Abstract
In recent times, robust green technological developments have advanced the goal of a circular economy by minimizing waste generation. The study was undertaken to explore the keratinolytic activity of chicken feather-degrading bacteria from South African soil. Isolates coded as SSN-01 and HSN-01 were identified as Bacillus sp. NFH5 and Bacillus sp. FHNM and their sequences were deposited in GenBank, with accession numbers MW165830.1 and MW165831.1, respectively. Extracellular enzyme production and thiol group generation by Bacillus sp. NFH5 peaked at 120 h with 1879.09 ± 88.70 U/mL and 9.49 ± 0.78 mM, respectively. Glutamic acid (4.44%), aspartic acid (3.50%), arginine (3.23%), glycine (2.61%), serine (2.08%), and proline (2.08%) were relatively higher in concentration. Keratinase (KerBAN) activity was highest at pH 8.0 and 90 °C but was inhibited by both EDTA and 1,10-phenanthroline. In addition, the keratinase-encoding gene (kerBAN) accessioned OK033360 had 362 amino acid residues, with molecular weight and theoretical isoelectric point of 39 kDa and 8.81, respectively. Findings from this study highlight the significance of Bacillus sp. NFH5 in the bio-recycling of recalcitrant keratinous wastes to protein hydrolysates – potential dietary supplements for livestock feeds. The properties of KerBAN underscore its application potential in green biotechnological processes.
Keywords: Amino acids, Bio-recycling, Keratinolytic protease, Livestock feed, Protein hydrolysate
1. Introduction
Recent times have seen significant agricultural productivity worldwide to sustain the teeming global population. On the other hand, the processing of agricultural produce engenders the generation of a wide variety of waste materials, many of which may serve as cheap industrial feedstocks, from the cleaner production and sustainable development viewpoints. However, a few agro-industrial wastes are not adequately exploited due to their structural stability [1]. Lignocellulosic, chitinous, and keratinous biomass are the most ubiquitous biopolymers contributing to the rise in carbon footprint [2], with keratinous materials being unusually recalcitrant to natural degradability because of the high keratin contents [3].
Keratin is a structural polypeptide with a high degree of insolubility occasioned by excess cross-linkages from hydrophobic interactions, hydrogen, and disulfide bonds [4]. Keratin-rich materials, such as avian feathers, generated in large amounts from poultry-processing farms often constitute serious environmental concerns due to their potential for litter, the generation of foul odors, and the ability to harbor pathogens [5]. Mishandling of this type of waste has drawn the attention of environmentalists and other stakeholders worldwide, with the enactment of necessary acts to regulate their disposal and management and promote a cleaner environment [6]. Keratinous wastes have been converted into valuable products using physicochemical treatment methods to propagate resource efficiency and a globally competitive economy [7]. In recent times, robust green technological developments have advanced the goal of a circular economy by minimizing waste [8].
Microbial keratinases can dismember keratinous polymers into peptides, amino acids, and non-protein nitrogenous compounds [9]. Keratinolytic microbes differ considerably in their capability to degrade keratin, and these variations have been attributed to their genetic diversity [10]. The expression of a battery of genes has been implicated in the complete decomposition of recalcitrant keratinous biomass [11]. Most significantly, the secretory-enzymatic machinery of keratinolytic microbes allows their growth in media formulated with the recalcitrant keratinous substrate as a sole source of carbon and nitrogen [12].
Previous studies have indicated that keratinolytic proteases from diverse microbial strains are predominantly serine- and metalloproteases [13]. Among bacterial species, Bacillus licheniformis keratinases have been the most extensively studied and used, as bioadditives, in commercial keratinase-based formulations [7]. More recently, however, keratinases from other Bacillus species showing high levels of variability have grown in importance, owing to their ability to degrade keratin within short timelines [14]. The uniqueness of keratinases from different bacterial sources could be due to variations in protein sequence identity, molecular configuration, and the nature of conserved residues. This study was aimed at characterizing Bacillus sp., a novel keratinolytic isolate from a local municipal dumpsite, for its keratin-degrading potentials and analyzing the amino acids profile of keratin protein hydrolysate generated. An additional aim of the study was to characterize the keratinase of the isolate for its biochemical properties. Furthermore, the structural identity of the Bacillus sp. keratinase was expatiated through molecular cloning, gene sequencing, and homology modeling.
2. Materials and methods
2.1. Preparation of keratin substrate
The keratin substrate used in the study was prepared from chicken feathers collected from a poultry farm. The chicken feathers were collected from a local poultry farm and processed to serve as keratin substrates. The blood stains associated with the feathers were removed by rinsing three times with tap water and then air-dried. The feathers were further oven-dried (60 °C for 48 h) to achieve constant weight before pulverizing to about 2 mm particle sizes and stored in air-tight containers at 25 °C.
2.2. Bacterial isolates and fresh inoculum preparation
Bacterial isolates (SSN-01 and HSN-01) employed for the present study were recovered from municipal dumpsites' soil samples and stored in the AEMREG culture repository. These isolates were thoroughly passaged in basal salt feather media (BSFM), formulated using (g/L) of MgCl2 (0.2), KH2PO4 (0.4), K2HPO4 (0.3), CaCl2 (0.22) and chicken feather powder (10). Erlenmeyer flasks of 100 mL capacity with 20 mL working volume were inoculated with 10 μL of each axenic culture, and the flasks were incubated for 48 h at 30 °C with constant agitation (140 rpm). A loopful of each culture broth was inoculated on basal salts feather agar (BSFA) plate and incubated for 24 h at 30°Ϲ. BSFA was the same as BSFM, except that it contained 15 g/L of agar bacteriological as a gelling agent. Bacterial colonies were transferred from the plates into sterile saline microtubes and vortexed. The optical density (OD) of each tube content was determined at 600 nm using a spectrophotometer to ensure uniform OD of 0.1. The resulting freshly standardized inoculum was used to initiate subsequent fermentation processes.
2.3. Keratinase production
The fermentation medium was constituted with BSFM in 250 mL capacity Erlenmeyer flasks. Medium sterilization was done by autoclaving (121 °C, at 15psi, 15min). Before inoculating with the freshly prepared starter culture (2%, v/v), the pH of the fermentation media was adjusted to 6.0 in an aseptic condition. Inoculated flasks were incubated with shaking (150 rpm) for 96 h at 30 °C. The fermentation broth was centrifuged at 15,000 rpm for 10 min to separate the filtrate from cell biomass and undegraded substrates. The supernatant extracted post-centrifugation served as crude keratinase.
2.3.1. Assay for keratinase activity
Keratinase activity was assayed as previously described [15]. Briefly, 0.5 mL of enzyme solution was thoroughly mixed with 0.5 mL of keratin azure dissolved in tris buffer of pH 8.0. The tube containing the mixture was allowed to stand for 1 h in a water bath at 50 °C. After incubation, the assay mixture's vials were immersed in ice-cold water for 10 min. Afterward, the filtrate was separated from the undegraded substrate by centrifuging the mixture at 15,000 rpm for 10 min. Aliquots (0.3 mL) were added to the microtiter plate, and the absorbance was determined at 595 nm using the SYNERGYMx 96 wells microplate reader (BioTek, USA). The keratinase-buffer solution incubated along the test experiments served as the control. One keratinase unit (U) was defined as any amount of enzyme causing an absorbance increase of 0.01 at the specified assay conditions.
2.3.2. Total protein determination
Bradford's [16] method was employed. A protein standard curve was prepared with bovine serum albumin (BSA) (0.06–2.0 mg/mL). Total protein was read off the standard curve.
2.3.3. Thiol concentration determination
Ellman's [17] method was employed. The crude enzyme solution (250 μL) was thoroughly mixed with 500 μL of distilled water and 50 μL of Ellman's reagent (5,5-dithiol-bis-(2-nitrobenzoic acid); DTNB) prepared by solubilizing 4 mg DTNB in 1 mL of phosphate buffer (pH 8), at 0.1 M. The mixture was incubated for 5 min at 25 °C, and the absorbance was read off at 412 nm.
2.4. Molecular identification of chicken feather-degrading bacteria
Extraction of genomic DNA from the two isolates (SSN-01 and HSN-01) under investigation was done using the Fungal/Bacterial Miniprep Kit, following Zymo Research's instructions. The universal target region for identifying bacterial species (16S region) was amplified using the universal oligonucleotides 27F: 5′-AGAGTTTGATCMTGGCTCAG-3′ and 1492R: 5′- CGGTTACCTTGTTACGACTT-3'. After PCR analysis, the products were loaded on agarose gel and electrophoresed. DNA Recovery Kit was used to extract the amplicons from the gel, and purification was carried out using Zymo Research Kit. Pure fragments were analyzed on ABI 3500xl Genetic Analyzer (Applied Biosystems, Thermofisher Scientific). CLC Bio Main Workbench v7.6 was used to compare the generated data against the NCBI database. Furthermore, related sequences were retrieved from the GenBank, and the evolutionary relationship was compared using MEGA X [18].
