Abstract
Endothelins are a family of biologically active peptides that are critical for development and function of neural crest-derived and cardiovascular cells. These effects are mediated by two G-protein-coupled receptors and involve transcriptional regulation of growth-responsive and/or tissue-specific genes. We have used the cardiac ANF promoter, which represents the best-studied tissue-specific endothelin target, to elucidate the nuclear pathways responsible for the transcriptional effects of endothelins. We found that cardiac-specific response to endothelin 1 (ET-1) requires the combined action of the serum response factor (SRF) and the tissue-restricted GATA proteins which bind over their adjacent sites, within a 30-bp ET-1 response element. We show that SRF and GATA proteins form a novel ternary complex reminiscent of the well-characterized SRF-ternary complex factor interaction required for transcriptional induction of c-fos in response to growth factors. In transient cotransfections, GATA factors and SRF synergistically activate atrial natriuretic factor and other ET-1-inducible promoters that contain both GATA and SRF binding sites. Thus, GATA factors may represent a new class of tissue-specific SRF accessory factors that account for muscle- and other cell-specific SRF actions.
Endothelins are a family of closely related peptide hormones (ET-1, -2, and -3) with essential functions for mammalian organogenesis and postnatal homeostasis (5, 39, 60). The founding member of the family, ET-1, was first identified as a potent endothelium-released vasoconstrictor (61). However, it is now well established that endothelins are synthesized in several tissues, where they act locally to promote cell growth and/or differentiation via two G-protein-coupled transmembrane domain receptors, ETA and ETB (1, 51). For example, ET-3–ETB interaction appears to be essential for development of melanocytes and myenteric ganglion neurons, since targeted or natural mutations of the ETB or ET-3 gene produce pigment abnormalities and aganglionic megacolon in mice and rats (6, 20, 29) and are associated with Hirschprung disease in humans (19, 47). On the other hand, ET-1-ETA interaction is evidently required for development of the heart and of specific neural crest-derived structures, and disruption of the ET-1 or ETA gene results in craniofacial and cardiovascular abnormalities that are incompatible with postnatal life (14, 33). Analysis of a number of markers suggests that, at least in the case of craniofacial development, ET-1–ETA signaling is required for normal cell proliferation and proper differentiation (15).
In addition to its essential role in embryonic development, ET-1 plays important functions in postnatal cardiovascular homeostasis, including regulation of cardiac and smooth muscle growth and contractility. In fact, the ET-1 pathway is dysregulated in several cardiovascular diseases, such as hypertension and heart failure, and ETA receptor antagonists prevent development of cardiac hypertrophy and increase survival in animal models of congestive heart failure (49, 50), confirming the potential relevance of targeting the ET-1/ETA pathway in human diseases (64).
The profound cellular effects elicited by ET-1 activation of ETA involve transcriptional modulation of growth-responsive and tissue-specific genes. For example, in cardiac and smooth muscle cells, where ET-1 has growth-promoting effects, several immediate-early genes like egr-1, c-jun, and c-fos are transiently induced in response to ET-1 treatment (24, 45, 52). Moreover, in cardiomyocytes, ET-1 modulates transcription of various cardiac-specific genes, including atrial natriuretic factor (ANF), the major heart secretory product (54, 55). The intracellular signaling cascades that are activated by ET1-ETA interaction have been extensively analyzed (7, 13, 25, 30, 43) and suggest that ETA stimulation can activate multiple signaling cascades via coupling to different G-proteins; for example, ETA coupling to Gq activates Ras-dependent pathways leading to stimulation of mitogen-activated protein kinases (MAPKs), whereas ETA coupling to Gi inhibits adenylate cyclase. Which of these cascades links ETA activation to the profound cellular and genetic changes induced by ET-1 remains undefined.
In order to elucidate the nuclear pathways that mediate cell-specific transcriptional responses to ET-1, we have used the cardiac ANF promoter, which at present is the best-studied tissue-specific ET-1 target. We present evidence showing that ET-1 responsiveness of the promoter requires the combined action of serum response factor (SRF) and tissue-specific GATA protein, which form a ternary complex over a 30-bp cis element harboring juxtaposed SRF and GATA binding sites. Formation of this complex requires both GATA and SRF binding sites and is mediated by physical interaction between the C-terminal zinc finger of GATA-4 and the DNA-binding domain of SRF. In transient-transfection assays, GATA factors and SRF synergistically activate several ET-1-inducible cardiac promoters. Thus, GATA proteins may represent a new class of tissue-specific SRF accessory factors that cooperate with SRF to mediate cell-specific nuclear signaling by extracellular stimuli.
MATERIALS AND METHODS
Cell cultures and transfections.
Neonatal cardiomyocytes were prepared from 4-day-old Sprague-Dawley rats and plated at a density of 125,000 cells/9.5-cm2 culture dish in six-well plates as previously described (11). For the ET-1 response element mapping assay, cardiomyocytes were transfected by calcium phosphate precipitation with 1.5 μg of wild-type or mutant ANF-luciferase reporter plasmids per 9.5-cm2 culture dish. From 16 to 20 h later, cardiomyocytes were washed twice with Dulbecco's modified Eagle's medium (DMEM; Canadian Life Technologies Inc.) and serum-free hormone-free medium was added (4). SFHF was supplemented with ET-1 or vehicle for 6 to 48 h. HeLa cells were plated at a density of 100,000 cells/9.5-cm2 dish in six-well plates (Falcon) in DMEM supplemented with 10% fetal bovine serum (qualified grade; Canadian Life Technologies Inc.). Transfections were carried out as in cardiomyocytes. For cotransfections, a total of 4 μg of expression vectors was used, generally 1 μg of SRF and 3 μg of GATA-4. The amount of DNA was kept constant by using the empty expression vector. Cardiomyocytes were harvested on the fifth day after plating, HeLa cells were harvested at 36 h posttransfection, and luciferase activity was assayed as previously described with a Berthold LB 953 luminometer (11).
Plasmids.
The mutations and deletions of the ANF promoter and the various GATA constructs have been described previously (3, 11, 18, 22, 42). For in vitro translation, GATA-4 constructs were subcloned into the pRSET plasmid as previously described (17). The luciferase reporter driven by the chicken −394αSkA has been described previously (44); the mouse −360c-fos and the rat −613αMHC luciferase constructs were kindly provided by T. Hoang (48) and P. M. Buttrick (10), respectively. The human SRF eukaryotic expression vectors used were either cytomegalovirus (CMV) (12) or simian virus 40 (SV40) driven (44). Human SRF was bacterially produced as a maltose-binding basic protein (MBP) fusion protein by subcloning a 1.6-kb XbaI-BamHI cDNA fragment from pCGN-SRF into the MBP coding region of the pMALC-2 vector (New England Biolabs). DNA fragments corresponding to the SRF DNA-binding domain including the MADS-box (amino acids 130 to 280) and the SRF MADS-box pm1, which contains point mutations at Arg143, Lys145, and Ile146, that abolish DNA binding were also subcloned into the pMALC-2 and pcDNA-3 vectors. All constructs were confirmed by sequencing.
Recombinant protein production.
SRF was bacterially produced as previously described (18). GATA-4 and SRF constructs were produced in vitro with or without [35S]methionine using the TNT coupled in vitro transcription-translation system (Promega Corp., Madison, Wis.).
Protein-protein binding assays.
