Abstract
Chlorantraniliprole (CAP) is an insecticide with low toxicity and high efficiency, which is widely used in agriculture in China. However, its potential ecological risks remain unknown. In this study, we investigated the impact of different CAP concentrations on bacterial and fungal communities in soil based on high-throughput sequencing. The results showed that CAP application had no significant effect on soil bacterial and fungal diversity, but altered the bacterial and fungal community structure. In particular, the soil bacterial and fungal community structure in the low CAP concentration treatment group exhibited large variability. Compared with 0 day, the phylum level of bacteria changed at 115 days, and fungi changed at 175 days, indicating that soil microbial community might have significant correlation with CAP degradation in soil. Correlation analysis between soil properties and microbial communities showed that TN, TP, and NO3–N were three key factors that significantly influenced microbial community structure. These results provide basic data for studying the effects of pesticides on ecosystem and potential remediation strategies of polluted soil.
Keywords: Chlorantraniliprole (CAP), Microbial communities, Soil, High-throughput sequencing
1. Introduction
Approximately 1–2.5 billion tons of pesticides are sprayed on farmland and towns every year around the world, which has become an important source of diffusion of chemical pollutants in the environment [1]. It has been estimated that, except for less than 1% of pesticides that act on target organisms, most of the pesticides enter the ecosystem and cause adverse effects on human health and the ecological environment [2]. Toxic pesticides that remain in the environment exhibit good fat-soluble properties, which seriously affects the growth, development, and reproduction of non-target organisms [49], especially in the soil environment.
One gram of soil may contain thousands of distinct microbial classification groups, and soil microbial groups have a direct or indirect impact on the health of plants in terrestrial ecosystems [3]. At the same time, the physical and chemical properties of the soil surface layer, including pH, soil organic carbon content, salinity, and available nitrogen content, are significantly correlated with the composition of soil microbial community. In addition, temperature and redox state can also affect the soil bacterial community [[4], [5], [6], [7]]. With the widespread use of pesticides, the effects of pesticides on soil microorganisms have gradually emerged. Pesticides are the main stressors to soil microorganisms [8], and some studies have shown that pesticides can also change non-target organisms. For example, the fungicide thiabendazole (TBZ) had been observed to alter the soil bacterial α-diversity [9], and cluster analysis of PCR-denaturing gradient gel electrophoresis banding patterns indicated significant difference in the microbial community structure between pesticide-treated and control soils [10]. Furthermore, Fournier et al. found that although both synthetic pesticide and biopesticide can change the co-occurrences of microbial taxa and community composition [11], their impact on microbial α-diversity is weak and transient. Several studies have shown that some pesticides may have little influence on the soil microorganisms. For instance, when compared with the control, the herbicide dichlorprop had no significant effect on microbial community structure at the phylum level, but enhanced the bacterial diversity and dichlorprop degradation related taxa in soil [12]. Vasileiadis et al. found that the main transformation product of iprodione, namely, 3,5-dichloroaniline, was not only a key factor for the observed structural changes in the soil bacterial and fungal community [13], but can also influence the abundance of certain bacterial and fungal community. These studies show that pesticides have different effects on soil microorganisms.
Chlorantraniliprole (CAP) is a type of anthranilamide pesticide developed by DuPont in 2007. It has a unique chemical structure and a novel mode of action. It not only has a broad insecticidal spectrum, but also has excellent control performance, shows no cross-resistance, etc. However, CAP is difficult to photolyze and hydrolyze in water, and can easily pollute water sources, harming the growth, development, and reproduction of aquatic organisms. It has been reported that most of the initially applied CAP (67% of CAP) can remain in a paddy field during flooding with only 20 days of holding time [14]. Under simulated sunlight or long-wave ultraviolet irradiation, the photolysis half-life of CAP in tap water and deionized water is 4.1 and 5.1 d, respectively, and the photolysis rate is hardly affected by humic acid and nitrate [15]. In soil-related experiments, the CAP degradation rate has been noted to vary with soil type. At field doses, CAP has been observed to exhibit only short-term effects on soil respiration, enzyme activity, and microbial phospholipid fatty acids (PLFA) profile in paddy soil, indicating that CAP does not pose a long-term threat to soil [16]. However, the use of high doses of CAP has been found to exert potential ecotoxic effects on soil bacterial community composition and microbial metabolism [17], and the root-associated microbiomes have been observed to be significantly altered by CAP application [18]. To date, research on the effects of CAP on the diversity and community composition of soil fungi is limited, with only a few related studies available on the combination of soil-related environmental factors. In the present study, the effects of CAP on the diversity and community structure of bacteria and fungi in soil as well as on the physical and chemical properties of the soil were examined by CAP treatments at three different concentrations under the same sampling time and co-cultivation for 235 days. The aims of this study were to: 1) evaluate the degradation kinetics of CAP in the soil; 2) determine the changes in the microbial diversity and community structure following treatment with different CAP concentrations; and 3) understand the influence of soil environment containing CAP on bacterial and fungal communities. The findings of this study can clarify the relationship between CAP and microorganisms in soil.
2. Materials and methods
2.1. Soil sample collection and pretreatment method
Soil samples were Fluvo-aquic soil which were collected from the surface layer (0–20 cm) of a wheat field in Shangshui County, Zhoukou City, Henan Province (33°54’25″N,114°61’16″E), China, and transported to the laboratory of Beijing Agricultural of University (40°09’39″N,116°30’ 80″E, Beijing) and the Eco-environment Research Center of Chinese Academy of Sciences (40°01’47″N,116°35’ 07″E, Beijing) in May 2017 for analyses. After the soil samples were naturally dried, they were ground and passed through a 20-mesh sieve.
The effects of CAP on soil microbes were studied using four different doses: 0 (CK, as the control), 0.8 mg/kg (C1,0.2 applicate rate), 4.0 mg/kg (C2,1 applicate rate), and 20.0 mg/kg (C3,5 applicate rate). CAP was dissolved in acetone (chromatography grade) to obtain a stock solution of 1000 mg/L and it was added to the soils to give a final concentration of 0.8, 4.0 and 20.0 mg/kg, respectively. Control samples (CK) were prepared by adding acetone without CAP. Each treatment was prepared in triplicate. After acetone volatilization, the soil samples were sieved again to homogenize, and then 500 g soil samples were added to a plastic flowerpot (height, 10 cm; top diameter, 12 cm; and bottom diameter, 8.5 cm) and incubated at 25 °C and 60% moisture content under natural light condition.
A total of 72 soil samples were collected on days 0, 20, 50, 115, 175, and 235, freeze-dried, and stored at −80 °C in a refrigerator for DNA extraction and investigation of soil physical and chemical properties. The physical and chemical properties of the soil were analyzed by the Institute of Soil Science, Chinese Academy of Sciences (Nanjing, China) with standard protocols. Briefly, the total phosphorous (TP) content was determined colorimetrically after wet digestion with H2SO4 and HClO4, respectively [19], while soil total nitrogen (TN) content was ascertained by using potassium persulfate oxidation method. Soil ammonium nitrogen (NH3–N) and nitrate nitrogen (NO3–N) contents were determined by SEAL Auto Analyzer 3 HR (Seal Analytical, UK) after 2 M KCl extraction. The total soil organic matter (SOM) and available phosphorous (AP) were measured according to previously published methods [20,21], and available potassium (AK) content was determined by ammonium acetate extraction method. The physicochemical properties of the collected soil samples are listed in Table 1.
