Abstract
Slt11p is a new splicing factor identified on the basis of synthetic lethality with a mutation in the 5′ end of U2 snRNA, a region that is involved in intermolecular U2/U6 helix II interaction. Slt11p is required for spliceosome assembly. Our genetic results suggest that Slt11p is involved in the base-pairing interaction of U2/U6 helix II in vivo. We showed that the recombinant protein binds to RNAs with some degree of structural specificity. Slt11p also anneals RNA and binds to the resulting duplexes, which contain two separated helical regions. These RNA structures are reminiscent of U2/U6 helix II, which is formed concomitantly with U4/U6 stem II, and suggest that Slt11p facilitates the cooperative formation of helix II in association with stem II in the spliceosome. We show that Slt11p and Slu7p, a second-step factor, interact with each other both in vivo and in vitro and that the binding of Slu7p to Slt11p impairs the RNA-binding activity of the latter. These results suggest that the function of Slt11p is regulated by Slu7p in the spliceosome.
Pre-mRNA splicing proceeds in a large RNA-protein complex, the spliceosome. During assembly, activation, and remodeling of the spliceosome, protein factors play essential roles in establishment, maintenance, regulation, and coordination of a series of dynamic RNA-RNA interactions which are directly involved in recognition of splice sites and formation of RNA structures that are important for the two-step splicing reaction (13, 30). Among these factors, RNA-binding proteins and RNA-dependent ATPases/RNA helicases (DEXD/H proteins) have been demonstrated or implicated in several RNA conformational changes in the splicing pathway (30). In yeasts, recognition of the 5′ splice site (5′-SS) is mediated directly through its base-pairing interaction with U1 small nuclear RNA (snRNA) in the formation of commitment complexes (29). The highly conserved branch point site (BPS) is first recognized by an RNA-binding protein, BBP (1). BBP is also involved in direct interactions with Prp40p, a component of U1 small nuclear ribonucleoprotein particle (snRNP) bound at the 5′-SS, and Mud2p, another RNA-binding protein bound to the polypyrimidine tract which is important for the selection of the 3′-SS. The BPS is then recognized through direct base-pairing interaction with a region in U2 snRNA (25). One component of the U2 snRNP, Prp11p, interacts with Mud2p (2). These protein-protein interactions provide a temporal and spatial framework for a cross-intron coordination of different RNA-RNA interactions (1). The function of Prp5p (a DEAD protein) has been associated with the recognition of BPS sequence by U2 snRNA (22).
Formation of the prespliceosome (containing U1 and U2 snRNPs) is followed immediately by recruitment of the preformed tri-snRNP consisting of U4/U6.U5, in which U4 and U6 snRNAs are complexed through extensive base-pairing interactions (stems I and II) (18). A series of conformational RNA changes is triggered through displacement of U1 snRNA by U6 snRNA at the 5′-SS. Another DEAD protein, Prp28p, facilitates this process (31). Recognition of the 5′-SS involves a highly conserved ACAGAGA sequence in U6 snRNA (18). The relative position of this motif with respect to U4/U6 stem I is critical to the recognition of the 5′-SS (15). Prp8p also exerts its function at this step (14, 15). Other RNA conformational changes during activation of the spliceosome include disruption of the U4/U6 duplex and formation of intermolecular U2/U6 interactions and an intramolecular Brow stem, near the 3′ end of U6 snRNA. Some of these RNA interactions presuppose the disruption of U4/U6 duplex, since regions in U6 snRNA that form U2/U6 helix I and the Brow stem are initially paired with U4 snRNA in U4/U6 stems I and II, respectively. Formation of U2/U6 helix Ia brings the BPS (recognized by a sequence in U2 snRNA immediately adjacent to helix Ia) close to the vicinity of the 5′-SS, which is recognized by the ACAGAGA sequence in U6 snRNA, also adjacent to helix Ia (see reference 18 for a review). Two RNA-dependent ATPases, Slt22p (or Brr2p), a large DEIH protein (21, 37), and Prp2p, a DEAH protein (33), are involved in spliceosomal activation prior to the first step. While the RNA target of Prp2p is not yet clear, Slt22p has been implicated in disruption of U2/U6 helix II (37) and U4/U6 duplex (26).
Following the first step, the spliceosome undergoes remodeling in order to form RNA structures that are important for the second splicing step. Loop 1 of U5 snRNA is important for tethering the two exons (23). However, a number of trans- and cis-acting factors are involved in the selection and recognition of the 3′-SS (34). RNA structures important for the second step usually involve noncanonical tertiary interactions (9, 17, 19). Two DEAH proteins, Prp16p (7) and Prp22p (28), are required for the second splicing step.
The U2/U6 helix II interaction, between the 5′ end of U2 snRNA and the 3′ end of U6 snRNA, is highly conserved (18). In yeast an 11-nucleotide (nt) substitution in U2 snRNA that disrupts the proposed helix II interaction confers mild growth defects (10). This 11-nt substitution in U2 snRNA (see Fig. 2A) was used as the starting mutation in a genetic screen for additional splicing mutants that are synthetically lethal with it (38). This screen yielded two new factors, Slt11p, an RNA-binding protein, and Slt22p, an RNA-dependent ATPase (37), and a number of previously characterized factors, Slt15p (or Prp17p), Slt16p (or Smd3p), Slt17p (or Slu7p), and Slt21p (or Prp8p) (38). In this study, we characterized the function of Slt11p in pre-mRNA splicing. We provide both genetic and biochemical data that Slt11p is involved in the RNA base-pairing interaction of U2/U6 helix II and that Slt11p interacts with Slu7p, a factor that is required for the recognition of the 3′-SS (5). Our results indicate that the RNA-annealing and -binding activities of Slt11p, which forms a dimer, facilitate the formation of U2/U6 helix II in association with U4/U6 stem II. We suggest that U2/U6 helix II plays a regulatory role in activation of the spliceosome. Furthermore, the effects of Slu7p on Slt11p suggest that the RNA-binding activity of the latter is regulated in spliceosomal activation.
FIG. 2.
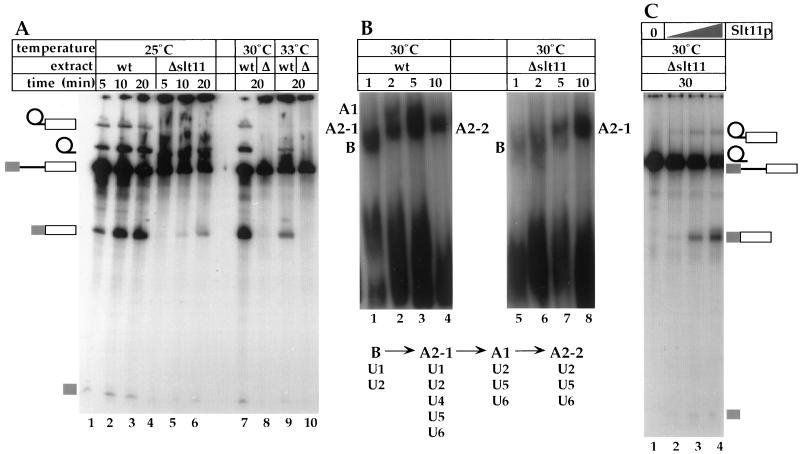
Slt11p is a splicing factor. (A) In vitro splicing assay of yeast pre-actin RNA with wild type (lanes 1 to 3, 7, and 9) and Δslt11 (lanes 4 to 6, 8, and 10) at 25°C (lanes 1 to 6), 30°C (lanes 7 and 8) and 33°C (lanes 9 and 10) for the indicated incubation times. (B) Spliceosome assembly in wild-type (lanes 1 to 4) and Δslt11 (lanes 6 to 8) extracts at 30°C and schematic of spliceosome assembly and snRNA content (based on data in reference 32). (C) Rescue of splicing activity of Δslt11 extract by recombinant Slt11p. Increasing amounts of His6-Slt11p (1, 10, and 100 ng [lanes 2, 3, and 4, respectively]) were added to a Δslt11 extract (lane 1), which was preincubated for 10 min at 25°C prior to a splicing reaction at 30°C. Note that the lariat intron ran very close to precursor RNA in this gel.
MATERIALS AND METHODS
Genetic manipulation of yeast and tests of synthetic lethality and genetic suppression.
