Abstract
Introduction
Sasang Constitutional Medicine (SCM) is a type of traditional Korean medicine where patients are classified as one of four Sasang constitution types (Sasang type) and medications consisting of medicinal herbs are prescribed according to the Sasang type. Despite the importance of personalized medicine, the operation mechanism is largely unknown. To gain a better understanding, we investigated the compound information that composes Sasang type-specific personalized herbal medicines on both multivariate and univariate levels.
Methods
Five machine learning classifiers including extremely randomized trees (ERT) were trained to investigate whether the Sasang type can be explained by compound information at the multivariate level. Hierarchical clustering was conducted to determine whether compounds are processed distributedly or specifically. Taxonomic and biosynthetic analyses were conducted on these compounds. A univariate level statistical test was conducted to provide more robust Sasang type-specific compound information.
Results
Using the trained ERT classifier, sixty important compounds were extracted. The sixty compounds were clustered into three groups, corresponding to each Sasang type-prominent compounds, suggesting that most compounds have specific preference for the Sasang type. Structural and biosynthetic characteristics of these Sasang type-prominent compounds were determined based on taxonomy and pathway analyses. Fourteen compounds showed statistically significant relevance with the Sasang type. Additionally, we predicted the Sasang type of unknown herbs, which were confirmed by their biological effects in functional assays.
Conclusion
This study investigated the personalized herbal medicines of the SCM using compound information. This study provided information on the chemical characteristics of the compounds that are essential for classifying the Sasang type of medicinal herbs, as well as predictions regarding the Sasang type of the commonly used but unidentified medicinal herbs.
Keywords: Personalized herbal medicine, Sasang constitutional medicine, Chemical characteristics, Compound information
1. Introduction
Sasang Constitutional Medicine (SCM) is a type of traditional Korean medicine, in which patients are classified into one of four Sasang constitution types (Sasang type): So-Eum (SE), So-Yang (SY), Tae-Eum (TE), and Tae-Yang (TY) [1]. The herbs are classified into four groups corresponding to the four Sasang types. For example, Panax Ginseng is used only for SE patients and Ephedra herba is used for TE patients. In addition, SCM is well-known for its personalized medicine characteristics, which means that despite the similar symptoms, the medications consisting of various medicinal herbs are prescribed differently depending on the patient's Sasang type.
Despite the importance of personalized medicine, however, it is only possible to assume the classification principle of SCM because the classification criteria for the herbs into four Sasang type groups or the meaning of each herb belonging to each Sasang type are largely unknown. Thus far, although various studies have been conducted to find the criteria and the meaning of herbal classification [[2], [3], [4]], most studies were conducted by applying the theoretical concepts of herbs used in conventional traditional medicine or by narratively reviewing the results for each Sasang type [3, 5]. Recently, chemical property-based various machine learning approaches have been applied to investigate natural products including herbal medicines [6, 7].
To gain a better understanding of the personalized medicine characteristics of SCM and its operation, we conducted a detailed investigation of the compound information that composes the Sasang type-specific personalized herbal medicines on both multivariate and univariate levels in this study. Using machine learning (ML) techniques and statistical analyses, compound patterns and specific compounds that enable Sasang type classification were found, and the chemical characteristics of the important compounds contributing to the classification were analyzed. The Sasang types of medicinal herbs, whose Sasang types were unidentified, were predicted based on compound information, and the prediction results were confirmed by a simple functional assay.
2. Methods
2.1. Identification of herbs
The list of herbs for each Sasang constitution type was obtained from 『Donguisusebowon Sinchukbon』 (『東醫壽世保元 辛丑本』; Longevity and Life Preservation of the Eastern Medicine), a book in which Jema Lee, the founder of SCM, presented the final result of his categorization of herbs by Sasang type. Previous studies [8, 9] that identified the medicinal herbs belonging to each Sasang type were also referenced. As a result, 144 herbs (47 for SE type, 37 for SY type, 44 for TE type, and 16 for TY type) were included in this study.
2.2. Construction of an herb-compound matrix dataset
Traditional Chinese Medicine Systems Pharmacology and Analysis Platform (TCMSP) (https://old.tcmsp-e.com/tcmsp.php), a database containing information on 499 herbs and 29,384 compounds, was used to extract compound information for each herb [10]. TCMSP was selected because the number of compounds included in the database has a greater advantage than other databases. When 144 herbs were analyzed using the database, 92 medicinal herbs (31 for the SE type, 30 for the SY type, 27 for the TE type, and 4 for the TY type) were included, allowing further analysis (Table 1).
