Abstract
Huntington’s disease (HD) is an incurable and progressive neurodegenerative disease affecting the basal ganglia of the brain. HD is caused due to expansion of the polyglutamine tract in the protein Huntingtin resulting in aggregates. The increased PolyQ length results in aggregation of protein Huntingtin leading to neuronal cell death. Vitamin B6, B12 and folate are deficient in many neurodegenerative diseases. We performed an integrated analysis of transcriptomic, metabolomic and cofactor-protein network of vitamin B6, B12 and folate was performed. Our results show considerable overlap of pathways modulated by Vitamin B6, B12 and folate with those obtained from transcriptomic and metabolomic data of HD patients and model systems. Further, in yeast model of HD we showed treatment of B6, B12 or folate either alone or in combination showed impaired aggregate formation. Transcriptomic analysis of yeast model treated with B6, B12 and folate showed upregulation of pathways like ubiquitin mediated proteolysis, autophagy, peroxisome, fatty acid, lipid and nitrogen metabolism. Metabolomic analysis of yeast model shows deregulation of pathways like aminoacyl-tRNA biosynthesis, metabolism of various amino acids, nitrogen metabolism and glutathione metabolism. Integrated transcriptomic and metabolomic analysis of yeast model showed concordance in the pathways obtained. Knockout of Peroxisomal (PXP1 and PEX7) and Autophagy (ATG5) genes in yeast increased aggregates which is mitigated by vitamin B6, B12 and folate treatment. Taken together our results show a role for Vitamin B6, B12 and folate mediated modulation of pathways important for preventing protein aggregation with potential implications for HD.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-023-03525-y.
Keywords: Huntingtin, Huntington disease, Metabolomics, Transcriptomics, Neurodegeneration, Autophagy, Cofactor protein interaction
Introduction
Huntington’s disease is a progressive incurable inherited neurodegenerative disease caused due to the expansion of CAG repeat in the gene Huntingtin (Htt). The increased CAG repeat, in turn, leads to increased polyglutamine (PolyQ) repeat in the protein resulting in protein aggregation (Davies and Scherzinger 1997). The disease manifestation includes motor, behavioural and cognitive dysfunction (Shannon 2011). The length of expanded CAG repeat is inversely correlated with age of onset while it is positively correlated with severity of disease (Duyao et al. 1993; Finkbeiner 2011). The protein aggregates are thought to induce neuronal cell death (Helmuth 2001; Arrasate and Finkbeiner 2012). HD is associated with brain volume changes, atrophy in caudate nucleus and putamen (Negi et al. 2014), which are also observed in presymptomatic HD patients (Mascalchi et al. 2004; Liu et al. 2016; Thota et al. 2021). Currently, the treatment is directed towards managing the symptoms (Novak and Tabrizi 2010). Hence understanding the factors that modulate aggregate formation or its degradation might help in better management of disease with a favourable outcome.
HD is associated with mitochondrial dysfunction and impaired energy metabolism (Browne and Beal 2004; Gu et al. 1996). Impaired functioning of enzymes like pyruvate and succinate dehydrogenase is shown to be associated with the disease (Sorbi et al. 1983; Saulle et al. 2004; Arning and Epplen 2012). Magnetic resonance spectroscopy (MRS) study shows changes in the levels of metabolites like N-acetylaspartate (NAA) and choline (Thota et al. 2021) while Positron emission tomography (PET) scan shows changes in glucose utilization (Thota et al. 2021). Previous transcriptomic, proteomic and metabolomic studies have shown deregulation of signalling and metabolic pathways (Sorolla et al. 2008; Graham et al. 2016b; Chaves et al. 2017; McGarry et al. 2020). The enzyme Pyridoxal Kinase (PDXK) which activate Pyridoxine (Vitamin B6) to Pyridoxal 5’ Phosphate is downregulated in postmortem human brain of HD patients while its activity is reduced in mice model of HD (Sorolla et al. 2016). In addition, studies have shown that in HD patients reduced vitamin B6 lead to increased protein oxidation and exacerbation of disease (Sorolla et al. 2010). Further, changes in levels of various metabolites like alanine, glutamate, tryptophan and deregulation of associated pathways modulated by vitamin B are also reported in HD (Rosas et al. 2015; Graham et al. 2016a; Herman et al. 2019). For many genetic diseases with non-synonymous single nucleotide polymorphisms (SNPs) in the genes coding for enzymes, vitamin/cofactor supplementation is used as the standard of care treatment (Ames et al. 2002). Hence vitamins might help to activate various enzymes by ensuring metabolic homeostasis which might have relevance for managing HD (Johri and Beal 2012). In addition, B12 and folate are important co-factors for many enzymatic reactions. In particular, low levels of B12 is associated with many neurological disorders (Nawaz et al. 2020). Low levels of B12 also results in Huntington’s like chorea (McCaddon 2013). Folate supplementation is recommended for HD along with B12 (Brennan et al. 1981). However, studies have shown that in the absence of B12, elevated levels of methyl tetrahydrofolate accumulate (Brennan et al. 1981). The methyl tetrahydrofolate was found to interact with kainic acid receptors (Brennan et al. 1981). Injection of kainic acid into the basal ganglia in animal models results in damage similar to patients affected with HD (Brennan et al. 1981). Similarly, homocysteine is associated with many neurodegenerative diseases (Obeid and Herrmann 2006; Herrmann and Obeid 2011; Sharma et al. 2015). Vitamin B12 and folate are essential for the reconversion of homocysteine into methionine while B6 is essential for converting homocysteine into cysteine through the trans-sulfuration pathway (Miller 2003). Taken together vitamin B6, B12 and folate could be potential disease modifiers which might help to achieve favorable prognosis in HD.
HD is also known to be associated with elevated lactate to pyruvate ratio (Jenkins et al. 1998; Ferreira et al. 2011). This in turn leads to a high NADH/NAD ratio resulting in the activation of mTOR and impaired autophagy (Maiese 2020; Xie et al. 2020). Autophagy is the major pathway that helps to clear protein aggregates formed by mutant Huntingtin (Sarkar and Rubinsztein 2008). Further, activation of autophagy is shown to confer neuroprotection (Cherra and Chu 2008). Modulating metabolic profile might help to activate autophagy with potential implications for disease prognosis.
Various model systems like yeast, C. elegans, zebrafish, mice, sheep have been used as a model system to understand the disease (Crook and Housman 2011; Morton and Howland 2013; Alexander et al. 2014; Das and Rajanikant 2014; Farshim and Bates 2018; Hofer et al. 2018; Morton 2018; Surguchov 2021; Sai Swaroop et al. 2022; Pradhan et al. 2022; Swaroop et al. 2022). Invertebrate models have helped to untangle the mechanisms associated with neurodegeneration in Parkinson’s Disease, Huntington’s disease and ALS (Link 2001; Ramaswamy et al. 2007; Van Damme et al. 2017; Surguchov 2021). Omic analysis and validation using cell culture models have been used to elucidate mechanisms in many diseases (Naik et al. 2020; Pradhan et al. 2022). The mice model of HD captures the tenets of the pathophysiology of the disease, hence is a very good model to study HD (Farshim and Bates 2018). The yeast model of HD has been used to understand factors modulating protein aggregation (Mason and Giorgini 2011; Rencus-Lazar et al. 2019). Previous unbiased screening studies using yeast Knockout (KO) libraries have conjured genes that modulate the toxicity of mutant Huntingtin in yeast (Tauber et al. 2011). Yeast and cell culture models have shown a potential role for tryptophan metabolism in disease (Perkins and Stone 1982; Beal et al. 1990; Giorgini et al. 2005; Mazarei and Leavitt 2015). In the yeast model of Alzheimer’s disease, riboflavin was shown to reduce aggregates and improve mitochondrial performance (Chen et al. 2020). Taken together the results show that genes in metabolic pathways and vitamin supplementation could modulate huntingtin protein aggregation and in turn help to achieve better prognosis and management of HD.
In the current study, we have analyzed the GEO transcriptomic datasets of HD patients, mice and yeast model of HD and binned the significantly differential genes into pathways. We demonstrate both from transcriptomic data and literature that B6, B12 and folate interacting proteins required for transport or their activation are downregulated. Further, a B6, B12 and folate cofactor-protein interaction network (cofactor network) was constructed. The cofactor-protein interaction network was merged with the transcriptomic network to obtain the common genes using the merge network function in Cytoscape. The gene expression profile from transcriptomic datasets were then imported into the merged cofactor-protein-transcriptomic network. The results showed downregulation of key enzymes in deregulated metabolomic pathways which are modulated by B6, B12 and folate. Analysis of published metabolomic data also showed the deregulation of metabolite levels which are in turn regulated by enzymes that require B6, B12 or folate as cofactors. To evaluate the role of B6, B12 and folate we carried out experiments using the yeast model of HD. B6, B12 and folate either individually or in combination attenuated protein aggregation in the yeast model of HD. Further, transcriptomic and metabolomic analysis was carried out in the yeast model treated with a combination of B6, B12 and folate and compared to controls. There was considerable concordance in the pathways obtained between our yeast transcriptomic and metabolomic datasets which show B6, B12 and folate modulate key pathways that might influence protein aggregation. Further, B6, B12 and folate also increased the activity of alanine amino transferase a B6 dependent enzyme and catalase a marker for peroxysomes. The results are discussed in light of these findings. B6, B12 and folate might help to achieve a potentially favourable prognosis in HD.
Material and methods
Bioinformatics analysis
Transcriptomics data
Datasets for differential gene expression (DGE) analysis were obtained from the Gene expression omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) (Edgar et al. 2002; Barrett et al. 2013). The datasets selected for analysis represent the following organisms- Saccharomyces cerevisiae, Mus musculus and Homo sapiens. The GEO series IDs analyzed are GSE18644 (Tauber et al. 2011) [S. cerevisiae]; GSE3621 (Hodges et al. 2008a), GSE10202 (Kuhn et al. 2007) [M. musculus] and GSE26927 (Durrenberger et al. 2012), GSE1751 (Borovecki et al. 2005) [H. sapiens] {transcriptomics datasets summary provided in Supplementary data S1}. The sample group selected represent only the differentially expressed genes (DEG) of the control and the disease excluding treatments or knockout studies. All human samples for DGE analysis are age and gender-matched. DGE analysis of the group set- control and disease were done using NetworkAnalyst (https://www.networkanalyst.ca/) (Xia et al. 2014, 2015; Zhou et al. 2019). NetworkAnalyst uses Linear Models for Microarray Analysis (limma) R/Bioconductor software package module that aids in analyzing gene expression datasets.
Differential gene expression analysis
The pre-normalized series matrix files for the GEO series IDs (GSE ID) selected were downloaded from the individual GSE ID page. A.txt file was compiled that included the sample data (GSM data) from the series matrix file which was grouped under two categories- Disease and Control. The group defined matrix file (.txt file) constitute the gene accession number as the identifier and intensity values for respective genes for each sample. This.txt matrix file so prepared was then uploaded to NetworkAnalyst which uses limma for DGE analysis. Parameters for data filtering was set at- variance filter = 15 and low abundance = 5 (Default settings) and no normalization. Study design specific comparison was set as Disease versus Control. The threshold for significant differentially expressed genes was set at log2 fold change = 1 and adjusted p-value ≤ 0.05 (Supplementary data S1).
Pathway enrichment using Enrichr and Cytoscape pathway network visualization
The biological pathway implications of the significantly expressing genes acquired from DGE analysis prior was investigated using enrichment analysis. The significant DEG were listed and run for enrichment analysis using Enrichr [https://maayanlab.cloud/Enrichr/] (Chen et al. 2013; Kuleshov et al. 2016; Xie et al. 2021). The list of DEG were uploaded to Enrichr, KEGG pathway was selected as a gene set reference. Pathways with an adjusted p-value ≤ 0.05 qualify as significant (Supplementary data S3). These pathways were then represented as pathway-gene interacting networks in Cytoscape 3.8.2 (Christmas et al. 2005).
Cofactor-protein interacting network and its overlapping with enriched pathways from transcriptomics datasets
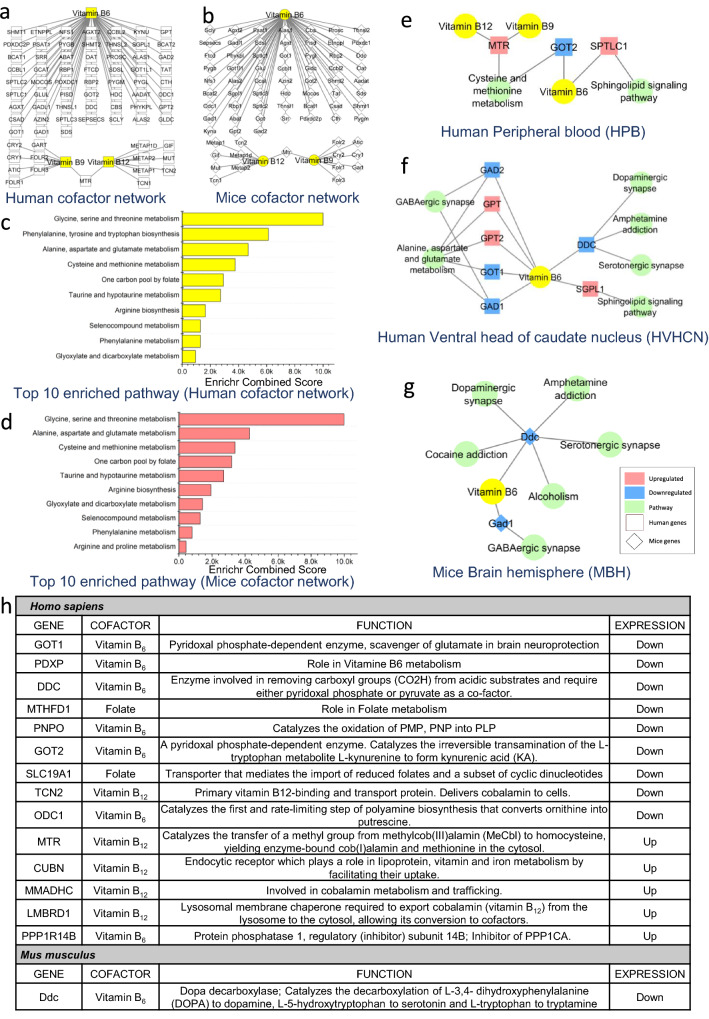
A cofactor-protein interaction network for Vitamin B6, B12 and folate and its interacting proteins were generated using pre-published data in Cytoscape 3.8.2 as described previously (Scott-Boyer et al. 2016; Naik et al. 2020) (Fig. 1a, b). An online tool DIOPT- DRSC Integrative Ortholog Prediction Tool (Hu et al. 2011) (https://www.flyrnai.org/cgi-bin/DRSC_orthologs.pl#), was used to predict the orthologs for mice and was used for generating cofactor-protein interacting network in Cytoscape 3.8.2 (Fig. 1b) [Supplementary data S4]. A comparative analysis was carried out for common genes between the cofactor-protein network and those involved in the enriched pathways from transcriptomic dataset analysis. Further, Cytoscape 3.8.2 was used to generate the integrated Transcriptomic-Cofactor-protein interaction network using the network merge tool for visualization (Fig. 1e–g). The generated network highlights key overlapping genes that are represented in both the cofactor-protein network and enriched pathway network from transcriptomics as shown in Fig. 1e–g.
