Abstract
Alternative splicing of cardiac troponin T (cTNT) exon 5 undergoes a developmentally regulated switch such that exon inclusion predominates in embryonic, but not adult, striated muscle. We previously described four muscle-specific splicing enhancers (MSEs) within introns flanking exon 5 in chicken cTNT that are both necessary and sufficient for exon inclusion in embryonic muscle. We also demonstrated that CUG-binding protein (CUG-BP) binds a conserved CUG motif within a human cTNT MSE and positively regulates MSE-dependent exon inclusion. Here we report that CUG-BP is one of a novel family of developmentally regulated RNA binding proteins that includes embryonically lethal abnormal vision-type RNA binding protein 3 (ETR-3). This family, which we call CELF proteins for CUG-BP- and ETR-3-like factors, specifically bound MSE-containing RNAs in vitro and activated MSE-dependent exon inclusion of cTNT minigenes in vivo. The expression of two CELF proteins is highly restricted to brain. CUG-BP, ETR-3, and CELF4 are more broadly expressed, and expression is developmentally regulated in striated muscle and brain. Changes in the level of expression and isoforms of ETR-3 in two different developmental systems correlated with regulated changes in cTNT splicing. A switch from cTNT exon skipping to inclusion tightly correlated with induction of ETR-3 protein expression during differentiation of C2C12 myoblasts. During heart development, the switch in cTNT splicing correlated with a transition in ETR-3 protein isoforms. We propose that ETR-3 is a major regulator of cTNT alternative splicing and that the CELF family plays an important regulatory role in cell-specific alternative splicing during normal development and disease.
The generation of multiple, functionally distinct protein isoforms from a single gene via alternative splicing is a common means of regulating gene expression. It has been estimated that more than one-third of human genes are alternatively spliced (21). Despite the prevalence of alternative splicing, the mechanisms by which it is regulated are not well understood. Cis-acting elements that mediate alternative splicing specific to different cell types have been identified in a few experimental systems (1, 3, 18, 19, 50, 66), and progress in identifying trans-acting factors involved in tissue-specific regulation, particularly neuron-specific splicing, has been made. KH-type splicing regulatory protein is enriched in neurons and has been isolated as a component of a complex that activates inclusion of the c-src neuronal N1 exon (43). The KH-type RNA binding protein Nova-1 is expressed exclusively in neurons of the central nervous system (4) and activates inclusion of exons in the glycine receptor and GABAA receptor pre-mRNAs (27). Brain polypyrimidine tract binding protein (brPTB), a protein related to the more ubiquitous PTB but enriched in brain, binds to the same sequence recognized by PTB and antagonizes the effects of Nova-1 (48).
In addition to the use of tissue-restricted factors, cell-specific alternative splicing may also be regulated by altering the abundance or activities of constitutive pre-mRNA splicing factors such as the SR protein family and hnRNP proteins (6, 31, 34, 58). These factors participate in constitutive splicing, but their expression is not entirely ubiquitous and undergoes tissue-specific regulation (14, 20, 30, 57, 65, 68). Regulation of alternative splice site choices by natural variation in the expression of these proteins, however, remains to be directly demonstrated.
For many genes, alternative splicing is also modulated in a developmental-stage-specific manner. Alternative splicing is determinative for cell fate decisions during development in Drosophila melanogaster, such as the regulation of sexual differentiation by the RNA binding protein Sex-lethal (reviewed in reference 35). Changes in the constitutive splicing machinery in nematodes also suggest a developmental role for alternative splicing regulation (54). Factors involved in the developmental regulation of alternative splicing in vertebrates, however, have not yet been reported.
Many proteins essential for striated muscle development exist in multiple isoforms generated by alternative splicing, including myogenic transcription factors, metabolic enzymes, and components of the myofibril. Skeletal-muscle-specific splicing patterns are induced during differentiation and can be induced in fibroblasts by expression of MyoD and myogenin (26, 50, 52). Alternative splicing of exon 5 of the cardiac troponin T (cTNT) gene undergoes a developmentally regulated switch such that mRNAs in embryonic striated muscle include the exon, but mRNAs in the adult do not (11). The developmental regulation of cTNT splicing is conserved in chickens, mice, rats, and humans (11, 28, 39, 60). The two cTNT isoforms generated by alternative splicing of exon 5 confer different levels of calcium sensitivity to the myofilament, thereby affecting the contractile properties of maturing muscle (16, 40).
Previously, we identified four cis-acting elements within the introns flanking exon 5 in the chicken cTNT that are both necessary and sufficient to promote exon inclusion specifically in embryonic striated muscle (10, 46, 50). These muscle-specific splicing enhancers (MSEs) were defined in cTNT minigenes by deletions or substitutions that prevented the activation of exon 5 inclusion in embryonic skeletal muscle cultures but that had little effect on the low level of exon inclusion observed in nonmuscle cultures (10, 46, 50). One of these MSEs, MSE2, is conserved in sequence and position in chicken and human cTNT (46). MSE2 is necessary and sufficient to promote muscle-specific cTNT exon 5 inclusion in embryonic skeletal muscle cultures when present in multiple copies (10). A conserved CUG motif found within MSE2 is found adjacent to alternative exons in several genes that undergo regulated splicing in striated muscle and is required for enhancer activity, suggesting that these elements play a common role in muscle-specific splicing (10, 46).
CUG-binding protein (CUG-BP) is a highly conserved protein that was purified based on its sequence-specific binding to RNA containing CUG repeats (59). CUG-BP is proposed to play a role in the pathogenesis of the trinucleotide expansion disease, myotonic dystrophy (DM1), in which nuclear accumulation of transcripts of the myotonin protein kinase (DMPK) gene containing expanded CUG repeats is proposed to create an RNA gain-of-function mutation. We have shown that CUG-BP binds to the conserved CUG motif within cTNT MSE2 and positively regulates MSE-dependent inclusion of exon 5 (46). Embryonically lethal abnormal vision (ELAV)-type RNA binding protein 3 (ETR-3) (25), which has high sequence similarity to CUG-BP, has also recently been reported to bind to RNAs containing CUG triplet repeats (36), suggesting that it may also regulate the alternative splicing of MSE-containing transcripts.
Here we report the identification of a novel family of splicing regulators that includes CUG-BP and ETR-3, which we call CELF proteins for CUG-BP- and ETR-3-like factors. Individual CELF family members are preferentially expressed in different cell types, and at least two CELF genes express multiple isoforms via alternative splicing. We demonstrate that CELF proteins bind to MSE-containing RNAs and activate MSE-dependent exon inclusion in fibroblasts when coexpressed with cTNT minigenes. Expression of these proteins is developmentally regulated. ETR-3 protein expression in particular correlates with changes in cTNT alternative splicing during heart development in both chickens and mice and during myogenic differentiation of the mouse C2C12 myoblast cell line. We propose that ETR-3 is a major regulator of cTNT alternative splicing. Our results strongly support the hypothesis that the CELF family plays a critical role in the regulation of cell-specific alternative splicing during normal development and disease.
MATERIALS AND METHODS
Identification and analysis of CELF sequences.
ETR3, CELF3, CELF4, and CELF5 were identified in searches of expressed sequence tag (EST) and high-throughput genomic sequence databases using a CUG-BP sequence query. ETR-3 was first identified in a screen of genes expressed in human fetal heart (25). Alternatively spliced forms of ETR-3 were recently found in a screen of genes induced during apoptosis in a neuroblastoma cell line and were called NAPOR-1, -2, and -3 (7); they were also found by a screen of a liver cDNA library using a CUG-BP probe (36). A truncated version of CELF3 called CAGH4 is also found in the database (38). A potential sixth family member was identified in cosmids from chromosome 15 (accession no. AC009690 and AC009524), but corresponding ESTs could not be found.