2.5. Optimization of physicochemical conditions for enhanced keratinase production
Extracellular keratinase production by the isolates was studied using the one-factor-at-a-time method. The physicochemical variables were optimized to improve the bacterial enzyme biosynthesis and secretion. At first, the pH for cultivating the bacteria was changed from 3.0 to 11.0. Likewise, the fermentation flasks were incubated at different temperatures (25 °C, 30°Ϲ, 35 °C, 40 °C and 45 °C). The inoculum size influence on enzyme activity by the isolates was investigated by kick-starting the fermentation process with different concentrations of the starter culture, ranging from 1% to 6% (v/v). Additionally, chicken feathers as the primary substrate for keratinase production varied (0.5–3%, w/v) to determine its concentration effect.
2.5.1. Time-course study
The kinetics of keratinase production by the study isolate was investigated at optimized physicochemical conditions. The submerged fermentation was carried out in 250 mL Erlenmeyer flasks containing improved fermentation media as optimized, and aliquots (4 mL) were aseptically withdrawn every 24 h for analysis. Cell-free extracts recovered by centrifugation were then evaluated for keratinase activity, pH, free thiol concentration, and total protein content.
2.6. Sample preparation and profiling of amino acid contents of feather hydrolysate
Chicken feathers were degraded using isolate SSN-01 in BSM, containing 1.5% (w/v) feathers, and inoculated with 2% (v/v) standardized inoculum. Fermentation was carried out under agitation (150 rpm) for 120 h at 30 °C. Subsequently, the medium was filtered through Whatman No. 1 filter paper (GE Healthcare, UK). After centrifugation (15,000 rpm for 15 min, at 4 °C), the resulting supernatant was freeze-dried in a vacuum concentrator (Martin Christ Gefriertrocknungsanlagen GmbH, Germany). The freeze-dried sample was then used for analysis. Amino acids analysis was carried out by high-performance liquid chromatography (HPLC) and fluorescence detection, following previously reported methods [[19], [20], [21]].
2.7. Effect of pH and temperature on the keratinase activity
The effect of pH (5.0–10.0) on enzyme activity was studied using the following buffer solutions (100 mM): sodium citrate (pH 5.0), potassium phosphate (pH 6.0–7.0), Tris- HCl (pH 8.0–9.0), and sodium bicarbonate-NaOH (pH 10) at 37 °C. The effect of temperature on keratinase activity was assessed from 30 °C to 100 °C, with a constant temperature interval of 10 °C.
2.8. Effect of protease inhibitors, metal ions, and laundry detergents on keratinase stability
The effect of protease inhibitors (phenylmethylsulfonyl fluoride (PMSF), ethylene diamine tetraacetic acid (EDTA), 1,10-phenanthroline), and metal ions (NaCl, KCl, CaCl2, MgCl2, ZnCl2, CuCl2, FeCl2, HgCl2, BaCl2, CoCl2, AlCl3 and FeCl3) was evaluated at 5 mM. Furthermore, the impact of selected solid laundry detergents (7 mg/mL), including Sunlight, Omo, Ariel, Maq, Surf, Sky, Freshwave, Evaklin, and Prowash, was studied following a previously described method [22].
2.9. Genomic DNA extraction and amplification of keratinase-encoding gene (kerBAN)
The genomic DNA of Bacillus sp. NFH5 was extracted, as reported previously [23]. DNA fragment coding for keratinase was amplified by conventional polymerase chain reaction (PCR) using a set of forward primer (kerBANF: TCATCTACTGATTACGTTCC) and reverse primer (kerBANR: TTAAGAAGCTTTATTTTCTTG). The primer pair was designed based on the nucleotide sequence of Bacillus thuringiensis keratinase gene (accession number KX155576) and the isolates' phylogenetic relatedness. The 25 μL reaction mixture, which consisted of 12.5 μL of OneTaq® Quick-Load® 2X master mix (New England Biolabs Inc., South Africa), 5.5 μL of nuclease-free water, 1 μL each of both forward and reverse primers, and 5 μL of DNA template was used for the PCR. The amplification of kerBAN was carried out using T100™ Thermal Cycler (Bio-Rad Laboratories Inc., Singapore), with the following conditions: initial denaturation at 95 °C for 5 min, 35 cycles of denaturation at 95 °C for 30 s, annealing at 50 °C for 1 min, extension at 72 °C for 1 min, and then final extension at 72 °C for 5 min. Subsequently, the amplified DNA fragment was electrophoresed on ethidium bromide-stained 1.2% (w/v) agarose gel (Merck chemicals (Pty) Ltd., South Africa), and the visualization was carried out using an ultraviolet transilluminator (Uvitec, UK).
2.9.1. DNA sequencing and phylogenetic analysis
The putative keratinase-encoding gene was analyzed using the dideoxynucleotide chain termination method [24]. Briefly, the amplicons were purified using the Nucleofast 96 well post-PCR clean-up plate (Macherey Nagel GmbH & Co., Düren, Germany) following the manufacturer's guidelines. Subsequently, the pure amplicons were sequenced with the BigDye Terminator V3.1 sequencing kit (Applied Biosystems) on an ABI3730XL DNA analyzer using a 50 cm capillary array and POP7 (Applied Biosystems) following the supplied manufacturer's protocols. The sequencing was carried out bi-directionally in order to obtain a reliable sequence. Geneious Prime V2020.1.1 (Biomatters Ltd., Auckland, NewZealand) was used to analyze the nucleotide sequences, while BLAST online tool was carried out to compare the sequence similarity with other sequences in the database. The nucleotide sequence of the keratinase-coding gene (kerBAN) was submitted to GenBank with the accession number OK033360.
2.9.2. Analysis of physicochemical parameters of the enzyme (KerBAN)
The physicochemical parameters include the number of amino acids, molecular weight, theoretical isoelectric point (pI), aliphatic index, instability index, and grand average of hydrophobicity (GRAVY) of strain NFH5 keratinase (KerBAN) were determined using Expasy-ProtParam (http://web.expasy.org/protparam) [25]. The presence of signal peptides in the protein and the sites of cleavage were determined using SignalP 6.0 Server (https://services.healthtech.dtu.dk/service.php?SignalP), while propeptide sequence identification was carried out by ProP 1.0 Server search (http://www.cbs.dtu.dk/services/ProP/).
2.9.3. Homology modeling and active site prediction
The keratinase structural homology modeling was carried out using SWISS-MODEL via the Expasy web server (https://swissmodel.expasy.org/) [26]. The active site was predicted using GASS-WEB (https://gass.unifei.edu.br/) online tool. After modeling, the quality of the model was determined using other structure validation methods, including QMEAN Z-Score [27] and Ramachandran plot analysis (https://zlab.umassmed.edu/bu/rama/index.pl).
2.10. Statistical analysis
The datasets generated from the triplicate experiments were submitted to analysis of variance (ANOVA), and the degree of freedom was set at P < 0.05 significance level. The analysis was conducted in Statistical Package for Social Science (SPSS) version 23.
3. Results
3.1. Bacterial isolates identification
Feather-degrading bacterial isolates coded as SSN-01 and HSN-01, previously isolated from the municipal dumpsite (Supplementary Fig. 1) and maintained in the AEMREG culture repository, were further explored for their keratinolytic potentials. The phylogenetic analysis of the 16S rRNA gene sequence indicated that both isolates belonged to Bacillus spp., with 100% similarity with Bacillus cereus group (sensu lato). Hits from the BLAST search showed that isolates SSN-01 and HSN-01 sequences are closely related to B. cereus S8 (NCBI: MT611946.1), Bacillus toyonensis SA04 (NCBI: OK350064.1), and Bacillus thuringiensis VMSES01 (NCBI: MZ234580.1); therefore, they were identified as Bacillus sp. NFH5 and Bacillus sp. FHNM, respectively (Supplementary Fig. 2). The nucleotide sequences have been deposited in the GenBank and are available under the respective accession numbers MW165830.1 and MW165831.1 for Bacillus sp. NFH5 and Bacillus sp. FHNM.
3.2. Optimization of fermentation process conditions
Three significant variables, including incubation temperature, initial medium pH, and chicken feather concentration, were optimized for enhanced keratinase production by Bacillus sp. NFH5. The isolate maximally secreted keratinolytic protease at 30 °C, with initial fermentation pH of 4 and substrate concentration of 1.5% (w/v) (see Supplementary Fig. 3).
