Abstract
Purpose:
To develop an end-to-end deep learning (DL) framework to segment ventilation defects on pulmonary hyperpolarized MRI.
Materials and methods:
The Multi-Ethnic Study of Atherosclerosis Chronic Obstructive Pulmonary Disease (COPD) study is a nested longitudinal case-control study in older smokers. Between February 2016 and July 2017, 56 participants (age, mean ± SD, 74 ± 8 years; 34 men) underwent same breath-hold proton (1H) and helium (3He) MRI, which were annotated for non-ventilated, hypo-ventilated, and normal-ventilated lungs. In this retrospective DL study, 820 1H and 3He slices from 42/56 (75%) participants were randomly selected for training, with the remaining 14/56 (25%) for test. Full lung masks were segmented using a traditional U-Net on 1H MRI and were imported into a cascaded U-Net, which were used to segment ventilation defects on 3He MRI. Models were trained with conventional data augmentation (DA) and generative adversarial networks (GAN)-DA.
Results:
Conventional-DA improved 1H and 3He MRI segmentation over the non-DA model (P = 0.007 to 0.03) but GAN-DA did not yield further improvement. The cascaded U-Net improved non-ventilated lung segmentation (P < 0.005). Dice similarity coefficients (DSC) between manually and DL-segmented full lung, non-ventilated, hypo-ventilated, and normal-ventilated regions were 0.965 ± 0.010, 0.840 ± 0.057, 0.715 ± 0.175, and 0.883 ± 0.060, respectively. We observed no statistically significant difference in DCSs between participants with and without COPD (P = 0.41, 0.06, and 0.18 for non-ventilated, hypo-ventilated, and normal-ventilated regions, respectively).
Conclusion:
The proposed cascaded U-Net framework generated fully-automated segmentation of ventilation defects on 3He MRI among older smokers with and without COPD that is consistent with our reference method.
Keywords: MRI, Deep learning, COPD, Ventilation defects, Hyperpolarized gas
1. Introduction
Hyperpolarized gas MRI is a functional pulmonary imaging modality that is used to assess ventilation defects in chronic obstructive pulmonary disease (COPD) and other lung diseases [1–4]. A strength of hyperpolarized gas (e.g., helium and xenon) MRI is the visual contrast between ventilated and non-ventilated regions in lung, which are often expressed as a percentage of the lung volume and have been shown to correlate with pulmonary function [1–4]. Due to the high cost and long polarization time (12–14 h) of helium (3He) gas [2], the sample size of 3He MRI cohorts is often limited [5–8].
Unfortunately, there is no consensus ‘gold standard’ method for quantifying ventilation defects via segmentation on 3He MRI as this has been only reported in few exploratory studies [5,6]. Quantification of ventilation defects was achieved by two linked steps: full lung mask was segmented on 1H MRI and was subsequently utilized for ventilation defects segmentation on 3He MRI [7]. Most of previous studies have utilized manual or semi-automatic methods (e.g., region growing and thresholding with manual editing) for full lung segmentation [5,7,9–12]. Segmentation of ventilation defects also often requires expert’s input. (e.g., to define cluster hierarchy or cluster number) [5–7,9,10,13,14]. Deep learning (DL) has the potential to provide increased reproducibility, efficiency, and robustness in both full lung and ventilation defects segmentation without operator / user input. One recent DL study segmented normal-ventilated regions but did not segment ventilation defects [15]. Another DL study segmented ventilation defects but did not segment hypo-ventilated lung regions (i.e., pathological progression from normal-ventilated to non-ventilated lung regions) [8]. As lung damage in COPD progresses from subclinical to exacerbation over decades [16], it is likely that affected lung regions initially manifest decreased ventilation and progress to completely non-ventilated over time. Quantification of hypo-ventilated lung regions can potentially serve as an imaging marker for COPD progression as well as for evaluation of potential early intervention.
The U-Net model is one of the most popular DL architectures in medical image segmentation [17]. One recent optimization proposed for the U-Net model is a cascaded U-Net, also known as a hierarchical U-Net [18–20], which can improve multi-label segmentation through first segmenting major classes with large interclass difference and then subdividing the major classes into tissue sub-classes [18–20]. Previous studies reported that cascaded U-Net could improve accuracy in multiclass brain and liver lesion segmentation over traditional U-Net models.
Data augmentation (DA) is critical for training DL models on small datasets. Conventional-DA (e.g., geometric transformations, deformations, and noise) has been widely used for medical image segmentation [21,22] applications. Recently, a novel DA method [23,24] was proposed using a pre-trained generative adversarial network (GAN) to generate realistic synthetic data by learning the training data distribution [25]. Previous studies have demonstrated improved performance of GAN-based DA methods over conventional DA methods on various imaging modalities including lung CT, brain MRI, and cell nuclei image [26–28].
The present study used inherently registered 1H MRI scans and hyperpolarized 3He MRI scans from the Multi-Ethnic Study of Atherosclerosis (MESA) COPD study [29]. Our goal was to develop an end-to-end DL framework to segment pulmonary ventilation defects including hypo-ventilated and non-ventilated regions on 3He MRI without any human intervention, eliminating operator dependence. To train the DL lung ventilation defect segmentation tool, GAN-based DA was compared with conventional-DA, and cascaded U-Net was compared with traditional U-Net.
2. Materials and methods
2.1. Participants and datasets
The MESA COPD Study is a nested longitudinal case-control study of 10+ pack-year smokers, and the recruitment criteria have been described previously [29]. A subset of MESA COPD participants underwent same breath-hold 1H and 3He MRI (n = 56) between February 2016 and July 2017 [age, mean ± standard deviation (SD), 74 ± 8 years, 34 men, Table 1]. For each participant, we included 9–10 inherently registered 1H and 3He MRI coronal slices that contain lungs. The paired 1H and 3He MRI scans were randomly split into training and test datasets with a 3:1 ratio at the participant-level (i.e., scans from 42 participants were used for training/validation and the remaining scans from 14 participants were used for testing). 820/1088 (75.37%) of slices were included in the training dataset; 164/820 (20%) of slices from the training dataset were randomly withheld and were used as validation dataset for parameter tuning during training; 268/1088 (24.63%) of slices from 14 participants were used for testing. This study was approved by the institutional review board (AAAO1456) and complied with HIPAA rules, and participants’ written informed consents were obtained. The 3He MRI were acquired under Food and Drug Administration-approved Investigational New Drug application for hyperpolarized gas (i.e., 3He).
Table 1.
Characteristics of the training and test sets of participants (n = 56).
| Variables | Participants in training dataset (n = 42) | Participants in test dataset (n = 14) | P-value* |
|---|---|---|---|
| Age, years | 73.5 ± 7.5 | 71.9 ± 7.0 | 0.43 |
| Male, N (%) | 25 (59.5%) | 9 (64.3%) | 0.75 |
| Race/Ethnicity, N (%) | 0.60 | ||
| White | 21 (50.0%) | 8 (57.1%) | |
| Black | 12 (28.6%) | 5 (35.7%) | |
| Hispanic | 9 (21.4%) | 1 (7.1%) | |
| Current smokers, N (%) | 10 (23.8%) | 6 (42.9%) | 0.19 |
| Pack years | 38.2 ± 20.9 | 46.8 ± 28.9 | 0.47 |
| COPD, N (%) | 22 (52.4%) | 8 (57.1%) | 0.76 |
| None | 20 (47.6%) | 6 (42.9%) | |
| Mild | 11 (26.2%) | 3 (21.4%) | |
| Moderate | 11 (26.2%) | 5 (35.7%) | |
| % normal-ventilated | 63.3 ± 21.3 | 59.7 ± 13.9 | 0.27 |
| % hypo-ventilated | 25.0 ± 14.8 | 26.8 ± 11.6 | 0.36 |
| % non-ventilated | 11.7 ± 10.1 | 13.5 ± 6.3 | 0.13 |
Note. — For quantitative vsariables, data are presented as mean ± standard deviation unless otherwise noted. COPD = chronic obstructive pulmonary disease.
