Abstract
Spinal muscular atrophy (SMA) is among the most common autosomal recessive disorders with different incidence rates in different ethnic groups. In the current study, we have determined SMN1, SMN2 and NAIP copy numbers in an Iranian population using MLPA assay. Cases were recruited from Genome-Nilou Laboratory, Tehran, Iran and Pars-Genome Laboratory, Karaj, Iran during 2012–2022. All enrolled cases had a homozygous deletion of exon 7 of SMN1. Moreover, except for 11 cases, all other cases had a homozygous deletion of exon 8 of SMN1. Out of 186 patients, 177 (95.16%) patients showed the same copy numbers of exons 7 and 8 of SMN2 gene. In addition, 53 patients (28.49%) showed 2 copies, 71 (38.17%) showed 3 copies and 53 patients (28.49%) showed 4 copies of SMN2 gene exons 7 and 8. The remaining 9 patients showed different copy numbers of exons 7 and 8 of SMN2 gene. The proportions of SMA patients with different numbers of normal NAIP were 0 copy in 73 patients (39.24%), 1 copy in 59 patients (31.72%), 2 copies in 53 patients (28.49%) and 4 copies in one patient (0.5%). These values are different from values reported in other populations. Integration of the data of the SMN1/2 and NAIP genes showed 17 genotypes. Patients with genotype 0-0-3-3-1 (0 copies of SMN1 (E7,8), 3 copies of SMN2 (E7,8) and 1 copy of NAIP (E5)) were the most common genotype in this study. Patients with 0-0-2-2-0 genotype were more likely to have type I SMA. The results of the current study have practical significance, particularly in the genetic counseling of at-risk families.
Subject terms: Cell biology, Genetics, Molecular biology
Introduction
Spinal muscular atrophy (SMA) is among the most common autosomal recessive disorders with an incidence rate of about 1 in 6000–10,000 live births. This disorder is described by degeneration of alpha motor neurons in the spinal cord and the medulla oblongata, leading to symmetrical proximal muscular atrophy. Heterozygous healthy carriers for this disorder have a frequency of 1 in 35 in the general population1. Based on the age of onset and reached motor functions, this disorder is classified into four clinical types, namely severe, intermediate, mild and adult-onset types being enumerated as types I to IV, respectively2. From a genetics point of view, this autosomal recessive disorder is caused by the dysfunction of the survival motor neuron (SMN) gene which is located on chromosome 5q13.2. This gene has two versions, namely SMN1 and SMN2. The former produces a full-length transcript. These two versions are different from each other in only five nucleotides. Homozygous deletion of SMN1 exon 7 is responsible for clinical disorder in approximately 94% of cases3. SMN2 has a partial function and can compensate homozygous deletions of SMN1 to some extent4.Therefore, copy numbers of SMN2 affect severity of SMA. Copy number of another gene located on chromosome 5q13.2, namely the neuronal apoptosis inhibitory protein (NAIP) gene has also been shown to be associated with severity of SMA5.
Variations in copy numbers of SMN1 and SMN2 have been reported in SMA patients from different populations. Moreover, different deletions and rearrangements have been detected in different ethnic groups6–8. Thus, identification of SMN1, SMN2 and NAIP copy numbers in SMA patients in each population has a practical significance, particularly in the genetic counseling of at risk families. In the current study, we have determined SMN1, SMN2 and NAIP copy numbers in an Iranian population of SMA patients using MLPA assay.
Methods and patients
Patients
A total of 186 SMA cases were enrolled in this study. Patients were referred to Genome-Nilou Laboratory, Tehran, Iran and Pars-Genome Laboratory, Karaj, Iran during 2012–2022. They were referred to Genome-Nilou laboratory by Iranian SMA Association and neurologists. They came from Tehran and other cities of Iran. All of them were genetically analyzed in Genome-Nilou laboratory. None of the patients used disease modifying therapies. All enrolled cases had a homozygous deletion of exon 7 of SMN1 gene, as confirmed by MLPA assay. Ethical approval for this study has been obtained from the Ethical Committee of Tehran University of Medical Sciences. All methods were carried out in accordance with relevant guidelines and regulations. Informed consent forms were signed by all patients or their parents.
MLPA assay
MLPA was performed using the SALSA MLPA Probemix P021-B1 for detection of deletions or duplication in the exons 7 and 8 of the SMN1, SMN2 and exon 5 of the NAIP genes (MRC-Holland, Amsterdam, Netherlands) as per the manufacturer’s instructions. The resulting fragments were separated using ABI PRISM 3100 (ThermoFisher Scientific, USA) and analyzed by GeneMarker software version 1.959. Peak heights were normalized to control healthy individuals in a similar method to a previous study10, and a deletion or duplication was expected when the normalized peak ratio value was 0 (homozygous deletion), 1 (heterozygous deletion), 3 (heterozygous duplication) and occasionally 4 (heterozygous triplication or homozygous duplication). Each experiment included 4 controls; 2 normal controls, 1 carrier and 1 affected person. All of the controls had been confirmed in an external genetic laboratory.
Statistical analyses
GraphPad Prism version 9.0 (GraphPad Software, La Jolla, CA, USA) (https://www.graphpad.com/guides/prism/latest/statistics/stat_checklist_kw.htm) was used for statistical analysis. Kruskal–Wallis test was performed to detect the relationship between copy number of exons 7 and 8 of SMN2 and deletion in exon 5 of NAIP gene, and SMA subtypes and age at onset. The quantitative data was expressed as mean ± standard deviation. The count data was expressed as the rate and frequency. P value less than 0.05 was considered statistically significant. To compare the distribution of clinical phenotypes (SMA subtypes) between patient groups with/without parental relationship, we used Chi-square (2 × 3 contingency table) (https://www.graphpad.com/quickcalcs/chisquared1.Chi-square/).
Results
General information
Based on the age of disease onset and clinical manifestations, 35, 47, 94 and 10 cases were classified as SMA types I–IV, respectively. A total of 114 cases (61.29%) were born to non-consanguineous parents. Others were born to first cousin (55 cases), first cousin once removed (14 cases) and second cousin (3 cases) parents. The study cohort included 83 females and 103 males.