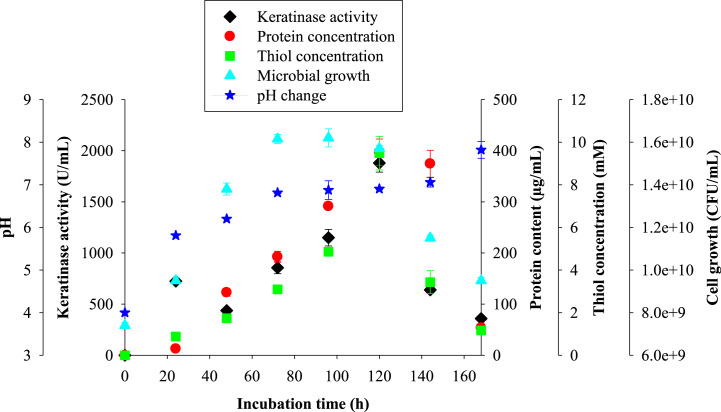
At the optimal process conditions, the keratinase production by NFH5 reached 724.55 ± 16.71 U/mL at 24 h of fermentation (Fig. 1) but declined after 48 h (437.27 ± 1.29 U/mL). Subsequently, the enzyme production increased, reaching a maximum at 120 h with the keratinase activity of 1879.09 ± 88.70 U/mL, representing a 3.4-fold increase in enzyme activity compared to 546 ± 20.63 U/mL obtained in an unoptimized condition (Supplementary Fig. 1). Further incubation resulted in a drastic decrease in enzyme activity. Microbial growth was highest at 72 h of incubation but reduced after 96 h of fermentation (Fig. 1). Medium total protein and thiol contents steadily rose with increasing incubation period, reaching respective maximum values of 397.05 ± 25.64 μg/mL and 9.49 ± 0.78 mM at 120 h (Fig. 1). Medium thiol concentration dropped after 120 h likewise the protein concentration that dramatically declined after 144 h. Furthermore, the pH of the fermentation media increased from the initial pH of 4.0 to a final value of 7.82 ± 0.19 after 168 h of incubation (Fig. 1).
Fig. 1.
Time-course profile of keratinase production by Bacillus sp. NFH5 in optimized medium conditions. Each point represents the average and standard deviation of triplicate experiments.
3.3. Amino acids analysis of chicken feather hydrolysate
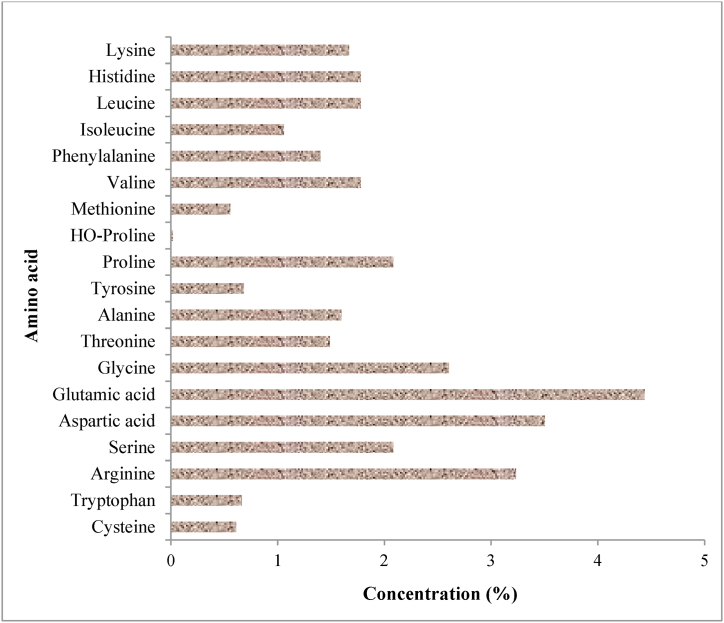
The depolymerized feathers' keratin was evaluated for various amino acids. The feather hydrolysate (with a total protein value of 55.67%) contained various amino acids at different concentrations (Fig. 2). The most abundant amino acids in the digest were glutamic acid (4.44%), aspartic acid (3.50%), arginine (3.23%), glycine (2.61%), serine (2.08%), and proline (2.08%). These occurred at far higher concentrations than methionine, tyrosine, tryptophan, and cysteine, which occurred at 0.56, 0.68, 0.67, and 0.61 (%), respectively. HO-proline was barely detected (Fig. 2).
Fig. 2.
Amino acid composition of feather hydrolysate from Bacillus sp. NFH5 fermentation medium.
3.4. The effect of pH and temperature on keratinase (kerBAN) activity
KerBAN was maximally active at pH 8.0 (Fig. 3). Notably, enzyme activity dropped at pH 7.0 (30.29 ± 0.28%) compared to activity (55.74 ± 6.12%) at pH 6.0. Beyond pH 8.0, enzyme activity steadily fell as alkalinity rose.
Fig. 3.
Effects of assay pH and temperature on KerBAN activity. Each point represents the average and standard deviation of triplicate experiments.
KerBAN activity rose steadily with assay temperature, attaining the highest value at 90 °C (Fig. 3). KerBAN also showed relatively high activity at 100 °C, with only a marginal decrease (or 6.2 ± 0.15%), compared to activity at optimum temperature.
3.5. The impact of chemical agents and laundry detergents on keratinase stability
KerBAN activity was markedly affected by the two metal ion chelators, EDTA and 1,10-phenanthroline, resulting in residual activity of 22 ± 1.72% and 27 ± 7.12%, respectively, at 5 mM (Table 1). In contrast, the serine protease inhibitor – PMSF did not considerably affect the enzyme activity (Table 1). Furthermore, KerBAN biocatalytic efficiency was not promoted by any metal ions evaluated; but overall maintained >80% residual activity (Table 1).
Table 1.
Effect of chemical agents and metal ions on KerBAN stability.
| Chemical agents | Concentration (mM) | Residual activity (%)a |
|---|---|---|
| Control | – | 100 ± 4.36 |
| PMSF | 5 | 93 ± 5.81 |
| EDTA | 5 | 22 ± 1.72 |
| 1,10-Phenanthroline | 5 | 27 ± 7.12 |
| Fe2⁺ | 5 | 79 ± 1.44 |
| Co2⁺ | 5 | 89 ± 3.26 |
| Fe³⁺ | 5 | 93 ± 0.70 |
| K⁺ | 5 | 89 ± 4.54 |
| Ca2⁺ | 5 | 88 ± 3.65 |
| Mg2⁺ | 5 | 88 ± 1.47 |
| Cu2⁺ | 5 | 83 ± 9.92 |
| Zn2⁺ | 5 | 84 ± 2.31 |
| Na⁺ | 5 | 92 ± 4.77 |
| Ba2⁺ | 5 | 92 ± 0.53 |
| Al³⁺ | 5 | 82 ± 5.91 |
Each treatment value indicates the average and standard deviation of triplicate assays.
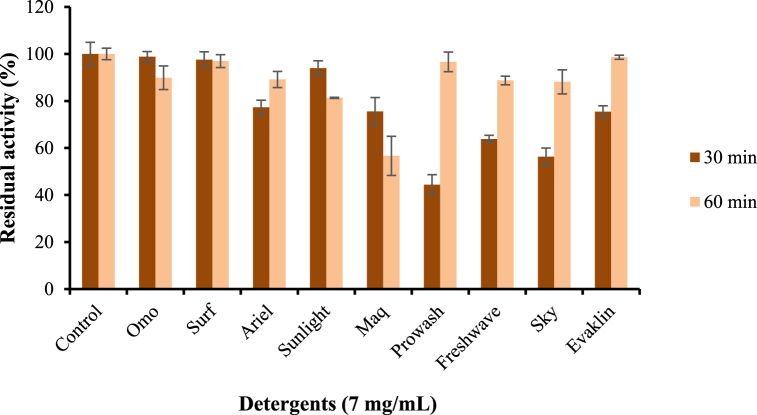
Fig. 4 presents the effects of various detergents on KerBAN stability. For most of the detergents, except Maq, Prowash, and Freshwave, enzyme stability was always high (≥80%) after both 30 and 60 min preincubation contact times. KerBAN was, however least stable (60 min pretreatment) with Maq, showing residual activity of 56.67% after 60 min (Fig. 4). Lowest stability value (44.4%) after 30 min pretreatment was, however, given by Prowash detergent. Most noteworthy, however, is the observation that residual KerBAN activity, with Ariel, Prowash, Freshwave, Sky, and Evaklin, was always more diminished, on 30 min pretreatment, than after 60 min pretreatment (Fig. 4).
Fig. 4.
The effect of laundry detergents on KerBAN stability. Each bar represents the average and standard deviation of triplicate experiments.
3.6. Keratinase-encoding gene (kerBAN) sequencing and physicochemical analysis
The putative keratinase-encoding gene in Bacillus sp. NFH5 was amplified and sequenced. The amplified DNA fragment coding keratinase (kerBAN) had a size of 1104 bp (Supplementary Fig. 4) as well as displayed a high percentage similarity with the keratinase and other peptidase genes from members of the B. cereus group. The gene sequence translation gave 362 amino acid residues with a molecular weight of ≈39 kDa. Other physicochemical properties of KerBAN, including its theoretical pI, GRAVY, aliphatic, and instability indexes, were deduced to be 8.81, −0.428, 70.61, and −18.79, respectively. The signal peptide and propeptide predictions indicated that the amino acid sequence lacked both peptides (Supplementary Figs. 5 and 6).