The Mann-Whitney U, Chi-squared, and Fisher’s exact tests were used as appropriate to compare differences between the training and test subjects.
2.2. MRI acquisition and annotation
3He MRI were acquired using a 3 T Achieva Philips MR scanner (Philips, Eindhoven, Netherlands) with a flexible wrap-around 3He radio frequency coil. 3He gas was polarized to 29 ± 5.7% using a 3He polarizer (GE Healthcare, Princeton, NJ). Participants were coached to exhale to their residual volume and then inhale 300 mL of 3He mixed with 700 mL N2 in order to image at approximately functional residual volume. Inherently registered coronal 1H MRI and 3He MRI were acquired at the same voxel size and matrix size (1.76 × 1.76 × 16.5 mm3, 256 × 256 × 12) during the same breath-hold [30]. 1H and 3He MRI was acquired using a fast section select gradient reversal (SSGR), TE/TR = 1.5/48 ms protocol. The 5 s 3He MRI was acquired first. It took ~3 s to switch the scanner from 3He to 1H acquisition. A 6 s 1H MRI scan was then acquired afterwards within the same breath hold. The total breath hold time was 15 s, which can be comfortably performed by all patients.
A total of 544 slices of 1H pulmonary MRI and 544 slices of 3He pulmonary MRI of 56 participants were annotated (i.e., 1088 slices were semi-automatically segmented) [31]. We developed a “ground-truth” method leveraging the experience of our Image Analysis Core Lab [32–35] as well as the combined experience of image analysts, radiologists, physiologists, and physicians. The image analysts have 15–20 years of experience in MRI analysis at Image Analysis Core Lab and the radiologist has over 35 years of experience in MRI reading and evaluation.
2.2.1. Full lung mask annotation
Lung masks were segmented on coronal 1H MRI images of each participant to define the lung boundaries (Fig. 1). A region of interest (ROI) was drawn on each coronal slice within each lung excluding regions with partial volume effects (i.e., loss of contrast between two adjacent tissues in a slice caused by insufficient resolution so that more than one tissue type occupies the same voxel or pixel). The mean and SD of the signal intensity (SI) of the lung ROI were calculated for each coronal slice of lungs. The lungs were then segmented on 1H MRI images using a region growing method with threshold values adaptively tied to mean SIlung ROI ± 2.576 SDlung ROI (Fig. 1). Manual corrections were applied if necessary. The corrections were mostly small anatomical corrections such as removing low intensity regions including cortical bones, central airways and background.
Fig. 1.
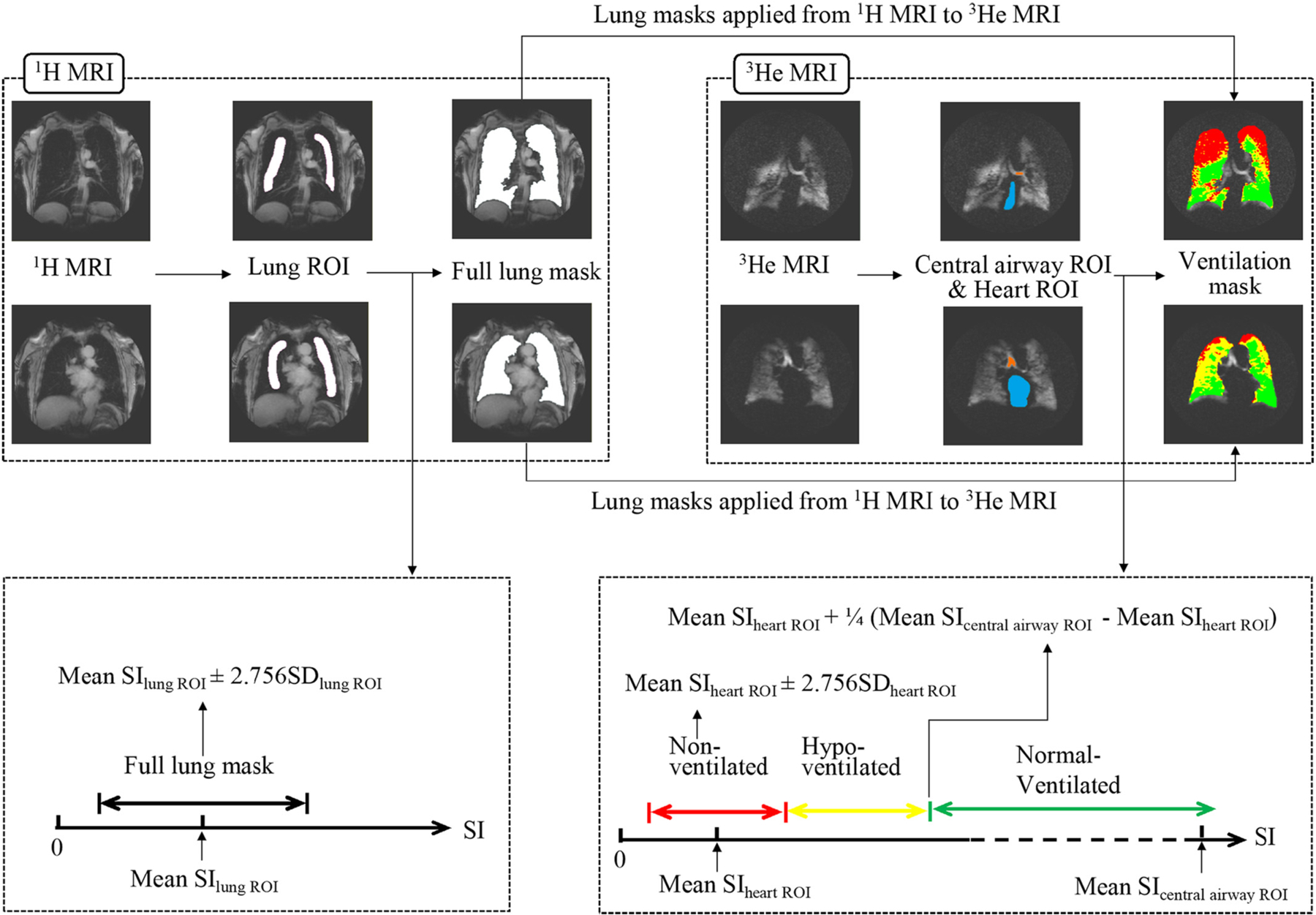
Illustration of ground truth segmentation. Full lung masks were segmented on 1H MRI and subsequently applied to 3He MRI. To determine appropriate thresholds for full lung masks and ventilation segmentations, ROIs were manually drawn inside the lungs on 1H MRI, and inside the central airways and the heart on 3He MRI. SD = standard deviation; SI = signal intensity; ROI = region of interest. Colour coding: Orange = central airway ROI; blue = heart ROI; red = non-ventilated lung regions; yellow = hypo-ventilated lung regions; green = normal-ventilated lung regions. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.2.2. Ventilation annotation
The full lung masks from 1H MRI images were applied to each corresponding 3He slice to define the lung boundaries on 3He MRI (Fig. 1). SI of heart was used as a reference for non-ventilated regions and SI of central airways was used as a reference for ventilated regions. ROIs of the heart and central airways were manually defined on the 3He MRI images excluding regions with partial volume effect. Non-ventilated regions were segmented via region growing with a threshold defined as the mean SIheart ROI ± 2.576 SDheart ROI. After non-ventilated regions were annotated, ventilated regions were further divided into normal-ventilated regions and hypo-ventilated regions. Normal-ventilated regions were segmented by region growing with a threshold as mean SIheart ROI + 0.25 (mean SIcentral airway ROI – mean SIheart ROI). The remaining lung regions (i.e., the first quartile of the range of mean SIheart ROI and mean SIcentral airway ROI) were defined as hypo-ventilated regions (Fig. 1).
