Abstract
In an ever-growing need for data storage capacity, the Deoxyribonucleic Acid (DNA) molecule gains traction as a new storage medium with a larger capacity, higher density, and a longer lifespan over conventional storage media. To effectively use DNA for data storage, it is important to understand the different methods of encoding information in DNA and compare their effectiveness. This requires evaluating which decoded DNA sequences carry the most encoded information based on various attributes. However, navigating the field of coding theory requires years of experience and domain expertise. For instance, domain experts rely on various mathematical functions and attributes to score and evaluate their encodings. To enable such analytical tasks, we provide an interactive and visual analytical framework for multi-attribute ranking in DNA storage systems. Our framework follows a three-step view with user-settable parameters. It enables users to find the optimal en-/de-coding approaches by setting different weights and combining multiple attributes. We assess the validity of our work through a task-specific user study on domain experts by relying on three tasks. Results indicate that all participants completed their tasks successfully under two minutes, then rated the framework for design choices, perceived usefulness, and intuitiveness. In addition, two real-world use cases are shared and analyzed as direct applications of the proposed tool. DNAsmart enables the ranking of decoded sequences based on multiple attributes. In sum, this work unveils the evaluation of en-/de-coding approaches accessible and tractable through visualization and interactivity to solve comparison and ranking tasks.
Keywords: Data storage, DNA, Medium, Attribute, Ranking, Visual analytic
Graphical Abstract
Nomenclature
- DNA:
Deoxyribonucleic Acid.
- ECC:
Error Correction Code.
- PCR:
Polymerase Chain Reaction.
- NGS:
Next Generation Sequence.
- MOSLA:
Molecular Storage for Long Term Archiving.
- SUS:
System Usability Scale.
1. Introduction
Existing data storage technologies cannot keep up with the exponential growth of modern data [1]. With advances in digital data storage, researchers have been seeking out alternative means to store more information in smaller media to bridge the gap between conventional storage media and the future data storage demands of mankind [2]. The uniqueness of DNA molecules as information carriers make them highly advantageous. When used as a data storage device, DNA as a molecular medium offers the benefits of high storage density, low maintenance costs, energy efficiency, and a long shelf-life. Compared to conventional storage media, these characteristics make DNA an up-and-coming storage medium for data storage and long-term archiving [3], [4], [5], [6]. Thanks to rapid advances in synthetic biology and next-generation sequencing (NGS), writing (synthesis) and reading (sequencing) have placed synthetic DNA in the lead position of alternative and novel storage media. When considering DNA for data storage, coding theorems for transmitting information from one source to a receiver over a channel becomes essential. These theorems model and evaluate the properties of one or more channels and their suitability to a specific en-/de-coding approach. The DNA data storage channel consists of three fundamental steps:
-
1.
Synthesis consists of producing short synthetic DNA sequences, or oligonucleotides, that contain the data as payload.
-
2.
Storage consists of a solution in which the synthetic DNA is stored.
-
3.
Sequencing, to read the short DNA sequences from storage.
Once sequencing is completed, the goal is to retrieve the original (encoded) sequence from the sequencing output using the decoder. Thanks to NGS technologies, these strands contain hundreds of characters [7]. However, DNA synthesis, storage, and sequencing may lead to errors in the resulting nucleotide sequence. The errors adhere to the following typology: insertion, deletion, and substitution of nucleotides. For example, when the GC content is lower than 40% and higher than 60%, the probability of synthesis and sequencing errors increases. Among many criteria, this motivates encoding the strands in a fashion that satisfies different conditions and constraints to optimize DNA storage.
Several researchers have demonstrated the feasibility of storing digital information and correcting errors in DNA molecules. Early works by Church et al. [8] and Goldman et al. [9] brought DNA storage to limelight. Alongside the development of DNA synthesis and sequencing technologies, newer and better methodologies for using DNA as a storage medium are published, which brought DNA storage to practical applications [10], [11], [49]. Aside from promises of a larger capacity and a longer lifespan, several efforts have been made to address the information and coding theoretic aspects of DNA data storage. These aspects specifically target the capacity of the storage channel [12], [13] and the design of error correction codes (ECC) [14], [15] for the specific errors [16] that arise inside a DNA data storage system. Additionally, in the field of ECC development, efforts have been made to provide solutions for other areas of DNA data storage, for example, constrained coding for DNA data storage [17], random access in DNA storage systems [18] and image storage solutions for DNA [19], [20]. Although ECCs have different approaches to correct information loss, one, if not the most important characteristic of an ECC is the ability to successfully recover the input data from the channel output. Traditionally, domain experts rely on time-consuming and non-visual approaches to evaluate and compare codes according to metrics to decide which sequences carry most of the encoded information. Navigating such a space takes years of experience and domain expertise.
Many approaches are concerned with reordering sequences. A noteworthy mention is for a specific scenario where sequences are reordered based upon the metric distances between every pair of decoded sequences [21]. The solution of the problem in this scenario is considered NP-hard [22]. There is no exact general solution; however, there are approximations. A naive approach to identifying similar sequences is not scalable, requiring every sequence to be tested against all other sequences. However, the related work has not yet considered relying on the interactive analysis or the visual exploration of such data, let alone combining multiple attributes and their weights. On the one hand, visually ranking the decoded sequences could speed up the time to determine the best encoding approach. On the other hand, visually finding out how much the ranking changes based on an attribute may shed further light on the robustness of one attribute to another.
To help domain experts navigate the parameter space, track potential errors, error sources, and evaluate the en-/de-coding approaches, we developed a visual analytical tool to rank the considered sequences by relying on information and coding theoretic metrics or attributes. We propose to employ a three-step workflow capable of handling multiple scenarios. Possible optimization scenarios in which the results of DNAsmart can be used comprise a combination of (a) the stored information, (b) the en-/de-coding approach(es) or code(s), (c) the parameter space of one code, (d) the ability of codes to adhere to DNA data storage specific characteristics (GC content, clustering of redundant sequencing output) and (e) other medium-based properties (e.g., lifespan). Three example scenarios are detailed. First, a comparative evaluation of aged synthetic DNA molecules pooled and stored in different conditions and locations. Recovering all information corresponds to finding the optimal (in the sense of lowest probability) subset of synthetic DNA molecules in a given sequencing pool. Second, a sensitivity analysis of the parameter space of a single coding approach results in a different set of synthetic DNAs. The recovery of all information corresponds to the search for the optimal parameter set. In both scenarios, all-information recovery is defined as minimizing the distance between a set of unordered sequences and a target reference. Finally, an indirect comparison of different ECC is made possible by considering user-specific attributes such as mutual information, which maximizes information recovery.
Motivated by creating better and more efficient DNA data storage systems and addressing these practical scenarios, the unordered nature of a set of decoded sequences, and the absence of a visualization method to rank such sequences, DNAsmart interactively evaluates DNA (s)torage sequences using (m)ultiple (a)ttributes and a (r)anking paradigm (t)ool. It leverages LineUp while adapting its main functionalities to the domain knowledge and specificity of DNA data storage [23]. To prioritize an attribute with a certain weight or multiple attributes with their corresponding weights over all other attributes, a set of decoded sequences is used as input data, and a ranking is provided by interactive sorting and grouping. Users have complete control over selecting attributes, sorting, merging, and adding weights to the chosen attributes. This flexibility is required to fit specific task requirements. Modifications are displayed live, in turn changing the ranking of a set of DNA sequences used in a data storage system. To evaluate DNAsmart, we performed a qualitative assessment with domain experts. We structured the visual analytical tasks by focusing on filling gaps related to the understanding and easy transition of the modules and the domain adaptability. We report the Top 3 decoded sequences based on three tasks, namely:
-
1.
when unsorted
-
2.
when sorted by a single attribute
-
3.
when merged and sorted by two or more attributes
Our results demonstrate that DNAsmart could leverage the subject-specific ranking task by allowing experts to interactively select attributes and rank the decoded sequences from DNA data storage systems.
