Abstract
Shewanella putrefaciens MR-1 has emerged as a good model to study anaerobic respiration and electron transport-linked metal reduction. Its remarkable respiratory plasticity suggests the potential for a complex regulatory system to coordinate electron acceptor use in the absence of O2. It had previously been suggested that EtrA (electron transport regulator A), an analog of Fnr (fumarate nitrate regulator) from Escherichia coli, may regulate gene expression for anaerobic electron transport. An etrA knockout strain (ETRA-153) was isolated from MR-1 using a gene replacement strategy. Reverse transcription-PCR analysis of total RNA demonstrated the loss of the etrA mRNA in ETRA-153. ETRA-153 cells retained the ability to grow on all electron acceptors tested, including fumarate, trimethylamine N-oxide (TMAO), thiosulfate, dimethyl sulfoxide, ferric citrate, nitrate, and O2, as well as the ability to reduce ferric citrate, manganese(IV), nitrate, and nitrite. EtrA is therefore not necessary for growth on, or the reduction of, these electron acceptors. However, ETRA-153 had reduced initial growth rates on fumarate and nitrate but not on TMAO. The activities for fumarate and nitrate reductase were lower in ETRA-153, as were the levels of fumarate reductase protein and transcript. ETRA-153 was also deficient in one type of ubiquinone. These results are in contrast to those previously reported for the putative etrA mutant METR-1. Molecular analysis of METR-1 indicated that its etrA gene is not interrupted; its reported phenotype was likely due to the use of inappropriate anaerobic growth conditions.
Shewanella putrefaciens MR-1 is a nonfermentative, facultatively anaerobic bacterium originally isolated from anaerobic sediments in Oneida Lake, N.Y. (32). In the absence of O2, MR-1 can utilize several electron acceptors for anaerobic respiration, including iron(III), manganese(IV), thiosulfate, fumarate, nitrate, trimethylamine N-oxide (TMAO), dimethyl sulfoxide (DMSO), and others. The ability to utilize many of these anaerobic electron acceptors is absent in aerobically grown cells (23, 25, 32). Several of MR-1's cytochromes are synthesized only under anaerobic conditions (24), and menaquinone levels are increased in cells grown under anaerobic conditions (26). These results imply that MR-1 has the ability to sense the presence or absence of O2 and to regulate the expression of multiple electron transport components necessary for anaerobic respiration. However, little is known about the regulation of these components in MR-1.
The ability to regulate electron transport components is essential to enable rapid adaptation to changing environments and to prevent the waste of cellular resources. For example, Fnr (fumarate nitrate regulator) from Escherichia coli transcriptionally activates genes necessary for anaerobic growth while repressing others needed for aerobic growth (5, 6). Fnr is a positive regulator for the transcription of fumarate and nitrate reductase genes in E. coli (9, 12, 20, 46). Several analogs to Fnr have been described, including (i) CAP (catabolite gene activator protein), which regulates transcription in response to glucose availability in E. coli (5, 17, 43); (ii) ANR (anaerobic regulation of arginine deiminase and nitrate reduction) from Pseudomonas aeruginosa (4); (iii) and HlyX (hemolysin regulator) from Actinobacillus pleuropneumoniae (18). One possible regulator in MR-1 is EtrA (electron transport regulator A), which has 73.6% identity to E. coli Fnr on the basis of amino acid alignments (41). A study describing the isolation of a putative etrA mutant, METR-1, concluded that the loss of EtrA rendered the cells unable to grow under anaerobic conditions on several electron acceptors, including nitrite, thiosulfate, sulfite, TMAO, DMSO, Fe(III), and fumarate (41). METR-1 was isolated using a strategy in which a vector derived from pACYC184 was used to interrupt etrA in the chromosome (41); this strategy relied on the presumption that pACYC184 would not replicate freely in MR-1. A subsequent report clearly demonstrated that pACYC184 does freely replicate in MR-1 (31) and is therefore not suitable for gene replacement or gene interruption strategies. This finding calls into question the phenotype reported for METR-1 (41) and prompted the reinvestigation of the role of EtrA in MR-1.
All materials used in this study were from sources previously described (35). A list of the bacteria and plasmids that were used in this study is presented in Table 1. For molecular biology experiments, S. putrefaciens strains were grown aerobically at room temperature (23 to 25°C) or at 30°C on Luria-Bertani (LB) medium, pH 7.4 (42). E. coli strains were grown aerobically at 37°C on LB medium. Growth media were supplemented with appropriate antibiotics when required, including chloramphenicol at 35 μg ml−1, tetracycline at 20 μg ml−1, and kanamycin at 50 μg ml−1; for some experiments, other concentrations were used where indicated. For other applications, S. putrefaciens was grown at room temperature either aerobically or anaerobically as previously described (24) in M1 defined medium (33) supplemented with 15 mM lactate, vitamin-free Casamino Acids (0.1 g liter−1), and an appropriate electron acceptor. Growth on ferric citrate was done using LM medium (27) supplemented with 15 mM lactate, 2 mM sodium bicarbonate, and 10 mM ferric citrate.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Descriptiona | Reference | Source |
|---|---|---|---|
| Strains | |||
| S. putrefaciens | |||
| MR-1 | Manganese-reducing strain from sediments of Lake Oneida, N.Y. | 32 | Previous study |
| ETRA-153 | etrA mutant derived from MR-1; etrA::Kmr | This work | |
| METR-1 | Original study: MR-1 but etrA::pACYC184 Tcr; this study: MR-1 containing freely replicating pACYC184 plus partial etrA sequence from MR-1 | 41 | K. H. Nealson |
| E. coli | |||
| JM109 | recA1 F′ traD36 lacIq Δ(lacZ)M15 proA+B+/e14−(McrA−) Δ(lac-proAB) thi gyrA96 (Nalr) endA1 hsdR17(rK− mK+) relA1 supE44 | 49 | Promega |
| S17-1λpir | λ(pir) hsdR pro thi; chromosomally integrated RP4-2 Tc::Mu Km::Tn7; donor strain to mate pEP185.2-derived plasmids into MR-1 | 45 | V. L. Miller |
| S17-1 | C600::RP-4 2-Tc::Mu-Km::Tn7 hsdR hsdM+recA thi pro; donor strain to mate pVK100-derived plasmids into MR-1 | 45 | M. L. P. Collins |
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Smr) endA1 nupG; used as host for plasmids derived from pCR2.1-TOPO | Invitrogen | |
| Plasmids | |||
| pVK100 | 23-kb broad-host-range cosmid; Tcr Kmr Tra+ | 11 | ATCC 37156 |
| pVKetrA | pVK100 with the MR-1 etrA open reading frame plus associated 5′ and 3′ regions | This work | |
| pCR2.1-TOPO | 3.9-kb vector for cloning PCR products; Kmr Apr | Invitrogen | |
| pUT/mini-Tn5Km | Apr; Tn5-based delivery plasmid; used as source of Kmr gene | 3 | D. Frank |
| pTOPO/etrA | pCR2.1-TOPO with the MR-1 etrA open reading frame plus associated 5′ and 3′ regions; 5.4 kb | This work | |
| pTOPO/etrA:Km | pTOPO-etrA with the Kmr gene replacing bases 255–501 of etrA; 7.3 kb | This work | |
| pEP185.2 | 4.28-kb mobilizable suicide vector derived from pEP184; oriR6K mobRP4 Cmr | 10 | V. L. Miller |
| pDSEPetrA | Kmr gene-interrupted etrA gene cloned into the SmaI site of pEP185.2; Cmr Kmr; 7.7 kb; used for gene replacement to generate strain ETRA-153 | This work | |
| pACYC184 | 4.24-kb vector; Cmr Tcr | 1 | New England Biolabs |
| pCALc | 5.8-kb vector used for cloning and sequencing; Apr | 47 | Stratagene |
Nalr, Smr, Apr, and Cmr, resistance to nalidixic acid, streptomycin, ampicillin, and chloramphenicol, respectively.
