Abstract
Hypoxic stress, like DNA damage, induces p53 protein accumulation and p53-dependent apoptosis in oncogenically transformed cells. Unlike DNA damage, hypoxia does not induce p53-dependent cell cycle arrest, suggesting that p53 activity is differentially regulated by these two stresses. Here we report that hypoxia induces p53 protein accumulation, but in contrast to DNA damage, hypoxia fails to induce endogenous downstream p53 effector mRNAs and proteins. Hypoxia does not inhibit the induction of p53 target genes by ionizing radiation, indicating that p53-dependent transactivation requires a DNA damage-inducible signal that is lacking under hypoxic treatment alone. At the molecular level, DNA damage induces the interaction of p53 with the transcriptional activator p300 as well as with the transcriptional corepressor mSin3A. In contrast, hypoxia primarily induces an interaction of p53 with mSin3A, but not with p300. Pretreatment of cells with an inhibitor of histone deacetylases that relieves transcriptional repression resulted in a significant reduction of p53-dependent transrepression and hypoxia-induced apoptosis. These results led us to propose a model in which different cellular pools of p53 can modulate transcriptional activity through interactions with transcriptional coactivators or corepressors. Genotoxic stress induces both kinds of interactions, whereas stresses that lack a DNA damage component as exemplified by hypoxia primarily induce interaction with corepressors. However, inhibition of either type of interaction can result in diminished apoptotic activity.
The critical role of p53 in tumor suppression is underscored by the findings that the p53 gene is mutated in over 50% of human cancers (17) and that mice nullizigous for the p53 gene develop tumors early in their lifetime (10). Two functions of p53 that have been proposed to be responsible for its role as a tumor suppressor are the induction of cell cycle arrest in response to DNA damage and the induction of apoptosis. Following DNA damage, p53 binds to DNA in a sequence-dependent manner, and through interactions with the transcriptional coactivator p300 (also called CBP) (4, 27) as well as basal transcription factors like TFIID (13), it induces the transcription of downstream effector genes whose products interact with and inhibit proteins involved in cell-cycle regulation (for a review, see 26).
The molecular events that lead to p53-dependent apoptosis are less clear. p53-dependent transactivation has been reported to induce apoptosis in some experimental systems (9, 53, 55). However, under other conditions it appears that macromolecular synthesis is completely dispensable for the induction of p53-dependent apoptosis (7, 51). Furthermore, expression of p53 mutants that lack transactivation capability is able to induce apoptosis in certain cell lines (9, 21, 40), and deletion of the polyproline-rich domain of p53 which is located between the transactivation and DNA binding domains of p53 abrogates the apoptotic properties of p53 but does not affect DNA binding, bax induction, or cell cycle inhibition (43). There is also increasing evidence that p53-dependent transrepression may also contribute to the induction of apoptosis (35, 36, 39, 41, 50). In addition, the interaction of p53 with two proteins, XPB and XPD, that are components of the basal transcription factor TFIIH, has been suggested to be essential for UV-induced apoptosis of human fibroblasts (52). The consensus from the above findings is that different mechanisms of p53-mediated apoptosis may exist which may function coordinately or independently in different experimental systems depending on factors such as cell type, type of stress, levels of p53, and oncogenic activity.
Accumulation of the p53 protein following genotoxic stress involves posttranscriptional mechanisms such as enhanced translation of p53 mRNA and decreased proteolytic degradation of the protein (26, 28, 33). Activation of p53 following genotoxic damage is achieved by induction of p53 levels and by modifications of the p53 protein such as phosphorylation and acetylation (reviewed in references 14 and 38). Phosphorylation of serines 15 and 20 following genotoxic stress (8, 29, 44–46, 48) has been shown to impair interaction between p53 and Mdm-2, resulting in enhanced p53 accumulation (38, 44, 48), suggesting another means of modulating p53-dependent apoptosis. p53 is also extensively phosphorylated at other sites in vitro and in vivo in response to genotoxic damage (reviewed in references 14, 29, and 38). Although some of these posttranslational modifications increase the sequence-specific DNA binding activity of p53 and its transactivation properties in vitro, the physiological significance of these modifications in vivo remains to be determined.
Tumor hypoxia develops in most solid tumors as a result of inefficient vascular development, or abnormal vascular architecture (6). Previous studies have demonstrated that hypoxia is an independent prognostic factor of survival independent of other factors, including tumor grade or treatment modality (surgery or radiotherapy) (22). In contrast to DNA damage, hypoxia is a physiological inducer of the p53 tumor suppressor gene product (15) and can act as a selective pressure during tumor growth for the elimination of cells with wild-type (wt) p53 and the clonal expansion of cells with mutant or otherwise inactive p53 protein (16). This observation provides a possible explanation for the more aggressive nature of hypoxic tumors compared to well-oxygenated ones and for the frequent occurrence of p53 mutations in advanced stages of tumor development.
Little is known about the regulation of p53 function in oxygen-deprived cells and the role of known endogenous downstream effectors of p53 in this process. In this study we present evidence indicating that similar to genotoxic damage, hypoxia induces p53 protein accumulation and apoptosis. Unlike DNA damage, hypoxia fails to enhance the transactivation properties of p53. However, hypoxia does induce p53 transrepression activity, and inhibition of this activity significantly decreases p53-dependent apoptosis under hypoxic conditions.
MATERIALS AND METHODS
Cell lines.
Human breast carcinoma MCF-7, human colorectal carcinoma RKO, Epstein-Barr virus-transformed human lymphocytes GM2184B, and HT1299 cell lines were obtained from the American Type Culture Collection (Manassas, Va.). p53+/+ and p53-null mouse embryonic fibroblasts (MEFs) transformed with E1A and ras (15) were a gift from Scott Lowe (Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.).
Hypoxic treatments.
Treatment of cells with hypoxia was performed either in a hypoxic chamber (<0.2% O2) (Sheldon Corp., Cornelius, Oreg.) or in specially designed aluminum chambers for dose-response experiments (2 to 0.02% O2) and for experiments in which cells were irradiated under hypoxia.
Determination of apoptotic cells.
Cells with apoptotic morphology were identified by staining with Hoechst dye 33342 (blue) (5 μg/ml) for changes in nuclear characteristics and with propidium iodide (pink) (5 μg/ml) for loss of membrane integrity. Apoptotic values were calculated as the percentage of apoptotic cells relative to the total number of cells in each field (>100 cells) and represent the average of four randomly selected fields per 60-mm-diameter dish.
Immunoblotting and immunoprecipitation.
Primary antibodies used for immunoblotting were DO-1 mouse monoclonal for human p53, M-19 rabbit polyclonal for human p21, and SMP14 mouse monoclonal for human Mdm-2 (Santa Cruz Biotechnology, Santa Cruz, Calif.). The anti-mSin3A (K-20) and anti-p300 (N-15) rabbit polyclonal antibodies used for coimmunoprecipitations of p53 protein were purchased from Santa Cruz Biotechnology. For immunoprecipitation and immunoblotting, cell extracts were prepared by lysing 107 to 108 cells in buffer A, which contained 10 mM Tris (pH. 7.5), 1 mM EDTA, 400 mM NaCl, 10% glycerol, 0.5% NP-40, 5 mM NaF, 0.5 M Na3VO5, 1 mM dithiothreitol (DTT), and 1 mM phenylmethylsulfonyl fluoride (PMSF). Lysates were incubated on ice for 10 min and then centrifuged at 6,000 × g to remove cellular debris. Forty to fifty micrograms of protein was used for Western transfer and immunoblotting. For immunoprecipitations cell lysates (2 to 4 mg) were mixed with an equal volume of TEG buffer, which contained 10 mM Tris (pH 7.5), 1 mM EDTA, 20% glycerol, 5 mM NaF, 0.5 mM Na3VO5, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 3 μg of polyclonal antibody 1801 and rocked at 4°C overnight. Antibody-protein complexes were captured by addition of 40 μl of protein A–G-Plus beads (Santa Cruz Biotechnology) for 2 h. Immunoprecipitates were washed three times with buffer B, which contained 0.5 M Tris (pH 7.9), 250 mM KCl, 0.3% NP-40, 0.1% Triton X-100, 0.01% sodium dodecyl sulfate, and 1 mM DTT; resuspended in 3X SDS gel loading buffer, and analyzed on 9% polyacrylamide gels.
Immunofluorescent staining.
