Abstract
gadd153, also known as chop, is a highly stress-inducible gene that is robustly expressed following disruption of homeostasis in the endoplasmic reticulum (ER) (so-called ER stress). Although all reported types of ER stress induce expression of Gadd153, its role in the stress response has remained largely undefined. Several studies have correlated Gadd153 expression with cell death, but a mechanistic link between Gadd153 and apoptosis has never been demonstrated. To address this issue we employed a cell model system in which Gadd153 is constitutively overexpressed, as well as two cell lines in which Gadd153 expression is conditional. In all cell lines, overexpression of Gadd153 sensitized cells to ER stress. Investigation of the mechanisms contributing to this effect revealed that elevated Gadd153 expression results in the down-regulation of Bcl2 expression, depletion of cellular glutathione, and exaggerated production of reactive oxygen species. Restoration of Bcl2 expression in Gadd153-overexpressing cells led to replenishment of glutathione and a reduction in levels of reactive oxygen species, and it protected cells from ER stress-induced cell death. We conclude that Gadd153 sensitizes cells to ER stress through mechanisms that involve down-regulation of Bcl2 and enhanced oxidant injury.
Gadd153, also known as Chop, is a leucine zipper transcription factor that is present at low levels under normal conditions but is robustly expressed in response to stress (5, 14, 40). gadd153 was originally identified based on its induction following treatment of cells with growth arresting and DNA damaging agents, though induced expression of the gene has also been strongly tied to perturbation of homeostasis in the endoplasmic reticulum (ER).
Proteins destined for transport to the cell membrane or to the cell exterior are synthesized on the ER surface and are then translocated to the ER lumen, where they are extensively modified by glycosylation and the addition of disulfide bonds. It is in the lumen of the ER, which provides a unique environment for protein folding, that proteins assume their mature, tertiary conformation. Disruption of homeostasis in the ER, which can occur, for example, as a result of nutrient deprivation or alteration of the organelle's calcium-rich oxidizing environment, can have devastating effects on the cell. Protein misfolding compromises cell function because essential polypeptides never exit the ER and are thus unable to perform their normal roles (reviewed in references 4 and 22). Additionally, accumulation of misfolded proteins in the ER triggers a unique signaling cascade referred to as the unfolded protein response (UPR). In the mammalian UPR a signal is transduced from the stressed ER to the nucleus, where transcription of a number of genes, including gadd153 and genes encoding ER resident proteins such as the glucose-regulated proteins (grp genes), is activated (reviewed in references 21 and 32).
The Grp's function as chaperones that guide proteins through the folding process, and their up-regulation in response to ER stress increases the cell's capacity to cope with the accumulation of immature, misfolded proteins in the ER. Indeed, if Grp78 induction is prevented, cell survival diminishes greatly following treatment with agents that stress the ER (24, 25). Following induction of the UPR, the kinetics of Gadd153 induction parallel exactly those seen for Grp78 (40). However, the effect of up-regulating Gadd153 in response to protein misfolding is much less intuitive than the effect of up-regulating expression of ER chaperones, and few studies have expressly addressed what function Gadd153 has in the ER stress response.
Several groups have reported that induction of Gadd153 correlates with the onset of apoptosis (12, 15). More recently it was reported that cell killing in response to toxins that perturb the ER was delayed in embryonic fibroblasts derived from mice lacking the gadd153 gene (45). These studies suggest that Gadd153 may be involved in the regulation of apoptosis, but to date no study has addressed the mechanistic relationship between expression of Gadd153 and cell death. Despite this, the assumption is commonly made in the literature that while UPR induction of Grp78 provides a survival signal, induction of Gadd153 serves as a death signal. Clearly, there is great need for more definitive evidence of the involvement of Gadd153 in apoptosis.
In the present study we sought to define in greater detail what role, if any, Gadd153 plays in the regulation of cell death. To accomplish this task we employed one cell line that constitutively overexpresses Gadd153 and several others that conditionally overexpress the protein. We report that while Gadd153 expression alone does not trigger cell death, it does sensitize cells to killing by agents that stress the ER. Importantly, overexpression of Gadd153 does not sensitize cells nonspecifically to all types of stress, since γ-irradiation, which induces cell death through a Gadd153-independent mechanism (29), kills cells irrespective of their Gadd153 status. Furthermore, we demonstrate that elevated Gadd153 expression down-regulates expression of the antiapoptotic protein Bcl2 and drastically depletes cells of glutathione, the primary intracellular scavenger of reactive oxygen species (ROS). Replenishment of Bcl2 returns cellular redox status to normal and protects cells from ER stress-induced death. Thus, elevated Gadd153 expression is tied to dramatic disruption of redox homeostasis, which readies cells for apoptosis.
MATERIALS AND METHODS
Cell culture, treatments, and generation of cell lines.
HeLa, Rat1, Rat-Myc, and gadd153−/− MEF (45) cell lines were grown in Dulbecco's modified essential medium (Life Technologies, Gaithersburg, Md.), supplemented with 10% fetal bovine serum (Hyclone, Logan, Utah), 100 U of penicillin per ml and 100 μg of streptomycin (Life Technologies) per ml, and were maintained in a humidified atmosphere containing 5% CO2. Rat1-Myc-Gadd153 (A94) cells were maintained as described above in medium containing 300 μg of hygromycin B (Sigma, St. Louis, Mo.) per ml (6).
Tunicamycin and thapsigargin were purchased from Sigma. Cells were γ-irradiated using a Gammacell 40 Exactor low-dose irradiator (Nordion International Inc., Ontario, Canada) with a 137Cs source at a specific dose rate of 1.22 Gy/min.
For the stable introduction of bcl2 into A94 cells, pSFFV-bcl2 (44) was cotransfected along with the puromycin-resistance pPur plasmid (Clontech Laboratories, Inc., Palo Alto, Calif.) into A94 cells by standard calcium phosphate transfection methods. Stable transformants were selected in 1 μg of puromycin per ml. Single colonies were expanded and screened by Western blot analysis for expression of Bcl2 and Gadd153. To generate clones that conditionally express Gadd153, the LacSwitch-inducible expression system (Stratagene, La Jolla, Calif.) was used. Both Rat1 and gadd153−/− cells were transfected with the LacI repressor expression plasmid P3′SS and the pOPRSVI-CHOPFlag plasmid generated by Matsumoto et al. (30). gadd153−/− cells were also transfected with P3′SS and CMVneo to generate the gadd153−/− Neo cell line. Transfected cells were placed under selection with 600 μg of geneticin (Life Technologies) and 300 μg of hygromycin B (Rat1, Gadd153i) per ml or 300 μg of geneticin and 100 μg of hygromycin B (gadd153−/− Gadd153i and gadd153−/− Neo) per ml. Single colonies were expanded and screened by Western blot analysis for inducible expression of Flag-tagged Gadd153 in the presence of 5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 8 to 24 h.
Survival assay.
Cell survival was determined using a standard clonogenic assay. Cells were initially plated in six-well cluster plates at a density of 30,000 cells/well. Following treatment, cells were trypsinized and serially diluted. Four dilutions were prepared from each treatment and ranged from 1:10 to 1:1,000,000. Colonies were allowed to grow for 10 to 14 days and were then stained with crystal violet (0.1% [wt/vol] crystal violet in 10% ethanol) and counted. Colony-forming efficiency was calculated based on the number of colonies that grew and the number of cells plated into each well and was expressed relative to control (untreated) cells.
Flow cytometric analysis of cell death and ROS production.
