Abstract
Kinetoplastid RNA editing is a posttranscriptional insertion and deletion of U residues in mitochondrial transcripts that involves RNA ligase. A complex of seven different polypeptides purified from Trypanosoma brucei mitochondria that catalyzes accurate RNA editing contains RNA ligases of ∼57 kDa (band IV) and ∼50 kDa (band V). From a partial amino acid sequence, cDNA and genomic clones of band IV were isolated, making it the first cloned component of the minimal RNA editing complex. It is indeed an RNA ligase, for when expressed in Escherichia coli, the protein autoadenylylates and catalyzes RNA joining. Overexpression studies revealed that T. brucei can regulate of total band IV protein at the level of translation or protein stability, even upon massively increased mRNA levels. The protein's mitochondrial targeting was confirmed by its location, size when expressed in T. brucei and E. coli, and N-terminal sequence. Importantly, genetic knockout studies demonstrated that the gene for band IV is essential in procyclic trypanosomes. The band IV and band V RNA ligases of the RNA editing complex therefore serve different functions. We also identified the gene for band V RNA ligase, a protein much more homologous to band IV than to other known ligases.
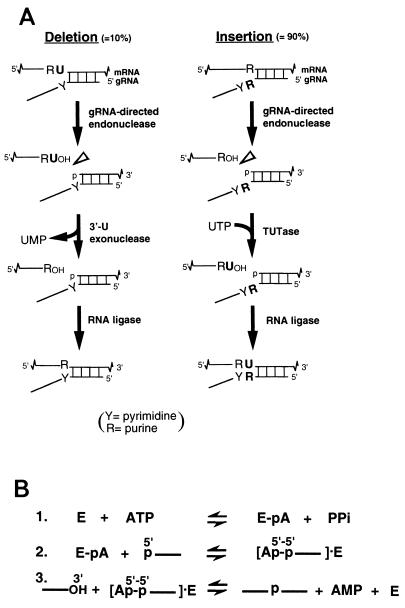
In kinetoplastid protozoans, many mitochondrial transcripts undergo RNA editing, a specific insertion and deletion of U residues at multiple sites, directed by guide RNAs (reviewed in references 2, 13, 15, 38, and 39). Both U deletional and U insertional editing cycles have been reproduced in vitro and shown to involve three enzymatic steps (Fig. 1A) (9, 20, 34, 35; see also reference 7). First, the mRNA is cleaved by a guide RNA (gRNA)-directed endonuclease, U residues are then added to or removed from the 3′ end of the upstream cleavage product by a terminal-U-transferase or 3′-U-exonuclease, and the mRNA is then rejoined by RNA ligase. A complex consisting of seven different polypeptides that contains all these activities and catalyzes both U-deletional and U-insertional editing rounds has been purified from Trypanosoma brucei mitochondria (10, 29). We have undertaken the cloning and characterization of the polypeptides that make up this complex, beginning with one identified as an RNA ligase.
FIG. 1.
Process of RNA editing and mechanism of RNA ligase action. (A) RNA editing has been found to involve the indicated enzymatic steps, as described in the introduction. TUTase, terminal-U-transferase. (B) Mechanism of many RNA and DNA ligases, including T4 and yeast tRNA ligase and evidently trypanosome RNA ligase. E, exonuclease.
RNA ligases are used by many cells in tRNA splicing (e.g., see references 5, 14, 45, and 49) and by bacteriophage T4 in tRNA repair (reviewed in reference 41), and they are also present in trypanosome mitochondria (3, 17, 46). These enzymes join RNA 3′ hydroxyl and 5′ phosphate termini, evidently by a common mechanism (30, 31; Fig. 1B). First, the ligase autoadenylylates, using ATP to form a covalent protein-AMP intermediate while releasing pyrophosphate (PPi). This reaction occurs in the absence of RNA and reverses with high concentrations of PPi. The AMP is then transferred to the 5′ phosphate of a donor RNA, generating a 5′-5′ linkage, and the 3′ hydroxyl of the acceptor RNA finally displaces this 5′ AMP, forming the new phosphodiester bond. T. brucei mitochondrial extract contains adenylylatable proteins of ∼57 and ∼50 kDa that deadenylylate when incubated with PPi or ligatable RNA but not when incubated with nonligatable RNA or ligatable DNA (29–31), indicating that they are RNA ligases. While some T. brucei lines show one ∼57-kDa polypeptide (8, 31), the TREU 667 line our laboratory works with has two closely migrating ∼57-kDa forms, as well as the ∼50-kDa species (29). These three polypeptides are all constituents of the minimal editing complex (29), indicating their roles in RNA editing.
To facilitate characterization of the enzymology and biological requirement for the ∼57-kDa RNA ligase, we cloned its cDNA, expressed it in Escherichia coli and trypanosomes, and performed genetic knockout analysis. These studies show that we have cloned the gene for a mitochondrially targeted RNA ligase that is essential in procyclic trypanosomes, regulated in abundance, and highly related to another predicted protein, evidently the ∼50-kDa ligase of the RNA editing complex.
MATERIALS AND METHODS
Procedures involving commercial reagents generally followed the manufacturer's recommendations.
Trypanosome propagation and preparation of mitochondrial extracts, DNA, and RNA.
Procyclic trypanosomes (strain TREU 667 or strain 427-derived transgenics of T. brucei brucei) were grown and mitochondrial extract was prepared and stored long term as described in reference 32, except that the extract was at 3 × 1010 cell equivalents/ml and used MRB (25 mM Tris-HCl [pH 8.0], 60 mM KCl, 10 mM magnesium acetate, 1 mM EDTA, 5% glycerol) supplemented with 5 mM dithiothreitol (DTT) and protease inhibitors (Pefabloc SC [Roche Molecular Biochemicals] at 1 mg/ml, Antipain [Sigma] at 50 μg/ml; E-64 [Sigma] at 10 μg/ml). Small-scale preparations from transformed trypanosomes were similar, except that 1 × 108 to 2 × 108 cells were lysed by vortexing rather than Dounce homogenizing, the percoll gradient was omitted, and the vesicles were suspended in 100 μl of MRB for the Triton X-100 treatment. DNA isolation (33) from 2 × 109 trypanosomes yielded ∼0.2 μg of DNA. RNA isolation, from ∼109 cells, utilized 75 ml of TRIZOL Reagent (Bethesda Research Laboratories [BRL]).
Protein purification and peptide sequencing.
For purification of the editing complex (basically scaling up of the protocol in reference 29), we used buffer P (25 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 1 mM EDTA, 5 mM DTT, 10% glycerol) supplemented with KCl to the millimolar concentrations indicated. We applied 7.5 × 1011 cell equivalents of extract (∼5 mg of protein per ml), diluted to 85 ml in buffer P (50 mM), at 2 ml/min to a 20-ml Q-Sepharose column in a 30-ml syringe, pre-equilibrated in buffer P (50 mM). After washing with 80 ml of buffer P (50 mM), a 130-ml buffer P (50 to 350 mM) linear gradient was run. Other purification runs suggest that buffer P (100 mM) may be preferable to buffer P (50 mM) above. The 7-ml fractions were analyzed by adenylylation. The seven peak fractions (∼170 to 200 mM KCl; <0.1 mg of protein per ml) were dialyzed for a few hours against buffer P (40 mM) and applied (at 0.5 ml/min) to a 3-ml DNA-cellulose column in a 5-ml syringe pre-equilibrated as described above. After a 12-ml wash, a 24-ml buffer P (50 to 350 mM) linear gradient was run, with the adenylylation activity eluting at ∼85 to 120 mM KCl.
For protein sequencing, the DNA-cellulose-purified peak fractions (4.5 ml) were treated with 4 mM PPi for 10 min on ice to deadenylylate the ligase polypeptides, precipitated, subjected to sodium dodecyl sulfate (SDS)–10% polyacrylamide gel electrophoresis, electroblotted onto a Bio-Rad TransBlot polyvinylidene difluoride membrane, and stained with 0.2% amido black. The Wistar protein analysis facility (Philadelphia, Pa.) performed trypsin digestion, high-pressure liquid chromatography (HPLC) separation of tryptic peptides, and sequencing by Edman degradation under the supervision of David Reim.
