Abstract
Escherichia coli (E. coli) bloodstream infections (BSIs) are among the most predominant causes of death in infants and children worldwide. NDM-5 (New Delhi Metallo-lactamase-5) is responsible for one of the main mechanisms of carbapenem resistance in E. coli. To analyze the phenotypic and genomic characteristics of NDM-5-producing E. coli from bloodstream infections (BSIs), a total of 114 E. coli strains was collected from a children’s hospital in Jiangsu province, China. Eight blaNDM-5-carrying E. coli strains were identified which were all carbapenem-resistant and carried diverse antimicrobial resistance genes apart from blaNDM-5. They belonged to six distinct sequence types (STs) and serotypes including one each for ST38/O7:H8, ST58/O?:H37, ST131/O25:H4, ST156/O11:H25 and ST361/O9:H30 and three strains are originating from a single clone belonging to ST410/O?:H9. Apart from blaNDM-5, the E. coli strains isolated from BSIs also carried other β-lactamase genes, including blaCMY-2 (n = 4), blaCTX-M-14 (n = 2), blaCTX-M-15 (n = 3), blaCTX-M-65 (n = 1), blaOXA-1 (n = 4) and blaTEM-1B (n = 5). The blaNDM-5 genes were located on three different types of plasmids, which were IncFII/I1 (n = 1), IncX3 (n = 4) and IncFIA/FIB/FII/Q1 (n = 3). The former two types were conjugatively transferable at frequencies of 10−3 and 10−6, respectively. The dissemination of NDM-producing strains, which exhibit resistance to the last-line antibiotics, carbapenems, may increase the muti-antimicrobial resistance burden among E. coli BSIs and further threaten public health.
Keywords: NDM-5, carbapenemase, Escherichia coli, multidrug resistance, bloodstream infections, whole-genome sequencing
1. Introduction
Bloodstream infections (BSIs) are considered among the most serious nosocomial pathologies with limited choices of effective therapeutic options. BSIs are frequently associated with high morbidity and mortality in hospitalized patients, which increases treatment costs, patients’ burden and diagnostic uncertainties [1]. According to the published data, 48.9 million incident cases have taken place worldwide, and 11.0 million deaths were associated with BSIs, which represented 19.7% of all global deaths [2]. Of note, BSIs remain a major problem for neonates and young children [3,4], because the diagnosis of BSIs in children is relatively difficult as they could manifest as serious infections in other sites of the body. Neonatal BSIs are still among the major reasons of morbidity and mortality in spite of the remarkable progress in neonatal medicine [5]. It was estimated that 3 million neonates were affected by BSIs, with mortality between 11% and 19%, worldwide [6].
BSIs are increasing globally, and in addition, the causative organisms are becoming resistant to clinical front-line antibiotics, which increases in-hospital mortality and length of stay [7]. BSIs are identified by positive blood cultures in patients with systemic signs of infection such as acute organ dysfunction, respiratory failure and even septic shock [8]. It was reported that sepsis was responsible for 60–80% of lost lives per year in children [9]. With rapid evolution and the acquisition of antibiotic resistance, Gram-negative strains are greatly responsible for BSIs [10]. According to previous studies, BSIs caused by MDR Enterobacterales increased significantly from 6.2% to 15.8% from 1997 to 2016 [11]. Escherichia coli was among the most common pathogens responsible for nosocomial BSIs [12,13,14]. In addition, E. coli BSIs are among the most predominant cause of death in infants and children worldwide [15]. The incidence of E. coli BSIs has significantly increased in England [16] and Australia [17] in recent years. A systematic review on sub-Saharan Africa found that E. coli accounted for 10% of blood culture positive for bacteriaemia or sepsis [18]. Meanwhile, BSIs are common among children cancer patients under five years old, and the most identified microorganisms in Egypt were Gram-negative bacteria, with E. coli being the dominant species (27.8%) [19]. Early-onset BSIs caused by E. coli are potentially fatal diseases (36.6%) among preterm infants [20].
As a member of the ESKAPE group of pathogens, E. coli strains that contain extended-spectrum β-lactamases (ESBLs) and carbapenemases could usually develop resistance to a variety of antimicrobial agents [21,22,23]. Resistance to third-generation cephalosporin and carbapenems in E. coli bloodstream isolates has rapidly increased [24,25]. ESBLs-producing Enterobacteriaceae (especially, E. coli) are important human pathogens and a cause of bloodstream infections that appeared multidrug resistant in developed and developing countries [26]. E. coli strains encoding CTX-M-β-lactamases are considered the widely distributed species relevant to globally disseminated ESBLs [27]. To date, 220 different variants of CTX-M-β-lactamases have been identified and clustered into five subfamilies based on their amino acid residues [28]. Recent molecular epidemiology studies in China have showed that CTX-M-14, CTX-M-15 and CTX-M-55 were the most common ESBLs; according to the Multi-Locus Sequence Typing (MLST) analysis, these ESBLs were often found in the sequence types ST131, ST405 and ST69 [29]. The first Indian report on a genome-wide comparison displayed the distribution of blaNDM-5 among ST131, ST405 and ST410, whereas blaCTX-M-15 was associated with cephalosporin resistance in ST131 and ST405 [30]. Whole-genome analysis in Australia, New Zealand and Singapore showed that Clade C1/C2 (30.2% and 53.5% of ST131, respectively) carrying CTX-M-type (CTX-M-14, CTX-M-15 (mainly) and CTX-M-27) ESBLs was clearly predominant [31]. The ST131 and ST410 clones are important human E. coli lineage among extraintestinal pathogenic E. coli (ExPEC) isolates worldwide [32,33]. The majority of ST131 isolates belong to the O25b:H4 serotype; other isolates in some clinical collections appeared to belong to the serotype O16:H5 [34]. Unlike ST131, ST410 has a wide distribution among different populations (humans, animals and livestock) [35]. The ST131 and ST410 clones are frequently resistant to most β-lactams, aminoglycosides and fluoroquinolones and could cause a great variety of infections, such as bloodstream infections, intra-abdominal infections and septic shock.
Carbapenems are the ‘last-resort’ antibiotics for treating severe infections caused by multidrug-resistant bacteria, yet the acquisition of carbapenemase genes by Gram-negative strains, particularly by Enterobacteriaceae, significantly compromises their efficacy [36]. Accordingly, about 32% of patients died within 14 days from bloodstream infections caused by carbapenem-resistant Enterobacteriaceae (CRE) [25]. The carbapenemase genes are often located on mobile genetic elements such as plasmids, and horizontal gene transfer can accelerate the spread of CRE between species worldwide [37]. Since research in 2008 showed that a novel carbapenem resistance gene, NDM , a carbapenem-hydrolyzing enzyme, was found in Klebsiella pneumoniae isolated from a patient in India, this gene has disseminated over many countries [38]. NDM-type carbapenemases mediate resistance to most β-lactams antibiotic except aztreonam [39]. NDM variants are increasingly becoming one of the main effectors of carbapenem resistance among E. coli isolates in clinical settings in China [40]. NDM-positive E. coli isolated from BSIs were reported from different countries across the world, including India [41], China [42], Sweden [43], Cuba [44] and several Latin American countries [45]. Despite their prevalence being relatively low, such strains could cause considerable mortality and pose great challenges to the treatment of bloodstream infections [46].
At the time of writing, a total of 44 NDM variants, namely, NDM-1 to NDM-44, were deposited in the database (https://www.ncbi.nlm.nih.gov/pathogens/refgene/#gene_family: (blaNDM) (accessed on 1 November 2022)). Among these, NDM-5 was firstly detected in E. coli isolated from the throat of a patient with a history of travel to India in 2011 [47] and subsequently reported in different countries including India [48], South Korea [49], Australia [50], America [51] and Egypt [52]. NDM-5 was also reported to be the dominant NDM variant produced by E. coli strains from different hospitals in Jiangsu province, China [53,54].
Yet, the phenotypic and genetic characteristics of the clinical NDM-5-producing E. coli strains remain largely unknown. To fill this gap, we collected a total of eight NDM-5-producing E. coli strains isolated from between years 2016 and 2020 from BSIs in a children’s hospital in Jiangsu, China. The clinical information of the studied strains was also collected. The antimicrobial resistance phenotypes and the genetic characteristics of all strains were comprehensively studied. In this study, we conducted a bioinformatics analysis to determine the molecular characteristics of NDM-5-positive E. coli carried by pediatric patients and assessed the transmission ability of the plasmid harboring blaNDM-5. Our findings shall increase the understanding of how to control of untreatable BSIs caused by carbapenem-resistant E. coli.
2. Materials and Methods
2.1. Bacteria Isolates and Clinical Data Collection
A total of 114 non-duplicated E. coli strains isolated from blood specimens from 114 patients from the Children’s Hospital of Soochow University (Suzhou, China) were collected during the period from September 2016 to October 2020. The total numbers of E. coli isolates during these periods were 1 in 2016 and 2019, respectively, 3 in 2017 and 3 in 2020. The strains were isolated in accordance with standard procedures. Briefly, the blood samples were first cultured with the BD BACTECTM FX Blood Culture System (BD Diagnostics, Sparks, MD, USA). The positive cultures were sub-cultured on blood agar and in Mueller–Hinton broth at 37 °C for 16–24 h to obtain the E. coli isolates. All E. coli isolates from patients meeting the inclusion criteria of the study were stored in the laboratory at –80 °C in cryovials containing 20% glycerol and the nutrient broth for further analysis. The clinical information of the patients from whom the blaNDM-5-positive E. coli isolates were isolated was obtained from the clinicians. The relevant clinical information and the origin of the strains are presented in Table 1.
