Abstract
The transport of metabolites, coenzymes, and ions across the mitochondrial inner membrane is still poorly understood. In most cases, membrane transport is facilitated by the so-called mitochondrial carrier proteins. The yeast Saccharomyces cerevisiae contains 35 members of the carrier family, but a function has been identified for only 13 proteins. Here, we investigated the yeast carrier Leu5p (encoded by the gene YHR002w) and its close human homologue Graves' disease protein. Leu5p is inserted into the mitochondrial inner membrane along the specialized import pathway used by carrier proteins. Deletion of LEU5 (strain Δleu5) was accompanied by a 15-fold reduction of mitochondrial coenzyme A (CoA) levels but did not affect the cytosolic CoA content. As a consequence, the activities of several mitochondrial CoA-dependent enzymes were strongly decreased in Δleu5 cells. Our in vitro and in vivo analyses assign a function to Leu5p in the accumulation of CoA in mitochondria, presumably by serving as a transporter of CoA or a precursor thereof. Expression of the Graves' disease protein in Δleu5 cells can replace the function of Leu5p, demonstrating that the human protein represents the orthologue of yeast Leu5p. The function of the human protein might not be directly linked to the disease, as antisera derived from patients with active Graves' disease do not recognize the protein after expression in yeast, suggesting that it does not represent a major autoantigen. The two carrier proteins characterized herein are the first components for which a role in the subcellular distribution of CoA has been identified.
Mitochondria perform a variety of processes, such as oxidative phosphorylation, the citric acid cycle, the β-oxidation of fatty acids, parts of the urea cycle, and the biosynthesis of heme and certain amino acids (13, 14, 61). The metabolic activity of mitochondria requires the rapid and highly specific exchange of molecules between the cytosol and the mitochondrial matrix space. To a large extent, this is facilitated by a family of transport proteins of the inner membrane, the so-called mitochondrial carrier proteins (for reviews, see references 20, 22, 31, 44, 45, and 67). Members of this family include proteins responsible for the exchange of ADP and ATP (termed AAC or ANT) and for the transport of, e.g., phosphate, citrate, carnitine, dicarboxylates, amino acids, flavin adenine dinucleotide (FAD), or protons. The biogenesis of carriers differs in various aspects from that of most other mitochondrial proteins (reviewed in references 2, 32, and 60). They lack an N-terminal targeting sequence (presequence) and they follow a unique import pathway involving the interaction with specialized import components in the outer membrane (Tom70), the intermembrane space (Tim8, Tim9, Tim10, Tim12, and Tim13), and the inner membrane (Tim18, Tim22, and Tim54).
In their transport-competent form, carrier proteins are dimeric (50). Each monomer is comprised of three homologous modules containing two transmembrane segments each. Both the N and C termini of carrier proteins face the intermembrane space. The primary sequences of carrier proteins typically share between 20 and 40% of amino acid residues, including a characteristic carrier signature motif in each of the matrix-exposed loops of the three modules. This motif is required for proper function of the carriers (see, e.g., reference 42) and for their insertion into the inner membrane (52).
The importance of mitochondrial carrier proteins for a living cell has been most impressively demonstrated by mouse mutants in, e.g., the ADP/ATP carrier ANT1 or the ornithine transporter ORNT1 (8, 25). Mutant mice exhibit the hallmarks of well-characterized diseases such as mitochondrial myopathies. For the identification of the function of the individual carrier proteins, the yeast Saccharomyces cerevisiae has proven to be an excellent model system. In the genome of this yeast, 35 members of the carrier family have been identified based on characteristic features in their primary structures (20, 44, 45). To date, the substrates of only 13 proteins have been elucidated, including ADP/ATP, phosphate, various citric acid cycle metabolites, carnitine, amino acids, and FAD.
The S. cerevisiae gene YHR002w encodes a protein that shares the characteristic features of mitochondrial carrier proteins such as the carrier signature motif, the tripartite structure, the presence of six transmembrane segments, and the lack of a typical N-terminal mitochondrial targeting sequence (presequence [44]). Highest sequence homology exists to human Graves' disease protein (hGP) (35% identical amino acid residues [69]), its bovine homologue (37% [21]), and to a protein of Saccharomyces pombe (46%; accession no. O 13805). The S. cerevisiae gene YHR002w, in an attempt to isolate the second α-isopropylmalate synthase (IPMS) in addition to the well-characterized Leu4p (9), has been cloned previously and termed LEU5 (19). This attempt was based on the observation that mutants in LEU4 were not leucine auxotrophic. Only double mutants in both LEU4 and LEU5 required the addition of leucine for growth. However, analysis of a partial sequence of LEU5 revealed that the protein might be membrane integrated and not directly involved in the biosynthesis of leucine (18, 19). The specific function of Leu5p remained elusive, though. We therefore sought to determine the subcellular localization of Leu5p and to define its function. Further, we intended to investigate the functional relationship between Leu5p and the human homologue hGP. Our biochemical and genetic results assign a crucial function to both proteins in the accumulation of coenzyme A (CoA) in the mitochondrial matrix.
MATERIALS AND METHODS
Yeast strains and growth of yeast.
The following S. cerevisiae strains were used: strain W303-1A or W303-1B (MATa or MATα, ade2-1 leu2-3,112 his311 his3-15 trp1-1 ura3-1) was used as the wild type; Δleu5 (W303-1A leu5::HIS3) (this study); Δcit2 (W303-1A cit2::URA3) (this study); Δleu5Δcit2 (W303-1A leu5::HIS3 cit2::URA3) (this study); Δcor1 (W303-1A cor1::HIS3) (11); Δcox6 (W303-1B cox6::URA3) (33); Δflx1 (W303-1A flx1::LEU2) (64); Δmir1 (W303-1B mir1::LEU2) (17). Cells were grown on 1% yeast extract, 2% Bacto Peptone supplemented with either 2% glucose (YPD), 3% glycerol (YPG), or 2% galactose (YPGal) unless stated otherwise. For selective growth, yeast cells were cultivated in 0.7% yeast nitrogen base, a carbon source as listed above, and 0.5% ammonium sulfate. Leucine (30 mg/liter), adenine, histidine, lysine, tryptophan, and uracil (20 mg/liter each) were added according to the auxotrophic requirements of the various strains.
Expression of hGP in yeast.
The gene encoding hGP was isolated from a cDNA library prepared from a Jurkat lymphoma cell line by PCR. A 1.3-kb DNA fragment was inserted into a yeast expression vector (pYES2; Invitrogen) under the control of the GAL10 promoter. The resulting plasmid, pYES2/hGP, was used for transformation into yeast strains. Expression of hGP was induced by inclusion of 0.2% galactose in the growth medium.
Fluorimetric measurement of α-IPMS activity.
Either intact or detergent-lysed mitochondria (50 μg each) were used to synthesize α-isopropylmalate (α-IPM) by incubation in 0.5 ml of SoH buffer (0.6 M sorbitol, 20 mM HEPES-KOH [pH 7.4]) for 15 min at 25°C. Intact mitochondria were supplemented with 2 mM α-ketoisovalerate and 2 mM pyruvate. Mitochondria lysed in 0.05% Triton X-100 detergent were centrifuged (10 min at 12,000 × g), and the supernatant was supplemented with 1 mM acetyl-CoA and 2 mM α-ketoisovalerate. Analysis of α-IPM by conversion to its umbelliferone derivative and fluorimetric detection was essentially performed as described previously (7). In brief, concentrated sulfuric acid (50 μl) and 2.5 ml of diethyl ether were added to the samples, followed by vortexing for 1 min. The ether phase was removed and dried under a stream of air. Concentrated sulfuric acid (0.2 ml) was added to the dried extract and the solution was left at room temperature for 15 min. Then 0.12 ml of 1.84 M resorcinol was added and samples were incubated at 37°C for 15 min. After addition of 2.5 ml of H2O, 50 μl of this mixture was added to 450 μl of a borate-carbonate buffer (pH 10). The fluorescence was measured in an M4 QII Zeiss fluorimeter (excitation, 360 nm; emission, 415 nm). The fluorescence recorded relative to 0.5 μg of quinine sulfate per ml (in 0.1 N sulfuric acid) was set to 40, and the fluorescence measured with 0.1 N sulfuric acid was adjusted to zero.
