Abstract
p105 (NFKB1) acts in a dual way as a cytoplasmic IκB molecule and as the source of the NF-κB p50 subunit upon processing. p105 can form various heterodimers with other NF-κB subunits, including its own processing product, p50, and these complexes are signal responsive. Signaling through the IκB kinase (IKK) complex invokes p105 degradation and p50 homodimer formation, involving p105 phosphorylation at a C-terminal destruction box. We show here that IKKβ phosphorylation of p105 is direct and does not require kinases downstream of IKK. p105 contains an IKK docking site located in a death domain, which is separate from the substrate site. The substrate residues were identified as serines 923 and 927, the latter of which was previously assumed to be a threonine. S927 is part of a conserved DSGΨ motif and is functionally most critical. The region containing both serines is homologous to the N-terminal destruction box of IκBα, -β, and -ɛ. Upon phosphorylation by IKK, p105 attracts the SCF E3 ubiquitin ligase substrate recognition molecules βTrCP1 and βTrCP2, resulting in polyubiquitination and complete degradation by the proteasome. However, processing of p105 is independent of IKK signaling. In line with this and as a physiologically relevant model, lipopolysaccharide (LPS) induced degradation of endogenous p105 and p50 homodimer formation, but not processing in pre-B cells. In mutant pre-B cells lacking IKKγ, processing was unaffected, but LPS-induced p105 degradation was abolished. Thus, a functional endogenous IKK complex is required for signal-induced p105 degradation but not for processing.
The NF-κB transcription factor family plays an evolutionarily conserved role in innate and adaptive immune responses, and its members are essential regulators of proinflammatory processes (12, 18, 33). NF-κB is also an important regulator in cell fate decisions, such as programmed cell death and proliferation control, and is critical in tumorigenesis (32, 42). The vertebrate NF-κB/Rel factors p50, p52, p65 (RelA), c-Rel, and RelB form various dimers and are under the control of a coevolved family of cytoplasmic IκB molecules, IκBα, -β, and -ɛ, and the nuclear IκB homologue Bcl-3 (1). p50 and p52 are formed by processing from the precursor molecules p105 and p100, respectively. Prototypic heterodimeric NF-κB p50-p65 is rapidly released from cytoplasmic complexes with IκBs upon cellular stimulation with diverse agents, including tumor necrosis factor alpha (TNF-α), interleukin-1β, bacterial lipopolysaccharides (LPS), and phorbol myristate acetate, following viral infection or exposure to γ-irradiation (31). All these agents induce an IκB kinase (IKK) complex to phosphorylate IκBα, -β, and -ɛ. The IKK complex contains the kinases IKKα and IKKβ and the noncatalytic IKKγ subunit, which is essential for signal responsiveness of the complex (see reference 17 for a review). Once phosphorylated by the IKK complex, the IκBs are ubiquitinated and degraded by the 26S proteasome, resulting in nuclear translocation of NF-κB. The ubiquitin ligase complex specific for IκBs, SCF (Skp1, Cullin, F-box protein), has recently been identified and contains as substrate recognition molecules the F-box and WD40 domain protein βTrCP1 (Slimb) (46, 53, 59) or its homologue βTrCP2 (HOS) (10). Further components in this complex are Skp1, Cdc53/Cul1, and ROC1 (49). The βTrCP component accounts for recognition of an IKK-phosphorylated DS(P)GLDS(P) motif containing serines 32 and 36 in IκBα and serines 19 and 23 in IκBβ (45, 54). βTrCP1 and βTrCP2 bind as homodimers to IκBα (48). A similar motif in the β-catenin (Armadillo) proteins of the Wnt/Wg pathways and in the human immunodeficiency virus type 1 (HIV-1)-encoded Vpu protein is phosphorylated by glycogen synthase kinase 3β (GSK3β) and casein kinase II (CKII), respectively. It then confers βTrCP1 binding and degradation of β-catenin and the Vpu-interacting HIV receptor CD4 by the ubiquitin-proteasome system (20, 30, 53).
The NF-κB precursor protein p105 (NFKB1) and its paralogue p100 (NFKB2) have unique roles in the control of NF-κB activity, since both act as IκB molecules when unprocessed and provide the subunits p50 and p52, respectively, upon processing. The expression of both genes is strongly autoregulated by NF-κB, providing a means to replace the proteolysed molecules, similar to the situation with IκBα (see reference 55 for a review). Different levels of proteolytic maturation of p105 by the 26S proteasome have been described. Posttranslational p50 production from p105 was shown in mammalian and yeast cells (8, 41) and requires ubiquitination (5, 40). A glycine-rich region at the end of the amino-terminal half of p105 is required for p50 generation, perhaps by limiting the processive action of the proteasome (27). Several groups have reported that processing of p105 may be enhanced by signal-induced phosphorylation caused by agents like phorbol ester, okadaic acid, TNF-α, double-stranded RNA, and hydrogen peroxide (7, 29, 34, 35, 37). In line with this, in cells stimulated with NF-κB-activating agents, including TNF-α, phorbol ester, and hydrogen peroxide, p105 and p100 were phosphorylated with the same kinetics as IκBα (37). However, the fold increase in p50 relative to p105 induced by the various conditions in these studies was modest compared to the clear decline in p105 levels.
The concept of posttranslational processing was challenged by a report that p105 processing occurs mainly as a constitutive process at the cotranslational level, leading to the production of p105-p50 heterodimers (25). This process involves a cotranslational dimerization of the Rel homology domain of the nascent polypeptides (26). Similarly, p52 production from p100 was shown to occur by a cotranslational process (15). The conditions which determine cotranslational versus posttranslational p50 formation remain to be determined. Recently, we and others demonstrated that p105 is subject to signal-induced complete degradation rather than processing (2, 13). TNF-α-induced p105 degradation rapidly releases p105-bound p50, which is detected in the nucleus as dimers complexed with Bcl-3 (13). Transfected IKKα and IKKβ cause efficient phosphorylation of p105 at a carboxy-terminal region of 18 amino acids, resulting in degradation of p105 by the proteasome. The phosphorylation sites were essential for TNF-α-induced p105 degradation, as shown by combined mutation of all potential phosphoacceptor sites (13). Furthermore, in cells transfected with Tpl-2 (Cot), this kinase interacts with a carboxy-terminal region of p105 and causes p105 phosphorylation and degradation, but it does not phosphorylate p105 directly (2). Since Cot acts upstream of the IKK complex (28), induction of IKK may be the mechanism by which Cot triggers p105 degradation. In contrast to these studies and after completion of this work, it was recently proposed that IKK phosphorylation of p105 primarily induces enhanced processing. It was reported that IKK-induced processing is promoted by sequestration of βTrCP and subsequent polyubiquitination of p105 (38).
In this work we have analyzed the molecular determinants of p105 phosphorylation by IKK and identified the major substrate residues and a separate kinase-docking site, which is part of a death domain. We show that IKK phosphorylation of p105 is direct and that it creates a recognition site for βTrCP proteins critically dependent on one of the determined phosphoacceptor serines. IKK-induced ubiquitination results in complete degradation but not enhanced processing of p105. This conclusion is supported by analysis of endogenous p105 in LPS-treated pre-B cells.
MATERIALS AND METHODS
Cell culture.
293 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 1 mM sodium pyruvate, and 100 U of penicillin-streptomycin per ml. 70Z/3 and 1.3E2 cells were maintained in RPMI medium supplemented with 10% fetal calf serum, 50 μM β-mercaptoethanol, and 100 U of penicillin-streptomycin per ml. 1.3E2 cell lines were electroporated with hemagglutinin (HA)-tagged IKKγ (IKKγ-HA) pcDNA3 expression vector, and clones stably expressing IKKγ-HA were selected and grown in medium containing G418 (1 mg/ml; Gibco). For stimulation, cells were treated with cycloheximide at 50 μg/ml for 293 cells and 25 μg/ml for 70Z/3 and 1.3E2 cells. N-acetyl-Leu-Leu-norlencinal (ALLN) (Calbiochem) and bacterial LPS (Sigma) were added at 50 and 10 μg/ml, respectively, for the indicated periods of time prior to preparation of extracts.
Plasmids.