All yeast strains were derived from W303-1A and -1B (Mata or Matα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100). Procedures for manipulation of yeast have been described by Adams et al. (3). A yeast strain containing chromosomal deletions of both U2 and U6 genes (see Fig. 3C) was described previously (38). A strain with a chromosomal deletion of SLT11 was constructed by replacing the entire open reading frame (ORF) with the yeast HIS3 gene. A yeast strain containing chromosomal deletion of U2 and U6 snRNA genes and SLT11 (see Fig. 3C) was constructed as described elsewhere (38). In all experiments, synthetic complete media were used. The plasmid shuffling method was used to test phenotypic defects of and genetic interactions between U2 and U6 snRNA mutations in SLT11 and Δslt11 backgrounds (see Fig. 3C). These strains were transformed with plasmids carrying U2 and U6 snRNA genes. The resultant transformants were grown on medium containing uracil prior to being transferred to 5-fluoroorotic acid (5-FOA) medium at 25 and 30°C. Yeast cells which were resistant to 5-FOA were then tested for additional phenotypic defects. Genetic interaction between Δslt11 and slu7-1 and slt17/slu7-100 was tested as described by Xu et al. (38).
FIG. 3.
Slt11p is involved in U2/U6 helix II interaction in vivo. (A) Inter- and intramolecular RNA interactions in the yeast spliceosome. Two conserved intron elements, the 5′-SS and the BPS (BPS int.), are recognized by U6 and U2 snRNAs, respectively. Both snRNAs can form two intermolecular base-pairing interactions (helices I and II) and an intramolecular interaction (Brow stem). (B) Substitution mutations in the helix II regions of U2 and U6 snRNAs. U2-γ is a control mutation. (C) Yeast strains with a double deletion of U2 and U6 genes (top) and a triple deletion of U2, U6 and SLT11 genes (bottom) used in genetic tests. Since SLT11 is not essential at ≤30°C, the maintenance plasmid for both strains contains only wild-type U2 and U6 genes. (D) Genetic interactions between U2 and U6 snRNA mutations in SLT11 (rows 1, 3, and 5) and Δslt11 (rows 2, 4, and 6) backgrounds. Panels a and b show 2-day growth (at 30 and 37°C, respectively) of the strain with the combination of U2 and U6 snRNA mutations in the SLT11 background on selective medium in the absence of the maintenance plasmid. Panels c and d show 2-day growth on 5-FOA-containing medium (at 25 and 30°C, respectively) of the strain with the combination of U2 and U6 snRNA mutations in the Δslt11 background. WT, wild type.
In vitro splicing assays and nondenaturing gel electrophoresis of spliceosome assembly.
Preparation of whole-cell yeast splicing extracts and in vitro splicing assays were performed as described previously (37). Synthetic 32P-labeled yeast pre-actin RNA was used in these assays and spliceosome assembly. Splicing complexes were analyzed by nondenaturing gel electrophoresis as described by Cheng and Abelson (8) and Tarn et al. (32), with modifications (37).
Site-directed mutagenesis.
Quick-change site-directed mutagenesis (Stratagene) was used to construct 5-nt, 4-nt, and substitution mutations in both U2 (U2-α, -β, and -γ) and U6 (U6-E, -F, -G, and -H) snRNA genes (see Fig. 3B). The 9-nt-substitution mutants of both U2 (U2-αβ) and U6 (U6-EF) (see Fig. 3B) mutations were constructed based on U2-α and U6-E, respectively.
Expression and purification of His-Slt11p and GST-Slu7p.
The ORF of Slt11p was cloned in pET16b (Novagen). The bacterial host cell BL21(DE3) was used to produce His6-Slt11p. Recombinant protein was found to be soluble and was thus purified using Ni2+-affinity chromatography with nickel-nitrilotriacetic acid resin (Qiagen). To construct glutathione S-transferase (GST)–Slu7p fusion protein expression vectors, the entire SLU7 ORF was cloned in pGEX-4T-1 (Pharmacia Biotech). The host XL1-Blue cells (Stratagene) were transformed with the GST-Slu7p constructs and pGEX-4T-1. GST-Slu7p protein (soluble) and GST were purified using chromatography affinity with glutathione Sepharose 4B (Pharmacia Biotech). RNase A and DNase I were added to both His6-Slt11p and GST-Slu7p samples prior to affinity chromatography to remove nucleic acids. All the purified proteins were dialyzed against 1× ACB (10 mM HEPES [pH 7.6], 100 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, and 10% [vol/vol] glycerol). The His6-Slt11p was used to produce rabbit antibodies at Comparative Animal Study Center of University of Toronto.
RNA-binding assay.
T7 templates for various RNA substrates were constructed by annealing two DNA oligonucleotides containing the promoter sequence of T7 RNA polymerase. Synthetic 32P-labeled RNA substrates were made from these DNA templates using T7 RNA polymerase with [α-32P]UTP. Both labeled and unlabeled substrates were gel purified. The RNA-binding assay was performed in a solution containing 1× ACB and 0.1% Triton in a final volume of 10 μl. Appropriate amounts (5 to 100 ng) of His6-Slt11p were mixed with 32P-labeled RNA (approximately 1 fmol, with different amounts of unlabeled RNA, if applicable), and the mixtures were kept at 25°C for 20 min and transferred to ice for another 20 min. The protein-RNA mixtures were then loaded directly on nondenaturing gels (4% acrylamide-bisacrylamide [60:1], 0.5× Tris-borate-EDTA [pH 7.5]) and run (in 0.5× Tris-borate-EDTA and 5 μM β-mercaptoethanol) for 2 h at 150 V at 4°C.
Glycerol gradient sedimentation.
A total of 100 μg of His6-Slt11p was layered onto a 10-to-40% glycerol step (5% increments) gradient (in 1× ACB) which was subsequently centrifuged at 46,000 rpm for 16 h in an SW50 Ti rotor (Beckman). Fractions (approximately 100 μl each) from the gradients were collected and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by blotting with rabbit anti-Slt11p antibodies and mouse anti-rabbit immunoglobulin G conjugated to horseradish peroxidase using an enhanced chemiluminescence kit (Kirkegaard & Perry Laboratories). For size standards, parallel gradients were run on samples containing bovine serum albumin (BSA) (68 kDa), albumin (45 kDa), and cytochrome c (12.5 kDa).
RNA-annealing assay.
Approximately 1 fmol of 32P-labeled RNA was mixed with various amounts of unlabeled RNA and various amounts of His6-Slt11p in a final volume of 10 μl (1× ACB), followed by 20 min of incubation at 25°C; 5 ng of proteinase K (with 0.1% SDS, final concentration) was then added to the mixture and incubated for another 15 min (after complete digestion of His6-Slt11p, as determined by immunoblotting [data not shown]). The resulting mixtures were immediately loaded on nondenaturing gels (6%; see above) and run for 2.5 h at 200 V at 4°C.
Affinity chromatography.
Recombinant GST-Slu7p and GST were incubated (for 60 min) with 200 μl of glutathione Sepharose 4B (50% slurry, equilibrated with 1× ACB containing 100 mM NaCl [1× ACB-100]) at final (ligand) concentrations of 1, 2, and 4 μg/μl (for GST-Slu7p) and 4 μg/μl (for GST). Minicolumns were assembled with proteins bound to the Sepharose. After one wash with 100 μl of 1× ACB-100), 100 μg of His6-Slt11 (mixed with 50 μg of BSA in 200 μl of 1× ACB-100) was applied to each column. The flowthrough was collected. After washes with 500 μl of 1× ACB-100, each column was eluted with 200 μl of 1× ACB with 500 mM NaCl, followed by 200 μl of 1× ACB with 1,000 mM NaCl and 200 μl of 1× ACB-100 with 1% SDS. Samples corresponding to 1% of the loading sample, 5% of the flowthrough fraction, and 10% of eluates were analyzed by SDS-PAGE followed by immunoblotting.
RESULTS
Slt11p is a new splicing factor involved in spliceosome activation.