The open reading frames for all five CELF proteins were cloned into the pcDNA3.1HisC vector (Invitrogen) in frame with the N-terminal Xpress epitope tag using 5′ primers containing the initiation codon and 3′ primers containing the termination codon. CUG-BP primers were GTTAGTGGATCCATGAACGGCACCCTGGACCA and GGCCGAAGCTTTCAGTAGGGCTTGCTGT. ETR-3 primers were CGGTGAGATCTATGAACGGAGCTTTGGA and CGGTCAAGCTTTCAGTAAGGTTTGCTGTCGT. CELF3 primers were GTTAGAGGATCCATGAAGGAGCCGGATGCCATCA and AATTACCTCGAGTCAGTAGGGCCGGTTGGCATCCTTA. CELF4 primers were GTTAGAGGATCCATGAAGGACCACGATGCCATC and AATTACCTCGAGTCAGTACGGGCGATTGGCGTCTTTG. CELF5 primers were CATCATCGGATCCATGGCCCGCCTGACGGAGAG and CATCATCTCTAGATCACGGGTCTTTGGGCCGCT. For all CELF proteins except CELF4, the expressed open reading frames begin at the codon for the first methionine shown in Fig. 1. The open reading frame for CELF4 in the expression vector starts at the codon for the methionine at position 48, as this was the first known methionine at the time of cloning; additional ESTs that contain the putative upstream translation presented in Fig. 1 have subsequently been entered in the database. The templates for PCR included a CUG-BP cDNA clone provided by L. Timchenko (Baylor College of Medicine, Houston, Tex.) and an ETR-3 cDNA provided by C. C. Liew (University of Toronto, Toronto, Ontario, Canada). The 5′ ends of CELF3 and CELF4 were not available in the EST database and were cloned by rapid amplification of cDNA ends (RACE) using the Marathon RACE kit (Clontech Laboratories, Inc.) on adult brain cDNA (Clontech). The 5′ RACE products provided the sequence of initiation codons, which were then used to amplify full-length CELF3 and CELF4 from commercial cDNA from adult brain (Clontech). The 5′ end of CELF5 mRNA (including the putative initiation codon) was found in cosmid R31341, and the 3′ end of CELF5 mRNA (including the termination codon and 3′ untranslated region [UTR]) was found in in several ESTs. CELF5 was amplified using cDNA generated from human total brain RNA (Clontech) in a 20-μl reaction mixture containing 2.5 μg of RNA, 50 ng of oligo (dT)12–18 primer (Life Technologies), First Strand buffer (Life Technologies), 0.01 M dithiothreitol [DTT], 0.5 mM deoxynucleoside triphosphate (dNTP) mixture, and 100 U of avian myeloblastosis virus reverse transcriptase (Life Technologies). The RNA and primers were heated to 65°C for 10 min and chilled on ice, and then the remaining reagents were added and the reaction mixture was incubated at 42°C for 1 h and at 70°C for 15 min and stored at −20°C. All PCRs were performed in a 50-μl reaction volume containing Taq Plus Precise buffer (Stratagene, La Jolla, Calif.), 0.25 mM dNTP mixture, 2.5 μl of dimethyl sulfoxide, 200 ng of each of the 5′ and 3′ primers, and 2.5 U of Taq Plus Precise enzyme (Stratagene) on an MJ Research PTC-100 using 30 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 2 min, followed by 1 cycle at 72°C for 10 min. PCR products were isolated from 1% agarose gels using the QIAquick gel extraction kit (Qiagen) and cloned directly into pcDNA3.1HisC. Sequences were confirmed by overlapping reads of both strands.
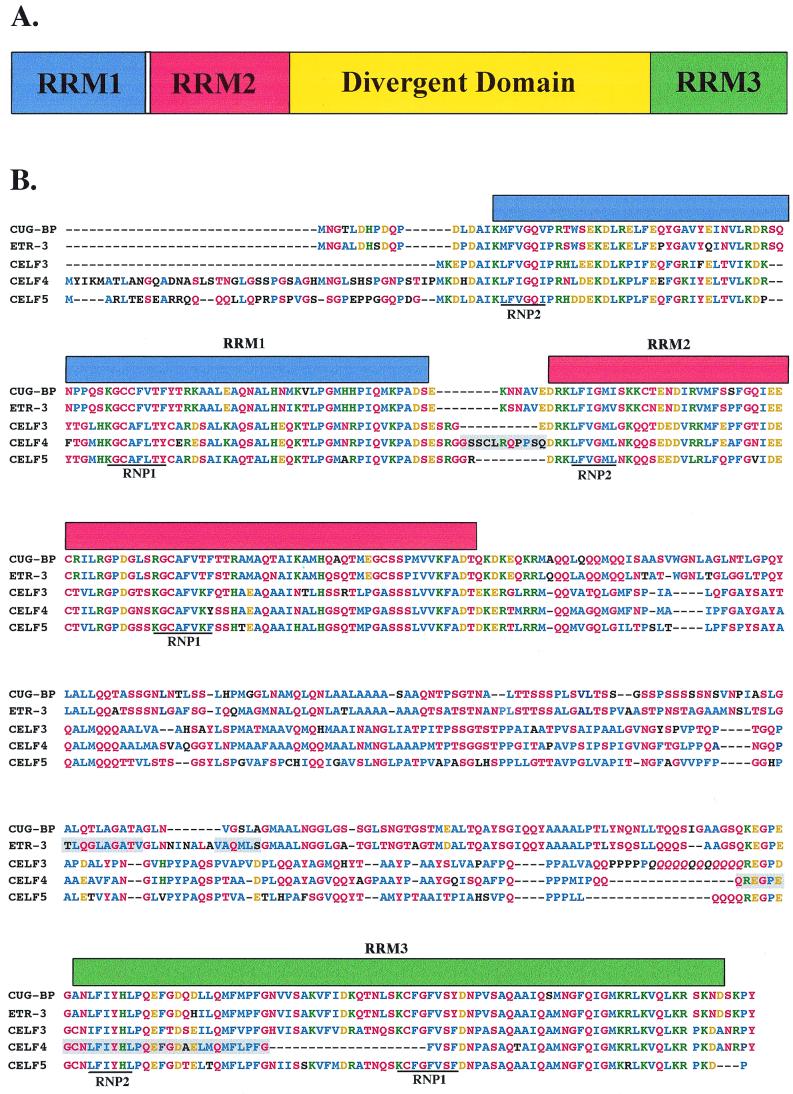
FIG. 1.
The CELF family members are closely related. (A) All of the CELF proteins possess the same domain structure: three RRMs and a divergent domain of unknown function between RRM2 and RRM3. Nomenclature is designed to consider CUG-BP and ETR-3 as CELF1 and CELF2, respectively. (B) Sequences of five human CELF proteins derived from PCR-amplified cDNAs (see Materials and Methods). Blue, conserved nonpolar amino acids; red, conserved uncharged polar residues; green, conserved positively charged residues; gold, conserved negatively charged residues; black, nonconserved residues. Known alternatively spliced regions are shaded. Italic's, a region of allelic variation in CELF3 due to a variable number of CAG repeats. (C) Phylogenetic relationship of human CELF proteins.
Sequence profile analyses were performed using PROSITE (24) and PSORTII (44). Nuclear localization predictions were made using both Reinhardt's NCNN and the k -NN nearest-neighbor prediction algorithms. The simple modular architecture research tool (SMART) (56) was used to identify proteins with a domain structure similar to that of the CELF proteins. Alignments and phylogenetic analyses were performed using DNASTAR software, which utilizes the Clustal V method (23).
Western blots.
Antipeptide antibodies against ETR-3 (against a 17-amino-acid peptide [QTSATSTNANPLSTGC] conjugated to keyhole limpet hemocyanin) were generated in rabbits by Cocalico Biologicals. Antipeptide antibodies against CELF4 (against a 15-amino-acid peptide [CIHPYPAQSPTAADP] conjugated to keyhole limpet hemocyanin) were generated in rabbits by Anaspec, Inc. The 3B1 mouse monoclonal antibody against CUG-BP was provided by M. Swanson (University of Florida, Gainsville). Tissues were dissected from embryonic-day-14, newborn, 4-day-old, and pregnant or 0- to 4-day-postpartum adult female Swiss Webster mice (Taconic Farms, Inc.) and from embryonic-day-8 to -20 and adult White Leghorn chickens (Texas A&M University) and homogenized directly in protein loading buffer (0.64 M Tris-HCl [pH 6.8], 10% glycerol, 2% sodium dodecyl sulfate [SDS], 5% β-mercaptoethanol). C2C12 murine myoblast cells were maintained at a density of 20 to 60% confluency in 100-mm-diameter tissue culture dishes in 10 ml of growth medium (low-glucose Dulbecco modified Eagle medium [DMEM] supplemented with 20% fetal bovine serum and 0.5% chick embryo extract) and cultured at 37°C in 5% CO2. To induce differentiation, growth medium was replaced with differentiation medium (low-glucose DMEM supplemented with 5% horse serum) when cells had reached 60% confluency. To collect total protein samples from C2C12 cultures, cells were rinsed with phosphate-buffered saline and then scraped directly into protein loading buffer and sonicated. Protein concentrations were determined using a noninterfering protein assay (Geno Technology, Inc.). Total protein samples were resolved on 10% denaturing polyacrylamide gels, with 50 to 60 μg loaded per lane, and transferred to Immobilon-P membranes (Millipore). Membranes were incubated for 1 h in blocking solution (5% milk in phosphate-buffered saline plus 0.05% Tween-20 [PBT]) at room temperature with shaking. The primary antibody was added, the mixture was incubated for 1 h, and then the membrane was washed twice for 20 min with PBT and blocked again for 1 h. Goat anti-rabbit or goat anti-mouse horseradish peroxidase-conjugated secondary antibody (Jackson ImmunoResearch Laboratories, Inc.) was added, and the mixture was incubated for 1 h; then the membrane was washed three times for 20 min with PBT, and bands were visualized with SuperSignal chemiluminescent substrate (Pierce) on Kodak film (Eastman Kodak).
