Abstract
It has been reported that inhibition of GPR65 may be effective for the treatment of certain cancers. Nevertheless, the role of GPR65 in various cancers remains unknown. We conducted an exhaustive pan-cancer analysis of GPR65 using multiple databases, including TCGA, GTEx, BioGPS, HPA, cBioPortal, and GeneCards. GPR65 was found to be differentially expressed in various cancers and linked to tumor mutational burden (TMB), microsatellite instability (MSI), and Ploidy, playing a key function in the tumor microenvironment (TME). It is closely linked to the development of Th17 cells as well as Th1 and Th2 cells in certain cancers. Our findings indicate that the expression of GPR65 is highly linked with clinical prognosis, mutations, and immune cell infiltration. It was revealed as an indicator of patient prognosis as well as a possible immunomodulatory role. As a possible new immunological checkpoint, GPR65 could be a target for tumor immunotherapy.
Keywords: TCGA, Immune microenvironment, GPR65, Immune checkpoints, Immunotherapy
1. Introduction
The immune system plays an active role in cancer and can inhibit tumor growth by destroying cancer cells or inhibiting their growth [1]. However, in the context of malignancy, there may be multiple immunosuppressive mechanisms to prevent effective antitumor immunity [2]. These immune checkpoint pathways, which usually remain self-tolerant and limit collateral tissue damage during the immune response, can be exploited by cancer to avoid immune destruction [3]. In recent years, immune checkpoint blockade has demonstrated promise in the treatment of a variety of malignancies [4].
GPR65 is an extracellular pH sensitive G protein-coupled receptor that has been found to be overexpressed in a variety of tumors and tumor cell lines [5]. Increased acid production caused by the up-regulation of glycolysis results in acidosis in the tumor microenvironment [6]. GPR65 promotes adaptation to an acidic environment to enhance cell survival and proliferation, thereby promoting tumor development [5,7]. GPR65 has been shown to inhibit the development of colorectal cancer in experimental mouse models [8], and overexpression of GPR65 in human tumors plays a role in the driving or maintaining tumor formation [9]. However, systematic pan-cancer analysis for GPR65 is still lacking.
Finding and verifying new pan-cancer genes is crucial to acquiring a deeper knowledge of the extraordinarily complicated process of carcinogenesis [10]. Cancer genomics databases, like The Cancer Genome Atlas (TCGA) [11], provides large datasets of tumor-associated functional genomics data for in-depth pan-cancer analysis. Genotype-Tissue Expression (GTEx) is a database for analyzing the relationship between human gene expression and regulation and genetic variation [12]. Copy number alterations (CNAs) are a common characteristic of human cancers, affecting multiple genes concurrently and contributing to the progression of cancer [13,14]. The Human Protein Atlas (HPA) database is based on proteomics, transcriptomics, and systems biology data to map tissues, cells, and organs [15]. It contains protein expression profiles from tumor tissues and normal tissues. The cBioPortal database is used to explore, visualize, and analyze multidimensional cancer genomics data [16]. GeneCards and BioGPS are human genetic information databases [17,18]. Genecards automatically integrates resources from approximately 150 gene-centric databases, including genomics, transcriptomics, proteomics, genetics, clinical, and functional. These databases can provide a wealth of information about the function of GPR65 in many cancers. In this research, we assessed the levels of GPR65 expression in pan-cancerous and normal control tissues and assessed the prognostic value of GPR65. We also considered the genetic alterations and protein levels of GPR65. The association between GPR65 and stromal infiltration score, immune infiltration score, immune-related genes, and immune checkpoints was investigated further. In addition, gene set enrichment analysis (GSEA) was utilized to determine the biological role of GPR65 in pan-cancer. Our findings demonstrate the role of GPR65 in pan-cancer, reveal its potential to influence the tumor microenvironment, and suggest that GPR65 may be a new immunotherapy target for cancer.
2. Materials and methods
2.1. Data source and availability
Exploring the potential role of GPR65 gene in cancer using multiple databases. RNA expression and clinical data of TCGA and GTEx were obtained from the UCSC Xena database [19]. The database cBioPortal was consulted for information on DNA copy number and methylation. The expression data were converted to log2 (x + 0.001). TIMER2 was utilized to compare the expression profile of GPR65 between several tumor types and nearby normal tissues [20]. BioGPS was utilized to examine the expression of GPR65 in numerous cancer cell lines and normal cells. TISIDB was utilized to examine the expression of GPR65 in immune and molecular subtypes [21].
2.2. Protein level analysis of GPR65 in multiple cancers
The HPA database was utilized to investigate the protein concentration of GPR65 in human tumor and normal tissues. The string database was utilized to build the protein-protein interaction (PPI) network of GPR65 [22]. The subcellular locations of GPR65 were visualized using GeneCards. Webgestalt database was used for GO enrichment analysis [23].
2.3. Relationship between GPR65 expression and survival prognosis
Cox proportional hazards model and Kaplan-Meier analysis were employed to examine the association among GPR65 mRNA expression and survival outcomes. R-packages such as “survival” [24] and “maxstat” [25] were used. The optimal cut-off values were calculated using the R package maxstat.
2.4. Tumor immune microenvironment and GPR65 expression
Genes related to chemokines, receptors, MHC, immunosuppressants, immunostimulants, and immune checkpoint pathways including inhibitory and stimulatory genes were retrieved from each cancer sample. Using the R package ESTIMATE, we estimated the patient's tumor stroma score on gene expression [26]. The infiltration score of immune-related cells in patients was assessed using the EPIC, Timer and quanTIseq methods in the R package IOBR [27].
2.5. GSEA of pathways associated with GPR65 gene expression in cancer patients
The correlation analysis of GPR65 with all genes using TCGA data was conducted using Pearson correlation coefficients. For gene set enrichment analysis, GPR65-correlated genes were chosen [28]. GSEA is performed using the clusterProfiler R package as well as the KEGG pathway database.
2.6. Statistical analyses
Expression differences were calculated using R software (version 4.1.2) [29] and p-values were used to determine the significance of expression differences. Log-rank test was used to analyze the survival outcome of Kaplan-Meier survival curves. P < 0.05 was deemed statistical significance. (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001).