Table 1.
List of SCM medicinal herbs.
| Sasang type | Chinese name | Pronunciation | English name | |
|---|---|---|---|---|
| 1 | SE | 白芍藥 | Baishao | Paeoniae Radix Alba |
| 2 | SE | 白朮 | Baizhu | Atractylodes Macrocephala Koidz. |
| 3 | SE | 半夏 | Banxia | Arum Ternatum Thunb. |
| 4 | SE | 陳皮 | Chenpi | Citrus Reticulata |
| 5 | SE | 川芎 | Chuanxiong | Chuanxiong Rhizoma |
| 6 | SE | 葱白 | Congbai | Allii Fistulost Bulbus |
| 7 | SE | 大腹皮 | Dafupi | Areca Catechu L. |
| 8 | SE | 當歸 | Danggui | Angelicae Sinensis Radix |
| 9 | SE | 大蒜 | Dasuan | Allii Sativi Bulbus |
| 10 | SE | 大棗 | Dazao | Jujubae Fructus |
| 11 | SE | 付子 | Fuzi | Aconiti Lateralis Radix Praeparata |
| 12 | SE | 甘草 | Gancao | Licorice |
| 13 | SE | 乾薑 | Ganjiang | Zingiberis Rhizoma |
| 14 | SE | 良薑 | Gaoliangjiang | Alpiniae Officinarum Rhizome |
| 15 | SE | 藿香 | Guanghuoxiang | Pogostemon Cablin (Blanco) Benth. |
| 16 | SE | 桂枝 | Guizhi | Cinnamomi Ramulus |
| 17 | SE | 厚朴 | Houpu | Magnolia Officinalis Rehd Et Wils |
| 18 | SE | 胡椒 | Hujiao | Piperis Fructus |
| 19 | SE | 靑皮 | Qingpi | Citri Reticulatae Pericarpium Viride |
| 20 | SE | 人蔘 | Renshen | Panax Ginseng C. A. Mey. |
| 21 | SE | 官桂 | Rougui | Cinnanmomi Cortex |
| 22 | SE | 生薑 | Shengjiang | Zingiber Officinale Roscoe |
| 23 | SE | 吳茱萸 | Wuzhuyu | Evodiae Fructus |
| 24 | SE | 香付子 | Xiangfu | Cyperi Rhizoma |
| 25 | SE | 茵蔯 | Yinchen | Artemisiae Scopariae Herba |
| 26 | SE | 鸎粟殼 | Yingsuke | Papaveris Pericarpium |
| 27 | SE | 益智仁 | Yizhi | Alpiniae Oxyphyliae Fructus |
| 28 | SE | 枳實 | Zhishi | Aurantii Fructus Immaturus |
| 29 | SE | 蒼朮 | Cangzhu | Atractylodes Lancea (Thunb.)Dc. |
| 30 | SE | 黃芪 | Huangqi | Hedysarum Multijugum Maxim. |
| 31 | SE | 砂仁 | Sharen | Amomum Aurantiacum H. T. Tsai Et S. W. Zhao |
| 32 | SY | 薄荷 | Bohe | Menthae Herba |
| 33 | SY | 柴胡 | Chaihu | Radix Bupleuri |
| 34 | SY | 車前子 | Cheqianzi | Plantaginis Semen |
| 35 | SY | 地骨皮 | Digupi | Lycii Cortex |
| 36 | SY | 獨活 | Duhuo | Radix Angelicae Biseratae |
| 37 | SY | 防風 | Fangfeng | Saposhnikoviae Radix |
| 38 | SY | 茯笭 | Fuling | Poria Cocos (Schw.) Wolf. |
| 39 | SY | 覆盆子 | Fupenzi | Rubi Fructus |
| 40 | SY | 甘遂 | Gansui | Kansui Radix |
| 41 | SY | 枸杞子 | Gouqizi | Lycii Fructus |
| 42 | SY | 荊芥 | Jingjie | Schizonepetae Herba |
| 43 | SY | 金銀花 | Jinyinhua | Lonicerae Japonicae Flos |
| 44 | SY | 苦蔘 | Kushen | Sophorae Flavescentis Radix |
| 45 | SY | 連翹 | Lianqiao | Forsythiae Fructus |
| 46 | SY | 牧丹皮 | Mudanpi | Cortex Moutan |
| 47 | SY | 木通 | Mutong | Caulis Akebiae |
| 48 | SY | 牛蒡子 | Niubangzi | Fructus Arctii |
| 49 | SY | 前胡 | Qianhu | Peucedani Radix |
| 50 | SY | 羌活 | Qianghuo | Notopterygii Rhizoma Et Radix |
| 51 | SY | 山茱萸 | Shanzhuyu | Cornus Officinalis Sieb. Et Zucc. |
| 52 | SY | 熟地黃 | Shudihuang | Rehmanniae Radix Praeparata |
| 53 | SY | 玄參 | Xuanshen | Figwort Root |
| 54 | SY | 澤瀉 | Zexie | Alisma Orientale (Sam.) Juz. |
| 55 | SY | 知母 | Zhimu | Anemarrhenae Rhizoma |
| 56 | SY | 山梔子 | Zhizi | Gardeniae Fructus |
| 57 | SY | 豬苓 | Zhuling | Polyporus Umbellatus (Pers)Fr. |
| 58 | SY | 黃栢 | Huangbo | Phellodendri Chinrnsis Cortex |
| 59 | SY | 川黃連 | Huanglian | Coptidis Rhizoma |
| 60 | SY | 沒藥 | Moyao | Myrrha |
| 61 | SY | 乳香 | Ruxiang | Olibanun |
| 62 | TE | 白果 | Baiguo | Ginkgo Semen |
| 63 | TE | 白芷 | Baizhi | A. Dahurica (Fisch.) Benth. Et Hook |
| 64 | TE | 栢子仁 | Baiziren | Platycladi Semen |
| 65 | TE | 五味子 | Beiwuweizi | Schisandrae Chinensis Fructus |
| 66 | TE | 大黃 | Dahuang | Radix Rhei Et Rhizome |
| 67 | TE | 浮萍 | Fuping | Spirodelae Herba |
| 68 | TE | 藁本 | Gaoben | Ligustici Rhizoma Et Radix |
| 69 | TE | 葛根 | Gegen | Radix Puerariae |
| 70 | TE | 苽蔕 | Guadi | Calyx Cucumis |
| 71 | TE | 黃芩 | Huangqin | Scutellariae Radix |
| 72 | TE | 桔梗 | Jiegeng | Platycodon Grandiforus |
| 73 | TE | 款冬花 | Kuandonghua | Farfarae Flos |
| 74 | TE | 蘿葍子 | Laifuzi | Raphani Semen |
| 75 | TE | 麻黃 | Mahuang | Ephedra Herba |
| 76 | TE | 牛黃 | Niuhuang | Bovis Calculus |
| 77 | TE | 蒲黄 | Puhuang | Pollen Typhae |
| 78 | TE | 桑白皮 | Sangbaipi | Mori Cortex |
| 79 | TE | 山藥 | Shanyao | Rhizoma Dioscoreae |
| 80 | TE | 升麻 | Shengma | Cimicifugae Rhizoma |
| 81 | TE | 石菖蒲 | Shichangpu | Acoritataninowii Rhizoma |
| 82 | TE | 使君子 | Shijunzi | Quisqualis Indica |
| 83 | TE | 酸枣仁 | Suanzaoren | Ziziphi Spinosae Semen |
| 84 | TE | 天門冬 | Tiandong | Asparagi Radix |
| 85 | TE | 烏梅 | Wumei | Mume Fructus |
| 86 | TE | 甘菊花 | Yejuhua | Chrysanthemi Indici Flos |
| 87 | TE | 薏苡仁 | Yiyiren | Coicis Semen |
| 88 | TE | 皂角 | Zaojiaoci | Gleditsiae Spina |
| 89 | TY | 蘆根 | Lugen | Phragmitis Rhizoma |
| 90 | TY | 木苽 | Mugau | Chaenomeles Sinensis (Thouin) Koehne |
| 91 | TY | 松花 | Songhuafen | Pine Pollen |
| 92 | TY | 松節 | Songjie | Lignum Pini Nodi |
SE, So-Eum type; SY, So-Yang type; TE, Tae-Eum type.
The compound information for each herb was extracted from the database and transformed into a simplified molecular-input line-entry system (SMILES) string [11]. A vector containing all compounds of 92 herbs was constructed using one-hot encoding (size of 1 × 4745). As a result, a herb-compound matrix (size of 92 × 4745) was constructed and used in analyses in this study. All data preprocessing and analysis were performed using Pandas, a Python library for data manipulation and analysis, and Scikit-learn, a Python module that integrates a broad range of machine learning algorithms [12].