Table 1.
Physicochemical properties of the soil samples.
| pH | TP(mg/kg) | TN(mg/kg) | NH3–N(mg/kg) | NO3–N(mg/kg) | AP(mg/kg) | AK(mg/kg) | SOM(%) |
|---|---|---|---|---|---|---|---|
| 5.21 | 355.80 | 1994.48 | 75.85 | 182.80 | 58.80 | 161.06 | 2.41 |
a Values represent means based on soil samples. TN: total nitrogen; TP: total phosphorus; NO3–N: nitrate nitrogen; NH3–N: ammonia nitrogen; AP: available phosphorus; AK: available potassium; SOM: soil organic matter.
2.2. CAP extraction and high-performance liquid chromatography-tandem mass spectrometry
CAP in soil samples was extracted and quantified as follows: The residual concentrations of CAP were measured through high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS). The supernatant was transferred to a 2-mL EP tube containing 50 mg of PSA, 150 mg of anhydrous magnesium sulfate, and 30 mg of C18, and vortexed for 1 min. After static precipitation, the supernatant was filtered through a 0.22-μm organic filter membrane for HPLC-MS/MS.
The HPLC-MS/MS analysis was performed on an Agilent system equipped with an Agilent ZORBA SB-C18 column (5 μm, 4.6 mm × 150 mm), quaternary pump, autosampler, and ultraviolet–visible detector under the following conditions: flow rate of the mobile phase (water/acetonitrile = 30/70, v/v), 0.2 mL/min; injection volume, 5 μL; and column temperature, 25 °C. The use of triple quadrupole mass spectrometer allowed working in multiple reaction monitoring mode. The electrospray ionization source was operated in positive ionization mode. The m/z transition pairs used for quantitation and confirmation were 484/452.7 (most sensitive) and 484/285.6, respectively. The detection limit of the method was 0.01–1.0 mg/kg and the recoveries were 94.1%–102.0% (RSD: 4.0%–6.9%, n = 5).
2.3. DNA extraction, PCR amplification, and high-throughput sequencing
The total genomic DNA of the soil samples was extracted with Fast DNA® SPIN Kit for Soil (MP Biomedicals LLC, USA) according to the manufacturer's instructions. After since, the V4 region of the 16 S rRNA region was amplified with primers 515 F (5′-GTGYCAGCMGCCGCGGTAA-3′) and 806 R (5′-GGACTACNVGGGTWTCTAAT-3′). The primers contained unique barcode (12 bp in length), which can sequence multiple samples at the same time in a single sequence. The 16 S PCR amplification system (total volume of 50 μL) included 37.5 μL of ddH2O, 5 μL of 10 × loading buffer, 4 μL of dNTPs, 0.5 μL of Taq DNA polymerase, 1 μL each of forward and reverse primers, and 1 μL of DNA template (20–30 ng/μL). The 16 S PCR reaction condition was as follows: 95 °C for 5 min, followed by 30 cycles of 94 °C for 20 s, 57 °C for 25 s, and 72 °C for 45 s, and a final extension at 72 °C for 10 min [22].
The ITS region was amplified with primers gITS7 (5′-GTGARTCATCGARTCTTTG-3′) and ITS4R (5′-TCCTCCGCTTATTGATATGC-3′). The ITS PCR amplification system (total volume of 50 μL) included 37.5 μL of ddH2O, 5 μL of 10 × loading buffer, 4 μL of dNTPs, 0.5 μL of Taq DNA polymerase, 1.5 μL each of forward and reverse primers, and 1.5 μL of DNA template (20–30 ng/μL). The ITS PCR reaction condition was as follows: 94 °C for 1 min, followed by 35 cycles of 94 °C for 20 s, 57 °C for 25 s, and 72 °C for 45 s, and a final extension at 72 °C for 10 min.
The PCR products were observed on 1% agarose gel and the target bands were extracted by E. Z.N.A.™ Gel Extraction Kit (Omega Bio-tek, Norcross, GA, USA) and purified. The purified product was mixed according to the fluorescence quantitative results, and sequencing library was prepared and sent to the Illumina Miseq high-throughput sequencing platform of MAGIGEN Gene Technology Company (Guangdong, China).
Amplicon data are available on NCBI Sequence Read Archive (SRA) under project number: PRJNA836153.
2.4. Sequence processing and data analysis
All the sequences obtained were clustered using the Galaxy generation platform (http://mem.rcees.ac.cn:8080) which includes a series of bioinformatic tools [23,24]. The database of 16 S is SILVA 138.1 and the database of ITS is UNITE 8.3. In a brief, raw sequences were spilt into samples according to their specific barcodes. The raw data of forward and reverse primer sequence with low-quality sequences (QC score<30), and short sequences [<290 bp (16 S rRNA), <276 bp (ITS)] were deleted for effective data operational taxonomic units (OTUs) classification analysis. UPARSE [50] was used to remove chimeras and an OTU table was generated at 97% similarity level. The taxonomy annotation was conducted using the RDP Classifier database [25]. In order to decrease the effects of sampling effort on the analysis, the OTU table was rarified for downstream analysis, which including calculations of Shannon index, inverse Simpson index, observed OTU number, and richness index (Chao1). The microbial community composition and structure at each classification level was investigated by statistical analysis, and principal component analysis (PCA) and canonical correspondence analysis (CCA) were employed to explore the effects of soil environmental factors on the populations of microbial community. Microsoft Excel 2016 was used for basic data processing, SPSS21.0 was utilized for one-way analysis of variance (one-way ANOVA) and significance analysis, and Origin 8.0 was employed for mapping.
3. Results
3.1. Dissipation of CAP in soil
The degradation dynamics of CAP in the soil samples (treated with 0.8, 4.0, and 20.0 mg/kg CAP) are shown in Fig. 1. The results indicated that 71%–75% CAP in soil was degraded after 235 days. A first-order kinetic model adequately described the dissipation kinetics of CAP at the three concentration levels. The half-lives of CAP treatments of 0.8, 4.0, and 20.0 mg/kg were 86.7, 99.0, and 115.0 d, respectively. The CAP degradation rate decreased with the increase in initial CAP concentration. This result suggested that CAP exhibits certain toxicity to the microbiota, and that soil microorganisms play an important role in the dissipation of CAP in soil.
Fig. 1.
Dissipation dynamics of CAP in soil at different initial CAP concentrations. Data are the means of triplicate measurements.
3.2. Effect of CAP on the α-diversity of soil bacterial communities
The soil samples treated with different concentrations of CAP were collected in the early, middle, and later stages of CAP degradation. After DNA extraction and high-throughput sequencing, a large number of 16 S rRNA gene sequences were obtained. After quality control, the high-quality sequence was divided into 25,738 OTUs (97% similarity). The rarefaction curve tended to be flat, indicating that the sequencing data were acceptable and the sequence depth of all the samples was sufficient. The changes in the bacterial community richness (observed OTU number) (Fig. 2-C), Chao1(Fig. 2-D), Shannon index(Fig. 2-A), and inverse Simpson index(Fig. 2-B) in the soil ecosystem are shown in Fig. 2.