In vitro binding studies were performed as previously described with minor modifications (17). In brief, 2 to 6 μl of 35S-labeled GATA proteins was incubated with 300 ng of immobilized SRF fusion proteins in 500 μl of binding buffer (150 mM NaCl, 50 mM Tris-Cl [pH 7.5], 0.3% Nonidet P-40, 1 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 0.25% bovine serum albumin [BSA]) for 2 h at 4°C with agitation and then centrifuged for 2 min at 15,000 rpm at room temperature (RT). Beads were washed three times by vortexing in 500 μl of binding buffer at RT and three times by vortexing in 500 μl of binding buffer without BSA. The protein complexes were released after boiling in Laemmli buffer and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Labeled proteins were visualized and quantified using a PhosphorImager screen and a STORM system (Molecular Dynamics).
Immunoprecipitations and immunoblots.
Coimmunoprecipitations of Flag-GATA-4 and hemagglutinin (HA)-SRF were carried out using nuclear extracts of 293T cells overexpressing the relevant proteins. Nuclear extracts were prepared as previously described (42). Coimmunoprecipitation reactions were carried out on 50 μg of nuclear extracts using 1 μl of 12CA5 antibody in 500 μl of binding buffer without BSA, and bound immunocomplexes were washed and subjected to SDS-PAGE as described previously (17). Proteins were transferred on Hybond polyvinylidene difluoride membrane and subjected to immunoblotting. Anti-Flag M5 (Sigma) and 12CA5 anti-HA monoclonal antibodies were used at a dilution of 1:8,000, revealed with an anti-mouse immunoglobulin-horseradish peroxidase conjugate (Sigma) at a dilution of 1:50,000, and visualized using ECL Plus (Amersham Pharmacia Biotechnology).
EMSAs.
For electrophoretic mobility shift assays (EMSAs), nuclear extracts were prepared from cardiomyocytes or HeLa cells overexpressing various recombinant proteins as previously described (22) with minor modifications. From 5 × 106 to 10 × 106 cells were washed with ice-cold phosphate-buffered saline (PBS) containing 1 mM sodium orthovanadate and scraped in 1 ml of ice-cold PBS containing 1 mM EDTA. The cells were resuspended in 400 μl of buffer A (20 mM HEPES [pH 7.9], 20 mM NaF, 1 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 1 mM EDTA, 1 mM EGTA, 0.25 mM sodium molybdate, 2 mM 10 μg DTT, 0.5 mM PMSF, 100 nM okadaic acid, plus 10 μg of leupeptin, 10 μg of aprotinin, and 10 μg of pepstatin per ml) and swell on ice for 15 min. Then 25 μl of 10% NP-40 was added, and microtubes were vortexed vigorously. The nuclei were then collected by centrifugation at 15,000 rpm at 4°C for 2 min. The pellets were resuspended in 5 volumes of buffer C (buffer A supplemented with 20% glycerol and 0.4 M NaCl) and shaken vigorously at 4°C for 1 h. The nuclear extracts were cleared by centrifugation at 15,000 rpm for 15 min at 4°C, and the protein concentration was assayed by the Bradford method. Binding reactions were performed according to Charron et al. (11), except that the reaction mixtures contained 1 or 10 μg of nuclear extracts from cardiomyocytes or 7.5 μg of nuclear extracts from HeLa cells overexpressing GATA-4 or SRF or recombinant SRF (rSRF) as specified in the figures. Reactions were loaded on a 4% polyacrylamide gel and run at 200 V at RT or 4°C in 0.25× Tris-borate-EDTA. Probes used were, from 5′ to 3′ (only the coding strand is shown), rat ANF proximal GATA-SRE-like (GATCCACTGATAACTTTAAAAGGGCATCTTCA), rat ANF proximal mutated-GATA-SRE-like (GATCCACTCCTAACTTTAAAAGGGCATCTTCA), rat ANF proximal GATA-mutated-SRE-like (GATCCACTGATAACGGGAAAAGGGCATCTTCA), and c-fos SRE (ACAGGATGTCCATATTAGGACATCTGCG). The GATA (WGATAR) and the SRE (CCW6GG) consensus motifs are underlined, and the mutations are shown in boldface. W and R denote, respectively, A or T and A or G.
RESULTS
To study the nuclear signaling of ETA, we used primary cardiomyocyte cultures in which we monitored the response of the ANF promoter to ET-1 stimulation. In these cells, ETA is the predominant endothelin receptor isoform, and it has been previously shown to mediate all ET-1 responses in cardiomyocytes, including changes in gene expression and cell shape which mimic those observed during in vivo cardiac hypertrophy (25, 55).
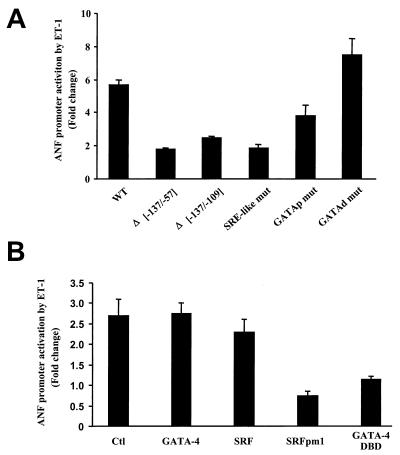
Consistent with previous reports (54), a luciferase reporter driven by a 695-bp ANF fragment is induced threefold following a 6-h treatment with ET-1 (10−9 M), with maximal response reaching sixfold after 48 h of treatment (Fig. 1). Deletion analysis revealed that a 30-bp proximal promoter region (−109 to −137 bp) containing a GATA binding site (11) juxtaposed to an A/T-rich sequence with strong homology to the binding site for SRF(s) is essential for transcriptional activation by ET-1 (Fig. 1A). Mutation of the SRE-like motif (SRE-like mut) or the GATA motif (GATAp mut) markedly inhibits the ET-1 responsiveness of the bp −695 promoter (by 50 to 75%), whereas mutation of the distal (bp −300) GATA binding site (GATAd mut) leads to a consistent, though small, enhancement of ET-1 responsiveness (Fig. 1A). These results suggest that the interaction of endogenous cardiac GATA proteins and SRF over their cognate binding sites within the proximal ANF promoter is essential for nuclear signaling by ET-1. Consistent with this, coexpression in cardiomyocytes of the −695ANF-luciferase reporter with a dominant negative form of SRF (SRFpm1) (46) or with a truncated GATA-4 protein that retains the DNA-binding region but removes all transcriptional activation domains (GATA-4 DBD) abrogates ET-1 responsiveness (Fig. 1B).
FIG. 1.
Mapping ET-1 response elements on the ANF promoter. ANF-luciferase reporter constructs were transfected in cardiomyocytes, which were then treated with ET-1 (1 nM) or vehicle for 48 h (A) or 6 h (B) as described in Materials and Methods. Promoter activity is expressed as the ratio of the luciferase activity recorded in the presence of ET-1 to the activity in the absence of ET-1 (fold change). The data shown represent the mean ± standard deviation of at least three different experiments, each carried out in duplicate. (A) All reporter constructs were driven by the wild type −695 rat ANF promoter (WT) or mutants thereof containing either internal deletions (Δ−137/−57 and Δ−137/−109) or point mutations in the proximal SRE-like sequence (SRE-like mut), the proximal GATA site (bp −120, GATAp mut), or the distal GATA element at bp −280 (GATAd mut). All mutants are described in Materials and Methods. (B) The wild-type ANF-luciferase construct was cotransfected in cardiomyocytes with wild-type or mutant GATA-4 or SRF expression vectors and stimulated or not with ET-1. Note that a dominant negative SRF form, SRFpm1, which no longer binds DNA but retains the ability to interact with GATA-4 (Fig. 3), and a dominant negative GATA-4 mutant, GATA-4 DBD, which contains the DNA-binding region (residues 200 to 332) but not the activation domains, abrogate ET-1 induction.