The analysts were blinded to the clinical information of the participants. One analyst performed the image analysis. A second analyst was included to evaluate inter-reader agreement. Two readings were included for intra-reader agreement. A total of 10 randomly selected MRI scans were used the intra-reader and inter-reader agreement assessment and were read twice. The intra-reader and inter-reader % CV was 1.7% and 2.6% for non-ventilated, 3.8% and 3.4% for hypo-ventilated, and 3.4% and 4.0% for normal-ventilated lung regions, respectively. SliceOmatic (TomoVision, Magog, Canada) was used to perform the manual ROI selection, region growing and manual corrections.
2.3. Deep learning framework and cascaded U-Net
The proposed DL framework consisted of 1H and 3He segmentation models trained separately on the 1H and 3He MRI training datasets (Fig. 2). Using supervised training, annotated full lung masks from 1H MRI were fed into the 3He model concatenated with the 3He MRI data to perform ventilation defect segmentation. After training, an end-to-end framework was built combining the two models for testing: the 1H MRI was first segmented by the 1H segmentation model, and the segmented full lung masks were concatenated with 3He MRI as the input of the 3He segmentation model. Such an end-to-end framework took only the original 1H and 3He MR images as input and required no human intervention.
Fig. 2.
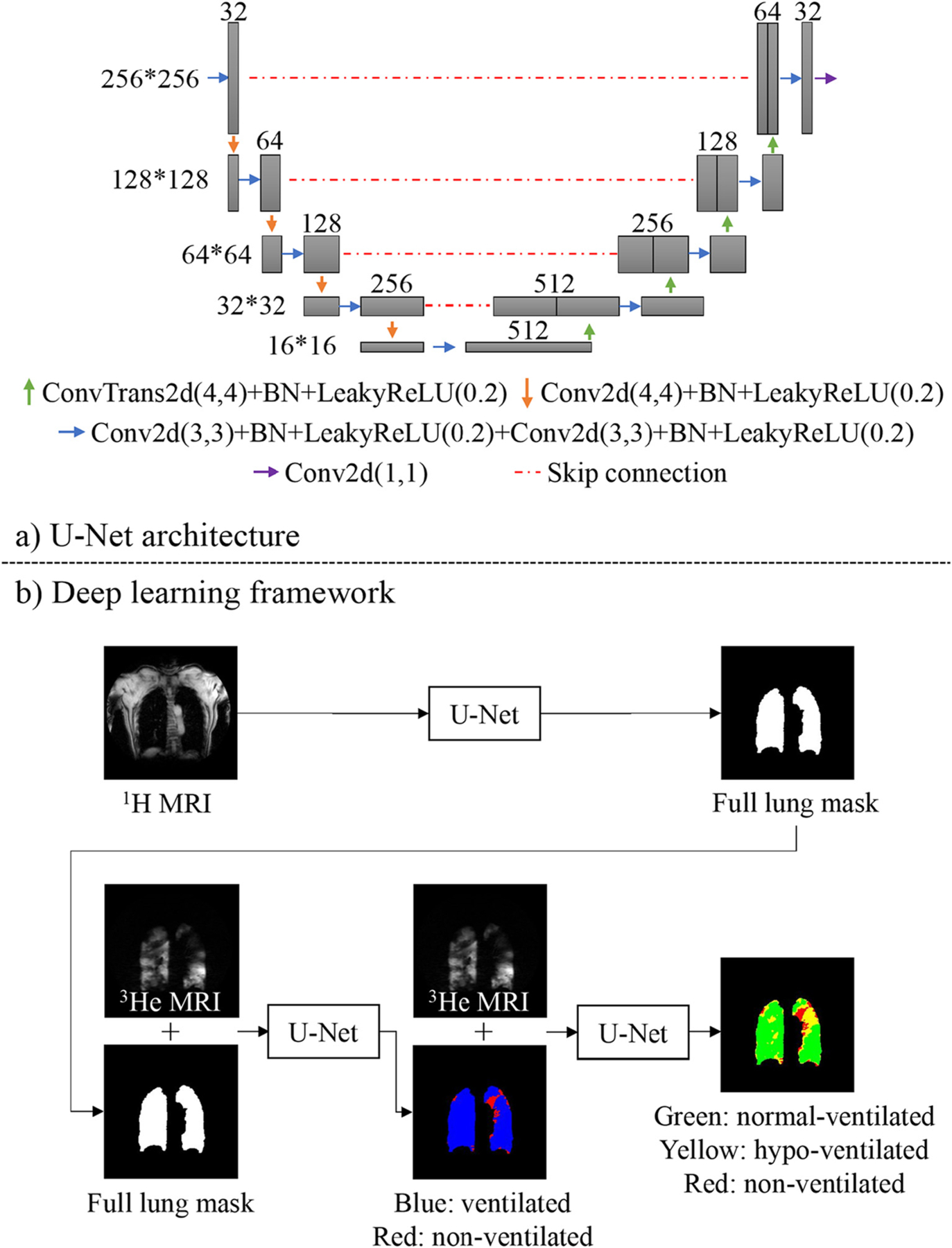
Conv2d = 2D convolution, BN = batch normalization, LeakyReLU = leaky rectified linear unit, ConvTrans2d = 2D transpose convolution.
a): U-Net architecture, in which we substituted the max pooling layer with a 2D convolution layer (Conv2d) with a 4*4 kernel.
b): The proposed end-to-end deep learning framework. Full lung mask was segmented from 1H MRI through a traditional U-Net. For 3He segmentation, we applied a two-layer cascaded U-Net which first segmented 3He MRI into ventilated (blue) and non-ventilated (red) lung regions. The ventilated regions were then segmented into normal-(green) and hypo-(yellow) ventilated lung regions. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We used a U-Net architecture (Fig. 2a) to build the 1H and 3He segmentation models (Fig. 2b). In the 1H segmentation model, we first used a traditional U-Net to generate full lung masks and replaced the max pooling layer with a 2D convolution layer. In the 3He segmentation model, we utilized a cascaded U-Net which consisted of two U-Nets. The cascaded U-Net utilized the full lung masks and 3He MRI scans as a two-channel input. The first step was to segment non-ventilated and ventilated regions. The segmented ventilated regions were subsequently imported into a second layer, which subdivided the ventilated regions into hypo-ventilated and normal-ventilated regions. We normalized the image intensity of 1H and 3He MRI to [−1,1] range.
2.4. Data augmentation
For conventional-DA, we included random scaling (scaled by 80–120% along each axis separately), random translation (translated by −10 to 10% pixels per axis), random rotation (−45° to 45 °), random shearing (−15 ° to 15 °), random horizontal flip (with probability P = 0.5), random vertical flip (with probability P= 0.5). Though flip is not a common source of variability in MRI scans, we utilized them to increase the model generalization as much as possible (e.g., to ensure that the model was not biased by a prevalence of certain lung patterns in left versus right lungs and in apical versus basal or central versus pleural regions). Implementation was performed using the Python library imgaug (Fig. 3).
Fig. 3.