Oligonucleotides, PCR, library screening, subcloning, and analysis.
PCR of genomic DNA used the following oligonucleotides with 192- to 512-fold degeneracy: IV.1(→), 5′-GGNATHATGG AYCCNAAYGA-3′; IV.1(←), 5′-ATYTGNGCNG TRAAYTCRTC-3′; IV.2(→), 5′-ATGYTNCCNC ARGTNGARGC-3′; IV.2(←), 5′-GCYTCNACYT GNGGNARCAT-3′ (R is A or G, Y is C or T, and H is A, C, or T). Subcloning into the untagged and tagged E. coli expression vectors used oligonucleotides IV.3(→) (5′-CCATGCCATG GAACTCCAAA GGTTGGGTGC TCCAC-3′) and IV.3(←) (5′-GCCCAAGCTT GTGACGCGTA GTGAATCACT ACC-3′) and oligonucleotides Nde-5′-IV (GATCCATATGCAACTCCAAA GGTTGGG) and Nde-3′-IV (GATCCATATG TTCGCCCTTT GTGGGGGC), respectively. Identification of the trans-splice site was done with oligonucleotides IV.4(→) (5′-CGCTATTATT AGAACAGTTTCTGTACTATA TTG-3′ [miniexon sequence]) and IV.4(←) (5′-GGCGCCGTTG ATTGGCGTAT GC-3′). Genomic knockout constructs used oligonucleotides pLew13/NotI (CACCGCGGTG GCGGCCGC), pLew13/MluI (GATCACGCGT AAAGAAATAT TCGACCTTC), pLew13/XbaI (GATCTCTAGA CTAAGCGGTTAGTGGAGC) and pLew13/StuI (GATCAGGCCT ACCCTTATGC AAAAAAG); their PCR analysis used the latter oligonucleotide and Upper-IV (TTCTCAGCAG TACATGAAGG G).
PCRs used Taq polymerase (BRL) and hot-start amplification for genomic DNA (∼2 ng, 54°C annealing) and for subcloning (0.2 ng of template, 41°C annealings). PCR of large fragments used Taq Extender (Stratagene). Reverse transcription (RT)-PCR to identify the trans-splice site used rTth polymerase (Perkin-Elmer), ∼1 ng of poly(A)+ RNA, a 95°C hot start, and a PCR with 65°C annealing.
Subclonings were performed using standard procedures (33). The PCR product from genomic DNA, generated using oligonucleotides IV.1(→) and IV.2(←) and blunted with T4 DNA polymerase, was cloned into EcoRV-cleaved pBluescript II KS (Stratagene). RT-PCR fragments were cloned into pCR2.1 (Invitrogen). For procaryotic expression of untagged protein, cDNA clone IV.13 (see below) was amplified using oligonucleotides IV.3(→) and IV.3(←) and cloned into pTrc 99A (Pharmacia) with NcoI and HindIII (sites in the primer oligonucleotides), forming pTrc-IV. For procaryotic expression of C-terminal His6-tagged protein, cDNA clone IV.13 was amplified using oligonucleotides Nde-5′-IV and Nde-3′-IV and cloned into that site of pRSETB (Invitrogen), forming pRSETB-IV. For trypanosome expression of untagged protein, the cDNA of clone IV.13, excised with EcoRI (far 5′ end) and MluI [30 bp upstream of the poly(A) site] and BamHI-HindIII-digested pLew82 (47), were blunted and ligated. A tetracycline (TC)-responsive T7 promoter drives the expression of a phleomycin resistance marker and the test gene in the sense (piT7LigIV) or antisense (piT7αLigIV) orientation (see Fig. 6A). For trypanosome genetic knockouts, a 3.6-kb upstream BamHI fragment was first cloned from genomic DNA into that site of pBluescript II KS and partly sequenced, and then a ∼1.1-kb upstream fragment overlapping the open reading frame (ORF) was amplified using oligonucleotides pLew13/NotI and pLew13/MluI and cloned into pLew13 (48) at those sites. Next, ∼0.3 kb of the 3′ region, amplified from genomic DNA using oligonucleotides pLew13/XbaI and pLew13/StuI, was inserted between those sites. This plasmid, pLew13-IV-k/o(G418), has band IV region sequences flanking the neomycin resistance gene. pLew13-IV-k/o(hygro) was made by replacing the neomycin resistance gene with the hygromycin resistance gene from pLew128 using BamHI and SalI. Upon homologous integration, these genes should be transcribed instead of the band IV ORF from upstream band IV sequences.
FIG. 6.
Sequence comparisons of T. brucei RNA ligase. Sequence alignment of the T. brucei band IV RNA ligase protein [Tb(IV); upper lines] with a predicted protein from an ORF of L. major [Lm(IV); middle lines] and a predicted protein from an ORF of T. brucei [Tb(V?); lower lines]. Dashes indicate identity, lowercase letters indicate similarities, uppercase letters indicate nonconservative differences, and online dots indicate spaces inserted for alignment.
A cDNA library from bloodstream T. brucei rhodesiense WRATat serodeme, clone MVAT4 in λ ZAPII (12; a kind gift of J. Donelson and N. El-Sayed) was screened (Stratagene) using 12 plates (150-mm diameter). Plaques were lifted on Colony/Plaque Screen membranes (NEN Research Products), denatured (33), and cross-linked (Stratalinker; default setting). Membranes were hybridized to a random primed probe of the partial genomic clone (above) and washed (much as described in reference 33). All analyzed plaques remained positive through the tertiary screen. cDNA-containing phagemids in pBluescript were excised from λ ZAPII and grown in SOLR cells (Stratagene). The clone with the longest insert, IV.13, was sequenced. It was used to screen the GenBank and Institute for Genomic Research databases using BLAST.
E. coli expression, protein purification, adenylylation, and RNA ligase assays.
For untagged procaryotic expression, pTrc-IV transformed into E. coli HR171prr+ (a gift of L. Snyder) was induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 30°C for ∼3 h. After being washed in phosphate-buffered saline, cells were sonicated in 50 mM Tris-HCl (pH 8.0)–1 mM EDTA–3 mM DTT with protease inhibitors (described above), and cleared lysates were dialyzed against this buffer at 4°C. For His-tagged protein, 200-ml cultures of pRSETB-IV-transformed BL21(DE3) cells (Invitrogen) were induced with 1 mM IPTG at 37°C for 3 to 4 h. Cells were resuspended in 20 ml of 50 mM Tris-HCl–2 mM EDTA (pH 7.8), freeze-thawed, incubated at 22°C for 30 min with 5 mM MgCl2–100 μg of lysozyme per ml–10 U of DNaseI per ml, and then resuspended and sonicated in 6 M guanidine-HCl–20 mM sodium phosphate–500 mM NaCl (pH 7.8). A Probond nickel column (Invitrogen) in 8 M urea–20 mM sodium phosphate–500 mM NaCl (pH 7.8) was loaded with cleared lysate and washed with this buffer adjusted with HCl to pHs 7.8, 6.0, and 5.3; the pH 4.0 eluant was dialyzed into 2 M urea–25 mM Tris-HCl (pH 7.6)–5 mM MgCl2–1 mM EDTA–5% glycerol–50 mM NaCl–0.1% β-mercaptoethanol.
Adenylylation and ligase assays were performed as previously described (29), using 4 μg of E. coli extract protein, 6 μg of trypanosome extract protein, or 0.8 μg of purified protein (its 2 M urea storage buffer was diluted 10-fold); the ligase assay also used 10 to 100 fmol of 5′-end-labeled pIBI30 transcript (29; 0.5 × 105 to 1 × 105 cpm). Assays using PPi pretreatment followed by pyrophosphatase can reduce adenylylation of band IV relative to band V. Protein concentration was determined with the Bio-Rad protein assay.
Generation of transgenic trypanosome cell lines for overexpression and knockout analysis.
For trypanosome ectopic expression, piT7LigIV or piT7αLigIV (10 μg) linearized with NotI within the rRNA gene nontranscribed spacer adjoining the transgene (see Fig. 6A) was electroporated (47, 48) into 2 × 107 procyclic 29.13 cells. Line 29.13 (48), generated from T. b. brucei strain 427, expresses T7 RNA polymerase and TC repressor (TetR) along with linked hygromycin and neomycin resistance genes. At 24 h, 5 or 50 ng of TC per ml was added for low or intermediate induction and 2.5 μg of phleomycin (Kayla) per ml was added for selection; cell lines were established and then propagated without selection (47). Four lines from each transfection-induction condition were induced with 500 ng of TC per ml, and after various times, mitochondrial extract was prepared. Clones selected at the two TC levels showed similar characteristics.