Miscellaneous procedures.
Previously published methods were used for the manipulation of DNA and for PCR (49), transformation of yeast cells (23), isolation of plasmids from yeast (49), isolation of yeast mitochondria and postmitochondrial supernatants (PMS) (12), protein import into mitochondria (15, 16, 55), whole-cell lysates by breaking the cells with glass beads (68), enzyme activities of citrate synthase (54), and determination of cellular citrate concentrations (41). For the enzymatic determination of CoA (40), mitochondria and PMS were deproteinized by the addition of 4% perchloric acid. After 5 min on ice, 150 mM potassium phosphate (pH 7) was added and the pH was adjusted to 7 with KOH. Samples were frozen in liquid nitrogen, thawed, and centrifuged for 10 min at 12,000 × g. The supernatant was used for detection of CoA.
RESULTS
Insertion of Leu5p into the mitochondrial inner membrane occurs along a carrier-specific pathway.
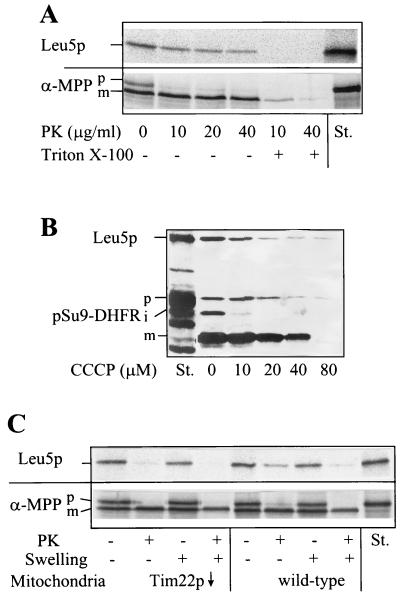
To identify Leu5p as a constituent of the mitochondrial inner membrane, we studied its import and localization in mitochondria. Leu5p was synthesized by in vitro translation in the presence of radioactive [35S]methionine. The radiolabeled protein was incubated with isolated yeast mitochondria in the presence of a membrane potential and ATP, and then samples were treated with proteinase K. A substantial fraction of Leu5p became resistant to digestion by proteinase K, indicating its uptake by the organelles (Fig. 1A). Consistent with the lack of a mitochondrial presequence, no proteolytic processing was observed. After lysis of the mitochondria with detergent, Leu5p was completely digested, indicating that the protease resistance was not caused by aggregation of Leu5p. Import of Leu5p was largely diminished after removing the surface receptors by treatment of the mitochondria with trypsin before the import reaction (data not shown). Depletion of the membrane potential, ΔΨ, by addition of the uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP) inhibited import strongly (Fig. 1B). Inhibition of import occurred in a fashion similar to that observed for import of a model precursor protein (pSu9-DHFR) into the matrix.
FIG. 1.
Leu5p is imported into the mitochondrial inner membrane. (A) [35S]methionine-labeled Leu5p and the precursor of α-MPP were incubated with isolated wild-type mitochondria (50 μg/sample) in import buffer containing 1 mM ATP and 2 mM NADH for 10 min at 25°C (55). Samples were chilled on ice, and mitochondria were reisolated and resuspended in SoH buffer (0.6 M sorbitol, 20 mM HEPES-KOH [pH 7.4]). Proteinase K (PK) was added at the indicated concentrations in the presence or absence of 0.1% Triton X-100. After 15 min protease digestion was halted by the addition of 1 mM phenylmethylsulfonyl fluoride, and proteins were precipitated with trichloroacetic acid and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The radiolabeled proteins were visualized by fluorography. (B) Import of Leu5p requires a membrane potential. Mitochondria in import buffer were treated with the indicated concentrations of CCCP for 3 min at 25°C. [35S]methionine-labeled Leu5p and the precursor of pSu9-DHFR were added and samples were incubated for 20 min at 25°C. After treatment with 50 μg of proteinase K per ml, samples were analyzed as described for panel A. (C) Leu5p is protease sensitive after the opening of the outer membrane and requires Tim22p for import. Import of radioactive Leu5p and α-MPP precursors was performed as for panel A using mitochondria isolated from wild-type and tim22 mutant strains Tim22 (Gal10) (51). The mitochondria were reisolated and subjected to a swelling procedure (15) in the presence or absence of 50 μg of proteinase K (PK) per ml. Protease digestion was halted by the addition of 1 mM phenylmethylsulfonyl fluoride. Proteins were precipitated with trichloroacetic acid and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and fluorography. Radioactive proteins were quantitated by phosphorimager analysis. Abbreviations: p, i, and m, precursor, intermediate, and mature forms of imported proteins; St., standard containing 50% of input precursor protein.
To analyze the submitochondrial localization of imported Leu5p, wild-type mitochondria were subjected to hypotonic swelling after the import reaction, a procedure causing the rupture of the outer, but not the inner, membrane (15, 24). The majority of imported Leu5p (>85%) became accessible to protease after the opening of the outer membrane, suggesting that Leu5p was exposed to the intermembrane space (Fig. 1C, right). The imported protein was resistant to extraction by treatment with alkaline buffers, indicating integration into the membrane (not shown). Together, these data suggest that Leu5p was imported into the mitochondrial inner membrane in a ΔΨ-dependent fashion.
We next tested whether Leu5p follows the import pathway of mitochondrial carrier proteins by employing a yeast mutant in which Tim22p can be depleted by regulated gene expression. Import of Leu5p into Tim22p-depleted mitochondria (51) was reduced by 70% compared to wild-type organelles (Fig. 1C). In contrast, import of the α-subunit of matrix processing peptidase (α-MPP) occurred at wild-type efficiency. This protein uses the Tim17p/Tim23p complex for its import into the matrix. In summary, Leu5p requires Tim22p for its import into the mitochondrial inner membrane and thus appears to follow the carrier-specific protein import pathway.
Phenotypical consequences of the deletion of LEU5.
