Abstract
A central theme in learning from image data is to develop appropriate representations for the specific task at hand. Thus, a practical challenge is to determine what features are appropriate for specific tasks. For example, in the study of gene expression patterns in Drosophila, texture features were particularly effective for determining the developmental stages from in situ hybridization images. Such image representation is however not suitable for controlled vocabulary term annotation. Here, we developed feature extraction methods to generate hierarchical representations for ISH images. Our approach is based on the deep convolutional neural networks that can act on image pixels directly. To make the extracted features generic, the models were trained using a natural image set with millions of labeled examples. These models were transferred to the ISH image domain. To account for the differences between the source and target domains, we proposed a partial transfer learning scheme in which only part of the source model is transferred. We employed multi-task learning method to fine-tune the pre-trained models with labeled ISH images. Results showed that feature representations computed by deep models based on transfer and multi-task learning significantly outperformed other methods for annotating gene expression patterns at different stage ranges.
Keywords: Deep learning, transfer learning, multi-task learning, image analysis, bioinformatics
1. INTRODUCTION
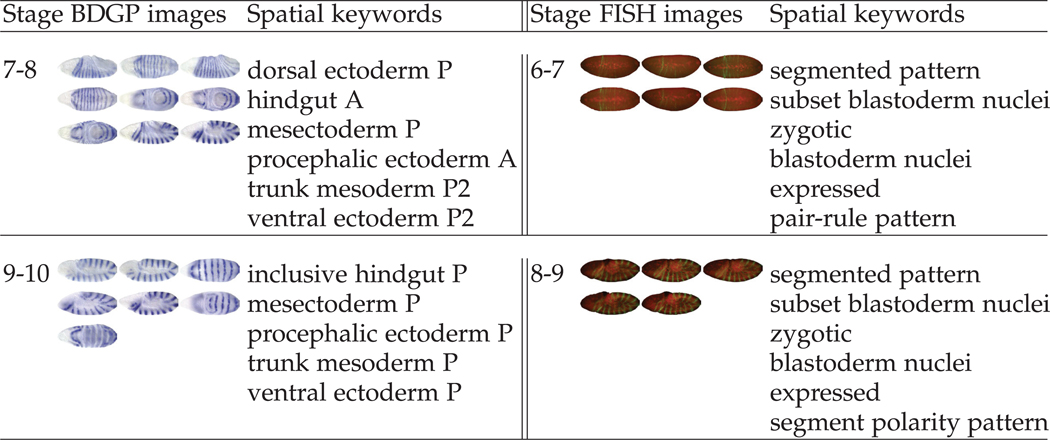
A General consensus in image-related research is that different recognition and learning tasks may require different image representations. Thus, a central challenge in learning from image data is to develop appropriate representations for the specific task at hand. Traditionally, a common practice is to hand-tune features for specific tasks, which is time-consuming and requires substantial domain knowledge. For example, in the study of gene expression patterns in Drosophila melanogaster, texture features based on wavelets, such as Gabor filters, were particularly effective for determining the developmental stages from in situ hybridization (ISH) images [29]. Such image representation, often referred to as “global visual features”, is not suitable for controlled vocabulary (CV) term annotation because each CV term is often associated with only a part of an image, thereby requiring an image representation of local visual features [11]. Examples of gene expression patterns and the associated CV terms are showed in Figure 1. Current state-of-the-art systems for CV term annotation first extracted local patches of an image and computed local features which are invariant to certain geometric transformations (e.g., scaling and translation). Each image was then represented as a bag of “visual words”, known as the “bag-of-words” representation [10], or a set of “sparse codes”, known as the “sparse coding” representation [12], [24], [30].
Fig. 1.
Gene expression patterns and the associated temporal stages and body part keywords in the BDGP [26] (left) and Fly-FISH [17] (right) databases for the gene engrailed in two stage-ranges.
In addition to being problem-dependent, a common property of traditional feature extraction methods is that they are “shallow”, because only one or two levels of feature extraction was applied, and the parameters for computing features are usually not trained using supervised algorithms. Given the complexity of patterns captured by biological images, these shallow models of feature extraction may not be sufficient. Therefore, it is desirable to develop a multi-layer feature extractor [7], [32], [33], alleviating the tedious process of manual feature engineering and enhancing the representation power.
In this work, we proposed to employ the deep learning methods to generate representations of ISH images. Deep learning models are a class of multi-level systems that can act on the raw input images directly to compute increasingly high-level representations. One particular type of deep learning models that have achieved practical success is the deep convolutional neural networks (CNNs) [16]. These models stack many layers of trainable convolutional filters and pooling operations on top of each other, thereby computing increasingly abstract representations of the inputs. Deep CNNs trained with millions of labeled natural images using supervised learning algorithms have led to dramatic performance improvement in natural image recognition and detection tasks [6], [13], [23].
However, learning a deep CNN is usually associated with the estimation of millions of parameters, and this requires a large number of labeled image samples. This bottleneck currently prevents the application of CNNs to many biological problems due to the limited amount of labeled training data. To overcome this difficulty, we proposed to develop generic and problem-independent feature extraction methods, which involves applying previously obtained knowledge to solve different but related problems. This is made possible by the initial success of transferring features among different natural image data sets [3], [22], [31]. These studies trained the models on the ImageNet data set that contains millions of labeled natural images with thousands of categories. The learned models were then applied to other image data sets for feature extraction, since layers of the deep models are expected to capture the intrinsic characteristics of visual objects.
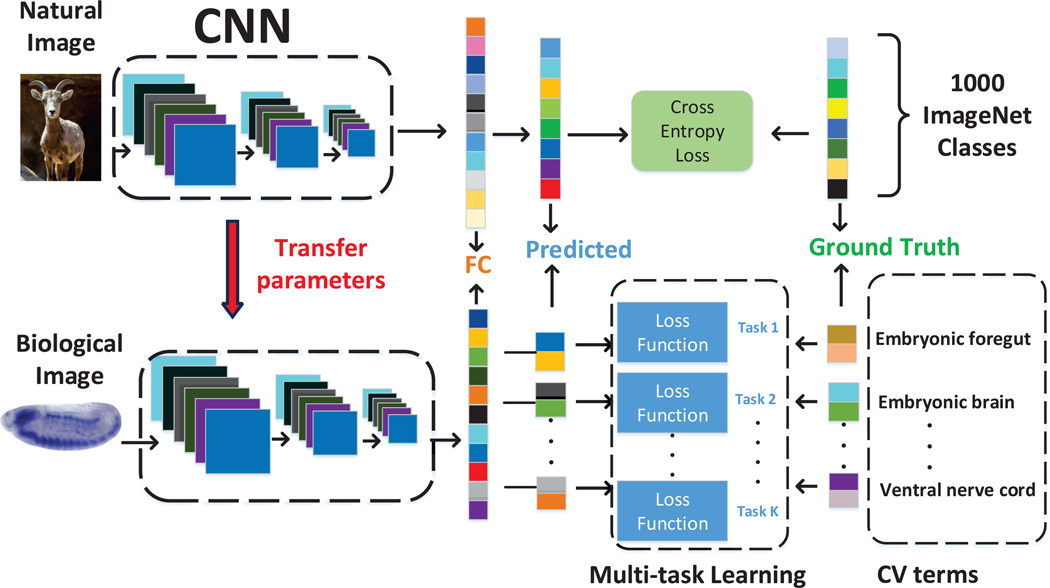
In this article, we explored whether the transfer learning property of CNNs can be generalized to compute features for biological images. We proposed to transfer knowledge from natural images by training CNNs on the ImageNet data set. We then proposed to fine-tune the trained model with labeled ISH images, and resumed training from already learned weights using multi-task learning schemes. To take this transfer learning idea one step further, we proposed another approach with partial transfer of parameters from pre-trained VGG model to be fine-tuned on the labeled ISH images. Specifically, we truncated the pre-trained VGG model at some intermediate layer followed by one max pooling and two fully connected layers to obtain the new CNN model. The three models were then all used as a feature extractors to compute image features from Drosophila gene expression pattern images. The resulting features were subsequently used to train and validate our machine learning method for annotating gene expression patterns. The overall pipeline of this work is given in Figure 2.