3.7. Homology modeling and model analysis
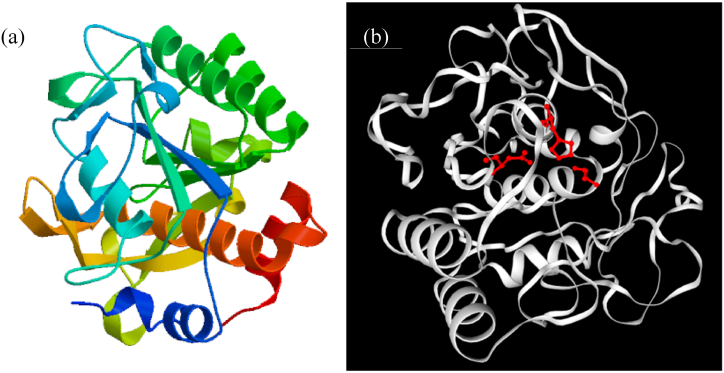
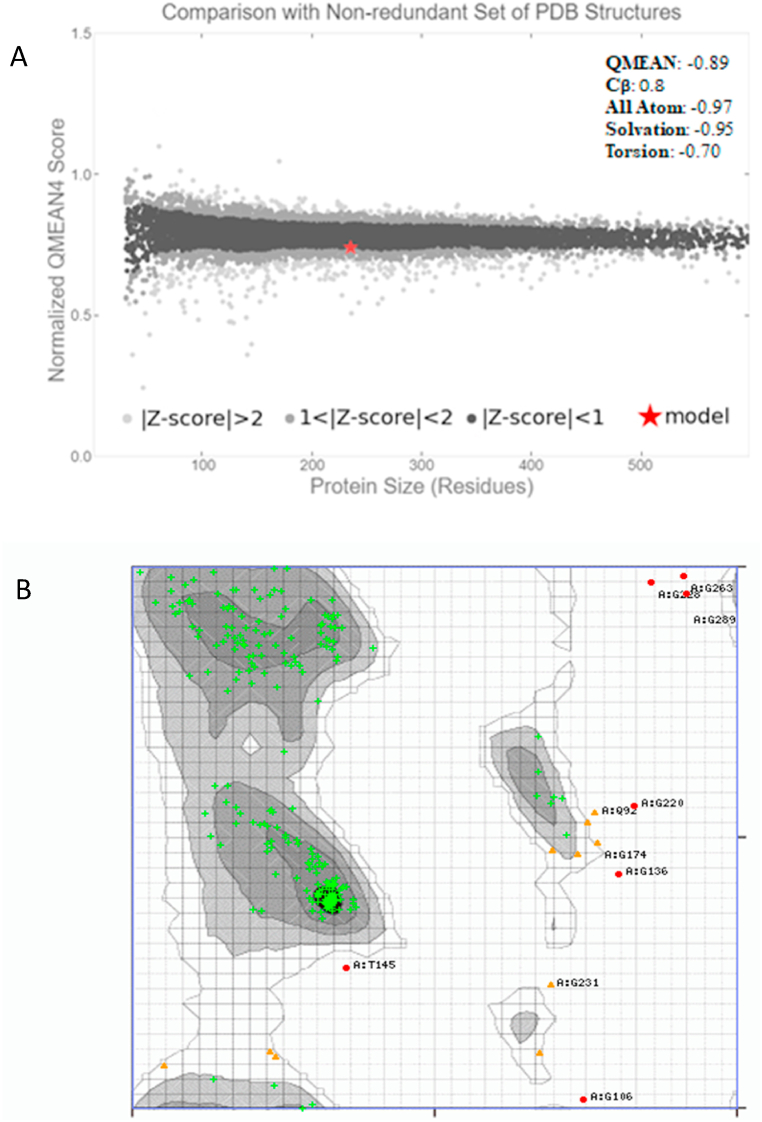
The three-dimensional (3D) protein structure was generated (Fig. 5a), while peptidase with a sequence identity score of 74.36% served as a template (Protein Data Bank (PDB) ID: 1thm). Subsequently, the active site prediction with GASS-WEB showed the presence of HIS 137 A; ASP 104 A; SER 291 A (red balls and sticks) at the catalytic cleft (Fig. 5b). Global model quality estimate (GMQE) and QMEANDisCo global score estimated for KerBAN were 0.59 and 0.86, respectively. The QMEAN Z-score was computed to be −0.89 (Fig. 6a). The Ramachandran plot showed that 93.227% of the residues were highly preferred observations, while 3.984% and 2.789% of the observations constituted the preferred and questionable (outliers) residues, respectively (Fig. 6b).
Fig. 5.
Homology three-dimensional (3D) structure of keratinase (KerBAN) from Bacillus sp. NFH5 generated by Swiss-Modelling using PDB ID: 1thm as a template (a); and active site prediction by GASS-WEB (b).
Fig. 6.
(A) QMEAN PDB structure of KerBAN showing Z-score values. (B) Ramachandran plot of the observed conformations. The numbers of highly preferred residues (green crosses; 93.227%), preferred residues (brown triangles; 3.984%), and questionable residues (red circles; 2.789%) are colour-coded for convenience.
4. Discussion
Keratinous waste-generating processes are among man's most environmentally impacting industrial activities [[28], [29], [30]]. Tannery wastes, potentially highly polluting [28], constitute up to 75% of the main raw material, animal hides, with only 25% going into leather manufacturing. Global poultry production was over 130 million tons in 2019 [31], generating tons of slow-to-degrade keratinous wastes with overwhelmingly high pollution potential. Keratinous wastes have also been considered the world's third most abundant natural biopolymer [2,30]. In recent times, the acute need to mitigate the deleterious effects of keratinous wastes on the environment has engendered an explosion in research targeted at identifying new and potent keratinolytic microbes and peptidases with potential for environmental and other biotechnological applications. Both keratinolytic isolates studied in this work originated from poultry feather-enriched natural composting sites. Isolation of these bacteria from this area is not particularly surprising, as it strengthens previous observations [[32], [33], [34], [35]] of isolating strongly keratinolytic bacteria from keratinous wastes-rich environments. Natural environments enriched in keratinous materials have often served as good sources for isolating keratin-eating microbes [36], being naturally enriched in keratinolytic organisms.
Bacillus species are industrial enzymes' most important microbial sources with excellent physicochemical properties. In recent times, keratinases, peptidases with robust physicochemical and biochemical properties, as well as multifaceted applications [14], have become identified among the sundry enzymes secreted in high proportions by members of the Bacillus genus [37]. The confirmatory identification, therefore, of the two chicken feather-degrading bacteria, isolates SSN-01 and HSN-01, as belonging to the Bacillus genus, was not surprising. Bacillus species have been described as the leading keratinase producers [38]. Both isolates SSN-01 and HSN-01 belonged to the B. cereus group, but because their identification process did not wholly discriminate among this sensu lato group, SSN-01 and HSN-01 became identified as Bacillus sp. NFH5 and Bacillus sp. FHNM, respectively. Alongside B. subtilis and B. licheniformis strains, the B. cereus group members have been described as some of the most effective microbial producers of keratinases. Keratinase production by members of the B. cereus group has been previously reported by several researchers [[39], [40], [41]]. Therefore, identifying both keratinolytic isolates as members of the B. cereus group is also not unexpected. The fact that the current isolates appear to be unknown strains, however, extends the number of strains of the B. cereus group from which keratinolytic proteases could be obtained. As new strains often have been associated with the production of enzymes with often varied characteristics, the continued discovery of new keratinolytic strains holds the potential for discovering new keratinases with desirable properties for novel vistas of keratinous biomass exploitation. Previously, such enzymes have been developed only through the cost-intensive process of protein engineering [42].
Keratinase titres, during fermentation by Bacillus sp. NFH5 was highest during the early stationary phase. This might be due to the accumulation of dead cells and their subsequent rupture and release of their intracellular constituent. Significantly lower maximum keratinase titres of 74.66 ± 1.52 U/mL, 420 ± 1.63 U/mL, and 292 U/g have been reported for B. cereus [40], B. cereus strain 151,007-R3_I05_21_26 F [38], and B. cereus [39], respectively, indicating markedly higher productivity by Bacillus sp. NFH5. Peak thiol concentration corresponded to maximum keratinase production at 120 h. This suggests the following: (1) that disulfide reductase activity was probably a significant component of Bacillus sp. NFH5 keratinolytic system, and (2) that it contributed markedly to the dismemberment of keratin polymers into smaller units of proteins with high contents of sulfhydryl group. Previously, Bacillus subtilis [43], Bacillus sp. CSK2 [44], and Bacillus sp. MBRL 575 [45], growing on keratinous material, engendered maximum liberation of thiols at medium concentrations of 15.5 ± 0.2 μM, 492.98 ± 82.99 μM, and 82 μM, respectively. Detection of high contents of thiol groups in the fermentation medium indicates efficient hydrolysis of disulfide linkages contributing to complete keratinolysis [9]. This observation thus highlights the relevance of strain NFH5 in converting keratinous residues into bio-accessible protein units. The protein concentration pattern suggests that microbial keratinase liberated more soluble proteins from chicken feather keratin during fermentation [46]. The drift of medium pH from the acidic to an alkaline range indicates the possible presence of ammonia in the production medium [47]. Ammonia, a major product of the deamination of proteolytic degradation products, has been associated with the growth of proteolytic bacteria on proteinaceous substrates [48].