Gene copy numbers in SMA patients
All enrolled cases had a homozygous deletion of exon 7 of SMN1. Moreover, except for 11 cases, all other cases had a homozygous deletion of exon 8 of SMN1. Out of 186 patients, 177 (95.16%) patients showed the same copy numbers of exons 7 and 8 of SMN2 gene. In addition, 53 patients (28.49%) showed 2 copies, 71 (38.17%) showed 3 copies and 53 patients (28.49%) showed 4 copies of SMN2 gene exons 7 and 8. The remaining 9 patients showed different copy numbers of exons 7 and 8 of SMN2 gene. The proportions of SMA patients with different numbers of normal NAIP were 0 copy in 73 patients (39.24%), 1 copy in 59 patients (31.72%), 2 copies in 53 patients (28.49%) and 4 copies in one patient (0.5%). Table 1 shows detailed characteristics of patients cohort.
Table 1.
Detailed characteristics of patients cohort.
| Case | Type | Sex | Age of onset | Age of diagnosis | Genes copy number | Parental relationship | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Exon7 SMN1 | Exon8 SMN1 | Exon7 SMN2 | Exon8 SMN2 | Exon5 NAIP | ||||||
| 1 | I | Female | 0M | 3M | 0 | 0 | 2 | 2 | 2 | First cousins |
| 2 | I | Female | 1M | 3M | 0 | 0 | 2 | 2 | 0 | Not related |
| 3 | I | Female | 2M | 12M | 0 | 0 | 2 | 2 | 0 | First cousins |
| 4 | I | Female | 2M | 12M | 0 | 0 | 2 | 2 | 0 | First cousin once removed |
| 5 | I | Female | 3M | 6M | 0 | 0 | 2 | 2 | 0 | Not related |
| 6 | I | Female | 3M | 12M | 0 | 0 | 2 | 2 | 0 | First cousins |
| 7 | I | Female | 3M | 18M | 0 | 0 | 2 | 2 | 0 | First cousins |
| 8 | I | Female | 4M | 4M | 0 | 0 | 2 | 2 | 0 | First cousins |
| 9 | I | Female | 4M | 16M | 0 | 0 | 2 | 2 | 0 | First cousins |
| 10 | I | Female | 4M | 2Y | 0 | 0 | 2 | 2 | 0 | Not related |
| 11 | I | Female | 5M | 9M | 0 | 0 | 2 | 2 | 0 | First cousins |
| 12 | I | Female | 5M | 12M | 0 | 0 | 2 | 2 | 0 | Not related |
| 13 | I | Female | 6M | 6M | 0 | 0 | 2 | 2 | 0 | First cousins |
| 14 | I | Female | 6M | 8M | 0 | 0 | 2 | 2 | 0 | Not related |
| 15 | I | Male | 0M | 1M | 0 | 0 | 2 | 2 | 0 | First cousins |
| 16 | I | Male | 1M | 1M | 0 | 0 | 2 | 2 | 0 | First cousin once removed |
| 17 | I | Male | 1M | 2M | 0 | 0 | 2 | 2 | 0 | Not related |
| 18 | I | Male | 1M | 3M | 0 | 0 | 2 | 2 | 2 | First cousins |
| 19 | I | Male | 1M | 4M | 0 | 0 | 2 | 2 | 0 | Not related |
| 20 | I | Male | 1M | 6M | 0 | 0 | 2 | 2 | 0 | Not related |
| 21 | I | Male | 2M | 2M | 0 | 0 | 2 | 2 | 1 | First cousins |
| 22 | I | Male | 2M | 2M | 0 | 0 | 2 | 2 | 0 | Not related |
| 23 | I | Male | 2M | 4M | 0 | 0 | 2 | 2 | 0 | First cousin once removed |
| 24 | I | Male | 2M | 9M | 0 | 0 | 2 | 2 | 0 | First cousin once removed |
| 25 | I | Male | 3M | 4M | 0 | 0 | 2 | 2 | 0 | First cousin once removed |
| 26 | I | Male | 3M | 10M | 0 | 0 | 2 | 2 | 0 | Not related |
| 27 | I | Male | 3M | 10M | 0 | 0 | 2 | 2 | 0 | Not related |
| 28 | I | Male | 3M | 3Y | 0 | 0 | 2 | 2 | 1 | First cousins |
| 29 | I | Male | 4M | 4M | 0 | 0 | 2 | 2 | 0 | First cousins |
| 30 | I | Male | 4M | 6M | 0 | 0 | 3 | 3 | 0 | First cousins |
| 31 | I | Male | 4M | 9M | 0 | 0 | 2 | 2 | 0 | Not related |
| 32 | I | Male | 5M | 12M | 0 | 0 | 2 | 2 | 0 | First cousins |
| 33 | I | Male | 6M | 8M | 0 | 0 | 3 | 3 | 2 | First cousins |
| 34 | I | Male | 6M | 8M | 0 | 0 | 4 | 4 | 0 | First cousins |
| 35 | I | Male | 6M | 12M | 0 | 0 | 2 | 2 | 1 | First cousins |
| 36 | II | Female | 6M | 7Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 37 | II | Female | 6M | 12Y | 0 | 0 | 2 | 2 | 0 | Not related |
| 38 | II | Female | 9M | 6Y | 0 | 1 | 3 | 2 | 1 | Not related |
| 39 | II | Female | 10M | 4Y | 0 | 0 | 2 | 2 | 0 | Not related |
| 40 | II | Female | 11M | 2Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 41 | II | Female | 11M | 7Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 42 | II | Female | 11M | 12Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 43 | II | Female | 12M | 4Y | 0 | 0 | 2 | 2 | 0 | First cousins |
| 44 | II | Female | 12M | 5Y | 0 | 0 | 2 | 2 | 0 | First cousins |
| 45 | II | Female | 12M | 5Y | 0 | 1 | 3 | 2 | 1 | Not related |
| 46 | II | Female | 12M | 11Y | 0 | 0 | 2 | 2 | 0 | Not related |
| 47 | II | Female | 12M | 12Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 48 | II | Female | 12M | 23Y | 0 | 0 | 2 | 2 | 0 | Not related |
| 49 | II | Female | 12M | 4Y | 0 | 0 | 2 | 2 | 0 | First cousins |
| 50 | II | Female | 14M | 3Y | 0 | 0 | 3 | 3 | 1 | First cousins |
| 51 | II | Female | 14M | 9Y | 0 | 0 | 3 | 3 | 1 | First cousin once removed |
| 52 | II | Female | 14M | 10Y | 0 | 0 | 3 | 3 | 0 | Not related |
| 53 | II | Female | 14M | 24Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 54 | II | Female | 15M | 2Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 55 | II | Female | 15M | 9Y | 0 | 1 | 4 | 4 | 2 | Second cousins |