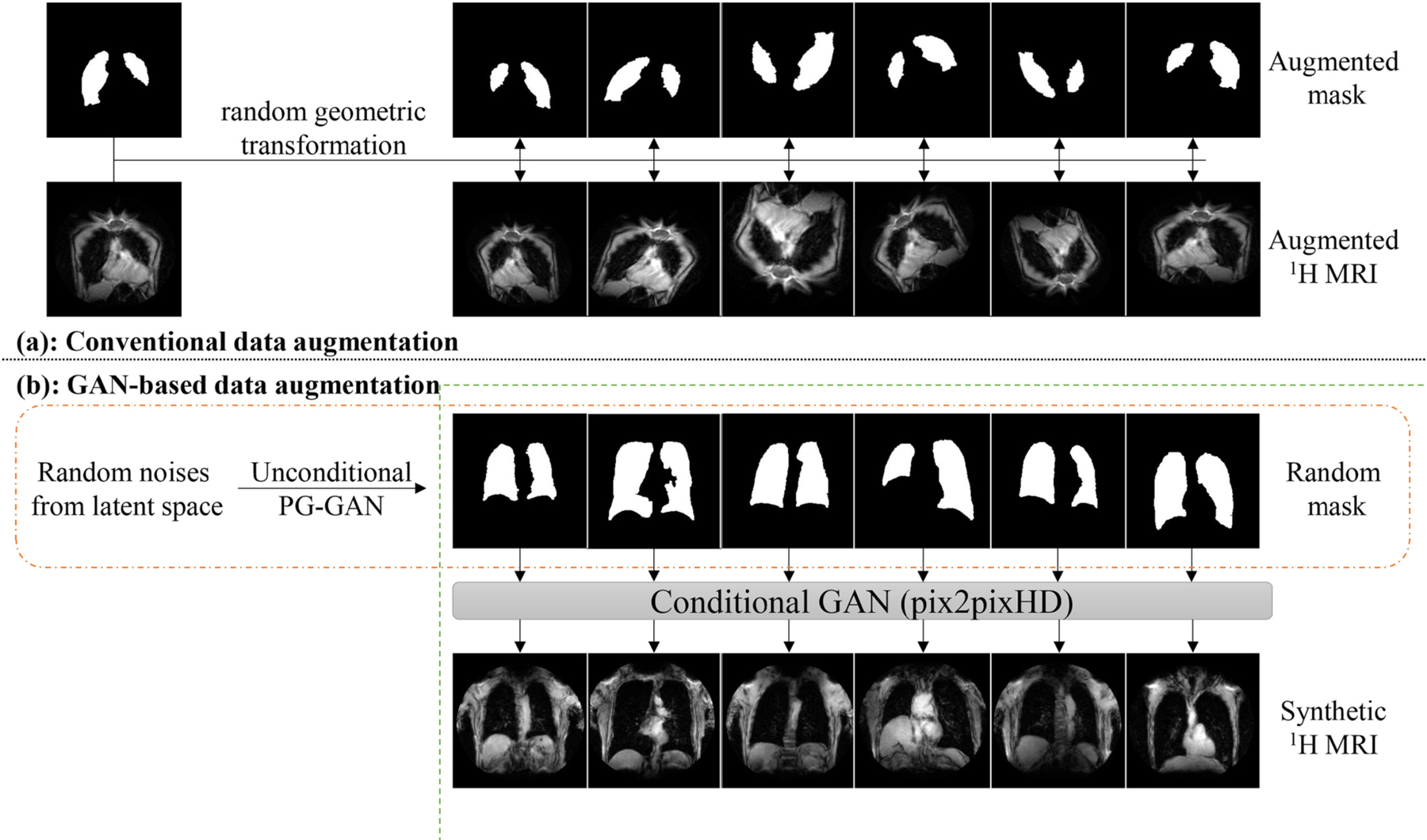
PG = progressively-growing. Data augmentation (DA). a): Conventional-DA includes random transformations: scaling (scaled by 80–120% along each axis separately), translation (translated by −10 to 10% pixels per axis), rotation (−45° to 45 °), shearing (−15 ° to 15 °), horizontal flip (probability P = 0.5), vertical flip (probability P = 0.5); b): GAN-based DA used two separate generative adversarial networks (GANs). The first unconditional GAN generated random synthetic full lung masks, and the second conditional GAN translated the random masks into corresponding synthetic 1H MR image.
For GAN-DA, we used a dual-GAN network consisting of two separate GANs, in which the first GAN [36] was trained to generate 1H lung masks from random latent noise, and the second conditional GAN (using pix2pixHD [37]) was trained to synthesize corresponding realistic 1H MRI images conditioned by the lung mask. Similarly, the first GAN was trained to generate 3He lung masks from random latent noise, and the second conditional GAN was trained to synthesize corresponding realistic 3He MRI images conditioned by the lung mask (Fig. 3).
Four different U-Net + DA segmentation models were trained:
Non-DA model: trained on (n = 410 × 0.8 = 328 for 1H MRI and n = 410 × 0.8 = 328 for 3He MRI, respectively) original coronal slices without any DA;
Conventional-DA model: trained on original coronal slices plus 10-fold slices generated using conventional-DA (n = 3608 for 1H MRI and n = 3608 for 3He MRI, respectively);
GAN-DA model: trained on original coronal slices plus 10-fold slices generated using dual-GAN DA (n = 3608 for 1H MRI and n = 3608 for 3He MRI, respectively);
Combined-DA model: trained on original coronal slices plus 5-fold slices generated using conventional-DA plus 5-fold slices generated using GAN-DA (n = 3608 for 1H MRI and n = 3608 for 3He MRI, respectively).
2.5. Training strategies
The training of DL models was performed using Python (version 3.6; Python Software Foundation, Wilmington, Del) and PyTorch library [38]. Random initialization was applied to each model. The models were trained with the following parameters: 15 epochs with a batch size of 16 for 1H lung segmentation and 100 epochs with a batch size of 4 for the 3He ventilated regions segmentation. A linear learning rate decay strategy was used [37], decreasing the learning rate from starting value 0.0001 to 0 starting from the 5th epoch for 1H segmentation model and the 50th epoch for the 3He segmentation model. The best model on the validation set was used to evaluate the performance on the test set. Detailed training and validation curves can be found in Supplemental Fig. 5. We used a hybrid loss function combining cross-entropy (CE) and Dice loss. For a sample image X ∈ RW×H (W, width; H, height), the segmentation model takes X as input and generates a probability map P, in which P(w, h, c) serves as the probability distribution at pixel (w, h) over C classes. The ground truth label for each pixel in X was provided by Y(w, h, c) as a one-hot vector, and the Y′(w, h, c) was the one-hot vector of predicted label. CE was defined as:
The CE compares the probability distributions of predictions and true labels. In addition, we used the Generalized Dice Loss (GDL) [39], an extension of the Dice similarity coefficient (DSC) which is an established evaluation metric in semantic segmentation, as a constraint on the spatial regularity of the prediction. DSC and GDL are defined as:
where Weightc is a class-wise weight to address data imbalance, and equals to the reciprocal of the population prevalence of that class [39]. A previous study found that the CE and GDL improved model training most when they have the same weight [40]. Therefore, both 1H and 3He models were trained with the Adam optimizer [41] to optimize the overall loss function L:
To test if there was overfitting in our model, we performed a 4-fold cross-validation by randomly splitting our dataset into 4 groups, each as a test dataset and the remaining 3 as a training dataset.
2.6. Statistical analysis
We evaluated the performance of our segmentation models using two metrics: DSC and mean surface distance (MSD). MSD is a surface-based metric and measures the average distance between the surfaces of predicted and ground-truth regions. DSC and MSD were calculated based on the entire 3D volume of full lungs or lung regions as we aimed to evaluate our algorithm at participant-level rather than slice-level. MSD was calculated through a public package provided by DeepMind (London, UK) [42]. Different DA methods were compared using DSCs and MSDs. Cascaded U-Net’s performance was compared with traditional U-Net (using directly 3 ventilation level classes) on 3He ventilation defect segmentation.
We used Wilcoxon signed-rank test to compare DSCs and MSDs between GAN-DA and conventional-DA methods (1H segmentation model), as well as cascaded and traditional U-Nets (3He segmentation model); the Wilcoxon signed-rank test was used to compare ground-truth values to inferred values for the total lung volume returned by the 1H model, and volumes of non-ventilated, hypo-ventilated, and normal-ventilated regions returned by the 3He model. Bonferroni-Holm correction was used when applicable. Mann-Whitney U test was used to compare models’ performance among participants with and without COPD. Kruskal-Wallis H Test was used to compare model performance in 4-fold cross-validation. Statistical analysis was performed using the SAS 9.4 package (SAS Institute. Inc., Cary, NC, USA). Two-tailed (α = 0.05) tests of significance were used.