For genetic knockout analysis, pLew13-IV-k/o(G418) or pLew13-IV-k/o(hygro) was cleaved with NotI and StuI to liberate the cassette and electroporated into wild-type 427 procyclic cells (48) as described above. After 24 h, G418 (15 μg/ml) or hygromycin (50 μg/ml) was added and cell lines were established (47). DNA isolated from these lines was analyzed by PCR using oligonucleotides Upper-IV (priming from genomic sequences 1.3 kb upstream of the band IV coding region, not present in the pLew 13-IV-k/o constructs) and pLew13/StuI (priming band IV 3′ untranslated region [UTR]). Amplification of the genomic locus yields a 2.9-kb product, while homologous integration of either knockout cassette yields a novel ∼6-kb product. Correct single-allele knockout lines were then electroporated with the alternate knockout construct or control constructs. Other genetic approaches (48) were also examined.
Northern blot analysis.
Poly(A)+ mRNA [10 or 30 μg, isolated using an oligo(dT) column (BRL)] was run on a 1% agarose-formaldehyde gel (33) and electroblotted onto Zetaprobe (Bio-Rad). Prehybridized membranes were treated as described above, using a random primed PCR product of the band IV ORF (described above) or the band I or band II cDNA clones (K.J.P. and M.H., unpublished data). Selective probing for endogenous band IV RNA used 42°C annealing and 5′-end-labeled TTACCCTTAT GCAAAAAAGA TGTGTTTGTGTGACGCGTAG to the 3′ UTR beyond the region present in the transgene.
RESULTS
Bands IVa and IVb are isoforms, while band V is a different RNA ligase.
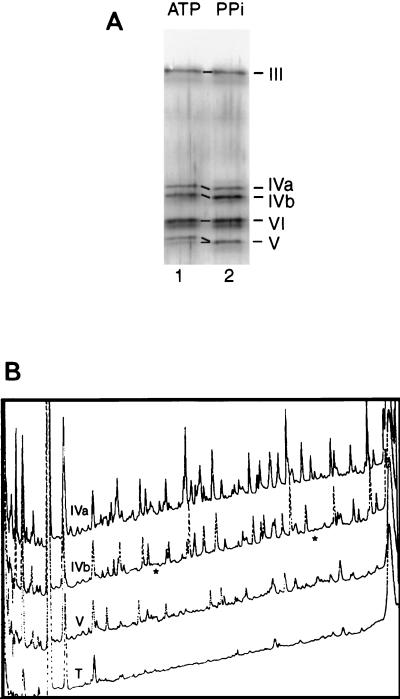
The three major adenylylatable polypeptides in mitochondrial extract of T. brucei line TREU 667, a doublet at ∼57 kDa and a single species at ∼50 kDa, represent RNA ligases. These adenylylatable polypeptides deadenylylate specifically in the presence of ligatable RNA and copurify with one another and with RNA ligation activity in all of the fractionation schemes examined (29; data not shown). Notably, they also copurify with gRNA-directed endonuclease, 3′-U-exonuclease, and terminal-U-transferase activities, as well as with catalysis of full-cycle U deletion and U insertion editing (10, 29). Purification using Q-Sepharose and DNA cellulose achieves ∼500-fold enrichment of ligase and full-cycle editing activities (10, 29) and yields only eight major silver-stainable bands (29). They are designated by apparent size, bands I through VII, with two closely migrating species that have virtually identical tryptic digest patterns (see Fig. 2B) designated IVa and IVb. Bands IVa, IVb, and V comigrate with the radiolabeled adenylylatable polypeptides, on both one- and two-dimensional gels (29). To confirm that the ligase is the observed protein and not a minor contaminant, the purified editing complex was incubated with ATP to adenylylate or PPi to deadenylylate ligases (Fig. 1B) and proteins were resolved on SDS-gels. Silver staining revealed that the electrophoretic mobility of band IV and band V, but not the other polypeptides, was altered by the treatment (Fig. 2A). Thus, the ligase proteins are the major silver-stainable species designated band IV and band V.
FIG. 2.
Polypeptides representing RNA ligase. (A) Silver stain of an SDS–8% polyacrylamide gel resolving T. brucei RNA ligase purified through Q-Sepharose and DNA-cellulose and then either adenylylated by incubation with 1 mM nonradioactive ATP (lane 1) or deadenylylated with 8 mM PPi, (lane 2). Samples were acetone precipitated prior to electrophoresis. The central portion of the gel is shown, including bands IVa, IVb, and V, which migrate faster when deadenylylated than when adenylylated, and bands III and VI, which do not change in migration upon these treatments. (In this preparation, band V was not fully adenylylated in lane 1). Bands I, II, and VII are off the top and bottom of this gel region. On such ≤8% polyacrylamide gels, band V (which stains brown and labels with [α-32P]ATP) migrates faster than band VI (which stains gray and does not label). On 10% polyacrylamide gels, the adenylylation-deadenylylation mobility difference can be readily observed for band V but not for band IV (27). (B) HPLC profile of tryptic peptides derived from bands IVa, IVb, and V and a trypsin control (T), representing elution volume on the x axis and signal intensity on the y axis. The asterisks indicate peaks present from band IVa but not from band IVb.
Following electrophoresis of the purified and deadenylylated editing complex, the excised bands were trypsin digested and resolved by HPLC. Bands IVa and IVb have virtually identical tryptic profiles (Fig. 2B) and thus are variants of one another. Band V has a very different tryptic profile (Fig. 2B) and different adenylylation properties (29; data not shown) and thus represents a distinct protein.
Band IV cDNA cloning reveals alternate trans splicing.
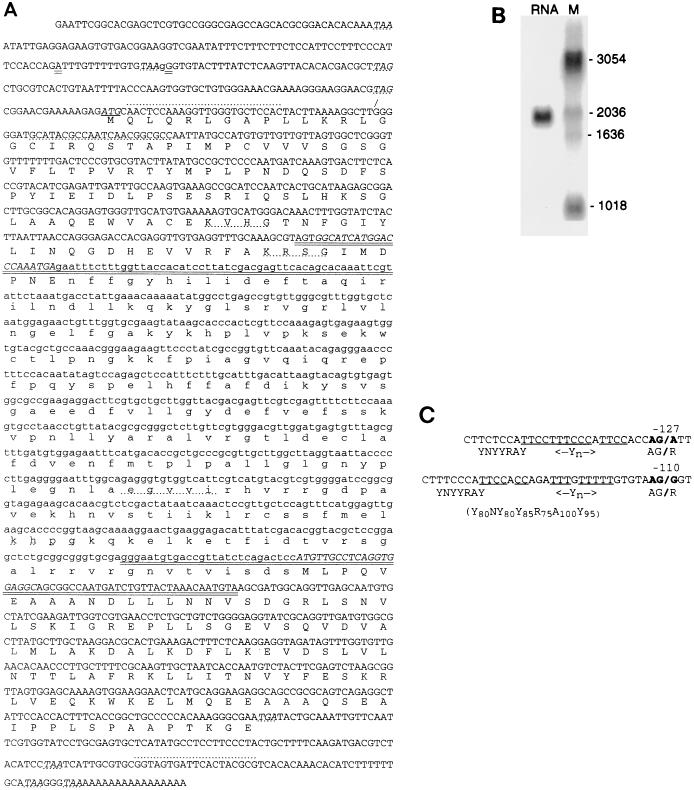
Two tryptic peptides from band IVa were subjected to Edman degradation, yielding sequences of 25 and 26 amino acids (the double-overlined amino acids in Fig. 3A). Degenerate oligonucleotides designed from their internal portions in both orientations (see Materials and Methods) were used for PCR of trypanosome genomic DNA, which should generate a contiguous protein-coding sequence since virtually no trypanosome genes contain introns. Oligonucleotides IV.1(→) and IV.2(←) generated a 777-nucleotide fragment (Fig. 3A; the new residues are in lowercase, and the primers are in italicized uppercase). The tryptic peptide sequences are in phase and not separated by termination codons. Verifying that the PCR product derives from the gene corresponding to the isolated protein, the sequences immediately interior to the primers (the lowercase double-overlined amino acids in Fig. 3A) encode the adjacent 17 and 9 amino acids of the sequenced tryptic peptides.