| 56 | II | Female | 15M | 11Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 57 | II | Female | 15M | 17Y | 0 | 0 | 3 | 3 | 0 | Not related |
| 58 | II | Female | 15M | 32Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 59 | II | Female | 17M | 18Y | 0 | 0 | 3 | 3 | 1 | First cousins |
| 60 | II | Male | 6M | 4Y | 0 | 0 | 3 | 3 | 0 | Not related |
| 61 | II | Male | 7M | 9M | 0 | 0 | 2 | 2 | 0 | Not related |
| 62 | II | Male | 7M | 7Y | 0 | 0 | 3 | 3 | 0 | Not related |
| 63 | II | Male | 7M | 12Y | 0 | 0 | 3 | 3 | 0 | Not related |
| 64 | II | Male | 7M | 18Y | 0 | 0 | 3 | 3 | 0 | Not related |
| 65 | II | Male | 8M | 3Y | 0 | 0 | 4 | 4 | 1 | Not related |
| 66 | II | Male | 8M | 15Y | 0 | 0 | 3 | 3 | 0 | Not related |
| 67 | II | Male | 9M | 12M | 0 | 0 | 3 | 3 | 1 | Not related |
| 68 | II | Male | 10M | 9Y | 0 | 1 | 3 | 3 | 1 | Not related |
| 69 | II | Male | 10M | 13Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 70 | II | Male | 10M | 14Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 71 | II | Male | 12M | 12M | 0 | 0 | 3 | 3 | 1 | Not related |
| 72 | II | Male | 12M | 9Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 73 | II | Male | 12M | 10Y | 0 | 0 | 2 | 2 | 0 | Not related |
| 74 | II | Male | 12M | 11Y | 0 | 0 | 4 | 4 | 1 | Not related |
| 75 | II | Male | 13M | 7Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 76 | II | Male | 14M | 29Y | 0 | 0 | 4 | 4 | 2 | Not related |
| 77 | II | Male | 15M | 2Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 78 | II | Male | 15M | 7Y | 0 | 0 | 3 | 3 | 0 | Not related |
| 79 | II | Male | 16M | 13Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 80 | II | Male | 17M | 10Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 81 | II | Male | 18M | 4Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 82 | II | Male | 22M | 13Y | 0 | 0 | 2 | 2 | 0 | Not related |
| 83 | III | Female | 20M | 16Y | 0 | 0 | 4 | 4 | 0 | Not related |
| 84 | III | Female | 2Y | 2Y | 0 | 0 | 3 | 3 | 0 | Not related |
| 85 | III | Female | 2Y | 7Y | 0 | 0 | 2 | 2 | 1 | First cousins |
| 86 | III | Female | 2Y | 23Y | 0 | 0 | 4 | 4 | 2 | First cousins |
| 87 | III | Female | 2Y | 25Y | 0 | 0 | 4 | 4 | 0 | Not related |
| 88 | III | Female | 2Y | 27Y | 0 | 0 | 2 | 2 | 0 | Not related |
| 89 | III | Female | 2Y | 31Y | 0 | 0 | 4 | 4 | 2 | First cousins |
| 90 | III | Female | 2Y | 49Y | 0 | 0 | 4 | 4 | 0 | First cousin once removed |
| 91 | III | Female | 3Y | 4Y | 0 | 0 | 4 | 4 | 0 | Not related |
| 92 | III | Female | 3Y | 5Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 93 | III | Female | 3Y | 9Y | 0 | 0 | 4 | 4 | 1 | Not related |
| 94 | III | Female | 3Y | 13Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 95 | III | Female | 3Y | 15Y | 0 | 0 | 3 | 3 | 0 | Not related |
| 96 | III | Female | 3Y | 17Y | 0 | 0 | 4 | 4 | 0 | First cousins |
| 97 | III | Female | 3Y | 18Y | 0 | 0 | 4 | 4 | 2 | Not related |
| 98 | III | Female | 3Y | 19Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 99 | III | Female | 3Y | 27Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 100 | III | Female | 3Y | 31Y | 0 | 0 | 4 | 4 | 2 | First cousins |
| 101 | III | Female | 4Y | 4Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 102 | III | Female | 4Y | 10Y | 0 | 0 | 3 | 3 | 0 | First cousins |
| 103 | III | Female | 4Y | 26Y | 0 | 0 | 4 | 4 | 2 | First cousins |
| 104 | III | Female | 4Y | 27Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 105 | III | Female | 4Y | 32Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 106 | III | Female | 4Y | 32Y | 0 | 0 | 3 | 3 | 2 | Not related |
| 107 | III | Female | 5Y | 6Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 108 | III | Female | 5Y | 9Y | 0 | 0 | 4 | 4 | 2 | First cousins |
| 109 | III | Female | 6Y | 23Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 110 | III | Female | 7Y | 13Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 111 | III | Female | 7Y | 32Y | 0 | 0 | 4 | 4 | 2 | First cousin once removed |
| 112 | III | Female | 7Y | 38Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 113 | III | Female | 8Y | 22Y | 0 | 0 | 4 | 4 | 2 | First cousins |
| 114 | III | Female | 8Y | 24Y | 0 | 0 | 3 | 3 | 0 | Not related |
| 115 | III | Female | 8Y | 39Y | 0 | 0 | 2 | 2 | 0 | Not related |
| 116 | III | Female | 9Y | 16Y | 0 | 0 | 4 | 4 | 2 | Not related |
| 117 | III | Female | 9Y | 21Y | 0 | 0 | 4 | 4 | 2 | Not related |
| 118 | III | Female | 9Y | 36Y | 0 | 0 | 2 | 2 | 0 | First cousins |
| 119 | III | Female | 9Y | 36Y | 0 | 0 | 4 | 4 | 2 | First cousins |