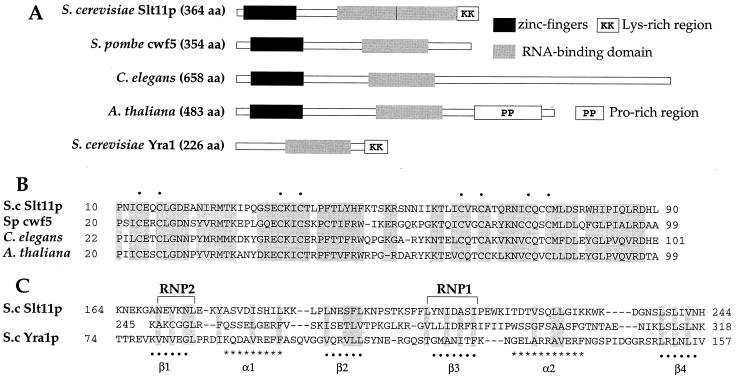
SLT11 (YBR065) was identified on the basis of synthetic lethality with a mutation near the 5′ end of the U2 snRNA that disrupts U2/U6 helix II (38). The protein can be divided into three regions (Fig. 1A). The N-terminal region (amino acids 1 to 150) contains two conserved zinc finger motifs that are found in three other proteins from Schizosaccharomyces pombe (cwf5), Caenorhabditis elegans, and Arabidopsis thaliana and several expressed sequence tags in the databases (Fig. 1B and data not shown). The cwf5 protein was identified on the basis of its association with a large complex that is involved in cell cycle control and pre-mRNA splicing (20). Slt11p contains two regions which are reminiscent of the RNA-binding domains (RBDs) of a class of RNA-binding proteins (6). However, the two putative RBDs of Slt11p contain divergent RNP1 and RNP2 subdomains that show homology to each other and to Yra1p (Fig. 1C), which possesses RNA-annealing activity (24). SLT11 was also identified as ECM2, which, in its mutant form, confers hypersensitivity to Calcofluor white, an indication of defects in cell surface biosynthesis and architecture (16). However, it is unclear whether its effect on the functions of extracellular matrix is direct or indirect.
FIG. 1.
Slt11p protein sequence and motifs. (A) Schematic of Slt11p and related proteins from S. pombe cwf5 (AL023592), C. elegans (Z69384), A. thaliana (AC004561), and S. cerevisiae Yra1p (U72633). (B) Alignment of the Slt11p zinc fingers with other proteins with similar motifs. Dots indicate conserved cysteine residues. (C) Alignment of the two potential RBDs in Slt11p with the RBD in Yra1p. The RNP core and other structural motifs are shown according to the work of Burd and Dreyfuss (6). In panels B and C, the conservation at a particular position among the aligned proteins is indicated by grey shading.
SLT11 is not essential for cell viability at ≤30°C. However, a chromosomal deletion of the entire ORF (Δslt11) confers temperature-sensitive growth at ≥33°C (data not shown). Although a splicing extract made from Δslt11 cells was active at 25°C, the overall splicing activity was significantly impaired (Fig. 2A, lanes 4 to 6), suggesting that Slt11p is required for maximum efficiency of splicing. The Δslt11 extract was completely inactive at ≥30°C (Fig. 2A, lanes 8 and 10). Spliceosome assembly at 30°C was assessed using native gel electrophoresis as described by Cheng and Abelson (8). Spliceosome assembly in the wild-type extract was largely consistent with kinetics described in references 8 and 32: complex B was detected in the initial stage of assembly (Fig. 2B, lane 1), followed by transient appearance of complexes A2-1 and A1 (Fig. 2B, lanes 2 and 3). After 10 min of incubation, accumulation of active spliceosome (i.e., complex A2-2, which ran with a mobility similar to that of complex A2-1 [8]) was observed (Fig. 2B, lane 4). The Δslt11 extract was defective in spliceosome assembly at 30°C: assembly of complex B was delayed, and complex A1 was not detected (Fig. 2B, lanes 5 to 8). The complex that accumulated after 10 min of incubation was likely A2-1 (Fig. 2B, lane 8). Similar results were observed with the wild-type extract at 25°C, whereas minimum formation of complex A1, in addition to A2-2, was detected in the Δslt11 extract (data not shown). The transition from complex A2-1 (containing all five spliceosomal snRNAs) to complex A1 (functional spliceosome, containing U2, U5, and U6 snRNAs) represents activation of the spliceosome (32). Our preliminary results suggest that Slt11p is required for spliceosome assembly, in particular, activation of the spliceosome.
Recombinant His6-Slt11p was produced in Escherichia coli and purified by Ni2+ affinity chromatography (data not shown). When added to the Δslt11 extract, the recombinant protein was able to rescue the splicing activity at 30°C (Fig. 2C, lanes 2 to 4), suggesting that the defect associated with the Δslt11 extract can be attributed solely to genetic depletion of Slt11p and that the recombinant protein is functionally competent.
Slt11 is involved in the RNA base-pairing interaction of U2/U6 helix II in vivo.
We showed previously that slt11-1 is synthetically lethal with mutations in the helix II region of both U2 and U6 snRNAs (38). A genetic approach was used to explore the role of SLT11 in the helix II interaction (Fig. 3). The effects on growth of partial disruption of helix II by substitutions of 5 nt (U2-α and U6-E) and 4 nt (U2-β and U6-F) in both snRNAs (Fig. 3B) were tested in SLT11 and Δslt11 yeast strains (Fig. 3C). None of these helix II mutations was found to confer temperature-sensitive or cold-sensitive defects by itself in the SLT11 background (Fig. 3D, panels R1/C1, R1/C2, R1/C3, R3/C1, and R5/C1; also data not shown). However, when tested in the Δslt11 background, each conferred synthetic lethality (Fig. 3D, panels R2/C2, R2/C3, R4/C1, and R6/C1 [partial lethality]), whereas a 3-nt substitution in a region outside helix II in U2 snRNA (U2-γ) showed no such lethality (Fig. 3D, panel R2/C5). In the Δslt11 background the synthetic lethality of both U6-E and U6-F was suppressed fully by U2-α and U2-β, respectively (Fig. 3D, R4/C2 and R6/C3); both combinations restored the helix II base-pairing interaction. This genetic complementation is allele specific; i.e., the other two combinations, U6-E–U2-γ and U6-F–U2-α, remained lethal in the Δslt11 background (Fig. 3D, panels R4/C3 and R6/C2, ). The control mutation, U2-γ, was unable to complement U6-E (Fig. 3D, panel R4/C5) but partially suppressed U2-F (Fig. 3D, panel R6/C5); this is probably due to extension of helix II by 3 bp in U2 (GGA) and U6 (UUU) (Fig. 3D, panel R6/C5). The suggestion that Slt11p is involved in the helix II base-pairing interaction was supported further by the observation that none of the combinations of U2-α, -β, and -γ with U6-E and -F mutations in the SLT11 background conferred detectable growth defects (Fig. 3D, panels R3/C2, R3/C3, R5/C2, and R5/C3).
When U2-α and U2-β mutations were combined, the resulting 9-nt substitution, U2-αβ, conferred a growth defect in the SLT11 strain at 37 and 16°C (10) (Fig. 3D, panel R1/C4) and was synthetically lethal with Δslt11 (Fig. 3D, panel R2/C4). The synthetic lethality was suppressed by U6-E (Fig. 3D, panel R4/C4), although not by U6-F (Fig. 3D, panel R6/C4). The corresponding mutation in U6 snRNA, U6-EF, in contrast, conferred lethality by itself (10). This phenotypic asymmetry is due to an additional intramolecular RNA-RNA interaction in which the helix II region of U6 snRNA is involved (unpublished observations). We note that of four combinations in which helix II is partially disrupted by 4 bp, i.e., U2-β–U6-wt (Fig. 3D, panel R2/C3), U2-wt–U6-F (panel R6/C1), U2-αβ–U6-E (panel R4/C4), and U2-γ–U6-F (panel R6/C5), three (all but the first) resulted in partial synthetic lethality with Δslt11. This indicates that the remaining RNA base-pairing interaction in these three cases may be sufficient, to various extents, to maintain the helix II interaction in the absence of Slt11p.
Slt11p binds to RNA in vitro.