Flow cytometry was performed on a FACScan flow cytometer equipped with an argon laser at 488 nm excitation (Becton Dickinson, San Jose, Calif.). To assay for apoptotic cells on the basis of sub-G1 content, cells were trypsinized after treatments and fixed in 70% ethanol. Following incubation with RNase (0.1%; Boehringer Mannheim/Roche Diagnostic Corporation, Indianapolis, Ind.), cells were stained with propidium iodide (PI; 50 μg/ml; Roche Diagnostic Corporation), and fluorescence signals from the stained cells were collected in the FL-2 detector using a 585/42 band-pass filter. A total of 100,000 events were collected. Cell cycle profiles were analyzed using MultiCycle software (Phoenix Flow Systems, San Diego, Calif.), and the sub-G1 peak was quantified to estimate apoptosis in the population. To examine the production of ROS, cells were harvested in phosphate-buffered saline and incubated in the presence of 20 μM 2′7′-dichlorodihydrofluorescein (H2DCF) diacetate (Molecular Probes, Eugene, Oreg.) in the dark at 37°C for 30 min. The shift in green fluorescence as measured in the FL-1 detector with a 530/30 band-pass filter is associated with ROS production and was determined from histogram data using CellQuest software (Becton Dickinson). A total of 20,000 events was collected for each histogram.
Western blot analysis.
Total cell lysates were prepared and 40 to 70 μg of protein was size fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to polyvinylidene difluoride membranes by standard techniques. Blots were probed with the following antibodies: monoclonal anti-Bcl2 (Transduction Laboratories, San Diego, Calif.), polyclonal anti-Gadd153 and anti-Grp78 (Santa Cruz Biotechnology Inc., Santa Cruz, Calif.), horseradish peroxidase-conjugated goat anti-mouse antibody (Amersham, Arlington Heights, Ill.), and horseradish peroxidase-conjugated goat anti-rabbit antibody (Amersham). Bands were detected with the enhanced chemiluminescence (ECL) system (Amersham).
CAT and β-galactosidase assays.
Subconfluent cells growing in 100-mm tissue culture dishes were transiently transfected using Superfect transfection reagent (Qiagen, Valencia, Calif.) according to the manufacturer's specifications. Rat1, Rat-Myc, and A94 cells were transfected with 30 μg of plasmid containing the bcl2 promoter P1 linked to the chloramphenicol acetyltransferase (cat) reporter gene, or a control vector that contained the cat gene but lacked the bcl2 promoter, along with 3 μg of a β-galactosidase expression plasmid. The same transfection method was used to introduce bcl2 promoter-reporter constructs and β-galactosidase expression plasmids into HeLa cells, along with 0.1 to 1.0 μg of either a CMV-gadd153 expression plasmid (14) or an empty vector control (CMVneo). Cells were harvested in a buffer containing 100 mM potassium phosphate buffer, and CAT assays were performed as previously described using 14C-labeled chloramphenicol (20). Percentages of chloramphenicol acetylation were obtained over the linear range of the assay (3 to 70% conversion) and were quantified using a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). Conversion values were normalized to β-galactosidase activity, which was measured using the Galactolight Plus kit (Tropix, Bedford, Mass.).
Site-directed mutagenesis.
The QuikChange site-directed mutagenesis kit (Stratagene) was used to mutate the CMV-gadd153 expression vector. The primer used to mutate two of the leucine residues in the leucine zipper region of gadd153 (L134A/L141A) was 5′-AGTGGCACAGGCAGCTGAAGAGAATGAACGGGCCAAG CAGGAAATC (the mutated nucleotides are underlined). The oligonucleotide used to mutate two serine residues within the activation domain of GADD153 (S79A/S82A) was 5′-AGCACCTCCCAGGCCCCTCACGCTCCAGATTCCAGT. The PCR conditions used were those recommended by the manufacturer: 95°C for 30 s followed by 17 cycles of 95°C for 30 s, 55°C for 1 min, and 68°C for 10 min. The resulting clones were sequenced to verify the mutations.
RT-PCR and Northern blot analysis.
To analyze RNA expression by reverse transcription-PCR (RT-PCR), 1 μg of total RNA from each sample was treated with DNase I and then 10% of this treated sample was used for RT-PCR with SuperScript One-Step RT-PCR with Platinum Taq System (Life Technologies). The primers for the gene-specific RT-PCR analysis were as follows: bcl2 primer 1, CTGGCATCTTCTCCTTCCAGC; bcl2 primer 2, ACCTA-CCCAGCCTCCGTTATC; gapdh primer 1, ACATCAAGAAGGTGGTGAAGCAGG; and gapdh primer 2, CTCTTGCTCTCAGATCCTTGCTGG. The conditions for the RT-PCR were 1 cycle of 50°C for 30 min, 94°C for 2 min, and then 35 cycles (bcl2) or 25 cycles (gapdh) of 94°C for 30 s, 55°C for 1 min, and 70°C for 1 min. Equal aliquots of the PCR products were electrophoresed through a 2% agarose gel in 1× TAE buffer.
To analyze RNA expression by Northern blot analysis, total RNA was harvested using STAT-60 (Tel-Test B, Friendswood, Tex.) according to the manufacturer's recommendations. Ten micrograms of total RNA was run on an agarose-formaldehyde gel and transferred to a GeneScreen Plus membrane (NEN Life Science Products). The grp78 cDNA, a generous gift from Amy S. Lee, was labeled by the random primer method and used as a probe. A 24-base oligonucleotide (3′-ACGGTATCTGATCGTCTTCGAACC-5′) complementary to 18S rRNA was also used as a probe to verify RNA integrity and loading differences.
Quantification of GSH and GSSG.
Cell glutathione (GSH) and the oxidized form of GSH (GSSG) were determined either using high-performance liquid chromatography (HPLC) or colorimetric assays. For HPLC determinations the method of Reed et al. (33) was employed. Briefly, cells were treated with ice-cold 5% trichloroacetic acid (TCA) followed by centrifugation to remove TCA-insoluble proteins. The acid supernatant was derivatized with 6 mM iodoacetic acid and 1% 2, 4-dinitrofluorobenzene to yield the S-carboxymethyl and 2, 4-dinitrophenyl derivatives of GSH and GSSG. Separation of GSH and GSSG derivatives was achieved on a 15- by 4.6-mm 10-μm C18 reversed-phase ion-exchange column. Colorimetric assessments of GSH and GSSG were done according to the method described by Anderson (2) with minor modifications due to adaptations of the assay to microtiterplate dimensions.
RESULTS
Enforced expression of Gadd153 sensitizes cells to ER stress.