Enzymatic digestion of keratinous materials has become one of the preferred approaches for processing keratin-rich waste materials into valuable hydrolysates [49,50]. According to Łaba et al. [50], such digestion of keratinous materials allows for the sustainable conversion of cheap and readily available keratin-rich agricultural wastes into far more valuable feather meal and protein hydrolysate products of improved dietary value, amino acid balance, and digestibility [51]. Amino acid analysis of the hydrolysate produced from feathers by Bacillus sp. NFH5 keratinolytic system shows that it was markedly enriched in free amino acids. On the basis of its free amino nitrogen content, the protein hydrolysate under investigation may serve as a cost-effective dietary supplement for livestock feed formulation, as feed cost is the major expenditure affecting poultry production globally [52]. Hydroxyproline was not detected in the protein hydrolysate as it plays no critical role in keratin protein; however, it is a significant component of collagen, where it provides mechanical support in tissues [53].
The Bacillus sp. NFH5 keratinase's (KerBAN) optimal pH of 8 agrees with other reports [54,55] that keratinolytic proteases, irrespective of the microbial source, are optimally active under neutral to alkaline conditions. This property has been said to promote their application in industrial productions, like leather processing and detergent formulation [22,56]. The decrease in enzyme activity at neutral pH may be attributed to the orientation of catalytic cleft or side chains at this pH condition, affecting substrate binding and product formation [4]. Vieille and Zeikus [57] have defined enzymes that retain maximum activity at >80 °C as hyperthermophilic enzymes. KerBAN was most active at the elevated temperature of 90 °C and can thus be regarded as hyperthermoactive. Enzyme hyperthermoactivity has previously been associated with keratinases from Fervidobacterium islandicum AW-1 (100 °C) [58], Bacillus circulans DZ100 (85 °C) [59], and Fervidobacterium pennavorans (80 °C) [60]. This property of KerBAN highlights the prospects of this novel enzyme for industrial and biotechnological processes requiring hyperthermoactive proteases. Some properties of Bacillus sp. NFH5 keratinase with other characterized bacterial keratinases presented in Table 2 further spotlight the merit KerBAN will enjoy in the protease market if its commercialization is put into perspective.
Table 2.
Some properties of KerBAN in comparison with other characterized bacterial keratinases.
| Microbial source | Max. prod. keratinase (U/mL)* | Optimum pH | Optimum temp. (oC)* | Keratinase type | Mol. Wt. (kDa)* | Ref.* |
|---|---|---|---|---|---|---|
| Bacillus sp. NFH5 | 1879.09 | 8.0 | 90 | Metallo | 39 | This study |
| Fervidobacterium islandicum AW-1 | – | 9 | 100 | – | >200, 97 | [58] |
| Bacillus circulans DZ100 | 15,000 | 12.5 | 85 | Serine | 32 | [59] |
| Pedobacter sp. 3.14.7 | 33.04 | 7.5 | 55 | Metallo | 25 | [61] |
| Pseudomonas aeruginosa KS-1 | 1200 | 9 | 60 | Serine | 45 | [62] |
| Bacillus cereus YQ15 | 925 | 10 | 60 | Serine-metallo | – | [63] |
| Acinetobacter sp. R-1 | 188 | 11 | 50 | Metallo | 25 | [64] |
| Chryseobacterium L99 sp. nov. | 213.8 | 8 | 40 | Serine | 33 | [65] |
| Streptomyces sp. | – | 11 | 45 | Serine-metallo | 44 | [66] |
| Brevibacillus brevis US575 | 7500 | 8 | 40 | Serine | 29.12 | [67] |
| Microbacterium sp. kr10 | – | 7.5 | 50 | Metallo | 42 | [68] |
(*) Max. prod. = Maximum produced; Temp. = Temperature; Mol. Wt. = Molecular weight; Ref. = Reference; (−) Not reported.
The removal of metal ions by ion chelators stalled the activity of KerBAN, suggesting that inherent metal ion(s) are crucial for the catalytic integrity of the enzyme [69]. A similar inhibition pattern was observed among other bacterial keratinases [4,64]. All the metal ions tested did not enhance KerBAN activity, which shows that this metallo-keratinase does not require extra cofactors to attain optimal catalytic efficiency. Likewise, previous reports of metallo-keratinase from other bacterial strains indicated that all the metal ions assessed negatively impacted the enzyme stability [70,71].
KerBAN showed an excellent stability profile in the presence of laundry detergents, indicating that it could be a good candidate for detergent formulation. During the study, KerBAN was less active when tested in the presence of some of the detergents at 30 min but showed higher activity at 1 h. While we do not have an immediate explanation for this, it is, however, quite possible that the following could have contributed to the observed phenomenon. Enzymes are proteins with complex three-dimensional structures, with native, active proteins being held together by a delicate balance of non-covalent forces. It is well known that detergents, especially if they contain anionic surfactants (e.g., alcohol sulfates (AS), alpha olefins (AOS), secondary alkane sulfonates (SAS), and linear alkene benzene sulfonates (LAS)), pose special stability problems for enzymes used in their formulations [[72], [73], [74], [75], [76]], often through engendering the oxidation, deamination, surfactant-induced unfolding and aggregation/precipitation of the latter (proteases). The surfactant-induced unfolding of enzymes by nonionic surfactants has particularly been associated with detergent protease inactivation [[73], [74], [75], [76]]. On the other hand, other chemical constituents of detergents, like chelators [72], patented stabilizers (e.g., alcohol ethers), polyols, alkanolamines, short-chain carboxylic acids, boron compounds, and some cations, especially of alkali metal salts like Ca2+ [75,76] have been shown to counteract the deleterious effects of anionic surfactants. KerBAN's recovery of activity at 1 h, after inhibition at 30 min exposure to some of the commercial detergents, may thus be due to the complex interactions between the various constituents of those detergents and the keratinolytic enzyme. Interactions between proteins and protein-denaturing surfactants found in commercial detergents may or may not, depending on the protein, nature of surfactants, and the other chemical components of the detergents (pH controlling agents, chelators, colorants, perfume, etc.), change a protein's secondary or tertiary structure (thus lead to its denaturation) [72,[74], [75], [76]]. In some instances, protein unfolding and denaturation have been found to be reversible [57]. It is additionally possible that, because of the peculiar composition of the detergents, interaction with KerBAN resulted in a reversible unfolding of the enzyme protein, with the protein being restored to its active conformation, following possible changes in the chemical environment of the reaction medium as the reaction was incubated for a longer time. There have been reports of possible precipitation of anionic surfactant molecules by chelators in commercial detergents [72,76]. Such anionic surfactant precipitation, if it is found to occur in those detergents (Prowash, Freshwave, and Sky), could probably partially explain enzyme activity recovery. A group of detergents did not exhibit any decrease in KerBAN activity at 30 min incubation. This could be explained by the fact that the peculiar chemical compositions of those detergents. It is a well-known fact that to ensure enzyme stabilization; several enzyme-containing commercial detergents are formulated with alcohol ethoxylates (AE) and relatively high levels of alcohol ethoxysulfates (AES), the only anionic surfactant not known to pose stability problems for enzymes in detergents [72]. It is possible that those detergents that did not inhibit KerBAN were formulated as above or similarly.
The molecular weight estimation of KerBAN is consistent with that of another keratinase from Bacillus spp [55]. The theoretical isoelectric point of the enzyme suggested that it is mostly active at an alkaline condition, as was corroborated by the assay-determined optimal activity of KerBAN. GRAVY is a physicochemical parameter used to predict a protein's hydrophobic or hydrophilic nature computed from the hydropathy values [77]. The negative value indicates that KerBAN is a hydrophilic protein [78]. The instability index measures protein stability experimentally, and any protein with a value less than 40 is considered stable [79]. In the case of KerBAN, the instability index at a negative value indicates its high stability. Furthermore, a high aliphatic index value indicates the thermostable nature of the keratinolytic protease over a wide temperature range. This parameter has been increasingly employed as an indicator for assessing the thermostability of globular proteins [80]. The absence of signal peptide and propeptide in the study amino acid sequence shows that the pair of primers used for gene cloning specifically targeted functional peptidase sequence in the bacterial genome.