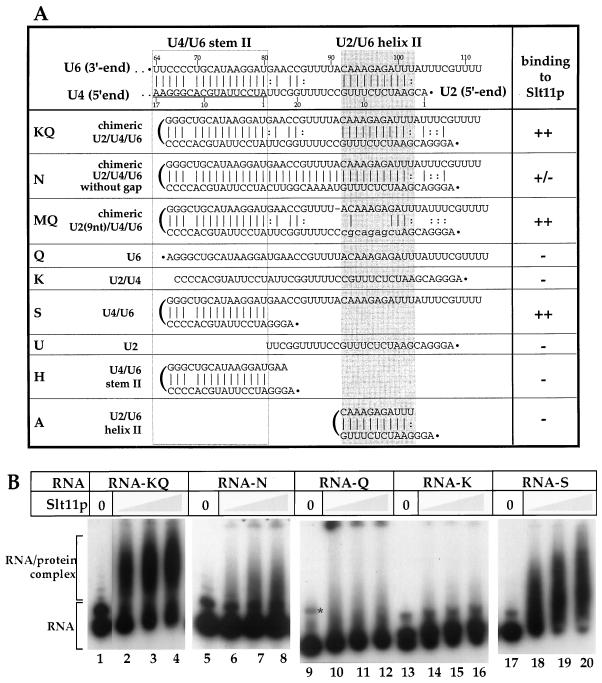
The genetic results (Fig. 3D) suggest that Slt11p is involved in the base-pairing interaction of U2/U6 helix II. We tested binding of the recombinant Slt11p to a synthetic RNA that corresponds to U2/U6 helix II (RNA-A) (Fig. 4A) but were unable to detect any significant binding activity by gel mobility shift (data not shown). Instead, we found that Slt11p binds to a synthetic RNA that contains two separate stems (Fig. 4B, lanes 2 to 4, and data not shown). These results suggested that Slt11p does not bind to helix II per se but that it binds to helix II in the presence of another stem or helix. We tested this hypothesis with an array of RNAs in which components of U2/U6 helix II and U4/U6 stem II were contained either in a single covalently linked molecule or in separate molecules (Fig. 4A).
FIG. 4.
RNA-binding activities of Slt11p. (A) Summary of RNA-binding activities of Slt11p to different synthetic RNAs. Shown on the top is the base-pairing interaction between the 3′ end of U6 snRNA with the 5′-end regions of U2 and U4 (underlined) snRNAs. The U2/U6 helix II-corresponding interaction is indicated by a grey box and U4/U6 stem II is indicated by an open box. All the synthetic RNAs were made according to the sequence shown on the top, and potential secondary structures are drawn on the basis of primary sequence. The 5′ end is indicated by a dot. ++, binding; +/-, weak binding; −, no binding. (B) RNA-binding activities of Slt11p to RNA-KQ, -N, -Q, -K, and -S. Three amounts (5, 10, and 20 ng) of Slt11p were tested for binding to 32P-labeled RNA (approximately 1 fmol). In the absence of Slt11p, all native RNAs ran as two bands in a nondenaturing gel. They likely represent different RNA conformations. The asterisk in lane 9 indicates a small portion of RNA-Q with a different conformation resulting from repeated freezing and thawing.
RNA-KQ (Fig. 4A) contains sequences derived from the 5′ end of U2 snRNA (nt 1 to 24), the 5′ end of U4 snRNA (nt 1 to 15), and the 3′ end of U6 snRNA (nt 66 to 112), thus corresponding to U2/U6 helix II and U4/U6 stem II (separated by an unpaired region). Binding of [32P]RNA-KQ to Slt11p was observed (Fig. 4B, lanes 1 to 4). This binding was competed efficiently by unlabeled cognate RNA (data not shown). When base pairing was introduced in the central region that separated the two helical elements (Fig. 4B) (RNA-N), Slt11p binding was almost entirely abolished (Fig. 4B, lanes 5 to 8). However, the individual introduction of 9-nt substitutions into the U2- or U6-corresponding portions of the chimeric RNA (e.g., RNA-MQ) (Fig. 4A) had a relatively small effect in reducing the binding of Slt11p (data not shown). For further characterization of the RNA requirements for Slt11p recognition and binding, RNA-KQ was divided into several constituent components (Fig. 4A). Neither RNA-Q nor -K, corresponding to the individual U6 and U2/U4 portions of RNA-KQ, respectively, bound to Slt11p sufficiently to be detected by gel shift (Fig. 4B, lanes 9 to 16). On the other hand, Slt11p bound nearly as well to RNA-S (corresponding to U4/U6 stem II and to the 3′ end of U6 snRNA) (Fig. 4B, lanes 17 to 20). Slt11p did not bind to RNA-U (corresponding to the 5′ end of U2 snRNA) (data not shown). The U4/U6 stem II portion of RNA-KQ and the U2/U6 helix II portion (RNA-H and -A, respectively) (Fig. 4A) alone also failed to bind to Slt11p (data not shown).
Although Slt11p contains domains that are characteristic of RNA-binding proteins (Fig. 1C), it does not bind to RNA nondiscriminatingly. Slt11p does not bind to RNA-N, -Q, -K, -U, -H, and -A (Fig. 4A), some of which may form single helices (RNA-A and H). It binds strongly to RNAs that contain two helical regions (RNA-KQ) (Fig. 4A) or one helical region with another unpaired sequences (RNA-MQ and -S) (Fig. 4A). Although the identity of nucleotides in the helical region(s) is not important for Slt11p binding (unpublished observations), the region that separates two helices seems to be important for Slt11p binding (Fig. 4B, lanes 5 to 8). These results suggest some degree of (structural) specificity for the RNA-binding activity of Slt11p. It is possible that Slt11p binds to helix II in the context of another helical element, i.e., U4/U6 stem II (Fig. 5A).
FIG. 5.
RNA-annealing activities of Slt11p. (A) Schematic of the U2/U6 helix II interaction in association with U4/U6 stem II in spliceosome assembly. (Top) While the 5′-SS and the BPS are recognized by conserved elements in U6 (black box) and U2 (grey box) snRNAs, the 5′ end of U2 snRNA can interact with the 3′ end of U6 snRNA, forming helix II, prior to the disruption of U4/U6 stem II. (Bottom) Concomitant formation of U2/U6 helix II and U4/U6 stem II. (B) Schematic of RNA-annealing assay. Slt11p was first mixed with 32P-labeled RNA (black) and unlabeled RNA (grey) and kept at 25°C for 20 min; then proteinase K was added, and incubation was continued for another 15 min. The resulting RNA duplex was then resolved on a nondenaturing gel. (C) Annealing of RNA-Q–RNA-K duplex. [32P]RNA-Q was mixed with increasing amounts of unlabeled RNA-K in the presence or absence of Slt11p in the annealing assay. The two controls were [32P]RNA-Q alone and mixed with Slt11p. Increasing amounts of unlabeled RNA-K were mixed with [32P]RNA-Q in the thermal annealing reaction (65°C for15 min → 42°C for15 min → 25°C) (lanes 8 to 11). The RNA sequences and potential base pairing are shown at the bottom. (D) Competition of annealing activities. Increasing amounts of unlabeled RNA-R and -Q or excess amounts of yeast tRNA (t) or control RNA (transcribed from pBluescript) (p) were added to the annealing assay (with [32P]RNA-Q, RNA-K and Slt11p) in the beginning. Lanes 2 to 6, 20-min annealing assay with increasing amounts of Slt11p; lane 1, [32P]RNA-Q alone.
Slt11p promotes efficient annealing of cRNAs in vitro.
During spliceosome assembly, U4 and U6 snRNAs are recruited into the prespliceosome as a duplex. It is thus unlikely that U4/U6 stem II is formed de novo in the spliceosome. However, U2 snRNA binds to the BPS independently of U4/U6 stem II. It is possible that U2/U6 helix II may form prior to disruption of U4/U6 stem II (Fig. 5A). One possible role for Slt11p, as suggested by genetic suppression (Fig. 3) and in vitro RNA binding (Fig. 4), is to facilitate the efficient formation of helix II in association with U4/U6 stem II. We determined if Slt11p is able to anneal cRNAs that contain these two elements.
The two RNAs used in the annealing assay, K and Q, can form two intermolecular helical structures corresponding to U2/U6 helix II and U4/U6 stem II (Fig. 4A). In this assay, [32P]RNA-Q was mixed with unlabeled RNA-K and Slt11p. Proteinase K was added following 20 min of incubation to remove Slt11p. The formation of RNA duplexes was determined by nondenaturing gel electrophoresis (Fig. 5B). When increasing amounts of RNA-K were mixed with [32P]RNA-Q in the presence of 20 ng of Slt11p, efficient annealing was observed (Fig. 5C, lanes 3 to 6). Although a small amount of spontaneous annealing was observed at a K/Q ratio of 4:1 (Fig. 5C, lane 7), at a K/Q ratio of 2:1, in the presence of Slt11p, more than 95% of [32P]RNA-Q was converted to the Q-K duplex (Fig. 5C, lane 5). This annealing was nearly complete within 5 min of incubation with Slt11p (data not shown). The RNA annealing promoted by Slt11p was significantly more efficient than thermal annealing (Fig. 3B, compare lanes 9 through 11 with lanes 3 through 6; also see Fig. 5D, lanes 3 to 6). This RNA-annealing activity was confirmed by strong competition for formation of the duplex by unlabeled cognate RNA-Q, mixed at the start of the assay (Fig. 5D, lanes 10 to 12). RNA-R, which contains a 9-nt substitution in the U2-corresponding region (Fig. 5D, lanes 7 to 9), was a marginally poorer competitor than RNA-Q. Nonspecific RNAs did not compete (Fig. 5D, lanes 13 and 14, and data not shown). Additional experiments indicated that the RNA-annealing activity of Slt11p requires a minimum of 11 consecutive base pairs and that this activity is not sequence specific (unpublished observations).