The remaining sections of this paper discuss related work on the DNA storage and visual ranking system, domain characterization for multi-attribute selection, explain the three DNAsmart workflows, show the usability of DNAsmart through use cases, and discuss the results.
2. Related work
This section reviews related work on DNA-based data storage, error-correcting codes, and interactive sorting techniques that aid in ranking and analytical systems to make sense of multi-attribute data.
2.1. DNA data storage
A representation of information into DNA is focused on storage and error-free retrieval of data encoded in the four DNA nucleotides. In recent years, considerable efforts have been invested in demonstrating the potential of using DNA as a storage system [24]. DNA has already been used as an information carrier in recent decades [25], [26]. However, the first large-scale attempts of encoding information into a synthetic DNA molecule were recorded in 2012 [8], [9], after which then emerged the usage of an Error-Correcting Code (ECC) for storing more significant amounts of data and more efficiently [5], [11], [14], [27].
With the rise of reasonably practical solutions that employ DNA as a storage medium, information-theoretic aspects were considered. A recent comprehensive survey outlined design considerations for advancing data storage using DNA. Furthermore, information theory measures, as well as distance measures, were presented as evaluation metrics. These evaluation metrics provide sufficient knowledge and techniques to identify and compare several coding systems [28]. However, most of the attention was directed to error correction and how the DNA channel should be modeled. Furthermore, different distance measures in information theory were used to design ECCs for the DNA storage channel, and different coding models have been derived [29], [30]. An erroneous collection of sub-strings of the original sequence using ECCs was obtained [31]. A particular focus was directed to the impact of insertions, deletions and substitution errors which affect, if not impair the whole data storage process [32], [33].
Additional research on information encoding into the DNA provides sufficient knowledge and techniques for identifying and grouping these unordered sets of sequences based on a single attribute. This fact has been explored for capacity analysis where the Shannon entropy helps users establish the upper limit on the amount of information that can be reliably stored in DNA under a given error rate [34], [35]. Also, techniques based on the Hamming distance have been employed to ensure sequence differences between DNA used for sequence identification (DNA barcodes). This work is established by preserving minimal distance and error-correcting properties among the barcodes [26], [36]. Furthermore, the fact of grouping unordered sets of sequences have been employed in the DNA reconstruction problem; where the goal is to minimize the Levenshtein distance between the original sequence and decoded sequences [37].
However, these methods do not address how to order these sequences based on multiple attributes. Rankings are popular for structuring unordered items by computing each item based on the value of one or more attributes. In addition, domain experts must compare and rank codes for specific tasks in the domain knowledge of coding theory. It is even more complicated to do so without a good command of mathematical concepts applied in coding theory. For example, finding the optimal code for a given task in a given scenario is time-consuming and difficult. The rationale of this work is to adapt the visual ranking paradigm presented by previous work to the specificity of the coding theory domain. While visualization of a ranking is straightforward, its interpretation is not because the rank of an item is only a summary of a complicated relationship between its attributes and those of other items. Therefore, this work aims to visually represent, analyze, and rank these sets of unordered sequences based on multiple attributes.
2.2. Visual ranking system
Interactive ranking and sorting is an active research area. Many systems exist for the visualization of multi-attribute rankings [38]. These systems focus on providing capabilities to generate, modify, and present tabular data and are not intended solely for ranking. For example, rows (data points) can be sorted by some column (attribute), but such systems do not natively support sorting based on combinations of columns or attributes. Consequently, these systems require a high level of formalism to define a ranking. Our system is built upon LineUp [23], a visual analytic system that performs ranking visualization of multi-attribute data and allows users to flexibly adjust weights to identify potential relationships. LineUp uses bar charts to facilitate ranking comparison while relying on tabular layouts to compare data attributes. It allows users to create custom rankings by clicking and dragging columns to adjust interactively the attribute weights used for the ranking. Users can see how changing the attribute weights affects the ranking of the data points. Additionally, DNAsmart allows users to select attributes of their choice by directly leveraging domain-specific knowledge for ranking tasks. DNAsmart is designed for information theory-specific tasks, requiring users to have a good knowledge of the attributes. The latter should be chosen with care as their integration significantly affects the ranking results of the set of sequences decoded from a DNA data storage system.
3. Domain characterization
In data storage, DNA as a storage medium comes with many challenges during synthesis, storage, and sequencing. One challenge is determining whether the complete reconstruction of the original data is possible from the given set of decoded sequences after storage. This task poses the coded string reconstruction problem, which requires reconstructing strings from their substrings satisfying predefined constraints. We describe our method based on: (a) transforming a set of decoded sequences into descriptors and (b) transforming information-theoretic metrics to qualitative measurements of attributes. To demonstrate the data extraction stage, we use the information-theoretic perspective of DNA storage as an example and detail our choice of multi-attributes. The encoding and decoding stages are external to the storage that even in the presence of error, it is possible to reconstruct the original data. The input sequences for the decoding entail processing short DNA molecules from the solution in which the DNA medium is held, then sequencing them.
3.1. Multi-Attributes
An important parameter, which could affect the final decoding result, is the choice of an attribute for the ranking of decoded sequences. Applying a default ranking attribute may lead to poor results. Assessing one sequence from a set of sequences that involves considering and integrating multiple attributes could enable accurate comparisons among the given sequences. The latter assumes user knowledge of good practices to choose said attributes. For instance, combining the mutual information and the entropy in one column is wrong. Example combinations are reported in Table 1. To satisfy the diverse requirements of different en-/de-coding approaches and user preferences, we implement and integrate extendable attributes for users to dynamically choose their preferred attributes with their preferred weights. By default, all attributes are given no weight. Choosing the suitable attribute is often challenging; such an analytical choice is specifically carried out depending on the en-/de-coding approach and the task. We integrate six domain-specific attributes and propose a new one by weighing in the domain-specific knowledge for DNA data storage and information theory. We identify and borrow specific attributes from distance-based, information theory, and biological domain namely:
-
1.
Hamming
-
2.
Levenshtein
-
3.
Damerau-Levenshtein
-
4.
Conditional entropy
-
5.
Mutual information
-
6.
GC content
Table 1.
Possible attribute-based combinations. Two types exist to optimize the results; either minimization or maximization.
| Minimization | Maximization |
|---|---|
| Hamming | Mutual Information |
| Levenshtein | Number of errors |
| Damerau-Levenshtein | – |
| Conditional entropy | – |
Additionally, the related work in question designed source codes for DNA storage to include attributes [10], [21], [29].
First, Entropy leads to considering the essential function of coding theory for the transmission of information from one source to another over a given channel. That is to say, a DNA molecule conceptually corresponds to an information carrier in a channel. Since more than one decoded sequence is involved, we define the concept of conditional entropy which measures the amount of information that exists in the decoded sequences given that the encoded sequence is known.
Second, the shared or mutual information among channels is essential for determining the information capacity [39]. Mutual information measures the accuracy with which the outputs of the channel, i.e., decoding sequence, represent the input of the channel or the preset DNA sequence.
3.2. Cumulative-based error weight
Analyzing the number of errors present in DNA data storage is an essential step in assessing the reliability and accuracy of the stored information. It can aid in identifying specific issues with the storage methods and techniques, allowing adjustments to improve the overall storage process. Furthermore, knowledge of error rates can help ensure data integrity and security and inform future DNA data storage technology advancements.