3. Results
3.1. Analysis of GPR65 expression in normal tissues and pan-cancer
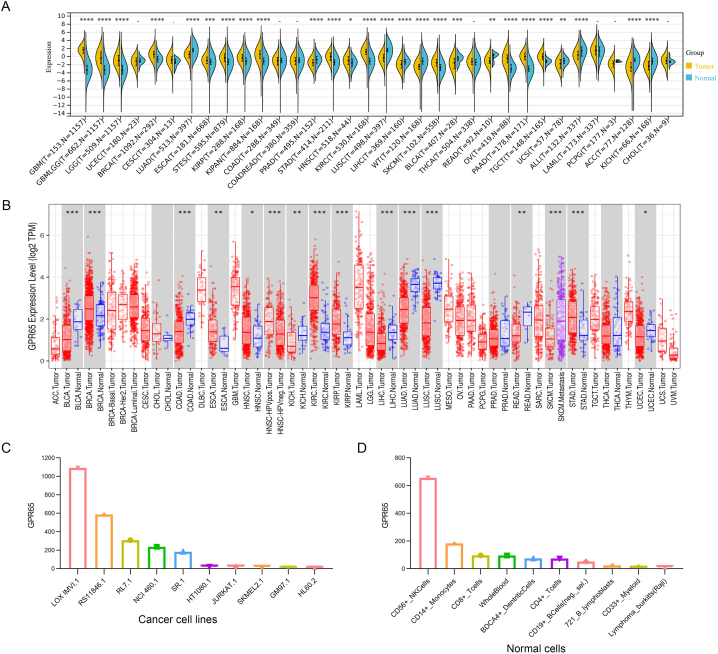
RNA-seq data from the TCGA and GTEx databases were used to examine the expression of GPR65 in pan-cancer. A significant difference in GPR65 expression was observed in 34 types of cancer, excluding those for which normal tissue data was unavailable. Compared to control tissues, GPR65 expression was elevated in glioblastoma multiforme (GBM), glioma (GBMLGG), brain lower grade glioma (LGG), breast invasive carcinoma (BRCA), esophageal carcinoma (ESCA), stomach and esophageal carcinoma (STES), Kidney renal papillary cell carcinoma (KIRP), pan-kidney cohort (KICH + KIRC + KIRP) (KIPAN), stomach adenocarcinoma (STAD), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), skin cutaneous melanoma (SKCM), ovarian serous cystadenocarcinoma (OV), pancreatic adenocarcinoma (PAAD), and testicular germ cell tumors (TGCT). In contrast to control tissues, GPR65 expression was decreased in lung adenocarcinoma (LUAD), prostate adenocarcinoma (PRAD), lung squamous cell carcinoma (LUSC), liver hepatocellular carcinoma (LIHC), high-risk wilms tumor (WT), bladder urothelial carcinoma (BLCA), rectum adenocarcinoma (READ), uterine carcinosarcoma (UCS), acute lymphoblastic leukemia (ALL), adrenocortical carcinoma (ACC), and kidney chromophobe (KICH) (Fig. 1A).
Fig. 1.
Differential expression of GPR65. (A) Expression of GPR65 in normal and cancerous tissues. (B) TIMER shows the level of GPR65 expression in TCGA tumors and nearby tissues, if available. (C) Expression of GPR65 in cancer cell lines. (D) Expression of GPR65 in normal cells.
Next, we used TIMER to conduct differential expression studies of GPR65 between tumors and adjacent normal tissues from the TCGA dataset. In tumor tissues of BRCA, KIRC, KIRP, STAD (P < 0.001), ESCA (P < 0.01), and HNSC (P < 0.05), GPR65 expression is greater than in control tissues. On the contrary, GPR65 expression was reduced in BLCA, colon adenocarcinoma (COAD), LIHC, LUAD, LUSC (P < 0.001), KICH, READ (P < 0.01), and uterine corpus endometrial carcinoma (UCEC) (P < 0.05) compared to the respective control tissues. In particular, some tumor types (eg cholangiocarcinoma (CHOL), PRAD, and thyroid carcinoma (THCA)) exhibited no differential expression (Fig. 1B).
Furthermore, we analyze the expression of GPR65 in cell lines with the BioGPS database. The results revealed that GPR65 was expressed in most cancer cell lines and was highly expressed in LOX IMUI.1, RS11846.1, and RL7.1. The top ten cancer cell lines with high expression of GPR65 are shown in Fig. 1C. And for normal cells, immune cells are where GPR65 expression is highest (Fig. 1D). Details are shown in Supplementary Figure S1A-B. Also, we found that GPR65 is mostly expressed in cancer cell lines from mesenchymal, bone marrow, lymphoid, and other tissues in the HPA database (Supplementary Figure S1C). We also investigated the mRNA expression of GPR65 in normal tissues and found that its expression was higher in the spleen, lung and small intestine (Supplementary Figure S1D).
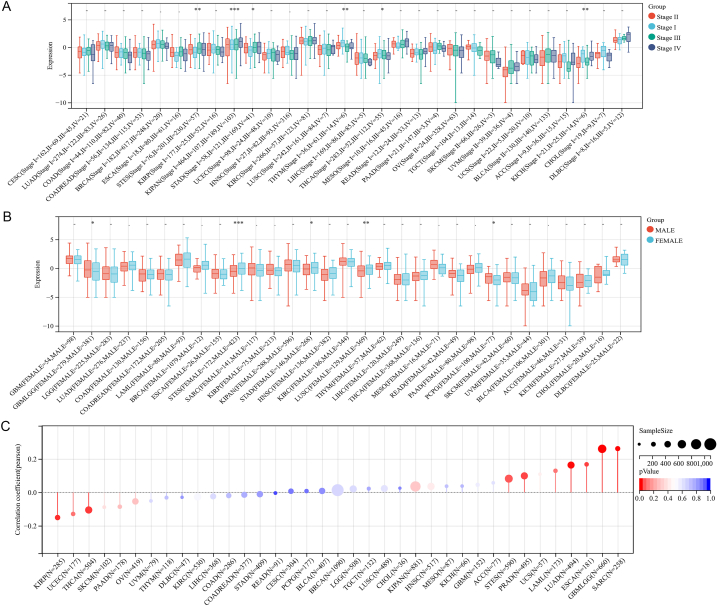
Next, we analyzed GPR65 expression in relation to World Health Organization (WHO) pathological tumor staging and found that GPR65 expression was stage-specific in some tumors (Fig. 2A). For example, it was lower in higher stages of thymoma (THYM), THCA, and KICH. On the contrary, higher was observed of GPR65 expression in higher stages in STES and KIPAN. For the relationship between GPR65 expression and gender in pan-cancer, we discovered that GBMLGG and pheochromocytoma and paraganglioma (PCPG) were higher in males, while STES, STAD, and LUSC were higher in females (Fig. 2B).
Fig. 2.
GPR65 expression and pan-cancer. (A) Expression of GPR65 in different cancer patients in pathological stages classified by WHO. (B) Expression of GPR65 in cancer patients of different genders.. (C) Analysis of the correlation between GPR65 and age of cancer patients.
We further calculated the Pearson correlation coefficient between age and GPR65 expression in each tumor (Fig. 2C) and observed it was prominently correlated in 6 tumors, with a significant positive correlation in 4 tumors: COAD, colon adenocarcinoma/rectum adenocarcinoma esophageal carcinoma (COADREAD), acute myeloid leukemia (LAML), and UCEC (P < 0.05). In comparison, a negative correlation was observed in 2 tumors: LIHC and CHOL (P < 0.05).
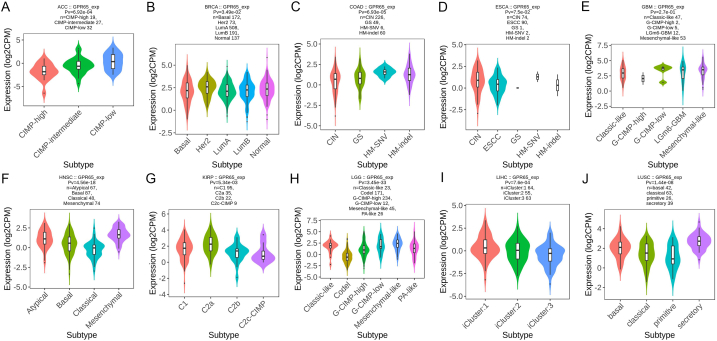
3.2. GPR65 expression is related to immune and molecular subtypes in human cancers
The TISIDB website was then used to investigate the impact of GPR65 expression on immunological and molecular subgroups among patients with malignancies. Six categories of immune subtypes were identified: C1 (wound healing), C2 (IFN-gamma dominant), C3 (inflammatory), C4 (lymphocyte deficient), C5 (immunologically silent), and C6 (TGF-b dominant). The findings demonstrated a relationship between GPR65 expression and several immunological subtypes in ACC, BLCA, BRCA, CESC, CHOL, COAD, ESCA, GBM, HNSC, and KICH (Fig. 3A–J). Furthermore, different immunological subgroups of the same cancer type expressed GPR65 in different ways. GPR65 displayed high expression in C6 kinds and low expression in C1 kinds, using COAD as an example. The expression of GPR65 was significantly correlated with ACC, BRCA, COAD, ESCA, GBM, HNSC, KIRP, LGG, LIHC, and LUSC for several molecular subtypes of cancer (Fig. 4A–J). The expression of GPR65 in other cancer subtypes is shown in Supplementary Figure S2, 3. The above results suggest that GPR65 is differentially expressed in cancer immune and molecular subtypes.
