Abstract
Sorafenib, which inhibits multiple kinases, is an effective frontline therapy for hepatocellular carcinoma (HCC). Ferroptosis is a form of iron-dependent programmed cell death regulated by lipid peroxidation, which can be induced by sorafenib treatment. Oncoprotein hepatitis B X-interacting protein (HBXIP) participates in multiple biological pro-tumor processes, including growth, metastasis, drug resistance, and metabolic reprogramming. However, the role of HBXIP in sorafenib-induced ferroptotic cell death remains unclear. In this study, we demonstrated that HBXIP prevents sorafenib-induced ferroptosis in HCC cells. Sorafenib decreased HBXIP expression, and overexpression of HBXIP blocked sorafenib-induced HCC cell death. Interestingly, suppression of HBXIP increased malondialdehyde (MDA) production and glutathione (GSH) depletion to promote sorafenib-mediated ferroptosis and cell death. Ferrostatin-1, a ferroptosis inhibitor, reversed the enhanced anticancer effect of sorafenib caused by HBXIP silencing in HCC cells. Regarding the molecular mechanism, HBXIP transcriptionally induced the expression of stearoyl-CoA desaturase (SCD) via coactivating the transcriptional factor ZNF263, resulting in the accumulation of free fatty acids and suppression of ferroptosis. Functionally, activation of the HBXIP/SCD axis reduced the anticancer activity of sorafenib and suppressed ferroptotic cell death in vivo and in vitro. HBXIP/SCD axis-mediated ferroptosis can serve as a novel downstream effector of sorafenib. Our results provide new evidence for clinical decisions in HCC therapy.
Keywords: HBXIP, SCD, ferroptosis, sorafenib, ZNF263, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) accounts for approximately 90% of primary liver cancers [1]. With the poorest five-year survival of 18%, liver cancer is the second most lethal cancer in the world. Sorafenib, an inhibitor of multiple kinases approved by the Food and Drug Administration, is an effective medication used as a frontline HCC therapy [1]. The safety and modest efficacy of sorafenib remain an effective option for therapy, except for lenvatinib. However, the mechanism of action of sorafenib remains unclear.
Ferroptosis is a form of cell death that differs from apoptosis and autophagy and is characterized by reactive oxygen species (ROS) and iron-dependent accumulation of lipid oxidates [2]. ROS is simultaneously controlled by the accumulation of fatty acids (FAs) and glutathione peroxidase 4 (GPX4). GPX4 inactivation accelerates the accumulation of glutathione (GSH), which further increases ROS production. In addition, inhibition of SLC7A11 (a key functional subunit of system Xc) can cause GPX4 inactivation and GSH depletion, subsequently triggering ferroptosis [3]. Erastin was the first ferroptosis inducer found to irreversibly inactivate SLC7A11. The combination of erastin and sorafenib remarkably enhances the anticancer activity of both drugs [4]. It was recently shown that NRF2 and MT-1G are critical for protecting cells against ferroptosis and inducing the sorafenib resistance in HCC cells [5, 6]. Thus, in-depth studies of the molecular mechanisms of ferroptosis-associated sorafenib efficacy in HCC may provide new treatment strategies.
As a conserved 18-kDa protein, hepatitis B X-interacting protein (HBXIP) acts as an oncoprotein and participates in multiple biological pro-tumor processes, including carcinogenesis, proliferation, migration, and drug resistance [7–9]. Aberrant HBXIP expression is associated with poor survival and prognosis in patients with HCC. HBXIP is a regulator that signals amino acid levels to mTORC1, acting as a guanine nucleotide exchange factor in Rag GTPases [10]. In addition, HBXIP coactivates several transcription factors, such as c-Myc, FOXO1, and c-Myb, which govern the transcription of tumor-related genes to accelerate tumorigenesis and metastasis in HCC [11–13]. HBXIP also drives metabolic reprogramming involving gluconeogenesis and the Warburg effect in HCC [12, 14]. However, whether HBXIP is involved in modulating sorafenib-associated ferroptosis in HCC requires further analysis.
Lipid metabolism is important in tumorigenesis and cancer development. Stearoyl-CoA desaturase (SCD) catalyzes the rate-limiting step in the conversion of saturated FAs into unsaturated FAs. Increased SCD expression and cancer aggressiveness are closely related in HCC, anaplastic thyroid carcinoma and lung cancer [15–17]. SCD increases coenzyme Q10 and unsaturated FAs in the membrane, thereby blocking ferroptosis and apoptosis in ovarian cancer cells [18]. However, whether SCD participates in HBXIP-accelerated metabolic reprogramming remains unclear.
In this study, we found that HBXIP suppresses ferroptosis by inducing abnormal free FA accumulation and blocks the anti-cancer activity of sorafenib in HCC cells. Mechanistic investigation revealed that HBXIP acts as a coactivator to induce SCD expression via coactivating transcription factor ZNF263, leading to upregulation of FA biosynthesis. Overexpression of HBXIP prevents ferroptosis and reduces the anti-tumor effect of sorafenib in vivo and in vitro. Our findings indicate that HBXIP is a potential downstream effector of sorafenib, providing new evidence for clinical decision-making in patients with HCC.
Materials and methods
Cell lines and cell culture
The human normal hepatocyte cell line LO2; the five different HCC cell lines HepG2, Hep3B, SMMC-7721, Huh7, and Bel-7402; and the human embryonic kidney cell line 293 T (HEK293T) were obtained from ATCC (Manassas, VA, USA). The HCC cell lines and HEK293T cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Gibco). HepG2 and Hep3B cells stably expressing green fluorescent protein tagged HBXIP were prepared through lentiviral infection. To obtain positive single and mixed clones, the cells were selected using puromycin (Invitrogen, Carlsbad, CA, USA) for 2 weeks and expanded for further selection. Two clones of HepG2 and Hep3B stably expressing HBXIP, HepG2-OE-HBXIP #10, HepG2-OE-HBXIP #12, Hep3B-OE-HBXIP #2, and Hep3B-OE-HBXIP #12, were used in subsequent experiments. Lipofectamine 3000 reagent (Invitrogen) was used for transfection.
Reagents
The following reagents were used: ferroptosis inhibitor ferrostatin-1 (MedChemExpress, Monmouth Junction, NJ, USA), ferroptosis activators erastin (Macklin, Shanghai, China, catalog #E872563) and RSL3 (Macklin, catalog #R873890) and sorafenib (Nexavar, Bayer Health Care, Leverkusen, Germany).
Plasmids and small interfering RNAs
The pcDNA3.1 (+), pcDNA3.1 (+)-HBXIP, pCMV-tag2B, pCMV-tag2B-HBXIP, pGL3-Basic, pRL-TK, pMG2.G, psPAX, and pCDH-RFP-HBXIP plasmids were maintained in our laboratory. To generate pGL3-SCD, the SCD promoter region was amplified from the genomic DNA of HepG2 cells using polymerase chain reaction (PCR) and cloned into the pGL3-Basic vector. All small interfering RNAs (siRNAs) and control siRNAs were purchased from RiboBio (Guangzhou, China).
5-Ethynyl-2′-deoxyuridine assay
Cell proliferation was assessed using the Cell-Light™ 5-Ethynyl-2′-deoxyuridine (EdU) imaging detection kit (RiboBio). The cells were transfected for 24 h or 48 h. A total of 104 cells were seeded into 24-well plates and evaluated according to the EdU detection kit protocol.
MTT assay
Cell viability was assessed using MTT reagent (Sigma, St. Louis, MO, USA) as described previously [19]. The cells were transfected with plasmids for 24 h. The cells (3000 cells/well) were seeded into 96-well plates and treated with indicated chemicals for 48 or 72 h. Absorbance was measured at 490 nm using a Multiskan FC system (Thermo Fisher Scientific, Waltham, MA, USA).
Colony formation assay
Stable cells were seeded into 6-well plates and re-treated with the chemicals every 3 days for 2 weeks. The cells were fixed in methanol at 25 °C for 10 min and then stained with crystal violet for 15 min and photographed.
Malondialdehyde and glutathione analysis
Transfected cells were seeded into 6-well plates and treated with the chemicals for 48 h. Cells or tissues were digested with RIPA for 25 min. A lipid peroxidation malondialdehyde (MDA) assay kit (Beyotime, Shanghai, China) was used to detect MDA levels, and a glutathione (GSH) assay kit (Beyotime) was used to measure GSH levels. A BCA protein assay kit (Beyotime) was used to assess the protein concentrations of cells or tissues. The data were normalized to protein concentrations.
Free FA detection and triglyceride analysis
Transfected cells were seeded into 6-well plates and treated with the chemicals for 48 h. Cells or tissues were digested with RIPA for 25 min. A free FA detection kit (Solarbio, Beijing, China, catalog #BC0595) and triglyceride (TG) assay kit (Solarbio, catalog #BC0620) were used to detect free FA and TG levels, respectively. A BCA protein assay kit (Beyotime) was used to assess the protein concentrations of cells or tissues. The data were normalized to protein concentrations.
Oil Red O staining
Oil Red O staining was performed as previously described [20]. Frozen xenograft sections (10 µm) or HCC cells were stained with Oil Red O working solution for 15 min after fixation in 4% paraformaldehyde. Positive Oil Red O staining was quantified using ImageJ software (NIH, Bethesda, MD, USA).