UV cross-linking.
The open reading frames for CUG-BP, ETR-3, and CELF4 were cloned downstream of glutathione S-transferase (GST) in the pGEX-2T GST fusion expression plasmid (Novagen). BL21(DE3)pLysS competent bacteria (Stratagene) were transformed and tested for the presence of the plasmid. One colony was grown overnight at 37°C in Terrific broth (53) plus 80 μg of Timentin/ml, diluted 1:10, and grown for an additional 2 h. IPTG (0.1 mM) was added, and cells were grown for 3 h and pelleted. The cell pellet was resuspended in 0.2 culture volumes of phosphate-buffered saline and sonicated on ice six times for 10 s each. Triton X-100 was added to a final concentration of 1%, and the total lysate was spun for 10 min at 12,000 × g. Recombinant GST fusion proteins were purified from the supernatant according to the GST-Bind buffer kit (Novagen) and quantitated using a protein assay kit (Bio-Rad), and the quantity and purity were checked on SDS-polyacrylamide gel electrophoresis gels by Coomassie staining (53).
The templates for in vitro RNA synthesis were derived from the MSE-containing and MSE-lacking minigenes used for transfection (see below). The MSE-containing substrate contains 30 nucleotides of the pBluescript vector (Stratagene) downstream of the T3 promoter, 93 nucleotides of cTNT intron 4 including MSE1, a 63-nucleotide artificial exon derived from skeletal troponin I (GGTTCACAACCATCTACGCATTCGAAGAGGCATTGGATCCGAACCAAGCAAGATGTCTGACAG), and 142 nucleotides of cTNT intron 5 including MSE2 to -4. The plasmid was linearized using SpeI (Roche Molecular Biochemicals). The substrate containing MSEs 2 to 4 was obtained by removing a SexAI-BamHI (Roche Molecular Biochemicals) fragment situated between the T3 promoter and the exon which encompassed MSE1. For the substrates containing the full-length intron and MSEs 2 to 4, the plasmids were linearized using SpeI (Roche Molecular Biochemicals); the MSE1 substrate was obtained by linearizing the MSE1 to 4 template with BstBI (Roche Molecular Biochemicals), which cuts within the exon. The linearized plasmids were transcribed with T3 RNA polymerase (Life Technologies). In vitro synthesis of uniformly labeled RNA was performed as described previously (9).
UV cross-linking was performed using 1 pmol of substrate (0.4 × 106 to 1.0 × 106 cpm) labeled with [α-32P]GTP and [α-32P]UTP and 500 ng of recombinant GST fusion proteins. Proteins and RNA were incubated at 30°C for 10 min with a mixture containing 30 μg of HeLa nuclear extract (13), 2 mM magnesium acetate, 2 mM ATP, 16 mM HEPES (pH 7.9) 65 mM potassium glutamate, 0.16 mM EDTA, 0.4 mM DTT, and 16% glycerol. In some experiments, 1 μg of tRNA (Ambion), 1 μg of bovine serum albumin, and 0.1 μg of heparin were substituted for HeLa nuclear extract without affecting the results. Incubation with bulk RNA and protein is necessary to decrease the nonspecific binding of the recombinant proteins. Reaction mixtures were placed on an aluminum block prechilled in ice water and irradiated 4 cm from a Philips G15T8 germicidal lamp for 8 min. Samples were digested with 0.5 μg of each RNase A and RNase T1 (Sigma) at 37°C for 20 min. An equal volume of protein loading buffer was added, and samples were denatured at 90°C and run on a SDS–12% polyacrylamide gel. Sizes were determined using prestained markers (Bio-Rad).
Cotransfection experiments.
R35C was derived from RTB33.51 (50) by filling in a BstBI site located 86 nucleotides upstream of the insertion site for the cTNT genomic fragment in the skeletal troponin I minigene. M2/M2TB has been described previously (10). RTBPSRAX was derived from RTBPSR (50) by removing all of the cTNT intron sequence (68 nucleotides between Asp718 and XbaI sites).
QT35 quail fibroblast cells were plated at a density of 1.8 × 106 cells/60-mm-diameter tissue culture dish in 3 to 5 ml of growth medium (F10 medium supplemented with 5% fetal bovine serum, 1% chick serum, 10% tryptose phosphate, and 2 mM l-glutamine) and cultured overnight at 37°C in 5% CO2. The medium was then changed to transfection medium (low-glucose DMEM supplemented with 10% fetal bovine serum and 2 mM l-glutamine), and cells were transfected with 2 μg of minigene DNA and 0 to 10 μg of CELF expression plasmid DNA using FuGene 6 (Roche Molecular Biochemicals). Medium was replaced with growth medium at 24 h, and total RNA was harvested 48 h following transfection by the method of Chomczynski and Sacchi (8) as modified by Xie and Rothblum (63). Total protein samples were collected from parallel plates by scraping cells into 250 μl of protein loading buffer and sonicating. Fifty microliters of each sample was run on a 10% denaturing polyacrylamide gel, and Western blot analysis was performed as described above using the anti-Xpress antibody (Invitrogen) at 1:5,000. To monitor cTNT splicing during muscle differentiation, C2C12 cells were transfected with 2 μg of R35C minigene DNA using FuGene 6 (Roche Molecular Biochemicals) at 30% confluency. Growth medium was replaced with differentiation medium the next day, and RNA was harvested every 24 h.
The extent of minigene exon inclusion was determined by reverse transcription-PCR (RT-PCR) analysis. cDNA was generated as described above. PCR was performed in a 40-μl volume containing Taq MgCl2-free buffer (Promega), 1.75 mM MgCl2, 0.2 mM dNTPs, 200 ng of each oligonucleotide (CATTCACCACATTGGTGTGC and AGGTGCTGCCGCCGGGCGGTGGCTG), 1.5 ng of γ-32P-labeled 5′ primer, and 2.5 U of Taq (Promega) for 18 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min, followed by 1 cycle at 72°C for 5 min. The 5′ primer was labeled with γ-32P by incubation with a mixture containing kinase buffer (70 mM Tris [pH 7.5], 1 mM MgCl2, 0.5 mM DTT), 0.7 U of polynucleotide kinase (USB Corporation), enough oligonucleotide for 1.5 ng per PCR, and equal molar amounts of [γ-32P]ATP in a 10-μl volume at 37°C for 30 min. PCR products were resolved on 5% nondenaturing polyacrylamide gels, and bands were quantitated using a PhosphorImager (Molecular Dynamics). The extent of endogenous cTNT exon inclusion in mouse heart was determined by RT-PCR analysis using primers AGCCGAGAGCATGTCTGACGCCGAGGAGGTGGT (in exon 3) and CAGCATCTTTGGCTTCATCAGGACCAACCT (in exon 6) and 30 cycles of amplification. Primer extension of chicken heart RNA was performed using a 5′-end-labeled oligo nucleotide complementary to cTNT exon 6 as described previously (64), and the ratios of primer extension products were quantitated by a PhosphorImager.
Nucleotide sequence accession numbers.
Accession numbers for the sequences of CELF3, CELF4, and CELF5 are AF329264, AF329265, and AF329266, respectively.
RESULTS
Identification of the CELF protein family.
A screen of human EST databases was performed to identify CUG-BP paralogs based on sequence similarity. Four proteins closely related to CUG-BP were found, including ELAV-type RNA binding protein 3 (ETR-3) and three novel proteins we call CELF3, CELF4, and CELF5 (Fig. 1B). A potential sixth family member was identified in cosmid sequences from chromosome 15 (see Materials and Methods), but no corresponding ESTs were found and this family member was not studied further. The three novel protein sequences presented are derived from open reading frames amplified by PCR from human brain cDNA. All five proteins have the same domain structure (Fig. 1A): three RNA binding domains composed of the RNP-containing RNA recognition motif (RRM) (reviewed in reference 5) and a domain of unknown function separating RRM2 and RRM3. We have termed the region separating RRM2 and RRM3 the divergent domain, because sequence similarities within this domain split the CELF family into two subgroups of closely related proteins (Fig. 1C), the first containing CUG-BP and ETR-3 (78% identical) and the second containing CELF3, CELF4, and CELF5 (CELF3 and CELF5 have 60.8 and 63.8% identity with CELF4, respectively). BLAST searches reveal that the divergent domains are unique. No known protein-protein, protein-RNA, or protein-DNA interaction motifs, targeting signals, or predicted secondary structure were identified within the divergent domain. Variable regions most likely due to alternative splicing were identified in both ETR-3 and CELF4 (Fig. 1B). CELF3 contains a region of allelic variation in which a variable number of CAG repeats encode a stretch of glutamines of unfixed length (Fig. 1B). All of the CELF proteins contain multiple potential protein kinase C and casein kinase II phosphorylation sites. All are predicted to have predominantly nuclear localization, and CELF3, CELF4, and CELF5 each possess a consensus nuclear localization signal sequence near the C terminus. CUG-BP is known to be distributed within both the nucleus and cytoplasm and is detectable by Western blot analysis and gel shift assays in two isoforms that differ in phosphorylation state and intracellular distribution (49, 59). Among vertebrate proteins, the CELF family is most closely related to the Hu family of RNA binding proteins, homologs of Drosophila nuclear protein ELAV that have been implicated in the regulation of mRNA stability and translation (reviewed in reference 32).