Immunofluorescent staining of RKO cells was performed as described by Graeber et al. (15). Cells were placed on Lab-Tek slides (Nunc, Naperville, Ill.), treated, and then fixed for 5 min with methanol previously stored at −20°C. After fixation, the slides were incubated for 20 min with 10% goat serum in phosphate-buffered saline (PBS). The slides were washed with PBS and then incubated with mouse anti-53 antibody (DO-1 at 1 μg/ml) for 60 min. After another wash, the slides were incubated in fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin (secondary) antibody (20 μg/ml; Caltag, South San Francisco, Calif.) for 60 min before a final wash. Coverslips were then mounted onto the slides, and the slides were photographed at a X600 magnification. As controls, slides that were stained with secondary antibody alone showed no significant fluorescence. All incubations were done at room temperature.
RNA isolation, Northern hybridization, and RPA.
Total RNA was isolated from 106 to 107 cells grown in monolayers using a modified guanidine isothiocyanate protocol (Trizol reagent; Gibco Life Sciences) according to the manufacturer's protocol. Northern hybridization assays were performed using 20 μg of total RNA. Radiolabeled probes to human and mouse α-tubulin, β-tubulin, and 18S RNA were synthesized with a random priming kit (Gibco BRL) from DNA fragments obtained by PCR amplification of mouse or human cDNA and gel purification of the DNA product. Ribonuclease protection assay (RPA) was performed with 10 μg of total RNA by using the RiboQuant kit and the human Stress-1 multiprobe template set (Pharmingen, San Diego, Calif.). Protected RNA-RNA hybrids were resolved on a 5% denaturing polyacrylamide gel and subjected to autoradiography. Quantitation was performed on a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Comet assay.
The comet assay was performed with RKO cells using a CometAssay kit (Trevigen, Gaithersburg, Md.) according to the manufacturer's instructions. Briefly, at the end of the treatment, cells were placed on ice, washed three times in ice-cold PBS, and counted. Approximately 250,000 cells were mixed with low-melting-point agarose (1%, wt/vol) and were placed on specially treated coverslips. Cells were lysed in a solution (2.5 M NaCl, 100 mM EDTA [pH 10], 10 mM Tris base, 1% sodium lauryl sarcosinate, 1% Triton X-100) for 30 min. Following cell lysis, the slides were placed in alkali buffer (NaOH, pH > 13) for 60 min, rinsed in Tris-borate-EDTA (TBE) buffer, and electrophoresed in 1× TBE buffer at 25 V for 20 min. The agarose was dehydrated by immersing it in 100% ethanol for 10 min and air dried. The DNA was visualized using SYBR-green dissolved in antifade solution and comets were analyzed on an epifluorescence microscope by capturing images with a charge-coupled device-cooled camera and analyzing individual comets with the Comet Analysis System (Loats Associates, Westminster, Md). Tail moment (percentage of DNA in the tail × length of migration) was automatically calculated after background correction for 75 comets in each experimental group. Results are reported as absolute tail moment measurement ± standard error of the mean (SEM).
RESULTS
Hypoxia induces p53 accumulation without increasing p21 protein levels.
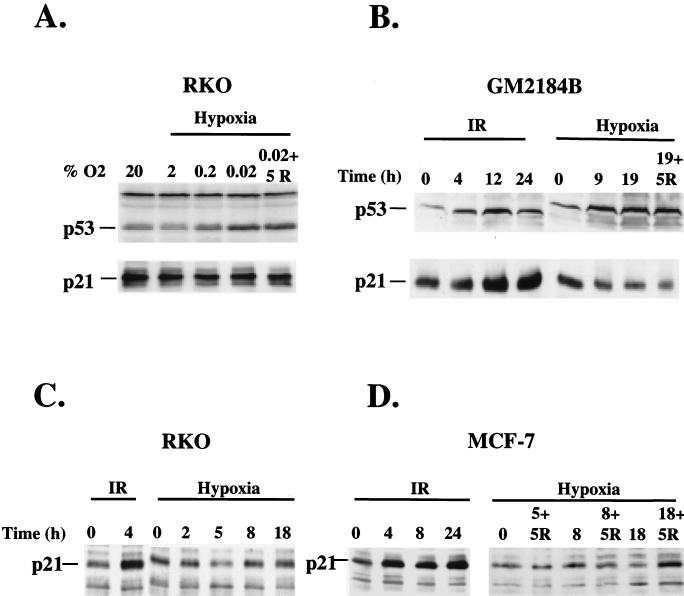
Activation of p53 protein by DNA damage is followed by increased synthesis of downstream effector proteins. The product of the p21–WAF-1–cip-1 gene (hereafter referred to as p21) exhibits the most consistent upregulation by p53 in response to DNA damage among various in vitro and in vivo systems (11, 37). Although hypoxia has been previously shown to induce p53 accumulation, it is not known whether p53 accumulation is followed by upregulation of p21 levels. To address this question we investigated the oxygen dependency of p53 and p21 accumulation in the wt p53 cell line RKO, which exhibits a p53-dependent upregulation of p21 after infrared (IR) treatment. Lowering oxygen levels from 20% to 0.02% by serial evacuations increased p53 protein levels (Fig. 1A). This increase was evident at O2 concentrations of 0.2%. However, none of the decreased oxygen levels resulted in increased p21 protein levels. In the human lymphocyte cell line GM2184, both IR treatment and hypoxia caused an increase in p53 levels of comparable magnitudes (Fig. 1B). However, while IR treatment of cells increased p21 levels, hypoxia decreased p21 levels. The same difference in the regulation of p21 levels by IR treatment and hypoxia was found in RKO and MCF-7 cells (Fig. 1C and D). Again, IR but not hypoxia increased p21 levels. These results demonstrate that the inability of hypoxia-induced p53 to induce p21 accumulation is not cell line specific but occurs in cell lines of different lineage.
FIG. 1.
Hypoxia induces p53 accumulation but uncouples it from p21 accumulation. (A) Western blot analysis of p53 and p21 levels from RKO exposed to different levels of hypoxia for 6 h or following 5 h of reoxygenation after hypoxia treatment. (B) Western blot analysis of p53 and p21 protein levels in GM2184B lymphoblasts with wt p53 following treatment with 6 Gy of IR or hypoxia for the indicated times. Also shown are western blot analyses of p21 levels in RKO cells (C) and in MCF-7 cells (D) after treatment with 6 Gy of IR or with hypoxia for the indicated times.
Hypoxia downregulates Mdm-2 protein levels.
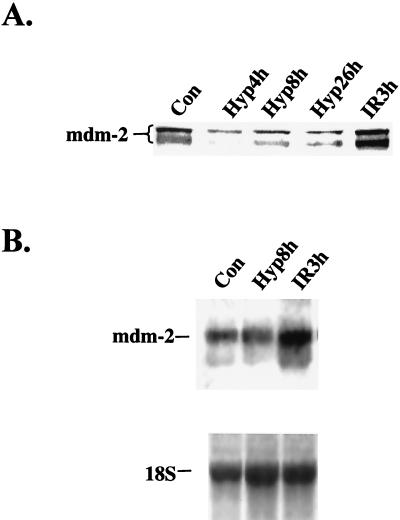
Another important p53-regulated effector is the product of the Mdm-2 oncogene. This transcriptional target of p53 participates in an autoregulatory feedback loop that regulates p53 levels by binding to the N terminus of p53, inhibiting its transactivation properties, and promoting its proteolytic degradation (20, 25, 33). In cells treated with hypoxia, Mdm-2 levels were markedly repressed at 4 h and remained repressed up to 26 h under hypoxia, while IR treatment induced a substantial increase in Mdm-2 levels (Fig. 2A). The decrease in Mdm-2 levels was not accompanied by a decrease in Mdm-2 mRNA levels (Fig. 2B), indicating that the decrease in Mdm-2 levels by hypoxia is due to a posttranscriptional mechanism. This decrease in Mdm-2 protein levels observed under hypoxia also suggests a likely mechanism for the increased accumulation of p53 by hypoxia (see Discussion).
FIG. 2.
IR but not hypoxia induces Mdm-2 protein and mRNA accumulation. (A) Time course of the regulation of Mdm-2 levels in cell extracts prepared at the indicated times under hypoxia or 3 h following treatment with 6 Gy of IR. (B) Northern blot analysis of Mdm-2 mRNA levels following treatment with hypoxia or 6 Gy of IR. Hybridization to 18S rRNA was used as a loading control (Con).
p53 induced by hypoxia fails to induce transactivation of endogenous downstream effectors.