Therefore, the cumulative-based error weight further supports sequence-based errors. The weight is defined as identifying the number of local regions that differ in a yet-to-be-decoded DNA sequence compared to an encoded one. The attribute is derived using a dynamic programming approach where the length of the decoded sequence is amplified against a reference sequence. To calculate an overall number of errors for each decoded sequence, we use a linear weighted sum model according to: ∑n=1ne; with e denoting an error region of size n for a pair of sequences. The three types of sequence-based errors are substitution, insertion, and deletion. Error regions for each sequence are computed according to its overall score. The score is the weighted sum of each error type based on its occurrence. To facilitate the interpretation of the error type and the overall number of errors, we constrain the normalization of the error type weights according to: ; denoting the occurrence of i insertions, d deletions, and s substitutions in a given sequence, respectively. DNAsmart employs the metrics mentioned above as multi-attributes to enable visual analysis and ranking of an unordered set of sequences. The framework transforms these multi-attributes into quantitative measurements to which weights are assigned. End-users provide further weight adjustments.
4. Workflow
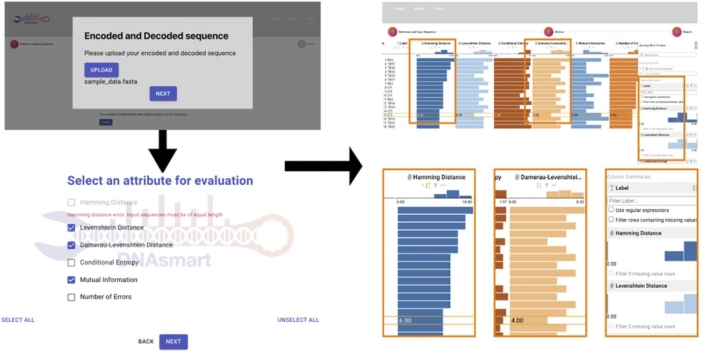
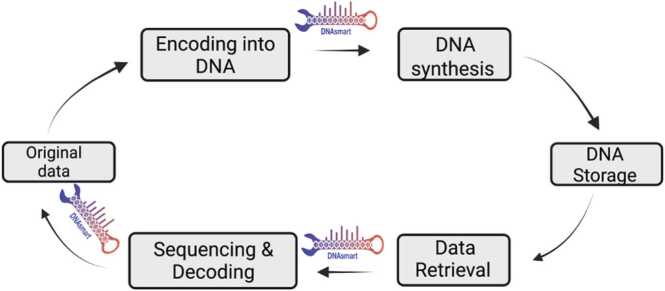
DNAsmart is a visual analytical method that combines lightweight data analytic and a visual technique to assist users with ranking tasks by weighing multiple attributes. The web-based implementation takes as input a multi-FASTA file type that accommodates sequences from either the synthesis or sequencing process. Fig. 1 shows the flexible use of DNAsmart in different steps of the DNA data storage process. A keyword extraction module analyzes all sequences and outputs a set of objects for the reference and each sequence, respectively. The User interface (UI) allows users to explore the sequences. As the user selects attributes of interest, the ranking view is displayed. Applying analytical tasks brings related sequences to the top and pushes down the less relevant ones. The layout of the framework is arranged in a step-wise fashion with three views: sequence, selection, and ranking.
Fig. 1.
Overview process of DNA data storage system with positions where DNAsmart can be used. DNAsmart accommodates sequences from either the synthesis or sequencing process.
4.1. Sequence view
DNAsmart automatically extracts sequences from a multi-FASTA file uploaded by the user with a two-fold purpose. It provides manipulable elements that serve as input for the ranking view. Upon uploading a file, the entire set of sequences is represented as a key and value pair.
4.2. Selection view
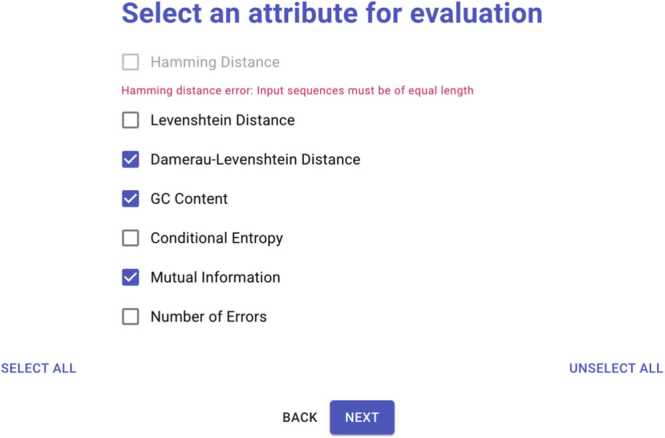
DNAsmart allows a flexible choice of attributes according to user task requirements. The Selection view lists different attributes with individual check-boxes for selection. The seven attributes are Hamming distance, Levenshtein distance, Damerau-Levenshtein distance, GC content, Conditional Entropy, Mutual Information, and Number of errors. Conditions are placed on the Hamming distance attribute as shown in Fig. 2 when the length of the sequences uploaded from the sequence view is not equal, prompting the user to upload sequences of equal length or to refrain from selecting this attribute. Users can select specific or all attributes based on their task requirements.
Fig. 2.
The selection view. The second step of the workflow. Attribute selection consists of selecting the right attributes for the right domain task.
4.3. Ranking view
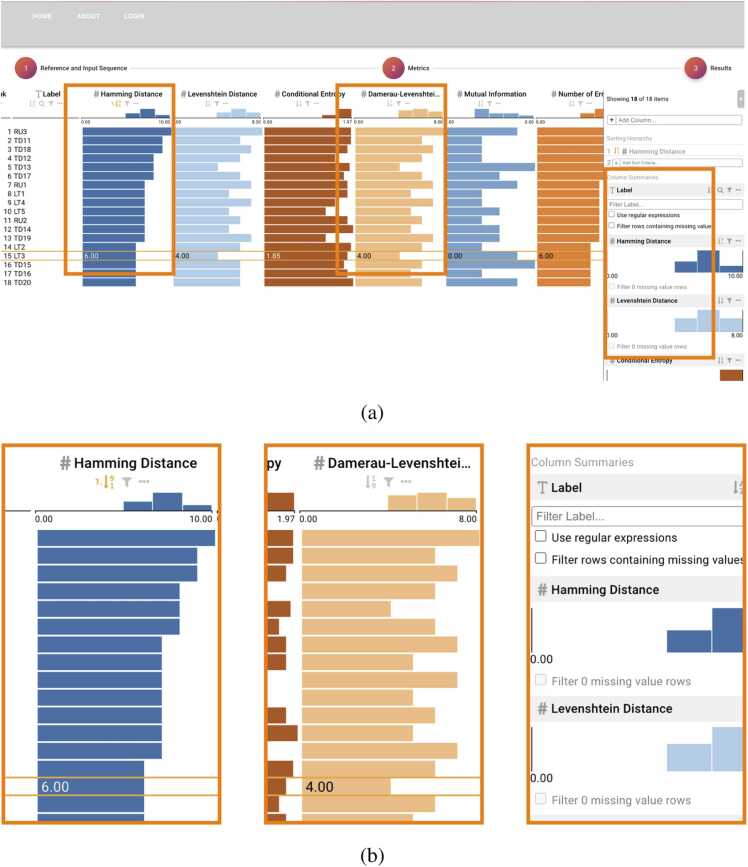
DNAsmart allows the interactive exploration of rankings based on multiple attributes computed for a given data set. As represented in Fig. 3, it shows a table where each row is a decoded sequence represented with its name, and each column represents an attribute. Their ranking orders the data and the rank scores are initially computed based on the arrangement of the given sequences. Users can interact with the tabular data by clicking and selecting the sort button for every relevant attribute. This corresponds to having an active sort functionality. Users may also implement weights on the attributes, which prompts the system to derive a new set of attribute weights based on the user interaction with the columns in said table.