We used four pairs of RNAs in various combinations. They represent wild-type helix II (RNA-K–RNA-Q), disrupted helix II due to a 9-nt substitution in either U2 snRNA (RNA-M–RNA-Q) or U6 snRNA (RNA-K–RNA-R), and restored helix II (RNA-M–RNA-R) (Fig. 6A). The intermolecular interaction corresponding to U4/U6 stem II in all four cases remained intact. The presence of this interaction was preponderant on the overall annealing activities of Slt11p (Fig. 6B, compare lanes 7 through 9 and lanes 13 through 15 with lanes 3 through 5 and lanes 17 through 19). When [32P]RNA-Q was mixed with RNA-M, corresponding to disrupted U2/U6 helix II interaction caused by the 9-nt substitution in U2 snRNA, the annealing efficiency was reduced marginally (Fig. 6B, lanes 7 to 9). Furthermore, three complexes, a, b, and c, were formed by Slt11p (Fig. 6B, lanes 7 to 9). These complexes probably represent RNA-M–RNA-Q duplexes with different conformations. They were largely eliminated when helix II-corresponding interaction was restored (Fig. 6B, lanes 17 to 19). These results suggest that the 9-nt substitution in U2 snRNA not only disrupts the U2/U6 helix II interaction but also introduces a conformational change(s) into the overall RNA structure(s) in vivo.
FIG. 6.
Characterization of the RNA-annealing and -binding activities of Slt11p. (A and D) Sequences and potential base-pairing of synthetic RNAs used in panels B and C and panels E and F, respectively. (B and E) Annealing of RNAs by Slt11p. Approximately 1 fmol of 32P-labeled RNAs was mixed with increasing amounts of unlabeled RNA and 10 ng of Slt11p in the RNA-annealing assay (Fig. 5C). The resulting RNA duplexes, after proteinase K digestion, were analyzed in 6% nondenaturing gels. The controls include [32P]RNA alone, with 10 ng of Slt11p, and with unlabeled RNAs in the absence of Slt11p. a, b, and c (panel B, lanes 7 to 9) indicate the complexes formed between RNA-Q and -M with altered configurations. In panel E, U′ indicates RNA-U with a different conformation and mobility. (C and F) Binding of Slt11p to RNA duplexes formed in B and E, respectively. Approximately 1 fmol of 32P-labeled RNAs was mixed with increasing amounts of unlabeled RNAs and 100 ng of Slt11p. The mixtures were first incubated at 25°C for 20 min and kept on ice for another 20 min prior to loading onto 4% nondenaturing gels. The controls include [32P]RNAs with unlabeled RNAs only and with 100 ng of Slt11p only. Note that RNA-K and -M ran as triplets and doublets, respectively, on nondenaturing gels. The asterisk indicates a protein-RNA-RNA ternary complex. In panel C, the arrow indicates an RNA duplex (lanes 1 to 4) observed in lanes 3 to 6 in panel B. Similar duplexes were not observed in lanes 7 to 9, 12 to 14, or 17 to 19, due partially to smearing. The formation of the Slt11p–RNA-M–RNA-R ternary complex is further indicated by the reduction of free RNA-M (lanes 17 to 19).
We sought to determine if Slt11p is able to bind to RNA duplexes that resulted from its own RNA-annealing activity. Although neither RNA-K nor RNA-Q bound individually to Slt11p (Fig. 4B, lanes 9 to 16), they formed an RNA duplex in the presence of Slt11p (Fig. 6B, lanes 3 to 6). When the labeled RNA-K or -M was mixed with unlabeled RNA-Q and Slt11p, large amounts of labeled RNAs were trapped in the wells (Fig. 6C, lanes 2 to 4, 7 to 9, 12 to 14, and 17 to 19). This is likely due to aggregation. However, formation of a possible protein-RNA-RNA tripartite complex (at a relatively low efficiency) was observed when [32P]RNA-K, RNA-Q, and Slt11p were mixed (Fig. 6C, lanes 2 to 4), since it did not occur in the absence of either Slt11p (Fig. 6C, lane 1) or unlabeled RNA (Fig. 6C, lane 5). RNA molecules in which the U2/U6 helix II-corresponding interaction was disrupted, [32P]RNA-M and RNA-Q, were unable to form this complex (Fig. 6C, lanes 7 to 9). Failure of RNA-M–RNA-Q and Slt11p to form this ternary complex is likely due to the alternative conformations introduced by the mutation in RNA-M (Fig. 6B, lanes 7 to 9). Consistent with this explanation, we observed that restoration of the helix II-corresponding interaction in RNA-M–RNA-R duplex, which eliminated complexes a and b (Fig. 6B, lanes 17 to 19), restored (at least partially) the formation of stable RNA-M–RNA-R–Slt11p complex (Fig. 6C, lanes 17 to 19). On the other hand, when the helix II-corresponding interaction was disrupted by mutation in the U6-corresponding RNA (RNA-R), no stable tripartite complex was observed (Fig. 6C, lanes 12 to 14).
We then tested another set of RNAs for the annealing activity of Slt11p (Fig. 6D and E). RNA-S contained a helical element corresponding to U4/U6 stem II and another region corresponding to the 3′ end of U6 snRNA (Fig. 6D). When it was mixed with labeled RNA-U, corresponding to the 5′ end of U2 snRNA, in the presence of Slt11p, a stable duplex was formed (Fig. 6E, lanes 3 to 5). However, when the helix II-corresponding interaction was disrupted by the 9-nt substitution in U6 snRNA (in RNA-T), such an RNA duplex was not detected (Fig. 6E, lanes 7 to 9). These results indicate that Slt11p is able to form a helical interaction (corresponding to U2/U6 helix II) in the presence of another preformed helical element (corresponding to U4/U6 stem II). Furthermore, a tripartite Slt11p–RNA-S–RNA-U complex was detected when the two RNAs were present in comparable amounts (i.e., at an S/U ratio of 1/1) (Fig. 6F, lane 3). At a higher S/U ratio, most of the labeled RNA-U was trapped in the wells (Fig. 6F, lanes 4 and 5). This aggregation might be caused by the binding of Slt11p to RNA-S (Fig. 4B, lanes 18 to 20, Fig. 4B) and annealing of labeled RNA-U to excess RNA-S associated with Slt11p. However, when the helix II-corresponding interaction was disrupted, only a small amount of tripartite Slt11p–RNA-T–RNA-U was detected in the presence of excess RNA-T (unlabeled) (Fig. 6F, lanes 8 to 10). The RNA substrates shown in Fig. 6D can form two helical elements in a different configuration than those shown in Fig. 6A. In both configurations, Slt11p is able to anneal two RNAs and to bind (under the experimental conditions tested) to the resulting duplex that contains two helical elements. The integrity of both helical elements seems to be important for the Slt11p-RNA interaction.
Slt11p forms a homodimer in the absence of RNA.
The formation of Slt11p-RNA-RNA ternary complexes (Fig. 6C) that contain two separated helical elements suggests that Slt11p may act as a dimer, with each subunit interacting with one of the two elements. We used glycerol gradient sedimentation to determine if the recombinant Slt11p forms a dimer. We used a mixture of three proteins (BSA, albumin, and cytochrome c) as molecular weight markers. Following centrifugation, the sedimentation profile indicated that these proteins are sufficiently separated to allow detection of an Slt11p dimer. Slt11p was found in two locations of the gradient, most likely representing, according to molecular weights, monomer (fractions 30 to 34) and dimer (fractions 20 to 22) forms (Fig. 7). We conclude that Slt11p forms a homodimer in the absence of RNA. It is possible that in the spliceosome, one subunit of Slt11p dimer binds to U4/U6 stem II, while the other subunit (with its RNA annealing activity) promotes the formation of U2/U6 helix II. The Slt11p dimer then binds to the resulting U2/U6/U4 complex (see Discussion).