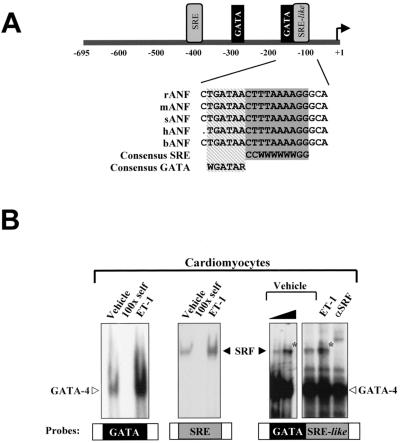
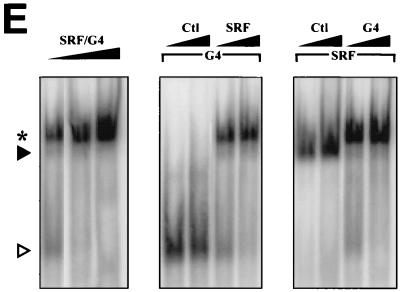
Given that both SRF and GATA proteins interact with evolutionarily conserved juxtaposed binding sites (Fig. 2A), we tested the possibility that they may form a DNA-binding ternary complex analogous to the well-studied ternary complex factor (TCF)-SRF ternary complex (16, 21, 28, 36, 53). Incubation of a 32-bp ANF probe containing the GATA-SRE region with cardiac extracts from cardiomyocytes treated or not with ET-1 revealed, in addition to the expected GATA and SRF complexes, the presence of a slower-migrating DNA-binding complex (marked by asterisks in Fig. 2B). This complex is not observed over a 30-bp c-fos SRE probe (Fig. 2B) but is efficiently inhibited by unlabeled SRE or GATA oligonucleotides and is displaced by incubation with an anti-SRF antibody (Fig. 2B and data not shown). Moreover, this SRF-containing complex is clearly observed in cells overexpressing SRF and GATA-4 or -6 and is dependent on the presence of both GATA and SRE elements (Fig. 2C). The SRF-GATA complex is abrogated in the presence of excess unlabeled oligonucleotides containing either GATA binding sites or well-characterized SREs from the c-fos or α-skeletal actin promoter (Fig. 2D); the complex is also eliminated in the presence of SRF or GATA-4 (or -6) antibodies (Fig. 2C, right panel, and 2D). N-terminally deleted GATA-4 protein (G4Δ) retains the ability to form a ternary complex with SRF that migrates faster than the ternary complex formed with wild-type GATA-4 (Fig. 2D, middle panel). In contrast, both SRF binding and ternary complex formation are severely reduced when a DNA-binding-defective mutant of GATA-4 (G4m) is coexpressed with SRF instead of native GATA-4, suggesting that DNA binding stabilizes SRF–GATA-4 interaction and that the ability of GATA-4 to interact with SRF is dissociable from its DNA-binding capacity. Similarly, a DNA-binding-defective SRF mutant (SRFpm1) fails to form a detectable ternary complex with GATA-4 (Fig. 2D, right panel), consistent with the requirement of both GATA and SRE elements for complex formation (Fig. 2C); this mutant decreases GATA-4 binding, suggesting that it retains the ability to physically contact GATA-4 (Fig. 2D, right panel). Next, we tested the effect of increased levels of GATA-4 and/or SRF on ternary complex formation. As shown in Fig. 2E, increased levels of SRF, GATA-4, or both enhance the formation of the ternary complex, suggesting that this may represent a possible mechanism of ET-1 regulation. Together, these data demonstrate that GATA proteins and SRF interact physically to form a DNA-binding ternary complex over a cardiac promoter through cooperative binding.
FIG. 2.
SRF and GATA factors form a ternary complex over a DNA element containing juxtaposed GATA and SRE motifs. (A) Schematic representation of the ANF promoter, focusing on the proximal ET-1 response element. Note the evolutionary conservation of the GATA/SRE-like element. (B to E) EMSAs were performed on c-fos-SRE (SRE), ANF-GATA (GATA), or ANF-GATA-SRE-like probes using nuclear extracts prepared from cardiomyocytes treated with ET-1 (100 nM) or vehicle for 6 h (B) or from HeLa cells overexpressing SRF, GATA-4 or -6, or both (C and D). Gels were exposed to a phosphorimager screen and developed in the STORM (Molecular Dynamics). GATA and SRF complexes are denoted by open and solid arrowheads, respectively, and the GATA-SRF complex is indicated by an asterisk. All probes and antibodies used are described in Materials and Methods. (B) ET-1 treatment (6 h) enhances GATA and SRF binding on the GATA and SRE probes and formation of the ternary complex. (C) The GATA-SRF ternary complex is present in HeLa cell extracts overexpressing SRF and GATA-4 or GATA-6 (left and right panels) and requires intact GATA and SRF binding sites. Please note the faint ternary complex band seen when only GATA factors are overexpressed, reflecting interaction of exogenous GATA proteins with endogenous HeLa cell SRF. (D) Both wild-type GATA-4 (G4) and an N-terminally truncated form (G4Δ, residues 200 to 440) form a ternary complex that is abrogated by unlabeled c-fos-SRE (SRE 1), αSKA-SRE (SRE2), or GATA probes and is eliminated in presence of either GATA-4 or SRF antibodies (Ab). Please note the different mobilities of the G4- and G4Δ-containing ternary complexes (left panels) and the reduced ability of a DNA-binding-defective GATA-4 mutant (G4m) to form a stable ternary complex while eliminating SRF-SRE binding. The right panel illustrates the inability of a DNA-binding-defective SRF mutant, SRFpm1, to form a ternary complex, although this mutant inhibits GATA-4 binding to its site. (E) Effects of increased amounts of GATA-4 and/or SRF on ternary complex formation. Binding was carried out using 4, 8, or 16 μg of extracts expressing GATA-4 and SRF (left panel) or using 2 μg of extracts overexpressing GATA-4 in presence of 2 or 4 μg of control (Ctl) or SRF-expressing extracts (middle panel) or using 2 μg of SRF-expressing extracts in the presence of 2 or 4 μg of extracts from control or GATA-4-expressing cells (right panel).
