Summary
Spatial targeting in transcranial magnetic stimulation protocols does not typically account for the idiosyncratic functional organization of individual human brains. Here, we provide a protocol for implementing targeted functional network stimulation (TANS), which accounts for each individual’s unique functional neuroanatomy and cortical folding patterns. Using an example dataset, we describe how to create a head model and estimate the best coil placement and stimulation intensity to minimize off-target effects.
For complete details on the use and execution of this protocol, please refer to Lynch et al. (2022).1
Subject areas: Bioinformatics, Clinical Protocol, Neuroscience, Cognitive Neuroscience
Graphical abstract
Highlights
-
•
Protocol for selective stimulation of individual-specific functional networks
-
•
Improved stimulation specificity with atargeted functional network stimulation (TANS)
-
•
Code for implementing TANS with an example precision functional mapping dataset
-
•
Steps for finding the best coil placement and estimation of the optimal stimulation dose
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Spatial targeting in transcranial magnetic stimulation protocols does not typically account for the idiosyncratic functional organization of individual human brains. Here, we provide a protocol for implementing targeted functional network stimulation, which accounts for each individual’s unique functional neuroanatomy and cortical folding patterns. Using an example dataset, we describe how to create a head model and estimate the best coil placement and stimulation intensity to minimize off-target effects.
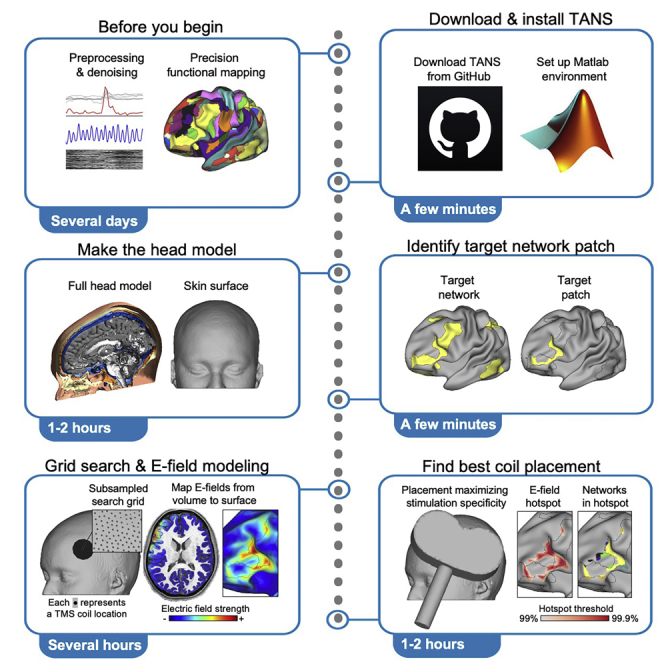
Before you begin
Precision functional mapping (PFM) is the practice of delineating functional brain organization at the individual level,2,3 typically using resting-state functional magnetic resonance imaging (fMRI) functional connectivity (FC). This approach stands in contrast to how FC analyses are typically performed – specifically, by pooling and averaging relatively small quantities of fMRI data from many persons. Precise delineation of functional networks at the individual patient level may be an important step towards making functional MRI a more clinically useful tool.4
One of the clinical applications that PFM is well-suited for is identifying patient-specific targets in brain stimulation interventions,4,5 including transcranial magnetic stimulation (TMS). Currently, spatial targeting in conventional TMS protocols is not informed by the patient’s functional neuroanatomy. Instead, generic coil placements based on scalp heuristics6,7 or stereotaxic coordinates derived from group-average functional maps8,9 have been used. When TMS is applied in this way, however, different functional networks are stimulated across individuals, and often multiple functionally discrete networks are simultaneously targeted within an individual.1 If the efficacy of TMS depends on engaging specific functional networks underlying a patient’s symptoms, widely used “one-size-fits-all” targeting approaches are unlikely to selectively engage any specific functional network across patients, which might prevent TMS from reaching its full therapeutic potential.
With this in mind, we recently developed an individualized TMS targeting approach, called Targeted Functional Brain Network Stimulation (hereafter referred to simply as “TANS”). TANS leverages precise fMRI-derived maps of an individual’s unique functional network topology and cortical folding patterns to find a coil placement and stimulation intensity for engaging the target functional network with minimal off-target effects. In this STAR protocol, we detail step-by-step how to implement TANS using an example PFM dataset that we have made publicly available.
Preprocessing and denoising of neuroimaging data
Timing: Dependent on amount of data per individual, typically several days
Note: This section describes the major preprocessing and denoising steps our group performs before implementing TANS. In principle, there are other equally valid preprocessing and denoising strategies that would also be compatible with TANS.
-
1.Acquire high-resolution (< 1 mm isotropic) anatomical data (T1-weighted and T2-weighted anatomical images) to help minimize partial volume effects and improve the accuracy of cortical surfaces.Note: The field-of-view (FOV) should be large enough to capture key facial features (ears, nose, etc.) that will be used later on during neuronavigation (see troubleshooting Problem #1).
-
a.Anatomical data can be preprocessed and cortical surfaces generated using the Human Connectome Project (HCP) PreFreeSurfer, FreeSurfer, and PostFreeSurfer pipelines.12
-
a.
-
2.Acquire at least 30-min of multi-echo fMRI data13,14 per-subject to improve the reliability of FC measurements.
-
a.Preprocessing of functional data should correct for head motion, geometric distortions in the phase-encoding direction, and precisely co-register functional data with anatomical data used to generate the cortical surfaces.
-
a.
Note: Avoid spatial smoothing in the volume when processing functional data (see15).
-
3.Discard S0-dependent (“not BOLD-like”) artifacts and retain T2∗-weighted (“BOLD-like”) signals of interest using multi-echo independent component analysis denoising (“ME-ICA”) or some other validated denoising algorithm.Note: ME-ICA (now implemented as a part of Tedana; DuPre et al.16) was empirically validated by17 and found by our group to improve the reliability of resting-state FC measurements and functional network maps.1,18 Other denoising algorithms, such as ICA-AROMA,19 can be used instead in the case of single-echo data.
-
a.Manually review and revise ICA component classifications if necessary following the criteria described in.20
-
b.Create and inspect grayplots21 to confirm appropriate separation of S0-dependent (artifacts including but not limited to the effects of head movement) and T2∗-weighted signals.
-
c.Perform mean gray matter time-series regression on the residual ME-ICA denoised time-series to remove spatially diffuse T2∗-weighted signals related to respiration.
-
a.
-
4.
Map the denoised fMRI time-series to the midthickness surfaces (“wb_command -volume-to-surface-mapping”, using the “-ribbon-constrained” method) and create a Connectivity Informatics Technology Initiative (CIFTI) file using Connectome Workbench command line utilities.22
Note: Code for preprocessing and denoising multi-echo fMRI data in this way is maintained in a separate online repository (https://github.com/cjl2007/Liston-Laboratory-MultiEchofMRI-Pipeline).
Precision functional mapping (PFM)
Timing: Typically several hours
Note: The TANS approach is intended to target functional brain networks mapped using PFM in individual subjects. In principle, however, any surface-registered spatial map, such as an ICA component, functional connectivity or task activation map can be used instead.
-
5.
Create a concatenated resting-state fMRI CIFTI (after temporal demeaning of each individual run).
-
6.