Fig. 3.
Expression of GPR65 in pan-cancer immune subtypes. (A) ACC, (B) BLCA, (C) BRCA, (D) CESC, (E) CHOL, (F) COAD, (G) ESCA, (H) GBM, (I) HNSC, (J) KICH.
Fig. 4.
Expression of GPR65 in pan-cancer molecular subtypes. (A) ACC, (B) BRCA, (C) COAD, (D) ESCA, (E) GBM, (F) HNSC, (G) KIRP, (H) LGG, (I) LIHC, (J) LUSC.
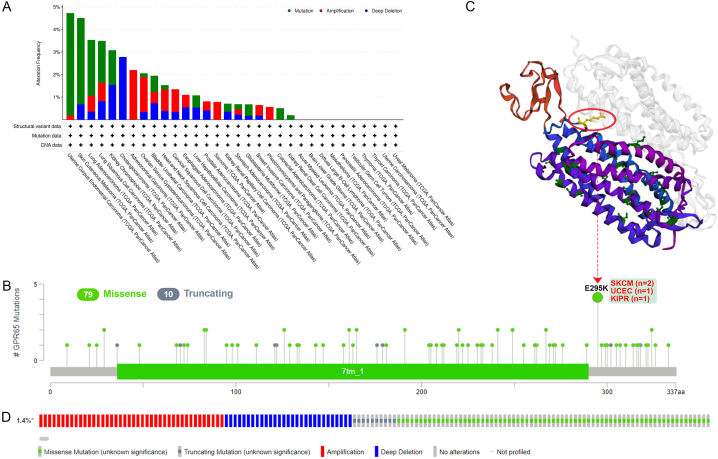
3.3. Genetic alterations of GPR65
Human malignancies originate as a result of a buildup of genetic mutations. Therefore, we intend to investigate GPR65 gene changes in human tumor tissues. According to our findings, endometrial cancer and skin melanoma had the highest incidence of GPR65 changes (>4%), with the “mutation” being the most common form. The frequency of “deep deletion” type CNA was the highest in cholangiocarcinoma (about 3%) and “amplification” type CNA in adrenocortical carcinoma (about 2%), as shown in Fig. 5A. We also display the different kinds, locations, and numbers of GPR65 gene mutations in Fig. 5B. Missense mutations made up the majority of the genetic changes (79 loci), followed by truncation mutations (10 loci), with the most common E mutation occurring at 295 loci close to the 7tm 1 domain and switching from E to K. An alteration in the E295K missense gene located close to the 7tm 1 structural domain was found in two cases of SKCM, one case of UCEC, and one case of KIPR. The E295K locus was acquired and made visible in the 3D structure of the GPR65 protein (Fig. 5C).
Fig. 5.
Genetic Alterations of GPR65. (A) Alterations of GPR65 in TCGA database. (B) The mutation types, sites, and number of cases of the GPR65 genetic alterations. (C) Mutation site (E295K) within the 7tm_1 domain is shown in the 3D structure of GPR65. (D) Mutations, structural variant, and copy-number alterations of GPR65.
The most common genetic changes in GPR65 were deep deletions, deep mutations, and amplifications (Fig. 5D). There are several types of GPR65 gene alterations in cancer patients, such as missense, amplification, gain, shallow deletion, and deep deletion in lung adenocarcinoma, which results in altered gene expression. Most putative copy number alterations of GPR65 are diploid, shallow deletion, and gain (Supplementary Fig. S4A and B). This suggests that the above cancers may be driven by changes in the GPR65 gene.
3.4. Expression of GPR65 and immune-related biomarkers
Then, we investigated the association of GPR65 expression with TMB and MSI, both of which are highly correlated with cancer sensitivity to immune checkpoint inhibitors (ICIs). The results showed that GPR65 expression was significantly positively correlated with TMB in UCEC, COADREAD, LAML and COAD, and negatively correlated with CHOL, DLBC, lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), and UVM (Supplementary Figure S5A). GPR65 expression positively associated with MSI in COADREAD, COAD, READ, and LAML and negatively with DLBC, GBMLGG, KIPAN, and TGCT (Supplementary Figure S5B). Analyzing the relationship between GPR65 expression and Ploidy, we found that GPR65 expression was positively correlated with Ploidy in LGG, KIPAN, PRAD, UCS, and SARC, and negatively correlated with Ploidy in PAAD, ACC, etc. (Supplementary Figure S5C).
3.5. Protein expression of GPR65
We found in the HPA database that GPR65 protein was highly expressed in prostate cancer and lower in skin cancer (Supplementary Figure S6A). GPR65 protein levels were higher in lymph nodes and tonsils, moderate in the appendix and bone marrow, and lower in the cervix, esophagus, and other normal human tissues (Supplementary Figure S6B). Localization on the cell membrane is essential to be a possible immune checkpoint. Using the GeneCards database, we analyze the subcellular localization of GPR65. As shown in Supplementary Figure S6C, the protein localization of GPR65 is mainly at the plasma membrane. Moreover, GPR65 is shown by the established PPI network to be strongly correlated with LY9, PTPN22, IKZF1, TLR1, TAGAP, PLEK, PTAFR, IGSF6, GPR183, and GPR39 proteins (Supplementary Figure S6D). Through the GO biological process enrichment analysis of the above genes in the Webgestalt database, we were able to determine their functions, which play an important role in cell activation, positive regulation of interleukin-6 biosynthetic process, leukocyte activation, regulation of inositol phosphate biosynthetic process, lymphocyte differentiation, regulation of interleukin-6 biosynthetic process, immune response, regulation of interleukin-6 production, interleukin-6 biosynthetic process, interleukin-6 production, and other different pathways involved (Supplementary Figure S6E).
