Abstract
Background:
In the United States, genetically admixed populations have the highest asthma prevalence and severe asthma exacerbations rates. This could be explained by environmental factors, but also by genetic variants that exert ethnic-specific effects. However, no admixture mapping has been performed for severe asthma exacerbations.
Objective:
We sought to identify genetic variants associated with severe asthma exacerbations in Hispanic/Latino subgroups by means of admixture mapping analyses and fine-mapping, and to assess their transferability to other populations and potential functional roles.
Methods:
We performed an admixture mapping in 1,124 Puerto Rican and 625 Mexican American children with asthma. Fine-mapping of the significant peaks was performed via allelic testing of common and rare variants. We performed replication across Hispanic/Latino subgroups, and the transferability to non-Hispanic/Latino populations was assessed in 1,001 African Americans, 1,250 Singaporeans, and 941 Europeans with asthma. The effects of the variants on gene expression and DNA methylation from whole blood were also evaluated in participants with asthma and in silico with data obtained through public databases.
Results:
Genome-wide significant associations of Indigenous American ancestry with severe asthma exacerbations were found at 5q32 in Mexican Americans as well as at 13q13-q13.2 and 3p13 in Puerto Ricans. The SNP rs1144986 (C5orf46) showed consistent effects for severe asthma exacerbations across Hispanic/Latino subgroups, but it was not validated in non-Hispanics/Latinos. This SNP was associated with DPYSL3 DNA methylation and SCGB3A2 gene expression levels.
Conclusions:
Admixture mapping study of asthma exacerbations revealed a novel locus that exhibited Hispanic/Latino-specific effects and regulated DPYSL3 and SCGB3A2.
Keywords: Admixture mapping, Hispanics/Latinos, ethnic subgroups, asthma, exacerbations, ancestry
INTRODUCTION
Asthma is a chronic inflammatory disease that shows remarkable heterogeneity by ethnicity, and may result from the complex interplay between environmental, behavioural, and genetic factors.[1] In fact, genetic ancestry is a critical factor for asthma susceptibility.[2] In the United States (U.S.), asthma prevalence is the highest in Puerto Ricans (14.0%), followed by African Americans (10.7%), and the lowest in Mexican Americans (5.4%) and Asian Americans (4.5%).[3] However, racial/ethnic disparities are not limited to asthma prevalence, since Puerto Ricans and African Americans also show the highest rates of asthma-related emergency room or urgent care visits.[4] Asthma exacerbations, defined as those events requiring urgent asthma care, hospitalizations, or administration of systemic corticosteroids, contribute to the large healthcare expenditures of asthma [5] and asthma-related deaths.[6] Moreover, exacerbations impair individuals’ quality of life [7] and long-term lung function.[8] Although environmental exposures, such as viral infections or air pollution, are known triggers for asthma exacerbations, several genetic loci for asthma exacerbations have been successfully uncovered through genome-wide association studies (GWAS) in Hispanic/Latino populations.[9–11]
Hispanics/Latinos are American admixed individuals with variable influences from Indigenous American, African and European populations (i.e., African ancestry is higher in Puerto Ricans, whereas Native American ancestry is higher in Mexicans).[12] In a scenario where a trait shows differential rates in admixed individuals, admixture mapping in the most affected populations can identify genetic variants associated with that trait. In fact, mapping causal variants by long-range linkage disequilibrium decreases multiple testing burden and can be coupled with functional approaches to prioritize genetic variants that could have population-specific effects.
Given the disparities in asthma prevalence and outcomes, multiple studies have leveraged locus-specific genetic ancestry to identify genetic variants associated with asthma-related outcomes.[12–15] However, no admixture mapping of severe asthma exacerbations has been performed to date. We hypothesized that genetic variation might partially explain the ethnic differences in severe asthma exacerbations among Hispanics/Latinos. Hence, we aimed to identify novel genetic variants associated with severe asthma exacerbations by admixture mapping in Hispanic/Latino children and youth with asthma. We then sought to evaluate the potential functional consequences of those genetic variants and attempted to validate them in non-Hispanic/Latino populations.
METHODS
A full description of the methods with a graphical summary of the workflow (Figure E1) can be found in the Supplemental material.
Discovery population
We analysed genotypes from Puerto Ricans (n=1,124) and Mexican Americans (n=625) from the study of Genes-Environment and Admixture in Latino Americans (GALA II), a case-control study of asthma in Hispanics/Latinos aged 8–21 years old recruited in the US and Puerto Rico between 2006 and 2014. GALA II was approved by the Human Research Protection Program Institutional Review Board of the University of California, San Francisco (US), and participants/parents provided written assent/consent, respectively. Participants were eligible if they reported four Hispanic/Latino grandparents. Asthma cases were diagnosed by a physician, had asthma symptoms, and reported use of controller or rescue medication in the two years preceding enrolment.[16] Severe asthma exacerbations were defined as the presence of any of the following events in the past 12 months: use of oral corticosteroids, asthma-related hospitalizations, or unscheduled asthma care.[17] Treatment category, defined following the Expert Panel Report 3 (EPR-3) guidelines for the diagnosis and management of asthma,[18] was used as a proxy for asthma severity. Three levels were defined based on the use of short beta-agonists (step 1); one inhaled corticosteroid, leukotriene inhibitor, or theophylline tablet (step 2); or more than one or a combination of an inhaled corticosteroid, leukotriene inhibitor, or theophylline tablet or a combination of inhaled corticosteroids and long-beta agonists (step 3).
Whole-genome sequencing data generation and processing
Whole-genome sequencing (WGS) was conducted on a HiSeq X system (Illumina, San Diego, CA). Data generation and quality control (QC) are described in the Supplemental material. Genotypes used in this study were based on TOPMed freeze 8 data.
Local and global ancestry estimation
The ancestral reference panel for ancestry estimation included 90 HapMap phase II Europeans (CEU), 90 Africans (YRI), and 71 Native Americans (NAM).[19] Local ancestry was estimated with RFMIX v2[20] based on biallelic single nucleotide polymorphisms (SNPs) that passed the quality control procedures of the WGS pipeline and that overlapped with the reference panel (402,838 SNPs). Global ancestry was calculated as the genome-wide average of local ancestry.
Admixture mapping in Hispanics/Latinos
The association of asthma exacerbations and the number of copies of African, European, and Indigenous American local ancestry (0, 1, or 2 copies) at each SNP analysed was tested separately in Hispanic/Latino subgroups. This was performed through logistic regression models with correction for global ancestry, age, sex, and treatment category (as a proxy for asthma severity). To account for multiple comparison testing, the final effective number of tests was defined as the sum of the two largest effective numbers of ancestry blocks, consistent with Horimoto et al. (2021),[21] and as detailed in the Supplemental material.
Fine-mapping and replication
Fine-mapping of variants with minor allele frequency (MAF)≥1% within the genome-wide significant admixture mapping peaks was performed with correction for the same covariates used in the admixture mapping. To correct for multiple comparison, an adjusted threshold of significance was defined as p =<0.05/number of effective tests. To evaluate whether independent SNPs account for the admixture mapping signal, we performed step-wise conditional regression analyses adjusting the association of local ancestry by the allele dosage of associated SNPs. Interaction of local ancestry on the association of the SNPs with exacerbations was evaluated in a regression model including the same covariates as in the main model with an interaction term SNP × local ancestry.
