Abstract
In the work reported here we have undertaken a functional dissection of a Polycomb response element (PRE) from the iab-7 cis-regulatory domain of the Drosophila melanogaster bithorax complex (BX-C). Previous studies mapped the iab-7 PRE to an 860-bp fragment located just distal to the Fab-7 boundary. Located within this fragment is an ∼230-bp chromatin-specific nuclease-hypersensitive region called HS3. We have shown that HS3 is capable of functioning as a Polycomb-dependent silencer in vivo, inducing pairing-dependent silencing of a mini-white reporter. The HS3 sequence contains consensus binding sites for the GAGA factor, a protein implicated in the formation of nucleosome-free regions of chromatin, and Pleiohomeotic (Pho), a Polycomb group protein that is related to the mammalian transcription factor YY1. We show that GAGA and Pho interact with these sequences in vitro and that the consensus binding sites for the two proteins are critical for the silencing activity of the iab-7 PRE in vivo.
Segment identity in the posterior two-thirds of the Drosophila melanogaster embryo, from parasegment 5 (PS5) to PS14, is determined by the pattern of expression of the bithorax complex (BX-C) homeotic genes, Ultrabithorax (Ubx), abdominal-A (abd-A), and Abdominal-B (Abd-B) (13, 32, 39, 46). These three homeotic genes are regulated by an elaborate cis-regulatory region that spans a DNA segment of over 300 kb. This large cis-regulatory region is subdivided into nine functionally autonomous domains, abx/bx, bxd/pbx, and iab-2 to iab-8 (2, 9, 29, 32). Each domain specifies the identity of a specific parasegment by activating one of the BX-C homeotic genes in a pattern appropriate for that parasegment. For example, the iab-5 cis-regulatory domain regulates Abd-B expression in a pattern that confers PS10 identity to the cells in this parasegment. Similarly, the iab-6, iab-7, and iab-8 cis-regulatory domains activate Abd-B expression in patterns appropriate for PS11, PS12, and PS13 identity, respectively (5, 9, 45). When one of the BX-C cis-regulatory domains is inactivated, the parasegment specified by the affected regulatory domain is transformed into a copy of the parasegment immediately anterior. Thus, in a deletion that inactivates iab-7, iab-7Sz, PS12 is transformed into a duplicate copy of PS11 (16). In this case, Abd-B expression in both PS11 and PS12 is driven by the iab-6 cis-regulatory domain.
The regulation of the BX-C homeotic genes during embryogenesis is subdivided into two phases: initiation and maintenance. In the initiation phase, the products of the gap and pair rule segmentation genes are responsible for initiating the parasegment-specific expression of the BX-C homeotic genes. These proteins are thought to interact with target sequences in the nine cis-regulatory domains (27, 36, 43, 48, 57, 59). However, the products of the segmentation genes are present only transiently in the early embryo, and regulation switches to a maintenance mode that recognizes and propagates the initial pattern during subsequent stages of development. Maintenance requires the trithorax-Group (trx-G) and Polycomb-Group (Pc-G) genes (reviewed in references 22, 38, 40, 50). The trx-G genes function to keep the homeotic genes on, while the Pc-G genes are negative regulators and function to maintain the inactive state of the homeotic genes. Experiments with homeotic reporter constructs have identified elements, called Polycomb response elements (PREs), in several of the BX-C cis-regulatory domains which appear to be targets for Pc-G action. When these PREs are combined with a parasegment-specific initiator, they maintain the segmentally restricted pattern of expression conferred on the reporter by the initiation element (7, 8, 10, 11, 23, 42, 51). In addition to this maintenance activity, the PREs have an unusual pairing-sensitive silencing activity (10, 19, 23, 30, 31, 37, 49). When they are included in a mini-white transgene, the PREs repress or even eliminate mini-white expression when the animals are homozygous for the transgene insert. Like classical transvection, pairing-sensitive silencing typically depends on whether these elements can pair. Silencing is observed in animals homozygous for the same mini-white insertion but is usually not found in animals which have the mini-white transposon inserted at two different locations.
One of the best-characterized BX-C PREs is the iab-7 PRE. This PRE is located in the iab-7 cis-regulatory domain, just distal to the Fab-7 boundary. When a restriction fragment containing the iab-7 PRE is combined with a bxd initiation element, it maintains the appropriate anterior limit (PS6) of reporter gene expression (23). The same iab-7 PRE fragment also functions as a pairing-sensitive silencer of mini-white. Like those of other PREs, both the maintenance and the pairing-sensitive silencing activities of the iab-7 PRE fragment depend on products of the Pc-G genes.
Insights into the function of the iab-7 PRE have also come from an analysis of the phenotypic effects of deletions that remove this PRE and/or the adjacent Fab-7 boundary in BX-C itself (16, 35). The simplest case involves deletions that remove both the PRE and the boundary, such as Fab-71. When the Fab-7 boundary is absent, the normally autonomous iab-6 and iab-7 cis-regulatory domains fuse into a single domain. In the fused domain, the positive elements in iab-6 ectopically activate iab-7 in PS11. As a consequence, Abd-B expression in PS11 is driven by the iab-7, not the iab-6, cis-regulatory domain, and these deletions produce a dominant gain-of-function phenotype, completely transforming PS11 into a duplicate copy of PS12. Deletions that remove only the boundary also produce a dominant gain-of-function transformation of PS11; however, they differ from the larger deletions such as Fab-71 in that there are often small clones of cells in PS11 which exhibit a loss-of-function phenotype and assume PS10 identity. In these clones, iab-6 is ectopically silenced in PS11 and Abd-B expression is controlled by iab-5. The mixed gain- and loss-of-function phenotypes due to deletions that remove just the boundary arise because there is a competition between positive elements in iab-6 that ectopically activate iab-7 and negative elements in iab-7 that ectopically silence iab-6. While this competition between positive and negative elements also occurs in the larger deletions, the silenced state is thought to be unstable because the iab-7 PRE is absent, leading to the constitutive activation of iab-7. Finally, deletions that remove only the iab-7 PRE produce no phenotype in heterozygotes; however, when homozygous or trans to a deficiency, a low percentage of the flies exhibit a gain-of-function transformation in which small clones of cells in PS11 have a PS12 identity (35). The poorly penetrant gain-of-function phenotype produced by the iab-7 PRE deletions suggests that the iab-7 cis-regulatory domain contains ancillary PRE-like elements that can help maintain the determined state.