3.5.1. Prognostic value of GPR65 in pan-cancer
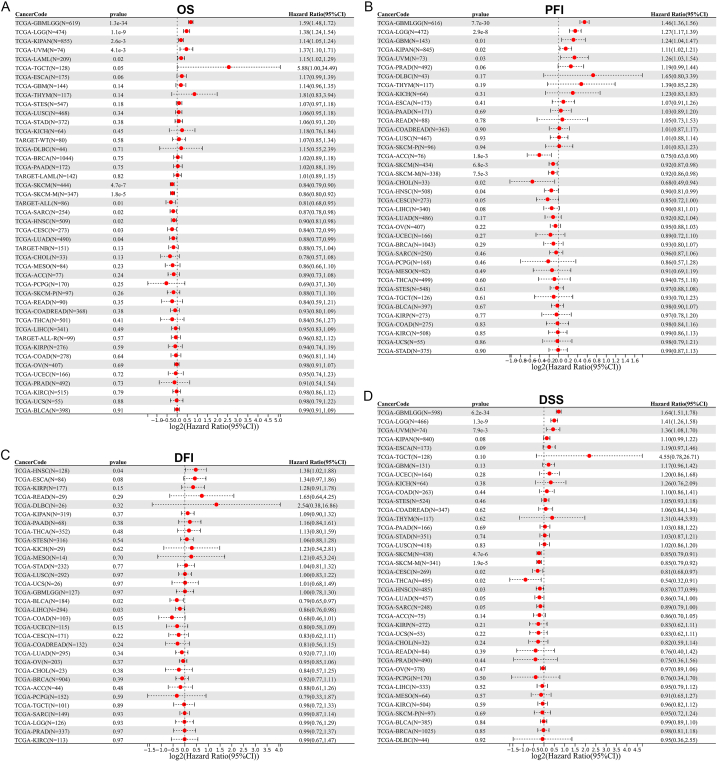
Moreover, we evaluated the prognostic significance of GPR65 in cancer patients. The link among GPR65 expression and tumor prognosis was analyzed using a Cox proportional hazards model. GPR65 was a low risk factor for patients with LUAD, CESC, HNSC, SARC, SKCM and SKCM-M, but a high-risk factor for patients with LAML, UVM, KIPAN, LGG and GBMLGG, as determined by an analysis of OS (Fig. 6A). In addition, PFI results demonstrated that GPR65 was a low-risk factor for patients with HNSC, CHOL, ACC, SKCM, and SKCM-M, but a high-risk factor for patients with UVM, KIPAN, GBM, LGG, and GBMLGG (Fig. 6B). GPR65 is a low-risk factor for patients with LIHC and BLCA, but a high-risk factor for patients with HNSC, according to a DFI analysis (Fig. 6C). Finally, analysis of DSS data demonstrated that GPR65 was a low-risk factor for patients with HNSC, THCA, CESC, SKCM, and SKCM-M, but a high-risk factor for patients with UVM, LGG, and GBMLGG (Fig. 6D).
Fig. 6.
Cox proportional hazards regression model analysis forest plot for GPR65. A Cox proportional hazards regression model with red dot coordinates greater than 0 for a certain cancer suggests that higher GPR65 expression has a better survival rate. (A) Results of GPR65 OS analysis. (B) Results of GPR65 PFI analysis. (C) Results of GPR65 DFI analysis. (D) Results of GPR65 DSS analysis. OS, overall survival; PFI, Progression Free Interval; DFI, Disease Free Interval; DSS, Disease Specific Survival. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
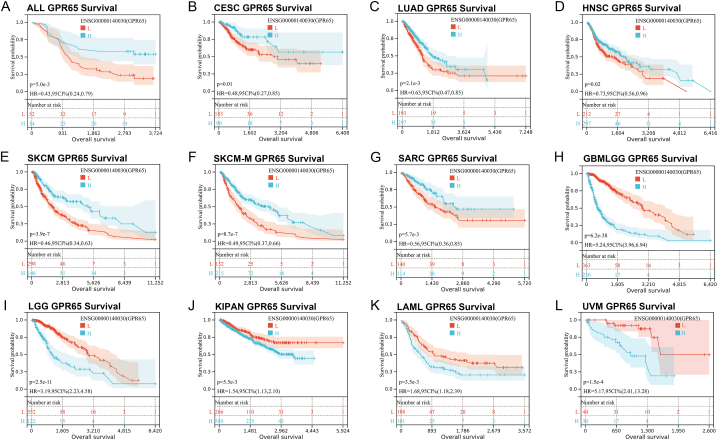
In Kaplan-Meier analysis, when the curve of high expression (H) was lower than the curve of low expression (L), it represents a lower survival rate for patients with high GPR65 expression. When the curve of low expression (L) was lower than the curve of high expression (H), represents a lower survival rate for patients with low GPR65 expression. The results of the Kaplan-Meier analysis of overall survival (OS) demonstrated that GPR65 is a positive factor in patients with ALL, CESC, HNSC, LUAD, SARC, SKCM, and SKCM-M (Fig. 7A–G), whereas it is a negative factor in patients with GBMLGG, KIPAN, LAML, LGG, and UVM (Fig. 7H–L). This suggests that treatment of various cancers in clinical practice should be concerned with high or low expression of GPR65, improving prognosis in various cancers by inhibiting or promoting GPR65 expression.
Fig. 7.
Kaplan-Meier OS of GPR65. Pan-cancer OS of GPR65 in the specified types of tumors from the TCGA database (A–L). In ALL, CESC, HNSC, LUAD, SARC, SKCM, and SKCM-M, high GPR65 expression was associated with improved OS. In GBMLGG, KIPAN, LAML, and UVM, high GPR65 expression was associated with worse OS.
3.5.2. Analysis of immune-related genes
Considering that it is advantageous for tumor immunotherapy to stimulate or mobilize the body's immune system and enhance the antitumor immunity of the tumor microenvironment. We analyzed the connection between GPR65 expression and chemokine, chemokine receptor, major histocompatibility complex (MHC), immunostimulator, and immunoinhibitor. Fig. 8A depicts the positive correlation between GPR65 expression and almost all of the 150 immune-related genes, including CCL4, CCL5, CCR1, CCR2, CCR5, HLA-DRA, HLA-DMB, HAVCR2, CSF1R, CD48, and CD86, among others. Furthermore, we explored the association of GPR65 with immune checkpoint gene expression. GPR65 expression was closely associated with 60 immune checkpoint genes involved in immune stimulation and inhibition, according to the findings. (Fig. 8B).
Fig. 8.
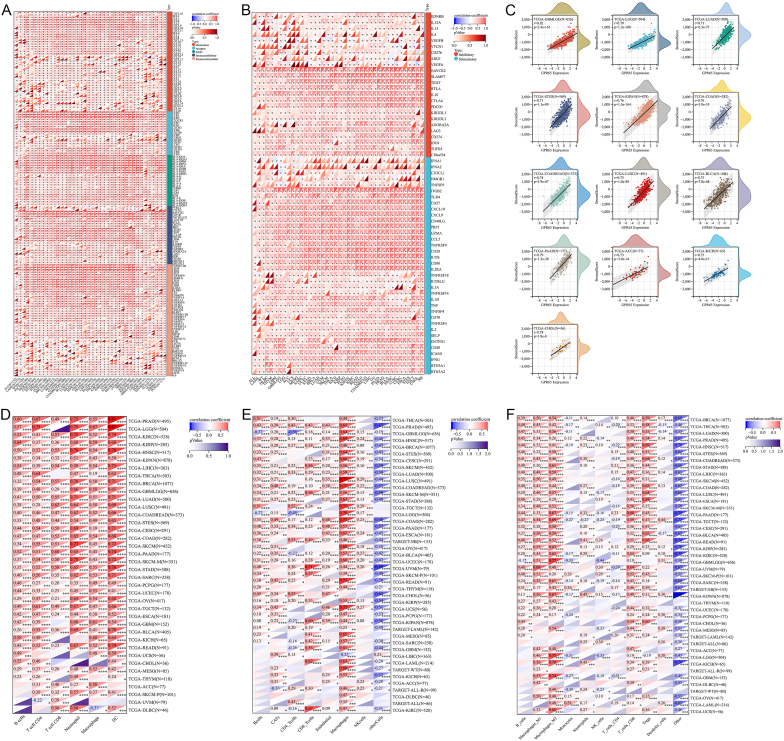
Correlationship between GPR65 expression and tumor immune microenvironment and infiltration. (A) Connection between the expression of GPR65 and immune stimulatory and inhibitory. (B) Correlation between the expression of GPR65 and chemokine, chemokine receptor, MHC, immunostimulator, and immunoinhibitor. (C) Relationship between GPR65 expression and stromal score. (D) Correlation of GPR65 with B cell, CD4 T cells, CD8 T cells, neutrophil, macrophage, and DC infiltration levels using the TIMER database. (E) Correlation of GPR65 with infiltration levels of B cells, CAFs, CD4 T cells, CD8 T cells, endothelial cells, macrophages, and NK cells using the EPIC database. (F) Correlation between GPR65 and infiltration levels of B cells, M1 macrophage, M2 macrophage, Monocytes, neutrophils, NK cells, CD4 T cells, CD8 T cells, Tregs, and dendritic cells using the quanTIseq database.
