Abstract
The capacity for nonself recognition is a ubiquitous and essential aspect of biology. In filamentous fungi, nonself recognition during vegetative growth is believed to be mediated by genetic differences at heterokaryon incompatibility (het) loci. Filamentous fungi are capable of undergoing hyphal fusion to form mycelial networks and with other individuals to form vegetative heterokaryons, in which genetically distinct nuclei occupy a common cytoplasm. In Neurospora crassa, 11 het loci have been identified that affect the viability of such vegetative heterokaryons. The het-c locus has at least three mutually incompatible alleles, termed het-cOR, het-cPA, and het-cGR. Hyphal fusion between strains that are of alternative het-c specificity results in vegetative heterokaryons that are aconidial and which show growth inhibition and hyphal compartmentation and death. A 34- to 48-amino-acid variable domain, which is dissimilar in HET-COR, HET-CPA, and HET-CGR, confers allelic specificity. To assess requirements for allelic specificity, we constructed chimeras between the het-c variable domain from 24 different isolates that displayed amino acid and insertion or deletion variations and determined their het-c specificity by introduction into N. crassa. We also constructed a number of artificial alleles that contained novel het-c specificity domains. By this method, we identified four additional and novel het-c specificities. Our results indicate that amino acid and length variations within the insertion or deletion motif are the primary determinants for conferring het-c allelic specificity. These results provide a molecular model for nonself recognition in multicellular eucaryotes.
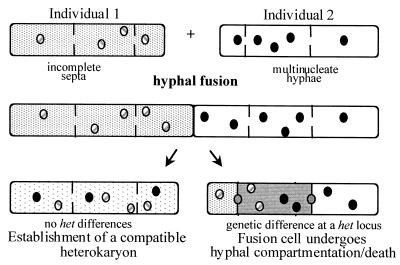
The ability to distinguish self from nonself is a critical feature in most multicellular eucaryotic organisms for maintenance of integrity and individuality. In filamentous fungi, nonself recognition during vegetative growth is mediated through a system known as vegetative or heterokaryon incompatibility (22, 29, 37). A filamentous fungal individual grows as an interconnected network of multinuclear hyphal filaments that are formed via hyphal self-fusion. Filamentous fungi also possess the remarkable attribute of being able to undergo hyphal fusion between different individuals to form vegetative heterokaryons (genetically different nuclei in a common cytoplasm). However, if fungal individuals undergo hyphal fusion but differ in allelic specificity at any one of a number of heterokaryon incompatibility loci (het; sometimes referred to as vic for vegetative incompatibility), the hyphal fusion cell is compartmentalized and dies (Fig. 1). Heterokaryon incompatibility is believed to play a role in filamentous fungi to prevent the spread of mycoviruses and debilitated organelles throughout fungal populations and to restrict resource plundering between individuals (7, 11, 12).
FIG. 1.
Diagrammatic representation of the consequences of hyphal fusion between fungal individuals that do, or do not, differ in specificity at heterokaryon incompatibility (het) loci, such as het-c in N. crassa.
In Neurospora crassa, 11 het loci have been genetically characterized (33, 35); the het-c locus was one of the first het loci identified (17). Vegetative heterokaryons forced by auxotrophic markers between strains that differ in het-c specificity or partial diploids that are heterozygous at het-c are aconidial, show greatly reduced growth rates, and exhibit hyphal compartmentation and death (17, 18, 25, 31, 34). Wild-type isolates fall into three het-c specificity groups (het-cOR compatible, het-cPA compatible, or het-cGR compatible) based on results from crosses using translocation strains (32) that generate het-c partial diploid progeny (24, 39). Genetic differences at het loci in N. crassa do not interfere with sexual fertility (34). Representatives from the three distinct and mutually incompatible allele types (het-cOR, het-cPA, and het-cGR) have been molecularly characterized (39, 40). The het-c locus encodes a polypeptide containing a consensus signal peptide sequence and a glycine-rich carboxyl-terminal domain (40). By chimeric construction between the three het-c allele types, it was determined that a 34- to 48-amino-acid (aa) variable domain (which is dissimilar in HET-COR, HET-CPA, and HET-CGR) confers allelic specificity (39). The het-c specificity domain of het-cOR, het-cPA, and het-cGR differs in both predicted amino acid sequence and in the pattern of insertion and deletion (indel) (Fig. 2).
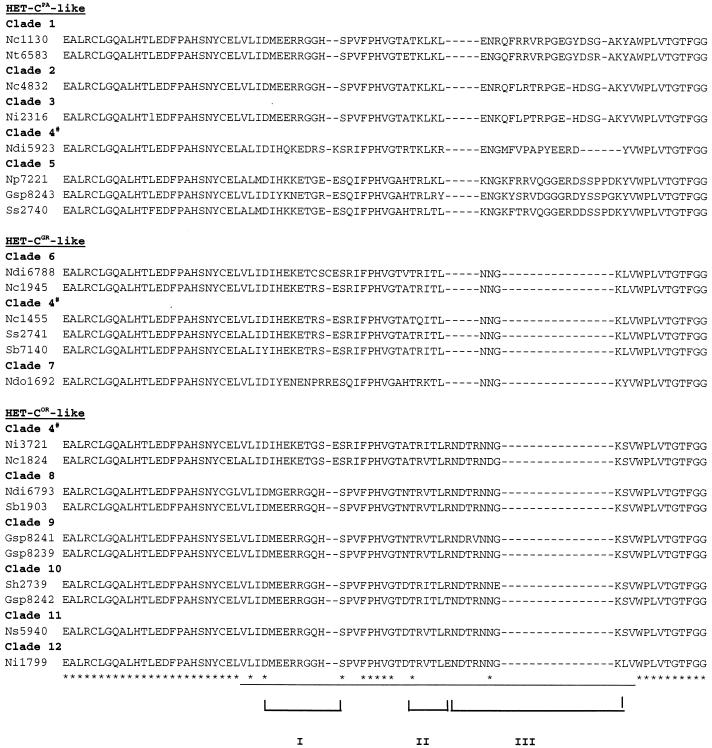
FIG. 2.
Predicted peptide sequences of the variable domain in naturally occurring het-c alleles. The underlined area is the het-c variable domain that is necessary and sufficient to confer het-c allelic specificity in the N. crassa het-cOR (FGSC 2489), het-cPA (FGSC 1130), and het-cGR (FGSC1945) alleles (39, 40). ∗, conserved sites. Dashes represent deletions. Regions I and II are polymorphic blocks; region III encompasses the indel motif. Each clade is supported by over 94% bootstrap values (48). HET-CPA-like, HET-COR-like, and HET-CGR-like refers to the indel pattern observed in the genetically characterized N. crassa het-cPA, het-cOR and het-cGR alleles, respectively (39). Clade 4 has representatives in all three HET-C-like classes based on the indel motif; separation of these three het-c types is supported by bootstrap values of less than 94%. GenBank accession numbers for nucleotide sequences of these alleles were previously cited (48); accession numbers are AF092695 to AF092732 and AF093679.
A molecular survey of the het-c variable domain in 15 N. crassa isolates and 25 isolates from related species and genera showed that all could be placed into one of the three previously identified het-c specificities based on indel motif (39, 48). Phylogenetic analyses based on DNA sequences of the het-c variable domain from N. crassa and related species showed trans species polymorphisms, i.e., isolates did not group according to genus or species but rather grouped according to DNA sequence type of the het-c specificity domain (48). Additional clades within each het-c specificity group were observed, suggesting that diversity within each het-c indel type could represent additional het-c specificities. Analysis of other loci that mediate nonself recognition, such as loci in the major histocompatibility complex (MHC) and the S locus in plants, also show multiple allelic polymorphisms that display transspecies polymorphisms (reviewed in reference 27). These data indicate that, like the MHC and the S locus, the het-c locus is subject to balancing selection and suggests that its function in mediating heterokaryon incompatibility is biologically significant as a nonself recognition system in this group of fungi.
Common molecular features are apparent among loci that mediate self and nonself recognition. Alleles that confer specificity are polymorphic and recognition is generally mediated by protein-protein interactions. The mechanism of allelic specificity has been examined in several fungal nonself recognition systems by the construction of chimeric (or hybrid) alleles. In Podospora anserina, a single amino acid difference in the alternative proteins encoded by the vegetative incompatibility locus, het-s, was sufficient to confer allelic specificity (14). In Ustilago maydis, a region composed of 30 to 48 amino acid residues was identified that regulates specificity at the b mating locus (49); artificial hybrid b alleles with novel specificity were generated by chimeric construction within this border region (50). In Coprinus cinereus, specificity of the homeodomain mating proteins HD1 and HD2 was determined by the N-terminal 160 to 170 aa (1). In N. crassa, amino acid sequence differences within the het-c variable domain could be the critical determinant for het-c specificity, or spatial differences within the indel motif could be the most important factor. To differentiate these two possibilities, we took advantage of the natural amino acid variation and indel motifs observed in the het-c variable domain among natural isolates (48) to construct chimeric alleles. We first asked whether the het-c specificity of these alleles, as assayed in N. crassa, was consistent with their grouping based on phylogenetic analyses. Second, we asked whether het-c specificity could be affected either by amino acid sequence and/or indel pattern within the het-c specificity domain. Third, we constructed a number of artificial het-c alleles that contained combinations of amino acid sequences and indel motifs that were not observed in our survey of naturally occurring het-c alleles. By this method, we were able to identify four additional, novel het-c specificities. Our results indicate that amino acid and spatial characteristics of the indel motif are the primary determinant for conferring het-c allelic specificity. These findings provide insight into understanding the molecular mechanism of nonself recognition during heterokaryon incompatibility in filamentous fungi and provide a molecular model for allelic specificity and nonself recognition mechanisms in multicellular eucaryotes.