2.3. Machine learning (ML) experimental details
2.3.1. ML model selection
Five well-known supervised machine-learning algorithms for classification were applied in this study. The models compared are as follows: extremely randomized trees (ERT), extreme gradient boosting (XGBoost), linear and nonlinear support vector machine (SVM), and multinomial logistic regression (Mlogit).
The ERT classifier is a decision tree-based ensemble method that is similar to random forests but uses randomly selected cut-off values rather than the optimal one. The strength of the ERT classifier is that it is robust to noise and can thus lead to a further decrease in overall variance while performing largely equal to or better than other tree-based classifiers [13]. Furthermore, the ensemble method can rank the importance of features used in a classification problem [14]. The XGBoost is a kind of Machine Learning algorithm belonging to a decision-tree-based ensemble and enrolls an advanced framework of gradient boosting [15]. The SVM classifier searches for the optimal hyperplane that maximizes the margin between classes in high-dimensional space [16]. The SVM classifier can be used as linear or nonlinear classifiers according to the applied kernel. The radial basis function (Gaussian kernel) was applied for the nonlinear SVM classifier. The one-vs.-rest scheme was used to apply the SVM, a binary classifier, into a multi-class problem. The Mlogit classifier is used to predict a nominal dependent variable with more than two categories [17]. The strength of the Mlogit model is that it measures how relevant a predictor (coefficient size) is and the direction of association (positive or negative) of the predictor.
2.3.2. Feature selection
The optimal number of features was selected by calculating the performance (accuracy) of the model while increasing the number of features from 10 to 4700 in increments of 10. The features were included from the feature (compound) with the highest feature importance score. It was calculated by a double nested cross-validation. This procedure was repeated ten times to avoid inconsistent results caused by randomness. Sixty features showed the best performance. The sixty features (compounds) showing the highest feature importance scores were selected by a nested cross-validation performance. All the feature selection procedures were conducted in the nested training set of each fold to avoid data leakage.
2.3.3. Hyperparameter optimization
For hyperparameter optimization, a randomized search on hyperparameters with nested cross-validation to avoid data leakage was conducted. The hyperparameter configuration can be varied across the folds because the hyperparameters were tuned for each fold. Supplementary Table S1summarizes the searched hyperparameters and their range for each ML model.
2.4. Model performance assessment
2.4.1. K-fold cross-validation
Each model was trained with stratified k-fold cross-validation (k = 4), in which the dataset was divided randomly into k disjoint subsets of approximately equal size according to the Sasang type.
2.4.2. AUROC, precision, recall, f1 score, and accuracy
The AUC was used to evaluate how well the ML model distinguishes the Sasang types with the compounds configuration of each herb. The area under the receiving operating characteristic (AUROC) curve was calculated using the implementation in the Scikit-learn Python package. The precision, recall, f1 score, and accuracy (equations (1), (2), (3), (4))) were used to evaluate the performance of the machine-learning model.
| (1) |
| (2) |
| (3) |
| (4) |
In this equation, tp denotes the true positive; fp, false positive; tn, true negative; fn, false negative. Macro-average methods that treat all classes equally were used to calculate the average values in multi-class classification settings.
2.5. Cell-based analysis
2.5.1. Cell culture and reagents
AGS (derived from the stomach), HepG2 (derived from the liver) and NRK-52E (derived from the kidney) cells were obtained from American Type Culture Collection (ATCC, Rockville, MD). The cells were maintained in Dulbecco's modified Eagle's medium liquid (DMEM) with high glucose levels, 10% fetal bovine serum (FBS), 50 units/ml penicillin, and 50 μg/ml streptomycin at 37 °C in a humidified atmosphere containing 5% CO2. For all experiments, the cells were starved for 12 h in FBS-free media [18]. Curcuma longa Radix, Houttuynia cordata and Leonurus japonicus Houtt were extracted using the medicinal standard herbs, which are guaranteed by Korea FDA and produced by the pharmaceutical company (Daewon pharmacy, Korea) approved as the good manufacturing practice (GMP) system as previously described [19, 20].
2.5.2. MTT assay
The cells were plated at a density of 1 × 10⁵ cells per well in 48-well culture plates and incubated in an FBS-free medium for 12 h. AGS and NRK-52E cell were incubated with drugs for 24 h [18]. HepG2 cells were incubated with drugs for 1 h, followed by a treatment with AA (10 μM) for 12 h and then iron (5 μM) for 6 h. The cell viability was defined as relative to the untreated control [i.e., viability (% of control) = 100 × (absorbance of the treated sample)/(absorbance of control)] as previously described [19,20].
2.6. Statistical analysis
Analysis of variance (ANOVA) test was used to assess the differences in the number of compounds belonging to the four Sasang types. Fisher's exact test was used to determine if there are non-random associations between each compound and each Sasang type, at a univariate level. Multiple comparison correction was not conducted because the purpose of Fisher's exact test was to suggest relevant compound candidates.
3. Results
3.1. Basic characteristics of the compounds comprising the SCM medicinal herbs
Ninety-two medicinal herbs were analyzed (31 for the SE type, 30 for the SY type, 27 for the TE type, and four for the TY type) in the current study (Table 1). Before examining whether it was possible to discriminate the Sasang type using the compound information composing each herb, we first described the basic characteristics of the compound information. The average numbers of compounds were 99.3, 95.2, 65.0, and 34.5 for SE, SY, TE, and TY types, respectively (Fig. 1A). We examined the difference between the number of compounds among the different Sasang type groups. Although the difference of compound number among the Sasang types was marginally significant (F-stat = 2.68, p-val = 0.052), it seems that the TY type is the primary factor, as the compound number of the other types appears comparable. (Fig. 1B). The overlap of compound lists belonging to each Sasang type was also examined using a Venn diagram (Fig. 1C).
Fig. 1.
Basic characteristics of Sasang herb data (A) Box plot of the compounds data used in the analysis. The number of compounds for each Sasang type was analyzed using a box plot, and the average number of compounds for each type was calculated. SE, So-Eum type; SY, So-Yang type; TE, Tae-Eum type; TY, Tae-Yang type. (B) Result of one-way ANOVA. The data were plotted as a function of the means and 95% confidence intervals. Tukey was conducted as a post-hoc analysis. (C) Venn diagram of the compounds belonging to each Sasang type.
3.2. Classification of each herb to a specific sasang type being explained by the multivariate level compound information
ML classifiers were trained for Sasang type and their performance of the classifiers was assessed to find out whether the Sasang type information can be explained by compound combination at the multivariate level, i.e., whether it is possible to discriminate the corresponding Sasang type using the compound information composing each herb. Four TY-type herbs were excluded because of their small sample size [21,22]. Four samples were too small for the model to learn the generalizable pattern. As a result, 88 herbs were used for subsequent analyses.