Fig. 2.
Dynamics of soil bacterial community diversity. C1, C2, and C3 correspond to 0.8, 4.0, and 20.0 mg/kg CAP treatment, respectively; CK indicates blank control group (acetone only); D0, D20, D50, D115, D175, and D235 denote soil samples collected on days 0, 20, 50, 115, 175, and 235, respectively. (Means with different small letters are significantly different from one another under different CAP concentration treatments.)
Soil microorganisms are important performers of soil ecological functions, and soil CAP pollution may affect the composition and structure of local microbial community. Therefore, microbial community analysis can be used to evaluate the changes in soil properties. With regard to the bacterial diversity, there were no significant differences (P>0.05) in Shannon index, inverse Simpson index, observed richness, and Chao1 among the different CAP treatment groups. The overall bacterial community diversity decreased with time and there were significant differences in microbial diversity in samples collected at different times (on days 0, 20, 50, 115, and 175, Fig. 2). The ANOVA for bacterial diversity indicated that there was significant interaction between time and CAP concentration. On day 235, Shannon index and inverse Simpson index of soil samples treated with 20.0 mg/kg CAP were significantly higher than those of the control, 0.8 mg/kg CAP treatment, and 4.0 mg/kg CAP treatment, indicating that high CAP concentration may increase bacterial diversity. This might be owing to the existence of a small number of CAP-degrading bacterial strains in the soil samples, which were stressed by CAP, with the death of some bacterial strains because of their inability to tolerate CAP and survival of a small quantity of strains that could use CAP as a source of energy and carbon for growth.
According to the correlation analysis between the α-diversity of bacterial communities and environmental factors (Table 2), Shannon index, inverse Simpson index, and observed richness were strongly significantly correlated with TN, TP, NO3–N, and organic matter (P < 0.01). Chao1 was strongly significantly negatively correlated with TN and NO3–N, and significantly negatively correlated with TN (P < 0.05). However, CAP concentration and other environmental factors had no correlation with Shannon index, inverse Simpson index, observed richness, and Chao1. These results indicated that the TN, TP, and NO3–N contents in the soil could change under CAP stress.
Table 2.
Correlation between the α-diversity of bacterial communities and environmental factors.
| Factors | Shannon | Inv-Simpson | Observed-richness | chao1 |
|---|---|---|---|---|
| TN | −0.421a | −0.322a | −0.466a | −0.489b |
| TP | −0.464a | 0.404a | 0.471a | −0.518a |
| NH3–N | 0.173 | 0.163 | 0.145 | −0.026 |
| NO3–N | −0.726a | −0.629a | −0.755a | −0.701a |
| AP | −0.147 | −0.130 | −0.208 | −0.275 |
| AK | 0.067 | 0.012 | 0.131 | 0.399 |
| SOM | −0.328a | −0.35a | −0.308a | −0.007 |
| c (CAP) | 0.070 | 0.058 | 0.079 | 0.086 |
a Values represent means based on soil samples. TN: total nitrogen; TP: total phosphorus; NO3–N: nitrate nitrogen; NH3–N: ammonia nitrogen; AP: available phosphorus; AK: available potassium; SOM: soil organic matter; c(CAP): concentration of CAP.
Correlation is significant at 0.01 level (2-tailed).
Correlation is significant at 0.05 level (2-tailed).
3.3. Effect of CAP on the α-diversity of soil fungal communities
After passing quality control, the high-quality sequences were divided into 5421 OTUs (97% similarity). The rarefaction curve tended to remain flat, indicating that the sequencing data were acceptable and the sequence depth of all samples was sufficient. The changes in the abundance of fungal communities [observed richness(Fig. 2-C), Chao1(Fig. 2-D), Shannon index(Fig. 2-A), and inverse Simpson index(Fig. 2-B)]in soil samples treated with different CAP concentrations at different sampling time are shown in Fig. 3.
Fig. 3.
Dynamics of soil fungal community diversity. C1, C2, and C3 correspond to 0.8, 4.0, and 20.0 mg/kg CAP treatment, respectively; CK indicates blank control group (acetone only); D0, D20, D50, D115, D175, and D235 denote soil samples collected on days 0, 20, 50, 115, 175, and 235, respectively. (Means with different small letters are significantly different from one another under different CAP concentration treatments.)
The results of significant difference analysis revealed no significant difference in the soil fungal diversity among all treated samples collected at the same sampling time, indicating that CAP concentration did not considerably alter the soil fungal diversity. This might be owing to the relatively isolated pot experiment with lack of animals and plants in the natural environment and inability to maintain original ecological environment (rainfall, temperature, etc.). The overall bacterial community diversity decreased with time and there were significant differences in microbial diversity in samples collected at different times. The ANOVA for fungal diversity indicated that there was significant interaction between time and CAP concentration. In addition, compared with day 0, a high amount of CAP was degraded in the soil, along with fungal diversity decreasing significantly on day 235.
The results of correlation analysis between fungal community α-diversity and environmental factors (Table 3) revealed that Shannon index, Simpson index, and observed richness were strongly negatively correlated with TN and NO3–N; Shannon index and Simpson index were significantly positively correlated with NH3–N; observed richness was strongly significantly positively correlated with TP; and Chao1 was strongly significantly negatively correlated with TN and significantly negatively correlated with NO3–N. The CAP concentration and other environmental factors had no correlation with the fungal diversity indices (Shannon index, Simpson index, observed richness, and Chao1). These results indicated that TN and NO3–N in soil have an effect on the α-diversity of soil fungal community.
Table 3.
Correlation between α-diversity of fungal community and environmental factors.
| Factors | Shannon | Inv-Simpson | Observed-richness | chao1 |
|---|---|---|---|---|
| TN | −0.296a | −0.260a | −0.459a | −0.411a |
| TP | 0.201 | 0.157 | 0.302a | 0.207 |
| NH3–N | 0.270b | 0.253b | 0.153 | −0.005 |
| NO3–N | −0.449a | −0.391a | −0.645a | −0.455b |
| AP | −0.056 | −0.051 | −0.219 | −0.392 |
| AK | 0.085 | 0.098 | 0.154 | 0.330 |
| SOM | −0.118 | −0.108 | −0.088 | −0.271 |
| c(CAP) | −0.017 | −0.050 | −0.039 | 0.048 |
Correlation is significant at 0.01 level (2-tailed).
Correlation is significant at 0.05 level (2-tailed).