Fig. 2.
Pipeline of deep models for transfer learning and multi-task learning. The network was trained on the ImageNet data containing millions of labeled natural images with thousands of categories (top row). The pre-trained parameters are then transferred to the target domain of biological images. We first directly used the pre-trained model to extract features from Drosophila gene expression pattern images. We then fne-tuned the trained model with labeled ISH images. We then employed the fine-tuned model to extract features to capture CV term-specific discriminative information (bottom row).
Experimental results showed that our approach of using CNNs outperformed the sparse coding methods [24] for annotating gene expression patterns at different stage ranges. In addition, our results indicated that the transfer and fine-tuning of knowledge by CNNs from natural images is very beneficial for producing high-level representations of biological images. Furthermore, we showed that the intermediate layers of CNNs produced the best gene expression pattern representations. This is because the early layers encode very primitive image features that are not enough to capture gene expression patterns. Meanwhile, the later layers captured features that are specific to the training natural image set, and these features may not be relevant to gene expression pattern images. Our result also showed that partial transfer of parameters led to improved performance, as compared to the complete transfer scheme.
2. DEEP MODELS FOR TRANSFER LEARNING AND FEATURE EXTRACTION
Deep learning models are a class of methods that are capable of learning hierarchy of features from raw input images. Convolutional neural networks (CNNs) are a class of deep learning models that were designed to simulate the visual signal processing in central nervous systems [1], [13], [16]. These models usually consist of alternating combination of convolutional layers with trainable filters and local neighborhood pooling layers, resulting in a complex hierarchical representations of the inputs. CNNs are intrinsically capable of capturing highly nonlinear mappings between inputs and outputs. When trained with millions of labeled images, they have achieved superior performance on many image-related tasks [13], [16], [23].
A key challenge in applying CNNs to biological problems is that the available labeled training samples are very limited. To overcome this difficulty and develop a universal representation for biological image informatics, we proposed to employ transfer learning to transfer knowledge from labeled image data that are problem-independent. The idea of transfer learning is to improve the performance of a task by applying knowledge acquired from different but related task with a lot of training samples. This approach of transfer learning has already yielded superior performance on natural image recognition tasks [3], [19], [22], [31].
In this work, we explored whether this transfer learning property of CNNs can be generalized to biological images. Specifically, the CNN model was trained on the ImageNet data containing millions of labeled natural images with thousands of categories and used directly as feature extractors to compute representations for ISH images. Although the pre-trained model is obtained from a different data source, the internal layers in the model act as generic representations of different levels of abstraction, which vary from simple object components like edges or corners to complicated structures like shapes. This property of the pre-trained model guarantees the feasibility of transfer learning in our study of gene expression patterns in Drosophila ISH images. In this work, we applied the pre-trained VGG model [23] that was trained on the ImageNet data to perform several computer vision tasks, such as localization, detection and classification. There are two pre-trained models in [23], which are “16” and “19” weight layers models. Since these two models generated similar performance on our ISH images, we used the “16” weight layers model in our experiment. The VGG architecture contains 36 layers. This network includes convolutional layers with fixed filter sizes and different numbers of feature maps. It also applied rectified non-linearity, max pooling to different layers.
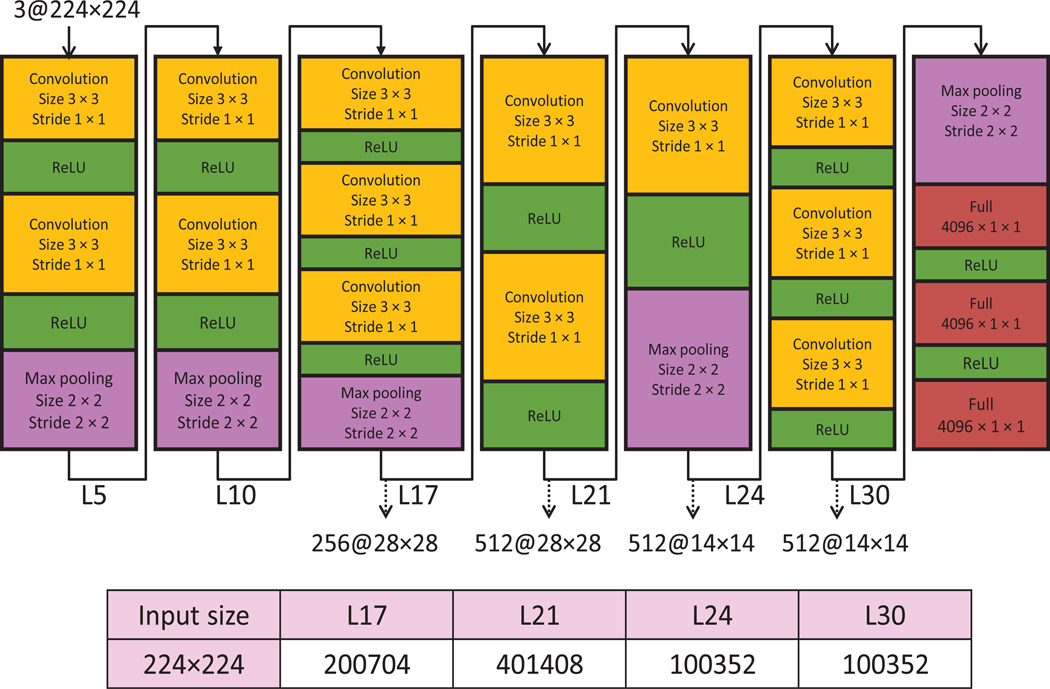
More details on various layers in the VGG weight layer model are given in Figure 3. Since the output feature representations of layers before the third max pooling layer involve larger feature vectors, we used each Drosophila ISH image as input to the VGG model and extracted features from layers 17, 21, 24, and 30 to reduce the computational cost. We then flattened all the feature maps and concatenated them into a single feature vector. For example, the number of feature maps in layer 21 is 512, and the corresponding size of feature maps is 28 × 28. Thus, the corresponding size of feature vector for this layer is 401,408.
Fig. 3.
Detailed architecture of the VGG model. “Convolution”, “Max pooling” and “ReLU” denote convolutional layer, max pooling layer and rectified linear unit function layer, respectively. This model consists of 36 layers. We extracted features from layers 71, 21, 24, and 30.