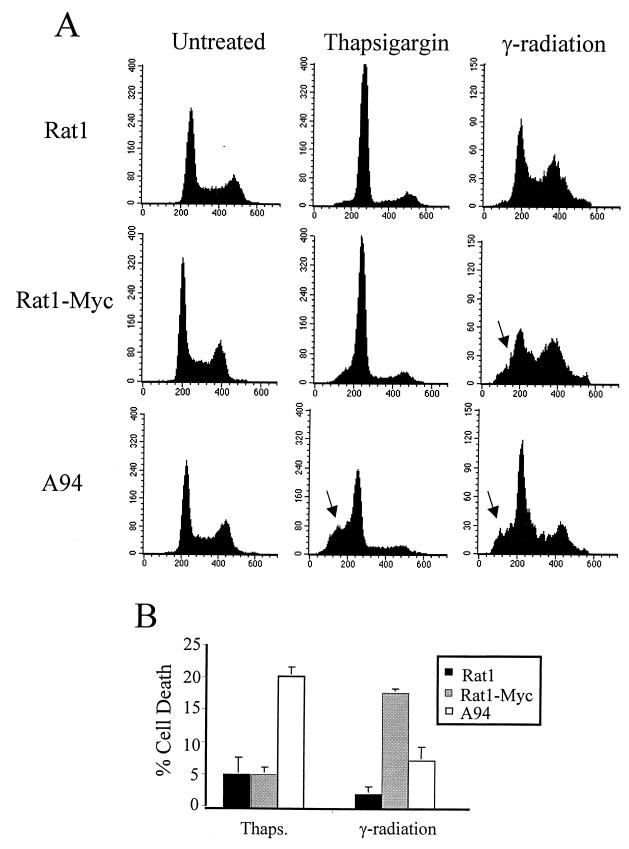
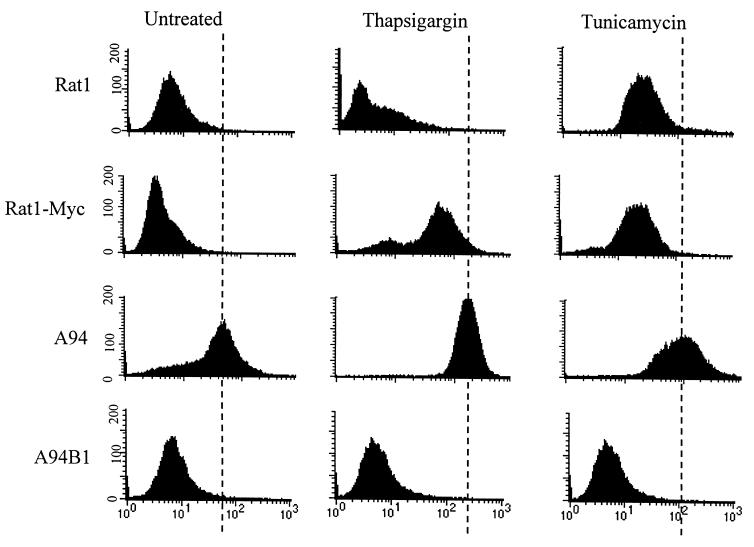
To better define Gadd153 function in the stress response, we investigated the responsiveness of A94 cells, which constitutively overexpress Gadd153, to several ER stress-inducing agents. A94 cells were derived from Rat1-Myc fibroblasts. A Rat1 cell line in which Gadd153 is constitutively overexpressed could not be established, presumably due to the growth-arresting properties of Gadd153. However, constitutive expression of the gene was achieved in Rat1-Myc cells, which overexpress c-Myc. A more detailed description of these cell lines can be found elsewhere (6). To examine the influence of Gadd153 on survival of cells exposed to ER stress, clonogenic assays were carried out with Rat1, Rat1-Myc, and A94 cells following their treatment with tunicamycin and thapsigargin, two classic ER stress agents. As seen in Fig. 1, constitutive expression of Gadd153 greatly reduced long-term survival following ER stress. Interestingly, c-Myc expression alone increased the sensitivity of Rat1 cells to thapsigargin and tunicamycin by two- and fivefold, respectively. However, with both agents overexpression of Gadd153 led to a further 10-fold reduction in survival compared to Rat1-Myc cells. To determine the specificity of these findings, we compared survival of Rat1, Rat1-Myc, and A94 cells following exposure to γ-irradiation, one of the few DNA-damaging treatments that does not induce Gadd153 expression (29). Importantly, elevated Gadd153 expression did not significantly alter the sensitivity of cells to γ-irradiation (Fig. 1). To confirm our findings that Gadd153-overexpressing cells have a heightened sensitivity to ER stress agents and to further investigate the mechanism involved, we examined cytotoxicity in a short-term assay using flow cytometric analysis for the detection of apoptotic cells. As shown in Fig. 2 for thapsigargin, apoptosis as measured by the appearance of a sub-G1 peak using flow cytometry was negligible in Rat1 and Rat1-Myc cells following 24-h treatment but was significantly elevated in A94 cells. Interestingly, despite the fact that clonogenic survival following γ-irradiation did not differ between the Rat1 and Rat1-Myc cells, flow cytometric analysis revealed a significant increase in apoptosis in Rat1-Myc cells compared to that in Rat1 cells. This is consistent with previous findings by others (6). Gadd153 expression, however, did not enhance but actually reduced the level of γ-irradiation-induced apoptosis.
FIG. 1.
Constitutive expression of Gadd153 sensitizes cells to ER stress as demonstrated in colony formation assays. Rat1, Rat-Myc, and A94 (Rat1-Myc-Gadd153) cell lines were treated with the indicated doses of thapsigargin or tunicamycin for 24 h or were exposed to ionizing radiation and allowed to recover for 24 h. Following treatments, cells were split into 100-mm tissue culture plates and colonies were counted 8 to 12 days later. Data are reported as percent survival relative to control (untreated) cells and are an average of at least three independent experiments. Error bars represent standard errors of the means.
FIG. 2.
Constitutive expression of Gadd153 sensitizes cells to ER stress-induced apoptosis. Cell lines were treated with thapsigargin (1 μM, 24 h) or γ-irradiation (5.0 rad, 48 h recovery). Following staining with PI, DNA content was measured using flow cytometry. (A) Histograms from a representative experiment. Arrows denote the appearance of a sub-G1 peak, which is indicative of apoptosis. (B) The sub-G1 content of each cell population was quantified. Data shown are the means ± standard errors of at least three independent experiments.
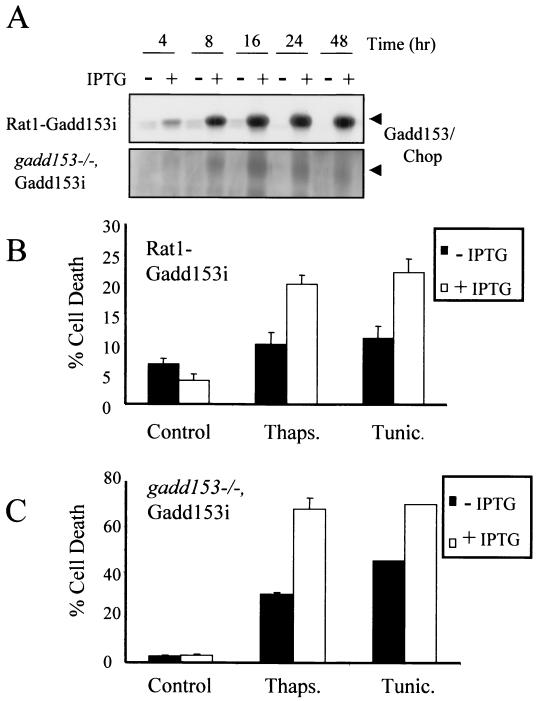
A complication of the A94 model system is that Gadd153 is overexpressed against a background of elevated c-Myc, and we clearly see an effect of c-Myc expression on survival following treatment with agents that perturb the ER (Fig. 1). We have confirmed this observation in another rat fibroblast cell line, Rat1a (19). While Rat1a cells were found to be relatively resistant to tunicamycin and thapsigargin, Rat1a-Myc, which constitutively overexpresses c-Myc, showed much more sensitivity to these agents (data not shown). These data are intriguing and demonstrate for the first time that expression of c-Myc sensitizes cells to ER stress. Further experiments are being carried out to address the involvement of c-Myc in ER stress signaling pathways. However, for the purpose of defining the role of Gadd153 in the ER stress response, it was essential that we examine the effects of Gadd153 independent of c-Myc overexpression to determine if Gadd153 expression alone sensitizes cells to ER stress. We therefore developed two cell lines in which overexpression of Gadd153 is conditional. A Gadd153 expression vector, where the gadd153 gene is under the control of the Lac repressor, was introduced into Rat1 fibroblasts and MEFs derived from mice in which both gadd153/chop gene alleles have been disrupted (45). The cell lines with inducible Gadd153 are referred to as Rat1-Gadd153i and gadd153−/− Gadd153i. That inclusion of IPTG in the culture medium results in accumulation of Gadd153 in both cell lines is demonstrated in Fig. 3A. Induction of Gadd153 alone did not induce cell death in either the MEF or Rat1 cell lines, even after 96 h or more of Gadd153 expression. However, cells pretreated for 8 to 16 h with IPTG were sensitized to killing by the ER stress agents thapsigargin and tunicamycin, as measured by flow cytometry (Fig. 3B and C).