Peak-wise independent significant SNPs identified in Mexican Americans were replicated in Puerto Ricans, and those identified in Puerto Ricans were assessed in Mexican Americans. Additional validation was sought in non-Hispanic/Latino participants with asthma, including 1,101 African Americans from the Study of African Americans, Asthma, Genes, and Environments (SAGE), 1,250 Singaporean Chinese from the Singapore Cross Sectional Genetic Epidemiology Study (SCSGES), and 941 Europeans from several studies: the MEchanism underlying the Genesis and evolution of Asthma (MEGA) study, the Genomics and Metagenomics of Asthma Severity (GEMAS) study, the Children, Allergy, Milieu, Stockholm, Epidemiology (BAMSE) study, and the Infancia and Medio Ambiente (INMA) study. Details of studies included in the replication are described in the Supplemental material and Table E1.
Fine-mapping of rare variants (MAF<1%) was conducted using the SKAT-O test[22] analysing 1-kilobase (kb) sliding windows with 500-bp increments within the admixture mapping peak limits. The threshold of significance was defined based on Storey q[23]<0.05.
Methylation profiling and quality control
DNA methylation from whole blood was profiled using the Illumina Infinium HumanMethylation450 BeadChip or the Infinium EPIC BeadChip arrays. QC is detailed in the Supplemental material. Briefly, low-quality probes and samples, outliers of DNA methylation, and samples with sex discordance or mixed genotype distributions on the control SNP probes were excluded. Standard background correction, dye-bias correction, inter-array normalization, and probe-type bias adjustment were conducted, and beta values were transformed to M-values.
RNA sample processing, sequencing, and quality control
Total RNA was quantified using the Quant-iT™ RiboGreen® RNA Assay Kit and normalized for library preparation with the Illumina TruSeq™ Stranded mRNA Sample Preparation Kit. Libraries were sequenced according to the manufacturer’s protocols using the HiSeq 4000 system (Illumina). Sample processing and quality control are detailed in the Supplemental material. Expression values were normalized across samples using an inverse normal transformation.
Functional assessment of associated SNPs
Quantitative trait loci (QTL) analyses in data from whole blood were conducted separately in Mexican American and Puerto Rican participants with asthma from GALA II for those CpG sites or genes with a transcription start site located within 1 Mb of the significant SNPs. Linear regression models were adjusted for exacerbations status, age, sex, treatment category, and genetic ancestry. Cis-expression QTL (eQTL) analyses were adjusted by the top 60 PEER factors[24], and cis-methylation QTL (meQTL) models by cell-type heterogeneity and methylation batch. The population sub-group results were then meta-analysed. In silico evidence of functional effects of the variant on gene expression and DNA methylation was assessed using the Genotype-Tissue Expression (GTEx) v8 Portal,[25] PhenoScanner v2,[26] Open Targets Genetics,[27] and the Genetics of DNA Methylation Consortium (GoDMC).[28] Chromatin interactions were determined using CHiCP.[29] Gene expression was inspected using the GTEx Portal[25] and REALGAR.[30]
RESULTS
Discovery sample characteristics
A total of 820 out of 1,124 Puerto Ricans had exacerbations in the last 12 months (72.9%), while 223 out of 625 Mexican Americans reported exacerbations for the same period (35.7%). Most of the exacerbators among Puerto Ricans and non-exacerbators among both ethnic subgroups were in the lowest treatment category, while the distribution in Mexican exacerbators was more balanced. Moreover, no significant differences in global ancestry were detected between exacerbators and non-exacerbators (p>0.05) (Table 1).
Table 1.
Characteristics of the asthma participants recruited between 2006 and 2014 for the Genes-Environments and Admixture in Latino Asthmatics (GALA II) study and included in the current study
| Puerto Ricans (N=1,124) | Mexican Americans (N=625) | |||||
|---|---|---|---|---|---|---|
| Exacerbators | Non-exacerbators | p-value | Exacerbators | Non-exacerbators | p-value | |
|
| ||||||
| Number of individuals | 820 | 304 | 223 | 402 | ||
| Sex (% male) | 443 (54.0) | 160 (52.6) | 7.27×10−1 | 129 (57.8) | 226 (56.2) | 7.57×10−1 |
| Age, mean+SD (years) | 12.2±3.2 | 13.8±3.6 | 1.83×10−11 | 12.2±3.2 | 13.2±3.4 | 6.16×10−4 |
| European ancestry, mean±SD (%) | 62.6±10.7 | 63.2±10.5 | 5.45×10−1 | 38.6±14.7 | 40.0±13.5 | 3.03×10−1 |
| African ancestry, mean±SD (%) | 23.5±11.7 | 26.7±11.4 | 1.57×10−1 | 4.6±2.2 | 5.0±3.3 | 3.03×10−1 |
| Indigenous American ancestry, mean±SD (%) | 13.8±3.93 | 14.3±4.8 | 1.16×10−1 | 56.7±15.2 | 55.0±14.2 | 2.00×10−1 |
| Treatment category, n (%) | ||||||
| Step 1 | 353 (43.0) | 227 (74.7) | 8.27×10−21 | 53 (23.7) | 185 (46.0) | 6.57×10−8 |
| Step 2 | 239 (29.1) | 41 (13.5) | 1.07×10−7 | 86 (38.6) | 131 (32.6) | 1.57×10−1 |
| Step 3 | 228 (27.9) | 36 (11.8) | 3.23×10−8 | 84 (37.7) | 86 (21.4) | 1.82×10−5 |
Abbreviation: SD: Standard deviation. For continuous variables, the mean and standard deviation are displayed and Mann-Whitney-Wilcoxon test was applied for the comparison of exacerbators versus non-exacerbators. For categorical variables, the number and proportion of subjects in each category are shown and a χ2 test was applied for the comparison of exacerbators versus non-exacerbators.
Stratified admixture mapping
Genome-wide significant associations were found between Indigenous American ancestry and severe asthma exacerbations at chromosomes 3p13 and 13q13.2 in Puerto Ricans (Table 2, Figure 1A, Figure E2 in the Supplemental material) (p≤4.33×10−5, accounting for 1,154 ancestry blocks). The most significant association at chromosome 3p13 corresponded to rs4677148, where Indigenous American ancestry was associated with lower odds of exacerbations (odds ratio [OR]: 0.57, 95% confidence interval [CI]: 0.44–0.74, p=2.55×10−5), while European ancestry was associated with a higher odds of having exacerbations (OR: 1.40, 95% CI: 01.16–1.94, p = 1.82×10−3) but no significant association with African ancestry was detected (p>0.05). The strongest association with the odds of severe exacerbations at 13q13.2 was located at rs10514839 (OR: 2.18, 95% CI: 1.55–3.06, p=6.56 × 10−6), whereby European or African ancestry showed no association.
Table 2.