FIG. 7.
Slt11p forms a homodimer in vitro in the absence of RNA. An immunoblot of fractions from the glycerol gradient (10 to 40%) sedimentation of Slt11p probed with rabbit anti-Slt11p antibodies is shown.
Slu7p interacts with Slt11p and impairs the RNA binding but not the annealing activities of Slt11p.
We showed previously that the original slt11-1 allele is synthetically lethal with several factors involved in the second splicing step, including Slu7p (38). Chromosomal deletion of SLT11 also caused synthetic lethality with slu7-1 and slt17/slu7-100 (data not shown). In order to determine direct protein-protein interaction, GST-Slu7p was produced in E. coli and purified (data not shown). We used affinity chromatography to determine if Slt11p binds to Slu7p in the absence of RNA (see Materials and Methods). While it was not detected in the 0.5 M and 1.0 M NaCl eluates of the GST column (Fig. 8A, lanes 15 and 16), Slt11p, bound to GST-Slu7, was detected in these eluates of GST-Slu7p columns with ligand concentrations of 4 and 2 μg/μl (Fig. 8A, lanes 3, 4, 7, and 8). At a lower ligand concentration (1 μg/μl), most of the bound Slt11p was eluted with 0.5 M NaCl (Fig. 8A, lanes 11 and 12). These results demonstrate a direct Slt11p-Slu7p binding that is ligand concentration dependent and susceptible to high salt concentrations. Mutations in the Zn finger region of Slt11p that failed to complement the synthetic lethality of Δslt11 with slt17/slu7-100 did not bind to GST-Slu7p in vitro (unpublished observations). These results suggest that the Slt11p-Slu7p interaction is important for pre-mRNA splicing.
FIG. 8.
Slt11p-Slu7p interaction and its effects on RNA-binding and -annealing activities of Slt11p. (A) Protein-protein interaction between Slt11p and Slu7p as determined by affinity chromatography. Western blots of flowthrough (lanes 2, 6, 10, and 14), 0.5 M NaCl (lanes 3, 7, 11, and 15), 1 M NaCl (lanes 4, 8, 12, and 16), and 1% SDS (S, lanes 5, 9, 13, and 17) eluates of four minicolumns with the indicated ligand concentrations were probed with rabbit anti-Slt11p antibody. (B) Effects of Slu7p on the RNA-binding activity of Slt11p. Approximately 1 fmol of [32P]RNA-KQ was mixed with increasing amounts of Slt11p and incubated for 20 min at 25°C; 20 or 40 ng of GST-Slu7p and 20 ng of GST were then added to the mixture and incubated for another 20 min. The mixtures were then loaded onto 4% nondenaturing gels. The controls include [32P]RNA alone, with 20 and 40 ng of GST-Slu7p, and with 20 ng of GST. (For binding of Slt11p to RNA-KQ, see Fig. 4B, lanes 2 to 4.) (C) Effects of Slu7p on the RNA-annealing activity of Slt11p. Twenty nanograms of Slt11p was mixed with 20, 40, or 60 ng of GST-Slu7p and 20 ng of GST and incubated for 20 min at 25°C. Approximately 1 fmol of [32P]RNA-Q and 2 mol of RNA-K were then added, and the mixtures were incubated for another 20 min, after which proteinase K was added for 15 min. The mixtures were then loaded onto 6% nondenaturing gels. The controls include [32P]RNA-Q alone, mixed with RNA-K only, or mixed with RNA-K and 40 or 60 ng of GST-Slu7, 20 ng of GST, and 20 ng of Slt11p.
In order to probe the function of this direct protein-protein interaction, we examined the effects of Slu7p on RNA-binding and -annealing activities of Slt11p. Slt11p was first incubated with [32P]RNA-KQ for 20 min, which resulted in Slt11p binding to the RNA (Fig. 4B, lanes 2 to 4). Different amounts of GST-Slu7p and GST were then added, and the mixture was incubated for another 20 min. While GST had no discernible effect on the formation of the Slt11p-RNA complex (Fig. 8B, lanes 8 to 10), GST-Slu7p almost completely abolished the protein-RNA complex (Fig. 8B, lanes 2 to 7), suggesting that Slu7p is able to interact with RNA-bound Slt11p and that this interaction impairs the binding of Slt11p to the RNA. Similar results were observed when GST-Slu7p was mixed with Slt11p prior to addition of RNA-KQ (data not shown). In contrast, when [32P]RNA-Q and RNA-K were added to the GST-Slu7p–Slt11p mixture (preincubated for 20 min), the annealing of these two RNAs was not affected by the presence of GST-Slu7p (Fig. 8C, lanes 8 to 10). The different effects of GST-Slu7p on Slt11p activities suggest that RNA-annealing and -binding activities are two distinct biochemical properties associated with different (i.e., monomer and dimer) (Fig. 9) forms of Slt11p and that Slu7p binds preferentially to one form of Slt11p that is active in RNA-binding (see Discussion).
FIG. 9.
Model for Slt11p function. Slt11p forms a homodimer in the absence of RNA. One of the subunits recognizes and binds to U4/U6 stem II. The other subunit facilitates the formation of U2/U6 helix II through its RNA-annealing function. The Slt11p dimer then binds to the resulting RNA structures with both helix II and stem II to maintain structural integrity. Stem II is divided by an unpaired bulge into two parts, including the helix II-proximal part (U4 nt 1 to 11 with U6 nt 70 to 80), which consists of 11 bp. Helix II also contains 11 bp.
DISCUSSION
Slt11p, a new splicing factor, is involved in activation of the spliceosome (Fig. 2). We have shown in this study that recombinant Slt11p binds to RNA (Fig. 4) and that it is able to anneal two RNAs (Fig. 6B) and, in particular, to form a helical interaction in the presence of another preformed helical element (Fig. 6E). The protein can bind to the resulting duplexes if they contain two separated helical regions (Fig. 6C and F). Furthermore, our genetic results suggest that Slt11p is involved in the RNA base-pairing interaction of U2/U6 helix II in vivo (Fig. 3). Taken together, these data suggest that Slt11p acts in the spliceosome to facilitate the formation of U2/U6 helix II in association with another helical element, likely U4/U6 stem II (Fig. 9). This is supported further by the (structural) requirements for Slt11p binding to RNAs (Fig. 4) and dimerization of Slt11p (Fig. 7). We also demonstrated that Slt11p and Slu7p interact with each other both in vivo and in vitro (Fig. 8A) and that Slu7p exerts different effects on the RNA-binding and -annealing activities of Slt11p (Fig. 8B and C). We suggest that the function of Slt11p is regulated by Slu7p in the spliceosome.
The RNA-annealing and -binding activities of Slt11p are two distinct biochemical properties.
Slt11p is able to anneal a wide range of cRNAs (Fig. 6B and data not shown), provided that a minimum of 11 consecutive base pairs can be formed (likely to allow detection of the resulting duplexes in nondenaturing gels). However, its RNA-binding activity has some degree of (structural) specificity (Fig. 4A). It requires one helical element (≥11 bp) and a second single-stranded region (e.g., RNA-S) (Fig. 4) or helical region (e.g., RNA-KQ) (Fig. 4). Although Slt11p did not bind RNA-K and RNA-Q individually (Fig. 4B, lanes 9 to 16), it was able to promote the efficient annealing of these RNAs (Fig. 6B, lanes 3 to 5) and to bind (weakly, under the conditions tested) to the resulting duplex (Fig. 6C, lanes 2 to 4). Similar results were observed with another set of RNAs (Fig. 6D, E, and F). In contrast, Slt11p was able to anneal RNA-M and -Q (resulting in RNA duplexes with altered configurations(Fig. 6B, lanes 7 to 9) but failed to bind stably to any of the duplexes (Fig. 6C, lanes 7 to 9). These results suggest that the RNA-annealing and -binding activities of Slt11p are two different biochemical properties with different substrate requirements. It seems that the structural aspects (the integrity of the two helical elements) (Fig. 6) of the RNA substrates rather than sequence are the determining factor for the Slt11p-RNA interactions. Our genetic results suggest the importance of RNA base pairing rather than sequence per se for SLT11-mediated helix II interaction in vivo (Fig. 3). Perhaps this is reflected in the lack of strict specificity for Slt11p activities in vitro (Fig. 4 and 6). However, it is possible that an additional factor(s) is required in vivo to maintain the specific SLT11-U2/U6 helix II interaction and that this factor(s) is absent in vitro.