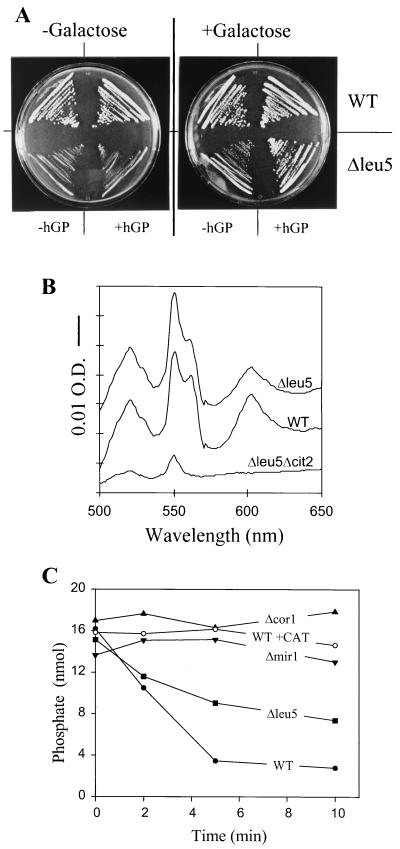
To initiate the functional investigation of Leu5p, the entire coding region of LEU5 was deleted using a single-step gene disruption procedure (1). The deletion of LEU5 was verified by PCR (data not shown). In comparison to wild-type cells, mutant cells lacking LEU5 (strain Δleu5) showed similar growth on rich media containing glucose (data not shown) but displayed strongly retarded growth on rich media containing glycerol (Fig. 2A, left part of left plate). On minimal media, Δleu5 cells did not exhibit an auxotrophy for leucine (19 and data not shown). Thus, Δleu5 cells display a leaky pet phenotype (63), suggesting that Leu5p performs a crucial but not essential function within mitochondria.
FIG. 2.
Leu5p-deficient cells are functional in oxidative phosphorylation. (A) Wild-type and Δleu5 cells were grown on YPG agar plates for 4 days at 30°C. The plate on the right contained an additional 0.2% galactose. Where indicated cells harbored the plasmid pYES2/hGP containing the cDNA of hGP. (B) Cytochrome spectra were recorded using isolated mitochondria from wild-type (WT) and Δleu5 cells which were grown overnight in YPD medium (30, 62). The bar represents an absorption difference of 0.01 (O.D.). (C) Membrane potential-driven formation of ATP. Mitochondria (10 μg) isolated from the indicated strains were incubated in buffer P (20 mM morpholinepropanesulfonic acid [MOPS]-KOH [pH 7.2], 0.25 M sucrose, 0.3 mM potassium phosphate, 5 mM MgCl2, 1 mg of fatty acid-free bovine serum albumin per ml, and 1 mM ADP) at 25°C. A membrane potential was generated by the addition of 2 mM NADH. The formation of ATP is accompanied by the decrease in free inorganic phosphate, which was measured by the malachite green assay (37, 43). One of the samples contained 50 nM carboxyatractyloside (CAT) to block the exchange of ADP and ATP between mitochondria and cytosol.
The reduced growth of Δleu5 cells was not caused by a defect in respiration. No significant differences in the cytochrome spectra of Δleu5 mitochondria were observed compared to wild-type organelles (Fig. 2B). In addition, Δleu5 mitochondria were active in oxidative phosphorylation as measured by the NADH-dependent incorporation of inorganic phosphate into ADP catalyzed by mitochondrial F1F0-ATP synthase (Fig. 2C) (43). The slight reduction in the rate of ATP production compared to wild-type cells cannot account for the severe impairment of growth on nonfermentable carbon sources. Organelles isolated from yeast strains defective in the phosphate carrier (Δmir1) or in complex III (Δcor1) were inactive in this assay. Furthermore, no incorporation of phosphate into ADP was observed when the ADP/ATP carrier was blocked by the addition of carboxyatractyloside (Fig. 2C) or when NADH was omitted (data not shown). These data suggest that the retarded growth of Δleu5 cells observed under nonfermentative conditions is due to the impairment of a process other than oxidative phosphorylation. Presumably, Leu5p, as a member of the carrier family, mediates the transport of a substrate required for proper function of an intramitochondrial biosynthetic process.
Leu5p is required for accumulation of CoA inside mitochondria.
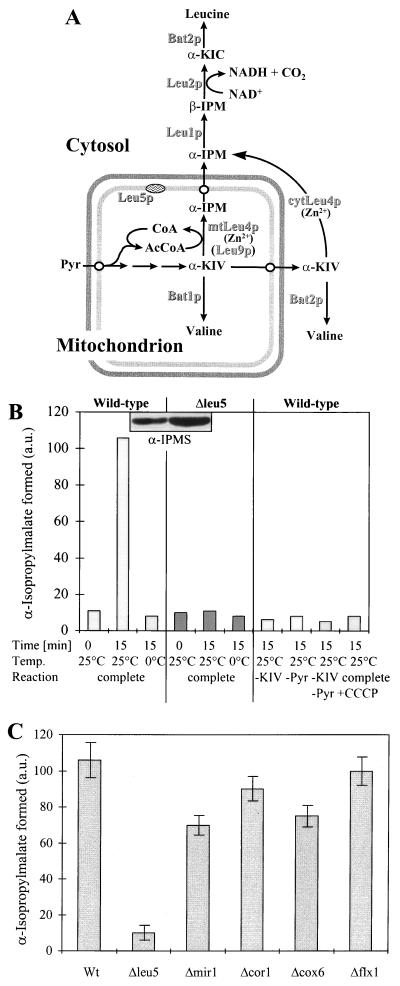
To identify the substrate of Leu5p, we took advantage of the phenotypical observations made for the inactivation of the LEU4 (coding for an IPMS) and LEU5 genes. The combined, but not the single, mutation of the two genes was reported to result in an auxotrophy for leucine (9, 19). In leu4 mutant cells, Chang and coworkers found 25% residual activity of IPMS (9), suggesting the existence of a second enzyme with IPMS activity. Initially, Leu5p was suspected to represent the second IPMS enzyme. However, to date this activity is attributed to a protein termed Leu9p (encoded by the gene YOR108w), which exhibits high sequence similarity to Leu4p (W. Pelzer, unpublished data). Leu9p is located in the mitochondrial matrix, in contrast to Leu4p, which residues in both mitochondria and the cytosol (see Fig. 3A) (3). Thus, in Δleu4 mutant cells, α-IPM is synthesized exclusively in mitochondria.
FIG. 3.
Intact Δleu5 mitochondria are defective in the synthesis of α-IPM. (A) The final steps of leucine biosynthesis in yeast. The locations of the various enzymes are indicated. Pyr, pyruvate; α-KIV, α-ketoisovalerate; β-IPM, β-isopropylmalate; α-KIC, α-ketoisocaproate. mtLeu4p and cytLeu4p are the mitochondrial and cytosolic forms of Leu4p, respectively. (B) Mitochondria were isolated from wild-type and Δleu5 cells grown in YPGal medium. They were incubated in SoH buffer for 15 min at 0 or 25°C with 2 mM α-ketoisovalerate (KIV) and 2 mM pyruvate unless indicated otherwise. One sample also contained 20 μM CCCP to deplete the membrane potential. α-IPM formed was converted to its umbelliferone derivative, which then was determined by fluorimetry. a.u., arbitrary units. In the insert, an immunostaining analysis of mitochondria isolated from wild-type and Δleu5 cells is shown using an antibody raised against purified IPMS (27). Loading of an equivalent amount of protein in both lanes was confirmed by staining with Ponceau S dye before immunodecoration. (C) Mitochondria isolated from the indicated strains were incubated with 2 mM α-ketoisovalerate and 2 mM pyruvate at 25°C as described for panel B and α-IPM was determined. The bars show the standard error of three independent experiments.
What might then lead to the leucine auxotrophy upon inactivation of LEU5 in a leu4 mutant background? We reasoned that Leu5p, as a mitochondrial carrier, may transport a compound related to the reaction catalyzed by mitochondrial IPMS (Fig. 3A). Potential candidates include α-ketoisovalerate, CoA as the precursor of the substrate acetyl-CoA, Zn2+ as a cofactor of Leu4p/Leu9p, or α-IPM as the product of the IPMS reaction (34). α-Ketoisovalerate is unlikely to be the substrate of Leu5p, since this metabolite is synthesized within the mitochondria (Fig. 3A) and Δleu5 cells show no auxotrophy for valine (data not shown).