3. DEEP MODELS FOR PARTIAL PARAMETER TRANSFER
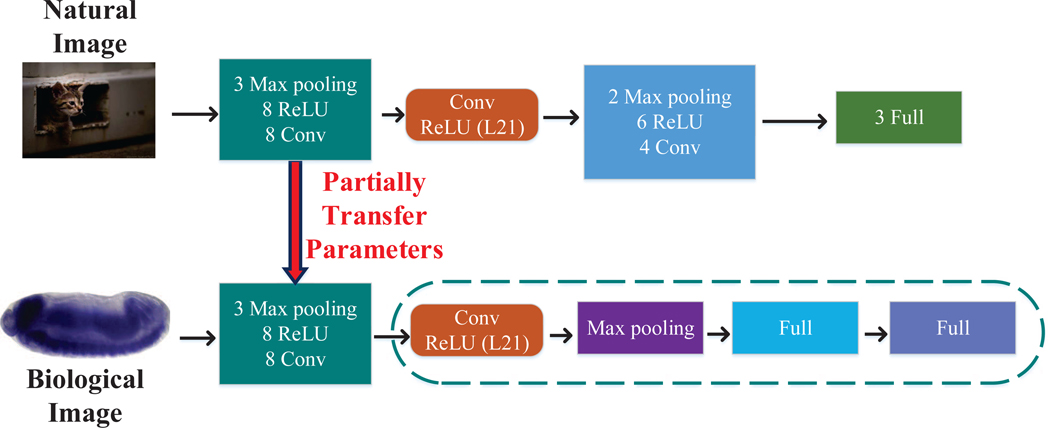
To account for the differences between natural and biological images, we proposed a new transfer learning scheme, known as partial parameter transfer, to only transfer part of the parameters learned from natural images to biological images. To be specific, we started from a pre-trained VGG model, and then truncated this VGG model at some intermediate layer. We then stacked one max pooling and two fully connected layers to obtain the new CNN model. The multi-task learning strategy was then used to fine-tune the modified CNN model from labeled ISH images. We first identified the most discriminative intermediate layer for gene expression annotation as a pivot layer. Then we kept the layers below this pivot layer, and added max pooling and fully connected layers, where the parameters are optimized during the fine-tuning stage using multi-task learning [25]. The pipeline of our method is illustrated in Figure 4.
Fig. 4.
Illustration of the partial transfer learning scheme. “Conv”, “Max pooling”, “ReLU” and “Full” denote the convolutional layer, max pooling layer, rectified linear unit layer and fully connected layer, respectively. The network was trained on the ImageNet data containing millions of labeled natural images with thousands of categories (left). The pre-trained parameters are partially transferred to the target domain of biological images. In particular, we truncated the pre-trained CNN model at layer 21, and attached one max pooling and two fully connected layers to obtain the new CNN model. Then we used multi-task learning approach to fine-tune the modified CNN model using labeled ISH images (right).
Note that this method is different from the fine-tuning method with multi-task learning we proposed before. We previously used the whole set of pre-trained architecture and parameters to obtain representations of ISH images. However, in this new scheme, we only retained several lower layers, which are mainly representative of local information. The intuition behind this method is that the higher layers of the pre-trained CNN model are usually more adjusted to label information of the training natural images, and these layers may not be informative enough for reflecting label information of gene expression pattern images. The proposed model with partial transfer learning could not only capture the common characteristics of images by pre-training, but also be representative for ISH images specifically from fine-tuning.
4. DEEP MODELS FOR MULTI-TASK LEARNING
In addition to the transfer learning scheme described above, we also proposed a multi-task learning strategy in which a CNN is first trained in the supervised mode using the ImageNet data and then fine-tuned on the labeled ISH Drosophila images. This strategy is different from the pre-trained model we used above. To be specific, the pre-trained model is designed to recognize objects in natural images while we studied the CV term annotation of Drosophila images instead. Although the leveraged knowledge from the source task could reflect some common characteristics shared in these two types of images such as corners or edges, extra efforts are also needed to capture the specific properties of ISH images. The Drosophila gene expression pattern images are organized into groups, and multiple CV term annotations are assigned to multiple images in the same group. This multi-image multi-label nature poses significant challenges to traditional image annotation methodologies. This is partially due to the fact that there are ambiguous multiple-to-multiple relationships between images and CV term annotations, since each group of images are associated with multiple CV term annotations.
In this paper, we proposed to use multi-task learning strategy [4], [8], [9] to overcome the above difficulty. To be specific, we first employed a CNN model that is pre-trained on natural images to initialize the parameters of a deep network. Then, we fine-tuned this network using multiple annotation term prediction tasks to obtain CV term-specific discriminative representation. The pipeline of our method is illustrated in Figure 2. We have a single pre-trained network with the same inputs but with multiple outputs, each of which corresponds to a term annotation task. These outputs are fully connected to a hidden layer that they share. Because all outputs share a common layer, the internal representations learned by one task could be used by other tasks. Note that the back-propagation is done in parallel on these outputs in the network. For each task, we used its individual loss function to measure the difference between outputs and the ground truth. In particular, we are given a training set of k tasks , , where denotes the i-th training sample, m denotes the total number of training samples. Note that we used the same groups of samples for different tasks, which is a simplified version of traditional multi-task learning. The output label denotes the CV term annotation status of training sample, which is binary with the form:
To quantitatively measure the difference between the predicted annotation results and ground truth from human experts, we used a loss function in the following form:
where
and denotes the ground truth label matrix over different tasks, and is the output matrix of our network through feedforward propagation. Note that denotes the network output before the softmax activation function. This loss function is a special case of the cross entropy loss function by using sigmoid function to induce probability representation [2]. Note that our multi-task loss function is the summation of multiple loss functions, and all of them are optimized simultaneously during training.
5. BIOLOGICAL IMAGE ANALYSIS
The Drosophila melanogaster has been widely used as a model organism for the study of genetics and developmental biology. To determine the gene expression patterns during Drosophila embryogenesis, the Berkeley Drosophila Genome Project (BDGP) used high throughput RNA in situ hybridization (ISH) to generate a systematic gene expression image database [26], [27]. In BDGP, each image captures the gene expression patterns of a single gene in an embryo. Each gene expression image is annotated with a collection of anatomical and developmental ontology terms using a CV term annotation to identify the characteristic structures in embryogenesis. This annotation work is now mainly carried out manually by human experts, which makes the whole process time-consuming and costly. In addition, the number of available images is now increasing rapidly. Therefore, it is desirable to design an automatic and systematic annotation approach to increase the efficiency and accelerate biological discovery [5], [11], [14], [15], [20], [21], [34].
Prior studies have employed machine learning and computer vision techniques to automate this task. Due to the effects of stochastic process in development, every embryo develops differently. In addition, the shape and position of the same embryonic part may vary from image to image. Thus, how to handle local distortions on the images is crucial for building robust annotation methods. The seminal work in [35] employed the wavelet-embryo features by using the wavelet transformation to project the original pixel-based embryonic images onto a new feature domain. In subsequent work, local patches were first extracted from an image and local features which are invariant to certain geometric transformations (e.g., scaling and translation) were then computed from each patch. Each image was then represented as a bag of “visual words”, known as the “bag-of-words” representation [10], or a set of “sparse codes”, known as the “sparse coding” representation [24], [30]. All prior methods used handcrafted local features combined with high-level methods, such as the bag-of-words or sparse coding schemes, to obtain image representations. These methods can be viewed as two-layer feature extractors. In this work, we proposed to employ the deep CNNs as a multi-layer feature extractor to generate image representations for CV term annotation.
We showed here that a universal feature extractor trained on problem-independent data set can be used to compute feature representations for CV term annotation. Furthermore, the model trained on problem-independent data set, such as the ImageNet data, can be fine-tuned on labeled data from specific domains using the error back propagation algorithm. This will ensure that the knowledge transferred from problem-independent images is adapted and tuned to capture domain-specific features in biological images. Since generating manually annotated biological images is both time-consuming and costly, the transfer of knowledge from other domains, such as the natural image world, is essential in achieving competitive performance.
6. EXPERIMENTS
6.1. Experimental setup
In this study, we used the Drosophila ISH gene expression pattern images provided by the FlyExpress database [15], [28], which contains genome-wide, standardized images from multiple sources, including the Berkeley Drosophila Genome Project (BDGP). For each Drosophila embryo, a set of high-resolution, two-dimensional image series were taken from different views (lateral, dorsal, and lateral-dorsal and other intermediate views). These images were then subsequently standardized semi-manually. In this study, we focused on the lateral-view images only, since most of images in FlyExpress are in lateral view.