RNA extraction, reverse transcription and RT-qPCR
The cells were transfected or treated with plasmids or the chemicals for 48 h. Cells or tissues from each group were treated with TRIzol reagent (Invitrogen). cDNA was extracted using a Transcriptor First Strand cDNA Synthesis Kit (TransGene Biotech, China). Reverse transcription quantitative polymerase chain reaction (RT-qPCR) was performed in triplicates using SYBR Premix Ex Taq (TaKaRa, Shiga, Japan). Primer sequences for the genes are listed in Supplementary Table S1.
Co-immunoprecipitation assay (Co-IP)
After 48 h of the transfection with plasmids, cells were lysed in a lysis buffer as previously described [21]. The products were incubated with Flag M2 affinity gel (Sigma-Aldrich) for 4 h at 4 °C. To explore endogenous protein interactions in HCC cells, protein G-Sepharoses (Santa Cruz Biotechnology, Dallas, TX, USA) was incubated with the negative control IgG or anti-ZNF263 and anti-HBXIP, followed by incubation with the cell lysate. After washing with washing buffer eight times, the binding proteins were washed out from the gel with washing buffer and then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting.
Dual-luciferase report gene system
The pGL3-Basic vector containing the wild-type or mutant SCD promoter along with pRL-TK and other plasmids was transfected into the cells. Transfected cells were incubated and analyzed using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA).
Chromatin immunoprecipitation assay
The EpiQuikTM ChIP Kit (EpiGentek Group, Farmingdale, NY, USA) coupled with RT-qPCR was used to evaluate the binding between HBXIP or ZNF263 and the SCD promoter. HBXIP or ZNF263 antibodies, as well as a negative control antibody IgG and positive control antibody anti-RNA polymerase II, were immunoprecipitated with protein-DNA complexes. The soluble fraction was used as the input control.
Western blotting analysis
Western blotting analysis was performed according to standard protocols [22]. All primary antibodies and their dilution ratios are provided in Supplementary Table S2. The bands on the polyvinylidene fluoride membranes were photographed using Glyco Band-Scan software version 4.50 (http://www.Glyco.com).
Immunohistochemistry staining
Immunohistochemical staining was performed as previously described [23]. The slides of xenografts were incubated with primary antibodies, including anti-HBXIP, anti-SCD, or anti-Ki67, at 4 °C overnight, followed by incubation with a secondary antibody at 37 °C for 30 min. After DAB and hematoxylin staining, the slides were dehydrated and sealed with neutral gum.
Animal experiments
All experimental procedures involving animals were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Six-week-old male BALB/c athymic nude mice were purchased from the Experimental Animal Center of Peking (Beijing, China). Stable cells (5 × 106) were seeded into the right flanks of the mice. After the xenografts had grown to 200 mm3, saline as a vehicle or sorafenib (30 mg/kg) was administered by gavage every day, and the mice were euthanized by the cervical dislocation method five weeks later. Before sacrifice, the tumor sizes and body weights were measured twice per week. The tumor volume (V) was calculated as follows: (L × W2)/2 (length, L, and width, W). The xenografts were excised and further assessed.
Statistical analysis
Statistical significance was quantified as the average of three independent replicates ± standard deviation via two-tailed Student’s t test, one-way analysis of variance or two-way analysis of variance (ANOVA), with the results indicated as ***P < 0.001, **P < 0.01, and *P < 0.05, respectively, and not significant (ns). The transcripts per million (TPM) and proteomics sequencing data with sorafenib drug sensitivity (IC50) information for several HCC cell lines were obtained from the Depmap database (https://depmap.org/portal/depmap/). The RNA-sequencing dataset of patients with HCC was obtained from The Cancer Genome Atlas (TCGA; https://tcga-data.nci.nih.gov/tcga/) or GSE10921 and GSE112790 in the Gene Expression Omnius (GEO) database (https://www.ncbi.nlm.nih.gov/geo/); and the data format was MINIML. R Foundation (2020) version 4.0.3 (The R Project for Statistical Computing, Vienna, Austria), GraphPad Prism 9 (GraphPad, Inc., La Jolla, CA, USA) and SPSS 22.0 (SPSS, Inc., Chicago, IL, USA) were used all the above analysis.
Results
Oncoprotein HBXIP is decreased by treatment with sorafenib in HCC
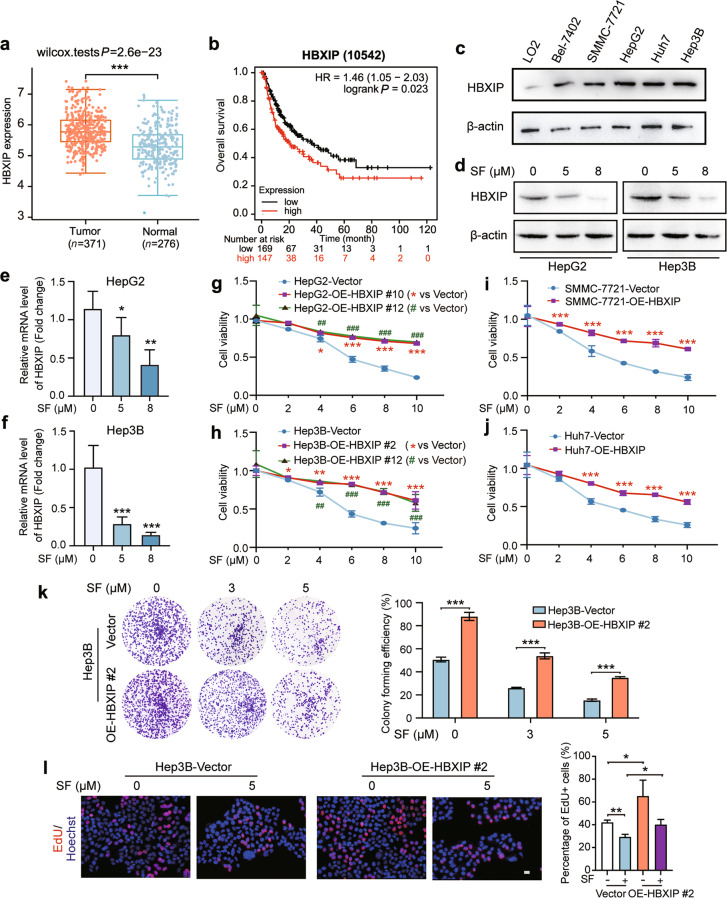
HBXIP is an oncoprotein that participates in tumorigenesis, proliferation, migration, drug resistance, and metabolic reprogramming in multiple cancers [7–9]. The mRNA level of HBXIP was higher in the 371 human HCC tissues than in the 276 normal tissues obtained from TCGA (https://tcga-data.nci.nih.gov) (Fig. 1a). In addition, Kaplan-Meier plotter data showed that higher HBXIP levels were associated with shorter overall survival in the 316 patients with HCC from TCGA dataset (http://kmplot.com/analysis/) (Fig. 1b). We next examined HBXIP expression in the human normal hepatocyte cell line LO2, as well as in five different human HCC cell lines: Bel-7402, SMMC-7721, HepG2, Huh7, and Hep3B. Western blotting showed that HBXIP was increased in these five HCC cell lines compared with that in the normal liver cell line LO2 (Fig. 1c). These results support the pro-tumor role of HBXIP in HCC development. We investigated whether sorafenib regulates the expression of the oncoprotein HBXIP. Interestingly, the mRNA and protein levels of HBXIP were robustly and dose-dependently inhibited by sorafenib in the HCC cell lines HepG2, Hep3B, SMMC-7721, and Huh7 (Fig. 1d–f; Supplementary Fig. S1b–d).
Fig. 1. Oncoprotein HBXIP is decreased by treatment with sorafenib in HCC.
a The HBXIP expression level in 371 HCC patients’ tumor tissues and 276 normal tissues from the TCGA dataset. b Kaplan–Meier plots of the overall survival of 316 HCC patients grouped by HBXIP mRNA levels from the TCGA dataset. c Western blotting (WB) analysis was performed to analyze the expression of HBXIP in LO2, Bel-7402, SMMC-7721, HepG2, Huh7, and Hep3B cells. d WB analysis was used to test the expression of HBXIP in HepG2 and Hep3B cells dose-dependently treated with DMSO or sorafenib (SF) for 48 h. e, f Fold change in mRNA levels of HBXIP determined by RT-qPCR assays in HepG2 and Hep3B cells dose-dependently treated with DMSO or sorafenib for 48 h. g, h MTT assay was utilized to examine cell viability in the indicated stable cells under the treatment of sorafenib. *P < 0.05, ***P < 0.01 or ##P < 0.01, ###P < 0.001 represented the P value of the cell viability in different OE-HBXIP group compared to the Vector group. i, j MTT assay was applied to evaluate the cell viability in SMMC-7721 and Huh7 cells transfected with pcDNA-Vector or pcDNA-HBXIP and treated with the indicated concentrations of sorafenib. ***P < 0.001. k A colony photograph of Hep3B and stable Hep3B-OE-HBXIP #2 cells dose-dependently treated with the indicated concentrations of sorafenib. l EdU incorporation assay in the indicated stable cells treated with DMSO or 5 μM sorafenib for 48 h. EdU positive cells were quantified by percentage of EdU+ (Red) /Hoechst+ (Blue). Scale bar = 20 μm. Columns with error bars symbolize the average of three independent replicates ± SD. Three experiments with consistent results tendency were analyzed by one-way ANOVA (e, f, k and l) or two-way ANOVA (g–j). ***P < 0.001, **P < 0.01, *P < 0.05.