We first investigated the IPMS reaction in intact isolated mitochondria using a fluorimetric assay which detects the fluorescent umbelliferone derivative of the product α-IPM. Wild-type mitochondria gave rise to the time- and temperature-dependent formation of α-IPM (Fig. 3B, left). Synthesis was fully dependent upon the addition of both α-ketoisovalerate and pyruvate (Fig. 3B, right). The latter is converted by pyruvate dehydrogenase to acetyl-CoA, which is unable to pass across the mitochondrial inner membrane (26). Generation of α-IPM was inhibited by the addition of the uncoupler CCCP, which presumably interferes with the potential-dependent uptake of α-ketoisovalerate and/or pyruvate into mitochondria. Thus, our assay system faithfully measured the synthesis of α-IPM in intact wild-type mitochondria. In contrast, organelles isolated from Δleu5 cells did not synthesize α-IPM (Fig. 3B, middle). This was surprising, as these mitochondria contained two- to threefold higher levels of IPMS protein as determined by immunostaining (Fig. 3B, insert). Pyruvate dehydrogenase was present at wild-type activities in detergent-lysed mitochondria of Δleu5 cells (data not shown). The defect in the synthesis of α-IPM was specific for Δleu5 cells, as wild-type levels of α-IPM were generated by mitochondria purified from cells defective in the phosphate carrier (Δmir1) and the FAD carrier (Δflx1) and cells defective in either complex III (Δcor1) or complex IV (Δcox6) (Fig. 3C). In conclusion, the IPMS enzyme present in intact Δleu5 mitochondria is incapable of synthesizing α-IPM.
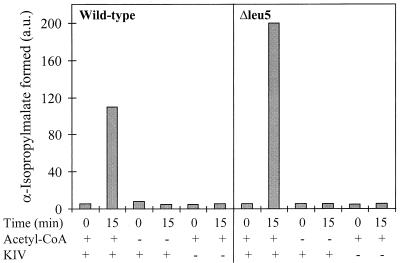
An entirely different result for the IPMS enzyme activity was obtained after detergent lysis of Δleu5 mitochondria. An almost twofold increase compared to wild-type organelles was detected (Fig. 4). Similar data were obtained using the CoA release assay previously used to measure the IPMS activity in cellular extracts (34 and data not shown). The formation of α-IPM was time and temperature dependent and required the addition of the substrates of IPMS, α-ketoisovalerate, and acetyl-CoA (Fig. 4 and data not shown). Addition of Zn2+ ions was not required to stimulate α-IPM synthesis, excluding the possibility that a lack of Zn2+ in the mitochondrial matrix was the reason for the defective IPMS reaction in intact Δleu5 mitochondria. In support of this conclusion, the Zn2+-dependent mitochondrial alcohol dehydrogenase exhibited wild-type activity in Δleu5 organelles (data not shown). In summary, Δleu5 mitochondria contain active IPMS. However, the enzyme activity is only detectable after the opening of the inner membrane.
FIG. 4.
Detergent extracts of Δleu5 mitochondria contain wild-type levels of α-IPMS activity. Mitochondria were isolated from wild-type and Δleu5 cells grown in YPGal medium. They were lysed in SoH buffer containing 0.05% Triton X-100 and centrifuged for 10 min at 12,000 × g. The clarified extract was incubated for 0 or 15 min with 2 mM α-ketoisovalerate (KIV) and 1 mM acetyl-CoA as indicated. α-IPM was determined as described for Fig. 3B. a.u., arbitrary units.
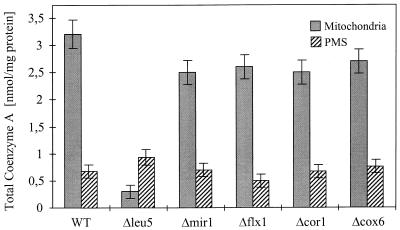
The results presented above excluded α-ketoisovalerate, Zn2+, and α-IPM as potential substrates of Leu5p. This rendered it likely that the defect in IPMS activity in intact Δleu5 mitochondria was due to a deficiency of CoA in the matrix. We therefore measured the content of CoA present in isolated mitochondria or in PMS of various yeast cells. In mitochondria and PMS of wild-type cells, we found 3.2 and 0.7 nmol CoA/mg of protein, respectively (Fig. 5). Using an estimated protein concentration of 300 mg/ml for both mitochondria and the cytoplasm, these numbers correspond to 0.96 and 0.21 mM CoA, respectively. Similar ratios were reported previously (46). Strikingly, mitochondria from Δleu5 cells contained about 15-fold lower levels of CoA compared to wild-type organelles, whereas a slight increase was found for PMS. No major variations in the CoA levels relative to wild-type controls were detected for either mitochondria or PMS derived from cells defective in the phosphate carrier (Δmir1), the FAD carrier (Δflx1), or the pet cells Δcor1 and Δcox6 (Fig. 5). Thus, Leu5p appears to be specifically required for the accumulation of CoA in the mitochondrial matrix.
FIG. 5.
Mitochondria isolated from Δleu5 cells contain strongly decreased levels of CoA. Mitochondria and PMS were prepared from the indicated yeast strains. The amount of CoA and its acylated derivatives was measured by a three-step enzyme assay (40). The bars show the standard error of three independent experiments. WT, wild-type strain.
LEU5 genetically interacts with CIT2 encoding the peroxisomal citrate synthase.
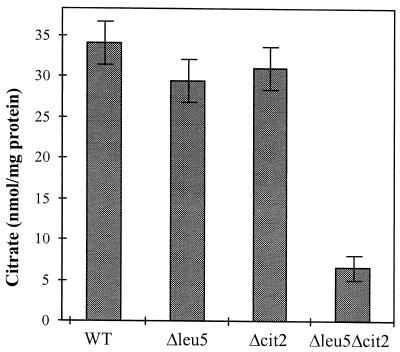
We further sought to obtain in vivo evidence for a deficiency of mitochondrial CoA in Δleu5 cells. First, we expected a low activity of mitochondrial citrate synthase (Cit1p) in these cells. Mitochondria lacking Cit1p activity can satisfy their needs for citrate by importing it from the cytosolic pool of citrate that is produced by the peroxisomal citrate synthase (Cit2p) (35). A Δcit1Δcit2 double mutant, on the other hand, does not grow on nonfermentable carbon sources due to low citrate levels, and the cells exhibit an auxotrophy for glutamate, a classical phenotype of citrate synthase deficiency (29). Therefore, deletion of the CIT2 gene in the Δleu5 background was anticipated to evoke a phenotype similar to that observed for Δcit1Δcit2 cells. Deletion of CIT2 alone is not associated with any phenotypical consequences (29, 66). Indeed, Δleu5Δcit2 cells contained fivefold lower levels of citrate compared to wild-type cells or mutant cells with single deletions of either LEU5 or CIT2 genes (Fig. 6). Further, the double mutant cells did not grow on rich media containing glycerol, i.e., they displayed a strict pet phenotype, unlike Δleu5 cells (data not shown). Finally, Δleu5Δcit2 cells showed a striking deficiency in cytochromes (Fig. 2B) and an auxotrophy for glutamate (not shown).
FIG. 6.
Low citrate levels in a cell lacking both mitochondrial Leu5p and peroxisomal Cit2p. Cell extracts were prepared from the indicated yeast cells and the content of citrate was measured. The bars show the standard error of three independent experiments. WT, wild-type strain.