Fig. 1.
a, b show vitamin B6, B12 and folate interacting protein networks in humans and mice. Results showing top ten pathways enriched for vitamin B6, B12 and folate interacting protein in c human and d mice. Network showing deregulated pathway in HD and overlapping of pathway genes associated with vitamin B6, B12 and folate as cofactors in following datasets. e Human peripheral blood-HPB [GSE1751], f Human ventral head of caudate nucleus-HVHCN [GSE26927] and g mice brain hemisphere-MBH [GSE3621] datasets. h Vitamin B6, B12 and folate interacting genes and their expression in HD patients and mice model of HD. The networks a, b, e, f and g were generated in Cytoscape 3.8.2. The plots c, d were made in Origin software
Cofactor interacting protein enrichment analysis
The list of genes from the cofactor-protein interaction network constructed in the earlier section (Cofactor-protein interacting network and its overlapping with enriched pathways from transcriptomics datasets) were investigated for enrichment analysis to elucidate its biological implication and function. The resulting pathways were then selected for significance at an adjusted p-value ≤ 0.05 (Fig. 1c, d). The enrichment analysis was carried out for yeast, mice and human pathways. Yeast and mice orthologs were predicted from human data using DIOPT—DRSC Integrative Ortholog Prediction Tool (Hu et al. 2011) [https://www.flyrnai.org/cgi-bin/DRSC_orthologs.pl#], to which enrichment analysis was performed (Supplementary data S4).
Metabolomics data
Quantitative differential metabolomics data of HD was collected from the literature. The metabolite profiles from these published data were used for metabolite set enrichment analysis to elucidate the role of the deregulated metabolites and their implication for the disease (Supplementary data S5).
Metabolite set enrichment analysis (MSEA)
The deregulated statistically significant differential metabolites from different profiling experiments of S. cerevisiae (Joyner et al. 2010), M musculus (Tsang et al. 2006) and H. sapiens (Rosas et al. 2015; Graham et al. 2016a, 2018; Herman et al. 2019) were collected. These metabolites were then converted to their HMDB annotations and the MSEA for individual datasets was carried out using MetaboAnalyst 5.0 (https://www.metaboanalyst.ca/). KEGG metabolite database was set as a reference for the enrichment of overlapping metabolites. The selection criteria for significant pathways were set at a FDR ≤ 0.25 (Supplementary data S5).
Common pathways analysis between Cofactor-protein interaction network and Metabolomics
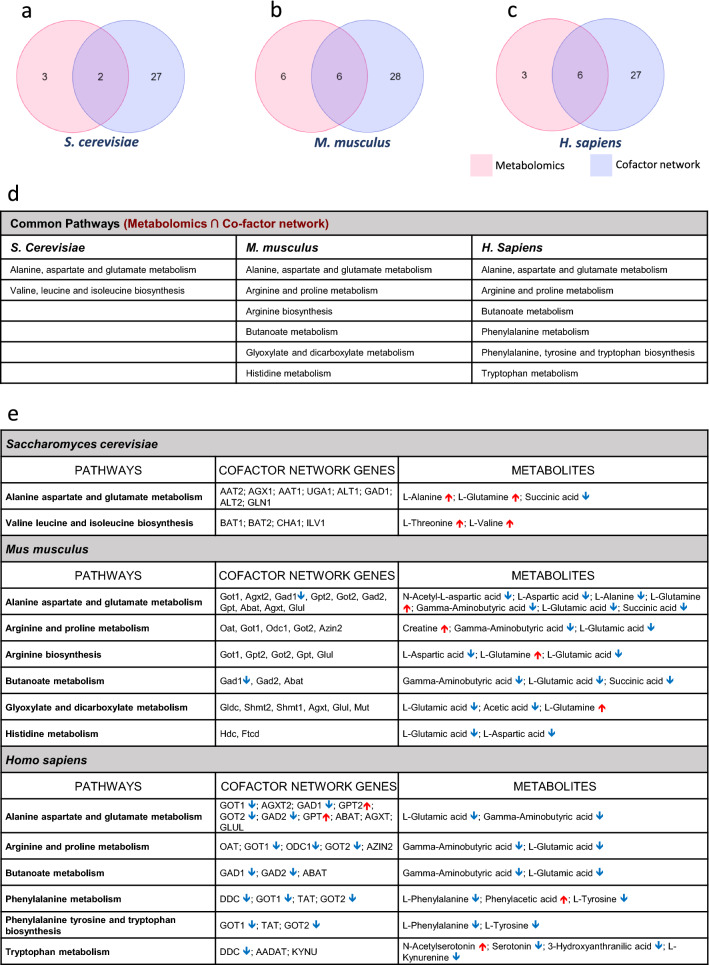
Commonality analysis was performed for pathways obtained by MSEA of metabolomic data and cofactor-protein interaction network (Fig. 2a–c). The proteins and metabolites representing these pathways were listed along with the gene expression values and metabolite levels respectively (Fig. 2d, e). (Venn diagrams was generated in Molbiotools Multiple list comparator- https://molbiotools.com/listcompare.php).
Fig. 2.
Showing overlap of deregulated metabolic pathways in HD (literature) and cofactor interacting protein-enriched pathway (cofactor network pathway) for B6, B12 and folate in a Saccharomyces cerevisiae b Mus musculus and c Homo sapiens. d The intersection of metabolic pathways deregulated (literature) in HD and pathways enriched by the cofactor-protein interacting proteins (cofactor network). (e) Common metabolic pathways deregulated showing genes expression and metabolite levels from transcriptomics and metabolomics datasets of HD. The Venn diagrams (a–c) was created using an online tool for multiple list comparator-https://molbiotools.com/listcompare.php
Experimental (Present Study)
Yeast Strain and plasmid
The yeast strain used for the study is Saccharomyces cerevisiae BY4741 and BY4742. The plasmid used in the study is p426 GPD (Addgene No. 1181 and 1183) which harbor the N-Terminal Huntingtin with a 25 or 72 CAG repeat attached by a linker to EGFP, henceforth mentioned as 25Q and 72Q. The selection marker for bacteria is ampicillin and the yeast selection marker is URA. The Yeast KO strain library was procured from Dharmacon (Product No. YSC1054). The KO used in the study PXP1, PEX7 and ATG5 are in the BY4742 background with G418 as the selection marker. The stain used for transcriptomic and metabolomic analysis is the BY4741 strain transformed with either 25Q or 72Q plasmid. Appropriate controls were used for the respective experiments performed.
Yeast culture, transformation and supplementation study
Yeast strain BY4741 and BY4742 were revived and transformed with p426 25Q GPD and p426 72Q GPD plasmid (Purchased from addgene, Addgene plasmid # 1181, 1183; http://n2t.net/addgene:1181/1183; RRID: Addgene_1181/1183) (Krobitsch and Lindquist 2000). Standard protocols were used for plating, culture, transformation and experiments as described in a previous study (Pradhan et al. 2022).
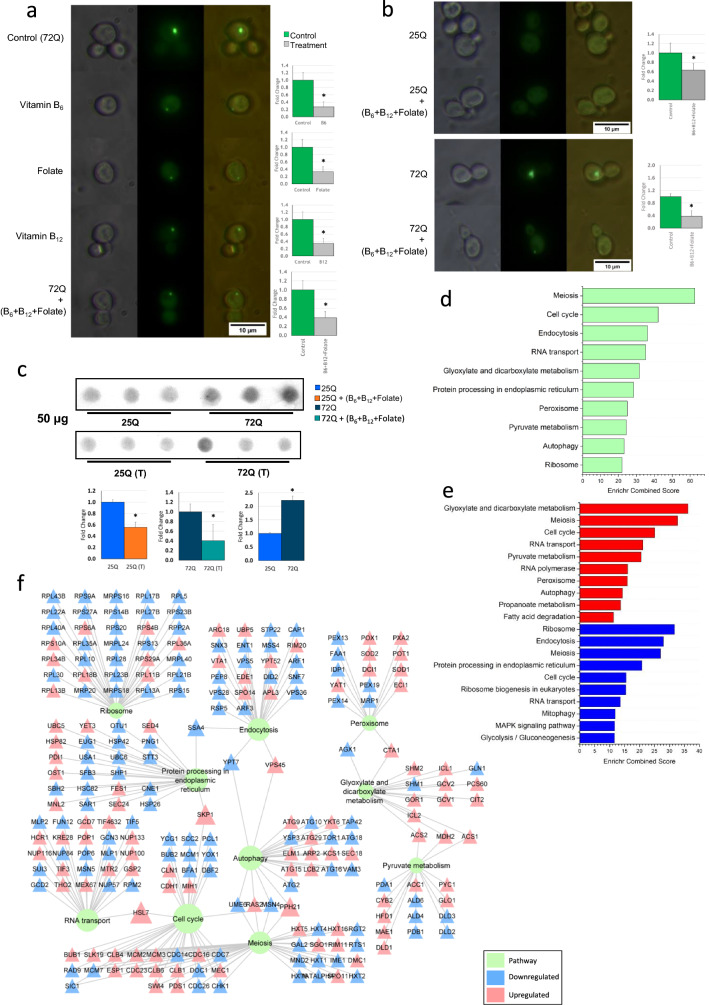
The 25Q/72Q transformed cells (BY4742) were cultured overnight to an OD600 of 0.8 or till the culture reached the logarithmic phase. These cells were then supplemented with vitamin B6 (SRL chemicals, 164,755), folate (Himedia, CMS175) and vitamin B12 (Sigma, V2876) individually or in combination (20 μg/ml each). To arrive at the optimum concentration of 20 µg/ml for each of B6, B12 and folate, we had titrated with different concentration of these vitamins (5, 10, 20 and 50 µg/ml for each of the vitamin) [Supplementary Data S2g]. The pre-formed protein aggregate in Htt 72Q transformed yeast were treated with vitamin B6, B12 and folate (20 μg/ml each) in combination, fluorescence imaging and quantification was done at a time period of 3 h. and 6 h. from treatment (Supplementary Data S2h). Fluorescence imaging was carried out using Laben BM-3000 FLT fluorescence microscope with an excitation wavelength of 470 nm and an emission wavelength of 509 nm after 12 h. from treatment (Fig. 3a, b).
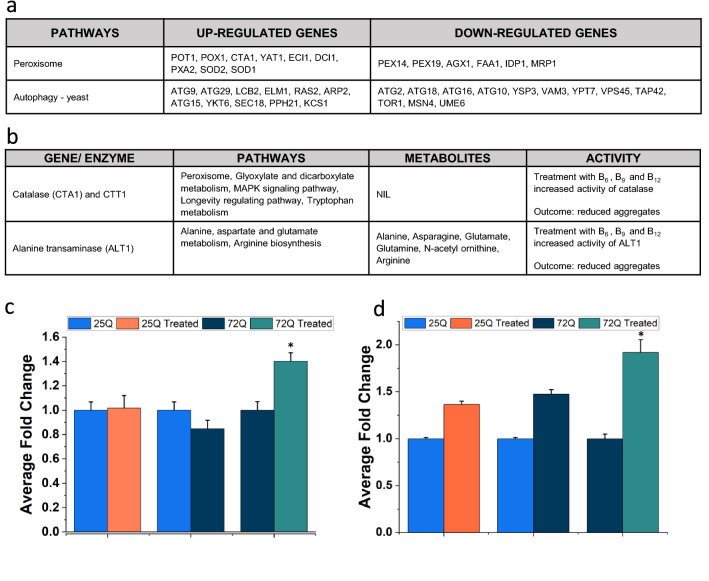
Fig. 3.
a Yeast transformed with Htt 72Q, GFP showing the aggregation as foci. Fluorescence data show a reduction in aggregate when yeast model of HD was treated with vitamin B6, B12 and folate individually as well in combination (concentration used 20 μg/ml) when compared to control. Fluorescence quantification showing significant reduction of aggregation in treatment, t-test: *p-value ≤ 0.05. (n ≥ 10). b Treatment of B6, B12 and folate on 25Q and 72Q transformed yeast showed significant reduction in protein aggregates in 72Q, *p-value ≤ 0.05. c Filter retardation assay for yeast Htt 25Q/72Q and B6, B12 and folate treated samples. The intensity of the blot shows aggregation in control and its attenuation in B6, B12 and folate treated yeast samples. The intensity of the blot was calculated and shown to be significantly reduced in the treated sample with a *p-value < 0.05 and n = 3. d Transcriptomics analysis (present study) showing top 10 significantly deregulated pathways (adjusted p-value ≤ 0.05) in 72Q vitamin B6, B12 and folate treated sample and d expression of genes involved in these deregulated pathways (only genes with adjusted p-value ≤ 0.05) shown in form of a network. e Enrichment analysis showing deregulated pathways based on upregulated (shown in red) and downregulated genes (shown in blue) in 72Q B6, B12 and folate treated samples. f The pathway-gene interaction networks. Figure (f) was generated in Cytoscape 3.8.2. The plots d and e was made using Origin software
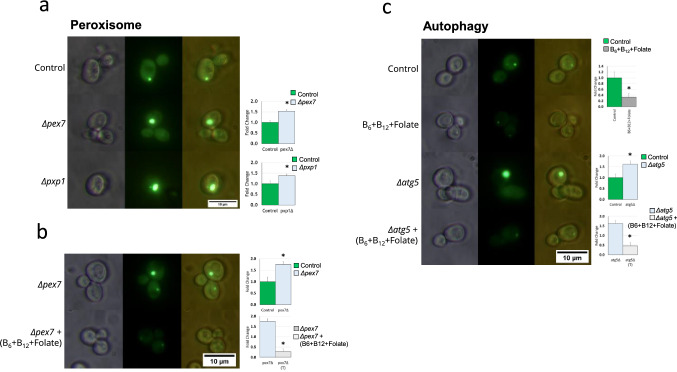
The knockout yeast strains used were procured from Dharmacon yeast knockout library (Product No. YSC1054). Yeast knockouts for the peroxisome pathway (PXP1 and PEX7) and autophagy pathway (ATG5) were used for experiments. The KO cells were grown with G418 (Santa Cruz Biotechnology, sc-29065A) selection and transformed with 72Q and selected using a double selection with URA- and G418 following standard procedures. The transformed KO cells were cultured following standard procedures. Further fluorescence imaging of the cells was carried out and the images were quantified in ImageJ software using Corrected total cell fluorescence (CTCF) analysis (Fig. 6a–c). To avoid bias the samples were blinded and randomized. All experiments were performed in three biological replicates (n = 3) and two technical replicates.