CELF proteins are widely expressed in adult tissues.
To examine the expression of CELF family members, commercial RNA dot blots containing samples from a variety of human tissues were probed with 3′-UTR probes. CELF3 and CELF5 mRNAs were restricted to brain, whereas CUG-BP, ETR-3, and CELF4 displayed broader patterns of expression (data not shown). Consistent with these results, CELF3 and CELF5 ESTs were identified only in brain-derived libraries.
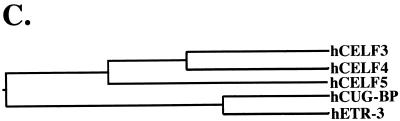
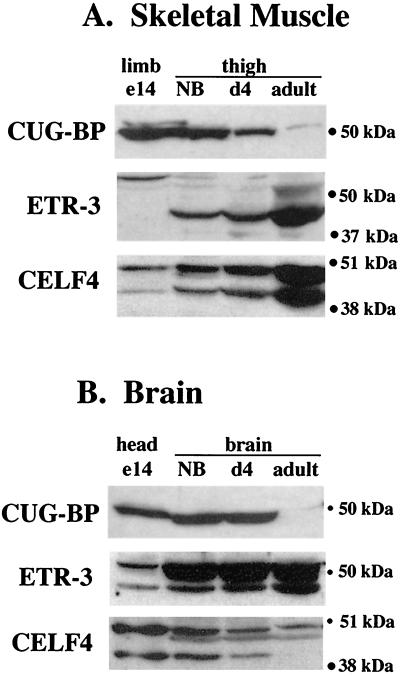
To examine the distribution of the three CELF family members that are not restricted to brain, Western blot analyses were performed on total protein samples extracted from a variety of adult mouse tissues (Fig. 2). A mouse monoclonal antibody against CUG-BP and rabbit polyclonal antipeptide antibodies against unique epitopes in ETR-3 and CELF4 were used. Each antipeptide antibody failed to recognize other CELF proteins that were transiently expressed in QT35 cells or as recombinant proteins (data not shown). Every tissue examined expressed one or more of the three CELF proteins, with skeletal muscle, heart, and brain showing the highest levels of CELF proteins overall. Each CELF family member was expressed in multiple tissues with a distinct pattern of tissue distribution. The multiple bands observed for ETR-3 and CELF4 suggest the presence of multiple isoforms, with different cell types showing different combinations of isoforms. At least some of these isoforms are likely to be generated by alternative splicing (Fig. 1B and data not shown). ETR-3 and CELF4 were particularly abundant in striated muscle and brain, where multiple isoforms were observed for each. In skeletal muscle, the pattern of these isoforms differed depending on the source of the tissue, perhaps due to differences in fiber type composition.
FIG. 2.
CELF proteins are widely expressed. Western blot analysis was performed on total protein samples extracted from adult, pregnant or postpartum female Swiss Webster mice. Fifty micrograms of total protein was loaded in each lane. ETR-3 and CELF4 polyclonal antipeptide antibodies failed to recognize other CELF proteins as transiently expressed or recombinant proteins. An abundant 30-kDa protein was also detected in brain samples on CELF4 blots (data not shown). The relative intensity of this band was reduced somewhat by the addition of a protease inhibitor cocktail to the homogenization buffer, suggesting abundant CELF4 protein expression and cleavage or degradation in brain.
CELF family members bind to cTNT MSEs in vitro and positively regulate MSE-dependent splicing in vivo.
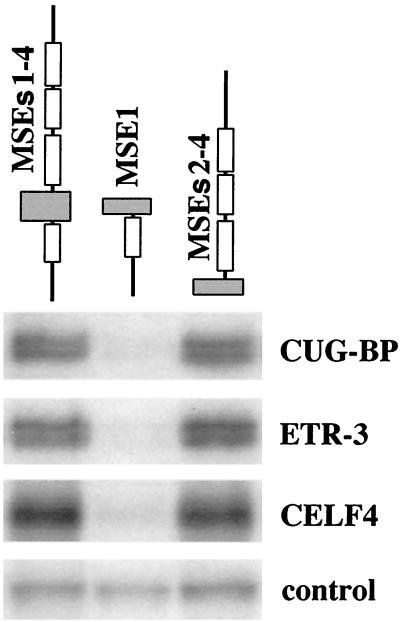
We have previously shown that CUG-BP binds to a human MSE2-like sequence flanking the human cTNT alternative exon 5 and promotes MSE-dependent exon inclusion (46). To determine whether CELF proteins differ in their ability to bind MSEs, we tested the binding of CUG-BP, ETR-3, and CELF4, the three CELF proteins expressed in striated muscle, to chicken cTNT MSEs. Sequence-specific binding of purified recombinant GST fusion proteins was tested by UV cross-linking using uniformly labeled RNAs containing MSEs 1 to 4, MSE1, or MSEs 2 to 4 from the chicken cTNT gene. All three CELF proteins bound to MSEs 1 to 4 and to MSEs 2 to 4 but not to an upstream fragment that contains MSE1 alone (Fig. 3), demonstrating that the binding of CELF proteins is restricted to the downstream intron. This binding is not due to the GST portion of the fusion proteins, as CELF fusion proteins made with other epitope tags demonstrate the same sequence-specific binding (data not shown). In experiments performed in the presence of HeLa nuclear extracts, a nonspecific 100-kDa RNA binding protein demonstrated equivalent levels of binding to all substrates, indicating that all three RNAs were intact and competent for binding. Therefore, the CELF proteins bind to a 142-nucleotide intronic region of the chicken cTNT gene that is sufficient for activation of exon inclusion in striated muscle (10, 50). Analysis using scanning mutations has identified multiple CELF protein binding sites within this region (unpublished data).
FIG. 3.
CELF proteins bind specifically to a downstream MSE-containing region. UV cross-linking experiments were performed using purified recombinant GST-CELF fusion proteins and uniformly 32P-labeled RNA (G and U) containing partial or full-length segments of the cTNT-regulated region. Proteins and RNA were preincubated in HeLa nuclear extract to increase the specificity of binding. Similar results were obtained when tRNA, bovine serum albumin, and heparin were used instead of HeLa nuclear extract (not shown). Each lane contains equal molar amounts of RNA. An approximately 100-kDa protein that bound nonspecifically to all three RNA substrates in HeLa cell nuclear extracts is also shown.
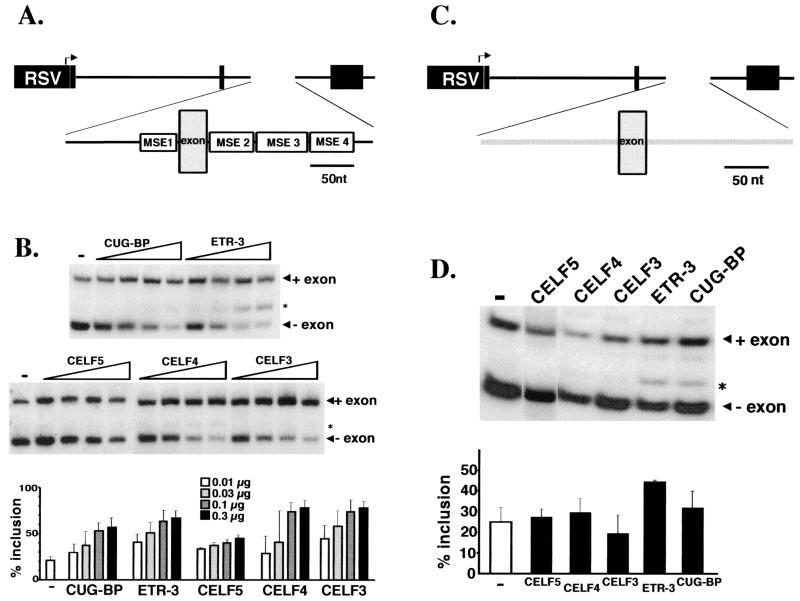
To determine whether different CELF family members can activate MSE-dependent exon inclusion, expression vectors for all five CELF proteins were cotransfected with the R35C minigene that contains a heterologous alternative exon flanked by MSEs 1 to 4 into QT35 quail fibroblast cells (Fig. 4A). This exon is appropriately regulated in embryonic skeletal muscle cultures and is predominantly skipped in QT35 cells, as in other nonmuscle cells (10, 50). Western blots probed with antibodies against the N-terminal epitope tag demonstrated that all of the expression vectors express proteins of the expected sizes at comparable levels and that increasing amounts of expression plasmid resulted in increasing amounts of expressed proteins (data not shown). Cotransfection with each of the five CELF proteins enhanced the level of exon inclusion in a dose-responsive manner (Fig. 4B). To determine whether enhanced exon inclusion by CELF proteins is MSE dependent, 300 ng of each CELF expression plasmid was cotransfected with a minigene containing the same alternative exon flanked by human β-globin intron 1 sequences which lack MSEs (Fig. 4C). The extent of exon inclusion in the presence of 300 ng of CUG-BP, CELF5, CELF4, or CELF3 expression plasmid did not differ from the level observed for the minigene alone (Fig. 4D). Exon inclusion was elevated by the addition of 300 ng of ETR-3 expression plasmid but remained well below levels observed with the MSE-containing minigene. Therefore, the CELF family members promote the MSE-dependent exon in vivo on RNA substrates to which they bind in a sequence-specific manner in vitro.