| 120 | III | Female | 10Y | 23Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 121 | III | Female | 11Y | 28Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 122 | III | Female | 13Y | 41Y | 0 | 0 | 2 | 2 | 0 | Not related |
| 123 | III | Female | 14Y | 34Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 124 | III | Female | 16Y | 41Y | 0 | 0 | 3 | 3 | 0 | First cousin once removed |
| 125 | III | Female | 17Y | 34Y | 0 | 0 | 2 | 2 | 0 | Not related |
| 126 | III | Female | 21Y | 32Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 127 | III | Male | 18M | 38Y | 0 | 0 | 3 | 3 | 1 | First cousin once removed |
| 128 | III | Male | 18M | 39Y | 0 | 0 | 4 | 4 | 2 | First cousins |
| 129 | III | Male | 19M | 7Y | 0 | 0 | 2 | 2 | 0 | Not related |
| 130 | III | Male | 2Y | 5Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 131 | III | Male | 2Y | 12Y | 0 | 0 | 4 | 4 | 2 | First cousins |
| 132 | III | Male | 2Y | 16Y | 0 | 0 | 3 | 3 | 0 | Not related |
| 133 | III | Male | 2Y | 30Y | 0 | 1 | 4 | 3 | 2 | Not related |
| 134 | III | Male | 2Y | 30Y | 0 | 0 | 4 | 4 | 2 | First cousins |
| 135 | III | Male | 2Y | 33Y | 0 | 1 | 4 | 3 | 2 | Not related |
| 136 | III | Male | 2Y | 37Y | 0 | 0 | 4 | 4 | 2 | First cousins |
| 137 | III | Male | 2Y | 40Y | 0 | 1 | 4 | 3 | 2 | Not related |
| 138 | III | Male | 2Y | 43Y | 0 | 1 | 4 | 3 | 2 | Not related |
| 139 | III | Male | 3Y | 5Y | 0 | 0 | 3 | 4 | 2 | Not related |
| 140 | III | Male | 3Y | 7Y | 0 | 0 | 2 | 2 | 0 | Not related |
| 141 | III | Male | 3Y | 8Y | 0 | 0 | 3 | 3 | 0 | Not related |
| 142 | III | Male | 3Y | 10Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 143 | III | Male | 3Y | 14Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 144 | III | Male | 3Y | 14Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 145 | III | Male | 3Y | 15Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 146 | III | Male | 3Y | 17Y | 0 | 0 | 4 | 4 | 1 | First cousins |
| 147 | III | Male | 3Y | 24Y | 0 | 0 | 4 | 4 | 2 | First cousins |
| 148 | III | Male | 3Y | 26Y | 0 | 0 | 4 | 4 | 2 | First cousins |
| 149 | III | Male | 3Y | 32Y | 0 | 1 | 3 | 2 | 2 | Not related |
| 150 | III | Male | 3Y | 41Y | 0 | 0 | 4 | 4 | 2 | Not related |
| 151 | III | Male | 3Y | 43Y | 0 | 0 | 4 | 4 | 2 | First cousin once removed |
| 152 | III | Male | 4Y | 4Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 153 | III | Male | 4Y | 10Y | 0 | 0 | 4 | 4 | 2 | Not related |
| 154 | III | Male | 4Y | 29Y | 0 | 0 | 4 | 4 | 2 | Not related |
| 155 | III | Male | 5Y | 7Y | 0 | 0 | 4 | 4 | 2 | First cousins |
| 156 | III | Male | 5Y | 15Y | 0 | 0 | 2 | 2 | 0 | First cousins |
| 157 | III | Male | 6Y | 20Y | 0 | 0 | 3 | 3 | 2 | Second cousins |
| 158 | III | Male | 6Y | 30Y | 0 | 0 | 2 | 2 | 0 | Not related |
| 159 | III | Male | 7Y | 29Y | 0 | 0 | 3 | 3 | 0 | Not related |
| 160 | III | Male | 7Y | 31Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 161 | III | Male | 7Y | 31Y | 0 | 0 | 4 | 4 | 2 | First cousins |
| 162 | III | Male | 11Y | 12Y | 0 | 0 | 3 | 3 | 2 | Not related |
| 163 | III | Male | 11Y | 33Y | 0 | 0 | 4 | 4 | 2 | First cousins |
| 164 | III | Male | 12Y | 22Y | 0 | 1 | 4 | 3 | 2 | Not related |
| 165 | III | Male | 12Y | 35Y | 0 | 0 | 4 | 4 | 2 | First cousins |
| 166 | III | Male | 12Y | 40Y | 0 | 0 | 3 | 3 | 0 | Not related |
| 167 | III | Male | 12Y | 50Y | 0 | 0 | 4 | 4 | 2 | Not related |
| 168 | III | Male | 13Y | 16Y | 0 | 0 | 3 | 3 | 2 | First cousins |
| 169 | III | Male | 13Y | 23Y | 0 | 0 | 4 | 4 | 2 | First cousins |
| 170 | III | Male | 13Y | 26Y | 0 | 0 | 4 | 4 | 0 | Not related |
| 171 | III | Male | 15y | 27Y | 0 | 0 | 4 | 4 | 2 | First cousins |
| 172 | III | Male | 15Y | 30Y | 0 | 0 | 4 | 4 | 0 | First cousin once removed |
| 173 | III | Male | 15Y | 43Y | 0 | 0 | 4 | 4 | 0 | First cousin once removed |
| 174 | III | Male | 17Y | 32Y | 0 | 0 | 4 | 4 | 2 | First cousins |
| 175 | III | Male | 18Y | 33Y | 0 | 0 | 4 | 4 | 2 | Not related |
| 176 | III | Male | 22Y | 29Y | 0 | 0 | 4 | 4 | 2 | First cousin once removed |
| 177 | IV | Female | 30Y | 41Y | 0 | 0 | 3 | 3 | 1 | Not related |
| 178 | IV | Male | 19Y | 29Y | 0 | 0 | 4 | 4 | 4 | First cousins |
| 179 | IV | Male | 20Y | 33Y | 0 | 0 | 4 | 4 | 2 | First cousins |
| 180 | IV | Male | 20Y | 40Y | 0 | 0 | 2 | 2 | 1 | Not related |
| 181 | IV | Male | 25Y | 45Y | 0 | 0 | 4 | 4 | 2 | Second cousins |
| 182 | IV | Male | 25Y | 49Y | 0 | 0 | 4 | 4 | 2 | Not related |
| 183 | IV | Male | 27Y | 42Y | 0 | 2 | 4 | 4 | 2 | Not related |
| 184 | IV | Male | 28Y | 40Y | 0 | 0 | 3 | 3 | 1 | First cousins |
| 185 | IV | Male | 28Y | 40Y | 0 | 0 | 4 | 4 | 2 | First cousins |
| 186 | IV | Male | 28Y | 40Y | 0 | 0 | 4 | 4 | 2 | First cousins |
Distribution of SMA patients in different groups of SMA is shown in Table 2. Type III SMA accounts for 50.53% of total cases.