Since hypoxia failed to increase p21 protein levels following p53 induction, we examined the ability of hypoxia-induced p53 to upregulate transcription of downstream effector genes. To address this question, we assayed for the levels of various mRNAs of known downstream p53 effectors after hypoxia or IR treatments. Such effectors include the cyclin-cdk inhibitor p21, the proapoptotic protein Bax, and the product of the growth arrest and DNA damage-inducible gene GADD-45. These effector genes have p53-responsive elements in their promoter regions and are induced in a p53-dependent manner following treatment with IR in a wide variety of cell lines and tissues (11, 23, 30, 31). To assay the levels of these products, we employed a multiribonuclease assay that allows the simultaneous detection of these mRNAs as well as the mRNA levels of housekeeping genes as an internal control in a single reaction.
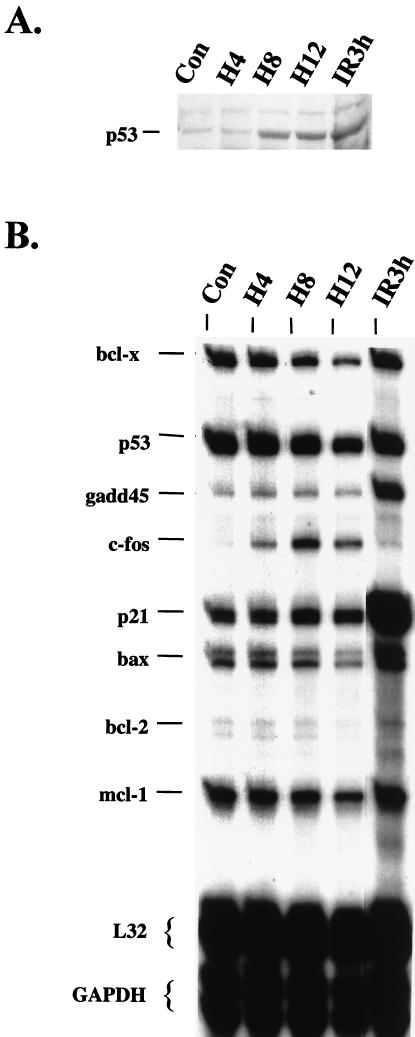
wt p53 MCF-7 cells were irradiated or exposed for different durations to hypoxia before protein and total RNA extraction. Induction of p53 occurred 8 h following hypoxia and reached levels comparable to those obtained 3 h after treatment with IR (Fig. 3A). However, hypoxia-induced p53 failed to increase the mRNA levels of the p21, GADD-45, or Bax genes (Fig. 3B). From the mRNAs assayed in this set, only the mRNA of the c-fos proto-oncogene increased under hypoxia, confirming previous observations that hypoxia is an inducer of this immediate-early gene (34). No change in p53 mRNA levels was observed under hypoxia, indicating that as with IR, accumulation of p53 protein by hypoxia is due to posttranscriptional mechanisms. These results indicate that hypoxia does not induce the transactivation of the same endogenous downstream effectors of p53 that DNA damage induces. Similar results were obtained in normal cervical epithelial cells, indicating that the lack of transactivation by hypoxia-induced p53 is not restricted to the transformed cell phenotype (Denko et al., unpublished results).
FIG. 3.
Hypoxia-induced p53 fails to transactivate endogenous effector gene mRNAs. MCF-7 cells were treated with hypoxia or with 6 Gy of IR, and at the times indicated whole-cell lysates and total RNA was prepared. (A) Western blot analysis of p53 levels following treatment of MCF-7 cells. (B) Multi-RPAs of mRNA levels of various p53 effectors and other genes involved in cellular stress and apoptotic responses. RPAs were performed in MCF-7 cells using RNA probes synthesized from a set containing various stress-induced genes (hStress set). Probes that are complementary to two housekeeping genes (ribosomal protein L32 and GAPDH) serve as normalization controls (Con).
A DNA damage-inducible signal activates p53 transactivation under hypoxia.
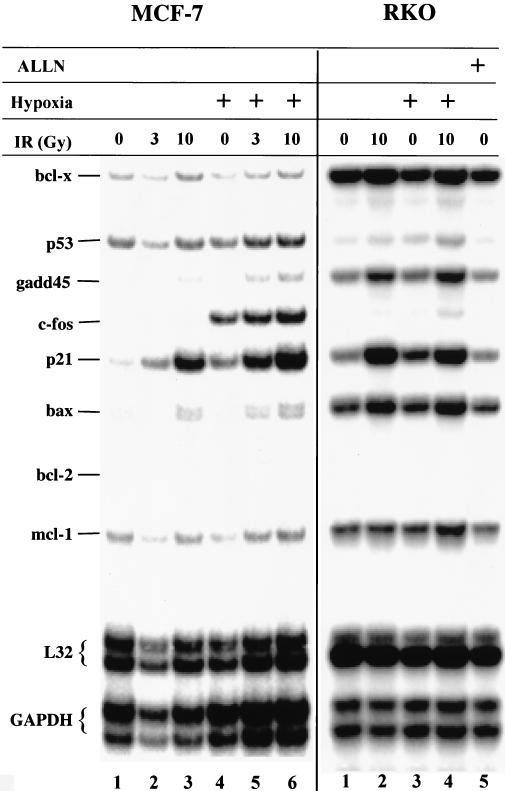
The lack of significant transactivation by hypoxia-inducible p53 could be explained by the following hypotheses: (i) hypoxia downregulates or inhibits the interaction of p53 with an accessory factor required for p53 transactivation, (ii) hypoxia-induced p53 or an accessory factor lacks a modification which is present in DNA damage-induced cells and which is necessary for transactivation, or (iii) both i and ii. To distinguish between these possibilities, we examined the transactivation properties of p53 in MCF-7 and RKO cells treated with IR or hypoxia or treated with IR while exposed to hypoxia. In MCF-7 cells, IR caused a significant dose-dependent induction of GADD-45, p21, and Bax mRNA levels (Fig. 4). Note that based on the levels of L32 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNAs, lane 2 is underloaded compared to control (lane 1) and the other lanes, by a factor of 2.2 Treatment with hypoxia for 9 h failed to increase GADD-45 or Bax levels, and increased p21 levels only 2- to 3-fold compared to 6- to 10-fold induction by 10 Gy of IR. However, in cells treated with IR while under hypoxia, a substantial increase in the levels of these same mRNAs was observed. Similar results to those found with MCF-7 cells were obtained in RKO cells, although the increases in p21 and Bax mRNA levels after combined hypoxia and IR treatments were less pronounced than those observed in MCF-7 cells (Fig. 4). This result suggests that hypoxia induces the accumulation of transcriptionally latent p53 that upon DNA damage becomes transcriptionally active. These results support the hypothesis that lack of p53-dependent transactivation is due not to a dominant, hypoxia-repressible event but rather to a lack of modification(s) of p53 and/or of accessory protein(s), which is induced by DNA damage.
FIG. 4.
Irradiation of MCF7 and RKO cells under hypoxia induces p53-dependent transactivation. Cells were grown in normoxic conditions (lanes 1 to 3, MCF-7, and lanes 1, 2, and 5, RKO) or exposed to hypoxia (lanes 4 to 6, MCF-7, and lanes 3 and 4, RKO). Cells in lanes 2, 3, 5, and 6 (MCF-7) and lanes 2 and 4 (RKO) received 3 or 10 Gy of IR, as indicated, 3 h before cell lysis. Cells in lane 5 (RKO) were treated with 20 μM ALLN for 3 h before cell lysis. Following cell lysis, total RNA was isolated and RPAs were performed as described in Materials and Methods.
Hypoxia induces nuclear accumulation of p53.