Table 1.
Clinical information of the cohort.
| Strain | Age * | Gender | Underlying Diseases | Clinical Department |
Antibiotic Treatment | Outcome | Year of Isolation |
|---|---|---|---|---|---|---|---|
| E1 | 3 Y | male | acute myeloid leukemia | hematology | meropenem + amikacin | recovery | 2019 |
| E2 | 13 Y | male | acute lymphoblastic leukemia, post allo-HSCT | hematology | meropenem + amikacin, tigecycline | automatically discharged | 2017 |
| E3 | 1 M | female | neonatal sepsis, premature infant | neonatology | meropenem | recovery | 2020 |
| E4 | 9 D | female | necrotizing enterocolitis, neonatal sepsis | neonatology | meropenem | recovery | 2020 |
| E5 | 4 M | male | congenital atresia of biliary tract | gastroenterology | imipenem | recovery | 2016 |
| E6 | 11 Y | male | T lymphoblastic leukemia/lymphoma | hematology | tigecycline + amikacin | recovery | 2020 |
| E7 | 3 Y | female | acute lymphoblastic leukemia | hematology | meropenem + amikacin | recovery | 2017 |
| E8 | 12 D | male | neonatal sepsis, premature infant | neonatology | meropenem | recovery | 2017 |
* Y, M and D in this column represent years, months and days, respectively.
2.2. Strain Identification and β-Lactamase Genes Confirmation
The species of all strains were identified using MALDI-TOF MS (Bruker Daltonik GmbH, Bremen, Germany) and 16S rRNA gene sequencing. The presence of the β-lactamase genes, including carbapenemase (blaNDM-5) and ESBLs genes (blaCMY-2, blaCTX-M-15, blaOXA-1, and blaTEM-1B) was detected by PCR targeting the entire blaNDM gene and other β-lactamase genes, which were validated with Sanger sequencing [55].
2.3. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was conducted for all isolates against 14 commonly used antibiotics including extended-spectrum β-lactam (ampicillin), aminoglycoside (gentamicin, tobramycin), monobactam (aztreonam), β-lactam/β-lactamase inhibitor combinations (ampicillin–sulbactam, piperacillin–tazobactam), carbapenem (imipenem), cephalosporins (cefepime, ceftriaxone, ceftazidime, cefotetan), fluoroquinolones (ciprofloxacin, levofloxacin) and trimethoprim–sulfamethoxazole. Antimicrobial susceptibility testing was conducted using the micro-broth dilution method. The minimum inhibitory concentrations (MICs) were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines [56]. E. coli ATCC 25922 was used as a quality control strain.
2.4. Conjugation Assay
A conjugation assay was performed using the filter-mating method [57]. Briefly, each blaNDM-5-carrying E. coli strain was used as the donor, and rifampicin-resistant E. coli EC600 was used as the recipient to test the transferability of the carbapenem resistance gene, blaNDM-5. The donor and the recipient were mixed in the ratio of 1:1 and cultured overnight on a nitrocellulose filter (HA type; pore size, 0.22 µm; Millipore Corp, Billerica, MA, USA) placed on LB agar. The filters were removed from the plates and placed in 1.0 mL of phosphate buffer. The cells were removed from the filter with a Vortex mixer. Dilutions were made, and all transconjugants were selected on LB agar plates supplemented with 1 µg/mL of meropenem and 600 µg/mL of rifampicin. The transconjugants were confirmed by PCR targeting the blaNDM-5 gene and by antimicrobial susceptibility testing. The conjugation frequencies were estimated by dividing the number of colony-forming units (CFUs) of the transconjugants by the number of CFUs of the recipient strains E. coli EC600.
2.5. DNA Extraction
The genomic DNA of each blaNDM-5-carrying E. coli was extracted from overnight cultures on BHI using a Genomic DNA Extraction Kit (QIAGEN, Valencia, CA, USA) according to the manufacturer’s instructions. The purity and concentrations of DNA were measured by a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA, USA).
2.6. Whole-Genome Sequencing and Bioinformatics Analysis
The extracted genomic DNA was subjected to whole-genome sequencing using the HiSeq platform (Illumina, San Diego, CA, USA). Trimmomatic was used to trim the raw reads in order to remove the adaptors and low-quality sequences [58]. De novo genome assembly was conducted with SPAdes v3.15.5 [59]. The draft genome sequences were annotated using RAST [60]. Acquired antimicrobial resistance genes (ARGs) were identified with ResFinder 4.1 [61]. Insertion sequences (ISs) were identified with the online software ISfinder [62]. Plasmid replicon types were identified with PlasmidFinder [63]. The comparison of the plasmids was performed using BLASTn, and the results were analyzed with the R tool [64]. The O:H serotype was identified with ECTyper v0.8.1 [65]. MLST v2.11 was used to determine the variety of STs [66]. Variant calling and core genome alignment of the strains E3, E4 and E5 was conducted using Snippy 3.1 [67]. Pairwise SNP was calculated using snp-dists 0.8.2 [68]. The harvest suite v.1.2 was used to remove recombination sequences and conduct the phylogenetic analysis using the assembled genome sequences as the input [69]. The generated phylogenetic tree was visualized and modified with iTOL v6 [70]. A heatmap of antimicrobial resistance genes was obtained using TBtools [71]. Plasmid alignment was visualized with BRIG [72].
2.7. Ethical Statements
For the study was obtained through the Children’s Hospital of Soochow University (Suzhou, China).
3. Results
3.1. Clinical Information and Overview of the NDM-5-Producing E. coli Strains
Among the 114 E. coli strains from BSIs collected from 2016 to 2020 in a children’s hospital in China, 8 (7.02%) strains designated E1–E8 carried blaNDM-5. The age of the eight patients ranged from 9 days to 13 years. Three (37.5%) were male patients, and five (62.5%) were female patients. The patients suffered from different diseases including acute myeloid leukemia, acute lymphoblastic leukemia, neonatal sepsis, congenital atresia of the biliary tract, etc., and were admitted to different departments in the hospital including hematology (n = 4), neonatology (n = 3) and gastroenterology (n = 1). The patients were treated with different antibiotics such as meropenem, tigecycline and amikacin. Seven patients recovered, and one patient was discharged automatically (Table 1).
3.2. Antimicrobial Resistance Profiles of the NDM-5-Producing E. coli Strains
The minimum inhibitory concentrations of commonly used antibiotics for the NDM-5-producing E. coli strains are listed in Table 2. All these strains exhibited multidrug resistance phenotypes. All eight strains were resistant to ampicillin, imipenem, cefepime, ceftriaxone, ceftazidime, cefotetan, ampicillin–sulbactam, piperacillin–tazobactam and trimethoprim–sulfamethoxazole. The proportion of E. coli strains that were non-susceptible to aztreonam, gentamycin and tobramycin were 62.5%, 50% and 75%, respectively. In addition, 87.5% of the eight strains were resistant to ciprofloxacin, and the remaining 12.5% strains exhibited intermediate resistance to ciprofloxacin. We found that 62.5% and 37.5% of the strains were resistant and intermediately resistant to levofloxacin, respectively.
Table 2.
MLST type and antibiotic resistance characteristics of Escherichia coli strains and their corresponding transconjugants.