Significant admixture mapping peaks identified in Mexican Americans and Puerto Ricans from GALA II (2006–2014).
| Ethnicity/Ancestry | Chr. band | Start (Bp)a | End (Bp)a | Peak size (Kb) | rsIDb | OR (95%CI) | p-value |
|---|---|---|---|---|---|---|---|
| Puerto Rican/ Indigenous American | 3p13 | 72283579 | 72585953 | 302 | rs4677148 | 0.57 (0.44–0.74) | 2.55×10−5 |
| 13q13.2 | 34471415 | 35919337 | 1,448 | rs10514839 | 2.18 (1.55–3.06) | 6.56×10−6 | |
| Mexican American/ Indigenous American | 5q32 | 147809331 | 147911637 | 102 | rs10477350 | 1.73 (1.34–2.24) | 3.23×10−5 |
Coordinates are referred to the human reference genome assembly GRCh38.
Genetic variant with the minimum association p-value within the peak.
Abbreviations: Bp: Base pairs; Chr: Chromosome; CI: confidence interval; Kb: Kilobases; OR: Odds ratio; rsID: reference single nucleotide polymorphism identifier; iSNPs: independent single nucleotide polymorphisms.
Figure 1. Manhattan plot for the admixture mapping results of severe exacerbations in participants with asthma from GALA II (2006–2014). A) Association results for Indigenous American ancestry in Puerto Ricans B) Association results for Indigenous American ancestry in Mexican Americans.
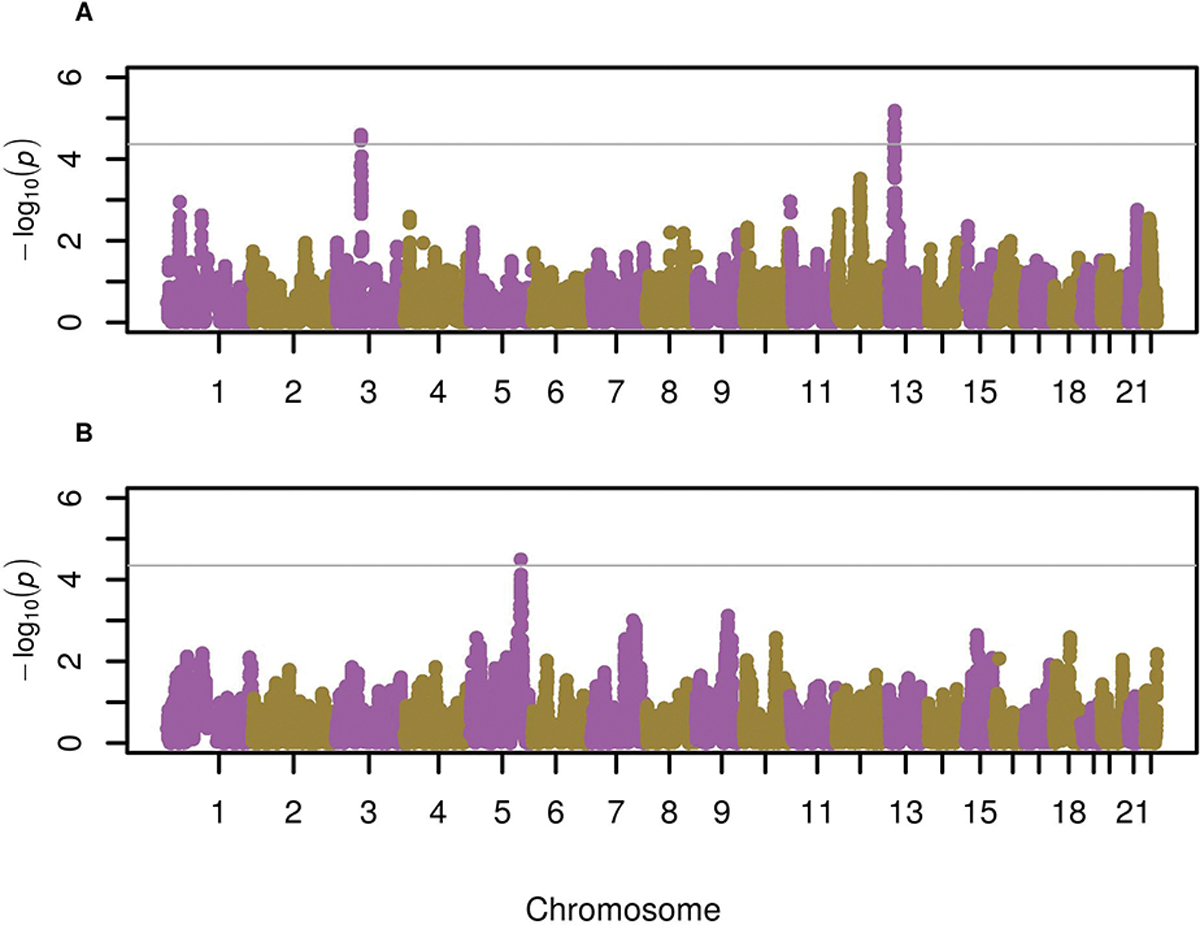
The association with local ancestry is represented as −log10 p-value on the y-axis along the chromosomes (x-axis). The threshold for significance (grey line) was defined as p<0.05/sum of the two largest values for the effective number of ancestry blocks: p≤4.33×10−5 in Puerto Ricans, accounting for 1,154 ancestry blocks (African: 560, European: 594, Native American: 432); and p≤4.51×10−5 in Mexican Americans, accounting for 1,107 ancestry blocks (African: 637, European: 470, Native American: 431).
Among Mexican Americans, we identified one admixture mapping peak at chromosome 5q32 whereby Indigenous American ancestry was genome-wide significantly associated with the odds of severe exacerbations (p≤4.51×10−5, accounting for 1,107 ancestry blocks) (Table 2, Figure 1B, Figure E3). The most significant association in the region was located at rs10477350, where Indigenous American was associated with greater odds of exacerbations (OR: 1.73, 95% CI: 1.34–2.24, p=3.23×10−5) and European ancestry with lower odds (OR: 0.60, 95% CI: 0.46–0.78, p=1.46×10−4).
No genome-wide significant associations were found between severe asthma exacerbations and African or European ancestry in both ethnic subgroups (Figure E4). Moreover, a meta-analysis of both ethnic subgroups did not yield any additional admixture mapping peak at genome-wide significant level.
Fine-mapping analyses
We next performed fine-mapping of common variants via allelic association testing (Table 3, Table E2). We found a region in the admixture mapping peak at 5q32 where genetic variation was significantly associated with severe asthma exacerbations in Mexican Americans after accounting for the number of variants tested within the peak. Specifically, four independent variants were associated with severe exacerbations (Figure 2A). The minor alleles of SNPs rs1144986 (C5orf46) and rs35439318 (SCGB3A2/CTC-327F10.1) were associated with lower odds of having exacerbations: OR for G allele: 0.43, 95% CI: 0.28–0.66, p=9.45×10−5 and OR for C allele: 0.51, 95% CI: 0.34–0.74, p=5.26×10−4, respectively. Moreover, the A alleles of rs7704889 (C5orf46/EEF1GP2) and rs10035432 (SCGB3A2/CTC-327F10.1) were associated with higher odds of exacerbations: OR: 1.58, 95% CI: 1.23–2.11, p=4.00×10−4 and OR: 1.61, 95% CI: 1.23–2.11, p=4.82×10−4, respectively. No significant associations were detected in any of the regions in Puerto Ricans.