Like other PREs from the Abd-B region of BX-C, the iab-7 PRE is associated with a prominent chromatin-specific nuclease-hypersensitive site called HS3 (16, 28). HS3 is approximately 230 bp in length and contains several recognizable sequence motifs. These include potential binding sites for the GAGA factor and the Zeste (z) protein. The GAGA factor is encoded by the Trithorax-like (Trl) gene and is thought to function in the formation and/or maintenance of nucleosome-free regions of chromatin (14, 33, 56). The Zeste protein has previously been implicated in pairing-dependent interactions at the white locus (41, 58). The idea that these sequence motifs might be important for the function of the iab-7 PRE is supported by the finding that mutations in both Trl and z reduce silencing activity (23). The HS3 nuclease-hypersensitive site also contains two copies of a 14-bp motif (KCRGCCATYDNNGD). This motif has a five-nucleotide core, GCCAT, with two additional conserved residues at position −3 (C) and +9 (G). Copies of this motif are found in the nuclease-hypersensitive sites associated with PREs located in three other Abd-B cis-regulatory domains, iab-5, iab-6, and iab-8 (1, 34; unpublished data). The same motif is also found in PREs from the Ubx region of BX-C and in PREs from both the Sex combs reduced and engrailed regulatory regions. Recent studies by Brown et al. (6) on a PRE from the engrailed (en) gene indicate that this motif is a binding site for the Pleiohomeotic protein (Pho), which is the Drosophila homolog of the mammalian transcription factor YY1.
In the studies reported here, we have analyzed the sequences required for the silencing activity of the iab-7 PRE in transgene assays. We show that sequences conferring silencing activity map to HS3. We also present evidence that the GAGA and Pho proteins bind to target sites in this hypersensitive region and that the consensus recognition sequences for these proteins are required for silencing activity in vivo.
MATERIALS AND METHODS
P-element transformation.
All transgenic flies were produced using a w1 strain.
Pairing-sensitive lines in pho background.
To examine the eye colors of flies containing the wild-type iab-7 PRE mini-white constructs in a pho mutant background, w−;TM6B,Sb,Tb/+;CiD/+ males were crossed to w−;6-22A T(3;4)69BC;101EF/TM3 virgins [the 6-22A T(3;4)69BC;101EF chromosome was a very kind gift from M. Müller). From the progeny of this cross, we selected w−; 6-22A T(3;4)69BC; 101EF/TM6B,Sb,Tb;CiD virgins, which were crossed either to w−;pho1/CiD or to w+;phoCV/CiD males (the pho alleles were a kind gift from J. Kassis). After these crosses, we collected w;TM6B, Sb,Tb/+;pho1/CiD and w−;TM6B,Sb,Tb/+;phoCV/CiD males, which were mated to virgins homozygous for one of the five third-chromosome pairing-sensitive iab-7 PRE inserts (w−;Ins-3X/Ins-3X). From the offspring of these crosses w−;Ins-3X/TM6B,Sb,Tb;pho1/+ virgins were crossed to w−;Ins-3X/TM6B,Sb,Tb; phoCV/+ males. In the progeny, pho1/phoCV pharate adults were recognized by their homeotic phenotype while the hetero- or homozygous state of the third-chromosome inserts was monitored by the dominant markers (Sb and Tb) on the TM6B balancer chromosome.
Construction of the 410- and 260-bp iab-7 PRE fragments.
The 410- and the 260-bp iab-7 PRE fragments were amplified by PCR from a 3.35-kb HindIII-to-XbaI Fab-7 fragment inserted into Bluescript. This Fab-7 region was originally isolated from phage lambda 8053 and spans bp 163 to 3517 as indicated previously (28). The 410-bp fragment was generated using two primers, FABP-1 (TGCTCTAGAGCAACTTCCTTCGTCCGTC) and FABP-4 (TGCTCTAGATGTCGGCAATTCGGATTC). The 260-bp fragment was also generated using two primers, FABP-2 (TGCTCTAGAGTTTCGTCGCTCACG) and FABP-4. These fragments were cloned into Bluescript at the XbaI site. An XhoI/NotI fragment was excised and inserted into the whiteenhancer-mini-white vector (23) in between the white enhancer and the mini-white gene. Both fragments were confirmed by sequencing.
Construction of the 260-bp HS3 fragment with mutations in the two GAGA sites.
Mutations in the two consensus GAGA binding sites were introduced using primers SES16 (CATGGATGTGAAACTTAACGTGCTCTTCGCGCATTGCGCTCGCGCTC) and SES17 (GAGCGCGAGCGCAATGCGCGAAGAGCACGTTAAGTTTCACATCCATG). The point mutations are indicated in boldface. A proximal fragment was amplified by PCR using primers FABP-2 and SES16, and a distal fragment was generated using primers SES17 and FABP-4. These two fragments were combined together for a second round of PCR amplification using primers FABP-2 and FABP-4. The larger fragment was cloned into Bluescript at the XbaI site. An XhoI/NotI fragment was excised and inserted into the whiteenhancer-mini-white vector (23) in between the white enhancer and the mini-white gene. The mutated fragment was confirmed by sequencing.
Gel mobility shift assay.
Oligonucleotides containing the wild-type and mutated Pho recognition motif were kinased with [α-32P]ATP. The complementary oligonucleotides were annealed by mixing them at equal molar concentrations, heating them at 65°C for 10 min, and then allowing them to cool to room temperature slowly before incubating them at 4°C for 30 min. The probes were then stored at −20°C.
The full-length Pho-expressing construct was a kind gift from Judy Kassis. Pho was expressed in a BL21 strain of Escherichia coli. Extraction of proteins in native form was carried out as described previously (52). The total protein extract was used directly for the gel mobility shift experiments. Nuclear extract was prepared from 0- to 16-h Drosophila embryos according to published procedures with minor modifications (25).
In a typical gel mobility shift experiment, 2 to 5 μg of protein extract was incubated with a mixture consisting of 105 cpm of end-labeled DNA (around 1 fmol), 1 μg of poly(dI-dC), and 5 μg of tRNA in binding buffer (25 mM HEPES [pH 7.6], 100 mM NaCl, 0.1 mM EDTA, 1 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride, and 10% glycerol) at room temperature for 30 min. The mixture was then analyzed on a 4% acrylamide-bisacrylamide (80:1) gel in 0.5× Tris-borate-EDTA (TBE) containing 2.5% glycerol. The gel was run in 0.5× TBE at 10 V/cm at room temperature, dried and exposed to X-ray film. The following oligonucleotide pairs were used to make the double-stranded probes: wt, CAGCTCGGCCATCATGGGG and CCCCATGATGGCCGAGCTG; mc, CAGCTCGGCACGCATGGGG and CCCCATGCGTGCCGAGCTG; m9, CAGCTCGGCCATCATGTGG and CCACATGATGGCCGAGCTG.