In the FlyExpress database, the embryogenesis of Drosophila has been divided into six discrete stage ranges (stages 1–3, 4–6, 7–8, 9–10, 11–12, and 13–17). We used those images in the later 5 stage ranges in the CV term annotation, since only a very small number of keywords were used in the first stage range. One characteristic of these images is that a group of images from the same stage and same gene are assigned with the same set of keywords. Prior work in [24] has shown that image-level annotation outperformed group-level annotation using the BDGP images. In this work, we focused on the image-level annotation only and used the same top 10 keywords that are most frequently annotated for each stage range as in [24]. The statistics of the numbers of images and most frequent 10 annotation terms for each stage range are given in Table 1.
TABLE 1.
Statistics of the data set used in this work. The table shows the total number of images for each stage range and the numbers of positive samples for each term.
| Stages | Number of images | # of positive samples for each term | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. 1 | No. 2 | No. 3 | No. 4 | No. 5 | No. 6 | No. 7 | No. 8 | No. 9 | No. 10 | ||
| 4–6 | 4173 | 953 | 438 | 1631 | 1270 | 1383 | 1351 | 351 | 568 | 582 | 500 |
| 7–8 | 1953 | 782 | 741 | 748 | 723 | 753 | 668 | 510 | 340 | 165 | 209 |
| 9–10 | 2153 | 899 | 787 | 778 | 744 | 694 | 496 | 559 | 452 | 350 | 264 |
| 11–12 | 7441 | 2945 | 2721 | 2056 | 1932 | 1847 | 1741 | 1400 | 1129 | 767 | 1152 |
| 13–17 | 7564 | 2572 | 2169 | 2062 | 1753 | 1840 | 1699 | 1273 | 1261 | 891 | 1061 |
For CV term annotation, our image data set is highly imbalanced with much more negative samples than positive ones. For example, there are 7564 images in stages 13–17, but only 891 of them are annotated the term “dorsal prothoracic pharyngeal muscle”. The commonly-used classification algorithms might not work well for our specific problem, because they usually aimed to minimizing the overall error rate without paying special attention to the positive class. Prior work in [24] has shown that using under-sampling with ensemble learning could produce better prediction performance. In particular, we selectively under-sampled the majority class to obtain the same number of samples as the minority class and built a model for each sampling. This process was performed many times for each keyword to obtain a robust prediction. Following [24], we employed classifier ensembles built on biased samples to train robust models for annotation. In order to further improve the performance, we produced the final prediction by using majority voting, since this sample scheme is one of the widely used methods for fusion of multiple classifiers. For comparison purpose, we also implemented the existing sparse coding image representation method studied in [24]. The annotation performance was measured using accuracy, specificity, sensitivity and area under the ROC curve (AUC) for CV term annotation. For all of these measures, a higher value indicates better annotation performance. All classifiers used in this work are the -norm regularized logistic regression.
6.2. Comparison of features extracted from different layers
The deep learning model consists of multiple layer of feature maps for representing the input images. With this hierarchical representation, a natural question is which layer has the most discriminative power to capture the characteristics of input images. When such networks were trained on natural image data set such as the ImageNet data, the features computed in lower layers usually correspond to local features of objects such as edges, corners or edge/color conjunctions. In contrast, the features encoded at higher layers mainly represent class-specific information of the training data. Therefore, for the task of natural object recognition, the features extracted from higher layers usually yielded better discriminative power [31].
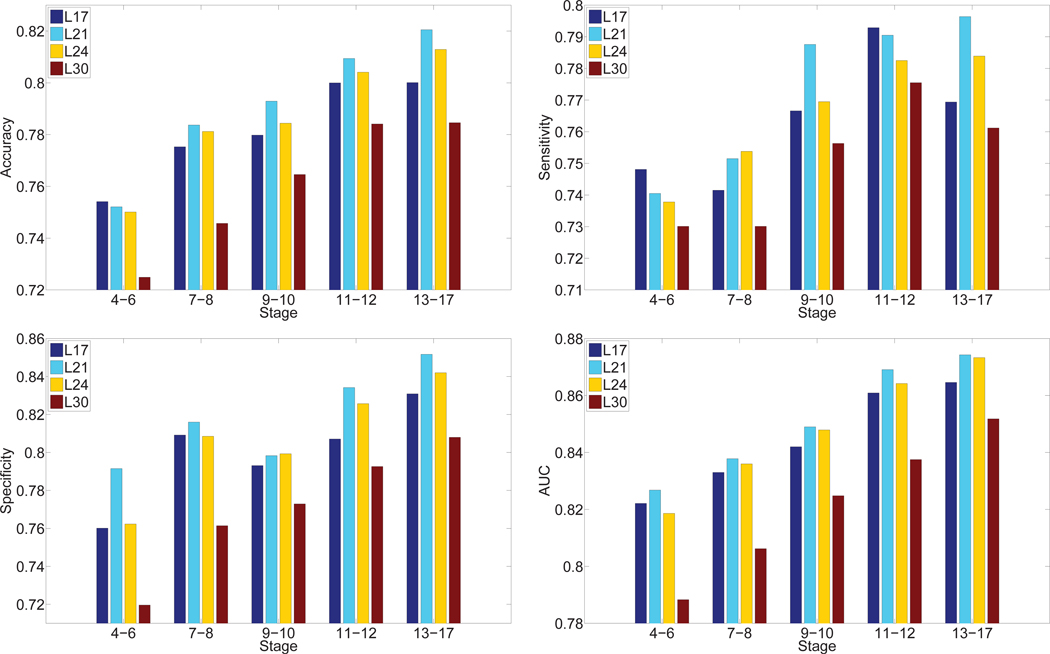
In order to identify the most discriminative features for the gene expression pattern annotation tasks, we compared the features extracted from various layers of the VGG network. Specifically, we used the ISH images as inputs to the pre-trained VGG network and extracted features from layers 17, 21, 24, and 30 for each ISH image. These features were used for the annotation tasks, and the results are given in Figure 5. We can observe that for all stage ranges, layer 21 features outperformed other features in terms of overall performance. Specifically, the discriminative power increased from layer 17 to layer 21, and then dropped afterwards as the depth of network increased. This indicates that gene expression features are best represented in the intermediate layers of CNN that was trained on natural image data set. One reasonable explanation about this observation is the lower layers compute very primitive image features that are not enough to capture gene expression patterns. Meanwhile, the higher layers captured features that are specific to the training natural image set, and these features may not be relevant for gene expression pattern images.
Fig. 5.
Comparison of annotation performance achieved by features extracted from different layers of deep models for transfer learning over five stage ranges. “Lx” denotes the hidden layer from which the features were extracted.