Eucaryotic expression vectors for p105 (p105pcDNA3Flag, p105ΔC1-ΔC5pcDNA3Flag, and p105SSS-AAApcDNA3Flag) and IκBα (IκBαpcDNA3 and IκBαΔNpcDNA3) as well as prokaryotic expression vectors (p105ΔNpRSET, p105ΔNSSS-AAApRSET, IκBαpGEX6P, and IκBαS32/36pGEX6P) were described previously (13). p105ΔC6 (amino acids 18 to 809) was generated by PCR, cloned via BamHI and SacI into pRSETB or, to generate p105ΔC6+SRD, a p105 construct with internal deletion of amino acids 809 to 916, into p105pRSETB. The inserts were excised via BamHI and KpnI and religated into pcDNA3Flag. p105ΔC7 (aa 18 to 776) was constructed via partial XbaI digestion, and p105ΔC8 (amino acids 18 to 751) was constructed by PCR. Note that due to these cloning procedures, ΔC6, ΔC7, and ΔC8 contain the artifical C-terminal additions ELEICSLVPPLEGPIL, EGPIL, and VVPLEGPIL, respectively. Mutant p105 (M1-M9) was generated by standard PCR techniques and cloned via SacI-KpnI digestion into procaryotic expression vector pRSETA or p105 (amino acids 18 to 968) pRSETB. The p105 cDNAs (18 to 968) coding for mutant proteins were religated after BamHI-KpnI digestion into pcDNA3, which was modified with an amino-terminal Flag tag inserted via HindIII and BamHI and a KpnI restriction site between XhoI and XbaI. All constructs were sequenced on a LiCor 4000L or with an ABI 377 sequencer (Invitek, Berlin, Germany). The eukaryotic p100 expression vector was constructed via BamHI-EcoRI digestion of p100pRSETC (13) and religation into pcDNA3Flag. Human p105 EST clones were from RZPD (Resource Center German Human Genome Project, accession numbers AL118961, AA258085, AA134618 and AW612589); human IκBβ1 and IκBβ2 cDNAs were described previously (16, 23). Human β-TrCP1 was PCR amplified from the hE3RSIκB cDNA (59) and inserted via NotI and XbaI into pcDNA3.1 modified with an amino-terminal HA tag.
Protein purification and in vitro translation.
p105 fragments (917 to 968) cloned into pRSETA were expressed in BL21/pLysS bacteria. Cell pellets were resuspended in 50 mM Tris (pH 7.4)–100 mM NaCl–5 mM β-mercaptoethanol–0.4 mM Pefabloc and lysed by sonication. Expressed proteins in the lysates were bound to Ni+-agarose (Qiagen) for 3 h at 4°C, washed three times with lysis buffer, and eluted for 1 h at 4°C in 300 mM imidazol–300 mM NaCl–50 mM NaPO4 (pH 6)–10% glycerol. Coupled in vitro transcription-translation was performed according to the manufacturer's protocol (Promega).
Antibodies.
The IKKγ (FL-419), IKKα (H-744), IκBα (C-21), HA (Y-11), p50 (D-17), and Cot (M-20) antibodies were obtained from Santa Cruz. Monoclonal antiubiquitin antibody (1B3; MoBiTec) and IKKβ antibody 10AG2 (Biosource) were used for Western blotting. Anti-Flag tag antibody M2 or M5 (Sigma) and anti-T7 tag antibody (Novagen) were used for immunoprecipitation or immunoblotting.
Extracts, EMSA, and Western blotting.
Whole-cell extracts were analyzed by electrophoretic mobility shift assay (EMSA) and Western blotting essentially as described previously (21).
293 cell transfection and preparation of extracts.
Plates (10 cm) of 293 cells were transiently transfected using the calcium phosphate precipitation method with 10 μg of total DNA. Cells were lysed 24 to 36 h after transfection. For detection of ubiquitinated proteins, one plate was lysed in 1 ml of lysis buffer containing 50 mM Tris (pH 7.6)–300 mM NaCl–0.5% Triton X-100–10 mM iodocetamide–0.4 mM NaVO4–0.4 mM EDTA–10 mM NaF–10 mM sodium pyrophosphate–1 mM dithiothreitol (DTT)–8 mM β-glycerophosphate and the complete protease inhibitor cocktail (Boehringer). Equal volumes of lysates (20 μl) were used for detection in Western blots and (1 ml) immunoprecipitation.
Immunoprecipitation.
Immunoprecipitation with cell extract equivalents of 10-cm plates were performed with anti-Flag antibody (M2; Sigma) or anti-gene 10 antibody (Novagen) in 1 ml of the indicated lysis buffers. The cell extracts were cleared for 1 h at 4°C with 20 μl of protein A-Sepharose (Pharmacia) and precipitated with antibodies and protein A-Sepharose for 3 h at 4°C. The precipitates were washed three times with 1 ml of the individual buffer used for immunoprecipitation, boiled in sodium dodecyl sulfate (SDS) loading buffer, and separated on 8 to 10% SDS–polyacrylamide gel electrophoresis (PAGE).
IKK assay.
Kinase assays were performed as described previously (13) using 1 μg of recombinant substrate protein and 25 ng of purified IKKβ.
RESULTS
IKKβ phosphorylates p105 directly at residues located within a DSXXDS motif.
We previously identified IKK phosphoacceptor sites between amino acids 920 and 936 of the p105 precursor molecule. Less efficient IKK phosphorylation was also observed in a region between amino acids 850 and 891, but deletion of this site had no effect on p105 stability (13). Phosphorylation was analyzed using transfected IKKs after immunoprecipitation in an in vitro kinase reaction. To formally rule out the possibility that p105 is not a direct target of IKKs, we now used recombinant IKKβ purified from baculovirus-infected insect cells in an in vitro kinase reaction (Fig. 1A). Recombinant IKKβ (Fig. 1B) phosphorylated p105 and IκBα with similar efficiency, while phosphorylation of IκBαS32,36A and p105ΔNAAA was abolished and strongly reduced, respectively. Similar results were obtained for recombinant IKKα (data not shown). Residual phosphorylation of the p105 mutant is likely due to the cryptic phosphorylation site between amino acids 850 and 891 (13). Thus, phosphorylation of p105 by IKKs is direct and does not need further kinases acting downstream of IKK. Next, we wanted to examine the phosphoacceptor sites in p105. Intriguingly, the published human p105 sequences contain either a threonine or a serine at position 927 (3, 14, 19, 36), but serine in rat and mouse p105 and either serine or threonine in chicken p105 (various cDNA and expressed sequence tag [EST] sequences in the database [not shown]). Similarly, human IκBβ was reported to contain a threonine at position 19, while in murine IκBβ the first potential acceptor position was determined to be a serine (23, 50). Importantly, it was shown that a threonine instead of a serine is not tolerated as a phosphoacceptor site in IκBα (6, 24). We resequenced several human p105 EST and cDNA clones and IκBβ1 and IκBβ2 cDNA clones in this region to determine whether the obvious differences in the published sequences are due to polymorphisms or arise from sequencing problems in this region, which indeed revealed compression effects upon sequencing. We found that both human p105 and human IκBβ in fact contain a serine instead of a threonine at positions 927 and 19, respectively (data not shown).
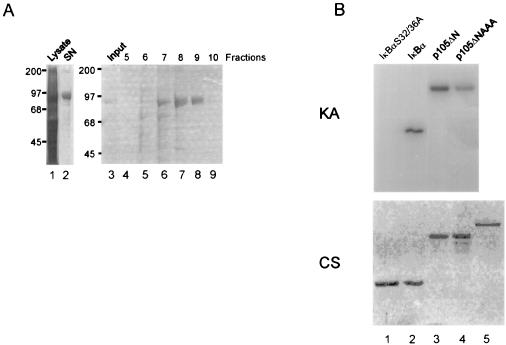
FIG. 1.
Purified recombinant IKKβ phosphorylates the signal response domain of p105 directly. (A) Coomassie brilliant blue-stained SDS-PAGE showing the purification of IKKβ. His-tagged IKKβ was expressed in SF9 cells with the baculovirus system. IKKβ protein from the lysates of infected cells (lane 1) was bound to Ni+ -agarose and cleaved off by thrombin digestion (lane 2). The supernatant was bound in 50 mM Tris (pH 7)–1 mM DTT–1 mM EDTA to a MonoQ column and eluted with a 10-ml gradient of 0 to 1 M NaCl (lanes 4 to 9). The protein eluted at about 500 mM NaCl. Sizes are shown in kilodaltons. (B) The purified protein (fraction 9) (lane 5, lower panel) was used in an in vitro kinase assay with IκBα S32/36A, wild-type IκBα, p105ΔN, and p105ΔNAAA, as indicated. p105ΔNAAA contains serine-to-alanine mutations at amino acids 921, 923, and 932 and a serine-to-threonine mutation at residue 927. Top, kinase assay (KA); bottom, Coomassie brilliant blue-stained (CS) SDS-PAGE of IκBα, p105, and IKKβ proteins.