Slt11p exists in two forms in the absence of RNA (Fig. 7). It is plausible that the dimer form binds to RNAs with two helical elements (Fig. 9). We note that the native U4/U6 stem II is divided into two regions with a single bulge. The region close to the 3′ end of U6 snRNA consists of 11 bp. U2/U6 helix II also contains 11 bp (Fig. 9). It is possible that the two subunits bind to each element in a similar or identical fashion. We suggest that the RNA-binding activity of Slt11p is attributed to its dimer form and the annealing activity to the monomer (and, perhaps, each subunit of the dimer). The different substrate specificities of these two activities can be explained as follows. As an RNA-binding protein (Fig. 1), the Slt11p monomer is able to anneal RNAs without strong specificity and to bind weakly to RNAs. Strong and cooperative RNA binding is established when both subunits of the dimer bind to two helical elements or one helical element along with a single-stranded region. In particular, Slt11p is able to form, at low efficiency, a tripartite complex with two RNAs that do not form proper base pairing (Fig. 6F, lanes 8 to 10). This tolerance of disrupted base-pairing interaction is consistent with cooperativity of RNA-binding activity of Slt11p. The different effects of Slu7p on two activities of Slt11p (Fig. 8B and C) are also consistent with the interpretation that the RNA-binding and -annealing activities of Slt11p are two distinct biochemical properties (see below). However, in order to dissect these two activities, it is necessary to test mutant Slt11p proteins that fail to form dimers for their RNA-binding and -annealing activities.
Slt11p is involved in formation of U2/U6 helix II in association with U4/U6 stem II.
During spliceosome assembly, after the U4/U6-U5 tri-snRNP is recruited into the prespliceosome, the resulting holospliceosome (corresponding to complex A2-1) (32) (Fig. 2B) undergoes a series of RNA conformational rearrangements. The switch at the 5′-SS (31) is followed immediately by disruption of U4/U6 stem I (15). The U6 portion of stem I is involved in U2/U6 helix I interaction (18). The mutual exclusion of U4/U6 stem I and U2/U6 helix I indicates that formation of the latter must follow unwinding of the former. By the same reasoning, the Brow stem forms following the disruption of U4/U6 stem II. On the other hand, it is possible to form U4/U6 stem II and U2/U6 helix II interactions concomitantly (Fig. 5A and 9). Our genetic study (Fig. 3) suggests that Slt11p is involved in the base-pairing interaction of U2/U6 helix II. The biochemical properties of recombinant Slt11p (see above) suggest that the protein binds to an RNA that resembles preformed U4/U6 stem II with the 3′ end of U6 snRNA (RNA-S) (Fig. 4A and B, lanes 18 to 20) and that anneals another short RNA (corresponding to the 5′ end of U2 snRNA, RNA-T) to this RNA (Fig. 6E, lanes 3 to 5). The outcome of this process is reminiscent of formation of U2/U6 helix II in association with (preformed) U4/U6 stem II (Fig. 9). We suggest that Slt11p acts as a dimer in the splicing process. During spliceosome assembly, one subunit of the Slt11p dimer may recognize U4/U6 stem II and anchor the dimer to the site of its action. The RNA-annealing activity of the other subunit may facilitate the formation of U2/U6 helix II. The dimer may then bind to the resulting RNA structure (Fig. 9).
Our preliminary biochemical data also suggest that Slt11p (dimer) binds to two separated helical elements cooperatively (Fig. 6F and unpublished observations). The genetic data (Fig. 3) are consistent with this notion of cooperativity. In the presence of Slt11p, most mutations in the helix II regions of both U2 and U6 snRNAs confer no or mild growth defects (Fig. 3) (10, 38). The only exceptions are the 9-nt (U6-E+F) (Fig. 3B) and 11-nt substitutions in the helix II region of U6 snRNA; these confer lethality by themselves (10). This is due to disruption of helix II and an intramolecular U6 interaction, in both of which the 3′ end of U6 snRNA is involved (unpublished observations). The tolerance of 4- and 5-nt substitutions of both U2 and U6 snRNAs and a 9-nt substitution of U2 snRNA (Fig. 3) suggests that in the presence of a proper (i.e., wild-type) U4/U6 stem II, dimeric Slt11p is able to form an imperfect U2/U6 helix II (in association with U4/U6 stem II) (Fig. 6F). Slt11p can be viewed as an RNA chaperone that maintains the structural integrity of RNA interactions, in addition to other activities. Its function is not essential for viability at permissive temperatures but is manifested when the integrity of the helix II interaction is compromised. This is demonstrated by synthetic lethality of Δslt11 with all these 4-nt and 5-nt substitution mutations in both U2 and U6 snRNAs and by allele-specific and mutual genetic suppression when helix II is restored (Fig. 3). However, it remains to be determined if mutations in the stem II region of U4 snRNA interact genetically with Δslt11 and mutations in the helix II region of both U2 and U6 snRNAs.
Wassarman and Steitz (35) detected a psoralen-cross-linked trimolecular U2/U4/U6 snRNA complex in HeLa cell nuclear extract. Two forms of the U2/U6 helix II interaction were observed. The first occurs in the relatively abundant snRNP complexes that sediment at >150S (containing all five spliceosomal snRNAs) in the absence of pre-mRNA substrate. This corresponds to the concomitant formation of the two intermolecular interactions of U2/U6 helix II and U4/U6 stem II, as suggested by Brow and Vidaver (4). The second form occurs in the S100 fraction and is dependent on the splicing reaction (36), which corresponds to de novo formation of helix II in the spliceosome assembly. It remains to be determined if helix II forms with stem II in the absence of pre-mRNA substrate in yeast. If it does, the existence of such a preassembled U2/U6/U4 snRNA complex (with or without U5 snRNA) may indicate an alternative pathway for spliceosome assembly in which the multi-snRNP complexes are recruited into the commitment complex in a single step. A large native complex containing all five snRNPs has been identified in yeast extracts in the absence of pre-mRNA (26). Regardless of the physiological relevance of this large complex to pre-mRNA splicing in vivo, the concomitant formation of U2/U6 helix II and U4/U6 stem II is topologically possible in the yeast spliceosome (Fig. 5A and 9). We note that Prp24p, another RNA-binding protein, is involved in formation of U4/U6 di-snRNP (11, 12, 27). Prp24p may also facilitate the formation of U2/U6 helix II. If so, it may explain why Slt11p is not essential at ≤30°C but is required, as an RNA chaperone (see above), for maximum efficiency of splicing and spliceosomal activation (Fig. 2).
Slu7p regulates the function of Slt11p in the spliceosome.
The concomitant formation of helix II and stem II raises the question of how they are unwound prior to the splicing reaction. Another factor identified in our genetic screen, Slt22p (or Brr2p), has been implicated in the unwinding of the U4/U6 duplex (26) and U2/U6 helix II (reference 37 and unpublished observations). Another question of concern is the dissociation of Slt11p from stem II and helix II. Slu7p may be involved in regulating this process.
Slu7p was identified in our genetic screen as Slt17p. Both slu7-1 and slt17/slu7-100 are synthetically lethal with mutations in the helix II region of U2 snRNA (38) and Δslt11 (see Results). The direct protein-protein interactions between the Slt11p dimer and Slu7p (Fig. 8A) (unpublished observations) and the inhibitory effect of this on the RNA-binding activity of Slt11p (Fig. 8B) suggest that one of the functions of Slu7p is to dissociate the Slt11p dimer from stem II and helix II. As discussed above, the RNA-annealing and -binding activities of Slt11p might be attributed to the monomeric and dimeric forms of Slt11p, respectively. If so, the inhibition of Slt11p RNA binding by Slu7p is due to interaction with the dimer form of Slt11p, whereas the RNA annealing activity of the Slt11p monomer is largely unaffected by Slu7p. We have observed that mutant Slt11p proteins that fail to form dimers also fail to bind the RNA substrates tested in this study. When tested, they do not interact with Slu7p in vitro (unpublished observations). These results are consistent with our interpretations. Although Slu7p is a second-step splicing factor (5), its interaction with Slt11p may occur prior to the first step. If so, the Slt11p dimer dissociates from helix II and stem II after Slu7p binds to it during spliceosomal activation. The two RNA duplexes can then be unwound by Slt22p. The Slt11p-Slu7p interaction may persist through both steps of splicing. It is possible that Slt11p plays an additional role(s) in both the first and second steps.