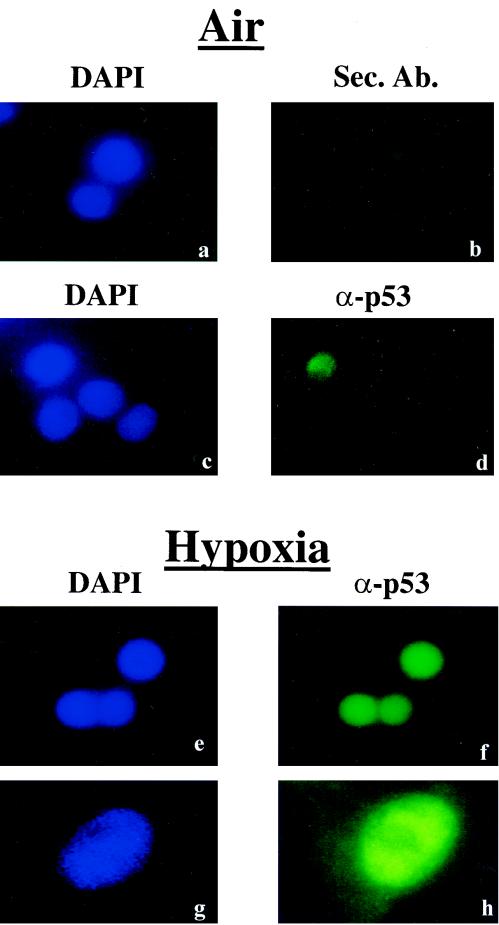
One mechanism that could explain the lack of p53-dependent transactivation of endogenous effector gene products under hypoxia is the nuclear exclusion of hypoxia-induced p53. This possibility is even more likely since the recent report by Ashcroft et al. (3) that p53 stabilized by hypoxia-mimicking agents like deferoxamine (DFO) was excluded from the nucleus. Although it was previously shown that hypoxia induced nuclear accumulation of p53 in untransformed human fibroblasts (15), it is still possible that in transformed cell lines like RKO, nuclear accumulation or transport of hypoxia-induced p53 is compromised. To address this question, we exposed RKO cells to hypoxia and subjected them to immunocytochemistry using the DO-1 monoclonal antibody. Under normoxia, a very low level of p53 immunoreactivity was present in these cells (Fig. 5d). After 12 h of hypoxia, a strong p53 signal that localized to the cell nuclei was evident (Fig. 5 f and h). These results indicate that as was the case in untransformed human fibroblasts, hypoxia-induced p53 accumulates in the nuclei of transformed cells and suggest that the mechanism of induction of p53 levels by DFO and physiological hypoxia are different.
FIG. 5.
Hypoxia-induced p53 is mainly localized to the nucleus. RKO cells were grown under normoxia (a to d), or exposed to hypoxia (e to h). Nuclei were visualized with DAPI (4′,6′-diamidino-2-phenylindole) counterstaining (a, c, e, and g), while p53 was visualized using the DO-1 monoclonal antibody and a fluorescein-conjugated mouse secondary antibody (d, f, and h). Panel b depicts fluorescence due to binding of the secondary antibody alone. Images in panels g and h were taken using a higher magnification objective (×60) than the other panels (×20).
Hypoxia induces p53-dependent transrepression.
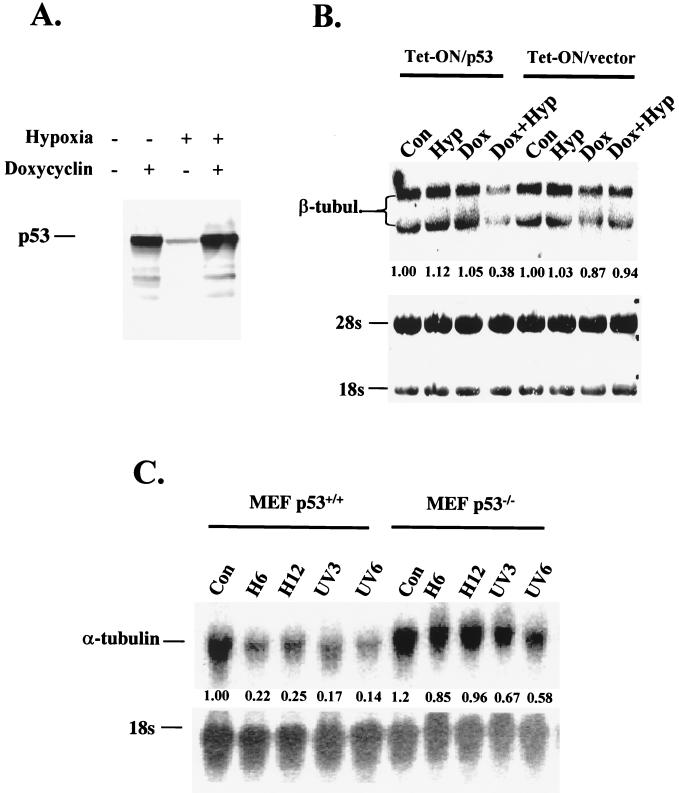
While p53 has been demonstrated to act as a transcriptional activator in response to DNA damage, it can also repress transcription of several downstream genes (1, 35–37, 41). Therefore, we examined the effect of hypoxia on the regulation of expression of genes that have been shown to be repressed in a p53-dependent manner. The mRNA for β-tubulin is repressed by p53 (35). To investigate whether the repression of β-tubulin is p53 dependent, we examined the effects of hypoxia on expression of β-tubulin mRNA in the p53-null cell line H1299, which was stably transfected with a p53-inducible gene construct under the control of a tetracycline-regulated promoter (Fig. 6A). A relatively low dose of doxycycline (200 ng/ml) was used to moderately induce p53 in these experiments. Under these conditions, we have found that p53 protein levels increase without any substantial induction of apoptosis compared to uninduced cells (data not shown). When H1299 cells were exposed to hypoxia in the presence of doxycycline, the levels of β-tubulin mRNA decreased to 38% of control levels. This decrease was p53 dependent, as in the absence of the p53-inducible gene hypoxia and doxycycline caused only a 6% decrease (Fig. 6B).
FIG. 6.
Hypoxia induces p53-dependent transrepression. (A) Doxycycline (200 ng/ml) induces p53 expression in H1299 cells transfected with a tet-inducible construct (clone 30) under normoxic and hypoxic conditions. The slight increase in p53 levels under hypoxia in the absence of doxycycline is probably due to stabilization of residual p53 expressed due to the “leakiness” of the construct. (B) Northern analysis of β-tubulin (β-tubul.) mRNA levels in an H1299 clone expressing a tet-inducible p53 construct and the parental clone transfected with empty vector. Cells were treated with hypoxia, doxycycline, or doxycycline for 2 h followed by treatment with hypoxia. In this cell line, hybridization with the β-tubulin probe results in two signals, probably because of alternative splicing of the β-tubulin. At the bottom, a methylene blue-stained nitrocellulose membrane, indicating levels of 18S and 28S RNA is shown. (C) Northern blot analysis of the p53-transrepressed gene coding for α-tubulin after treatment of p53+/+ and p53−/− MEFs with hypoxia or with 10 J of UV radiation/m2. Hybridization to 18S rRNA was used as a control. This is a light PhosphorImager scan that better represents the repression levels of α-tubulin. Values represent relative β-tubulin levels.
Another gene whose expression is repressed in a p53-dependent manner is α-tubulin. mRNA levels of this gene decrease in response to p53 induction that leads to growth arrest (36), or to apoptosis (37). We assayed for changes in α-tubulin mRNA levels in p53+/+ and p53−/− MEFs after treatment with hypoxia and UV. Hypoxia decreased α-tubulin mRNA levels to 22 and 25% of control levels after 8 and 16 h of treatment in the p53+/+ MEFs (Fig. 6C). UV treatment also caused a significant decrease in α-tubulin levels. In p53−/− cells, however, α-tubulin mRNA levels decreased only slightly in response to hypoxia (85% at 6 h and 96% at 12 h) and moderately in response to UV (67% at 3 h and 58% at 6 h). These results indicate that while p53-dependent transactivation is absent in cells exposed to hypoxia, p53 transrepression is still present.
Hypoxia fails to induce acetylation of Lys382 but induces phosphorylation of Ser15.