FIG. 3.
Nucleotide and amino acid sequences of T. brucei RNA ligase band IVa. (A) Nucleotide sequence and predicted amino acid sequence of the cloned trypanosome cDNA. The two sequenced tryptic peptides are indicated by the double-overlined amino acids. The lowercase residues correspond to the new sequences obtained from the PCR using primers IV.1(→) and IV.2(←), and the uppercase residues correspond to those additionally obtained from cDNA clone IV.13. The first AUG and in-frame termination codons are in italics with solid or dotted underlining, respectively. Other conserved amino acid motifs discussed in the text are indicated by dotted underlining. The oligonucleotides used as PCR primers the entire expressed protein region and for the 5′ region of the mRNA and are indicated by dotted overlining on lines 5 and 30 (oligonucleotides IV.3) and by dotted underlining on line 6 (oligonucleotide IV.4(←)). The latter clones showed miniexon trans splicing to either of the double-underlined purine residues on line 3 (see panel C). The longer cDNAs derived from strain TREU 667 contain a G residue just upstream from the position of the trans-splice site used in the shorter cDNA (the lowercase g residue on line 3), while cDNA IV.13 isolated from the library generated from WRATat (12) has an A at that position; a second apparent polymorphism is that the two former cDNA kinds both contain an extra GTG (valine) codon inserted after codon 14 (indicated by the diagonal overline). (B) Northern blot of 2 μg of strain TREU 667 poly(A)+ RNA probed with the PCR product generated using oligonucleotides IV.1(→) and IV.2(←). The sizes of the molecular size markers in lane M are indicated in bases on the right. (C) The two splice acceptor sites are numbered relative to the ATG initiation sequence. The miniexon sequence becomes joined to the mRNA at the indicated boldface purine, replacing the sequence upstream of the diagonal line. Also indicated are matches to the splice consensus acceptor sequence; below is shown the percent conservation of mammalian branch site consensus sequences. Y, pyrimidine; R, purine; N, is any residue.
Screening of a trypanosome cDNA library using this PCR product yielded several related clones. Double-strand sequencing of the longest cDNA, ∼1.8 kb, confirmed that it contains the PCR product and the entirety of both sequenced peptides, providing 11 amino acids of additional confirmation that the cloned gene corresponds to the purified protein (Fig. 3A). Its predicted 468-amino-acid ORF (indicated below the nucleotide sequence in Fig. 3A) begins ∼250 nucleotides from the 5′ end of this cDNA and extends to ∼100 nucleotides before the 3′ poly(A) site. In frame, four translation termination codons (dot-underlined italic residues) are upstream and four are downstream of the identified ORF; termination codons are also throughout the other two reading frames. The predicted protein is 52 kDa, close to the 57 kDa (29, 31) or 54 kDa (9) estimated from the SDS-gel mobility of the adenylylatable trypanosome protein.
Surprisingly, the 5′ end of this cDNA lacks the 39-nucleotide miniexon that is trans spliced onto all mature nucleus-encoded trypanosome mRNAs (2), even though this cDNA is of at least the expected length [Fig. 3B; band IV mRNA is ∼1.9 kb, while the cloned cDNA plus an average-size trypanosome poly(A) tail would be ∼2 kb]. This suggests that the cDNA could correspond to unspliced pre-mRNA. To determine the miniexon addition site, we performed RT-PCR of poly(A)+ RNA using a miniexon upstream primer and products of two sizes were obtained. Both extend upstream less far than the initial cDNA, with the miniexon joined to the pre-mRNA at a consensus splice acceptor site 127 or 110 nucleotides upstream of the noted ATG (the residues indicated by double underlining in Fig. 3A, line 3). Specifically, both miniexon addition sites are at a purine residue, immediately following an AG dinucleotide, preceded within a few nucleotides by a polypyrimidine tract (28 of 30 and 8 of 9 residues, respectively), preceded within 20 to 30 nucleotides by a good match to the YNYYRAY consensus branch site (Fig. 3C). Thus, band IV RNA ligase mRNA has at least two miniexon addition sites that generate the ∼1.9-kb mRNAs.
Expressed band IV protein is a mitochondrially targeted functional RNA ligase.
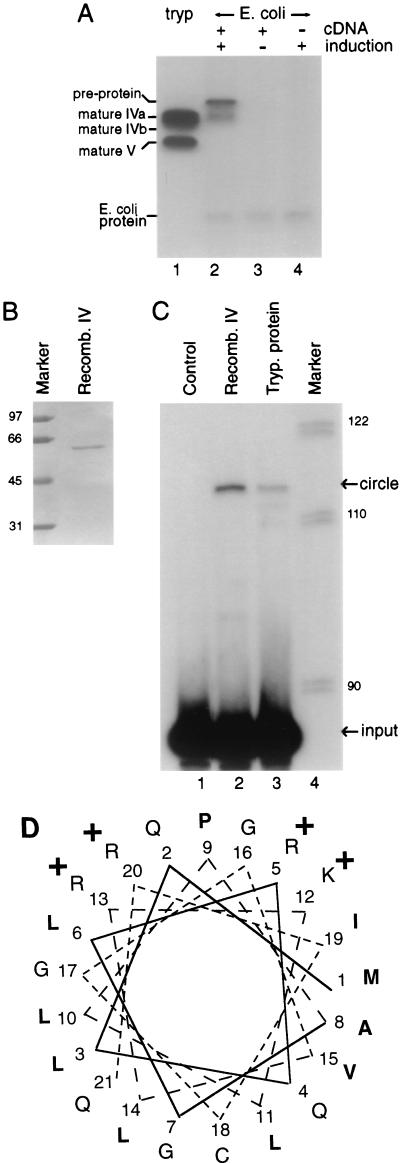
When untagged band IV was expressed in E. coli, induced cell extracts showed a novel adenylylatable protein (Fig. 4A, lanes 2 to 4). Limited RNA ligase activity could also be observed in these induced extracts, although the protein is almost entirely in inclusion bodies (data not shown). Therefore, we examined His6-tagged recombinant protein purified after guanidine solubilization of the inclusion bodies (Fig. 4B). It had substantial adenylylation and RNA ligase activity, while similarly isolated control proteins showed none (Fig. 4C and data not shown). We concluded that the cloned cDNA encodes a functional RNA ligase.
FIG. 4.
T. brucei RNA ligase expressed in E. coli. (A) Adenylylation of the trypanosome (tryp) mitochondrial editing complex showing the ∼57-kDa band IV doublet and ∼50-kDa band V (lane 1) or E. coli extracts made from cells with (lanes 2 and 3) or without (lane 4) pTrc99A containing the untagged T. brucei RNA ligase ORF and induced with IPTG (lanes 2 and 4) or uninduced (lane 3). The adenylylatable protein comigrates with the induced polypeptide observed upon silver staining (not shown). A lighter exposure of this gel shows distinct bands IVa and IVb in lane 1. (B) Coomassie stain of an SDS-gel of the nickel column-purified, C-terminally His6-tagged band IV protein expressed in E. coli. (C) RNA ligase assay using the His6-tagged band IV protein expressed in E. coli and purified on a nickel column (lane 2) using partly purified editing complex from trypanosome mitochondria (lane 3) or a buffer control (lane 1). (D) Helical-wheel representation of the predicted N-terminal amino acid sequence of T. brucei RNA ligase band IVa. Basic residues are indicated by plus signs, and hydrophobic residues are in boldface. The valine at position 15 in the clones from TREU 667 is shown. Molecular sizes (kilodaltons) of markers are shown to the left and right of panels B and C, respectively.