Construct a functional connectivity matrix summarizing the temporal correlation between the time-courses of all cortical vertices and subcortical voxels.
Note: Correlations between nodes less than 10 mm apart (geodesic and Euclidean space used for cortico-cortical and subcortical-cortical distance, respectively) should be set to zero to avoid contamination of correlation values from spatial smoothing. Correlations between voxels belonging to subcortical structures should also be set to zero.
-
7.Functional networks can then be identified by subjecting the FC matrix to the InfoMap community detection algorithm.23
-
a.Threshold the functional connectivity matrix to retain the strongest X% correlations (for example, 0.01, 0.02, 0.05, 0.1, 0.2, 0.5, 1, 2, and 5%) to each vertex and voxel.Note: The optimal graph threshold will maximize size-weighted average homogeneity relative to the median of the size-weighted average homogeneity calculated from randomly rotated networks, as done in.24
-
b.Assign functional network identities to communities based on their topology (e.g., spatial pattern of representation in different cortical and subcortical zones) and functional connectivity properties either manually or via a template-matching algorithm.
 CRITICAL: Reliable and accurate mapping of functional brain networks at the single-subject level requires a sufficient amount of high quality data per-subject. Previous work indicates that acquiring 30 or more minutes of resting-state multi-echo fMRI data per-subject can help ensure reliable mapping of individual-specific functional neuroanatomy for individualized TMS targeting.1,18
Note: Code for performing PFM in this way is maintained in the same online repository containing TANS (https://github.com/cjl2007/Targeted-Functional-Network-Stimulation/tree/main/PFM).
CRITICAL: Reliable and accurate mapping of functional brain networks at the single-subject level requires a sufficient amount of high quality data per-subject. Previous work indicates that acquiring 30 or more minutes of resting-state multi-echo fMRI data per-subject can help ensure reliable mapping of individual-specific functional neuroanatomy for individualized TMS targeting.1,18
Note: Code for performing PFM in this way is maintained in the same online repository containing TANS (https://github.com/cjl2007/Targeted-Functional-Network-Stimulation/tree/main/PFM).
-
a.
Key resources table
Materials and equipment
-
•
Anatomical data and surface-registered functional brain network maps formatted per before you begin.
-
•
Linux or MacOS operating system. No specific computer configuration is required. Timing estimates in this protocol are based on a Linux machine running Ubuntu 20.04.5 with 2 × Xeon Silver 12-core 2.220 GHz processors and 400 GB RAM.
-
•
SimNIBS27 version 4.0 is required.
-
•
Matlab version 2017b or higher is required.
-
•
Utilities for reading and writing CIFTI and GIFTI files in Matlab. Specifically, the utilities from the Midnight Scan Club codebase (https://github.com/MidnightScanClub/MSCcodebase).
Step-by-step method details
Below, we provide step-by-step instructions for how to use TANS. Example data from a highly-sampled individual subject (“ME01”, study author CJL) is used for demonstration purposes. The entire example dataset (n = 6, including ME01) can be obtained from Box (https://wcm.box.com/v/TANS-ExampleData).
Download the Targeted Functional Network Stimulation (TANS) code.
Timing: A few minutes
This section describes how to obtain the TANS code from GitHub and to set up the Matlab environment.
Install TANS by downloading the code from GitHub (https://github.com/cjl2007/Targeted-Functional-Network-Stimulation) and adding the folder to the Matlab search path.
-
1.
Open a Matlab terminal and add the TANS, SimNIBS, and CIFTI/GIFTI utility folders to the Matlab search path.
% Define directories
Paths{1} = '/SimNIBS-4.0/';
Paths{2} = '/TANS/';
Paths{3} = '/Utilities/';
% Add the directories
addpath(genpath(Paths{1}));
addpath(genpath(Paths{2}));
addpath(genpath(Paths{3}));
CRITICAL: Before proceeding, confirm that FSL, FreeSurfer, Connectome Workbench, and SimNIBS command line utilities are available when using system commands in Matlab.
If successful, each of the commands below should return status as 0.
% Confirm various software is available
[status,∼] = system('mris_convert -version'); % freesurfer
[status,∼] = system('charm -version'); % simnibs v4.0
[status,∼] = system('wb_command -version'); % connectome workbench
[status,∼] = system('flirt -version'); % fsl
If successful, each of the commands below should return status as 2.
% Confirm that functions for reading and writing
% CIFTI and GIFTI files are also available
status = exist('ft_read_cifti_mod','file');
status = exist('gifti','file');
Note: If status returns an unexpected value, this indicates that either the operating system environment variable or a path to a critical software package is incorrect or missing. This must be fixed before proceeding (see troubleshooting Problem #2).
Create the head model
Timing: Typically 1–2 h
This section describes how to make the head model used later on for E-field modeling.
-
2.Use the tans_headmodels function to create a head model for E-field modeling using the CHARM method.11 In addition, this function generates skin surface geometry files (∗.surf.gii) that can be used for visualizing certain results later on.
-
a.The inputs to the function are:
-
i.Subject: The subject ID (string).
-
ii.T1w: Path to T1w-weighted anatomical image (string).
-
iii.T2w: Path to T2w-weighted anatomical image (string).
-
iv.OutDir : Path to the output folder (string).
-
v.Paths: Paths to folders that must be added to Matlab search path (cell array of strings).
-
i.
-
a.
% Define inputs
Subject = 'ME01';
T1w = ['/example_data/' Subject '/anat/T1w/T1w_acpc_dc_restore.nii.gz'];
T2w = ['/example_data/' Subject '/anat/T1w/T2w_acpc_dc_restore.nii.gz'];
OutDir = ['/example_data/' Subject '/tans'];
% run the tans_headmodels function
tans_headmodels(Subject,T1w,T2w,OutDir,Paths);
Note: The T2-weighted image should be already co-registered to the T1-weighted anatomical image (“T1w”). If a T2-weighted image is not available, T2w can be set to [];
Note: Visually examine the key outputs of the tans_headmodels function located in the [OutDir '/HeadModel/m2m_’ Subject ‘/’] folder to confirm tissue segmentations (final_tissues.nii.gz, skin.nii.gz) and skin surface geometry files (Skin.surf.gii) look correct. Manually edit tissue segmentations, if needed.
Identify the target network patch
Timing: Typically less than 1 min
Functional networks usually consist of multiple nodes spanning different cortical zones in both hemispheres. This section describes how to identify which part of the network TMS can be targeted to maximize stimulation specificity.
-
3.Use the tans_roi function to find the largest patch of the target network that is accessible to TMS (i.e., on a gyral crown) within a specified search space.
-
a.The inputs to the function are:
-
i.TargetNetwork: A CIFTI file containing the functional network of interest (structure array). Non-zero values in TargetNetwork.data are considered target network vertices.
-
ii.MidthickSurfs: Paths to low (32k) dimensional FS_LR midthickness surfaces (Cell array of strings, MidthickSurfs{1} = path to LH, MidthickSurfs{2} = path to RH).
-
iii.VertexSurfaceArea: A CIFTI file containing the vertex surface areas (structure array).
-
iv.Sulc: CIFTI file containing sulcal depth information (structure array).
-
v.SearchSpace: A CIFTI file containing a binarized mask representing the search space (structure array). If there is no particular region of interest, SearchSpace can be set to []to consider the entire cortical surface.
-
vi.OutDir: Path to the output folder (string).
