Abstract
In recent years, the human virome has gained importance, especially after the SARS-CoV-2 pandemic, due to its possible involvement in autoimmune, inflammatory diseases, and cancer. Characterization of the human virome can be carried out by shotgun next-generation sequencing (metagenomics), which allows the identification of all viral communities in an environmental sample and the discovery of new viral families not previously described. Variations in viral quantity and diversity have been associated with disease development, mainly due to their effect on gut bacterial microbiota. Phages can regulate bacterial flora through lysogeny; this is associated with increased susceptibility to infections, chronic inflammation, or cancer. The virome characterization in different human body ecological niches could help elucidate these particles' role in disease. Hence, it is important to understand the virome's influence on human health and disease. The present review highlights the significance of the human virome and how it is associated with disease, focusing on virome composition, characterization, and its association with cancer.
Keywords: Human virome, Microbiota, Metagenomics, Cancer
1. Introduction
The human microbiome includes all the microorganisms (protozoa, fungi, bacteria, archaea, viruses), their genetic material, and metabolites, which inhabit and interact in the different ecological human body niches [[1], [2], [3], [4]]. An imbalance in the proportion of microorganism species has been associated with disease development [5]. Several studies have focused mainly on the characterization of the bacterial microbiome and its metabolites in cancer, autoimmune, neurological, and metabolic diseases, among others [[5], [6], [7]]. All this evidence has made it possible to recognize the role played by the bacterial microbiome in human health and immunity.
Moreover, in recent years, especially after the SARS-CoV-2 pandemic, efforts have been made to characterize not only the bacteria that inhabit our body but also the viruses (human virome) [8,9]. The human virome is composed of viruses that infect eukaryotic cells, bacteria (phages), other microorganisms such as protozoa, fungi, or archaea, and genetic elements derived from viruses anchored to the host chromosomes [10]. Furthermore, the imbalance of the human virome has been associated with the development of cancer, metabolic, and inflammatory diseases [[11], [12], [13]]. For example, Chen F. et al. (2022) compared the gut virome of colorectal cancer patients and healthy subjects. The authors found that the virome was drastically altered, identifying viruses that were only present in cancer patients [14].
Moreover, phages can be lysogenic or lytic. Lysogenic phages do not kill the cell; they integrate into the host genome without interfering with the replication [15]. On the other hand, lytic phages bind to specific receptors in the cell and then penetrate and invade the host; by doing so, they hijack the cell’s machinery to make copies of the virus and release their genetic material to the environment. Lytic phages have been used as alternatives to antibiotic therapies and to treat infections [15]. Furthermore, phages can act as vectors by transferring genetic material to other bacteria (Transduction), improving their virulence, production of virulence factors, toxic synthesis, and antibiotic resistance [12,16]. The order Caudovirales (Myoviridae, Siphoviridae, and Podoviridae), which are double-stranded DNA phages, is the most prevalent [8,17].
A metagenomics approach using shotgun-type next-generation sequencing (NGS) technology has made it possible to characterize the human virome [5,6,12]. The main advantage of NGS over other methods is that it does not require prior knowledge of the viruses in the samples; hence all the genetic material can be identified and characterized [15,18]. Breitbart M. et al. (2002) started the virome study with the characterization of viruses present in a seawater sample, in which a wide diversity of viral communities was found and a high number of viral genomes that had not been previously characterized [19]. Since then, the number of publications to describe the virome in different ecological niches has increased using Metaviromics approaches for its characterization [20].
The adult gut virome is highly stable and may be dominated by one virotype over different time periods [8,21]; however, the interindividual variability is significant. For instance, Reyes A. et al. (2010) compared the virome of 88 different subjects and found that only eight genomes were found in more than one individual [21]. Furthermore, an imbalance of the human virome could be promoted by different factors, such as diet, breastfeeding, medications (antibiotics, chemotherapy, immunosuppressants), host genetics, geographic region, underlying diseases, and age [13]. Moreover, based on the virus ability to kill bacterial hosts, the human virome could potentially modulate the proliferation or decrease of specific bacterial communities, and their production of metabolites, leading to an imbalanced microbiome [12,16,17].
In addition to the bacterial microbiome regulation, the virome may be involved in the human immune system development by directly interacting with the host’s mucosa at an early age. In this way, the immune system produces antibodies and cytokines that protect the host against viral or bacterial infections, thus proposing a mutualistic symbiotic relationship between the human being and the virome [10].
Understanding how the human virome is associated with disease is of the utmost importance to acknowledge the influence that it has on human health. The present review highlights the importance of the human virome and how it is associated with disease, focusing on virome composition, characterization, and its association with cancer.
2. Human virome composition
The human virome is composed of viral genetic elements anchored to the host chromosomes (human endogenous retroviruses) and single and double-stranded DNA and RNA viruses (ssDNA, dsDNA, ssRNA, dsRNA) that inhabit the different ecological niches of the human body, such as skin, mucous membranes, respiratory system, genitourinary system, gastrointestinal system, and even blood or cerebrospinal fluid [10,22]. It is estimated that the human body has 1012 virus-like particles (VLP), with the highest proportion in the gastrointestinal tract (∼109–10 [10] VLP). However, the quantity and diversity of the intestinal virome will depend on individual and environmental factors such as diet, geographical location, age, and lifestyle that influence its composition [[22], [23], [24], [25]]. Most of the studies have focused on the description of the intestinal virome; however, the characterization of other ecological niches like the oral cavity, nostrils, ears, genitourinary system, and respiratory system, among others, could help to generate associations between the bacterial microbiome composition, host pathogenesis, and immunity [22]. Table 1 shows viruses present in various ecological niches of the human body [18,23,[25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37]].