DNA manipulations performed in this study were done according to standard techniques (42) as described previously (35). Electroporation of pACYC184 into MR-1 was done as described previously (31) with the following exceptions: the voltage was 1.15 kV and, immediately after electroporation, the cells were resuspended in 0.5 ml of LB broth, incubated at room temperature or 30°C on a gyrotary shaker at 180 rpm for 2 h prior to being plated on appropriate media. Primers used in PCR, inverse PCR, and colony PCR are listed in Table 2. The primers were designed using the genomic sequence for MR-1 (The Institute for Genomic Research [TIGR]). We assigned base numbers to etrA so that +1 corresponds to the first base of the start codon and 753 corresponds to the last base of its stop codon; negative numbers refer to sequence upstream of etrA. DNA sequencing was performed either by thermal cycle DNA sequencing with custom primers as previously described (35) or with an ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit by an automated single-capillary method (Applied Biosystems ABI Prism 310). Computer-assisted sequence analysis and comparisons were done with MacVector software (Accelrys, San Diego, Calif.).
TABLE 2.
Synthetic oligonucleotides used in this study
| Name | Oligonucleotide sequencea |
|---|---|
| Oligonucleotides based on etrA | |
| E1 | 5′-CATAGCCCAGCGTCTTGTG-3′ |
| E2 | 5′-ACGATTATGCCAAATGACTTGAGT-3′ |
| E3 | 5′-ATTGCGATCGATGAACGTCTCGCCGCCTTTA-3′ |
| E4 | 5′-ATTGCGATCGATCCTTGTTCGGTGATGGTGTAACTT-3′ |
| E5 | 5′-CTTTGTATCCCGTTCACCCTCAAT-3′ |
| E6 | 5′-ATCCTAGCATTACCCGCCAAGA-3′ |
| E7 | 5′-GGCTGCCCTGTGGACTGGTA-3′ |
| E8 | 5′-CGAAGCGGTTAGCAAGGTTACTGA-3′ |
| Oligonucleotide based on Kmr gene: K1 | 5′-ATTGCGATCGATTTATGCTTGTAAACC GTT-3′ |
| Oligonucleotides based on cat gene | |
| C1 | 5′-CAGACGGCATGATGAACCTGAATC-3′ |
| C2 | 5′-CCACCGTTGATATATCCCAATGGC-3′ |
| Oligonucleotides based on pCALc | |
| S1 | 5′-ATACGACTCACTATAGGGGAATTG-3′ |
| S2 | 5′-TGGCTGCTGAGACGGCTATGAAAT-3′ |
Underlined regions indicate the ClaI site engineered into oligonucleotides E3, E4, and K1.
Construction of an etrA insertion mutation.
Construction of a plasmid for gene replacement of etrA was completed using the strategy described by Myers and Myers (35). A 1,532-bp fragment containing the entire etrA gene plus 5′ and 3′ flanking sequences was generated by PCR of MR-1 genomic DNA using primers E1 and E2 (Table 2). This PCR product was cloned into pCR2.1-TOPO, generating pTOPO/etrA. Inverse PCR (37) of pTOPO/etrA using primers E3 and E4 (Table 2) generated TOPO/etrA(Δ246), a 5.2-kb fragment that is missing 246 bp of internal etrA sequence. The 2.1-kb kanamycin resistance (Kmr) gene from pUT/mini-Tn5Km was generated by PCR with primer K1. Following digestion with ClaI, the Kmr gene was ligated to the TOPO/etrA(Δ246) fragment, generating pTOPO/etrA:Km. A 3.4-kb DNA fragment containing the etrA gene interrupted by the Kmr gene was generated by PCR using pTOPO/etrA:Km as the template and primers E1 and E2. This fragment was blunt ended and ligated into the SmaI site of the suicide vector pEP185.2, generating pDSEPetrA, which was electroporated into the donor strain E. coli S17-1λpir. Throughout, appropriate analyses (restriction digests, PCR, DNA sequencing) were done to verify that the expected constructs were obtained.