All of these phenotypical observations made for Δleu5Δcit2 cells conform well to the idea that Leu5p facilitates the accumulation of CoA in the mitochondrial matrix. The deficiency of mitochondrial CoA in addition to the lack of peroxisomal Cit2p results in a reduced cellular content of citrate (Fig. 6). As a consequence, synthesis of α-ketoglutarate (by the citric acid cycle) and its transamination product, glutamate, the precursor of C5 amino acids, is decreased. Further, production of succinyl-CoA, a substrate of the first step of heme biosynthesis, is hampered, explaining the defect in cytochromes in Δleu5Δcit2 cells. Hence, the surprising connection between mitochondrial Leu5p and peroxisomal Cit2p provides in vivo evidence for Leu5p performing a function in the accumulation of CoA in mitochondria.
The hGP functionally complements the defect of Leu5p.
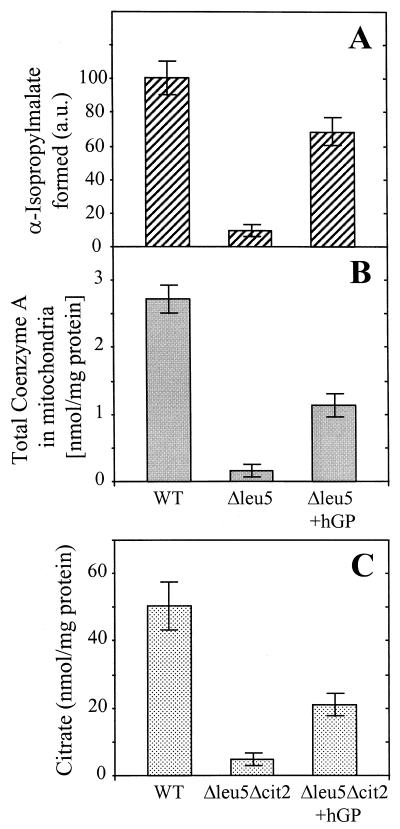
The significant homology between hGP and yeast Leu5p suggests a similar function of the two proteins. To test this idea, Δleu5 cells were transformed with a plasmid carrying the hGP gene under the control of a galactose-inducible promoter. The resulting Δleu5/hGP cells were analyzed for growth on nonfermentable carbon sources. When expression of hGP was induced by the addition of galactose, Δleu5/hGP cells grew at wild-type rates (Fig. 2A, right plate). Δleu5 cells grew much slower under these conditions, suggesting that hGP could replace the function of Leu5p. In keeping with this interpretation, hardly any differences to wild-type cells were obtained for the activity of IPMS enzymes in intact mitochondria (Fig. 7A). Further, the levels of mitochondrial CoA were almost fully restored in Δleu5/hGP cells (sixfold increase compared to Δleu5 cells) (Fig. 7B). Finally, expression of hGP in Δleu5Δcit2 cells partially restored the cellular citrate concentration to the levels of wild-type cells (Fig. 7C). Partial replacement of the function of yeast proteins has been observed for a number of other mammalian carriers. Our data demonstrate that the human protein can at least partially replace Leu5p function and suggest a role of hGP in the accumulation of CoA in mitochondria.
FIG. 7.
hGP can functionally replace yeast Leu5p. The formation of α-IPM (A), the total content of CoA in mitochondria (B), and the cellular amount of citrate (C) in wild-type (WT) and Δleu5Δcit2 mutant cells with and without expressed hGP was measured as described for Fig. 3B, 5, and 6. The bars show the standard error of at least three independent experiments.
DISCUSSION
In this study, we used biochemical and genetic approaches to identify a function of the mitochondrial carriers Leu5p and hGP in the accumulation of CoA in the matrix. First, the CoA levels in Δleu5 mitochondria were 15-fold lower than those in wild-type or various mutant organelles. No corresponding decrease in the CoA concentration was detectable in the PMS of Δleu5 cells (see below). Second, the CoA-dependent enzyme IPMS was largely defective in intact Δleu5 mitochondria. This was solely due to a shortage in the substrate acetyl-CoA, since the enzyme was fully active when detergent extracts of Δleu5 mitochondria were supplied with acetyl-CoA. Third, the surprising interference between mitochondrial Leu5p and peroxisomal citrate synthase Cit2p is best explained on the basis of a role of Leu5p in CoA accumulation in the mitochondrial matrix. The decreased CoA levels in Δleu5 mitochondria lead to impaired synthesis of citrate by mitochondrial Cit1p. In Δleu5Δcit2 cells, citrate can no longer be supplied to mitochondria by peroxisomal Cit2p-catalyzed synthesis. This results in a mutant cell that closely mimics the double deletion of the CIT1 and CIT2 genes in that the cellular citrate concentration is largely decreased. As a further consequence, the flux through the citric acid cycle in Δleu5Δcit2 cells is low, and cells are impaired in generating glutamate from α-ketoglutarate. This defect may be further enhanced by the low activity of the other CoA-dependent enzymes of the citric acid cycle. In passing, our data nicely support the functional communication between mitochondria and peroxisomes by retrograde regulation (see, e.g., references 10 and 36). A final point supporting the role of Leu5p in mitochondrial CoA accumulation is the dramatic defect in heme-containing proteins in Δleu5Δcit2 cells. The heme deficiency is a result of the lowered activity of δ-aminolevulinate synthase, the first enzyme of heme biosynthesis using succinyl-CoA as a substrate. The impairment of heme biosynthesis in Δleu5Δcit2 cells may be the major reason why these cells do not grow on the nonfermentable carbon source glycerol.
One may wonder why Δleu5 cells, as opposed to Δleu5Δcit2 cells, are not auxotrophic for glutamate or defective in heme-containing proteins. First, the Km values for acetyl-CoA are 10-fold higher for IPMS than for citrate synthase, rendering IPMS most sensitive to reduced CoA concentrations (38, 48). A second obvious reason may be the relatively high amounts of leucine required for cellular protein biosynthesis.
To our knowledge, Leu5p and hGP are the first proteins for which a function in the cellular distribution of CoA has been demonstrated. The cellular compartmentation of CoA is an important but still poorly understood aspect of metabolism (47). In wild-type cells, the highest CoA concentrations are found in mitochondria and the peroxisomes (46, 65), where the coenzyme participates in numerous pathways, such as the citric acid cycle, the biosynthesis of heme, the β-oxidation of fatty acids, and the glyoxylate cycle. About 5- to 10-fold lower levels of CoA as compared to mitochondria are found in the cytosol of wild-type cells (Fig. 5) (46). Strikingly, in a mutant lacking Leu5p the relative concentrations of CoA in mitochondria and the cytosol were reversed. These data suggest that either CoA or a precursor of CoA is the substrate of the Leu5p carrier protein. A decision between these possibilities cannot be made presently, since little is known about the compartmentation and the molecular identity of the five enzymes participating in CoA biosynthesis. Only for the first enzymatic step has the gene been identified. Pantothenate kinase (gene YDR531w in S. cerevisiae [6]) catalyzes the committed step of biosynthesis and is tightly regulated in its activity by acetyl-CoA (47). While pantothenate kinase is known to reside in the cytosol, the location of the enzymes completing biosynthesis of CoA has not been determined with certainty. Dephospho-CoA kinase mediating the final reaction of CoA biosynthesis has been reported to be associated with mitochondria but is believed to be located outside the inner membrane (53, 58). These findings render it likely that CoA is synthesized externally to the mitochondrial inner membrane and must be transported into the matrix space. In support of this view, an in vitro transport system for CoA uptake into isolated mitochondria of rat liver has been reported (59). Mitochondrial import of CoA required an electrical gradient (56), but the transporter has not been identified (57). Using isolated yeast mitochondria, we were unable to apply these findings for setting up a transport assay for CoA. This was mainly due to the fact that radiolabeled CoA was rapidly metabolized when added to isolated yeast mitochondria, presumably by cleavage of CoA to 4-phosphopantetheine, a reaction catalyzed by CoA hydrolase (5). The data presented here fit nicely with the cytosolic synthesis of CoA, as Δleu5 cells were capable of producing normal levels of cytosolic CoA. On that basis, the most likely substrate of Leu5p is CoA. Clearly, definitive identification of the substrate of Leu5p will have to await the purification of Leu5p and the reconstitution of the transport reaction, as has been recently achieved for several mitochondrial carrier proteins (28, 45).