We generated stable HBXIP-overexpressing HCC cell lines in HepG2 and Hep3B cells, and two of 13 clones were further evaluated (Supplementary Fig. S1a). HepG2-OE-HBXIP #10, HepG2-OE-HBXIP #12, Hep3B-OE-HBXIP #2, and Hep3B-OE-HBXIP #12 cells were used to determine the inhibitory effect of sorafenib on cell proliferation. MTT and colony formation assays indicated that stable HBXIP overexpression increased the survival of HepG2 and Hep3B cells treated with sorafenib (Fig. 1g, h, and k; Supplementary Fig. S1e). Similar to the results observed in HepG2 and Hep3B cells, transient overexpression of HBXIP in SMMC-7721 and Huh7 cells decreased sorafenib-mediated cell death in the MTT assay (Fig. 1i, j). Similarly, treatment with 5 μM sorafenib decreased EdU-positive Hep3B cells, whereas HBXIP overexpression increased EdU-positive Hep3B cells. Compared to the vector group treated with sorafenib, HBXIP overexpression abolished the ability of sorafenib to inhibit HCC cell proliferation (Fig. 1l). A positive correlation between HBXIP mRNA or protein levels and sorafenib sensitivity (IC50) was also found in the Depmap database (R(TPM) = 0.574, P < 0.001 and R(protein expression) = 0.986, P < 0.001) (Supplementary Fig. S1f and S1g) (https://depmap.org/portal/depmap/). These results indicate that sorafenib downregulates HBXIP, and HBXIP overexpression affects the efficiency of sorafenib therapy in HCC.
HBXIP is involved in ferroptosis suppression in sorafenib-treated HCC cells
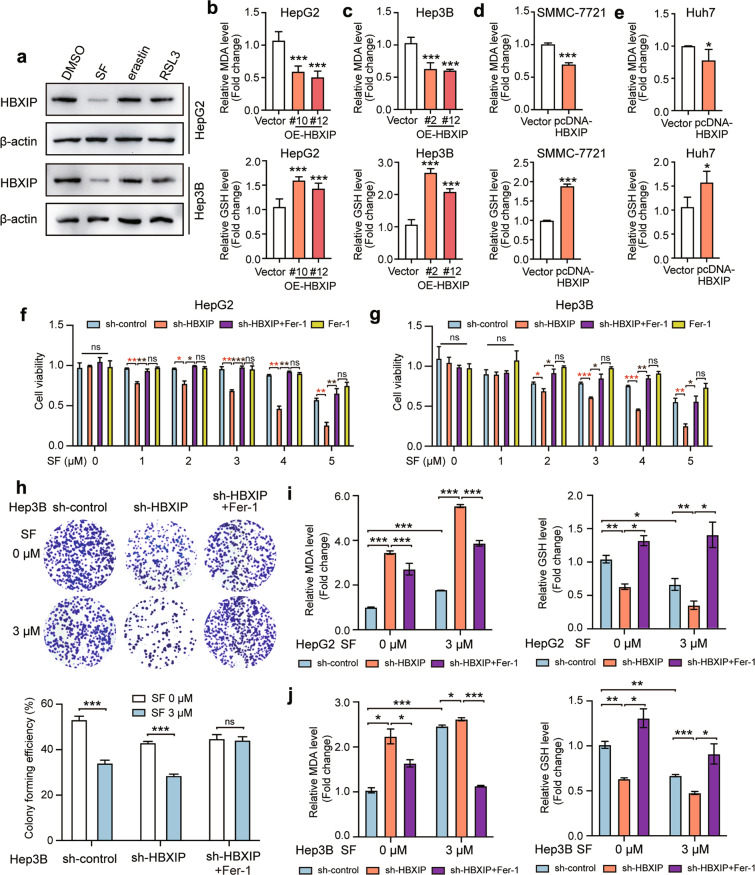
A recent study has revealed that ferroptosis is essential for sorafenib-induced cell death in HCC [24]. Here, we investigated whether HBXIP is involved in inhibiting ferroptosis. Two ferroptosis activators, erastin and RSL3, along with sorafenib, were used to treat HepG2 and Hep3B cell lines for 48 h. The Western blotting results showed that erastin and RSL3 did not decrease the protein levels of HBXIP, suggesting that the decrease in HBXIP was sorafenib-specific in HCC cells (Fig. 2a). The main characteristic of ferroptosis is the accumulation of lipid peroxidation products, such as MDA [25]. Moreover, lipid peroxidation mediated by GSH depletion can induce ferroptosis [26]. An increase in MDA and GSH depletion indicates ferroptosis. We evaluated the levels of MDA and GSH in HepG2 and Hep3B cells stably expressing HBXIP. MDA deletion and GSH accumulation were observed in HBXIP-overexpressing cells compared to in vector-transfected HepG2 and Hep3B cells, indicating that ferroptosis was inhibited (Fig. 2b, c). Similar results were observed in SMMC-7721 and Huh7 cells that were transiently transfected with the HBXIP expression plasmid (pcDNA-HBXIP) (Fig. 2d, e). The MTT assay showed that silencing of HBXIP increased cell death after treatment with sorafenib for 48 h. Sorafenib-induced cell death was blocked by ferrostatin-1 (an inhibitor of ferroptosis, Fer-1). The rate of cell death induced by sorafenib following HBXIP knockdown in the Fer-1 group was lower than that in the group without Fer-1, whereas it did not significantly differ from that in the sh-control+Fer-1 group (Fig. 2f, g). Similar results were observed in the colony formation assay (Fig. 2h). These results confirm that the increase in HBXIP silencing-induced cell death following sorafenib treatment was associated with ferroptosis. MDA was profoundly increased in HBXIP-knockdown HCC cells; this effect was reversed by Fer-1. Knockdown of HBXIP promoted the depletion of intracellular GSH in HCC cells, which was reversed by Fer-1, similar to the results observed for MDA (Fig. 2i, j). The above results indicate that ferroptosis can be induced by silencing of HBXIP, which promotes sorafenib-induced the cell death.
Fig. 2. HBXIP is involved in ferroptosis suppression in sorafenib-treated HCC cells.
a WB analysis was applied to analyze the expression of HBXIP in HepG2 and Hep3B cells with DMSO, SF, erastin or RSL3 (the activators of ferroptosis) for 48 h. b, c MDA and GSH were detected in HepG2, Hep3B cells and two stable HBXIP overexpression clones. d, e MDA and GSH were detected in SMMC-7721 and Huh7 cells transfected with pcDNA-Vector or pcDNA-HBXIP for 48 h. f, g MTT assay was performed to test the cell viability of HepG2 and Hep3B cells transfected with sh-control or sh-HBXIP. After 48 h of the transfection, the gradient concentrations of sorafenib (SF) were used to treat the indicated cells. 10 μM ferrostatin-1 (a ferroptosis inhibitor, Fer-1) or DMSO along with sorafenib was added into sh-HBXIP and sh-control group. h A colony photograph of Hep3B cells transfected with sh-control or sh-HBXIP. After 48 h of the transfection, DMSO or 3 μM sorafenib was used to treat the indicated stable cells for at least 2 weeks. 10 μM Fer-1 or DMSO along with sorafenib was added into sh-HBXIP group. i, j MDA and GSH were detected in HepG2 and Hep3B cells transfected with sh-control or sh-HBXIP. After 48 h of the transfection, the gradient concentrations of sorafenib were utilized to treat the indicated cells. 10 μM Fer-1 or DMSO along with sorafenib was added into sh-HBXIP group. Columns with error bars symbolize the average of three independent replicates ± SD. Three experiments with consistent results tendency were analyzed by two-tailed Student’s t test (b–e) or two-way ANOVA (f–j). ***P < 0.001, **P < 0.01, *P < 0.05, ns not significant.