FIG. 4.
CELF proteins promote MSE-dependent exon inclusion. (A) The MSE-containing R35C minigene contains an alternative exon flanked upstream by the last 99 nucleotides of cTNT intron 4, which includes MSE1, and downstream by the first 142 nucleotides of cTNT intron 5, which includes MSEs 2 to 4. This intron-exon-intron cassette is inserted between exons 2 and 4 of the constitutively spliced chicken skeletal troponin I gene. RSV, Rous sarcoma virus. (B) Quail QT35 fibroblasts were cotransfected with 2 μg of R35C and 0 (−), 10, 30, 100, or 300 ng of CUG-BP, ETR-3, CELF5, CELF4, or CELF3 expression plasmid. Increasing concentrations of expression vector showed increasing amounts of protein expression (data not shown). RNA was harvested after 48 h, and RT-PCR analysis was performed to determine the extent of exon inclusion. (C) The MSE-lacking substrate RTBPSRAX contains the same alternative exon as R35C flanked upstream by the last 78 nucleotides of human β-globin intron 1 and downstream by the first 96 nucleotides of the same intron. (D) QT35 cells were cotransfected with 2 μg of RTBPSRAX alone (−) and with 300 ng of CELF5, CELF4, CELF3, ETR-3, or CUG-BP expression plasmid. Asterisk, cryptic splice observed in some PCRs in which the exon is included but the 30-nucleotide exon upstream of the alternative exon is skipped. The appearance of this cryptic splice is inconsistent and represents at most 15% of spliced mRNAs. It is unknown why this cryptic splice is enhanced most efficiently by ETR-3, but enhancement does not appear to be MSE dependent as it is observed for both MSE-containing and MSE-lacking minigenes. Loading was adjusted so that approximately equal counts were loaded in each lane.
Differential ability of CELF proteins to promote exon inclusion via MSE2 alone.
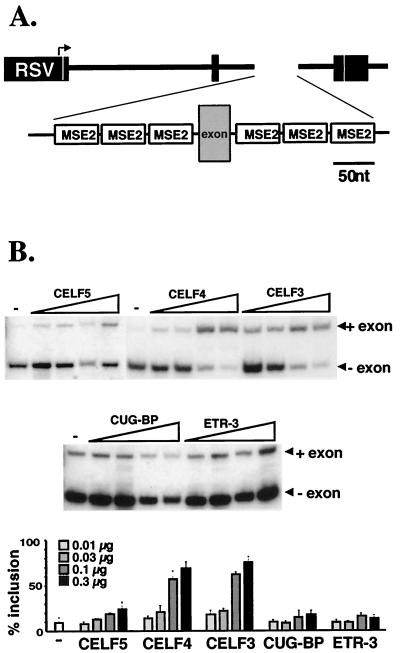
We previously reported that multiple copies of a single, 39-nucleotide MSE, MSE2, is sufficient to drive robust muscle-specific inclusion of a heterologous exon (10). To determine whether the CELF proteins can promote exon inclusion via this element alone, CUG-BP, ETR-3, CELF3, CELF4, and CELF5 were cotransfected in QT35 fibroblasts with the M2/M2TB minigene, which contains a 59-nucleotide alternative exon flanked on both sides by three copies of MSE2 (Fig. 5A). Both CELF3 and CELF4 promoted exon inclusion (Fig. 5B). CELF5 also promoted exon inclusion, but to a much lesser extent. In contrast, neither CUG-BP nor ETR-3 affected M2/M2TB exon inclusion (Fig. 5B), even when up to 10 μg of expression plasmid was cotransfected with the minigene (data not shown). This difference in ability to promote exon inclusion via MSE2 alone indicates a functional division of the CELF protein family into two subgroups, consistent with the division suggested by the sequence analysis discussed above.
FIG. 5.
CELF3, CELF4, and to a lesser extent CELF5, but not CUG-BP or ETR-3, can promote exon inclusion via MSE2 alone. (A) The M2/M2TB minigene contains a 52-nucleotide exon flanked upstream and downstream by three copies of the 39-nucleotide cTNT MSE2 from intron 5. RSV, Rous sarcoma virus. (B) QT35 cells were cotranfected with 2 μg of the M2/M2TB minigene and 0 (−), 10, 30, 100, or 300 ng of CELF5, CELF4, CELF3, CUG-BP, or ETR-3 expression plasmid. RT-PCR analysis was performed on RNA harvested 48 h posttransfection. Loading was adjusted so that approximately equal counts were loaded in each lane.
CELF protein expression is dynamic during striated muscle and brain development.
The expression of CUG-BP, ETR-3, and CELF4 in striated muscle and brain and the ability of these proteins to regulate the splicing of cTNT, a transcript that undergoes developmental stage-specific alternative splicing, prompted us to investigate whether the expression of these CELF proteins is developmentally regulated in muscle and brain. To examine the developmental profiles of the three widely expressed CELF proteins, Western blot analyses were performed on total protein samples collected from mice at different stages of embryonic and postnatal life using the antibodies against CUG-BP, ETR-3, and CELF4 (Fig. 6). All three family members displayed changes during development. The abundance of CUG-BP decreased significantly throughout skeletal muscle development to low levels in adult thigh muscle. CUG-BP remained constant during early stages of brain development and was reduced to a very low but detectable level in the adult brain. ETR-3 increased in abundance in skeletal muscle and brain and underwent a transition from high-molecular-mass isoforms (primarily a band at approximately 52 kDa) in embryonic limb to predominantly lower-molecular-mass isoforms (approximately 42 and 50 kDa) in adult thigh. CELF4 levels increased in adult skeletal muscle without any apparent change in isoform distribution, while a low-molecular-mass isoform (approximately 42 kDa) is lost in adult brain.
FIG. 6.
Developmental profile of CELF proteins in skeletal muscle and brain. Western blot analysis was performed on total protein samples extracted from embryonic-day-14 (e14), newborn (NB), postnatal-day-4 (d4), and adult pregnant or 0- to 4-day-postpartum female Swiss Webster mice. Fifty micrograms of total protein was loaded in each lane. In brain samples, high levels of a 30-kDa protein are observed on CELF4 blots (data not shown), suggesting that a high degree of tissue-specific cleavage or degradation of CELF4 may be occurring in brain as described for Fig. 2.
Changes in ETR-3 protein correlate with developmental changes in cTNT splicing in cardiac and skeletal muscle.
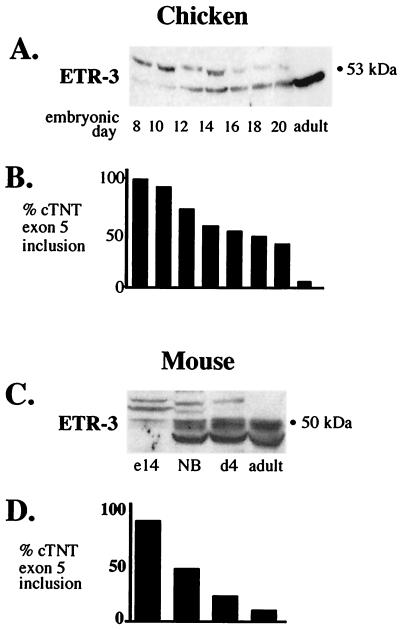
To test whether changes in CELF proteins occur concomitantly with changes in cTNT splicing during development, Western blot analyses were performed on total protein extracts from heart at different stages of development using antibodies against CUG-BP, ETR-3, and CELF4. Because the developmental switch in cTNT splicing is conserved among species, whole-tissue extracts were collected from both chickens and mice at various stages of embryonic and postnatal life. Primer extension or RT-PCR analysis was performed on parallel tissue samples to determine the extent of cTNT exon 5 inclusion over the same time course. In both chickens and mice, a dramatic transition from high- to low-molecular-weight ETR-3 isoforms concomitant with a switch from cTNT exon inclusion to exon skipping was observed (Fig. 7). Preliminary experiments indicate that this transition is not due to changes in phosphorylation, and we have cloned several alternatively spliced forms of ETR-3 from mouse heart tissue (data not shown). CUG-BP protein levels are somewhat reduced in the adult heart in both chickens and mice but do not change concomitantly with changes in cTNT splicing (data not shown). CELF4 is also slightly reduced in the adult chicken heart but does not undergo any apparent changes in the avian embryo or in the developing or adult mouse heart (data not shown). Therefore, the conserved developmental switch in cTNT splicing occurs concomitantly with a conserved switch in ETR-3 isoforms.