Table 1.
Viruses present in several ecological niches of the human body.
| Human body site | VLP Approximate amount | Eukaryotic viruses | Phages |
|---|---|---|---|
| Gastrointestinal tract | ∼109 |
Siphoviridae Podoviridae Myoviridae Microviridae Inoviridae Caliciviridae, Picornaviridae Reoviridae Virgaviridae |
Microviridae Siphoviridae Podoviridae Myoviridae Inoviridae |
| Oral cavity | ∼108 |
Herpesviridae, Papillomaviridae, Anelloviridae Redondoviridae |
Siphoviridae Podoviridae Myoviridae |
| Respiratory tract | n/a |
Anelloviridae Redondoviridae Adenoviridae, Herpesviridae Papillomaviridae |
Siphoviridae Podoviridae Myoviridae Microviridae Inoviridae |
| Blood | n/a |
Anelloviridae Herpesviridae, Marseilleviridae, Mimiviridae, Phycodnaviridae Picornaviridae |
Myoviridae, Siphoviridae, Podoviridae, Microviridae Inoviridae |
| Nervous system | ∼104 | Herpesviridae | Myoviridae, Siphoviridae Podoviridae |
| Skin | n/a |
Polyomaviridae, Papillomaviridae Circoviridae Adenoviridae, Anelloviridae, Herpesviridae, |
Siphoviridae Podoviridae Myoviridae |
| Genitourinary system | ∼107 |
Anelloviridae Papillomaviridae Polyomaviridae Herpesviridae |
Siphoviridae Podoviridae Myoviridae |
n/a: Not available; Modified from Liang and Bushman, 2021.
Regarding the intestinal virome, the viruses that infect eukaryotic cells (eukaryotic viruses) are less than 10% of the total virome, which can be in a latency state until a host immunosuppression condition occurs [22]. Eukaryotic viruses can be ssDNA (Anelloviridae, Circoviridae) and dsDNA (Adenoviridae, Herpesviridae, Papillomaviridae, Polyomaviridae), which cause several host diseases [13,21,23,27]. Few RNA viruses infect eukaryotes and may induce an infectious process. RNA viruses such as Reoviridae (rotavirus), Caliciviridae (norovirus), and Picornaviridae (enterovirus) are examples of viruses that cause gut infection [13,28,37]. RNA viruses may be underestimated, as the stability of their genetic material is low, and not all of these viruses can be detected in a sample [22].
Most of the human virome is composed of phages (∼90%), which can infect and lyse the bacteria in the gut [13,22,23,28,37]. However, these phages can also interact differently with bacteria. For example, phages can insert their genome into the bacterial chromosome and remain dormant until an induction signal leads to virus multiplication and bacterial lysis (lysogeny) [15]. An interaction by pseudolysogeny can also occur, in which the viral genome is free in the host without the lytic or lysogeny process. Finally, phages can insert their genome into the host and produce new viral particles by budding, keeping the bacterial cell intact [13,25]. In the human gut, Myoviridae, Podoviridae, Siphoviridae, and Microviridae are the most predominant phages [21,28,29,38].
Human endogenous retroviruses (viral genetic elements), coupled to the host chromosome, represent approximately 8% of the human genome and can be transferred to offspring [13]. These genetic elements do not produce viral particles, but they can generate proteins associated with the development of various autoimmune, neurodegenerative, and inflammatory diseases or cancer [11,13,29,32,39,40]. The viral genome harbors replication, transcription, and translation promoter regions, which could explain how these viral genetic elements regulate gene expression when anchored to the host cell genome and may promote disease [41].
3. Virome and cancer
Several viruses have been classified as causal agents of carcinogenesis, such as the Epstein Barr virus (EBV), human papillomavirus (HPV), hepatitis B virus (HBV), hepatitis C virus (HCV), and human T-cell lymphotropic virus type 1 (HTLV-1) [11,13]. The effect of these viruses on carcinogenesis is based on the direct transformation of infected cells with virus-derived oncogenes. Cancer could also be generated indirectly when a virus-induced infection causes chronic inflammation [11]. Immune system development is related to gut microbiota composition; for instance, it has been described that the immune system of animal models was dysregulated in the lack of microbial communities [42]. Therefore, virome composition could be implicated in immune system development by regulating bacterial communities; moreover, a depressed immune system is associated with inflammatory diseases and cancer development [43].
Moreover, recent studies have highlighted the virome’s role in carcinogenesis (especially in CRC) [[44], [45], [46], [47]]. The principal associated mechanism is the modulation of bacterial communities or direct host cell transformation [45,47]. The molecular and functional role is still under research. However, these new insights allow the potential use of virome biomarkers as diagnostic, prognostic, or treatment tools in several cancer types.