3. Results
3.1. Participant characteristics
Characteristics of the study participants, stratified by training and test datasets, are shown in Table 1. The training dataset contains 42/56 (75%) participants (73.5 ± 7.5 years; 25 men) and the test dataset includes the remaining 14/56 (25%) participants (71.9 ± 7.0 years; 9 men). There were 22 participants with mild and moderate COPD in the training datasets and 8 participants with mild and moderate COPD in the test dataset. There were no large differences between the training and test datasets, as would be expected by the allocation due to chance (Table 1).
3.2. 1H MRI segmentation
Compared with the non-DA model (0.955 ± 0.012), improvement in DSCs was observed with combined-DA model (0.965 ± 0.010, P = 0.0007) and conventional-DA model (0.961 ± 0.011, P = 0.03), but not GAN-DA model (0.959 ± 0.010, P = 0.13) (Table 2). There was no significant difference between DSC of combined-DA model (0.965 ± 0.010) and DSC of conventional-DA model (0.961 ± 0.011, P = 0.13). Additionally, combined-DA yielded higher DSC than GAN-DA model (0.959 ± 0.010, P = 0.007).
Table 2.
Comparison of different data augmentation (DA) strategies for full lung mask segmentation on 1H MRI test set (n = 14).
| Models | DSC | P value (vs. non-DA model) | MSD (mm) | P value (vs. non-DA model) |
|---|---|---|---|---|
| Non-DA model | 0.955 ± 0.012 (0.947, 0.953, 0.964) |
– | 1.055 ± 0.953 (0.632, 0.775, 0.916) |
– |
| Conventional-DA model | 0.961 ± 0.011 (0.955, 0.963, 0.968) |
0.03 | 0.594 ± 0.137 (0.536, 0.601, 0.680) |
0.03 |
| GAN-DA model | 0.959 ± 0.011 (0.949, 0.959, 0.967) |
0.13 | 0.928 ± 0.690 (0.550, 0.726, 0.883) |
0.31 |
| Combined-DA model | 0.965 ± 0.010* (0.955, 0.967, 0.971) |
0.0007 | 0.657 ± 0.254* (0.477, 0.630, 0.715) |
0.005 |
Note. —Data are presented as mean ± standard deviation (25th percentile, median, 75th percentile). DA = data augmentation, GAN = generative adversarial network, DSC = Dice Similarity Coefficient, MSD = mean surface distance. Wilcoxon signed-rank tests with the Bonferroni-Holm correction were used to account for multiple comparisons. Adjusted P-values are reported.
Combined-DA model has statistically significantly higher DSC (P = 0.0007) and lower MSD (P = 0.01) compared with GAN-DA model.
DSCs are not statistically significantly different between conventional-DA and GAN-DA, or between conventional-DA and combined-DA.
MSDs are not statistically significantly different between conventional-DA and GAN-DA, or between conventional-DA and combined-DA.
Similarly, conventional-DA model (0.594 ± 0.137 mm) had smaller MSD than non-DA model (1.055 ± 0.953 mm, P = 0.005) (Table 2). Combined-DA models also reduced the surface distance to 0.657 ± 0.254 mm (P = 0.03) when compared to the non-DA model. MSD of GAN-DA model (0.928 ± 0.690 mm) was not significantly different from that of the non-DA model (1.055 ± 0.953 mm, P = 0.31). MSD of conventional-DA model (0.594 ± 0.137 mm) was not significantly different from combined-DA model (0.657 ± 0.254 mm, P = 0.86) but was significantly lower than GAN-DA model (0.928 ± 0.690 mm, P = 0.01).
There was no significant difference observed between DL-predicted total lung volume and ground truth total lung volume (3.727 ± 1.075 L vs. 3.721 ± 1.109 L, P = 0.86).
3.3. 3He MRI segmentation
GAN- and combined-DA did not improve 3He segmentation (Supplemental Tables 1–3). Therefore, we only applied conventional-DA in 3He model training.
In the cascaded U-Net 3He model using an end-to-end framework, DSCs of non-ventilated, hypo-ventilated, and normal-ventilated regions were 0.840 ± 0.057, 0.715 ± 0.175, and 0.883 ± 0.060, respectively. The DSC of cascaded U-Net for non-ventilated regions was significantly higher than traditional U-Net (0.840 ± 0.055 vs. 0.818 ± 0.055, P < 0.001, Table 3). Cascaded U-Net also had lower MSD for non-ventilated regions than traditional U-Net (1.275 ± 1.396 mm vs. 1.432 ± 1.343 mm, P < 0.005, Table 3). DSCs and MSDs for the cascaded U-Net on 3He MRI were not significantly different between COPD and non-COPD participants (P = 0.06–0.75, Table 4). For non-ventilated, hypo-ventilated, and normal-ventilated regions, DL-predicted volumes were not significantly different from ground truth volumes (0.528 ± 0.334 L vs. 0.556 ± 0.370 L, 1.049 ± 0.584 L vs. 1.012 ± 0.610 L, and 2.150 ± 0.345 L vs. 2.153 ± 0.674 L, respectively, P = 0.12–0.36). Fig. 4 showed examples of a higher agreement with cascaded U-Net segmentation than with traditional U-Net segmentation when compared to the ground truth.
Table 3.
Comparison of traditional U-Net and cascaded U-Net on ventilation defect segmentation in the test set (n = 14).
| Traditional U-Net | Cascaded U-Net | P value | |
|---|---|---|---|
| Non-ventilated (3He) | |||
| DSC | 0.818 ± 0.055 | 0.840 ± 0.057 | <0.001 |
| MSD (mm) | 1.432 ± 1.343 | 1.275 ± 1.396 | <0.005 |
| Hypo-ventilated (3He) | |||
| DSC | 0.697 ± 0.192 | 0.715 ± 0.175 | 0.24 |
| MSD (mm) | 1.288 ± 1.357 | 1.166 ± 1.237 | 0.30 |
| Normal ventilated (3He) | |||
| DSC | 0.875 ± 0.070 | 0.883 ± 0.060 | 0.50 |
| MSD (mm) | 1.359 ± 0.845 | 1.506 ± 1.054 | 0.76 |
Table 4.
Comparison between ventilation defect segmentation in participants with and without COPD using cascaded U-Net in the test set (n = 14).
| COPD |
No COPD |
P value | |
|---|---|---|---|
| n = 8) | (n = 6) | ||
| Non-ventilated (3He) | |||
| DSC | 0.851 ± 0.056 | 0.826 ± 0.061 | 0.41 |
| MSD (mm) | 1.228 ± 1.630 | 1.338 ± 1.158 | 0.75 |
| Hypo-ventilated (3He) | |||
| DSC | 0.653 ± 0.208 | 0.799 ± 0.066 | 0.06 |
| MSD (mm) | 1.516 ± 1.561 | 0.700 ± 0.331 | 0.66 |
| Normal ventilated (3He) | |||
| DSC | 0.861 ± 0.071 | 0.912 ± 0.024 | 0.18 |
| MSD (mm) | 1.855 ± 1.158 | 1.042 ± 0.748 | 0.08 |
Note. —Data are presented as mean ± standard deviation. DSC = Dice similarity coefficient, MSD = mean surface distance. The Mann-Whitney U test was used to compare participants with and without COPD.
Fig. 4.