Table 2.
Results of genetic diagnosis of SMA patients.
| Clinical type | Type I | Type II | Type III | Type IV | Total |
|---|---|---|---|---|---|
| Number | 35 | 47 | 94 | 10 | 186 |
| Proportion | 18.81% | 25.26% | 50.53% | 5.37% | 100% |
While exon 7 was absent in all SMA patients of all classes, exon 8 was present in 4 Type II, 6 Type III and 1 type IV SMA cases. In fact, in 94.08% (175/186) of the patients, homozygous deletion of both exons 7 and 8 of the SMN1 gene was reported. Among these, 18.81% (35/186) of patients were diagnosed with SMA Type I, 25.26% (47/186) with Type II, 50.53% (94/186) with Type III, and 5.37% (10/186) with Type IV. In 5.9% (11/186) of the patients, homozygous deletion of the 7th exon and heterozygous deletion of 8th exon of the SMN1 gene were detected (Table 3). There was no correlation between different SMA types and deletion types of exons 7 and 8 of SMN1 gene (P value = 0.31).
Table 3.
SMN1 Exons copy numbers in patients with different clinical types of SMA.
| Clinical types | Type I | Type II | Type III | Type IV | Total | |
|---|---|---|---|---|---|---|
| Copy number | ||||||
| EXON 7 | 0 | 35 (18.81%) | 47 (25.26%) | 94 (50.53%) | 10 (5.37%) | 186 (100%) |
| 1 | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| Total | 35 (18.81%) | 47 (25.26%) | 94 (50.53%) | 10 (5.37%) | 186 (100%) | |
| EXON 8 | 0 | 35 (18.81%) | 43 (23.11%) | 88 (47.31%) | 9 (4.83%) | 175 (94.08%) |
| 1 | 0 (0%) | 4 (2.15%) | 6 (3.22%) | 0 (0%) | 10 (5.37%) | |
| 2 | 0 (0%) | 0 (0%) | 0 (0%) | 1 (0.53%) | 1 (0.53%) | |
| Total | 35 (18.81%) | 47 (25.269%) | 94 (50.53%) | 10 (5.37%) | 186 (100%) | |
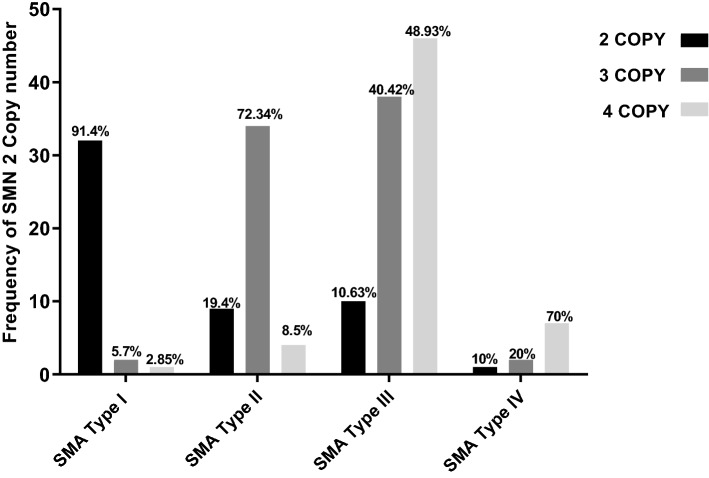
Totally, 27.95% (52/186), 38.7% (72/186), and 28.49% (53/186) of patients had 2, 3, and 4 copies of exons 7 and 8 of the SMN2 gene, respectively (Fig. 1). However, 9 patients showed different normal copy numbers of exons 7 and 8 of SMN2 gene. Three out of nine patients showed 3 copies of exon 7 and 2 copies of exon 8, five out of nine patients showed 4 copies of exon 7 and 3 copies of exon 8 and one patient showed 3 copies of exon 7 and 4 copies of exon 8 of SMN2 gene. In addition, 39.24% (73/186), 32.73% (59/186), 31.72% (53/186) and 0.53% (1/186) of patients had 0, 1, 2 and 4 copies of the exon 5 of the NAIP gene, respectively. The presence of two copies of SMN2 gene was most common in type I patients, accounting for 91.42% (32/35) of these patients. The presence of 3 copies of SMN2 was most common in type II patients, accounting for 72.34% (34/47) of patients. Finally, having 4 copies of this gene was most common in type III and type IV patients, accounting for 48.93% (46/94) and 70% (7/10) of patients, respectively.
Figure 1.
The percentage of individuals with various numbers of exon 7 of the SMN2 gene.
Figure 1 shows the percentage of individuals with various number of SMN2 gene copies.