Fig. 6.
a, b Peroxisome pathway and c autophagy pathway knockout showing increase in aggregating and subsequently B6, B12 and folate treatment leads to attenuation of aggregates
Filer retardation assay
25Q/72Q transformed BY4742 yeast strain were cultured overnight to an OD600 of 0.8. An aliquot of 16 × 106 cells was resuspended in lysis buffer [30 mM Tris–HCl (Himedia, MB030), pH 8.0, 200 mM NaCl (Himedia, MB023), 2 mM EDTA (Himedia, RM1370), 5% glycerol, 10 mM Phenylmethylsulfonyl fluoride (Himedia, RM1592)]. Further, the cells were lysed using Precellys evolution homogenizer (P000062-PEVO0-A). The filter retardation assay procedure was adapted from the previously described methodology (Cohen et al. 2012) using Bio-Dot microfiltration apparatus (Bio-Rad). The protein lysate was diluted in TBST- Tris Buffered Saline with Tween-20 [50 mM Tris–HCL (Himedia, MB030), pH 8.0, 250 mM NaCl (Himedia, MB023), 0.1% Tween-20 (Himedia, MB067)]. Either 50 or 100 μg of protein sample in Tris Buffered Saline (TBS) with 2% SDS (Himedia, MB010) were heat-treated at 100 °C for 10 min and then loaded onto the apparatus, housing conditioned nitrocellulose membrane (0.2 µm, Cat. No: 10600016, GE Health care) in 2% (w/v) SDS containing TBS buffer. Vacuum was applied for 5 min or till the sample was filtered. The 25Q/72Q EGFP protein on the blot was immune-detected with mice anti-GFP antibody (Santa Cruz Biotechnology, B-2, sc-9996), as primary antibody and anti-mice streptavidin-HRP conjugate antibody as the secondary antibody (Invitrogen, SNN404Y). This was then visualized with a stabilized chromogen (TMB) substrate (Invitrogen, SB01) in Syngene Gbox F3. The mean quantification of the blot was done using ImageJ software (Fig. 3c). The experiment was performed in three biological replicates (n = 3).
Catalase and ALT activity assay
The whole cell lysate of 25Q/72Q and 25Q/72Q cells treated with B6, B12 and folate each containing the soluble proteins 250 ug was used for the catalase and ALT assays. The catalase assay was performed previously described (Martins and English 2014). An endpoint reading was taken at the end of 10 min at 240 nm. For ALT activity assay, HiPer SGPT (ALAT) Estimation Kit (Himedia, HTBC009) was used. Assay procedure was followed as per the manufacturer’s instruction. The activity of catalase and ALT was calculated as fold change with respect to respective controls. The graphs were generated using Origin 2021 (Fig. 5c, d).
Fig. 5.
Integrated analysis of transcriptomics and metabolomics data (present study). a Representing two key pathways altered in 72Q treated with B6, B12 and folate compared to 72Q, showing upregulated and downregulated genes. b Table summarising Catalase and ALT1 associated pathways and metabolites if any and assay activity. c Results showing significantly increased ALT activity in 72Q treated samples, *p-value < 0.05 and n = 3. d Results showing significantly increased Catalase activity in 72Q treated samples, *p-value < 0.05 and n = 3. The graphs were made in Origin software
RNA isolation, library preparation, sequencing, and analysis
Total RNA was extracted using the standard Trizol method from control cells [72Q (BY4741) transformed] and 72Q cells supplemented with Vitamin B6, B12 and folate. The experiments were performed in three biological replicates (n = 3). Tapestation 2100 from Agilent Biosystems was used to evaluate the quality of total RNA. mRNA libraries were prepared using NEBNext® mRNA Library Prep Reagent Set for Illumina. Briefly, mRNA was isolated from total RNA using oligodT beads, heat fragmented and converted into cDNA. Then end repair of fragmented ends was performed to which Illumina adapter was ligated at both the ends. Size selection of adapter-ligated cDNA was made to obtain the final mRNA library. The libraries were sequenced using Illumina® Hiseq2500 to get 100 bp paired-end reads. The sequencing depth for each sample was > 10 million reads. The reads were checked for quality using FastQC ([CSL STYLE ERROR: reference with no printed form.]) and then aligned to the reference Saccharomyces cerevisiae S288C strain (Downloaded from The University of California, Santa Cruz (UCSC) genome browser) using bowtie2(Langmead 2010) with default parameters. Samtools (Li et al. 2009) was used to obtain a binary alignment map file (BAM file). An annotation file for Saccharomyces cerevisiae S288C strain was downloaded from UCSC and read counts were obtained using BEDTools (Quinlan and Hall 2010). DESeq package from R(Anders and Huber 2010) was used to compute differential gene expression between 72 and 72Q supplemented with Vitamin B6, B12 and folate (Supplementary data S6).
Significant genes with an adjusted p-value ≤ 0.05 were considered for enrichment analysis. Enrichment was carried out using Enrichr, only the pathways having adjusted p-value ≤ 0.05 were considered to be significant (Fig. 3d). The upregulated and downregulated genes were analyzed separately and binned into pathways separately. Pathways having an adjusted p-value ≤ 0.05 were considered to be significant (Fig. 4e). Further, a pathway network and gene enriching the pathway along with their expression was constructed using Cytoscape 3.8.2 (Fig. 4f).
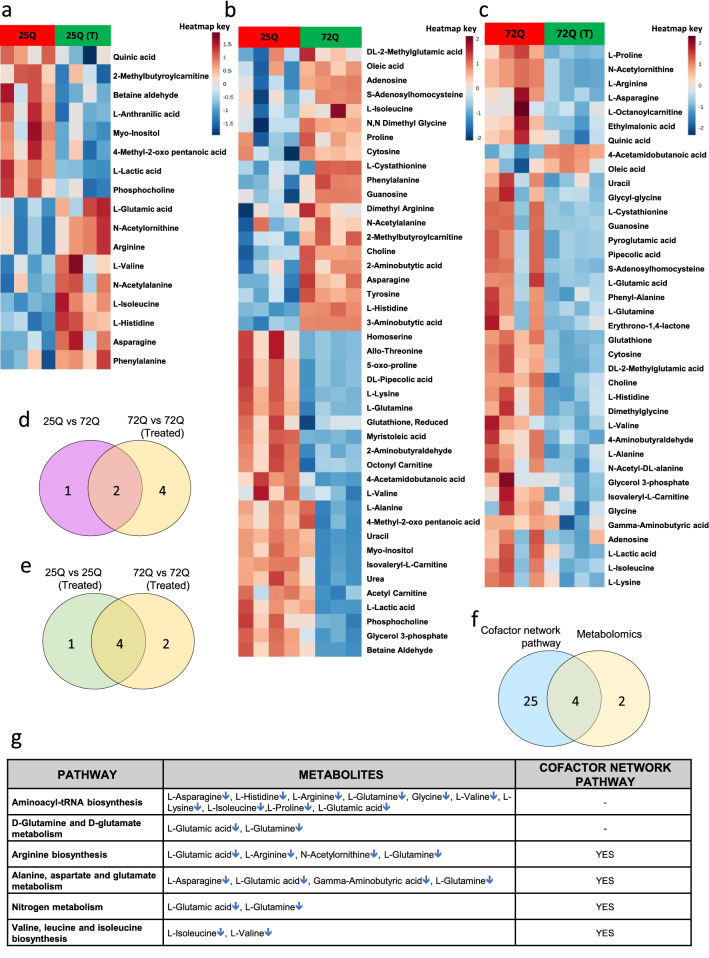
Fig. 4.
Yeast metabolomics (present study). Heatmap for metabolites altered in B6, B12 and folate treated sample. a 25Q vs 25Q + vitamin B6, B12 and folate, b 25Q vs 72Q and c 72Q vs 72Q + vitamin B6, B12 and folate. Venn diagram showing overlap of pathways between d 25Q vs 72Q and 72Q vs 72Q + vitamin B6, B12 and folate [72Q vs 72Q (Treated)], e 25Q vs 25Q + vitamin B6, B12 and folate [25Q vs 25Q (Treated)] and 72Q vs 72Q + vitamin B6, B12 and folate [72Q vs 72Q (Treated)] g Showing overlap of cofactor-protein network pathway and altered metabolomics pathways in vitamin B6, B12 and folate treated sample. e MSEA results showing pathways deregulated in 72Q vs 72Q (Treated) and those common to cofactor-protein interaction network pathways. representing regulation of metabolites for the deregulated pathways. Figures a–c was created using an online tool- MetaboAnalyst 5.0 (https://www.metaboanalyst.ca/). The Venn diagrams d–f was created using an online tool for multiple list comparator-https://molbiotools.com/listcompare.php
The sequencing data has been submitted to the GEO database with a Submission ID: SUB11437723 and BioProject ID: PRJNA835609 (http://www.ncbi.nlm.nih.gov/bioproject/835609).
Yeast metabolomics, sample preparation and analysis
Yeast cells (BY4741) expressing the 25Q or 72Q served as controls or those treated with a combination of B6, B12 and folate served as experimental sets. The experiments were performed in four biological replicates (n = 4). An aliquot of 8 × 106 yeast cells each of control and experimental sets were snap-frozen in liquid nitrogen and stored in -80 °C until further use. The metabolites were extracted from these aliquots using following procedure. The pellet was subjected to a freeze–thaw procedure (three cycles). Further, it was resuspended in 750 μl of extraction buffer (EB) [4:1 methanol: water] spiked with labelled internal standard, 2.5 μM each of Jasmonic acid (Sigma, 14,631), L-Tryptophan 15N2, 98% (Cambridge Isotope Laboratories, Inc, NLM-800), Sodium Pyruvate 13C3, 99% (Cambridge Isotope Laboratories, Inc, CLM-2440) and D-Glucose, U-13C6, 99% (Cambridge Isotope Laboratories, Inc, CLM-1396). The samples were sonicated in ice for 15 s with 5–10 pulses. This is followed by the addition of 450 μl of cold chloroform. After vortexing for 10 min, 150 μl of cold Millipore Type 1 water was added and the mixture was then incubated at – 20 °C for 20 min followed by centrifugation at 4000 rpm for 10 min at 4 °C to get separation of the two layers. The different layers were then pooled into a new tube and dried in a speed vac (miVac) for about 40 min. The tubes were resuspended in a 50:50 methanol–water solution after drying, followed by vortexing and ice sonication for 5 min. This was transferred to an Amicon filter column (Amicon Ultra 0.5 ml filter unit, Cat. No. UFC500396, Merck Millipore, Massachusetts, USA), which was pre-conditioned with 50:50 methanol–water. The Amicon filter with the resuspended pooled metabolite mixture was centrifuged at 13,000 rpm at 4 °C for 6 h. and the filtrate was collected in a collecting tube and quantitative metabolomics was performed.
Quantitative targeted metabolomics was carried out in positive ionization mode using Agilent 6490 iFunnel triple quadrupole LC/MS with a 2 μl injection volume of filtrate. The samples were separated using a Waters X-Bridge amide 3.5 μm, 4.6 × 100 mm (part no. 186004868, Waters, Milford, USA) column and injected into Agilent 6490 QQQ. The liquid chromatography (LC) and mass spectrometry (MS) systems were configured as described previously (Pulukool et al. 2021; Bhagavatham et al. 2021; Pradhan et al. 2022).
All the data processing were performed as described in a previous study (Pradhan et al. 2022). Briefly, processing of acquired targeted metabolomics data with abundances of metabolites, including peak integration with relevant retention time for each metabolite was done using Agilent Mass Hunter- Quantitative analysis software (Version No. B.07.00). From the labelled inter standards used, L-Tryptophan with the lowest CV (Coefficient of Variation) was used for data normalization. Only those metabolites with CV ≤ 20% after normalization was used for further analysis. An unbiased PCA analysis clustered the samples into two separate groups which are represented by a 2D score plot (Supplementary data S2i–k). A two-way t-test analysis was carried out with FDR ≤ 0.25 to determine the differential metabolites between the control and experimental groups using MetaboAnalyst 5.0 [https://www.metaboanalyst.ca]. The significant differential metabolites are represented in the form of a heatmap (Fig. 4a–c).
Metabolite set enrichment analysis (MSEA)
The significant differential metabolites identified between controls and the B6, B12 and folate treated sample sets were analyzed using metabolite set enrichment analysis (MSEA). MSEA was performed with KEGG metabolite set library as a reference for enrichment. Significant pathways were selected with FDR ≤ 0.25 (Supplementary Data S2l–n). A chord diagram was used to illustrate the link between the significant pathways and the metabolites altered using Origin 2021(Supplementary Data S2m). Further, a Venn diagram was used to show the overlap of the pathways obtained by metabolic analysis and those obtained from cofactor-protein interaction network analysis (Fig. 4f).