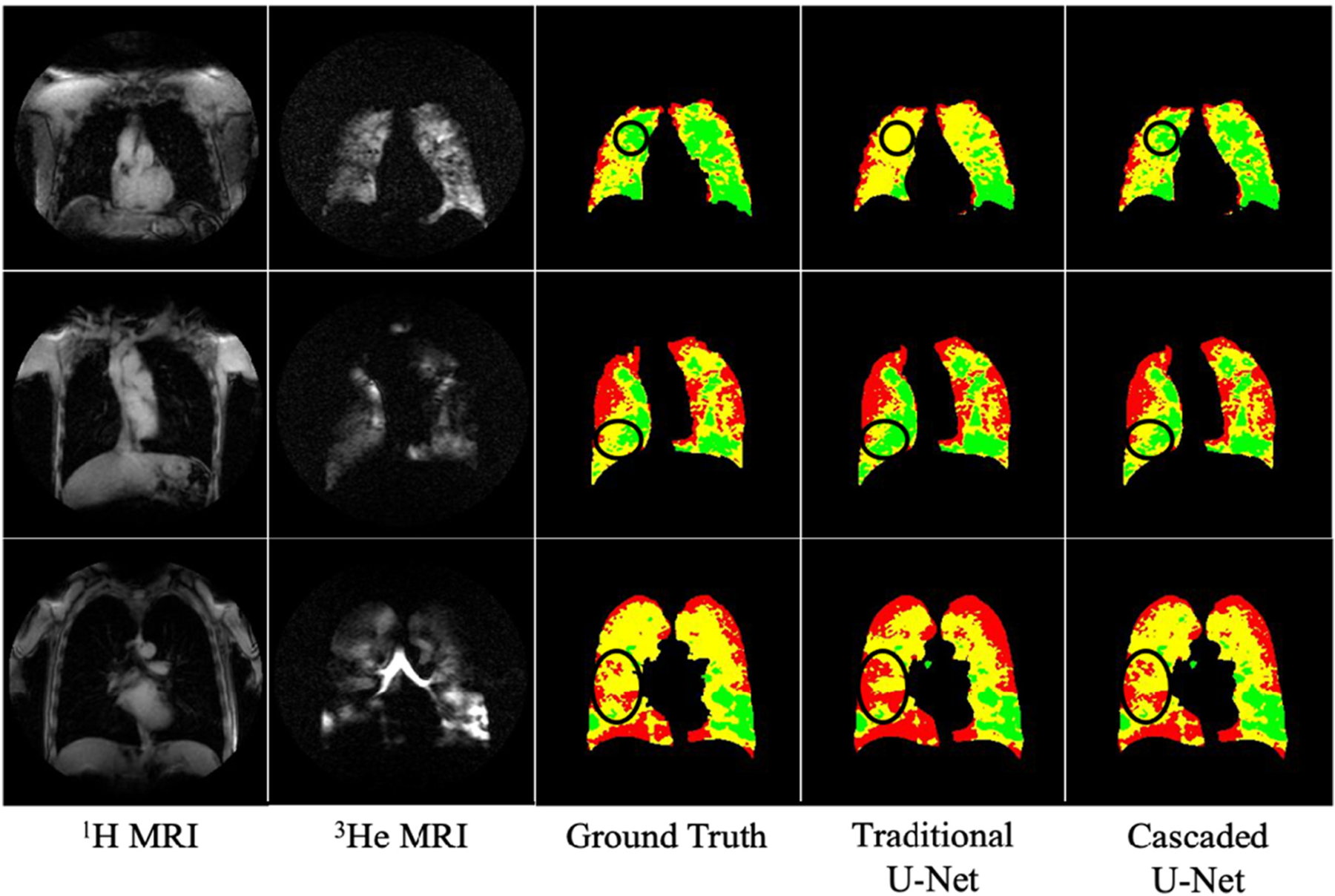
Comparison of ventilation segmentation results between traditional U-Net and cascaded U-Net. Red = non-ventilated lung region; yellow = hypo-ventilated lung region; green = normal-ventilated lung regions. The circled regions highlight higher agreement with cascaded U-Net segmentation than with traditional U-Net segmentation when compared to the ground truth. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In 4-fold cross-validation, there was no overfitting in traditional U-Net and cascaded U-Net, as there was no statistically significant difference among DSCs from 4 folds validation (Data not shown).
4. Discussion
4.1. Cascaded U-Net for multi-class lung ventilation segmentation
Our cascaded U-Net framework provided a fully automatic segmentation of pulmonary ventilation defects on 3He MRI among participants with and without COPD. This end-to-end framework accelerated the process of ventilation segmentation and yielded results consistent with our semi-automatic reference standard. This approach with no human input, provided quantification results equivalent to human annotation. One strength of our study is the use of inherently registered and fully annotated 1H and 3He MRI, which enabled us to validate a fully automated DL pipeline in an end-to-end manner.
Compared with a previous DL study which segmented ventilated and non-ventilated lung regions, our cascaded U-Net additionally segmented a hypo-ventilated class to distinguish the lung regions with impaired ventilation from the non-ventilated lung regions with no airflow [8]. Furthermore, our model had better performance in non-ventilated region segmentation than a previous DL study (DSC, 0.84 ± 0.06 vs. 0.70 ± 0.30) [8]. We found that conventional-DA and combined-DA models improved full lung mask segmentation from baseline performance (i.e., no DA applied) with statistical significance.
Likewise, the Cascaded U-Net improved segmentation accuracy with statistical significance for non-ventilated regions compared with traditional U-Net. The finding is in agreement with previous studies that utilized cascaded U-Net in liver tumor and brain tumor segmentation [18,19]. Multi-layer cascaded U-Net addressed multi-class segmentation through applying a hierarchical strategy [18,19]. The Cascaded U-Net took advantage of hierarchical distribution of lung ventilation. In the present study, the first layer U-Net learned the features that best differentiate ventilated and non-ventilated regions; the second layer learned the features to distinguish normal-from hypo-ventilated regions. In the present study, we utilized this multi-scale model to improve the segmentation for lung ventilation defects regions (Fig. 4).
Prior the development and application of DL models in medical image analysis, clustering methods were prevalent in pulmonary ventilation defect segmentation on 3He MRI [5–7,10,13]. Although clustering methods can segment ventilation defects on hyperpolarized gas MRI, the segmentation of full lung mask from 1H MRI could still be time-consuming. The end-to-end DL model in the present study would tremendously decrease analysis time. Our DL framework provided accurate full lung segmentation from 1H MRI and multi-categorical ventilation segmentation from 3He MRI through a multi-channel scheme.
4.2. GAN-DA for data-limited learning
In the present study, we explored a novel data augmentation (i.e., GAN-DA). Our results showed that both conventional-DA and combined-DA statistically significantly improved the 1H MRI segmentation while GAN-DA did not. Combined-DA had higher mean DSCs, and lower mean MSDs than conventional-DA but these differences did not reach statistical significance. Our finding was in agreement with a previous study [27] which suggested that GAN-DA and conventional-DA provided additional information independently. GAN-DA interpolated and enriched the diversity of training data in latent space, while conventional-DA could only extrapolated beyond the observed original scans through geometric transformations [27].
In contrast to a previous report that GAN-DA using a dual-GAN approach improved nuclei segmentation [26], we found that GAN-DA impaired the performance of 3He MRI segmentation. A potential explanation is that GAN-DA failed to generate high-quality synthetic 3He MRI images (Supplemental Fig. 2), in contrast to the high-quality synthetic 1H MRI images (Supplemental Fig. 1). Using a pre-trained VGG16 network (Supplemental Fig. 3) [43] and t-Distributed Stochastic Neighbor Embedding [44], we extracted and visualized feature distributions of synthetic and real masks and MRI images for 1H MRI (Supplemental Fig. 4a and b), and for 3He MRI (Supplemental Fig. 4c and d). As noted in Supplemental Fig. 4c, we observed large discrepancies between synthetic and real 3He MRI masks. Future studies are needed to further investigate the utility of GAN-based DA to improve DL segmentation of biomedical images with complex textures rather than well-contrasted anatomical or biological structures.
4.3. Limitations and conclusions
We noticed that the difference between COPD and non-COPD participants is borderline significant for DSC of hypo-ventilated regions, and for MSD of normal-ventilated regions (i.e., P= 0.06, P = 0.08, respectively). With the small sample size of the COPD and non-COPD participants (i.e., n = 8, n = 6, respectively) in the test dataset, the present study could not provide a definitive conclusion on our DL model’s consistency on hypo- and normal-ventilated regions between COPD and non-COPD participants. Future large-scale studies are needed to further evaluate our DL model on the segmentation of different disease conditions.