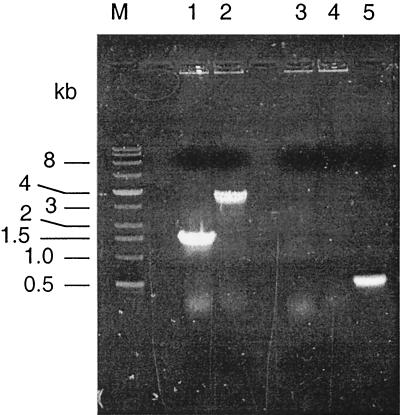
E. coli S17-1λpir(pDSEPetrA) was mated with MR-1, and MR-1 exconjugants were selected using kanamycin under aerobic conditions on defined medium with 15 mM lactate as the electron donor. Colonies were screened by colony PCR (48) using primers E1 and E2; those lacking the expected wild-type 1.5-kb PCR product were pursued as putative insertional mutants. One strain, designated ETRA-153, met all criteria expected for an etrA knockout: (i) it lacked the expected 1.5-kb wild-type PCR product for etrA; (ii) it was positive for a 3.4-kb PCR product, consistent with Kmr gene-interrupted etrA (Fig. 1); (iii) it was negative for a PCR product using primers C1 and C2 (Table 2), specific to the cat gene of pEP185.2 (Fig. 1); and (iv) it grew in the presence of kanamycin but not in the presence of chloramphenicol. The lack of the cat gene and sensitivity to chloramphenicol are consistent with a double-crossover gene replacement. The etrA gene from ETRA-153 interrupted by the Kmr gene was cloned into pCR2.1TOPO for sequencing. The sequence obtained (using primers E5 and E6 [Table 2]) confirmed that the etrA gene in ETRA-153 was interrupted by the Kmr gene at the expected position (data not shown by request).
FIG. 1.
Colony PCRs with primers specific for the etrA gene or the chloramphenicol acetyltransferase gene (cat) from pEP185.2. Lanes 1 and 2 show reactions with the etrA primers E1 and E2 (Table 2), and lanes 3 to 5 show reactions with the cat primers C1 and C2 (Table 2). The templates for PCR were as follows: MR-1 (lanes 1 and 3), ETRA-153 (lanes 2 and 4), and pDSEPetrA (lane 5). The sizes of the DNA markers (lane M) in kilobases are indicated on the left.
Characterization and complementation of the etrA insertion mutant.
Expression of etrA was analyzed by reverse transcription-PCR (RT-PCR) of total RNA using the Titan One Tube RT-PCR system (Roche Molecular Biochemicals, Indianapolis, Ind.). Total RNA was isolated from aerobically grown cells using a hot-phenol method followed by treatment with RNase-free DNase as previously described (28). All standard precautions to prevent RNase contamination were taken (42). Total RNA (2 μg) from each strain served as the template, using primers E5 and E6. The expected 662-bp RT-PCR product was present in MR-1 and MR-1(pVK100) but was absent in ETRA-153(pVK100) (Fig. 2). Complementation of ETRA-153 with pVKetrA restored the etrA transcript to ETRA-153 (Fig. 2).
FIG. 2.
Expression of etrA in S. putrefaciens cells grown aerobically as determined by RT-PCR of 2 μg of total RNA from the following strains: MR-1 (lane 1), MR-1(pVK100) (lane 2), ETRA-153(pVK100) (lane 3), and ETRA-153(pVKetrA) (lane 4). The etrA product is indicated by the arrow at the right. Primers E5 and E6 (Table 2) for RT-PCR were based on the etrA sequence from the MR-1 genome project (TIGR). The sizes of the DNA markers (lane M) in kilobases are indicated on the left.
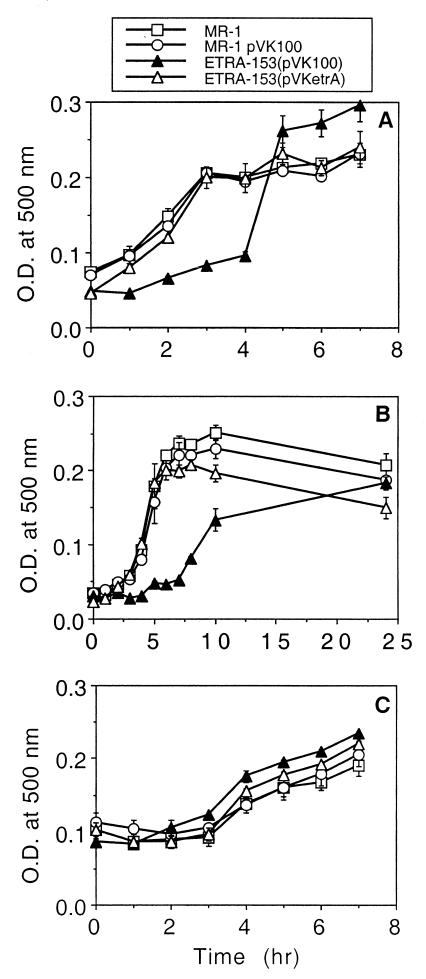
The ability of ETRA-153 to grow on and reduce various electron acceptors was investigated in defined medium, sampled once daily over a period of 4 days. For most electron acceptors, growth was assessed by measuring culture turbidity at 500 nm using a Beckman DU-64 spectrophotometer. Growth on Fe(III) was assessed by measuring total cellular protein (27). Concentrations of nitrate, nitrite, Fe(II), and Mn(II) were determined as previously described (36). The growth rates and maximal growth yields of ETRA-153(pVK100) were essentially the same as those of MR-1 using 20 mM fumarate, 20 mM TMAO, 5 mM DMSO, 10 mM thiosulfate, 10 mM ferric citrate, 2 mM nitrate, or O2 as the terminal electron acceptor (data not shown by request). ETRA-153(pVK100) was able to reduce 10 mM ferric citrate, 2 mM nitrate, and 5 mM MnO2 at rates that were essentially identical to those of MR-1 (data not shown by request). However, closer examination of short-term growth, examined hourly, demonstrated that ETRA-153(pVK100) has a lower initial rate of growth on fumarate and nitrate but that its rate of growth on TMAO is identical to that of MR-1 (Fig. 3).
FIG. 3.
Comparison of levels of short-term growth of various strains of S. putrefaciens with fumarate (A), nitrate (B), and TMAO (C). Medium was inoculated with cultures pregrown anaerobically on the tested substrate. Culture turbidity was measured over the course of 1 day until ETRA-153(pVK100) reached growth comparable to that of MR-1. Values are mean ± high and low values from two parallel but independent experiments for each strain. O.D., optical density.