Protein three-dimensional structure provides clues regarding protein molecular orientation and functional properties. Model evaluation with GMQE and QMEANDisCo global overall suggested a good model quality. The relatively lower score (although above the threshold) of GMQE could be attributed to the target-template sequence coverage, which differs from QMEANDisCo, that is not dependent on explicit sequence coverage [81]. Similarly, QMEAN is another important parameter for predicting model quality. The QMEAN Z-score close to zero suggests a native-like structure in terms of the geometric arrangement of various protein residues on a global scale, while below −4 signifies a poor-quality model [27]. The QMEAN Z-score obtained in this study is consistent with that reported for other enzymes [82]. With the percentage of residues in the highly preferred and preferred regions, the Ramachandran plot provides stereochemical quality information on the enzyme stability and indicates that the model is acceptable [79,83].
5. Conclusion
The identity of chicken feather-degrading bacteria from a local municipal dumpsite was confirmed as Bacillus sp. NFH5 and Bacillus sp. FHNM. NFH5 grew and utilized chicken feathers as a carbon and nitrogen source, maximally secreted keratinase, and directed free thiol group liberation at 120 h of fermentation. The analysis of the feather hydrolysates showed the presence of various amino acids at different concentrations suggesting the prospect of the protein hydrolysate in livestock dietary feed formulation. The Metallo-keratinase showed optimal catalytic efficiency at pH 8.0 and 90 °C. The activity of KerBAN after treatment with solid laundry detergent indicates its significance in the detergent industry. Detection of the putative keratinase-coding genes confirms the keratinolytic potentials demonstrated by the isolate. An optimal enzyme production at a more extended fermentation period is unsustainable from an industrial perspective. Hence, the kerBAN elucidation is a motivation for future study, which will include gene cloning and heterologous expression in a competent commercial host for higher keratinase yield, which would address the problem of the extended incubation period for maximum productivity associated with wild bacterial isolates.
Declarations
Author contribution statement
Lupho Kokwe: Performed the experiments; Wrote the paper. Nonso E. Nnolim: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper. Lewis I. Ezeogu, Bruce Sithole: Analyzed and interpreted the data; Wrote the paper. Uchechukwu U. Nwodo: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by the Industrial Biocatalysis Hub, funded by the Department of Science and Innovation and the Technology Innovation Agency.
Data availability statement
Data associated with this study has been deposited at “NCBI” under the accession number [MW165830.1, MW165831.1 and OK033360].
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e13635.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Shestakova A., Timorshina S., Osmolovskiy A. Biodegradation of keratin-rich husbandry waste as a path to sustainable agriculture. Sustainability. 2021;13:8691. doi: 10.3390/su13168691. [DOI] [Google Scholar]
- 2.Lange L., Huang Y., Busk P.K. Microbial decomposition of keratin in nature—a new hypothesis of industrial relevance. Appl. Microbiol. Biotechnol. 2016;100:2083–2096. doi: 10.1007/s00253-015-7262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falade A.O. Valorization of agricultural wastes for production of biocatalysts of environmental significance: towards a sustainable environment. Environ. Sustain. 2021;4:317–328. doi: 10.1007/s42398-021-00183-9. [DOI] [Google Scholar]
- 4.Bokveld A., Nnolim N.E., Digban T.O., Okoh A.I., Nwodo U.U. Chryseobacterium aquifrigidense keratinase liberated essential and nonessential amino acids from chicken feather degradation. Environ. Technol. 2021 doi: 10.1080/09593330.2021.1969597. [DOI] [PubMed] [Google Scholar]
- 5.Biswas I., Mitra D., Senapati A., Mitra D., Chattaraj S., Ali M., Basak G., Panneerselvam P., Das Mohapatra P.K. Valorization of vermicompost with bacterial fermented chicken feather hydrolysate for the yield improvement of tomato plant: a novel organic combination. Int. J. Recycl. Org. Waste Agric. 2021;10:29–42. doi: 10.30486/ijrowa.2020.1904599.1104. [DOI] [Google Scholar]
- 6.Devi S., Chauhan A., Bishist R., Sankhyan N., Rana K., Sharma N. Production, partial purification and efficacy of keratinase from Bacillus halotolerans L2EN1 isolated from the poultry farm of Himachal Pradesh as a potential laundry additive. Biocatal. Biotransform. 2022 doi: 10.1080/10242422.2022.2029851. [DOI] [Google Scholar]
- 7.Nnolim N.E., Nwodo U.U. Microbial keratinase and the bio-economy: a three-decade meta-analysis of research exploit. Amb. Express. 2021;11:12. doi: 10.1186/s13568-020-01155-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sypka M., Jodłowska I., Białkowska A.M. Keratinases as versatile enzymatic tools for sustainable development. Biomolecules. 2021;11:1900. doi: 10.3390/biom11121900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hendrick Q., Nnolim N.E., Nwodo U.U. Chryseobacterium cucumeris FHN1 keratinolytic enzyme valorized chicken feathers to amino acids with polar, anionic and non-polar imino side chain characteristics. Biocatal. Agric. Biotechnol. 2021;35 doi: 10.1016/j.bcab.2021.102109. [DOI] [Google Scholar]
- 10.Kang D., Shoaie S., Jacquiod S., Sørensen S.J., Ledesma-Amaro R. Comparative genomics analysis of keratin-degrading Chryseobacterium species reveals their keratinolytic potential for secondary metabolite production. Microorganisms. 2021;9:1042. doi: 10.3390/microorganisms9051042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang Z., Zhang J., Liu B., Du G., Chen J. Biochemical characterization of three keratinolytic enzymes from Stenotrophomonas maltophilia BBE11-1 for biodegrading keratin wastes. Int. Biodeterior. Biodegrad. 2013;82:166–172. doi: 10.1016/j.ibiod.2013.03.008. [DOI] [Google Scholar]
- 12.Kowalczyk P., Mahdi-Oraibi S., Misiewicz A., Gabzdyl N., Miskiewicz A., Szparecki G. Feather-degrading bacteria: their biochemical and genetic characteristics. Arabian J. Sci. Eng. 2018;43:33–41. doi: 10.1007/s13369-017-2700-2. [DOI] [Google Scholar]
- 13.Gurunathan R., Huang B., Ponnusamy V.K., Hwang J.S., Dahms H.U. Novel recombinant keratin degrading subtilisin like serine alkaline protease from Bacillus cereus isolated from marine hydrothermal vent crabs. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-90375-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Almahasheer A.A., Mahmoud A., El-Komy H., Alqosaibi A.I., Aktar S., AbdulAzeez S., Borgio J.F. Novel feather degrading keratinases from Bacillus cereus group: biochemical, genetic and bioinformatics analysis. Microorganisms. 2022;10:93. doi: 10.3390/microorganisms10010093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nnolim N.E., Okoh A.I., Nwodo U.U. Proteolytic bacteria isolated from agro-waste dumpsites produced keratinolytic enzymes. Biotechnol. Rep. 2020;27 doi: 10.1016/j.btre.2020.e00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 17.Ellman G.L. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 18.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. Mega X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeVries J.W., Koski C.M., Egberg D.C., Larson P.A. Comparison between a spectrophotometric and a high-pressure liquid chromatography method for determining tryptophan in food products. J. Agric. Food Chem. 1980;28:896–898. doi: 10.1021/jf60231a025. [DOI] [PubMed] [Google Scholar]
- 20.Einarsson S., Josefsson B., Lagerkvist S. Determination of amino acids with 9-fluorenylmethyl chloroformate and reversed-phase high-performance liquid chromatography. J. Chromatogr. 1983;282:609–618. doi: 10.1016/S0021-9673(00)91638-8. [DOI] [Google Scholar]
- 21.Gehrke C.W., Wall L.L., Sr., Absheer J.S., Kaiser F.E., Zumwalt R.W. Sample preparation for chromatography of amino acids: acid hydrolysis of proteins. JAOAC (J. Assoc. Off. Anal. Chem.) 1985;68:811–821. doi: 10.1093/jaoac/68.5.811. [DOI] [Google Scholar]