3.4. Effect of CAP on soil microbial community structure
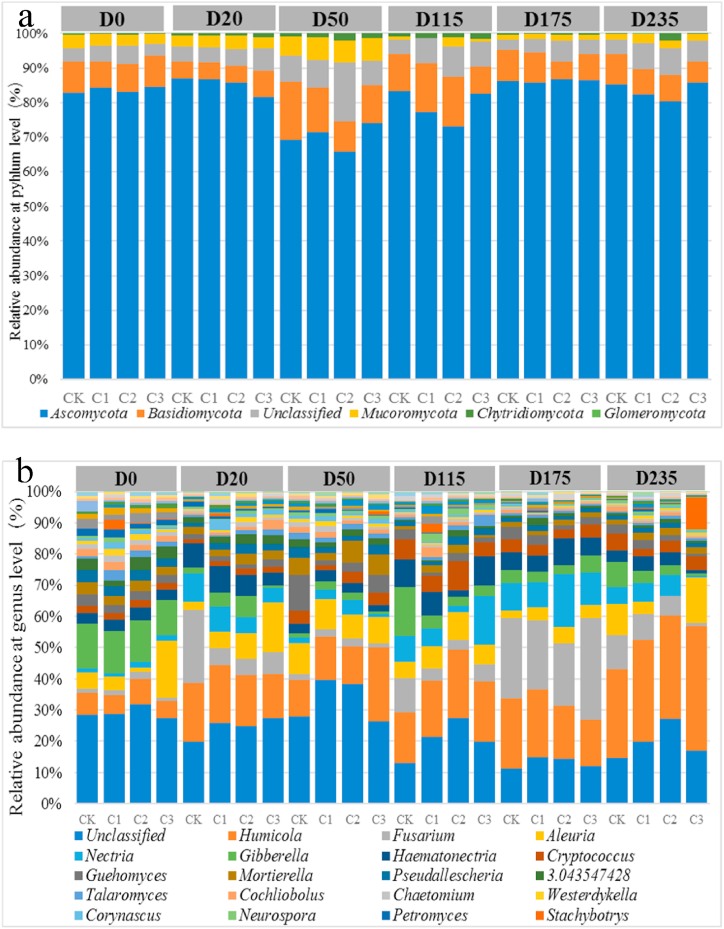
The relative abundances of the bacterial community at the phylum and genus levels in different soil samples are shown in Fig. 4a and 4b, respectively. A total of 48 bacterial phyla were detected in the soil samples, of which Proteobacteria, Actinobacteria, Acidobacteria, Thaumarchaeota, Bacteroidetes, Chloroflexi, Firmicutes, Planctomycetes, and Verrucomicrobia were predominant, with Proteobacteria being the most dominant phylum. Analysis of the bacterial community structure at different sampling points revealed that the abundance of Bacteroidetes significantly increased in all treatment groups from day 115, and the abundance of Acidobacteria in all treatment groups decreased on days 115 and 235, when compared with those on day 0. Furthermore, on day 235, with the increase in the concentration of CAP, the relative abundance of Acidobacteria accelerated, whereas the relative abundance of Thaumarchaeota slowed down.
Fig. 4.
Taxonomic classification of bacterial community at the phylum(a) and genus(b) levels.
Fig. 4b shows the bacterial genera with a relative abundance of more than 1%. The relative abundance of Unclassified genus significantly decreased from day 20 and reached the lowest on day 235, whereas the relative abundance of Janibacter increased from day 115 and that of Pseudomonas increased from day 20. Furthermore, the relative abundance of Nitrososphaera decreased in all treatment groups with the increase in CAP concentration.
For simplicity, only taxonomic groups with >1.0% abundance at the genus level in at least one sample are presented. The values shown are the means of triplicate measurements. C1, C2, and C3 correspond to 0.8, 4.0, and 20.0 mg/kg CAP treatment, respectively; CK indicates blank control group (acetone only); D0, D20, D50, D115, D175, and D235 denote soil samples collected on days 0, 20, 50, 115, 175, and 235, respectively.
The relative abundances of the fungal community at the phylum and genus levels in different soil samples are shown in Fig. 5a and b, respectively. Fig. 5 shows the comparison of the relative abundances of different fungal phyla and genera in each soil sample. A total of five fungal phyla, including Glomeromycota, Chytridiomycota, Mucoromycota, Basidiomycota, and Ascomycota, were detected in each soil sample, with Ascomycota being the dominant phylum. Comparison of the fungal community structure in samples collected at different sampling points revealed that the relative abundance of Mucoromycota in all treatment groups decreased on day 175, when compared with that on day 0, and decreased with the increase in CAP concentration.
Fig. 5.
Taxonomic classification of fungal community at the phylum(a) and genus(b) levels.
Fig. 5b illustrates the soil fungal community at the genus level with relative abundance of more than 1%. The dominant fungal genus was Humicola. The relative abundance of Nectria increased from day 20 and reached the highest on day 175. In contrast, the relative abundance of Gibberella decreased on days 20 and 50, when compared with that on day 0, but recovered to the initial level (noted on day 0) on day 115. On day 235, the relative abundance of Fusarium decreased with the increase in CAP concentration, and that of Humicola increased with the increase in CAP concentration.
For simplicity, only taxonomic groups with >1.0% abundance at the genus level in at least one sample are presented. The values shown are the means of triplicate measurements. C1, C2, and C3 correspond to 0.8, 4.0, and 20.0 mg/kg CAP treatment, respectively; CK indicates blank control group (acetone only); D0, D20, D50, D115, D175, and D235 denote soil samples collected on days 0, 20, 50, 115, 175, and 235, respectively.
3.5. Correlations between bacterial community structure and environment variables
PCA of soil bacterial community composition under different treatments at various times is shown in Fig. 6. The two principal coordinates, PC1 and PC2, explained 47% and 13% of the total variation, respectively. The samples collected on days 0, 20, and 50 and those collected on days 115 and 175 were relatively concentrated, indicating that the functional compositions of the two groups of soil bacteria were comparatively similar, respectively. However, samples collected on day 235 were far apart, implying that the functional composition of soil bacteria in these samples was quite different from the other samples. The different color spots in Fig. 6 denote different CAP concentrations, and black spots represent the control group. It can be noted from the figure that all the colors are dispersed, indicating that CAP concentration may have no significant effect on bacterial community composition.
Fig. 6.
PCA of soil bacterial community composition under different treatments at various times. C1, C2, and C3 correspond to 0.8, 4.0, and 20.0 mg/kg CAP treatment, respectively; CK indicates blank control group (acetone only); D0, D20, D50, D115, D175, and D235 denote soil samples collected on days 0, 20, 50, 115, 175, and 235, respectively.
Based on the Bray Curtis and Jaccard methods, the partial Mantel test (Table 4) was conducted on the β-diversity of soil bacterial community and environmental factors. The results showed that TN, TP, NH3–N, and NO3–N were positively correlated with the bacterial community structure, whereas CAP concentration, AP, AK, and SOM were not significantly correlated with the bacterial community structure.
Table 4.
Partial Mantel test of soil bacterial community structure diversity and environmental variables based on Bray Curtis and Jaccard methods.
| Factor | r.BC | p.BC | r.JC | p.JC |
|---|---|---|---|---|
| c(CAP) | −0.0176 | 0.676 | −0.0240 | 0.701 |
| TN | 0.1145 | 0.002 | 0.1319 | 0.008 |
| TP | 0.265 | 0.001 | 0.2588 | 0.001 |
| NH3–N | 0.0728 | 0.012 | 0.0586 | 0.051 |
| NO3–N | 0.2797 | 0.001 | 0.2985 | 0.001 |
| AP | −0.0023 | 0.514 | −4.91E-05 | 0.496 |
| AK | 0.0126 | 0.352 | 0.0333 | 0.818 |
| SOM | 0.0609 | 0.075 | 0.061 | 0.134 |
Bold numbers indicate that P values are significant at 0.05 level.