-
vii.Paths: Paths to folders that must be added to Matlab search path (cell array of strings).
-
i.
-
a.
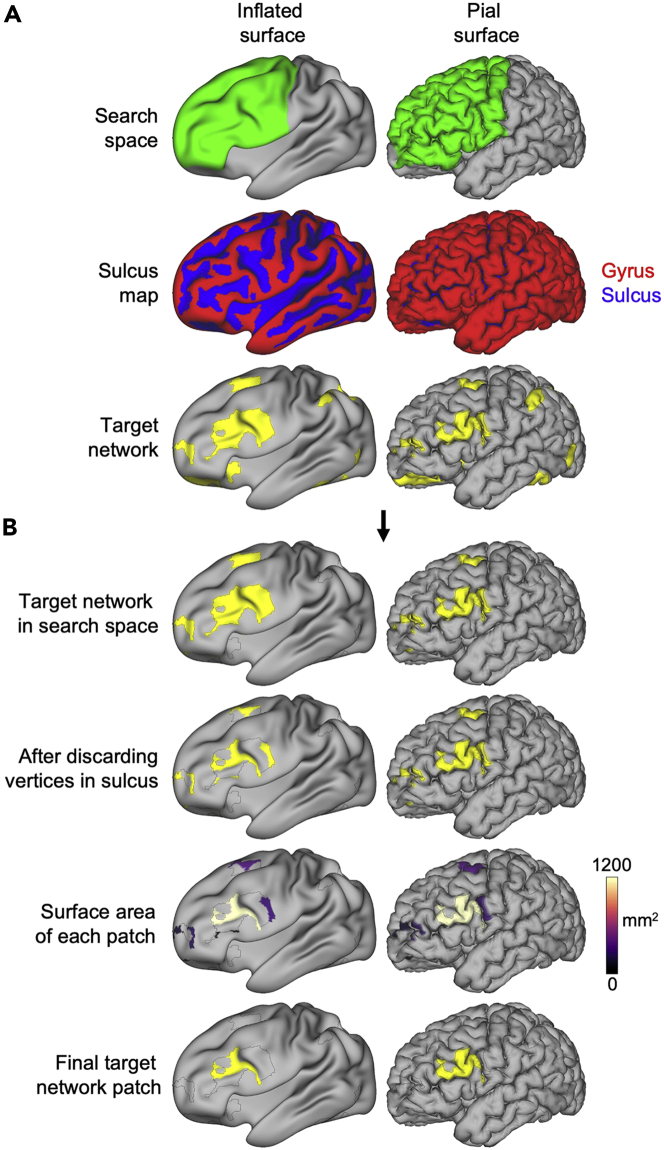
Note: Visually examine outputs of the tans_roi function located in the [OutDir '/ROI/’] folder.
Specifically, the target network vertices retained inside the search space (TargetNetwork+SearchSpace.dtseries.nii), the effect of discarding target network vertices in a sulcus (TargetNetwork+SearchSpace+SulcalMask.dtseries.nii), the size of each remaining network patch (TargetNetwork+SearchSpace+SulcalMask+Clusters.dtseries.nii), and the final target network patch selected (TargetNetworkPatch.dtseries.nii).
Note: All of these CIFTI files can be viewed on any of the fs_LR_32k surfaces (inflated and pial surfaces both shown in Figure 1).
CRITICAL: Confirm that the target network patch is amenable to TMS before proceeding (see troubleshooting Problem #3). For example, a patch that is located on the inferior surface of the brain is unlikely to produce good results.
% read in the functional networks for this individual;
FunctionalNetworks = ft_read_cifti_mod([‘/example_data/’ Subject '/pfm/' Subject '_FunctionalNetworks.dtseries.nii']);
% isolate the target network
TargetNetwork = FunctionalNetworks;
TargetNetwork.data(TargetNetwork.data∼=9) = 0; % note: 9 == frontoparietal network
TargetNetwork.data(TargetNetwork.data∼=0) = 1; % binarize.
% load pre-made mask representing the lateral PFC;
SearchSpace = ft_read_cifti_mod('LPFC_LH+RH.dtseries.nii');
% load sulcal depth information;
Sulc = ft_read_cifti_mod([‘/example_data/’ Subject…
'/anat/MNINonLinear/fsaverage_LR32k/' Subject '.sulc.32k_fs_LR.dscalar.nii']);
BrainStructure = SearchSpace.brainstructure; % extract brain structure index
Sulc.data(BrainStructure==-1) = []; % remove medial wall vertices
% define the other input variables;
MidthickSurfs{1} = ['/example_data/' Subject '/anat/T1w/fsaverage_LR32k/'…
Subject '.L.midthickness.32k_fs_LR.surf.gii']; % LH
MidthickSurfs{2} = ['/example_data/' Subject '/anat/T1w/fsaverage_LR32k/'…
Subject '.R.midthickness.32k_fs_LR.surf.gii']; % RH
VertexSurfaceArea = ft_read_cifti_mod([‘/example_data/’ Subject…
'/anat/T1w/fsaverage_LR32k/' Subject '.midthickness_va.32k_fs_LR.dscalar.nii']);
OutDir = [ '/example_data/' Subject '/tans/Network_Frontoparietal'];
% run the tans_roi function
tans_roi(TargetNetwork,MidthickSurfs,VertexSurfaceArea,Sulc,SearchSpace,OutDir,Path);
Figure 1.
Identifying the target network patch using the tans_roi function
(A and B) Inputs for the function include the search space, sulcus map, and target functional network map (A). The search space is a binarized mask that constrains which elements of the target functional network are considered eligible targets for TMS. In this example, the search space is the lateral prefrontal cortex (green). The sulcus map (generated by Freesurfer) quantifies how far each vertex is from a hypothetical middle point between gyri and sulci (values > 0 indicate the vertex is closer to a gyrus and values < 0 indicate vertex is closer to a sulcus). The final output of the function is the largest patch of the target network after discarding all network vertices outside of the search space or in a sulcus (B).
Make a search grid on the scalp above the target network patch centroid
Timing: Typically 5–10 min
This section describes how to create a “search grid” – a series of possible coil placements that will be evaluated.
-
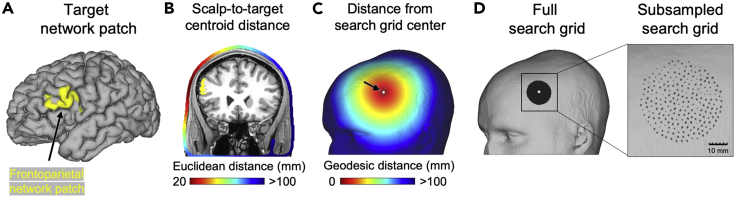
4.Use the tans_searchgrid function to generate an array of three-dimensional coordinates representing a search grid on the scalp directly above the centroid of the target network patch (see Figure 2).
-
a.The inputs to the function are:
-
i.TargetNetworkPatch: A CIFTI file indexing the functional network patch (structure array). Non-zero values in TargetNetwork.data are considered target network vertices.
-
ii.PialSurfs: Paths to low (32k) dimensional FS_LR pial surfaces (Cell array of strings, PialSurfs{1} = path to LH, PialSurfs{2} = path to RH).
-
iii.SkinSurf: Path to the skin surface geometry file (string).
-
iv.SearchGridRadius: Radius of the search grid. Recommended value is 20 mm.