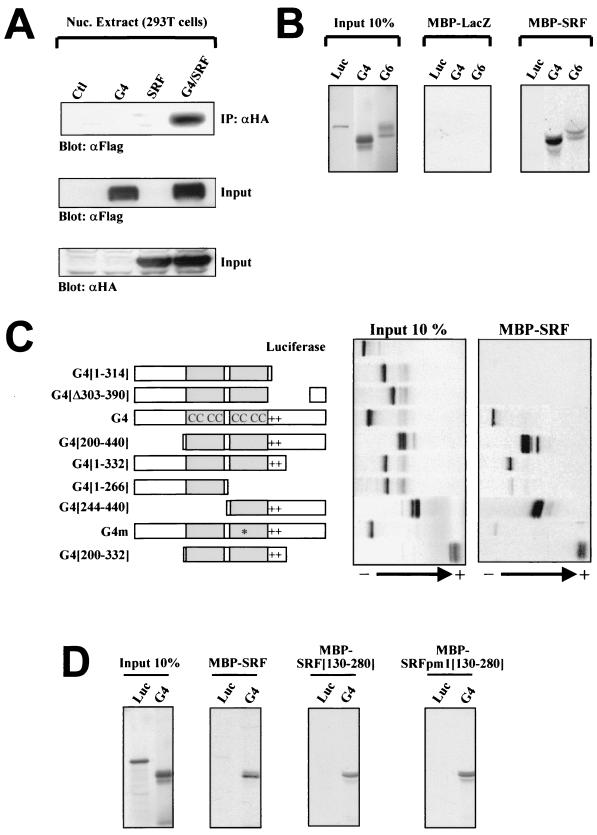
Physical interaction between GATA-4 and SRF was further analyzed using in vivo coimmunoprecipitations (Fig. 3A) and in vitro pull-down assays (Fig. 3B), which confirmed the ability of the two proteins to interact directly even in the absence of DNA. Structure-function studies revealed that the minimal DNA-binding domain of GATA-4, composed of the second zinc finger and the basic regions (amino acids 244 to 332), is necessary and sufficient for physical interaction with SRF; however, DNA binding is not essential for interaction with SRF, as revealed by the ability of G4m (which harbors a point mutation in the zinc finger) to interact, albeit with decreased efficiency, with SRF (Fig. 3C). Similarly, the core SRF DNA-binding domain (SRF[130–280]) is sufficient for GATA interaction. However, SRF's ability to bind DNA is not required for GATA interaction, as evidenced by the ability of SRF[130–280]pm1, which harbors mutations in the basic region that abolish DNA binding, to efficiently associate with GATA-4 (Fig. 3D). It is noteworthy that this structure-function analysis carried out in the absence of DNA is in general agreement with the data obtained in the presence of the GATA-SRF DNA fragment in gel shift experiments (Fig. 2D). Indeed, formation of the SRF-GATA ternary complex required only the DNA-binding domain of GATA-4 (200 to 332), and while DNA-binding-defective mutants of GATA-4 (G4m) and SRF (SRFpm1) did not produce ternary complexes, they nevertheless abrogated SRF and GATA binding, respectively; this is consistent with their ability to physically interact with each other in the absence of DNA binding.
FIG. 3.
SRF can interact physically with cardiac GATA factors in the absence of DNA. (A) SRF interacts in vivo with GATA-4. Nuclear extracts from 293T cells transfected with empty vectors (Ctl), Flag-GATA-4 (G4), and/or HA-SRF (SRF) were immunoprecipitated using an anti-HA antibody, separated by SDS–10% PAGE, transferred to polyvinylidene difluoride membranes, and subjected to immunoblotting using an anti-Flag antibody (top panel). The lower two panels are Western blots carried out on the same nuclear extracts using either HA (to reveal tagged SRF proteins) or Flag (to reveal tagged GATA-4 proteins) antibodies. (B and C) Luciferase (Luc), GATA-4 (G4), or GATA-6 (G6) and a series of deletion mutants of GATA-4 were translated and labeled as described in Materials and Methods. LacZ and SRF in fusion with MBP were produced in bacteria. (B) SRF directly interacts with both cardiac factors GATA-4 and -6. (C) In vitro mapping of the GATA-4 domain required for interaction with SRF to the second zinc finger and the basic region (244 to 332). (D) The SRF DNA-binding domain is sufficient for interaction with GATA-4. SRF (130–280)pm1 is a DNA-binding-deficient mutant containing three point mutations in the basic region. Protein complexes were separated by electrophoresis, exposed on a phosphorimager screen, and developed in the STORM.
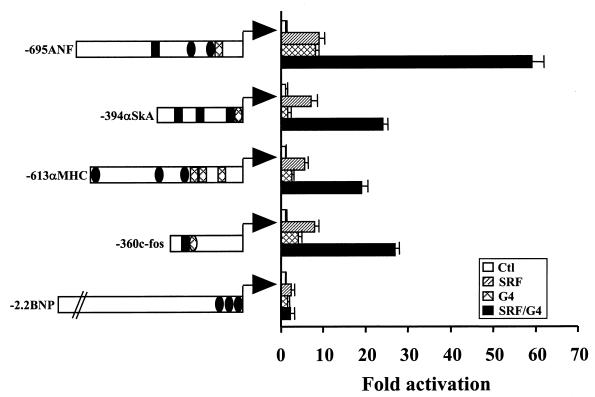
Next, we tested the transcriptional properties of a GATA-4–SRF–DNA complex. In cotransfection assays, both GATA-4 and SRF significantly activate the ANF promoter (up to 20-fold), and when they are added together, a synergistic 80-fold promoter activation is achieved (Fig. 4A). This synergy is dependent on the presence of both SRE and GATA elements, suggesting that, although the two proteins can physically associate in solution, DNA binding is indeed required for stable interaction and/or for formation of the transcriptionally competent complex. Other cardiac GATA factors, GATA-5 and -6, can also synergize with SRF over the ANF promoter; however, no synergy could be observed under the same conditions with GATA-1, -2, or -3 (Fig. 4B). Synergy is dependent on the DNA-binding domains and the DNA-binding ability of both SRF and GATA-4, as evidenced by the inability of SRFpm1 and G4m (which no longer bind their respective sites) to functionally cooperate (Fig. 4C and D); this finding is consistent with the requirement for intact SRE and GATA elements to observe cooperative SRF-GATA transactivation (Fig. 4A). However, although sufficient for physical interaction, neither the GATA-4 nor SRF DNA-binding domain is sufficient for transcriptional synergy, which requires the activation domains of both proteins (Fig. 4C and D). Finally, we tested whether the promoters of other genes that are transcriptionally activated by ET-1 in cardiomyocytes share the ability to respond synergistically to SRF and GATA-4. In addition to the c-fos promoter, two cardiac promoters, α-skeletal actin and α-myosin heavy chain, are activated by ET-1 (24, 59; unpublished data). All three promoters contain adjacent GATA and SRE elements, and all three are synergistically activated by SRF and GATA-4. In contrast, the B-type natriuretic peptide (BNP) promoter which is modestly induced by ET-1 (35) harbors GATA but no SRF binding sites and is unable to support GATA-SRF synergy (Fig. 5). Together, these data demonstrate that DNA-bound SRF can cooperate with members of the GATA family to form a transcriptionally active ternary complex that regulates expression of some tissue-specific and growth-responsive genes.
FIG. 4.
Synergistic activation of the cardiac ANF promoter by GATA-4 and SRF. (A) HeLa cells were transfected with the −695ANF-luc (WT) or ANF-luc vector containing point mutations in the GATA or SRE elements as described for Fig. 1 in the presence of GATA-4 (G4) or CMV-driven SRF expression vectors or both, or in presence of the backbone vector (Ctl). Please note that the 5′ deletion at bp −371 removes a consensus distal SRE that is not required for ET-1 response or GATA-SRF synergy, while the −135/−57 bp internal deletion removes the proximal GATA SRE but retains the distal GATA (bp-300) and SRE (bp-400) sites. (B) Other GATA factors were tested for their ability to synergize with SRF over the ANF promoter. Only the cardiac GATA proteins GATA-4, -5, and -6 (G4, G5, and G6) show synergy with SRF. (C) Mapping of GATA-4 (G4) and SRF functional domains required for transcriptional synergy. HeLa cells were cotransfected with the −695ANF-luciferase reporter and various GATA-4 mutants in the presence or absence of the CMV-driven SRF expression vector (C) or with expression vectors for different SRF mutants in the presence or absence of wild-type GATA-4 expression vector (D). Please note that in panel D, SRF proteins were produced using SV40-driven expression vectors. All plasmids used are described in Materials and Methods; the data shown are the means ± standard deviation of at least three different experiments, each carried out in duplicate.