Transcriptomics and metabolomics integration
The list of significant differential genes from our yeast transcriptomic dataset and significant differential metabolites obtained for our yeast metabolomic dataset were uploaded for a joint-pathway analysis in MetaboAnalyst 5.0. The integration was carried out using the option All pathways (integrated), which provided the metabolic and the gene only regulatory pathways. The significant pathways with FDR ≤ 0.25 were used for further discussions. All integrated significant pathways, with the expression levels of the differential genes and the levels of differential metabolites from our transcriptomic and metabolomic analysis of control and B6, B12 and folate treated samples are provided in Supplementary Data S7.
Results
Cofactor network analysis of B6, B12 folate shows deregulation of pathways with potential implications for HD in mice and human
We analyzed the expression levels of the transporters and enzymes which are essential for the uptake of B6, B12 and folate in HD patients as well as model systems using expression datasets from the GEO database (Data on DEG provided in Supplementary data S1). The cofactor-protein interaction network of B6, B12 and folate for human and mice were generated using Cytoscape 3.8.2 (Fig. 1a, b). The mice network was created from the human cofactor-protein interaction network data using the orthologue approach as mentioned in methods. The genes involved in the cofactor-protein interaction network were further binned into pathways. The top 10 enriched pathways are provided (Fig. 1c, d) [Find complete list of significant pathways in Supplementary data S2a–c]. The major pathways include metabolism of amino acids, one-carbon metabolism, selinocompound metabolism, taurine and hypotaurine pathways (Fig. 1c, d). To understand the expression levels of genes common to the cofactor-protein interaction network in HD an integrated analysis with significant differentially expressed genes from transcriptomic datasets of humans HD patients (Borovecki et al. 2005; Durrenberger et al. 2012) and mice model (Hodges et al. 2008b) of HD was performed.
Integration of cofactor network with transcriptomic datasets shows the deregulated metabolic pathways with the core enzymes which use Vitamin B6, B12 and folate as cofactors
The Vitamin B6, B12 and folate protein interaction pathway was analyzed and the genes were categorized into pathways. Further, the transcriptomic data from both human and mice (Supplementary data S3) was imported into this network. The Human peripheral blood (HPB) [GSE1751] dataset network showed enrichment of cysteine and methionine metabolism pathway. The gene 5-methyltetrahydrofolate-homocysteine methyltransferase (MTR) which requires B12 and folate as a cofactor for its activity enriched in the pathway is upregulated while glutamic-oxaloacetic transaminase 2 (GOT2) gene which requires vitamin B6 as a cofactor is downregulated (Fig. 2e). The sphingolipid signalling pathway involving serine palmitoyltransferase long chain base subunit 1 (SPTLC1) gene which uses B6 as a cofactor was also upregulated (Fig. 1e).
In the Human ventral head of caudate nucleus (HVHCN) [GSE26927] dataset network, the glutamate decarboxylase 1 (GAD1) gene and the glutamate decarboxylase 2 (GAD2) gene are involved in GABAergic synapse which uses B6 as cofactor were downregulated (Fig. 1f). The alanine, aspartate and glutamate metabolism pathway with three enzymes glutamic-oxaloacetic transaminase 1 (GOT1), glutamic-pyruvic transaminase (GPT) and glutamic-pyruvic transaminase 2 (GPT2) use B6 as a cofactor (Fig. 1f). The GOT1 was downregulated while the GPT were upregulated. The B6 dependent enzyme dopa decarboxylase (DDC) was downregulated and is involved in the pathways like dopaminergic synapse, amphetamine addiction and serotonergic synapse (Fig. 1f). Similarly, the B6 dependent enzyme sphingosine-1-phosphate lyase 1 (SGPL1) involved in the sphingolipid signalling pathway was upregulated (Fig. 1f).
To reiterate if our observations are also found in model systems, we analyzed the Mice brain hemisphere (MBH) [GSE3621] dataset. Our analysis showed that the B6 dependent enzyme glutamate decarboxylase 1 (Gad1) was downregulated and is involved in GABAergic synapse while the B6 dependent enzyme dopa decarboxylase (Ddc) which is downregulated is involved in cocaine addiction, dopaminergic synapse, amphetamine addiction, serotonergic synapse and alcoholism (Fig. 1g). Further, we also looked at the proteins that interact with B6, B12 or folate or use them as a cofactor and their expression levels in the transcriptomic data (Fig. 1h). Details on Enriched pathways have been provided in Supplementary data S3.
Further, our analysis shows increased expression levels of B12 transporters [lysosomal cobalamin transport escort protein LMBD1, cubilin (Intrinsic Factor-Cobalamin Receptor) CUBN] in HD patients compared to controls (Fig. 1h). However, the transport protein of B12, transcobalamin 2 (TCN2) was found to be downregulated in HD patients (Fig. 1h). Further, the pyridoxamine 5´-phosphate oxidase (PNPO) the enzyme involved in the activation of B6 by converting it into pyridoxal 5´ phosphate was downregulated (Fig. 1h). In addition, the enzyme involved in the import of reduced folate was also found to be downregulated (Fig. 1h). The expression levels of these genes might decide the availability of these cofactors in various other enzymatic pathways which might have relevance for the disease. Since genes associated with biosynthesis, metabolism or transport of B6, B12 and folate are deregulated it is conceived that HD patients might suffer from low levels of these vitamins. Hence supplementing B6, B12 and folate will help mitigate protein aggregation associated disease progression in HD. We have used the yeast model of HD to validate these findings.
Integrated analysis cofactor-protein interaction network with metabolomic pathway data and gene expression data shows genes and metabolites that might have potential implications for disease
Commonality analysis was performed for yeast, mice model of HD and HD patients by integrated analysis of pathways obtained from cofactor-protein interaction network with metabolic datasets from literature. The mice and yeast cofactor-protein interaction network were generated from the human network using the orthologue approach as described in methods (Supplementary data S4). To the common pathways obtained from cofactor-protein network and metabolomic datasets, gene expression profiles from transcriptomic datasets were integrated.
Commonality analysis of cofactor-protein interaction network and metabolomic pathway data analyzed from yeast model of HD from the literature shows alanine, aspartate and glutamate metabolism as well as valine leucine and isoleucine biosynthesis (Fig. 2a, d). The expression levels of genes and metabolites are provided (Fig. 2e). However, significant changes in the expression levels of genes that use B6, B12 and folate as cofactors in the deregulated metabolic pathways in yeast (Fig. 2e).
Commonality analysis was performed for pathways obtained for the mice metabolomics datasets from literature and cofactor-protein interaction network (Supplementary Data S5). The results show six common pathways between the mice datasets (Fig. 2b, d). These pathways include metabolism of various amino acids (alanine, aspartate, glutamate, histidine, arginine and proline), butanoate, glyoxylate and decarboxylate metabolism (Fig. 2d). In mice datasets, many of the metabolites belonging to these pathways are found to be downregulated while significant changes were not observed in most of the genes involved in these pathways except for Gad1 (B6 cofactor) as observed in the transcriptomics datasets from different studies (Fig. 2e).
Further, commonality analysis of human metabolomic pathway datasets from literature and cofactor-protein interaction network was carried out (Fig. 2c). The pathways that were commonly altered were the metabolism of various amino acids (alanine, aspartate, glutamate, arginine, proline, phenylalanine, tyrosine and tryptophan) (Fig. 2d). Further, we compared the expression levels of genes in these deregulated metabolic pathways using transcriptomic datasets from different studies (Supplementary Data S1). In the human datasets, most of the metabolites involved in these deregulated metabolic pathways were found to be downregulated. Consistent with the results of metabolic pathways, expression levels of most of the genes in these pathways were also found to be downregulated (Enriched pathway details for the cofactor-protein interacting network has been provided in Supplementary data S4 and for metabolomics data in Supplementary data S5). The enzymes which use B6 as a cofactor belonging to six metabolic pathways, which include GOT1/2, AGXT2, GAD1/2, ODC1, DDC were downregulated while GPT and GPT2 were upregulated (Fig. 2e). In addition, the low levels of B6, B12 and folate might also affect the levels of metabolites due to reduced enzyme activity.
Yeast model of HD treated with Vitamin B6, B12 or folate or its combination induced remodelling of many pathways as well as mitigated protein aggregation
Yeast model of HD expressing N-Terminal-Htt-25Q-EGFP and N-Terminal-Htt-72Q-EGFP was used for all the studies. Yeast model of HD expressing N-Terminal-Htt-25Q-EGFP or N-Terminal-Htt-72Q-EGFP showed diffused fluorescence and fluorescent foci respectively. The yeast controls and those which were incubated with either B6, B12 or folate or a combination of B6, B12 and folate did not show any significant difference in growth (Supplementary Data S2f). Yeast model of HD treated with B6, B12 and folate in 25Q did not show any change in diffused fluorescence. The yeast model of HD expressing 72Q treated with either B6, B12 or folate showed significantly decreased aggregates compared to 72Q controls (Fig. 3a). Further, treatment with a combination of B6, B12 and folate completely abrogated aggregation in 72Q (Fig. 3b). The effective concentration for a combination of B6, B12 and folate were performed by titrating different concentrations of these vitamins to arrive at the most effective concentration (Supplementary Data S2g). Maximal decrease of protein aggregates was observed at a vitamin concentration (B6, B12 and folate) of 20 µg/ml each. Based on our results since the genes associated with biosynthesis, metabolism or transport associated with B6, B12 and folate are deregulated all subsequent were performed by using a combination of these vitamins. The fluorescent images were quantified using ImageJ as given in methods (Fig. 3a). The results showed that the fluorescent aggregate is significantly lower in the vitamin B6, B12 and folate treated sets compared to control. Further, filter retardation assay was carried out on the control and experimental sets treated with a combination of B6, B12 and folate (Fig. 3c). The results of filter retardation assay performed with a protein concentration of 50 μg and 100 μg protein concentration is provided in Fig. 3c and Supplementary Data S2q. The images were quantified using ImageJ as given in methods (Fig. 3c). A significant increase in intensity was observed in the 72Q sets as compared to 25Q sets (Fig. 3b). No Significant changes in intensity were observed between 25 and 25Q treated B6, B12 and folate (Fig. 3b). However, a significant decrease in protein aggregates was observed in the vitamin B6, B12 and folate treated sets compared to control (Fig. 3b). The results of fluorescence image data and those obtained by filter retardation assay shows concurrence between them (Fig. 3a–c). The results show significant decrease in the treated sets compared to controls. A previous study on yeast model of 25Q and 46Q has shown that after 24 h significant signals were observed both for 25Q and 46Q (Cohen et al. 2012).
To elucidate the mechanistic aspects of our findings, transcriptomic analysis of control (72Q) and experimental sets treated with a combination of B6, B12 and folate was carried out using adjusted p-value of ≤ 0.05 as described in methods. The transcriptomic analysis binned genes into 63 significant pathways which is provided in Supplementary Data S2d. Our analysis shows the significant deregulated pathways include metabolic pathways, peroxisome, autophagy and pathways involved in the protein life cycle (Fig. 3d). The metabolic pathways include fatty acid degradation, nitrogen metabolism, glycerolipid metabolism, biosynthesis of unsaturated fatty acids and purine metabolism. (Fig. 3d and Supplementary Data S2d). Further, we also analyzed the deregulated pathways with the upregulated and downregulated genes separately which yielded 46 and 36 pathways respectively (Supplementary Data S2e). The top 10 deregulated pathways obtained from upregulated and downregulated genes are provided in Fig. 3e. Furthermore, using the genes enriched in the top 10 deregulated pathways obtained from our yeast transcriptomics data, a gene interaction network was generated. The expression levels of the genes were imported into the network from our yeast transcriptomic data which has been illustrated in Fig. 3f (Details on differential expressed genes are shown in Supplementary data S6). The results show enrichment of Autophagy and Endocytosis, Glyoxylate and dicarboxylate metabolism with peroxisome and pyruvate metabolism (Fig. 3f). Our analysis also shows reduced levels of TOR1 which negatively regulate autophagy and increased expression levels of Catalase CTA1 (Fig. 3f). Taken together our results shows that B6, B12 and folate modulate metabolic pathways as well as peroxisomes, autophagy and ubiquitin mediated proteolysis which might mitigate Huntingtin protein aggregation in the yeast model of HD.
Yeast model of HD treated with a combination of Vitamin B6, B12 or folate modulated metabolomic pathways as well as mitigated protein aggregation
Further, we carried out the metabolomic analysis of the yeast model of HD expressing N-Terminal-Htt-25Q-EGFP and N-Terminal-Htt-72Q-EGFP which are treated with a combination of B6, B12 and folate. The experiments were performed in four biological replicates. All the procedures and analysis has been performed as described previously (Pulukool et al. 2021; Bhagavatham et al. 2021). Briefly, 8 million cells were homogenized after spiking internal standards. The homogenate was spun down and the resulting supernatant was passed through a 3 kDa cutoff filter as described in methods.
A total of 165 metabolites were targeted in the positive ionization mode for all the samples. In case of 25Q and 25Q treated with B6, B12 and folate 59 metabolites were detected. In case of 25Q and 72Q 58 metabolites were detected. In case of 72Q and 72Q treated with B6, B12 and folate 63 metabolites were detected. From the detected metabolites those having a CV of ≤ 20% after normalization with internal standard were used for further analysis using MetaboAnalyst 5.0. A principal component analysis (PCA) of each of the sample pairs analyzed, clustered the samples into two different groups, which is represented by a score plot (Supplementary data S2i–k). In case of 25Q and 25Q treated with B6, B12 and folate 17 significant differential metabolites were obtained (Fig. 4a). In case of 25Q with 72Q 43 significant differential metabolites were obtained (Fig. 4b). In case of 72Q with 72Q treated with B6, B12 and folate 37 significant differential metabolites were obtained (Fig. 4c). All significant differential metabolites were obtained at a False Discovery Rate (FDR) of ≤ 0.25. The significant differential metabolites were represented by a Heat map (Fig. 4a–c). Metabolite set enrichment analysis (MSEA) of the significant differential metabolites were carried out using MetaboAnalyst 5.0 as given in methods. The results are provided in the Supplementary Data S2n–p.