Although ETRA-153 maintained the ability to grow on and reduce all of the electron acceptors tested, its lower initial growth rate on fumarate and nitrate indicated that it may have lower levels of fumarate and nitrate reductase. Since ≥98% of the fumarate reductase activity in MR-1 is localized to the soluble fraction (23), soluble fractions isolated from cells grown anaerobically with 20 mM fumarate were analyzed for fumarate reductase activity by monitoring the fumarate-dependent oxidation of reduced benzyl viologen under anaerobic conditions as described previously (23). The nitrate reductase activity of washed cells grown anaerobically with 2 mM nitrate was determined under anaerobic conditions. Briefly, overnight cultures were centrifuged and resuspended in 0.4 M potassium phosphate buffer (pH 7.5), with cell densities equalized to an optical density of 0.10 at 500 nm. Lactate and formate were added to a final concentration of 15 mM each, and the reaction was started by adding NaNO3 (0.5 M) to a final concentration of 1 mM. Samples (1 ml) were removed at 0, 15, 30, 45, and 60 min and were immediately filtered through 0.2-μm-pore-size filters. The filtrates were immediately placed on ice until they were analyzed for nitrate. The fumarate reductase activity of ETRA-153(pVK100) was 42% less than that of MR-1 (pVK100) (Fig. 4A). Complementing the mutant with pVKetrA restored fumarate reductase activity to the level of the wild type (Fig. 4A). Similarly, nitrate reduction by whole cells of ETRA-153(pVK100) was 35% less than that catalyzed by MR-1(pVK100) (Fig. 4A); complementing the mutant with pVKetrA restored the rate of nitrate reduction to that of the wild type (Fig. 4A). The decreases in nitrate and fumarate reductase activities correlate with the lower initial rates of growth of ETRA-153 (Fig. 3), suggesting that EtrA positively, but only partially, regulates the use of these electron acceptors in the absence of O2. This is in contrast to results of studies of E. coli Fnr mutants that show nearly complete loss of fumarate and nitrate reductase activities under anaerobic conditions (2, 15). In one study of four different Fnr mutants, fumarate reductase activities were less than 12% of wild-type levels and nitrate reductase activities were less than 4% of wild-type levels (15). EtrA appears to play a more subtle role in regulation in MR-1 than Fnr in E. coli.
FIG. 4.
(A) Analysis of the fumarate reductase activities of soluble fractions and the nitrate reductase activity of whole cells. For each panel, the strains were MR-1(pVK100) (first set of bars), ETRA-153(pVK100) (second set of bars), and ETRA-153(pVKetrA) (third set of bars). The average activities for MR-1(pVK100) were 14.8 μmol min−1 mg of protein−1 and 9.4 nmol min−1 ml−1 for fumarate and nitrate reductase, respectively. Values are relative means ± standard deviations (SD) of results of independent experiments (n = 3 for fumarate reductase; n = 5 for nitrate reductase). ∗, statistically significant to a P of ≤0.03. (B) Analysis of fumarate reductase protein of soluble fractions using an antibody specific for fumarate reductase. Protein levels are relative to those of MR-1(pVK100) using NIH Image software. Values are relative means ± SD for soluble fractions isolated from three different cultures. §, statistically significant from strains 1 and 3 to a P of ≤0.005; ‡, statistically significant from strain 1 to a P of ≤0.03. (C) RNase protection of 10 μg of RNA isolated from cells grown anaerobically on fumarate with a probe specific for fumarate reductase. Values are relative to those for MR-1(pVK100) using NIH Image software and are the means ± SD of results for three independently grown cultures. †, statistically significant to a P of ≤0.02.
Polyclonal antibodies specific for fumarate reductase (FumR) were generated against an antigen produced by recombinant technology. Briefly, an internal fragment of fumR encoding a 121-residue fragment of FumR was cloned into pBAD/Thio-TOPO (Invitrogen, Carlsbad, Calif.). Following arabinose-induced expression in E. coli TOP10 cells, the FumR protein fusion was purified using HisBind Quick resin (Novagen, Madison, Wis.). The purified protein was used as an antigen to generate polyclonal antibodies in New Zealand White rabbits (Cocalico Biologicals, Reamstown, Pa.). Purified immunoglobulin G was obtained from rabbit sera, nonspecific antibodies were removed by absorption with autoclaved E. coli JM109 cells, and Western blotting was done as described previously (30). The specificity of the antibody was confirmed using CMTn-3, a fumarate reductase-minus mutant (29) as a negative control. The level of FumR protein was 60% lower in ETRA-153(pVK100) than in MR-1(pVK100) (Fig. 4B); complementation with pVKetrA increased the level of FumR protein, although it was 35% less than that seen with MR-1(pVK100) (Fig. 4B).
In E. coli, Fnr has a direct role in the transcriptional regulation of the genes for fumarate and nitrate reductase (9, 12, 20, 46). Since both the protein and activity levels for fumarate reductase were partially reduced in ETRA-153, the levels of fumarate reductase (fumR) mRNA were examined by RNase protection assay (RPA III kit; Ambion, Austin, Tex.). Since the nitrate reductase genes have not yet been identified in MR-1, the nitrate reductase transcript could not be examined. A 266-bp internal fragment of fumR was cloned into pCR2.1TOPO; the desired orientation was verified by PCR. Using M13 primers, a 509-bp fragment containing the fumR and flanking 5′ and 3′ vector DNAs was generated by PCR; using this fragment as a template, the antisense fumR RNA probe was generated using a MAXIscript kit (Ambion). The probe was gel purified on a Tris-borate-EDTA–urea polyacrylamide gel. RNase protection assays were done using total RNA (10 μg) and 400 pg of the probe. The level of the fumR transcript was 73% lower in ETRA-153(pVK100) than in MR-1(pVK100) (Fig. 4C); complementation with pVKetrA increased the fumR transcript to levels similar to those of MR-1(pVK100) (Fig. 4C).