3.6. Correlationship between GPR65 expression and tumor immune microenvironment
Immune and stromal cells in the microenvironment of a tumor are of great diagnostic and prognostic value. We discovered that infiltration of stromal and immune cells was highly linked to GPR65 expression. To explore the impact of GPR65 expression on the tumor microenvironment, the ESTIMATE algorithm was used to examine the association between GPR65 expression and stromal score. In 13 cancer types, including GBMLGG, LGG, LUAD, and STES, the expression of this gene had a significant positive correlation (r > 0.7) with the stromal score (Fig. 8C).
We analyzed the relationship between GPR65 expression and diverse immune cells (Timer, EPIC, and quanTIseq). Ultimately, this gene expression was significantly linked to immune-related cellular infiltration in 44 cancer types, 38 of which were co-targeted by the three approaches. The TIMER2 data demonstrated a correlation between GPR65 and the levels of B cell, CD4 T cell, CD8 T cell, neutrophil, macrophage, and dendritic cell (DC) infiltration in TCGA pan-cancer. Among them, neutrophils and DC exhibited strong positive correlation with multiple cancer types (Fig. 8D). According to EPIC findings, B cells, CAFs, CD8 T cells, endothelial cells, macrophages, and NK cells were all positively correlated with GPR65 expression levels in TCGA pan cancers, with macrophages exhibiting a statistically high positive association in nearly all types of cancer. (Fig. 8E). The results of quanTIseq showed that GPR65 was positively correlated with the infiltration levels of B cells, M1 macrophages, M2 macrophages, Neutrophils, CD8 T cells, Tregs and dendritic cells in TCGA pan-cancer, among which M2 macrophages showed a significant positive correlation in multiple cancer types, while GPR65 had a negative correlation with Monocyte infiltration levels (Fig. 8F).
3.7. GSEA of GPR65
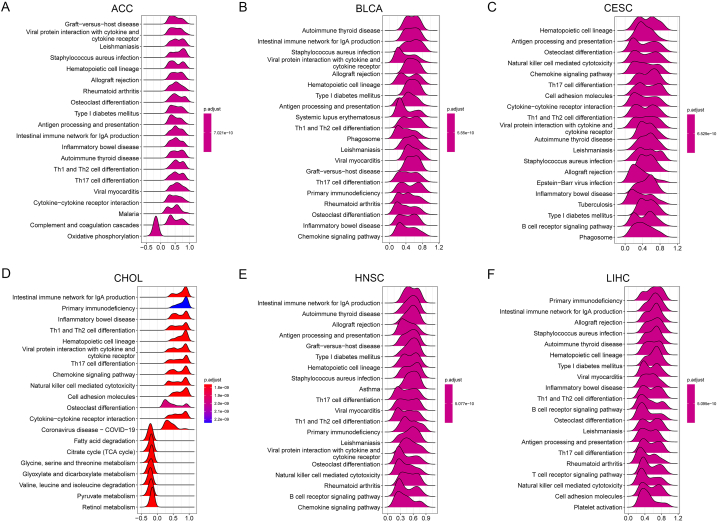
We evaluated the pathways by which GPR65 may be involved in the application of GSEA in 33 tumor types of TCGA. Immune-related pathways are significantly associated with GPR65, especially Th1 and Th2 cell differentiation, Th17 cell differentiation pathways such as ACC, BLCA, CESC, CHOL, HNSC, LIHC (Fig. 9A–F). The balance of Th1 and Th2 cell differentiation has a significant effect on the body's normal functioning, and a shift in this balance can result in the development of diseases. The main function of Th17 cells is to promote the mobilization, recruitment and activation of neutrophils and to mediate pro-inflammatory responses. It is closely associated with autoimmune diseases, infectious diseases, and tumorigenesis. These results suggest that GPR65 is closely associated with the regulation of the tumor immune microenvironment and tumor immune escape.
Fig. 9.
The GSEA analysis of GPR65 in pan-cancer(A–F). TOP20 GSEA terms in specific cancer types. Irregular graphs with colors show the relative expression intensity of different genes in the pathway. When the overall horizontal coordinate of the graph is greater than 0, it indicates positive regulation of the pathway by GPR65. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
GPR65, a member of the proton-sensing G protein-coupled receptor family, is closely related to the TME [30]. Extracellular matrix (ECM), stromal cells, and immune cells compose the TME (including T and B lymphocytes, tumor-associated macrophages, etc.) [31]. The composition of TME has been discovered to affect the response to immune checkpoint blockade (ICB) [[32], [33], [34]]. ICB has profoundly changed the landscape of cancer treatment, with antibodies targeting the inhibitory T cell receptors PD-1, PD-L1, and CTLA4 that vastly improve patient survival in many [[35], [36], [37], [38], [39], [40]]. Compared with healthy cells, the acidity outside of tumor cells is stronger, and extracellular tumor acidosis accelerates [41]. GPR65 is closely associated with the expression of several markers of immune cells, including T cells, and GPR65 expressed by immune cells can modulate responses in acidic microenvironments [42].
Previous research has already shown that GPR65 inhibits hematological malignancies and makes blood cancer cells more sensitive to acidotic stress in the microenvironment [43]. The reduction of c-myc oncogene expression by GPR65-mediated G13/Rho signaling contributes to the tumor suppressive effect of GPR65 [44]. This is similar to our study. After a comprehensive systematic pan-cancer analysis of GPR65, we found an important relationship between its expression level and the clinical prognosis of patients. Mutations in GPR65 also raise the risk of developing cancer. In addition, GPR65 was significantly associated with immune-related cellular infiltration, revealing its potential function as a possible participant in the regulation of the immune microenvironment. Therefore, as a new cancer immunotherapy, GPR65 could be coupled with established immune checkpoint inhibitors to increase immune infiltration and response in cancer. All such research shows that GPR65 may have been a promising immunotherapy target for the treatment of cancer.
In the first step of our study, we analyzed the expression of GPR65 in normal and pan-cancer tissues. Compared to normal tissues, GPR65 was found to be overexpressed in GBM, GBMLGG, LGG, BRCA, ESCA, STES, KIRP, KIPAN, STAD, HNSC, KIRC, SKCM, OV, PAAD and TGCT . This is consistent with a previous study in gastric cancer [45]. On the contrary, GPR65 expression was decreased in LUAD, PRAD, LUSC, LIHC, WT, BLCA, READ, UCS, ALL, ACC, and KICH. Different levels of GPR65 expression may reflect distinct underlying functions and mechanisms in distinct tumor types. Furthermore, we analyzed the expression of GPR65 in various cancer cell lines and normal cells. Per the findings, GPR65 was expressed in the majority of cancer cell lines and was abundantly expressed in LOX IMUI.1, RS11846.1, and RL7.1. And for normal cells, the highest GPR65 expression is found in immune cells. The study by Choi, J. W. et al. confirmed this as well [46].
Membrane localization is required as a possible immune checkpoint. Utilising GeneCards database, we investigated the subcellular localization of GPR65 and found that the protein is primarily localized to the plasma membrane. Furthermore, the constructed PPI network revealed that GPR65 is strongly affiliated with the LY9, PTPN22, IKZF1, TLR1, TAGAP, PLEK, PTAFR, IGSF6, GPR183, and GPR39 proteins.