| Strains | Description | MLST | Serotype a | Minimum Inhibitory Concentration (μg/mL) b | Conjugation Efficiency c | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | SAM | ATM | SXT | CIP | TZP | GEN | FEP | CRO | CAZ | CTT | TOB | IPM | LVX | |||||
| E1 | donor | ST38 | O7:H8 | ≥32 | ≥32/16 | 2 | ≥16/304 | 1 | 64/4 | ≤1 | ≥64 | ≥64 | ≥64 | ≥64 | ≤1 | ≥16 | 1 | NA |
| E2 | donor | ST58 | O?:H37 | ≥32 | ≥32/16 | ≤1 | ≥16/304 | 2 | 64/4 | ≥16 | 16 | ≥64 | ≥64 | ≥64 | ≥16 | ≥16 | 1 | NA |
| E3 | donor | ST410 | O?:H9 | ≥32 | ≥32/16 | ≥64 | ≥16/304 | ≥4 | ≥128/4 | ≤1 | ≥64 | ≥64 | ≥64 | ≥64 | ≥16 | 4 | ≥8 | NA |
| E4 | donor | ST410 | O?:H9 | ≥32 | ≥32/16 | ≥64 | ≥16/304 | ≥4 | ≥128/4 | ≤1 | ≥64 | ≥64 | ≥64 | ≥64 | ≥16 | 8 | ≥8 | NA |
| E5 | donor | ST410 | O?:H9 | ≥32 | ≥32/16 | 4 | ≥16/304 | 0.5 | ≥128/4 | ≥16 | ≥64 | ≥64 | ≥64 | ≥64 | 8 | ≥16 | 1 | NA |
| E6 | donor | ST131 | O25:H4 | ≥32 | ≥32/16 | ≥64 | ≥16/304 | ≥4 | ≥128/4 | ≥16 | ≥64 | ≥64 | ≥64 | ≥64 | ≥16 | ≥16 | ≥8 | NA |
| E7 | donor | ST361 | O9:H30 | ≥32 | ≥32/16 | 16 | ≥16/304 | ≥4 | ≥128/4 | ≥16 | ≥64 | ≥64 | ≥64 | ≥64 | ≥16 | ≥16 | ≥8 | NA |
| E8 | donor | ST156 | O11:H25 | ≥32 | ≥32/16 | 16 | ≥16/304 | 2 | ≥128/4 | ≤1 | ≥64 | ≥64 | ≥64 | ≥64 | ≤1 | ≥16 | 2 | NA |
| E1-TC | transconjugant | ST80 | O75:H7 | ≥32 | ≥32/16 | ≤1 | ≤1/19 | ≤0.25 | 64/4 | ≥16 | 16 | ≥64 | ≥64 | ≥64 | ≤1 | ≥16 | 0.5 | 2.5 × 10−3 |
| E6-TC | transconjugant | ST80 | O75:H7 | ≥32 | ≥32/16 | 16 | ≥16/304 | 1 | ≥128/4 | ≥16 | ≥64 | ≥64 | ≥64 | ≥64 | ≥16 | 16 | 1 | 2.9 × 10−6 |
| E7-TC | transconjugant | ST80 | O75:H7 | ≥32 | ≥32/16 | 16 | ≥16/304 | ≤0.25 | ≥128/4 | ≥16 | ≥64 | ≥64 | ≥64 | ≥64 | ≥16 | ≥16 | 0.5 | 3.5 × 10−6 |
| E8-TC | transconjugant | ST80 | O75:H7 | ≥32 | ≥32/16 | ≤1 | ≤1/19 | ≤0.25 | 64/4 | ≤1 | 16 | ≥64 | ≥64 | ≥64 | ≤1 | ≥16 | 0.5 | 1.6 × 10−6 |
| EC600 | recipient | ST80 | O75:H7 | 16 | 8/4 | ≤1 | ≤1/19 | ≤0.25 | ≤4/4 | ≤1 | ≤1 | ≤1 | ≤1 | ≤4 | ≤1 | ≤1 | 0.5 | NA |
a O? indicates that the O serotype of the strain was not typable using the current scheme. b Abbreviations: AMP, ampicillin; SAM, ampicillin–sulbactam; ATM, aztreonam; SXT, trimethoprim–sulfamethoxazole; CIP, ciprofloxacin; TZP, piperacillin–tazobactam; GEN, gentamicin; FEP, cefepime; CRO, ceftriaxone; CAZ, ceftazidime; CTT, cefotetan; TOB, tobramycin; IPM, imipenem; LVX, levofloxacin. c NA, not applicable.
3.3. Genomic Characteristics of the NDM-5-Producing E. coli Strains
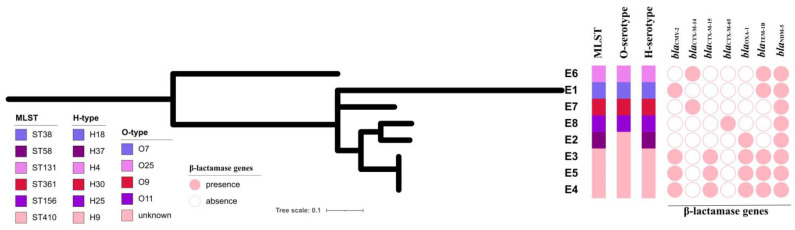
Draft genome sequences of the eight NDM-5-producing E. coli strains were obtained. These strains belonged to six different sequence types including ST410 (n = 3, strains E3, E4 and E5), ST38 (n = 1, strain E1), ST58 (n = 1, strain E2), ST131 (n = 1, strain E6), ST361 (n = 1, strain E7) and ST156 (n = 1, strain E8). The strains E3, E4 and E5 belonged to an unknown O serotype (O?) and to serotype H9. The strains E1, E2, E6, E7 and E8 belonged to the serotypes O7:H18, O?:H37, O25:H4, O9:H30 and O11:H25, respectively. The strains E3, E4 and E5 shared the same sequence type and serotype, and the number of pairwise SNPs among the three strains was <20, suggesting they belonged to a single clone (Figure 1).
Figure 1.
Phylogeny of blaNDM-5-positive E. coli isolates from bloodstream infections. MLST, O and H serotypes and the presence of β-lactamase genes are plotted.
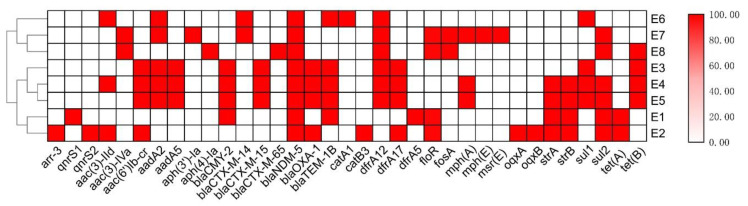
The E. coli strains carried different antimicrobial resistance genes conferring resistance to different classes of antibiotics including aminoglycoside, fosfomycin, fluoroquinolone, macrolides, chloramphenicol, rifampicin, sulphonamide, tetracycline, trimethoprim, extended-spectrum β-lactam and carbapenem. The number of acquired antimicrobial resistance genes carried by each E. coli strain ranged from 8 to 17. Two strains (E1 and E2) carried the tetracycline resistance gene, tet(A), while four strains (E3, E4, E5 and E8) harbored the tetracycline resistance gene tet(B). Apart from blaNDM-5, the E. coli strains isolated from BSIs also carried other β-lactamase genes, including blaCMY-2 (n = 4), blaCTX-M-14 (n = 2), blaCTX-M-15 (n = 3), blaCTX-M-65 (n = 1), blaOXA-1 (n = 4) and blaTEM-1B (n = 5). Particularly, the strains E3, E4 and E5 carried five β-lactamase genes, including blaNDM-5, blaCMY-2, blaCTX-M-15, blaOXA-1 and blaTEM-1B, which could pose a significant threat to public health. Despite belonging to a single clone, the resistance genes carried by the three strains were not identical, suggesting active genetic recombination could have occurred in this high-risk clone. The strain E1 carried blaCMY-2 and blaTEM-1B, the strain E2 carried blaOXA-1, the strain E6 carried blaCTX-M-14 and blaTEM-1B, the strain E7 carried blaCTX-M-14 and the strain E8 carried blaCTX-M-65 (Figure 2).
Figure 2.
Heatmap of antimicrobial resistance genes carried by E. coli isolates in this study. The horizontal axis represents the antimicrobial resistance genes, and the vertical axis represents the strain IDs. The red boxes represent the presence of the corresponding items among the sequenced isolates, and the white boxes represent their absence. The gradient identity bar indicates the percentage similarity of related genes. The similarity tree was constructed using agglomerative hierarchical clustering, with the degree of similarity between different clusters calculated by the average linkage method and the degree of similarity of different isolates (calculated using the Spearman’s rank correlation coefficient).The classes of antibiotics to which the genes confer resistance included aminoglycosides: strA, strB, aac(3)-IId, aac(6)-Ib-cr, aadA2, aadA5, aph3-Ia, aph(4)-Ia and aac(3)-IV; fosfomycin: fosA; fluoroquinolones: qnrS1 and qnrS2; macrolides: mph(A), mph(B), mph(E) and msr(E); chloramphenicol: catA1, catB3, and floR; rifampicin: arr-3; sulphonamide: sul1 and sul2; tetracycline: tet(A) and tet(B); trimethoprim: dfrA5, dfrA12 and dfrA17; extended-spectrum β-lactams: blaCMY-2, blaTEM-1D, blaOXA-1, blaCTX-M-14, blaCTX-M-15, and blaCTX-M-65; carbapenems: blaNDM-5.