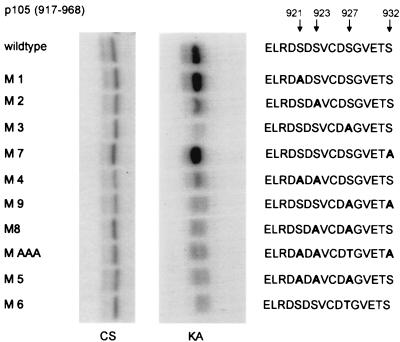
Since p105 contains a serine in position 927, there is one motif in this position (DSVCDS927) which matches the IKK sites found in all small IκBs (DSXXDS). To analyze the sequence requirement for phosphorylation of p105 by IKKs, serine residues in the destruction box were mutated to alanines (Fig. 2). Phosphorylation was analyzed using p105 peptides spanning amino acids 917 to 968 in an in vitro kinase reaction with purified recombinant IKKβ. While single mutations of serine 921 or 932 to alanine had no discernible effect, mutation of either serine 923 or serine 927 strongly reduced phosphorylation. Serine 927 was clearly the most sensitive residue. Any double mutation affecting one of these residues (in M4, M9, and M8) was also severely impaired, as were the mutants p105AAA and M5. Importantly, replacement of serine 927 by threonine (in M6) resulted equally in a strong reduction of phosphorylation, underscoring the strong serine-over-threonine preference of IKKs. Thus, while S921, which is not conserved phylogenetically, appears to play no role, two serines (S923 and 927) are critical for full activity.
FIG. 2.
Mutational analysis of IKK phosphorylation sites in p105. p105 cDNA fragments encoding amino acids 917 to 968 of p105 were cloned into pRSETA. Purified proteins (see Coomassie-stained SDS gel [CS], left panel) were used in an in vitro kinase assay with purified IKKβ (KA, right panel). Mutated amino acids in the p105 destruction box are shown in bold letters.
IKK phosphorylation triggers interaction of p105 with the F-box proteins βTrCP1 and HOS/βTrCP2.
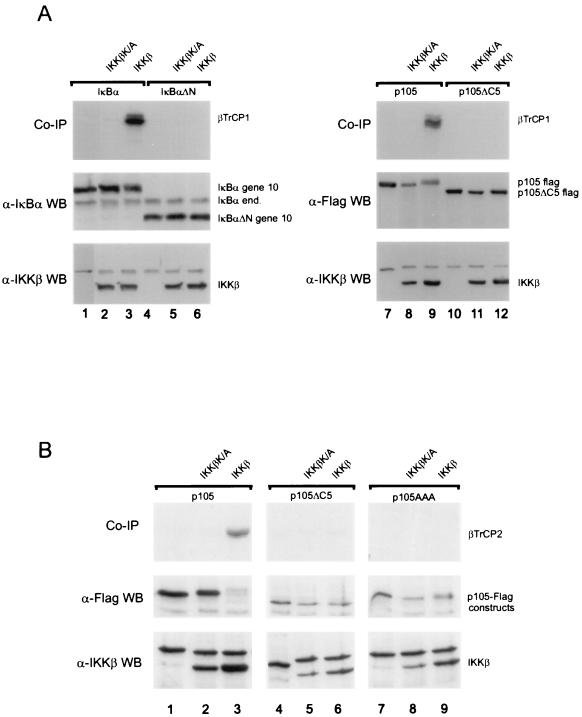
The IKK phosphorylated destruction box of IκBα is recognized by the F-box proteins βTrCP1 and βTrCP2 to trigger degradation (10, 45, 47, 53, 54, 59). Since IKK phosphorylation of p105 coincides with proteasome-dependent p105 proteolysis, a likely possibility is that phosphorylated p105 could associate with βTrCPs, as suggested earlier (13). As expected, in vitro-translated βTrCP1 could be coimmunoprecipitated with IκBα from extracts of cells expressing full-length IκBα and IKKβ (Fig. 3A, lane 3), but not when the destruction box was deleted (in IκBαΔN, lane 6) or the kinase was inactive (in IKKβK/A, lane 2). In a virtually identical manner, βTrCP1 could be precipitated with p105 (lane 9) strictly dependent on both the C-terminal destruction box (missing in p105ΔC5, lane 12) and active IKKβ (lane 8 versus 9). Likewise, βTrCP2 bound to p105 phosphorylated by IKKβ (Fig. 3B, lane 3), but did not bind to p105 mutants with deletion or point mutations of all serines in the destruction box (lanes 6 and 9). Thus, the similarity between the recognition sites in IκBα and p105 suggests that both proteins utilize the same βTrCP1- or -2-containing SCF complexes for signal-dependent ubiquitination.
FIG. 3.
SCF ligase receptor subunits βTrCP1 and βTrCP2 recognize IKK-phosphorylated p105. (A) Plates of 293 cells were transfected with 1 μg of IκBα (lanes 1 to 3), 1 μg of IκBαΔN (lanes 4 to 6), 2 μg of p105 (lanes 7 to 9), or 2 μg of p105ΔC5 (lanes 10 to 12) together with kinase inactive IKKβ (lanes 2, 5, 8, and 11) or wild-type IKKβ (lanes 3, 6, 9, and 12). Cells were extracted with 200 mM NaCl–50 mM HEPES (pH 7.5)–0.5% NP-40–1 mM EDTA–10% glycerol. The extracts were diluted to 100 mM NaCl–50 mM HEPES (pH 7.5)–0.5% NP-40–1 mM EDTA– 5% glycerol and incubated for preclearance and immunoprecipitation with 4.5 μl of in vitro-translated, 35S-labeled βTrCP1-HA. Immunoprecipitations were performed with anti-gene 10 (IκBα) or anti-Flag (p105) antibodies. Coprecipitated βTrCP1 (top panels), expression (Western blots) of IκBα constructs and endogenous IκBα (anti-IκBα antibody; middle panel, left), of p105 constructs (anti-Flag antibody, right panel) as well as of expressed wild-type and mutant IKKβ are shown as indicated. (B) A similar experiment was performed using p105 (lane 1 to 3) p105ΔC5 (lanes 4 to 6) and p105AAA (lanes 7 to 9) expression constructs cotransfected with IKKβ K/A (lanes 2, 5, and 8) or IKKβ (lanes 3, 6, and 9). Cell lysis and immunoprecipitation were performed as described above, using anti-Flag antibodies and in vitro translated βTrCP2-HA/Hos protein.
IKK and βTrCP induce ubiquitination and subsequent proteasomal degradation of p105.
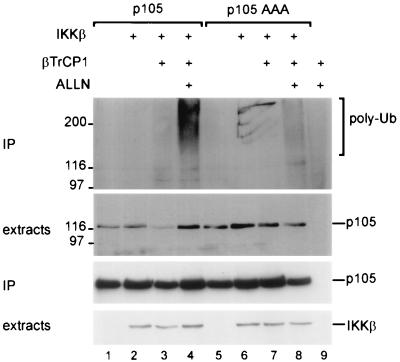
The effect on ubiquitination and proteolysis of p105 in response to IKK phosphorylation and βTrCP recognition was analyzed in intact cells. When p105 was cotransfected along with IKKβ and βTrCP1, polyubiquitination was observed (Fig. 4, lane 3, top panel). Ubiquitination was enhanced when degradation was blocked with a proteasome inhibitor (lane 4). Mutation of residues critical for p105 phosphorylation by IKKs greatly reduced ubiquitin conjugation (lanes 7 and 8). IKKβ and βTrCP1-mediated ubiquitination was paralleled by a loss of p105 (compare lane 3 with lanes 1 and 2, second panel), which was abolished by proteasome inhibition (lane 4) or by mutation of IKK phosphorylation sites in p105 (lane 7). Thus, in an IKK-dependent fashion, p105 is targeted by βTrCP/SCF, polyubiquitinated, and degraded by the proteasome.
FIG. 4.
Specific phosphorylation of p105 triggers βTrCP1-dependent ubiquitination and proteasomal degradation. Plates of 293 cells were transfected with 3 μg of p105 (lanes 1 to 4) or p105AAA (lanes 5 to 8) together with 2 μg of IKKβ (lanes 2, 3, 4, 6, 7, and 8) and 1 μg of β-TrCP1-HA (lanes 3, 4, 7, and 8) or with βTrCP1-HA alone (lane 9). Cells were treated for 2 h with ALLN as indicated, extracts were prepared and immunoprecipitation was performed with anti-Flag antibodies. Top panel, antiubiquitin Western blot of precipitated proteins. Second panel, anti-Flag Western blot of cell extracts; third panel, anti-Flag Western blot of precipitates, lower panel, Western blot detection of IKKβ in extracts.
IKK and βTrCP trigger posttranslational complete degradation but not processing of p105; critical role of serine 927.