The pathways obtained for 25Q and 25Q treated with B6, B12 and folate, 25Q with 72Q are provided in the Supplementary Data S2o, p. The analysis of 25Q and 25Q treated with B6, B12 and folate showed enrichment of pathways belonging to aminoacyl t-RNA, metabolism of amino acids (glutamate, glutamine, arginine, proline, leucine, isoleucine and valine as well as histidine metabolism). The analysis of 25Q and 72Q treated with B6, B12 and folate showed enrichment of pathways belonging to aminoacyl t-RNA, metabolism of amino acids (glutamate, glutamine, phenylalanine, tyrosine and tryptophan biosynthesis). The pathways obtained for 72Q and 72Q treated with B6, B12 and folate along with the upregulated and downregulated differential metabolites are provided in Fig. 4g. The analysis of 72Q and 72Q treated with B6, B12 and folate showed enrichment of aminoacyl t-RNA, metabolism of amino acids (glutamate, glutamine, arginine, proline, glycine, serine, threonine, leucine, isoleucine and valine, alanine, aspartate) and glutathione metabolism (Fig. 4g). The results of our analysis have been represented by a chord diagram that shows the link between the deregulated pathways with associated metabolites Supplementary Data S2m. Further, we have also provided the deregulated pathways with the levels of associated metabolites with respect to controls (Supplementary Data S2n).
Further, we performed a commonality analysis of pathways obtained in 25Q and 72Q with those obtained from 72 and 72Q treated with B6, B12 and folate (Fig. 4d). Our analysis shows 2 pathways Glutamine and glutamate metabolism and aminoacyl-tRNA biosynthesis were common between the two sets (Supplementary Data S2o). Comparative analysis for common pathways obtained for 25Q and 25Q treated with B6, B12 and folate as well as 72Q and 72Q treated with B6, B12 and folate showed 4 common pathways between the two sets (Fig. 4e). These 4 pathways include Valine, leucine isoleucine biosynthesis, Arginine biosynthesis, Glutamine and glutamate metabolism and aminoacyl-tRNA biosynthesis (Supplementary Data S2p). Furthermore, to understand the role of cofactors (B6, B12 and folate) we generated a cofactor-protein interaction network using yeast orthologues using Cytoscape. The genes in the cofactor-protein interaction network were further binned into pathways. We then compared the metabolic pathways obtained in our yeast model system (72Q with 72Q treated with B6, B12 and folate) with the pathways obtained from the cofactor-protein interaction network. We find an overlap of six pathways between the two datasets which include metabolism of amino acids (arginine, proline, glycine, serine, threonine, valine, leucine and isoleucine) and nitrogen metabolism (Fig. 4f). These results show that B6, B12 and folate influence the functioning of metabolic pathways and modulate protein aggregation in the yeast model of HD. To further understand if there is concurrence between the transcriptomic and metabolomic datasets of the yeast model system, an integrated analysis was carried out using MetaboAnalyst 5.0.
Integrated analysis of transcriptomic and metabolomic data from the yeast model of HD shows deregulated pathways with potential implications for disease
Our integrated analysis showed that there is concordance between the metabolomic data and the transcriptomic data. A total of 35 significant pathways were found to be enriched in the integrated analysis. The pathways that were enriched include metabolism of various amino acids, glycerolipids, glutathione metabolism, fatty acid degradation, glyoxylate and dicarboxylate metabolism, autophagy, mitophagy, ubiquitin mediated proteolysis, pyruvate metabolism, glycolysis gluconeogenesis etc. (Supplementary Data S7). The significant pathways along with the genes and metabolites are provided in Supplementary Data S7. The peroxisome and autophagy pathway along with the upregulated and downregulated genes are provided (Fig. 5a). To probe the relevance of deregulated pathways to protein aggregation we employed yeast knockout for the genes from these pathways.
The enzyme activity correlates with the gene expression levels and pathways enriched in transcriptomic and metabolomic analysis in yeast model of HD
Further we asked if the changes in gene expression or the product of the enzymatic reactions correlate with the activity of marker enzymes from pathways obtained in the yeast model of HD treated with B6, B12 and folate compared to appropriate controls. Our transcriptomic, metabolomic and integrated analysis shows enrichment of alanine, aspartate and glutamate metabolism (Fig. 5b). Consistent with this the activity of ALT was significantly higher in 72Q treated with B6, B12 and folate compared to controls (Fig. 5c). The results of transcriptomic and integrated analysis also showed enrichment of peroxisome pathway and consistent with this Catalase (CTA1) activity was significantly higher in 72Q treated with B6, B12 and folate compared to controls (Fig. 5d). Taken together our results show considerable concordance between the changes in pathways observed with expression and activity of marker genes and enzymes in yeast model of HD.
Experiments using yeast knock out of genes from the deregulated pathways confirm their role in protein aggregation
Both transcriptomic and metabolomic integrated omic analysis show peroxisome and autophagy pathways having an association with the 72Q and 72Q treated with B6, B12 and folate (Fig. 5a). Yeast knockout (KO) for the peroxisomal protein- PEX7 shows elevated protein aggregate formation compared to controls (Fig. 6a, b). PEX7 is involved in the transport of peroxisomal proteins into peroxisomes (Rodrigues et al. 2015, p. 7). Further, KO of the peroxisomal protein PXP1 also leads to significantly elevated levels (1.38 fold, p ≤ 0.0012) of protein aggregation (Fig. 6a, b). PXP1 is involved in thiamine pyrophosphate containing enzyme with 2-hydroxy acyl-CoA lyase activity. Previous studies have shown that Htt aggregates are shown to be cleared out through the autophagy pathway and has implicated a role for ATG5 is demonstrated in the process (Sarkar and Rubinsztein 2008; Cortes and La Spada 2014). Consistent with previous observations yeast KO for ATG5 showed significantly elevated levels (1.62 fold, p ≤ 0.044) of protein aggregate formation (Fig. 6c). Further, we probed if the addition of a combination of B6, B12 and folate could mitigate protein aggregate formation in yeast KO for peroxisomal and autophagy genes (Fig. 6a–c). The addition of vitamin B significantly reduced protein aggregate formation in PEX7 (0.28 fold, p ≤ 3.223E−06) and ATG5 Kos (0.46 fold, p ≤ 0.0026). The absorbance of the experimental samples (Htt 72Q transformed control strain and knockout strain) with/without treatment of vitamin B6, B12 and folate showed no significant change in OD600 (Supplementary Data S2r). The results indicate that metabolic remodelling could potentially modulate protein aggregate formation in HD. For the yeast model of HD expressing 72Q controls, PEX7 and ATG5 KO treated with a combination B6, B12 and Folate or those without treatment, the fluorescence was quantified using ImageJ (Fig. 6a–c).
Discussion
Previous studies have identified B6 and B12 as major cofactors involved in many neurodegenerative diseases, highlighting their role in reducing homocysteine and its derivatives (Sharma et al. 2015). Homocysteine was shown to be elevated in HD patients (Zoccolella et al. 2006). Similarly, deregulation of cysteine metabolism and redox stress is also associated with neurodegenerative diseases including HD (Paul et al. 2018). Mutated Huntingtin protein influences homocysteine metabolism by modulating cystathionine-beta-synthase (CBS) activity (Andrich et al. 2004; Zoccolella et al. 2006). Notwithstanding this, B6 is an essential cofactor of CBS and low levels of B6 is associated with elevated homocysteine and decreased cysteine (Sharma et al. 2015).
Our transcriptomic, metabolomic and cofactor-protein interaction network analysis shows deregulation of metabolic pathways of various amino acid pathways and neurotransmitters. Previous, studies on HD patients and mice model of HD have shown low expression levels and activity of the enzyme PDXK which is involved in the activation of vitamin B6 to pyridoxal phosphate (Sorolla et al. 2016). In addition, the transcriptomic analysis of humans shows that PNPO which catalyze the synthesis of pyridoxal phosphate is downregulated (Fig. 1h). Pyridoxal phosphate is a cofactor for various reactions which include transamination, decarboxylation of amino acids and racemization (Liang et al. 2019). Consistent with this, analysis of published metabolomic datasets show deregulation of pathways modulated by B6. Reduced alanine, compromised GABAergic, serotonergic and dopaminergic signalling are reported in HD patients, which is consistent with reduced pyridoxal phosphate activity (Reilmann et al. 1995; Sorolla et al. 2016; Hsu et al. 2018; Du et al. 2013) (Cepeda et al. 2014). Our transcriptomic datasets show elevated expression levels of ALT1 and increased activity of ALT (GPT) in B6, B12 and folate treated 72Q yeast compared to 72Q controls. HD is associated with defects in the sphingolipid metabolism pathway (Di Pardo and Maglione 2018). SPTLC1 is a B6 dependent enzyme, subunit of serine palmitoyltransferase involved in the synthesis of 3-oxosphinganine. Sphingolipid-metabolizing enzymes expression levels are perturbed in both early and later stages of the disease in mice model systems and HD patients (Di Pardo and Maglione 2018). The levels of SGPL1 were increased in the brain of mice model of HD (Di Pardo et al. 2017). Taken together our results show that supplementation of B6 might mitigate defects in sphingolipid and amino acid metabolism in HD with potentially favourable consequences.
The reduced expression levels of TCN2 gene a Vitamin B12 binding and transporter protein might have potential implications for HD. Previous studies have also shown that B12 is important for neuronal health (Gröber et al. 2013). Studies have shown that B12 deficiency results in reversible chorea (Pacchetti et al. 2002). It is interesting to note that chorea is characteristic of HD(Roos 2010). Our transcriptomic and cofactor-protein interaction network shows deregulation of multiple pathways involving B12 with potential implications for HD. A previous study of the levels of B12 in the Indian population showed a deficiency of this vitamin (Singla et al. 2019), which is especially pronounced in the vegetarians compared to non-vegetarians in the Indian population (Singla et al. 2019). Taken together it is apparent that the low levels of B12 might lead to impaired neuronal function in HD patients especially in the Indian population.
Further, in our yeast model system B6, B12 or folate either alone or in combination abrogated protein aggregation. To understand the mechanistic aspects in the yeast model of HD, transcriptomic analysis of B6, B12 and folate treated sets were compared with control. Our analysis shows upregulation of ubiquitin mediated proteolysis and autophagy pathway. Further metabolomic analysis of yeast model of HD treated with a combination of B6, B12 and folate shows changes in metabolic pathways which considerably overlap with the pathways obtained from the cofactor-protein interaction network. Integrative analysis between transcriptomic and metabolomic analysis shows considerable concordance between the datasets. Previous studies on yeast and mammalian cell culture models have shown that both ubiquitin mediated proteolysis and autophagy pathway are important for the degradation of mutant Huntingtin (Seo et al. 2007; Li et al. 2010; Thibaudeau et al. 2018; Ravikumar et al. 2002; Ravikumar and Rubinsztein 2006). In our study on ATG5 KO as well as previous studies, it is evident that KO of this gene in the autophagy pathway lead to Htt protein aggregation (Komatsu et al. 2006; Metzger et al. 2010, p. 7). In both Parkinson’s disease (PD) and HD, the damaged mitochondria and the aggregation of causative protein are associated with impaired autophagy and the ubiquitin–proteasome function (Li and Li 2011; Bloom 2014; Ortega and Lucas 2014; Atik et al. 2016; Zhao et al. 2016; Harrison et al. 2019; Finkbeiner 2020). Upregulation of autophagy by drugs targeting autophagy like rapamycin/CCI-779, lithium, trehalose and rilmenidine are of interest in the treatment of HD (Ravikumar et al. 2002; Panda et al. 2019). mTOR a negative regulator of autophagy when inhibited by rapamycin, helps to clear protein aggregation in HD(Ravikumar et al. 2002, 2004; Jung et al. 2010). Our yeast transcriptomic data shows downregulation of TOR1 in 72Q treated with B6, B12 and folate. Our study also demonstrates a reduction in protein aggregation in ATG5 KO yeast model treatment with a combination of B6, B12 and folate. The Vitamin B6, B12 and folate treatment effects were shown to be independent of KO of ATG5. Hence, an involvement of autophagy in the clearance of aggregate after treatment with the three B vitamins could not be confirmed in this study. Despite this the vitamin B levels were found to modulate Htt protein aggregation or its clearance which might help to achieve a favourable prognosis and better management of HD.
Our transcriptomic analysis, as well as the integrated omic analysis, shows upregulation of peroxisomes in B6, B12 and folate treated yeast model of HD. Peroxisomes are ubiquitous organelles present in almost all tissue types of all organisms and are involved in lipid biosynthesis (Kassmann 2014; Uzor et al. 2020). Peroxisomal dysfunction is associated with various neurodegenerative diseases (Jo et al. 2020). Peroxisome biogenesis is modulated by peroxisome proliferator-activated receptor (PPAR) activation due to PPAR gamma coactivator 1a (PGC-1a) (Bagattin et al. 2010). PPARG was shown to be downregulated in mice model of HD and lymphocytes of HD patients and PPARG agonist was shown to result in favourable prognosis in mice model of HD (Chiang et al. 2010). The downregulation of PPARG leads to dysregulation of energy homeostasis (Chiang et al. 2012). 3-Nitropropionate (3NP) mice model of HD shows downregulation of oxidative stress and peroxisome markers indicate compromised peroxisome function (Moslemi et al. 2019). Consistent with the importance of peroxisomes our study shows that KO of PEX7 which aid the insertion of peroxisomal proteins into the peroxisomal membrane (Purdue et al. 1999) cause increased protein aggregation in the yeast model of HD. Mutations in PEX7b is associated with severe neurological symptoms like epilepsy (Purdue et al. 1999; Malheiro et al. 2015; Landino et al. 2017). Further, our analysis shows upregulation of fatty acid oxidation and biosynthesis of unsaturated acids in the yeast model of HD treated with B6, B12 and folate. Our transcriptomic data also shows that CTA1 expression is upregulated and catalase assay shows elevated activity in B6, B12 and folate treated 72Q compared to control 72Q set. The CTA1 is also a peroxisome marker (Horiguchi et al. 2001). From our transcriptomic analysis it is evident that the CTA1 is involved in pathways like peroxisome, glyoxylate and dicarboxylate metabolism, MAPK signaling pathway, Longevity regulating pathway and tryptophan pathway. Metabolism of cholesterol and fatty acids are deregulated in HD (Block et al. 2011). Peroxisomes are the sites of fatty acid degradation and unsaturated fatty acid biosynthesis in all organisms including yeast (Wanders et al. 2016; Schrader et al. 2016). KO of PXP1 which is a thiamine pyrophosphate containing enzyme with 2-hydroxy acyl-CoA lyase activity significantly increased protein aggregation (Fig. 6a). Deficiency of docosahexaenoic acid is the major polyunsaturated fatty acid (PUFA) in the brain and retina is associated with memory loss, learning disability and impaired vision is coordinately synthesized by endoplasmic reticulum and peroxisomes (Wanders et al. 2016; Jump 2002). Supplementation of essential fatty acids in HD was found to be beneficial as it prevents polyQ aggregation, inhibits HDAC and activates the ubiquitin–proteasome system (Das and Vaddadi 2004). Eicosapentaenoic acid and docosahexaenoic acid downregulate genes that stimulate lipid synthesis and upregulate genes that stimulate fatty oxidation (Block et al. 2011). Unsaturated fatty acids modulate membrane fluidity (Leekumjorn et al. 2009) and freshly isolated cells from HD patients have altered membrane fluidity (Sameni et al. 2018). Taken together our results show B6, B12 and folate upregulate peroxisome pathway and might play a critical role to achieve a favourable prognosis in HD.