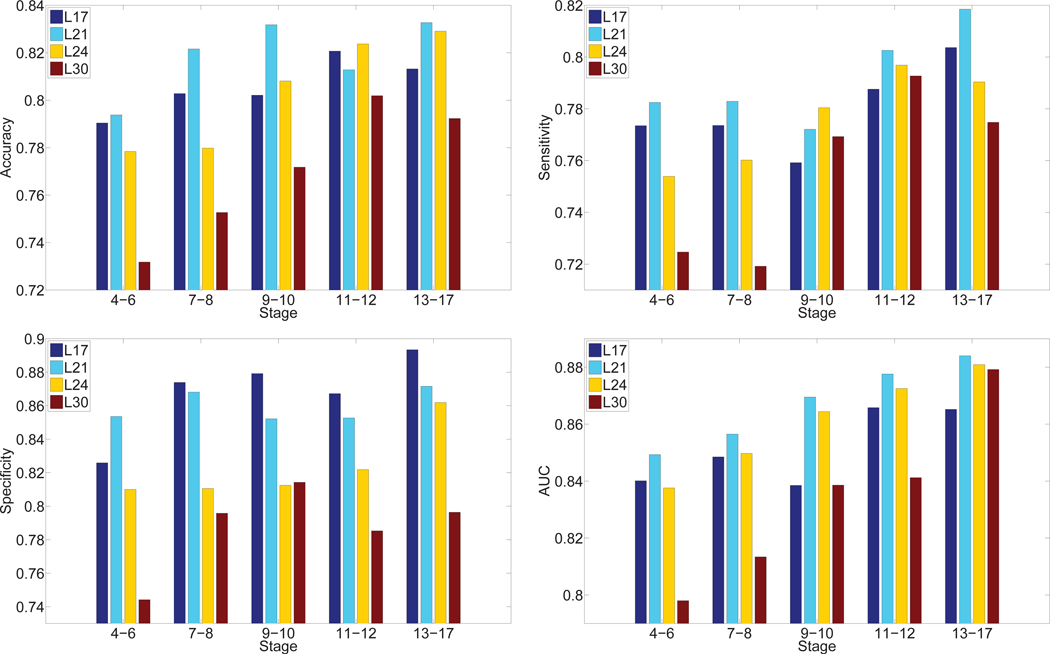
Then we proposed to use multi-task learning strategy to fine-tune the pre-trained network with labeled ISH images. In order to show the gains through fine-tuning on pre-trained model, we extracted features from the same hidden layers that are used for the pre-trained model. We reported the predictive performance achieved by features of different layers in the proposed fine-tuned model in Figure 6. It can be observed from the results that the predictive performance was generally higher on middle layers in the deep architecture. In particular, layer 21 outperformed other layers significantly. This result is consistent with the observation found on the pre-trained model.
Fig. 6.
Comparison of annotation performance achieved by features extracted from different layers of the deep models for multi-task learning over five stage ranges. “Lx” denotes the hidden layer from which the features were extracted.
6.3. Comparison with prior methods
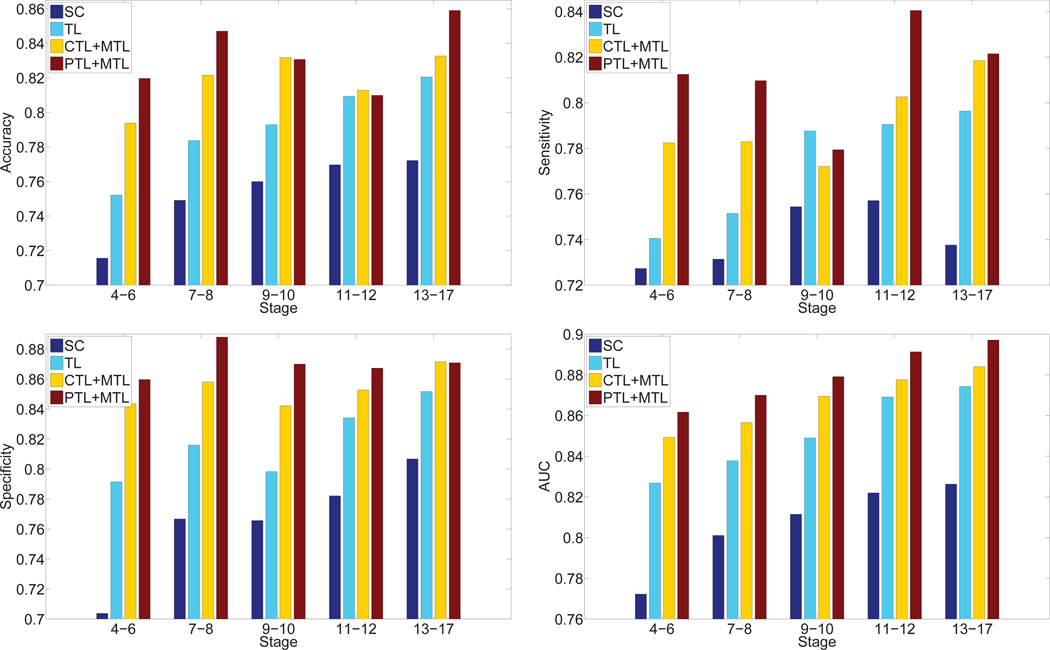
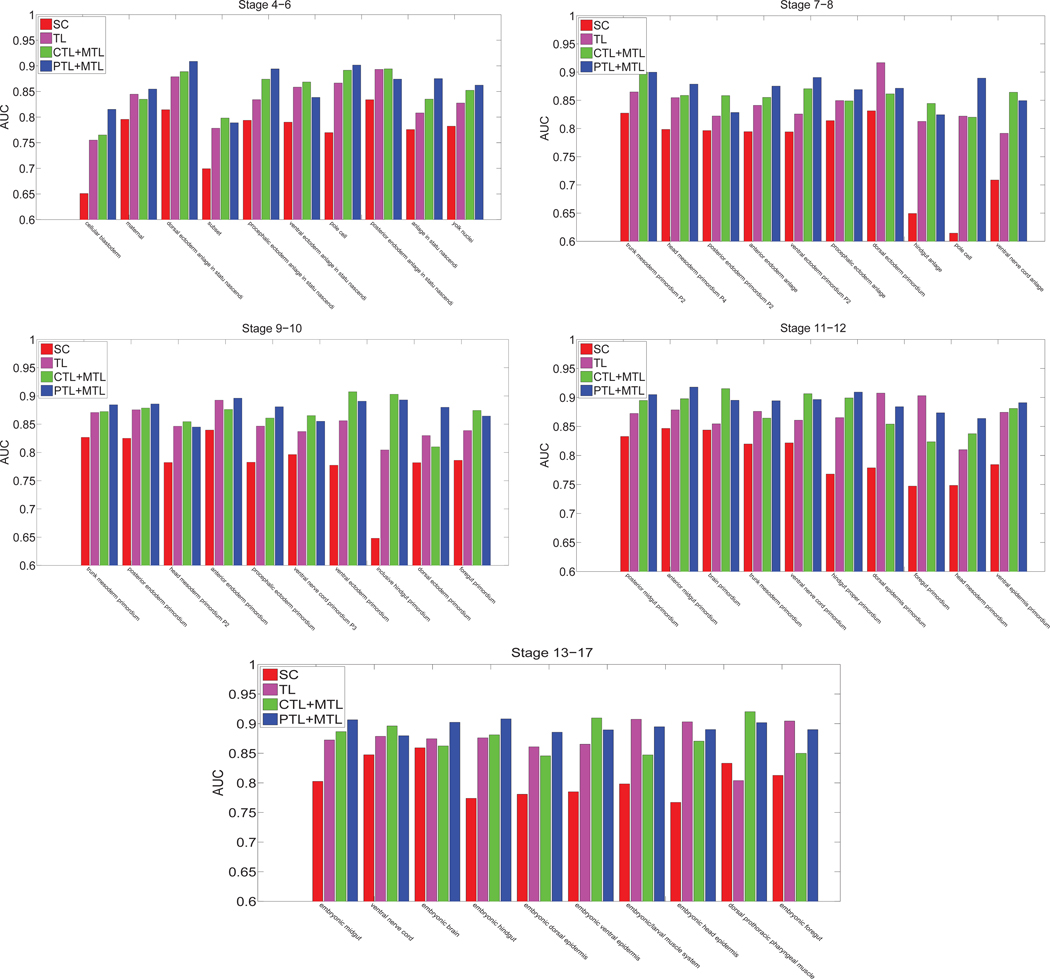
We also compared the performance achieved by different methods including sparse coding, transfer learning model and multi-task learning. These results demonstrated that our deep models with multi-task learning were able to accurately annotate gene expression images over all embryogenesis stage ranges. To compare our generic features with the domain-specific features used in [24], we compared the annotation performance of our deep learning features with that achieved by the domain-specific sparse coding features [24]. For the sparse coding method, they first extracted a sequence of patches from each image, and applied the scale invariant feature transform (SIFT) descriptor [18] to represent each patch. They constructed the codebook based on these SIFT feature vectors by applying the k-means algorithm. The cluster centers were treated as visual words. They also set the number of visual words to 2000 as in [30]. They then employed lasso type regularization to obtain different weights of multiple visual words in the codebook for each SIFT feature vector. The average pooling function was used to summarize the final representations of images. The regularization parameter λ was tuned through cross-validation on a subset of images. Deep learning models include transfer learning, multi-task learning combined with complete transfer learning or partial transfer learning. In this experiment, we only considered the features extracted from layer 21 since they yielded the best performance among different layers. For the multi-task learning model with partial transfer learning, we truncated the pre-trained CNN model at layer 21, and immediately stacked one max pooling and two fully connected layers to obtain the new CNN model. Then we used multi-task learning approach to fine-tune the modified CNN model from labeled ISH images. The performance of these four types of features averaged over all terms is given in Figure 7 and Table 2. We can observe that the deep models for multi-task learning features outperformed the sparse coding features and transfer learning features consistently and significantly in all cases. To examine the performance differences on individual anatomical terms, we showed the AUC values on each term in Figure 8 for different stage ranges. We can observe that our features extracted from layer 21 of the VGG networks for transfer learning and multi-task learning outperformed the sparse coding features over all stage ranges for all terms consistently. These results demonstrated that our generic features of deep models were better at representing gene expression pattern images than the problem-specific features based on sparse coding. In addition, we observed that partial transfer of parameters from models trained on natural images led to better performance than the complete transfer scheme. This showed that our new partial transfer learning method is effective in transferring knowledge from natural images to biological images.