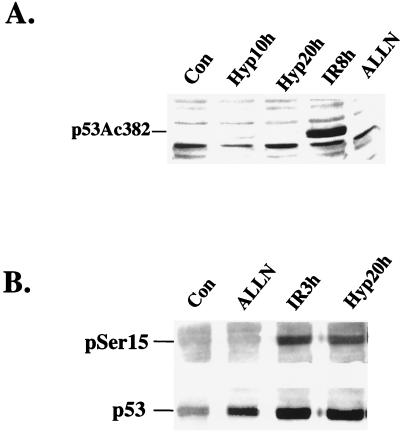
Activation of p53-dependent transcriptional activity by DNA-damaging agents has been associated with specific posttranscriptional modifications of the protein. The transcriptional coactivator p300 has been shown to interact with p53 and induce acetylation of its C terminus, at Lys320 and Lys382 (18, 42). Furthermore, acetylation of p53 is proposed to unmask the DNA binding domain of p53 and to increase its transactivation ability. We investigated the effects of hypoxia and IR on p53 acetylation using a rabbit polyclonal antibody that recognizes p53 acetylated at Lys382. RKO cells treated with hypoxia, IR, and the proteasome inhibitor ALLN were lysed, and extracts were subjected to immunoblotting using this antibody. ALLN induces accumulation of p53 protein by inhibiting its proteolytic degradation and does not induce p53-dependent transactivation (Fig. 4). As seen in Fig. 7A, only IR caused a significant increase in p53 protein acetylation, while 10 or 20 h of hypoxia did not result in an increase in p53 acetylation at this site. ALLN also failed to induce this modification.
FIG. 7.
Differences in p53 modifications induced by IR and hypoxia. (A) IR but not hypoxia induces acetylation of Lys382. RKO cells were treated with hypoxia, 6 Gy of IR, or 20 μM ALLN for the times indicated. Immunoblotting was performed with a rabbit polyclonal antibody raised against Lys382. (B) Both IR and hypoxia induce Ser15 phosphorylation of p53. Treatment times are indicated. The top panel shows an immunoblot using a rabbit polyclonal that recognizes p53 phosphorylated at Ser15. The bottom panel shows the same immunoblot after being stripped and reprobed with the DO-1 monoclonal antibody.
In contrast, hypoxia exhibited an ability similar to that of IR in stimulating phosphorylation of Ser15 (Fig. 7B). Human p53 is phosphorylated on at least two sites near the N terminus in response to DNA damage: at Ser15 and Ser20 (8, 29, 45, 46, 48). Phosphorylation on Ser15 has been shown to decrease the affinity of Mdm-2 for binding at the N terminus of the protein (44). Treatment of RKO cells with hypoxia for 20 h induced robust phosphorylation on Ser15 as determined by immunoblotting with a polyclonal antibody specific for phosphorylated Ser15. IR also caused Ser15 phosphorylation 3 h after treatment, while treatment with ALLN, a calpain I inhibitor that induces p53 accumulation by decreasing its proteolytic degradation (46), did not result in Ser15 phosphorylation. When the same immunoblot was stripped and reprobed with the DO-1 monoclonal antibody that recognizes wt p53 protein, a significant induction of p53 protein levels was observed in cells treated with ALLN, IR, and hypoxia, compared to untreated control cells. The phosphorylation of p53 on Ser15 was induced as early as 4 h of hypoxia and was present at comparable levels after 20 h of continuous exposure to hypoxia (Koumenis et al., unpublished observations). Therefore, phosphorylation of p53 on Ser15 is similar for both DNA damage and hypoxia.
Hypoxia fails to induce a p53-p300 interaction but promotes a strong interaction between p53 and the corepressor mSin3A.
The lack of p53-dependent transactivation and acetylation of p53 on Lys382 and the induction of p53-dependent transrepression by p53 under hypoxia suggest that hypoxia must influence the interaction of p53 with transcriptional accessory proteins in a unique manner. To further elucidate the mechanism of selective regulation of p53 function by hypoxia, we examined the interactions between p53 and transcriptional coactivators and corepressors. The transcriptional coactivator p300 has been shown to interact with a number of transcriptional activators and enhancers, including p53, and its function has been demonstrated to be indispensable for p53-dependent transactivation (4, 18, 27, 42). The transcriptional corepressor mSin3A, a member of the NCoR transcriptional repressor complex, is utilized by transcriptional repressors like the Mad-Max complex and nuclear hormone receptors and has recently been shown to interact strongly with p53 (36). mSin3A also interacts with histone deacetylases (HDACs), which promote deacetylation of core histones at the promoter region and induce transcriptional repression of specific genes (19).
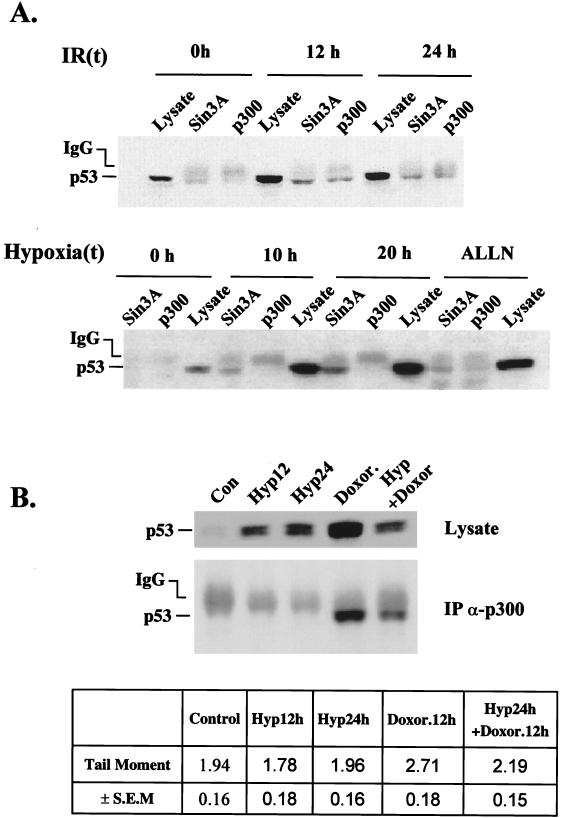
The interaction between p53 and p300 or p53 and mSin3A was examined in RKO cells treated with hypoxia, ALLN, or IR. Accumulation of p53 protein was strongly induced in these cells by hypoxia, ALLN, and IR in a dose-dependent manner (Fig. 8A). In extracts immunoprecipitated with an anti-p300 polyclonal antibody, p53 was present only in cells treated with IR, with maximal levels occurring at 8 h post-IR treatment. In contrast, in immunoprecipitates using an anti-mSin3A antibody, p53 was detected in cells treated with both hypoxia and IR and to a lesser extent in cells treated with ALLN. The strong interaction between p53 and mSin3A under hypoxic conditions was time dependent and followed kinetics similar to those of p53 induction. More significantly, higher levels of p53 were coimmunoprecipitated with mSin3A after 20 h of hypoxia compared to the levels following treatment with ALLN, suggesting that hypoxia induces a stress-specific modification that leads to a strong interaction between p53 and mSin3A. No change in the levels of p300 or mSin3A was observed following treatments with hypoxia (Koumenis et al., unpublished data).
FIG. 8.
Interactions of p53, p300, and mSin3A after hypoxia and ionizing radiation exposure. (A) Immunoprecipitation of mSin3a or p300 was performed on extracts from cells treated with 3 Gy of IR (top) or hypoxia-ALLN (bottom) using polyclonal antibodies against the corresponding proteins. One-tenth of the cell extract was loaded in a lane of each set to assay for p53 levels. The blot was probed with the DO-1 anti-p53 monoclonal antibody. (B) Immunoblot on cell lysate (top) and immunoprecipitation (IP) followed by immunoblotting (bottom) of p53 levels in extracts of cells treated with doxorubicin (100 ng/ml) for 12 h, hypoxia for 12 and 24 h, or both doxorubicin (12 h) and hypoxia (24 h). In the third case, doxorubicin was added during the last 12 h of hypoxia. The same anti-p300 polyclonal antibody (N-15) that was used in the top panel of B was used here. The table at the bottom of the figure indicates the tail moment values from cells treated as described above as determined by the Comet assay. Values are averages of 75 cells per treatment group and are reported along with SEMs for each group.
Since DNA damage is able to induce transactivation by hypoxia-inducible, transcriptionally latent p53, we hypothesized that DNA damage should be able to induce an interaction between hypoxia-induced p53 and p300. We treated RKO cells with doxorubicin, hypoxia, or a combination of both treatments (Fig. 8B). Doxorubicin was used as the DNA-damaging agent because we found that it induces higher levels of p53 accumulation and a stronger interaction between p53 and p300. Both doxorubicin and hypoxia induced p53 accumulation. The combination of the two treatments induced p53 accumulation that was no greater than the induction found by doxorubicin or hypoxia alone. When lysates were immunoprecipitated with an anti-p300 antibody, p53 was present in the extracts treated with doxorubicin but not with hypoxia. However, in cells treated with both doxorubicin and hypoxia, the anti-p300 antibody was able to coimmunoprecipitate p53, indicating that DNA damage induced an interaction between p53 and p300 even under hypoxia. The lower levels of p53 in this lane compared to those in extracts treated with doxorubicin alone may reflect the difference in DNA damage induced by doxorubicin under oxic versus hypoxic conditions as detected by the comet assay (Fig. 8B). This assay measures DNA damage in the genome through an increase in the electrophoretic mobility of nuclear DNA in an electrofield (12). The increase in DNA mobility (seen as migrating DNA comet) is a direct measure of the extent of total DNA damage (single- and double-strand breaks). Hypoxia itself did not induce any measurable DNA damage and reduced the net damage induced by doxorubicin after a 12-h exposure.