-
v.GridSpacing: The approximate distance between pairs of vertices in the search grid. Recommended value is 2 mm.
-
vi.OutDir: Path to the output folder (string).
-
vii.Paths: Paths to folders that must be added to Matlab search path (cell array of strings).
-
i.
-
a.
% define inputs
PialSurfs{1} = [‘/example_data/' Subject '/anat/T1w/fsaverage_LR32k/' Subject…
'.L.pial.32k_fs_LR.surf.gii'];
PialSurfs{2} = [‘/example_data/' Subject '/anat/T1w/fsaverage_LR32k/'…
Subject '.R.pial.32k_fs_LR.surf.gii'];
TargetPatch = [OutDir '/ROI/TargetNetworkPatch.dtseries.nii’]
SkinSurf = [‘/example_data/' Subject '/tans/HeadModel/m2m_' Subject '/Skin.surf.gii'];
SearchGridRadius = 20; % in mm
GridSpacing = 2; % in mm
% run the tans_searchgrid function
[SubSampledSearchGrid,∼] = tans_searchgrid(TargetPatch,PialSurfs,SkinSurf,…
GridSpacing,SearchGridRadius,OutDir,Paths);
Note: Both the size (radius) and density of the search grid influence the total number of simulations performed. The effect of subsampling is shown in Figure 2C.
Figure 2.
Constructing a search grid of coil positions on the scalp directly above the centroid of the target network patch using the tans_searchgrid function
(A) The target network patch is identified with tans_roi.m.
(B) The distance in Euclidean space between the centroid of the target network patch and each point on the scalp is calculated.
(C) The nearest point on the scalp (black arrow pointing to white foci) is identified and serves as the center point in the search grid.
(D) The geodesic distance from the center of the search grid is calculated. All skin vertices within a distance specified by the SearchGridRadius variable are identified (SearchGridRadius is set to the default value of 20 mm in this example) and compromise the initial search grid. Subsampling (set by the GridDensity variable) is used to reduce the total number of simulations that will need to be performed. In this example, the spacing between points in the search grid is approximately 2 mm.
Perform electric field modeling at each point in search grid
Timing: Depends on the total number of simulations performed and available parallel pools in Matlab, typically several hours
This section describes how to perform E-field simulations at each point in the search grid.
-
5.Use the tans_simnibs function to perform electric field simulations at each point in the search grid using SimNIBS.27 A number of coil orientations determined by an angle (in degrees) specified by the user are performed.
-
a.The inputs to the function are:
-
i.SearchGridCoords: Number of coil positions × 3 numeric array generated by tans_searchgrid function.
-
ii.HeadMesh: Path to head model (string).
-
iii.CoilModel: Path to the coil model (.cdd file, string). Avoid using non-planar coil models, if possible (see troubleshooting Problem #4).
-
iv.AngleResolution: Inter angle resolution used in the search grid (numeric).
-
v.DistanceToScalp: Distance from scalp to TMS coil (numeric).
-
vi.SkinSurf: Path to the skin surface geometry file (string).
-
vii.MidthickSurfs: Paths to low (32k) dimensional FS_LR midthickness surfaces (Cell array of strings, MidthickSurfs{1} = path to LH, MidthickSurfs{2} = path to RH).
-
viii.WhiteSurfs: Paths to low (32k) dimensional FS_LR white surfaces (Cell array of strings, WhiteSurfs{1} = path to LH, WhiteSurfs{2} = path to RH).
-
ix.PialSurfs: Paths to low (32k) dimensional FS_LR pial surfaces (Cell array of strings, PialSurfs{1} = path to LH, PialSurfs{2} = path to RH).
-
x.MedialWallMasks: Paths to low (32k) dimensional FS_LR medial wall masks (Cell array of strings, MedialWallMasks{1} = path to LH, MedialWallMasks{2} = path to RH).
-
xi.nThreads: The number of parallel pools that will be used in Matlab (numeric).
-
xii.OutDir: Path to the output folder (string).
-
xiii.Paths: Paths to folders that must be added to Matlab search path (cell array of strings).
-
i.
-
a.
% define inputs
WhiteSurfs{1} = ['/example_data/' Subject '/anat/T1w/fsaverage_LR32k/'…
Subject '.L.white.32k_fs_LR.surf.gii'];
WhiteSurfs{2} = ['/example_data/' Subject '/anat/T1w/fsaverage_LR32k/'…
Subject '.R.white.32k_fs_LR.surf.gii'];
MedialWallMasks{1} = ['/example_data/' Subject '/anat/MNINonLinear/fsaverage_LR32k/'…
Subject '.L.atlasroi.32k_fs_LR.shape.gii'];
MedialWallMasks{2} = ['/example_data/' Subject '/anat/MNINonLinear/fsaverage_LR32k/'…
Subject '.R.atlasroi.32k_fs_LR.shape.gii'];
HeadMesh = ['/example_data/' Subject '/tans/HeadModel/' Subject '.msh'];
CoilModel = [Paths{1} '/…/coil_models/Drakaki_BrainStim_2022/MagVenture_MCF-B65.ccd'];
DistanceToScalp = 2;
AngleResolution = 30; % in degrees
nThreads = 20; %
% run the tans_simnibs function;
tans_simnibs(SearchGridCoords,HeadMesh,CoilModel,AngleResolution,DistanceToScalp,…
SkinSurf,MidthickSurfs,WhiteSurfs,PialSurfs,MedialWallMasks,nThreads,OutDir,Paths);
Note: The strength of the electric field associated with each coil placement is mapped to the individual’s cortical surfaces.
Note: A fixed stimulation intensity (dI/dt = 1 A/μs) is modeled at this point because the strength of the E-field has no effect on its spatial distribution (including where it is maximal relative to the target network, which is what will be optimized in subsequent steps, see Figure 3).
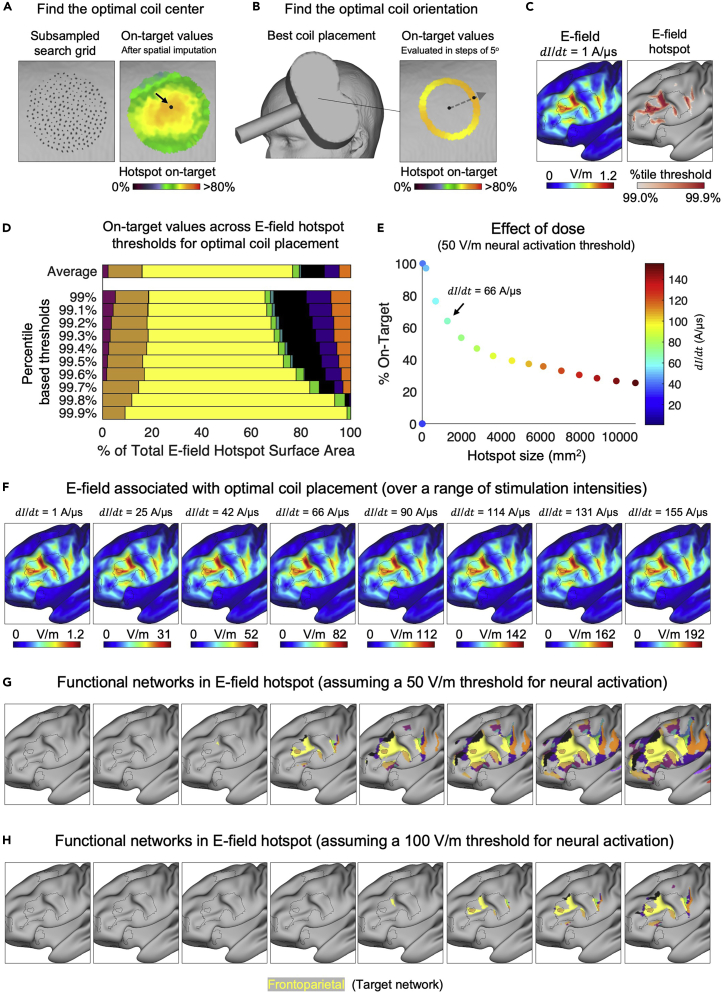
Figure 3.