Fig. 7.
Performance comparison of different methods. “SC” and “TL” denote sparse coding and transfer learning, respectively. “CTL + MTL” and “PTL + MTL” denote the performance achieved by complete and partial transfer learning, respectively, with multi-task learning models. We only consider the features extracted from layer 21 of these two deep models.
TABLE 2.
Performance comparison in terms of accuracy, sensitivity, specificity, and AUC achieved by CNN models and sparse coding features for all stage ranges. “PTL+MTL” and “CTL+MTL” denote the features extracted from layer 21 of the deep models for multi-task learning with complete and partial transfer learning, respectively. “SC” and “TL” denote the performance of the sparse coding and transfer learning features.
| Measures | Methods | Stage 4–6 | Stage 7–8 | Stage 9–10 | Stage 11–12 | Stage 13–17 |
|---|---|---|---|---|---|---|
| Accuracy | PTL+MTL | 0.8197±0.0279 | 0.8471±0.0225 | 0.8307±0.0291 | 0.8099±0.0318 | 0.8591±0.0301 |
| CTL+MTL | 0.7938±0.0381 | 0.8216±0.0231 | 0.8318±0.0216 | 0.8128±0.0325 | 0.8327±0.0256 | |
| TL | 0.7521±0.0326 | 0.7837±0.0269 | 0.7929±0.0231 | 0.8094±0.0331 | 0.8205±0.0304 | |
| SC | 0.7217±0.0352 | 0.7401±0.0351 | 0.7549±0.0303 | 0.7659±0.0326 | 0.7681±0.0231 | |
| Sensitivity | PTL+MTL | 0.8104±0.0391 | 0.8014±0.0317 | 0.7794±0.0327 | 0.8312±0.0297 | 0.8207±0.0331 |
| CTL+MTL | 0.7825±0.0372 | 0.7829±0.0368 | 0.7721±0.0412 | 0.8026±0.0401 | 0.8185±0.0259 | |
| TL | 0.7405±0.0293 | 0.7515±0.0342 | 0.7876±0.0401 | 0.7905±0.0389 | 0.7964±0.0317 | |
| SC | 0.7321±0.0408 | 0.7190±0.0331 | 0.7468±0.0298 | 0.7576±0.0329 | 0.7328±0.0235 | |
| Specificity | PTL+MTL | 0.8591±0.0291 | 0.8779±0.0206 | 0.8617±0.0318 | 0.8673±0.0332 | 0.8709±0.0317 |
| CTL + MTL | 0.8436±0.0376 | 0.8581±0.0380 | 0.8422±0.0284 | 0.8527±0.0252 | 0.8716±0.0256 | |
| TL | 0.7915±0.0247 | 0.8160±0.0316 | 0.7983±0.0315 | 0.8342±0.0237 | 0.8517±0.0306 | |
| SC | 0.7140±0.0389 | 0.7605±0.0392 | 0.7629±0.0298 | 0.7749±0.0329 | 0.8005±0.0298 | |
| AUC | PTL+MTL | 0.8607±0.0415 | 0.8671±0.0341 | 0.8736±0.0302 | 0.8913±0.0246 | 0.8972±0.0231 |
| CTL + MTL | 0.8493±0.0427 | 0.8565±0.0279 | 0.8695±0.0276 | 0.8776±0.0291 | 0.8824±0.0197 | |
| TL | 0.8344±0.0439 | 0.8401±0.0346 | 0.8508±0.0257 | 0.8702±0.0271 | 0.8746±0.0299 | |
| SC | 0.7687±0.0432 | 0.7834±0.0358 | 0.7921±0.0294 | 0.8061±0.0342 | 0.8105±0.0280 |
Fig. 8.
Performance comparison of different methods for all stage ranges. “SC”, ‘TL”, “CTL + MTL” and “PTL + MTL” denote sparse coding, transfer learning, complete transfer learning and partial transfer learning with multi-task learning models, respectively.
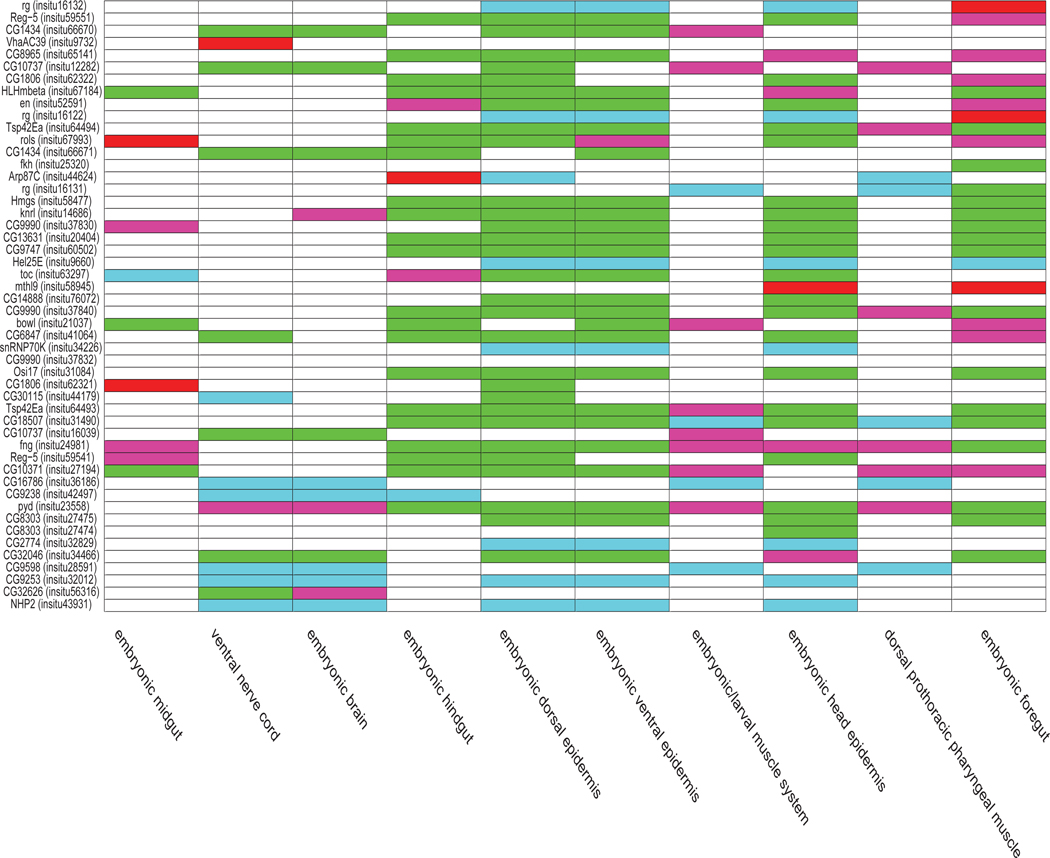
In Figure 9, we provided a term-by-term and image-by-image comparison between the results of the deep model for multi-task learning with partially transfer parameters and the sparse coding features for the 10 terms in stages 13–17. The x-axis corresponds to the 10 terms. The y-axis corresponds to a subset of 50 images in stages 13–17 with the largest numbers of annotated terms. Overall, it is clear that the total number of green and blue entries is much more than the number of red and pink entries, indicating that, among all predictions disagreed by these two methods, the predictions by the multi-task learning features were correct most of the time.