FIG. 3.
Inducible expression of Gadd153 sensitizes cells to ER stress-induced death. (A) gadd153, under control of an IPTG-inducible promoter, was introduced into Rat1 fibroblasts and MEFs derived from gadd153/chop−/− mice. Accumulation of Gadd153 is evident in Western blots following introduction of 5 mM IPTG to the culture medium. (B and C) Rat1-Gadd153i (B) and gadd153−/− Gadd153i (C) cells were treated with thapsigargin (1 μM, 24 h) or tunicamycin (1 μg/ml, 24 h) in the presence or absence of 5 mM IPTG, and apoptosis was quantified using flow cytometry. Data are averages of at least three independent experiments. Error bars represent standard errors of the means.
Down-regulation of Bcl2 contributes to Gadd153-mediated sensitization of cells to ER stress.
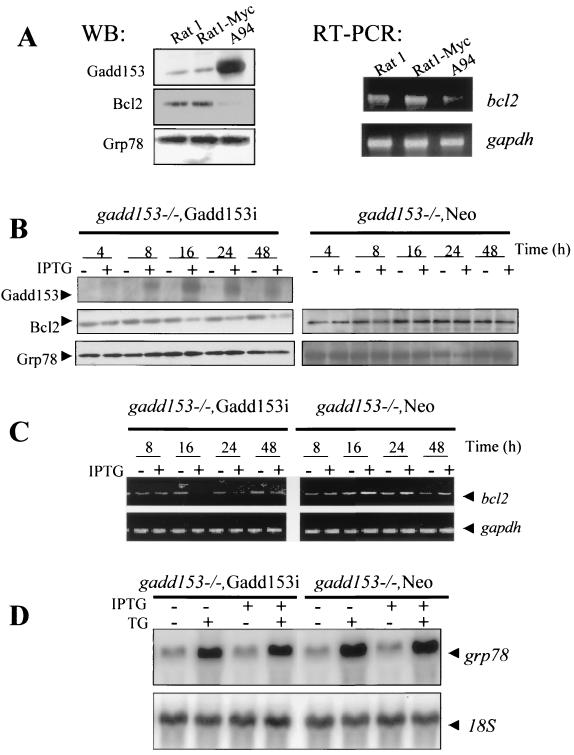
We next sought to determine how Gadd153 sensitizes cells to ER stress, and we first compared Bcl2 expression in parental and Gadd153-overexpressing cell lines. As shown in Fig. 4A, c-Myc expression did not alter Bcl2 expression relative to the parental Rat1 cells. However, in the Gadd153-expressing A94 cells, the level of Bcl2 protein was dramatically reduced, as was the bcl2 RNA level as determined by RT-PCR. Grp78 levels were also assessed and found to be equivalent in the Rat1, Rat1-Myc, and A94 cell lines (Fig. 4A), indicating that ER stress was not basally elevated as a consequence of Gadd153 overexpression. When challenged with a stressful treatment, all three cell lines showed Grp78 expression induced to similar levels (data not shown). Similar results were obtained with gadd153−/− Gadd153i cells; concomitant with IPTG induction of Gadd153, Bcl2 protein levels declined (Fig. 4B). Treatment with IPTG did not, however, result in increased ER stress, since Grp78 levels remained constant (Fig. 4B). This was further demonstrated with gadd153−/− Neo cells, which lack Gadd153 completely. Treatment of gadd153−/− Neo cells with IPTG had no effect on the expression of Grp78 or Bcl2, indicating that the down-regulation of bcl2 observed in gadd153−/− Gadd153i cells was the result of induced Gadd153 expression and not a nonspecific effect of IPTG on the cells. Down-regulation of bcl2 mRNA expression by Gadd153 was also demonstrated using RT-PCR (Fig. 4C). Inclusion of IPTG in the culture medium of gadd153−/− Gadd153i cells resulted in down-regulation of bcl2 mRNA but had no effect on expression of bcl2 in gadd153−/− Neo cells. Furthermore, these gadd153−/− cell lines had similar levels of basal and thapsigargin-induced grp78 RNA expression, regardless of the presence of IPTG or IPTG-induced Gadd153 expression (Fig. 4D).
FIG. 4.
Gadd153 down-regulates Bcl2. (A) Western blot demonstrating that Bcl2 protein levels are diminished in cells constitutively overexpressing Gadd153, while Grp78 expression is unaffected by constitutively elevated Gadd153 levels. The RT-PCR shows that the bcl2 mRNA level is also decreased basally in A94 cells. The gapdh RT-PCR was performed as a control for RNA integrity and loading. (B) Western blot demonstrating down-regulation of Bcl2 in gadd153−/− Gadd153i cells but not in gadd153−/− Neo cells. Concomitant with induction of Gadd153 expression by inclusion of 5 mM IPTG in the culture medium, Bcl2 expression decreases, while expression of Grp78 remains constant. In contrast, IPTG had no effect on Bcl2 protein levels in the vector control cells. There was no Gadd153 expression in the gadd153−/− Neo cells, so no blot is shown for this protein. (C) RT-PCR demonstrating that inclusion of IPTG in the culture medium of gadd153−/− Neo cells has no effect on bcl2 mRNA levels. In contrast, IPTG-mediated induction of Gadd153 in gadd153−/−, Gadd153i cells results in diminished abundance of bcl2 mRNA. (D) Northern blot analysis showing that 5 mM IPTG pretreatment for 18 h, which induces Gadd153 expression in gadd153−/− Gadd153i cells, does not influence the induction of grp78 RNA levels in response to ER stress incurred by 1 μM thapsigargin (TG) for 6 h.
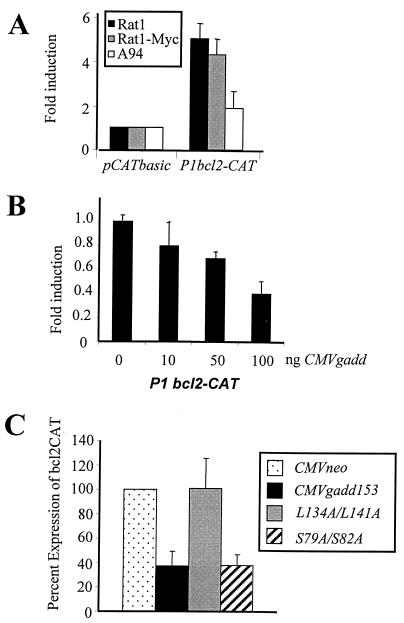
To determine how Gadd153 acts to down-regulate Bcl2, we examined the effects of Gadd153 expression on bcl2 promoter activity using a CAT reporter system. A construct containing the P1 promoter region of bcl2 linked to the cat coding region (44) was transfected into Rat1, Rat1-Myc, and A94 cells. bcl2 promoter activity was similar in Rat1 and Rat1-Myc cells. However, the activity of the bcl2 promoter was significantly repressed in Gadd153-overexpressing A94 cells (Fig. 5A). To verify that the decreased bcl2 promoter expression indeed was related to Gadd153 expression, we performed transient-transfection experiments in which HeLa cells were cotransfected with the bcl2-cat reporter plasmid along with increasing concentrations of a gadd153 expression vector. As shown in Fig. 5B, Gadd153 expression inhibited bcl2 promoter activity in a dose-dependent manner. Similar cotransfection experiments performed with the bcl2 promoter-reporter construct and an empty vector control (CMVneo) demonstrated no effect of the control vector on the bcl2 promoter (data not shown). These data suggest that Gadd153 down-regulates Bcl2 expression by inhibiting bcl2 transcription. We next tested repression of the bcl2 promoter by several Gadd153 mutants. When mutations were made in the leucine zipper region of the protein (L134A/L141A), Gadd153 was no longer able to repress the bcl2 promoter (Fig. 5C). In contrast, Gadd153 harboring two mutations in the protein's transactivation domain (S79A/S82A) repressed the bcl2 promoter as well as wild-type Gadd153 did (Fig. 5C).