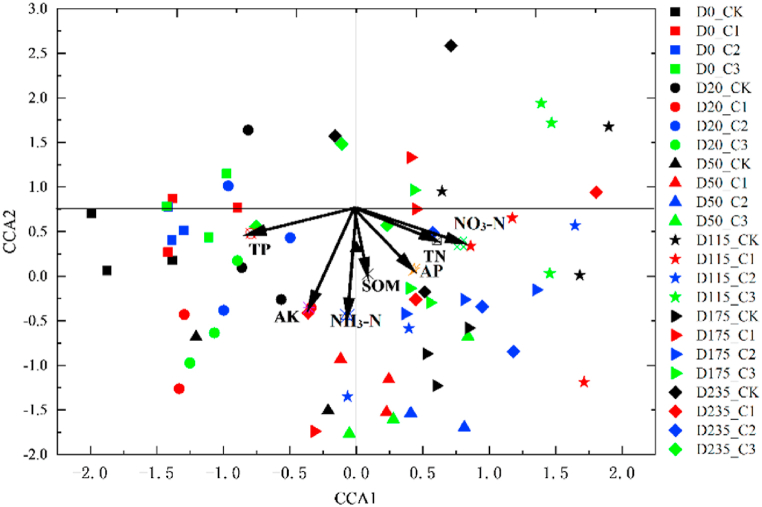
It can be observed from Fig. 7 that under CAP stress, soil TP, AK, AN, and NO3–N had a strong influence on soil bacterial community, followed by AP and TN; in contrast, TP, AK, NH3–N, and NO3–N had no significant influence on soil bacterial community.
Fig. 7.
CCA of soil bacterial community and environmental variables under different treatments at various sampling times. The length of the arrow represents the intensity of the influence of the environmental factor on the community. The longer the arrow is, the greater is the influence of the environmental factor. C1, C2, and C3 correspond to 0.8, 4.0, and 20.0 mg/kg CAP treatment, respectively; CK indicates blank control group (acetone only); D0, D20, D50, D115, D175, and D235 denote soil samples collected on days 0, 20, 50, 115, 175, and 235, respectively.
3.6. Correlations between soil fungal community structure and environment variables
PCA of soil fungal community composition in different soil samples is shown in Fig. 8. The two principal coordinates, PC1 and PC2, explained 42% and 14% of the total variation, respectively. The distances between the samples on days 0 and 115 as well as on days 175 and 235 were far, and the distance from the samples on day 235 was the farthest, indicating significant difference in the functional composition of soil fungi. The samples collected on day 50 were relatively clustered, implying that the functional composition of soil fungi was similar. It can be observed from Fig. 8 that the black spots are relatively concentrated, when compared with the color spots, implying that CAP treatment of soil has a certain influence on the functional composition of fungi in soil.
Fig. 8.
PCA of fungal community composition in different soil samples. C1, C2, and C3 correspond to 0.8, 4.0, and 20.0 mg/kg CAP treatment, respectively; CK indicates blank control group (acetone only); D0, D20, D50, D115, D175, and D235 denote soil samples collected on days 0, 20, 50, 115, 175, and 235, respectively.
Based on Bray Curtis and Jaccard methods, partial Mantel test (Table 5) was conducted on the β-diversity of soil fungal community and environmental factors. The results revealed that TN, TP, NH3–N, AP, NO3–N, and SOM were positively correlated with the soil fungal community structure, with TP and NO3–N exhibiting significant correlation with the soil fungal community structure (P < 0.05). In contrast, no significant correlation was observed between CAP concentration and soil fungal community structure.
Table 5.
Partial Mantel test of soil fungal community structure diversity and environmental variables based on Bray Curtis and Jaccard methods.
| Factors | r.BC | p.BC | r.JC | p.JC |
|---|---|---|---|---|
| c(CAP) |
0.0030 |
0.438 |
−0.0428 |
0.81 |
| TN | 0.0336 | 0.271 | −0.0123 | 0.57 |
| TP | 0.1085 | 0.005 | 0.1825 | 0.001 |
| NH3–N | 0.0492 | 0.064 | −0.0143 | 0.642 |
| NO3–N | 0.1155 | 0.021 | 0.114 | 0.035 |
| AP | 0.0315 | 0.268 | 0.059 | 0.14 |
| AK | −0.0155 | 0.628 | −0.0122 | 0.589 |
| SOM | −0.0107 | 0.503 | 0.0463 | 0.227 |
Bold numbers indicate P values are significant at 0.05 level.
CCA was performed to further identify the relationship between environmental factors and soil fungal community structure (Fig. 9). The first two axes represented 7.32% and 3.2% of the variation in the soil fungal communities, respectively, indicating that the patterns of soil fungal community composition cannot be predominantly explained by the environmental factors examined in this study. In contrast, TP, TN, and NO3–N exhibited the highest correlation with the soil fungal community structure, followed by AK and AP.
Fig. 9.
CCA of soil fungal community and environmental variables under different treatments at various times. C1, C2, and C3 correspond to 0.8, 4.0, and 20.0 mg/kg CAP treatment, respectively; CK indicates blank control group (acetone only); D0, D20, D50, D115, D175, and D235 denote soil samples collected on days 0, 20, 50, 115, 175, and 235, respectively.
4. Discussion
Land cultivation is still one of the important planting modes in agriculture. Soil provides crops with essential nutrients for growth, while crops uptake water, inorganic salts, and nutrients needed for growth from soil, along with some toxic and harmful substances that remain in the soil. The absorbed pesticide residues are transmitted to each organ of the plant along with the nutrients and are enriched in the plant body. CAP is a type of anthranilamide pesticide, which is widely used and sold in more than 100 countries around the world. However, some studies have found that CAP may have side effects on non-target plants and animals, especially on some arthropods [26]. For instance, when systemic insecticide is applied to seed treatments, it can cause negative effects on non-target organisms. Furthermore, CAP may have side effects on beneficial arthropods (such as Paederus fuscipes and honey bees) [26,27], and higher concentrations of CAP can easily be detected in soil and vegetables, indicating longer persistence of CAP in the environment [28]. However, information about the stress of CAP is still limited. Hence, it is necessary to determine the effect of CAP on soil microbial community. Soil microorganisms can directly affect the soil environment and play an important role in the ecological functions related to soil sustainability. In the present study, by using high-throughput 16 S rDNA sequencing, the effects of three different concentrations of CAP on the microbial communities in soils were characterized. The results demonstrated that the half-lives of treatments with 0.8, 4.0, and 20.0 mg/kg CAP were 86.7, 99.0, and 115.0 d, respectively, indicating that the CAP degradation rate decreased with the increase in the initial concentration of CAP. These findings are similar to those determined in soil dissipation experiments for the fungicide fluopyram [29] and herbicide halosulfuron methyl [30], and may be owing to the fact that high concentration of pesticides hinders the pesticides degradation ability of soil microorganisms.