Our transcriptomic analysis shows enrichment of glycerolipid metabolism in B6, B12 and folate treated yeast model of HD. In particular, the genes TGL3 (triglyceride lipase), PAH1 (phosphatidyl phosphatase) and ATG15 are found to be upregulated. The gene ATG15 also connect glycerolipid metabolism with autophagy (van Zutphen et al. 2014). PAH1 is involved in the conversion of phosphatidate into diacylglycerol and is important for the biogenesis of lipid droplets as well as the synthesis of other lipids (Pascual et al. 2013; Adeyo et al. 2011). TGL3 is associated with lipid droplets that help to convert triacylglycerol into diacylglycerol and then to phosphatidylcholine or phosphatidylethanolamine (Schmidt et al. 2013; de Kroon 2007; Rajakumari and Daum 2010). Axotomized retinal ganglion cells are shown to hydrolyze triacylglycerol and synthesize phospholipids which induced axon regeneration (Yang et al. 2020). Lipid droplets are utilized by vacuoles or lysosomes in a process that resembles microautophagy (van Zutphen et al. 2014), involving ATG15 a phospholipase (van Zutphen et al. 2014; Ramya and Rajasekharan 2016). In Drosophila expression of mutant Huntingtin in fat body cells or neurons lead to weight loss as well as loss of intracellular lipid stores (Lakra et al. 2019; Aditi et al. 2016). However, in fibroblasts, hepatocytes, striatal and primary neurons from mice model of HD showed compromised macroautophagy leading to accumulation of lipids in cells (Martinez-Vicente et al. 2010). In addition, the transcriptomic data also shows down regulation of TOR1 which might help to upregulate autophagy. Overall, our results from transcriptomic analysis in the yeast model of HD show changes in glycerolipid metabolism and autophagy pathway which might have implications for protein aggregate clearance.
Taken together our present results of cofactor-protein, GEO dataset transcriptomic and published metabolomic data of mice, human and yeast combined with experiments using yeast model show a critical role for B6, B12 and folate in modulating protein aggregation. Combination of B6, B12 and folate could modulate sphingolipid metabolism, peroxisomes pathway, beta-oxidation and unsaturated fatty acid biosynthesis as well as upregulating ubiquitin–proteasome system and autophagy. The results from the present study show that a combination of B6, B12 and folate could clear preformed protein aggregates in yeast model of HD has relevance for neuronal protection. The present findings concomitant with our previous results of atrophy of caudate nucleus and putamen as well as changes in brain volume in presymptomatic HD patients (Thota et al. 2021) shows that supplementation of vitamin B before the onset of symptoms in genetically susceptible individuals might help to achieve better management of HD. Hence supplementation of B6, B12 and folate might help to achieve a favourable prognosis in HD.
Conclusions
Our integrated transcriptomic and vitamin B6, B12 and folate cofactor-protein interaction network shows deregulated pathways like alanine, aspartate and glutamate metabolism, cysteine and methionine metabolism, sphingolipid signalling pathway, GABAergic, dopaminergic and serotonergic synapse, amphetamine addiction. Low levels of alanine are associated with HD patients and elevated alanine reduced the flux through glycolytic pathway which is a potential targets in the disease (Pradhan et al. 2022). The glutamine and glutamate pathway was shown to be important and over expression of glutamine synthase was protective in Drosophila (Vernizzi et al. 2020). Death of GABAergic neurons is associated with HD chorea (Lloyd 1980; Hsu et al. 2018) while many HD patients show depression associated with dysfunction of serotonin. Elevated homocysteine observed in HD leads to mitochondrial dysfunction and lipid biosynthesis (Zoccolella et al. 2006; Browne and Beal 2004; Gu et al. 1996; Block et al. 2011). Further integration of metabolic data with cofactor network and superimposing gene expression data from transcriptomic data over it shows deregulated metabolic pathways which might have potential implications for disease. Furthermore, the yeast model of HD treated with B6, B12 or folate either alone or in combination showed clearance of protein aggregates. Transcriptomic analysis of yeast model treated with a combination of B6, B12 and folate shows upregulation of autophagy, ubiquitin mediated proteolysis, peroxisome, glycerolipid metabolism, fatty acid oxidation and unsaturated fatty acid biosynthesis while purine metabolism, porphyrin and chlorophyll metabolism and ribosome are downregulated. Metabolomic analysis of yeast model treated with B6, B12 and folate in the present study show changes in aminoacyl tRNA biosynthesis, metabolism of amino acids. Integrated transcriptomic and metabolomic analysis of our yeast model showed considerable concordance between the pathways obtained. Consistent with the expression levels observed in transcriptomic data, the activity of marker enzymes ALT and CAT1 was found to be elevated in B6, B12 and folate treated 72Q compared to 72Q controls. Further yeast KO was used to validate the findings. Yeast KO for PEX7 a protein involved in peroxisomal protein import significantly increased protein aggregates, reiterating the role of peroxisome in HD. Similarly, yeast KO for PXP1 also showed increased protein aggregation. Yeast KO for ATG5 showed increased protein aggregation. The increased protein aggregation observed in all the yeast KO was mitigated by treating with a combination of B6, B12 and folate. Taken together our results show B6, B12 and folate modulate the activation of the peroxisomal pathway and upregulate autophagy and ubiquitin mediated proteolysis which might help in the clearance of protein aggregates with potential implications in HD. The results from our present study concomitant with our previously published results of atrophy and volume change in HD brain in presymptomatic patients shows that initiating therapeutic interventions before the onset of symptoms might help to achieve favourable prognosis in genetically susceptible individuals.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We acknowledge our institute Sri Sathya Sai Institute of Higher Learning, Central Research Instruments Facility (CRIF), Prasanthi Nilayam and Institute of Bioinformatics and Applied Biotechnology. We acknowledge the grant support from the Department of Biotechnology-Basic Research in Modern Biology DBT (BRB): BT/PR8226/BRB/10/1224/2013, Department of Science and Technology-The Science and Engineering Research Board–Extra Mural Research DST-SERB-EMR: EMR/2017/005381, Department of Biotechnology- Bioinformatics Infrastructure facility DBT-BIF: BT/BI/25/063/2012, Department of Science and Technology- Fund for improvement of Science and Technology Infrastructure in Higher Educational Institutions (DST-FIST): SR/FST/LSI-616/2014, University Grants Commission-Special Assistance Program (UGC-SAP III): F.3-19 /2018/DRS-III(SAP-II) for infrastructure funding. Financial support was provided by The Department of Science & Technology Fund for Improvement of S&T Infrastructure in Higher Educational Institutions (grant no. SR/FST/LSI- 5361/2012), The Department of Biotechnology, India, Glue grant (BTIPR23078/MED/29/1253/2017), and The Departments Information Technology, Biotechnology and Science and Technology, Government of Karnataka, India. Senior Research Fellowship- Department of Science and Technology‐Innovation in Science Pursuit for Inspired Research, India (DST/INSPIRE Fellowship/2016/IF160535).
Data availability
The sequencing data has been submitted to the GEO database with a Submission ID: SUB11437723 and BioProject ID: PRJNA835609 (http://www.ncbi.nlm.nih.gov/bioproject/835609).
Declarations
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Research involving Human Participants and/or Animals
We confirm that the research does not involve human participants and/or animals.
Informed consent
Since the study does not involve any human participants, informed consent is not applicable for the study.
Footnotes
Bibha Choudhary and Venketesh Sivaramakrishnan are contributed equally.
Contributor Information
Sai Sanwid Pradhan, Email: saisanwidpradhan@sssihl.edu.in.
K. Raksha Rao, Email: raksharaok@gmail.com.
Meghana Manjunath, Email: meghanamanjunath91@gmail.com.
R. Saiswaroop, Email: saiswaroopr@sssihl.edu.in
Durga Prasad Patnana, Email: patnanadurgaprasad@sssihl.edu.in.
Kanikaram Sai Phalguna, Email: saiphalguna5@gmail.com.
Bibha Choudhary, Email: vibha@ibab.ac.in.
Venketesh Sivaramakrishnan, Email: s.venketesh@gmail.com.
References
- Adeyo O, Horn PJ, Lee S, et al. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J Cell Biol. 2011;192:1043–1055. doi: 10.1083/jcb.201010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aditi K, Shakarad MN, Agrawal N. Altered lipid metabolism in Drosophila model of Huntington’s disease. Sci Rep. 2016;6:31411. doi: 10.1038/srep31411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander AG, Marfil V, Li C. Use of Caenorhabditis elegans as a model to study Alzheimer’s disease and other neurodegenerative diseases. Front Genet. 2014 doi: 10.3389/fgene.2014.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames BN, Elson-Schwab I, Silver EA. High-dose vitamin therapy stimulates variant enzymes with decreased coenzyme binding affinity (increased Km): relevance to genetic disease and polymorphisms. Am J Clin Nutr. 2002;75:616–658. doi: 10.1093/ajcn/75.4.616. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrich J, Saft C, Arz A, et al. Hyperhomocysteinaemia in treated patients with Huntington’s disease homocysteine in HD. Mov Disord. 2004;19:226–228. doi: 10.1002/mds.10629. [DOI] [PubMed] [Google Scholar]
- Arning L, Epplen JT. Genetic modifiers of Huntington’s disease: beyond CAG. Future Neurol. 2012;7:93–109. doi: 10.2217/fnl.11.65. [DOI] [Google Scholar]
- Arrasate M, Finkbeiner S. Protein aggregates in Huntington’s disease. Exp Neurol. 2012;238:1–11. doi: 10.1016/j.expneurol.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atik A, Stewart T, Zhang J. Alpha-synuclein as a biomarker for Parkinson’s disease. Brain Pathol. 2016;26:410–418. doi: 10.1111/bpa.12370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagattin A, Hugendubler L, Mueller E. Transcriptional coactivator PGC-1α promotes peroxisomal remodeling and biogenesis. PNAS. 2010;107:20376–20381. doi: 10.1073/pnas.1009176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal MF, Matson WR, Swartz KJ, et al. Kynurenine Pathway measurements in Huntington’s disease striatum: evidence for reduced formation of kynurenic acid. J Neurochem. 1990;55:1327–1339. doi: 10.1111/j.1471-4159.1990.tb03143.x. [DOI] [PubMed] [Google Scholar]
- Bhagavatham SKS, Khanchandani P, Kannan V, et al. Adenosine deaminase modulates metabolic remodeling and orchestrates joint destruction in rheumatoid arthritis. Sci Rep. 2021;11:15129. doi: 10.1038/s41598-021-94607-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block RC, Dorsey ER, Beck CA, et al. Altered cholesterol and fatty acid metabolism in Huntington disease. J Clin Lipidol. 2011;4:17–23. doi: 10.1016/j.jacl.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom GS. Amyloid-β and tau: the trigger and bullet in Alzheimer disease pathogenesis. JAMA Neurol. 2014;71:505–508. doi: 10.1001/jamaneurol.2013.5847. [DOI] [PubMed] [Google Scholar]
- Borovecki F, Lovrecic L, Zhou J, et al. Genome-wide expression profiling of human blood reveals biomarkers for Huntington’s disease. Proc Natl Acad Sci U S A. 2005;102:11023–11028. doi: 10.1073/pnas.0504921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan MJW, van der Westhuyzen J, Kramer S, Metz J. Neurotoxicity of folates: Implications for vitamin B12 deficiency and Huntington’s chorea. Med Hypotheses. 1981;7:919–929. doi: 10.1016/0306-9877(81)90046-3. [DOI] [PubMed] [Google Scholar]
- Browne SE, Beal MF. The energetics of Huntington’s disease. Neurochem Res. 2004;29:531–546. doi: 10.1023/B:NERE.0000014824.04728.dd. [DOI] [PubMed] [Google Scholar]
- Cepeda C, Murphy KPS, Parent M, Levine MS. The Role of Dopamine in Huntington’s Disease. Prog Brain Res. 2014;211:235–254. doi: 10.1016/B978-0-444-63425-2.00010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves G, Özel RE, Rao NV, et al. Metabolic and transcriptomic analysis of Huntington’s disease model reveal changes in intracellular glucose levels and related genes. Heliyon. 2017;3:e00381. doi: 10.1016/j.heliyon.2017.e00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ji B, Hao X, et al. FMN reduces Amyloid-β toxicity in yeast by regulating redox status and cellular metabolism. Nat Commun. 2020;11:867. doi: 10.1038/s41467-020-14525-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherra SJ, Chu CT. Autophagy in neuroprotection and neurodegeneration: a question of balance. Future Neurol. 2008;3:309–323. doi: 10.2217/14796708.3.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang M-C, Chen C-M, Lee M-R, et al. Modulation of energy deficiency in Huntington’s disease via activation of the peroxisome proliferator-activated receptor gamma. Hum Mol Genet. 2010;19:4043–4058. doi: 10.1093/hmg/ddq322. [DOI] [PubMed] [Google Scholar]
- Chiang M-C, Chern Y, Huang R-N. PPARgamma rescue of the mitochondrial dysfunction in Huntington’s disease. Neurobiol Dis. 2012;45:322–328. doi: 10.1016/j.nbd.2011.08.016. [DOI] [PubMed] [Google Scholar]