To further investigate the phenotype of ETRA-153, subcellular fractions of fumarate-grown cells were prepared and analyzed. Cytoplasmic membrane, intermediate membrane, outer membrane (OM), and soluble (cytoplasm plus periplasm) fractions were purified by an EDTA-lysozyme-Brij protocol as previously described (24). The intermediate membrane is a hybrid of the cytoplasmic membrane and the OM (24). The separation and purity of these subcellular fractions were assessed by spectral cytochrome content analysis (24), by membrane buoyant-density analysis (24), and with sodium dodecyl sulfate (SDS)-polyacrylamide gels (14, 22) stained for protein with Pro-Blue or heme as previously described (34). Protein was determined by a modified Lowry method (28). SDS-polyacrylamide gel electrophoresis patterns confirmed a prominent separation of the various fractions and demonstrated that they were comparable to those of previous experiments (23–25, 28, 30, 34). Protein and heme stains of these fractions were similar for all strains (data not shown by request). Cytochrome spectra of the various subcellular fractions indicated that the cytochrome content and distribution of ETRA-153(pVK100) were similar to those of MR-1(pVK100) (data not shown). Using an antibody specific for the OM cytochrome OmcA (30), Western blotting of OM fractions of MR-1(pVK100), ETRA-153(pVK100), and ETRA-153(pVKetrA) showed no significant differences among these strains in the levels of OmcA (data not shown). CymA, a tetraheme cytochrome c, is required for growth on and reduction of nitrate, fumarate, Fe(III), and Mn(IV) (28, 35); since ETRA-153 grew well on these electron acceptors, EtrA is not required for the expression of cymA. A more subtle role for EtrA in the expression of cymA cannot be excluded at this time.
Since the levels of various quinones are affected by growth under aerobic versus anaerobic conditions (26), the quinone contents of the various strains were examined by thin-layer chromatography (TLC). Quinones were extracted from cells by a method adapted from the work of Kröger and Dadák (13) as previously described (35). Quinones were resolved by TLC as described previously (7) on Merck Kieselgel 60 F254 plates (20 by 20 cm, 0.25 mm thick) developed with petroleum ether-diethyl ether (9/1 [vol/vol]). The plates were examined under reflective UV light (wavelength, 254 nm). All strains were similar in their contents of menaquinones and most ubiquinone species. However, ETRA-153(pVK100) was deficient in one of the ubiquinone species that was present in all other strains tested (Fig. 5). The deficiency in one type of ubiquinone seen in ETRA-153 (Fig. 5) indicates that EtrA may play a role in the regulation of ubiquinone synthesis. However, this role appears to be minor since the levels of menaquinones and the other ubiquinones were similar to those of the wild type. The presence of multiple ubiquinone bands on TLC is due to variation in the lengths of the membrane-anchoring isoprenyl side chains. The length of a side chain is determined by the enzyme polyprenyl diphosphate synthase, and p-hydroxybenzoate polyprenyltransferase catalyzes the attachment of various isoprenyl chain substrates to p-hydroxybenzoate (40). However, it has been shown that the exact length of the isoprenyl side chain is not crucial for function (38, 39). This implies that a deficiency in one species of ubiquinone will not likely affect growth since other forms can effectively substitute. This may explain why ETRA-153 can grow on all tested electron acceptors despite being deficient in one particular ubiquinone. In E. coli, the ratios of aerobic to anaerobic quinone were not altered in an Fnr deletion mutant, indicating that Fnr is not involved in the regulation of quinone synthesis (44).
FIG. 5.
Thin-layer chromatogram of quinones isolated from S. putrefaciens cells grown anaerobically with fumarate as the electron acceptor. Lanes 1 and 2 were loaded with ubiquinone-10 and menaquinone-4 standards, respectively. The other lanes were loaded with quinone extracts (equivalent to 0.10 g [wet weight] of cells) isolated from cells of the following strains: MR-1 (lane 3), MR-1(pVK100) (lane 4), ETRA-153(pVK100) (lane 5), ETRA-153(pVKetrA) (lane 6), MR-1(pACYC184) (lane 7), and METR-1 (lane 8). The samples were loaded at the bottom, and migration was upward. The positions of various quinones are indicated at the right. I, methylmenaquinone; II, menaquinone; III–VI, various ubiquinones. The identities of these quinones were previously verified by high-pressure liquid chromatography with diode array detection (35).
Reexamination of the putative etrA mutant METR-1.
In 1993, Saffarini and Nealson (41) described the putative etrA mutant METR-1 and concluded that etrA was necessary for anaerobic growth on nitrite, thiosulfate, sulfite, TMAO, DMSO, Fe(III), and fumarate (41). This conclusion is in contrast to the results reported here for ETRA-153, which grew readily on all of these electron acceptors. METR-1 was putatively constructed by site-specific insertion through a single crossover between etrA in the genome and plasmid pSETR-3 (a small fragment of etrA in the vector pACYC184) (41). This approach presumed that pACYC184 could not replicate in MR-1 and could be maintained only by insertion into the genome. However, it has since been shown that pACYC184 can readily replicate in MR-1 (31); this finding raised the question as to how the strategy of Saffarini and Nealson could have been successfully used for site-specific insertional mutagenesis.
To investigate these discrepancies, further experiments were done with METR-1. Colony PCR with primers E7 and E8 (Table 2) designed to amplify etrA indicated that METR-1 does not have an interrupted etrA gene; it yielded the same 844-bp product as MR-1 (data not shown by request). Plasmid DNA was isolated from METR-1, and restriction digests with SalI and EcoRV indicated that the strain contains a 7.8-kb plasmid. An NcoI digest yielded two fragments of approximately 3.6 and 4.1 kb (data not shown). No plasmid should be present if METR-1 is a true single-crossover mutant; MR-1 does not contain this 7.8-kb plasmid. The plasmid in METR-1 is almost twice as large as expected for pACYC184 (4.2 kb) or pSETR3 (4 kb). Its two NcoI fragments were subcloned into pCALc (Stratagene, La Jolla, Calif.), and the DNA sequences of the ends of these fragments were determined using primers S1 and S2 (Table 2). The larger NcoI fragment contained sequence that matched pACYC184, whereas the smaller NcoI fragment contained sequence that matched etrA of MR-1 (TIGR MR-1 genome project) (data not shown). Since only the ends of the fragments were sequenced, the identity of the “extra” DNA is unknown. These results are consistent with the demonstrated ability of pACYC184 to replicate freely in MR-1 (31).