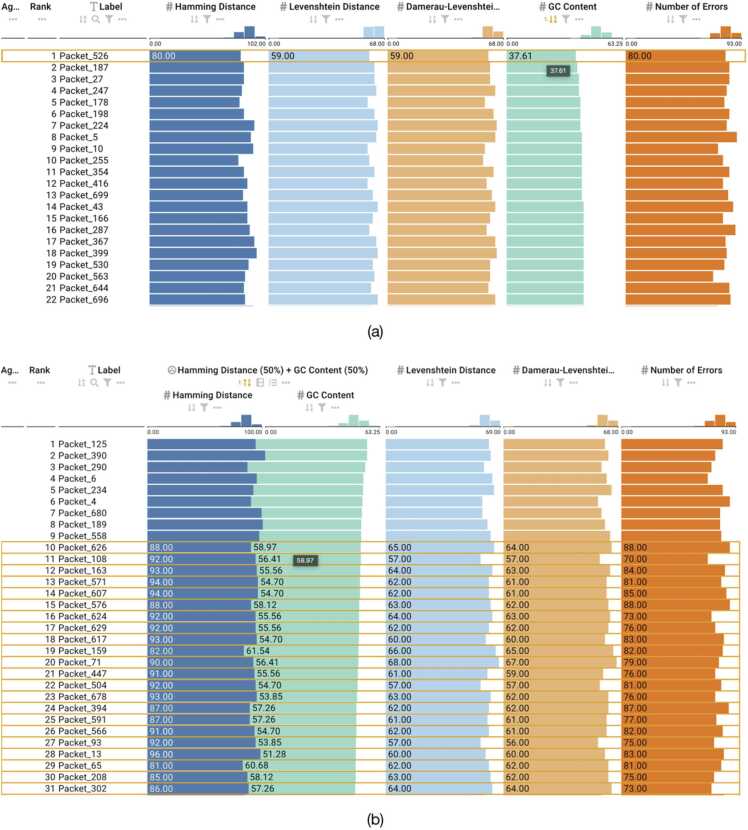
Fig. 3.
The ranking view. The third and last step of the workflow. Area-based visual idioms provided by the bar charts effectively depict larger and smaller values and enable interactive functionalities. (a) Screenshot overview. (b) Detail from left to right: The Hamming column with an active sort functionality, the Damerau-Levenshtein column with a highlighted value for the selected sequence, and the sidebar.
Clicking and dragging these columns merges two or more attributes, then automatically sets equal weights to the sequences in the data set. Thus, ranking all the rows in the table according to their resulting rank scores. Specific weight adjustments are possible thanks to user-settable functionalities specific to attribute weights. As presented in the LineUp API, DNAsmart inherits the full interactive functionalities to explore the unordered set of sequences. Ranking enables sorting sequences by each attribute in the columns or by user-defined combinations of attributes. Filtering is possible for one or a combination of columns. In addition, regular expressions may be used for advanced filtering. Grouping and aggregating enable a display mode where the height of each row is reduced to a minimum height of a single pixel. Our tool also inherits all interaction and encoding idioms. We decided to mention only the idioms that we have changed. Moreover, since the default API does not use a colorblind-friendly scheme for the ranking visualization, we replaced the default with the Tableau10 colorblind-safe palette. This renders the color encoding more accessible [40], [41].
4.4. Design considerations
DNAsmart is built around five fundamental design considerations. First, DNAsmart supports decision-making and provides the optimal set of sequences and the ranking system that benefits users by allowing them to navigate through the sequences or even different separate attributes. Second, the tool allows low computational overhead. DNAsmart uses a randomized approach to determine the similarity between sequences based on multi-attributes. Third, DNAsmart supports the traditional minimization problem when comparing a set of sequences. That is to say, minimizing the computational expense of calculating different metrics between all input sequences. Fourth, although assumptions are considered for the input format, the input can be arbitrary and does not necessarily belong to a particular error-correction code. The input format follows multiple sequences in one FASTA file format. Fifth and last, the workflow does not presuppose a definite number of sequences in one given set.
5. Results
To demonstrate the usability of DNAsmart, we show several features of DNAsmart with two exemplary use cases - (I) barcode validation and selection and (II) parameter space of an encoding scheme.
The case study overview can be seen in Figs. 4, 5, 6, 7, 8 and 9.
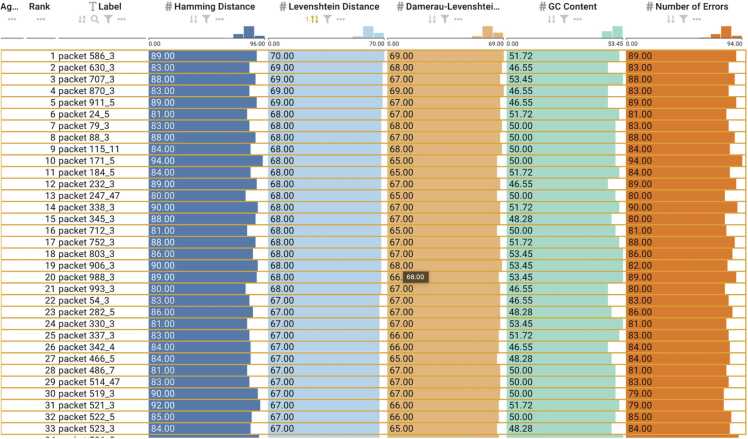
Fig. 4.
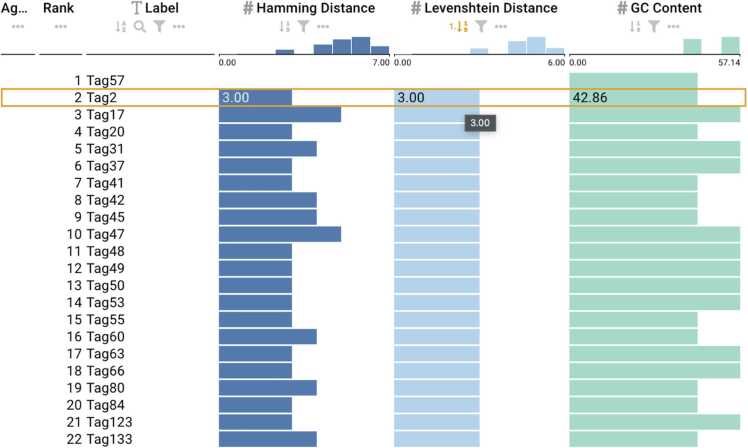
Barcodes from Faircloth et al[43] with an active sort functionality on the Levenshtein column depicting the minimum distance and GC content.
Fig. 5.
Barcodes from Adey et al. [44] with an active sort functionality on the Levenshtein column depicting the minimum distance and GC content.
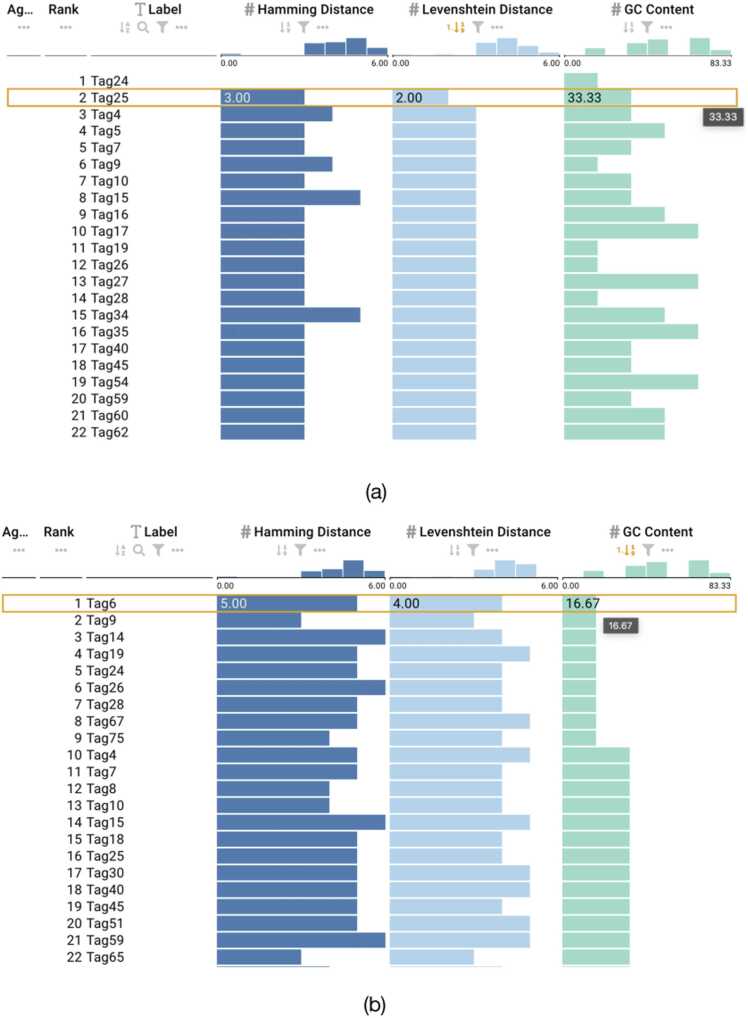
Fig. 6.