Searching for a TMS coil placement that best aligns the strongest portion of E-field (the “hotspot”) with the target network using the tans_optimize function
(A) The maximum on-target value across all orientations is calculated at each point in the search grid. Spatial smoothing is used to impute on-target values for non-sampled points in the search grid.
(B) The skin surface vertex maximizing the on-target value (on average within a distance specified by the Uncertainty variable) when it is set as the coil center is considered to be the optimal choice (black foci in A marked by arrow). Next, the exact coil orientation maximizing the on-target value is identified.
(C) The magnitude of the E-field (C, left) and various percentile based definitions of the hotspot (C, right) associated with the optimal coil placement.
(D) The average on-target value across E-field hotspot thresholds.
(E) A range of stimulation intensities can be modeled using the tans_dose function to estimate stimulation specificity achieved given an assumed neural activation threshold and minimum hotspot size (50 V/m and 1,000 mm2 are used in the example shown in E).
(F) Note how the stimulation intensity has no effect on the spatial distribution of the E-field (F), which is why a fixed stimulation intensity of dI/dt = 1 A/μs can be used to identify the optimal coil placement in the first place.
(G and H) The functional networks represented inside the suprathreshold portion of the E-field assuming a 50 V/m (G) and 100 V/m (H) threshold for neural activation.
Find the coil placement that best aligns the electric field hotspot with the target network
Timing: Depends on the total number of simulations performed, typically 1–2 h
This section describes how the best coil placement is identified.
-
6.Use the tans_optimize function to find which coil placement has the greatest on-target value (calculated as the proportion of the E-field hotspot inside the target network). In other words, this function identifies the coil placement that maximizes stimulation specificity.
-
a.The inputs to the function are:
-
i.Subject: The subject ID (string).
-
ii.TargetNetwork: A CIFTI file containing the functional network of interest (structure array). Non-zero values in TargetNetwork.data are considered target network vertices.
-
iii.AvoidanceRegion: A CIFTI file indexing the brain regions or networks of no interest (structure array). Non-zero values in AvoidanceRegion.data are considered non-target network vertices.
-
iv.PercentileThresholds: A range of percentiles used for operationalizing the E-field hotspot (numeric). For example, linspace(99.9,99,10).
-
v.SearchGrid: Path to Search Grid GIFTI file (string).
-
vi.DistanceToScalp: DistanceToScalp: Distance from scalp to TMS coil (numeric).
-
vii.SkinSurf: Path to the skin surface geometry file (string).
-
viii.VertexSurfaceArea: A CIFTI file containing the vertex surface areas (structure array).
-
ix.MidthickSurfs: Paths to low (32k) dimensional FS_LR midthickness surfaces (Cell array of strings, MidthickSurfs{1} = path to LH, MidthickSurfs{2} = path to RH).
-
x.WhiteSurfs: Paths to low (32k) dimensional FS_LR white surfaces (Cell array of strings, WhiteSurfs{1} = path to LH, WhiteSurfs{2} = path to RH).
-
xi.PialSurfs: Paths to low (32k) dimensional FS_LR pial surfaces (Cell array of strings, PialSurfs{1} = path to LH, PialSurfs{2} = path to RH).
-
xii.MedialWallMasks: Paths to low (32k) dimensional FS_LR medial wall masks (Cell array of strings, MedialWallMasks{1} = path to LH, MedialWallMasks{2} = path to RH).
-
xiii.HeadMesh: Path to head model (string).
-
xiv.AngleResolution: Inter angle resolution used when fine tuning coil orientation (numeric).
-
xv.Uncertainty: Coil center position uncertainty (in mm, numeric).
-
xvi.CoilModel: Path to the coil model (string).
-
xvii.OutDir: Path to the output folder (string).
-
xviii.Paths: Paths to folders that must be added to Matlab search path (cell array of strings).
-
i.
-
a.
% define input variables
PercentileThresholds = linspace(99.9,99,10);
Uncertainty = 5; % in mm
% isolate the target network again
TargetNetwork = FunctionalNetworks;
TargetNetwork.data(TargetNetwork.data∼=9) = 0; % 9 == frontoparietal network
TargetNetwork.data(TargetNetwork.data∼=0) = 1; % binarize.
Note: The entire target network is typically used at this stage, not just the target patch identified earlier by the tans_roi function. Main outputs of the tans_optimize function are shown in Figures 3A–3D.
% run the "tans_optimize.m" module;
tans_optimize(Subject,TargetNetwork,[],PercentileThresholds,SearchGrid,DistanceToScalp,…
SkinSurf,VertexSurfaceArea,MidthickSurfs,WhiteSurfs,PialSurfs,MedialWallMasks,HeadMesh,…
AngleResolution,PositionUncertainty,CoilModel,OutDir,Paths);
Optional: Users can specify an avoidance region or network to minimize certain kinds of off-target effects. This may be desirable, for example, when targeting network A, and avoiding stimulation of network B is more important than avoiding stimulation of network C.
% specify an avoidance region;
AvoidanceRegion = FunctionalNetworks;
AvoidanceRegion.data(AvoidanceRegion.data∼=17) = 0; % 17 == Somatomotor-Hand
AvoidanceRegion.data(AvoidanceRegion.data∼=0) = 1; % binarize.
% run the tans_optimize function;
tans_optimize(Subject,TargetNetwork,AvoidanceRegion,PercentileThresholds,SearchGrid,…
DistanceToScalp,SkinSurf,VertexSurfaceArea,MidthickSurfs,WhiteSurfs,PialSurfs,…
MedialWallMasks,HeadMesh,AngleResolution,PositionUncertainty,CoilModel,OutDir,Paths);
Estimate the optimal dose
Timing: Typically less than 1 min
This section describes how the optimal stimulation intensity (dose) for target engagement with minimal off-target stimulation can be estimated with an assumed neural activation threshold.
-
7.Evaluate the effect of stimulation intensity on the on-target value when using the coil placement prescribed by tans_optimize using the tans_dose function (see Figures 3E–3H).
-
a.The inputs to the function are:
-
i.magnE: A CIFTI file containing the E-field associated with the coil placement prescribed by tans_optimize (can be either a string path or a structure array).
-
ii.VertexSurfaceArea: A CIFTI file containing the vertex surface areas (structure array).
-
iii.DiDt: A range of stimulation intensities (the speed of variation of the current throughout the stimulating coil, in units of A/us, numeric).
-
iv.AbsoluteThreshold: This value (in V/m units) represents an assumed neural activation threshold (numeric).
-
v.MinHotSpotSize: The minimum size (in surface area, mm2) of the E-field hotspot (numeric).