One limitation of our study is that the ROIs for determining thresholds for lung MRI segmentation were manually selected. We excluded partial volume effects in ROI selection for threshold calculation and this can be a source of error during inference. Additionally, some limitations in the original data might be propagated in the augmented data and could be a potential source of error. It should be noted that there is currently no consensus on a reference method for MRI lung mask segmentation and ventilation defects segmentation [5,7–9]. Both previously reported methods and our method rely on experts’ judgment and experience to some degree. We believe that expert panels and cross-validation among different institutes are needed to establish a consensus on lung ventilation segmentation. It should also be pointed out that for investigators who prefer other reference lung segmentation methods, our pre-trained model may not be directly generalized without fine-tuning. With appropriate labels, future studies will test if this DL algorithm can potentially generate end-to-end lung segmentation model that may tremendously cut down analysis time for different reference methods [5,7–9].
In conclusion, we developed a reliable end-to-end DL framework using a cascaded U-Net to automatically segment 1H and 3He MRI into non-ventilated, hypo-ventilated, and normal-ventilated regions. The performance of this DL model was found to be similar for participants with and without COPD. Conventional-DA and cascaded U-Net improved the robustness of quantifying ventilation defects.
Note. —Data are presented as mean ± standard deviation. DSC = Dice similarity coefficient, MSD = mean surface distance. Wilcoxon signed-rank test was used to compare traditional U-Net and cascaded U-Net.
Supplementary Material
Acknowledgements
This research was supported by grants R01-HL093081, R01-HL077612, and R01-HL121270 from the National Heart, Lung, and Blood Institute and contracts 75N92020D00001, HHSN268201500003I, N01-HC-95159, 75N92020D00005, N01-HC-95160, 75N92020D00002, N01-HC-95161, 75N92020D00003, N01-HC-95162, 75N92020D00006, N01-HC-95163, 75N92020D00004, N01-HC-95164, 75N92020D00007, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, and N01-HC-95169 and by grants UL1-TR-000040, UL1-TR-001079, and UL1-TR-001420 from the National Center for Advancing Translational Sciences (NCATS) and P30 DK26687 from National Institute of Diabetes and Digestive and Kidney Diseases and Medical Research Council grant MR/M008894/1. The authors thank the other investigators, the staff, and the participants of the MESA study for their valuable contributions. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S. Department of Health and Human Services.
Abbreviations:
- 1H
proton
- 3He
hyperpolarized helium
- CE
cross-entropy
- CNN
convolutional neural networks
- COPD
chronic obstructive pulmonary disease
- DA
data augmentation
- DL
deep learning
- DSC
Dice similarity coefficient
- MESA
Multi-Ethnic Study of Atherosclerosis
- MSD
mean surface distance
- ReLU
Rectified Linear Unit
- ROI
region of interest
- SI
signal intensity
- SD
standard deviation
Footnotes
CRediT authorship contribution statement
Xuzhe Zhang: Methodology, Software, Visualization, Writing – original draft. Elsa D. Angelini: Conceptualization, Methodology, Supervision, Writing – review & editing. Fateme S. Haghpanah: Software, Writing – review & editing. Andrew F. Laine: Methodology, Supervision, Writing – review & editing. Yanping Sun: Resources, Investigation, Writing – review & editing. Grant T. Hiura: Formal analysis, Writing – review & editing. Stephen M. Dashnaw: Methodology, Resources, Investigation, Writing – review & editing. Martin R. Prince: Methodology, Data curation, Investigation, Writing – review & editing. Eric A. Hoffman: Resources, Investigation, Writing – review & editing. Bharath Ambale-Venkatesh: Resources, Investigation, Writing – review & editing. Joao A. Lima: Resources, Investigation, Writing – review & editing. Jim M. Wild: Resources, Investigation, Writing – review & editing. Emlyn W. Hughes: Resources, Methodology, Investigation, Writing – review & editing. R. Graham Barr: Methodology, Conceptualization, Supervision, Project administration, Funding acquisition, Writing – review & editing. Wei Shen: Conceptualization, Methodology, Resources, Data curation, Writing – review & editing, Visualization, Supervision, Project administration.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mri.2022.06.016.
References
- [1].de Lange EE, Altes TA, Patrie JT, Gaare JD, Knake JJ, Mugler JP 3rd, et al. Evaluation of asthma with hyperpolarized helium-3 MRI: correlation with clinical severity and spirometry. Chest 2006;130(4):1055–62. [DOI] [PubMed] [Google Scholar]
- [2].Fain S, Schiebler ML, McCormack DG, Parraga G. Imaging of lung function using hyperpolarized helium-3 magnetic resonance imaging: review of current and emerging translational methods and applications. J Magn Reson Imaging 2010;32 (6):1398–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kirby M, Pike D, Coxson HO, McCormack DG, Parraga G. Hyperpolarized (3)He ventilation defects used to predict pulmonary exacerbations in mild to moderate chronic obstructive pulmonary disease. Radiology 2014;273(3):887–96. [DOI] [PubMed] [Google Scholar]
- [4].Woodhouse N, Wild JM, Paley MN, Fichele S, Said Z, Swift AJ, et al. Combined helium-3/proton magnetic resonance imaging measurement of ventilated lung volumes in smokers compared to never-smokers. J Magn Reson Imaging 2005;21 (4):365–9. [DOI] [PubMed] [Google Scholar]
- [5].Kirby M, Heydarian M, Svenningsen S, Wheatley A, McCormack DG, Etemad-Rezai R, et al. Hyperpolarized 3He magnetic resonance functional imaging semiautomated segmentation. Acad Radiol 2012;19(2):141–52. [DOI] [PubMed] [Google Scholar]
- [6].Lui JK, LaPrad AS, Parameswaran H, Sun Y, Albert MS, Lutchen KR. Semiautomatic segmentation of ventilated airspaces in healthy and asthmatic subjects using hyperpolarized 3He MRI. Comput Math Methods Med 2013;2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zha W, Niles DJ, Kruger SJ, Dardzinski BJ, Cadman RV, Mummy DG, et al. Semiautomated ventilation defect quantification in exercise-induced bronchoconstriction using hyperpolarized helium-3 magnetic resonance imaging: a repeatability study. Acad Radiol 2016;23(9):1104–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tustison NJ, Avants BB, Lin Z, Feng X, Cullen N, Mata JF, et al. Convolutional neural networks with template-based data augmentation for functional lung image quantification. Acad Radiol 2019;26(3):412–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hughes PJC, Horn FC, Collier GJ, Biancardi A, Marshall H, Wild JM. Spatial fuzzy c-means thresholding for semiautomated calculation of percentage lung ventilated volume from hyperpolarized gas and (1) H MRI. J Magn Reson Imaging 2018;47 (3):640–6. [DOI] [PubMed] [Google Scholar]
- [10].Mathew L, Kirby M, Etemad-Rezai R, Wheatley A, McCormack DG, Parraga G. Hyperpolarized 3He magnetic resonance imaging: preliminary evaluation of phenotyping potential in chronic obstructive pulmonary disease. Eur J Radiol 2011;79(1):140–6. [DOI] [PubMed] [Google Scholar]
- [11].Ray N, Acton ST, Altes T, de Lange EE, Brookeman JR. Merging parametric active contours within homogeneous image regions for MRI-based lung segmentation. IEEE Trans Med Imaging 2003;22(2):189–99. [DOI] [PubMed] [Google Scholar]
- [12].Zha W, Fain SB, Schiebler ML, Evans MD, Nagle SK, Liu F. Deep convolutional neural networks with multiplane consensus labeling for lung function quantification using UTE proton MRI. J Magn Reson Imaging 2019;50(4):1169–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tustison NJ, Avants BB, Flors L, Altes TA, Lange Eed, Mugler JP, et al. Ventilation-based segmentation of the lungs using hyperpolarized 3He MRI. J Magn Reson Imaging 2011;34(4):831–41. [DOI] [PubMed] [Google Scholar]