3.4. Transferability and Genetic Analysis of blaNDM-5-Carrying Plasmids
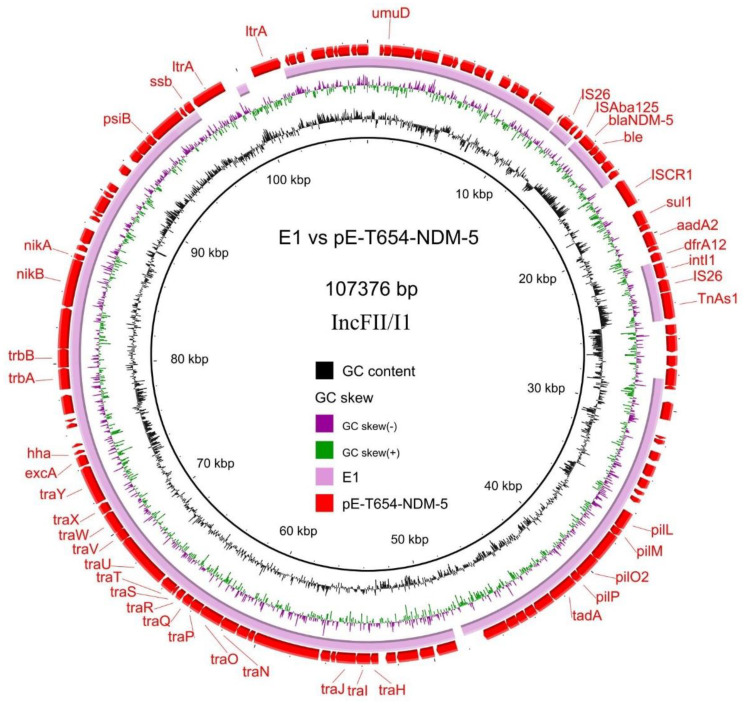
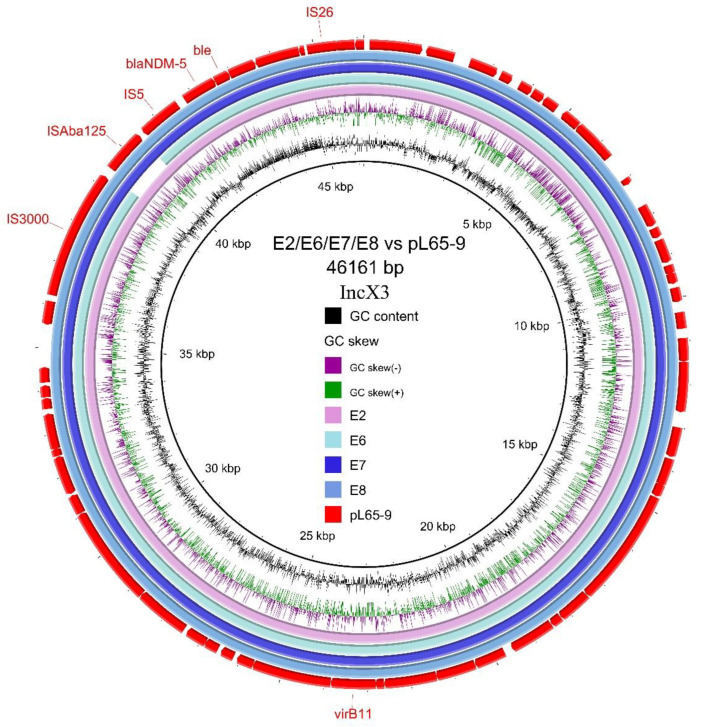
To further investigate the role of NDM-5-producing E. coli strains in bacterial dissemination, we analyzed the drafts of the whole-genome sequences of all strains and found that the blaNDM-5 genes were located in contigs ranging from 3 Kb to 43 Kb. The blaNDM-5 gene in strain E2 was located on a 43,563 bp contig, and the blaNDM-5 genes in other strains were all located on contigs < 9 Kb. The blaNDM-5-carrying contigs were generally too short to accurately resolve the genetic context of blaNDM-5. The blaNDM-5-bearing contigs in these strains aligned well to different plasmids, including the 107kb IncFII/I1 plasmid pE−T654−NDM-5 (accession no.: CP090291, aligned strain: E1; Figure 3), the 46 kb IncX3 plasmid pL65-9 (accession no.: CP034744; aligned strains: E2, E6, E7 and E8; Figure 4) and the 108 kb IncFIA/FIB/FII/Q1 plasmid pC405−NDM5 (accession no.: LC521844; aligned strains: E3, E4 and E5; Figure 5). The blaNDM-5 genes in the strains E1, E6, E7 and E8 were conjugatively transferable to E. coli EC600 with conjugation efficiencies of 2.5 × 10−3, 2.9 × 10−6, 3.5 × 10−6 and 1.6 × 10−6, respectively. The transconjugants were all resistant to carbapenems and cephalosporins (Table 2). blaNDM-5 in the strains E2, E3, E4 and E5 were non-conjugatively transferable under the experimental conditions of this study.
Figure 3.
Circular alignments of the reference plasmid sequence (pE−T654−NDM−5, GenBank accession: CP090291) with homologous blaNDM-5−carrying contigs from the E. coli strain E1 in this study. Representative genes such as antimicrobial resistance genes, conjugation−associated genes and mobile genetic elements are labelled in the outermost circle.
Figure 4.
Circular alignments of the reference plasmid sequence (pL65−9, GenBank accession: CP034744) with homologous blaNDM-5-carrying contigs from the E. coli strains E2, E6, E7 and E8 in this study. Representative genes such as antimicrobial resistance genes and mobile genetic elements are labelled in the outermost circle.
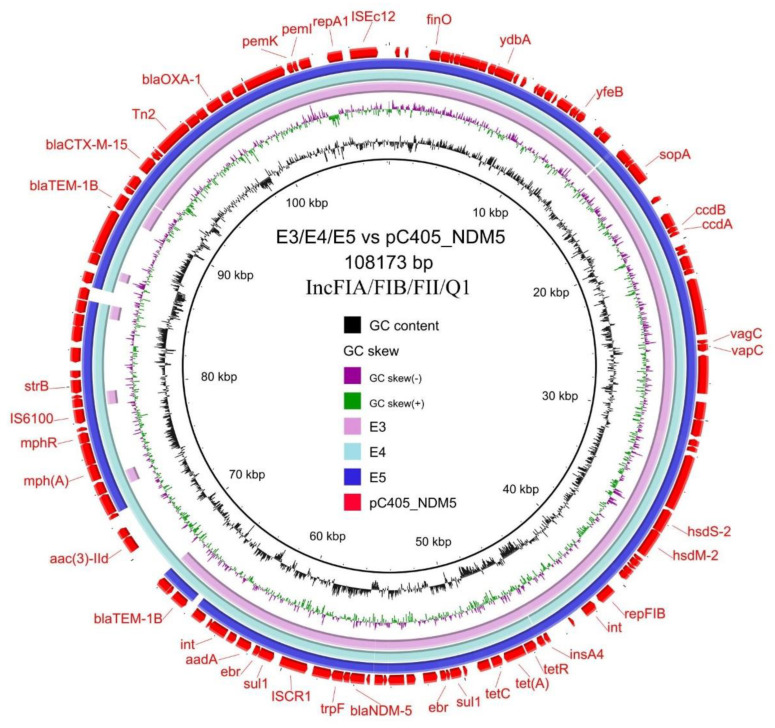
Figure 5.
Circular alignments of the reference plasmid sequence (pC405−NDM5, GenBank accession: LC521844) with homologous blaNDM-5−carrying contigs from the E. coli strain E3, E4 and E5 in this study. Representative genes such as antimicrobial resistance genes, conjugation-associated genes and mobile genetic elements are labelled in the outermost circle.
4. Discussion
Carbapenems are used as the last-resort antibiotics to treat Gram-negative bacterial infections. The emergence of carbapenem-resistant organisms is considered one of the most urgent public health concerns [73]. The acquisition of carbapenemase genes by Gram-negative strains is one of the major mechanisms of carbapenem resistance, and the horizontal gene transfer of carbapenemase genes located on mobile genetic elements, including transposons and plasmids, is causing a widespread dissemination of CRE [37]. E. coli is among the most frequently isolated CRE worldwide which could colonize both elderly patients (aged >60 years) and the pediatric population [74,75]. Carbapenem-resistant E. coli isolated from blood samples has been reported across the world [76]. Carbapenem-resistant E. coli strains carrying different blaNDM gene variants were found in North America, Africa, Asia and Europe [77]. A study from hospitals in Chongqing, China, on strains from BSIs suggested that the infection ratio of E. coli in children was significantly higher than in adults, and most of the E. coli strains from BSIs in children expressed blaNDM, particularly blaNDM-5 [78]. Furthermore, the spread of ESBL-producing E. coli is one of the most important driving factors of the abuse of carbapenems, which indirectly exacerbates the selective pressure of carbapenemase producers [79]. It is well known that CTX-M-15 and CTX-M-14 are the most common CTX-Ms worldwide, often reported in South-East Asia [27]. Our study also demonstrated that carbapenemase-producing E. coli are often multidrug resistant and usually express other antibiotic resistance genes.
The genomic and phenotypic characteristics of blaNDM-5/blaCTX-M-positive E. coli from BSIs still remain poorly understood. In this study, a total of eight NDM-5-producing strains were obtained from 114 E. coli strains from BSIs in pediatric patients. The number of NDM-5-producing E. coli isolates was higher in 2017 (n = 3) and 2020 (n = 3) than in 2016 (n = 1) and 2019 (n = 1). No NDM-5-producing E. coli was isolated from blood samples in 2018. This could be associated with the different number of samples collected each year. After treatment with different antibiotics such as meropenem, tigecycline and amikacin, seven patients recovered, and one patient was discharged automatically. The E. coli strains causing BSIs carry genes responsible for resistance to aminoglycosides [strA, strB, aac(3)-IId, aac(6)-Ib-cr, aac(3)-IV, aadA2, aadA5, aph(3)-la, aph(4)-la], extended-spectrum β-lactams (blaCMY-2, blaCTX-M-15, blaOXA-1, blaTEM-1B and blaNDM-5) and fluoroquinolones (qnrS1, qnrS2). These findings are well in accordance with the multidrug resistance to aminoglycosides, third-generation cephalosporins, carbapenems and quinolones determined in this study. A previous study reported the first genome sequence of an MDR E. coli carrying blaNDM-5 and two copies of blaCTX-M-14 from a BSI in China [80]. Similarly, our research showed that both an ESBL gene and a carbapenemase gene were detected in six isolates, including blaCTX-M-15/blaNDM-5 in strains E3, E4 and E5, blaCTX-M-14/blaNDM-5 in strains E6 and E7 and blaCTX-M-65/blaNDM-5 in strain E8. Additionally, a previous study firstly reported an E. coli ST410 strain coharboring the blaNDM-5, blaOXA-1, blaCTX-M-15, blaCMY-2, aac(3)-IIa and aac(6)-Ib-cr genes that was collected from a bloodstream infection in China [81]. We also found that an E. coli ST410 strain coharbored the blaNDM-5, blaOXA-1, blaCTX-M-15, blaCMY-2, blaTEM-1D, blaAmpC1, aac(3)-IId and aac(6)-Ib-cr genes; it was collected from a bloodstream infection in Suzhou, China.