Binding of GAGA to the iab-7 PRE.
DNA to prepare the HS3 iab-7 PRE affinity matrix was made by PCR amplification using the same primers used to generate the 260-bp iab-7 PRE fragment (see above) except that one of the primers had a 5′ biotin modification. The 260-bp PCR product was purified from a 2% agarose gel using a Qiagen kit for extraction of DNA from agarose gels. The purified DNA was incubated with a 10-μl suspension of streptavidin-coated magnetic beads (from Dynal) by following the supplier's protocol. Saturated binding of DNA to the beads was monitored by comparing the input and unbound DNA on a 2% agarose gel. DNA matrices were also prepared using a 260-bp segment of Fab-7 which does not contain any consensus GAGA binding sites and a 270-bp segment from the Ubx promoter region (−251 to +19) which contains four consensus GAGA binding sites.
The nuclear extract was incubated with the affinity matrix under reaction conditions identical to those used for the gel shift assays. After a 30-min incubation at room temperature the affinity matrix beads were separated from the unbound proteins and washed with 0.1 M KCl in elution buffer containing 25 mM HEPES (pH 7.6), 1 mM dithiothreitol, 20% glycerol, and 0.1% Nonidet P-40. Bound proteins were then eluted by steps of increasing KCl concentration. Eluted fractions were separated by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS–10% PAGE), blotted onto an Immobilon polyvinylidene difluoride (PVDF) transfer membrane (from Millipore), and processed for Western analysis using an anti-GAGA antibody kindly provided by Jordan Raff (44).
RESULTS
Silencing activity maps to the iab-7 PRE nuclease-hypersensitive site.
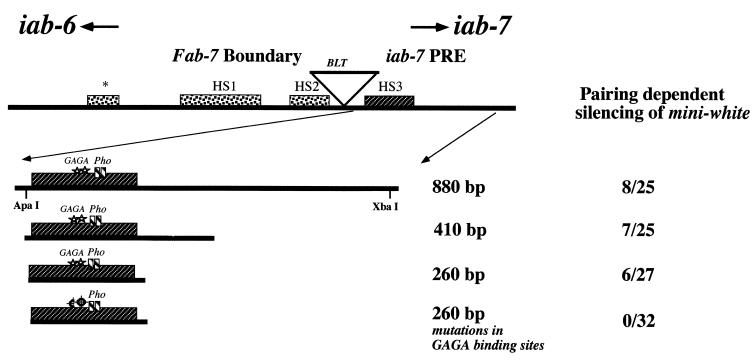
Shown in Fig. 1 is a map of the Fab-7 boundary and iab-7 PRE DNA segment from BX-C. Previous studies indicate that the Fab-7 boundary is defined by the three proximal nuclease-hypersensitive regions, HS∗, HS1, and HS2, while the iab-7 PRE maps to an 860-bp ApaI-XbaI fragment just distal to the boundary (23, 24, 35). This 860-bp fragment functions as a Pc-G-dependent silencer in two different assays, a mini-white silencing assay and a bxd-Ubx maintenance assay (23). One of the notable features of the 860-bp iab-7 PRE fragment is the ∼230-bp nuclease-hypersensitive region, HS3. Although chromatin-based silencing mechanisms are generally thought to be associated with decreased rather than increased accessibility, we have found that PREs from elsewhere in the Abd-B cis-regulatory region also map to restriction fragments which contain nuclease-hypersensitive regions in chromatin digests (1, 28; unpublished data). This correlation suggested that HS3 may contain target sites for proteins that function to recruit Pc-G proteins and hence might be essential for establishing and maintaining Pc-G-dependent silencing.
FIG. 1.
Defining the minimal PRE and mutations in GAGA binding sites. The Fab-7 boundary and iab-7 PRE region of the BX-C are shown at the top. These two elements lie adjacent to one another, and both map between the PS11 regulatory domain, iab-6, on the left and the PS12 regulatory domain, iab-7, on the right. The Bluetail transposon, BLT, which is subject to the control of iab-7 but not iab-6, lies between hypersensitive sites HS2 and HS3 (16). The 860-, 410-, and 260-bp fragments containing HS3 are shown. The number of pairing-sensitive lines out of the total number of transgenic lines of each construct is shown to the right. Stars, GAGA consensus binding sites; small boxes, Pho consensus binding sites. The bottom construct depicts the 260-bp fragment containing mutations in both of the GAGA consensus binding sites.
To explore this possibility, we asked whether sequences spanning HS3 are sufficient to recapitulate the silencing activity of the 860-bp iab-7 PRE fragment. We first tested a 410-bp fragment which extends from the ApaI site to near the middle of the 860-bp fragment and which includes HS3. The 410-bp fragment was placed in the same whiteenhancer-mini-white vector as that used previously to test the silencing activity of the full-length fragment (23). As indicated in Fig. 1, the frequency of wenhancer-mini-white transformants exhibiting pairing-dependent silencing for the 410-bp fragment is close to that for the full-length fragment. (An example of pairing-sensitive silencing for another iab-7 PRE mini-white transgene is shown in Fig. 4. Note that the eye color of flies hemizygous for this transgene insert is dark orange, while the eye color of flies homozygous for this transgene is white.) Since the precise endpoints of HS3 are not known (+/− 30 to 40 bp), we next tested a 260-bp fragment that is estimated to be slightly larger than the nucleosome-free region and that should extend only slightly beyond its likely limits (Fig. 1). As with the 410-bp fragment, the frequency of wenhancer-mini-white transformants exhibiting pairing-dependent silencing with the 260-bp fragment is close to that of the intact ApaI-XbaI fragment.
FIG. 4.
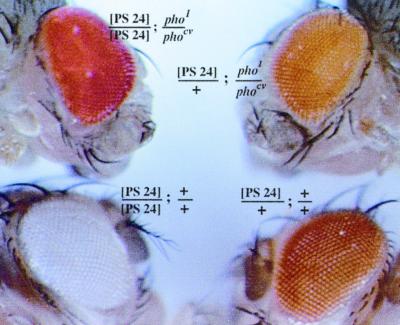
Pho is required for the silencing activity of the iab-7 PRE. Shown is the eye color phenotype of one of the 860-bp iab-7 PRE mini-white transgenic lines, P24, in different genetic backgrounds. As a hemizygote in a wild-type background, this insert produces a light orange eye color phenotype. When the transgene is homozygous, mini-white expression is repressed, and the eye color of the P24/P24 animals is almost white. The pho gene is required for the iab-7 PRE-dependent silencing of mini-white. This can be seen by comparing the eye color of P24/P24; pho1/phocv animals with that of wild-type P24/P24 animals.