Figure 4A also shows that the major adenylylatable product expressed in E. coli from the untagged band IV construct, which is presumably the same as the primary trypanosome translation product, migrates to a position equivalent to that of a protein ∼2 to 3 kDa larger than the mature trypanosome band IVa ligase protein. However, when this same ORF is expressed in trypanosomes, its protein product is the size of the mature band IVa protein and is found in mitochondria (see Fig. 5B). These results suggest that the trypanosome band IV protein is cleaved to the mature size upon mitochondrial import, consistent with it naturally being a mitochondrial protein not encoded in mitochondrial DNA. Accordingly, the N-terminal portion of the predicted protein conforms to trypanosome mitochondrial signal sequences (16, 28, 42, 43), being rich in basic and hydrophobic but devoid of acidic residues and able to form an amphipathic α-helix (Fig. 4D). Therefore, the band IV primary translation product begins with a signal sequence and is the cytoplasmic precursor to band IV mitochondrial RNA ligase.
FIG. 5.
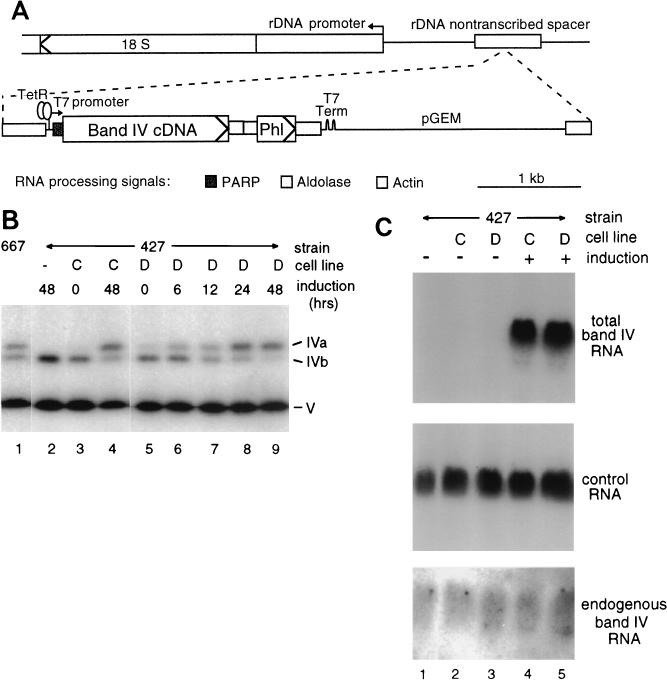
Expression of the band IV construct in trypanosomes. (A) Plasmid piT7LigIV, used for inducible overexpression of band IV cDNA in transfected trypanosomes, has a T7 promoter with a tet operator driving expression of the band IV ORF and a linked phleomycin resistance gene (Phl, the ble gene from Streptoalloteichus hindustanus) plus the indicated 5′ trans-splicing and 3′ polyadenylation signals to allow normal trypanosome mRNA processing (47). Transcription stops at the T7 terminator before reading into the pGEM vector sequences. Linearization within the adjoining segment derived from the transcriptionally silent rRNA gene (rDNA) intergenic spacer allows homologous targeting to this region of the host chromosome, with the desired ORF reading in the reverse orientation of the rRNA gene. This DNA was electroporated into procyclic cell line 29.13, which expresses T7 RNA polymerase and TetR to allow regulation of the TC-responsive T7 promoter (47). In plasmid piT7αLigIV, the band IV ORF cassette was, instead, inserted in the antisense orientation. Term, termination signal; PARP, procyclic acid repeat protein (procyclin). (B) Adenylylation assay of extracts prepared from our original cell line (lane 1, TREU 667; reference 29), from the parent cell line used for the transfections (lane 2, line 29.13, in the 427 background), and from two representative stably transformed lines expressing the cloned band IVa gene (cell lines C and D), without and at various times following TC induction. To score molecules that were already adenylylated in the extract, the assays were performed following PPi treatment (29). (C) Northern blotting of RNAs from similarly uninduced and induced cultures of the untransformed and transformed cells used a probe for total band IV mRNA (top panel), a probe for a different cloned mitochondrial cDNA (middle panel; band I; K.J.P., unpublished results), and an oligonucleotide probe specific for the endogenous band IV mRNA (bottom panel).
Total band IV protein is regulated in trypanosomes.
Band IV protein was expressed from the cloned cDNA in trypanosomes using the TC-regulatable piT7LigIV construct ectopically integrated into 29.13 cells (Fig. 5A; see Materials and Methods). These parental 29.13 cells show adenylylatable polypeptides corresponding to band IVb and band V (Fig. 5B, lane 2) but none the size of band IVa, which was present in our original TREU 667 strain (lane 1). Notably, all eight of the piT7LigIV-transformed 29.13 cell lines examined also show an adenylylatable band IVa (lanes 3 to 9; data not shown). Since its abundance increases markedly upon TC induction (lanes 3 to 9; data not shown), it is evidently encoded by the cloned cDNA. From its size, we conclude that the cloned gene encodes the band IVa isoform of this RNA ligase.
As the amount of adenylylatable band IVa protein from the cloned gene increases following induction, the amount of adenylylatable band IVb protein from the cells' endogenous gene correspondingly decreases, maintaining an approximately constant amount of band IVa plus band IVb (Fig. 5B). In contrast, the amount of adenylylatable band V RNA ligase protein, which is encoded by a different gene (Fig. 2B; also see reference 29 and Fig. 6), is unchanged upon band IVa induction (Fig. 5B). These abundances of adenylylatable bands IVa, IVb, and V are similarly observed in whole-cell extracts, mitochondrial extracts, and extracts deadenylylated prior to assay (data not shown). This demonstrates that band IVa induction affects the actual amount of band IVb protein and not merely its localization or extent of preadenylylation. Thus, the amount of total band IV ligase protein in the trypanosome is regulated to not appreciably exceed that present in the normal editing complex. Furthermore, band IV and band V RNA ligases are treated as nonequivalent species by this regulatory process.
Northern blotting was used to examine whether the observed band IV protein regulation is due to modulation in band IV mRNA abundance (Fig. 5C). A coding region probe showed that transgene RNA is induced to very high levels relative to endogenous band IV mRNA (Fig. 5C, top; long exposures reveal a band in lanes 1 to 3 that is ∼1/1,000 of the intensity seen upon induction). Yet endogenous band IV mRNA levels, specifically detected with an oligonucleotide for the distal 3′ UTR not present in the transgene (see Materials and Methods; Fig. 3B), remain constant upon induction (Fig. 5C, bottom). Controls analyzing two other mitochondrial mRNAs confirmed approximately equal loading of the lanes (Fig. 5C, middle; data not shown). Hence, the observed regulation of band IV protein occurs beyond the level of mRNA abundance, evidently translational or posttranslational.
Band IV RNA ligase is an essential gene.
If this ligase is needed for RNA editing, it should be critical in procyclic cells, which require mitochondrial genes and therefore presumably editing of their transcripts. Southern blotting indicated that the band IV gene is single copy (restriction enzymes that do not cut within the probe region generate a single band, while ones that cut once generate two bands; data not shown), so there should be two alleles in the diploid genome to knock out. Making use of the trypanosome's homologous recombination of introduced DNA (11, 19, 40), procyclic 427 cells were transfected with either of two otherwise identical constructs designed to replace a band IV coding region with a hygromycin or G418 resistance gene [from pLew13-IV-k/o(hygro) or pLew13-IV-k/o(G418); see Materials and Methods]. Individually, both of these constructs can readily replace an allele of band IV since their transfection yielded numerous cell lines and all of those analyzed showed proper integration (Table 1). In all cases, the PCR analysis (see Materials and Methods) generated the novel ∼6-kb product diagnostic of homologous integration into one band IV allele and the 2.9-kb product diagnostic of the remaining band IV allele. These resultant single-allele knockout cell lines were readily transfectable with control constructs that target the other drug resistance gene to the β-tubulin locus using plasmids pLew128 and pLew13 (Table 1). However, when we instead attempted to transfect these single-allele knockout cell lines with the alternate band IV knockout construct, only a few cell lines grew and none showed only the intended double replacement (Table 1). Critically, in multiple such second-transfection attempts, all of the resulting cell lines retained a copy of the intact band IV gene, since their PCR analysis still showed the diagnostic 2.9-kb product (Table 1). Additional analyses using other PCR primers showed that such cell lines result from events which normally occur only extremely rarely in trypanosome transfections: mistargeting of the second drug resistance cassette or acquisition of an extra copy of the locus (data not shown). These data demonstrate that true double-knockout cells are inviable. We concluded that the band IV gene is essential for procyclic trypanosomes.