FIG. 5.
(A) Plasmid constructs containing the bcl2 promoter region P1 linked to the cat reporter gene were transfected into Rat1, Rat1-Myc, and A94 cell lines. Acetylation of 14C-chloramphenicol, which reflects activity of the bcl2 promoter, was quantified. Data are expressed as fold induction of the promoter activity relative to that of the vector control, which harbored the cat gene but lacked the bcl2 promoter. (B) bcl2-cat promoter-reporter plasmids were introduced into HeLa cells along with increasing concentrations of a gadd153 expression vector, and bcl2 promoter activity was determined. Data are reported as fold induction relative to the activity of the bcl2 promoter in HeLa cells transfected with a vector lacking the gadd153 gene (CMVneo). (C) bcl2-cat promoter-reporter plasmids were introduced in HeLa cells along with 1 μg of expression vector for CMVneo, wild-type Gadd153, Gadd153 harboring mutations in the leucine zipper domain (L134A/L141A), or Gadd153 harboring mutations in the transactivation domain (S79A/S82A). bcl2 promoter activity was determined 48 h later. Data are expressed relative to the activity of the bcl2 promoter-transfected empty CMVneo vector. All data are averages of at least three independent experiments and bars represent standard errors of the means.
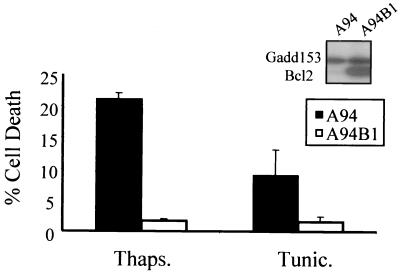
To determine whether down-regulation of Bcl2 contributes to sensitization to ER stress in Gadd153-overexpressing cells, expression of Bcl2 in A94 cells was restored by stable introduction of a bcl2 expression plasmid, and the sensitivity of the resulting cell line, A94B1, to ER stress-induced apoptosis was examined using flow cytometry. Cell death was negligible in A94B1 cells treated with thapsigargin and tunicamycin (Fig. 6), clearly demonstrating a protective effect of Bcl2 in Gadd153-overexpressing cells.
FIG. 6.
Restoration of Bcl2 expression in A94 cells protects against ER stress-induced death. A94 cells transfected with bcl2 expression constructs show greatly enhanced Bcl2 levels (Western blot, insert). Following treatment with either thapsigargin (1 μM, 24 h) or tunicamycin (1 μg/ml, 24 h), cells were harvested, stained with PI, and analyzed using flow cytometry. The sub-G1 content of each cell population was quantified. Data are averages of at least three independent experiments and error bars represent standard errors of the means.
Intracellular GSH content is decreased in cells expressing Gadd153.
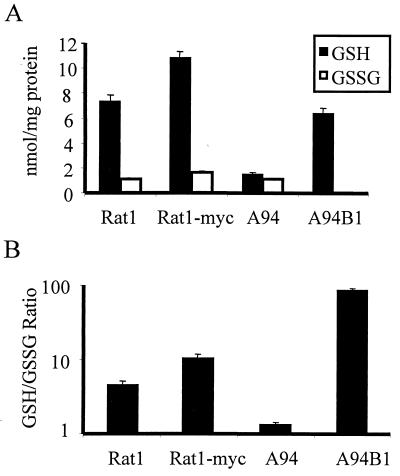
Reduced Bcl2 expression has been linked to lowered levels of cellular GSH, the tripeptide thiol that is an essential component of cellular defenses against oxidative stress and reactive electrophiles. In addition, we have previously implicated Gadd153 in the cellular response to oxidative stress (19). We therefore reasoned that Gadd153 might perturb cellular redox states through a Bcl2-dependent mechanism. Using HPLC and colorimetric assays, GSH and GSSG levels were quantified in the Gadd153-overexpressing cell line, A94. Compared to GSH levels in Rat1 and Rat1-Myc, GSH levels were dramatically reduced in A94 cells, while GSSG levels remained unchanged (Fig. 7A). The ratio of GSH to GSSG, which gives an indication of the overall redox status of the cell, was five- and eightfold less in A94 cells relative to that in the Rat1 and Rat1-Myc lines, respectively (Fig. 7B). These data indicate that A94 cells are unable to maintain the normal reducing environment found in Rat1 and Rat1-Myc cells. Furthermore, restoration of Bcl2 expression in A94 cells, which protects against ER stress-induced death (Fig. 5), returned GSH levels in the Gadd153-overexpressing cells to normal (Fig. 7A) and increased the GSH/GSSG ratio above that seen in parental cells (Fig. 7B). Differences between GSH/GSSG ratios in A94B1 cells and Rat1 parental cells probably reflect the abundance of Bcl2 in A94B1 cells, which exceeds the endogenous levels observed in either the Rat1 or Rat1-Myc lines.
FIG. 7.
GSH is depleted from Gadd153-overexpressing cells in a Bcl2-dependent manner. (A) HPLC was used to measure cellular GSH and GSSG levels. (B) The ratio of GSH to GSSG, which gives an estimate of the overall redox state of the cell, is reported. All data are reported as averages of three or more independent experiments and bars represent standard errors of the means.
The observations described above suggest that Gadd153-overexpressing cells have a greater oxidant burden than those that do not express Gadd153 constitutively. To determine more directly if this is so, production of reactive oxygen species (ROS) was measured in Rat1, Rat1-Myc, and A94 cells. Cells were loaded with the cell-permeable fluorescent dye, H2DCF diacetate. Upon entry into cells, the dye is oxidized by ROS generating a fluorescent product. Fluorescence, measured by flow cytometry, gives an estimate of intracellular ROS production. Using this approach we found that oxidant production was low in untreated Rat1 and Rat1-Myc cells. In sharp contrast, ROS production was significantly elevated in untreated A94 cells (Fig. 8), consistent with the reduced GSH levels in the Gadd153-overexpressing cells. Furthermore, ROS production in A94B1 cells was low and comparable to that observed in Rat1 and Rat1-Myc cells. ROS production increased somewhat in Rat1 and Rat1-Myc cells following thapsigargin and tunicamycin treatment but was greatest in A94 cells treated with thapsigargin and tunicamycin (Fig. 8). Consistent with their elevated GSH levels, oxidant production following ER stress was not observed in A94B1 cells, which overexpress Bcl2 and are resistant to ER stress (Fig. 8).
FIG. 8.