FIG. 5.
Functional cooperation between GATA-4 and SRF is not restricted to the ANF promoter. HeLa cells were cotransfected with the luciferase reporter under the control of the promoter of the rat ANF (−695ANF), the chicken α-skeletal actin (−394αSKA), the rat α-myosin heavy chain (−613αMHC), the mouse c-fos (−360c-fos), or rat BNP (−2.2BNP) genes. Note that except for the BNP promoter, which contains no recognizable SRE motif, all other promoters contain one or more SRE (depicted by a rectangle) and GATA (depicted by an ellipse) sites; on each of these promoters, at least one SRE is in close proximity to a GATA site. High-affinity binding sites are shown as black areas, and weak affinity sites (based on published reports or on the present work) are hatched. Only promoters containing both GATA and SRE sites are synergistically activated by transient overexpression of GATA-4 and SRE. Transfections were carried out in HeLa cells as described for Fig. 4. All reporter constructs are described in Materials and Methods. The data are from one representative experiment carried out in duplicate. Similar results were obtained on at least two other occasions.
DISCUSSION
We have used the ANF promoter to elucidate the transcription pathways that link activation of the G-protein-coupled endothelin ETA receptor to cell-specific changes in gene expression and, consequently, cell fate. We found that transcriptional regulation of several ET-1-responsive cardiac genes involves combinatorial interaction between the tissue-specific GATA family of transcription factors and SRF through formation of a novel DNA-bound ternary complex reminiscent of the well-studied TCF-SRF ternary complex, identified over the c-fos promoter and involving SRF and the growth factor-regulated, ubiquitous TCFs. The results obtained identify for the first time an endothelin response element that could account for cell-specific endothelin—and possibly other growth and differentiation factor—actions. Moreover, given the coexpression of SRF and members of the GATA family in various endothelin target tissues such as ovaries and vascular smooth muscles (discussed below), the transcription pathway described herein may represent a paradigm for elucidating cell-specific nuclear signaling by ETA and possibly other G-protein-coupled receptors.
ET-1 is a potent growth promoter of vascular and cardiac myocytes and a survival-differentiation factor for neural crest cells; although these effects clearly involve transcriptional regulation of specific genes, the identities of these target genes are only starting to be unraveled (15). Moreover, while numerous Ras-dependent intracellular signaling cascades are transiently activated by ET-1–ETA association, the pathway linking ETA stimulation to nuclear events has remained undefined. In fact, few promoters have been reported to be regulated by ET-1, and their precise ET response elements were not mapped; they include cardiac α- and β-myosin heavy chain (59), ANF (54), BNP (35), and c-Fos (24). Interestingly, it was shown that ET-1 enhanced the activity of a c-fos SRE-driven reporter and that this required an intact SRE; however, mutations that abolished formation of the TCF-SRF ternary complex, the presumed mediator of growth factor signaling, did not affect ET-1 responsiveness, indicating that ET-1 action is SRF dependent but TCF independent (24). In agreement with this, our results suggest that a GATA-SRF, not a TCF-SRF, ternary complex likely mediates ET-1 regulation of cardiac promoters which contain SREs but no adjacent TCF sites. This does not exclude the possibility that other elements and factors may also contribute to ET-1 response in a cell- and promoter-dependent manner. Indeed, as discussed in the introduction, several intracellular pathways known to activate various transcription factors are induced by ET-1; they include, among others, p38 MAPK, which phosphorylates and activates MEF2 proteins (23, 62), and ATF6 (57), an SRF-interacting protein (63). Whether ATF6 and MEF2 are part of a larger ET-1 response complex on the ANF promoter would be worth investigating. At present, several lines of evidence suggest that the presence of GATA and SRF factors is essential for the ET-1 responsiveness of ANF: (i) dominant negative GATA or SRF forms abrogate ET-1 response (Fig. 1) and (ii) cotransfection of the ANF promoter with an ETA expression vector renders the ANF promoter responsive to ET-1 only in cells containing both GATA and SRF proteins, likely through formation of the GATA-SRF ternary complex (J. Wang and M. Nemer, unpublished data).
The mechanism(s) by which the GATA-SRF ternary complex mediates ET-1 response is presently undefined. Binding of both GATA and SRF is induced by ET-1, and this may in turn enhance ternary complex formation. Additionally, ET-1 induces GATA-4 phosphorylation (F. Charron and M. Nemer, unpublished data); whether postranslational modifications of GATA-4 modulate ternary complex formation or activity is presently being investigated. Preliminary evidence indicates that the GATA-4–SRF synergy can be further enhanced by some MAPK forms. The identification for the first time of an ET regulatory element and effector transcription complexes will pave the way for elucidation of the signaling cascades and molecules more proximal to transcription in which specificity may lie.
In addition to its growth-promoting effects on cardiac myocytes, ET-1 is a well-known mitogen for smooth muscle cells, including vascular and mesangial cells (39), which express high levels of GATA-6 and SRF (9, 56). Moreover, SRF binding sites are required for cardiac and smooth muscle-specific expression of several genes, such as ANF, α-myosin heavy chain, α-skeletal actin, SM22, and α-smooth muscle actin (3, 32, 34). In the case of the ANF promoter, two SRF binding sites have been characterized; the high-affinity site, centered around bp −400, was shown to be essential for ANF promoter activity in postnatal ventricles (3); in addition to SRF, the element binds a cardiac-enriched nuclear protein whose identity remains to be elucidated (3). The proximal ANF SRE discussed in this paper is a low-affinity binding site for SRF that was shown to contribute to ANF promoter activation in response to α1-adrenergic stimulation (27), although in our own hands this site has no effect on α1-adrenergic activation of the promoter. The mechanism by which SRF acts to regulate smooth and cardiac muscle-specific genes has remained enigmatic ever since it was noted that the CArG box, present on actin promoters and required for their muscle-specific expression, was identical to the serum response element mapped on the c-fos promoter (38, 41) and that both elements interacted equally well with SRF (8). Based on the present data, it is tempting to speculate on the role of GATA proteins as tissue-specific cofactors for SRF. The present work also suggests that a GATA-SRF ternary complex likely contributes to promoter architecture, allowing proteins bound to distal enhancer elements to interact with those present on the proximal promoter and with the basal transcription machinery. Indeed, molecular modeling based on the crystal structure of SRF and the nuclear magnetic resonance structure of GATA-1 bound to their respective sites (data not shown) reveal that binding of the SRF-GATA complex to the proximal ANF promoter induces a DNA bend that may bring distal enhancers into closer interactions with downstream elements. This may in turn explain the modest contribution of the proximal SRE to α1-adrenergic responsiveness, given that the ANF α1-adrenergic response elements are present on both sides of the GATA SRE (at bp −80 and −450) (2).