FIG. 7.
An ETR-3 isoform switch that correlates with cTNT splicing is conserved in avian and mammalian heart development. (A) ETR-3 Western blot of total protein samples extracted from embryonic and adult chicken hearts. Sixty micrograms of total proteins was loaded in each lane. (B) Results from primer extension of endogenous cTNT mRNA from the same time course using a 32P-labeled oligonucleotide complementary to exon 6. (C) ETR-3 Western blot of total protein samples from embryonic-day-14 (e14), newborn (NB), postnatal-day-4 (d4), and adult mouse hearts. Fifty micrograms of total protein was loaded per lane. (D) RT-PCR of endogenous cTNT was performed over the same time course and quantitated by PhosphorImager.
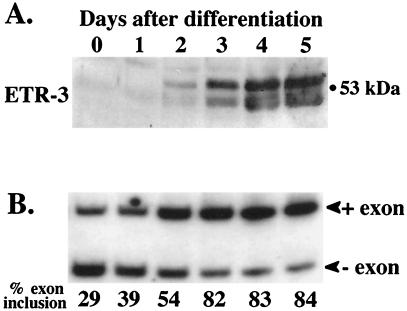
Muscle-specific splicing is induced as part of the myogenic program, since it can be induced in fibroblasts by expression of the myogenic regulators MyoD and myogenin (50). To monitor cTNT splicing during muscle differentiation, we transfected the R35C minigene containing MSEs 1 to 4 into C2C12 mouse myoblast cells induced to differentiate and monitored the extent of exon inclusion over time. An exogenous minigene was used to examine splicing during myoblast differentiation because endogenous cTNT mRNA cannot be detected until differentiation is well under way. As shown in Fig. 8B, cTNT exon inclusion is strongly induced during C2C12 differentiation. This effect is MSE dependent as alternative exons flanked by introns that lack MSEs maintain a constant level of exon inclusion (data not shown). Western blot analysis on total protein extracts from differentiating C2C12 cultures demonstrates a dramatic increase in ETR-3 protein expression during the same time course (Fig. 8A). CUG-BP and CELF4 protein levels were invariant during differentiation (41; data not shown).
FIG. 8.
Induction of ETR-3 protein expression and cTNT exon inclusion occur simultaneously during skeletal muscle differentiation. (A) ETR-3 Western blot of total protein extracts from C2C12 murine myoblast cells induced to differentiate. Fifty micrograms of total protein was loaded in each lane. (B) RT-PCR analysis of cTNT minigene RNA from similar cultures transfected with R35C harvested over the same time course. Loading was adjusted so that approximately equal counts were loaded in each lane.
DISCUSSION
A novel family of splicing regulators.
In this study, we provide evidence that members of the CELF family of RNA binding proteins are regulators of cell-specific alternative splicing. This family includes CUG-BP, a protein that has previously been reported to bind to a conserved intronic splicing element and regulate alternative splicing in a sequence-dependent manner (46). By screening human EST databases, we identified four paralogs of CUG-BP including ETR-3, originally identified in a fetal heart library (25), and three novel factors we called CELF3, CELF4, and CELF5. Following submission of the manuscript of this report, another group reported the identification of these proteins and called them Bruno-like (BRUNOL) proteins (17). CUG-BP, ETR-3, and CELF4 all bind to RNA substrates containing chicken cTNT MSEs, and all five CELF proteins promote MSE-dependent exon inclusion in chicken cTNT minigene pre-mRNAs in quail fibroblast cells. The ability of the human CELF proteins to regulate MSE-dependent alternative splicing of avian cTNT pre-mRNAs, the conservation in sequence and position of MSE2 in chicken and human cTNT (46), and the high degree of conservation in the splicing pattern of cTNT transcripts in birds and mammals (11, 28, 39, 60) all indicate that MSE-dependent regulation of alternative splicing by the CELF family is a conserved mechanism of regulating gene expression during development in vertebrates.
Several muscle-specific genes whose pre-mRNAs undergo regulated alternative splicing contain potential MSE sequences (10), suggesting that the CELF family may be involved in the regulation of the alternative splicing of a subset of genes in muscle. Although the total abundance of the three widely expressed CELF proteins was particularly high in striated muscle tissues, every tissue examined by Western blot analysis contains at least one CELF family member. Dot blots indicate that CELF3 and CELF5 are restricted to brain. Together, these results suggest that each CELF protein may play a distinct role in alternative splicing in different tissues or act on different target pre-mRNAs within tissues where more than one family member is present.
The CELF proteins differ in their abilities to activate splicing by MSE2 alone. We have previously shown that CUG-BP binds to an MSE2-like sequence downstream of human cTNT exon 5 in correlation with regulation in vivo (46). CUG-BP (and ETR-3) failed, however, to activate inclusion of an exon flanked by multiple copies of MSE2 from the chicken cTNT gene. It should be noted that, in the previous study, MSE2 was in the context of the natural introns, which presumably contain additional regulatory elements. Although the binding of CUG-BP to the human MSE2-like sequence is clearly required for regulation, the results presented in Fig. 5 indicate that MSE2 binding is not sufficient. Additional elements flanking MSE2, different spacing between elements, or elements within human MSE2 not found in chicken MSE2 are required for CUG-BP function.
CELF proteins as regulators of alternative splicing during normal development and disease.
The shift in alternative splicing of cTNT exon 5 from exon inclusion in the embryo to exon skipping in adult striated muscle is highly conserved in birds and mammals (11, 28, 39, 60). Western blot analysis demonstrates that in both chicken and mouse heart, ETR-3 undergoes a developmental transition from high- to low-molecular-weight isoforms concomitant with the switch to cTNT exon 5 skipping. CUG-BP and CELF4 remain largely invariant over this same time course, strongly suggesting that ETR-3 may be the primary regulator of cTNT alternative splicing in vivo. Preliminary evidence suggests that the embryonic and adult ETR-3 isoforms observed in heart are generated by alternative splicing, and at least some of the larger ETR-3 isoforms expressed in embryonic mouse heart are also present in differentiated C2C12 cells (A. N. Ladd and T. A. Cooper, unpublished observations). Different splice forms of ETR-3 may differ in their abilities to recognize or bind cTNT RNA, their abilities to promote exon 5 inclusion, or their nuclear and cytoplasmic localizations. We have cloned several different ETR-3 splice variants and are currently addressing this issue. It is possible that a different splicing factor is responsible for regulating both the switch in cTNT splicing and the transition in ETR-3 isoforms during heart development. ETR-3 mRNAs contain potential binding sites for CUG-BP and ETR-3 (36), which raises the possibility of CELF protein autoregulation or cross talk in the developing heart, where CUG-BP, ETR-3, and CELF4 are all highly expressed. There is also a striking correlation, however, between the level of ETR-3 protein (but not CUG-BP or CELF4) and the extent of cTNT exon 5 inclusion during differentiation of myoblasts into myotubes. Preliminary studies of variants cloned from mouse heart indicate that high-molecular-weight isoforms promote cTNT exon inclusion (A. N. Ladd and T. A. Cooper, unpublished observations). Both high- and low-molecular-weight isoforms of ETR-3 protein are induced concomitantly with the increase in cTNT exon inclusion during C2C12 differentiation. Therefore, in two independent systems (heart development and skeletal muscle differentiation), changes in cTNT alternative splicing directly correlate with changes in the expression of high-molecular-weight ETR-3 protein isoforms, ETR-3 shows sequence-specific binding to a conserved intronic element demonstrated to be required for regulated splicing of cTNT exon 5, and coexpression of ETR-3 with cTNT minigenes promotes element-dependent exon inclusion in fibroblasts. Together, these results strongly suggest that ETR-3 plays a major role in cTNT alternative splicing regulation.
cTNT is likely to be just one of multiple targets of CELF protein regulation. Although neither CUG-BP nor CELF4 expression correlated with changes in cTNT splicing in the heart, both underwent developmental changes in skeletal muscle. The abundance of CUG-BP decreased significantly during thigh muscle development, while CELF4 levels increased dramatically during this same period. CUG-BP, ETR-3, and CELF4 also displayed changes in expression during brain development. This is consistent with their involvement in the developmental-stage-specific regulation of other target pre-mRNAs. CUG-BP has been suggested to play a role in neuron-specific splicing of an exon in the γ2 of GABAA transcripts; GABAA gene expression is known to undergo changes during brain development (67). The identification of other CELF family target genes will be an important goal for future studies.