Herpes simplex virus-like particles were first observed in Burkitt’s lymphoma cells [48]. After several studies, these viral particles were identified as EBV, and their gene products have been detected in different human cancers like nasopharyngeal carcinoma, Hodgkin’s disease, gastric carcinomas, and lymphoproliferative disorders [49]. The principal EBV oncogenic mechanism is epigenetic regulation (histone, DNA, nucleosome, and chromatin modification) that control viral gene expression [50].
Similarly, HPV has been identified as a high or low-malignant risk factor; its association with cervical, oral, oropharyngeal, and penile carcinomas has been well-documented [[51], [52], [53]]. HPV induces carcinogenesis by integrating into the host cells and triggering an altered gene expression [54].
Likewise, Hepatocellular carcinoma (HCC) has been associated with HBV and HCV infections [55,56]. HCC develops from HCV chronic infection that leads to hepatic fibrosis and cirrhosis by upregulating profibrogenic signaling pathways [57] and cell cycle stimulation [58]. On the other hand, HBV can integrate its viral genome into the host cells, producing viral gene products which act as transcriptional activators of cell growth-associated genes [59].
Additionally, HTLV-1 has been associated as an etiological agent of Adult T-cell leukemia, transforming infected cells by integrating into HTLV-1 regulatory and accessory genes, which trigger CD4+ T-lymphocytes malignancy [60]. Moreover, this virus has also been found in cutaneous T-cell lymphoma (mycosis fungoides) [61].
Virome’s role in carcinogenesis is still under study since no direct or causal associations have been reported. For example, an imbalanced gut virome has been reported in patients with colorectal cancer (CRC) [[11], [12], [13],22]; this imbalance is characterized by presenting a greater viral quantity and diversity in the feces of individuals with CRC compared to healthy individuals [40]. Moreover, individuals with CRC may have a lower diversity of butyrate-producing bacteria (Firmicutes like Faecalibacterium prausnitzii, Eubacterium rectale) or a decrease in bacteria that supports a healthy gut state (R. intestinalis) [62].
Similarly, an enrichment of Bacteroides, Parabacteroides sp., Alistipes putredinis, Bilophila wadsworthia, Lachnospiraceae bacterium and Escherichia coli, Gemella, Peptostreptococcus, Parvimonashave, and Fusobacterium, has been observed in CRC patients [63,64]. Among the phages involved in this imbalance are Inovirus, Tunalikevirus, Siphoviridae, Myoviridae, Streptococcus phage SpSL1, Streptococcus phage 5093, Streptococcus phage K13, Vibrio phage pYD38-A, Enterobacteria phage HK544 [40]. Phages have been associated with antimicrobial activity when lysing gut bacteria [65]. Hence, virome gut alteration could promote pathologic bacteria enrichment and trigger gut inflammation [66]. Research suggests that phages may be involved in the “leaky gut” development that could lead to pathogenic bacteria entry into intestinal cells, inducing a chronic gut inflammatory process [67]. A decrease in beneficial bacteria may be due to a greater quantity of bacterial lysis by specific phages, as observed in environmental media [68].
In a metagenomic analysis of colorectal carcinomas, phages were the most predominant viral species. Moreover human endogenous retrovirus K113, human herpesviruses 7-6B, Megavirus chilensis, Cytomegalovirus (CMV) and Epstein-Barr virus (EBV), were the most frequently detected viruses in primary CRC and metastases patients [69].
Moreover, persistent human CMV infections have also been associated with altered outcomes in cancer patients promoted by an upregulation of the WNT signaling pathway (associated with CRC cell proliferation and migration) in CRC-derived HT29 and SW480 ″stem-like” cells [70].
Similarly, JC virus (polyomavirus family) infections have been associated with CRC by genetic and epigenetic instability promotion. This chromosomal instability (CIN) and the CpG island methylator phenotype (CIMP) are promoted by oncogenic T-antigen (T-Ag) JVC virus expression [71,72]. Moreover, DNA viruses can infect the host cell and promote long-term persistent infections, which could be an important aspect of cell transformation and oncogenesis [73].
On the other hand, phages serve as mediators of horizontal gene transfer, promoting the exchange of virulence, metabolic, or antimicrobial resistance genes between bacterial species in the gut [16]. As mentioned above, virome imbalance could induce an increase in pathogenic bacteria such as enterobacteria. Hence, the exchange of genetic material in these bacteria would cause a greater infective capacity, greater survival rate, and resistance to antibiotic treatments.
Nowadays, the public health problem caused by multidrug-resistant bacteria has been broadly described [74]. The surveillance of viruses as mediators of antimicrobial resistance gene transfer could help to develop new strategies against infectious diseases.