Five well-known classification ML models were applied for this study. The ML models compared in this study were as follows: ERT, XGBoost, linear and nonlinear (RBF) SVM, and Mlogit (see Materials and Methods for more details). Since the ERT model outperformed the other models (Table 2), the ERT classifier was selected. The macro-averaged accuracy and f1 score of the ERT classifier were 0.511 ± 0.121 and 0.498 ± 0.129, respectively (mean ± SD).
Table 2.
Overall classification performance for each model.
|
Model |
Precision | Recall | F1 score | Accuracy |
|---|---|---|---|---|
| ERT | 0.505 0.125 | 0.522 0.139 | 0.498 0.129 | 0.511 0.121 |
| XGBoost | 0.394 0.064 | 0.436 0.078 | 0.374 0.077 | 0.397 0.067 |
| SVM (RBF) | 0.475 0.066 | 0.482 0.062 | 0.467 0.070 | 0.489 0.067 |
| SVM(Linear) | 0.441 0.087 | 0.514 0.109 | 0.420 0.092 | 0.455 0.085 |
| Mlogit | 0.464 0.061 | 0.446 0.061 | 0.422 0.049 | 0.466 0.067 |
Mean SD. ERT, extremely randomized trees; XGBoost, extreme gradient boosting; SVM, support vector machine; Mlogit, multinomial logistic regression.
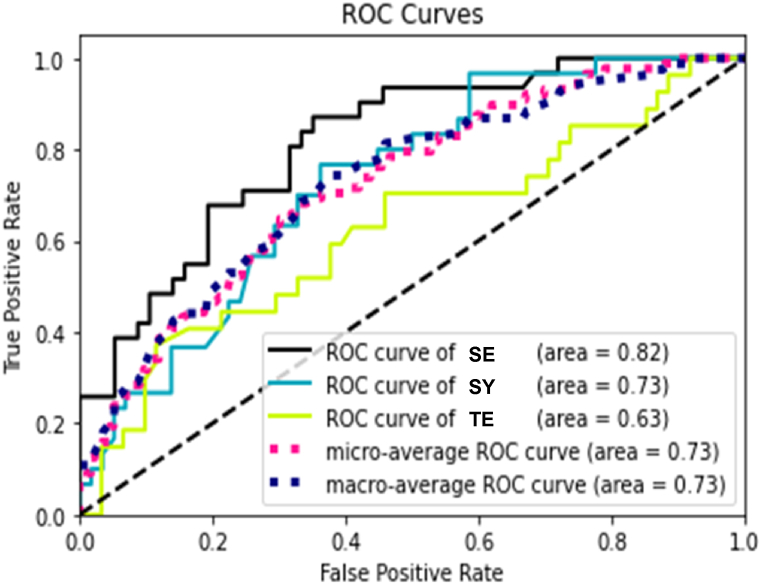
The ERT classifier was investigated more thoroughly for a detailed analysis of the classifier performance. The macro-average AUROC of the classifier was 0.73 (Fig. 2). The classification performance of the ERT model for the individual Sasang type was as follows: the average precision, recall, and f1 score for the SE type were 0.612, 0.671, and 0.621, respectively; 0.464, 0.451, and 0.434, respectively, for the SY type; 0.440, 0.443, and 0.439, respectively, for TE type (Table 3, Supplementary Figure S1). The result showed statistically significant classification performance for the SE and SY type, suggesting that the configuration of the compound composing each herb has information to discriminate the Sasang type of each herb. In other words, the classification of each herb to a specific type can be explained by the compound configuration of the herbs.
Fig. 2.
Classification performance of the Sasang type decoder. Receiver operating characteristic (ROC) curve of 88 herbs with 60 compound features. SE, So-Eum type; SY, So-Yang type; TE, Tae-Eum type.
Table 3.
Classification performance of the extremely randomized trees classifier for individual Sasang type.
| Precision | Recall | F1 score | Accuracy | |
|---|---|---|---|---|
| SE | 0.612 0.155 | 0.671 0.132 | 0.621 0.076 | 0.511 0.121 |
| SY | 0.464 0.271 | 0.451 0.127 | 0.434 0.189 | |
| TE | 0.440 0.132 | 0.443 0.185 | 0.439 0.159 | |
| Average | 0.505 | 0.522 | 0.498 | 0.511 |
Mean SD. SE, So-Eum type; SY, So-Yang type; TE, Tae-Eum type.
3.3. Most compounds showing selectivity for particular sasang type
To avoid curse of dimensionality, feature selection was conducted. The optimal number of features was chosen by the double nested cross-validation performance (see the Methods 2.3.2). Sixty turned out to be the optimal number and the sixty features (compounds) that have the highest feature importance scores were selected by the nested cross-validation performance. The sixty features (compounds) were analyzed to understand which compounds are processed in a distributed manner and which are processed in a labeled-line manner for the purpose of classifying the Sasang type information. The sixty compounds were clustered based on the cosine similarity between each vector of the sixty compounds (size of 88 × 1) and representative Sasang type vector for each type (size of 88 × 1), representing the distribution of each compound and each Sasang type within 88 herbs, respectively. To define the clusters, the dendrogram was cut at the second level, resulting in three clusters and one compound (Fig. 3). We found that the three clusters corresponded to TE-prominent (n = 8), SY-prominent (n = 9), and SE-prominent compound groups (n = 42), which suggests that the majority of compounds have selectivity for particular Sasang type.
Fig. 3.
Clustered heatmap of the 60 compounds showing SC-prominent compounds. Each row represents each compound, and the column represents each SC type. The color represents the similarity (cosine similarity) between the distribution of each compound and that of each SC type. The data was clustered with respect to rows. The compounds were clustered into three groups at the second level dendrogram, resulting in TE-prominent, SY-prominent, and SE-prominent compounds, respectively. SE, So-Eum type; SY, So-Yang type; TE, Tae-Eum type.
3.4. Chemical characteristics of the sasasng type-prominent compounds
To identify the chemical characteristics of the Sasang type-prominent compounds, firstly, the chemical taxonomy of each cluster, i.e., the structural classification of chemical entities using ClassyFire [23], was identified (Supplementary Tables S2, S3, and S4). Many of the TE-prominent compounds showed a class of fatty acyls (7/8). Unlike TE-prominent compounds, SY-prominent compounds showed heterogeneous composition with various classes: prenol lipids (2/9), fatty acyls (2/9), coumarins and derivatives (2/9), carboxyl acids and derivatives (1/9), cinnamic acids and derivatives (1/9), and organooxygen compounds (1/9). The SE-prominent compounds were identified as prenol lipids (21/42), flavonoids (9/42), and benzene and substituted derivatives (4/42).