MATERIALS AND METHODS
Strains and media.
Escherichia coli strain DH5α (F− endA1 hsdR17 supE44 lacZM15) (Bethesda Research Laboratories, Gaithersburg, Md.) was used for routine DNA manipulation work. Strains used for chimeric construction and transformation recipients are listed in Table 1, along with their origin and het-c specificity. All strains were grown on Vogel's vegetative growth media (VM) (46). A cross to construct het-c deletion strain CJ44 was performed between X22-2 and Xa-3 (Table 1; Q. Xiang and N. L. Glass, unpublished results) by using Westergaard's media and standard crossing conditions (10) and selecting for progeny that formed heterokaryons compatible with both het-cOR and het-cPA strains.
TABLE 1.
List of N. crassa strains
| Strain | Genotype | Source or reference |
|---|---|---|
| C2-2-9 | het-cOR thr-2 A | 43 |
| C9-2 | het-cPA thr-2 a | 43 |
| FGSC2193 | NM149 T(II;V) het-cGR A | 32; D. D. Perkins |
| C15-1 | het-cOR arg-5; pan-2; inl a | R. Todd and L. Glass |
| Xa-3 | het-cPA arg-5; pan-2 A | Q. Xiang and L. Glass |
| X22-2 | Δhet-c thr-2 a | Q. Xiang and L. Glass |
| CJ44 | Δhet-c arg-5; pan-2 A | This study |
Construction of chimeric alleles.
Construction and identification of het-c chimeric alleles were as previously described (39). Previous results indicated that the het-c variable domain was necessary and sufficient to confer allelic specificity; amino acid differences outside of this region between HET-COR, HET-CPA, and HET-CGR did not affect allelic specificity (underlined region in Fig. 2) (39). Plasmids carrying N. crassa het-cOR and het-cPA alleles were used for chimeric allele construction (39), depending on availability of restriction sites. A unique DNA 220-bp StuI-SalI or 650-bp EcoRV-SalI fragment encompassing the het-c variable domain from naturally occurring and artificial alleles was exchanged in frame with an otherwise het-cOR or het-cPA allele. Chimeric alleles were identified by restriction digests using conserved XhoI or ApaI restriction site differences located within the exchanged fragment. The chimeric alleles were cloned into pCB1004 (6) or pOKE103 vector (gift of R. L. Metzenberg) to test for het-c specificity in N. crassa by transformation assays.
Generation of artificial het-c specificity regions by PCR mutagenesis.
The specificity domains for artificial alleles were generated by a recombinant PCR technique (23, 45) that requires two sets of primers and two rounds of PCR. The oligonucleotides used in the construction of artificial het-c specificity domains are listed in Table 2. Plasmids containing het-c were used as first-round PCR templates. The first round of PCR was performed by standard protocol: initial 5-min denaturation at 94°C, followed by 30 step cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C, and a final extension at 72°C for 10 min. Reactions were carried out in volumes of 50 μl containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2.0 mM MgCl2, 100 mM (each) dATP, dCTP, dGTP, and dTTP, 0.2 mM concentrations of each primer, 1.25 U of Taq polymerase, and 50 ng of plasmid DNA as template. The first-round PCR products were purified from agarose gel using a QIAEXII Gel Purifying kit (Qiagen Inc., Mississauga, Ontario, Canada). Two corresponding first-round PCR products were combined and used as templates for the second round of PCR to construct recombinant products. The PCR conditions resembled those of the first round except that annealing was done at 51°C. PCR products from the second round were cloned into pCRII vector (Invitrogen, San Diego, Calif.) for further manipulations. The predicted DNA sequence of the artificial constructs was confirmed by DNA sequence analysis (Nucleic Acid and Protein Synthesis Unit; The University of British Columbia).
TABLE 2.
Oligonucleotide primers used for the construction of artificial het-c alleles
| Primer name | Sequencea |
|---|---|
| Red | GGA GAC ATG GCG ATA TCG |
| SP1.3 | GCT CAT GCC AGG AAC AAC |
| del3.3 | AGC GTA CTT GAG TTT GAG TTT AGT CGC AGT GC |
| del3.5 | ACT CAA ACT CAA GTA CGC TTG GCC CCT GGT |
| po1.3 | CTG AAC TGT CCA TTG TTA CGT GTG TCA |
| po1.5 | CAC GTA ACA ATG GAC AGT TCA GGA GCG TAA GA |
| po2.3 | GTC TAT TCT CAC GTG TGT CAT TCT TGA G |
| po2.5 | GAC ACA CGT GAG AAT AGA CAG TTC AGG CGC GTA AGA |
| po3.3 | CTG TCT ATT CTC GAG TGT GAT TCG CGT ATC |
| po3.5 | CAC ACT CGA GAA TAG ACA GTT CAG GCG CGT AAG A |
| po4.3 | GTG TCA TTC TTG AGT TTG AGT TTA GTC GC |
| po4.5 | CTC AAA CTC AAG AAT GAC ACA CGT AAC |
| p26m-3 | CGC CCC AGA ATC ATC TCT TTC CTC ATA TGG |
| p26m-5 | GAA AGA GAT GAT TCT GGG GCG AAG TAC ACG |
| pd1.3 | AGC GTA CTT TCC TTC TCC GGG TCT TAC |
| pd1.5 | GGA GAA GGA AAG TAC GCT TGG CCC CTG |
| pd2.3 | AGC ATA CTT TAC GCG CCT GAA CTG TCT |
| pd2.5 | AGG CGC GTA AAG TAT GCT TGG CCC CTG |
| pd3.3 | AAG CGT ACT TCG GTC TTA CGC GCC TGA A |
| pd3.5 | GTA AGA CCG AAG TAC GCT TGG CCC CTG |
| pd4.3 | AGC GTA CTT TCT ATT CTC GAG TTT GAG |
| pd4.5 | CTC GAG AAT AGA AAG TAC GCT TGG CCC CTG |
| od-3 | TCC ATT GTT GAG TGT GAT TCG AGT ATC TG |
| od-5 | ATC ACA CTC AAC AAT GGA AAG TCG GTT |
| go-3 | CTT ACG TGT GTC ATT CTT GAG TGT GAT TCG AGT AGC |
| go-5 | AAG AAT GAC ACA CGT AAG TTG GTT TGG CCC TTG G |
The primer sequences are shown in the 5′-to-3′ orientation.
Secondary-structure predictions.
Secondary-structure predictions of the HET-C variable domain were examined using Gibrat (21), Levin (30), DPM (13), and SOPMA (19, 20) prediction programs (http://www.bcp.fr) and the proteomics tools at the ExPASy (Expert Protein Analysis System) proteomics server of the Swiss Institute of Bioinformatics (SIB) (http://www.expasy.ch/).
Transformation assays.
N. crassa spheroplasts were prepared as described by Schweizer et al. (41). Strains C9-2, C2-2-9, and FGSC2193 (Table 1) were used as recipients for transformation assays with the pCB1004 (hygR) (6) vector constructs. Strain CJ44 (Table 1) was used for cotransformation with pCB1004 and pOKE103 (pan-2+; gift of R. L. Metzenberg) vector constructs. For transformation experiments, a modified procedure from P. anserina transformation (4) was applied to N. crassa. Fifty microliters of spheroplasts was thawed on ice and subsequently heat shocked at 48°C for 5 min, followed by a 30-s incubation on ice. The spheroplasts were then placed at room temperature for 10 min before DNA was added. One microgram of DNA construct was used for each 50 μl of spheroplasts. The mixture of spheroplasts and DNA was incubated at room temperature for an additional 10 min and then added to 1 ml of 40% polyethylene glycol (3350)–10 mM MOPS (morpholine propanesulfonic acid)–50 mM CaCl2. After 10 min, 7 ml of prewarmed top agar was added into the mixture and poured onto agar plates containing 250 μg of hygromycin (Calbiochem, San Diego, Calif.)/ml or media lacking pantothenic acid. For each transformation experiment, 15 individual transformants were transferred to separate VM plates containing 200 μg of hygromycin/ml or lacking pantothenic acid and were incubated at 30°C. Transformants were inspected for up to a week for growth inhibition or altered morphology that is characteristic of het-c incompatibility (18). For cotransformation experiments, 0.5 μg of each plasmid was used per 50 μl of spheroplasts.