-
vi.TargetNetwork: A CIFTI file containing the functional network of interest (structure array). Non-zero values in TargetNetwork.data are considered target network vertices.
-
vii.AvoidanceRegion: A CIFTI file indexing the brain regions or networks of no interest (structure array). Non-zero values in AvoidanceRegion.data are considered non-target network vertices.
-
viii.FunctionalNetworks: A CIFTI file indexing all functional networks (string or structure array)
-
ix.NetworkLabels: Path to a .xls file containing the names and desired colors of each functional network (string).
-
x.OutDir: Path to the output folder (string).
-
xi.Paths: Paths to folders that must be added to Matlab search path (cell array of strings).
-
i.
-
a.
% define some new input variables;
NetworkLabels = [DataDir '/' Subject '/pfm/' Subject '_FunctionalNetworks.xlsx'];
magnE = [OutDir '/Optimize/magnE_BestCoilCenter+BestOrientation.dtseries.nii'];
DiDt = linspace(1,155,100) ∗ 1e6; % A/us
AbsoluteThreshold = 50; % V/m
MinHotSpotSize = 500; % surface area (mmˆ2)
% run the "tans_dose.m" module; tans_dose(magnE,VertexSurfaceArea,DiDt,AbsoluteThreshold,MinHotSpotSize,TargetNetwork,[],…
FunctionalNetworks,NetworkLabels,[OutDir '/Optimize/'],Paths);
Enter coil placement information into neuronavigation software
Timing: Typically 10–15 min
The tans_optimize function generates two files relevant for neuronavigation. Specifically, a pair of text files in [OutDir '/Optimize/’] containing three-dimensional coordinates representing the TMS coil’s center (CoilCenterCoordinates.txt) and orientation (CoilOrientationCoordinates.txt) on the scalp.
Note: These instructions are specific to the Brainsight software, but in principle these coordinates should be compatible with other neuronavigation software.
-
8.
Open the Brainsight software.
-
9.Load the anatomical data (the T1w image in native volume space).
-
a.Generate the skin and full brain curvilinear reconstructions.
-
a.
-
10.Select the “Configure Targets” option within the “Targets” panel (see Methods video S1).
-
a.First, enter the coil orientation coordinates (“CoilOrientationCoordinates.txt”) into the “Crosshairs Origin” field and create a Marker by opening the “New…” dropdown menu and selecting “Marker”.
-
b.Next, enter the coil center coordinates (“CoilCenterCoordinates.txt”) into the “Crosshairs Origin” field.
-
c.Open the “Crosshairs:” dropdown menu and select “Mini TMS Coil” (see better visualize the orientation of the coil) and select “Optimize Now” so that the coil lays flat against the skin surface.
-
d.Use the “Twist” angle adjustment tool to find the orientation that is in line with the Coil Orientation marker.
-
a.
Expected outcomes
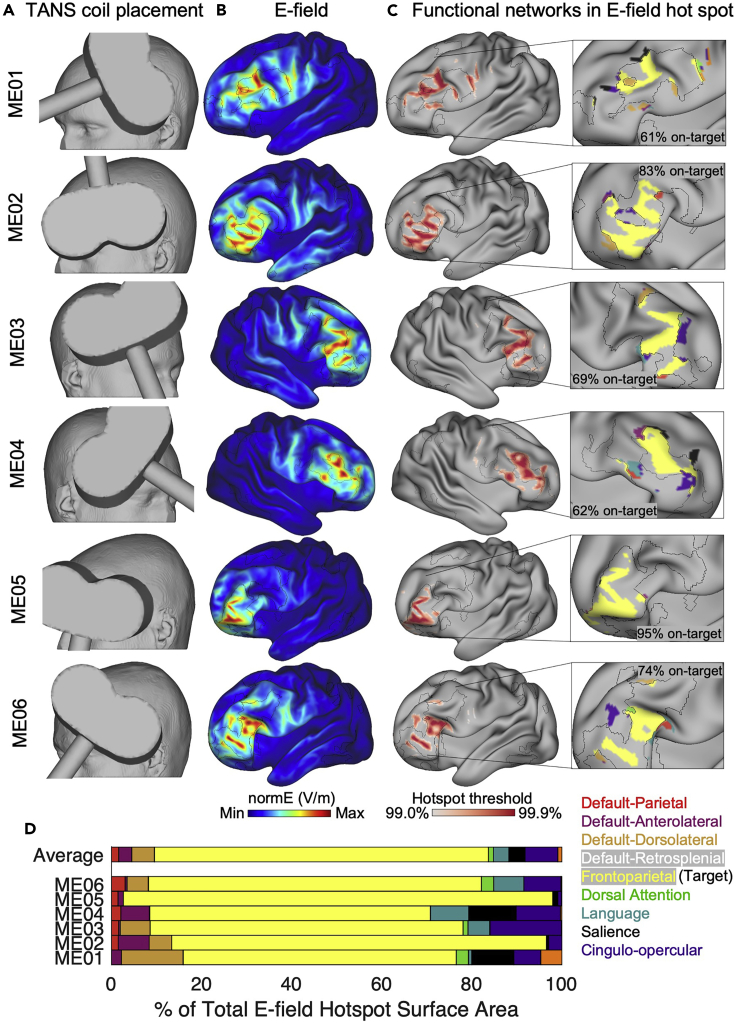
It is expected that TANS will find a coil placement for stimulating the target functional brain network with minimal stimulation of non-target functional networks (to the extent that is possible given the size of the target functional network relative to the size of the E-field hotspot). To provide context for what the results of TANS are expected to look like, Figure 4A shows the optimal coil placement found using TANS for stimulating the frontoparietal network in ME01 and the other five example PFM datasets available online. The E-field associated with the optimal coil placement and boundaries of the target functional network (black borders) in each individual is shown in Figure 4B. All functional networks in the E-field hotspot are shown in Figure 4C (the 99.5% threshold is used for visualization purposes). A stacked horizontal bar plot (Figure 4D) summarizes the percentage of the total E-field hotspot surface area that is occupied by each functional brain network.
Figure 4.
Example outcomes when targeting the frontoparietal network in 6 healthy individuals
(A) The coil placement that maximized the on-target value in each patient.
(B) The electric field (E-field) generated by the optimal TMS coil placement.
(C) The E-field hotspots are defined using percentile based thresholds (99%–99.9%, in 0.1% steps, in this case). The middle (99.5%) threshold is selected for visualizing functional networks inside the E-field hotspot in the inset.
(D) Overall variability in functional network stimulation across subjects summarized in a horizontal stacked bar graph.
Limitations
The main limitation of the TANS approach (specifically, the tans_simnibs function) is that it involves a grid-search procedure during which many E-field simulations are performed. This can take multiple hours to complete, which can be undesirable when time is a factor. To reduce runtime, we recommend setting the nThreads variable to a number that will fully utilize the parallel processing capabilities of your Matlab environment (to increase the number of simulations performed at once). Another limitation of TANS (and of TMS more generally) is that smaller functional networks or those more often represented in a fissure or on the medial surface of the brain will be less amenable to selective stimulation than larger networks with prominent representation on the lateral surface of the brain.
Troubleshooting
Problem 1
Anatomical landmarks needed for neuronavigation co-registration procedures are corrupted or missing.