Mitochondria isolated from Δleu5 cells contain low yet significant amounts of CoA. Thus, an alternative pathway must exist to supply the organelles with CoA. Most likely, another member of the mitochondrial carrier family takes over the function of Leu5p in accumulating CoA in the mitochondrial matrix, even though it does so at low efficiency. The best candidates for such a supplementary task are the three ADP/ATP carrier proteins (AAC) of yeast. In support of this suggestion, Leu5p shares highest sequence similarity to the AAC subgroup of the yeast carriers (44, 45). Further, based on their substrate specificity, AAC proteins seem to be optimally suited for the transport of the adenine nucleotide CoA across the mitochondrial inner membrane.
Our study answers the long-standing question of the connection between the IPMS Leu4p and the membrane protein Leu5p (9, 18, 19). Only Δleu4Δleu5 double mutant cells, but not the single mutants, exhibit an auxotrophy for leucine. Now, this finding can be easily understood on the basis of the low content of mitochondrial CoA upon deletion of LEU5. In the absence of Leu4p, α-IPM is synthesized by Leu9p (Fig. 3A), an isoenzyme of Leu4p that exhibits 83% amino acid identity and is localized to the mitochondrial matrix (W. Pelzer, unpublished). Since Leu4p is located in both the matrix and the cytosol (3), in cells lacking LEU5, α-IPM can be generated by cytosolic Leu4p. In the Δleu4Δleu5 double mutant, however, synthesis of α-IPM can only be mediated by mitochondrial Leu9p, which functions poorly due to the low CoA concentration.
Graves' disease is a multifactorial autoimmune disorder in which hyperthyroidism is caused by the production of autoantibodies against the thyrotropin receptor and other thyroid proteins (reviewed in references 4 and 39). A cDNA for hGP has been identified by expression cloning in an immunoscreen using antisera from patients with active Graves' disease (69). The similarity of hGP to mitochondrial carrier proteins has been noted earlier, but a function has not been assigned yet. As shown here, hGP can replace the yeast carrier Leu5p, demonstrating that both proteins are functional orthologues required for the accumulation of CoA in the mitochondrial matrix. We were unable to detect hGP after functional expression in yeast by immunostaining with antisera derived from several patients with hyperthyroidism (data not shown). These findings indicate that hGP is not a major autoantigen of Graves' disease. Further, our insights into the function of hGP in CoA transport suggest no direct involvement of the mitochondrial carrier in this disorder.
The identification of the function of Leu5p and hGP in the accumulation of CoA in the mitochondrial matrix is a seminal step in the understanding of the mechanisms underlying proper subcellular distribution of CoA. For a comprehensive knowledge of CoA metabolism our studies will have to be followed up by a search for components involved in the biosynthesis of CoA. Moreover, membrane transporters facilitating supply of, e.g, peroxisomes, with this important cofactor will have to be identified.
ACKNOWLEDGMENTS
The expert technical assistance of B. Niggemeyer, M. Dienst, and M. Weidgans is gratefully acknowledged. We are indebted to G. Kohlhaw for his invaluable advice throughout this study. We thank G. Kohlhaw, W. Neupert, N. Pfanner, P. A. Srere, and A. Tzagoloff for kindly providing yeast strains and antisera, M. Grussendorf for antisera of patients with Graves' disease, and M. Brunner and coworkers for providing mitochondria depleted in Tim22p.
Our work was supported by grants of the Sonderforschungsbereich 286 of the Deutsche Forschungsgemeinschaft, the Volkswagen-Stiftung, the Fonds der Chemischen Industrie, and the Hungarian Funds OKTA. H.K. acknowledges a fellowship from Deutscher Akademischer Auslandsdienst DAAD.
Footnotes
We wish to dedicate this publication to the memory of the late Paul A. Srere.
REFERENCES
- 1.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer M F, Hofmann S, Neupert W, Brunner M. Protein translocation into mitochondria: the role of the TIM complexes. Trends Cell Biol. 2000;10:25–31. doi: 10.1016/s0962-8924(99)01684-0. [DOI] [PubMed] [Google Scholar]
- 3.Beltzer J P, Morris S R, Kohlhaw G B. Yeast LEU4 encodes mitochondrial and nonmitochondrial forms of alpha-isopropylmalate synthase. J Biol Chem. 1988;263:368–374. [PubMed] [Google Scholar]
- 4.Brix T H, Kyvik K O, Hegedus L. What is the evidence of genetic factors in the etiology of Graves' disease? A brief review. Thyroid. 1998;8:727–734. doi: 10.1089/thy.1998.8.727. [DOI] [PubMed] [Google Scholar]
- 5.Bucovaz E T, MacLeod R M, Morrison J C, Whybrew W D. The coenzyme A-synthesizing protein complex and its proposed role in CoA biosynthesis in bakers' yeast. Biochimie. 1997;79:787–798. doi: 10.1016/s0300-9084(97)86938-6. [DOI] [PubMed] [Google Scholar]
- 6.Calder R B, Williams R S B, Ramaswamy G, Rock C O, Campbell E, Unkles S E, Kinghorn J R, Jackowski S. Cloning and characterisation of a eukaryotic pantothenate kinase gene (panK) from Aspergillus nidulans. J Biol Chem. 1999;274:2014–2020. doi: 10.1074/jbc.274.4.2014. [DOI] [PubMed] [Google Scholar]
- 7.Calvo J M, Bartholomew J C, Stieglitz B I. Fluorometric assay of enzymatic reactions involving acetyl Coenzyme A in aldol condensations. Anal Biochem. 1969;28:164–181. doi: 10.1016/0003-2697(69)90168-7. [DOI] [PubMed] [Google Scholar]
- 8.Camacho J A, Obie C, Biery B, Goodman B K, Hu C, Almashanu S, Steel G, Casey R, Lambert M, Mitchell G A, Valle D. Hyperonithinaemia-hyperammonaemia-homocitrullinuria syndrome is caused by mutations in a gene encoding a mitochondrial ornithine transporter. Nat Genet. 1999;22:151–158. doi: 10.1038/9658. [DOI] [PubMed] [Google Scholar]
- 9.Chang L F, Cunningham T S, Gatzek P R, Chen W J, Kohlhaw G B. Cloning and characterization of yeast LEU4, one of two genes responsible for alpha-isopropylmalate synthesis. Genetics. 1984;108:91–106. doi: 10.1093/genetics/108.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chelstowska A, Butow R A. RTG genes in yeast that function in communication between mitochondria and the nucleus are also required for expression of genes encoding peroxisomal proteins. J Biol Chem. 1995;270:18141–18146. doi: 10.1074/jbc.270.30.18141. [DOI] [PubMed] [Google Scholar]