- Christmas RA-CI, Bolouri, H, Schwikowski, B, Anderson, M, Kelley, R, Landys, N, Workman, C, Ideker, T, Cerami, E, Sheridan, R, Bader, GD, Sander, C (2005) Cytoscape: a software environment for integrated models of biomolecular interaction networks. American Association for Cancer Research Education Book, pp 12–16, 10.1101/gr.1239303.metabolite [DOI] [PMC free article] [PubMed]
- Cohen A, Ross L, Nachman I, Bar-Nun S. Aggregation of PolyQ proteins is increased upon yeast aging and affected by Sir2 and Hsf1: novel quantitative biochemical and microscopic assays. PLoS ONE. 2012;7:4785. doi: 10.1371/journal.pone.0044785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes CJ, La Spada AR. The many faces of autophagy dysfunction in Huntington’s disease: from mechanistic pathways to therapeutic opportunities. Drug Discov Today. 2014;19:963–971. doi: 10.1016/j.drudis.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook ZR, Housman D. Huntington’s disease: can mice lead the way to treatment? Neuron. 2011;69:423–435. doi: 10.1016/j.neuron.2010.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Rajanikant GK. Huntington disease: can a zebrafish trail leave more than a ripple? Neurosci Biobehav Rev. 2014;45:258–261. doi: 10.1016/j.neubiorev.2014.06.013. [DOI] [PubMed] [Google Scholar]
- Das UN, Vaddadi KS. Essential fatty acids in Huntington’s disease. Nutrition. 2004;20:942–947. doi: 10.1016/j.nut.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Davies SW, Scherzinger E. Nuclear inclusions in Huntington’s disease. Trends Cell Biol. 1997;7:422. doi: 10.1016/S0962-8924(97)88136-6. [DOI] [PubMed] [Google Scholar]
- de Kroon AIPM. Metabolism of phosphatidylcholine and its implications for lipid acyl chain composition in Saccharomyces cerevisiae. Biochim Biophys Acta. 2007;1771:343–352. doi: 10.1016/j.bbalip.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Di Pardo A, Maglione V. Sphingolipid metabolism: a new therapeutic opportunity for brain degenerative disorders. Front Neurosci. 2018;12:249. doi: 10.3389/fnins.2018.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pardo A, Amico E, Basit A, et al. Defective Sphingosine-1-phosphate metabolism is a druggable target in Huntington’s disease. Sci Rep. 2017;7:5280. doi: 10.1038/s41598-017-05709-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Pang TYC, Hannan AJ. A tale of two maladies? Pathogenesis of depression with and without the Huntington’s disease gene mutation. Front Neurol. 2013 doi: 10.3389/fneur.2013.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrenberger PF, Fernando FS, Magliozzi R, et al. Selection of novel reference genes for use in the human central nervous system: a BrainNet Europe Study. Acta Neuropathol. 2012;124:893–903. doi: 10.1007/s00401-012-1027-z. [DOI] [PubMed] [Google Scholar]
- Duyao M, Ambrose C, Myers R, et al. Trinucleotide repeat length instability and age of onset in Huntington’s disease. Nat Genet. 1993;4:387–392. doi: 10.1038/ng0893-387. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farshim PP, Bates GP. Mouse models of Huntington’s disease. Methods Mol Biol. 2018;1780:97–120. doi: 10.1007/978-1-4939-7825-0_6. [DOI] [PubMed] [Google Scholar]
- Ferreira IL, Cunha-Oliveira T, Nascimento MV, et al. Bioenergetic dysfunction in Huntington’s disease human cybrids. Exp Neurol. 2011;231:127–134. doi: 10.1016/j.expneurol.2011.05.024. [DOI] [PubMed] [Google Scholar]
- Finkbeiner S. Huntington’s disease. Cold Spring Harb Perspect Biol. 2011 doi: 10.1101/cshperspect.a007476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner S. The autophagy lysosomal pathway and neurodegeneration. Cold Spring Harb Perspect Biol. 2020;12:a033993. doi: 10.1101/cshperspect.a033993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgini F, Guidetti P, Nguyen Q, et al. A genomic screen in yeast implicates kynurenine 3-monooxygenase as a therapeutic target for Huntington disease. Nat Genet. 2005;37:526–531. doi: 10.1038/ng1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham SF, Kumar P, Bahado-Singh RO, et al. Novel metabolite biomarkers of Huntington’s disease as detected by high-resolution mass spectrometry. J Proteome Res. 2016;15:1592–1601. doi: 10.1021/acs.jproteome.6b00049. [DOI] [PubMed] [Google Scholar]
- Graham SF, Kumar PK, Bjorndahl T, et al. Metabolic signatures of Huntington’s disease (HD): 1H NMR analysis of the polar metabolome in post-mortem human brain. Biochim Biophys Acta (BBA) Mol Basis Dis. 2016;1862:1675–1684. doi: 10.1016/j.bbadis.2016.06.007. [DOI] [PubMed] [Google Scholar]
- Graham SF, Pan X, Yilmaz A, et al. Targeted biochemical profiling of brain from Huntington’s disease patients reveals novel metabolic pathways of interest. Biochim Et Biophys Acta Mol Basis Dis. 2018;1864:2430–2437. doi: 10.1016/j.bbadis.2018.04.012. [DOI] [PubMed] [Google Scholar]
- Gröber U, Kisters K, Schmidt J. Neuroenhancement with vitamin B12—underestimated neurological significance. Nutrients. 2013;5:5031–5045. doi: 10.3390/nu5125031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Gash MT, Mann VM, et al. Mitochondrial defect in Huntington’s disease caudate nucleus. Ann Neurol. 1996;39:385–389. doi: 10.1002/ana.410390317. [DOI] [PubMed] [Google Scholar]
- Harrison TM, La Joie R, Maass A, et al. Longitudinal tau accumulation and atrophy in aging and Alzheimer disease. Ann Neurol. 2019;85:229–240. doi: 10.1002/ana.25406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmuth L. Protein clumps Hijack cell’s clearance system. Science. 2001;292:1467–1468. doi: 10.1126/science.292.5521.1467a. [DOI] [PubMed] [Google Scholar]
- Herman S, Niemelä V, Emami Khoonsari P, et al. Alterations in the tyrosine and phenylalanine pathways revealed by biochemical profiling in cerebrospinal fluid of Huntington’s disease subjects. Sci Rep. 2019 doi: 10.1038/s41598-019-40186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann W, Obeid R. Homocysteine: a biomarker in neurodegenerative diseases. Clin Chem Lab Med. 2011;49:435–441. doi: 10.1515/CCLM.2011.084. [DOI] [PubMed] [Google Scholar]
- Hodges A, Hughes G, Brooks S, et al. Brain gene expression correlates with changes in behavior in the R6/1 mouse model of Huntington’s disease. Genes Brain Behav. 2008;7:288–299. doi: 10.1111/j.1601-183X.2007.00350.x. [DOI] [PubMed] [Google Scholar]
- Hofer S, Kainz K, Zimmermann A, et al. Studying Huntington’s disease in yeast: from mechanisms to pharmacological approaches. Front Mol Neurosci. 2018 doi: 10.3389/fnmol.2018.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi H, Yurimoto H, Goh T-K, et al. Peroxisomal catalase in the methylotrophic yeast Candida boidinii: transport efficiency and metabolic significance. J Bacteriol. 2001;183:6372–6383. doi: 10.1128/JB.183.21.6372-6383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y-T, Chang Y-G, Chern Y. Insights into GABAAergic system alteration in Huntington’s disease. Open Biol. 2018;8:180165. doi: 10.1098/rsob.180165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Flockhart I, Vinayagam A, et al. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics. 2011;12:357. doi: 10.1186/1471-2105-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins BG, Rosas HD, Chen Y-CI, et al. 1H NMR spectroscopy studies of Huntington’s disease: correlations with CAG repeat numbers. Neurology. 1998;50:1357–1365. doi: 10.1212/WNL.50.5.1357. [DOI] [PubMed] [Google Scholar]
- Jo DS, Park NY, Cho D-H. Peroxisome quality control and dysregulated lipid metabolism in neurodegenerative diseases. Exp Mol Med. 2020;52:1486–1495. doi: 10.1038/s12276-020-00503-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johri A, Beal MF. Antioxidants in Huntington’s disease. Biochim Biophys Acta. 2012;1822:664–674. doi: 10.1016/j.bbadis.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner PM, Matheke RM, Smith LM, Cichewicz RH. Probing the metabolic aberrations underlying mutant huntingtin toxicity in yeast and assessing their degree of preservation in humans and mice. J Proteome Res. 2010;9:404–412. doi: 10.1021/pr900734g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jump DB. Dietary polyunsaturated fatty acids and regulation of gene transcription. Curr Opin Lipidol. 2002;13:155–164. doi: 10.1097/00041433-200204000-00007. [DOI] [PubMed] [Google Scholar]
- Jung CH, Ro S-H, Cao J, et al. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassmann CM. Myelin peroxisomes – Essential organelles for the maintenance of white matter in the nervous system. Biochimie. 2014;98:111–118. doi: 10.1016/j.biochi.2013.09.020. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Krobitsch S, Lindquist S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc Natl Acad Sci USA. 2000;97(4):1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A, Goldstein DR, Hodges A, et al. Mutant huntingtin’s effects on striatal gene expression in mice recapitulate changes observed in human Huntington’s disease brain and do not differ with mutant huntingtin length or wild-type huntingtin dosage. Hum Mol Genet. 2007;16:1845–1861. doi: 10.1093/hmg/ddm133. [DOI] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakra P, Aditi K, Agrawal N. Peripheral expression of mutant Huntingtin is a critical determinant of weight loss and metabolic disturbances in Huntington’s disease. Sci Rep. 2019;9:10127. doi: 10.1038/s41598-019-46470-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landino J, Jnah AJ, Newberry DM, Iben SC. neonatal rhizomelic chondrodysplasia punctata type 1: weaving evidence into clinical practice. J Perinat Neonatal Nurs. 2017;31:350–357. doi: 10.1097/JPN.0000000000000282. [DOI] [PubMed] [Google Scholar]
- Langmead B (2010) Aligning short sequencing reads with Bowtie. In: Curr Protoc Bioinformatics Chapter 11:Unit 11.7. 10.1002/0471250953.bi1107s32 [DOI] [PMC free article] [PubMed]
- Leekumjorn S, Cho HJ, Wu Y, et al. The role of fatty acid unsaturation in minimizing biophysical changes on the structure and local effects of bilayer membranes. Biochim Biophys Acta. 2009;1788:1508–1516. doi: 10.1016/j.bbamem.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X-J, Li S. Proteasomal dysfunction in aging and Huntington disease. Neurobiol Dis. 2011;43:4. doi: 10.1016/j.nbd.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang C-E, Huang S, et al. Inhibiting the ubiquitin–proteasome system leads to preferential accumulation of toxic N-terminal mutant huntingtin fragments. Hum Mol Genet. 2010;19:2445–2455. doi: 10.1093/hmg/ddq127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Han Q, Tan Y, et al. Current advances on structure-function relationships of pyridoxal 5′-phosphate-dependent enzymes. Front Mol Biosci. 2019 doi: 10.3389/fmolb.2019.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link CD. Transgenic invertebrate models of age-associated neurodegenerative diseases. Mech Ageing Dev. 2001;122:1639–1649. doi: 10.1016/s0047-6374(01)00291-3. [DOI] [PubMed] [Google Scholar]
- Liu W, Yang J, Burgunder J, et al. Diffusion imaging studies of Huntington’s disease: a meta-analysis. Parkinsonism Relat Disord. 2016;32:94–101. doi: 10.1016/j.parkreldis.2016.09.005. [DOI] [PubMed] [Google Scholar]
- Lloyd KG. The neuropathology of GABA neurons in extrapyramidal disorders. J Neural Transm Suppl. 1980 doi: 10.1007/978-3-7091-8582-7_25. [DOI] [PubMed] [Google Scholar]
- Maiese K. New Insights for nicotinamide: metabolic disease, autophagy, and mTOR. Front Biosci (landmark Ed) 2020;25:1925–1973. doi: 10.2741/4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malheiro AR, da Silva TF, Brites P. Plasmalogens and fatty alcohols in rhizomelic chondrodysplasia punctata and Sjögren-Larsson syndrome. J Inherit Metab Dis. 2015;38:111–121. doi: 10.1007/s10545-014-9795-3. [DOI] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, Wong E, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins D, English AM. Catalase activity is stimulated by H2O2 in rich culture medium and is required for H2O2 resistance and adaptation in yeast. Redox Biol. 2014;2:308–313. doi: 10.1016/j.redox.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascalchi M, Lolli F, Della Nave R, et al. Huntington disease: volumetric, diffusion-weighted, and magnetization transfer MR imaging of brain. Radiology. 2004;232:867–873. doi: 10.1148/radiol.2322030820. [DOI] [PubMed] [Google Scholar]
- Mason RP, Giorgini F. Modeling Huntington disease in yeast. Prion. 2011;5:269–276. doi: 10.4161/pri.5.4.18005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazarei G, Leavitt BR. Indoleamine 2,3 dioxygenase as a potential therapeutic target in Huntington’s disease. JHD. 2015;4:109–118. doi: 10.3233/JHD-159003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaddon A. Vitamin B12 in neurology and ageing; clinical and genetic aspects. Biochimie. 2013;95:1066–1076. doi: 10.1016/j.biochi.2012.11.017. [DOI] [PubMed] [Google Scholar]
- McGarry A, Gaughan J, Hackmyer C, et al. Cross-sectional analysis of plasma and CSF metabolomic markers in Huntington’s disease for participants of varying functional disability: a pilot study. Sci Rep. 2020;10:20490. doi: 10.1038/s41598-020-77526-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger S, Saukko M, Van Che H, et al. Age at onset in Huntington’s disease is modified by the autophagy pathway: implication of the V471A polymorphism in Atg7. Hum Genet. 2010;128:453–459. doi: 10.1007/s00439-010-0873-9. [DOI] [PubMed] [Google Scholar]
- Miller AL. The methionine-homocysteine cycle and its effects on cognitive diseases. Altern Med Rev. 2003;8:7–19. [PubMed] [Google Scholar]
- Morton AJ. Large-brained animal models of Huntington’s disease: Sheep. Methods Mol Biol. 2018;1780:221–239. doi: 10.1007/978-1-4939-7825-0_12. [DOI] [PubMed] [Google Scholar]
- Morton AJ, Howland DS. Large genetic animal models of Huntington’s disease. J Huntingtons Dis. 2013;2:3–19. doi: 10.3233/JHD-130050. [DOI] [PubMed] [Google Scholar]