Growth rate determinations.
The linear growth rate (LGR) of transformants was measured in race tubes as previously described (10). Individual colonies cut from a transformation plate were placed at one end of a 40-ml glass tube containing 25 ml of VM, plus supplements and/or hygromycin. All tubes were incubated at 25°C. The starting point was marked as the leading edge of the colony after overnight growth; subsequent growth was recorded as the distance to the leading edge of the colony at 24-h intervals. Each experiment contained at least three replicate transformants for each construct, plus controls.
Light microscopy.
The stain Evan's Blue (Direct Blue 53, CI23860; Aldrich Chemical Co., Milwaukee, Wis.) is excluded by cells with intact plasma membranes (16) and was used to identify dead hyphal compartments (25). Transformants were inoculated on cellophane membrane layered on top of petri dishes containing selective media and incubated at 30°C. Cellophane membranes with adherent mycelium were removed from the medium, placed on glass slides, and flooded with 1% (wt/vol) Evans blue in water. After 5 min, mycelia were rinsed in distilled water, floated off cellophane onto the slide, and mounted in glycerol-phosphate buffer under a coverslip. Samples were examined under bright-field illumination on an Olympus microscope.
RESULTS
Phylogenetic and structural features of the het-c variable domain.
Allelic specificity of het-c is dependent upon a 34- to 48-aa region (Fig. 2), which is variable between HET-COR, HET-CPA, and HET-CGR; exchange of this region by chimeric construction is sufficient to switch allelic specificity (39). The variable domain differs in both predicted amino acid sequence and in indel motif between het-cOR, het-cPA, and het-cGR alleles. Phylogenetic analysis of the het-c variable domain among N. crassa isolates and related species and genera showed 12 major clades, which were supported by bootstrap levels of over 94% (48) (Fig. 2). Isolates within each clade group by het-c indel motif, with the exception of clade 4, which includes het-cOR-like, het-cGR-like, and het-cPA-like isolates.
Two regions that show high amino acid diversity are designated “I” and “II” in Fig. 2. The 24 isolates fall into two groups based on amino acid variations within region I. The first group contains a consensus sequence of M(G)EERRGG(Q)H and includes isolates that have either a HET-cPA-like or HET-cOR-like indel motif. The second group of alleles is much more variable in amino acid sequence in region I but has a consensus of IH(Y)E(Q/K)K(N)ET(N/D)G(R/P/C)S(E/R). All of the HET-cGR-like peptides have this consensus sequence. Secondary-structure predictions indicate that both amino acid variations found in region I would form an α-helix. The second variable region (II; Fig. 2) has a consensus sequence of T(A)XTR(Q)L(I/V/K)T(R/K)L(Y/R); only the second T residue is conserved among all 24 alleles. Although this region is variable, it is predicted to form or be part of a β-sheet (antiparallel) structure.
Two regions that are highly conserved among all 24 predicted peptides immediately bracket the indel region, I(V)FPHVG and WPLVTGTF (Fig. 2). The I(V)FPHVG region is predicted to be part of a β-sheet with adjacent sequences that are part of region II. The second highly conserved region flanking the indel region, WPLVTGTF, is also predicted to form a β-sheet (antiparallel) structure. The indel region (III; Fig. 2) is predicted to form a loop or coil. These secondary-structure predictions indicate that the variable indel loop region is flanked by the two conserved regions predicted to form antiparallel β-strands.
Introduction of different het-c constructs yields three phenotypic classes of transformants.
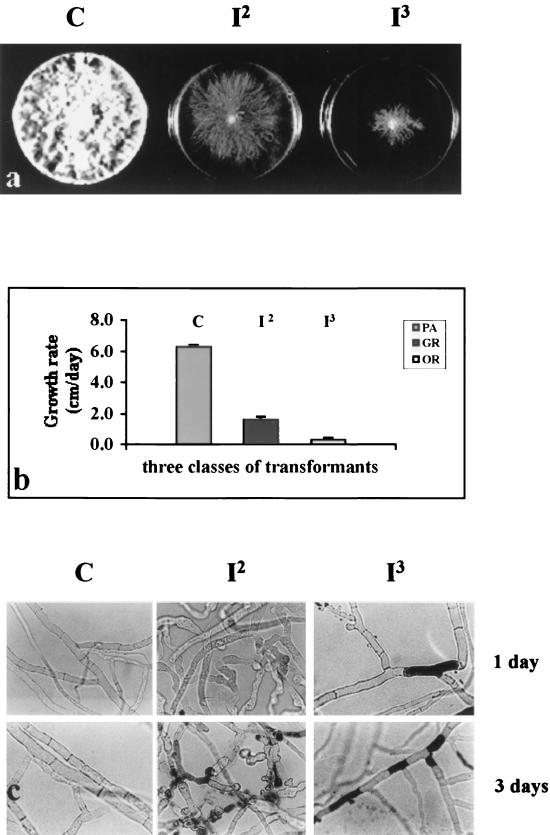
To determine whether the 12 het-c phylogenetic branches each confer a different het-c allelic specificity (Fig. 2) or whether they fall into one of the three het-c specificities previously identified (het-cOR, het-cPA, or het-cGR compatible), we assessed the het-c specificity of chimeric alleles by transformation assays with N. crassa. Chimeric constructs between the het-c variable domain from the 24 naturally occurring alleles (Fig. 2) and an otherwise het-cOR and/or het-cPA allele were constructed (see Materials and Methods). Each chimeric construct was introduced into strains that differed in het-c specificity, C2-2-9 (het-cOR), C9-2 (het-cPA), and FGSC2193 (het-cGR) (Table 1). Three phenotypic classes could be distinguished by the morphology of colonies, LGR, and occurrence of hyphal compartmentation and death (HCD) in the transformants. Compatible (class 1) transformants [for example, introduction of a het-cPA allele into C9-2 (PA); Table 3] displayed vigorous growth and conidiation with an LGR of 4.0 to 6.5 cm/day (Fig. 3). Less than 1% HCD was observed in compatible transformants. Class 2 (intermediate incompatible) transformants [for example, introduction of a het-cPA allele into a C2-2-9 (OR); Table 3] had an LGR of 1.0 to 2.5 cm/day, were aconidial, and exhibited a swollen hyphal morphology (Fig. 3). HCD (approximately 20%) was observed after 2 days of growth. Class 3 transformants [for example, the introduction of a het-cOR allele into C9-2 (PA); Table 3] displayed severely inhibited growth with an LGR of <1.0 cm/day. These transformants showed a flat, curling, aconidial morphology and approximately 20 to 30% dead hyphal compartments (Fig. 3).
TABLE 3.
Phenotypes of transformants containing constructs of naturally occurring het-c allelesa
| Allele | C9-2 (PA) | C2-2-9 (OR) | FGSC2193 (GR) |
|---|---|---|---|
| het-cPA-like | |||
| Nc1130 | C | I2 | Cb |
| Nc6583 | C | I2 | C |
| Ni4832 | C | I2 | C |
| Nt2316 | C | I2 | C |
| Ndi5923 | I2 | I2 | I2 |
| Np7221 | C | I2 | C |
| Gsp8243 | C | I2 | C |
| Ss2740 | C | I2 | C |
| het-cGR-like | |||
| Ndi6788 | I2 | I2 | C |
| Nc1945 | I2 | I2 | C |
| Nc1455 | I2 | I2 | C |
| Ss2741 | I2 | I2 | C |
| Sh7140 | I2 | I2 | C |
| Ndo1692 | I2 | I2 | C |
| het-cOR-like | |||
| Ni3721 | I3 | C | I3 |
| Nc1824 | I3 | C | I3 |
| Ndi6793 | I3 | C | I3 |
| Sb1903 | I3 | C | I3 |
| Gsp8241 | I3 | C | I3 |
| Gsp8239 | I3 | C | I3 |
| Sh2739 | I3 | C | I3 |
| Gsp8242 | I3 | C | I3 |
| Ns5940 | I3 | C | I3 |
| Ni1799 | I3 | C | I3 |
I2, incompatible transformants with an LGR of 1 to 2.5 cm/day showing ∼20 to 30% HCD; I3, severely incompatible transformants with an LGR of <1 cm/day showing 20 to 30% HCD; C, fully compatible transformants with an LGR of 4 to 6.5 cm/day with <1% HCD.
Transformation results (39) and analysis of partial diploid progeny (24) indicate that strain FGSC2193 has a recessive allele-specific suppressor for het-c incompatibility. FGSC2193 and C9-2 are not isogenic strains and probably differ not only in het genotype but also at a large number of other loci that may affect the morphological phenotype associated with het-c vegetative incompatibility.
FIG. 3.