The eight NDM-5-producing strains belonged to six different sequence types including ST410, ST38, ST58, ST131, ST361 and ST156, all of which have been reported to be associated with multidrug resistance phenotypes. Among the sequence types in this study, ST410, ST38, ST361 and ST156 E. coli were identified as NDM-5-producers in previous studies [53,82,83,84]. ST131 and ST58 E. coli have been reported to be NDM-1 producers [85,86]. Previous epidemiological studies demonstrated that the dominant E. coli sequence type associated with BSIs in China was ST131, followed by ST69 and ST38 [87,88]. ST131 is the predominant E. coli lineage among extraintestinal pathogenic E. coli (ExPEC) isolates worldwide and is commonly reported to produce extended-spectrum β-lactamases [32]. Previous studies reported that CTX-M-14 and CTX-M-15 were the most common ESBLs in E. coli ST131 isolates [32], which is in line with the results of our study. Two serotypes of ST131, O16:H5 and O25b:H4 have been identified [34]. In 2008, the serogroup O25b and the sequence type ST131were detected in many countries from analyses of the population biology of ESBL-producing E. coli [89]. The strain E6 in this study belongs to O25b:H4, ST131. Due to the limited number of strains included in this study, we were unable to compare the prevalence of different sequence types that we calculated with that reported in the literature.
Three strains, belonging to a single clone with the sequence type ST410 and the serotype O?:H9, were collected in this study. ST410 has been reported worldwide as an extraintestinal pathogen associated with resistance to different classes of antibiotics such as fluoroquinolones, polymyxins, third-generation cephalosporins and carbapenems [33,90]. ST410 was further classified into several clades, each of which is associated with a different serotype, including clades A and B (serotype O8:H21), clade C (O8:H9), and clade D and E (O?:H9) [35]. We found that the strains E3, E4 and E5 recovered from bloodstream infection belonged to the serotype clades D and E, which has not been published before. Strains belonging to the clades D and E could be recovered from human and animal samples, posing threat to both populations [35]. The clone ST410 E. coli undergoes constant evolution, with its resistome changing with time. Around 2014, a clade of ST410 acquired the carbapenemase gene, blaNDM-5, on a conserved IncFII plasmid; this could be the ancestral source of the blaNDM-5-carrying plasmids in the ST410 strains E3, E4 and E5 analyzed in this study [33]. A previous study reported the isolation of an ST410 E. coli strain a carrying blaNDM-5-encoding IncX3 plasmid from a 59-year-old patient with BSI in China, yet the ST410 E. coli carrying a blaNDM-5-encoding IncFII plasmid isolated from blood samples of patients with BSIs was not reported before this study [91]. In addition, we reported the first ST410 clone which co-harbored five β-lactamase genes, including blaNDM-5, blaCMY-2, blaCTX-M-15, blaOXA-1 and blaTEM-1B. Considering that the ST410 clone isolated in this study carries a cassette of important resistance genes, has the potential of transmission between patients and could cause BSIs, it should be monitored closely in the future.
Plasmids play a pivotal role in the dissemination of antimicrobial resistance genes. blaNDM-5 has been reported to be closely associated with different types of plasmids such as IncX3, IncFII and IncHI2 [91,92,93]. Among these, the IncF plasmid, with a low copy number, has a narrow host range (Enterobacteriaceae) and has greatly improved the fitness cost through its antimicrobial resistance determinants by horizontal gene transfer [94]. This is the most common plasmid type in the ST131 clone and is widely distributed in clinically Enterobacteriaceae. The IncX3 plasmid containing blaNDM-5 has showed a varied geographical distribution in China, as it has been isolated in Jiangsu province [95], Shanghai [96] and Zhejiang province [97]. The blaNDM-5 genes in the strains E1−E8 were all plasmid-borne. Three different types of blaNDM-5-carrying plasmids were identified among the strains, including the IncFII/I1 pE−T654−NDM-5−like plasmids, the IncX3 pL65−9−like plasmids and the IncFIA/FIB/FII/Q1 pC405−NDM5−like plasmids. The former two types carry conjugation-associated genes and are conjugatively transferable at different frequencies. The pC405−NDM5−like plasmids are non-self-transmissible. The reason that blaNDM-5 in the strains E3, E4 and E5 are non-conjugative transferable could be explained by the fact that the IncFIA/FIB/FII/Q1 pC405−NDM5−like plasmid does not carry conjugation-associated genes. The fact that no transconjugants were detected for strain E2 could be explained by a low conjugation frequency (~10−6) of the IncX3 plasmids. The transferability of blaNDM-5-carrying plasmids may promote the rapid dissemination of resistance-encoding elements among Gram-negative bacterial pathogens. Controlled measures such as optimizing antibiotic usage and the development of new antibiotics or antibiotic adjuvants are urgently needed to prevent the emergence and transmission of resistant bacteria such as carbapenem-resistant E. coli.
5. Conclusions
In summary, this study collected 8 blaNDM-5-carrying E. coli strains from a total of 114 E. coli strains from bloodstream infections in a children’s hospital in Jiangsu province, China. These strains were carbapenem-resistant and carried diverse antimicrobial resistance genes apart from blaNDM-5. The eight strains belonged to six distinct STs and serotypes including one each for ST38/O7:H8, ST58/O?:H37, ST131/O25:H4, ST156/O11:H25 and ST361/O9:H30, and three strains are originating from a single clone belonging to ST410/O?:H9. Five β-lactamase genes were identified in the ST410 E. coli strains, i.e., blaNDM-5, blaCMY-2, blaCTX-M-15, blaOXA-1 and blaTEM-1B. The blaNDM-5 gene was located on three different types of plasmids, which were IncFII/I1 (n = 1), IncX3 (n = 4) and IncFIA/FIB/FII/Q1 (n = 3). The former two types were conjugatively transferable at frequencies of 10−3 and 10−6, respectively. The results of this study could increase our understanding of carbapenem-resistant E. coli from BSIs. Effective strategies such as the development of new antibiotics or antibiotic adjuvants and the optimization of antibiotic usage are needed for the control of untreatable BSIs caused by carbapenem-resistant E. coli.