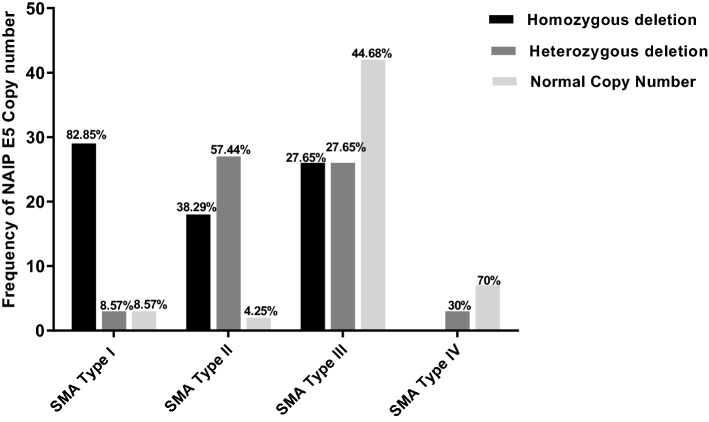
Figure 2 shows the percentage of individuals with various numbers of NAIP gene copies. There was a significant difference in the distribution of NAIP gene copy numbers among different types of SMA (× 2 = 69, P < 0.0001). All patients carrying deletion of two copies of NAIP gene had severe (type I) SMA, accounting for 82.85% (29/35) of patients. Having one copy of this gene was most common in type II patients, accounting for 57.44% (27/47) of patients. The presence of two copies of NAIP gene was most common in type III and type IV patients, accounting for 44.68% (42/94) and 60% (6/10), respectively (Fig. 2).
Figure 2.
The percentage of individuals with various numbers of NAIP gene.
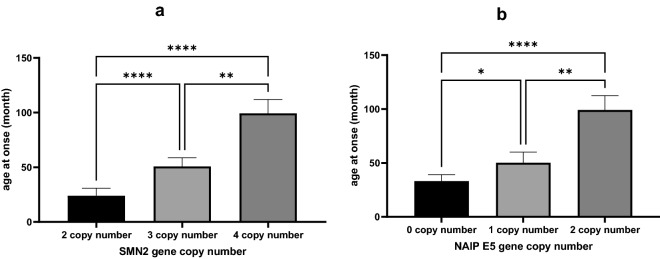
The average age of onset for patients with 2 copies of SMN2 gene (23.86 ± 50.14 month) was significantly lower than that of patients with 3 (50.75 ± 69 month) or 4 (99.25 ± 96.56 month) copies of SMN2 (P < 0.0001) (Fig. 3a).
Figure 3.
Relationship between copy numbers of exon 7 of SMN2 (a) and NAIP (b) genes and age at onset of patients. A non-parametric Kruskal–Wallis test was used to identify significant association between the age at onset of patients and SMN2 and NAIP genes copy number (* P value < 0.05, *** P value < 0.001 and **** P value < 0.0001).
The average age of onset of SMA in patients with 0 copy of the NAIP gene (33.1 ± 51.98) was also less than that of patients with 1 (50.15 ± 75.81) and 2 (99 ± 96.65) copies (P < 0.0001) (Fig. 3b).
There was a significant difference in the distribution of NAIP gene copy numbers among different types of SMA. All patients carrying deletion of two copies of SMN2 and NAIP genes had severe (type I) SMA.
Chi-square (2 × 3 contingency table) was performed to compare the distribution of NAIP E5 copy numbers between patient’s groups with/without parental relationship (not related vs. related groups). The analysis showed that there was significant difference (× 2 = 25.36, P < 0.0001) in the distribution of NAIP E5 copy numbers in patient’s groups regarding the parental relationship. In fact, 95 out of 115 (82.6%) of patients with no parental relationship had no or one NAIP E5 copy number and 20 (17.4%) of patients with no parental relationship had two NAIP E5 copy numbers. However, among the 71 patients with parental relationships, 27 patients (38%) had no NAIP E5 copy number, 34 patients (47.9%) had two NAIP E5 copy numbers and 10 patients (14.1%) had one NAIP E5 copy number.
There was also a strong significant correlation between copy numbers of SMN2 and NAIP genes (R = 0.68, P < 0.0001) and the copy numbers of SMN2 and NAIP genes had synergistic effect on SMA phenotype.
Integration of the data of the SMN1/2 and NAIP genes showed 17 genotypes. Patients with genotype 0-0-3-3-1 (0 copies of SMN1 (E7,8), 3 copies of SMN2 (E7,8) and 1 copy of NAIP (E5)) were the most common genotype in this study (Table 4). Patients with 0-0-2-2-0 genotype were more likely to have type I SMA.
Table 4.
Relationship between SMN1 (E7and E8) -SMN2 (E7and E8) -NAIP (E5) genotype and clinical phenotype of SMA.
| Genotype | Case numbers (%) | Age of onset, month (mean ± SD) | Clinical classification (%) | Sex | |||
|---|---|---|---|---|---|---|---|
| Type I | Type II | Type III | Type IV | ||||
| 0-0-3-3-1 | 47 (25.26%) | 54.48 ± 78.54 | 0 (0%) | 22 (46.8%) | 23 (48.9%) | 2 (4.25%) |
28 Female 19 Male |
| 0-0-2-2-0 | 45 (24.19%) | 21.42 ± 42.32 | 27 (60%) | 9 (20%) | 9 (20%) | 0 (0%) |
24 Female 21 Male |
| 0-0-4-4-2 | 37 (19.89%) | 113 ± 99 | 0 (0%) | 1 (2.7%) | 31 (83.78%) | 5 (13.5%) |
11 Female 26 Male |
| 0-0-3-3-0 | 19 (10.21%) | 41 ± 52.1 | 1 (5.26%) | 9 (47.36%) | 9 (47.36%) | 0 (0%) |
8 Female 11 Male |
| 0-0-4-4-0 | 9 (4.83%) | 74.75 ± 77.33 | 1(11.11%) | 0 (0%) | 8 (88.88) | 0 (0%) |
5 Female 4 Male |
| 0-0-2-2-1 | 5 (2.68%) | 55 ± 103.8 | 3 (60%) | 0 (0%) | 1(20%) | 1(20%) |
1 Female 4 Male |
| 0-0-3-3-2 | 4 (2.15%) | 82.5 ± 61.19 | 1 (25%) | 0 (0%) | 4 (75%) | 0 (0%) |
1 Female 4 Male |
| 0-0-4-4-1 | 4 (2.15%) | 23 ± 15.09 | 0 (0%) | 2 (50%) | 2 (50%) | 0 (0%) |
1 Female 3 Male |
| 0-1-3-2-1 | 2 (1.07%) | 10.5 ± 2.12 | 0 (0%) | 2 (100%) | 0 (0%) | 0 (0%) |
2 Female 0 Male |
| 0-0-2-2-2 | 2 (1.07%) | 1 ± 0 | 2 (100%) | 0 (0%) | 0 (0%) | 0 (0%) |
1 Female 1 Male |
| 0-1-3-2-2 | 1 (0.53%) | 36 ± 0 | 0 (0%) | 1 (100%) | 0 (0%) | 0 (0%) |
0 Female 1 Male |
| 0-1-4-3-2 | 5 (2.68%) | 48 ± 53 | 0 (0%) | 0 (100%) | 5 (100%) | 0 (0%) |
0 Female 5 Male |
| 0-0-4-4-4 | 1(0.53%) | 228 ± 0 | 0 (0%) | 0 (0%) | 0 (0%) | 1 (100%) |
0 Female 1 Male |
| 0-1-4-4-2 | 1 (0.53%) | 15 ± 0 | 0 (0%) | 1 (0%) | 0 (0%) | 0 (0%) |
1 Female 0 Male |
| 0-2-4-4-2 | 1 (0.53%) | 324 ± 0 | 0 (0%) | 0 (0%) | 0 (0%) | 1 (0%) |
0 Female 1 Male |
| 0-1-3-3-1 | 1 (0.53%) | 10 ± 0 | 0 (0%) | 1 (0%) | 0 (0%) | 0 (0%) |
0 Female 1 Male |
| 0-0-3-4-2 | 1 (0.53%) | 36 ± 0 | 0 (0%) | 0 (0%) | 1 (0%) | 0 (0%) |
0 Female 1 Male |