Barcodes from Meyer et al. [42] with a selected sequence. (a) Active sort functionality on the Levenshtein column showing the minimum distance within the barcodes. (b) Active sort functionality on the GC content showing the lowest value.
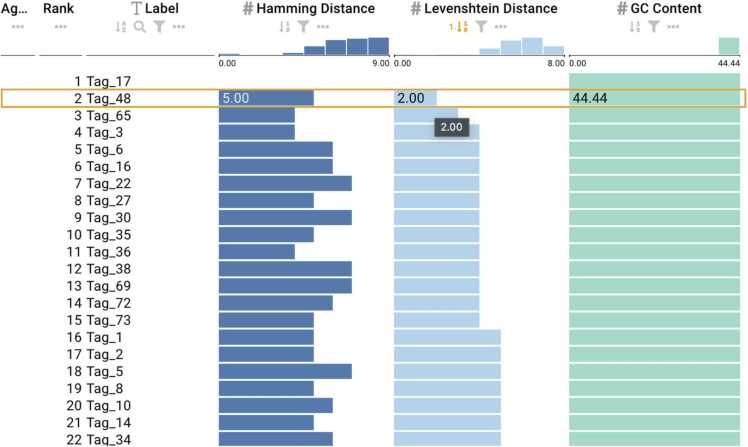
Fig. 7.
Barcodes from Meyer et al. [45] based on Hamming distance with a selected sequence. (a) Active sort functionality on the Hamming column ranking the barcodes from smallest to largest. (b) Active sort functionality on the GC content showing the least GC content value.
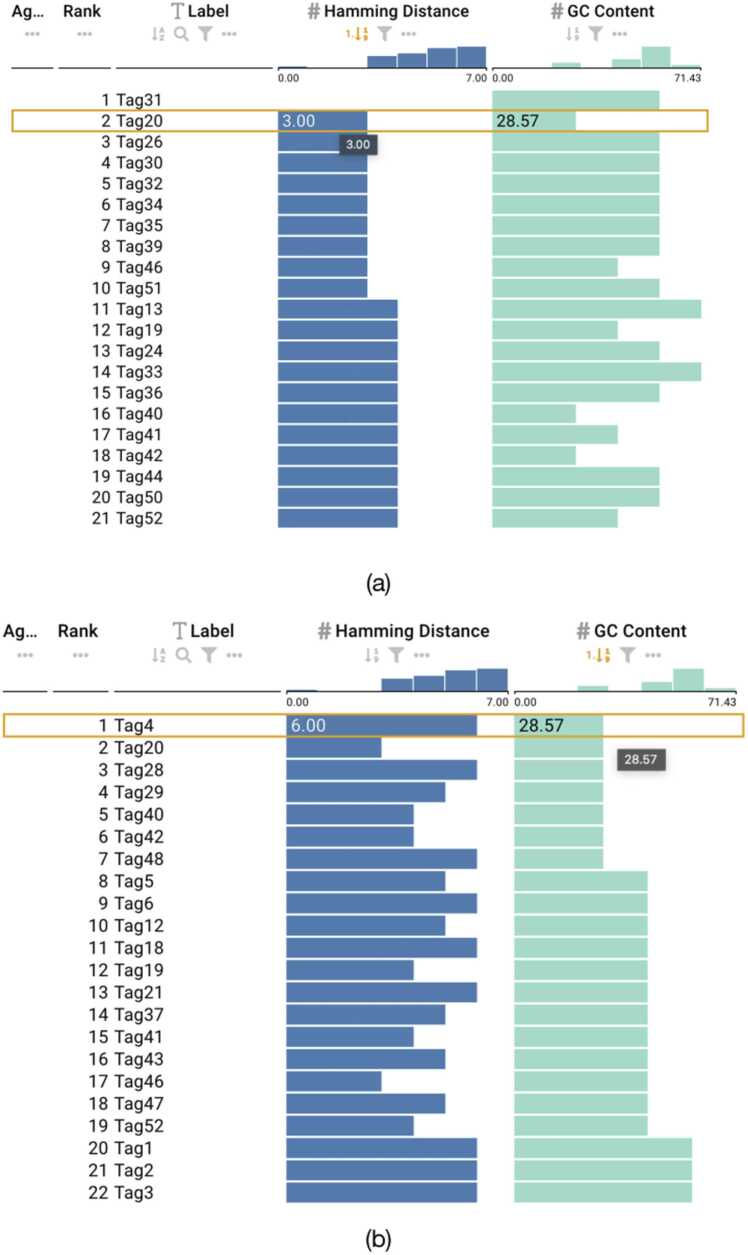
Fig. 8.
Sequences from Grass encoding scheme. (a) Active sort functionality on the GC content column showing the least GC content value. (b) Two merged attributes (Hamming and GC content) of equal weight with an active sort functionality. Selected sequences depicting sequences with maximum hamming distance and values that conform to GC content constraints.
Fig. 9.
Sequences from Fountain encoding scheme. Active sort functionality on the Levenshtein column. Selected sequences depict the ordering of sequences based on Levenshtein distance that is separated in the distance metric space.
5.1. Use case 1: barcode validation and selection
To demonstrate the function and utility of DNAsmart in validating the conformance of these barcodes, we uploaded the independent FASTA files containing barcodes for each of the sets of codes, respectively. We visualized the set of codes using DNAsmart based on three attributes- Hamming distance, Levenshtein distance, and GC content.
We validated five sets of pre-existing barcodes to ensure that all pairwise comparisons within these barcodes were greater than the minimum expected Levenshtein or Hamming distance. A summary of the sets of barcodes is detailed in Table 2. We also validated that the barcodes conform to constraints such as GC content. GC content of< 40% and> 60% can be problematic during their synthesis and sequencing process.
Table 2.
Summary of barcodes designed based on three attributes. The values are the observed minimum distance and GC content percentage using DNAsmart. Meyer et al.* [42] barcodes were designed based on Levenshtein distance.
| Barcodes | Number of barcodes | Hamming distance | Levenshtein distance | GC content |
|---|---|---|---|---|
| Adey et al. | 96 | – | 2 | 40–60% |
| Hamady et al. | 80 | 2 | – | 50% |
| Meyer et al. | 52 | 3 | – | < 40% – > 60% |
| Meyer et al.* | 75 | – | 2 | < 40% – > 60% |
| Faircloth et al. | 211 | – | 3 | 40–60% |
To ascertain barcode conformance, we appropriately formatted an input file for these barcodes and uploaded the file to DNAsmart using the sequence view. Then in the selection view, we selected the Hamming distance, Levenshtein distance, and GC content, respectively. We used these attributes to test and validate the barcodes. Then we visualized our data using the ranking view to provide an interactive exploration of rankings of the barcodes based on the three attributes we selected.