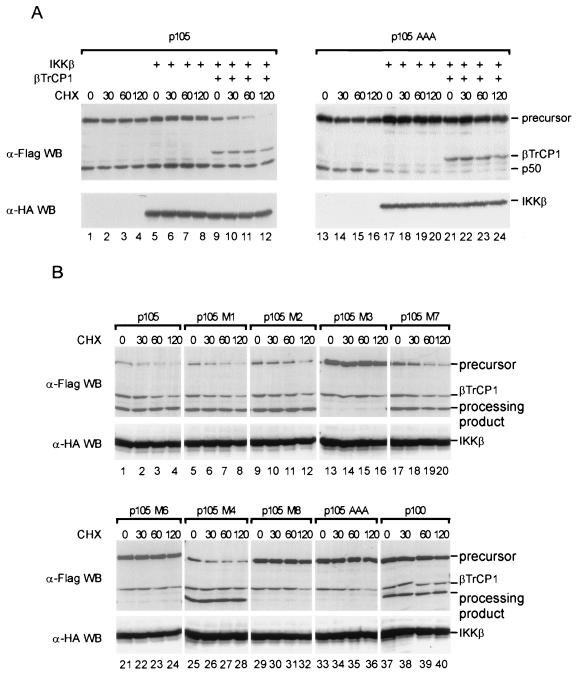
A matter of debate is the relation of p105 processing versus complete p105 degradation in response to IKK signaling (13, 38). In order to uncouple de novo synthesis and cotranslational processes from posttranslational events, the effect of IKKβ and βTrCP on p105 processing and degradation was analyzed in the presence of cycloheximide (Fig. 5A). When p105 was expressed alone, the total amounts of p105 and p50 as well as the precursor-product ratio did not change over time following translation inhibition (lanes 1 to 4). Coexpression of IKKβ resulted in increased production of both p105 and p50, perhaps by enhancing transcription of the p105 expression vector, but did not change the precursor-product ratio (lanes 5 to 8). However, when βTrCP1 was coexpressed with p105 and IKKβ, p105 amounts were gradually diminished after cycloheximide addition and nearly disappeared after 2 h (lanes 9 to 12). In stark contrast, the amounts of p50 were unaffected and did not increase, as expected if any posttranslational processing were regulated by a concerted action of IKKβ and βTrCP. Taken together, these results strongly suggest that complete degradation but not posttranslational processing of p105 is regulated by IκB kinases and βTrCP/SCF complexes.
FIG. 5.
IKKβ and βTrCP1 trigger serine 927-dependent degradation of p105. (A) Transfections were performed in quadruplicate. One fourth of the CaPO4 DNA precipitate was used to transfect a 6-cm plate of 293 cells. Each transfection contained a total of 20 μg of DNA with 2 μg of p105 expression vector (lanes 1 to 12) or 2 μg of p105AAA (lanes 13 to 24). For cotransfection, 4 μg of IKKβ (lanes 5 to 12 and 17 to 24) and 2 μg of βTrCP1-Flag (lanes 9 to 12 and 21 to 24) were used. Cells were treated with cycloheximide (CHX) for the indicated times, and extracts were prepared in RIPA buffer containing the complete protease inhibitor cocktail (Boehringer). (B) 293 cells were cotransfected with IKKβ, βTrCP1, and wild-type p105 or various p105 proteins (residues 18 to 968) carrying point mutations as shown in Fig. 2 or with an expression vector encoding p100. Transfection, stimulation, and lysis of cells were performed as described for panel A.
Intriguing observations were made when the IKK phosphorylation sites in p105 were inactivated in the same type of experiment: Here, the product-precursor ratio again was unaffected when the p105 mutant was expressed alone and analyzed at different time points after cycloheximide administration (lanes 13 to 16). However, slightly less p50 than p105 was produced. In contrast to wild-type p105, p105AAA was stable and was not degraded when coexpressed along with IKKβ and βTrCP (lanes 21 to 24), as predicted for a process dependent on IKK phosphorylation. However, surprisingly, a strong reduction in relative p50 amounts, and thus in processing, was obtained when the p105 mutant was coexpressed with IKKβ, and this reduction was independent of transfected βTrCP (lanes 17 to 24). These lower amounts of p50 did not change further after translation inhibition (lanes 17 to 20 and 21 to 24), and thus any interference of IKKβ with processing of mutant p105 must occur at or very shortly after translation (see also below).
We next assessed the effect of the single phosphorylation site mutants (Fig. 2) for IKKβ- and βTrCP-mediated posttranslational degradation and for relative levels of p105 processing (Fig. 5B). Both mutants in which phosphorylation by IKK was unaffected (M1 and M7) showed a time course of precursor degradation like that of wild-type p105, with a half-life of about 30 to 60 min. All proteins with a mutation of serine 923 but not of serine 927 (M2 and M4) displayed delayed degradation. Mutants M3, M6, M8, and p105AAA were completely stable and present at elevated levels. These proteins have in common a mutation of serine 927 to either alanine or threonine and are almost inactive IKK substrates. Thus, phosphorylation of S927 is the major prerequisite for functional βTrCP interaction with p105, in agreement with the positioning of this residue in the central DSG motif shared by all SCF substrates. The same mutants all revealed strongly reduced processing when coexpressed with IKKβ, while proteins defective in serine 923 but with serine 927 intact revealed processing levels like the wild-type protein (Fig. 5A and below).
In the same assay, we also tested p100. It was shown previously that following TNF-α stimulation, endogenous p100 was phosphorylated with the same kinetics as p105 or IκBα in orthophosphate-labeled cells; however, IKKα did not phosphorylate p100 in vitro (13, 37). In fact, p100 was not degraded in the presence of IKKβ and βTrCP after inhibition of translation, and processing to p52 was also unaffected (Fig. 5, lanes 37 to 40). Thus, the inducible in vivo phosphorylation of p100 observed earlier must be due to a kinase different from IKK but not necessarily independent of IKK.
IKKβ physically interacts with a docking site on p105 which is part of a death domain and separate from the substrate site.
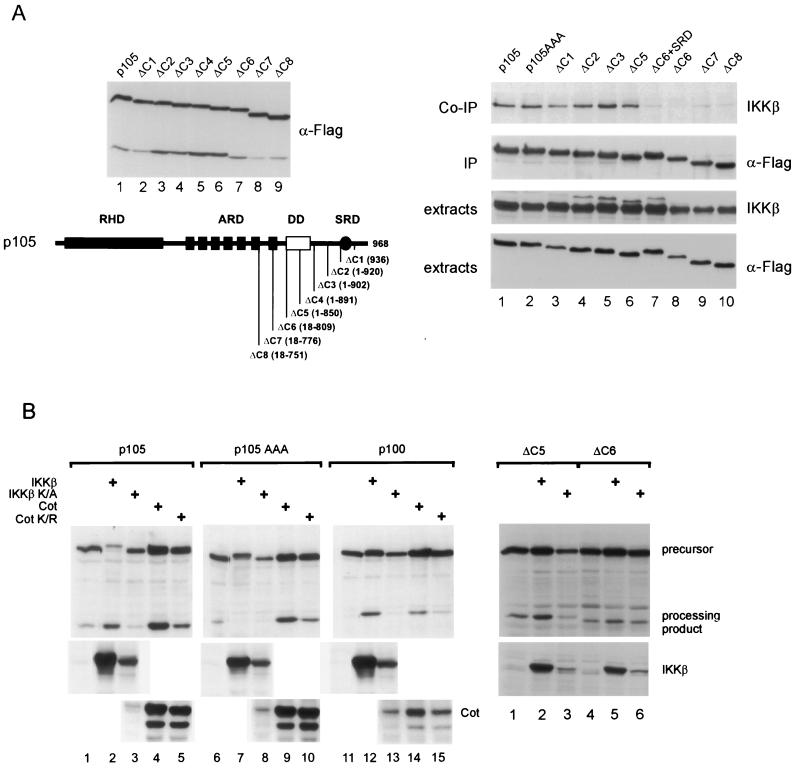
We previously showed that both IKKα and IKKβ can physically associate with p105 (13). To test the requirement of the phosphorylation sites for binding of IKKβ and to delineate the interacting region, p105 constructs with progressively deleted C-terminal sequences were generated (Fig. 6A, left panels). All deletion mutants underwent processing (Fig. 6A) and interacted with p65 (not shown). p105 wild-type and mutant proteins were coexpressed with IKKβ and subjected to coimmunoprecipitation (Fig. 6A, right panel). Intriguingly, point mutation of the IKK substrate serine residues in p105AAA did not affect the robust physical interaction of IKKβ (lanes 1 and 2). The p105 deletion mutants ΔC1 to ΔC5, either still containing all phosphorylation sites (ΔC1), lacking the major IKK phosphorylation sites (ΔC2), or devoid of all IKK sites, including cryptic sites (ΔC5), all interacted with IKKβ with the same efficiency as the wild-type protein (lanes 1 compared to 2 to 6). However, further deletion of sequences N-terminal to amino acid 850 (ΔC6, ΔC7, and ΔC8) resulted in an almost complete loss of interaction (lanes 1 to 6 compared to 8 to 10). The signal response domain, containing the IKK phosphorylation sites, could not restore IKKβ interaction when fused to ΔC6 (lane 7 versus 8). These results reveal that IKKβ binds to sequences in p105 located more than 70 amino acids amino terminal to the major phosphorylation sites and that the substrate serines do not contribute to efficient physical interaction. Similar findings were obtained for IKKα (data not shown). Thus, substrate recognition of p105, and probably of other substrates, by IKKs is not determined solely by specificity-determining residues in close proximity to the substrate serines, but also by the quality of a separate docking site on the substrate.
FIG. 6.