Furthermore, a review of the biosynthetic characteristics of the major secondary metabolites in sixty compounds that are crucial in the Sasang type classification could suggest an interesting point of view [24]. As shown in Supplementary Table S2, the majority of the components in the medicinal herbs classified as TE type were fatty acid-based substances synthesized via a polyketide biosynthetic pathway that requires various polyketide synthases. In the case of compounds from the medicinal herbs for the SY type, the shikimate, mevalonate, and polyketide pathways were involved in biosynthesizing these metabolites. As in the case of SY type, the compound list for the SE type showed that various biosynthetic pathways were involved for these compounds, but terpenoids appeared most commonly, basically biosynthesized through the mevalonic (isoprenoid) pathway [25].
3.5. Identification of sasasng type-specific compounds from a univariate level analysis
A univariate level statistical test (Fisher's exact test) was conducted on the sixty compounds showing a high feature importance score to find the compounds that are more relevant to the Sasang type, i.e., the Sasang type-specific compounds. Fourteen Sasang type-specific compounds were found accordingly, and the relative ratio of each compound for each Sasang type was analyzed (Fig. 4). Among them, ten, three, and one compound are relevant to the SE, SY, and TE types, respectively. Guaiene, -cis-.beta.-Elemene diastereomer, naringin, 3691-11-0, o-cymol, cadalin, cadinene, alpha-terpineol, germacrene, l-limonen are found to be SE-specific compounds (Fig. 4A); FER, marmesin, decyl acetate to be SY-specific compounds (Fig. 4B); and arachidonic acid to be TE-specific compounds (Fig. 4C).
Fig. 4.
SC type-specific compounds. Fourteen compounds showed significant relevance with the SC type (according to Fisher's exact test). The bar graph represents the relative frequency of each compound, indicating which SC type compound shows relevance. The relative frequency is . (A) SE-specific compounds. (B) SY-specific compounds. (C) TE-specific compound. SE, So-Eum type; SY, So-Yang type; TE, Tae-Eum type.
3.6. Sasang type prediction of unidentified medicinal herbs based on compound level information, and their investigations by functional assay
The trained ML classifier was applied to predict the Sasang type of medicinal herbs whose Sasang types are unidentified as a further application. Thirty medicinal herbs were selected according to the amount of the usage [26].
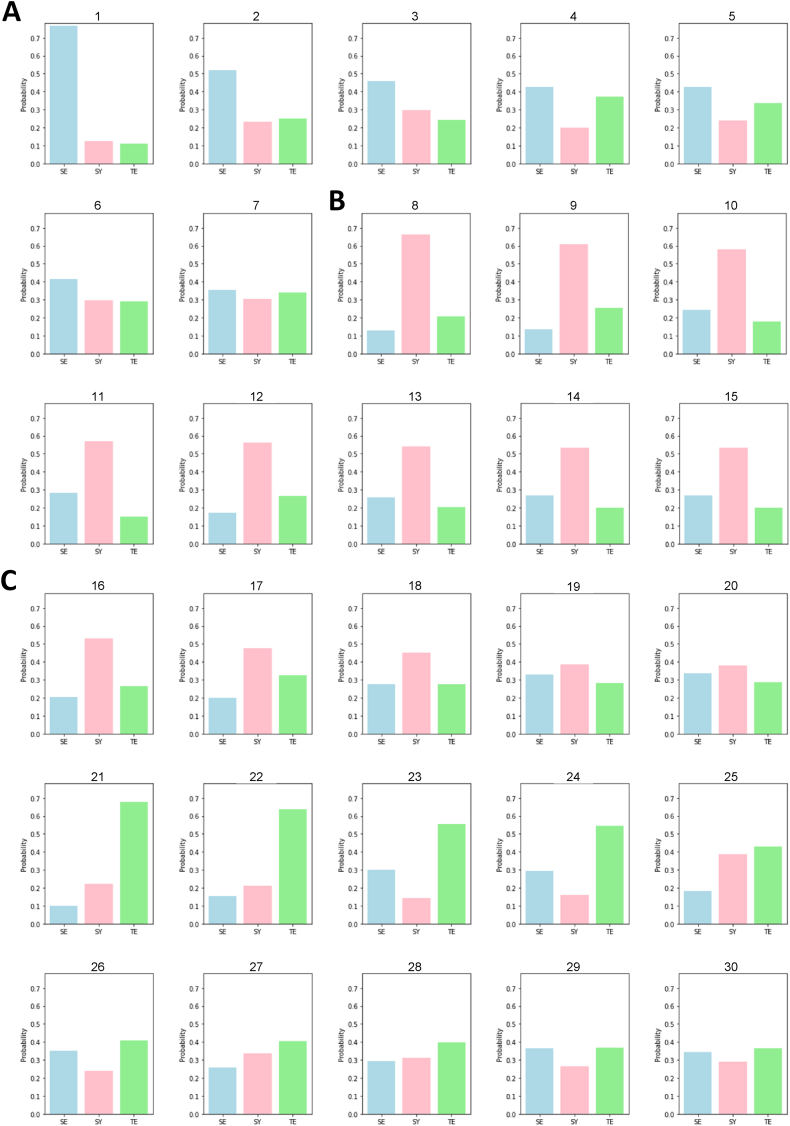
The trained ML classifier successfully predicted the Sasang type of the thirty herbs with the probability of which Sasang type each herb belongs. The prediction procedure was repeated 10 times for each herb, and the mean probability was presented to provide stability (Fig. 5 and Table 4). As a result, seven kinds of herbs (from number 1 to number 7), including Curcuma longa Radix and Eriobotrya japonica Lindley corresponded to SE type (Fig. 5A), and thirteen herbs (from number 8 to number 20), such as Houttuynia cordata, Cistanche deserticola, and Lindera strychnifolia Vill. are related to the SY type (Fig. 5B). Ten herbs (from number 21 to number 30) containing Leonurus japonicus Houtt. and Benincasa hispida Cogniaux were categorized as the TE type (Fig. 5C).
Fig. 5.
Predicted result of top 30 unidentified medicinal herbs using trained ERT model. The Sasang type of the 30 medicinal herbs was predicted using the trained ERT model. The bar represents the mean probability (ten trials repeated) of which constitution each medicinal herb belongs. The predicted Sasang type of each herb is visualized using background colors, and the herbs are clustered based on their predicted Sasang type. In each herbal group, the herbs were sorted according to their probability. The numbers indicate the name of herbs in Table 4 (A) Compounds 1–7 predicted as SE type. (B) Compounds 8–15 predicted as SY type. (C) Compounds 16–30 predicted as TE type. SE, So-Eum type; SY, So-Yang type; TE, Tae-Eum type.
Table 4.