Table 3.
Association results of independent genetic variants at chromosome 5 that were peak-wise significantly associated with severe asthma exacerbations in Hispanics/Latinos from GALA II (2006–2014)
| Mexican Americans | Puerto Ricans | Meta-analysis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||
| rsID | Position (Bp)a | A1/A2 | Closest gene | EAF | OR (95% CI) | p-value | EAF | OR (95% CI) | p-value | OR (95%CI) | p-value |
|
| |||||||||||
| rs1144986 | 147883415 | G/A | C5orf46 | 0.13 | 0.43 (0.28–0.66) | 9.45×10−5 | 0.22 | 0.79 (0.62–1.00) | 4.94×10−2 | 0.60 (0.33–1.08) | 1.61×10−4 b |
| rs7704889 | 147911637 | A/G | C5orf46/EEF1GP2 | 0.54 | 1.58 (1.23–2.03) | 4.00×10−4 | 0.42 | 0.97 (0.79–1.20) | 7.94×10−1 | 1.23 (0.77–1.98) | 6.57×10−3 b |
| rs10035432 | 147855193 | A/G | SCGB3A2/CTC-327F10.1 | 0.26 | 1.61 (1.23–2.11) | 4.82×10−4 | 0.21 | 1.02 (0.81–1.29) | 8.72×10−1 | 1.28 (0.81–2.00) | 5.93×10−3 b |
| rs35439318 | 147862111 | C/T | SCGB3A2/CTC-327F10.1 | 0.15 | 0.51 (0.34–0.74) | 5.26×10−4 | 0.22 | 0.83 (0.65–1.05) | 1.27×10−1 | 0.66 (0.41–1.08) | 1.45×10−3 b |
Coordinates are referred to the human reference genome assembly GRCh38
A random effect model was applied since heterogeneity was found between populations (Cochran’s Q p-value<0.05). Association p-values<0.05 are shown in bold.
Abbreviations: A1: Effect allele; A2: Non-effect allele; Bp: Base pairs; CI: Confidence interval; EAF: Effect allele frequency; OR: Odds ratio; rsID: reference single nucleotide polymorphism identifier.
Figure 2. Locus zoom for the fine-mapping results displaying the region around the most significant signals at 5q32 in Mexican Americans with asthma from GALA II (2006–2014).

The statistical significance of association results (−log10 p-value) is represented for each SNP as a dot (left y-axis) by chromosome position (x-axis). SNPs are color-coded to show their linkage disequilibrium (LD) with the most significant SNP based on the pairwise r2 values from Mexican Americans. The peak-wise significance threshold is represented as a dashed blue line.
The step-wise addition of the independent SNPs as covariates to the regression model that tested the association of the lead ancestry SNP with severe asthma exacerbations revealed that the SNPs accounted for the 5q32 peak since the association of local ancestry became non significant (Table E3). Moreover, the association of the SNPs revealed by fine-mapping and severe exacerbations remained significant after additional adjustment by local ancestry, and no significant interaction with local ancestry was found (Table E4). To assess the robustness of the associations to socio-economic and clinical factors, we performed sensitivity analyses with adjustment by second-hand smoking exposure, insurance status, income quartile, maternal education, and obesity, confirming that the effects were not explained by these factors (Table E5).
We next assessed whether the independent variant replicated among Hispanics/Latino subgroups. In Puerto Ricans, the SNP rs1144986 (OR for G allele: 0.79, 95% CI: 0.62–1.00, p=4.94×10−2) had consistent effects compared with Mexican Americans. In the meta-analysis of Hispanics/Latinos, the G allele of rs1144986 showed a protective effect over exacerbations (OR: 0.60, 95% CI: 0.33–1.08, p=1.61×10−4, Cochran’s Q p=1.40×10−2). Moreover, an analysis in 1,462 Mexican Americans and 2,346 Puerto Ricans revealed that this SNP was not associated with the underlying asthma susceptibility at p<0.05 (Table E6).
The SNP that showed significant and consistent effects in Hispanic/Latino subgroups (rs1144986) was assessed for validation in non-Hispanic/Latino populations. However, no association was found with severe exacerbations (p>0.05) in Singaporean Chinese, African Americans, or Spanish individuals, or with less severe exacerbations, including school absences in 421 Swedish children or wheezing in the last year in 100 Spanish individuals (Table E7).
We next tested the association of rare variants with severe asthma exacerbations. No significant associations were found within the admixture mapping peaks identified in either Puerto Ricans or Mexican Americans (q>0.05).
Assessment of functional effects of the genetic variants
The effect of rs1144986 on DNA methylation in whole blood from Hispanic/Latino participants with asthma was evaluated for 196 CpG sites located within ±1 Megabase (Mb) of the SNP. The SNP rs1144986 was significantly associated with three probes annotated to DPYSL3, being cg04833034, at 57.4 kb of DPYSL3, the most significant CpG (p=5.17×10−6, q=3.41×10−3). Moreover, associations for CpGs at PPP2R2B and C5orf46 were also observed) (Table 4). Notably, the associations with cg24686270 and cg10930901 were replicated in the GoDMC results[28] (p=1.1×10−139 and 7.1×10−29, respectively). DPYSL3 expression in bronchial epithelial cells was found to be increased in severe asthma in publicly available gene expression datasets of asthma participants (Figure E5). Additionally, the meQTL rs1144986 showed evidence of chromatin interaction with C5orf46 in lymphoblastoid cells (CHiCAGO score=11.7) and association with H3K4me1 histone marks in blood (Table E4). Notably, none of the significant CpGs regulated the expression of nearby genes in whole blood in a subset of 126 Puerto Ricans and 40 Mexicans with DNA methylation and gene expression data available at a false discovery rate-adjusted p<0.05 (Table E8).
Table 4.
Significant results from the QTL mapping in Hispanics/Latinos from GALA II (2006–2014)
| Puerto Ricans | Mexican Americans | Meta-analysisa | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||||
| SNP - analysis | Target | Positionb | Gene | Coef (SE) | p-value | PQ | Coef (SE) | p-value | PQ | Coef (SE) | p-value | Q |
|
| ||||||||||||
| rs1144986 -meQTL | cg04833034 | 147452619 | DPYSL3 | 0.18 (0.04) | 1.58×10−5 | 0.66 | 0.11 (0.07) | 1.28×10−1 | NAc | 0.16 (0.04) | 6.36×10−6 | 8.13×10−4 |
|
| ||||||||||||
| cg09639133 | 147451646 | DPYSL3 | 0.22 (0.05) | 5.10×10−5 | 0.53 | 0.14 (0.07) | 5.25×10−2 | NAc | 0.19 (0.04) | 1.13×10−5 | 8.13×10−4 | |
|
| ||||||||||||
| cg10930901 | 147505871 | DPYSL3 | −0.18 (0.05) | 7.96×10−5 | 0.26 | −0.05 (0.07) | 4.56×10−1 | NAc | −0.14 (0.04) | 1.84×10−4 | 8.79×10−3 | |
|
| ||||||||||||
| cg01112778 | 147081424 | PPP2R2B | 0.14 (0.05) | 4.41×10−3 | 0.53 | 0.17 (0.09) | 5.29×10−2 | NAc | 0.15 (0.04) | 5.56×10−4 | 2.03×10−2 | |
|
| ||||||||||||
| cg24686270 | 147901750 | C5orf46 | 0.10 (0.03) | 4.25×10−3 | 0.47 | 0.10 (0.06) | 1.21×10−1 | NAc | 0.10 (0.04) | 1.04×10−4 | 2.98×10−2 | |
|
| ||||||||||||
| rs1144986 - eQTL | ENSG00000164265 | 147870682 | SCGB3A2 | 0.68 (0.08) | 1.01×10−15 | NA | 0.47 (0.10) | 1.85×10−6 | NA | 0.60 (0.06) | 6.12×10−21 | 3.06×10−20 |
A fixed effect model was applied since no heterogeneity was found between populations (Cochran’s Q p-value>0.05).