Author Contributions
Conceptualization, L.H., S.W. and N.D.; methodology, L.H. and H.H.; validation, C.X., M.Z., Y.L. (Yuanyuan Li) and Y.L. (Yunbing Li); formal analysis, H.H. and N.D.; data curation, L.H., H.H. and N.D.; writing—original draft preparation, H.H. and L.H.; writing—review and editing, L.H., S.W. and N.D.; project administration, N.D.; funding acquisition, N.D. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by an independent ethics committee and the Institutional Review Board of Children’s hospital of Soochow University.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The assembled genome sequences of all E. coli strains from bloodstream infections in this study were deposited in the NBCI database under BioProject accession number PRJNA890582.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study was funded by the Natural Science Foundation of Jiangsu Province (No. BK20220493) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Di Franco S., Alfieri A., Pace M.C., Sansone P., Pota V., Fittipaldi C., Fiore M., Passavanti M.B. Blood Stream Infections from MDR Bacteria. Life. 2021;11:575. doi: 10.3390/life11060575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudd K.E., Johnson S.C., Agesa K.M., Shackelford K.A., Tsoi D., Kievlan D.R., Colombara D.V., Ikuta K.S., Kissoon N., Finfer S., et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet. 2020;395:200–211. doi: 10.1016/S0140-6736(19)32989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shakur S.M., Whitehall J., Mudgil P. Pediatric bloodstream infections in metropolitan Australia. World J. Pediatr. WJP. 2019;15:161–167. doi: 10.1007/s12519-018-00221-3. [DOI] [PubMed] [Google Scholar]
- 4.Malande O.O., Nuttall J., Pillay V., Bamford C., Eley B. A ten-year review of ESBL and non-ESBL Escherichia coli bloodstream infections among children at a tertiary referral hospital in South Africa. PloS ONE. 2019;14:e0222675. doi: 10.1371/journal.pone.0222675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaidi A.K., Ganatra H.A., Syed S., Cousens S., Lee A.C., Black R., Bhutta Z.A., Lawn J.E. Effect of case management on neonatal mortality due to sepsis and pneumonia. BMC Public Health. 2011;11:S13. doi: 10.1186/1471-2458-11-S3-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleischmann-Struzek C., Goldfarb D.M., Schlattmann P., Schlapbach L.J., Reinhart K., Kissoon N. The global burden of paediatric and neonatal sepsis: A systematic review. Lancet Respir. Med. 2018;6:223–230. doi: 10.1016/S2213-2600(18)30063-8. [DOI] [PubMed] [Google Scholar]
- 7.Naylor N.R., Pouwels K.B., Hope R., Green N., Henderson K.L., Knight G.M., Atun R., Robotham J.V., Deeny S.R. The health and cost burden of antibiotic resistant and susceptible Escherichia coli bacteraemia in the English hospital setting: A national retrospective cohort study. PLoS ONE. 2019;14:e0221944. doi: 10.1371/journal.pone.0221944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dellinger R.P., Levy M.M., Rhodes A., Annane D., Gerlach H., Opal S.M., Sevransky J.E., Sprung C.L., Douglas I.S., Jaeschke R., et al. Surviving Sepsis Campaign: International guidelines for management of severe sepsis and septic shock, 2012. Intensive Care Med. 2013;39:165–228. doi: 10.1007/s00134-012-2769-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kissoon N., Carcillo J.A., Espinosa V., Argent A., Devictor D., Madden M., Singhi S., van der Voort E., Latour J. World Federation of Pediatric Intensive Care and Critical Care Societies: Global Sepsis Initiative. Pediatr. Crit. Care Med. A J. Soc. Crit. Care Med. World Fed. Pediatr. Intensive Crit. Care Soc. 2011;12:494–503. doi: 10.1097/PCC.0b013e318207096c. [DOI] [PubMed] [Google Scholar]
- 10.Holmes C.L., Anderson M.T., Mobley H.L.T., Bachman M.A. Pathogenesis of Gram-Negative Bacteremia. Clin. Microbiol. Rev. 2021;34:e00234-20. doi: 10.1128/CMR.00234-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diekema D.J., Hsueh P.R., Mendes R.E., Pfaller M.A., Rolston K.V., Sader H.S., Jones R.N. The Microbiology of Bloodstream Infection: 20-Year Trends from the SENTRY Antimicrobial Surveillance Program. Antimicrob. Agents Chemother. 2019;63:e00355-19. doi: 10.1128/AAC.00355-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim D., Yoon E.J., Hong J.S., Choi M.H., Kim H.S., Kim Y.R., Kim Y.A., Uh Y., Shin K.S., Shin J.H., et al. Major Bloodstream Infection-Causing Bacterial Pathogens and Their Antimicrobial Resistance in South Korea, 2017–2019: Phase I Report From Kor-GLASS. Front. Microbiol. 2021;12:799084. doi: 10.3389/fmicb.2021.799084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian L., Sun Z., Zhang Z. Antimicrobial resistance of pathogens causing nosocomial bloodstream infection in Hubei Province, China, from 2014 to 2016: A multicenter retrospective study. BMC Public Health. 2018;18:1121. doi: 10.1186/s12889-018-6013-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian L., Zhang Z., Sun Z. Antimicrobial resistance trends in bloodstream infections at a large teaching hospital in China: A 20-year surveillance study (1998–2017) Antimicrob. Resist. Infect. Control. 2019;8:86. doi: 10.1186/s13756-019-0545-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Randolph A.G., McCulloh R.J. Pediatric sepsis: Important considerations for diagnosing and managing severe infections in infants, children, and adolescents. Virulence. 2014;5:179–189. doi: 10.4161/viru.27045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abernethy J., Guy R., Sheridan E.A., Hopkins S., Kiernan M., Wilcox M.H., Johnson A.P., Hope R. Epidemiology of Escherichia coli bacteraemia in England: Results of an enhanced sentinel surveillance programme. J. Hosp. Infect. 2017;95:365–375. doi: 10.1016/j.jhin.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 17.Williamson D.A., Lim A., Wiles S., Roberts S.A., Freeman J.T. Population-based incidence and comparative demographics of community-associated and healthcare-associated Escherichia coli bloodstream infection in Auckland, New Zealand, 2005–2011. BMC Infect. Dis. 2013;13:385. doi: 10.1186/1471-2334-13-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okomo U., Akpalu E.N.K., Le Doare K., Roca A., Cousens S., Jarde A., Sharland M., Kampmann B., Lawn J.E. Aetiology of invasive bacterial infection and antimicrobial resistance in neonates in sub-Saharan Africa: A systematic review and meta-analysis in line with the STROBE-NI reporting guidelines. Lancet Infect. Dis. 2019;19:1219–1234. doi: 10.1016/S1473-3099(19)30414-1. [DOI] [PubMed] [Google Scholar]
- 19.Elseady N.S.M., Khamis N., AbdelGhani S., Rabea H.M., Elanany M.G., Nashat Alsheshtawi K., Abdelrahim M.E.A. Antibiotic sensitivity/resistance pattern of hospital acquired blood stream infection in children cancer patients: A retrospective study. Int. J. Clin. Pract. 2021;75:e14617. doi: 10.1111/ijcp.14617. [DOI] [PubMed] [Google Scholar]
- 20.Stoll B.J., Puopolo K.M., Hansen N.I., Sánchez P.J., Bell E.F., Carlo W.A., Cotten C.M., D’Angio C.T., Kazzi S.N.J., Poindexter B.B., et al. Early-Onset Neonatal Sepsis 2015 to 2017, the Rise of Escherichia coli, and the Need for Novel Prevention Strategies. JAMA Pediatr. 2020;174:e200593. doi: 10.1001/jamapediatrics.2020.0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Oliveira D.M.P., Forde B.M., Kidd T.J., Harris P.N.A., Schembri M.A., Beatson S.A., Paterson D.L., Walker M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020;33:e00181-19. doi: 10.1128/CMR.00181-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saber A., Sebbar N.K., Sert Y., Alzaqri N., Hökelek T., El Ghayati L., Talbaoui A., Mague J.T., Baba Y.F., Urrutigoîty M., et al. Syntheses of N-substituted benzimidazolone derivatives: DFT calculations, Hirshfeld surface analysis, molecular docking studies and antibacterial activities. J. Mol. Struct. 2020;1200:127174. doi: 10.1016/j.molstruc.2019.127174. [DOI] [Google Scholar]
- 23.Bouzian Y., Karrouchi K., Sert Y., Lai C.-H., Mahi L., Ahabchane N.H., Talbaoui A., Mague J.T., Essassi E.M. Synthesis, spectroscopic characterization, crystal structure, DFT, molecular docking and in vitro antibacterial potential of novel quinoline derivatives. J. Mol. Struct. 2020;1209:127940. doi: 10.1016/j.molstruc.2020.127940. [DOI] [Google Scholar]
- 24.Skjøt-Rasmussen L., Olsen S.S., Jensen U.S., Hammerum A.M. Increasing consumption of antimicrobial agents in Denmark parallels increasing resistance in Escherichia coli bloodstream isolates. Int. J. Antimicrob. Agents. 2012;40:86–88. doi: 10.1016/j.ijantimicag.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 25.Tamma P.D., Goodman K.E., Harris A.D., Tekle T., Roberts A., Taiwo A., Simner P.J. Comparing the Outcomes of Patients With Carbapenemase-Producing and Non-Carbapenemase-Producing Carbapenem-Resistant Enterobacteriaceae Bacteremia. Clin. Infect. Dis. 2017;64:257–264. doi: 10.1093/cid/ciw741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pitout J.D. Extraintestinal Pathogenic Escherichia coli: A Combination of Virulence with Antibiotic Resistance. Front. Microbiol. 2012;3:9. doi: 10.3389/fmicb.2012.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peirano G., Pitout J.D.D. Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae: Update on Molecular Epidemiology and Treatment Options. Drugs. 2019;79:1529–1541. doi: 10.1007/s40265-019-01180-3. [DOI] [PubMed] [Google Scholar]