SCD is the downstream effector of HBXIP in sorafenib-treated HCC cells
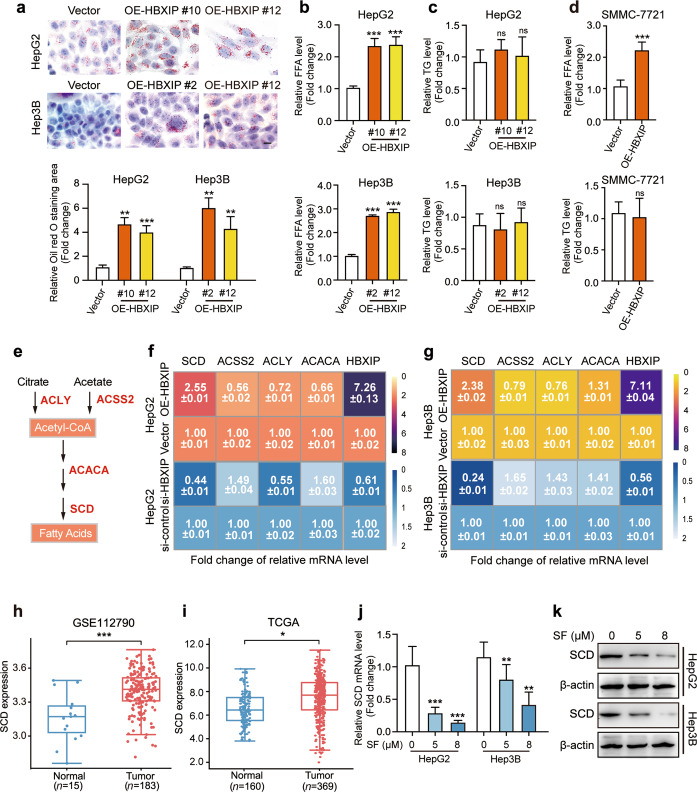
Given the inhibitory function of HBXIP in ferroptosis, we examined the mechanism by which HBXIP regulates ferroptosis. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (https://www.genome.jp/kegg/) and Gene Ontology (GO) enrichment analysis were performed to summarize the differentially expressed genes between the two groups based on the HBXIP expression level in 371 patients with HCC from TCGA dataset. The results revealed that differentially expressed genes associated with HBXIP expression were mainly related to lipid metabolism, such as FA metabolic process, PPAR signaling pathway, and TG metabolic process (Supplementary Fig. S2a, b). Accumulation of FAs that control ROS is important for ferroptosis; we examined whether HBXIP can upregulate FA metabolism to inhibit ferroptosis. Oil Red O staining showed that stable HBXIP overexpression increased the accumulation of lipid metabolism in HCC cell lines (Fig. 3a). Analysis of free FA and TG in HepG2, Hep3B and SMMC-7721 cells revealed that overexpression of HBXIP promoted the accumulation of free FA, but not esterifiable TG (Fig. 3b–d). Thus, we evaluated whether HBXIP participates in the progression of FA generation. ATP citrate lyase (ACLY), acyl-CoA synthetase short chain family member 2 (ACSS2), acetyl-CoA carboxylase alpha (ACACA), and SCD were the major FA synthesizing rate-limiting enzymes and have been shown play vital roles in HCC fatty acid metabolism [27–30] (Fig. 3e). The RT-qPCR results verified that only SCD was positively associated with HBXIP overexpression or knockdown (Fig. 3f, g).
Fig. 3. SCD is the downstream effector of HBXIP in sorafenib-treated HCC cells.
a Oil red O staining of the indicated stable cells (HepG2-Vector, HepG2-OE-HBXIP #10 #12, Hep3B-Vector, Hep3B-OE-HBXIP #2 #12). Scale bar, 20 μm. b, c Free fatty acids and TG were detected in HepG2, Hep3B cells and two HBXIP stable expression clones. d Free fatty acids and TG were detected in SMMC-7721 cells transfected with pcDNA-Vector or pcDNA-HBXIP for 48 h. e Schematic diagram of several fatty acids key enzymes. f, g The average of three times fold changes ± SD in mRNA levels of HBXIP determined by RT-qPCR assays in HepG2 or Hep3B cells transfected with pcDNA-Vector and pcDNA-HBXIP or si-control and si-HBXIP. h SCD expression level in 183 HCC patients’ tumor tissues and 15 normal tissues from GSE112790. i SCD expression level in 369 HCC patients’ tumor tissues and 160 normal tissues from TCGA dataset. j Fold change in mRNA levels of SCD determined by RT-qPCR assays in the indicated cells dose-dependently treated with sorafenib for 48 h. k WB analysis was used to analyze the expression of SCD in HepG2 cells or Hep3B cells dose-dependently treated with sorafenib for 48 h. Columns with error bars symbolize the average of three independent replicates ± SD. Three experiments with consistent results tendency were analyzed by two-tailed Student’s t test (a–d) or one-way ANOVA (j). ***P < 0.001, **P < 0.01, *P < 0.05, ns not significant.
SCD catalyzes the synthesis of unsaturated FAs and is a rate-limiting kinase. Several studies have demonstrated the tumor-promoting role of SCD in multiple cancers, including in colorectal cancer [31], lung cancer [32], clear cell renal cell carcinoma [33], and prostate cancer [34]. Specifically, SCD suppresses lipid oxidation and induces cell death by regulating sorafenib sensitivity [18]. Compared to in normal tissues, SCD expression was significantly increased in tumor tissues from GSE112790 in the GEO and TCGA databases, suggesting that SCD serves as a pro-tumor gene in HCC (Fig. 3h, i). However, the regulatory mechanism underlying SCD overexpression in HCC is poorly understood. Furthermore, we evaluated the effect of HBXIP on SCD using RT-qPCR and Western blotting in HepG2 and Hep3B cells stably overexpressing HBXIP. We found that SCD mRNA and protein expression were upregulated by stable HBXIP overexpression (Supplementary Fig. S2c–f). To verify whether SCD is the downstream effector of HBXIP in sorafenib treatment, RT-qPCR and Western blotting analysis were performed. Sorafenib treatment decreased the mRNA and protein levels of SCD in the HepG2 and Hep3B cell lines (Fig. 3j, k; Supplementary Fig. S2g). Taken together, these results indicate that HBXIP induces FA accumulation in HCC cells and that SCD is the downstream effector of HBXIP in response to sorafenib treatment.
SCD expression is transcriptionally activated by HBXIP in an SREBP-independent manner
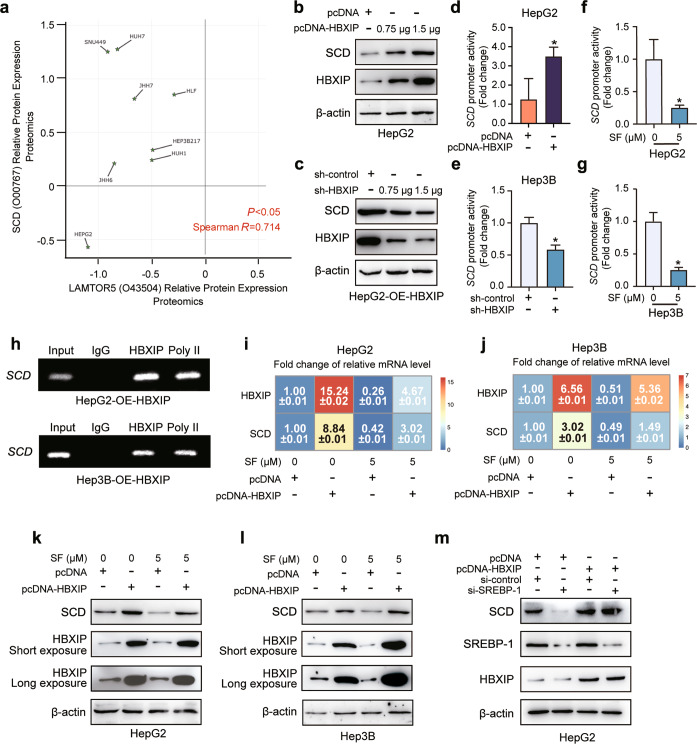
To further explore the mechanism by which HBXIP modulates SCD expression in HCC cells, we analyzed the proteomic data of several HCC cell lines using the Depmap database. We found a positive correlation between HBXIP protein levels and SCD (R = 0.714, P < 0.05) (Fig. 4a). We transfected the overexpression or interference plasmid of HBXIP in a dose-dependent manner into HepG2 and HepG2-OE-HBXIP stable cells, followed by western blotting. Combined with the results shown in Fig. 3f, g, dose-dependent overexpression and silencing of HBXIP affected the mRNA and protein levels of SCD (Fig. 4b, c; Supplementary Fig. S2h, i). As HBXIP can act as a transcriptional coactivator, we hypothesized that it can induce the transcription of the SCD promoter. The dual-luciferase reporter gene assay showed that HBXIP promoted the activity of the full-length SCD promoter (−920 to +55) (Fig. 4d, e). Furthermore, the full-length SCD promoter activity was inhibited by sorafenib treatment (Fig. 4f, g). In HepG2 and Hep3B cells stably overexpressing-HBXIP, we observed binding of HBXIP to the SCD promoter using the chromatin immunoprecipitation (ChIP) assay (Fig. 4h). We also found that sorafenib-induced suppression of HBXIP and that SCD was rescued by exogenous HBXIP overexpression at the mRNA and protein levels in HCC cells (Fig. 4i–l). These results indicate that HBXIP transcriptionally upregulates SCD expression in HCC cells.