CELF family members are likely to play a critical role during development. ETR-1, a putative homolog of CUG-BP in Caenorhabditis elegans, is essential for muscle differentiation (42). CUG-BP has also been implicated in the pathogenesis of myotonic dystrophy (59), a neuromuscular disease caused by an expansion of an unstable region of CTG repeats in the 3′ UTR of the DMPK gene (2, 15, 37). Congenital patients have numerous developmental problems, including arrested or delayed muscle maturation (55), the absence or reduction of some muscle fiber types, and mental retardation (22). Overexpression of DMPK transcripts containing expanded CUG repeats suppresses myogenic differentiation in culture (51, 61), suggesting a direct link between CUG expansion and the developmental defects observed in congenital patients.
The importance of CELF family function is probably not diminished in adulthood. DM1 is the most common form of adult onset muscular dystrophy and is associated with manifestations in several different tissues. The most affected are striated muscle and brain, the tissues that demonstrate the highest levels of CELF protein expression. It has been proposed that the expanded CUG repeats in the DMPK transcripts of affected individuals create a gain-of-function mutation that affects the processing of other mRNAs by disrupting RNA processing proteins (62). Consistent with this model, DMPK is actively transcribed and both expanded DMPK transcripts (12) and the hypophosphorylated isoform of CUG-BP (59) accumulate in the nuclei of DM1 cells. The splicing of cTNT is also disrupted in striated-muscle cultures from DM1 patients and in normal cells expressing transcripts with large CUG repeats (46), consistent with the altered function of CELF proteins. If CELF family function is disrupted in DM1, the missplicing of a battery of target pre-mRNAs including cTNT could explain many of the different manifestations of the disease in affected individuals.
Other potential functions of the CELF proteins.
Phylogenetic analysis indicates that the CELF family is most closely related to the Hu family, which regulates mRNA stability and translation (reviewed in reference 32). Embryo deadenylation element binding protein, a homolog of CUG-BP identified in Xenopus laevis, has been shown to regulate mRNA deadenylation (45). Bruno, a Drosophila protein similar to CUG-BP, regulates translation of oskar mRNAs not localized to the posterior pole of the oocyte (33). Based on the similarities of the mammalian CELF proteins to these factors, it is likely that CELF protein function is not limited to regulation of alternative splicing. Other splicing regulatory factors have been implicated in transport or processing of mRNAs within the cytoplasm, including PTB (29), hnRNP A1 (47), and several SR proteins (6). CUG-BP is known to be localized in both the nucleus and cytoplasm (49), consistent with its having roles in both nuclear and cytoplasmic RNA processing events such as splicing, translation, regulation of mRNA stability, and mRNA shuttling. Indeed, the loss of cytoplasmic CUG-BP in DM1 cells may contribute as much to the pathogenesis of DM1 through changes in translation or mRNA stability as the changes in alternative splicing caused by its accumulation in the nucleus.
ACKNOWLEDGMENTS
We thank Lubov Timchenko, C. C. Liew, and Maurice Swanson for kind gifts of reagents or clones, Claire Lo, Gopal Singh, and Wade Haaland for their technical assistance, and Sue Berget, Rajesh Savkur, Chris Smith, and Maurice Swanson for helpful discussions on the manuscript.
This work was supported by grants to T.A.C. from the Muscular Dystrophy Association and NIH (AR45653). A.N.L. was supported by a postdoctoral NRSA fellowship from the NIH.
REFERENCES
- 1.Black D L. Activation of c-src neuron-specific splicing by an unusual RNA element in vivo and in vitro. Cell. 1992;69:795–807. doi: 10.1016/0092-8674(92)90291-j. [DOI] [PubMed] [Google Scholar]
- 2.Brook J D, McCurrach M E, Harley H G, Buckler A J, Church D, Aburatani H, Hunter K, Stanton V P, Thirion J P, Hudson T, Sohn R, Zemelman B, Snell R G, Rundle S A, Crow S, Davies J, Shelbourne P, Buxton J, Jones C, Juvonen V, Johnson K, Harper P S, Shaw D J, Housman D E. Molecular basis of myotonic dystrophy—expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 3.Buckanovich R J, Darnell R B. The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol Cell Biol. 1997;17:3194–3201. doi: 10.1128/mcb.17.6.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckanovich R J, Posner J B, Darnell R B. Nova, the paraneoplastic Ri antigen, is homologous to an RNA-binding protein and is specifically expressed in the developing motor system. Neuron. 1993;11:657–672. doi: 10.1016/0896-6273(93)90077-5. [DOI] [PubMed] [Google Scholar]
- 5.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 6.Cáceres J F, Screaton G R, Krainer A R. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi D K, Ito T, Mitsui Y, Sakaki Y. Fluorescent differential display analysis of gene expression in apoptotic neuroblastoma cells. Gene. 1998;223:21–31. doi: 10.1016/s0378-1119(98)00364-3. [DOI] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.Cooper T A. In vitro splicing of cardiac troponin T precursors. Exon mutations disrupt splicing of the upstream intron. J Biol Chem. 1992;267:5330–5338. [PubMed] [Google Scholar]
- 10.Cooper T A. Muscle-specific splicing of a heterologous exon mediated by a single muscle-specific splicing enhancer from the cardiac troponin T gene. Mol Cell Biol. 1998;18:4519–4525. doi: 10.1128/mcb.18.8.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper T A, Ordahl C P. A single cardiac troponin T gene generates embryonic and adult isoforms via developmentally regulated alternate splicing. J Biol Chem. 1985;260:11140–11148. [PubMed] [Google Scholar]
- 12.Davis B M, McCurrach M E, Taneja K L, Singer R H, Housman D E. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc Natl Acad Sci USA. 1997;94:7388–7393. doi: 10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu X D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 15.Fu Y H, Pizzuti A, Fenwick R G, King J, Rajnarayan S, Dunne P W, Dubel J, Nasser G A, Ashizawa T, Dejong P, Wieringa B, Korneluk R, Perryman M B, Epstein H F, Caskey C T. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science. 1992;255:1256–1258. doi: 10.1126/science.1546326. [DOI] [PubMed] [Google Scholar]
- 16.Godt R E, Fogaca R T H, Silva I K, Nosek T M. Contraction of developing avian heart muscle. Comp Biochem Physiol [A] 1993;105:213–218. doi: 10.1016/0300-9629(93)90197-c. [DOI] [PubMed] [Google Scholar]
- 17.Good P J, Chen Q, Warner S J, Herring D. A family of human RNA-binding proteins related to the Drosophila Bruno translational regulator. J Biol Chem. 2000;275:28583–28592. doi: 10.1074/jbc.M003083200. [DOI] [PubMed] [Google Scholar]
- 18.Gooding C, Roberts G C, Moreau G, Nadal-Ginard B, Smith C W. Smooth muscle-specific switching of alpha-tropomyosin mutually exclusive exon selection by specific inhibition of the strong default exon. EMBO J. 1994;13:3861–3872. doi: 10.1002/j.1460-2075.1994.tb06697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo W, Mulligan G J, Wormsley S, Helfman D M. Alternative splicing of beta-tropomyosin pre-mRNA: cis-acting elements and cellular factors that block the use of a skeletal muscle exon in nonmuscle cells. Genes Dev. 1991;5:2096–2107. doi: 10.1101/gad.5.11.2096. [DOI] [PubMed] [Google Scholar]
- 20.Hanamura A, Caceres J F, Mayeda A, Franza B R, Jr, Krainer A R. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- 21.Hanke J, Brett D, Zastrow I, Aydin A, Delbruck S, Lehmann G, Luft F, Reich J, Bork P. Alternative splicing of human genes: more the rule than the exception? Trends Genet. 1999;15:389–390. doi: 10.1016/s0168-9525(99)01830-2. [DOI] [PubMed] [Google Scholar]
- 22.Harper P S. Myotonic dystrophy as a trinucleotide repeat disorder—a clinical perspective. In: Wells R D, Warren S T, editors. Genetic instabilities and hereditary neurological diseases. Boston, Mass: Academic Press; 1998. pp. 115–130. [Google Scholar]
- 23.Higgins D G, Sharp P M. Fast and sensitive multiple sequence alignments on a microcomputer. Comput Appl Biosci. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- 24.Hofmann K, Bucher P, Falquet L, Bairoch A. The PROSITE database, its status in 1999. Nuceic Acids Res. 1999;27:215–219. doi: 10.1093/nar/27.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang D M, Hwang W S, Liew C C. Single pass sequencing of a unidirectional human fetal heart cDNA library to discover novel genes of the cardiovascular system. J Mol Cell Card. 1994;26:1329–1333. doi: 10.1006/jmcc.1994.1151. [DOI] [PubMed] [Google Scholar]