To compare the distribution of clinical phenotypes (SMA subtypes) between patient’s groups with/without parental relationship, patients’ group was divided into three subtypes I, II and III &IV. Chi square test in the 2 × 3 contingency table analysis provided evidence that there was significant difference (× 2 = 26.12, P < 0.0001) in the distribution of clinical phenotypes (SMA subtypes) in patient’s groups regarding the parental relationship. The frequency of type I patients was higher in patients with parental relationship (first cousins or first cousin once removed) while the frequency of patients with types II and III subtypes was higher in patients with non-consanguineous families (Fig. 4).
Figure 4.
The distribution of clinical phenotypes (SMA subtypes) between patient’s groups with/without parental relationship. Chi-square (2 × 3 contingency table) was performed to compare the distribution of clinical phenotypes between patient’s groups (not related and related groups).
Discussion
In the current study, we assessed SMN1, SMN2 and NAIP copy numbers in a large population of Iranian patients with SMA. The majority of enrolled patients were born in non-consanguineous families which is consistent with high rate of normal carriers in Iranian population. A previous study in Iranian population estimated a carrier frequency of 5% in this population11. Consistent with this report, a more recent literature review has suggested higher frequency of heterozygous carriers of the SMN1 mutations among Caucasian and Asian populations compared to the Black population12.
In our cohort of patients, all patients except for 11 cases had a homozygous deletion of exon 8 of SMN1. This finding is comparable with the findings in Chinese population2 and some other populations13.
The proportions of SMA cases with different numbers of normal SMN2 copies were 2 copies in 53 (28.49%), 3 copies in 71 (38.17%) and 4 copies in 53 (28.49%). These values are significantly different from those reported by Fang et al. in Chinese population2. They reported the presence of 1–4 normal SMN2 copies in 2 patients (4.8%), 14 (33.3%), 24 (57.1%) and 2 (4.8%) patients in their cohort, respectively2. Amara et al. have reported that 31.3% of Tunisian type I SMA patients carry one copy of SMN2, though all patients of other forms had a minimum of 2 copies14.
The proportions of SMA patients with different numbers of normal NAIP were 0 copy in 73 patients (39.24%), one copy in 59 patients (31.72%), 2 copies in 53 patients (28.49%) and 4 copies in one patient (0.5%). These figures are also different from Fang et al. report in Chinese population as authors reported 0–2 copies in 4 (9.5%), 26 (61.9%) and 12 patients (28.6%), respectively2. Moreover, NAIP has been reported to be absent in the majority of Tunisian SMA type 1 patients14.
Thus, there is significant difference in the copy number of mentioned genes among SMA patients of different populations. This difference might be due to the presence of some founder mutations in each population.
We also compared the copy numbers of SMN2 and NAIP between four classes of SMA patients. These disease-modifying genes have been shown to influence age of onset of SMA patients. These two genes have been shown to be the most important modifier genes whose copy numbers can influence clinical course of SMA. Hassan et al. have shown that the combination of these genes has better performance in prediction of patients' prognosis than using CNVs of exon 7 of SMN2 gene only. While CNVs of exon 7 of SMN2 gene could predict response of patients to genetic therapy, deletion of exon 5 of NAIP gene alone could not predict severity of SMA15. Another study has shown that NAIP deletion is significantly related to the clinical severity of SMA and is a marker for prediction of SMA prognosis16. This finding has also been confirmed in our study, since all patients carrying deletion of two copies of NAIP gene had severe (type I) SMA.
Zhang et al.17 have determined five combined SMN1-SMN2-NAIP genotypes in their cohort of SMA patients with 0-3-1 genotype being the commonest one. Similarly, in our cohort of patients, 0-3-1 genotype had the highest frequency accounting for 26.19% of cases. Moreover, Zhang et al., have reported the synergistic effect of copy numbers of SMN2 and NAIP genes on clinical course of SMA. They have demonstrated association between the combined SMN1-SMN2-NAIP genotypes with fewer copies and earlier disease onset and higher mortality in SMA patients17. Another study in Vietnamese population has shown association between copy numbers of SMN2 and clinical severity of SMA. However, heterozygous NAIP deletion has been found commonly in SMA patients of this population in an independent manner from the clinical phenotype18. The latter finding is not consistent with our study, since we found association between copy numbers of both SMN2 and NAIP genes and age of disease onset in Iranian population. Similar finding has been reported among Malaysian SMA patient19.
Taken together, the current study is the largest and the most comprehensive genetic analysis of Iranian patients that analyzed SMN1, SMN2 and NAIP copy numbers simultaneously. This study also shows the spectrum of SMN2 and NAIP copy numbers in Iranian SMA patients.
Acknowledgements
We would like to thank Iranian SMA Association and Iranian SMA Registry of TUMS for their kind supports in the current study.