(A) Physical interaction of IKKβ with p105 is conferred by the N-terminal half of a death domain. (Left panel) p105 and C-terminal deletion constructs were expressed in 293 cells and analyzed in a Western blot for expression and basal processing levels. Bottom, schematic summary of deletion constructs. RHD, Rel homology domain; ARD, ankyrin repeat domain; DD, death domain (residues 805 to 892); SRD, signal response domain. Amino acids encoded by the deletion mutants are indicated. Right panel, 293 cells were transfected with 4 μg of p105, p105AAA, p105ΔC1-p105ΔC8, or p105ΔC6+SRD, as indicated, along with 3 μg of IKKβ expression vector (lanes 1 to 10). IKKβ, coprecipitated with Flag-tagged p105 proteins, is shown in an anti-IKKβ Western blot (top panel), expression of p105 proteins and IKKβ is shown by Western blotting of precipitated proteins or extracts with the respective antibodies, as indicated (lower panels). Cell lysis and immunoprecipitation was performed with RIPA buffer. Extracts or precipitated proteins were resolved by SDS-PAGE, probed with anti-IKKβ antibody, stripped, and reprobed with anti-Flag antibody. (B) Left panel, effect of kinases on processing efficiencies of p105, p105 mutants, and p100. In a total of 10 μg of DNA, Flag-tagged p105, p105AAA, and p100 (2 μg of each) were transfected alone (lanes 1, 6, and 11) or along with 4 μg of the different kinase expression vectors for IKKβ (lanes 2, 7, and 12), IKKβK/A (lanes 3, 8, and 13), Cot (lanes 4, 9, and 14), or CotK/R (lanes 5, 10, and 15), as indicated. Top panels, precursor and processing products in Western blots revealed by anti-Flag antibody. Middle and bottom panels, Western blots of extracts with anti-IKKβ and anti-Cot antibodies. Right panel, Flag-p105ΔC5 and -p105ΔC6 were transfected either alone (lanes 1 and 4) or together with IKKβ (lanes 2 and 5) or IKKβK/A (lanes 3 and 6) and analyzed as described above.
Interestingly, the IKK docking site is located in the N-terminal half of a death domain which is phylogenetically conserved in p105 (9, 44) (Fig. 6A, left panel).
The demonstration of an IKK docking site also provides an explanation of why IKKβ overexpression represses processing of p105 mutants lacking the phosphorylation sites. By physically interacting with the p105 mutants at an early step in the biogenesis of p105 and p50, unable to dissociate upon substrate phosphorylation due to lack of phosphorylation sites, it could hinder access of the proteasome or other processing components or affect p105 folding. This would predict that an enzymatically inactive IKK mutant should have the same effect.
We have investigated the consequence of kinase expression on p105 processing in more detail (Fig. 6B). While expression of wild-type IKKβ led to increased p50, presumably by increasing p105 expression and concomitant p105 degradation, inactive IKKβ reduced the amounts of p50 (Fig. 6B, left panel, compare lane 1 with 2 and 3). We also tested the mitogen-activated protein kinase kinase kinase-related kinase Cot, which does not directly phosphorylate p105. Like IKKβ, Cot increased p105 expression and thus also the amount of p50, while inactive Cot had no discernible effect (lanes 4 and 5). However, coexpression of either wild-type or kinase-inactive IKKβ with p105AAA strongly impaired p50 production while having no effect on the p105 amounts (lanes 6 to 8). The very similar effect of wild-type and kinase-inactive IKKβ on p105 mutants clearly demonstrates that the kinase activity of IKKβ was not required for processing inhibition, which is thus likely due to physical interaction. As with wild-type p105, Cot and its kinase-dead variant did not repress p50 production from p105AAA; instead, amounts of p105 and p50 were slightly enhanced. Furthermore, these experiments also make clear that the IKK sites are in principle not required for basal processing to occur (lanes 1 and 6) (see also Fig. 5A, lanes 13 to 16, and Fig. 6A, left panel). We have also analyzed the role of the IKK docking site for processing inhibition of p105 mutants by IKKβ (Fig. 6B, right panel). p50 formation from ΔC5, containing the docking site, was reduced by coexpressed inactive IKKβ (lane 1 compared to 3). In contrast, no change in p50 levels was observed for ΔC6, lacking the docking site (lanes 4 and 6), when coexpressed with IKKβK/A.
When p100, which is not an IKK phosphorylation substrate, was tested in the same experimental setting, only a very low level of processing was observed under these conditions (Fig. 6B, left panel). However, cotransfection of IKKβ, but not of IKKβK/A, led to strong accumulation of the processing product (lanes 11 to 13). Likewise, Cot but not kinase-inactive Cot led to an apparent increase in p100 processing (compare lane 11 with 14 and 15). The increase in the processing product p52 with either IKKβ or Cot was paralleled in both cases by increased p100 levels, suggesting that these kinases indeed primarily act on the expression of p100 and that processing is secondary to this effect.
LPS induces degradation but not processing of cellular p105; requirement for a functional IKK complex containing IKKγ.
The data obtained from in vitro experiments and from transfected cells strongly suggest a model for activation of cytoplasmic p105 complexes to release sequestered NF-κB subunits, including the p105 processing product p50. IKKs phosphorylate a destruction box equivalent to that of the small IκBs and generate a recognition site for βTrCP-containing SCF ubiquitin ligases. Subsequent polyubiquitination of p105 leads to complete degradation.
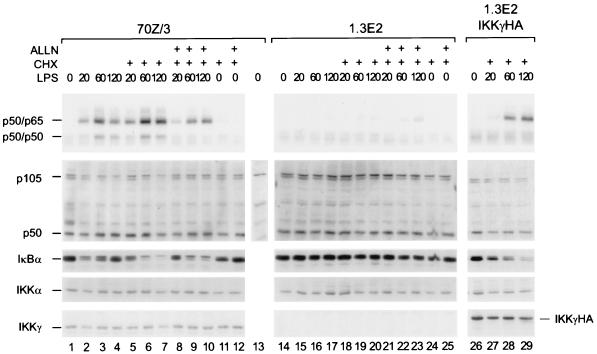
To further prove that degradation of endogenous p105 is observed with a physiological NF-κB inducer and that an intact endogenous IKK complex is required, we tested the pre-B-cell line 70Z/3 and its mutant derivative 1.3E2. These mutant cells lack IKKγ (NEMO), the only regular noncatalytic subunit of the tripartite IKK complex (22, 43, 57). LPS treatment of 70Z/3 cells in fact led to a rapid decrease in p105 amounts which was more pronounced when de novo protein synthesis was blocked with cycloheximide (Fig. 7, left panel, lanes 1 to 4 and 5 to 7). This decrease was completely blocked by the proteasome inhibitor ALLN (lanes 8 to 10). Despite the decrease in p105, no changes in p50 amounts were observed. Thus, LPS primarily induced p105 degradation rather than processing. Similarly, as expected, LPS treatment resulted in degradation of IκBα at 20 min and the appearance of a phosphoform with retarded migration. IκBα de novo synthesis was observed at later time points and was inhibited by cycloheximide. In striking contrast, LPS did not affect the stability of either p105 or IκBα in 1.3E2 mutant cells (Fig. 7, center panels). The LPS-induced decreases in steady-state amounts of IκBα and p105 in wild-type cells resulted in induced NF-κB DNA-binding activities (upper left panel). Release of p50-p65 complexes paralleled IκBα degradation, as expected, and p50 homodimers were released upon degradation of p105, but not when p105 degradation was blocked with ALLN. No activation of hetero- or homodimers was observed in the mutant cells. LPS induction of the DNA-binding species p50-p65 and p50-p50 as well as of IκBα and p105 degradation could be partially rescued by stable expression in 1.3E2 cells of IKKγ (right panels). Taken together, these data demonstrate that a functional endogenous IKK complex containing IKKγ is required for LPS-induced p105 degradation and p50 release. Since the relative levels of p50 are unchanged in the absence of IKKγ, processing in these cells is a constitutive mechanism, independent of IKK signaling.
FIG. 7.
Requirement of a functional cellular IKK complex for rapid LPS-induced proteolysis of p105. 70Z/3 cells (lanes 1 to 13), 1.3E2 cells (lanes 14 to 25), or 1.3E2 cells stably expressing HA-tagged IKKγ (lanes 26 to 29) were LPS stimulated for the indicated times in either the presence or absence of cycloheximide (CHX) or CHX and ALLN (30 min of incubation), as indicated. As a control, cells were treated with CHX or CHX and ALLN alone for 150 min (lanes 11, 12, 24, and 25). An EMSA with NF-κB DNA-binding activities is shown in the top panel. Migration of p50-p65 as well as of p50-p50 complexes is indicated. Detection of p105, p50, IκBα, IKKα, and IKKγ by Western blotting is shown below, and the migration of the specific bands is indicated. Please note that a nonspecific band migrates slightly slower than p105, as determined by peptide competition (lane 13).