- 26.Ichida M, Endo H, Ikeda U, Matsuda C, Ueno E, Shimada K, Kagawa Y. MyoD is indispensable for muscle-specific alternative splicing in mouse mitochondrial ATP synthase gamma-subunit pre-mRNA. J Biol Chem. 1998;273:8492–8501. doi: 10.1074/jbc.273.14.8492. [DOI] [PubMed] [Google Scholar]
- 27.Jensen K B, Dredge B K, Stefani G, Zhong R, Buckanovich R J, Okano H J, Yang Y Y, Darnell R B. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000;25:359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 28.Jin J P, Lin J J. Isolation and characterization of cDNA clones encoding embryonic and adult isoforms of rat cardiac troponin T. J Biol Chem. 1989;264:14471–14477. [PubMed] [Google Scholar]
- 29.Kaminski A, Hunt S L, Patton J G, Jackson R J. Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encepthalomyocarditis virus RNA. RNA. 1995;1:924–938. [PMC free article] [PubMed] [Google Scholar]
- 30.Kamma H, Portman D S, Dreyfuss G. Cell type-specific expression of hnRNP proteins. Exp Cell Res. 1995;221:187–196. doi: 10.1006/excr.1995.1366. [DOI] [PubMed] [Google Scholar]
- 31.Kanopka A, Muhlemann O, Petersen-Mahrt S, Estmer C, Ohrmalm C, Akusjarvi G. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature. 1998;393:185–187. doi: 10.1038/30277. [DOI] [PubMed] [Google Scholar]
- 32.Keene J D. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc Natl Acad Sci USA. 1999;96:5–7. doi: 10.1073/pnas.96.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim-Ha J, Kerr K, Macdonald P M. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell. 1995;81:403–412. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- 34.Lin C H, Patton J G. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA. 1995;1:234–245. [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez A J. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu Rev Genet. 1998;32:279–305. doi: 10.1146/annurev.genet.32.1.279. [DOI] [PubMed] [Google Scholar]
- 36.Lu X, Timchenko N A, Timchenko L T. Cardiac elav-type RNA-binding protein (ETR-3) binds to RNA CUG repeats expanded in myotonic dystrophy. Hum Mol Genet. 1999;8:53–60. doi: 10.1093/hmg/8.1.53. [DOI] [PubMed] [Google Scholar]
- 37.Mahadevan M, Tsilfidis C, Sabourin L, Shutler G, Amemiya C, Jansen G, Neville C, Narang M, Barcelo J, Ohoy K, Leblond S, Earlemacdonald J, Dejong P J, Wieringa B, Korneluk R G. Myotonic dystrophy mutation—an unstable CTG repeat in the 3′ untranslated region of the gene. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- 38.Margolis R L, Abraham M R, Gatchell S B, Li S H, Kidwai A S, Breschel T S, Stine O C, Callahan C, McInnis M G, Ross C A. cDNAs with long CAG trinucleotide repeats from human brain. Hum Genet. 1997;100:114–122. doi: 10.1007/s004390050476. [DOI] [PubMed] [Google Scholar]
- 39.McAuliffe J J. Delineation of the cardiac troponin T expression pattern during murine development. J Cell Biochem. 1994;18(Suppl.):W347. [Google Scholar]
- 40.McAuliffe J J, Gao L Z, Solaro R J. Changes in myofibrillar activation and troponin C Ca2+ binding associated with troponin T isoform switching in developing rabbit heart. Circ Res. 1990;66:1204–1216. doi: 10.1161/01.res.66.5.1204. [DOI] [PubMed] [Google Scholar]
- 41.Miller J W, Urbinati C R, Teng-Umnuay P, Stenberg M G, Byrne B J, Thornton C A, Swanson M S. Recruitment of human muscleblind proteins to (CUG)n expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milne C A, Hodgkin J. ETR-1, a homologue of a protein linked to myotonic dystropy, is essential for muscle development in Caenorhabditis elegans. Curr Biol. 1999;9:1243–1246. doi: 10.1016/s0960-9822(99)80504-1. [DOI] [PubMed] [Google Scholar]
- 43.Min H, Turck C W, Nikolic J M, Black D L. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- 44.Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paillard L, Omilli F, Legagneux V, Bassez T, Maniey D, Osborne H B. EDEN and EDEN-BP, a cis element and an associated factor that mediate sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 1998;17:278–287. doi: 10.1093/emboj/17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Philips A V, Timchenko L T, Cooper T A. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 47.Piñol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 48.Polydorides A D, Okano H J, Yang Y Y, Stefani G, Darnell R B. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc Natl Acad Sci USA. 2000;97:6350–6355. doi: 10.1073/pnas.110128397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roberts R, Timchenko N A, Miller J W, Reddy S, Caskey C T, Swanson M S, Timchenko L T. Altered phosphorylation and intracellular distribution of a (CUG)n triplet repeat RNA-binding protein in patients with myotonic dystrophy and in myotonin protein kinase knockout mice. Proc Natl Acad Sci USA. 1997;94:13221–13226. doi: 10.1073/pnas.94.24.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ryan K J, Cooper T A. Muscle-specific splicing enhancers regulate inclusion of the cardiac troponin T alternative exon in embryonic skeletal muscle. Mol Cell Biol. 1996;16:4014–4023. doi: 10.1128/mcb.16.8.4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sabourin L A, Tamai K, Narang M A, Korneluk R G. Overexpression of 3′-untranslated region of the myotonic dystrophy kinase cDNA inhibits myoblast differentiation in vitro. J Biol Chem. 1997;272:29626–29635. doi: 10.1074/jbc.272.47.29626. [DOI] [PubMed] [Google Scholar]
- 52.Saitoh O, Olson E N, Periasamy M. Muscle-specific RNA processing continues in the absence of myogenin expression. J Biol Chem. 1990;265:19381–19384. [PubMed] [Google Scholar]
- 53.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 54.Sanford J R, Bruzik J P. Developmental regulation of SR protein phosphorylation and activity. Genes Dev. 1999;13:1513–1518. doi: 10.1101/gad.13.12.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sarnat H B, Silbert S W. Maturational arrest of fetal muscle in neonatal myotonic dystrophy. A pathologic study of four cases. Arch Neurol. 1976;33:466–474. doi: 10.1001/archneur.1976.00500070008002. [DOI] [PubMed] [Google Scholar]
- 56.Schultz J, Milpetz F, Bork P, Ponting C P. SMART, a simple modular architecture research tool: identification of signalling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Screaton G R, Caceres J F, Mayeda A, Bell M V, Plebanski M, Jackson D G, Bell J I, Krainer A R. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 1995;14:4336–4349. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh R, Valcarcel J, Green M R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 59.Timchenko L T, Miller J W, Timchenko N A, Devore D R, Datar K V, Lin L J, Roberts R, Caskey C T, Swanson M S. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Townsend P J, Barton P J R, Yacoub M H, Farza H. Molecular cloning of human cardiac troponin T isoforms: expression in developing and failing heart. J Mol Cell Card. 1995;27:2223–2236. doi: 10.1016/s0022-2828(95)91587-7. [DOI] [PubMed] [Google Scholar]
- 61.Usuki F, Ishiura S, Saitoh N, Sasagawa N, Sorimachi H, Kuzume H, Maruyama K, Terao T, Suzuki K. Expanded CTG repeats in myotonin protein kinase suppresses myogenic differentiation. Neuroreport. 1997;8:3749–3753. doi: 10.1097/00001756-199712010-00018. [DOI] [PubMed] [Google Scholar]
- 62.Wang J, Pegoraro E, Menegazzo E, Gennarelli M, Hoop R C, Angelini C, Hoffman E P. Myotonic dystrophy: evidence for a possible dominant-negative RNA mutation. Hum Mol Genet. 1995;4:599–606. doi: 10.1093/hmg/4.4.599. [DOI] [PubMed] [Google Scholar]
- 63.Xie W Q, Rothblum L I. Rapid, small-scale RNA isolation from tissue culture cells. BioTechniques. 1991;11:324–327. [PubMed] [Google Scholar]
- 64.Xu R, Teng J, Cooper T A. The cardiac troponin T alternative exon contains a novel purine-rich positive splicing element. Mol Cell Biol. 1993;13:3660–3674. doi: 10.1128/mcb.13.6.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zahler A M, Neugebauer K M, Lane W S, Roth M B. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science. 1993;260:219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- 66.Zhang L, Askiya M, Sherman T G, Grabowski P J. Essential nucleotides direct neuron-specific splicing of gamma-2 pre-mRNA. RNA. 1996;2:682–698. [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang L, Liu W, Grabowski P J. Coordinate repression of a trio of neuron-specific splicing events by the splicing regulator PTB. RNA. 1999;5:117–130. doi: 10.1017/s1355838299981530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang W-J, Wu J Y. Functional properties of p54, a novel SR protein active in constitutive and alternative splicing. Mol Cell Biol. 1996;16:5400–5408. doi: 10.1128/mcb.16.10.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]