Fig. 4. SCD expression is transcriptionally activated by HBXIP in an SREBP-independent manner.
a The proteomics data of positive correlation between HBXIP and SCD in 8 HCC cell lines from Depmap database (https://depmap.org/portal/depmap/). The correlation coefficient (R) and P value (P) are based on Spearman correlation. b WB analysis exhibited the expression of SCD and HBXIP in HepG2 cells transfected with pcDNA-HBXIP and pcDNA-Vector in a dose-dependent manner. c WB analysis exhibited the expression of SCD and HBXIP in HepG2-OE-HBXIP #10 stable cells transfected with sh-control and sh-HBXIP in a dose-dependent manner. d, e SCD promoter activity was detected by DLR gene assay. pcDNA-Vector and pcDNA-HBXIP or sh-control and sh-HBXIP were transiently transfected into the indicated cells for 48 h. f, g SCD promoter activity was detected by DLR gene assay. DMSO or 5 μM sorafenib was added into HepG2 or Hep3B cells. After 48 h of treatment, luciferase activities were measured in the dark. h ChIP analysis of the SCD promoter in stable HepG2-OE-HBXIP #10 or Hep3B-OE-HBXIP #2 cells immunoprecipitated with negative control IgG, positive control polymerase II (Poly II) or anti-HBXIP. i, j The average of three times fold changes ± SD in mRNA levels of HBXIP and SCD determined by RT-qPCR assays in the indicated cells. After the transfection with pcDNA-Vector and pcDNA-HBXIP for 48 h, DMSO or 5 μM sorafenib was used to treat the indicated cells. k, l WB analysis exhibited the expression levels of SCD and HBXIP in the indicated cells. After the transfection with pcDNA-Vector and pcDNA-HBXIP for 48 h, DMSO or 5 μM sorafenib was applied to treat the indicated cells. m WB analysis exhibited the expression levels of SCD and HBXIP in HepG2 cells transfected with pcDNA-Vector and pcDNA-HBXIP with or without si-control or si-SREBP-1. Columns with error bars symbolize the average of three independent replicates ± SD. Three experiments with consistent results tendency were analyzed by two-tailed Student’s t test (d–g). *P < 0.05.
According to a previous report, SREBP-1 can serve as a transcription factor for SCD following sorafenib treatment [35]. SREBP-1/SCD1-mediated lipogenesis protects cancer cells from ROS and ferroptotic death [36]. Moreover, HBXIP can upregulate SREBP-1c in breast cancer [9]. Thus, we investigated whether SREBP-1c is involved in HBXIP-activated SCD transcription in HCC cells. Although SREBP-1 silencing decreased SCD protein levels, its knockdown did not affect the upregulation of SCD mediated by exogenous overexpression of HBXIP in HepG2 cells (Fig. 4m). Taken together, HBXIP stimulates SCD transcription and expression in an SREBP-1-independent manner.
HBXIP stimulates SCD transcription via coactivating transcription factor ZNF263
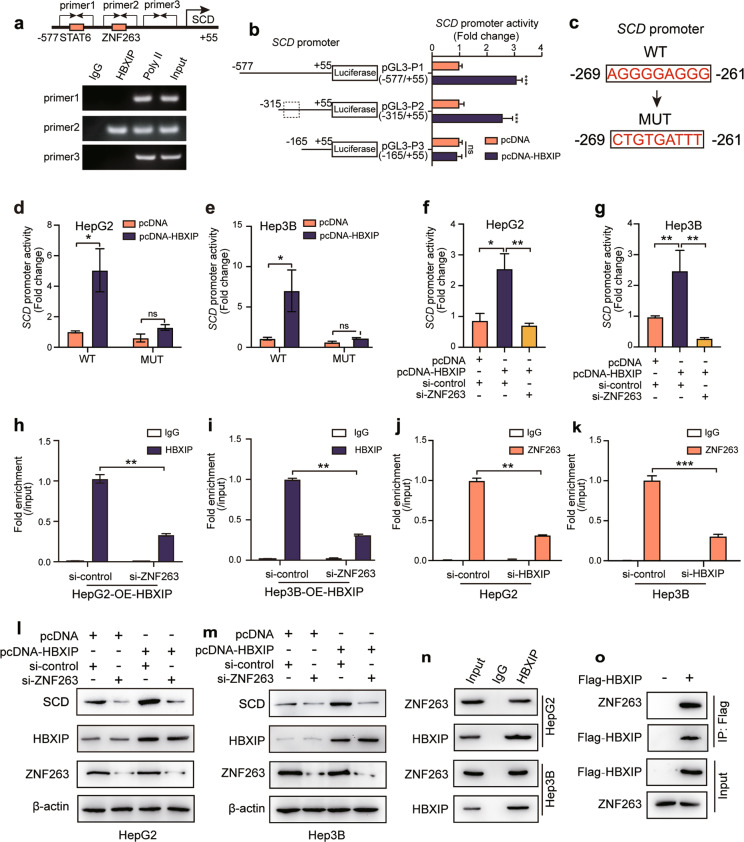
We next performed bioinformatics analysis using a transcription factor binding prediction server (JASPAR, http://jaspar.genereg.net/) to predict SCD transcriptional factors coactivated by HBXIP. We identified two putative binding sites for the transcription factors STAT6 (−469 to −455) and ZNF263 (−293 to −246) within the SCD promoter region. A ChIP assay followed by PCR was conducted to verify binding of HBXIP to the SCD promoter in HCC cells by incubation with an HBXIP antibody. Our data revealed HBXIP enrichment in the SCD promoter using primer 2 (Fig. 5a). Three truncated promoter regions shorter than the full-length promoter (−920/+55), including −577/+55 (pGL3-P1), −315/+55 (pGL3-P2), and −165/+55 (pGL3-P3), were constructed to test the binding specificity of HBXIP to the SCD promoter. Dual-luciferase reporter gene assays indicated that pGL3-P3 lost its response to HBXIP overexpression compared with pGL3-P1 and pGL3-P2, suggesting that the −315/−165 fragment is the key region in the HBXIP-regulated SCD promoter (Fig. 5b).
Fig. 5. HBXIP stimulates SCD transcription via coactivating transcription factor ZNF263.
a ChIP analysis of the SCD promoter in HepG2 cells immunoprecipitated with negative control IgG, positive control polymerase II or anti-HBXIP detected by three different primers of SCD promoter. Schematic diagram (up) shows the potential binding site of transcriptional factors on SCD promoter. b The activities of different SCD promoter regions were detected by DLR gene assay in HepG2 cells transfected with pcDNA-Vector or pcDNA-HBXIP for 48 h. c The diagram shows the sequences of the SCD promoter (WT) and the ZNF263 binding sites mutation (MUT). d, e WT and MUT SCD promoter activities were detected by DLR gene assay. pcDNA-Vector and pcDNA-HBXIP were transiently transfected into HepG2 or Hep3B cells for 24 h. f, g SCD promoter activity was detected by DLR gene assay. si-control or si-ZNF263 along with pcDNA-Vector or pcDNA-HBXIP were transiently transfected into HepG2 or Hep3B cells for 24 h. h, i ChIP analysis of the SCD promoter followed by RT-qPCR. si-control or si-ZNF263 were transiently transfected into stable HepG2-OE-HBXIP #10 or Hep3B-OE-HBXIP #2 cells immunoprecipitated with negative control IgG or anti-HBXIP. j, k ChIP analysis of the SCD promoter followed by RT-qPCR. si-control or si-HBXIP were transiently transfected into HepG2 or Hep3B cells immunoprecipitated with negative control IgG or anti-ZNF263. l, m WB analysis exhibited the expression levels of SCD, HBXIP and ZNF263 in HepG2 or Hep3B cells transiently transfected with pcDNA-Vector or pcDNA-HBXIP together with si-control or si-ZNF263. n Co-IP analysis of endogenous HBXIP and ZNF263 in the indicated cells. o Co-IP analysis of exogenous Flag-HBXIP and ZNF263. pCMV-Vector and pCMV-HBXIP were transiently transfected into HEK293T cells. Columns with error bars symbolize the average of three independent replicates ± SD. Three experiments with consistent results tendency were analyzed by two-tailed Student’s t test (b, d–k). ***P < 0.001, **P < 0.01, *P < 0.05, ns not significant.
Based on the predicted ZNF263 binding site in the SCD promoter, we mutated the ZNF263 binding site (ZNF263-MUT) using site-directed mutagenesis and tested the promoter activity of SCD (Fig. 5c). The promoter activity of mutated SCD was lower than that in the wild-type and was not altered by HBXIP overexpression (Fig. 5d, e). Similar results were observed when ZNF263 was knocked down in HBXIP-overexpressing HCC cells (Fig. 5f, g). In addition, ChIP assays confirmed that knocking down of ZNF263 in stable HBXIP-overexpressing HCC cells led to weaker affinity between HBXIP and the SCD promoter (Fig. 5h, i). Enrichment of ZNF263 in the SCD promoter was evaluated in a ChIP assay following silencing of HBXIP in HepG2 and Hep3B cells. RT-qPCR of the ChIP product showed that HBXIP silencing reduced the interaction between ZNF263 and the SCD promoter, supporting that HBXIP coactivates ZNF263 and promotes SCD transcription (Fig. 5j, k). Furthermore, in ZNF263-silenced HepG2 and Hep3B cells, overexpression of HBXIP did not upregulate SCD protein levels (Fig. 5l, m). Co-immunoprecipitation assays further revealed that both endogenous and exogenous HBXIP interacted with ZNF263 in HCC cells and HEK293T cells (Fig. 5n, o). Taken together, HBXIP stimulates SCD transcription and expression by coactivating ZNF263 in HCC cells.