Related to preprocessing and denoising of neuroimaging data. Administering TMS precisely requires use of a neuronavigation software (for example, Rogue Research’s Brainsight program). The accuracy of neuronavigation procedure depends on co-registration of multiple key facial landmarks (e.g., the tragus of the ears, bridge and tip of the nose) with the anatomical image. These facial landmarks can be corrupted or missing entirely in anatomical images with a smaller FOV, which can decrease the accuracy of the neuronavigation procedure and in turn limit the impact of the TANS approach.
Potential solution
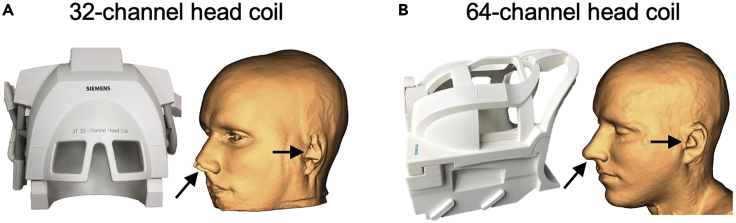
If possible, acquire a new T1w image using a head coil designed for large FOV acquisitions and full head coverage. For example, compare the two skin reconstructions for the same individual when using a 32-channel (Figure 5A) versus 64-channel head coil (Figure 5B). Inform the scan technician that capturing these facial landmarks is a priority. If any non-rigid facial landmark will be used for neuronavigation (the tragus of the ear), we recommend that they be in their “natural” position during MRI scanning. For example, we recommend avoiding compression of non-rigid facial features with the cushions often used to immobilize the head during MRI scanning. The original target coordinates produced by the tans_optimize function can be transformed into this image space and used for neuronavigation instead.
Figure 5.
Potential solution for low quality skin reconstructions
(A and B) Head coils designed for larger field of view acquisitions can better capture facial landmarks needed for neuronavigation. Comparison of skin reconstructions for the same individual when using a 32-channel (A) versus 64-channel head coil (B).
Problem 2
The Matlab environment is not configured properly.
Related to step 1. TANS requires Matlab to make calls to the operating system in order to execute certain FreeSurfer, Connectome Workbench, and FSL commands. Both the system environment variable and all paths to critical software packages must be properly specified before running TANS.
Potential solution
If Matlab cannot call all of the required software packages (FreeSurfer, SimNIBS, Connectome Workbench, and FSL), use Matlab’s “setenv” function to update the PATH variable so that it includes the correct paths to all required programs.
Problem 3
The target network patch identified is not accessible to TMS.
Related to step 3. The tans_roi function automatically excludes from consideration all parts of the target functional network that are located within a sulcus or on the medial wall of the brain. In addition, users have the option to specify a search space to further constrain which parts of the functional network are considered potential stimulation targets. If no search space is specified, the target network patch can be located in a brain area less amenable to TMS (for example, in inferior temporal or insular cortex).
Potential solution
If the target network patch identified by tans_roi is located in a brain region not accessible to TMS, users can re-run tans_roi with an updated search space mask that excludes these regions before proceeding to the other steps.
Problem 4
The coil placement prescribed by TANS is not physically possible.
Related to step 5. When using a non-planar (bent) TMS coil model, it is possible that TANS will identify a coil placement that is not physically possible to achieve because part of the stimulating coil is located within the head.
Potential solution
After running the tans_optimize function, users are encouraged to visually inspect both the skin and coil surface geometry files. Users can consider switching to a planar (not bent) TMS coil and re-running tans_simnibs and tans_optimize if there is overlap between the skin and coil surface files.
Problem 5
The coil handle is oriented in the direction of the face and interferes with neuronavigation tracking equipment.
Related to step 10. In practice, it is usually desirable to have the TMS coil handle and all the cables attached to it oriented posteriorly away from the subject’s face and neuronavigation tracking equipment. TANS does not automatically consider whether or not the prescribed coil placement will be inconvenient in this way.
Potential solution
If TANS identifies a coil orientation with the handle pointed anteriorly towards the subject’s face as the best choice, we recommend considering flipping the orientation 180° and reversing the current direction electronically to achieve the intended E-field but with a more convenient approach.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by either the lead contact, Conor Liston (col2004@med.cornell.edu) , or lead technical contact, Charles J. Lynch (cjl2007@med.cornell.edu).
Materials availability
-
•
This study did not generate new unique reagents.
Acknowledgments
We thank the staff at the Citigroup Biomedical Imaging Center for assistance with data collection. This work was supported by grants to C.L. from the National Institute of Mental Health, the National Institute on Drug Addiction, the Hope for Depression Research Foundation, and the Foundation for OCD Research. C.J.L. was supported by an NIMH F32 National Research Service Award (F32MH120989).
Author contributions
Conceptualization, C.J.L., C.L.; Methodology, C.J.L., I.E., H.B.; Investigation, C.J.L., I.E., J.D.P.; Funding Acquisition, C.L.; Resources, C.L., J.D.P; Writing – Original Draft, C.J.L., C.L.; Writing – Review & Editing, C.J.L., I.E., J.D.P., C.L.; Supervision, C.L.
Declaration of interests
C.L. is listed as an inventor for Cornell University patent applications on neuroimaging biomarkers for depression that are pending or in preparation.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2023.102118.
Contributor Information
Charles J. Lynch, Email: cjl2007@med.cornell.edu.
Conor Liston, Email: col2004@med.cornell.edu.
Data and code availability
An example dataset (Database: https://wcm.box.com/v/TANS-ExampleData) and code for implementing precision functional mapping and TANS is available online (https://github.com/cjl2007/Targeted-Functional-Network-Stimulation; https://doi.org/10.5281/zenodo.7566548). Code for processing multi-echo fMRI data is also available online (https://github.com/cjl2007/Liston-Laboratory-MultiEchofMRI-Pipeline,; https://doi.org/10.5281/zenodo.6958611).