TABLE 1.
Knockout analysis of band IV with different transfecting plasmids
| Recipient cell line | % of cell lines exhibiting targeted knockout
with:
|
No. of cell lines obtaineda
with:
|
||||
|---|---|---|---|---|---|---|
| pLew13-IV- k/o(hygro) | pLew13-IV- k/o(G418) | pLew13-IV- k/o(hygro) | Control construct (hygro)b | pLew13-IV- k/o(G418) | Control construct (G418)c | |
| Wild typed | 100 | 100 | 19e | 23 | 12f | 17 |
| First allele IV knockout (with G418r gene) | 0 | 4g | 24 | |||
| First allele IV knockout (with hygror gene) | 0 | 8g | 17 | |||
Number of lines obtained out of 24 possible (number of microtiter wells seeded with transfected cells).
Plasmid pLew128.
Plasmid pLew13.
Line 427.
Ten of these lines were analyzed by PCR.
Five of these lines were analyzed by PCR.
All of these lines were analyzed by PCR.
We turned to this knockout protocol because initial attempts to generate cell lines which survive on only a regulatable ectopic copy of the band IV gene (48) yielded no complete knockout lines that were appropriately down-regulatable, despite several attempts (data not shown). This protocol (48), initially demonstrated using nonessential genes, has recently been reported to be similarly unsuccessful in knocking out several other essential gene functions in procyclic trypanosomes (E. Ullu and C. Tschudi, personal communication). We had also attempted to deplete band IV through ectopic expression of antisense RNA using piT7αLigIV, but band IV protein abundance was not detectably affected and band IV mRNA decreased only very modestly, despite the antisense RNA being induced to >1,000-fold excess (data not shown).
A leishmania band IV homologue.
Only two predicted proteins with significant homology to T. brucei band IV were detected in the GenBank database. One is a previously unidentified 490-amino-acid ORF from Leishmania major (gb/AAC24666.1; chromosome 1 cosmid L5701.8) with 85% sequence identity (92% sequence similarity) to the central 306 amino acids of the T. brucei protein and >50% identity (>80% similarity) to the adjoining 40 N-terminal and 46 C-terminal amino acids (Fig. 6). The remaining N-terminal region of the predicted L. major protein, although showing virtually no primary sequence homology with the T. brucei protein, appears to be a mitochondrial targeting sequence (16, 28, 42, 43) since its first ∼17 amino acids are almost all hydrophobic or basic, could form an amphipathic helix, have no acidic residues, and begin with MRRL. This suggests that the leishmania ORF is a homologue of the trypanosome band IV RNA ligase and is also mitochondrial.
The T. brucei band V RNA ligase gene.
The other predicted protein detected in the GenBank database is a T. brucei ORF (CAB95523; chromosome 1) that has 55% identity (70% similarity) to the central 208 amino acids of band IV (Fig. 6). It also has significant homology (∼40% identity; ∼60% similarity) throughout its length. The probability that a hit of this similarity will occur by chance in the database is 10−86, so most likely they are functionally related proteins. Further suggesting that this ORF encodes a ligase is the fact that it conserves the KXXG and EGφφφ motifs (X is any residue; φ is a hydrophobic residue) that are typical of RNA ligases (36, 37), including the band IV sequences from T. brucei and L. major (underlined residues in Fig. 6). In fact, another RNA ligase is known in T. brucei. It is band V of the minimal RNA editing complex, reported to be 47 kDa (8) or 50 kDa (29, 31) and mitochondrially imported. Importantly, the protein predicted from this homologous T. brucei ORF is 416 amino acids, with a calculated molecular mass 47.5 kDa. Furthermore, its N terminus is a typical mitochondrial targeting sequence. Their common predicted length, location, and function strongly imply that this T. brucei ORF encodes band V, the other RNA ligase of the minimal RNA editing complex.
T. brucei subspecies show minimal differences in band IV.
The Institute for Genomic Research T. brucei database, representing terminal sequences of sheared DNA from T. brucei strain TREU 927, includes five fragments of band IV. They have 1 to 3% sequence difference from the band IV cDNA clone (Fig. 3) isolated from T. brucei rhodesiense (12), and most do not affect the amino acid sequence.
DISCUSSION
We report cloning the first component of the minimal trypanosome mitochondrial RNA editing complex, an essential RNA ligase. The clone's identity was verified because it encodes the entirety of both sequenced peptides from the purified protein (Fig. 3), which was confirmed to be an RNA ligase (Fig. 2A; 29), and the expressed protein is adenylylatable and catalyzes RNA joining (Fig. 4A and C). A larger primary translation product (Fig. 4A) and its N-terminal sequence are consistent with its mitochondrial targeting. Furthermore, the adenylylatable product ectopically expressed in T. brucei is mitochondrial and has the same size as the trypanosome band IVa RNA ligase (Fig. 5B). Importantly, this expressed protein coregulates with the endogenously encoded band IV RNA ligase isoform (Fig. 5B), suggesting that it becomes part of the RNA editing complex.
Genetic knockout analyses show that band IV is critical in procyclic cells (Table 1), as might be expected for a component vital in RNA editing. Either of two band IV replacement cassettes readily integrate homologously to knock out one of the diploid band IV gene copies, and the resultant single-knockout out lines were readily transfectable with analogous cassettes targeting different genetic loci (Table 1). However, in multiple controlled experiments, transfection of those single-knockout lines with the alternate band IV knockout cassettes yielded no viable products in which the remaining band IV gene was replaced (Table 1). This inability to attain double-knockout cells in light of substantial positive controls demonstrated the requirement for band IV RNA ligase.
We used this genetic approach because an alternate procedure (48) has repeatedly failed to generate cell lines which survive solely on an appropriately down-regulatable ectopic copy of several essential genes in procyclic T. brucei (Ullu and Tschudi, personal communication), including band IV (unpublished data). Furthermore, band IV protein was not diminished by ectopic expression of antisense band IV RNA, despite a ∼1,000-fold excess over endogenous band IV mRNA.
The essential nature of the band IV RNA ligase indicates that its function cannot be replaced by the band V RNA ligase. Thus, the two ligases of the editing complex serve different enzymatic and/or structural roles. Although the present data do not define these roles, separate studies using biochemical analyses and dominant negative cell lines indicate that band IV RNA ligase serves in U deletion while the band V RNA ligase serves in U insertion (C.E.H. et al., unpublished data; Cruz Reyes et al., unpublished data).
Band IV RNA ligase.
The two ∼57-kDa adenylylatable polypeptides present in the purified editing complex and crude extract of our TREU 667 cells (29) are virtually identical proteins (Fig. 2B). The few tryptic peptides that derive only from band IVa (starred positions in Fig. 2B) could reflect their small difference in length. Indicating that bands IVa and IVb represent two natural alleles, strain 427 has only the IVb variant while the transfected band IV cDNA, obtained from another trypanosome subspecies (12), generates the IVa variant (Fig. 5). Cells used in two other studies (8, 31) also appear homogenic, although for which form remains unclear. Since the two band IV isoforms transfer their adenylylated AMP to ligatable RNA with equal efficiency (29), they appear to be similarly active in RNA ligation.
Our transfections detect a regulatory process through which the trypanosome can prevent excess band IV protein from accumulating. Upon induction of the ectopic gene expressing the IVa isoform, the level of the endogenous IVb isoform proportionately decreases, keeping the total amount of band IV protein approximately constant (Fig. 5B). This compensatory reduction in the endogenously encoded ligase is selective for band IV protein and does not affect the level of the band V protein, even though it is encoded by a related gene and comprises another RNA ligase of the editing complex, indicating that this process treats these two ligases separately. Additional experiments showed that this regulation occurs at the level of band IV translation or protein stability, not at transcription or mRNA stability (Fig. 5C), and not at subcellular protein distribution or adenylylation efficiency. Such regulation might be useful for members of a multiprotein complex needed in stoichiometric amounts. Indeed, in yeast, an analogous form of regulation serves to prevent overaccumulation of ribosomal proteins; ones made in excess and not assembled into ribosomes are subject to rapid degradation (27, 44).