For barcodes designed based on Levenshtein distance, we used DNAsmart to interact with the barcodes by clicking and selecting the sort button for the Levenshtein attribute. Only those barcodes provided by Faircloth et al [43] maintained a minimum Levenshtein distance sufficient to correct one error (Levenshtein distance ≥3) across all pairwise comparisons. As shown in Fig. 4, the barcodes also conformed to appropriate GC content constraints within the range of 40% and 60%. However, the barcodes provided by Adey et al. [44] and Meyer et al. [42] contained pairwise comparisons below the minimum expected Levenshtein distance reported as shown in Figs. 5 and 6. The minimum Levenshtein distance reported for the barcodes was 4 for Adey et al. [44] and 3 for Meyer et al. [42], respectively. Using the ranking view of DNAsmart, we clicked and selected the Levenshtein distance for both sets of barcodes and observed a minimum distance of 2 for each set of barcodes. This is shown in Figs. 5 and Figs. 6a. Additionally, we evaluated the barcodes based on GC content, and we found that the barcodes from Adey et al. conformed to appropriate GC content compatible for synthesis and sequencing with an exact GC content of 44.44% as also shown in Fig. 5. However, we found that some barcodes did not satisfy the constraints for the barcodes from Meyer et al., using DNAsmart and ranking based on GC content. As shown in Fig. 6b, some barcodes from this set had GC content as low as 16.67% and as high as 83.33%. These barcodes would be problematic for the synthesis and sequencing technology.
For barcodes designed based on Hamming distance, only substitution errors can be corrected by constructing codes with a larger minimum distance between barcodes. The minimum Hamming distance between barcodes needs to be at least 2k+ 1 to correct K errors, as this significantly reduces the probability of reading errors interfering with accurate barcode identification. To ascertain whether barcodes satisfy the constraints of minimum Hamming distance and GC content, we selected the input file for barcodes designed based on Hamming distance. Using the ranking view from DNAsmart, we found that the observed minimum Hamming distance of 3 shown in Fig. 7a is the same as the expected Hamming distance for the barcodes provided by Meyer et al. [45]. However, the GC content in Fig. 7b of the barcodes do not conform to the appropriate GC content as the set of barcodes contains GC content as low as 28.57% and as high as 71.43%. For barcodes from Hamady et al. [46], the observed minimum Hamming distance of 2 conformed to their expected Hamming distance, and the barcodes satisfy the GC content constraint.
Barcodes aid in information indexing when the quantity of digital information surpasses the capacity of a single storage pool. Barcodes can be randomly generated, yet selecting and validating them requires avoiding sequences that do not conform to established constraints. DNAsmart provide visual feedback on how these constraints affects the ranking of the barcodes. Additionally, DNAsmart also supports merging two or more attributes, which could affect the resulting rank of barcodes. For the sake of brevity, we did not show this option for this use case.
5.2. Use case 2: parameter space of an encoding scheme
To ensure proper information retrieval, one requires that sequences at minimum Hamming or Levenshtein distance be avoided in the information string. Furthermore, due to synthesis and sequencing constraints, sequences with improper GC content are to be avoided. This poses a challenging question of combining relevant distance attributes with GC content and other relevant attributes. For that purpose, we visually explored data using DNAsmart based on five attributes- Hamming, Levenshtein, Damerau-Levenshtein, GC content, and Number of errors. The dataset considered for this use case was derived using the Grass encoding scheme [5] and the Fountain encoding scheme [11] containing about 713 and 1000 sequences respectively.
For the sequences from the Grass encoding, we uploaded the FASTA file of the encoding scheme and selected the five attributes. Then we visualized the dataset using the ranking view of DNAsmart to provide an interactive exploration for combining and ranking of the sequences based on the attributes we selected. When sorted by the GC content as shown in 8a, we found that the dataset contains inappropriate GC content as low as 37.61% and as high as 63.25%. However, since DNAsmart supports merging two or more attributes, we prioritized and merged the Hamming and GC content attributes with a corresponding equal weight of 50% for each attribute. We clicked and sorted the sequences based on this combination. We found that sequences with maximum Hamming distance also have appropriate GC content, thus satisfying the GC content constraints and leading to proper information retrieval. This is shown in Fig. 8b.
For the sequences from the Fountain encoding, we performed an ordering task based on the Levenshtein attributes. We selected five attributes, as earlier mentioned, and interactively explored the dataset using DNAsmart. We confirmed that the encoding scheme satisfies the appropriate GC content constraints as it contains GC content within 40% and 60%. Additionally, sorting and ranking the dataset based on Levenshtein distance result in a significant ordering of the sequences that are well separated in the distance metric space as shown in Fig. 9.
In summary, using DNAsmart, the recovery of information corresponds to overviewing and finding an optimal set of DNA sequences which could provide knowledge for further domain-specific analysis. The information provided from these explorations can lead to further clustering tasks in the domain. DNAsmart allows the exploration of any dataset that satisfies several properties, such as those coming from DNA data storage systems.
6. User evaluation
DNAsmart enables the influential ranking of an unordered set of sequences by exploring multi-attributes derived from digitized data (quantitative and qualitative improvement of decoded information).
6.1. Analytical tasks
We have structured the visual analysis tasks we want to address with the ranking framework to be based on a single attribute or set of attributes. The goal of each task is to find the TOP 3 ranking of the decoded sequences: 1. when unsorted (default view, no interaction needed), 2. when sorted by a single attribute (one interaction needed), and 3. when three attributes are merged and sorted by specific weights (three interactions needed).
6.2. User study
A user study was performed to investigate the effectiveness of the tool. Nine participants (3 female and 6 male) were recruited from the Molecular Storage for Long-Term Archiving (MOSLA) consortium. More than half of them indicated that they have experience with information visualization, and the rest of them had considerable experience in data storage. Eight participants indicated having experience with DNA Storage systems, and only one reported having none. To train the domain experts to use the tool, a randomly generated set of sequences was used as a toy example data set. The second data set was derived from simulation using an error simulation tool that simulates errors during synthesis, Polymerase Chain Reaction (PCR), storage, and sequencing [16]. The user evaluation started with a written description and instructions to proceed. The evaluation was divided into two steps: the training step and the testing step. For each step, there were a series of questions regarding the use of the tool. The questions were divided into two training questions and three testing questions. There was no time limit for both steps. The training involved selecting the Top 3 decoded sequences in two instances when sorted by one attribute and when merged and sorted by two attributes, respectively. Participants were provided with hints on how to sort, drag and merge attributes. Participants were given the correct answer to both training questions. Testing involved selecting the Top 3 decoded sequences in three instances when unsorted, sorted by one attribute, merged, and sorted by three attributes, respectively. Participants answer these questions using the previously learned interactive features (e.g., sorting) to visualize the desired ranking. At the end of the study, participants were required to fill out a post-hoc questionnaire comprising: a demographic form, an experience form, and a system usability scale or SUS form.
6.3. Findings
We outlined 3 tasks that the participants had to perform using DNAsmart. The findings covered the three views of the tool: sequence, selection, and ranking. The tasks had slightly different levels of complexity. We measured task completion times to approximate the time it took each participant to complete a task. All participants completed the training. In testing, 7 out of 9 participants provided correct answers. Training and testing took about 4 min (μ = 240, σ = 15.5) and 2 min (μ = 113, σ = 49.6), respectively.
We found that participants spent more time in task 3 because they were required to use more than one interaction. They had to recall different functionalities to merge, add weights and sort results and use 3 attributes. In the post-hoc questionnaire, the 5-point based Likert scale results were categorized into three main sections: design choices, usefulness, and intuitiveness as shown in Tables 3, 4, 5, respectively.
Table 3.