DISCUSSION
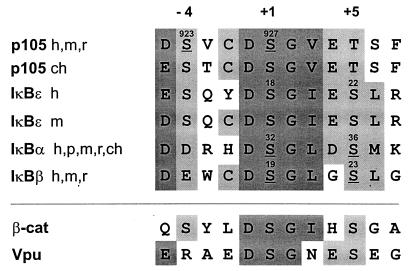
The NF-κB protein p105 acts in a dual fashion as a cytoplasmic IκB molecule, able to associate with other NF-κB subunits, and as the precursor for the p50 subunit. We have shown previously that p105 is subject to signal-dependent proteolysis, which gives rise to induced release of p50 homodimers and is mediated by the IKK complex (13). In this work we have investigated specificity-determining residues for the action of IKK and provide evidence for IKK-dependent ubiquitination and complete degradation of p105. By using purified recombinant IKKβ, we have shown first that IKK directly phosphorylates p105. This experiment is important, since it excludes the formal possibility that unknown kinases downstream of IKK could account for the observed p105 phosphorylation. Using this purified kinase, we have determined that serines 923 and 927 are the major substrate sites. Serines 923 and 927 are in the same spacing (SXXXS) as the phosphorylation sites in IκBα, -β, and -ɛ and are both preceded by acidic amino acids (Fig. 8). The p105 sequence in fact bears extended similarity with the sequences surrounding IKK sites in the small IκBs. Serine 927 is part of a conserved DSGΨ (where Ψ is a hydrophobic residue) motif. All small IκBs and p105 share at the −5 position of this serine an acidic amino acid and at −4 an acidic residue or serine. The −2 and +3 positions bear further IκB-specific preferences, cysteine and acidic residues, respectively. However, compared to the small IκBs, the phosphorylated residues in p105 (Fig. 8, underlined) are in nonequivalent positions, −4 and +1 versus +1 and +5 relative to the generally conserved central DSGψ motif (Fig. 8). The sequence conservation suggests that IKK may utilize solely serines in the +4 spacing, preferentially when both are preceded by acidic residues. This, however, is not sufficient to create a bona fide IKK substrate site, since Vpu is not phosphorylated by IKK (D. Krappmann, not shown). Perhaps the −4 and −5 positions of the IκBs are specificity-determining residues.
FIG. 8.
Phylogenetic conservation (h, human; m, murine; r, rat; ch, chicken; p, porcine) of destruction boxes in p105, in IκBα,-β, and -ɛ, in β-catenin, and in HIV-1 Vpu. Position numbers of the critical serine residues in the respective full-length proteins are indicated. Serines where stimulated phosphorylation has been shown experimentally are underlined.
Functionally, the conserved serines differ in the various IκBs. Unlike the situation in IκBα, where single mutation of either serine 32 or 36 did not fully abrogate inducible phosphorylation (6, 51), mutation of S927 almost completely abrogated phosphorylation of p105. In IκBɛ, any mutations of serines in the conserved motif had little or no effect on phosphorylation (45), although degradation was abolished by mutation of serines 18 and 22 to alanine (52). Perhaps the relatively high number of serines close to the conserved substrate site in IκBɛ provide alternative phosphorylation sites for the IKK complex in the mutant proteins.
Our analysis of physical interaction of IKKβ with p105 revealed that the kinase binds to a region that is nonoverlapping with the destruction box and that mutation or deletion of the substrate serines does not affect interaction strength. The docking site was delineated to the N-terminal half of a death domain and is separated by 70 amino acids from the destruction box (Fig. 6A). The interaction of IKK with a docking site may contribute to substrate recognition in addition to specificity-determining residues flanking the substrate serines.
The death domain confers homo- and heterotypic protein interactions and is mostly found in receptors or adaptors that signal cell death. It is also conserved in signaling molecules which are not associated with apoptotic pathways but regulate NF-κB activation, such as Drosophila Pelle and Tube, which are part of the Toll-Dorsal pathway (9, 56). The fact that the death domain is phylogenetically conserved in p105 and in p100 (reference 44 and data not shown) indicates an essential function for these molecules. The physical IKK-death domain interaction may indicate that IKKs also interact with death domains of other molecules and that this interaction could be relevant for recruitment of IKKs to activated receptor-adapter complexes. Similarly, the death domain could engage the precursors into heterotypic complexes with other signaling molecules.
We have also analyzed the physical interaction of IKKs with IκBα and IκBβ, which do not contain a death domain. Compared to the robust IKK-p105 interaction, IκBα was only weakly bound by IKKβ (V. Heissmeyer, unpublished data). However, human IκBβ revealed a stronger interaction with IKKβ, which was conferred by a C-terminal PEST sequence shared by the IκBβ1 and IκBβ2 splicing isoforms (V. Heissmeyer, unpublished data).
In contrast to p105, p100 is not phosphorylated by IKKs (13), consistent with the fact that the carboxy-terminal amino acids of p100, downstream of the death domain, show no conservation with p105. Accordingly, p100 was not degraded upon coexpression with IKKβ and βTrCP (Fig. 5B and 6A). Thus, p100 is the only cytoplasmic IκB protein not directly phosphorylated by IKKs. However, IKKβ binds to p100 (V. Heissmeyer, data not shown). It is thus possible that IKK, once bound to the death domain of p100, activates a further, unknown kinase to phosphorylate p100.
The overall similarity of the sequence context of IKK phosphorylation sites in p105 and IκBα suggested that p105 should interact with the same type of ubiquitin ligase as IκBα. In fact, the interaction efficiency of βTrCP1 with IκBα and p105 was virtually identical. Furthermore, when comparing the related F-box proteins βTrCP1 and βTrCP2, which bind equally well to phosphorylated IκBα (47), both also interact with phosphorylated p105 with comparable efficiency. The interaction with βTrCP2 was lost completely when the major IKK sites were mutated (p105AAA), indicating that the phosphorylated minor sites in p105 (between residues 850 and 891) cannot attract the F-box protein. The binding of both βTrCPs again underscores the similarity of the destruction boxes in p105 and the small IκBs and discriminates these proteins from β-catenin, which, upon GSK3β phosphorylation, can attract βTrCP1 but not βTrCP2 (11). Our data also reveal that the last residue in the DSGΨXS consensus sequence for βTrCP recognition is not maintained for p105, which contains a threonine, a very poor IKK substrate. This is intriguing, since the DSGΨXS motif is strictly conserved in all other proven and potential βTrCP substrates (Fig. 8), including armadillo and plakoglobin (not shown). The last serine in the motif is functionally important in IκBα, since single mutation of this residue (serine 36) completely abolishes induced degradation (4, 6). Yaron et al. (58) have shown that short IκBα competitor peptides with singly phosphorylated serine 36 or 32 have strongly impaired inhibitory effects on IκBα ubiquitination compared to their doubly phosphorylated counterparts. It is therefore possible that βTrCPs recognize the phosphorylated signal sequences in IκBα and p105 in a slightly different manner.
We have shown that coexpression of IKKβ and βTrCP1 triggers p105 polyubiquitination which results in complete proteasomal degradation but not in enhanced processing of p105 (Fig. 4 and 5). This result is in contrast to the conclusions drawn by Orian et al. (38), who reported that IKK predominantly enhanced processing. We demonstrated that the expression of IKKβ alone led to an increase in p50, but this effect is ascribed to IKKβ-induced expression of p105, resulting in increased amounts of p50 produced by processing and loss of p105 by simultaneous IKKβ-induced degradation. This conclusion is also supported by the observation that IKKβ, which does not phosphorylate p100, enhances p52 production along with p100 expression. Likewise, Cot, a kinase which does not phosphorylate p105, enhanced production of p50 and p52 as well as of p105 and p100, most likely by acting on the expression vector. Importantly, by the use of cycloheximide, we have shown that at the posttranslational level, and thus independent of any effects of the kinase on the expression vector, IKKβ (βTrCP) triggered complete degradation but not processing of p105. The observed degradation was fully dependent on serine 927, in agreement with the pivotal role of this residue as an IKK phosphoacceptor site. We also showed that in mouse pre-B cells, LPS triggered degradation but not processing of endogenous p105. LPS-induced degradation but not basal processing required a functional endogenous IKK complex.
Our data fully support the notion that p105 contains a carboxy-terminal destruction box that, like the N-terminal domain in IκBα, upon IKK phosphorylation, is recognized by an SCFβTrCP E3 ubiquitin ligase which mediates polyubiquitination and complete degradation by the proteasome. Thus, p105 is degraded by the same mechanism as IκBα. It is also interesting to note that both proteins can obviously be degraded when complexed with Rel factors (p50-p65 and the processing product of p105 or other p105-associated Rel factors, respectively).
The basal processing reaction, in contrast, has been shown to require a glycine-rich region (residues 372 to 394) and an acidic domain (residues 446 to 454) (27, 39), both located at the end of the first half of the precursor. That the basal processing reaction does not require carboxy-terminal sequences containing the destruction box described here is also supported by the fact that deletion or mutation of the IKK phosphorylation sites in p105 does not affect basal processing and that processing was not reduced in cells lacking a functional IKK complex.