Phenotypic and growth characteristics of the three classes of transformants. Shown are the phenotypes of compatible (class 1 [C]) transformants, such as the introduction of a het-cPA allele into C9-2 (PA) spheroplasts, incompatible class 2 (I2) transformants, such as introduction of a het-cGR allele into C9-2 (PA) spheroplasts, and class 3 (I3) transformants, such as introduction of a het-cOR allele into C9-2 (PA) spheroplasts. The genotypes of strains are given in Table 1. (a) Representative colony growth of the three classes of transformants. All are shown after 3 days of incubation at 30°C on solid VM (see Materials and Methods). (b) Average growth rate (centimeters/day) of three classes of transformants represented by the introduction of het-cPA (C), het-cGR (I2), and het-cOR (I3) alleles into a C9-2 (PA) strain (see above). (c) HCD in the three classes of transformants represented by the introduction of het-cPA (C), het-cGR (I2), and het-cOR (I3) alleles into a C9-2 (PA) strain (see above) after 1 and 3 days of vegetative growth. Evan's blue (16) was used to stain dead hyphal compartments as described in Materials and Methods. Magnification, ×61.
Chimeric alleles containing het-cOR-like variable domains conferred het-cOR allelic specificity.
The het-cOR-like alleles fell into six distinct clades by phylogenetic analysis (48) (Fig. 2). All of the het-c chimeric constructs containing a het-cOR-like indel motif produced class 3, severely incompatible transformants when introduced into C9-2 (PA) and FGSC2193 (GR) (Table 3). Fully compatible transformants were obtained when these constructs were introduced into C2-2-9 (OR). The chimeric het-cOR-like alleles thus displayed an identical het-c specificity to a het-cOR allele, even though amino acid variability occurred in both regions I and II (Fig. 2). In particular, het-cOR-like chimeric alleles containing either of the two motifs in region I displayed identical het-c specificity, for example, Ni3721 and Sb1903 (Table 3).
Chimeric het-cGR-like alleles conferred a het-c specificity identical to that of N. crassa het-cGR.
The het-cGR-like alleles group together based on indel motif within the het-c variable domain but show three distinct clades by phylogenetic analyses (48). The het-cGR-like alleles lack the het-cPA-like and het-cOR-like insertions and have a similar motif in regions I and II, although some amino acid variability is present (Fig. 2). The introduction of all of the het-cGR-like chimeric constructs into C9-2 (PA), C2-2-9 (OR), and FGSC2193 (GR) yielded identical transformation results as a canonical het-cGR allele (Table 3). Thus, the observed variations in the predicted amino acid sequence among the het-cGR-like alleles did not affect het-c allelic specificity. In particular, although phylogenetic analysis showed a relationship between het-cGR-like alleles Nc1455, Ss2741, and Sb7140 and het-cOR-like alleles Ni3721 and Nc1824 (clade 4, Fig. 2), Nc1455, Ss2741, and Sb7140 conferred het-cGR specificity while Ni3721 and Nc1824 conferred het-cOR specificity.
Chimeric het-cPA-like alleles conferred an identical het-c specificity as a het-cPA allele with the exception of Ndi5923.
The het-cPA-like alleles contain a 42- to 48-bp insertion (14 to 16 aa) compared to het-cGR-like and het-cOR-like alleles. Phylogenetic analysis showed that these het-cPA-like alleles fell into five distinct clades (Fig. 2). A high degree of nonsynonymous substitutions among these alleles results in amino acid variation in regions I and II and especially in the het-cPA-specific insertion (III; Fig. 2). Secondary-structure predictions of the indel motif in the HET-CPA-like peptides showed an additional short β-sheet, plus a variable loop between the conserved antiparallel β-strands. When the het-cPA-like chimeric constructs were introduced into C2-2-9 (OR), C9-2 (PA), and FGSC2193 (GR), all of the het-cPA-like chimeric constructs displayed a het-c specificity pattern identical to those of transformants containing the N. crassa canonical het-cPA allele (Table 3), with the single exception of the Ndi5923 construct.
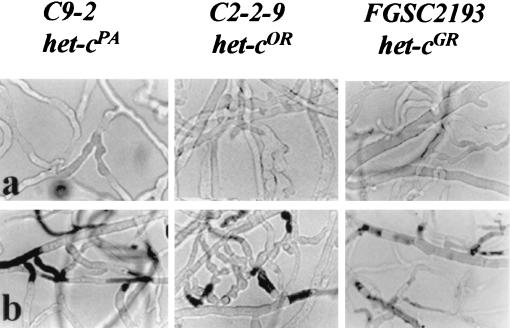
Unlike the other het-cPA-like chimeric constructs, the Ndi5923 construct yielded incompatible transformants when introduced into C2-2-9 (OR), C9-2 (PA), and FGSC2193 (GR) strains (Table 3 and Fig. 4). The class 2-incompatible transformants in all three recipient strains displayed a similar phenotype, characterized by an LGR of approximately 1.3 to 1.8 cm per day, abnormal swollen hyphal morphology, and dead hyphal compartments (Fig. 4). The Ndi5923 allele has a het-cPA-like insertion that is 12 to 18 bp (four to six amino acids) shorter than other het-cPA-like alleles (depending on the reference allele), in addition to amino acid differences.
FIG. 4.
Phenotypes of the class-2-incompatible transformants caused by the introduction of chimeric allele Ndi5923 into C9-2 (PA), C2-2-9 (OR), and FGSC2193 (GR) after 1 day of growth (a) and 3 days of growth (b). Hyphae were treated with the vital dye Evan's blue (16), which stains dead hyphal compartments (25). Magnification, ×68.
The transformation results using the chimeric alleles showed that grouping of alleles by indel motif was a good predictor of het-c specificity. We did not observe het-c specificity differences among the 12 different clades within each het-c specificity group that were observed by phylogenetic analysis (48). However, we did identify a chimeric allele, Ndi5923, which showed length and amino acid differences within the het-cPA-like insertion and conferred a novel het-c specificity.
Artificially constructed amino acid variations in regions I and II do not affect het-c specificity.
The transformation results using the chimeric alleles showed that naturally occurring amino acid variations in regions I and II (Fig. 2) did not affect het-c allelic specificity. However, some variations in amino acid composition in region II were specific for a het-c type, and therefore, the role of amino acid variability on het-c specificity could not be completely addressed by our chimeric constructs using naturally occurring het-c alleles. We therefore constructed a number of artificial het-c alleles with novel combinations of region II and indel motifs (Fig. 2, region III).
The first type of allele (Fig. 5, po3 and po4) had a mosaic combination of predicted amino acid sequence and indel motif between a het-cPA and a het-cOR allele. The po3 allele has a het-cPA-specific insertion in an otherwise het-cOR allele. The introduction of po3 into C9-2 (PA), C2-2-9 (OR), and FGSC2193 (GR) yielded an incompatibility-compatibility spectrum identical to that of a canonical het-cPA allele (Table 4). The po4 allele has a het-cOR indel motif but is otherwise identical to a het-cPA allele in predicted amino acid sequence (Fig. 5). When introduced into C9-2 (PA), C2-2-9 (OR), and FGSC2193 (GR), po4 displayed a het-c specificity identical to that of a canonical het-cOR allele (Table 4).
FIG. 5.
Predicted amino acid sequences in the variable domain of artificially constructed het-c alleles (for details on construction, see Materials and Methods).
TABLE 4.
Phenotypes of transformants containing artificially constructed het-c alleles in recipient strains that differ in het-c specificitya
| Allele | C9-2 (PA) | C2-2-9 (OR) | FGSC2193 (GR) |
|---|---|---|---|
| het-cPA | C | I2 | Cb |
| het-cGR | I2 | I2 | C |
| het-cOR | I3 | C | I3 |
| po3 | C | I2 | C |
| po4 | I3 | C | I3 |
| od1 | I2 | I2 | C |
| go1 | I3 | C | I3 |
| del3 | C | C | C |
| Ndi5923 | I2 | I2 | I2 |
| Ndi5923m | C | I2 | C |
| pd1 | I2 | I3 | I2 |
| pd2 | I2 | I2 | I2 |
| pd3 | I2 | I3 | I2 |
| pd4 | I2 | I3 | C |
| po1 | I3 | I2 | I3 |
| po2 | I3 | I2 | I3 |
C, fully compatible transformants with an LGR of 4 to 6.5 cm/day with <1% HCD; I2, incompatible transformants with an LGR of 1 to 2.5 cm/day which show ∼20 to 30% HCD; I3, severely incompatible transformants that have an LGR of <1 cm/day and show 20 to 30% HCD.
The second type of construct (od1 and go1) contains mosaic combinations between region II and the indel motif between a het-cGR and a het-cOR allele (Fig. 5). Construct od1 was generated from het-cOR by a 15-bp deletion of the het-cOR-specific insertion sequence (encoding RNDTR), thus creating an allele that is HET-COR for amino acid sequences but het-cGR-like for the indel motif. The go1 construct was generated from a het-cGR allele by the addition of 15 bp encoding the het-cOR-specific insertion sequence (RNDTR) and is thus het-cGR for predicted amino acid sequence within the het-c variable region, with the exception of the het-cOR-like insertion. The od1 and go1 chimeric constructs displayed a het-c specificity that was indistinguishable from the specificities of het-cGR and het-cOR alleles, respectively (Table 4). Thus, these data support what was observed in transformation experiments with the naturally occurring het-c chimeric constructs: allelic specificity is completely dependent upon the indel motif and amino acid variability outside of the indel motif does not contribute to het-c allelic specificity.