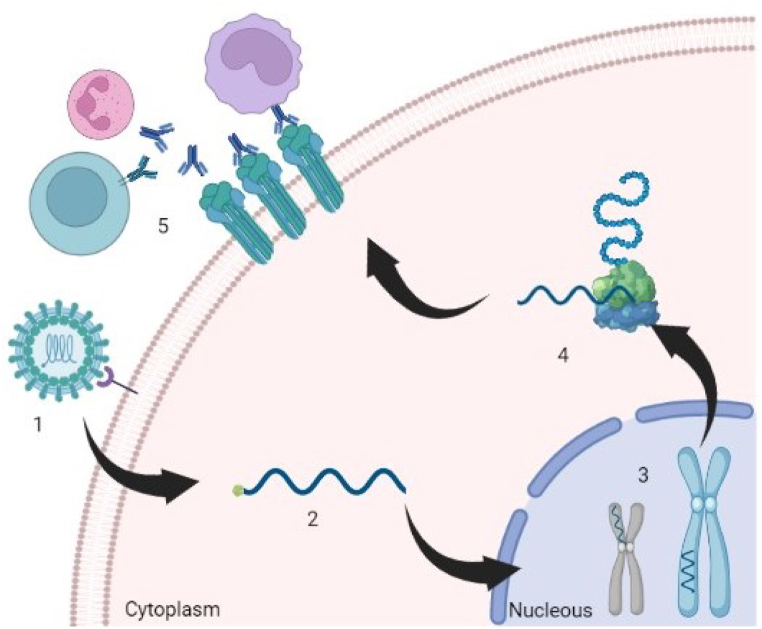
In this context, it is proposed that eukaryotic viruses could insert viral genetic elements into the host genome, encoding viral envelope proteins which will be expressed in the host cell membranes. These proteins would later be recognized by the immune system cells (lymphocytes, neutrophils, macrophages), inducing chronic inflammation and subsequent cancer development (Fig. 1).
Fig. 1.
Chronic inflammation induced by viral proteins anchored to plasma membranes. 1: Recognition of the viral particle by the membrane receptor. 2: Entry of viral genetic material into the cytoplasm. 3: Integration of the viral genome into the genome (chromosome) of the host. 4: Expression of viral genetic elements. 5: Anchoring of viral proteins in the plasma membrane and stimulation of the immune response.
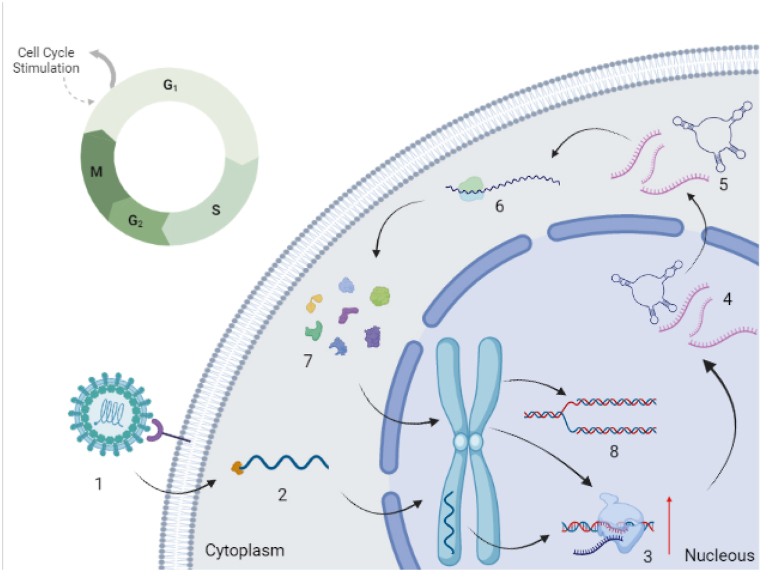
Viruses can transform cells (prokaryotes and eukaryotes) by inserting viral genetic elements into the host genome, which could be involved in gene expression regulation by modulating viral particle replication, transcription, or translation. Therefore, if those genetic elements are inserted into the host genome, there could be a continuous stimulation of replication, transcription, or translation processes of genes in which that fragment has been inserted. In this context, gene overexpression could lead to chronic inflammation and cancer development (Fig. 2) [75,76].
Fig. 2.
Cell cycle stimulation by viral protein factors. 1: Recognition of the viral particle by the membrane receptor. 2: Entry of viral genetic material into the cytoplasm and integration of the viral genome into the genome (chromosome) of the host. 3: Transcription of viral genetic elements. 4: Maturation of mRNA, miRNAs, rRNA of viral genetic elements. 5–6: Expression of viral genetic elements associated with viral replication/transcription stimulating factors. 7: Entry of viral replication/transcription stimulating factors into the nucleus. 8: Stimulation of replication and transcription of the host genome.
The viral genetic elements characterization in the human genome could clarify the quantity in which these elements are found and evaluate their ability to regulate gene expression. It would allow the generation of research projects to determine whether these elements are involved in stimulating the cell cycle and gene expression in different types of cancer, and autoimmune/chronic inflammatory diseases, among others.
Therefore, a continuous virome characterization in several environmental ecological niches is necessary, especially in those in direct contact with the human population. Furthermore, it could compare whether DNA/RNA fragments of the viral particles, found in the environment, are present in the human host genome.
4. Virome characterization
The virome characterization in environmental and human ecological niches has been possible thanks to the use of high throughput metagenomic sequencing techniques; this technology allows the identification of viral communities and the characterization of new viruses or viruses that cannot be cultured [77].
Shotgun metagenomic analysis allows us to classify, at a taxonomic level, the raw data obtained in metagenomics sequencing. This technology includes several steps like 1) sample collection, processing, and sequencing, 2) sequence raw data processing, 3) taxonomic profile and functional-genomic features sequence analysis, 4) statistical and biological analysis, and 5) validation [78].
Metagenomics has been used in several environmental or human complex microbiomes [[79], [80], [81], [82], [83]]. The advantage of shotgun metagenomics over 16S rRNA NGS sequencing is the more powerful taxonomic identification in less abundant microbiome taxa [84]. Hence, this technology could characterize the global microbiome community organization.