To confirm that the silencing activity of the 410- and 260-bp fragments is mediated by Pc-G proteins, we tested the effects of mutations in Pc-G genes. As observed for the full-length iab-7 PRE fragment (23), the silencing of mini-white by the two smaller fragments is suppressed by mutations in the Pc-G group genes (Table 1 and data not shown). The results for Polycomb and Sex combs midleg are shown in Table 1. These findings provided further evidence that HS3 contains target sites for proteins that function to establish and maintain Pc-G-silencing complexes.
TABLE 1.
Interaction of the iab-7 PRE with Pc-G and trxG mutationsa
| Gene testedb | Allele | Interaction with:
|
|||||
|---|---|---|---|---|---|---|---|
| Pairing-sensitive line
|
Non-pairing-sensitive line
|
||||||
| 410.1 | 410.2 | 260.1 | 260.2 | 410.3 | 260.3 | ||
| Polycomb | Pc1 | 0 | + | + | + | 0 | 0 |
| Sex Combs Midleg | ScmET50 | + | + | + | + | 0 | 0 |
| ScmSuz302 | + | + | + | + | 0 | 0 | |
| Trithorax-like | Trl13c | + | 0 | ||||
Interactions of homozygous 410- and 260-bp iab-7 transgenic lines with Polycomb, Sex Combs Midleg, and Trithorax-like alleles. +, increased mini-white expression; 0, no change in expression.
Polycomb and Sex Combs Midleg are PcG genes; Trithorax-like is a trxG gene.
The silencing activity of the HS3 fragment requires the GAGA protein.
Chromatin immunoprecipitation experiments by Strutt et al. (55) indicate that the GAGA factor interacts with sequences from the iab-7 PRE DNA segment in vivo. They found that DNA fragments spanning the iab-7 PRE region could be immunoprecipitated from formaldehyde-cross-linked chromatin with antibodies directed against the GAGA protein. There are two sequences in the 860-bp ApaI-XbaI iab-7 PRE fragment that closely match the consensus GAGA protein binding site. These potential binding sites are located near the middle of HS3 (Fig. 1). They are arranged in opposite orientations and are separated by 12 bp (see Fig. 3).
FIG. 3.
Mutations in Pho consensus binding sites. Black bar, 860-bp iab-7 PRE ApaI-XbaI fragment; large box, hypersensitive site 3; stars and small boxes, GAGA and Pho consensus binding sites, respectively. The 860-bp iab-7 PRE fragment was inserted into an enhancer-less mini-white transgene. Portions of the sequence for each of the fragments tested are shown. The number of pairing-sensitive lines out of the total number of transgenic lines is shown for each construct. (A) Wild-type sequence; (B) 15-bp deletion that removes the proximal Pho consensus binding site; (C) 60-bp deletion that removes both Pho consensus binding sites and the intervening sequence; (D) point mutations generated in the Pho core consensus sequence; (E) point mutations generated in both of the +9 conserved residues. Boldface, conserved sequences within the Pho consensus binding site; italics, GAGA consensus sites. For the point mutations (D and E), the mutated sequence is shown in boldface above the residues that were mutated.
We have previously shown that the silencing activity of the 860-bp iab-7 PRE fragment is reduced by mutations in the gene encoding the GAGA factor, Trl. Since the GAGA binding sites in the 860-bp iab-7 fragment map to HS3, one would predict that the silencing activity of the small 260-bp HS3 fragment would also require the GAGA factor. This seems to be the case. As shown in Table 1, mini-white silencing by the 260-bp fragment is suppressed by a Trl mutation.
GAGA protein interacts with target sites in HS3.
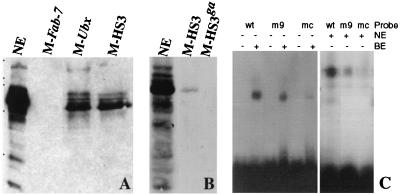
To demonstrate that the GAGA factor interacts with the two recognition sequences in HS3, we incubated nuclear extracts with magnetic beads containing the 260-bp HS3 fragment. After the beads were washed, bound protein was eluted from them with increasing concentrations of salt and analyzed by Western blotting. As a positive control we linked a fragment from the Ubx promoter, which is known to interact with GAGA protein in vitro, to the magnetic beads. As a negative control we used a fragment from a region of the Fab-7 boundary that has no consensus GAGA binding sites. As shown for the 0.4 M salt fraction in Fig. 2A, the GAGA protein is bound to beads containing the HS3 and Ubx promoter fragments but is not bound to the control Fab-7 beads. To confirm that GAGA binding to HS3 depends on the two consensus sites, we mutated the two sites. As shown in Fig. 2B, when nuclear extracts are incubated with magnetic beads containing the mutant HS3 fragment, the GAGA protein is not detected in the salt-eluted fractions.
FIG. 2.
GAGA and Pho proteins bind to recognition sequences in HS3. (A) Binding of GAGA protein to the iab-7 PRE. In this experiment nuclear extract was incubated with matrix containing different DNA sequences. The first was a control sequence from a region of Fab-7 that does not contain any consensus GAGA protein recognition sequences. The second was a fragment from the Ubx promoter, which contains four consensus GAGA recognition sequences. The third was the 260-bp iab-7 PRE fragment spanning HS3 (see text and Fig. 1). It contains two consensus GAGA recognition sequences. Proteins bound to each matrix were eluted with increasing salt concentrations. Shown is the 0.4 M KCl wash. The protein samples eluted from each matrix were separated by SDS–10% PAGE and transferred on a PVDF membrane for Western analysis with an anti-GAGA antibody. Similar quantities of nuclear extract and affinity matrix were used in order to compare the relative abundances of protein in each fraction. While multiple isoforms of GAGA (plus presumptive breakdown products) are detected by the antibody in nuclear extracts, only a subset of these bind to the matrix. For the HS3 matrix, the major species is the lowest band, which corresponds to the 70-kDa isoform, while larger isoforms are generally present in lower yield. Note that little or no GAGA protein appears to be bound to the Fab-7 matrix, which does not contain consensus GAGA binding sites. M-Fab-7, matrix containing Fab-7 DNA; M-Ubx, matrix containing Ubx promoter DNA; M-HS3, matrix containing HS3 DNA. (B) Binding of GAGA protein to the iab-7 PRE depends on the two consensus GAGA binding sites. We compared the binding of the GAGA factor to an affinity matrix containing the wild-type HS3 sequence and a matrix containing the HS3 sequence with mutations in the two consensus GAGA binding sites (see text and Fig. 1). The 70-kDa isoform binds to the matrix containing the wild-type HS3 sequence (M-HS3) but does not bind to the matrix containing the HS3 sequence with GAGA binding site mutations (M-HS3ga). (C) Labeled double-stranded oligonucleotide probes (see Materials and Methods). wt, wild type; m9, mutated at position 9 (G to T); mc, mutated at core positions 2, 3, and 4 (CAT to ACG). Two to five micrograms of protein extract (NE, nuclear extract; BE, bacterial extract expressing Pho) was incubated with the labeled DNA and cold carrier (poly[dI-dC] and tRNA) in binding buffer at room temperature for 30 min. The mixture was then analyzed on a 4% acrylamide-bisacrylamide (80:1) gel in 0.5× TBE containing 2.5% glycerol. Specificity was confirmed by competition experiments in which a 100-fold molar excess of either wild-type or mutant oligonucleotides was included in the binding mixture. The core mutant was found to be a much weaker competitor than the wild-type sequence. To a lesser extent this was also true of the +9 mutant (data not shown).