Chromosomal positions referred to the genome assembly GRCh38. For CpG sites, the location of the CpG is stated. For genes, the location of the transcription start site is shown.
The SNP did not meet the genotype inclusion criteria for the individuals profiled with the EPIC array.
The CpG was not available in the 450K array.
Abbreviations PQ: Cochran’s Q-test p-value; Coef: Regression coefficient estimate; NA: Not available/applicable; Q: Q-value; SE: Standard error of the regression coefficient estimate.
The effect of rs1144986 over the expression of genes with transcription start site within ±1Mb was assessed. From the 5 SNP-gene pairs, the SNP rs1144986 showed significant associations with gene expression of SCGB3A2 at q<0.05 in whole blood (p=6.62×10−22; q=6.62×10−20), and the association was also validated in blood according to the GTEx (p=5.1×10−10),[25] PhenoScanner and Open Targets Genetics (minimum p=3.3×10−310).[26,27]
DISCUSSION
Admixture mapping analysis of severe asthma exacerbations in Hispanic/Latino children with asthma identified several genomic regions in which local Indigenous American or European ancestry was associated with severe asthma exacerbations in Puerto Ricans and Mexican Americans. Although fine-mapping in Puerto Ricans revealed no significant variants, we found four independent SNPs explaining the admixture mapping peak for Indigenous American ancestry at chromosome 5q32 in Mexican Americans, including rs1144986 (C5orf46), which showed significant consistent effects among Hispanic/Latino subgroups. Interestingly, none of the four loci were associated with asthma exacerbations in non-Hispanic/Latino populations, suggesting ethnic-specific effects shared among Hispanic/Latino subgroups. This could be explained, at least partially, due to differences in allele frequency and ethnic background. For example, at 5q32, Native American ancestry was associated with higher odds of exacerbations in Mexican Americans, and the risk allele (A) of rs1144986 was more common among Native Americans (frequency=98%) and Mexicans (frequency=87%) than in Puerto Ricans (frequency=78%) or Europeans (frequency=67%) (Tables E4, E6). The fact that Indigenous American ancestry is around 2% in African Americans[31] could contribute to the lack of validation of these genetic variants despite the higher risk allele frequency (89%) observed.
An analysis of functional effects revealed that rs1144986 altered DNA methylation of several nearby genes, including DPYSL3. The protein DPYSL3 is involved in cytoskeleton remodeling by participating in the signaling of class 3 semaphorins. While the role of DPYSL3 in asthma is unclear, it is co-expressed with genes involved in airway type 2 inflammation[32]. However, DPYSL3 expression in whole blood data from Hispanics/Latinos could not be evaluated due to low levels or lack of expression, consistently with the GTEx data.[25] Additionally, the protective allele of rs1144986 increased SCGB3A2 expression in blood (Figure 3), although SCGB3A2 is predominantly expressed in the lung.[25] SCGB3A2 is involved in lung development[33] and has shown an anti-inflammatory role in mice.[34–37] More recently, the role of secretoglobins as airway immunoregulators is gaining interest.[38] Lung expression levels of pro-inflammatory cytokines, IL-4, IL-5, and IL-13, are lower in ovalbumin-induced mice pre-treated with SCGB3A2 compared with those not treated with it.[36,37] Plasma SCGB3A2 levels are decreased in severe asthma,[39] and SCGB3A2 polymorphisms have been associated with asthma susceptibility.[40] In human bronchial epithelial cells, SCGB3A2 decreased airway inflammation inhibiting ERK and JNK activation.[41] In lung cancer, SCGB3A2 induced pyroptotic cell death. Interestingly, participants with higher SCGB3A2 gene expression manifested higher survival rates.[42] Moreover, SCGB3A2 shows anti-fibrotic activity through increased expression of STAT1 phosphorylation and SMAD7 expression,[43,44] hence, decreasing SMAD2/3 phosphorylation, which attenuates the TGFβ signaling,[44] a key pathway implicated in airway remodeling,[45] allergic airway inflammation,[46] and drug response in asthma.[47] Taken together, this evidence suggests that these genes may be involved in susceptibility to asthma exacerbations and merit further exploration to understand their specific role.
Figure 3. Gene expression levels of SCGB3A2 in whole blood by genotype at rs1144986 in participants with asthma from GALA II (2006–2014). A) Mexican Americans and B) Puerto Ricans.

Our study has several strengths and limitations. We focused on minority populations at high risk of asthma that have undergone distinct historical processes and show differential asthma exacerbations rates. Although we identified several regions in which local ancestry was associated with severe exacerbations, analyzing two different Hispanic/Latino subgroups may have hindered our power to detect associations. Specifically, this could have affected the identification of SNPs in the regions where Indigenous American ancestry was associated with exacerbations in Puerto Ricans, as this ancestral population has a smaller contribution to their genetic background compared with Mexican Americans. Our study was especially underpowered to detect an association of rare variants. Specifically, assuming 30% of causal risk variants, we were only powered at >80% to detect rare variant signals with a maximum OR≥18 in Puerto Ricans, likely due to the larger sample size compared with Mexicans. Moreover, although genetic signals identified by fine-mapping exceeded the respective peak-wise threshold of significance (Table 2, Table E2), none would have exceeded a stringent correction for the total number of independent variants tested in the discovery stage (p=0.05/1,977SNPs=2.53×10−5). Furthermore, we also sought to determine if these genetic variants may exert the same effects in non-Hispanic/Latino populations. The fact that none of the variants was validated in non-Hispanics/Latinos suggests that they could exhibit ethnic-specific effects. We attempted to overcome these limitations by performing functional analyses to reveal the effects of the variant identified on whole blood gene expression and DNA methylation in Hispanics/Latinos. Although whole blood epigenetic and transcriptional signals may capture the inflammatory component related to specific blood cell types, future studies in airway tissues should be required due to the modest cross-tissue replication described in asthma.[48] However, given that we analysed a mixed cell type tissue, our analyses were adjusted by blood cell-type heterogeneity to overcome this limitation.[48] Despite this, other epigenetic mechanisms known to be involved in asthma (e.g., histone modifications or miRNAs[49]) could also underlie the functional role of the genetic variation. Moreover, chromatin interactions were evaluated in silico using a publicly available database due to the lack of experimental data from Hispanic/Latino participants with asthma. Therefore, the role of these loci in asthma exacerbations should be explored in future studies using cell lines from asthma participants or animal models of this disease.