The histone deacetylase inhibitor TSA reduces hypoxia-induced apoptosis and p53-dependent transrepression.
The p53-mSin3A interaction under hypoxia suggests that transcriptional repression mediated by this interaction may be important for p53-induced apoptosis by this stress. To investigate this possibility, we tested the effects of the HDAC inhibitor trichostatin A (TSA) on p53-dependent apoptosis following treatments with IR and hypoxia. TSA is an inhibitor of HDAC activity and has been shown to inhibit transcription by several transcriptional repressors, including p53 (24, 36, 54).
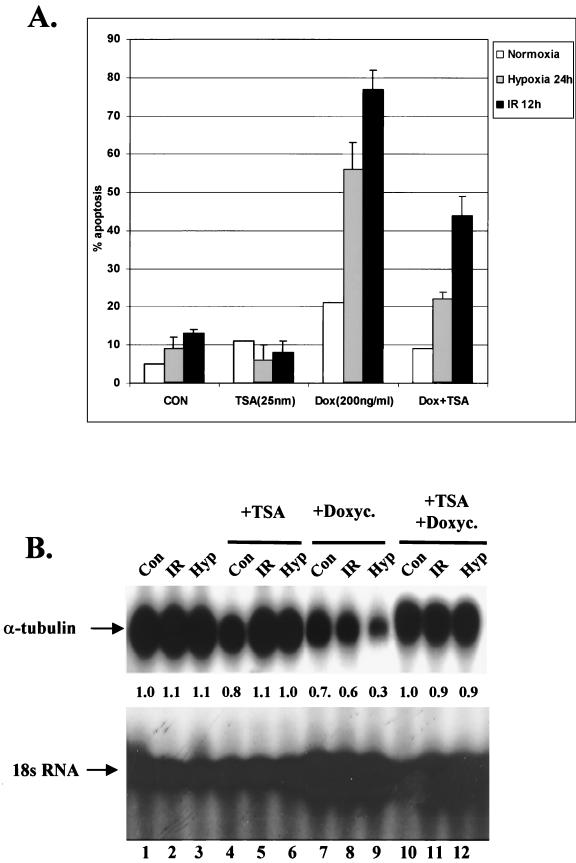
To test the effects of TSA on hypoxia- and IR-induced apoptosis, cells from tet-inducible H1299 cells were treated with TSA alone, doxycycline, or TSA followed by doxycycline, and cells with apoptotic morphology were visualized with Hoechst 33342 and propidium iodide staining and counted. As shown in Fig. 9A, TSA did not cause significant changes in apoptotic levels in cells under normoxic or hypoxic conditions or treated with IR. Addition of doxycycline prior to treatment induced a substantial increase in the apoptotic levels induced by hypoxia or IR. The level of apoptosis induced by hypoxia was reduced by TSA to the level of Dox treatment alone, suggesting that a trichostatin-sensitive transcriptional repression pathway mediates almost all p53-dependent apoptosis under hypoxia. In contrast, only 40% of radiation-induced apoptosis was inhibited by trichostatin, consistent with our data demonstrating the interactions of p53 with both transcriptional activation and repression pathways following IR treatments.
FIG. 9.
(A) TSA inhibits p53-dependent apoptosis under hypoxia. H1299 cells transfected with a p53 expression vector under the control of the tetracycline promoter were treated with IR or hypoxia. One group of cells did not receive any additional treatment (CON). TSA (25 nM) was added to a second group 1 h prior to treatment. A third group was treated with doxycycline 2 h prior to treatment, while both TSA and doxycycline were added to a fourth group prior to treatments. At the end of the treatment, cells with apoptotic morphology were visualized with Hoechst 3342 and phosphatidyl inositol staining. The total number of cells and the number of apoptotic cells in four different fields in 60-mm-diameter dishes were counted and expressed as the percentage of the total number of cells. Numbers represent the average of three independent experiments. Error bars represent SEM (B) TSA inhibits p53 transrepression under hypoxia. RKO cells were treated as described above, with the exception that cells were lysed 6 h after IR treatment and 12 h after hypoxia treatment. Northern blot analysis was performed on total RNA using a probe for human α-tubulin and 18S rRNA as described above.
To test whether the reduction of apoptosis by TSA correlated with a reduction in transcriptional repression, we assayed the levels of α-tubulin mRNA (Fig. 9B). p53 was induced by the addition of doxycycline, and RKO cells were treated in the same manner as described above, but cells were lysed at earlier time points to avoid effects due to apoptosis. Cells treated with IR were lysed at 6 h following treatments, while cells treated with hypoxia were lysed at 12 h after the onset of hypoxia. We found that pretreatment with TSA did not alter the mRNA levels of α-tubulin in untreated cells or cells treated with IR or hypoxia (lanes 4 to 6, respectively). In contrast, pretreatment with doxycycline caused a modest decrease of α-tubulin mRNA levels in untreated cells (lane 7) and cells treated with IR (lane 8), and a substantially bigger decrease in cells exposed to hypoxia (lane 9). When TSA was added 1 h prior to doxycycline, no decrease in α-tubulin mRNA levels was seen in IR-treated and hypoxia-treated cells compared to those in control cells. These results demonstrate that a strong correlation exists between the ability of TSA to reverse hypoxia-induced p53-dependent apoptosis and to inhibit p53-dependent transrepression.
DISCUSSION
The results described in this study demonstrate that hypoxia is unique among stresses that modulate p53, as it induces p53 protein accumulation and transcriptional repression but not the transcriptional induction of endogenous downstream effector genes. Furthermore, hypoxia is the first physiological stress that has been shown to lack a component of transactivation-competent p53 for the induction of apoptosis.
One of the most consistently transactivated genes in response to p53 induction is p21. Both p21 protein and mRNA failed to increase in response to hypoxia in human and rodent cells. The mRNA levels of other p53 effector genes, bax, GADD-45, and APO-1-Fas also failed to increase in response to hypoxia. Furthermore, we have shown that hypoxia not only fails to activate another important effector of p53, Mdm-2, but actually suppresses its protein levels. The lack of change in the mRNA levels of Mdm-2 by hypoxia points to a posttranscriptional mechanism for this decrease, like decreased mRNA translation or enhanced protein ubiquination. The results presented here, which show a marked reduction of Mdm-2 levels under hypoxia, provide a direct explanation for the observed accumulation of p53 under hypoxic conditions. Taken together, the findings listed above indicate that the dissociation between p53 accumulation and effector activation is not limited to one experimental system, nor does it apply to one effector, but instead it reflects a conserved, stress-specific cellular response.
The results from the experiments in which hypoxia is combined with IR treatments strongly suggest that the lack of p53 transactivation under hypoxia is due to a specific posttranslational modification(s) of p53 or an accessory protein which requires DNA damage for induction. Phosphorylation of p53 at different sites, including Ser15 and Ser20, has been proposed to induce the transcriptional activity of p53. However, at least for phosphorylation at the Ser15 position of human p53, this does not appear to be the case. Hypoxia was found to induce significant phosphorylation of Ser15, indicating that modification of this site alone is not sufficient for transcriptional activity. Furthermore, Ser15 phosphorylation does not appear to be required for p53 protein accumulation under hypoxia, since we have shown that the decrease in Mdm-2 levels under hypoxia is most likely the mechanism of p53 accumulation under these conditions (2).
Our immunocytochemistry experiments also rule out the possibility that the lack of p53 transactivation is due to a failure of hypoxia-induced p53 to accumulate in or be actively transported to the nucleus of RKO cells. These results extend our previous findings using untransformed human fibroblasts but also raise the interesting possibility that the mechanism for p53 accumulation by hypoxia is different from the one responsible for induction by DFO or other hypoxia-mimicking agents. While DFO and CoCl2 have been extensively used as a biochemical surrogate of hypoxia, we have found that at the molecular level these stresses exhibit significant differences in their profiles of induction of gene expression (Denko et al., unpublished observations). However, the finding by Ashcroft et al. that p53 accumulated by a nongenotoxic stress is not transcriptionally active is in agreement with our study (3).