The GAGA binding sites in HS3 are important for PRE function in vivo.
The experiments described in the previous section indicate that the binding of the GAGA protein in nuclear extracts to HS3 requires the two consensus recognition sequences. Since mutations in Trl disrupt the silencing activity of the 260-bp iab-7 PRE fragment (23), we predicted that the two GAGA recognition sequences are essential for the in vivo function of this PRE. This prediction is correct. As indicated in Fig. 1, when the GAGA binding sites in the 260-bp HS3 fragment are mutated, HS3 is unable to silence the wenhancer-mini-white transgene.
Interaction of Pho with the iab-7 PRE.
In addition to the GAGA protein binding sites, the iab-7 PRE contains two copies of a 14-bp sequence motif. These two motifs are located in the distal half of HS3, just beyond the two GAGA binding sites (see Fig. 3). They are arranged in opposite orientations and are separated from each other by 32 bp. A similar motif has been found in PREs from elsewhere in BX-C and in other Drosophila genes (34). As indicated in Fig. 3, this conserved motif consists of an 8-bp central core sequence, CRGCCATY. In addition there is a conserved G at position +9. Studies by Brown et al. (6) have shown that this motif is a binding site for the Pho protein in an en PRE. However, since the noncore sequences in the iab-7 PRE motifs are quite different from those in the en PRE (34), an important question is whether Pho can actually bind to the iab-7 PRE sequences.
To address this problem, we asked whether the Pho protein expressed in bacteria would gel shift a short double-stranded oligonucleotide probe spanning the proximal motif. As shown in Fig. 2C, the Pho protein expressed in bacteria binds to the oligonucleotide probe. To determine whether this interaction depends on the 8-bp core motif, the CRGCCATY core sequence was changed from CGGCCATC to CGGCACGC (mutated bases are in boldface). As shown in Fig. 2C, little or no shifting of the mutant probe is observed with the bacterial Pho protein. We also tested the effects of mutation in the G residue (G→T) at position +9. This mutation causes a small but reproducible reduction in the yield of the shifted probe. These findings indicate that the Pho protein can interact with the conserved sequence motif in the iab-7 PRE.
We also used the same set of oligonucleotide probes to assay for DNA binding activity in nuclear extracts. As shown in Fig. 2C, the wild-type probe gives a gel shift with nuclear extract similar to that observed for the bacterial Pho protein. Moreover, as was observed for bacterial Pho, the core sequence mutations essentially eliminate the shift with the nuclear extract, while the yield of the shifted probe is reduced by the G→T mutation at position +9. These results suggest that Pho protein in nuclear extracts (or a protein with similar sequence specificity) binds to the conserved iab-7 PRE sequence motif.
The conserved sequence motif is required for silencing activity in vivo.
If Pho protein binding to the conserved sequence motifs in the iab-7 PRE is important in vivo, then mutations in either one or both of the motifs should disrupt silencing activity. To address this question, we generated a number of mutations in the Pho protein binding sites. Since we expected that some of these mutations might have only a small effect on silencing activity, we tested the mutant PREs in a mini-white vector that does not have whiteenhancer. As can be seen in Fig. 3, about two-thirds of the mini-white lines containing the control 860-bp iab-7 PRE fragment are pairing sensitive. This figure is higher than the frequency observed with the mini-white vector containing whiteenhancer (Fig. 1).
In the first set of mutations, we deleted sequences spanning either one or both of the conserved Pho motifs. Removal of a 26-bp sequence spanning the proximal motif reduced the frequency of pairing-sensitive silencing from more than two-thirds of the lines to less than one-fourth. When both Pho motifs plus the intervening sequences were deleted (Fig. 3), the frequency of pairing-sensitive lines was less than 10%. Since these two deletions remove sequences besides the putative Pho binding sites, we generated an iab-7 PRE in which point mutations were introduced into seven of the eight bases in the core sequence of each motif (Fig. 3). As for the larger double deletion, there is dramatic reduction in the frequency of pairing-sensitive lines, and less than 5% of inserts containing the mutant PRE exhibit silencing activity. We also tested the effect of mutating the conserved G residue at position +9 in both motifs. Consistent with the in vitro gel shift assays, changing the conserved G residue has at most only a minor effect on silencing activity.
Pho is required for pairing-sensitive silencing in vivo.
The experiments described in the previous section show that both of the Pho binding sites in the iab-7 PRE are required for full silencing activity. If the binding of the Pho protein to these sites in vivo is critical for establishing and maintaining a silencing complex, then the silencing activity of the wild-type iab-7 PRE should depend on the pho gene. To test this prediction, we compared the eye color of a pairing-sensitive iab-7 PRE line of wild-type flies with that of flies carrying a semiviable pho mutant combination. As can be seen in Fig. 4, the silencing activity of the iab-7 PRE is abrogated when the pho function is compromised. When there is only a single copy of the P24 transgene insert, the eye colors of wild-type and pho mutant flies are very similar. However, the nearly complete repression of mini-white expression observed in wild-type flies carrying two copies of the P24 transgene insert is alleviated in pho mutant flies, and the eye color is red instead of pale yellow or white. These results provide strong support for the idea that the Pho protein is directly involved in the establishment and maintenance of functional silencing complexes at the iab-7 PRE.