It has been reported that microbial diversity is strongly negatively correlated with the contents of pesticide residues in the soil [31]. In the present study, the dynamic investigation of bacterial diversity in soil samples revealed that the Shannon index and inverse Simpson index of 20 mg/kg CAP treatment group were significantly higher than those of the control group on day 235, and the effect of low CAP concentration on soil microbial diversity was significantly higher than that of high CAP concentration, similar to that reported by Wu et al. [16] in paddy soil.
pH is one of the most important factors affecting the degradation of pesticides [32]. Analysis of a group of soil samples with a wide range of pH revealed that pH is an important factor affecting the composition of soil bacterial community [3]. However, in the present study, there was no significant difference in the effect of pH among treatments with different CAP concentrations and diverse sampling times. Therefore, it is still unclear whether soil pH could affect soil microbial community in the presence of CAP. In addition to soil pH, soil bacterial communities are also affected by many factors, such as soil available nitrogen [4], organic carbon content [5], temperature [6], and redox state [7]. However, there are still no relevant studies to confirm the factors or combinations of factors that can affect the composition of soil microorganisms. Most of the previous studies could only explain a small part of the changes in the soil microbial community through local soil types or environmental variables, and it is impossible to guarantee completely consistent objective conditions under laboratory settings with uniformly controlled variables. For example, when exploring the contribution of environmental and spatial parameters to the changes in the soil microbial community in North China Plain, soil environmental factors accounted for 19.7% and combined action of spatial and environmental variables accounted for 6.6%, and most of the parameters (76.1%) could not interpret the actual data results [33] through model parameters or variables affecting each other. Furthermore, investigation of the soil microbial community in a single city park in New York revealed that the soil microbial diversity was essentially the same as that found in soils worldwide [34].
Some studies have suggested that climate is the best factor for predicting global soil microbial richness and community composition, followed by soil and spatial model [35]. Furthermore, soil microbial communities are related to different soil characteristics, indicating that the composition of microbial communities is not a simple reflection of microorganisms in their environment, but is the result of selective environmental pressures such as insecticide behavior and metabolic pathways in soil [36], which further explains the changes in the bacterial and fungal community structure. The results of the present study revealed that the relative abundance of Janibacter increased from day 115, while that of Pseudomonas increased from day 20. Janibacter, a member of the Intrasporangiaceae family of Actinobacteria, is a Janus-faced bacterium that has both antibiotic resistance/pathogenicity and ability to degrade pollutants [37]. In the present study, on day 115, the increase Janibacter abundance in the soil may be due to the fact that the bacteria can use CAP as an energy source. Previous studies have demonstrated that Pseudomonas sp. could degrade high concentrations of the herbicide atrazine [38,39]. Gao et al. isolated Pseudomonas sp. GW13 which could degrade CAP rapidly [40]. Since Pseudomonas could affect the degradation of CAP, the soil microbial communities would exhibit distinct responses to different concentrations of CAP accordingly. The relative abundance of Nitrososphaera decreased in all CAP treatment groups, and the decrease enhanced with the increase in CAP concentration. Nitrososphaera, known for its ability to drive the global nitrogen cycle, is a kind of soil ammonia-oxidizing archaeon (AOA) [41]. AOA are widespread in nature and play an important role in soil nitrification [42], because of their ability to perform the first step of nitrification through the ammonia monooxygenase enzyme [43,44]. In the present study, the abundance of Nitrososphaera decreased with time, which may be owing to the soil samples preserved in flowerpots for a long time, far away from the agricultural production environment. Similar results have also been reported in a previous study in which successional plots analysis showed that the abundance of AOA declined with time [45]. These findings suggest that the effect of agriculture on the relative abundance of these soil microbial genera is reversible. Besides, the present study observed that the bacterial community composition changed with time in all the CAP treatment groups and the control, which might be owing to the variations in the moisture content at the initial time point as well as the addition of acetone in the experimental system.
With regard to the fungal community in soil under CAP stress, on day 235, the relative abundance of Fusarium decreased with the increase in CAP concentration, indicating that CAP inhibited the growth of Fusarium. On the contrary, the relative abundance of Humicola increased with the increase in CAP concentration, indicating that CAP promoted the growth of Humicola. Several species of this genus are thermophilic fungi, which can break down complex natural substrates [46]. With the treatment of pesticide formulations—imidacloprid, benomyl, andmetribuzin, the relative abundance of the genera Humicola changed [51]. Previous studies have confirmed that pathogens within the Fusarium genus are the primary agents of Fusarium head blight (FHB) of wheat, which causes yield reduction and deoxynivalenol contamination, attracting immense concern worldwide [47]. In addition, Fusarium oxysporum f. sp. lini is one of the major flax pathogens affecting the world's crop production [48]. Whether the increase of CAP content in soil will inhibit the decrease of Fusarium pathogens needs further study. PCA of the fungal community composition of different soil samples indicated that the soil fungal functional compositions on days 20 and 50 were similar, with significant differences between the initial sampling point and all other sampling points after day 115. On day 115, CAP in the soil was degraded by more than half, and hence, CAP stress may have an impact on the soil fungal community; nevertheless, time and physical and chemical properties were mainly the overall influencing factors. Partial Mantel tests revealed that the TN, TP, NH3–N, NO3–N, available phosphorus, and organic matter contents were positively correlated with the soil fungal community diversity.
5. Conclusion
In this study, the effects of different CAP concentrations on the bacterial and fungal communities in soil were examined. The results showed that the overall bacterial community diversity decreased with time and there were significant differences in microbial diversity in samples collected at different times. The CAP application had no significant effect on soil bacterial and fungal diversity, but the interaction between CAP application and time significantly affects microbial diversity. The CAP affected the relative abundance of soil microbial community composition. The relative abundance of Nectria, Janibacter and Nitrososphaera increased, the relative abundance of Fusarium decreased at late. In addition, TN, TP, and NO3–N were the three key factors that significantly influenced soil microbial community structure.
Author contribution statement
QIAN TANG: Performed the experiments; Analyzed and interpreted the data; Wrote the paper. Pingping Wang: Performed the experiments. Huijun Liu: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper. Decai Jin: Conceived and designed the experiments.Xiangning Chen and Lifei Zhu: Contributed reagents, materials, analysis tools or data.
Funding statement
Huijun Liu was supported by National Natural Science Foundation of China [31601658], CAS [kf2019009], Beijing municipal youth top-notch talent program [2018000021223ZK34].
Data availability statement
Data associated with this study has been deposited at NCBI (www.ncbi.nlm.nih.gov) under the accession number [SRR19137846, SRR19137845, SRR19137844 for fungus and SRR19137849, SRR19137848, SRR19137847 for bacteria].
Additional information
No additional information is available for this paper.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
We thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
Contributor Information
Huijun Liu, Email: huijunliu78@163.com.
Xiangning Chen, Email: cxn@bua.edu.cn.