Next, we investigated the association among GPR65 expression and gender in pan-cancer. We noticed that GBMLGG and PCPG were higher in males, while STES, STAD, and LUSC were higher in females. In addition, age showed a positive correlation with GPR65 expression in COAD, COADREAD, LAML and UCEC, and a negative correlation in LIHC and CHOL. These findings could have a significant impact on how immunotherapy plans are chosen for people of different ages and genders. Cox proportional hazards model analysis (including OS, DSS, PFI, and DFI) and Kaplan-Meier OS analysis were conducted to investigate the prognostic significance of GPR65 expression in pan-cancer. The analysis indicated a link between high GPR65 expression in GBMLGG, KIPAN, LAML, LGG, and UVM with poor OS. There was a link between high expression of GPR65 and poor DSS in UVM, LGG, and GBMLGG. Furthermore, the PFI results revealed that GPR65 was a high-risk aspect for UVM, KIPAN, GBM, LGG, and GBMLGG patients. In DFI analysis, we found that GPR65 was a high-risk aspect for HNSC patients. The poor prognosis of GBM is affected by GPR65 overexpression, and targeting GPR65 could be a potential therapy for GBM, according to a previous study [47]. Based on this, we conjecture that treatment against GPR65 could be a target for immunotherapy in many types of tumors.
The occurrence and development of cancer is closely related to the accumulation of genetic variation [48,49]; therefore, this study explored changes in the GPR65 gene in human cancer samples. The GPR65 gene is located on the long arm of chromosome 14 (14q31-q32.1) [50]. The role of GPR65 gene polymorphisms in the pathogenesis of systemic lupus erythematosus and type 2 diabetes has been revealed [51,52], however, there are no relevant studies on GPR65 gene alterations in human pancytopenia. We used the cBioPortal database to discover the fact that there are various types of GPR65 gene alterations in the cancer group, and mutation is the most frequent GPR65 change in pan-cancer. These findings suggest that GPR65 gene alterations can be considered as a possible driver of cancer.
TMB, an emerging biomarker, has been found to correlate with immune checkpoint inhibitor efficacy in numerous clinical studies, including PD-1/PD-L1 inhibition [53,54]. The clinical attention of predicting the efficacy of tumor immunotherapy is growing. Therefore, It is essential to study the linkage among TMB and GPR65 expression. In UCEC, COADREAD, LAML, and COAD, the expression of GPR65 was positively related to TMB, whereas it was significantly negatively related in CHOL, DLBC, UVM, etc. The expression of GPR65 was positively correlated with MSI in COADREAD, COAD, READ, and LAML, and negatively correlated with TMB in DLBC, GBMLGG, KIPAN and TGCT. We found that GPR65 expression was positively correlated with Ploidy in LGG, KIPAN, PRAD, UCS and SARC, and inversely associated with Ploidy in PAAD, ACC, etc. Ploidy is a characteristic of cancer and is related to the chromosomal instability that contributes to cancer development [55], and estimating the purity and ploidy of tumors facilitates the study of cancer genome evolution and heterogeneity within tumors.
After GSEA analysis of GPR65, we discovered that GPR65 is strongly linked with immune-related pathways. Th1 and Th2 cell differentiation pathways regulate each other antagonistically. When the ratio of Th1 and Th2 differentiation is imbalanced it is involved in the progression of multiple tumors [56]. Th17 cells are moldable, and Th17 cells can be activated to induce tumor immunity. However, it has also been suggested that Th17 cell-associated cytokines play an oncogenic role in cancer metastasis by enhancing angiogenesis and tumor immune evasion [57,58]. Th1 and Th2 cells, and Th17-derived cytokines are all involved in tumor regulation by affecting the tumor microenvironment [56,[59], [60], [61], [62]]. These results suggest that GPR65 is closely associated with the regulation of the tumor immune microenvironment and tumor immune escape.
Immediately thereafter, we conducted a correlation analysis between GPR65 expression and various immune cells using three databases (TIMER, EPIC and quanTIseq). 44 cancers exhibited a strong association between expression of this gene and immune-related cellular infiltration (38 of which were jointly indicated by all three databases). Intriguingly, we observed a negative correlation between GPR65 expression and CD4 T cell infiltration values in GBMLGG tissues, but a positive correlation in CHOL. Furthermore, in some tumor tissues, EPIC and quanTIseq results demonstrated a negative correlation between GPR65 expression and T cell CD4 infiltration levels, whereas Timer yielded positive results, such as GBM, KIRC, and KIPAN. This suggests that GPR65 plays distinct immunomodulatory roles in various cancer types. Furthermore, we evaluated the correlation between GPR65 expression and immune regulatory genes and discovered that GPR65 expression was remarkably and positively correlated with the majority of immune-related genes and immune checkpoint genes. These findings indicated that GPR65 functions as an immune checkpoint.
We conducted an exhaustive and systematic analysis of GPR65 and indirectly validated the results of the analysis using different databases. Nevertheless, our research remains with some limitations and more in vivo and clinical studies are necessary to examine the anti-tumor activity of GPR65 and to validate the immune checkpoint role of GPR65. This could improve the credibility of our results. In addition, we lack specific complete cases and data to determine the benefit of anti-GPR65-targeted drugs in terms of survival or inhibition of tumor growth in cancer models. Future research is required to determine the role of GPR65 expression in immune infiltration and human cancers, in addition to the development and testing of new GPR65-targeted antitumor agents.
In summary, this exhaustive pan-cancer analysis of GPR65 exhibited a significant association between GPR65 expression and clinical prognosis, mutations, and immune cell infiltration across a spectrum of human cancers. Its potential function as a patient prognostic indicator and immunomodulatory agent was uncovered. As the possible new immune checkpoint, GPR65 may become a target for immunotherapy against tumors.
Declarations
Funding
This study was supported by the National Natural Science Foundation of China (No. 81971795).
Author contributions statement
JHS, KR, and LLW conceived the research. The data analysis was conducted by LLW, LLS, and HS. LLW, LLS, HS, and YHX authored and contributed to the validation of the manuscript. SDZ, GSA, and JL performed some interpretation of the results. All authors contributed to the article and approved the submitted version.
Declaration of competing interest
The authors declare no competing interests.
Data availability
The data involved in this study are available in public databases as well as in supplementary materials. For further information, please contact the authors.
Acknowledgments
The authors thank the Department of Forensic Pathology, School of Forensic Medicine, Shanxi Medical University for providing the data analysis platform for this study.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.heliyon.2023.e13617.
Contributor Information
Kang Ren, Email: renkang@sxmu.edu.cn.
Junhong Sun, Email: junhong.sun@sxmu.edu.cn.