The failure of p53 to become acetylated on Lys382 or to associate with p300 under hypoxia indicates that the lack of p300-p53 interaction and C-terminal acetylation may be responsible for the lack of p53-dependent transactivation of endogenous effector genes under hypoxia. These findings also underscore the requirement for C-terminal acetylation for p53-dependent transactivation, irrespective of other modifications that may occur at the N terminus. Other modifications like phosphorylation of Ser20 or phosphorylations at the C terminus in response to hypoxia treatments are currently being investigated.
The lack of association between p53 and p300 and acetylation of p53 at Lys382 under hypoxia is intriguing and may provide a biochemical handle for understanding the relationship between posttranslational modifications and activity of p53. Alternatively, this lack of interaction between the two proteins may be due to functional competition of p53 with factors that limit the availability of p300. Interestingly, another hypoxia-inducible factor, p35srj, has been reported to associate with p300 and to inhibit Hif-1α activity (5). A similar mechanism could be responsible for disrupting the association of p300 with p53. Still, our results with the combined hypoxia–DNA-damaging treatments suggest that if any competitive inhibition is responsible for lack of p53 transactivation under hypoxia, then DNA damage must shift the dynamics of this competitive inhibition towards a functional p53-p300 interaction.
It has been proposed that while p53 requires a functional interaction with the transcriptional coactivator p300 and PCAF (both proteins have histone acetylase and chromatin decondensation activities), p53-dependent transrepression may require interactions with proteins that possess deacetylase and chromatin condensation properties, like HDACs (19, 36). Murphy et al. have shown that p53 can coprecipitate not only with mSin3A but also with HDAC, indicating that, by binding to an as yet unidentified factor, p53 can recruit the HDAC complex to the promoter. Using an immunoprecipitation-PCR-based assay, they also demonstrated that this interaction occurs at the promoters of the repressed genes like MAP4 (36). Our coimmunoprecipitation studies lend support to the finding that p53 interacts with the histone deacetylase complex in vivo through mSin3A. This interaction between p53 and mSin3A is particularly strong under hypoxia.
What is the biological role of p53-dependent transrepression under hypoxia? Murphy et al. have shown that overexpression of the p53-repressed gene MAP4 delayed the onset of apoptosis induced by activation of a temperature sensitive-p53 construct (35). More recently, work from the same investigators has shown that compromising transcriptional repression by inhibiting histone deacetylase activity results in a dramatic decrease in the levels of p53-dependent apoptosis (36). Here, we also demonstrate that TSA decreases p53-dependent apoptosis and transrepression of α-tubulin under hypoxic conditions, to the levels of doxycycline-treated cells alone, indicating that almost all of hypoxia-induced apoptosis occurs through this trichostatin-sensitive pathway. This result is consistent with our finding that hypoxia-inducible p53 only interacts with transcriptional corepressors such as mSin3A and does not interact with transcriptional cofactors such as p300. Furthermore, as DNA damage induces interactions between p53 and both transcriptional coactivators and repressors, trichostatin only reduced radiation-induced apoptosis by 40%. Thus, in the case of activation of p53 by DNA damage, both transcriptional activation and repression pathways appear to be involved, as has been reported in the literature.
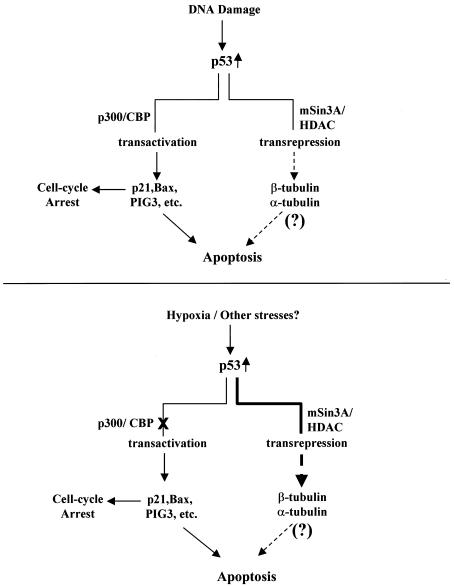
The results presented here describe a new paradigm for the regulation of p53 function by a physiological stress. As depicted in our model (Fig. 10), DNA damage induces the modification of p53 and interaction with accessory proteins that result in two pools of p53, one that possesses transactivation potential and one that possesses transrepression potential. Each pool of DNA damage-induced p53 is capable of inducing apoptosis. In contrast, hypoxia-induced p53 primarily interacts with transcriptional corepressors in signalling for apoptosis. Although a variety of genes exist that are transcriptionally repressed by p53, the identification of that gene(s) that modulates the critical effectors of apoptosis such as APAF-1 and caspase 9 (47) in a p53-dependent manner will be the next major contribution to understanding tumor suppression.
FIG. 10.
Model for the regulation of p53 function by genotoxic stress (top) and hypoxia (bottom). See text for details.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) grants CA67166, CA64489, and CA88480 to A.J.G. and ES05777 to M.B.K. C.K. was supported by National Research Service Award F32CA675754-02 from the NIH. M.B.K. is the Steven Birnbaum Scholar of the Leukemia Society of America and is also supported by the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital.
We thank Thea Tlsty for helpful suggestions and Nick Denko for critical review of the manuscript.
REFERENCES
- 1.Ahn J, Murphy M, Kratowicz S, Wang A, Levine A J, George D L. Down-regulation of the stathmin/Op18 and FKBP25 genes following p53 induction. Oncogene. 1999;18:5954–5958. doi: 10.1038/sj.onc.1202986. [DOI] [PubMed] [Google Scholar]
- 2.Alarcon R, Koumenis C, Geyer R K, Maki C G, Giaccia A J. Hypoxia induces p53 accumulation through MDM2 down-regulation and inhibition of E6-mediated degradation. Cancer Res. 1999;59:6046–6051. [PubMed] [Google Scholar]
- 3.Ashcroft M, Taya Y, Vousden K H. Stress signals utilize multiple pathways to stabilize p53. Mol Cell Biol. 2000;20:3224–3233. doi: 10.1128/mcb.20.9.3224-3233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharya S, Michels C L, Leung M K, Arany Z P, Kung A L, Livingston D M. Functional role of p35srj, a novel p300/CBP binding protein, during transactivation by HIF-1. Genes Dev. 1999;13:64–75. doi: 10.1101/gad.13.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown J M, Giaccia A J. The unique physiology of solid tumors: opportunities (and problems) for cancer therapy. Cancer Res. 1998;58:1408–1416. [PubMed] [Google Scholar]
- 7.Caelles C, Helmberg A, Karin M. p53-dependent apoptosis in the absence of transcriptional activation of p53-target genes. Nature. 1994;370:220–223. doi: 10.1038/370220a0. [DOI] [PubMed] [Google Scholar]
- 8.Canman C E, Lim D S, Cimprich K A, Taya T, Tamai K, Sakaguchi K E, Appella M B, Kastan M B, Siliciano J D. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, Ko L J, Jayaraman L, Prives C. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 1996;10:2438–2451. doi: 10.1101/gad.10.19.2438. [DOI] [PubMed] [Google Scholar]
- 10.Donehower L A, Harvey M, Slagle B L, McArthur M J, Montgomery C A J, Butel J S, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;19:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 11.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 12.Fairbairn D W, Olive P L, O'Neal K L. The comet assay: a comprehensive review. Mutat Res. 1995;339:37–59. doi: 10.1016/0165-1110(94)00013-3. [DOI] [PubMed] [Google Scholar]
- 13.Farmer G, Colgan J, Nakatani Y, Manley J L, Prives C. Functional interaction between p53, the TATA-binding protein (TBP), and TBP-associated factors in vivo. Mol Cell Biol. 1996;16:4295–4304. doi: 10.1128/mcb.16.8.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giaccia A J, Kastan M B. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 1998;12:2973–2983. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- 15.Graeber T G, Peterson J F, Tsai M, Monica K, Fornace A J, Jr, Giaccia A J. Hypoxia induces accumulation of p53 protein, but activation of a G1-phase checkpoint by low-oxygen conditions is independent of p53 status. Mol Cell Biol. 1994;14:6264–6277. doi: 10.1128/mcb.14.9.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graeber T G, Osmanian C, Jacks T, Housman D E, Koch C J, Lowe S W, Giaccia A J. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumors. Nature. 1996;379:88–91. doi: 10.1038/379088a0. [DOI] [PubMed] [Google Scholar]