- Moslemi M, Motamedi F, Asadi S, Khodagholi F. Peroxisomal malfunction caused by mitochondrial toxin 3-NP: protective role of oxytocin. Iran J Pharm Res. 2019;18:296–307. [PMC free article] [PubMed] [Google Scholar]
- Naik AA, Narayanan A, Khanchandani P, et al. Systems analysis of avascular necrosis of femoral head using integrative data analysis and literature mining delineates pathways associated with disease. Sci Rep. 2020;10:18099. doi: 10.1038/s41598-020-75197-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz A, Khattak NN, Khan MS, et al. Deficiency of vitamin B12 and its relation with neurological disorders: a critical review. J Basic Appl Zool. 2020;81:10. doi: 10.1186/s41936-020-00148-0. [DOI] [Google Scholar]
- Negi RS, Manchanda KL, Sanga S. Imaging of Huntington’s disease. Med J Armed Forces India. 2014;70:386–388. doi: 10.1016/j.mjafi.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak MJU, Tabrizi SJ. Huntington’s disease. BMJ. 2010;340:c3109–c3109. doi: 10.1136/bmj.c3109. [DOI] [PubMed] [Google Scholar]
- Obeid R, Herrmann W (2006) Mechanisms of homocysteine neurotoxicity in neurodegenerative diseases with special reference to dementia. 580:2994–3005. 10.1016/j.febslet.2006.04.088 [DOI] [PubMed]
- Ortega Z, Lucas JJ. Ubiquitin–proteasome system involvement in Huntington’s disease. Front Mol Neurosci. 2014;7:77. doi: 10.3389/fnmol.2014.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacchetti C, Cristina S, Nappi G. Reversible chorea and focal dystonia in vitamin B12 deficiency. N Engl J Med. 2002;347:295. doi: 10.1056/NEJM200207253470417. [DOI] [PubMed] [Google Scholar]
- Panda PK, Fahrner A, Vats S, et al. Chemical screening approaches enabling drug discovery of autophagy modulators for biomedical applications in human diseases. Front Cell Dev Biol. 2019;7:38. doi: 10.3389/fcell.2019.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual F, Soto-Cardalda A, Carman GM. PAH1-encoded phosphatidate phosphatase plays a role in the growth phase- and inositol-mediated regulation of lipid synthesis in Saccharomyces cerevisiae. J Biol Chem. 2013;288:35781–35792. doi: 10.1074/jbc.M113.525766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BD, Sbodio JI, Snyder SH. Cysteine metabolism in neuronal redox homeostasis. Trends Pharmacol Sci. 2018;39:513–524. doi: 10.1016/j.tips.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins MN, Stone TW. An iontophoretic investigation of the actions of convulsant kynurenines and their interaction with the endogenous excitant quinolinic acid. Brain Res. 1982;247:184–187. doi: 10.1016/0006-8993(82)91048-4. [DOI] [PubMed] [Google Scholar]
- Pradhan SS, Thota SM, Saiswaroop R, et al. Integrated multi-omic analysis of Huntington disease and yeast model delineates pathways modulating protein aggregation. Dis Models Mechan. 2022 doi: 10.1242/dmm.049492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulukool SK, Bhagavatham SKS, Kannan V, et al. Elevated dimethylarginine, ATP, cytokines, metabolic remodeling involving tryptophan metabolism and potential microglial inflammation characterize primary open angle glaucoma. Sci Rep. 2021;11:9766. doi: 10.1038/s41598-021-89137-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdue PE, Skoneczny M, Yang X, et al. Rhizomelic Chondrodysplasia punctata, a peroxisomal biogenesis disorder caused by defects in Pex7p, a peroxisomal protein import receptor: a mini review. Neurochem Res. 1999;24:581–586. doi: 10.1023/A:1023957110171. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S, Daum G. Janus-faced enzymes yeast Tgl3p and Tgl5p catalyze lipase and acyltransferase reactions. Mol Biol Cell. 2010;21:501–510. doi: 10.1091/mbc.E09-09-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, McBride JL, Kordower JH. Animal models of Huntington’s disease. ILAR J. 2007;48:356–373. doi: 10.1093/ilar.48.4.356. [DOI] [PubMed] [Google Scholar]
- Ramya V, Rajasekharan R. ATG15 encodes a phospholipase and is transcriptionally regulated by YAP1 in Saccharomyces cerevisiae. FEBS Lett. 2016;590:3155–3167. doi: 10.1002/1873-3468.12369. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Rubinsztein DC. Role of autophagy in the clearance of mutant huntingtin: a step towards therapy? Mol Aspects Med. 2006;27:520–527. doi: 10.1016/j.mam.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Vacher C, Berger Z, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Reilmann R, Rolf LH, Lange HW. Decreased plasma alanine and isoleucine in Huntington’s disease. Acta Neurol Scand. 1995;91:222–224. doi: 10.1111/j.1600-0404.1995.tb00438.x. [DOI] [PubMed] [Google Scholar]
- Rencus-Lazar S, DeRowe Y, Adsi H, et al. Yeast models for the study of amyloid-associated disorders and development of future therapy. Front Mol Biosci. 2019;6:15. doi: 10.3389/fmolb.2019.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues TA, Grou CP, Azevedo JE. Revisiting the intraperoxisomal pathway of mammalian PEX7. Sci Rep. 2015;5:11806. doi: 10.1038/srep11806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos RA. Huntington’s disease: a clinical review. Orphanet J Rare Dis. 2010;5:40. doi: 10.1186/1750-1172-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas HD, Doros G, Bhasin S, et al. A systems-level “misunderstanding”: the plasma metabolome in Huntington’s disease. Ann Clini Transl Neurol. 2015;2:756–768. doi: 10.1002/acn3.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sai Swaroop R, Akhil PS, Sai Sanwid P, et al. Integrated multi-omic data analysis and validation with yeast model show oxidative phosphorylation modulates protein aggregation in amyotrophic lateral sclerosis. J Biomol Struct Dyn. 2022 doi: 10.1080/07391102.2022.2090441. [DOI] [PubMed] [Google Scholar]
- Sameni S, Malacrida L, Tan Z, Digman MA. Alteration in fluidity of cell plasma membrane in Huntington disease revealed by spectral phasor analysis. Sci Rep. 2018;8:734. doi: 10.1038/s41598-018-19160-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Rubinsztein DC. Huntington’s disease: degradation of mutant huntingtin by autophagy. FEBS J. 2008;275:4263–4270. doi: 10.1111/j.1742-4658.2008.06562.x. [DOI] [PubMed] [Google Scholar]
- Saulle E, Gubellini P, Picconi B, et al. Neuronal vulnerability following inhibition of mitochondrial complex II: a possible ionic mechanism for Huntington’s disease. Mol Cell Neurosci. 2004;25:9–20. doi: 10.1016/j.mcn.2003.09.013. [DOI] [PubMed] [Google Scholar]
- Schmidt C, Athenstaedt K, Koch B, et al. Regulation of the yeast triacylglycerol lipase TGl3p by formation of nonpolar lipids. J Biol Chem. 2013;288:19939–19948. doi: 10.1074/jbc.M113.459610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader M, Costello JL, Godinho LF, et al. Proliferation and fission of peroxisomes—an update. Biochim Biophys Acta (BBA) Mol Cell Res. 2016;163:971–983. doi: 10.1016/j.bbamcr.2015.09.024. [DOI] [PubMed] [Google Scholar]
- Scott-Boyer MP, Lacroix S, Scotti M, et al. A network analysis of cofactor-protein interactions for analyzing associations between human nutrition and diseases. Sci Rep. 2016;6:19633. doi: 10.1038/srep19633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H, Sonntag K-C, Kim W, et al. Proteasome activator enhances survival of Huntington’s disease neuronal model cells. PLoS ONE. 2007;2:e238. doi: 10.1371/journal.pone.0000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon KM. Huntington’s disease—clinical signs, symptoms, presymptomatic diagnosis, and diagnosis. Handb Clin Neurol. 2011;100:3–13. doi: 10.1016/B978-0-444-52014-2.00001-X. [DOI] [PubMed] [Google Scholar]
- Sharma M, Tiwari M, Tiwari RK. Hyperhomocysteinemia: impact on neurodegenerative diseases. Basic Clin Pharmacol Toxicol. 2015;117:287–296. doi: 10.1111/bcpt.12424. [DOI] [PubMed] [Google Scholar]
- Singla R, Garg A, Surana V, et al. Vitamin b12 deficiency is endemic in Indian population: a perspective from North India. Indian J Endocrinol Metab. 2019;23:211–214. doi: 10.4103/ijem.IJEM_122_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorbi S, Bird ED, Blass JP. Decreased pyruvate dehydrogenase complex activity in Huntington and Alzheimer brain. Ann Neurol. 1983;13:72–78. doi: 10.1002/ana.410130116. [DOI] [PubMed] [Google Scholar]
- Sorolla MA, Reverter-Branchat G, Tamarit J, et al. Proteomic and oxidative stress analysis in human brain samples of Huntington disease. Free Radical Biol Med. 2008;45:667–678. doi: 10.1016/j.freeradbiomed.2008.05.014. [DOI] [PubMed] [Google Scholar]
- Sorolla MA, Rodríguez-Colman MJ, Tamarit J, et al. Protein oxidation in Huntington disease affects energy production and vitamin B6 metabolism. Free Radic Biol Med. 2010;49:612–621. doi: 10.1016/j.freeradbiomed.2010.05.016. [DOI] [PubMed] [Google Scholar]
- Sorolla MA, Rodríguez-Colman MJ, Vall-Llaura N, et al. Impaired PLP-dependent metabolism in brain samples from Huntington disease patients and transgenic R6/1 mice. Metab Brain Dis. 2016;31:579–586. doi: 10.1007/s11011-015-9777-7. [DOI] [PubMed] [Google Scholar]
- Surguchov A. Invertebrate models untangle the mechanism of neurodegeneration in Parkinson’s disease. Cells. 2021;10:407. doi: 10.3390/cells10020407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaroop RS, Pradhan SS, Darshan VMD, et al. Integrated network pharmacology approach shows a potential role of Ginseng catechins and ginsenosides in modulating protein aggregation in Amyotrophic Lateral Sclerosis. 3 Biotech. 2022;12:333. doi: 10.1007/s13205-022-03401-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauber E, Miller-fleming L, Mason RP, et al. Functional gene expression profiling in yeast implicates translational dysfunction in mutant huntingtin toxicity. J Bio Chem. 2011;286:410–419. doi: 10.1074/jbc.M110.101527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaudeau TA, Anderson RT, Smith DM. A common mechanism of proteasome impairment by neurodegenerative disease-associated oligomers. Nat Commun. 2018;9:1097. doi: 10.1038/s41467-018-03509-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thota SM, Chan KL, Pradhan SS, et al. Multimodal imaging and visual evoked potentials reveal key structural and functional features that distinguish symptomatic from presymptomatic Huntington’s disease brain. Neurol India. 2021;69:1247–1258. doi: 10.4103/0028-3886.329528. [DOI] [PubMed] [Google Scholar]
- Tsang TM, Woodman B, Mcloughlin GA, et al. Metabolic characterization of the R6/2 transgenic mouse model of Huntington’s disease by high-resolution MAS 1H NMR spectroscopy. J Proteome Res. 2006;5:483–492. doi: 10.1021/pr050244o. [DOI] [PubMed] [Google Scholar]
- Uzor N-E, McCullough LD, Tsvetkov AS. Peroxisomal DYSFUNCTION IN NEUROLOGICAL DISEASES AND BRAIN AGING. Front Cell Neurosci. 2020 doi: 10.3389/fncel.2020.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme P, Robberecht W, Van Den Bosch L. Modelling amyotrophic lateral sclerosis: progress and possibilities. Dis Model Mech. 2017;10:537–549. doi: 10.1242/dmm.029058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zutphen T, Todde V, de Boer R, et al. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2014;25:290–301. doi: 10.1091/mbc.E13-08-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernizzi L, Paiardi C, Licata G, et al. Glutamine synthetase 1 increases autophagy lysosomal degradation of mutant huntingtin aggregates in neurons, ameliorating motility in a drosophila model for Huntington’s disease. Cells. 2020;9:196. doi: 10.3390/cells9010196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanders RJA, Waterham HR, Ferdinandusse S. Metabolic interplay between peroxisomes and other subcellular organelles including mitochondria and the endoplasmic reticulum. Front Cell Dev Biol. 2016 doi: 10.3389/fcell.2015.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Benner MJ, Hancock REW. NetworkAnalyst–integrative approaches for protein-protein interaction network analysis and visual exploration. Nucleic Acids Res. 2014;42:W167–174. doi: 10.1093/nar/gku443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Gill EE, Hancock REW. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat Protoc. 2015;10:823–844. doi: 10.1038/nprot.2015.052. [DOI] [PubMed] [Google Scholar]
- Xie N, Zhang L, Gao W, et al. NAD+ metabolism: pathophysiologic mechanisms and therapeutic potential. Sig Transduct Target Ther. 2020;5:227. doi: 10.1038/s41392-020-00311-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Bailey A, Kuleshov MV, et al. Gene set knowledge discovery with Enrichr. Curr Protoc. 2021;1:e90. doi: 10.1002/cpz1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Wang X, Wang J, et al. Rewiring neuronal glycerolipid metabolism determines the extent of Axon Regeneration. Neuron. 2020;105:276–292.e5. doi: 10.1016/j.neuron.2019.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Hong Y, Li S, Li X-J. Compartment-dependent degradation of mutant huntingtin accounts for its preferential accumulation in neuronal processes. J Neurosci. 2016;36:8317–8328. doi: 10.1523/JNEUROSCI.0806-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Soufan O, Ewald J, et al. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47:W234–W241. doi: 10.1093/nar/gkz240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoccolella S, Martino D, Defazio G, et al. Hyperhomocysteinemia in movement disorders: current evidence and hypotheses. Curr Vasc Pharmacol. 2006;4:237–243. doi: 10.2174/157016106777698414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data has been submitted to the GEO database with a Submission ID: SUB11437723 and BioProject ID: PRJNA835609 (http://www.ncbi.nlm.nih.gov/bioproject/835609).