- 28.Naas T., Oueslati S., Bonnin R.A., Dabos M.L., Zavala A., Dortet L., Retailleau P., Iorga B.I. Beta-lactamase database (BLDB)—Structure and function. J. Enzym. Inhib. Med. Chem. 2017;32:917–919. doi: 10.1080/14756366.2017.1344235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S., Zhao S.Y., Xiao S.Z., Gu F.F., Liu Q.Z., Tang J., Guo X.K., Ni Y.X., Han L.Z. Antimicrobial Resistance and Molecular Epidemiology of Escherichia coli Causing Bloodstream Infections in Three Hospitals in Shanghai, China. PLoS ONE. 2016;11:e0147740. doi: 10.1371/journal.pone.0147740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Devanga Ragupathi N.K., Veeraraghavan B., Muthuirulandi Sethuvel D.P., Anandan S., Vasudevan K., Neeravi A.R., Daniel J.L.K., Sathyendra S., Iyadurai R., Mutreja A. First Indian report on genome-wide comparison of multidrug-resistant Escherichia coli from blood stream infections. PLoS ONE. 2020;15:e0220428. doi: 10.1371/journal.pone.0220428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris P.N.A., Ben Zakour N.L., Roberts L.W., Wailan A.M., Zowawi H.M., Tambyah P.A., Lye D.C., Jureen R., Lee T.H., Yin M., et al. Whole genome analysis of cephalosporin-resistant Escherichia coli from bloodstream infections in Australia, New Zealand and Singapore: High prevalence of CMY-2 producers and ST131 carrying blaCTX-M-15 and blaCTX-M-27. J. Antimicrob. Chemother. 2018;73:634–642. doi: 10.1093/jac/dkx466. [DOI] [PubMed] [Google Scholar]
- 32.Nicolas-Chanoine M.H., Bertrand X., Madec J.Y. Escherichia coli ST131, an intriguing clonal group. Clin. Microbiol. Rev. 2014;27:543–574. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roer L., Overballe-Petersen S., Hansen F., Schønning K., Wang M., Røder B.L., Hansen D.S., Justesen U.S., Andersen L.P., Fulgsang-Damgaard D., et al. Escherichia coli Sequence Type 410 Is Causing New International High-Risk Clones. mSphere. 2018;3:e00337-18. doi: 10.1128/mSphere.00337-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dahbi G., Mora A., Mamani R., López C., Alonso M.P., Marzoa J., Blanco M., Herrera A., Viso S., García-Garrote F., et al. Molecular epidemiology and virulence of Escherichia coli O16:H5-ST131: Comparison with H30 and H30-Rx subclones of O25b:H4-ST131. Int. J. Med. Microbiol. IJMM. 2014;304:1247–1257. doi: 10.1016/j.ijmm.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 35.Falgenhauer L., Imirzalioglu C., Ghosh H., Gwozdzinski K., Schmiedel J., Gentil K., Bauerfeind R., Kämpfer P., Seifert H., Michael G.B., et al. Circulation of clonal populations of fluoroquinolone-resistant CTX-M-15-producing Escherichia coli ST410 in humans and animals in Germany. Int. J. Antimicrob. Agents. 2016;47:457–465. doi: 10.1016/j.ijantimicag.2016.03.019. [DOI] [PubMed] [Google Scholar]
- 36.Rodríguez-Baño J., Gutiérrez-Gutiérrez B., Machuca I., Pascual A. Treatment of Infections Caused by Extended-Spectrum-Beta-Lactamase-, AmpC-, and Carbapenemase-Producing Enterobacteriaceae. Clin. Microbiol. Rev. 2018;31:e00079-17. doi: 10.1128/CMR.00079-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonomo R.A., Burd E.M., Conly J., Limbago B.M., Poirel L., Segre J.A., Westblade L.F. Carbapenemase-Producing Organisms: A Global Scourge. Clin. Infect. Dis. 2018;66:1290–1297. doi: 10.1093/cid/cix893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yong D., Toleman M.A., Giske C.G., Cho H.S., Sundman K., Lee K., Walsh T.R. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009;53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dortet L., Poirel L., Nordmann P. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. BioMed Res. Int. 2014;2014:249856. doi: 10.1155/2014/249856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sekar R., Mythreyee M., Srivani S., Sivakumaran D., Lallitha S., Saranya S. Carbapenem-resistant Enterobacteriaceae in Pediatric Bloodstream Infections in Rural Southern India. Indian Pediatr. 2017;54:1021–1024. doi: 10.1007/s13312-017-1204-1. [DOI] [PubMed] [Google Scholar]
- 42.Li X., Sun L., Zhu Y., Shen M., Tu Y. Draft genome sequence of Escherichia coli ST977: A clinical multidrug-resistant strain harbouring bla(NDM-3) isolated from a bloodstream infection. J. Glob. Antimicrob. Resist. 2018;13:121–122. doi: 10.1016/j.jgar.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Löfmark S., Sjöström K., Mäkitalo B., Edquist P., Tegmark Wisell K., Giske C.G. Carbapenemase-producing Enterobacteriaceae in Sweden 2007-2013: Experiences from seven years of systematic surveillance and mandatory reporting. Drug Resist. Updates Rev. Comment. Antimicrob. Anticancer. Chemother. 2015;20:29–38. doi: 10.1016/j.drup.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 44.Yu H., González Molina M.K., Carmona Cartaya Y., Hart Casares M., Aung M.S., Kobayashi N., Quiñones Pérez D. Multicenter Study of Carbapenemase-Producing Enterobacterales in Havana, Cuba, 2016-2021. Antibiotics. 2022;11:514. doi: 10.3390/antibiotics11040514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villegas M.V., Pallares C.J., Escandón-Vargas K., Hernández-Gómez C., Correa A., Álvarez C., Rosso F., Matta L., Luna C., Zurita J., et al. Characterization and Clinical Impact of Bloodstream Infection Caused by Carbapenemase-Producing Enterobacteriaceae in Seven Latin American Countries. PLoS ONE. 2016;11:e0154092. doi: 10.1371/journal.pone.0154092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lutgring J.D., Balbuena R., Reese N., Gilbert S.E., Ansari U., Bhatnagar A., Boyd S., Campbell D., Cochran J., Haynie J., et al. Antibiotic Susceptibility of NDM-Producing Enterobacterales Collected in the United States in 2017 and 2018. Antimicrob. Agents Chemother. 2020;64:e00499-20. doi: 10.1128/AAC.00499-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hornsey M., Phee L., Wareham D.W. A novel variant, NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob. Agents Chemother. 2011;55:5952–5954. doi: 10.1128/AAC.05108-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahmad N., Ali S.M., Khan A.U. Detection of New Delhi Metallo-β-Lactamase Variants NDM-4, NDM-5, and NDM-7 in Enterobacter aerogenes Isolated from a Neonatal Intensive Care Unit of a North India Hospital: A First Report. Microb. Drug Resist. 2018;24:161–165. doi: 10.1089/mdr.2017.0038. [DOI] [PubMed] [Google Scholar]
- 49.Cho S.Y., Huh H.J., Baek J.Y., Chung N.Y., Ryu J.G., Ki C.S., Chung D.R., Lee N.Y., Song J.H. Klebsiella pneumoniae co-producing NDM-5 and OXA-181 carbapenemases, South Korea. Emerg. Infect. Dis. 2015;21:1088–1089. doi: 10.3201/eid2106.150048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wailan A.M., Paterson D.L., Caffery M., Sowden D., Sidjabat H.E. Draft Genome Sequence of NDM-5-Producing Escherichia coli Sequence Type 648 and Genetic Context of blaNDM-5 in Australia. Genome Announc. 2015;3 doi: 10.1128/genomeA.00194-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rojas L.J., Hujer A.M., Rudin S.D., Wright M.S., Domitrovic T.N., Marshall S.H., Hujer K.M., Richter S.S., Cober E., Perez F., et al. NDM-5 and OXA-181 Beta-Lactamases, a Significant Threat Continues To Spread in the Americas. Antimicrob. Agents Chemother. 2017;61:e00454-17. doi: 10.1128/AAC.00454-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mohamed N.M., Zakaria A.S., Edward E.A. Genomic Characterization of International High-Risk Clone ST410 Escherichia coli Co-Harboring ESBL-Encoding Genes and bla(NDM-5) on IncFIA/IncFIB/IncFII/IncQ1 Multireplicon Plasmid and Carrying a Chromosome-Borne bla(CMY-2) from Egypt. Antibiotics. 2022;11:1031. doi: 10.3390/antibiotics11081031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun P., Xia W., Liu G., Huang X., Tang C., Liu C., Xu Y., Ni F., Mei Y., Pan S. Characterization Of bla (NDM-5)-Positive Escherichia coli Prevalent In A University Hospital In Eastern China. Infect. Drug Resist. 2019;12:3029–3038. doi: 10.2147/IDR.S225546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu M., Li X., Xie Y., Bi D., Sun J., Li J., Tai C., Deng Z., Ou H.Y. ICEberg 2.0: An updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 2019;47:D660–D665. doi: 10.1093/nar/gky1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zou D., Huang Y., Zhao X., Liu W., Dong D., Li H., Wang X., Huang S., Wei X., Yan X., et al. A novel New Delhi metallo-β-lactamase variant, NDM-14, isolated in a Chinese Hospital possesses increased enzymatic activity against carbapenems. Antimicrob. Agents Chemother. 2015;59:2450–2453. doi: 10.1128/AAC.05168-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.CLSI Performance Standards for Antimicrobial Susceptibility Testing. Approved Standard. CLSI Document M100. 2022. [(accessed on 1 November 2022)]. Available online: https://clsi.org/standards/products/microbiology/documents/m100/
- 57.Taylor D.E., De Grandis S.A., Karmali M.A., Fleming P.C. Transmissible plasmids from Campylobacter jejuni. Antimicrob. Agents Chemother. 1981;19:831–835. doi: 10.1128/AAC.19.5.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Overbeek R., Olson R., Pusch G.D., Olsen G.J., Davis J.J., Disz T., Edwards R.A., Gerdes S., Parrello B., Shukla M., et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kleinheinz K.A., Joensen K.G., Larsen M.V. Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage. 2014;4:e27943. doi: 10.4161/bact.27943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carattoli A., Zankari E., García-Fernández A., Voldby Larsen M., Lund O., Villa L., Møller Aarestrup F., Hasman H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.R Core Team R: A language and environment for statistical computing. 2014;1:201. [Google Scholar]
- 65.Bessonov K., Laing C., Robertson J., Yong I., Ziebell K., Gannon V.P.J., Nichani A., Arya G., Nash J.H.E., Christianson S. ECTyper: In silico Escherichia coli serotype and species prediction from raw and assembled whole-genome sequence data. Microb. Genom. 2021;7:000728. doi: 10.1099/mgen.0.000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Larsen M.V., Cosentino S., Rasmussen S., Friis C., Hasman H., Marvig R.L., Jelsbak L., Sicheritz-Pontén T., Ussery D.W., Aarestrup F.M., et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seemann T. Snippy: Rapid Haploid Variant Calling and Core Genome Alignment. [(accessed on 1 January 2022)]. Available online: https://github.com/tseemann/snippy.