SRF, initially isolated as the nuclear protein that mediates transcriptional response of c-fos and other immediate-early genes to growth factors, has been one of the most extensively characterized transcription factors (reviewed in reference 58). It is now well established that many SRF-dependent responses to growth factor stimulation are mediated by an SRF-containing ternary complex in which the TCF is the target of several MAPK cascades (21, 36, 53). At least three different but related TCFs have been identified; functional as well as structural analyses of the TCF-SRF-DNA ternary complex suggest that TCFs act as growth-regulated SRF cofactors (37, 40). Unexpectedly, while mutations that abolish TCF binding rendered the c-fos promoter unresponsive to some growth factors, they did not abolish serum regulation or endothelin stimulation (26, 31). This led to the speculation that an unidentified SRF cofactor that would interact with the DNA-binding SRF domain and form a ternary complex with SRF and DNA must exist. Our results suggest that GATA factors may fulfill these criteria. Indeed, GATA-4 and -6 interact with the DNA-binding domain of SRF and form a stable ternary complex, as evidenced by gel shift analysis and supported by molecular modeling (Fig. 2 and 3 and data not shown). Remarkably, we found that the well-studied c-fos SRE contains two inverted GATA motifs flanking the SRF binding sequences (see c-fos SRE sequence in Materials and Methods) which bound recombinant GATA factors, albeit with lower affinity than the ANF GATA sites (our unpublished work). Moreover, the c-fos promoter as well as a c-fos SRE heterologous promoter were synergistically activated by SRF and GATA factors in many cell types (Fig. 5 and data not shown). Whether a GATA-SRF ternary complex can substitute for the TCF-SRF complex over the c-fos promoter and mediate cell-specific serum or growth-differentiation responses in GATA-expressing cells deserves to be investigated.
Finally, we have recently shown that GATA factors can interact with other MADS-box-containing proteins, namely, the MEF2 factors (42). Although the ability to associate with MADS-box transcription factor through similar domains may be a general feature of GATA proteins, it is noteworthy that functional cooperativity between GATA and MEF2, on the one hand, and GATA and SRF, on the other hand, involves distinct mechanisms. In the case of MEF2, transcriptional synergy occurs via GATA-dependent recruitment of MEF2 to target promoters and does not require the DNA-binding ability of MEF2 (42). In contrast, transcriptional cooperativity between GATA and SRF requires both GATA and SRF DNA-binding abilities and sites. While the molecular basis for these differences is not clear at present, the ability of GATA and MADS factors to interact via different mechanisms underscores the potential relevance to various cellular processes of a functional interaction between the two evolutionarily conserved transcription factor families.
ACKNOWLEDGMENTS
We are grateful to Brian Wilkes for molecular modeling studies, to Lynda Robitaille for technical assistance, to Lise Laroche for secretarial help, and to members of the Nemer lab for helpful discussions.
This work was supported by grants from the Medical Research Council of Canada (MT-13056 and MOP-36382) and in part by a grant from the Société de recherches sur le cancer inc. S.M. was the recipient of an MRC studentship, and M.N. is a senior Scientist of the MRC.
The first two authors contributed equally to this work.
REFERENCES
- 1.Arai H, Hori S, Aramori I, Ohkubo H, Nakanishi S. Cloning and expression of a cDNA encoding an endothelin receptor. Nature. 1990;348:730–732. doi: 10.1038/348730a0. [DOI] [PubMed] [Google Scholar]
- 2.Ardati A, Nemer M. A nuclear pathway for a1-adrenergic receptor signaling in cardiac cells. EMBO J. 1993;12:5131–5139. doi: 10.1002/j.1460-2075.1993.tb06208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argentin S, Ardati A, Tremblay S, Lihrmann I, Robitaille L, Drouin J, Nemer M. Developmental stage-specific regulation of atrial natriuretic factor gene transcription in cardiac cells. Mol Cell Biol. 1994;14:777–790. doi: 10.1128/mcb.14.1.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Argentin S, Sun Y-L, Lihrmann I, Schmidt T J, Drouin J, Nemer M. Distal cis-acting promoter sequences mediate glucocorticoid stimulation of cardiac atrial natriuretic factor gene transcription. J Biol Chem. 1991;266:23315–23322. [PubMed] [Google Scholar]
- 5.Battistini B, Chailler P, D'Orleans-Juste P, Briere N, Sirois P. Growth regulatory properties of endothelins. Peptides. 1993;14:385–399. doi: 10.1016/0196-9781(93)90057-n. [DOI] [PubMed] [Google Scholar]
- 6.Baynash A G, Hosoda K, Giaid A, Richardson J A, Emoto N, Hammer R E, Yanagisawa M. Interaction of endothelin-3 with endothelin-B receptor is essential for development of epidermal melanocytes and enteric neurons. Cell. 1994;79:1277–1285. doi: 10.1016/0092-8674(94)90018-3. [DOI] [PubMed] [Google Scholar]
- 7.Bogoyevitch M A, Glennon P E, Andersson M B, Clerk A, Lazou A, Marshall C J, Parker P J, Sugden P H. Endothelin-1 and fibroblast growth factors stimulate the mitogen- activated protein kinase signaling cascade in cardiac myocytes: the potential role of the cascade in the integration of two signaling pathways leading to myocyte hypertrophy. J Biol Chem. 1994;269:1110–1119. [PubMed] [Google Scholar]
- 8.Boxer L M, Prywes R, Roeder R G, Kedes L. The sarcomeric actin CArG-binding factor is indistinguishable from the c-Fos serum response factor. Mol Cell Biol. 1989;9:515–522. doi: 10.1128/mcb.9.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Browning C L, Culberson D E, Aragon I V, Fillmore R A, Croissant J D, Schwartz R J, Zimmer W E. The developmentally regulated expression of serum response factor plays a key role in the control of smooth muscle-specific genes. Dev Biol. 1998;194:18–37. doi: 10.1006/dbio.1997.8808. [DOI] [PubMed] [Google Scholar]
- 10.Buttrick P M, Kaplan M L, Kitsis R N, Leinwand L A. Distinct behavior of cardiac myosin heavy chain gene constructs in vivo: discordance with in vitro results. Circ Res. 1993;72:1211–1217. doi: 10.1161/01.res.72.6.1211. [DOI] [PubMed] [Google Scholar]