References
- 1.Fenner K., Canonica S., Wackett L.P., et al. Evaluating pesticide degradation in the environment: blind spots and emerging opportunities. Science. 2013;341(6147):752–758. doi: 10.1126/science.1236281. [DOI] [PubMed] [Google Scholar]
- 2.Kuhad R.C., Johri A.K., Singh A., et al. vol. 1. U.S.A.; 2004. Applied Bioremediation and Phytoremediation. Springer Berlin Heidelberg; pp. 35–54. [Google Scholar]
- 3.Fierer N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017;15(10):579–590. doi: 10.1038/nrmicro.2017.87. [DOI] [PubMed] [Google Scholar]
- 4.Cederlund H., et al. Soil carbon quality and nitrogen fertilization structure bacterial communities with predictable responses of major bacterial phyla. Appl. Soil Ecol. 2014;84:62–68. [Google Scholar]
- 5.Sul W.J., Asuming-Brempong S., Wang Q., et al. Tropical agricultural land management influences on soil microbial communities through its effect on soil organic carbon. Soil Biol. Biochem. 2013;65:33–38. [Google Scholar]
- 6.Oliverio A.M., Bradford M.A., Fierer N. Identifying the microbial taxa that consistently respond to soil warming across time and space. Global Change Biol. 2017;21:17–2129. doi: 10.1111/gcb.13557. [DOI] [PubMed] [Google Scholar]
- 7.Pett-Ridge J., Firestone M.K. Redox fluctuation structures microbial communities in a wet tropical soil. Appl. Environ. Microbiol. 2005;71:6998–7007. doi: 10.1128/AEM.71.11.6998-7007.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siedt M., Schäffer A., Smith K.E.C., et al. Comparing straw, compost, and biochar regarding their suitability as agricultural soil amendments to affect soil structure, nutrient leaching, microbial communities, and the fate of pesticides. Sci. Total Environ. 2020;751 doi: 10.1016/j.scitotenv.2020.141607. [DOI] [PubMed] [Google Scholar]
- 9.Papadopoulou E.S., Genitsaris S., Omirou M., et al. Bioaugmentation of thiabendazole-contaminated soils from a wastewater disposal site: factors driving the efficacy of this strategy and the diversity of the indigenous soil bacterial community. Environ. Pollut. 2017;233:16–25. doi: 10.1016/j.envpol.2017.10.021. [DOI] [PubMed] [Google Scholar]
- 10.Walvekar V.A., Bajaj S., Singh D.K., et al. Ecotoxicological assessment of pesticides and their combination on rhizospheric microbial community structure and function of Vigna radiata. Environ. Sci. Pollut. Res. Int. 2017;24(20):17175–17186. doi: 10.1007/s11356-017-9284-y. [DOI] [PubMed] [Google Scholar]
- 11.Fournier B., Pereira Dos Santos S., Gustavsen J.A., et al. Impact of a synthetic fungicide (fosetyl-Al and propamocarb-hydrochloride) and a biopesticide (Clonostachys rosea) on soil bacterial, fungal, and protist communities. Sci. Total Environ. 2020;738 doi: 10.1016/j.scitotenv.2020.139635. [DOI] [PubMed] [Google Scholar]
- 12.Zhu Y., Guo J. Impact of dichlorprop on soil microbial community structure and diversity during its enantioselective biodegradation in agricultural soils. J Environ Sci Health B. 2020;55(11):974–982. doi: 10.1080/03601234.2020.1802186. [DOI] [PubMed] [Google Scholar]
- 13.Vasileiadis S., Puglisi E., Papadopoulou E.S., et al. Blame it on the metabolite: 3,5-Dichloroaniline rather than the parent compound is responsible for the decreasing diversity and function of soil microorganisms. Appl. Environ. Microbiol. 2018;84(22):e01536. doi: 10.1128/AEM.01536-18. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Redman Z.C., Anastasio C., Tjeerdema R.S. Quantum yield for the aqueous photochemical degradation of chlorantraniliprole and simulation of its environmental fate in a model California rice field. Environ. Toxicol. Chem. 2020;39(10):1929–1935. doi: 10.1002/etc.4827. [DOI] [PubMed] [Google Scholar]
- 15.Lavtižar V., Van Gestel C.A., Dolenc D., et al. Chemical and photochemical degradation of chlorantraniliprole and characterization of its transformation products. Chemosphere. 2014;95(1):408–414. doi: 10.1016/j.chemosphere.2013.09.057. [DOI] [PubMed] [Google Scholar]
- 16.Wu M., Liu J., Li W., et al. Temporal dynamics of the compositions and activities of soil microbial communities post-application of the insecticide chlorantraniliprole in paddy soils. Ecotoxicol. Environ. Saf. 2017;144:409–415. doi: 10.1016/j.ecoenv.2017.06.056. [DOI] [PubMed] [Google Scholar]
- 17.Wu M., Li G., Chen X., et al. Rational dose of insecticide chlorantraniliprole displays a transient impact on the microbial metabolic functions and bacterial community in a silty-loam paddy soil. Sci. Total Environ. 2018;616–617:236–244. doi: 10.1016/j.scitotenv.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 18.Wang S.S., Cui H., Chen M.Z., et al. Simultaneous quantitation of 3ADON and 15ADON chemotypes of DON-producing Fusarium species in Chinese wheat based on duplex droplet digital PCR assay. J. Microbiol. Methods. 2021;190 doi: 10.1016/j.mimet.2021.106319. [DOI] [PubMed] [Google Scholar]
- 19.Parkinson J.A., Allen S.E. A wet oxidation procedure suitable for the determination of nitrogen and mineral nutrients in biological material. Commun. Soil Sci. Plant Anal. 2008;6:1–11. [Google Scholar]
- 20.Wang S., Zhang B., Li T., et al. Soil vanadium(V)-reducing related bacteria drive community response to vanadium pollution from a smelting plant over multiple gradients. Environ. Int. 2020;138 doi: 10.1016/j.envint.2020.105630. [DOI] [PubMed] [Google Scholar]
- 21.Yeomans J.C., Bremner J.M. A rapid and precise method for routine determination of organic carbon in soil. Commun. Soil Sci. Plant Anal. 1988;19(13):1467–1476. [Google Scholar]
- 22.Kong X., Jin D., Tai X., et al. Bioremediation of dibutyl phthalate in a simulated agricultural ecosystem by Gordonia sp. strain QH-11 and the microbial ecological effects in soil. Sci. Total Environ. 2019;667:691–700. doi: 10.1016/j.scitotenv.2019.02.385. [DOI] [PubMed] [Google Scholar]
- 23.Feng K., Zhang Z., Cai W., et al. Biodiversity and species competition regulate the resilience of microbial biofilm community. Mol. Ecol. 2017;26(21):6170–6182. doi: 10.1111/mec.14356. [DOI] [PubMed] [Google Scholar]
- 24.Kong X., Jin D., Jin S., et al. Responses of bacterial community to dibutyl phthalate pollution in a soil-vegetable ecosystem. J. Hazard Mater. 2018;353:142–150. doi: 10.1016/j.jhazmat.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 25.Wang Q., Garrity G.M., Tiedje J.M., et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliveira R.L., Gontijo P.C., Sâmia R.R., et al. Long-term effects of chlorantraniliprole reduced risk insecticide applied as seed treatment on lady beetle Harmonia axyridis (Coleoptera: coccinellidae) Chemosphere. 2019;219:678–683. doi: 10.1016/j.chemosphere.2018.12.058. [DOI] [PubMed] [Google Scholar]