- 11.Crivellone M D, Wu M A, Tzagoloff A. Assembly of the mitochondrial membrane system. Analysis of structural mutants of the yeast coenzyme QH2-cytochrome c reductase complex. J Biol Chem. 1988;263:14323–14333. [PubMed] [Google Scholar]
- 12.Daum G, Böhni P C, Schatz G. Import of proteins into mitochondria: cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- 13.De Winde J H, Grivell L A. Global regulation of mitochondrial biogenesis in Saccharomyces cerevisiae. Prog Nucleic Acid Res Mol Biol. 1993;46:51–91. doi: 10.1016/s0079-6603(08)61018-1. [DOI] [PubMed] [Google Scholar]
- 14.Dickinson J R, Schweizer M. The metabolism and molecular physiology of Saccharomyces cerevisiae. London, United Kingdom: Taylor and Francis Ltd.; 1999. [Google Scholar]
- 15.Diekert, K., A. I. P. M. deKroon, G. Kispal, and R. Lill. Isolation and sub-fractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol., in press. [DOI] [PubMed]
- 16.Diekert K, Kispal G, Guiard B, Lill R. An internal targeting signal directing proteins into the mitochondrial intermembrane space. Proc Natl Acad Sci USA. 1999;96:11746–11751. doi: 10.1073/pnas.96.21.11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dietmeier K, Zara V, Palmisano A, Palmieri F, Voos W, Schlossmann J, Moczko M, Kispal G, Pfanner N. Targeting and translocation of the phosphate carrier/p32 to the inner membrane of yeast mitochondria. J Biol Chem. 1993;268:25958–25964. [PubMed] [Google Scholar]
- 18.Drain P, Schimmel P. Multiple new genes that determine activity for the first step of leucine biosynthesis in Saccharomyces cerevisiae. Genetics. 1988;119:13–20. doi: 10.1093/genetics/119.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drain P, Schimmel P. Yeast LEU5 is a PET-like gene that is not essential for leucine biosynthesis. Mol Gen Genet. 1986;204:397–403. doi: 10.1007/BF00331015. [DOI] [PubMed] [Google Scholar]
- 20.El Moualij B, Duyckaerts C, Lamotte-Brasseur J, Sluse F E. Phylogenetic classification of the mitochondrial carrier family of Saccharomyces cerevisiae. Yeast. 1997;13:573–581. doi: 10.1002/(SICI)1097-0061(199705)13:6<573::AID-YEA107>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 21.Fiermonte G, Runswick M J, Walker J E, Palmieri F. Sequence and pattern of expression of a bovine homologue of a human mitochondrial transport protein associated with Grave's disease. DNA Sequence. 1992;3:71–78. doi: 10.3109/10425179209033999. [DOI] [PubMed] [Google Scholar]
- 22.Fiore C, Trezeguet V, Le Saux A, Roux P, Schwimmer C, Dianoux A C, Noel F, Lauquin G J, Brandolin G, Vignais P V. The mitochondrial ADP/ATP carrier: structural, physiological and pathological aspects. Biochimie. 1998;80:137–150. doi: 10.1016/s0300-9084(98)80020-5. [DOI] [PubMed] [Google Scholar]
- 23.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glick B S, Brandt A, Cunningham K, Müller S, Hallberg R L, Schatz G. Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell. 1992;69:809–822. doi: 10.1016/0092-8674(92)90292-k. [DOI] [PubMed] [Google Scholar]
- 25.Graham B H, Waymire K G, Cottrell B, Trounce I A, MacGregor G R, Wallace D C. A mouse model for mitochondrial myopathy and cardiomyopathy resulting from a deficiency in the heart/muscle isoform of the adenine nucleotide translocator. Nat Genet. 1997;16:226–234. doi: 10.1038/ng0797-226. [DOI] [PubMed] [Google Scholar]
- 26.Haddock B A, Yates D W, Garland P B. The localization of some coenzyme A-dependent enzymes in rat liver mitochondria. Biochem J. 1970;119:565–573. doi: 10.1042/bj1190565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hampsey D M, Kohlhaw G. Inactivation of yeast alpha-isopropylmalate synthase by CoA. Antagonism between CoA and adenylates and the mechanism of CoA inactivation. J Biol Chem. 1981;256:3791–3796. [PubMed] [Google Scholar]
- 28.Kaplan R S. High-level bacterial expression of mitochondrial transport proteins. J Bioenerg Biomembr. 1996;28:41–47. [PubMed] [Google Scholar]
- 29.Kim K S, Rosenkrantz M S, Guarente L. Saccharomyces cerevisiae contains two functional citrate synthase genes. Mol Cell Biol. 1986;6:1936–1942. doi: 10.1128/mcb.6.6.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kispal G, Csere P, Guiard B, Lill R. The ABC transporter Atm1p is required for mitochondrial iron homeostasis. FEBS Lett. 1997;418:346–350. doi: 10.1016/s0014-5793(97)01414-2. [DOI] [PubMed] [Google Scholar]
- 31.Klingenberg M, Nelson D. Structure-function relationships in the mitochondrial carrier family. In: Papa S, Tager J M, editors. Biochemistry of cell membranes. Basel, Switzerland: Birkhäuser; 1995. pp. 191–219. [Google Scholar]
- 32.Koehler C M, Merchant S, Schatz G. How membrane proteins travel across the mitochondrial intermembrane space. Trends Biochem Sci. 1999;24:428–432. doi: 10.1016/s0968-0004(99)01462-0. [DOI] [PubMed] [Google Scholar]
- 33.Koerner T J, Homison G, Tzagoloff A. Nuclear mutants of Saccharomyces cerevisiae with altered subunits 4, 5, and 6 of cytochrome oxidase. J Biol Chem. 1985;260:5871–5874. [PubMed] [Google Scholar]
- 34.Kohlhaw G B. Alpha-isopropylmalate synthase from yeast. Methods Enzymol. 1988;166:414–423. doi: 10.1016/s0076-6879(88)66054-x. [DOI] [PubMed] [Google Scholar]
- 35.Lewin A S, Hines V, Small G M. Citrate synthase encoded by the CIT2 gene of Saccharomyces cerevisiae is peroxisomal. Mol Cell Biol. 1990;10:1399–1405. doi: 10.1128/mcb.10.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao X, Butow R A. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell. 1993;72:61–71. doi: 10.1016/0092-8674(93)90050-z. [DOI] [PubMed] [Google Scholar]
- 37.Lill R, Dowhan W, Wickner W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell. 1990;60:271–280. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- 38.Lindbladh C, Rault M, Hagglund C, Small W C, Mosbach K, Bulow L, Evans C, Srere P A. Preparation and kinetic characterization of a fusion protein of yeast mitochondrial citrate synthase and malate dehydrogenase. Biochemistry. 1994;33:11692–11698. doi: 10.1021/bi00205a004. [DOI] [PubMed] [Google Scholar]
- 39.McIver B, Morris J C. The pathogenesis of Graves' disease. Endocrinol Metab Clin N Am. 1998;27:73–89. doi: 10.1016/s0889-8529(05)70299-1. [DOI] [PubMed] [Google Scholar]