The T. brucei band IV RNA ligase shows >85% identity over the central 300 amino acids with the predicted protein from an unidentified ORF of L. major which also appears would mitochondrially localize (Fig. 6). Although no RNA ligase has been cloned and no RNA editing complex has been purified from leishmania, we speculate that this ORF encodes the leishmania homologue of the T. brucei band IV RNA ligase and that it is part of a mitochondrial RNA editing complex. Its relationship to an RNA ligase activity enriched from L. tarentolae (6) remains to be determined.
With the exception of the kinetoplastid RNA ligases, all other known 3′-5′ RNA ligases appear to function in tRNA remodeling. Cellular RNA ligases in many species, including yeast (references 5 and 45 and references therein), wheat germ (25), Candida albicans (4), and humans (24), act in tRNA splicing (49), while the enzyme in T4 (reference 23 and references therein) rejoins host tRNA molecules cleaved during the phage attack. In contrast, the trypanosome mitochondrial band IV and band V RNA ligases reside in a compartment where no tRNA cleavage-rejoining is known to occur and appear, instead, to rejoin pre-mRNAs after U insertions and deletions in RNA editing. Therefore, these ligases might be expected to have a different RNA specificity. Accordingly, while T4 RNA ligase joins tRNAs to pCp more efficiently than it dimerizes heterologous RNAs, the trypanosome ligases show the reverse preference (data not shown). Furthermore, while other RNA ligases are associated with a polynucleotide kinase and cyclic phosphodiesterase needed to remodel the cleaved tRNA termini, we have found no evidence of such activities with the trypanosome mitochondrial RNA ligases (data not shown). Indeed, these activities do not appear to be important in RNA editing since the gRNA-directed endonuclease, 3′-U-exonuclease, and terminal-U-transferase all generate 3′ hydroxyl and 5′ phosphate termini (Fig. 1A; 9, 20, 26, 30, 34). The disparate biological role of the trypanosome RNA ligases may help explain their low overall sequence similarity to other ligases (data not shown).
The T. brucei band IV mRNA uses either of two closely positioned trans-splice sites (Fig. 3A), both good matches to consensus splice acceptor sequences (Fig. 3C). Unlike cis-splicing sites of higher eucaryotes that are largely within protein coding regions, trypanosomes trans splice a common miniexon sequence to all nucleus-encoded pre-mRNAs within their 5′ UTRs, and thus the precise location of the acceptor site may not be critical. Other trypanosome mRNAs also utilize more than one trans-splice site (1, 22). Additionally, our trypanosome expression studies suggest that RNA transcribed from an ectopic construct can produce protein considerably less effectively than endogenous mRNA (Fig. 5B and C). This could arise since standard trypanosome expression vectors (47, 48) generate a chimeric 3′ UTR (Fig. 3A and 5A) and 3′ UTRs can affect RNA subcellular distribution (21).
Band V RNA ligase.
Intriguingly, a T. brucei ORF recently contributed from the Sanger Centre encodes a predicted protein that is >50% identical to the central region of band IV (Fig. 6). These proteins are related with a confidence over 75 orders of magnitude greater than that of the next most similar predicted sequence. This predicted protein is almost assuredly the second ligase of the RNA editing complex (29) because it is approximately the same size as the band V RNA ligase protein, conserves motifs characteristic of RNA ligases, and begins with a typical mitochondrial targeting sequence (Fig. 6). The high homology between band IV and the presumptive band V RNA ligase contrasts with their only minimal sequence homology to other known ligases (data not shown). However, the T. brucei band IV protein is markedly less similar to the presumed T. brucei band V protein than it is to the presumed L. major band IV protein, suggesting that these two RNA ligases assumed their distinct roles in editing well before these species diverged, ∼108 years ago (18). Evidence that the two RNA ligases of the RNA editing complex derived from a common ancestor in ancient times may prove useful in further understanding the evolution of RNA editing.
ACKNOWLEDGMENTS
We gratefully acknowledge John Donelson and Najib El-Sayed for providing the cDNA library, David Reim and the Wistar protein analysis facility for protein sequencing, Larry Snyder for the HR171prr+ cells, Elisabetta Ullu and Chris Tschudi for communicating to us their unpublished results, Nina Agabian for helpful discussions, and Alevtina Zhelonkina for technical assistance with Northern blots.
L.N.R. was a Howard Hughes predoctoral fellow. This work was supported by NIH grant GM34231.
The first two authors contributed equally to this work.
REFERENCES
- 1.Agabian N. Trans splicing of nuclear pre-mRNAs. Cell. 1990;61:1157–1160. doi: 10.1016/0092-8674(90)90674-4. [DOI] [PubMed] [Google Scholar]
- 2.Arts G J, Benne R. Mechanism and evolution of RNA editing in kinetoplastida. Biochim Biophys Acta. 1996;1307:39–59. doi: 10.1016/0167-4781(96)00021-8. [DOI] [PubMed] [Google Scholar]
- 3.Bakalara N, Simpson A M, Simpson L. The Leishmania kinetoplast-mitochondrion contains terminal uridylyl transferase and RNA ligase activities. J Biol Chem. 1989;264:18679–18686. [PubMed] [Google Scholar]
- 4.Baymiller J, Jennings S, Kienzle B, Gorman J A, Kelly R, McCullough J E. Isolation and sequence of the tRNA ligase-encoding gene of Candida albicans. Gene. 1994;142:129–134. doi: 10.1016/0378-1119(94)90367-0. [DOI] [PubMed] [Google Scholar]
- 5.Belford H G, Westaway S K, Abelson J, Greer C L. Multiple nucleotide cofactor use by yeast ligase in tRNA splicing: evidence for independent ATP- and GTP-binding sites. J Biol Chem. 1993;268:2444–2450. [PubMed] [Google Scholar]
- 6.Blanc V, Alfonzo J, Aphasizhev R, Simpson L. The mitochondrial RNA ligase from L. tarentolaecan join RNA molecules bridged by a complementary RNA. J Biol Chem. 1999;274:24289–24296. doi: 10.1074/jbc.274.34.24289. [DOI] [PubMed] [Google Scholar]
- 7.Blum B, Bakalara N, Simpson L. A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60:189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- 8.Corell R A, Read L K, Riley G R, Nellissery J K, Allen T E, Kable M L, Wachal M D, Seiwert S D, Myler P J, Stuart K D. Complexes from Trypanosoma bruceithat exhibit deletion editing and other editing-associated properties. Mol Cell Biol. 1996;16:1410–1418. doi: 10.1128/mcb.16.4.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruz-Reyes J, Sollner-Webb B. Trypanosome U-deletional RNA editing involves gRNA-directed endonuclease cleavage, terminal U exonuclease, and RNA ligase activities. Proc Natl Acad Sci USA. 1996;93:8901–8906. doi: 10.1073/pnas.93.17.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cruz-Reyes J, Rusché L, Sollner-Webb B. Trypanosoma bruceiU insertion and U deletion activities co-purify with an enzymatic editing complex but are differentially optimized. Nucleic Acids Res. 1998;26:3634–3639. doi: 10.1093/nar/26.16.3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eid J, Sollner-Webb B. Stable transformation of T. bruceithat occurs exclusively by homologous recombination. Proc Natl Acad Sci USA. 1991;88:2118–2121. doi: 10.1073/pnas.88.6.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Sayed N, Alarconb C, Beck J, Sheffield V, Donelson J. cDNA expressed sequence tags of T. brucei rhodesienseprovide new insights into the biology of the parasite. Mol Biochem Pharmacol. 1995;73:75–90. doi: 10.1016/0166-6851(95)00098-l. [DOI] [PubMed] [Google Scholar]