References
- 1.Lynch C.J., Elbau I.G., Ng T.H., Wolk D., Zhu S., Ayaz A., Power J.D., Zebley B., Gunning F.M., Liston C. Automated optimization of TMS coil placement for personalized functional network engagement. Neuron. 2022;110:3263–3277.e4. doi: 10.1016/j.neuron.2022.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laumann T.O., Gordon E.M., Adeyemo B., Snyder A.Z., Joo S.J., Chen M.-Y., Gilmore A.W., McDermott K.B., Nelson S.M., Dosenbach N.U.F., et al. Functional system and areal organization of a highly sampled individual human brain. Neuron. 2015;87:657–670. doi: 10.1016/j.neuron.2015.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon E.M., Laumann T.O., Gilmore A.W., Newbold D.J., Greene D.J., Berg J.J., Ortega M., Hoyt-Drazen C., Gratton C., Sun H., et al. Precision functional mapping of individual human brains. Neuron. 2017;95:791–807.e7. doi: 10.1016/j.neuron.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gratton C., Kraus B.T., Greene D.J., Gordon E.M., Laumann T.O., Nelson S.M., Dosenbach N.U.F., Petersen S.E. Defining individual-specific functional neuroanatomy for precision psychiatry. Biol. Psychiatry. 2020;88:28–39. doi: 10.1016/j.biopsych.2019.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greene D.J., Marek S., Gordon E.M., Siegel J.S., Gratton C., Laumann T.O., Gilmore A.W., Berg J.J., Nguyen A.L., Dierker D., et al. Integrative and network-specific connectivity of the basal ganglia and thalamus defined in individuals. Neuron. 2020;105:742–758.e6. doi: 10.1016/j.neuron.2019.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beam W., Borckardt J.J., Reeves S.T., George M.S. An efficient and accurate new method for locating the F3 position for prefrontal TMS applications. Brain Stimul. 2009;2:50–54. doi: 10.1016/j.brs.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mir-Moghtadaei A., Siddiqi S.H., Mir-Moghtadaei K., Blumberger D.M., Vila-Rodriguez F., Daskalakis Z.J., Fox M.D., Downar J. Updated scalp heuristics for localizing the dorsolateral prefrontal cortex based on convergent evidence of lesion and brain stimulation studies in depression. Brain Stimul. 2022;15:291–295. doi: 10.1016/j.brs.2022.01.013. [DOI] [PubMed] [Google Scholar]
- 8.Weigand A., Horn A., Caballero R., Cooke D., Stern A.P., Taylor S.F., Press D., Pascual-Leone A., Fox M.D. Prospective validation that subgenual connectivity predicts antidepressant efficacy of transcranial magnetic stimulation sites. Biol. Psychiatry. 2018;84:28–37. doi: 10.1016/j.biopsych.2017.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blumberger D.M., Vila-Rodriguez F., Thorpe K.E., Feffer K., Noda Y., Giacobbe P., Knyahnytska Y., Kennedy S.H., Lam R.W., Daskalakis Z.J., Downar J. Effectiveness of theta burst versus high-frequency repetitive transcranial magnetic stimulation in patients with depression (THREE-D): a randomised non-inferiority trial. Lancet. 2018;391:1683–1692. doi: 10.1016/S0140-6736(18)30295-2. [DOI] [PubMed] [Google Scholar]
- 10.Nielsen J.D., Madsen K.H., Puonti O., Siebner H.R., Bauer C., Madsen C.G., Saturnino G.B., Thielscher A. Automatic skull segmentation from MR images for realistic volume conductor models of the head: assessment of the state-of-the-art. Neuroimage. 2018;174:587–598. doi: 10.1016/j.neuroimage.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Puonti O., Van Leemput K., Saturnino G.B., Siebner H.R., Madsen K.H., Thielscher A. Accurate and robust whole-head segmentation from magnetic resonance images for individualized head modeling. Neuroimage. 2020;219:117044. doi: 10.1016/j.neuroimage.2020.117044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glasser M.F., Sotiropoulos S.N., Wilson J.A., Coalson T.S., Fischl B., Andersson J.L., Xu J., Jbabdi S., Webster M., Polimeni J.R., et al. The minimal preprocessing pipelines for the Human Connectome project. Neuroimage. 2013;80:105–124. doi: 10.1016/j.neuroimage.2013.04.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Posse S., Wiese S., Gembris D., Mathiak K., Kessler C., Grosse-Ruyken M.-L., Elghahwagi B., Richards T., Dager S.R., Kiselev V.G. Enhancement of BOLD-contrast sensitivity by single-shot multi-echo functional MR imaging. Magn. Reson. Med. 1999;42:87–97. doi: 10.1002/(sici)1522-2594(199907)42:1<87::aid-mrm13>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 14.Kundu P., Voon V., Balchandani P., Lombardo M.V., Poser B.A., Bandettini P.A. Multi-echo fMRI: a review of applications in fMRI denoising and analysis of BOLD signals. Neuroimage. 2017;154:59–80. doi: 10.1016/j.neuroimage.2017.03.033. [DOI] [PubMed] [Google Scholar]
- 15.Coalson T.S., Van Essen D.C., Glasser M.F. The impact of traditional neuroimaging methods on the spatial localization of cortical areas. Proc. Natl. Acad. Sci. USA. 2018;115:E6356–E6365. doi: 10.1073/pnas.1801582115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DuPre E., Salo T., Markello R., Kundu P., Whitaker K., Handwerker D. Zenodo; 2020. ME-ICA/Tedana: 0.0. 9a. [Google Scholar]
- 17.Kundu P., Inati S.J., Evans J.W., Luh W.-M., Bandettini P.A. Differentiating BOLD and non-BOLD signals in fMRI time series using multi-echo EPI. Neuroimage. 2012;60:1759–1770. doi: 10.1016/j.neuroimage.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lynch C.J., Power J.D., Scult M.A., Dubin M., Gunning F.M., Liston C. Rapid precision functional mapping of individuals using multi-echo fMRI. Cell Rep. 2020;33:108540. doi: 10.1016/j.celrep.2020.108540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pruim R.H.R., Mennes M., van Rooij D., Llera A., Buitelaar J.K., Beckmann C.F. ICA-AROMA: a robust ICA-based strategy for removing motion artifacts from fMRI data. Neuroimage. 2015;112:267–277. doi: 10.1016/j.neuroimage.2015.02.064. [DOI] [PubMed] [Google Scholar]
- 20.Griffanti L., Douaud G., Bijsterbosch J., Evangelisti S., Alfaro-Almagro F., Glasser M.F., Duff E.P., Fitzgibbon S., Westphal R., Carone D., et al. Hand classification of fMRI ICA noise components. Neuroimage. 2017;154:188–205. doi: 10.1016/j.neuroimage.2016.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Power J.D. A simple but useful way to assess fMRI scan qualities. Neuroimage. 2017;154:150–158. doi: 10.1016/j.neuroimage.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith S.M., Vidaurre D., Beckmann C.F., Glasser M.F., Jenkinson M., Miller K.L., Nichols T.E., Robinson E.C., Salimi-Khorshidi G., Woolrich M.W., et al. Functional connectomics from resting-state fMRI. Trends Cogn. Sci. 2013;17:666–682. doi: 10.1016/j.tics.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosvall M., Bergstrom C.T. Maps of random walks on complex networks reveal community structure. Proc. Natl. Acad. Sci. USA. 2008;105:1118–1123. doi: 10.1073/pnas.0706851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon E.M., Laumann T.O., Marek S., Raut R.V., Gratton C., Newbold D.J., Greene D.J., Coalson R.S., Snyder A.Z., Schlaggar B.L., et al. Default-mode network streams for coupling to language and control systems. Proc. Natl. Acad. Sci. USA. 2020;117:17308–17319. doi: 10.1073/pnas.2005238117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcus D.S., Harms M.P., Snyder A.Z., Jenkinson M., Wilson J.A., Glasser M.F., Barch D.M., Archie K.A., Burgess G.C., Ramaratnam M., et al. Human Connectome Project informatics: quality control, database services, and data visualization. Neuroimage. 2013;80:202–219. doi: 10.1016/j.neuroimage.2013.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischl B. FreeSurfer. Neuroimage. 2012;62:774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thielscher A., Antunes A., Saturnino G.B. Field modeling for transcranial magnetic stimulation: a useful tool to understand the physiological effects of TMS? Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2015;2015:222–225. doi: 10.1109/EMBC.2015.7318340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
An example dataset (Database: https://wcm.box.com/v/TANS-ExampleData) and code for implementing precision functional mapping and TANS is available online (https://github.com/cjl2007/Targeted-Functional-Network-Stimulation; https://doi.org/10.5281/zenodo.7566548). Code for processing multi-echo fMRI data is also available online (https://github.com/cjl2007/Liston-Laboratory-MultiEchofMRI-Pipeline,; https://doi.org/10.5281/zenodo.6958611).