- 68.Seemann T., Page A., Klotzl F. Snp-Dists: Pairwise SNP Distance Matrix from A FASTA Sequence Alignment. 2018. [(accessed on 1 January 2022)]. Available online: https://github.com/tseemann/snp-dists.
- 69.Treangen T.J., Ondov B.D., Koren S., Phillippy A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Letunic I., Bork P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen C., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., Xia R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 72.Alikhan N.F., Petty N.K., Ben Zakour N.L., Beatson S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kelly A.M., Mathema B., Larson E.L. Carbapenem-resistant Enterobacteriaceae in the community: A scoping review. Int. J. Antimicrob. Agents. 2017;50:127–134. doi: 10.1016/j.ijantimicag.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lipworth S., Vihta K.D., Davies T., Wright S., Tabirao M., Chau K., Vaughan A., Kavanagh J., Barker L., George S., et al. Molecular epidemiology and antimicrobial resistance phenotype of paediatric bloodstream infections caused by Gram-negative bacteria. Commun. Med. 2022;2:101. doi: 10.1038/s43856-022-00161-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu W., Zhao F., Chen J. Analysis of drug resistance genes of integrons in clinical isolates of Escherichia coli from elderly bloodstream infections. Cell. Mol. Biol. (Noisy-Le-Grand Fr.) 2022;68:67–72. doi: 10.14715/cmb/2022.68.6.11. [DOI] [PubMed] [Google Scholar]
- 76.Foglia F., Della Rocca M.T., Melardo C., Nastri B.M., Manfredini M., Montella F., De Filippis A., Finamore E., Galdiero M. Bloodstream infections and antibiotic resistance patterns: A six-year surveillance study from southern Italy. Pathog. Glob. Health. 2022:1–11. doi: 10.1080/20477724.2022.2129161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cuzon G., Bonnin R.A., Nordmann P. First identification of novel NDM carbapenemase, NDM-7, in Escherichia coli in France. PLoS ONE. 2013;8:e61322. doi: 10.1371/journal.pone.0061322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu H., Ma D., Liu B., Yang S., Lin Q., Yu R., Jia X., Niu S., Zhang Q., Huang S. Differences in the Distribution of Species, Carbapenemases, Sequence Types, Antimicrobial Heteroresistance and Mortality Rates Between Pediatric and Adult Carbapenemase-Producing Enterobacterales in Bloodstream Infections. Front. Med. 2022;9:827474. doi: 10.3389/fmed.2022.827474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dortet L., Cuzon G., Nordmann P. Dissemination of carbapenemase-producing Enterobacteriaceae in France, 2012. J. Antimicrob. Chemother. 2014;69:623–627. doi: 10.1093/jac/dkt433. [DOI] [PubMed] [Google Scholar]
- 80.Zheng W., Yue M., Zhang J., Ruan Z. Coexistence of two bla(CTX-M-14) genes in a bla(NDM-5)-carrying multidrug-resistant Escherichia coli strain recovered from a bloodstream infection in China. J. Glob. Antimicrob. Resist. 2021;26:11–14. doi: 10.1016/j.jgar.2021.05.002. [DOI] [PubMed] [Google Scholar]
- 81.Huang J., Ma S., Yu Q., Fu M., Shao L., Shan X., Li X. Whole genome sequence of an Escherichia coli ST410 isolate co-harbouring bla(NDM-5), bla(OXA-1), bla(CTX-M-15), bla(CMY-2), aac(3)-IIa and aac(6′)-Ib-cr genes isolated from a patient with bloodstream infection in China. J. Glob. Antimicrob. Resist. 2019;19:354–355. doi: 10.1016/j.jgar.2019.10.027. [DOI] [PubMed] [Google Scholar]
- 82.Bi R., Kong Z., Qian H., Jiang F., Kang H., Gu B., Ma P. High Prevalence of bla (NDM) Variants Among Carbapenem-Resistant Escherichia coli in Northern Jiangsu Province, China. Front Microbiol. 2018;9:2704. doi: 10.3389/fmicb.2018.02704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Park Y., Choi Q., Kwon G.C., Koo S.H. Emergence and transmission of New Delhi metallo-beta-lactamase-5-producing Escherichia coli Sequence Type 361 in a Tertiary Hospital in South Korea. J. Clin. Lab. Anal. 2020;34:e23041. doi: 10.1002/jcla.23041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tang B., Chang J., Cao L., Luo Q., Xu H., Lyu W., Qian M., Ji X., Zhang Q., Xia X., et al. Characterization of an NDM-5 carbapenemase-producing Escherichia coli ST156 isolate from a poultry farm in Zhejiang, China. BMC Microbiol. 2019;19:82. doi: 10.1186/s12866-019-1454-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bonnin R.A., Poirel L., Carattoli A., Nordmann P. Characterization of an IncFII plasmid encoding NDM-1 from Escherichia coli ST131. PLoS ONE. 2012;7:e34752. doi: 10.1371/journal.pone.0034752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Paskova V., Medvecky M., Skalova A., Chudejova K., Bitar I., Jakubu V., Bergerova T., Zemlickova H., Papagiannitsis C.C., Hrabak J. Characterization of NDM-Encoding Plasmids From Enterobacteriaceae Recovered From Czech Hospitals. Front. Microbiol. 2018;9:1549. doi: 10.3389/fmicb.2018.01549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang Y., Wang H., Li Y., Hou Y., Hao C. Drug susceptibility and molecular epidemiology of Escherichia coli in bloodstream infections in Shanxi, China. PeerJ. 2021;9:e12371. doi: 10.7717/peerj.12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Quan J., Zhao D., Liu L., Chen Y., Zhou J., Jiang Y., Du X., Zhou Z., Akova M., Yu Y. High prevalence of ESBL-producing Escherichia coli and Klebsiella pneumoniae in community-onset bloodstream infections in China. J. Antimicrob. Chemother. 2017;72:273–280. doi: 10.1093/jac/dkw372. [DOI] [PubMed] [Google Scholar]
- 89.Nicolas-Chanoine M.H., Blanco J., Leflon-Guibout V., Demarty R., Alonso M.P., Caniça M.M., Park Y.J., Lavigne J.P., Pitout J., Johnson J.R. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 2008;61:273–281. doi: 10.1093/jac/dkm464. [DOI] [PubMed] [Google Scholar]
- 90.Falgenhauer L., Waezsada S.E., Gwozdzinski K., Ghosh H., Doijad S., Bunk B., Spröer C., Imirzalioglu C., Seifert H., Irrgang A., et al. Chromosomal Locations of mcr-1 and bla CTX-M-15 in Fluoroquinolone-Resistant Escherichia coli ST410. Emerg. Infect. Dis. 2016;22:1689–1691. doi: 10.3201/eid2209.160692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gu J.N., Chen L., Weng X.B., Yang X.Y., Pan D.M. Clinical and Microbiological Characteristics of a Community-Acquired Carbapenem-Resistant Escherichia coli ST410 Isolate Harbouring blaNDM-5-Encoding IncX3-Type Plasmid From Blood. Front. Med. 2021;8:658058. doi: 10.3389/fmed.2021.658058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ma Z., Zeng Z., Liu J., Liu C., Pan Y., Zhang Y., Li Y. Emergence of IncHI2 Plasmid-Harboring blaNDM-5 from Porcine Escherichia coli Isolates in Guangdong, China. Pathogens. 2021;10:954. doi: 10.3390/pathogens10080954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xu L., Wang P., Cheng J., Qin S., Xie W. Characterization of a novel bla (NDM-5)-harboring IncFII plasmid and an mcr-1-bearing IncI2 plasmid in a single Escherichia coli ST167 clinical isolate. Infect. Drug Resist. 2019;12:511–519. doi: 10.2147/IDR.S192998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Villa L., García-Fernández A., Fortini D., Carattoli A. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 2010;65:2518–2529. doi: 10.1093/jac/dkq347. [DOI] [PubMed] [Google Scholar]
- 95.Liu Z., Xiao X., Li Y., Liu Y., Li R., Wang Z. Emergence of IncX3 Plasmid-Harboring bla (NDM-) (5) Dominated by Escherichia coli ST48 in a Goose Farm in Jiangsu, China. Front. Microbiol. 2019;10:2002. doi: 10.3389/fmicb.2019.02002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang F., Xie L., Wang X., Han L., Guo X., Ni Y., Qu H., Sun J. Further Spread of bla NDM-5 in Enterobacteriaceae via IncX3 Plasmids in Shanghai, China. Front. Microbiol. 2016;7:424. doi: 10.3389/fmicb.2016.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li X., Fu Y., Shen M., Huang D., Du X., Hu Q., Zhou Y., Wang D., Yu Y. Dissemination of bla(NDM-5) gene via an IncX3-type plasmid among non-clonal Escherichia coli in China. Antimicrob. Resist. Infect. Control. 2018;7:59. doi: 10.1186/s13756-018-0349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The assembled genome sequences of all E. coli strains from bloodstream infections in this study were deposited in the NBCI database under BioProject accession number PRJNA890582.