Despite the improvements in performance and sensitivity, challenges persist, for instance, the simultaneous characterization of the different types of viruses present in a sample (dsDNA, ssDNA, ssRNA, dsRNA). In addition, the newly identified viruses (mainly phages) are difficult to assign a specific host (bacteria) [77]. However, technological alternatives to solve these problems have already been proposed, such as commercial protocol development that allows the simultaneous extraction of DNA and RNA in several biological samples. Furthermore, there is an improvement in metagenomics protocol and bioinformatic tools for the species identification that makes up the virome [77,85]. Likewise, amplification methodologies are being improved, allowing the identification of longer sequences, improving genome coverage, and reducing the error rate [77]. The use of machine learning has also been implemented to identify the viral genome regions involved in host pathogenicity [86]. These methodologies will help to understand the interaction between viruses and their host and identify possible targets for drug development that prevent or limit the infectious/inflammatory process produced by viruses in the human body.
5. Conclusions
The study of the virome has gained importance due to its possible role in the development of several diseases including cancer. Virome biomarkers could be used as new diagnostic, prognostic, or treatment tools in several cancer types. Shotgun metagenomic technology becomes a powerful instrument to assess a trans-kingdom interaction (virome, bacterial microbiota, fungi, host cells) to search its role in carcinogenesis. Most studies have focused mainly on intestinal virome characterization due to its association with intestinal diseases and CRC. Viral genetic elements could be a new topic of investigation to evaluate their role in disease development. In conclusion, metagenomic technology is a powerful tool for human virome characterization. Knowledge obtained from metagenomic analysis could help to understand the genesis of several diseases and new treatment strategies development.
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Funding statement
This study is supported by Universidad UTE.
Data availability statement
Data included in article/supp. material/referenced in article.
Declaration of interest’s statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
No additional information is available for this paper.
Abbreviations
- CMV
Cytomegalovirus
- CRC
colorectal cancer
- dsDNA
double-stranded DNA
- dsRNA
double-stranded RNA
- EBV
Epstein Barr virus
- HBV
hepatitis B virus
- HCV
hepatitis C virus
- HCC
Hepatocellular carcinoma
- HPV
human papillomavirus
- HTLV-1
human T-cell lymphotropic virus type 1
- NGS
Next Generation Sequencing
- ssDNA
single-stranded DNA
- ssRNA
single-stranded RNA
- VLP
virus like particles
References
- 1.del Campo-Moreno R., Alarcón-Cavero T., D'Auria G., Delgado-Palacio S., Ferrer-Martínez M. Microbiota en la salud humana: técnicas de caracterización y transferencia. Enferm. Infecc. Microbiol. Clín. 2018;36(4):241–245. doi: 10.1016/j.eimc.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Gilbert J., Blaser M.J., Caporaso J.G., Jansson J., Lynch S.V., Knight R. Current understanding of the human microbiome. Nat. Med. 2018;24(4):392–400. doi: 10.1038/nm.4517.Current. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mousa W.K., Chehadeh F., Husband S. Recent advances in understanding the structure and function of the human microbiome. Front. Microbiol. 2022;13(825338):1–22. doi: 10.3389/fmicb.2022.825338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ariza-Andraca R., García-Ronqsuillo M. El microbioma humano. Su papel en la salud y en algunas enfermedades. Cir. Cir. 2016;84(Supl 1):31–35. [Google Scholar]
- 5.Durack J., Lynch S.V. The gut microbiome: relationships with disease and opportunities for therapy. J. Exp. Med. 2019;216(1):20–40. doi: 10.1084/jem.20180448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vijay A., Valdes A.M. Role of the gut microbiome in chronic diseases: a narrative review. Eur. J. Clin. Nutr. 2022;76(4):489–501. doi: 10.1038/s41430-021-00991-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Rahman M.M., Islam M.R., Shohag S., et al. Microbiome in cancer: role in carcinogenesis and impact in therapeutic strategies. Biomed. Pharmacother. 2022;149(112898):1–22. doi: 10.1016/j.biopha.2022.112898. [DOI] [PubMed] [Google Scholar]
- 8.Minot S., Sinha R., Chen J., et al. 2011. The Human Gut Virome : Inter-individual Variation and Dynamic Response to Diet; pp. 1616–1625. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wylie K.M., Weinstock G.M., Storch G.A. Emerging view of the human virome. Transl. Res. 2012;160(4):283–290. doi: 10.1016/j.trsl.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Virgin H. The virome in mammalian physiology and disease. Cell. 2014;157(1):142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santiago-Rodriguez T.M., Hollister E.B. Human virome and disease : high-throughput phage-bacteria dysbiosis and development of human gut. Viruses. 2019;11(7):656. doi: 10.3390/v11070656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spencer L., Olawuni B., Singh P. Gut virome: role and distribution in health and gastrointestinal diseases. Front. Cell. Infect. Microbiol. 