IKK-regulated and ubiquitin-mediated p105 degradation is an important bifurcation in NF-κB signaling downstream of the IKK complex. This bifurcation provides a means to regulate p50 homodimers, which may act as inhibitors to limit transcriptional responses of p50-p65 or as activators, depending on the availability of Bcl-3. To dissect the regulation of p50 and Bcl-3 is important for understanding the function of these molecules in the immune response and in oncogenesis.
ACKNOWLEDGMENTS
We thank Michael Karin for the gift of IKKα and -β expression vectors and baculovirus constructs, Yinon Ben-Neriah for the βTrCP1 expression vector, Serge Fuchs and Ze'ev Ronai for the HOS/βTrCP2 plasmid, and Ulf Rapp for Cot constructs. 1.3E2 cells were provided by Carol Sibley. We thank Erika Scharschmidt for expert technical assistance and Rudolf Dettmer for purification of IKKs.
RERERENCES
- 1.Baeuerle P A, Baltimore D. NF-κB: ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 2.Belich M P, Salmeron A, Johnston L H, Ley S C. TPL-2 kinase regulates the proteolysis of the NF-κB-inhibitory protein NF-κB1 p105. Nature. 1999;397:363–368. doi: 10.1038/16946. [DOI] [PubMed] [Google Scholar]
- 3.Bours V, Villalobos J, Burd P R, Kelly K, Siebenlist U. Cloning of a mitogen-inducible gene encoding a κB DNA-binding protein with homology to the rel oncogene and to cell-cycle motifs. Nature. 1990;348:76–80. doi: 10.1038/348076a0. [DOI] [PubMed] [Google Scholar]
- 4.Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlist U. Control of IκBα proteolysis by site-specific, signal-induced phosphorylation. Science. 1995;267:1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- 5.Coux O, Goldberg A L. Enzymes catalyzing ubiquitination and proteolytic processing of the p105 precursor of nuclear factor κB1. J Biol Chem. 1998;273:8820–8828. doi: 10.1074/jbc.273.15.8820. [DOI] [PubMed] [Google Scholar]
- 6.DiDonato J, Mercurio F, Rosette C, Li J W, Suyang H, Ghosh S, Karin M. Mapping of the inducible IκB phosphorylation sites that signal its ubiquitination and degration. Mol Cell Biol. 1996;16:1295–1304. doi: 10.1128/mcb.16.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donald R, Ballard D W, Hawiger J. Proteolytic processing of NF-κB/IκB in human monocytes. ATP-dependent induction by pro-inflammatory mediators. J Biol Chem. 1995;270:9–12. doi: 10.1074/jbc.270.1.9. [DOI] [PubMed] [Google Scholar]
- 8.Fan C M, Maniatis T. Generation of p50 subunit of NF-κB by processing of p105 through an ATP-dependent pathway. Nature. 1991;354:395–398. doi: 10.1038/354395a0. [DOI] [PubMed] [Google Scholar]
- 9.Feinstein E, Kimchi A, Wallach D, Boldin M, Varfolomeev E. The death domain: a module shared by proteins with diverse cellular functions. Trends Biochem Sci. 1995;20:342–344. doi: 10.1016/s0968-0004(00)89070-2. [DOI] [PubMed] [Google Scholar]
- 10.Fuchs S Y, Chen A, Xiong Y, Pan Z Q, Ronai Z. HOS, a human homolog of Slimb, forms an SCF complex with Skp1 and Cullin1 and targets the phosphorylation-dependent degradation of IκB and β-catenin. Oncogene. 1999;18:2039–2046. doi: 10.1038/sj.onc.1202760. [DOI] [PubMed] [Google Scholar]
- 11.Hart M, Concordet J P, Lassot I, Albert I, del los Santos R, Durand H, Perret C, Rubinfeld B, Margottin F, Benarous R, Polakis P. The F-box protein β-TrCP associates with phosphorylated β-catenin and regulates its activity in the cell. Curr Biol. 1999;9:207–210. doi: 10.1016/s0960-9822(99)80091-8. [DOI] [PubMed] [Google Scholar]
- 12.Hatada E N, Krappmann D, Scheidereit C. NF-κB and the innate immune response. Curr Opin Immunol. 2000;12:52–58. doi: 10.1016/s0952-7915(99)00050-3. [DOI] [PubMed] [Google Scholar]
- 13.Heissmeyer V, Krappmann D, Wulczyn F G, Scheidereit C. NF-κB p105 is a target of IκB kinases and controls signal induction of Bcl-3-p50 complexes. EMBO J. 1999;18:4766–4778. doi: 10.1093/emboj/18.17.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heron E, Deloukas P, van Loon A P. The complete exon-intron structure of the 156-kb human gene NFKB1, which encodes the p105 and p50 proteins of transcription factors NF-κB and IκBγ: implications for NF-κB-mediated signal transduction. Genomics. 1995;30:493–505. doi: 10.1006/geno.1995.1270. [DOI] [PubMed] [Google Scholar]
- 15.Heusch M, Lin L, Geleziunas R, Greene W C. The generation of nfkb2 p52: mechanism and efficiency. Oncogene. 1999;18:6201–6208. doi: 10.1038/sj.onc.1203022. [DOI] [PubMed] [Google Scholar]
- 16.Hirano F, Chung M, Tanaka H, Maruyama N, Makino I, Moore D D, Scheidereit C. Alternative splicing variants of IκBβ establish differential NF-κB signal responsiveness in human cells. Mol Cell Biol. 1998;18:2596–2607. doi: 10.1128/mcb.18.5.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 18.Karin M, Delhase M. The IκB kinase (IKK) and NF-κB: key elements of proinflammatory signalling. Semin Immunol. 2000;12:85–98. doi: 10.1006/smim.2000.0210. [DOI] [PubMed] [Google Scholar]
- 19.Kieran M, Blank V, Logeat F, Vandekerckhove J, Lottspeich F, Le Bail O, Urban M B, Kourilsky P, Baeuerle P A, Israel A. The DNA binding subunit of NF-κB is identical to factor KBF1 and homologous to the re1 oncogene product. Cell. 1990;62:1007–1018. doi: 10.1016/0092-8674(90)90275-j. [DOI] [PubMed] [Google Scholar]
- 20.Kitagawa M, Hatakeyama S, Shirane M, Matsumoto M, Ishida N, Hattori K, Nakamichi I, Kikuchi A, Nakayama K. An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of β-catenin. EMBO J. 1999;18:2401–2410. doi: 10.1093/emboj/18.9.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krappmann D, Emmerich F, Kordes U, Scharschmidt E, Dorken B, Scheidereit C. Molecular mechanisms of constitutive NF-κB/Rel activation in Hodgkin/Reed-Sternberg cells. Oncogene. 1999;18:943–953. doi: 10.1038/sj.onc.1202351. [DOI] [PubMed] [Google Scholar]
- 22.Krappmann D, Hatada E N, Tegethoff S, Li J, Klippel A, Giese K, Baeuerle P A, Scheidereit C. The IκB kinase (IKK) complex is tripartite and contains IKKγ but not IKAP as a regular component. J Biol Chem. 2000;275:29779–29787. doi: 10.1074/jbc.M003902200. [DOI] [PubMed] [Google Scholar]
- 23.Lee J W, Choi H S, Gyuris J, Brent R, Moore D D. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol Endocrinol. 1995;9:243–254. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Peet G W, Pullen S S, Schembri King J, Warren T C, Marcu K B, Kehry M R, Barton R, Jakes S. Recombinant IκB kinases α and β are direct kinases of IκB. J Biol Chem. 1998;273:30736–30741. doi: 10.1074/jbc.273.46.30736. [DOI] [PubMed] [Google Scholar]
- 25.Lin L, DeMartino G N, Greene W C. Cotranslational biogenesis of NF-κB p50 by the 26S proteasome. Cell. 1998;92:819–828. doi: 10.1016/s0092-8674(00)81409-9. [DOI] [PubMed] [Google Scholar]
- 26.Lin L, DeMartino G N, Greene W C. Cotranslational dimerization of the rel homology domain of NF-κB1 generates p50–p105 heterodimers and is required for effective p50 production. EMBO J. 2000;19:4712–4722. doi: 10.1093/emboj/19.17.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin L, Ghosh S. A glycine-rich region in NF-κB p105 functions as a processing signal for the generation of the p50 subunit. Mol Cell Biol. 1996;16:2248–2254. doi: 10.1128/mcb.16.5.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin X, Cunningham E T J, Mu Y, Geleziunas R, Greene W C. The proto-oncogene Cot kinase participates in CD3/CD28 induction of NF-κB acting through the NF-κB-inducing kinase and IκB kinases. Immunity. 1999;10:271–280. doi: 10.1016/s1074-7613(00)80027-8. [DOI] [PubMed] [Google Scholar]