- 11.Charron F, Paradis P, Bronchain O, Nemer G, Nemer M. Cooperative interaction between GATA-4 and GATA-6 regulates myocardial gene expression. Mol Cell Biol. 1999;19:4355–4365. doi: 10.1128/mcb.19.6.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C Y, Schwartz R J. Recruitment of the tinman homolog Nkx-2.5 by serum response factor activates cardiac α-actin gene transcription. Mol Biol Cell. 1996;16:6372–6384. doi: 10.1128/mcb.16.11.6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choukroun G, Hajjar R, Kyriakis J M, Bonventre J V, Rosenzweig A, Force T. Role of the stress-activated protein kinases in endothelin-induced cardiomyocyte hypertrophy. J Clin Investig. 1998;102:1311–1320. doi: 10.1172/JCI3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clouthier D E, Hosoda K, Richardson J A, Williams S C, Yanagisawa H, Kuwaki T, Kumada M, Hammer R E, Yanagisawa M. Cranial and cardiac neural crest defects in endothelin-A receptor-deficient mice. Development. 1998;125:813–824. doi: 10.1242/dev.125.5.813. [DOI] [PubMed] [Google Scholar]
- 15.Clouthier D E, Williams S C, Yanagisawa H, Wieduwilt M, Richardson J A, Yanagisawa M. Signaling pathways crucial for craniofacial development revealed by endothelin-A receptor-deficient mice. Dev Biol. 2000;217:10–24. doi: 10.1006/dbio.1999.9527. [DOI] [PubMed] [Google Scholar]
- 16.Dalton S, Treisman R. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell. 1992;68:597–612. doi: 10.1016/0092-8674(92)90194-h. [DOI] [PubMed] [Google Scholar]
- 17.Durocher D, Charron F, Warren R, Schwartz R J, Nemer M. The cardiac transcription factors Nkx2–5 and GATA-4 are mutual cofactors. EMBO J. 1997;16:5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durocher D, Chen C Y, Ardati A, Schwartz R J, Nemer M. The ANF promoter is a downstream target for Nkx-2.5 in the myocardium. Mol Cell Biol. 1996;16:4648–4655. doi: 10.1128/mcb.16.9.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edery P, Attie T, Amiel J, Pelet A, Eng C, Hofstra R M, Martelli H, Bidaud C, Munnich A, Lyonnet S. Mutation of the endothelin-3 gene in the Waardenburg-Hirschsprung disease (Shah-Waardenburg syndrome) Nat Genet. 1996;12:442–444. doi: 10.1038/ng0496-442. [DOI] [PubMed] [Google Scholar]
- 20.Gariepy C E, Cass D T, Yanagisawa M. Null mutation of endothelin receptor type B gene in spotting lethal rats causes aganglionic megacolon and white coat color. Proc Natl Acad Sci USA. 1996;93:867–872. doi: 10.1073/pnas.93.2.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gille H, Strahl T, Shaw P E. Activation of ternary complex factor Elk-1 by stress-activated protein kinases. Curr Biol. 1995;5:1191–1200. doi: 10.1016/s0960-9822(95)00235-1. [DOI] [PubMed] [Google Scholar]
- 22.Grépin C, Dagnino L, Robitaille L, Haberstroh L, Antakly T, Nemer M. A hormone-encoding gene identifies a pathway for cardiac but not skeletal muscle gene transcription. Mol Cell Biol. 1994;14:3115–3129. doi: 10.1128/mcb.14.5.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han J, Jiang Y, Li Z, Kravchenko V V, Ulevitch R J. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature. 1997;386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- 24.Herman W H, Simonson M S. Nuclear signaling by endothelin-1. A Ras pathway for activation of the c-fos serum response element. J Biol Chem. 1995;270:11654–11661. doi: 10.1074/jbc.270.19.11654. [DOI] [PubMed] [Google Scholar]
- 25.Hilal-Dandan R, Ramirez M T, Villegas S, Gonzalez A, Endo-Mochizuki Y, Brown J H, Brunton L L. Endothelin ETA receptor regulates signaling and ANF gene expression via multiple G protein-linked pathways. Am J Physiol. 1997;272:H130–H137. doi: 10.1152/ajpheart.1997.272.1.H130. [DOI] [PubMed] [Google Scholar]
- 26.Hill C S, Treisman R. Differential activation of c-fos promoter elements by serum, lysophosphatidic acid, G proteins and polypeptide growth factors. EMBO J. 1995;14:5037–5047. doi: 10.1002/j.1460-2075.1995.tb00186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hines W A, Thorburn J, Thorburn A. A low-affinity serum response element allows other transcription factors to activate inducible gene expression in cardiac myocytes. Mol Cell Biol. 1999;19:1841–1852. doi: 10.1128/mcb.19.3.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hipskind R A, Buscher D, Nordheim A, Baccarini M. Ras/MAP kinase-dependent and -independent signaling pathways target distinct ternary complex factors. Genes Dev. 1994;8:1803–1816. doi: 10.1101/gad.8.15.1803. [DOI] [PubMed] [Google Scholar]
- 29.Hosoda K, Hammer R E, Richardson J A, Baynash A G, Cheung J C, Giaid A, Yanagisawa M. Targeted and natural (piebald-lethal) mutations of endothelin-B receptor gene produce megacolon associated with spotted coat color in mice. Cell. 1994;79:1267–1276. doi: 10.1016/0092-8674(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 30.James A F, Xie L H, Fujitani Y, Hayashi S, Horie M. Inhibition of the cardiac protein kinase A-dependent chloride conductance by endothelin-1. Nature. 1994;370:297–300. doi: 10.1038/370297a0. [DOI] [PubMed] [Google Scholar]
- 31.Johansen F E, Prywes R. Two pathways for serum regulation of the c-Fos serum response element require specific sequence elements and a minimal domain of serum response factor. Mol Cell Biol. 1994;14:5920–5928. doi: 10.1128/mcb.14.9.5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim S, Ip H S, Lu M M, Clendenin C, Parmacek M S. A serum response factor-dependent transcriptional regulatory program identifies distinct smooth muscle cell sublineages. Mol Cell Biol. 1997;17:2266–2278. doi: 10.1128/mcb.17.4.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurihara Y, Kurihara H, Oda H, Maemura K, Nagai R, Ishikawa T, Yazaki Y. Aortic arch malformations and ventricular septal defect in mice deficient in endothelin-1. J Clin Investig. 1995;96:293–300. doi: 10.1172/JCI118033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li L, Liu Z, Mercer B, Overbeek P, Olson E N. Evidence for serum response factor-mediated regulatory networks governing SM22alpha transcription in smooth, skeletal, and cardiac muscle cells. Dev Biol. 1997;187:311–321. doi: 10.1006/dbio.1997.8621. [DOI] [PubMed] [Google Scholar]
- 35.Liang F, Lu S, Gardner D G. Endothelin-dependent and -independent components of strain-activated brain natriuretic peptide gene transcription require extracellular signal regulated kinase and p38 mitogen-activated protein kinase. Hypertension. 2000;35:188–192. doi: 10.1161/01.hyp.35.1.188. [DOI] [PubMed] [Google Scholar]
- 36.Marais R, Wynne J, Treisman R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell. 1993;73:381–393. doi: 10.1016/0092-8674(93)90237-k. [DOI] [PubMed] [Google Scholar]
- 37.Matsubara H, Kanasaki M, Murasawa S, Tsukaguchi Y, Nio Y, Inada M. Differential gene expression and regulation of angiotensin II receptor subtypes in rat cardiac fibroblasts and cardiomyocytes in culture. J Clin Investig. 1994;93:1592–1601. doi: 10.1172/JCI117139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miwa T, Kedes L. Duplicated CArG box domains have positive and mutually dependent regulatory roles in expression of the human alpha-cardiac actin genes. Mol Cell Biol. 1987;7:2803–2813. doi: 10.1128/mcb.7.8.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyauchi T, Masaki T. Pathophysiology of endothelin in the cardiovascular system. Annu Rev Physiol. 1999;61:391–415. doi: 10.1146/annurev.physiol.61.1.391. [DOI] [PubMed] [Google Scholar]