The indel motif of three amino acids that define a het-cGR allele is essential for vegetative incompatibility.
A het-cGR allele has the smallest variable domain, which is characterized by a 9-bp indel motif (encoding NNG), which is also conserved among almost all het-cOR alleles. In het-cPA-like alleles, the predicted amino acid sequence of this 9-bp indel is variable but includes the predicted asparagine residue (N) (Fig. 2). The indel motif of HET-CGR is predicted to form a short loop between the two conserved antiparallel β-strands. Removal of the codons for the NNG residues results in the predicted formation of a single β-sheet structure that includes both the conserved PHVGTRITL and WPLVTGTF regions (Fig. 2).
A het-cGR allele that contained a 9-bp deletion that removed the codons for NNG was generated (Fig. 5, del3). Only compatible transformants were obtained when del3 was introduced into C2-2-9 (OR), C9-2 (PA), and FGSC2193 (GR) strains by transformation. The del3 transformants displayed growth rates comparable to those of other compatible transformants and did not display HCD. These data suggest that the loop structure between the antiparallel β-strands is important in the structural maintenance of the het-c specificity domain and that loss of this region produces a nonfunctional allele.
The novel specificity of Ndi5923 could be converted to het-cPA specificity by increasing the length of the het-cPA-type insertion.
The results obtained with the artificial and naturally occurring chimeric constructs indicated that amino acid variations in regions I and II do not materially affect het-c specificity; het-c specificity is dependent upon the indel motif in region III (Fig. 2). The Ndi5923 allele displayed a novel het-c specificity and produced incompatible transformants in C9-2 (PA), C2-2-9 (OR), and FGSC2193 (GR) strains (Table 3). The het-cPA-like insertion in Ndi5923 shows both amino acid composition and length variations compared to those of other het-cPA-like alleles.
To determine whether the predicted amino acid sequence of the het-cPA-like insertion from Ndi5923 was the most important factor in het-c specificity or whether the length of the insertion mattered, we constructed an artificial allele, Ndi5923m. Fifteen base pairs were inserted into the Ndi5923 construct, thus making an allele with a het-cPA indel insertion size identical to that of the het-cPA allele from C9-2 (Fig. 5). The 15-bp addition encoded 5 aa that are observed in the C9-2 het-cPA allele. Of these five codons, only the serine (S) and lysine (K) residues are conserved among HET-CPA-like peptides (Fig. 2). When Ndi5923m was introduced into C2-2-9 (OR), C9-2 (PA), and FGSC2193 (GR) strains, it behaved identically to het-cPA and yielded wild-type transformants in C9-2 and FGSC2193 and class-2-incompatible transformants in C2-2-9 (Table 4). These data showed that the novel het-c specificity displayed by Ndi5923 could be converted to het-cPA specificity by altering the size and amino acid composition of the het-cPA-like insertion.
Variations in amino acid sequence in the indel motif affect het-c specificity and severity of incompatibility response.
A set of artificial het-c constructs, pd1, pd2, pd3, and pd4, which contained variations in length of the het-cPA insertion, was generated from the C9-2 het-cPA allele (Fig. 5). The pd1 construct has a deletion of 15 bp (5 aa) and thus has a het-cPA-like insertion of identical size but with a predicted amino acid composition different from that of Ndi5923. The lysine (K) residue that is conserved among all naturally occurring HET-C polypeptides, with the exception of Ndi5923 HET-C, is included in the pd1 construct (Fig. 2 and 5). The pd2 construct has a deletion of 21 bp (removing 7 aa) within the het-cPA-like insertion. The pd3 construct has a deletion of 30 bp (removing 10 aa) within the het-cPA-specific insertion and thus has a size identical to that of a het-cOR allele, although the 5-aa insertion is placed differently in respect to the NNG motif (Fig. 5). The pd4 construct is missing the entire het-cPA-specific insertion and thus has an indel motif that is identical in size to that of a het-cGR allele, although it has the predicted amino acid composition of a het-cPA allele (Fig. 5). In particular, instead of having the 9-bp indel motif (NNG) from het-cGR, the pd4 HET-C has ENR from het-cPA.
The pd4 construct displayed an identical het-c specificity to a canonical het-cGR allele in transformation experiments, although the severity of the incompatibility response was affected (Table 4). The introduction of pd4 into C2-2-9 (OR) yielded class 3, severely incompatible transformants, but the introduction of het-cGR into C2-2-9 (OR) yielded class-2-incompatible transformants. Similar to het-cGR, the introduction of pd4 into C9-2 (PA) yielded class-2-incompatible transformants and compatible transformants in FGSC2193 (GR). These data show that although het-c specificity is dependent upon indel motif, amino acid differences within the indel motif can affect the severity of the incompatibility phenotype.
Similar to Ndi5923, incompatible transformants were obtained when pd1, pd2, and pd3 were introduced into C9-2 (PA), C2-2-9 (OR), and FGSC2193 (GR) (Table 4 and Fig. 6). The pd1 allele has a size identical to but an amino acid composition different from those of Ndi5923 and produced class-2-incompatible transformants in C9-2 (PA) and FGSC2193 (GR) but produced severely incompatible transformants in C2-2-9 (OR). The pd2 construct, which has an indel size that is different from those of all other alleles, yielded transformants that displayed a class-2-incompatible phenotype. The pd3 construct has a variable domain that is identical in size to that of a het-cOR allele (Fig. 7). Secondary-structure predictions of the predicted variable region from PD3 and HET-COR showed different profiles in the loop region between the antiparallel β-strands. Unlike a standard het-cOR allele, the introduction of pd3 into C2-2-9 (OR), C9-2 (PA), and FGSC2193 yielded a spectrum of incompatible transformants identical to that of pd1 (Table 4). These data indicated that amino acid composition variation in the indel motif can confer novel het-c specificity, perhaps by altering spatial characteristics of the loop domain formed by the indel motif.
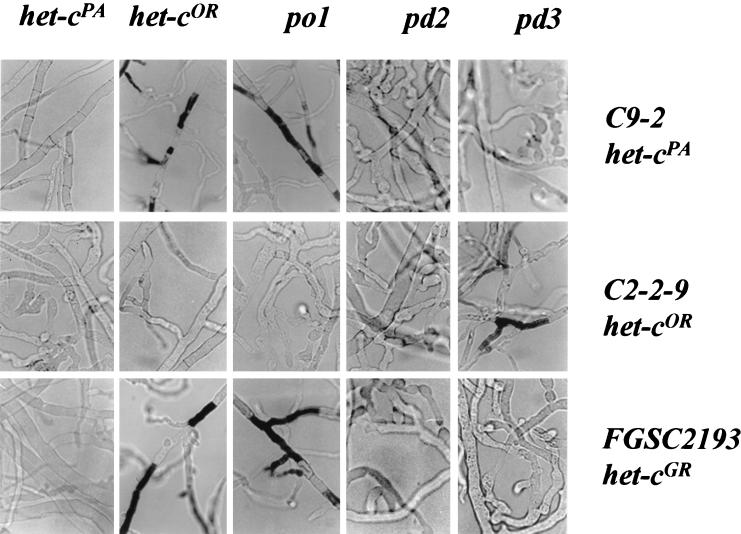
FIG. 6.
Phenotypes of C9-2 (het-cPA), C2-2-9 (het-cOR), and FGSC2193 (het-cGR) transformants containing het-cPA, het-cOR, po1, pd2, and pd3 constructs observed after 1 day of growth. Hyphae were treated with the vital dye Evan's blue (16), which stains dead hyphal compartments. Magnification, ×64.
FIG. 7.
Amino acid comparison between predicted products of alleles that confer identical het-c specificity, Ndi5923 and PD1 (A), and predicted products of alleles that confer different het-c specificity, HET-COR and PD3 (B). ∗, amino acid identity.
Novel het-c specificity can be generated by increasing length of indel motif.
Our results indicated that variations in the pattern and length of the het-cPA-like insertion could generate alleles with novel het-c specificity. To examine if increasing the length of the variable domain affected het-c specificity, two artificial alleles were constructed, po1 and po2, which contain both of the het-cOR-specific and het-cPA-specific insertions. These two constructs are 15 bp longer than a typical het-cPA allele. The po1 and po2 constructs are identical, except that po1 has the predicted NNG sequences from het-cOR while po2 has the predicted ENR sequences from het-cPA (Fig. 5).