Author contributions
S.G.F. and S.S. wrote the draft and revised it. MHH designed and supervised the study. M.R.A., A.R., H.S.A., G.Z., M.H. and S.A. performed the experiment. S.E. and S.Y. analysed the data. M.M.T.A. and P.S. collected the data. All the authors read and approved the submitted version.
Data availability
The datasets generated and/or analysed during the current study are available in the Clinvar repository (https://www.ncbi.nlm.nih.gov/clinvar/?gr=0&term=smn).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shahram Savad, Email: Shahram.savad@yahoo.com.
Soudeh Ghafouri-Fard, Email: s.ghafourifard@sbmu.ac.ir.
References
- 1.Ogino S, Wilson RB. Genetic testing and risk assessment for spinal muscular atrophy (SMA) Hum. Genet. 2002;111:477–500. doi: 10.1007/s00439-002-0828-x. [DOI] [PubMed] [Google Scholar]
- 2.Fang P, et al. Molecular characterization and copy number of SMN1, SMN2 and NAIP in Chinese patients with spinal muscular atrophy and unrelated healthy controls. BMC Musculoskelet. Disord. 2015;16:1–8. doi: 10.1186/s12891-015-0457-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogino S, Wilson RB. Spinal muscular atrophy: Molecular genetics and diagnostics. Expert Rev. Mol. Diagn. 2004;4:15–29. doi: 10.1586/14737159.4.1.15. [DOI] [PubMed] [Google Scholar]
- 4.Elsheikh B, et al. An analysis of disease severity based on SMN2 copy number in adults with spinal muscular atrophy. Muscle Nerve. 2009;40:652–656. doi: 10.1002/mus.21350. [DOI] [PubMed] [Google Scholar]
- 5.Al-Rajeh S, et al. Molecular analysis of the SMN and NAIP genes in Saudi spinal muscular atrophy patients. J. Neurol. Sci. 1998;158:43–46. doi: 10.1016/S0022-510X(98)00053-7. [DOI] [PubMed] [Google Scholar]
- 6.Watihayati MS, Zabidi-Hussin AM, Tang TH, Matsuo M, Nishio H. Deletion analyses of SMN1 and NAIP genes in Malaysian spinal muscular atrophy patients. Pediatr. Int. 2007;49:11–14. doi: 10.1111/j.1442-200X.2007.02302.x. [DOI] [PubMed] [Google Scholar]
- 7.Omrani O, Bonyadi M, Barzgar M. Molecular analysis of the SMN and NAIP genes in Iranian spinal muscular atrophy patients. Pediatr. Int. 2009;51:193–196. doi: 10.1111/j.1442-200X.2008.02665.x. [DOI] [PubMed] [Google Scholar]
- 8.Wang C-C, Jong Y-J, Chang J-G, Chen Y-L, Wu S-M. Universal fluorescent multiplex PCR and capillary electrophoresis for evaluation of gene conversion between SMN1 and SMN2 in spinal muscular atrophy. Anal. Bioanal. Chem. 2010;397:2375–2383. doi: 10.1007/s00216-010-3761-1. [DOI] [PubMed] [Google Scholar]
- 9.Hulce D, Li X, Snyder-Leiby T, Johathan-Liu CS. GeneMarker® genotyping software: Tools to increase the statistical power of DNA fragment analysis. J. Biomol. Tech. 2011;22:S35–S36. [Google Scholar]
- 10.Savad S, et al. Molecular genetic analysis of patients with duchenne/becker muscular dystrophy by multiplex ligation-dependent probe amplification and next-generation sequencing techniques. Precis. Med. Clin. OMICS. 2022;2:25. doi: 10.5812/pmco-123209. [DOI] [Google Scholar]
- 11.Hasanzad M, et al. Carrier frequency of SMA by quantitative analysis of the SMN1 deletion in the Iranian population. Eur. J. Neurol. 2010;17:160–162. doi: 10.1111/j.1468-1331.2009.02693.x. [DOI] [PubMed] [Google Scholar]
- 12.Verhaart IE, et al. Prevalence, incidence and carrier frequency of 5q–linked spinal muscular atrophy—a literature review. Orphanet. J. Rare Dis. 2017;12:1–15. doi: 10.1186/s13023-017-0671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jedrzejowska M, et al. Phenotype modifiers of spinal muscular atrophy: The number of SMN2 gene copies, deletion in the NAIP gene and probably gender influence the course of the disease. Acta Biochim. Polon. 2009;56:1. doi: 10.18388/abp.2009_2521. [DOI] [PubMed] [Google Scholar]
- 14.Amara A, et al. Correlation of SMN2, NAIP, p44, H4F5 and Occludin genes copy number with spinal muscular atrophy phenotype in Tunisian patients. Eur. J. Paediatr. Neurol. 2012;16:167–174. doi: 10.1016/j.ejpn.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Hassan HA, Zaki MS, Issa MY, El-Bagoury NM, Essawi ML. Genetic pattern of SMN1, SMN2, and NAIP genes in prognosis of SMA patients. Egypt. J. Med. Hum. Genet. 2020;21:1–7. doi: 10.1186/s43042-019-0044-z. [DOI] [Google Scholar]
- 16.Akutsu T, et al. Molecular genetics of spinal muscular atrophy: Contribution of the NAIP gene to clinical severity. Kobe J. Med. Sci. 2002;48:25–31. [PubMed] [Google Scholar]
- 17.Zhang Y, et al. The analysis of the association between the copy numbers of survival motor neuron gene 2 and neuronal apoptosis inhibitory protein genes and the clinical phenotypes in 40 patients with spinal muscular atrophy: Observational study. Medicine. 2020;99:e18809. doi: 10.1097/MD.0000000000018809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tran VK, et al. SMN2 and NAIP gene dosages in Vietnamese patients with spinal muscular atrophy. Pediatr. Int. 2008;50:346–351. doi: 10.1111/j.1442-200X.2008.02590.x. [DOI] [PubMed] [Google Scholar]
- 19.Watihayati MS, et al. Combination of SMN2 copy number and NAIP deletion predicts disease severity in spinal muscular atrophy. Brain Dev. 2009;31:42–45. doi: 10.1016/j.braindev.2008.08.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the Clinvar repository (https://www.ncbi.nlm.nih.gov/clinvar/?gr=0&term=smn).