Abbreviations
GPR65, G protein-coupled receptor 65; TCGA, the Cancer Genome Atlas; GTEx, Genotype Tissue Expression; HPA, Human Protein Atlas; TMB, tumor mutational burden; MSI, microsatellite instability; TME, tumor microenvironment; GSEA, Gene set enrichment analysis; ACC, Adrenocortical carcinoma; BLCA, Bladder Urothelial Carcinoma; BRCA, Breast invasive carcinoma; CESC, Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, Cholangiocarcinoma; COAD, Colon adenocarcinoma; COADREAD, Colon adenocarcinoma/Rectum adenocarcinoma Esophageal carcinoma; DLBC, Lymphoid Neoplasm Diffuse Large B-cell Lymphoma; ESCA, Esophageal carcinoma; GBM, glioblastoma multiforme; GBMLGG, Glioma; HNSC, Head and Neck squamous cell carcinoma; KICH, Kidney Chromophobe; KIPAN, Pan-kidney cohort (KICH + KIRC + KIRP); KIRC, Kidney renal clear cell carcinoma; KIRP, Kidney renal papillary cell carcinoma; LAML, Acute Myeloid Leukemia; LGG, Brain Lower Grade Glioma; LIHC, Liver hepatocellular carcinoma; LUAD, Lung adenocarcinoma; LUSC, Lung squamous cell carcinoma; OV, Ovarian serous cystadenocarcinoma; PAAD, Pancreatic adenocarcinoma; PCPG, Pheochromocytoma and Paraganglioma; PRAD, Prostate adenocarcinoma; READ, Rectum adenocarcinoma; SARC, Sarcoma; SKCM, Skin Cutaneous Melanoma; STAD, Stomach adenocarcinoma; HNSC, head and neck squamous cell carcinoma; STES, Stomach and Esophageal carcinoma; TGCT, Testicular Germ Cell Tumors; THCA, Thyroid carcinoma; THYM, Thymoma; UCEC, Uterine Corpus Endometrial Carcinoma; UCS, Uterine Carcinosarcoma; UVM, Uveal Melanoma; ALL, Acute Lymphoblastic Leukemia; WT, High-Risk Wilms Tumor; OS, overall survival; PFI, Progression Free Interval; DFI, Disease Free Interval; DSS, Disease Specific Survival; ICB, immune checkpoint blockade.
Appendix B. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Schreiber R.D., Old L.J., Smyth M.J. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 2.Andersen M.H. The targeting of immunosuppressive mechanisms in hematological malignancies. Leukemia. 2014;28:1784–1792. doi: 10.1038/leu.2014.108. [DOI] [PubMed] [Google Scholar]
- 3.Topalian S.L., Drake C.G., Pardoll D.M. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Topalian S.L., Taube J.M., Pardoll D.M. Neoadjuvant checkpoint blockade for cancer immunotherapy. Science. 2020;367 doi: 10.1126/science.aax0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ihara Y., Kihara Y., Hamano F., Yanagida K., Morishita Y., Kunita A., Yamori T., Fukayama M., Aburatani H., Shimizu T. The G protein-coupled receptor T-cell death-associated gene 8 (TDAG8) facilitates tumor development by serving as an extracellular pH sensor. Proc. Natl. Acad. Sci. USA. 2010;107:17309–17314. doi: 10.1073/pnas.1001165107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gatenby R.A., Gillies R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 7.Cipriani B., Miller D., Naylor A., Milne G., Young B., Satchell R., Sarkar S., Smith Z., McPherson R., Nika A. Inhibition of GPR65 counteracts low pH induced immunosuppressive polarization of macrophages: in vitro and in vivo characterization of potent, selective and orally bioavailable small molecule GPR65 antagonists. Cancer Res. 2022;82:2162. [Google Scholar]
- 8.Marie M.A., Sanderlin E.J., Satturwar S., Hong H., Lertpiriyapong K., Donthi D., Yang L.V. GPR65 (TDAG8) inhibits intestinal inflammation and colitis-associated colorectal cancer development in experimental mouse models. Biochim. Biophys. Acta, Mol. Basis Dis. 2022;1868 doi: 10.1016/j.bbadis.2021.166288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sin W.C., Zhang Y., Zhong W., Adhikarakunnathu S., Powers S., Hoey T., An S., Yang J. G protein-coupled receptors GPR4 and TDAG8 are oncogenic and overexpressed in human cancers. Oncogene. 2004;23:6299–6303. doi: 10.1038/sj.onc.1207838. [DOI] [PubMed] [Google Scholar]
- 10.Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomczak K., Czerwińska P., Wiznerowicz M. Review the cancer genome atlas (TCGA): an immeasurable source of knowledge. Contemporary Oncology/Współczesna Onkologia. 2015;2015:68–77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N. The genotype-tissue expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harbers L., Agostini F., Nicos M., Poddighe D., Bienko M., Crosetto N. Somatic copy number alterations in human cancers: an analysis of publicly available data from the cancer genome atlas. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.700568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X., Kim Y., Tsang E.K., Davis J.R., Damani F.N., Chiang C., Hess G.T., Zappala Z., Strober B.J., Scott A.J., et al. The impact of rare variation on gene expression across tissues. Nature. 2017;550:239–243. doi: 10.1038/nature24267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 16.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Stein T.I., Nudel R., Lieder I., Mazor Y., et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 2016;54:1. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 18.Wu C., Orozco C., Boyer J., Leglise M., Goodale J., Batalov S., Hodge C.L., Haase J., Janes J., Huss J.W. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:1–8. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldman M., Craft B., Hastie M., Repečka K., McDade F., Kamath A., Banerjee A., Luo Y., Rogers D., Brooks A.N. The UCSC Xena platform for public and private cancer genomics data visualization and interpretation. bioRxiv. 2019 [Google Scholar]
- 20.Li T., Fu J., Zeng Z., Cohen D., Li J., Chen Q., Li B., Liu X.S. TIMER2. 0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509–W514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ru B., Wong C.N., Tong Y., Zhong J.Y., Zhong S.S.W., Wu W.C., Chu K.C., Wong C.Y., Lau C.Y., Chen I. TISIDB: an integrated repository portal for tumor–immune system interactions. Bioinformatics. 2019;35:4200–4202. doi: 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- 22.Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., Doncheva N.T., Legeay M., Fang T., Bork P. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Therneau T. 2015. A Package for Survival Analysis in S. R Package Version 2. [Google Scholar]
- 25.Ogłuszka M., Orzechowska M., Jędroszka D., Witas P., Bednarek A.K. Evaluate Cutpoints: adaptable continuous data distribution system for determining survival in Kaplan-Meier estimator. Comput. Methods Progr. Biomed. 2019;177:133–139. doi: 10.1016/j.cmpb.2019.05.023. [DOI] [PubMed] [Google Scholar]
- 26.Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W., Treviño V., Shen H., Laird P.W., Levine D.A. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:1–11. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng D., Ye Z., Shen R., Yu G., Wu J., Xiong Y., Zhou R., Qiu W., Huang N., Sun L. IOBR: multi-omics Immuno-oncology biological research to decode tumor microenvironment and signatures. Front. Immunol. 2021:2547. doi: 10.3389/fimmu.2021.687975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian A., Kuehn H., Gould J., Tamayo P., Mesirov J.P. GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics. 2007;23:3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- 29.Chambers J.M. Springer; 2008. Software for Data Analysis: Programming with R. [Google Scholar]