2022;12(March:1–11. doi: 10.3389/fcimb.2022.836706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bai G.-H., Lin S.-C., Hsu Y.-H., Chen S.-Y. The human virome: viral metagenomics, relations with human diseases , and therapeutic applications. Viruses. 2022;14(278):1–29. doi: 10.3390/v14020278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen F., Li S., Guo R., et al. Meta-analysis of fecal viromes demonstrates high diagnostic potential of the gut viral signatures for colorectal cancer and adenoma risk assessment. J. Adv. Res. 2022;(xxxx) doi: 10.1016/j.jare.2022.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carding S.R., Hoyles N.D.L. 2017. Review Article : the Human Intestinal Virome in Health and Disease; pp. 800–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moon K., Cho J. Metaviromics coupled with phage-host identification to open the viral ‘ black box. J. Microbiol. 2021;59(3):311–323. doi: 10.1007/s12275-021-1016-9. [DOI] [PubMed] [Google Scholar]
- 17.Ogilvie L.A., Jones B.V. The human gut virome : form and function. Emerg Top life Sci. 2017:351–362. doi: 10.1042/ETLS20170039. [DOI] [PubMed] [Google Scholar]
- 18.Rascovan N., Duraisamy R., Desnues C. Metagenomics and the human virome in asymptomatic individuals. Annu. Rev. Microbiol. 2016;70:125–141. doi: 10.1146/annurev-micro-102215-095431. [DOI] [PubMed] [Google Scholar]
- 19.Breitbart M., Salamon P., Andresen B., et al. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. U. S. A. 2002;99(22):14250–14255. doi: 10.1073/pnas.202488399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raghavendra P., Thammineni P. 2018. Pathogen Identification Using Novel Sequencing Methods; pp. 161–202. (Advances in Cell and Molecular Diagnostics). [DOI] [Google Scholar]
- 21.Reyes A., Haynes M., Hanson N., et al. Viruses in the fecal microbiota of monozygotic twins and their mothers. Nature. 2010;466(7304):334–338. doi: 10.1038/nature09199.Viruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao Z., Sugimura N., Burgermeister E., Ebert M.P., Zuo T., Lan P. The gut virome: a new microbiome component in health and disease. EBioMedicine. 2022;81(104113):1–17. doi: 10.1016/j.ebiom.2022.104113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shkoporov A.N., Clooney A.G., Sutton T.D.S., et al. Article the human gut virome is highly diverse , stable, and individual specific. Cell Host Microbe. 2019;26(4):527–541.e5. doi: 10.1016/j.chom.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 24.Shkoporov A.N., Hill C. Review bacteriophages of the human gut : the “known unknown” of the microbiome. Cell Host Microbe. 2019;25(2):195–209. doi: 10.1016/j.chom.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 25.Bushman F.D., Liang G. The human virome : assembly , composition and host interactions. Nat. Rev. Microbiol. 2021;19(August) doi: 10.1038/s41579-021-00536-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abeles S.R., Ly M., Santiago-rodriguez T.M., Pride D.T. 2015. Effects of Long Term Antibiotic Therapy on Human Oral and Fecal Viromes; pp. 1–18. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dutilh B.E., Cassman N., Mcnair K., et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun. 2014:1–11. doi: 10.1038/ncomms5498. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guerin E., Shkoporov A., Stockdale S.R., et al. Biology and taxonomy of crAss-like bacteriophages , the most abundant virus in the human gut biology and taxonomy the most abundant virus in the human gut. Cell Host Microbe. 2018;24(5):653–664.e6. doi: 10.1016/j.chom.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 29.Edwards R.A., Vega A., Norman H., et al. Global phylogeography and ancient evolution of the widespread human gut virus crAssphage. Nat. Microbiol. 2019;4(10):1727–1736. doi: 10.1038/s41564-019-0494-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yutin N., Makarova K., Gussow A., et al. Discovery of an expansive bacteriophage family that includes the most abundant viruses from the human gut. Nat. Microbiol. 2018;3(1):38–46. doi: 10.1038/s41564-017-0053-y.Discovery. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang T., Breitbart M., Lee W.H., et al. RNA viral community in human feces : prevalence of plant pathogenic viruses. PLoS Biol. 2006;4(1) doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aggarwala V., Liang G., Bushman F.D. 2017. Viral Communities of the Human Gut : Metagenomic Analysis of Composition and Dynamics; pp. 1–10. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young J.C., Chehoud C., Bittinger K., et al. Viral metagenomics reveal blooms of anelloviruses in the respiratory tract of lung transplant recipients. Am. J. Transplant. 2015;15(1):200–209. doi: 10.1111/ajt.13031.Viral. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abbas A.A., Taylor L.J., Dothard M.I., et al. Redondoviridae, a family of small, circular DNA viruses of the human oro-respiratory tract that are associated with periodontitis and critical illness. Cell Host Microbe. 2019;25(5):719–729. doi: 10.1016/j.chom.2019.04.001. (Redondoviridae) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spezia P., Macera L., Mazzetti P., et al. Redondovirus DNA in human respiratory samples. J. Clin. Virol. 2020;131(January) doi: 10.1016/j.jcv.2020.104586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lázaro-Perona F., Dahdouh E., Román-Soto S., et al. Metagenomic detection of two vientoviruses in a human sputum sample. Viruses. 