In summary, we leveraged local ancestry to identify genomic regions that contribute to severe asthma exacerbations in Hispanic/Latinos. Indigenous American ancestry was associated with asthma exacerbation risk at 5q32-q33.1 and novel association of a genetic variant with severe asthma exacerbations with a potential population-specific effect were uncovered. Moreover, these variants had functional effects on SCGB3A2 gene expression and DPYSL3 DNA methylation, two genes that are plausibly implicated in severe exacerbations.
Supplementary Material
KEY MESSAGE.
What is already known on this topic – Admixed minorities in the United States, including Hispanic/Latino subgroups, show disproportionate rates of asthma and asthma exacerbations compared with European-descent populations, but no study has assessed the contribution of genetic variation to asthma exacerbations by admixture mapping.
What this study adds – Admixture mapping analysis of severe asthma exacerbations in Hispanics/Latinos revealed a functional genetic variant with ethnic-specific effect over two biologically plausibly genes implicated in this trait (DPYSL3 and SCGB3A2).
How this study might affect research, practice or policy – Our findings have prioritized novel gene targets for future research in asthma.
ACKNOWLEDGEMENTS
We acknowledge the participants, families, recruiters, health care providers and community clinics for their participation and the contribution of the high-performance compute cluster Wynton HPC underlying UCSF’s Research Computing Capability to the results of this research. We also thank Sandra Salazar for her support as GALA II and SAGE study coordinator and the New York Genome Center investigators and teams for whole-genome sequencing sample preparation, quality control, data generation, data processing, and initial joint genotyping. We gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed. We also gratefully acknowledge the contributions of the investigators of the NHLBI TOPMed Consortium (https://www.nhlbiwgs.org/topmed-banner-authorship). The genotyping of the GEMAS and MEGA studies was performed at the Spanish National Cancer Research Centre, in the Human Genotyping lab, a member of CeGen, PRB3, and is supported by grant PT17/0019, of the PE I+D+i 2013–2016, funded by ISCIII and ERDF. ISGlobal acknowledges support from the Spanish Ministry of Science and Innovation through the “Centro de Excelencia Severo Ochoa 2019–2023” Program (CEX2018-000806-S), and support from the Generalitat de Catalunya through the CERCA Program.
FUNDING
This work was funded by the Spanish Ministry of Science and Innovation MCIN/AEI/10.13039/501100011033 and the European Regional Development Fund “ERDF A way of making Europe” by the European Union grant [SAF2017-83417R], and by MCIN/AEI/10.13039/501100011033 [grant PID2020-116274RB-I00]. Whole-genome sequencing for the TOPMed (Trans-Omics in Precision Medicine) program was supported by the NHLBI. Whole-genome sequencing for GALA II (NHLBI TOPMed: Genes-environments and Admixture in Latino Americans) Study (phs000920) was performed at the New York Genome Center [3R01HL117004-01S3]. Centralized read mapping and genotype calling, along with variant quality metrics and filtering, were provided by the TOPMed Informatics Research Center [3R01HL117626-02S1]. Phenotype harmonization, data management, sample-identity quality control, and general study coordination were provided by the TOPMed Data Coordinating Center [3R01HL120393-02S1]. WGS of part of GALA II was performed by New York Genome Center under The Centers for Common Disease Genomics of the Genome Sequencing Program (GSP) Grant [UM1 HG008901]. The GSP Coordinating Center [U24 HG008956] contributed to cross-program scientific initiatives and provided logistical and general study coordination. GSP is funded by the National Human Genome Research Institute, the National Heart, Lung, and Blood Institute, and the National Eye Institute [NA]. This work was also supported by the Sandler Family Foundation[NA], the American Asthma Foundation [NA], the RWJF Amos Medical Faculty Development Program [NA], Harry Wm. and Diana V. Hind Distinguished Professor in Pharmaceutical Sciences II [NA], the National Heart, Lung, and Blood Institute of the National Institutes of Health [R01HL117004, R01HL128439, R01HL135156, X01HL134589, R01HL155024, R01AI079139, R01HL141845, R01HL118267 and R01DK113003], the National Institute of Health and Environmental Health Sciences [R01ES015794 and R21ES24844], the National Institute on Minority Health and Health Disparities [R01MD010443 and R56MD013312], the Tobacco-Related Disease Research Program [Award Number 24RT-0025 and 27IR-0030]. The BAMSE study was funded by the Swedish Heart-Lung Foundation and the Swedish Research Council and Region Stockholm (ALF project and database maintenance). The INMA study was funded by the Instituto de Salud Carlos III [Red INMA G03/176, CB06/02/0041], Spanish Ministry of Health [FIS-FEDER PI16/1288, FIS-FEDER PI19/1338; Miguel Servet FEDER 15/0025 and 20/0006], Generalitat Valenciana [BEST/2020/059]. This work was also partially funded by Fundación Canaria Instituto de Investigación Sanitaria de Canarias [PIFIISC19/17]. JV was funded by ISCIII and the European Regional Development Fund “ERDF A way of making Europe” by the European Union [PI16/00049 and PI19/00141]. MP-Y was funded by the Ramón y Cajal Program [RYC-2015-17205] by MCIN/AEI/10.13039/501100011033 and by the European Social Fund “ESF Investing in your future” and by the EAACI Allergopharma Award 2021. JV and M.P.-Y were funded Instituto de Salud Carlos III (ISCIII) [CB06/06/1088]. EH-L was supported by a fellowship awarded by MCIN/AEI/10.13039/501100011033 and by “ESF Investing in your future” [PRE2018-083837]. JP-G was supported by a fellowship awarded by the Spanish Ministry of Universities [FPU19/02175].
ROLE OF THE FUNDING SOURCE
The funders of the study had no role in study design, data collection, data analysis, data interpretation, writing of the report, or decision to submit the report for publication.