- 40.Mo Y, Vaessen B, Johnston K, Marmorstein R. Structures of SAP-1 bound to DNA targets from the E74 and c-fos promoters: insights into DNA sequence discrimination by Ets proteins. Mol Cell. 1998;2:201–212. doi: 10.1016/s1097-2765(00)80130-6. [DOI] [PubMed] [Google Scholar]
- 41.Mohun T, Garrett N, Treisman R. Xenopus cytoskeletal actin and human c-fos gene promoters share a conserved protein-binding site. EMBO J. 1987;6:667–673. doi: 10.1002/j.1460-2075.1987.tb04806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morin S, Charron F, Robitaille L, Nemer M. GATA-dependent recruitment of MEF2 proteins to target promoters. EMBO J. 2000;19:2046–2055. doi: 10.1093/emboj/19.9.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ono K, Tsujimoto G, Sakamoto A, Eto K, Masaki T, Ozaki Y, Satake M. Endothelin-A receptor mediates cardiac inhibition by regulating calcium and potassium currents. Nature. 1994;370:301–304. doi: 10.1038/370301a0. [DOI] [PubMed] [Google Scholar]
- 44.Paradis P, MacLellan W R, Belaguli N S, Schwartz R J, Schneider M D. Serum response factor mediates AP-1-dependent induction of the skeletal alpha-actin promoter in ventricular myocytes. J Biol Chem. 1996;271:10827–10833. doi: 10.1074/jbc.271.18.10827. [DOI] [PubMed] [Google Scholar]
- 45.Pribnow D, Muldoon L L, Fajardo M, Theodor L, Chen L Y, Magun B E. Endothelin induces transcription of fos/jun family genes: a prominent role for calcium ion. Mol Endocrinol. 1992;6:1003–1012. doi: 10.1210/mend.6.7.1508217. [DOI] [PubMed] [Google Scholar]
- 46.Prywes R, Zhu H. In vitro squelching of activated transcription by serum response factor: evidence for a common coactivator used by multiple transcriptional activators. Nucleic Acids Res. 1992;20:513–520. doi: 10.1093/nar/20.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puffenberger E G, Hosoda K, Washington S S, Nakao K, deWit D, Yanagisawa M, Chakravart A. A missense mutation of the endothelin-B receptor gene in multigenic Hirschsprung's disease. Cell. 1994;79:1257–1266. doi: 10.1016/0092-8674(94)90016-7. [DOI] [PubMed] [Google Scholar]
- 48.Rajotte D, Sadowski H B, Haman A, Gopalbhai K, Meloche S, Liu L, Krystal G, Hoang T. Contribution of both STAT and SRF/TCF to c-fos promoter activation by granulocyte-macrophage colony-stimulating factor. Blood. 1996;88:2906–2916. [PubMed] [Google Scholar]
- 49.Sakai S, Miyauchi T, Kobayashi M, Yamaguchi I, Goto K, Sugishita Y. Inhibition of myocardial endothelin pathway improves long-term survival in heart failure. Nature. 1996;384:353–355. doi: 10.1038/384353a0. [DOI] [PubMed] [Google Scholar]
- 50.Sakai S, Miyauchi T, Sakurai T, Kasuya Y, Ihara M, Yamaguchi I, Goto K, Sugishita Y. Endogenous endothelin-1 participates in the maintenance of cardiac function in rats with congestive heart failure. Marked increase in endothelin-1 production in the failing heart. Circulation. 1996;93:1214–1222. doi: 10.1161/01.cir.93.6.1214. [DOI] [PubMed] [Google Scholar]
- 51.Sakurai T, Yanagisawa M, Takuwa Y, Miyazaki H, Kimura S, Goto K, Masaki T. Cloning of a cDNA encoding a non-isopeptide-selective subtype of the endothelin receptor. Nature. 1990;348:732–735. doi: 10.1038/348732a0. [DOI] [PubMed] [Google Scholar]
- 52.Shamim A, Pelzer T, Grohe C, Neyses L. Induction of Egr-1 mRNA and protein by endothelin 1, angiotensin II and norepinephrine in neonatal cardiac myocytes. Mol Cell Biochem. 1999;195:11–17. doi: 10.1023/a:1006887307568. [DOI] [PubMed] [Google Scholar]
- 53.Shaw P E, Schroter H, Nordheim A. The ability of a ternary complex to form over the serum response element correlates with serum inducibility of the human c-fos promoter. Cell. 1989;56:563–572. doi: 10.1016/0092-8674(89)90579-5. [DOI] [PubMed] [Google Scholar]
- 54.Shubeita H E, McDonough P M, Harris A N, Knowlton K U, Glembotski C C, Brown J H, Chien K R. Endothelin induction of inositol phospholipid hydrolysis, sarcomere assembly, and cardiac gene expression in ventricular myocytes: a paracrine mechanism for myocardial cell hypertrophy. J Biol Chem. 1990;265:20555–20562. [PubMed] [Google Scholar]
- 55.Sugden P H, Bogoyevitch M A. Endothelin-1-dependent signaling pathways in the myocardium. Trends Cardiovasc Med. 1996;6:87–94. doi: 10.1016/1050-1738(96)00013-8. [DOI] [PubMed] [Google Scholar]
- 56.Suzuki E, Evans T, Lowry J, Truong L, Bell D W, Testa J R, Walsh K. The human GATA-6 gene: structure, chromosomal location, and regulation of expression by tissue-specific and mitogen-responsive signals. Genomics. 1996;38:283–290. doi: 10.1006/geno.1996.0630. [DOI] [PubMed] [Google Scholar]
- 57.Thuerauf D J, Arnold N D, Zechner D, Hanford D S, DeMartin K M, McDonough P M, Prywes R, Glembotski C C. p38 mitogen-activated protein kinase mediates the transcriptional induction of the atrial natriuretic factor gene through a serum response element: a potential role for the transcription factor ATF6. J Biol Chem. 1998;273:20636–20643. doi: 10.1074/jbc.273.32.20636. [DOI] [PubMed] [Google Scholar]
- 58.Treisman R. Journey to the surface of the cell: Fos regulation and the SRE. EMBO J. 1995;14:4905–4913. doi: 10.1002/j.1460-2075.1995.tb00173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang D L, Chen J J, Shin N L, Kao Y C, Hsu K H, Huang W Y, Liew C C. Endothelin stimulates cardiac alpha- and beta- myosin heavy chain gene expression. Biochem Biophys Res Commun. 1992;183:1260–1265. doi: 10.1016/s0006-291x(05)80326-2. [DOI] [PubMed] [Google Scholar]
- 60.Yanagisawa M. The endothelin system: a new target for therapeutic intervention. Circulation. 1994;89:1320–1322. doi: 10.1161/01.cir.89.3.1320. [DOI] [PubMed] [Google Scholar]
- 61.Yanagisawa M, Kurihara H, Kimura S, Tomobe Y, Kobayashi M, Mitsui Y, Yazaki Y, Goto K, Masaki T. A novel potent vasoconstrictor peptide produced by vascular endothelial cells. Nature. 1988;332:411–415. doi: 10.1038/332411a0. [DOI] [PubMed] [Google Scholar]
- 62.Zhao M, New L, Kravchenko V V, Kato Y, Gram H, di Padova F, Olson E N, Ulevitch R J, Han J. Regulation of the MEF2 family of transcription factors by p38. Mol Cell Biol. 1999;19:21–30. doi: 10.1128/mcb.19.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu C, Johansen F E, Prywes R. Interaction of ATF6 and serum response factor. Mol Cell Biol. 1997;17:4957–4966. doi: 10.1128/mcb.17.9.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zolk O, Quattek J, Sitzler G, Schrader T, Nickenig G, Schnabel P, Shimada K, Takahashi M, Bohm M. Expression of endothelin-1, endothelin-converting enzyme, and endothelin receptors in chronic heart failure. Circulation. 1999;99:2118–2123. doi: 10.1161/01.cir.99.16.2118. [DOI] [PubMed] [Google Scholar]