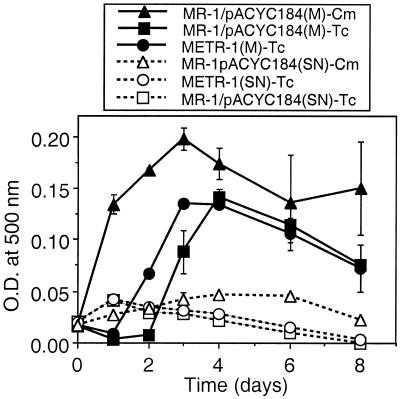
While this molecular analysis indicates that the etrA gene was not interrupted in METR-1, it does not provide a rationale for the reported phenotype of METR-1 (41). Four factors may have contributed to the reported phenotype: (i) no control strains were compared with METR-1; (ii) qualitative growth measurements, such as visual turbidity and end product odor formation, were used for some electron acceptors, which limits the detection of growth changes that fall between the extremes; (iii) low concentrations of electron acceptors (2 mM) were used, which may not have been sufficient to support robust growth; and (iv) tetracycline was used to select for the tetracycline resistance (Tcr) marker of pACYC184 (tetracycline may markedly compromise anaerobic growth because resistance is encoded by an energy-intensive class C tetracycline efflux pump) (8, 16, 19). Levels of growth of METR-1 and MR-1(pACYC184) were similar when fumarate (20 mM) was nonlimiting (Fig. 6). The growth of MR-1(pACYC184) was more rapid and reached a higher maximal density when the medium contained chloramphenicol (35 μg ml−1) than when it contained tetracycline (16 μg ml−1); this demonstrated that tetracycline can partially impede growth under anaerobic conditions. In contrast, all strains grew very poorly under the conditions used by Saffarini and Nealson (41); i.e., when fumarate was limiting (2 mM), no significant increase in turbidity was observed over 8 days (Fig. 6). These conditions are clearly inadequate to support growth, even of MR-1, but explain why METR-1 was reported as negative for growth on fumarate (41). Growth on TMAO, DMSO, and O2 was also investigated; similar to fumarate-grown cells, both METR-1 and MR-1(pACYC184) grew well and to similar extents on TMAO (20 mM) and DMSO (5 mM) but grew poorly when placed under limiting electron acceptor conditions (2 mM) (data not shown by request). Growth on O2 was evident under all tested conditions, but maximal turbidity was less when the strains were grown with tetracycline than when they were grown with chloramphenicol (data not shown by request). In summary, tetracycline-grown cells showed reduced growth, which is consistent with energy expenditure for the tetracycline efflux pump, leaving less energy for cell growth. Also, there was little or no growth over 8 days when electron acceptors were limiting (2 mM); therefore, an electron acceptor concentration of 2 mM is insufficient to adequately assess quantitative growth differences between various strains in liquid culture.
FIG. 6.
Anaerobic growth of METR-1 and MR-1(pACYC184) using fumarate as the electron acceptor. Two variations of media were compared: M1 medium (pH 7.4) with 15 mM lactate, 20 mM fumarate, and either tetracycline (16 μg ml−1) or chloramphenicol (35 μg ml−1) (M) and M1 medium (pH 7.9) with 30 mM lactate, 2 mM fumarate, and either tetracycline (8 μg ml−1) or chloramphenicol (30 μg ml−1) (SN). SN refers to conditions reported by Saffarini and Nealson (41). METR-1 was not examined with chloramphenicol because it is Tcr only.
Concluding remarks.
Although EtrA plays a subtle role in MR-1 anaerobic gene regulation, other regulatory components are likely involved as well. This is not surprising considering that themes of cooperative regulation and redundancy are common in living organisms. Given the complex respiratory diversity displayed by MR-1, it is possible that there are multiple regulatory elements involved in the expression of various electron transport components. For example, multiple cytochromes are localized to the outer membrane of MR-1 under anaerobic conditions (24), but little is known about the regulatory mechanisms involved in the expression or localization of these various cytochromes. However, cytochrome spectra and SDS-polyacrylamide gel electrophoresis analysis of subcellular fractions of ETRA-153 indicated that, relative to MR-1, there were no major changes in cytochrome content or distribution. This implies that EtrA plays no significant role in the global regulation of cytochrome production and that other as yet uncharacterized regulators are likely involved in the control of cytochromes expressed under anaerobic conditions.
Since it has been suggested that MR-1 may have more than one Fnr-like element (41), the MR-1 genomic fragments compiled by TIGR were searched using MacVector for other possible analogs to Fnr or EtrA. Both nucleotide and amino acid searches were completed. No other open reading frames with significant homology to etrA were identified in the MR-1 genome. Queries based on ANR (Pseudomonas aeruginosa, GenBank accession number M98276; Pseudomonas fluorescens, GenBank accession number AF053611), HlyX (Actinobacillus pleuropneumoniae, GenBank accession number M80712), and Fnr (Vibrio cholerae, GenBank accession number AF244992) in the MR-1 genome showed significant homology with EtrA, but no other convincing alignments were obtained. Although MR-1 inevitably has other regulatory elements, it appears that they are not obviously homologous with Fnr or EtrA.
Also, the sequence upstream of the fumarate reductase gene in MR-1 was searched (using MacVector software) for the presence of a possible Fnr consensus sequence (TTGATnnnnATCAA) (where n is any base) (4, 21), allowing for differences in the spacing of the half-sites; this consensus sequence was not found in the 8 kb immediately upstream of the fumarate reductase gene. At 3.9 kb upstream of the fumarate reductase gene, the sequence TCGATcttcCTCAA is present, which has two variations from the Fnr-binding consensus sequence (bold letters). At 7.2 kb upstream of the fumarate reductase gene, the sequence TTGACaataATTAA is present, which also has two variations from the Fnr consensus sequence (bold letters). Although these two sites may potentially be recognition sites for EtrA, this is only speculative. The consensus sequence for EtrA is not known and may be different than that for Fnr.