HBXIP/SCD axis affects the efficacy of sorafenib via ferroptosis
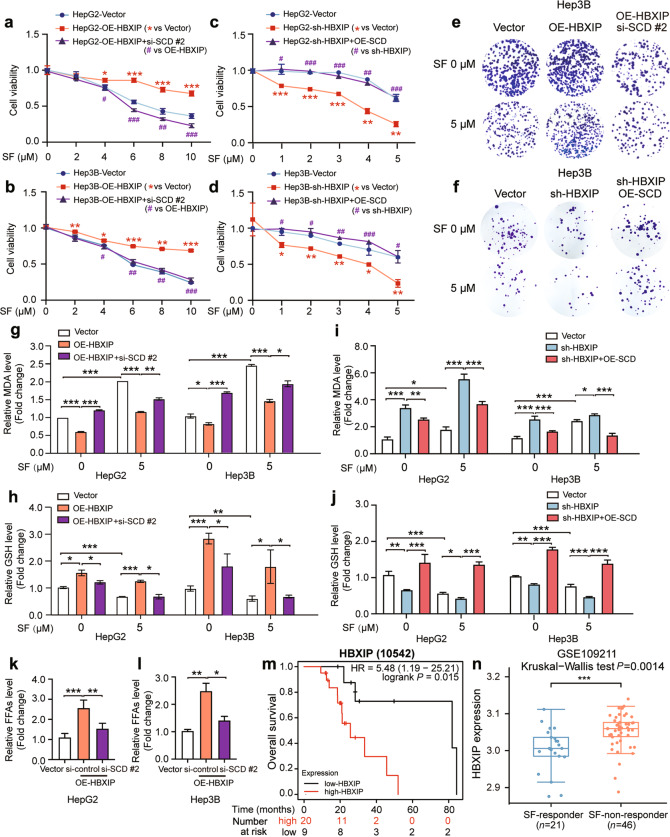
To verify whether the HBXIP/SCD axis is critical for regulating sorafenib-induced ferroptosis, two siRNAs of SCD were designed and the interference efficiency was evaluated (Supplementary Fig. S3a). First, we analyzed the proliferation of HCC cells using the MTT assay. Sorafenib inhibited the proliferation of HepG2 and Hep3B cells; this efficacy was disrupted by HBXIP overexpression. Moreover, SCD silencing in HBXIP-overexpressing HepG2 and Hep3B cells restored the efficacy of sorafenib (Fig. 6a, b). Similarly, HBXIP silencing enhanced the efficacy of sorafenib, and SCD overexpression abolished the increase in sorafenib efficacy mediated by HBXIP silencing in HCC cells (Fig. 6c, d). Consistent with the above findings, the colony formation assay indicated that inhibition of cell death triggered by HBXIP overexpression in Hep3B cells treated with sorafenib was robustly reversed by silencing of SCD (Fig. 6e; Supplementary Fig. S3b). The colony formation assay showed that the efficacy of sorafenib enhanced by HBXIP silencing was abolished by SCD overexpression (Fig. 6f; Supplementary Fig. S3c). To clarify the role of the HBXIP/SCD axis in sorafenib-mediated ferroptosis, we measured MDA and GSH levels in the HCC cell lines. In HBXIP-overexpressing HCC cells treated with sorafenib, SCD silencing increased MDA production and intracellular GSH depletion, and vice versa (Fig. 6g–j). Next, the accumulation of free FAs increased by HBXIP was reversed by SCD silencing in HepG2 and Hep3B cells (Fig. 6k, l). We investigated the overall survival of patients with HCC treated with sorafenib from TCGA datasets and found that high expression of HBXIP was associated with shorter overall survival (Fig. 6m). We also analyzed HBXIP expression in 21 sorafenib responders and 46 sorafenib non-responders with HCC from GSE109211 in the GEO database. The data showed that HBXIP expression was lower in sorafenib responders than in sorafenib non-responders (Fig. 6n). These data demonstrate that the HBXIP/SCD axis negatively regulates ferroptosis and can disrupt the efficacy of sorafenib in HCC.
Fig. 6. HBXIP/SCD axis affects the efficacy of sorafenib via ferroptosis.
a, b MTT assay showed the cell viability in the indicated stable cells treated with the gradient concentrations of sorafenib. *P < 0.05, **P < 0.01, ***P < 0.001 OE-HBXIP group compared to the Vector group. #P < 0.05, ##P < 0.01, ###P < 0.001 OE-HBXIP + si-SCD #2 group compared to the OE-HBXIP group. c, d MTT assay showed the cell viability in the indicated stable cells treated with the gradient concentrations of sorafenib. *P < 0.05, **P < 0.01, ***P < 0.001 sh-HBXIP group compared to the Vector group. #P < 0.05, ##P < 0.01, ###P < 0.001 sh-HBXIP + OE-SCD group compared to the sh-HBXIP group. e A colony photograph of Hep3B cells and stable Hep3B-OE-HBXIP #2 cells transfected with si-control or si-SCD #2. After 48 h of the transfection, DMSO or 5 μM sorafenib was used to treat the indicated stable cells for at least 2 weeks. f A colony photograph of Hep3B cells and stable Hep3B-sh-HBXIP cells transfected with pcDNA-Vector or pcDNA-SCD. After 48 h of the transfection, DMSO or 5 μM sorafenib was applied to treat the indicated stable cells for at least 2 weeks. g, h MDA and GSH were detected in HepG2 and Hep3B cells transfected with pcDNA-HBXIP or pcDNA-Vector together with or without si-control and si-SCD #2 following treatment of DMSO or 5 μM sorafenib. i, j MDA and GSH were detected in HepG2 and Hep3B cells transfected with pcDNA-SCD or pcDNA-Vector together with or without si-control and si-HBXIP following treatment of DMSO or 5 μM sorafenib. k, l Free fatty acids and TG were detected in HepG2 and Hep3B cells transfected with pcDNA-HBXIP or pcDNA-Vector together with or without si-control and si-SCD #2. m Kaplan-Meier plots of the overall survival of 29 HCC patients after the treatment of sorafenib grouped by HBXIP mRNA levels from the TCGA dataset. n HBXIP levels in 21 HCC sorafenib responders and 46 HCC sorafenib non-responders from the GSE109211. Columns with error bars symbolize the average of three independent replicates ± SD. Three experiments with consistent results tendency were analyzed by one-way ANOVA (k and l) or two-way ANOVA (a–d, g–j). ***P < 0.001, **P < 0.01, *P < 0.05.
Sorafenib promotes ferroptosis by inhibition of the HBXIP/SCD axis in vivo
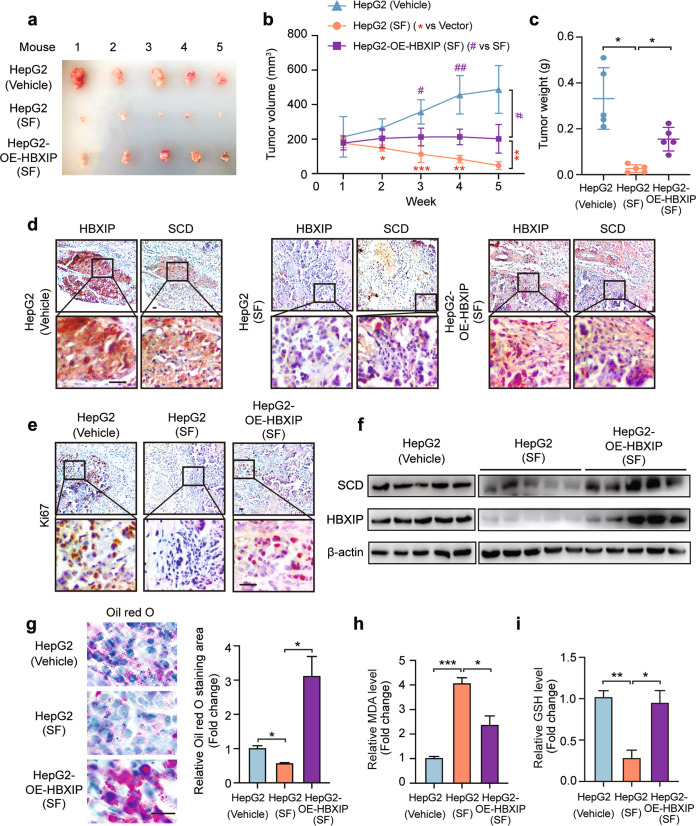
To verify the function of the HBXIP/SCD axis in sorafenib-induced ferroptosis and growth inhibition of HCC in vivo, HepG2 and stably overexpressing HBXIP HepG2 cells were transplanted into the right flank of BALB/c nude mice. Sorafenib obviously inhibited tumor growth, and HBXIP overexpression significantly disrupted the suppression of xenografts growth by sorafenib in vivo (Fig. 7a–c). Immunohistochemical analysis showed that the staining intensities of HBXIP, SCD, and Ki67 were decreased in sorafenib-treated mouse tissues compared with those in the vehicle group, and HBXIP overexpression restored the levels of SCD and Ki67 in sorafenib-treated mouse tissues (Fig. 7d, e). Western blotting revealed the same results as immunohistochemical staining (Fig. 7f). Oil Red O staining verified that the decrease in FA accumulation induced by sorafenib was reversed by HBXIP overexpression (Fig. 7g). Additionally, the changes in MDA and GSH levels were confirmed in vivo (Fig. 7h, i). Therefore, the HBXIP/SCD axis is implicated in sorafenib-mediated ferroptosis and tumor growth suppression in sorafenib-treated HCC.