Gadd153 increases cellular ROS production. Control (untreated) cells and cells treated with thapsigargin (1 μM, 24 h) or tunicamycin (1 μg/ml, 24 h) were incubated with H2DCF diacetate, which freely crosses cell membranes and is cleaved intracellularly to form the less membrane-permeative H2DCF. In the presence of ROS this dye is oxidized, producing a fluorescent product, DCF. Flow cytometry was used to analyze fluorescence in each cell population. ROS production, indicated as a rightward shift in peak fluorescence, was maximal in A94 cells, regardless of treatment. This experiment was repeated three times, and representative histograms are shown.
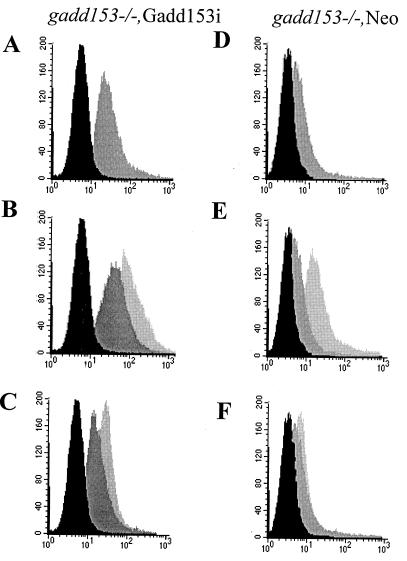
The finding that Gadd153 expression contributes to oxidant production was confirmed in gadd153−/− MEFs that conditionally express Gadd153. As measured by flow cytometry, ROS production increased substantially in gadd153−/− Gadd153i cells when Gadd153 expression was induced with IPTG (Fig. 9A). Furthermore, while thapsigargin and tunicamycin both induced ROS production in gadd153−/− Gadd153i cells, the oxidant burden was exaggerated in gadd153−/− Gadd153i cells in which Gadd153 expression was restored (Fig. 9B and C). In contrast, although inclusion of IPTG in the culture medium of gadd153−/− Neo cells did cause some increase in ROS (presumably a nonspecific effect), it was not nearly as great as that seen in the Gadd153-expressing cells (Fig. 9D, E, and F).
FIG. 9.
Conditional expression of Gadd153 increases oxidative stress. ROS production was examined in gadd153−/− Gadd153i cells, which conditionally express Gadd153 (A, B, and C), and in gadd153−/− Neo cells, which are vector control cells (D, E, and F), using H2DCF dye and flow cytometry. (A and D) Cells were treated with IPTG (dark gray peak) or with a vehicle control (black peak). Significant ROS production was observed in response to Gadd153 induction (A) but not in IPTG-treated control cells (D). (B and E) Following pretreatment with IPTG or vehicle control, cells were exposed to tunicamycin (TN; 1 μg/ml, 12 h). The black peak represents fluorescence in untreated control cells (−IPTG, −TN). The dark gray peak corresponds to tunicamycin-treated cells lacking Gadd153 (−IPTG, +TN), while the light gray peak corresponds to ROS production in IPTG-pretreated cells exposed to tunicamycin (+IPTG, +TN). Tunicamycin treatment increased ROS production in both cell lines, but ROS production was maximal in Gadd153-expressing cells treated with tunicamycin. (C and F) Following pretreatment with IPTG or vehicle control, cells were exposed to thapsigargin (TG; 1 μM, 12 h). The black peak represents fluorescence in untreated cells (−IPTG, −TG). The dark gray peak corresponds to thapsigargin-treated cells that were not pretreated with IPTG (−IPTG, +TG), while the light gray peak corresponds to cells receiving IPTG followed by thapsigargin (+IPTG, +TG). Maximal ROS production was again detected in Gadd153-expressing cells treated with thapsigargin. All experiments were repeated three times and representative histograms are shown.
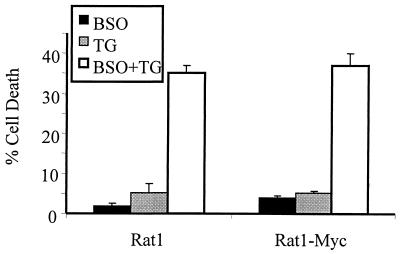
That disruption of redox equilibrium contributes to ER stress-induced cell death was confirmed in experiments using buthionine sulfoximine (BSO), an inhibitor of GSH neosynthesis. Pretreatment of Rat1 and Rat1-Myc cells with BSO depleted the cells of GSH (data not shown) and greatly sensitized them to tunicamycin and thapsigargin (Fig. 10). Interestingly, A94 cells were not protected from ER stress by pretreatment with N-acetylcysteine (NAC), which supplies cells with cysteine, a necessary precursor in the synthesis of GSH (data not shown). However, such NAC pretreatment also failed to increase significantly the GSH levels in the A94 cells (data not shown).
FIG. 10.
GSH depletion sensitizes cells to ER stress. Rat1 and Rat1-Myc cells were pretreated with BSO (100 μM, 24 h) followed by treatment with thapsigargin (TG; 1 μM, 24 h). Cell death was quantified using flow cytometry. Data represent averages from at least three independent experiments. Error bars denote standard errors of the means.
DISCUSSION
In this study we examined the influence of Gadd153 expression on cell survival following ER stress. Using a cell line that constitutively overexpresses Gadd153 and two additional cell lines that conditionally overexpress the protein, we have demonstrated that elevated Gadd153 expression sensitizes cells to agents that perturb ER function. Concomitant with Gadd153 expression, production of the Bcl2 protein is repressed. Furthermore, expression of Gadd153 results in severe depletion of intracellular thiols. As such a redox imbalance would predict, ROS production was markedly increased in association with overexpression of Gadd153. Restoration of Bcl2 in Gadd153-overexpressing cells returns redox homeostasis to the cells and protects them from ER stress-induced death.
In Gadd153-overexpressing cells, Bcl2 protein levels are dramatically reduced relative to parental cells, and we have provided evidence that this is due to reduced transcription of the bcl2 gene. While it has been suggested that Gadd153 can directly regulate gene transcription (37), there is no evidence of a Gadd153-responsive site in the bcl2 promoter. An alternative explanation is that Gadd153 exerts its effects on the bcl2 promoter indirectly by negatively regulating another transcription factor. Gadd153 has long been recognized as an inhibitor of gene transcription, primarily through its dimerization with and negative regulation of other leucine zipper transcription factors of the C/EBP (13, 34) and CREB (36, 43) families. Furthermore, there is precedence for repression of the bcl2 promoter through the action of transcription factor complexes (24). Our finding that mutation of the leucine zipper domain of Gadd153 abolishes its effects on the bcl2 promoter suggests that the ability of Gadd153 to influence bcl2 expression requires complex formation with other proteins.