Prediction probability for the top 30 unidentified medicinal herbs to belong to which constitution.
| Chinese name | English name | SE | SY | TE | |
|---|---|---|---|---|---|
| 1 | 鬱金 | Curcuma longa Radix | 0.766 | 0.125 | 0.110 |
| 2 | 枇杷葉 | Eriobotrya japonica Lindley | 0.519 | 0.231 | 0.250 |
| 3 | 防己 | Sinomenium acutum Rehder et Wilson | 0.459 | 0.298 | 0.243 |
| 4 | 丹蔘 | Salvia miltiorrhiza Bunge | 0.427 | 0.202 | 0.371 |
| 5 | 艾葉 | Artemisia princeps Pampanini | 0.426 | 0.239 | 0.335 |
| 6 | 丁香 | Syzygium aromaticum Merrill et Perry | 0.414 | 0.296 | 0.290 |
| 7 | 龍膽草 | Gentiana scabra Bunge | 0.354 | 0.304 | 0.342 |
| 8 | 魚腥草 | Houttuynia cordata | 0.129 | 0.665 | 0.206 |
| 9 | 肉蓯蓉 | Cistanche deserticola | 0.137 | 0.610 | 0.253 |
| 10 | 烏藥 | Lindera strychnifolia Vill. | 0.243 | 0.580 | 0.177 |
| 11 | 淫羊藿 | Epimedium koreanum Nakai | 0.282 | 0.569 | 0.149 |
| 12 | 肉荳蔲 | Myristica fragrans | 0.170 | 0.563 | 0.266 |
| 13 | 北沙參 | Glehnia littoralis Fr. Schmidt ex Miquel | 0.258 | 0.540 | 0.202 |
| 14 | 細⾟ | Asarum sieboldii Miq. | 0.268 | 0.532 | 0.199 |
| 15 | 辛夷 | Magnolia denudata Desrousseaux | 0.268 | 0.532 | 0.199 |
| 16 | 檳榔子 | Areca catechu Linné | 0.205 | 0.530 | 0.265 |
| 17 | 百合 | Lilium lancifolium Thunberg | 0.201 | 0.475 | 0.324 |
| 18 | 小茴香 | Foeniculum vulgare | 0.275 | 0.451 | 0.274 |
| 19 | 蛇床子 | Torilis japonica | 0.330 | 0.388 | 0.282 |
| 20 | 紫蘇葉 | Perillae Folium | 0.336 | 0.379 | 0.285 |
| 21 | 益母草 | Leonurusjaponicus Houtt | 0.101 | 0.220 | 0.679 |
| 22 | 冬瓜子 | Benincasa hispida Cogniaux | 0.153 | 0.210 | 0.637 |
| 23 | 決明子 | Cassia tora Linné | 0.301 | 0.143 | 0.556 |
| 24 | 杜仲 | Eucommia ulmoides Oliver | 0.294 | 0.162 | 0.543 |
| 25 | 白蒺藜 | Tribulus terrestris | 0.183 | 0.386 | 0.430 |
| 26 | 釣鉤藤 | Uncariae Ramulus cum Uncus | 0.352 | 0.239 | 0.409 |
| 27 | 麥芽 | Hordeum vulgare Linné | 0.257 | 0.337 | 0.405 |
| 28 | 川貝母 | Fritillariae Cirrhosae Bulbus | 0.293 | 0.310 | 0.397 |
| 29 | 牛膝 | Twotoothed Achyranthes | 0.366 | 0.266 | 0.368 |
| 30 | 威靈仙 | Chinese Clematis | 0.343 | 0.291 | 0.366 |
SE, So-Eum type; SY, So-Yang type; TE, Tae-Eum type.
Additionally, we wanted to know whether the predicted herbs with the highest rank in each type have a biological function in each organ corresponding to the Sasang type. According to SCM theory, each Sasang type is highly related to specific organs (i.e. SE, kidney; SY, stomach; TE, liver) [1, 2, 3]; Thus, we tested the anti-cancer effects of Curcuma longa Radix (SE) in the NRK-52E kidney cancer cell line and Houttuynia cordata (SY) in the AGS stomach cancer cell line as well as anti-oxidant effects of Leonurus japonicus Houtt (TE) in the HepG2 hepatocyte (Fig. 6A–C). The water extract of Curcuma longa Radix (the first ranking herb in SE) and Houttuynia cordata (the first ranking herb in SY) significantly inhibited the proliferation of cancer derived from kidney and stomach, respectively. Leonurus japonicus Houtt (the first ranking herb in TE) markedly inhibited the oxidative damage induced by AA + iron.
Fig. 6.
Cell viability in three types of cells. The cells were plated at a density of 1 × 10⁵ cells per well in 48-well culture plates and incubated in an FBS-free medium for 12 h. (A and B) AGS and NRK-52E cells were incubated with drugs for 24 h. (C) HepG2 cells were incubated with drugs for 1 h, followed by a treatment with AA (10 μM) for 12 h and then iron (5 μM) for 6 h. All data represent means ± SD of 4 independent experiments (*ρ < 0.05 and **ρ < 0.01 vs. control group; ##ρ < 0.01 vs. AA + iron-treated group). AA, arachidonic acid; con, vehicle-treated control.
4. Discussion
SCM is a unique form of personalized medicine in traditional Korean medicine, in which the patients are classified into one of four Sasang constitution types: SE, SY, TE, or TY. In SCM, herbal medicines (composed of various medicinal herbs) are prescribed based on the patients’ Sasang type, in addition to their symptoms, and the applied medicinal herbs were divided into four groups corresponding to the four Sasang types; the herbs themselves are inextricably linked to the concept of Sasang type. Furthermore, a study of the herbs could provide more objective insight than investigating the Sasang type-diagnosed patients by SCM experts to understand the intrinsic principle of SCM, in that the agreement rate for diagnosed Sasang type among three qualified SCM experts is between 52.5% and 68.4% [2,27]. Using a novel drug-centric approach, this study examined the compound patterns that enable Sasang type classification and the chemical characteristics of each herbal group that contributes to the medicinal effect of SCM via ML techniques.
A previous study examined the major botanical compounds, such as phenol, alkaloid, and terpenoid, contained in each herb from a biomedical point of view. Lim et al. reported that phenolics were dominant in the TY-type herbs, iridoids and triterpenes were in the SY-type herbs, saponins were in the TE-type herbs, and monoterpene and sesquiterpenes were in SE type herbs [3]. On the other hand, because this research was still limited as a review on the characteristics of each herb, more systematic and data-driven group-level characteristics of each constitutional herbal group consisting of corresponding medicinal herbs have not been identified.