Fig. 7. Sorafenib promotes ferroptosis by inhibition of the HBXIP/SCD axis in vivo.
a Imaging of the xenografts derived from BALB/c nude mice transplanted with 5 × 106 HepG2 and stable HepG2-OE-HBXIP cells with the treatment of saline or sorafenib (30 mg/kg, SF). b Volumes of tumors in each group (n = 5). #P < 0.05, ##P < 0.01 vs HepG2 (SF). **P < 0.01, ***P < 0.001 vs HepG2-OE-HBXIP. c Tumor weights in each group (n = 5). d IHC staining of the xenografts incubated with anti-SCD or anti-HBXIP, respectively. Scale bar, 20 μm. e IHC staining of xenografts incubated with anti-Ki67. Scale bar, 20 μm. f WB analysis exhibited the expression of SCD and HBXIP in xenografts. g Oil red O staining of the xenografts’ slides. h, i MDA and GSH were detected in the xenografts. Columns with error bars symbolize the average of three independent replicates ± SD. Three experiments with consistent results tendency were analyzed by one-way ANOVA (c, g–i) or two-way ANOVA (b). ***P < 0.001, **P < 0.01, *P < 0.05.
Discussion
The high mortality of HCC due to its asymptomatic state, diagnostic difficulty, and lack of effectively therapeutic methods has created challenges in the field of oncology [37]. Therefore, novel therapeutic targets and prognostic markers must be identified. In this study, we demonstrated that HBXIP is a novel target for HCC treatment. HBXIP was highly expressed in tumor tissues compared to in normal tissues and predicted shorter overall survival of patients with HCC in TCGA dataset. Sorafenib is a therapeutic drug approved for treating HCC; however, tumor heterogeneity and acquired resistance limit the therapeutic efficacy of sorafenib [38]. Here, we found that HBXIP dose-dependently responded to sorafenib treatment. HBXIP overexpression significantly blocked sorafenib-induced cell death in HCC cells. These results reveal the pivotal role of the oncoprotein HBXIP in HCC cell death regulation and sorafenib treatment.
Ferroptosis is a form of programmed cell death that is dependent on iron ions, differs from autophagy and apoptosis, and is regulated by lipid oxidation [39]. A recent study showed that sorafenib mainly promotes oxidative stress-induced ferroptosis in HCC by impairing mitochondrial function and promoting GSH depletion mediated by SLC7A11 activity inhibition [40]. Knockdown of MT-1G contributes to sorafenib-induced ferroptosis in HCC by increasing GSH depletion and ROS production [6]. These studies suggest that sorafenib induces ferroptosis in HCC treatment, which supports the relation between HBXIP and ferroptosis inhibition. We found that the ferroptosis activators, erastin and RSL3, did not decrease HBXIP expression, confirming that sorafenib-induced HBXIP expression loss was sorafenib-specific. Sorafenib-induced cell death was increased by silencing of HBXIP. Importantly, in HBXIP-silenced HCC cell lines, MDA production and intracellular GSH depletion were increased compared to in the control group, and inhibition of cell proliferation was reversed by ferrostatin-1 (a ferroptosis inhibitor). Our results present a new function for the oncoprotein HBXIP in cell death regulation.
Lipid synthesis, storage and degradation are associated with cell sensitivity to ferroptosis. Recently, ACSL4 and 15-LOX were shown to induce ferroptosis, catalyze acyl Co-A derivatives, and drive accumulation of oxidized phospholipids on intracellular membranes [41]. Thus, the mechanism of lipid metabolism and ferroptosis suggests a treatment strategy for cancer. Analysis of differentially expressed genes in the HBXIP expression-divided groups showed that HBXIP plays a regulatory role in lipid metabolism. After screening several key lipid synthesis kinases, SCD was identified as the downstream effector of HBXIP in sorafenib-induced imbalance in lipid metabolism and ferroptosis.
SCD is a major enzyme that catalyzes the transformation of saturated FAs into monounsaturated FAs. Protein dynamics and cell fluidity are influenced by the balance between unsaturated FAs and saturated FAs, which is critical for cancer cells [42]. The accumulation of unsaturated FAs not only accelerates tumor proliferation and metastasis but also inhibits cell apoptosis and ferroptosis. A previous study showed that inhibition of SCD activity can promote both apoptosis and ferroptosis by decreasing coenzyme Q10 and membrane-unsaturated FAs and increasing long-chain saturated ceramides [18]. Next, we examined the regulation of SCD by HBXIP in HCC cells. HBXIP was shown to bind to the SCD promoter and activate its transcriptional activity. Our previous study revealed that HBXIP co-activates the transcription factor SREBP-1c in breast cancer [9]. Given that SCD promoter activity is affected by SREBP-1c [35], we silenced SREBP-1 using siRNAs and tested SCD expression in HCC cells. Unexpectedly, upregulation of SCD triggered by HBXIP was independent of SREBP-1. Different mutations of SCD promoter were constructed, and a fragment (−315/−165) containing ZNF263 putative binding sites was found to be the core promoter of SCD for HBXIP coactivation. Sun et al. revealed that ZNF263 can induce resistance to apoptosis via activation of endoplasmic reticulum stress-dependent autophagy in HCC [43]. We found that ZNF263 is a transcriptional factor for the SCD promoter and activates its transcription. Our results revealed that HBXIP transcriptionally promoted the SCD expression by coactivating ZNF263.
Our animal experiments revealed that HBXIP abolished sorafenib-induced inhibition of tumor growth in vivo. According to analysis of the GSE109211 dataset, HBXIP expression was lower in sorafenib responders than in sorafenib non-responders, indicating that sorafenib efficacy is associated with HBXIP levels in HCC.
We identified the mechanism of action for HBXIP in sorafenib-induced ferroptosis. HBXIP can block ferroptotic cell death by disturbing free FA accumulation and upregulating SCD expression in HCC cells. The transcriptional factor ZNF263 is coactivated by HBXIP and then recruited to the SCD promoter. Finally, the HBXIP/SCD axis blocks the cell death triggered by sorafenib treatment in vivo and in vitro. These data provide insight into the important role of the HBXIP/SCD axis in HCC and support HBXIP as a therapeutic target of sorafenib.
Supplementary information
Acknowledgements
This study was supported by National Natural Science Foundation of China (82072943, China), National Natural Science Foundation of China (82072929, China) and National Natural Science Foundation of China (31870752, China).
Author contributions
All authors have participated in the article preparation. LZ designed the research methods. LZ, XML and XHS performed the experiments, analyzed the data, and prepared the manuscript. LZ, XHS, XML, KY, XLF, XW, HMS, QQL, FFX, SMG, JQM participated in the experiments. WYZ and LHY designed and revised the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Contributor Information
Wei-ying Zhang, Email: zhwybao@nankai.edu.cn.
Li-hong Ye, Email: yelihong@nankai.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41401-022-00981-9.