- 13.Estevez A, Simpson L. Uridine insertion/deletion RNA editing in trypanosome mitochondria—a review. Gene. 1999;240:247–260. doi: 10.1016/s0378-1119(99)00437-0. [DOI] [PubMed] [Google Scholar]
- 14.Greer C L, Peebles C L, Gegenheimer P, Abelson J. Mechanism of action of a yeast RNA ligase in tRNA splicing. Cell. 1983;32:537–546. doi: 10.1016/0092-8674(83)90473-7. [DOI] [PubMed] [Google Scholar]
- 15.Hajduk S. Defining the editing “reaction.”. Trends Microbiol. 1997;5:1–2. doi: 10.1016/S0966-842X(96)30039-5. [DOI] [PubMed] [Google Scholar]
- 16.Hausler T, Stierhof Y-D, Blattner J, Clayton C. Conservation of mitochondrial targeting sequence function in mitochondrial and hydrogenosomal proteins from the early-branching eukaryotes crithidia, trypanosoma and trichomonas. Eur J Cell Biol. 1997;73:240–251. [PubMed] [Google Scholar]
- 17.Huang J, Van der Ploeg L H T. A 5′ exoribonuclease and RNA ligase of T. brucei. Nucleic Acids Res. 1988;16:9737–9759. doi: 10.1093/nar/16.20.9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lake J, de la Cruz V, Ferrieira P, Morel C, Simpson L. Evolution of parasitism: kinetoplastid protozoan history reconstructed from mitochondrial rRNA gene sequences. Proc Nat Acad Sci USA. 1988;85:4779–47783. doi: 10.1073/pnas.85.13.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee M, Van der Ploeg L. Homologous recombination and stable transfection in the parasitic protozoan T. brucei. Science. 1990;250:1583–1586. doi: 10.1126/science.2177225. [DOI] [PubMed] [Google Scholar]
- 20.Kable M L, Seiwert S D, Heidmann S, Stuart K. RNA editing: a mechanism for gRNA-specified uridylate insertion into precursor mRNA. Science. 1996;273:1189–1195. doi: 10.1126/science.273.5279.1189. [DOI] [PubMed] [Google Scholar]
- 21.Long R M, Singer R H, Meng X, Gonzalez I, Nasmyth K, Jasen R-P. Mating type switching in yeast is controlled by asymmetrical localization of ASH1 mRNA. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- 22.Metzenberg S, Agabian N. Human and fungal 3′ splice sites are used by Trypanosoma bruceifor trans splicing. Mol Biochem Parasitol. 1996;83:11–23. doi: 10.1016/s0166-6851(96)02742-9. [DOI] [PubMed] [Google Scholar]
- 23.Penner M, Morad I, Snyder L, Kauffmann G. Phage T4-coded stp: double edged effector of coupled DNA and tRNA-restriction systems. J Mol Biol. 1995;249:857–868. doi: 10.1006/jmbi.1995.0343. [DOI] [PubMed] [Google Scholar]
- 24.Perkins K K, Furneaux H, Hurwitz J. Isolation and characterization of an RNA ligase from HeLa cells. Proc Natl Acad Sci USA. 1985;82:684–688. doi: 10.1073/pnas.82.3.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pick L, Furneaux H, Hurwitz J. Purification of wheat germ RNA ligase II: mechanism of action of wheat germ RNA ligase. J Biol Chem. 1986;261:6694–6704. [PubMed] [Google Scholar]
- 26.Piller K J, Rusché L N, Cruz-Reyes J, Sollner-Webb B. Resolution of the RNA editing gRNA-directed endonuclease from two other endonucleases of Trypanosoma bruceimitochondria. RNA. 1997;3:279–290. [PMC free article] [PubMed] [Google Scholar]
- 27.Planta R, Raué H. Control of ribosome biogenesis in yeast. Trends Genet. 1988;4:64–68. doi: 10.1016/0168-9525(88)90042-x. [DOI] [PubMed] [Google Scholar]
- 28.Roise D. Recognition and binding of mitochondrial presequences during the import of proteins into mitochondria. J Bioenerg Biomembr. 1997;29:19–27. doi: 10.1023/a:1022403604273. [DOI] [PubMed] [Google Scholar]
- 29.Rusché L, Cruz-Reyes J, Piller K J, Sollner-Webb B. Purification of a functional enzymatic editing complex from Trypanosoma bruceimitochondria. EMBO J. 1997;16:4069–4081. doi: 10.1093/emboj/16.13.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rusché L N, Piller K J, Sollner-Webb B. Guide RNA-mRNA chimeras, which are potential RNA editing intermediates, are formed by endonuclease and RNA ligase in a trypanosome mitochondrial extract. Mol Cell Biol. 1995;15:2933–2941. doi: 10.1128/mcb.15.6.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabatini R, Hajduk S L. RNA ligase and its involvement in guide RNA/mRNA chimera formation: evidence for a cleavage-ligation mechanism of Trypanosoma bruceimRNA editing. J Biol Chem. 1995;270:7233–7240. doi: 10.1074/jbc.270.13.7233. [DOI] [PubMed] [Google Scholar]
- 32.Sabatini R, Adler B, Madison-Antenucci S, McManus M, Hajduk S. Biochemical methods for analysis of kinetoplastid RNA editing. Methods Companion Methods Enzymol. 1998;15:15–26. doi: 10.1006/meth.1998.0602. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Seiwert S D, Heidmann S, Stuart K. Direct visualization of uridylate deletion in vitrosuggests a mechanism for kinetoplastid RNA editing. Cell. 1996;84:831–841. doi: 10.1016/s0092-8674(00)81062-4. [DOI] [PubMed] [Google Scholar]
- 35.Seiwert S, Stuart K. RNA editing: transfer of genetic information from gRNA to precursor mRNA in vitro. Science. 1994;266:114–117. doi: 10.1126/science.7524149. [DOI] [PubMed] [Google Scholar]
- 36.Shuman S, Ru X-M. Mutational analysis of vaccinia DNA ligase defines residues essential for covalent catalysis. Virology. 1995;211:73–83. doi: 10.1006/viro.1995.1380. [DOI] [PubMed] [Google Scholar]
- 37.Shuman S, Schwer B. RNA capping enzyme and DNA ligase: a superfamily of covalent nucleotidyl transferases. Mol Microbiol. 1995;17:405–410. doi: 10.1111/j.1365-2958.1995.mmi_17030405.x. [DOI] [PubMed] [Google Scholar]
- 38.Smith H, Gott J, Hanson M. A guide to RNA editing. RNA. 1997;3:1105–1123. [PMC free article] [PubMed] [Google Scholar]
- 39.Stuart K, Allen T, Heidmann S, Seiwert S. RNA editing in kinetoplastid protozoa. Microbiol Mol Biol Rev. 1997;61:105–120. doi: 10.1128/mmbr.61.1.105-120.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ten Ashroek A, Ouellette M, Borst P. Targeted insertion of the neomycin phosphotransferase gene into the tubulin gene cluster of T. brucei. Nature. 1990;348:174–175. doi: 10.1038/348174a0. [DOI] [PubMed] [Google Scholar]
- 41.Uhlenbeck O C, Gumport R I. T4 RNA ligase. In: Boyer P D, editor. The enzymes. XV. New York, N.Y: Academici Press; 1982. pp. 31–58. [Google Scholar]
- 42.von Heijne G. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 1986;5:1335–1342. doi: 10.1002/j.1460-2075.1986.tb04364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Heijne G. Protein sorting signals: simple peptides with complex functions. EXS. 1995;73:67–76. doi: 10.1007/978-3-0348-9061-8_4. [DOI] [PubMed] [Google Scholar]
- 44.Warner J R. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 45.Westaway S K, Belford H G, Apostol B L, Abelson J, Greer C L. Novel activity of a yeast ligase deletion polypeptide: evidence for GTP-dependent tRNA splicing. J Biol Chem. 1993;268:2435–2443. [PubMed] [Google Scholar]
- 46.White T C, Borst P. RNA end-labeling and RNA ligase activities can produce a circular rRNA in whole cell extracts from trypanosomes. Nucleic Acids Res. 1987;15:3275–3290. doi: 10.1093/nar/15.8.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wirtz E, Hoek M, Cross G. Regulated processive transcription of chromatin by T7 RNA polymerase in T. brucei. Nucleic Acids Res. 1998;26:4626–4634. doi: 10.1093/nar/26.20.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wirtz E, Leal S, Ochatt C, Cross G. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in T. brucei. Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 49.Zillmann M, Gorovsky M A, Phizicky E M. Conserved mechanism of tRNA splicing in eukaryotes. Mol Cell Biol. 1991;11:5410–5416. doi: 10.1128/mcb.11.11.5410. [DOI] [PMC free article] [PubMed] [Google Scholar]