ABBREVIATIONS
- 1KGP
1000 Genomes project
- 95% CI
95% confidence interval
- CEU
Utah residents with Northern and Western European ancestry
- eQTL
Expression quantitative trait loci
- EPR-3
Expert Panel Report 3
- FDR
False Discovery Rate
- GALA II
Genes-Environment and Admixture in Latino Americans II study
- GEMAS
Genomics and Metagenomics of Asthma Severity
- GTEx
Genotype-Tissue Expression
- GWAS
Genome-wide association study
- Kb
Kilobase
- MAF
Minor allele frequency
- Mb
Megabase
- MCMC
Markov Chain Monte Carlo
- MEGA
MEchanism underlying the Genesis and evolution of Asthma
- meQTL
Methylation quantitative trait loci
- NAM
Native American
- OR
Odds ratio
- QC
Quality control
- SAGE
Study of African Americans, Asthma, Genes and Environment
- SCSGES
Singapore Cross Sectional Genetic Epidemiology Study
- SNP
single nucleotide polymorphism
- TOPMed
Trans-Omics for Precision Medicine
- US
United States of America
- WGS
Whole-genome sequencing
- YRI
Yoruba in Ibadan, Nigeria
Footnotes
CONFLICTS OF INTEREST
EH-L, and M.P.-Y. report funding from the Spanish Ministry of Science and Innovation (MCIN/AEI/10.13039/501100011033) and by “ESF Investing in your future” by the European Union. JP-G reports funding from the Spanish Ministry of Universities. M.P.-Y. and F.L.D. report grants from MCIN/AEI/10.13039/501100011033 and the European Regional Development Fund “ERDF A way of making Europe” by the European Union. MP-Y reports grant support from GlaxoSmithKline, Spain paid to Fundación Canaria Instituto de Investigación Sanitaria de Canarias (FIISC) for a project outside the submitted work. MP-Y and JV reports grants from Instituto de Salud Carlos III, Madrid, Spain. JV also reports funding by ISCIII and the European Regional Development Fund “ERDF A way of making Europe”. JMH-P has received fees from CSL Behring, GSK, Astra-Zeneca, laboratorios Menarini, Boehringer Ingelheim, FAES, laboratorios Esteve, Laboratorios Ferrer, Mundipharma, Laboratorios Rovi, Roche, Novartis, GRIFOLS, Pfizer, Acthelion-Jansen, Chiesi y Laboratorios Bial for the realization of courses, talks, consultancies, and other activities related to his professional activity. FTC has received research support from the Singapore Ministry of Education Academic Research Fund, Singapore Immunology Network (SIgN), National Medical Research Council (NMRC) (Singapore), Biomedical Research Council (BMRC) (Singapore), and the Agency for Science Technology and Research (A*STAR) (Singapore). F.T.C. has received consulting fees from Sime Darby Technology Centre; First Resources Ltd; Genting Plantation, and Olam International, outside the submitted work. YYS has received research support from the NUS Resilience & Growth Postdoctoral Fellowships. The other authors declare no conflict of interest.
AVAILABILITY OF DATA AND MATERIALS
TOPMed WGS and RNA-seq data from GALA II are available on dbGaP under accession number phs000920.v4.p2. TOPMed WGS data from SAGE is available on dbGaP under accession number phs000921.v4.p1.
REFERENCES
- 1.Dharmage SC, Perret JL, Custovic A. Epidemiology of Asthma in Children and Adults. Front Pediatr 2019;7:246. doi: 10.3389/fped.2019.00246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hernandez-Pacheco N, Flores C, Oh SS, et al. What Ancestry Can Tell Us About the Genetic Origins of Inter-Ethnic Differences in Asthma Expression. Curr Allergy Asthma Rep 2016;16:53. doi: 10.1007/s11882-016-0635-4 [DOI] [PubMed] [Google Scholar]
- 3.National Health Interview Survey: Most Recent Asthma Data. Centers for Disease Control and Prevention. 2019.https://www.cdc.gov/asthma/most_recent_data.html [Google Scholar]
- 4.Oraka E, Iqbal S, Flanders WD, et al. Racial and ethnic disparities in current asthma and emergency department visits: findings from the National Health Interview Survey, 2001–2010. J Asthma 2013;50:488–96. doi: 10.3109/02770903.2013.790417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nurmagambetov T, Kuwahara R, Garbe P. The Economic Burden of Asthma in the United States, 2008–2013. Ann Am Thorac Soc 2018;15:348–56. doi: 10.1513/AnnalsATS.201703-259OC [DOI] [PubMed] [Google Scholar]
- 6.Krishnan V, Diette GB, Rand CS, et al. Mortality in patients hospitalized for asthma exacerbations in the United States. Am J Respir Crit Care Med 2006;174:633–8. doi: 10.1164/rccm.200601-007OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briggs A, Nasser S, Hammerby E, et al. The impact of moderate and severe asthma exacerbations on quality of life: a post hoc analysis of randomised controlled trial data. J patient-reported outcomes 2021;5:6. doi: 10.1186/s41687-020-00274-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calhoun WJ, Haselkorn T, Miller DP, et al. Asthma exacerbations and lung function in patients with severe or difficult-to-treat asthma. J Allergy Clin Immunol 2015;136:1125–7.e4. doi: 10.1016/j.jaci.2015.05.014 [DOI] [PubMed] [Google Scholar]
- 9.Yan Q, Forno E, Herrera-Luis E, et al. A genome-wide association study of severe asthma exacerbations in Latino children and adolescents. Eur Respir J 2021;57:2002693. doi: 10.1183/13993003.02693-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandez-Pacheco N, Farzan N, Francis B, et al. Genome-wide association study of inhaled corticosteroid response in admixed children with asthma. Clin Exp Allergy 2019;49:789–98. doi: 10.1111/cea.13354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrera-Luis E, Espuela-Ortiz A, Lorenzo-Diaz F, et al. Genome-wide association study reveals a novel locus for asthma with severe exacerbations in diverse populations. Pediatr Allergy Immunol 2021;32:106–15. doi: 10.1111/pai.13337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galanter JM, Gignoux CR, Torgerson DG, et al. Genome-wide association study and admixture mapping identify different asthma-associated loci in Latinos: the Genes-environments & Admixture in Latino Americans study. J Allergy Clin Immunol 2014;134:295–305. doi: 10.1016/j.jaci.2013.08.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gignoux CR, Torgerson DG, Pino-Yanes M, et al. An admixture mapping meta-analysis implicates genetic variation at 18q21 with asthma susceptibility in Latinos. J Allergy Clin Immunol 2019;143:957–69. doi: 10.1016/j.jaci.2016.08.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pino-Yanes M, Gignoux CR, Galanter JM, et al. Genome-wide association study and admixture mapping reveal new loci associated with total IgE levels in Latinos. J Allergy Clin Immunol 2015;135:1502–10. doi: 10.1016/j.jaci.2014.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goddard PC, Keys KL, Mak ACY, et al. Integrative genomic analysis in African American children with asthma finds three novel loci associated with lung function. Genet Epidemiol 2021;45:190–208. doi: 10.1002/gepi.22365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishimura KK, Galanter JM, Roth LA, et al. Early-life air pollution and asthma risk in minority children. The GALA II and SAGE II studies. Am J Respir Crit Care Med 2013;188:309–18. doi: 10.1164/rccm.201302-0264OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddel HK, Taylor DR, Bateman ED, et al. Asthma control and exacerbations - Standardizing endpoints for clinical asthma trials and clinical practice. Am J Respir Crit Care Med 2009;180:59–99. doi: 10.1164/rccm.200801-060ST [DOI] [PubMed] [Google Scholar]
- 18.U.S. Department of Health and Human Services NI of H. Expert Panel Report 3: Guidelines for the Diagnosis and Management of Asthma. National Institute of Health’s National Heart Lung and Blood Institute; 2007. [Google Scholar]