2020;12(327):3–7. doi: 10.3390/v12030327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abeles S.R., Robles-sikisaka R., Ly M., et al. Human oral viruses are personal , persistent and gender-consistent. ISME J. 2014;8(9):1753–1767. doi: 10.1038/ismej.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shkoporov A.N., Khokhlova E.V., Fitzgerald C.B., et al. ΦCrAss001 represents the most abundant bacteriophage family in the human gut and infects Bacteroides intestinalis. Nat. Commun. 2018:1–8. doi: 10.1038/s41467-018-07225-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandes M.A., Verstraete S.G., Phan T.G., et al. Enteric virome and bacterial microbiota in children with ulcerative colitis and crohn's disease. J. Pediatr. Gastroenterol. 2019;68(1):30–36. doi: 10.1097/MPG.0000000000002140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakatsu G., Zhou H., Ka W., et al. Alterations in enteric virome are associated with colorectal cancer and survival outcomes. Gastroenterology. 2018;155(2):529–541.e5. doi: 10.1053/j.gastro.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 41.Chaitanya K.V. Structure and organization of virus genomes. Genome and Genomics. 2019;25:1–30. doi: 10.1007/978-981-15-0702-1_1. [DOI] [Google Scholar]
- 42.Round J.L., Mazmanian S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009;9(5):313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grivennikov S.I., Greten F.R., Karin M. Immunity, inflammation, and cancer. Cell. 2011;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao R., Zhu Y., Kong C., et al. Alterations, interactions, and diagnostic potential of gut bacteria and viruses in colorectal cancer. Front. Cell. Infect. Microbiol. 2021;11(July):1–13. doi: 10.3389/fcimb.2021.657867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hannigan G.D., Duhaime M.B., Ruffin M.T., Koumpouras C.C., Schloss P.D. Diagnostic potential and interactive dynamics of the colorectal cancer virome. MBio. 2018;9(6):1–13. doi: 10.1128/mBio.02248-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z., Guo K., Liu Y., Huang C., Wu M. Dynamic impact of virome on colitis and colorectal cancer: immunity, inflammation, prevention and treatment. Semin. Cancer Biol. 2022;86(P2):943–954. doi: 10.1016/j.semcancer.2021.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emlet C., Ruffin M., Lamendella R. Enteric virome and carcinogenesis in the gut. Dig. Dis. Sci. 2020;65(3):852–864. doi: 10.1007/s10620-020-06126-4. [DOI] [PubMed] [Google Scholar]
- 48.Yasunaga J., Matsuoka M. Leukaemogenic mechanism of human T-cell leukaemia virus type I. Rev. Med. Virol. 2007;17:301–311. doi: 10.1002/rmv.548. [DOI] [PubMed] [Google Scholar]
- 49.Young L.S., Rickinson A.B. Epstein–barr virus: 40 years on. Nat. Rev. Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 50.Tempera I., Lieberman P.M. Epigenetic regulation of EBV persistence and oncogenesis. Semin. Cancer Biol. 2014:22–29. doi: 10.1016/j.semcancer.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boshartb M., Gissmann L., Ikenberg H., Kleinheinzl A., Scheurlen W., Hausen H. A new type of papillomavirus DNA, its presence in genital cancer biopsies and in cell lines derived from cervical cancer. EMBO J. 1984;3(5):1151–1157. doi: 10.1002/j.1460-2075.1984.tb01944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Durst M., Gissmann L., Ikenberg H., Hausen H.Z.U.R. A papillomavirus , DNA- from a cervical carcinoma and-its - prevalence in cancer biopsy samples from different geographic regions. Proc. Natl. Acad. Sci. U. S. A. 1983;80(12):3812–3815. doi: 10.1073/pnas.80.12.3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gillison M.L., Koch W.M., Capone R.B., et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J. Natl. Cancer Inst. 2000;92(9) doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 54.Münger K., Baldwin A., Edwards K.M., et al. Mechanisms of human papillomavirus-induced oncogenesis. J. Virol. 2004;78(21):11451–11460. doi: 10.1128/JVI.78.21.11451–11460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Donato F., Boffetta P., Uoti M. A meta-analysis of epidemiological studies on the combined effect of hepatitis B and C virus infections in causing hepatocellular carcinoma. Int. J. Cancer. 1998;75(3):347–354. doi: 10.1002/(sici)1097-0215(19980130)75:3<347::aid-ijc4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 56.Choo Q., Kuo G., Weiner A.M.Y.J., Overby L.R., Bradley D.W., Houghton M. Isolation of a cDNA clone derived from a blood- borne non-A , non-B viral hepatitis genome. Science (80-) 1988;244(4902):359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 57.Schulze-Krebs A., Preimel D., Popov Y., et al. Hepatitis C virus–replicating hepatocytes induce fibrogenic activation of hepatic stellate cells. Gastroenterology. 2005;129(1):246–258. doi: 10.1053/j.gastro.2005.03.089. [DOI] [PubMed] [Google Scholar]