- [14].He M, Driehuys B, Que LG, Huang YT. Using hyperpolarized (129)Xe MRI to quantify the pulmonary ventilation distribution. Acad Radiol 2016;23(12): 1521–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Astley JR, Biancardi AM, Hughes PJC, Smith LJ, Marshall H, Eaden J, et al. 3D deep convolutional neural network-based ventilated lung segmentation using multi-nuclear hyperpolarized gas MRI. In: Petersen J, San José Estépar R, Schmidt-Richberg A, Gerard S, Lassen-Schmidt B, Jacobs C, et al., editors. Thoracic Image Analysis Cham: Springer International Publishing; 2020. p. 24–35. [Google Scholar]
- [16].Young AL, Bragman FJS, Rangelov B, Han MK, Galban CJ, Lynch DA, et al. Disease progression modeling in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2020;201(3):294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ronneberger O, Fischer P, Brox T. In: Navab N, Hornegger J, Wells WM, Frangi AF, editors. U-Net: Convolutional Networks for Biomedical Image Segmentation Springer International Publishing; 2015. p. 234–41. [Google Scholar]
- [18].Christ PF, Elshaer MEA, Ettlinger F, Tatavarty S, Bickel M, Bilic P, et al. In: Ourselin S, Joskowicz L, Sabuncu MR, Unal G, Wells W, editors. Automatic Liver and Lesion Segmentation in CT Using Cascaded Fully Convolutional Neural Networks and 3D Conditional Random Fields Springer International Publishing; 2016. p. 415–23. [Google Scholar]
- [19].Liu H, Shen X, Shang F, Ge F, Wang F. In: Zhu D, Yan J, Huang H, Shen L, Thompson PM, Westin C-F, et al. , editors. CU-Net: Cascaded U-Net with Loss Weighted Sampling for Brain Tumor Segmentation Springer International Publishing; 2019. p. 102–11. [Google Scholar]
- [20].Wang L, Li G, Shi F, Cao X, Lian C, Nie D, et al. Volume-based analysis of 6-month-old infant brain MRI for autism biomarker identification and early diagnosis In: Medical Image Computing and Computer-Assisted Intervention : MICCAI International Conference on Medical Image Computing and Computer-Assisted Intervention. 11072; 2018. p. 411–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nalepa J, Marcinkiewicz M, Kawulok M. Data augmentation for brain-tumor segmentation: a review. Front Comput Neurosci 2019:13. [DOI] [PMC free article] [PubMed]
- [22].Shorten C, Khoshgoftaar TM. A survey on image data augmentation for deep learning. J Big Data 2019;6(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Shin H-C, Tenenholtz NA, Rogers JK, Schwarz CG, Senjem ML, Gunter JL, et al. In: Gooya A, Goksel O, Oguz I, Burgos N, editors. Medical Image Synthesis for Data Augmentation and Anonymization Using Generative Adversarial Networks Springer International Publishing; 2018. p. 1–11. [Google Scholar]
- [24].Frid-Adar M, Klang E, Amitai M, Goldberger J, Greenspan H. Synthetic data augmentation using GAN for improved liver lesion classification In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018; ); 2018. p. 289–93. [Google Scholar]
- [25].Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative adversarial nets. In: Ghahramani Z, Welling M, Cortes C, Lawrence ND, Weinberger KQ, editors. Advances in Neural Information Processing Systems 27. Curran Associates, Inc.; 2014. p. 2672–80. [Google Scholar]
- [26].Pandey S, Singh PR, Tian J. An image augmentation approach using two-stage generative adversarial network for nuclei image segmentation. Biomed Signal Proc Control 2020;57:101782. [Google Scholar]
- [27].Bowles C, Chen L, Guerrero R, Bentley P, Gunn R, Hammers A, et al. GAN Augmentation: Augmenting Training Data Using Generative Adversarial Networks. arXiv:181010863 [cs] 2018.
- [28].Han C, Kitamura Y, Kudo A, Ichinose A, Rundo L, Furukawa Y, et al. Synthesizing diverse lung nodules wherever massively: 3D multi-conditional GAN-based CT image augmentation for object detection In: 2019 International Conference on 3D Vision (3DV); 2019. p. 729–37. [Google Scholar]
- [29].Smith BM, Austin JH, Newell JD Jr, D’Souza BM, Rozenshtein A, Hoffman EA, et al. Pulmonary emphysema subtypes on computed tomography: the MESA COPD study. Am J Med 2014;127(1). 94 e7–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Horn FC, Tahir BA, Stewart NJ, Collier GJ, Norquay G, Leung G, et al. Lung ventilation volumetry with same-breath acquisition of hyperpolarized gas and proton MRI. NMR Biomed 2014;27(12):1461–7. [DOI] [PubMed] [Google Scholar]
- [31].Shen W, Sun Y, Balte P, Dashnaw S, Prince M, Hoffman EA, et al. Low and absent ventilation percent on He-3 MRI and cardiac function: the Mesa COPD Study. Am J Respir Crit Care Med 2018;197:A3919. [Google Scholar]
- [32].Geer EB, Shen W, Strohmayer E, Post KD, Freda PU. Body composition and cardiovascular risk markers after remission of Cushing’s disease: a prospective study using whole-body MRI. J Clin Endocrinol Metab 2012;97(5):1702–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Shen W, Chen J, Gantz M, Velasquez G, Punyanitya M, Heymsfield SB. A single MRI slice does not accurately predict visceral and subcutaneous adipose tissue changes during weight loss. Obesity (Silver Spring, Md) 2012;20(12):2458–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Shen W, Velasquez G, Chen J, Jin Y, Heymsfield SB, Gallagher D, et al. Comparison of the relationship between bone marrow adipose tissue and volumetric bone mineral density in children and adults. J Clin Densitom 2014;17(1):163–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Reyes-Vidal CM, Mojahed H, Shen W, Jin Z, Arias-Mendoza F, Fernandez JC, et al. Adipose tissue redistribution and ectopic lipid deposition in active acromegaly and effects of surgical treatment. J Clin Endocrinol Metab 2015;100(8):2946–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Karras T, Aila T, Laine S, Lehtinen J. Progressive growing of GANs for improved quality, stability, and variation International Conference on Learning Representations. 2018. https://openreview.net/forum?id=Hk99zCeAb. [Google Scholar]
- [37].Wang T-C, Liu M-Y, Zhu J-Y, Tao A, Kautz J, Catanzaro B. High-resolution image synthesis and semantic manipulation with conditional GANs In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2018. p. 8798–807. [Google Scholar]
- [38].Paszke A, Gross S, Chintala S, Chanan G, Yang E, DeVito Z, et al. Automatic Differentiation in PyTorch 2017.
- [39].Sudre CH, Li W, Vercauteren T, Ourselin S, Cardoso MJ. Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations In: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: Third International Workshop, DLMIA 2017, and 7th International Workshop, ML-CDS 2017, Held in Conjunction with MICCAI 2017 Quebec City, QC. Europe PMC Funders; 2017. p. 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Horváth A, Tsagkas C, Andermatt S, Pezold S, Parmar K, Cattin P. Spinal cord gray matter-white matter segmentation on magnetic resonance AMIRA images with MD-GRU In: International Workshop and Challenge on Computational Methods and Clinical Applications for Spine Imaging. Springer; 2018. p. 3–14. [Google Scholar]
- [41].Kingma DP, Ba J Adam: A Method for Stochastic Optimization. arXiv:14126980 [cs] 2017.
- [42].Michelle Livne LD, DeepMind Technologies. Surface Distance Available from: https://github.com/deepmind/surface-distance; 2018.
- [43].Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. In: Bengio Y, LeCun Y, editors. 3rd International Conference on Learning Representations San Diego, CA, USA: ICLR; 2015. [Google Scholar]
- [44].Maaten Lvd, Hinton G. Visualizing Data using t-SNE. J Mach Learn Res 2008;9(86): 2579–605. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