This study aimed to identify the principle/criteria for classifying medicinal herbs into the Sasang types using a data-driven approach. The current study examined whether the principle of distribution is explainable at the multi-compound-level by multivariate analyses, such as ML [28]. The Sasang type classification could be explained by multi-compound configuration, being confirmed by statistical significance in the classification performance of the ML classifier on the SE and SY type herbs. Various patterns made by multiple compounds would help classify different type groups at the multivariate level. Although the classification performance for the TE type was statistically insignificant (p value = 0.25), the herbs were only investigated at the compound level in this study. Other factors, in addition to the compound factor, would help account for the Sasang type classification principles. It is important how much the Sasang type classification can be explained by other characteristics, such as traditional theory-based taste and action information, in addition to the compound information in further analyses.
This study investigated which compounds play important roles in type discrimination to interpret the multi-compound patterns derived from ML analysis. The 60 compounds deducted by the ML pattern analysis were examined using hierarchical clustering, and 14 Sasang type-specific compounds were found by statistical analysis. On the other hand, because this result reflects only univariate-level investigation, much more remains to be investigated for multivariate-level interpretation.
The characteristics of these compounds were analyzed using chemical information. The secondary metabolites in the plants were biosynthesized by the action of various enzymes and generally constituted the active ingredients of medicinal plants [29,30]. It was also reported that the building blocks that make up the drug target of the bodies and a line of enzymes normally involved in the biosynthesis of secondary metabolites [31,32]. Although it is difficult to assume that the compounds listed in Fig. 4 have the representative activity of each herbal medicine and there is little scientific evidence to directly connect the biosynthetic pathway of some of the representative compounds to SCM, the potential and novelty of these attempts themselves cannot be ignored. In the TE type, fatty acid-based materials are normally synthesized through the polyketide biosynthetic pathway. On the other hand, the shikimate, mevalonate, and polyketide pathways were involved in the SY constitution. In the case of SE, one of the most abundant components, terpenoids, was related to the mevalonic (isoprenoid) pathway. Although this analysis alone is not enough to explain the characteristics of herbal medicines in the SCM, at least there are some differences for each Sasasng type in terms of the biosynthetic pathway.
This study has some limitations. Owing to the small sample size, medicinal herbs of the TY type were excluded from the analyses. The TY type frequently has a small sample size in other studies regarding Sasang type diagnosis because of the rarity of the TY type in the population distribution in SCM. The majority of studies excluded the TY type from their analyses. The same situation occurred in this study although we adopted a drug-centric approach. In addition, in this study, the herb-compound matrix was constructed using one-hot encoding, not considering the chemical similarity between compounds. It would be better to consider vector embedding in future research. In addition, in this study, we provided information about the predictive type of the medicinal herbs using a trained ML classifier. However, readers should note that this prediction is only a result of nonlinear pattern computation based on compound information, not based on the whole information comprising the Sasang type, implying that this predictive information was not recommended for direct clinical application. If there exists a discrepancy between the prediction result and the clinical application, this indicates that additional information is needed to the dataset. Refining the model remains to conduct additional research in the future.
5. Conclusions
Here, we comprehensively investigated the compounds composing the Sasang type-specific personalized herbal medicines. We confirmed that most of the 60 compounds showing high feature importance determined by the ERT classifier have selectivity for particular Sasang type. We found that most compounds showed selectivity for particular Sasang type. We also investigated taxonomic and biosynthetic characteristics of the 59 Sasang-prominent compounds (8, 9, and 42 compounds for TE, SY, and SE, respectively). Furthermore, we identified 14 Sasang type-specific compounds showing statistically significance with the Sasang type at a univariate level. Lastly, using a trained ERT classifier, we predicted the Sasang type of commonly used but unidentified medicinal herbs, and indirectly confirmed the prediction result with a simple in vitro experiment examining the biological function of herbs.
Declarations
Author contribution statement
Ji-Hwan Kim: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data. Chang-Eop Kim: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper; Analyzed and interpreted the data.Sa-Yoon Park, Young Woo Kim: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper. Yu Rim Song, Young Pyo Jang, Young Pyo Jang: Performed the experiments.
Funding statement
Prof. Ji-Hwan Kim was supported by National Research Foundation of Korea [NRF-2017R1C1B5017048 and NRF-2022R1I1A2066653]. Prof. Chang-Eop Kim was supported by National Research Foundation of Korea [2020M3A9E4103843]; Ministry of Health and Welfare [HF20C0087].
Data availability statement
Data will be made available on request.
Declaration of interest's statement
The authors declare no conflict of interest.
Additional information
Supplementary content related to this article has been published online at [URL].
Acknowledgement
Kim YW would like to thank to the Ph.D.’s program of Kyungpook National University for completing the thesis through this work.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.heliyon.2023.e13692.
Contributor Information
Ji-Hwan Kim, Email: jani77@gachon.ac.kr.