- 27.Williams J.R., Swale D.R., Anderson T.D. Comparative effects of technical-grade and formulated chlorantraniliprole to the survivorship and locomotor activity of the honey bee, Apis mellifera (L.) Pest Manag. Sci. 2020;76(8):2582–2588. doi: 10.1002/ps.5832. [DOI] [PubMed] [Google Scholar]
- 28.Xu B., Wang K., Vasylieva N., et al. Development of a nanobody-based ELISA for the detection of the insecticides cyantraniliprole and chlorantraniliprole in soil and the vegetable bok choy. Anal. Bioanal. Chem. 2021;413(9):2503–2511. doi: 10.1007/s00216-021-03205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y., Xu J., Dong F., et al. Response of microbial community to a new fungicide fluopyram in the silty-loam agricultural soil. Ecotoxicol. Environ. Saf. 2014;108:273–280. doi: 10.1016/j.ecoenv.2014.07.018. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y., Du L., Liu H., et al. Halosulfuron methyl did not have a significant effect on diversity and community of sugarcane rhizosphere microflora. J. Hazard Mater. 2020;399 doi: 10.1016/j.jhazmat.2020.123040. [DOI] [PubMed] [Google Scholar]
- 31.Lu X.M., Lu P.Z. Response of microbial communities to pesticide residues in soil restored with Azolla imbricata. Appl. Microbiol. Biotechnol. 2018;102(1):475–484. doi: 10.1007/s00253-017-8596-7. [DOI] [PubMed] [Google Scholar]
- 32.Baker K.L., Marshall S., Nicol G.W., et al. Degradation of metalaxyl-M in contrasting soils is influenced more by differences in physicochemical characteristics than in microbial community composition after re-inoculation of sterilised soils. Soil Biol. Biochem. 2010;42(7):1123–1131. [Google Scholar]
- 33.Shi Y., Li Y., Xiang X., et al. Spatial scale affects the relative role of stochasticity versus determinism in soil bacterial communities in wheat fields across the North China Plain. Microbiome. 2018;6(1):27. doi: 10.1186/s40168-018-0409-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramirez K.S., Leff J.W., Barberán A., et al. Biogeographic patterns in below-ground diversity in New York City's Central Park are similar to those observed globally. Proc. Biol. Sci. 2014;281(1795) doi: 10.1098/rspb.2014.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tedersoo L., Bahram M., Põlme S., et al. Fungal biogeography. Global diversity and geography of soil fungi. Science. 2014;346(6213) doi: 10.1126/science.1256688. [DOI] [PubMed] [Google Scholar]
- 36.Cai Z., Ma J., Wang J., et al. Impact of the novel neonicotinoid insecticide Paichongding on bacterial communities in yellow loam and Huangshi soils. Environ. Sci. Pollut. Res. Int. 2016;23(6):5134–5142. doi: 10.1007/s11356-015-5733-7. [DOI] [PubMed] [Google Scholar]
- 37.Zhao Q., Jin M., Zhou Z., et al. Complete genome sequence of Janibacter melonis M714, a janus-faced bacterium with both human health impact and industrial applications. Curr. Microbiol. 2020;77(8):1883–1889. doi: 10.1007/s00284-020-01951-2. [DOI] [PubMed] [Google Scholar]
- 38.Silva E., Fialho A.M., Sá-Correia I., et al. Combined bioaugmentation and biostimulation to cleanup soil contaminated with high concentrations of atrazine. Environ. Sci. Technol. 2004;38(2):632–637. doi: 10.1021/es0300822. [DOI] [PubMed] [Google Scholar]
- 39.Kerminen K., Le Moël R., Harju V., et al. Influence of organic matter, nutrients, and cyclodextrin on microbial and chemical herbicide and degradate dissipation in subsurface sediment slurries. Sci. Total Environ. 2018;618:1449–1458. doi: 10.1016/j.scitotenv.2017.09.302. [DOI] [PubMed] [Google Scholar]
- 40.Gao W., Li D.Y., You H. Functional characterization and genomic analysis of the chlorantraniliprole-degrading strain Pseudomonas sp. GW13. Bioeng. 2019;16(4):106. doi: 10.3390/bioengineering6040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kobayashi S., Hira D., Yoshida K., et al. Nitric oxide production from nitrite reduction and hydroxylamine oxidation by copper-containing dissimilatory nitrite reductase (NirK) from the aerobic ammonia-oxidizing archaeon, Nitrososphaera viennensis. Microb. Environ. 2018;33(4):428–434. doi: 10.1264/jsme2.ME18058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reyes C., Hodgskiss L.H., Kerou M., et al. Genome wide transcriptomic analysis of the soil ammonia oxidizing archaeon Nitrososphaera viennensis upon exposure to copper limitation. ISME J. 2020;14(11):2659–2674. doi: 10.1038/s41396-020-0715-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Könneke M., Bernhard A.E., de la Torre J.R., et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437(7058):543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 44.Treusch A.H., Leininger S., Kletzin A., et al. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ. Microbiol. 2005;7(12):1985–1995. doi: 10.1111/j.1462-2920.2005.00906.x. [DOI] [PubMed] [Google Scholar]
- 45.Zhalnina K., de Quadros P.D., Gano K.A., et al. Ca. Nitrososphaera and Bradyrhizobium are inversely correlated and related to agricultural practices in long-term field experiments. Front. Microbiol. 2013;1(4):104. doi: 10.3389/fmicb.2013.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sabrin R.M.I., Shaimaa G.A.M., Ahmed E.A., et al. Natural products of the fungal genus Humicola: diversity, Biological.Activity, and industrial importance. Curr. Microbiol. 2021;78:2488–2509. doi: 10.1007/s00284-021-02533-6. [DOI] [PubMed] [Google Scholar]
- 47.Wang C., Qin Y., Li Y., et al. Variations of root-associated bacterial cooccurrence relationships in paddy soils under chlorantraniliprole (CAP) stress. Sci. Total Environ. 2021;779 doi: 10.1016/j.scitotenv.2021.146247. [DOI] [PubMed] [Google Scholar]
- 48.Kanapin A., Bankin M., Rozhmina T., et al. Genomic regions associated with Fusarium wilt resistance in flax. Int. J. Mol. Sci. 2021;22(22) doi: 10.3390/ijms222212383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agrawal A., Pandey R.S., Sharma B. Water pollution with special reference to pesticide contamination in India. J. Water Resour. Protect. 2010;2(5):432–448. [Google Scholar]
- 50.Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10(10):996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 51.Streletskii R., Astaykina A., Krasnov G., et al. Changes in bacterial and fungal community of soil under treatment of pesticides. Agron. J. 2022;12(1):124. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data associated with this study has been deposited at NCBI (www.ncbi.nlm.nih.gov) under the accession number [SRR19137846, SRR19137845, SRR19137844 for fungus and SRR19137849, SRR19137848, SRR19137847 for bacteria].