- 40.Michal G, Bergmeyer H U. Methods of enzymatic analysis. New York, N.Y: Academic Press; 1974. pp. 1975–1981. [Google Scholar]
- 41.Moellering H, Gruber W. Determination of citrate with citrate lyase. Anal Biochem. 1966;17:369–376. doi: 10.1016/0003-2697(66)90172-2. [DOI] [PubMed] [Google Scholar]
- 42.Muller V, Heidkamper D, Nelson D R, Klingenberg M. Mutagenesis of some positive and negative residues occurring in repeat triad residues in the ADP/ATP carrier from yeast. Biochemistry. 1997;36:16008–16018. doi: 10.1021/bi971867l. [DOI] [PubMed] [Google Scholar]
- 43.Nargang F E, Künkele K-P, Mayer A, Ritzel R G, Neupert W, Lill R. “Sheltered disruption” of Neurospora crassa MOM22, an essential component of the mitochondrial protein import complex. EMBO J. 1995;14:1099–1108. doi: 10.1002/j.1460-2075.1995.tb07093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelson D R, Felix C M, Swanson J M. Highly conserved charge-pair networks in the mitochondrial carrier family. J Mol Biol. 1998;277:285–308. doi: 10.1006/jmbi.1997.1594. [DOI] [PubMed] [Google Scholar]
- 45.Palmieri L, Runswick M J, Fiermonte G, Walker J E, Palmieri F. Yeast mitochondrial carriers: bacterial expression, biochemical identification and metabolic significance. J Bioenerg Biomembr. 2000;32:67–77. doi: 10.1023/a:1005564429242. [DOI] [PubMed] [Google Scholar]
- 46.Robishaw J D, Berkich D, Neely J R. Rate-limiting step and control of coenzyme A synthesis in cardiac muscle. J Biol Chem. 1982;257:10967–10972. [PubMed] [Google Scholar]
- 47.Rock C O, Calder R B, Karim M A, Jackowski S. Pantothenate kinase regulation of the intracellular concentration of coenzyme A. J Biol Chem. 2000;275:1377–1383. doi: 10.1074/jbc.275.2.1377. [DOI] [PubMed] [Google Scholar]
- 48.Roeder P R, Kohlhaw G B. Alpha-isopropylmalate synthase from yeast. A zinc metalloenzyme. Biochim Biophys Acta. 1980;613:482–487. doi: 10.1016/0005-2744(80)90103-5. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 50.Schroers A, Burkovski A, Wohlrab H, Kramer R. The phosphate carrier from yeast mitochondria. Dimerization is a prerequisite for function. J Biol Chem. 1998;273:14269–14276. doi: 10.1074/jbc.273.23.14269. [DOI] [PubMed] [Google Scholar]
- 51.Sirrenberg C, Bauer M F, Guiard B, Neupert W, Brunner M. Import of carrier proteins into the mitochondrial inner membrane mediated by Tim22. Nature. 1996;384:582–585. doi: 10.1038/384582a0. [DOI] [PubMed] [Google Scholar]
- 52.Sirrenberg C, Endres M, Folsch H, Stuart R A, Neupert W, Brunner M. Carrier protein import into mitochondria mediated by the intermembrane proteins Tim10/Mrs11 and Tim12/Mrs5. Nature. 1998;391:912–915. doi: 10.1038/36136. [DOI] [PubMed] [Google Scholar]
- 53.Skrede S, Halvorson O. Mitochondrial pantetheinephosphate adenylyltransferase and dephospho-CoA kinase. Eur J Biochem. 1983;131:57–63. doi: 10.1111/j.1432-1033.1983.tb07231.x. [DOI] [PubMed] [Google Scholar]
- 54.Srere P A. Enzyme concentrations in tissues. Science. 1967;158:936–937. doi: 10.1126/science.158.3803.936. [DOI] [PubMed] [Google Scholar]
- 55.Steiner H, Zollner A, Haid A, Neupert W, Lill R. Biogenesis of mitochondrial heme lyases in yeast. Import and folding in the intermembrane space. J Biol Chem. 1995;270:22842–22849. doi: 10.1074/jbc.270.39.22842. [DOI] [PubMed] [Google Scholar]
- 56.Tahiliani A G. Dependence of mitochondrial coenzyme A uptake on the membrane electrical gradient. J Biol Chem. 1989;264:18426–18432. [PubMed] [Google Scholar]
- 57.Tahiliani A G, Keene T, Kaplan R S. Characterization of the inhibitor sensitivity of the coenzyme A transport system in isolated rat heart mitochondria. J Bioenerg Biomembr. 1992;24:635–640. doi: 10.1007/BF00762356. [DOI] [PubMed] [Google Scholar]
- 58.Tahiliani A G, Neely J R. Mitochondrial synthesis of coenzyme A is on the external surface. J Mol Cell Cardiol. 1987;19:1161–1167. doi: 10.1016/s0022-2828(87)80526-6. [DOI] [PubMed] [Google Scholar]
- 59.Tahiliani A G, Neely J R. A transport system for coenzyme A in isolated rat heart mitochondria. J Biol Chem. 1987;262:11607–11610. [PubMed] [Google Scholar]
- 60.Truscott K N, Pfanner N. Import of carrier proteins into mitochondria. Biol Chem. 1999;380:1151–1156. doi: 10.1515/BC.1999.146. [DOI] [PubMed] [Google Scholar]
- 61.Tzagoloff A. Mitochondria. New York, N.Y: Plenum Press; 1982. [Google Scholar]
- 62.Tzagoloff A, Akai A, Needleman R B. Assembly of the mitochondrial membrane system. Characterization of nuclear mutants of Saccharomyces cerevisiae with defects in mitochondrial ATPase and respiratory enzymes. J Biol Chem. 1975;250:8228–8235. [PubMed] [Google Scholar]
- 63.Tzagoloff A, Dieckmann C L. PET genes of Saccharomyces cerevisiae. Microbiol Rev. 1990;54:211–225. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tzagoloff A, Jang J, Glerum D M, Wu M. FLX1 codes for a carrier protein involved in maintaining a proper balance of flavin nucleotides in yeast mitochondria. J Biol Chem. 1996;271:7392–7397. doi: 10.1074/jbc.271.13.7392. [DOI] [PubMed] [Google Scholar]
- 65.Van Broekhoven A, Peeters M C, Debeer L J, Mannaerts G P. Subcellular distribution of coenzyme A: evidence for a separate coenzyme A pool in peroxisomes. Biochem Biophys Res Commun. 1981;100:305–312. doi: 10.1016/s0006-291x(81)80097-6. [DOI] [PubMed] [Google Scholar]
- 66.van Roermund C W, Elgersma Y, Singh N, Wanders R J, Tabak H F. The membrane of peroxisomes in Saccharomyces cerevisiae is impermeable to NAD(H) and acetyl-CoA under in vivo conditions. EMBO J. 1995;14:3480–3486. doi: 10.1002/j.1460-2075.1995.tb07354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walker J E, Runswick M J. The mitochondrial transport protein superfamily. J Bioenerg Biomembr. 1993;25:435–446. doi: 10.1007/BF01108401. [DOI] [PubMed] [Google Scholar]
- 68.Woonter M, Jaehning J A. Accurate initiation by RNA polymerase II in whole cell extracts from Saccharomyces cerevisiae. J Biol Chem. 1990;265:8979–8982. [PubMed] [Google Scholar]
- 69.Zarrilli R, Oates E L, McBride O W, Lerman M I, Chan J Y, Santisteban P, Ursini M V, Notkins A L, Kohn L D. Sequence and chromosomal assignment of a novel cDNA identified by immunoscreening of a thyroid expression library: similarity to a family of mitochondrial solute carrier proteins. Mol Endocrinol. 1989;3:1498–1505. doi: 10.1210/mend-3-9-1498. [DOI] [PubMed] [Google Scholar]