The introduction of both po1 and po2 into C9-2, C2-2-1, and FGSC2193 produced transformants that displayed incompatible phenotypes (Table 4 and Fig. 6). Both C9-2 (PA) and FGSC2193 (GR) strains that contained either po1 or po2 constructs displayed severely incompatible phenotypes (LGR of <0.5 cm/day); dead hyphal compartments could be observed after 24 h of growth. The C2-2-9 (OR) transformants that contained either po1 or po2 constructs showed a class-2-incompatible phenotype. Secondary-structure predictions showed that the indel region of PO1 and PO2 contained two loops with an additional short region predicted to form a β-sheet between the two conserved antiparallel β strands. The fact that po1 and po2 constructs showed an identical novel het-c specificity and similar spectrum of phenotypes suggested that they might also confer an identical het-c specificity.
Alleles conferring novel het-c specificity are not self-incompatible.
The po1, po2, pd1, pd2, pd3, and Ndi5923 constructs displayed a novel het-c specificity and produced incompatible transformants when introduced into C9-2 (PA), C2-2-9 (OR), and FGSC2193 (GR) strains. It is possible that the incompatible phenotypes that we observed were the result of self-incompatibility of these alleles rather than of incompatibility triggered by an interaction with the resident het-c allele. To determine whether po1, po2, pd1, pd2, pd3, and Ndi5923 conferred self-incompatibility, each construct was individually introduced into a het-c deletion strain, CJ44 (Table 1). In all cases, only compatible, wild-type transformants were obtained (Table 5). In particular, po1 and po2, which have both het-cOR-like and het-cPA-like insertions, were not self-incompatible. These data indicate that po1, po2, pd1, pd2, pd3, and pd4 confer a novel het-c specificity in N. crassa that is different from het-cPA, het-cOR, and het-cGR allelic specificities.
TABLE 5.
Phenotypes and growth rates of CJ44 transformants containing different combinations of het-c allele pairs
| Allele pair | Phenotypea | Growth rate (cm/day) (mean ± SD) | HCDb |
|---|---|---|---|
| pCB1004 + pOKE | C | 4.6 ± 0.20 | − |
| het-cOR + het-cOR | C | 4.7 ± 0.20 | − |
| het-cPA + het-cPA | C | 4.8 ± 0.20 | − |
| het-cOR + het-cPA | I | 0.3 ± 0.10 | +++ |
| het-cOR + het-cGR | I | 0.4 ± 0.10 | +++ |
| Ndi5923 + Ndi5923 | C | 4.6 ± 0.17 | − |
| pd1 + pd1 | C | 4.3 ± 0.30 | − |
| pd2 + pd2 | C | 5.4 ± 0.60 | − |
| pd3 + pd3 | C | 4.4 ± 0.30 | − |
| po1 + po1 | C | 4.7 ± 0.57 | − |
| po2 + po2 | C | 4.8 ± 0.46 | − |
| pd1 + Ndi5923 | C | 4.5 ± 0.35 | − |
| pd1 + pd2 | I | 0.5 ± 0.12 | +++ |
| pd1 + pd3 | I | 0.4 ± 0.05 | +++ |
| pd1 + po1 | I | 0.3 ± 0.10 | +++ |
| pd2 + pd3 | I | 0.6 ± 0.10 | +++ |
| pd2 + po1 | I | 0.4 ± 0.06 | +++ |
| pd3 + po1 | I | 0.4 ± 0.10 | +++ |
| pd3 + het-cOR | I | 0.6 ± 0.15 | +++ |
| po1 + po2 | C | 4.7 ± 0.30 | − |
Transformants were compatible (C) or incompatible (I).
−, <1% HCD was observed in the growing colony. +++, 20 to 30% HCD was observed in the colony after 1 day of growth.
Alleles with variable specificity domains confer four novel and different het-c specificities.
The introduction of po1, po2, pd1, pd2, pd3, and Ndi5923 into C2-2-9 (OR), C9-2 (PA), and FGSC2193 (GR) produced transformants that displayed all of the hallmarks of het-c incompatibility (growth inhibition, suppression of conidiation, and HCD), indicating that these constructs conferred a novel het-c specificity. However, it was unclear whether these constructs each conferred an identical het-c specificity or defined new and different het-c allelic specificities. To distinguish these two possibilities, po1, po2, pd1, pd2, pd3, and Ndi5923 were cotransformed in pairwise combinations into CJ44 (Table 5); the presence of alternative alleles was coselected by growth on hygromycin media which lacked pantothenic acid (see Materials and Methods). As expected, the introduction by cotransformation of constructs of identical type yielded only compatible transformants (e.g., het-cOR plus het-cOR, pd1 plus pd1, or po1 plus po1) (Table 5), while the cotransformation of het-cPA plus het-cOR and het-cGR plus het-cOR constructs produced severely incompatible transformants.
Only two pairwise combinations of novel alleles yielded compatible transformants. The pd1 construct has an indel motif of the same size as that of Ndi5923, although only four predicted amino acid positions are conserved (Fig. 7, E, N, F, and E). However, cotransformation of pd1 and Ndi5923 into CJ44 yielded only compatible transformants, indicating that these two constructs conferred an identical het-c specificity (Table 5). Similarly, po1 and po2 have an indel motif of identical size but differ in predicted amino acid sequence within the NNG or ENR motif (Fig. 5). The cotransformation of po1 and po2 alleles into CJ44 also yielded compatible transformants, indicating that these two alleles have identical het-c specificity (Table 5).
Pairwise cotransformation of all other allele combinations resulted in severely incompatible transformants (Table 5). These transformants displayed a typical het-c incompatibility phenotype with an LGR of approximately 0.5 cm/day and showed 20 to 30% HCD throughout the colony. In particular, the introduction of pd3 and het-cOR into CJ44 yielded incompatible transformants, even though they had specificity domains with identical size (Fig. 7). Thus, four additional novel het-c specificities were identified in this study. One, Ndi5923, was identified from a survey of naturally occurring alleles. The other three were identified from artificially constructed alleles that contained length variations in the indel motif (pd2 and po1/po2) or variations in amino acid sequence of the indel motif (pd3).
DISCUSSION
het-c specificity is dependent upon indel motif.
The het-c variable domain characterized from species and genera related to N. crassa showed that the alleles could be grouped into one of the three het-c specificities identified in N. crassa, based on the indel motif; the indel motif pattern exhibits transspecies polymorphisms (reference 48 and Fig. 2). Phylogenetic analysis (either including or excluding the indel motif) supported this grouping but also split each group into additional clades. Transspecies polymorphisms were also apparent in these clades; clade 5 contains a het-c allele from Neurospora pannonica, Sordaria sclerogenia, and a Gelasinospora sp. isolate. These data suggest that multiple specificities might occur at het-c, in addition to het-cOR, het-cPA, and het-cGR-types. However, the results of this study indicate that naturally occurring variations in amino acid sequences in the variable domain do not affect het-c specificity; het-c specificity is dependent upon the indel motif. The neutral amino acid variation surrounding the indel motif has presumably been retained in populations because of selection for alleles conferring alternative het-c specificity, which is dependent upon the indel motif. Transspecies polymorphism associated with two different indel motifs has also been reported in allelic lineages in the MHC Aβ1 locus of mice and rats, which have been preserved for over 10 million years (15). Interestingly, intragenic recombination surrounding the indel motif of the Aβ1 locus was also observed, thus creating hybrid genes. Occasional recombination within the het-c variable region could also link different neutral polymorphisms (such as those found in regions I and II) with identical indel motifs, resulting in apparent transspecies polymorphisms within an indel type. Phylogenetic analysis treats insertions and deletions as single events, and therefore, mutations that result in amino acid variability outside of the indel motif significantly affect tree topology (48). In this study, we determined that chimeric constructs containing variable domains from naturally occurring het-c alleles fell into one of three het-c allelic specificities identified in N. crassa. The only exception was a naturally occurring allele from Neurospora discreta. If this alternative specificity is also under balancing selection, it should increase in frequency within the N. discreta population, similar to what has been hypothesized for both S alleles and polymorphic loci in the MHC (27).
DNA sequences of various het loci from N. crassa and P. anserina show that alleles conferring alternative specificities are polymorphic. For example, the alternative het-s polypeptides of P. anserina differ by 12 aa substitutions, although only a single amino acid change is sufficient to switch allelic specificity (44). Similarly, 16 polymorphic positions were identified in predicted P. anserina het-c polypeptides (not related to N. crassa het-c) (36). As with het-s, allelic specificity at P. anserina het-c was dependent upon a single amino acid difference and constructs conferring novel het-c allelic specificities were generated by chimeric allele construction. In N. crassa, the predicted het-6 polypeptides show only 68% amino acid identity, with polymorphic positions scattered throughout the open reading frame (42). Although alternative alleles at het loci are polymorphic, it is unclear whether selection mechanisms are maintaining polymorphisms at het loci other than the N. crassa het-c locus. The introduction of the N. crassa het-cOR, het-cPA, and het-cGR alleles into P. anserina resulted in growth inhibition and HCD (38). These data indicate that the mechanism of het-c-mediated vegetative incompatibility is well conserved among filamentous fungi, but whether it occurs in a particular species may be dependent on the presence of polymorphisms within the indel motif.
Secondary-structure predictions and het-c specificity.