- 19.Pino-Yanes M, Thakur N, Gignoux CR, et al. Genetic ancestry influences asthma susceptibility and lung function among Latinos. J Allergy Clin Immunol 2015;135:228–35. doi: 10.1016/j.jaci.2014.07.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maples BK, Gravel S, Kenny EE, et al. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am J Hum Genet 2013;93:278–88. doi: 10.1016/j.ajhg.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horimoto ARVR, Xue D, Thornton TA, et al. Admixture mapping reveals the association between Native American ancestry at 3q13.11 and reduced risk of Alzheimer’s disease in Caribbean Hispanics. Alzheimers Res Ther 2021;13:122. doi: 10.1186/s13195-021-00866-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S, Wu MC, Lin X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics 2012;13:762–75. doi: 10.1093/biostatistics/kxs014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci 2003;100:9440–5. doi: 10.1073/pnas.1530509100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stegle O, Parts L, Durbin R, et al. A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput Biol 2010;6:e1000770. doi: 10.1371/journal.pcbi.1000770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Consortium GTEx. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020;369:1318–30. doi: 10.1126/science.aaz1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamat MA, Blackshaw JA, Young R, et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics 2019;35:4851–3. doi: 10.1093/bioinformatics/btz469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghoussaini M, Mountjoy E, Carmona M, et al. Open Targets Genetics: systematic identification of trait-associated genes using large-scale genetics and functional genomics. Nucleic Acids Res 2021;49:D1311–20. doi: 10.1093/nar/gkaa840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Min JL, Hemani G, Hannon E, et al. Genomic and phenotypic insights from an atlas of genetic effects on DNA methylation. Nat Genet 2021;53:1311–21. doi: 10.1038/s41588-021-00923-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schofield EC, Carver T, Achuthan P, et al. CHiCP: a web-based tool for the integrative and interactive visualization of promoter capture Hi-C datasets. Bioinformatics 2016;32:2511–3. doi: 10.1093/bioinformatics/btw173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shumyatcher M, Hong R, Levin J, et al. Disease-Specific Integration of Omics Data to Guide Functional Validation of Genetic Associations. AMIA. Annu Symp proceedings AMIA Symp 2017;2017:1589–96.http://www.ncbi.nlm.nih.gov/pubmed/29854229 [PMC free article] [PubMed] [Google Scholar]
- 31.Bryc K, Durand EY, Macpherson JM, et al. The genetic ancestry of African Americans, Latinos, and European Americans across the United States. Am J Hum Genet 2015;96:37–53. doi: 10.1016/j.ajhg.2014.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Modena BD, Bleecker ER, Busse WW, et al. Gene Expression Correlated with Severe Asthma Characteristics Reveals Heterogeneous Mechanisms of Severe Disease. Am J Respir Crit Care Med 2017;195:1449–63. doi: 10.1164/rccm.201607-1407OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurotani R, Tomita T, Yang Q, et al. Role of secretoglobin 3A2 in lung development. Am J Respir Crit Care Med 2008;178:389–98. doi: 10.1164/rccm.200707-1104OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kido T, Yoneda M, Cai Y, et al. Secretoglobin superfamily protein SCGB3A2 deficiency potentiates ovalbumin-induced allergic pulmonary inflammation. Mediators Inflamm 2014;2014. doi: 10.1155/2014/216465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoneda M, Xu L, Kajiyama H, et al. Secretoglobin Superfamily Protein SCGB3A2 Alleviates House Dust Mite-Induced Allergic Airway Inflammation in Mice. Int Arch Allergy Immunol 2016;171:36–44. doi: 10.1159/000450788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiba Y, Kurotani R, Kusakabe T, et al. Uteroglobin-related protein 1 expression suppresses allergic airway inflammation in mice. Am J Respir Crit Care Med 2006;173:958–64. doi: 10.1164/rccm.200503-456OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mandal AK, Zhang Z, Ray R, et al. Uteroglobin represses allergen-induced inflammatory response by blocking PGD2 receptor-mediated functions. J Exp Med 2004;199:1317–30. doi: 10.1084/jem.20031666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mootz M, Jakwerth CA, Schmidt-Weber CB, et al. Secretoglobins in the big picture of immunoregulation in airway diseases. Allergy 2021;:all.15033. doi: 10.1111/all.15033 [DOI] [PubMed] [Google Scholar]
- 39.Inoue K, Wang X, Saito J, et al. Plasma UGRP1 levels associate with promoter G-112A polymorphism and the severity of asthma. Allergol Int 2008;57:57–64. doi: 10.2332/allergolint.O-07-493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao G, Lin X, Zhou M, et al. Association between CC10 +38A/G polymorphism and asthma risk: A meta-analysis. Pakistan J Med Sci 2013;29:1439–43. doi: 10.12669/pjms.296.3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Tanino Y, Sato S, et al. Secretoglobin 3A2 attenuates lipopolysaccharide-induced inflammation through inhibition of ERK and JNK pathways in bronchial epithelial cells. Inflammation 2015;38:828–34. doi: 10.1007/s10753-014-9992-0 [DOI] [PubMed] [Google Scholar]
- 42.Yokoyama S, Nakayama S, Xu L, et al. Secretoglobin 3A2 eliminates human cancer cells through pyroptosis. Cell death Discov 2021;7:12. doi: 10.1038/s41420-020-00385-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cai Y, Kimura S. Secretoglobin 3A2 Exhibits Anti-Fibrotic Activity in Bleomycin-Induced Pulmonary Fibrosis Model Mice. PLoS One 2015;10:e0142497. doi: 10.1371/journal.pone.0142497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurotani R, Okumura S, Matsubara T, et al. Secretoglobin 3A2 suppresses bleomycin-induced pulmonary fibrosis by transforming growth factor beta signaling down-regulation. J Biol Chem 2011;286:19682–92. doi: 10.1074/jbc.M111.239046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sagara H, Okada T, Okumura K, et al. Activation of TGF-beta/Smad2 signaling is associated with airway remodeling in asthma. J Allergy Clin Immunol 2002;110:249–54. doi: 10.1067/mai.2002.126078 [DOI] [PubMed] [Google Scholar]
- 46.Hansen G, McIntire JJ, Yeung VP, et al. CD4(+) T helper cells engineered to produce latent TGF-beta1 reverse allergen-induced airway hyperreactivity and inflammation. J Clin Invest 2000;105:61–70. doi: 10.1172/JCI7589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burchard EG, Avila PC, Nazario S, et al. Lower bronchodilator responsiveness in Puerto Rican than in Mexican subjects with asthma. Am J Respir Crit Care Med 2004;169:386–92. doi: 10.1164/rccm.200309-1293OC [DOI] [PubMed] [Google Scholar]
- 48.Hudon Thibeault A-A, Laprise C. Cell-Specific DNA Methylation Signatures in Asthma. Genes (Basel) 2019;10:932. doi: 10.3390/genes10110932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alashkar Alhamwe B, Miethe S, Pogge von Strandmann E, et al. Epigenetic Regulation of Airway Epithelium Immune Functions in Asthma. Front Immunol 2020;11:1747. doi: 10.3389/fimmu.2020.01747 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
TOPMed WGS and RNA-seq data from GALA II are available on dbGaP under accession number phs000920.v4.p2. TOPMed WGS data from SAGE is available on dbGaP under accession number phs000921.v4.p1.