Fig. 9.
Comparison of prediction results between the deep models for multi-task learning with partial transfer learning and the sparse coding features for the 10 terms in stages 13–17. The x-axis shows the 10 terms. The y-axis corresponds to a subset of 50 images in stages 13–17 with the largest numbers of annotated terms. The gene names and the FlyExpress image IDs in parentheses are displayed. The prediction results of different methods compared with the ground truth are distinguished by different colors. The white entries correspond to predictions agreed upon by these two methods, while non-white entries were used to denote different types of disagreements. Specifically, the green and blue entries correspond to correct predictions by the multi-task learning features but incorrect predictions by the sparse coding features. Green and blue indicate positive and negative samples, respectively, in the ground truth. Similarly, the red and pink entries correspond to incorrect predictions by the multi-task learning features but correct predictions by the sparse coding features. Red and pink indicate positive and negative samples, respectively, in the ground truth.
7. CONCLUSIONS AND FUTURE WORK
In this work, we proposed to employ the deep convolutional neural networks as a multi-layer feature extractor to generate generic representations for ISH images. We used the deep convolutional neural network trained on large natural image set as feature extractors for ISH images. We first directly used the model trained on natural images as feature extractors. We then employed multi-task classification methods to fine-tune the pre-trained and modified model with labeled ISH images. Although the number of annotated ISH images is small, it nevertheless improved the pre-trained model. We compared the performance of our generic approach with the problem-specific methods. Results showed that our proposed approach significantly outperformed prior methods on ISH image annotation. We also showed that the intermediate layers of deep models produced the best gene expression pattern representations.
In the current study, we focus on using deep models for CV annotation. There are many other biological image analysis tasks that require appropriate image representations such as developmental stage prediction. We will consider broader applications in the future. In this work, we considered a simplified version of the problem in which each term is associated with all images in the same group. We will extend our model to incorporate the image group information in the future.
ACKNOWLEDGMENT
We would like to acknowledge support for this project from the National Science Foundation (NSF grant IIS-0953662, DBI-1147134 and DBI-1350258) and NIH (R01 LM010730, HG002516-09). We also gratefully acknowledge the support of NVIDIA Corporation with the donation of the Tesla K40 GPU used for this research.
Biography

Wenlu Zhang received the BS degree in Computer Science from Information Engineering University, Zhengzhou, China, in 2008, the MS degree in Computer Science from City College of New York, New York, NY, in 2010. Currently, she is a PhD candidate of Computer Science at Old Dominion University. Her research interests include machine learning, data mining, and bioinformatics

Rongjian Li received the BS and MS degrees in applied mathematics from Xi’an Jiaotong University, Xi’an, China, in 2007 and 2012, respectively. Currently, he is working towards the PhD degree in the Department of Computer Science at Old Dominion University. His research interests include machine learning, data mining, and bioinformatics.

Tao Zeng received the MS degree in Neuroscience from Chinese Academy of Sciences and MS degree in Computer Science from Old Dominion University, in 2008 and 2014, respectively. He is currently pursuing the PhD degree in Computer Science at Washington State University. His interests include biomedical image segmentation, data mining and deep learning.

Qian Sun is a software engineer at Google. She obtained her PhD degree in Computer Science from Arizona State University in 2015. She received her BS degree in Electrical Engineering and Automation from Nanjing University of Aeronautics and Astronautics, China, in 2008. Her research areas include data mining, machine learning with the applications in bioinformatics.

Sudhir Kumar received his bachelors degree in electrical and electronics engineering and masters degree in biology from the Birla Institute of Technology Science in India in 1990. He completed his PhD in genetics at Penn State University in 1996. He is currently a Laura H. Carnell Professor and the Director of the Institute for Genomics and Evolutionary Medicine at Temple University. His major research interests focus on the discovery of evolutionary patterns and processes underlying the diversity of life on earth and the uses of this knowledge in genomics, precision medicine, and developmental evolution. He is also interested in developing new methods and algorithms for big data (images and sequences) and translating them into widely used software packages and knowledge bases.

Jieping Ye received the PhD degree in computer science from the University of Minnesota, Twin Cities, MN, USA, in 2005. He is an Associate Professor of Department of Computational Medicine and Bioinformatics and Department of Electrical Engineering and Computer Science at the University of Michigan, Ann Arbor, MI, USA. His research interests include machine learning, data mining, and biomedical informatics. He serves as an Action Editor for Data Mining and Knowledge Discovery. Dr. Ye has served as Senior Program Committee/Area Chair/Program Committee Vice Chair of many conferences including NIPS, ICML, KDD, IJCAI, ICDM, SDM, ACML, and PAKDD. He serves as a PC Co-Chair of SDM 2015. He serves as an Associate Editor for IEEE Transactions On Knowledge and Data Engineering and IEEE Transactions On Pattern Analysis And Machine Intelligence. He won the NSF CAREER Award in 2010. His papers have been selected for the outstanding student paper at ICML in 2004, the KDD best research paper honorable mention in 2010, the KDD best research paper nomination in 2011 and 2012, the SDM best research paper runner up in 2013, the KDD best research paper runner up in 2013, and the KDD best student paper award in 2014.

Shuiwang Ji received the PhD degree in computer science from Arizona State University, Tempe, Arizona, in 2010. Currently, he is an Associate Professor in the School of Electrical Engineering and Computer Science, Washington State University, Pullman, Washington. His research interests include machine learning, data mining, computational neuroscience, and bioinformatics. He received the National Science Foundation CAREER Award in 2014. He is currently an Associate Editor for BMC Bioinformatics and IEEE Transactions on Neural Networks and Learning Systems and an editorial board member of Data Mining and Knowledge Discovery. He is a senior member of IEEE.
Contributor Information
Wenlu Zhang, Department of Computer Science, Old Dominion University, Norfolk, VA, 23529..
Rongjian Li, Department of Computer Science, Old Dominion University, Norfolk, VA, 23529..
Tao Zeng, School of Electrical Engineering and Computer Science, Washington State University, Pullman, WA, 99163..
Qian Sun, Department of Computer Science and Engineering, Arizona State University, Tempe, AZ, 85287..
Sudhir Kumar, Institute for Genomics and Evolutionary Medicine and the Department of Biology, Temple University, Philadelphia, PA 19122..