- 30.Justus C.R., Dong L., Yang L.V. Acidic tumor microenvironment and pH-sensing G protein-coupled receptors. Front. Physiol. 2013;4:354. doi: 10.3389/fphys.2013.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen F., Zhuang X., Lin L., Yu P., Wang Y., Shi Y., Hu G., Sun Y. New horizons in tumor microenvironment biology: challenges and opportunities. BMC Med. 2015;13:1–14. doi: 10.1186/s12916-015-0278-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petitprez F., Meylan M., de Reyniès A., Sautès-Fridman C., Fridman W.H. The tumor microenvironment in the response to immune checkpoint blockade therapies. Front. Immunol. 2020;11:784. doi: 10.3389/fimmu.2020.00784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fridman W.H., Sautès-Fridman C., Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat. Rev. Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 34.Guerra L., Bonetti L., Brenner D. Metabolic modulation of immunity: a new concept in cancer immunotherapy. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.107848. [DOI] [PubMed] [Google Scholar]
- 35.Yan Y., Zhang L., Zuo Y., Qian H., Liu C. Immune checkpoint blockade in cancer immunotherapy: mechanisms, clinical outcomes, and safety profiles of PD-1/PD-L1 inhibitors. Arch. Immunol. Ther. Exp. 2020;68:1–15. doi: 10.1007/s00005-020-00601-6. [DOI] [PubMed] [Google Scholar]
- 36.Fang J., Chen F., Liu D., Gu F., Chen Z., Wang Y. Prognostic value of immune checkpoint molecules in breast cancer. Biosci. Rep. 2020;40 doi: 10.1042/BSR20201054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye J., Ji X., Dennis P.A., Abdullah H., Mukhopadhyay P. Relationship between progression‐free survival, objective response rate, and overall survival in clinical trials of PD‐1/PD‐L1 immune checkpoint blockade: a meta‐analysis. Clin. Pharmacol. Ther. 2020;108:1274–1288. doi: 10.1002/cpt.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ansell S.M., Lesokhin A.M., Borrello I., Halwani A., Scott E.C., Gutierrez M., Schuster S.J., Millenson M.M., Cattry D., Freeman G.J. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin's lymphoma. N. Engl. J. Med. 2015;372:311–319. doi: 10.1056/NEJMoa1411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Topalian S.L., Hodi F.S., Brahmer J.R., Gettinger S.N., Smith D.C., McDermott D.F., Powderly J.D., Carvajal R.D., Sosman J.A., Atkins M.B. Safety, activity, and immune correlates of anti–PD-1 antibody in cancer. N. Engl. J. Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka A., Sakaguchi S. Regulatory T cells in cancer immunotherapy. Cell Res. 2017;27:109–118. doi: 10.1038/cr.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corbet C., Feron O. Tumour acidosis: from the passenger to the driver's seat. Nat. Rev. Cancer. 2017;17:577–593. doi: 10.1038/nrc.2017.77. [DOI] [PubMed] [Google Scholar]
- 42.Onozawa Y., Fujita Y., Kuwabara H., Nagasaki M., Komai T., Oda T. Activation of T cell death-associated gene 8 regulates the cytokine production of T cells and macrophages in vitro. Eur. J. Pharmacol. 2012;683:325–331. doi: 10.1016/j.ejphar.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 43.Justus C.R., Sanderlin E.J., Dong L., Sun T., Chi J.-T., Lertpiriyapong K., Yang L.V. Contextual tumor suppressor function of T cell death-associated gene 8 (TDAG8) in hematological malignancies. J. Transl. Med. 2017;15:1–14. doi: 10.1186/s12967-017-1305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Z., Dong L., Dean E., Yang L.V. Acidosis decreases c-Myc oncogene expression in human lymphoma cells: a role for the proton-sensing G protein-coupled receptor TDAG8. Int. J. Mol. Sci. 2013;14:20236–20255. doi: 10.3390/ijms141020236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y., Shen Z., Wang B., Ye C., Lai Z., Jiang H., Wang Z., Jiang K., Ye Y., Wang S. Long non-coding RNA GPR65-1 is up-regulated in gastric cancer and promotes tumor growth through the PTEN-AKT-slug signaling pathway. Cell Cycle. 2018;17:759–765. doi: 10.1080/15384101.2018.1426414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi J.-W., Lee S.Y., Choi Y. Identification of a putative G protein-coupled receptor induced during activation-induced apoptosis of T cells. Cell. Immunol. 1996;168:78–84. doi: 10.1006/cimm.1996.0051. [DOI] [PubMed] [Google Scholar]
- 47.Wang H.-X., Chen Y.-H., Zhou J.-X., Hu X.-Y., Tan C., Yan Y., Huang Q.-L., Shen J.-Y., Xu H.-C., Li F. Overexpression of G-protein-coupled receptors 65 in glioblastoma predicts poor patient prognosis. Clin. Neurol. Neurosurg. 2018;164:132–137. doi: 10.1016/j.clineuro.2017.11.017. [DOI] [PubMed] [Google Scholar]
- 48.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 49.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 50.Kyaw H., Zeng Z., Su K., Fan P., Shell B.K., Carter K.C., Li Y. Cloning, characterization, and mapping of human homolog of mouse T-cell death-associated gene. DNA Cell Biol. 1998;17:493–500. doi: 10.1089/dna.1998.17.493. [DOI] [PubMed] [Google Scholar]
- 51.Ke X. Presence of multiple independent effects in risk loci of common complex human diseases. Am. J. Hum. Genet. 2012;91:185–192. doi: 10.1016/j.ajhg.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ghalamkari S., Nasiri M., Ariannia S., Farrokhseresht R. G-protein coupled-receptor 65 5′ UTR gene polymorphism in the pathogenesis of systemic lupus erythematosus. Rheumatology Research. 2017;2:139–143. [Google Scholar]
- 53.Chalmers Z.R., Connelly C.F., Fabrizio D., Gay L., Ali S.M., Ennis R., Schrock A., Campbell B., Shlien A., Chmielecki J. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9:1–14. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yarchoan M., Hopkins A., Jaffee E.M. Tumor mutational burden and response rate to PD-1 inhibition. N. Engl. J. Med. 2017;377:2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duensing A., Duensing S. Centrosomes, polyploidy and cancer. Polyploidization and Cancer. 2010:93–103. doi: 10.1007/978-1-4419-6199-0_6. [DOI] [PubMed] [Google Scholar]
- 56.Lee H.L., Jang J.W., Lee S.W., Yoo S.H., Kwon J.H., Nam S.W., Bae S.H., Choi J.Y., Han N.I., Yoon S.K. Inflammatory cytokines and change of Th1/Th2 balance as prognostic indicators for hepatocellular carcinoma in patients treated with transarterial chemoembolization. Sci. Rep. 2019;9:1–8. doi: 10.1038/s41598-019-40078-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao S., Hsu T.-W., Li M.O. Immunity beyond cancer cells: perspective from tumor tissue. Trends Cancer. 2021;7:1010–1019. doi: 10.1016/j.trecan.2021.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shahid A., Bharadwaj M. The connection between the Th17 cell related cytokines and cancer stem cells in cancer: novel therapeutic targets. Immunol. Lett. 2019;213:9–20. doi: 10.1016/j.imlet.2019.07.001. [DOI] [PubMed] [Google Scholar]
- 59.Chang S.H. T helper 17 (Th17) cells and interleukin-17 (IL-17) in cancer. Arch Pharm. Res. (Seoul) 2019;42:549–559. doi: 10.1007/s12272-019-01146-9. [DOI] [PubMed] [Google Scholar]
- 60.Salazar Y., Zheng X., Brunn D., Raifer H., Picard F., Zhang Y., Winter H., Guenther S., Weigert A., Weigmann B. Microenvironmental Th9 and Th17 lymphocytes induce metastatic spreading in lung cancer. J. Clin. Invest. 2020;130:3560–3575. doi: 10.1172/JCI124037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin W., Zhang H.-l., Niu Z.-y., Wang Z., Kong Y., Yang X.-s., Yuan F. The disease stage-associated imbalance of Th1/Th2 and Th17/Treg in uterine cervical cancer patients and their recovery with the reduction of tumor burden. BMC Wom. Health. 2020;20:1–7. doi: 10.1186/s12905-020-00972-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benvenuto M., Focaccetti C., Ciuffa S., Fazi S., Bei A., Miele M.T., Albonici L., Cifaldi L., Masuelli L., Bei R. Polyphenols affect the humoral response in cancer, infectious and allergic diseases and autoimmunity by modulating the activity of TH1 and TH2 cells. Curr. Opin. Pharmacol. 2021;60:315–330. doi: 10.1016/j.coph.2021.08.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data involved in this study are available in public databases as well as in supplementary materials. For further information, please contact the authors.