An important finding of this study is that elevated Gadd153 expression results in thiol depletion. Direct measurement of cellular GSH levels indicated that GSH pools were severely depleted in Gadd153-overexpressing cells. We confirmed this finding by measuring ROS production and again found that Gadd153 expression, even in the absence of stress, perturbed redox equilibrium (Fig. 9A). GSH has a number of vital functions in the cell. In addition to its role in detoxifying electrophiles and scavenging free radicals, GSH maintains protein integrity by preventing oxidation of thiol groups and by reducing disulfide bonds induced by oxidative stress (reviewed in references 3 and 27). Thiol depletion has been shown to sensitize cells to death caused by a variety of stimuli, and in some cell types it is sufficient to induce apoptosis (8, 35, 41). It is clear from these studies and others (9, 16, 17, 38) that in many cell types control of GSH homeostasis is closely associated with regulation of apoptosis. An interesting question raised by this study is how Gadd153 expression and Bcl2 repression lead to redox imbalance. Bcl2 is known to influence GSH levels, but how this occurs remains controversial (39). Our glutathione measurements reveal that the total level of glutathione (GSH plus GSSG) is dramatically lower in Gadd153-overexpressing cells because GSH pools are depleted, while GSSG levels remain essentially unchanged. There are several possible explanations for these findings. First, GSH could be rapidly exported from the Gadd153-overexpressing cells. This does not appear to be the case, as measurement of thiols in the medium of parental cells and those that overexpress Gadd153 indicated that both had extruded equally low levels of GSH (data not shown). Another possible explanation is that Gadd153 interferes with GSH neosynthesis. Synthesis of GSH is a two-step process that is dependent on the sequential actions of γ-glutamyl cysteine synthetase, which is rate limiting, and glutathione synthetase. Regulation of these enzymes, particularly of γ-glutamyl cysteine synthetase, is complex and occurs transcriptionally (31, 42), posttranscriptionally (26), and posttranslationally (10, 28). Whether Gadd153 can influence GSH neosynthesis is currently under investigation.
While our data demonstrate that elevated Gadd153 levels sensitize cells to ER stress, we also found that overexpression of c-Myc increases susceptibility of cells to ER stress-induced death. We have confirmed these findings with another rat fibroblast cell line, Rat1a-Myc, which constitutively expresses c-Myc and displays enhanced sensitivity to thapsigargin and tunicamycin compared to its parental line, Rat1a (data not shown). While the mechanism whereby c-Myc sensitizes cells to ER stress remains to be determined, it appears to be distinct from that of Gadd153. For example, expression of c-Myc does not downregulate Bcl2, while Bcl2 expression is markedly depressed in all cells that express Gadd153, regardless of c-Myc expression. Furthermore, unlike our observations in Gadd153-overexpressing cells, GSH levels were not reduced in Rat1-Myc cells but were actually higher compared to those in the parental Rat1 cell line.
In summary, we have established an important mechanistic link between Gadd153 expression and cell survival following ER stress. By decreasing Bcl2 levels, primarily through transcriptional mechanisms, and by depleting cells of essential thiols, Gadd153 poises cells for death. Further studies investigating how Gadd153 represses the bcl2 promoter and perturbs cellular redox status should enhance our understanding of the role of Gadd153 in the stress response.
ACKNOWLEDGMENTS
We thank Robert Wersto, Joe Chrest, and Christa Morris for their assistance with the flow cytometry experiments described in the manuscript. We also thank David Ron for generously providing us with the gadd153/chop knockout cells, as well as M. Matsumoto and S. Akira for their gift of the pORSVI-CHOPFlag and P3′SS vectors used to generate the cell lines that conditionally express Gadd153.
Lars-O. Klotz is a recipient of a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft (DFG), Bonn, Germany (KL 1245/1-1).
REFERENCES
- 1.Alarcon R M, Rupnow B A, Graeber T G, Knox S J, Giaccia A J. Modulation of c-Myc activity and apoptosis in vivo. Cancer Res. 1996;56:4315–4319. [PubMed] [Google Scholar]
- 2.Anderson M E. Determination of glutathione and glutathione disulfide in biological samples. Methods Enzymol. 1985;113:548–555. doi: 10.1016/s0076-6879(85)13073-9. [DOI] [PubMed] [Google Scholar]
- 3.Anderson M E, Luo J L. Glutathione therapy: from prodrugs to genes. Semin Liver Dis. 1998;18:415–424. doi: 10.1055/s-2007-1007174. [DOI] [PubMed] [Google Scholar]
- 4.Aridor M, Balch W E. Integration of endoplasmic reticulum signaling in health and disease. Nat Med. 1999;5:745–751. doi: 10.1038/10466. [DOI] [PubMed] [Google Scholar]
- 5.Carlson S G, Fawcett T W, Bartlett J D, Bernier M, Holbrook N J. Regulation of the C/EBP-related gene gadd153 by glucose deprivation. Mol Cell Biol. 1993;13:4736–4744. doi: 10.1128/mcb.13.8.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C, Nussenzweig A, Guo M, Kim D, Li G C, Ling C C. Down-regulation of gadd153 by c-myc in rat fibroblasts and its effect on cell growth and radiation-induced apoptosis. Oncogene. 1996;13:1659–1665. [PubMed] [Google Scholar]
- 7.Cherney B W, Bhatia K, Tosato G. A role for deregulated c-Myc expression in apoptosis of Epstein-Barr virus-immortalized B cells. Proc Natl Acad Sci USA. 1994;91:12967–12971. doi: 10.1073/pnas.91.26.12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiba T, Takahashi S, Sato N, Ishii S, Kikuchi K. Fas-mediated apoptosis is modulated by intracellular glutathione in human T cells. Eur J Immunol. 1996;5:1164–1169. doi: 10.1002/eji.1830260530. [DOI] [PubMed] [Google Scholar]
- 9.Cotgreave I A, Gerdes R G. Recent trends in glutathione biochemistry—glutathione-protein interactions: a molecular link between oxidative stress and cell proliferation? Biochem Biophys Res Commun. 1998;242:1–9. doi: 10.1006/bbrc.1997.7812. [DOI] [PubMed] [Google Scholar]
- 10.Estrela J M, Gil F, Vila J M, Vina J. Alpha-adrenergic modulation of glutathione metabolism in isolated rat hepatocytes. Am J Physiol. 1988;255:E801–E805. doi: 10.1152/ajpendo.1988.255.6.E801. [DOI] [PubMed] [Google Scholar]
- 11.Evan G I, Wyllie A H, Gilbert C S, Littlewood T D, Land H, Brooks M, Waters C M, Penn L Z, Hancock D C. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 12.Eymin B, Dubrez L, Allouche M, Solary E. Increased gadd153 messenger RNA level is associated with apoptosis in human leukemic cells treated with etoposide. Cancer Res. 1997;57:686–695. [PubMed] [Google Scholar]
- 13.Fawcett T W, Eastman H B, Martindale J L, Holbrook N J. Physical and functional association between GADD153 and CCAAT/enhancer-binding protein beta during cellular stress. J Biol Chem. 1996;271:14285–14289. doi: 10.1074/jbc.271.24.14285. [DOI] [PubMed] [Google Scholar]