Acknowledgments
This work was supported by National Institutes of Health grant R01GM50786 to C.R.M. The graduate fellowship for T. M. Maier was provided in part by the Robert G. Zach Family Foundation.
We are grateful to V. L. Miller for graciously providing pEP185.2, to D. Frank for providing pUT/mini-Tn5Km, and to J. M. Myers for providing expertise and advice in the laboratory and for initial studies with anti-FumR. We thank the Protein & Nucleic Acid Shared Facility of the Medical College of Wisconsin for performing automated nucleic acid sequencing for this study.
The preliminary genomic sequence data was obtained from TIGR through their website at http://www.tigr.org. Sequencing of S. putrefaciens MR-1 genomic DNA by TIGR was accomplished with support from the Department of Energy.
REFERENCES
- 1.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chippaux M, Giudici D, Abou-Jaoudé A, Casse F, Pascal M C. A mutation leading to the total lack of nitrite reductase activity in Escherichia coli K12. Mol Gen Genet. 1978;160:225–229. doi: 10.1007/BF00267485. [DOI] [PubMed] [Google Scholar]
- 3.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertional mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galimand M, Gamper M, Zimmermann A, Haas D. Positive FNR-like control of anaerobic arginine degradation and nitrate respiration in Pseudomonas aeruginosa. J Bacteriol. 1991;173:1598–1606. doi: 10.1128/jb.173.5.1598-1606.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guest J R. Oxygen-regulated gene expression in Escherichia coli. The 1992 Marjory Stephenson Prize Lecture. J Gen Microbiol. 1992;138:2253–2263. doi: 10.1099/00221287-138-11-2253. [DOI] [PubMed] [Google Scholar]
- 6.Gunsalus R P. Control of electron flow in Escherichia coli: coordinated transcription of respiratory pathway genes. J Bacteriol. 1992;174:7069–7074. doi: 10.1128/jb.174.22.7069-7074.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Itoh T, Funabashi H, Katayama-Fujimura Y, Iwasaki S, Kuraishi H. Structure of methylmenaquinone-7 isolated from Alteromonas putrefaciens IAM 12079. Biochim Biophys Acta. 1985;840:51–55. [Google Scholar]
- 8.Johnson R, Adams J. The ecology and evolution of tetracycline resistance. Trends Ecol Evol. 1992;7:295–299. doi: 10.1016/0169-5347(92)90226-2. [DOI] [PubMed] [Google Scholar]
- 9.Jones H M, Gunsalus R P. Regulation of Escherichia coli fumarate reductase (frdABCD) operon expression by respiratory electron acceptors and the fnr gene product. J Bacteriol. 1987;169:3340–3349. doi: 10.1128/jb.169.7.3340-3349.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinder S A, Badger J L, Bryant G O, Pepe J C, Miller V L. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O8 and construction of a transformable R−M+ mutant. Gene. 1993;136:271–275. doi: 10.1016/0378-1119(93)90478-l. [DOI] [PubMed] [Google Scholar]
- 11.Knauf V C, Nester E W. Wide host range cloning vectors: cosmid clone bank of Agrobacterium Ti plasmids. Plasmid. 1982;8:45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- 12.Kolesnikow T, Schröder I, Gunsalus R P. Regulation of narK gene expression in Escherichia coli in response to anaerobiosis, nitrate, iron, and molybdenum. J Bacteriol. 1992;174:7104–7111. doi: 10.1128/jb.174.22.7104-7111.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kröger A, Dadák V. On the role of quinones in bacterial electron transport; the respiratory system of Bacillus megaterium. Eur J Biochem. 1969;11:328–340. doi: 10.1111/j.1432-1033.1969.tb00776.x. [DOI] [PubMed] [Google Scholar]
- 14.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 15.Lambden P R, Guest J R. Mutants of Escherichia coli K12 unable to use fumarate as an anaerobic electron acceptor. J Gen Microbiol. 1976;97:145–160. doi: 10.1099/00221287-97-2-145. [DOI] [PubMed] [Google Scholar]
- 16.Lenski R E, Simpson S C, Nguyen T T. Genetic analysis of a plasmid-encoded, host genotype-specific enhancement of bacterial fitness. J Bacteriol. 1994;176:3140–3147. doi: 10.1128/jb.176.11.3140-3147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, Wing H, Lee D, Wu H, Busby S. Transcription activation by Escherichia coli FNR protein: similarities to, and differences from, the CRP paradigm. Nucleic Acids Res. 1998;26:2075–2081. doi: 10.1093/nar/26.9.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacInnes J I, Kim J E, Lian C, Soltes G A. Actinobacillus pleuropneumoniae hlyX gene homology with the fnr gene of Escherichia coli. J Bacteriol. 1990;172:4587–4592. doi: 10.1128/jb.172.8.4587-4592.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McMurry L, Petrucci R E, Jr, Levy S B. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc Natl Acad Sci USA. 1980;77:3974–3977. doi: 10.1073/pnas.77.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melville S B, Gunsalus R P. Mutations in fnr that alter anaerobic regulation of electron transport-associated genes in Escherichia coli. J Biol Chem. 1990;265:18733–18736. [PubMed] [Google Scholar]
- 21.Melville S B, Gunsalus R P. Isolation of an oxygen-sensitive FNR protein of Escherichia coli: interaction at activator and repressor sites of FNR-controlled genes. Proc Natl Acad Sci USA. 1996;93:1226–1231. doi: 10.1073/pnas.93.3.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myers C R, Collins M L P. Cell-cycle-specific oscillation in the composition of chromatophore membrane in Rhodospirillum rubrum. J Bacteriol. 1986;166:818–823. doi: 10.1128/jb.166.3.818-823.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myers C R, Myers J M. Fumarate reductase is a soluble enzyme in anaerobically grown Shewanella putrefaciens MR-1. FEMS Microbiol Lett. 1992;98:13–20. [Google Scholar]