- 58.Axley P., Ahmed Z., Ravi S., Singal A.K. Review article hepatitis C virus and hepatocellular carcinoma: a narrative review. J. Clin. Transl. Hepatol. 2018;6(1):79–84. doi: 10.14218/JCTH.2017.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Di Bisceglie A.M. Hepatitis B and hepatocellular carcinoma adrian. Hepatology. 2019;49(5 suppl):S56–S60. doi: 10.1002/hep.22962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen Y., Williams V., Filippova M., Filippov V., Duerksen-hughes P. Viral carcinogenesis: factors inducing DNA damage and virus integration. Cancers (Basel) 2014;6:2155–2186. doi: 10.3390/cancers6042155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poiesz B.J., Ruscetti F.W., Gazdart A.D.I.F., Bunnt P.A., Minnat J.D., Gallo R.C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. U. S. A. 1980;77(12):7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gao R., Wang Z., Li H., et al. Gut microbiota dysbiosis signature is associated with the colorectal carcinogenesis sequence and improves the diagnosis of colorectal lesions. J. Gastroenterol. Hepatol. 2020;35(12):2109–2121. doi: 10.1111/jgh.15077. [DOI] [PubMed] [Google Scholar]
- 63.Nakatsu G., Li X., Zhou H., et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat. Commun. 2015;6(8727):1–9. doi: 10.1038/ncomms9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feng Q., Liang S., Jia H., et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat. Commun. 2015;6(6528):1–13. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- 65.Barr J.J., Auro R., Furlan M., et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc. Natl. Acad. Sci. U. S. A. 2013;110(26):10771–10776. doi: 10.1073/pnas.1305923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gogokhia L., Buhrke K., Bell R., et al. Expansion of bacteriophages is linked to aggravated intestinal inflammation and colitis Lasha. Cell Host Microbe. 2019;25(2):285–299. doi: 10.1016/j.chom.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tetz G., Tetz V. Bacteriophage infections of microbiota can lead to leaky gut in an experimental rodent model. Gut Pathog. 2016;8(1):1–4. doi: 10.1186/s13099-016-0109-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weinbauer M.G., Rassoulzadegan F. Are viruses driving microbial diversification and diversity? Environ. Microbiol. 2004;6(1):1–11. doi: 10.1046/j.1462-2920.2003.00539.x. [DOI] [PubMed] [Google Scholar]
- 69.Marongiu L., Landry J.J.M., Rausch T., et al. Metagenomic analysis of primary colorectal carcinomas and their metastases identifies potential microbial risk factors. Mol. Oncol. 2021;15(12):3363–3384. doi: 10.1002/1878-0261.13070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Teo W.H., Chen H.P., Huang J.C., Chan Y.J. Human cytomegalovirus infection enhances cell proliferation, migration and upregulation of EMT markers in colorectal cancer-derived stem cell-like cells. Int. J. Oncol. 2017;51(5):1415–1426. doi: 10.3892/ijo.2017.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goel A., Li M.S., Nagasaka T., et al. Association of JC virus T-antigen expression with the methylator phenotype in sporadic colorectal cancers. Gastroenterology. 2006;130(7):1950–1961. doi: 10.1053/j.gastro.2006.02.061. [DOI] [PubMed] [Google Scholar]
- 72.Jung W.T., Li M.S., Goel A., Boland C.R. JC virus T-antigen expression in sporadic adenomatous polyps of the colon. Cancer. 2008;112(5):1028–1036. doi: 10.1002/cncr.23266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McLaughlin-Drubin M.E.M.K. Viruses associated with human cancer margaret. Biochim. Biophys. Acta. 2008;1782(3):127‐150. doi: 10.1016/j.bbadis.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.World Health Organization. Antimicrobial Resistance. Antimicrobial Resistance.
- 75.Virgin H.W. The virome in mammalian physiology and disease. Cell. 2014;157(1):142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu X., Hong T., Parameswaran S., et al. Human virus transcriptional regulators xing. Cell. 2020;182(1):24–37. doi: 10.1016/j.cell.2020.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smith S.E., Huang W., Tiamani K., Unterer M., Khan Mirzaei M., Deng L. Emerging technologies in the study of the virome. Curr. Opin. Virol. 2022;54(101231):1–10. doi: 10.1016/j.coviro.2022.101231. [DOI] [PubMed] [Google Scholar]
- 78.Quince C., Walker A.W., Simpson J.T., Loman N.J., Segata N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017;35(9):833–844. doi: 10.1038/nbt.3935. [DOI] [PubMed] [Google Scholar]
- 79.Venter J.C., Remington K., Heidelberg J.F., et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304(5667):66–74. doi: 10.1126/SCIENCE.1093857. [DOI] [PubMed] [Google Scholar]
- 80.Qin J., Li R., Raes J., et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Oh J., Byrd A.L., Deming C., et al. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014;514(7520):59–64. doi: 10.1038/nature13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huttenhower C., Gevers D., Knight R., et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sunagawa S., Coelho L.P., Chaffron S., et al. Structure and function of the global ocean microbiome. Science (80-) 2015;(6237):348. doi: 10.1126/SCIENCE.1261359/SUPPL_FILE/SUNAGAWA_TABLES1.XLSX. [DOI] [PubMed] [Google Scholar]
- 84.Durazzi F., Sala C., Castellani G., Manfreda G., Remondini D., De Cesare A. Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci. Rep. 2021;11(1):1–10. doi: 10.1038/s41598-021-82726-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moya A., Pérez Broncal V. vol. 1838. 2018. (The Human Virome Methods and Protocols). [DOI] [Google Scholar]
- 86.Park J.J., Chen S. Metaviromic identification of discriminative genomic features in SARS-CoV-2 using machine learning. Patterns. 2022;3(100407):1–15. doi: 10.1016/j.patter.2021.100407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supp. material/referenced in article.