References
- 1.Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380:1450–62. doi: 10.1056/NEJMra1713263. [DOI] [PubMed] [Google Scholar]
- 2.Su Y, Zhao B, Zhou L, Zhang Z, Shen Y, Lv H, et al. Ferroptosis, a novel pharmacological mechanism of anti-cancer drugs. Cancer Lett. 2020;483:127–36. doi: 10.1016/j.canlet.2020.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Dolma S, Lessnick SL, Hahn WC, Stockwell BR. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell. 2003;3:285–96. doi: 10.1016/S1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
- 4.Yu Y, Xie Y, Cao L, Yang L, Yang M, Lotze MT, et al. The ferroptosis inducer erastin enhances sensitivity of acute myeloid leukemia cells to chemotherapeutic agents. Mol Cell Oncol. 2015;2:e1054549. doi: 10.1080/23723556.2015.1054549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun X, Ou Z, Chen R, Niu X, Chen D, Kang R, et al. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology. 2016;63:173–84. doi: 10.1002/hep.28251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun X, Niu X, Chen R, He W, Chen D, Kang R, et al. Metallothionein-1G facilitates sorafenib resistance through inhibition of ferroptosis. Hepatology. 2016;64:488–500. doi: 10.1002/hep.28574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L, Fang R, Liu B, Shi H, Wang Y, Zhang W, et al. Deacetylation of tumor-suppressor MST1 in Hippo pathway induces its degradation through HBXIP-elevated HDAC6 in promotion of breast cancer growth. Oncogene. 2016;35:4048–57. doi: 10.1038/onc.2015.476. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Zhang Z, Zhou X, Li L, Liu Q, Wang Z, et al. The oncoprotein HBXIP enhances migration of breast cancer cells through increasing filopodia formation involving MEKK2/ERK1/2/Capn4 signaling. Cancer Lett. 2014;355:288–96. doi: 10.1016/j.canlet.2014.09.047. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Y, Li H, Zhang Y, Li L, Fang R, Li Y, et al. Oncoprotein HBXIP modulates abnormal lipid metabolism and growth of breast cancer cells by activating the LXRs/SREBP-1c/FAS signaling cascade. Cancer Res. 2016;76:4696–707. doi: 10.1158/0008-5472.CAN-15-1734. [DOI] [PubMed] [Google Scholar]
- 10.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng S, Wu H, Wang F, Lv J, Lu J, Fang Q, et al. The oncoprotein HBXIP facilitates metastasis of hepatocellular carcinoma cells by activation of MMP15 expression. Cancer Manag Res. 2019;11:4529–40. doi: 10.2147/CMAR.S198783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi H, Fang R, Li Y, Li L, Zhang W, Wang H, et al. The oncoprotein HBXIP suppresses gluconeogenesis through modulating PCK1 to enhance the growth of hepatoma cells. Cancer Lett. 2016;382:147–56. doi: 10.1016/j.canlet.2016.08.025. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Fang R, Cui M, Zhang W, Bai X, Wang H, et al. The oncoprotein HBXIP up-regulates YAP through activation of transcription factor c-Myb to promote growth of liver cancer. Cancer Lett. 2017;385:234–42. doi: 10.1016/j.canlet.2016.10.018. [DOI] [PubMed] [Google Scholar]
- 14.Yang N, Wang T, Li Q, Han F, Wang Z, Zhu R, et al. HBXIP drives metabolic reprogramming in hepatocellular carcinoma cells via METTL3-mediated m6A modification of HIF-1α. J Cell Physiol. 2021;236:3863–80. doi: 10.1002/jcp.30128. [DOI] [PubMed] [Google Scholar]
- 15.Bansal S, Berk M, Alkhouri N, Partrick DA, Fung JJ, Feldstein A. Stearoyl-CoA desaturase plays an important role in proliferation and chemoresistance in human hepatocellular carcinoma. J Surg Res. 2014;186:29–38. doi: 10.1016/j.jss.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Roemeling CA, Marlow LA, Pinkerton AB, Crist A, Miller J, Tun HW, et al. Aberrant lipid metabolism in anaplastic thyroid carcinoma reveals stearoyl CoA desaturase 1 as a novel therapeutic target. J Clin Endocrinol Metab. 2015;100:E697–E709. doi: 10.1210/jc.2014-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noto A, De Vitis C, Pisanu ME, Roscilli G, Ricci G, Catizone A, et al. Stearoyl-CoA-desaturase 1 regulates lung cancer stemness via stabilization and nuclear localization of YAP/TAZ. Oncogene. 2017;36:4671–2. doi: 10.1038/onc.2017.212. [DOI] [PubMed] [Google Scholar]
- 18.Tesfay L, Paul BT, Konstorum A, Deng Z, Cox AO, Lee J, et al. Stearoyl-CoA desaturase 1 protects ovarian cancer cells from ferroptotic cell death. Cancer Res. 2019;79:5355–66. doi: 10.1158/0008-5472.CAN-19-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Meerloo J, Kaspers GJL, Cloos J. Cell sensitivity assays: the MTT assay. Methods Mol Biol. 2011;731:237–45. doi: 10.1007/978-1-61779-080-5_20. [DOI] [PubMed] [Google Scholar]
- 20.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–7. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 21.Liu N, Li H, Li S, Shen M, Xiao N, Chen Y, et al. The Fbw7/human CDC4 tumor suppressor targets proproliferative factor KLF5 for ubiquitination and degradation through multiple phosphodegron motifs. J Biol Chem. 2010;285:18858–67. doi: 10.1074/jbc.M109.099440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin B, Yu J, Nowsheen S, Zhao F, Wang L, Lou Z. STK38 promotes ATM activation by acting as a reader of histone H4 ufmylation. Sci Adv. 2020;6:eaax8214. doi: 10.1126/sciadv.aax8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z, Dou C, Jia Y, Li Q, Zheng X, Yao Y, et al. RIG-I suppresses the migration and invasion of hepatocellular carcinoma cells by regulating MMP9. Int J Oncol. 2015;46:1710–20. doi: 10.3892/ijo.2015.2853. [DOI] [PubMed] [Google Scholar]
- 24.Wang Q, Bin C, Xue Q, Gao Q, Huang A, Wang K, et al. GSTZ1 sensitizes hepatocellular carcinoma cells to sorafenib-induced ferroptosis via inhibition of NRF2/GPX4 axis. Cell Death Dis. 2021;12:426. doi: 10.1038/s41419-021-03718-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang WS, Stockwell BR. Ferroptosis: death by lipid peroxidation. Trends Cell Biol. 2016;26:165–76. doi: 10.1016/j.tcb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–31. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu L, Zhu Y, Lin X, Lu B, Zhou X, Zhou F, et al. The IKKβ-USP30-ACLY axis controls lipogenesis and tumorigenesis. Hepatology. 2021;73:160–74. doi: 10.1002/hep.31249. [DOI] [PubMed] [Google Scholar]
- 28.Sun L, Kong Y, Cao M, Zhou H, Li H, Cui Y, et al. Decreased expression of acetyl-CoA synthase 2 promotes metastasis and predicts poor prognosis in hepatocellular carcinoma. Cancer Sci. 2017;108:1338–46. doi: 10.1111/cas.13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lally JSV, Ghoshal S, DePeralta DK, Moaven O, Wei L, Masia R, et al. Inhibition of acetyl-coa carboxylase by phosphorylation or the inhibitor ND-654 suppresses lipogenesis and hepatocellular carcinoma. Cell Metab. 2019;29:174–82. doi: 10.1016/j.cmet.2018.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai KKY, Kweon S-M, Chi F, Hwang E, Kabe Y, Higashiyama R, et al. Stearoyl-CoA desaturase promotes liver fibrosis and tumor development in mice via a Wnt positive-signaling loop by stabilization of low-density lipoprotein-receptor-related proteins 5 and 6. Gastroenterology. 2017;152:1477–91. doi: 10.1053/j.gastro.2017.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen L, Ren J, Yang L, Li Y, Fu J, Li Y, et al. Stearoyl-CoA desaturase-1 mediated cell apoptosis in colorectal cancer by promoting ceramide synthesis. Sci Rep. 2016;6:19665. doi: 10.1038/srep19665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noto A, Raffa S, De Vitis C, Roscilli G, Malpicci D, Coluccia P, et al. Stearoyl-CoA desaturase-1 is a key factor for lung cancer-initiating cells. Cell Death Dis. 2013;4:e947. doi: 10.1038/cddis.2013.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von Roemeling CA, Marlow LA, Wei JJ, Cooper SJ, Caulfield TR, Wu K, et al. Stearoyl-CoA desaturase 1 is a novel molecular therapeutic target for clear cell renal cell carcinoma. Clin Cancer Res. 2013;19:2368–80. doi: 10.1158/1078-0432.CCR-12-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fritz V, Benfodda Z, Rodier G, Henriquet C, Iborra F, Avancès C, et al. Abrogation of de novo lipogenesis by stearoyl-CoA desaturase 1 inhibition interferes with oncogenic signaling and blocks prostate cancer progression in mice. Mol Cancer Ther. 2010;9:1740–54. doi: 10.1158/1535-7163.MCT-09-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu G, Kuang S, Cao R, Wang J, Peng Q, Sun C. Sorafenib kills liver cancer cells by disrupting SCD1-mediated synthesis of monounsaturated fatty acids the ATP-AMPK-mTOR-SREBP1 signaling pathway. FASEB J. 2019;33:10089–103. doi: 10.1096/fj.201802619RR. [DOI] [PubMed] [Google Scholar]
- 36.Yi J, Zhu J, Wu J, Thompson CB, Jiang X. Oncogenic activation of PI3K-AKT-mTOR signaling suppresses ferroptosis via SREBP-mediated lipogenesis. Proc Natl Acad Sci USA. 2020;117:31189–97. doi: 10.1073/pnas.2017152117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 38.Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S, Kim JS, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10:25–34. doi: 10.1016/S1470-2045(08)70285-7. [DOI] [PubMed] [Google Scholar]
- 39.Mou Y, Wang J, Wu J, He D, Zhang C, Duan C, et al. Ferroptosis, a new form of cell death: opportunities and challenges in cancer. J Hematol Oncol. 2019;12:34. doi: 10.1186/s13045-019-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li ZJ, Dai HQ, Huang XW, Feng J, Deng JH, Wang ZX, et al. Artesunate synergizes with sorafenib to induce ferroptosis in hepatocellular carcinoma. Acta Pharmacol Sin. 2021;42:301–10. doi: 10.1038/s41401-020-0478-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doll S, Proneth B, Tyurina YY, Panzilius E, Kobayashi S, Ingold I, et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat Chem Biol. 2017;13:91–98. doi: 10.1038/nchembio.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Viswanathan VS, Ryan MJ, Dhruv HD, Gill S, Eichhoff OM, Seashore-Ludlow B, et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature. 2017;547:453–7. doi: 10.1038/nature23007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cui J, Liu J, Fan L, Zhu Y, Zhou B, Wang Y, et al. A zinc finger family protein, ZNF263, promotes hepatocellular carcinoma resistance to apoptosis via activation of ER stress-dependent autophagy. Transl Oncol. 2020;13:100851. doi: 10.1016/j.tranon.2020.100851. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.