Jieping Ye, Department of Electrical Engineering and Computer Science and the Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, MI 48109..
Shuiwang Ji, School of Electrical Engineering and Computer Science, Washington State University, Pullman, WA, 99163..
REFERENCES
- [1].Bengio Y, Courville A, and Vincent P. Representation learning: A review and new perspectives. IEEE Transactions on Pattern Analysis and Machine Intelligence, 35(8):1798–1828, 2013. [DOI] [PubMed] [Google Scholar]
- [2].Bishop C. Neural Networks for Pattern Recognition. Oxford University Press, 1995. [Google Scholar]
- [3].Donahue J, Jia Y, Vinyals O, Hoffman J, Zhang N, Tzeng E, and Darrell T. DeCAF: A deep convolutional activation feature for generic visual recognition. In Proceedings of the 31st International Conference on Machine Learning, pages 647–655, 2014. [Google Scholar]
- [4].Fei H. and Huan J. Structured feature selection and task relationship inference for multi-task learning. Knowledge and information systems, 35(2):345–364, 2013. [Google Scholar]
- [5].Frise E, Hammonds AS, and Celniker SE Systematic imagedriven analysis of the spatial Drosophila embryonic expression landscape. Molecular Systems Biology, 6:345, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Girshick R, Donahue J, Darrell T, and Malik J. Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the 27th IEEE Conference on Computer Vision and Pattern Recognition, 2014. [Google Scholar]
- [7].Guo Y. and Schuurmans D. Multi-label classification with output kernels. In Machine Learning and Knowledge Discovery in Databases, pages 417–432. Springer, 2013. [Google Scholar]
- [8].Guo Y. and Xue W. Probabilistic multi-label classification with sparse feature learning. In Proceedings of the Twenty-Third international joint conference on Artificial Intelligence, pages 1373–1379. AAAI Press, 2013. [Google Scholar]
- [9].He J. and Zhu Y. Hierarchical multi-task learning with application to wafer quality prediction. In Data Mining (ICDMF, 2012 IEEE 12th International Conference on, pages 290–298. IEEE, 2012. [Google Scholar]
- [10].Ji S, Li Y-X, Zhou Z-H, Kumar S, and Ye J. A bag-of-words approach for Drosophila gene expression pattern annotation. BMC Bioinformatics, 10(1):119, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ji S, Sun L, Jin R, Kumar S, and Ye J. Automated annotation of Drosophila gene expression patterns using a controlled vocabulary. Bioinformatics, 24(17):1881–1888, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ji S, Yuan L, Li Y-X, Zhou Z-H, Kumar S, and Ye J. Drosophila gene expression pattern annotation using sparse features and term-term interactions. In Proceedings of the Fifteenth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pages 407–416, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Krizhevsky A, Sutskever I, and Hinton G. Imagenet classification with deep convolutional neural networks. In Bartlett P, Pereira F, Burges C, Bottou L, and Weinberger K, editors, Advances in Neural Information Processing Systems 25, pages 1106–1114. 2012. [Google Scholar]
- [14].Kumar S, Jayaraman K, Panchanathan S, Gurunathan R, Marti-Subirana A, and Newfeld SJ BEST: a novel computational approach for comparing gene expression patterns from early stages of Drosophila melanogaster develeopment. Genetics, 169:2037–2047, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kumar S, Konikoff C, Van Emden B, Busick C, Davis KT, Ji S, Wu L-W, Ramos H, Brody T, Panchanathan S, Ye J, Karr TL, Gerold K, McCutchan M, and Newfeld SJ Fly Express: visual mining of spatiotemporal patterns for genes and publications in Drosophila embryogenesis. Bioinformatics, 27(23):3319–3320, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].LeCun Y, Bottou L, Bengio Y, and Haffner P. Gradient-based learning applied to document recognition. Proceedings of the IEEE, 86(11):2278–2324, November 1998. [Google Scholar]
- [17].Lécuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes T, Tomancak P, and Krause H. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell, 131:174–187, 2007. [DOI] [PubMed] [Google Scholar]
- [18].Lowe DG Distinctive image features from scale-invariant keypoints. International journal of computer vision, pages 91–110, 2004. [Google Scholar]
- [19].Oquab M, Laptev I, Bottou L, and Sivic J. Learning and transferring mid-level image representations using convolutional neural networks. In Proceedings of the 27th IEEE Conference on Computer Vision and Pattern Recognition, 2014. [Google Scholar]
- [20].Pruteanu-Malinici I, Mace DL, and Ohler U. Automatic annotation of spatial expression patterns via sparse Bayesian factor models. PLoS Comput Biol, 7(7):e1002098, 07 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Puniyani K, Faloutsos C, and Xing EP SPEX2: automated concise extraction of spatial gene expression patterns from fly embryo ISH images. Bioinformatics, 26(12):i47–56, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Razavian AS, Azizpour H, Sullivan J, and Carlsson S. Cnn features off-the-shelf: an astounding baseline for recognition. In Proceedings of the 27th IEEE Conference on Computer Vision and Pattern Recognition Workshops, 2014. [Google Scholar]
- [23].Simonyan K. and Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556, 2014. [Google Scholar]
- [24].Sun Q, Muckatira S, Yuan L, Ji S, Newfeld S, Kumar S, and Ye J. Image-level and group-level models for Drosophila gene expression pattern annotation. BMC Bioinformatics, 14:350, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, and Rabinovich A. Going deeper with convolutions. arXiv preprint arXiv:1409.4842, 2014. [Google Scholar]
- [26].Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu S, Lewis SE, Richards S, Ashburner M, Hartenstein V, Celniker SE, and Rubin GM Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biology, 3(12):research0088.1–0088.14, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tomancak P, Berman B, Beaton A, Weiszmann R, Kwan E, Hartenstein V, Celniker S, and Rubin G. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biology, 8(7):R145, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Van Emden B, Ramos H, Panchanathan S, Newfeld S, and Kumar S. Flyexpress: an image-matching web-tool for finding genes with overlapping patterns of expression in drosophila embryos. Tempe, AZ, 85287530, 2006. [Google Scholar]
- [29].Yuan L, Pan C, Ji S, McCutchan M, Zhou Z-H, Newfeld S, Kumar S, and Ye J. Automated annotation of developmental stages of Drosophila embryos in images containing spatial patterns of expression. Bioinformatics, 30(2):266–273, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yuan L, Woodard A, Ji S, Jiang Y, Zhou Z-H, Kumar S, and Ye J. Learning sparse representations for fruit-fly gene expression pattern image annotation and retrieval. BMC Bioinformatics, 13:107, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zeiler MD and Fergus R. Visualizing and understanding convolutional networks. In Proceedings of the European Conference on Computer Vision, pages 818–833. Springer, 2014. [Google Scholar]
- [32].Zhang D, He J, Liu Y, Si L, and Lawrence R. Multi-view transfer learning with a large margin approach. In Proceedings of the 17th ACM SIGKDD international conference on Knowledge discovery and data mining, pages 1208–1216. ACM, 2011. [Google Scholar]
- [33].Zhang J. and Huan J. Inductive multi-task learning with multiple view data. In Proceedings of the 18th ACM SIGKDD international conference on Knowledge discovery and data mining, pages 543–551. ACM, 2012. [Google Scholar]
- [34].Zhang W, Feng D, Li R, Chernikov A, Chrisochoides N, Osgood C, Konikoff C, Newfeld S, Kumar S, and Ji S. A mesh generation and machine learning framework for Drosophila gene expression pattern image analysis. BMC Bioinformatics, 14:372, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhou J. and Peng H. Automatic recognition and annotation of gene expression patterns of fly embryos. Bioinformatics, 23(5):589–596, 2007. [DOI] [PubMed] [Google Scholar]