DISCUSSION
Polycomb-dependent silencing plays a central role in Drosophila development (22, 38, 49, 50). For the homeotic genes of the ANT-C and BX-C complexes, Pc-G silencing provides the mechanism for remembering segmental identity and consequently is critical for maintaining a commitment to the determined state. Pc-G silencing also plays a key role in the regulation of genes that function in other aspects of development including neurogenesis, oogenesis, and stem cell lineages. While the importance of Pc-G silencing in many developmental pathways has been amply documented, it is not yet understood how Pc-G proteins are recruited to appropriate target genes, how silencing complexes are established at these targets, and how the complexes are faithfully propagated from mother to daughter cell.
One approach for addressing these questions is the characterization of cis-acting elements, PREs, that can establish and maintain Pc-G silencing complexes. In the work presented here, we have defined the sequences important for iab-7 PRE function. We have also presented evidence indicating that two proteins, the GAGA factor and Pho, interact directly with this PRE and are required for its silencing activity in vivo.
The iab-7 PRE was initially identified in transgene assays using fragments from the iab-6 to -7 region of BX-C. These studies showed that an 860-bp iab-7 fragment can establish and maintain Pc-G-dependent silencing complexes in two different assays: the pairing-sensitive silencing of mini-white and the maintenance of parasegmentally restricted patterns of Ubx:LacZ expression (23). At the proximal end of this 860-bp fragment is the ∼230-bp nuclease-hypersensitive region, HS3. Since Pc-G-dependent silencing is generally believed to involve a marked reduction in DNA accessibility, not enhanced accessibility, it is important to determine whether this nucleosome-free region of chromatin plays any role in the silencing activity of the iab-7 PRE. Two lines of evidence argue that sequences in HS3 are critical for silencing activity. First, we have shown that a small 260-bp fragment spanning HS3 is sufficient to mediate Pc-G-dependent silencing activity in the mini-white assay. Second, site-directed mutagenesis experiments indicate that sequences essential for silencing activity map to HS3.
An attractive hypothesis is that HS3 provides accessible target sequences for one or more sequence-specific DNA binding proteins. In this model, these DNA binding proteins would interact with their cognate sequences in HS3 and nucleate the assembly of Pc-G silencing complexes by recruiting Pc-G proteins. It seems likely that nucleosome-free regions of chromatin play a similar role in the functioning of other PREs. For example, the three other known PREs in the Abd-B cis-regulatory region, the iab-8 PRE (1), the iab-6 PRE (unpublished data), and Mcp (8, 28, 37), all map to small DNA fragments that contain one or more prominent nuclease-hypersensitive sites. Of these, the Mcp PRE has been characterized in the most detail. Like the iab-7 PRE, the nuclease-hypersensitive region of Mcp is essential for its silencing activity. However, it is not sufficient on its own to direct the assembly of functional silencing complexes, and adjacent proximal or distal flanking sequences are required (37). The chromatin structure of the Mcp element at ectopic sites has also been examined. (A ftz-LacZ transgene was used in this analysis. Unfortunately, the mini-white transgenes are not suitable for examining the chromatin structure of the iab-7 PRE fragments.) The transgene Mcp element has a nuclease-hypersensitive region of approximately the same size and position as that of the endogenous element (37).
Our experiments also indicate that two DNA binding proteins, the GAGA factor and Pho, interact with target sites in HS3 and play a critical role in the silencing activity of the iab-7 PRE. The GAGA factor was initially identified as a potent activator of transcription in nuclear extracts (4, 53, 54) and has generally been thought to be involved in the activation rather than the repression of gene expression. The stimulatory activity of the GAGA factor appears to be due to its ability to prevent histones and other repressive proteins from associating with promoters that have GAGA binding sites (12). In in vitro chromatin assembly experiments the GAGA factor facilitates the formation of a nucleosome-free region of chromatin across the hsp70 promoter (56). In vivo, mutations in the GAGA binding sites of heat shock promoters reduce promoter accessibility and suppress transcription (18, 21, 33). Further support for a role in transcriptional activation comes from genetic studies on mutations in Trl, the gene encoding the GAGA protein. Trl mutations exhibit genetic interactions with homeotic genes in BX-C that are hallmarks of the trx-G genes, not the Pc-G genes (14). Additionally, the expression of several pair rule genes which have GAGA binding sites in their promoters is severely reduced in embryos from Trl mutant mothers (3).
Although it is now well established that the GAGA factor promotes the transcription of many different genes, our results argue that this protein must also play an essential role in the silencing activity of the iab-7 PRE. Several lines of evidence support this conclusion. First, the silencing activity of the iab-7 PRE is impaired by Trl mutations. Second, the GAGA protein binds to the iab-7 PRE both in vivo (55) and in vitro. Third, mutations in the GAGA binding sites of the iab-7 PRE eliminate GAGA protein binding in nuclear extracts and abrogate silencing activity in vivo.
What role does the GAGA factor play in the silencing activity of the iab-7 PRE? At this point the most plausible hypothesis is that the GAGA factor is required to generate a nucleosome-free region over HS3. In this view, the function of the GAGA factor would be analogous to its presumed role in gene activation, namely, to ensure that sequences in HS3 are accessible for the assembly of large multicomponent protein complexes. When the GAGA protein is reduced as in Trl mutants or when the GAGA binding sites are mutant, the HS3 nucleosome-free region would not be formed properly. As a consequence, target sequences for the DNA binding proteins (such as possibly Pho) that are actually responsible for recruiting the large Pc-G silencing complexes to the PRE would be unavailable. While this hypothesis is consistent with the well-documented activities of the GAGA factor at promoters both in vitro and in vivo, we cannot exclude the possibility that GAGA is not only required for the formation of HS3 but also plays a more active role in recruiting Pc-G proteins to the iab-7 PRE. Supporting this hypothesis, Horard et al. (26) found that GAGA binding is required for the in vitro association of Pc-G complexes with fragments from the bxd PRE.
The other protein that is critical for the silencing activity of the iab-7 PRE is Pho. Like the GAGA factor, Pho appears to function by directly interacting with target sequences in HS3. Several lines of evidence support this conclusion. First, the silencing activity of the iab-7 PRE in vivo depends on pho function and is eliminated by mutations in the pho gene. Second, the Pho protein binds to two conserved target sequences in the iab-7 PRE. Third, mutations in these two sites not only eliminate binding in vitro but also compromise silencing activity in vivo. Pho has also been directly implicated in the silencing activity of three other PREs, one from the en gene (6) and two from BX-C (15, 47). The Pho protein has been shown to bind to these PREs in vitro, while mutations in either the Pho binding sites or in the pho gene itself reduce or eliminate silencing.