- 17.Greenblatt M S, Bennett W P, Hollstein M, Harris C C. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 18.Gu W, Shi X L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 19.Hassig C A, Tong J K, Fleischer T C, Owa T, Grable P G, Ayer D E, Schreiber S L. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc Natl Acad Sci USA. 1998;95:3519–3524. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 21.Haupt Y, Rowan S, Shaulian E, Vousden K H, Oren M. Induction of apoptosis in HeLa cells by trans-activation-deficient p53. Genes Dev. 1995;9:2170–2183. doi: 10.1101/gad.9.17.2170. [DOI] [PubMed] [Google Scholar]
- 22.Hockel M, Knoop C, Schlenger K, Vorndran B, Baussmann E, Mitze M, Knapstein P G, Vaupel P. Intratumoral pO2 predicts survival in advanced cancer of the uterine cervix. Radiother Oncol. 1993;26:45–50. doi: 10.1016/0167-8140(93)90025-4. [DOI] [PubMed] [Google Scholar]
- 23.Kastan M B, Zhan Q, El-Diery W, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr A mammalian cell-cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 24.Kim YB, Yoshida M, Horinouchi S. Selective induction of cyclin-dependent kinase inhibitors and their roles in cell cycle arrest caused by trichostatin A, an inhibitor of histone deacetylase. Ann NY Acad Sci. 1999;886:200–203. doi: 10.1111/j.1749-6632.1999.tb09416.x. [DOI] [PubMed] [Google Scholar]
- 25.Kubbutat M H, Jones S N, Vousden K H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 26.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 27.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 28.Maki C G, Howley P M. Ubiquitination of p53 and p21 is differentially affected by ionizing and UV radiation. Mol Cell Biol. 1997;17:355–363. doi: 10.1128/mcb.17.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meek D W. Multisite phosphorylation and the integration of stress signals at p53. Cell Signal. 1998;10:159–166. doi: 10.1016/s0898-6568(97)00119-8. [DOI] [PubMed] [Google Scholar]
- 30.Miller M, Wilder S, Bannasch D, Israeli D, Lehlbach K, Li-Weber M, Friedman S L, Galle P R, Stremmel W, Oren M, Krammer P H. p53 activates the CD95 (APO-1/Fas) gene in response to DNA damage by anticancer drugs. J Exp Med. 1998;188:2033–2045. doi: 10.1084/jem.188.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyashita T, Reed J C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 32.Momand J, Zambetti G P, Olson D C, George D, Levine A J. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 33.Mosner J, Mummenbrauer T, Bauer C, Sczakiel G, Grosse F, Deppert W. Negative feedback regulation of wild-type p53 biosynthesis. EMBO J. 1995;14:4442–4449. doi: 10.1002/j.1460-2075.1995.tb00123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller J M, Krauss B, Kaltschmidt C, Baeuerle P A, Rupec R A. Hypoxia induces c-fos transcription via a mitogen-activated protein kinase-dependent pathway. J Biol Chem. 1997;272:23435–23439. doi: 10.1074/jbc.272.37.23435. [DOI] [PubMed] [Google Scholar]
- 35.Murphy M, Hinman A, Levine A J. Wild-type p53 negatively regulates the expression of a microtubule-associated protein. Genes Dev. 1996;10:2971–2980. doi: 10.1101/gad.10.23.2971. [DOI] [PubMed] [Google Scholar]
- 36.Murphy M, Ahn J, Walker K K, Hoffman W H, Evans R M, Levine A J, George D L. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 1999;13:2490–2501. doi: 10.1101/gad.13.19.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polyak K, Xia Y, Zweier J L, Kinzler K W, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 38.Prives C. Signaling to p53: breaking the MDM2–p53 circuit. Cell. 1998;95:5–8. doi: 10.1016/s0092-8674(00)81774-2. [DOI] [PubMed] [Google Scholar]
- 39.Roperch J P, Alvaro V, Prieur S, Tuynder M, Nemani M, Lethrosne F, Piouffre L, Gendron M C, Israeli D, Dausset J, Oren M, Amson R, Telerman A. Inhibition of presenilin 1 expression is promoted by p53 and p21WAF-1 and results in apoptosis and tumor suppression. Nat Med. 1998;4:835–838. doi: 10.1038/nm0798-835. [DOI] [PubMed] [Google Scholar]
- 40.Ryan K M, Vousden K H. Characterization of structural p53 mutants which show selective defects in apoptosis but not cell cycle arrest. Mol Cell Biol. 1998;18:3692–3698. doi: 10.1128/mcb.18.7.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sabbatini P, Chiou S K, Rao L, White E. Modulation of p53-mediated transcriptional repression and apoptosis by the adenovirus E1B 19K protein. Mol Cell Biol. 1995;15:1060–1070. doi: 10.1128/mcb.15.2.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakaguchi K, Herrera J E, Siato S, Miki T, Bustin M, Vassilev A, Anderson C W, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakamuro D, Sabbatini P, White E, Prendergast G C. The polyproline region of p53 is required to activate apoptosis but not growth arrest. Oncogene. 1997;15:887–898. doi: 10.1038/sj.onc.1201263. [DOI] [PubMed] [Google Scholar]
- 44.Shieh S Y, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 45.Shieh S Y, Taya Y, Prives C. DNA damage-inducible phosphorylation of p53 at N-terminal sites including a novel site, Ser20, requires tetramerization. EMBO J. 1999;18:1815–1823. doi: 10.1093/emboj/18.7.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siliciano J D, Canman C E, Taya Y, Sakaguchi K, Appella E, Kastan M B. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soengas M S, Alarcon R M, Yoshida H, Giaccia A J, Hakem R, Mak T W, Lowe S W. Apaf-1 and caspase-9 in p53-dependent apoptosis and tumor inhibition. Science. 1999;284:156–159. doi: 10.1126/science.284.5411.156. [DOI] [PubMed] [Google Scholar]
- 48.Unger T, Juven-Gershon T, Moallem E, Berger M, Sionov R V, Lozano G, Oren M, Haupt Y. Critical role for Ser20 of human p53 in the negative regulation of p53 by Mdm2. EMBO J. 1999;18:1805–1814. doi: 10.1093/emboj/18.7.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Unger T, Sionov R V, Moallem E, Yee C L, Howley P M, Oren M, Haupt Y. Mutations in serines 15 and 20 of human p53 impair its apoptotic activity. Oncogene. 1999;18:3205–3212. doi: 10.1038/sj.onc.1202656. [DOI] [PubMed] [Google Scholar]
- 50.Venot C, Maratrat M, Dureuil C, Conseiller E, Bracco L, Debussche L. The requirement for the p53 proline-rich functional domain for mediation of apoptosis is correlated with specific PIG3 gene transactivation and with transcriptional repression. EMBO J. 1998;17:4668–4679. doi: 10.1093/emboj/17.16.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner A J, Kokontis J M, Hay N. Myc-mediated apoptosis requires wild-type p53 in a manner independent of cell cycle arrest and the ability of p53 to induce p21/waf1/cip1. Genes Dev. 1994;8:2817–2830. doi: 10.1101/gad.8.23.2817. [DOI] [PubMed] [Google Scholar]
- 52.Wang X W, Vermeulen W, Coursen J D, Gibson M, Lupold S E, Forrester K, Xu G, Elmore L, Yeh H, Hoeijmakers J H, Harris C C. The XPB and XPD DNA helicases are components of the p53-mediated apoptosis pathway. Genes Dev. 1996;10:1219–1232. doi: 10.1101/gad.10.10.1219. [DOI] [PubMed] [Google Scholar]
- 53.Yonish-Rouach E, Deguin V, Zaitchouk T, Breugnot C, Mishal Z, Jenkins J R, May E. Transcriptional activation plays a role in the induction of apoptosis by transiently transfected wild-type p53. Oncogene. 1995;11:2197–2205. [PubMed] [Google Scholar]
- 54.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostain A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 55.Zhu J, Zhou W, Jiang J, Chen X. Identification of a novel p53 functional domain that is necessary for mediating apoptosis. J Biol Chem. 1998;273:13030–13036. doi: 10.1074/jbc.273.21.13030. [DOI] [PubMed] [Google Scholar]