- 22.Paul T., Das A., Mandal A., Halder S.K., Jana A., Maity C., DasMohapatra P.K., Pati B.R., Mondal K.C. An efficient cloth cleaning properties of a crude keratinase combined with detergent: towards industrial viewpoint. J. Clean. Prod. 2014;66:672–684. doi: 10.1016/j.jclepro.2013.10.054. [DOI] [Google Scholar]
- 23.Maugeri T.L., Carbone M., Fera M.T., Gugliandolo C. Detection and differentiation of Vibrio vulnificus in seawater and plankton of a coastal zone of the Mediterranean Sea. Res. Microbiol. 2006;157:194–200. doi: 10.1016/j.resmic.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 24.Sanger F., Nicklen S., Coulson A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gasteiger E., Hoogland C., Gattiker A., Wilkins M.R., Appel R.D., Bairoch A. In: The Proteomics Protocols Handbook. Walker J.M., editor. Springer Protocols Handbooks. Humana press; 2005. Protein identification and analysis tools on the ExPASy server; pp. 571–607. [DOI] [Google Scholar]
- 26.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., Lepore R. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296. doi: 10.1093/nar/gky427. –W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benkert P., Biasini M., Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dettmer A., Cavalli É., Ayub M.A., Gutterres M. Optimization of the unhairing leather processing with enzymes and the evaluation of inter-fibrillary proteins removal: an environment-friendly alternative. Bioproc. Biosyst. Eng. 2012;35:1317–1324. doi: 10.1007/s00449-012-0719-z. [DOI] [PubMed] [Google Scholar]
- 29.Gupta R., Sharma R., Beg Q.K. Revisiting microbial keratinases: next generation proteases for sustainable biotechnology. Crit. Rev. Biotechnol. 2013;33:216–228. doi: 10.3109/07388551.2012.685051. [DOI] [PubMed] [Google Scholar]
- 30.Bagewadi Z.K., Mulla S.I., Ninnekar H.Z. Response surface methodology based optimization of keratinase production from Trichoderma harzianum isolate HZN12 using chicken feather waste and its application in dehairing of hide. J. Environ. Chem. Eng. 2018;6:4828–4839. doi: 10.1016/j.jece.2018.07.007. [DOI] [Google Scholar]
- 31.Brockotter F. 2020. Popularity of poultry continues globally.https://www.poultryworld.net/poultry/popularity-of-poultry-continues-globally/ Available from: Accessed. [Google Scholar]
- 32.Kim J.M., Lim W.J., Suh H.J. Feather-degrading Bacillus species from poultry waste. Process Biochem. (Amsterdam, Neth.) 2001;37:287–291. doi: 10.1016/S0032-9592(01)00206-0. [DOI] [Google Scholar]
- 33.Tork S.E., Shahein Y.E., El-Hakim A.E., Abdel-Aty A.M., Aly M.M. Production and characterization of thermostable metallo-keratinase from newly isolated Bacillus subtilis NRC 3. Int. J. Biol. Macromol. 2013;55:169–175. doi: 10.1016/j.ijbiomac.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 34.Habbeche A., Saoudi B., Jaouadi B., Haberra S., Kerouaz B., Boudelaa M., Badis A., Ladjama A. Purification and biochemical characterization of a detergent-stable keratinase from a newly thermophilic actinomycete Actinomadura keratinilytica strain Cpt29 isolated from poultry compost. J. Biosci. Bioeng. 2014;117:413–421. doi: 10.1016/j.jbiosc.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 35.Jana A., Halder S.K., Dasgupta D., Hazra S., Mondal P., Bhaskar T., Ghosh D. Keratinase biosynthesis from waste poultry feathers for proteinaceous stain removal. ACS Sustain. Chem. Eng. 2020;8:17651–17663. doi: 10.1021/acssuschemeng.0c04378. [DOI] [Google Scholar]
- 36.Agrahari S., Wadhwa N. Degradation of chicken feather a poultry waste product by keratinolytic bacteria isolated from dumping site at Ghazipur poultry processing plant. Int. J. Poultry Sci. 2010;9:482–489. [Google Scholar]
- 37.Li Q. Structure, application, and biochemistry of microbial keratinases. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.674345. 10.3389%2Ffmicb.2021.674345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shalaby M.M., Samir R., Goma F.A.Z.M., Rammadan M.A. Enhanced fusidic acid transdermal delivery achieved by newly isolated and optimized Bacillus cereus Keratinase. Biotechnol. Rep. 2021;30 doi: 10.1016/j.btre.2021.e00620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arokiyaraj S., Varghese R., Ahmed B.A., Duraipandiyan V., Al-Dhabi N.A. Optimizing the fermentation conditions and enhanced production of keratinase from Bacillus cereus isolated from halophilic environment. Saudi J. Biol. Sci. 2019;26:378–381. doi: 10.1016/j.sjbs.2018.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akhter M., Wal Marzan L., Akter Y., Shimizu K. Microbial bioremediation of feather waste for keratinase production: an outstanding solution for leather dehairing in tanneries. Microbiol. Insights. 2020;13 doi: 10.1177/1178636120913280. 10.1177%2F1178636120913280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nie L., Zhang R., Zhang L., Ma M., Li C., Zhang Y., An Y., Xu H., Xiao S., Wang T. Mutations in the regulatory regions result in increased streptomycin resistance and keratinase synthesis in Bacillus thuringiensis. Arch. Microbiol. 2021;203:5387–5396. doi: 10.1007/s00203-021-02525-x. [DOI] [PubMed] [Google Scholar]
- 42.Sharma K.M., Kumar R., Panwar S., Kumar A. Microbial alkaline proteases: optimization of production parameters and their properties. J. Genet. Eng. Biotechnol. 2017;15:115–126. doi: 10.1016/j.jgeb.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeong J.H., Jeon Y.D., Lee O.M., Kim J.D., Lee N.R., Park G.T., Son H.J. Characterization of a multifunctional feather-degrading Bacillus subtilis isolated from forest soil. Biodegradation. 2010;21:61029–61040. doi: 10.1007/s10532-010-9363-y. [DOI] [PubMed] [Google Scholar]
- 44.Nnolim N.E., Nwodo U.U. Bacillus sp. CSK2 produced thermostable alkaline keratinase using agro-wastes: keratinolytic enzyme characterization. BMC Biotechnol. 2020;20:65. doi: 10.1186/s12896-020-00659-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kshetri P., Ningthoujam D.S. Keratinolytic activities of alkaliphilic Bacillus sp. MBRL 575 from a novel habitat, limestone deposit site in Manipur, India. SpringerPlus. 2016;5:595. doi: 10.1186/s40064-016-2239-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rieger T.J., De Oliveira C.T., Pereira J.Q., Brandelli A., Daroit D.J. Proteolytic system of Bacillus sp. CL18 is capable of extensive feather degradation and hydrolysis of diverse protein substrates. Br. Poultry Sci. 2017;58:329–335. doi: 10.1080/00071668.2017.1293229. [DOI] [PubMed] [Google Scholar]
- 47.Tiwary E., Gupta R. Medium optimization for a novel 58 kDa dimeric keratinase from Bacillus licheniformis ER-15: biochemical characterization and application in feather degradation and dehairing of hides. Bioresour. Technol. 2010;101:6103–6110. doi: 10.1016/j.biortech.2010.02.090. [DOI] [PubMed] [Google Scholar]
- 48.He Z., Sun R., Tang Z., Bu T., Wu Q., Li C., Chen H. Biodegradation of feather waste keratin by the keratin-degrading strain Bacillus subtilis 8. J. Microbiol. Biotechnol. 2018;28:314–322. doi: 10.4014/jmb.1708.08077. [DOI] [PubMed] [Google Scholar]
- 49.Grazziotin A., Pimentel F.A., De Jong E.V., Brandelli A. Nutritional improvement of feather protein by treatment with microbial keratinase. Anim. Feed Sci. Technol. 2006;126:135–144. doi: 10.1016/j.anifeedsci.2005.06.002. [DOI] [Google Scholar]
- 50.Łaba W., Żarowska B., Chorążyk D., Pudło A., Piegza M., Kancelista A., Kopeć W. New keratinolytic bacteria in valorization of chicken feather waste. Amb. Express. 2018;8:9. doi: 10.1186/s13568-018-0538-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paul T., Das A., Mandal A., Halder S.K., DasMohapatra P.K., Pati B.R., Mondal K.C. Valorization of chicken feather waste for concomitant production of keratinase, oligopeptides and essential amino acids under submerged fermentation by Paenibacillus woosongensis TKB2. Waste Biomass Valori. 2014;5:575–584. doi: 10.1007/s12649-013-9267-2. [DOI] [Google Scholar]
- 52.Kidd M.T., Maynard C.W., Mullenix G.J. Progress of amino acid nutrition for diet protein reduction in poultry. J. Anim. Sci. Biotechnol. 2021;12:45. doi: 10.1186/s40104-021-00568-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li P., Wu G. Roles of dietary glycine, proline, and hydroxyproline in collagen synthesis and animal growth. Amino Acids. 2018;50:29–38. doi: 10.1007/s00726-017-2490-6. [DOI] [PubMed] [Google Scholar]