Chang-Eop Kim, Email: eopchang@gachon.ac.kr.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Kim J.Y., Pham D.D. Sasang constitutional medicine as a holistic tailored medicine. Evid. base Compl. Alternative Med. 2009;6(S1):11–19. doi: 10.1093/ecam/nep100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park S.Y., Park M., Lee W.Y., Lee C.Y., Kim J.H., Lee S., Kim C.E. Machine learning-based prediction of Sasang constitution types using comprehensive clinical information and identification of key features for diagnosis. Integr. Med. Res. 2021;10(3) doi: 10.1016/j.imr.2020.100668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim S.H., Jeon E.S., Lee J., Han S.Y., Chae H. Pharmacognostic outlooks on medical herbs of Sasang typology. Integr. Med. Res. 2017 Sep;6(3):231–239. doi: 10.1016/j.imr.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim E.J., Hong Y.S., Seo S.H., Park S.E., Na C.S., Son H.S. Metabolite markers for characterizing Sasang constitution type through GC-MS and 1H NMR-based metabolomics study. Evid. base Compl. Alternative Med. 2019;2019 doi: 10.1155/2019/8783496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee W.Y., Lee C.Y., Kim C.E., Kim J.H. Investigating the biomarkers of the sasang constitution via network pharmacology approach. J. Evidence-Based Complementary Altern. Med. 2021;2021 doi: 10.1155/2021/6665130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim H.W., Wang M., Leber C.A., Nothias L.F., Reher R., Kang K.B., Cottrell G.W. NPClassifier: a deep neural network-based structural classification tool for natural products. J. Nat. Prod. 2021;84(11):2795–2807. doi: 10.1021/acs.jnatprod.1c00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang R., Li X., Zhang X., Qin H., Xiao W. Machine learning approaches for elucidating the biological effects of natural products. Nat. Prod. Rep. 2021;38(2):346–361. doi: 10.1039/d0np00043d. [DOI] [PubMed] [Google Scholar]
- 8.Kim J.H. The study on the selection of sasang constitution-specific herbs in 『dongyisusebowon sinchuk-bon』 from TCMID and TCMSP. Journal of Sasang Constitutional Medicine. 2019;31(3):19–33. [Google Scholar]
- 9.Kim K.Y., Kim J.Y. A research on the classification of herbal medicines based on the Sasang constitution (Taeumin and Taeyangin Part) Journal of Sasang Constitutional Medicine. 2002;14(1):1–9. [Google Scholar]
- 10.Zhao J., Lin F., Liang G., Han Y., Xu N., Pan J., Luo M., Yang W., Zeng L. Exploration of the molecular mechanism of polygonati rhizoma in the treatment of osteoporosis based on network Pharmacology and molecular docking. Front. Endocrinol. 2022 Jan 5;12 doi: 10.3389/fendo.2021.815891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Behery H., Attia A.F., El-Feshawy N., Torkey H. Efficient machine learning model for predicting drug-target interactions with case study for Covid-19. Comput. Biol. Chem. 2021;93 doi: 10.1016/j.compbiolchem.2021.107536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 13.Geurts P., Ernst D., Wehenkel LJMl. Extremely randomized trees. 2006;63(1):3–42. [Google Scholar]
- 14.Saeys Y., Abeel T., Van de Peer Y., editors. Joint European Conference on Machine Learning and Knowledge Discovery in Databases. Springer; 2008. Robust feature selection using ensemble feature selection techniques. [Google Scholar]
- 15.Wang X., You X., Zhang L., Huang D., Aramini B., Shabaturov L., Jiang G., Fan J. A radiomics model combined with XGBoost may improve the accuracy of distinguishing between mediastinal cysts and tumors: a multicenter validation analysis. Ann. Transl. Med. 2021;9(23):1737. doi: 10.21037/atm-21-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinwart I., Christmann A. Springer Science & Business Media; 2008. Support Vector Machines. [Google Scholar]
- 17.Bohning D.J., Aotio S.M. Multinomial logistic regression algorithm. Ann. inst. Stat. Math. 1992;44(1):197–200. [Google Scholar]
- 18.Yun U.J., Bae S.J., Song Y.R., Kim Y.W. A critical YAP in malignancy of HCC is regulated by evodiamine. Int. J. Mol. Sci. 2022;23(3):1855. doi: 10.3390/ijms23031855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song Y.R., Jang B., Lee S.M., Bae S.J., Bak S.B., Kim Y.W. Angelica gigas NAKAI and its active compound, decursin, inhibit cellular injury as an antioxidant by the regulation of AMP-activated protein kinase and YAP signaling. Molecules. 2022;27(6):1858. doi: 10.3390/molecules27061858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee E.H., Baek S.Y., Park J.Y., Kim Y.W. Emodin in Rheum undulatum inhibits oxidative stress in the liver via AMPK with Hippo/Yap signalling pathway. Pharm. Biol. 2020 Dec;58(1):333–341. doi: 10.1080/13880209.2020.1750658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park S.H., Kim M.G., Lee S.J., Kim J.Y., Chae H. Temperament and character profiles of sasang typology in an adult clinical sample. J. Evidence-Based Complementary Altern. Med. 2011;2011 doi: 10.1093/ecam/nep034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S.H., Hwang M., Choi S.H., Kim H.J., Lee E.J., Kwon C.Y., Chung S.Y., Kim J.W., Chang G.T. Analysis of the bio-psychological characteristics of Sasang typology in Korean preschool children using the ponderal index and the temperament and character inventory. J. Compl. Integr. Med. 2020;18(1):175–183. doi: 10.1515/jcim-2017-0171. [DOI] [PubMed] [Google Scholar]
- 23.Djoumbou Feunang Y., Eisner R., Knox C., Chepelev L., Hastings J., Owen G., Fahy E., Steinbeck C., Subramanian S., Bolton E., Greiner R., Wishart D.S. ClassyFire: automated chemical classification with a comprehensive, computable taxonomy. J. Cheminf. 2016;8:61. doi: 10.1186/s13321-016-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jan R., Asaf S., Numan M., Lubna, Kim K.M. Plant secondary metabolite biosynthesis and transcriptional regulation in response to biotic and abiotic stress conditions. Agronomy. 2021;11:968. [Google Scholar]
- 25.Zhou Y., Lu X., Chen L., Zhang P., Zhou J., Xiong Q., Shen Y., Tian W. Polysaccharides from Chrysanthemun indicum L. enhance the accumulation of polysaccharide and atractylenolide in Atractylodes macrocephala Koidz. Int. J. Biol. Macromol. 2021 Nov 1;190:649–659. doi: 10.1016/j.ijbiomac.2021.09.010. [DOI] [PubMed] [Google Scholar]
- 26.Park M., Lee C.Y., Lee T.H., Kim Y.S., Kim C.E. Identifying theoretical characteristics of traditional medicines in Korea, China, and Japan through the herb usage data. Journal of Physiology & Pathology in Korean Medicine. 2018;32(3):149–156. [Google Scholar]
- 27.Baek Y.H., Kim H.S., Lee S.W., Jang E.S. The concordance and validity assessment of diagnosis for the expert in sasang constitution. Journal of Sasang constitutional medicine. 2014;26(3):295–303. [Google Scholar]
- 28.Shi Y., Zhang L., Wang Z., Lu X., Wang T., Zhou D., Zhang Z. Multivariate machine learning analyses in identification of major depressive disorder using resting-state functional connectivity: a multicentral study. ACS Chem. Neurosci. 2021 Aug 4;12(15):2878–2886. doi: 10.1021/acschemneuro.1c00256. [DOI] [PubMed] [Google Scholar]
- 29.Wu T., Kerbler S.M., Fernie A.R., Zhang Y. Plant cell cultures as heterologous bio-factories for secondary metabolite production. Plant Commun. 2021;2(5) doi: 10.1016/j.xplc.2021.100235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bisht R., Bhattacharyya A., Shrivastava A., Saxena P. An overview of the medicinally important plant type III PKS derived polyketides. Front. Plant Sci. 2021;12 doi: 10.3389/fpls.2021.746908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gani U., Vishwakarma R.A., Misra P. Membrane transporters: the key drivers of transport of secondary metabolites in plants. Plant Cell Rep. 2021;40(1):1–18. doi: 10.1007/s00299-020-02599-9. [DOI] [PubMed] [Google Scholar]
- 32.Ancheeva E., Daletos G., Proksch P. Bioactive secondary metabolites from endophytic fungi. Curr. Med. Chem. 2020;27(11):1836–1854. doi: 10.2174/0929867326666190916144709. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.