Unlike that of Trl, the phenotypes of pho mutants are similar to those seen for other Pc-G genes (17, 20). Animals homozygous for loss-of-function alleles die at the pupal stage and exhibit homeotic transformations of legs and abdomen. The late lethal phase is due to a substantial maternal contribution, and mutant embryos lacking a maternal source of wild-type Pho die with severe homeotic transformations and other developmental defects. The homeotic transformations evident in mutant animals indicate that pho is likely to have a direct role in Pc-G silencing. For the iab-7 PRE, our results argue that silencing activity depends on the binding of the Pho protein to the two target sites in HS3. Both seem to be important, as silencing activity is compromised when one is deleted. Whereas we suppose that the major function of the GAGA factor is to ensure that sequences in HS3 are accessible to other proteins, the phenotypic effects of pho mutations suggest that it plays a more active role in silencing. A plausible hypothesis is that it functions (perhaps together with as yet unidentified factors) to recruit components of the silencing machinery to the PRE, such as Polycomb or Sex Combs Midleg, which do not appear to interact directly with DNA. Supporting the possibility that other factors besides Pho play a critical role in recruiting Polycomb group complexes, Shimell et al. (47) have found that a PRE fragment from iab-2, which contains Pho binding sites and which is able to silence mini-white, is not sufficient to confer full Pc-G maintenance activity. Moreover, we have found that mutations in the two Pho binding sites have only a minor effect on the maintenance activity of the 860-bp iab-7 PRE fragment in an iab-7 Ubx-LacZ assay system (unpublished data). Clearly it will be of interest to identify these other factors.
ACKNOWLEDGMENTS
R.K.M., J.M., K.H., and S.E.S. contributed equally to this work.
We thank members of the Karch and Schedl laboratory for advice and encouragement. Thanks also to H. Gyurkovics and J. Gausz for helpful discussions and comments on the work.
F.K. acknowledges support from the Swiss National Fund and the State of Geneva. R.K.M. was supported by a Swiss National Fund grant (no. 31-43432.95). P.S. acknowledges support from NIH. K.H. and S.E.S. were supported by an NIH predoctoral training grant.
REFERENCES
- 1.Barges S, Mihaly J, Galloni M, Hagstrom K, Müller M, Shanower G, Schedl P, Gyurkovics H, Karch F. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development. 2000;127:779–790. doi: 10.1242/dev.127.4.779. [DOI] [PubMed] [Google Scholar]
- 2.Bender W, Akam M, Karch F, Beachy P A, Peifer M, Spierer P, Lewis E B, Hogness D S. Molecular genetics of the bithorax complex in Drosophila melanogaster. Science. 1983;221:23–29. doi: 10.1126/science.221.4605.23. [DOI] [PubMed] [Google Scholar]
- 3.Bhat K M, Farkas G, Karch F, Gyurkovics H, Gausz J, Schedl P. The GAGA factor is required in the early Drosophila embryo not only for transcriptional regulation but also for nuclear division. Development. 1996;122:1113–1124. doi: 10.1242/dev.122.4.1113. [DOI] [PubMed] [Google Scholar]
- 4.Biggin M D, Tjian R. Transcription factors that activate the Ubx promoter in developmentally staged extracts. Cell. 1988;53:699–711. doi: 10.1016/0092-8674(88)90088-8. [DOI] [PubMed] [Google Scholar]
- 5.Boulet A, Lloyd A, Sakonju S. Molecular definition of the morphogenetic and regulatory functions and the cis-regulatory elements of the Drosophila Abd-B homeotic gene. Development. 1991;111:393–405. doi: 10.1242/dev.111.2.393. [DOI] [PubMed] [Google Scholar]
- 6.Brown J L, Mucci D, Whiteley M, Dirksen M L, Kassis J A. The Drosophila Polycomb-group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 7.Busturia A, Bienz M. Silencers in Abdominal-B, a homeotic Drosophila gene. EMBO J. 1993;12:1415–1425. doi: 10.1002/j.1460-2075.1993.tb05785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Busturia A, Wightman C D, Sakonju S. A silencer is required for maintenance of transcriptional repression throughout Drosophila development. Development. 1997;124:4343–4350. doi: 10.1242/dev.124.21.4343. [DOI] [PubMed] [Google Scholar]
- 9.Celniker S E, Sharma S, Keelan D J, Lewis E B. The molecular genetics of the bithorax complex of Drosophila: cis-regulation in the Abdominal-B domain. EMBO J. 1990;9:4277–4286. doi: 10.1002/j.1460-2075.1990.tb07876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan C S, Rastelli L, Pirrotta V. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 1994;13:2553–2564. doi: 10.1002/j.1460-2075.1994.tb06545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiang A, O'Connor M B, Paro R, Simon J, Bender W. Discrete Polycomb-binding sites in each parasegmental domain of the bithorax complex. Development. 1995;121:1681–1689. doi: 10.1242/dev.121.6.1681. [DOI] [PubMed] [Google Scholar]
- 12.Croston G E, Kerrigan L A, Lira L M, Marshak D R, Kadonaga J T. Sequence-specific antirepression of histone H1 mediated inhibition of basal RNA polymerase II transcription. Science. 1991;251:643–649. doi: 10.1126/science.1899487. [DOI] [PubMed] [Google Scholar]
- 13.Duncan I. The bithorax complex. Annu Rev Genet. 1987;21:285–319. doi: 10.1146/annurev.ge.21.120187.001441. [DOI] [PubMed] [Google Scholar]
- 14.Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Karch F. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature. 1994;371:806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- 15.Fritsch C, Brown J L, Kassis J A, Muller J. The DNA-binding Polycomb group protein Pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development. 1999;126:3905–3913. doi: 10.1242/dev.126.17.3905. [DOI] [PubMed] [Google Scholar]
- 16.Galloni M, Gyurkovics H, Schedl P, Karch F. The Bluetail transposon: evidence for independent cis-regulatory domains and domain boundaries in the bithorax complex. EMBO J. 1993;12:1087–1097. doi: 10.1002/j.1460-2075.1993.tb05750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gehring W J. A recessive lethal (l(4)29) with a homeotic effect in D. melanogaster. Drosophile Inf Serv. 1970;45:103. [Google Scholar]