- 24.Myers C R, Myers J M. Localization of cytochromes to the outer membrane of anaerobically grown Shewanella putrefaciens MR-1. J Bacteriol. 1992;174:3429–3438. doi: 10.1128/jb.174.11.3429-3438.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myers C R, Myers J M. Ferric reductase is associated with the membranes of anaerobically grown Shewanella putrefaciens MR-1. FEMS Microbiol Lett. 1993;108:15–22. [Google Scholar]
- 26.Myers C R, Myers J M. Role of menaquinone in the reduction of fumarate, nitrate, iron(III) and manganese(IV) by Shewanella putrefaciens MR-1. FEMS Microbiol Lett. 1993;114:215–222. [Google Scholar]
- 27.Myers C R, Myers J M. Ferric iron reduction-linked growth yields of Shewanella putrefaciens MR-1. J Appl Bacteriol. 1994;76:253–258. doi: 10.1111/j.1365-2672.1994.tb01624.x. [DOI] [PubMed] [Google Scholar]
- 28.Myers C R, Myers J M. Cloning and sequence of cymA, a gene encoding a tetraheme cytochrome c required for reduction of iron(III), fumarate, and nitrate by Shewanella putrefaciens MR-1. J Bacteriol. 1997;179:1143–1152. doi: 10.1128/jb.179.4.1143-1152.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myers C R, Myers J M. Isolation and characterization of a transposon mutant of Shewanella putrefaciens MR-1 deficient in fumarate reductase. Lett Appl Microbiol. 1997;25:162–168. doi: 10.1046/j.1472-765x.1997.00196.x. [DOI] [PubMed] [Google Scholar]
- 30.Myers C R, Myers J M. Outer membrane cytochromes of Shewanella putrefaciens MR-1: spectral analysis, and purification of the 83-kDa c-type cytochrome. Biochim Biophys Acta. 1997;1326:307–318. doi: 10.1016/s0005-2736(97)00034-5. [DOI] [PubMed] [Google Scholar]
- 31.Myers C R, Myers J M. Replication of plasmids with the p15A origin in Shewanella putrefaciens MR-1. Lett Appl Microbiol. 1997;24:221–225. doi: 10.1046/j.1472-765x.1997.00389.x. [DOI] [PubMed] [Google Scholar]
- 32.Myers C R, Nealson K H. Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science. 1988;240:1319–1321. doi: 10.1126/science.240.4857.1319. [DOI] [PubMed] [Google Scholar]
- 33.Myers C R, Nealson K H. Respiration-linked proton translocation coupled to anaerobic reduction of manganese(IV) and iron(III) in Shewanella putrefaciens MR-1. J Bacteriol. 1990;172:6232–6238. doi: 10.1128/jb.172.11.6232-6238.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myers J M, Myers C R. Isolation and sequence of omcA, a gene encoding a decaheme outer membrane cytochrome c of Shewanella putrefaciens MR-1, and detection of omcA homologs in other strains of S. putrefaciens. Biochim Biophys Acta. 1998;1373:237–251. doi: 10.1016/s0005-2736(98)00111-4. [DOI] [PubMed] [Google Scholar]
- 35.Myers J M, Myers C R. Role of the tetraheme cytochrome CymA in anaerobic electron transport in cells of Shewanella putrefaciens MR-1 with normal levels of menaquinone. J Bacteriol. 2000;182:67–75. doi: 10.1128/jb.182.1.67-75.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Myers J M, Myers C R. Role for outer membrane cytochromes OmcA and OmcB of Shewanella putrefaciens MR-1 in reduction of manganese dioxide. Appl Environ Microbiol. 2001;67:260–269. doi: 10.1128/AEM.67.1.260-269.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ochman H, Gerber A S, Hartl D L. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okada K, Kainou T, Matsuda H, Kawamukai M. Biological significance of the side chain length of ubiquinone in Saccharomyces cerevisiae. FEBS Lett. 1998;431:241–244. doi: 10.1016/s0014-5793(98)00753-4. [DOI] [PubMed] [Google Scholar]
- 39.Okada K, Kamiya Y, Zhu X, Suzuki K, Tanaka K, Nakagawa T, Matsuda H, Kawamukai M. Cloning of the sdsA gene encoding solanesyl diphosphate synthase from Rhodobacter capsulatus and its functional expression in Escherichia coli and Saccharomyces cerevisiae. J Bacteriol. 1997;179:5992–5998. doi: 10.1128/jb.179.19.5992-5998.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okada K, Suzuki K, Kamiya Y, Zhu X, Fujisaki S, Nishimura Y, Nishino T, Nakagawa T, Kawamukai M, Matsuda H. Polyprenyl diphosphate synthase essentially defines the length of the side chain of ubiquinone. Biochim Biophys Acta. 1996;1302:217–223. doi: 10.1016/0005-2760(96)00064-1. [DOI] [PubMed] [Google Scholar]
- 41.Saffarini D A, Nealson K H. Sequence and genetic characterization of etrA, an fnr analog that regulates anaerobic respiration in Shewanella putrefaciens MR-1. J Bacteriol. 1993;175:7938–7944. doi: 10.1128/jb.175.24.7938-7944.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T, editors. Molecular cloning: a laboratory manual. 2nd ed. 1 to 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 43.Shaw D J, Rice D W, Guest J R. Homology between CAP and Fnr, a regulator of anaerobic respiration in Escherichia coli. J Mol Biol. 1983;166:241–247. doi: 10.1016/s0022-2836(83)80011-4. [DOI] [PubMed] [Google Scholar]
- 44.Shestopalov A I, Bogachev A V, Murtazina R A, Viryasov M B, Skulachev V P. Aeration-dependent changes in composition of the quinone pool in Escherichia coli. Evidence of post-transcriptional regulation of the quinone biosynthesis. FEBS Lett. 1997;404:272–274. doi: 10.1016/s0014-5793(97)00143-9. [DOI] [PubMed] [Google Scholar]
- 45.Simon R, Priefer U, Pühler A. A broad host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:37–45. [Google Scholar]
- 46.Stewart V. Requirement of Fnr and NarL functions for nitrate reductase expression in Escherichia coli K-12. J Bacteriol. 1982;151:1320–1325. doi: 10.1128/jb.151.3.1320-1325.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 48.Townsend K M, Frost A J, Lee C W, Papadimitriou J M, Dawkins H J S. Development of PCR assays for species- and type-specific identification of Pasteurella multocida isolates. J Clin Microbiol. 1998;36:1096–1100. doi: 10.1128/jcm.36.4.1096-1100.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]