- 14.Fornace A J, Nebert D W, Hollander M C, Luethy J D, Papathanasiou M, Fargnoli J, Holbrook N J. Mammalian genes coordinately regulated by growth arrest signals and DNA-damaging agents. Mol Cell Biol. 1989;9:4196–4203. doi: 10.1128/mcb.9.10.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman A D. GADD153/CHOP, a DNA damage-inducible protein, reduced CAAT/enhancer binding protein activities and increased apoptosis in 32D c13 myeloid cells. Cancer Res. 1996;56:3250–3256. [PubMed] [Google Scholar]
- 16.Ghibelli L, Coppola S, Rotilio G, Lafavia E, Maresca V, Ciriolo M R. Non-oxidative loss of glutathione in apoptosis via GSH extrusion. Biochem Biophys Res Commun. 1995;216:313–320. doi: 10.1006/bbrc.1995.2626. [DOI] [PubMed] [Google Scholar]
- 17.Ghibelli L, Fanelli C, Rotilio G, Lafavia E, Coppola S, Colussi C, Civitareale P, Ciriolo M R. Rescue of cells from apoptosis by inhibition of active GSH extrusion. FASEB J. 1998;12:479–486. doi: 10.1096/fasebj.12.6.479. [DOI] [PubMed] [Google Scholar]
- 18.Guyton K Z, Xu Q, Holbrook N J. Induction of the mammalian stress response gene GADD153 by oxidative stress: role of AP-1 element. Biochem J. 1996;314:547–554. doi: 10.1042/bj3140547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoang A T, Cohen K J, Barrett J F, Bergstrom D A, Dang C V. Participation of cyclin A in Myc-induced apoptosis. Proc Natl Acad Sci USA. 1994;91:6875–6879. doi: 10.1073/pnas.91.15.6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holbrook N J, Luethy J D, Kin J. Transient expression of foreign genes in lymphoid cells is enhanced by phorbol ester. Mol Cell Biol. 1987;7:2610–2613. doi: 10.1128/mcb.7.7.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaufman R J. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 22.Kopito R R. ER quality control: the cytoplasmic connection. Cell. 1997;88:427–430. doi: 10.1016/s0092-8674(00)81881-4. [DOI] [PubMed] [Google Scholar]
- 23.Li R, Pei H, Watson D K, Papas T S. EAP1/Daxx interacts with ETS1 and represses transcriptional activation of ETS1 target genes. Oncogene. 2000;19:745–753. doi: 10.1038/sj.onc.1203385. [DOI] [PubMed] [Google Scholar]
- 24.Little E, Lee A S. Generation of a mammalian cell line deficient in glucose-regulated protein stress induction through targeted ribozyme driven by a stress-inducible promoter. J Biol Chem. 1995;270:9526–9534. [PubMed] [Google Scholar]
- 25.Liu H, Bowes R C, van de Water B, Sillence C, Nagelkerke J F, Stevens J L. Endoplasmic reticulum chaperones GRP78 and calreticulin prevent oxidative stress, Ca2+ disturbances, and cell death in renal epithelial cells. J Biol Chem. 1997;272:21751–21759. doi: 10.1074/jbc.272.35.21751. [DOI] [PubMed] [Google Scholar]
- 26.Liu R M, Gao L, Choi J, Forman H J. Gamma-glutamylcysteine synthetase: mRNA stabilization and independent subunit transcription by 4-hydroxy-2-nonenal. Am J Physiol. 1998;275:L861–L869. doi: 10.1152/ajplung.1998.275.5.L861. [DOI] [PubMed] [Google Scholar]
- 27.Lu S C. Regulation of hepatic glutathione synthesis: current concepts and controversies. FASEB J. 1999;13:1169–1183. [PubMed] [Google Scholar]
- 28.Lu S C, Kuhlenkamp J, Garcia-Ruiz C, Kaplowitz N. Hormone-mediated down-regulation of hepatic glutathione synthesis in the rat. J Clin Investig. 1991;88:260–269. doi: 10.1172/JCI115286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luethy J D, Holbrook N J. Activation of the gadd153 promoter by genotoxic agents: a rapid and specific response to DNA damage. Cancer Res. 1992;52:5–10. [PubMed] [Google Scholar]
- 30.Matsumoto M, Minami M, Takeda K, Sakao Y, Akira S. Ectopic expression of CHOP (GADD153) induces apoptosis in M1 myeloblastic leukemia cells. FEBS Lett. 1996;395:143–147. doi: 10.1016/0014-5793(96)01016-2. [DOI] [PubMed] [Google Scholar]
- 31.Mulcahy R T, Wartman M A, Bailey H H, Gipp J J. Constitutive and beta-naphthoflavone-induced expression of the human gamma-glutamylcysteine synthetase heavy subunit gene is regulated by a distal antioxidant response element/TRE sequence. J Biol Chem. 1997;272:7445–7454. doi: 10.1074/jbc.272.11.7445. [DOI] [PubMed] [Google Scholar]
- 32.Pahl H L. Signal transduction from the endoplasmic reticulum to the cell nucleus. Physiol Rev. 1999;79:683–701. doi: 10.1152/physrev.1999.79.3.683. [DOI] [PubMed] [Google Scholar]
- 33.Reed D J, Babson J R, Beatty P W, Brodie A E, Ellis W W, Potter D W. High-performance liquid chromatography analysis of nanomole levels of glutathione, glutathione disulfide, and related thiols and disulfides. Anal Biochem. 1980;106:55–62. doi: 10.1016/0003-2697(80)90118-9. [DOI] [PubMed] [Google Scholar]
- 34.Ron D, Habener J H. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 1992;6:439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- 35.Sato N, Iwata S, Nakamura K, Hori T, Mori K, Yodoi J. Thiol-mediated redox regulation of apoptosis. Possible roles of cellular thiols other than glutathione in T cell apoptosis J. Immunol. 1995;154:3194–3203. [PubMed] [Google Scholar]
- 36.Sylvester S L, ap Rhys C M J, Luethy-Martindale J D, Holbrook N J. Induction of GADD153, a CCAAT/enhancer-binding protein (C/EBP)-related gene, during the acute phase response in rats. J Biol Chem. 1994;269:20119–20125. [PubMed] [Google Scholar]
- 37.Ubeda M, Wang X Z, Zinszner H, Wu I, Habener J F, Ron D. Stress-induced binding of the transcriptional factor CHOP to a novel DNA control element. Mol Cell Biol. 1996;16:1479–1489. doi: 10.1128/mcb.16.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van den Dobbelsteen D J, Nobel C S I, Schlegel J, Cotgreave I A, Orrenius S, Slater A F. Rapid and specific efflux of reduced glutathione during apoptosis induced by anti-Fas/APO-1 antibody. J Biol Chem. 1996;271:15420–15427. doi: 10.1074/jbc.271.26.15420. [DOI] [PubMed] [Google Scholar]
- 39.Voehringer D W. BCL-2 and glutathione: alterations in cellular redox state that regulate apoptosis sensitivity. Free Radic Biol Med. 1999;27:945–950. doi: 10.1016/s0891-5849(99)00174-4. [DOI] [PubMed] [Google Scholar]
- 40.Wang X Z, Lawson B, Brewer J W, Zinszner H, Sanjay A, Mi L J, Boorstein R, Kreibich G, Hendershot L M, Ron D. Signals from the stressed endoplasmic reticulum induce C/EBP-homologous protein (CHOP/GADD153) Mol Cell Biol. 1996;16:4273–4280. doi: 10.1128/mcb.16.8.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watson R W, Rotstein O D, Nathens A B, Dackiw A P, Marshall J C. Thiol-mediated redox regulation of neutrophil apoptosis. Surgery. 1996;120:150–157. doi: 10.1016/s0039-6060(96)80282-0. [DOI] [PubMed] [Google Scholar]
- 42.Wild A C, Moinova H R, Mulcahy R T. Regulation of gamma-glutamylcysteine synthetase subunit gene expression by the transcription factor Nrf2. J Biol Chem. 1999;274:33627–33636. doi: 10.1074/jbc.274.47.33627. [DOI] [PubMed] [Google Scholar]
- 43.Wolfgang C D, Chen B P C, Martindale J L, Holbrook N J, Hai T. Gadd153/Chop10, a potential target gene of the transcriptional repressor ATF3. Mol Cell Biol. 1997;17:6700–6707. doi: 10.1128/mcb.17.11.6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zauli G, Gibellini D, Caputo A, Bassini A, Negrini M, Monne M, Mazzoni M, Capitani S. The human immunodeficiency virus type-1 Tat protein upregulates Bcl-2 gene expression in Jurkat T-cell lines and primary peripheral blood mononuclear cells. Blood. 1995;86:3823–3834. [PubMed] [Google Scholar]
- 45.Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot R T, Remotti H, Stevens J L, Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]