Questionnaire results showing how the participants rated the tool for intuitiveness.
| Description | Strongly Agree | Agree | Neutral | Disagree | Strongly Disagree |
|---|---|---|---|---|---|
| add weights | 3 | 2 | 2 | 2 | 0 |
| upload data | 5 | 4 | 0 | 0 | 0 |
| select attributes | 5 | 2 | 0 | 2 | 0 |
| how item are ranked | 5 | 2 | 0 | 2 | 0 |
| learn before using | 3 | 5 | 0 | 1 | 0 |
| confident | 1 | 3 | 2 | 2 | 1 |
| merge attributes | 5 | 2 | 0 | 2 | 0 |
| transition | 5 | 1 | 3 | 0 | 0 |
| need assistance | 2 | 1 | 0 | 4 | 2 |
| learn to use quickly | 2 | 0 | 1 | 2 | 4 |
Table 4.
Questionnaire results showing how the participants rated the tool for design choices.
| Description | Strongly Agree | Agree | Neutral | Disagree | Strongly Disagree |
|---|---|---|---|---|---|
| complex workflow | 1 | 1 | 2 | 4 | 1 |
| well integrated | 2 | 5 | 2 | 0 | 0 |
| inconsistency | 0 | 1 | 2 | 2 | 4 |
Table 5.
Questionnaire results showing how the participants rated the tool for usefulness.
| Description | Strongly Agree | Agree | Neutral | Disagree | Strongly Disagree |
|---|---|---|---|---|---|
| useful for DNA | 3 | 5 | 1 | 0 | 0 |
| will use frequently | 1 | 2 | 4 | 2 | 0 |
Results indicated that participants rated the tool as intuitive, easy to use (μ = 3.7, σ = 0.71), useful for ranking a set of unordered sequences from a DNA data storage system (μ = 3.7, σ = 0.5), and that the workflow was not complex (μ = 2.8, σ = 0.83). All participants indicated that the step-wise transitions were very well integrated and that they are willing to use the tool frequently. However, two participants indicated that they will need to learn or need some assistance before using the tool; hence they were neutral on its frequent use.
7. Discussion and future work
Selecting DNA sequences, or a set of DNA sequences that satisfy combinatorial constraints, is motivated by the tasks of storing information in DNA sequences used for data storage, computation, or as molecular barcodes in chemical libraries [22]. The selection of an optimal set of sequences is important to minimize errors due to cross-hybridization between different barcodes and their complements, to achieve higher information density for encoding algorithms, and to obtain large sets of barcodes for large-scale applications.
The global exploration of different sets of sequences using DNAsmart allows us to discover the discrepancies in the sequences, whether in the selection of the barcode, in the implicit comparison of the encoding scheme, or in the validation of the sequences able to resist aging. This further leads us to suggest that researchers evaluate sequences in use or before using them to ensure that they satisfy constraints based on multiple attributes, are valid across sets, and are robust to the suite of substitution, insertion, and deletion errors affecting massively parallel sequencing technologies.
This work opens the way for further discussion on integrating information visualization for a domain task such as the reconstruction of coded sequence, where experts rely exclusively on data analysis without visualization. This would provide a more detailed understanding of coded sequences, and a visual way to verify the integrity and accuracy of error-correcting codes. Moreover, there are known sources of noise present in various steps. For future work, we plan to address issues related to uncertainty visualization. Visualization of such uncertainties as part of our design is paramount to better assess, understand, and potentially mitigate the effects of strong noise.
Future use of DNAsmart in other areas may include evolutionary analysis of sequences (e.g., non-coding sequences for non-membrane folding), quality control, and validation of synthesis and sequencing methods (e.g., validation of synthetic DNA synthesized using the enzymatic synthesis method), and estimation of the mutual relationship between two positions in a protein family. Moreover, our tool may be extended with the addition of more attributes to benefit further domain-specific questions. The latter prompts the use of different visual encoding considerations.
8. Conclusions
In this work, first, we showed how stacked bar charts can be leveraged to guide the scientific exploration of decoded information from a DNA data storage system by ranking the set of decoded sequences. Second, The user study has shown that DNAsmart offers an intuitive way to visually understand multi-attribute rankings of a set of sequences that are not possible to know without a visual analytical tool. Third, although the number of participants was not high enough to conduct statistical analyses, we plan to share our tool with more domain experts to collect further feedback and data. In sum, our tool, DNAsmart, supported users in the customization of the ranking view by integrating a selection view to dynamically choose preferred attributes based on their needs and quickly learn how to use the interactive functionalities. To the best of our knowledge, this tool is the first application of visual techniques to the domain of DNA data storage.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
CRediT authorship contribution statement
Conceptualization: C.E. and G.H., implementation: C.E., writing manuscript: C.E. and G.H., proofreading: C.E., M.W., D.H. and G.H., visualization: C.E. and G.H., andsupervision: G.H.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to thank all members of the MOSLA consortium for participating in the user study.
Funding
This work has been financially supported by the Hessian Ministry for Science and the Arts (LOEWE) in the context of the Molecular Storage for Long-Term Archiving (MOSLA) consortium. The funders had no role in study design, data collection, and analyses, publication decision, or manuscript preparation.
Availability of data and materials
DNAsmart is implemented as a client-side web application and relies on React to support efficient visualization updates when data is modified [47]. The code and data for DNAsmart is publicly available at https://github.com/sombiri/DNAsmart. The evaluation is shared under its own branch. A deployed prototype of DNAsmart is available at https://dnasmart.mathematik.uni-marburg.de/.
References
- 1.Shrivastava S., Badlani R. Data storage in DNA. Int J Electr Energy. 2014:119–124. [Google Scholar]
- 2.Reinsel D., Gantz J., Rydning J. The digitization of the world from edge to core. IDC White Pap. 2018;13 [Google Scholar]
- 3.Cox J.P. Long-term data storage in DNA. TRENDS Biotechnol. 2001;19:247–250. doi: 10.1016/s0167-7799(01)01671-7. [DOI] [PubMed] [Google Scholar]
- 4.Anchordoquy T.J., Molina M.C. Preservation of DNA. Cell Preserv Technol. 2007;5:180–188. [Google Scholar]
- 5.Grass R.N., Heckel R., Puddu M., Paunescu D., Stark W.J. Robust chemical preservation of digital information on DNA in silica with error-correcting codes. Angew Chem Int Ed. 2015;54:2552–2555. doi: 10.1002/anie.201411378. [DOI] [PubMed] [Google Scholar]
- 6.Zhirnov V., Zadegan R.M., Sandhu G.S., Church G.M., Hughes W.L. Nucleic acid memory. Nat Mater. 2016;15:366–370. doi: 10.1038/nmat4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tabatabaei Y.S.H., Gabrys R., Olgica M. Portable and error-free DNA-Based data storage. Sci Rep. 2017;7 doi: 10.1038/s41598-017-05188-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Church G.M., Gao Y., Kosuri S. Next-generation digital information storage in DNA. Science. 2012;337 doi: 10.1126/science.1226355. 1628-1628. [DOI] [PubMed] [Google Scholar]
- 9.Goldman N., Bertone P., Chen S., Dessimoz C., LeProust E.M., Sipos B., et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature. 2013;494:77–80. doi: 10.1038/nature11875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Organick L., Ang S.D., Chen Y.-J., Lopez R., Yekhanin S., Makarychev K., et al. Random access in large-scale DNA data storage. Nat Biotechnol. 2018;36:242. doi: 10.1038/nbt.4079. [DOI] [PubMed] [Google Scholar]
- 11.Erlich Y., Zielinski D. DNA fountain enables a robust and efficient storage architecture. Science. 2017;355:950–954. doi: 10.1126/science.aaj2038. [DOI] [PubMed] [Google Scholar]
- 12.R. Heckel, I. Shomorony, K. Ramchandran, N. David, Fundamental limits of DNA storage systems, in: 2017 IEEE International Symposium on Information Theory (ISIT), IEEE, 2017, pp. 3130–3134.