ACKNOWLEDGMENTS
We thank Michael Costanza, Quoc Hugnh, and Shou-Jiang Tang for their technical assistance, B. Blencowe, D. Jansma, M. Kober, C. Köth, V. Lay, J. Li, A. MacMillan, and W. Rice for their advice and suggestions during the course of this study, and members of the Friesen lab, present and past, for their support. D.X. thanks Peggy M. Pasternak and Yiming Xu for their support and stimulating discussion.
This research was supported by the National Cancer Institute of Canada and the Medical Research Council of Canada.
REFERENCES
- 1.Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 2.Abovich N, Liao X C, Rosbash M. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 1994;8:843–854. doi: 10.1101/gad.8.7.843. [DOI] [PubMed] [Google Scholar]
- 3.Adams A, Gottschling D E, Kaiser C A, Stearns T. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- 4.Brow D A, Vidaver R M. An element in human U6 RNA destabilizes the U4/U6 spliceosomal RNA complex. RNA. 1995;1:122–131. [PMC free article] [PubMed] [Google Scholar]
- 5.Brys A, Schwer B. Requirement of SLU7 in yeast pre-mRNA splicing is dictated by the distance between the branchpoint and the 3′ splice site. RNA. 1996;2:707–717. [PMC free article] [PubMed] [Google Scholar]
- 6.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 7.Burgess S M, Guthrie C. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell. 1993;73:1377–1391. doi: 10.1016/0092-8674(93)90363-u. [DOI] [PubMed] [Google Scholar]
- 8.Cheng S C, Abelson J. Spliceosome assembly in yeast. Genes Dev. 1987;1:1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- 9.Collins C A, Guthrie C. Allele-specific genetic interactions between Prp8 and RNA active site residues suggest a function for Prp8 at the catalytic core of the spliceosome. Genes Dev. 1999;13:1970–1982. doi: 10.1101/gad.13.15.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Field J D, Friesen J D. Functionally redundant interactions between U2 and U6 spliceosomal snRNAs. Genes Dev. 1996;10:489–501. doi: 10.1101/gad.10.4.489. [DOI] [PubMed] [Google Scholar]
- 11.Ghetti A, Company M, Abelson J. Specificity of Prp24 binding to RNA: a role for Prp24 in the dynamic interaction of U4 and U6 snRNAs. RNA. 1995;1:132–145. [PMC free article] [PubMed] [Google Scholar]
- 12.Jandrositz A, Guthrie C. Evidence for a Prp24 binding site in U6 snRNA and in a putative intermediate in the annealing of U6 and U4 snRNAs. EMBO J. 1995;15:820–832. doi: 10.1002/j.1460-2075.1995.tb07060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krämer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 14.Kuhn A N, Li Z, Brow D A. Splicing factor Prp8 governs U4/U6 unwinding during activation of the spliceosome. Mol Cell. 1999;3:65–75. doi: 10.1016/s1097-2765(00)80175-6. [DOI] [PubMed] [Google Scholar]
- 15.Li Z, Brow D A. A spontaneous duplication in U6 spliceosomal RNA uncouples the early and late functions of the ACAGA element in vivo. RNA. 1996;2:879–894. [PMC free article] [PubMed] [Google Scholar]
- 16.Lussier M, White A M, Sheraton J, di Paolo T, Treadwell J, Southard S B, Horenstein C I, Chen-Weiner J, Ram A F, Kapteyn J C, Roemer T W, Vo D H, Bondoc D C, Hall J, Zhong W W, Sdicu A M, Davis J, Klis F M, Robbins P W, Bussey H. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics. 1997;147:435–450. doi: 10.1093/genetics/147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luukkonen B G M, Séraphin B. The role of branchpoint-3′ splicing site spacing and interaction between intron terminal nucleotides in 3′ splice site selection in Saccharomyces cerevisiae. EMBO J. 1997;16:779–892. doi: 10.1093/emboj/16.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madhani H D, Guthrie C. Dynamic RNA-RNA interactions in the spliceosome. Annu Rev Genet. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- 19.Madhani H D, Guthrie C. Randomization-selection analysis of snRNAs in vivo: evidence for a tertiary interaction in the spliceosome. Genes Dev. 1994;8:1071–1086. doi: 10.1101/gad.8.9.1071. [DOI] [PubMed] [Google Scholar]
- 20.McDonald W H, Ohi R, Smelkova N, Frendewey D, Gould K L. Myb-related fission yeast cdc5p is a component of a 40S snRNP-containing complex and is essential for pre-mRNA splicing. Mol Cell Biol. 1999;19:5352–5362. doi: 10.1128/mcb.19.8.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noble S M, Guthrie C. Identification of novel genes required for yeast pre-mRNA splicing by means of cold-sensitive mutations. Genetics. 1996;143:67–80. doi: 10.1093/genetics/143.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Day C L, Dalbadie-McFarland G, Abelson J. The Saccharomyces cerevisiae Prp5 protein has RNA-dependent ATPase activity with specificity for U2 small nuclear RNA. J Biol Chem. 1996;271:33261–33267. doi: 10.1074/jbc.271.52.33261. [DOI] [PubMed] [Google Scholar]
- 23.O'Keefe R T, Norman C, Newman A J. The invariant U5 snRNA loop 1 sequence is dispensable for the first catalytic step of pre-mRNA splicing in yeast. Cell. 1996;86:679–689. doi: 10.1016/s0092-8674(00)80140-3. [DOI] [PubMed] [Google Scholar]
- 24.Portman D S, O'Conner J P, Dreyfuss G. YRA1, an essential Saccharomyces cerevisiae gene, encodes a novel nuclear protein with RNA annealing activity. RNA. 1997;3:527–537. [PMC free article] [PubMed] [Google Scholar]
- 25.Query C C, Strobel S A, Sharp P A. Three recognition events at the branch site adenine. EMBO J. 1995;15:1392–1402. [PMC free article] [PubMed] [Google Scholar]
- 26.Raghunathan P L, Guthrie C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr Biol. 1998;16:847–855. doi: 10.1016/s0960-9822(07)00345-4. [DOI] [PubMed] [Google Scholar]
- 27.Raghunathan P L, Guthrie C. A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science. 1998;279:857–860. doi: 10.1126/science.279.5352.857. [DOI] [PubMed] [Google Scholar]
- 28.Schwer B, Gross C H. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 1998;17:2086–2094. doi: 10.1093/emboj/17.7.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Séraphin B, Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989;59:349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- 30.Staley J P, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 31.Staley J P, Guthrie C. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- 32.Tarn W Y, Lee K R, Cheng S C. Yeast precursor mRNA processing protein PRP19 associates with the spliceosome concomitant with or just after dissociation of U4 small nuclear RNA. Proc Natl Acad Sci USA. 1993;90:10821–10825. doi: 10.1073/pnas.90.22.10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teigelkamp S, McGarvey M, Plimpton M, Beggs J D. The splicing factor PRP2, a putative RNA helicase, interacts directly with pre-mRNA. EMBO J. 1994;13:888–896. doi: 10.1002/j.1460-2075.1994.tb06332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Umen J G, Guthrie C. The second catalytic step of pre-mRNA splicing. RNA. 1995;1:869–885. [PMC free article] [PubMed] [Google Scholar]
- 35.Wassarman D A, Steitz J A. Interactions of small nuclear RNA's with precursor messenger RNA during in vitro splicing. Science. 1992;257:1918–1925. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- 36.Wassarman D A, Steitz J A. A base-pairing interaction between U2 and U6 small nuclear RNAs occurs in >150S complexes in HeLa cell extract: implications for the spliceosome assembly pathway. Proc Natl Acad Sci USA. 1993;90:7139–7143. doi: 10.1073/pnas.90.15.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu D, Nouraini S, Field D, Tang S-J, Friesen J D. An RNA-dependent ATPase associated with U2/U6 snRNAs in pre-mRNA splicing. Nature. 1996;381:709–713. doi: 10.1038/381709a0. [DOI] [PubMed] [Google Scholar]
- 38.Xu D, Field D J, Tang S-J, Moris A, Bobechko B P, Friesen J D. Synthetic lethality of yeast slt mutations with U2 small nuclear RNA mutations suggests functional interactions between U2 and U5 snRNPs that are important for both steps of pre-mRNA splicing. Mol Cell Biol. 1998;18:2055–2066. doi: 10.1128/mcb.18.4.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]