The indel motif is predicted to form a coiled-loop structure between conserved antiparallel β-strands. The allele-specific indel motif may form a protruding loop that mediates het-c allele specificity, perhaps by protein-protein interactions (between alternative HET-C proteins or with other proteins). The predicted secondary structure of HET-C is reminiscent of immunoglobulin molecules; crystallographic studies with immunoglobulin molecules show that the antigen interface consists of three hypervariable loops known as the complementarity determining region (CDR) which are formed on nine antiparallel β-strands of the variable domain (9). Both length variations and amino acid substitutions occur in the CDRs and are correlated with antigen binding specificity (5). The large number of sequence variations in the loop region of antibodies shows that the structural framework of the variable domain is fairly insensitive to changes in amino acid composition and length of the CDR loop regions. In this study, novel het-c specificities were generated either by alterations in amino acid and/or length of the indel motif. Only one construct, del3, which removed the loop between the conserved antiparallel β-strands, destroyed het-c function. Although many of the alleles that confer alternative het-c specificities give significantly different secondary-structure profiles in the indel region (het-cOR, het-cPA, het-cGR and po1/po2), it is not obvious from such predictions what differences are crucial for conferring alternative het-c specificities. Future structural studies using the predicted products of the seven different het-c specificities identified in this study will provide useful tools to address this issue.
Relationship between allelic specificity and recognition.
An allelic specificity region functioning in heteromeric complex formation has been described for several fungal mating systems. In U. maydis, the b mating-type genes (bE and bW) have variable and constant regions (28). Analysis of chimeric bE and bW alleles showed that single-amino-acid alterations in the N-terminal variable region were sufficient to generate novel mating specificities. It was hypothesized that these amino acid differences affected the capacity of different bE and bW proteins to heterodimerize (26). In P. anserina, alternative het-s polypeptides (HET-S and HET-s) have been shown to form both heterodimers and homodimers via yeast two-hybrid experiments (8). The results in this study also support the possibility that vegetative incompatibility is mediated by HET-C heteromeric complex formation. In an immunoprecipitation study, alternative HET-C proteins formed a heteromeric complex during vegetative incompatibility (47; G. Iyer and N. L. Glass, unpublished data). From the results generated in this study, we would predict that the amino acid composition and length differences in the indel motif in the chimeric constructs that displayed novel specificity (Ndi5923/pd1, pd2, pd3, and po1/po2) may facilitate heterocomplex formation between alternative HET-C proteins. The specificity domain may directly mediate protein-protein interactions or, alternatively, affect the conformation of a different region of HET-C that mediates physical interactions between alternative HET-C polypeptides.
Models for het-c vegetative incompatibility.
Vegetative incompatibility reactions appear to have conserved features in filamentous fungi. In N. crassa and P. anserina, the vegetative incompatibility response involves common stages of hyphal compartmentation, vacuolization, and death (3, 18, 25). Forced heterokaryons or transformants containing alternative het-c alleles show growth inhibition, growth arrest, suppression of conidiation, and HCD. The morphological phenotype and severity of the incompatibility phenotype can be affected by the genetic background of recipient strains (reference 39 and this study). In addition, it is possible that variations in the indel motif affect the thermodynamics of heterocomplex formation and thus affect the phenotypic output of vegetative incompatibility. Further genetic experiments and experiments that assess the affinity of alternative HET-C proteins to form a heterocomplex will be required to differentiate these two possibilities.
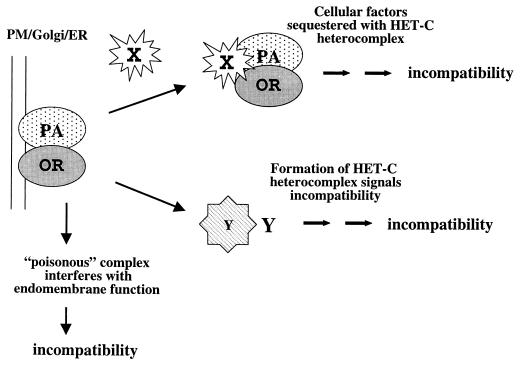
The HET-C protein is predicted to enter the endomembrane system and to ultimately reside within either the golgi or the plasma membrane. The presence of alternative HET-C proteins within a common cytoplasm results in growth inhibition and HCD, presumably by heterocomplex formation (Fig. 8). Chimeric het-c allele construction and secondary-structure prediction in this study suggested that the specificity domain of het-c may be involved in the mechanism of stable HET-C heteromeric complex formation. An interaction of alternative HET-C proteins may mediate vegetative incompatibility directly, perhaps by interfering with the normal functioning of HET-C within the cell, although mutational analysis has shown that het-c is not an essential gene; het-c mutants are indistinguishable from the wild type in morphology (40). Alternatively, heterocomplex formation between HET-C proteins may result in the formation of a “poison” complex that affects normal cellular function of either the plasma membrane or endomembrane system. In P. anserina, a poison heteromeric complex model, in which heteromeric complexes between the products of incompatible genes are lethal to the cell, has been proposed for mediating vegetative incompatibility (2). The formation of such a complex may result in general growth inhibition and morphological changes, until a threshold of HET-C heterocomplex is formed and HCD is triggered. Alternatively, HET-C heterocomplex formation may trigger vegetative incompatibility via signaling mechanisms that result in growth inhibition and HCD (Fig. 8). The results of this study have provided tools and information on the molecular basis of allelic specificity that will be extremely useful in deciphering the structural mechanism of recognition, which ultimately leads to the phenotypic manifestations of vegetative incompatibility in filamentous fungi.
FIG. 8.
Alternative models for het-c-mediated vegetative incompatibility. The model depicts the heteromeric complex formation between the products of incompatible het-c alleles, the formation of which is dependent upon variations in the indel motif in the variable domain. The HET-C heteromeric complex may trigger vegetative incompatibility by a poison effect on function of the endoplasmic reticulum (ER) and golgi or plasma membrane (PM) or by a dominant-negative effect on cell growth by an interaction with a modulator (X) or by regulating the activity or expression of a separate component (Y) that triggers the activation of the downstream effectors of vegetative incompatibility. OR, HET-COR; PA, HET-CPA.
ACKNOWLEDGMENTS
We thank S. Sarkar and G. Iyer and the members of the N. L. Glass laboratory for critical reading of the manuscript. We also thank the members of J. Wu's graduate committee, Michel Roberge, Jim Kronstad, and Sally Otto, for their advice and suggestions.
We gratefully acknowledge financial support from the Canadian Natural Sciences and Engineering Research Council (NSERC) and a National Institutes of Health GM60468 grant to N.L.G.