- 54.Daroit D.J., Brandelli A. A current assessment on the production of bacterial keratinases. Crit. Rev. Biotechnol. 2014;34:372–384. doi: 10.3109/07388551.2013.794768. [DOI] [PubMed] [Google Scholar]
- 55.Nnolim N.E., Udenigwe C.C., Okoh A.I., Nwodo U.U. Microbial keratinase: next generation green catalyst and prospective applications. Front. Microbiol. 2020;11:3280. doi: 10.3389/fmicb.2020.580164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fang Z., Yong Y.C., Zhang J., Du G., Chen J. Keratinolytic protease: a green biocatalyst for leather industry. Appl. Microbiol. Biotechnol. 2017;101:7771–7779. doi: 10.1007/s00253-017-8484-1. [DOI] [PubMed] [Google Scholar]
- 57.Vieille C., Zeikus G.J. Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 2001;65:1–43. doi: 10.1128/MMBR.65.1.1-43.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nam G.W., Lee D.W., Lee H.S., Lee N.J., Kim B.C., Choe E.A., Hwang J.K., Suhartono M.T., Pyun Y.R. Native-feather degradation by Fervidobacterium islandicum AW-1, a newly isolated keratinase-producing thermophilic anaerobe. Arch. Microbiol. 2002;178:538–547. doi: 10.1007/s00203-002-0489-0. [DOI] [PubMed] [Google Scholar]
- 59.Benkiar A., Nadia Z.J., Badis A., Rebzani F., Soraya B.T., Rekik H., Naili B., Ferradji F.Z., Bejar S., Jaouadi B. Biochemical and molecular characterization of a thermo-and detergent-stable alkaline serine keratinolytic protease from Bacillus circulans strain DZ100 for detergent formulations and feather-biodegradation process. Int. Biodeterior. Biodegrad. 2013;83:129–138. doi: 10.1016/j.ibiod.2013.05.014. [DOI] [Google Scholar]
- 60.Friedrich A.B., Antranikian G. Keratin degradation by Fervidobacterium pennavorans, a novel thermophilic anaerobic species of the order Thermotogales. Appl. Environ. Microbiol. 1996;62:2875–2882. doi: 10.1128/aem.62.8.2875-2882.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rios P., Bezus B., Cavalitto S., Cavello S.I. Production and characterization of a new detergent-stable keratinase expressed by Pedobacter sp. 3.14.7, a novel Antarctic psychrotolerant keratin-degrading bacterium. J. Genet. Eng. Biotechnol. 2022;20:81. doi: 10.1186/s43141-022-00356-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sharma R., Gupta R. Substrate specificity characterization of a thermostable keratinase from Pseudomonas aeruginosa KS-1. J. Ind. Microbiol. Biotechnol. 2010;37:785–792. doi: 10.1007/s10295-010-0723-8. [DOI] [PubMed] [Google Scholar]
- 63.Zhang R.X., Wu Z.W., Cui H.Y., Chai Y.N., Hua C.W., Wang P., Li L., Yang T.Y. Production of surfactant-stable keratinase from Bacillus cereus YQ15 and its application as detergent additive. BMC Biotechnol. 2022;22:26. doi: 10.1186/s12896-022-00757-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang R.X., Gong J.S., Zhang D.D., Su C., Hou Y.S., Li H., Shi J.S., Xu Z.H. A metallo-keratinase from a newly isolated Acinetobacter sp. R-1 with low collagenase activity and its biotechnological application potential in leather industry. Bioproc. Biosyst. Eng. 2016;39:193–204. doi: 10.1007/s00449-015-1503-7. [DOI] [PubMed] [Google Scholar]
- 65.Lv L.X., Sim M.H., Li Y.D., Min J., Feng W.H., Guan W.J., Li Y.Q. Production, characterization and application of a keratinase from Chryseobacterium L99 sp. nov. Process Biochem. (Amsterdam, Neth.) 2010;45:1236–1244. doi: 10.1016/j.procbio.2010.03.011. [DOI] [Google Scholar]
- 66.Tatineni R., Doddapaneni K.K., Potumarthi R.C., Vellanki R.N., Kandathil M.T., Kolli N., Mangamoori L.N. Purification and characterization of an alkaline keratinase from Streptomyces sp. Bioresour. Technol. 2008;99:1596–1602. doi: 10.1016/j.biortech.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 67.Jaouadi N.Z., Rekik H., Badis A., Trabelsi S., Belhoul M., Yahiaoui A.B., Aicha H.B., Toumi A., Bejar S., Jaouadi B. Biochemical and molecular characterization of a serine keratinase from Brevibacillus brevis US575 with promising keratin-biodegradation and hide-dehairing activities. PLoS One. 2013;8 doi: 10.1371/journal.pone.0076722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thys R.C.S., Brandelli A. Purification and properties of a keratinolytic metalloprotease from Microbacterium sp. J. Appl. Microbiol. 2006;101:1259–1268. doi: 10.1111/j.1365-2672.2006.03050.x. [DOI] [PubMed] [Google Scholar]
- 69.Hendrick Q., Nnolim N.E., Nontongana N., Nwodo U. Sphingobacterium multivorum HNFx produced thermotolerant and chemostable keratinase on chicken feathers. Biologia. 2022 doi: 10.1007/s11756-022-01126-3. [DOI] [Google Scholar]
- 70.Thys R.C.S., Lucas F.S., Riffel A., Heeb P., Brandelli A. Characterization of a protease of a feather‐degrading Microbacterium species. Lett. Appl. Microbiol. 2004;39:181–186. doi: 10.1111/j.1472-765X.2004.01558.x. [DOI] [PubMed] [Google Scholar]
- 71.Wang S.L., Hsu W.T., Liang T.W., Yen Y.H., Wang C.L. Purification and characterization of three novel keratinolytic metalloproteases produced by Chryseobacterium indologenes TKU014 in a shrimp shell powder medium. Bioresour. Technol. 2008;99:5679–5686. doi: 10.1016/j.biortech.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 72.Russell G.L., Britton L.N. Use of certain alcohol ethoxylates to maintain protease stability in the presence of anionic surfactants. J. Surfactants Deterg. 2002;5:5–10. doi: 10.1007/s11743-002-0198-9. [DOI] [Google Scholar]
- 73.Stoner M.R., Dale D.A., Gualfetti P.J., Becker T., Manning M.C., Carpenter J.F., Randolph T.W. Protease autolysis in heavy-duty liquid detergent formulations: effects of thermodynamic stabilizers and protease inhibitors. Enzym. Microb. Technol. 2004;34:114–125. doi: 10.1016/j.enzmictec.2003.09.008. [DOI] [Google Scholar]
- 74.Stoner M.R., Dale D.A., Gualfetti P.J., Becker T., Randolph T.W. Surfactant‐induced unfolding of cellulase: kinetic studies. Biotechnol. Prog. 2006;22:225–232. doi: 10.1021/bp0501468. [DOI] [PubMed] [Google Scholar]
- 75.Lund H., Kaasgaard S.G., Skagerlind P., Jorgensen L., Jorgensen C.I., van de Weert M. Correlation between enzyme activity and stability of a protease, an alpha-amylase and a lipase in a simplified liquid laundry detergent system, determined by differential scanning calorimetry. J. Surfactants Deterg. 2012;15:9–21. doi: 10.1007/s11743-011-1272-5. [DOI] [Google Scholar]
- 76.Lund H., Kaasgaard S.G., Skagerlind P., Jorgensen L., Jørgensen C.I., Van de Weert M. Protease and amylase stability in the presence of chelators used in laundry detergent applications: correlation between chelator properties and enzyme stability in liquid detergents. J. Surfactants Deterg. 2012;15:265–276. doi: 10.1007/s11743-011-1318-8. [DOI] [Google Scholar]
- 77.Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 78.Chang K.Y., Yang J.R. Analysis and prediction of highly effective antiviral peptides based on random forests. PLoS One. 2013;8 doi: 10.1371/journal.pone.0070166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dutta B., Banerjee A., Chakraborty P., Bandopadhyay R. Silico studies on bacterial xylanase enzyme: structural and functional insight. J. Genet. Eng. Biotechnol. 2018;16:749–756. doi: 10.1016/j.jgeb.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Enany S. Structural and functional analysis of hypothetical and conserved proteins of Clostridium tetani. J. Infect. Public Health. 2014;7:296–307. doi: 10.1016/j.jiph.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 81.Studer G., Rempfer C., Waterhouse A.M., Gumienny R., Haas J., Schwede T. QMEANDisCo—distance constraints applied on model quality estimation. Bioinformatics. 2020;36:1765–1771. doi: 10.1093/bioinformatics/btz828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Santhoshkumar R., Yusuf A. Silico structural modeling and analysis of physicochemical properties of curcumin synthase (CURS1, CURS2, and CURS3) proteins of Curcuma longa. J. Genet. Eng. Biotechnol. 2020;18:24. doi: 10.1186/s43141-020-00041-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Filiz E., Koç İ. In silico sequence analysis and homology modeling of predicted beta-amylase 7-like protein in Brachypodium distachyon L. J. BioSci. Biotechnol. 2014;3:61–67. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this study has been deposited at “NCBI” under the accession number [MW165830.1, MW165831.1 and OK033360].