- 13.A. Lenz, P.H. Siegel, A. Wachter-Zeh, E. Yaakobi, An upper bound on the capacity of the DNA storage channel, in: 2019 IEEE Information Theory Workshop (ITW), IEEE, 2019, pp. 1–5.
- 14.H.M. Kiah, G.J. Puleo, O. Milenkovic, Codes for DNA storage channels, in: 2015 IEEE Information Theory Workshop (ITW), IEEE, 2015, pp. 1–5.
- 15.Heckel R., Mikutis G., Grass R.N. A characterization of the DNA data storage channel. Sci Rep. 2019;9:1–12. doi: 10.1038/s41598-019-45832-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwarz M., Welzel M., Kabdullayeva T., Becker A., Freisleben B., Heider D. Mesa: automated assessment of synthetic DNA fragments and simulation of DNA synthesis, storage, sequencing and pcr errors. Bioinformatics. 2020;36:3322–3326. doi: 10.1093/bioinformatics/btaa140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Löchel H.F., Welzel M., Hattab G., Hauschild A.-C., Heider D. Fractal construction of constrained code words for DNA storage systems. Nucleic Acids Res. 2022;50 doi: 10.1093/nar/gkab1209. e30-e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El-Shaikh A., Welzel M., Heider D., Seeger B. High-scale random access on DNA storage systems. NAR Genom Bioinforma. 2022;(4 () doi: 10.1093/nargab/lqab126. lqab126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.M. Dimopoulou, E.G. SanAntonio, M. Antonini, A jpeg-based image coding solution for data storage on DNA, in: 2021 29th European Signal Processing Conference (EUSIPCO), IEEE, 2021, pp. 786–790.
- 20.X. Pic M. Antonini A constrained shannon-fano entropy coder for image storage in synthetic DNA 2022 30th Eur Signal Process Conf (EUSIPCO), IEEE 2022 1367 1371. (pp.).
- 21.Rashtchian C., Makarychev K., Racz M., Ang S., Jevdjic D., Yekhanin S., et al. Clustering billions of reads for DNA data storage. Adv Neural Inf Process Syst. 2017;30 [Google Scholar]
- 22.Tulpan D.C., Hoos H.H., Condon A.E. International Workshop on DNA-Based Computers. Springer; 2002. Stochastic local search algorithms for DNA word design; pp. 229–241. (pp.) [Google Scholar]
- 23.Gratzl S., Lex A., Gehlenborg N., Pfister H., Streit M. Lineup: Visual analysis of multi-attribute rankings. IEEE Trans Vis Comput Graph. 2013;19:2277–2286. doi: 10.1109/TVCG.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong Y., Sun F., Ping Z., Ouyang Q., Qian L. Dna storage: research landscape and future prospects. Natl Sci Rev. 2020;7:1092–1107. doi: 10.1093/nsr/nwaa007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clelland C.T., Risca V., Bancroft C. Hiding messages in DNA microdots. Nature. 1999;399:533–534. doi: 10.1038/21092. [DOI] [PubMed] [Google Scholar]
- 26.Heider D., Barnekow A. Dna-based watermarks using the DNA-crypt algorithm. BMC Bioinforma. 2007;8 doi: 10.1186/1471-2105-8-176. 176-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D. Limbachiya, V. Dhameliya, M. Khakhar, M.K. Gupta, On optimal family of codes for archival DNA storage, in: 2015 Seventh International Workshop on Signal Design and Its Applications in Communications (IWSDA), IEEE, 2015, pp. 123–127.
- 28.Ezekannagha C., Becker A., Heider D., Hattab G. Design considerations for advancing data storage with synthetic DNA for long-term archiving. Mater Today Bio. 2022 doi: 10.1016/j.mtbio.2022.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.R. Gabrys, E. Yaakobi, O. Milenkovic, Codes in the damerau distance for DNA storage, in: 2016 IEEE International Symposium on Information Theory (ISIT), IEEE, 2016, pp. 2644–2648.
- 30.Song W., Cai K., Immink K.A.S. Sequence-subset distance and coding for error control in DNA-based data storage. IEEE Trans Inf Theory. 2020;66:6048–6065. [Google Scholar]
- 31.Kiah H.M., Puleo G.J., Milenkovic O. Codes for DNA sequence profiles. IEEE Trans Inf Theory. 2016;62:3125–3146. doi: 10.1109/TIT.2016.2555321. [DOI] [Google Scholar]
- 32.Kovačević M., Tan V.Y.F. Codes in the space of multisets—coding for permutation channels with impairments. IEEE Trans Inf Theory. 2018;64:5156–5169. doi: 10.1109/TIT.2017.2789292. [DOI] [Google Scholar]
- 33.Lenz A., Siegel P.H., Wachter-Zeh A., Yaakobi E. Coding over sets for DNA storage. IEEE Trans Inf Theory. 2020;66:2331–2351. doi: 10.1109/TIT.2019.2961265. [DOI] [Google Scholar]
- 34.F. Balado, On the shannon capacity of DNA data embedding, in: 2010 IEEE International Conference on Acoustics, Speech and Signal Processing, IEEE, 2010, pp. 1766–1769.
- 35.Balado F. Capacity of DNA data embedding under substitution mutations. IEEE Trans Inf Theory. 2012;59:928–941. [Google Scholar]
- 36.Bystrykh L.V. Generalized DNA barcode design based on hamming codes. PloS One. 2012;7 doi: 10.1371/journal.pone.0036852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sabary O., Yucovich A., Shapira G., Yaakobi E. Reconstruction algorithms for DNA-storage systems. bioRxiv. 2020 doi: 10.1038/s41598-024-51730-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Few S. Show Me the Numbers: Designing Tables and Graphs to Enlighten. second ed. Analytics Press; Oakland, CA, USA: 2012. [Google Scholar]
- 39.Csiszár I., Körner J. Cambridge University Press; 2011. Information Theory: Coding Theorems for Discrete Memoryless Systems. [Google Scholar]
- 40.Setlur V., Stone M.C. A linguistic approach to categorical color assignment for data visualization. IEEE Trans Vis Comput Graph. 2015;22:698–707. doi: 10.1109/TVCG.2015.2467471. [DOI] [PubMed] [Google Scholar]
- 41.Hattab G., Rhyne T.-M., Heider D. Ten simple rules to colorize biological data visualization. PLOS Comput Biol. 2020;16 doi: 10.1371/journal.pcbi.1008259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer M., Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5448. pdb-prot5448. [DOI] [PubMed] [Google Scholar]
- 43.Faircloth B.C., Glenn T.C. Not all sequence tags are created equal: designing and validating sequence identification tags robust to indels. PLoS One. 2012;7 doi: 10.1371/journal.pone.0042543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adey A., Morrison H.G., Xun X., Kitzman J.O., Turner E.H., Stackhouse B., et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11:1–17. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meyer M., Stenzel U., Hofreiter M. Parallel tagged sequencing on the 454 platform. Nat Protoc. 2008;3:267–278. doi: 10.1038/nprot.2007.520. [DOI] [PubMed] [Google Scholar]
- 46.Hamady M., Walker J.J., Harris J.K., Gold N.J., Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fedosejev A. Packt Publishing Ltd; 2015. React. js essentials. [Google Scholar]
- 49.Welzel M., Schwarz P.M., Löchel H.F., Kabdullayeva T., Clemens S., Becker A., et al. DNA-Aeon provides flexible arith-metic coding for constraint adherence and error correction in dna stor-age. Nature Communications. 2023;14:628. doi: 10.1038/s41467-023-36297-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
DNAsmart is implemented as a client-side web application and relies on React to support efficient visualization updates when data is modified [47]. The code and data for DNAsmart is publicly available at https://github.com/sombiri/DNAsmart. The evaluation is shared under its own branch. A deployed prototype of DNAsmart is available at https://dnasmart.mathematik.uni-marburg.de/.