REFERENCES
- 1.Asante-Owusu R N, Banham A H, Böhnert H U, Mellor E J, Casselton L A. Heterodimerization between two classes of homeodomain proteins in the mushroom Coprinus cinereus brings together potential DNA-binding and activation domains. Gene. 1996;172:25–31. doi: 10.1016/0378-1119(96)00177-1. [DOI] [PubMed] [Google Scholar]
- 2.Begueret J, Turcq B, Clave C. Vegetative incompatibility in filamentous fungi—het genes begin to talk. Trends Genet. 1994;10:441–446. doi: 10.1016/0168-9525(94)90115-5. [DOI] [PubMed] [Google Scholar]
- 3.Beisson-Schecroun J. Incompatibilité cellulaire et interactions nucleo-cytoplasmiques dans les phénomènes de barrage chez Podospora anserina. Ann Genet. 1962;4:3–50. [PubMed] [Google Scholar]
- 4.Berges T, Barreau C. Heat shock at an elevated temperature improves transformation efficiency of protoplasts from Podospora anserina. J Gen Microbiol. 1989;135:601–604. doi: 10.1099/00221287-135-3-601. [DOI] [PubMed] [Google Scholar]
- 5.Branden C, Tooze J. Introduction to protein structure. New York, N.Y: Garland Publishing, Inc.; 1991. pp. 179–199. [Google Scholar]
- 6.Carroll A M, Sweigard J A, Valent B. Improved vectors for selecting resistance to hygromycin. Fungal Genet Newsl. 1994;41:22. [Google Scholar]
- 7.Caten C E. Vegetative incompatibility and cytoplasmic infection in fungi. J Gen Microbiol. 1972;72:221–229. doi: 10.1099/00221287-72-2-221. [DOI] [PubMed] [Google Scholar]
- 8.Coustou V, Deleu C, Saupe S, Bégueret J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc Natl Acad Sci USA. 1997;94:9773–9778. doi: 10.1073/pnas.94.18.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies D R, Padlan E A, Sheriff S. Antibody-antigen complexes. Annu Rev Biochem. 1990;59:439–473. doi: 10.1146/annurev.bi.59.070190.002255. [DOI] [PubMed] [Google Scholar]
- 10.Davis R H, De Serres F J. Genetic and microbial research techniques for Neurospora crassa. Methods Enzymol. 1970;17A:79–143. [Google Scholar]
- 11.Debets A J M, Griffiths A J F. Polymorphism of het-genes prevents resource plundering in Neurospora crassa. Mycol Res. 1998;102:1343–1349. [Google Scholar]
- 12.Debets F, Yang X, Griffiths A J F. Vegetative incompatibility in Neurospora—its effect on horizontal transfer of mitochondrial plasmids and senescence in natural populations. Curr Genet. 1994;26:113–119. doi: 10.1007/BF00313797. [DOI] [PubMed] [Google Scholar]
- 13.Deléage G, Roux B. An algorithm for protein secondary structure prediction based on class prediction. Protein Eng. 1987;1:289–294. doi: 10.1093/protein/1.4.289. [DOI] [PubMed] [Google Scholar]
- 14.Deleu C, Clavé C, Bégueret J. A single amino acid difference is sufficient to elicit vegetative incompatibility in the fungus Podospora anserina. Genetics. 1993;135:45–52. doi: 10.1093/genetics/135.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Figueroa F, Gunther E, Klein J. MHC polymorphisms pre-dating speciation. Nature. 1988;335:265–271. doi: 10.1038/335265a0. [DOI] [PubMed] [Google Scholar]
- 16.Gaff D F, Okong'O-Ogola O. The use of non-permeating pigments for testing the survival of cells. J Exp Bot. 1971;22:756–758. [Google Scholar]
- 17.Garnjobst L. Genetic control of heterocaryosis in Neurospora crassa. Am J Bot. 1953;40:607–614. [Google Scholar]
- 18.Garnjobst L, Wilson J F. Heterocaryosis and protoplasmic incompatibility in Neurospora crassa. Proc Natl Acad Sci USA. 1956;42:613–618. doi: 10.1073/pnas.42.9.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geourjon C, Deléage G. SOPM: a self-optimized method for protein secondary structure prediction. Protein Eng. 1994;7:157–164. doi: 10.1093/protein/7.2.157. [DOI] [PubMed] [Google Scholar]
- 20.Geourjon C, Deléage G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci. 1995;11:681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- 21.Gibrat J F, Garnier J, Robson B. Further developments of protein secondary structure prediction using information theory. New parameters and consideration of residue pairs. J Mol Biol. 1987;198:425–443. doi: 10.1016/0022-2836(87)90292-0. [DOI] [PubMed] [Google Scholar]
- 22.Glass N L, Jacobson D J, Shiu K T. The genetics of hyphal fusion and vegetative incompatibility in filamentous ascomycetes. Annu Rev Genet. 2000;34:165–186. doi: 10.1146/annurev.genet.34.1.165. [DOI] [PubMed] [Google Scholar]
- 23.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 24.Howlett B, Leslie J F, Perkins D D. Putative multiple alleles at the vegetative (heterokaryon) incompatibility loci het-c and het-8 in Neurospora crassa. Fungal Genet Newsl. 1993;40:40–42. [Google Scholar]
- 25.Jacobson D J, Beurkens K, Klomparens K L. Microscopic and ultrastructural examination of vegetative incompatibility in partial diploids heterozygous at het loci in Neurospora crassa. Fungal Genet Biol. 1998;23:45–56. doi: 10.1006/fgbi.1997.1020. [DOI] [PubMed] [Google Scholar]
- 26.Kämper J, Reichmann M, Romeis T, Bölker M, Kahmann R. Multiallelic recognition: nonself-dependent dimerization of the bE and bW homeodomain proteins in Ustilago maydis. Cell. 1995;81:73–83. doi: 10.1016/0092-8674(95)90372-0. [DOI] [PubMed] [Google Scholar]
- 27.Klein J, Sato A, Nagl S, O'hUigin C. Molecular trans-species polymorphism. Annu Rev Ecol Syst. 1998;29:1–21. [Google Scholar]
- 28.Kronstad J W, Leong S A. The b mating-type locus of Ustilago maydis contains variable and constant regions. Genes Dev. 1990;4:1384–1395. doi: 10.1101/gad.4.8.1384. [DOI] [PubMed] [Google Scholar]
- 29.Leslie J F. Fungal vegetative compatibility. Annu Rev Phytopathol. 1993;31:127–150. doi: 10.1146/annurev.py.31.090193.001015. [DOI] [PubMed] [Google Scholar]
- 30.Levin J M, Robson B, Garnier J. An algorithm for secondary structure determination in proteins based on sequence similarity. FEBS Lett. 1986;205:303–308. doi: 10.1016/0014-5793(86)80917-6. [DOI] [PubMed] [Google Scholar]
- 31.Mylyk O M. Heterokaryon incompatibility genes in Neurospora crassa detected using duplication-producing chromosome rearrangements. Genetics. 1975;80:107–124. doi: 10.1093/genetics/80.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perkins D D. Chromosome rearrangements in Neurospora and other filamentous fungi. Adv Genet. 1997;36:239–398. doi: 10.1016/s0065-2660(08)60311-9. [DOI] [PubMed] [Google Scholar]
- 33.Perkins D D. Main features of vegetative incompatibility in Neurospora crassa. Fungal Genet Newsl. 1982;35:44–46. [Google Scholar]
- 34.Perkins D D. The use of duplication-generating rearrangements for studying heterokaryon incompatibility genes in Neurospora. Genetics. 1975;80:87–105. doi: 10.1093/genetics/80.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perkins D D, Radford A, Sachs M S. The Neurospora compendium: chromosomal loci. San Diego, Calif: Academic Press; 2000. [Google Scholar]
- 36.Saupe S, Turcq B, Bégueret J. Sequence diversity and unusual variability at the het-c locus involved in vegetative incompatibility in the fungus Podospora anserina. Curr Genet. 1995;27:466–471. doi: 10.1007/BF00311217. [DOI] [PubMed] [Google Scholar]
- 37.Saupe S J. Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol Mol Biol Rev. 2000;64:489–502. doi: 10.1128/mmbr.64.3.489-502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saupe S J, Clavé C, Sabourin M, Bégueret J. Characterization of hch, the Podospora anserina homolog of the het-c heterokaryon incompatibility gene of Neurospora crassa. Curr Genet. 2000;38:39–47. doi: 10.1007/s002940000130. [DOI] [PubMed] [Google Scholar]
- 39.Saupe S J, Glass N L. Allelic specificity at the het-c heterokaryon incompatibility locus of Neurospora crassa is determined by a highly variable domain. Genetics. 1997;146:1299–1309. doi: 10.1093/genetics/146.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saupe S J, Kuldau G A, Smith M L, Glass N L. The product of the het-C heterokaryon incompatibility gene of Neurospora crassa has characteristics of a glycine-rich cell wall protein. Genetics. 1996;143:1589–1600. doi: 10.1093/genetics/143.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schweizer M, Case M E, Dykstra C C, Giles N H, Kushner S R. Identification and characterization of recombinant plasmids carrying the complete qa-2+ cluster from Neurospora crassa including the qa-1+ regulatory gene. Proc Natl Acad Sci USA. 1981;78:5086–5090. doi: 10.1073/pnas.78.8.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith M L, Micali O C, Hubbard S P, Mir-Rashed N, Jacobson D J, Glass N L. Vegetative incompatibility in the het-6 region of Neurospora crassa is mediated by two linked genes. Genetics. 2000;155:1095–1104. doi: 10.1093/genetics/155.3.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith M L, Yang C J, Metzenberg R L, Glass N L. Escape from het-6 incompatibility in Neurospora crassa partial diploids involves preferential deletion within the ectopic segment. Genetics. 1996;144:523–531. doi: 10.1093/genetics/144.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turcq B, Deleu C, Denayrolles M, Begueret J. Two allelic genes responsible for vegetative incompatibility in the fungus Podospora anserina are not essential for cell viability. Mol Gen Genet. 1991;228:265–269. doi: 10.1007/BF00282475. [DOI] [PubMed] [Google Scholar]
- 45.Vallette F, Mege E, Reiss A, Adesnik M. Construction of mutant and chimeric genes using polymerase chain reaction. Nucleic Acids Res. 1989;17:723–733. doi: 10.1093/nar/17.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vogel H J. Distribution of lysine pathways among fungi: evolutionary implications. Am Nat. 1964;98:435–446. [Google Scholar]
- 47.Wu J. Ph.D. thesis. Non-self recognition in filamentous fungi—the het-c mediated vegetative incompatibility in Neurospora crassa. Vancouver, Canada: University of British Columbia; 2000. [Google Scholar]
- 48.Wu J, Saupe S J, Glass N L. Evidence for balancing selection operating at the het-c heterokaryon incompatibility locus in a group of filamentous fungi. Proc Natl Acad Sci USA. 1998;95:12398–12403. doi: 10.1073/pnas.95.21.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yee A R, Kronstad J W. Construction of chimeric alleles with altered specificity at the b incompatibility locus of Ustilago maydis. Proc Natl Acad Sci USA. 1993;90:664–668. doi: 10.1073/pnas.90.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yee A R, Kronstad J W. Dual sets of chimeric alleles identify specificity sequences for the bE and bW mating and pathogenicity genes of Ustilago maydis. Mol Cell Biol. 1998;18:221–232. doi: 10.1128/mcb.18.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]