- 29.MacKichan M L, Logeat F, Israel A. Phosphorylation of p105 PEST sequence via a redox-insensitive pathway up-regulates processing of p50 NF-κB. J Biol Chem. 1996;271:6084–6091. doi: 10.1074/jbc.271.11.6084. [DOI] [PubMed] [Google Scholar]
- 30.Margottin F, Bour S P, Durand H, Selig L, Benichou S, Richard V, Thomas D, Strebel K, Benarous R. A novel human WD protein, h-βTrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol Cell. 1998;1:565–574. doi: 10.1016/s1097-2765(00)80056-8. [DOI] [PubMed] [Google Scholar]
- 31.May M J, Ghosh S. Signal transduction through NF-κB. Immunol Today. 1998;19:80–88. doi: 10.1016/s0167-5699(97)01197-3. [DOI] [PubMed] [Google Scholar]
- 32.Mayo M W, Baldwin A S. The transcription factor NF-κB: control of oncogenesis and cancer therapy resistance. Biochim Biophys Acta. 2000;1470:M55–M62. doi: 10.1016/s0304-419x(00)00002-0. [DOI] [PubMed] [Google Scholar]
- 33.Medzhitov R, Janeway C., Jr Innate immune recognition: mechanisms and pathways. Immunol Rev. 2000;173:89–97. doi: 10.1034/j.1600-065x.2000.917309.x. [DOI] [PubMed] [Google Scholar]
- 34.Mellits K H, Hay R T, Goodbourn S. Proteolytic degradation of MAD3 (IκBα) and enhanced processing of the NF-κB precursor p105 are obligatory steps in the activation of NF-κB. Nucleic Acids Res. 1993;21:5059–5066. doi: 10.1093/nar/21.22.5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mercurio F, DiDonato J A, Rosette C, Karin M. p105 and p98 precursor proteins play an active role in NF-κB-mediated signal transduction. Genes Dev. 1993;7:705–718. doi: 10.1101/gad.7.4.705. [DOI] [PubMed] [Google Scholar]
- 36.Meyer R, Hatada E N, Hohmann H P, Haiker M, Bartsch C, Rothlisberger U, Lahm H W, Schlaeger E J, van Loon A P, Scheidereit C. Cloning of the DNA-binding subunit of human nuclear factor κB: the level of its mRNA is strongly regulated by phorbol ester or tumor necrosis factor α. Proc Natl Acad Sci USA. 1991;88:966–970. doi: 10.1073/pnas.88.3.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naumann M, Scheidereit C. Activation of NF-κB in vivo is regulated by multiple phosphorylations. EMBO J. 1994;13:4597–4607. doi: 10.1002/j.1460-2075.1994.tb06781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orian A, Gonen H, Bercovich B, Fajerman I, Eytan E, Israel A, Mercurio F, Iwai K, Schwartz A L, Ciechanover A. SCFβ-TrCP ubiquitin ligase-mediated processing of NF-κB p105 requires phosphorylation of its C-terminus by IκB kinase. EMBO J. 2000;19:2580–2591. doi: 10.1093/emboj/19.11.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orian A, Schwartz A L, Israel A, Whiteside S, Kahana C, Ciechanover A. Structural motifs involved in ubiquitin-mediated processing of the NF-κB precursor p105: roles of the glycine-rich region and a downstream ubiquitination domain. Mol Cell Biol. 1999;19:3664–3673. doi: 10.1128/mcb.19.5.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orian A, Whiteside S, Israel A, Stancovski I, Schwartz A L, Ciechanover A. Ubiquitin-mediated processing of NF-κB transcriptional activator precursor p105: reconstitution of a cell-free system and identification of the ubiquitin-carrier protein, E2, and a novel ubiquitin-protein ligase, E3, involved in conjugation. J Biol Chem. 1995;270:21707–21714. doi: 10.1074/jbc.270.37.21707. [DOI] [PubMed] [Google Scholar]
- 41.Palombella V J, Rando O J, Goldberg A L, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-κB1 precursor protein and the activation of NF-κB. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 42.Rayet B, Gelinas C. Aberrant rel/nfkb genes and activity in human cancer. Oncogene. 1999;18:6938–6947. doi: 10.1038/sj.onc.1203221. [DOI] [PubMed] [Google Scholar]
- 43.Rooney J W, Emery D W, Sibley C H. 1.3E2, a variant of the B lymphoma 70Z/3, defective in activation of NF-κB and OTF-2. Immunogenetics. 1990;31:73–78. doi: 10.1007/BF00661216. [DOI] [PubMed] [Google Scholar]
- 44.Schultz J, Copley R R, Doerks T, Ponting C P, Bork P. SMART: a Web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shirane M, Hatakeyama S, Hattori K, Nakayama K. Common pathway for the ubiquitination of IκBα, IκBβ, and IκBɛ mediated by the F-box protein FWD1. J Biol Chem. 1999;274:28169–28174. doi: 10.1074/jbc.274.40.28169. [DOI] [PubMed] [Google Scholar]
- 46.Spencer E, Jiang J, Chen Z J. Signal-induced ubiquitination of IκBα by the F-box protein Slimb/β-TrCP. Genes Dev. 1999;13:284–294. doi: 10.1101/gad.13.3.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki H, Chiba T, Kobayashi M, Takeuchi M, Suzuki T, Ichiyama A, Ikenoue T, Omata M, Furuichi K, Tanaka K. IκBα ubiquitination is catalyzed by an SCF-like complex containing Skp1, cullin-1, and two F-box/WD40-repeat proteins, βTrCP1 and βTrCP2. Biochem Biophys Res Commun. 1999;256:127–132. doi: 10.1006/bbrc.1999.0289. [DOI] [PubMed] [Google Scholar]
- 48.Suzuki H, Chiba T, Suzuki T, Fujita T, Ikenoue T, Omata M, Furuichi K, Shikama H, Tanaka K. Homodimer of two F-box proteins βTrCP1 or βTrCP2 binds to IκBα for signal-dependent ubiquitination. J Biol Chem. 2000;275:2877–2884. doi: 10.1074/jbc.275.4.2877. [DOI] [PubMed] [Google Scholar]
- 49.Tan P, Fuchs S Y, Chen A, Wu K, Gomez C, Ronai Z, Pan Z Q. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IκBα. Mol Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- 50.Thompson J E, Phillips R J, Erdjument Bromage H, Tempst P, Ghosh S. IκBβ regulates the persistent response in a biphasic activation of NF-κB. Cell. 1995;80:573–582. doi: 10.1016/0092-8674(95)90511-1. [DOI] [PubMed] [Google Scholar]
- 51.Traenckner E B, Pahl H L, Henkel T, Schmidt K N, Wilk S, Baeuerle P A. Phosphorylation of human IκBα on serines 32 and 36 controls IκBα proteolysis and NF-κB activation in response to diverse stimuli. EMBO J. 1995;14:2876–2883. doi: 10.1002/j.1460-2075.1995.tb07287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whiteside S T, Epinat J C, Rice N R, Israel A. IκBɛ, a novel member of the IκB family, controls RelA and cRel NF-κB activity. EMBO J. 1997;16:1413–1426. doi: 10.1093/emboj/16.6.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winston J T, Strack P, Beer Romero P, Chu C Y, Elledge S J, Harper J W. The SCFβ-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu C, Ghosh S. β-TrCP mediates the signal-induced ubiquitination of IκBβ. J Biol Chem. 1999;274:29591–29594. doi: 10.1074/jbc.274.42.29591. [DOI] [PubMed] [Google Scholar]
- 55.Wulczyn F G, Krappmann D, Scheidereit C. The NF-κB/Rel and IκB gene families: mediators of immune response and inflammation. J Mol Med. 1996;74:749–769. doi: 10.1007/s001090050078. [DOI] [PubMed] [Google Scholar]
- 56.Xiao T, Towb P, Wasserman S A, Sprang S R. Three dimensional structure of a complex between the death domains of Pelle and Tube. Cell. 1999;99:545–555. doi: 10.1016/s0092-8674(00)81542-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamaoka S, Courtois G, Bessia C, Whiteside S T, Weil R, Agou F, Kirk H E, Kay R J, Israel A. Complementation cloning of NEMO, a component of the IκB kinase complex essential for NF-κB activation. Cell. 1998;93:1231–1240. doi: 10.1016/s0092-8674(00)81466-x. [DOI] [PubMed] [Google Scholar]
- 58.Yaron A, Gonen H, Alkalay I, Hatzubai A, Jung S, Beyth S, Mercurio F, Manning A M, Ciechanover A, Ben Neriah Y. Inhibition of NF-κB cellular function via specific targeting of the IκB-ubiquitin ligase. EMBO J. 1997;16:6486–6494. doi: 10.1093/emboj/16.21.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning A M, Andersen J S, Mann M, Mercurio F, Ben Neriah Y. Identification of the receptor component of the IκBα-ubiquitin ligase. Nature. 1998;396:590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]