- 18.Gilmour D S, Thomas G H, Elgin S C R. Drosophila nuclear proteins bind to regions of alternating C and T residues in gene promoters. Science. 1989;245:1487–1490. doi: 10.1126/science.2781290. [DOI] [PubMed] [Google Scholar]
- 19.Gindhart J G, Jr, Kaufman T C. Identification of Polycomb and trithorax group responsive elements in the regulatory region of the Drosophila homeotic gene Sex combs reduced. Genetics. 1995;139:797–814. doi: 10.1093/genetics/139.2.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Girton J R, Jeon S H. Novel embryonic and adult homeotic phenotypes are produced by pleiohomeotic mutations in Drosophila. Dev Biol. 1994;161:393–407. doi: 10.1006/dbio.1994.1040. [DOI] [PubMed] [Google Scholar]
- 21.Glaser R L, Thomas G, Siegfried E S, Elgin S C R, Lis J. Optimal heat-induced expression of the Drosophila hsp26 gene requires a promoter sequence containing (CT)n (GA)n repeats. J Mol Biol. 1990;211:751–761. doi: 10.1016/0022-2836(90)90075-W. [DOI] [PubMed] [Google Scholar]
- 22.Hagstrom K, Schedl P. Remembrance of things past: maintaining gene expression patterns with altered chromatin. Curr Opin Genet Dev. 1997;7:814–821. doi: 10.1016/s0959-437x(97)80045-7. [DOI] [PubMed] [Google Scholar]
- 23.Hagstrom K, Muller M, Schedl P. A Polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila bithorax complex. Genetics. 1997;146:1365–1380. doi: 10.1093/genetics/146.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagstrom K, Muller M, Schedl P. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 1996;15:3202–3215. doi: 10.1101/gad.10.24.3202. [DOI] [PubMed] [Google Scholar]
- 25.Han W, Yu Y, Alton N, Pick L. Multiple proteins interact with the fushi tarazu proximal enhancer. Mol Cell Biol. 1993;13:5549–5559. doi: 10.1128/mcb.13.9.5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horard B, Tatout C, Poux S, Pirrotta V. Structure of a Polycomb response element and in vitro binding of Polycomb group complexes containing GAGA factor. Mol Cell Biol. 2000;20:3187–3197. doi: 10.1128/mcb.20.9.3187-3197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ingham P W, Martinez-Arias A. The correct activation of Antennapedia and bithorax complex genes requires the fushi tarazu gene. Nature. 1986;324:592–597. doi: 10.1038/324592a0. [DOI] [PubMed] [Google Scholar]
- 28.Karch F, Galloni M, Sipos L, Gausz J, Gyurkovics H, Schedl P. Mcp and Fab-7: molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res. 1994;22:3138–3146. doi: 10.1093/nar/22.15.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karch F, Weiffenbach B, Peifer M, Bender W, Duncan I, Celniker S, Crosby M, Lewis E B. The abdominal region of the bithorax complex. Cell. 1985;43:81–96. doi: 10.1016/0092-8674(85)90014-5. [DOI] [PubMed] [Google Scholar]
- 30.Kassis J A, Vansickle E P, Sensabaugh S M. A fragment of engrailed regulatory DNA can mediate transvection of the white gene in Drosophila. Genetics. 1991;128:751–761. doi: 10.1093/genetics/128.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kassis J A. Unusual properties of regulatory DNA from the Drosophila engrailed gene: three “pairing-sensitive” sites within a 1.6-kb region. Genetics. 1994;136:1025–1038. doi: 10.1093/genetics/136.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis E B. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 33.Lu Q, Wallrath L L, Granok H, Elgin S C. (CT)n (GA)n repeats and heat shock elements have distinct roles in chromatin structure and transcriptional activation of the Drosophila hsp26 gene. Mol Cell Biol. 1993;13:2802–2814. doi: 10.1128/mcb.13.5.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mihaly J, Mishra R K, Karch F. A conserved sequence motif in Polycomb-response elements. Mol. Cell. 1998;1:1065–1066. doi: 10.1016/s1097-2765(00)80107-0. [DOI] [PubMed] [Google Scholar]
- 35.Mihaly J, Hogga I, Gausz J, Gyurkovics H, Karch K. In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development. 1997;124:1809–1820. doi: 10.1242/dev.124.9.1809. [DOI] [PubMed] [Google Scholar]
- 36.Muller J, Bienz M. Sharp anterior boundary of homeotic gene expression conferred by the fushi tarazu protein. EMBO J. 1992;11:3653–3661. doi: 10.1002/j.1460-2075.1992.tb05450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Müller M, Hagstrom K, Gyurkovics H, Pirrotta V, Schedl P. The Mcp element from the bithorax complex mediates long-distance regulatory interactions. Genetics. 1999;153:1333–1356. doi: 10.1093/genetics/153.3.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paro R. Imprinting a determined state into the chromatin of Drosophila. Trends Genet. 1990;6:416–421. doi: 10.1016/0168-9525(90)90303-n. [DOI] [PubMed] [Google Scholar]
- 39.Peifer M, Karch F, Bender W. The bithorax complex: control of segmental identity. Genes Dev. 1987;1:891–898. doi: 10.1101/gad.1.9.891. [DOI] [PubMed] [Google Scholar]
- 40.Pirrotta V. Polycomb silencing and the maintenance of stable chromatin states. Results Probl Cell Differ. 1999;25:205–228. doi: 10.1007/978-3-540-69111-2_10. [DOI] [PubMed] [Google Scholar]
- 41.Pirrotta V. The genetics and molecular biology of zeste in Drosophila melanogaster. Adv Genet. 1991;29:301–348. doi: 10.1016/s0065-2660(08)60110-8. [DOI] [PubMed] [Google Scholar]
- 42.Poux S, Kostic C, Pirrotta V. Hunchback-independent silencing of late Ubx enhancers by a Polycomb group response element. EMBO J. 1996;15:4713–4722. [PMC free article] [PubMed] [Google Scholar]
- 43.Qian S, Capovilla M, Pirrotta V. The bx region enhancer, a distant cis-control element of the Drosophila Ubx gene, and its regulation by hunchback and other segmentation genes. EMBO J. 1991;10:1415–1425. doi: 10.1002/j.1460-2075.1991.tb07662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raff J, Kellum R, Alberts B. The Drosophila GAGA transcription factor is associated with specific regions of heterochromatin throughout the cell cycle. EMBO J. 1994;13:5977–5983. doi: 10.1002/j.1460-2075.1994.tb06943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanchez-Herrero E. Control of the expression of the bithorax complex genes abdominal-A and Abdominal-B by cis-regulatory regions in Drosophila embryos. Development. 1991;111:437–449. doi: 10.1242/dev.111.2.437. [DOI] [PubMed] [Google Scholar]
- 46.Sanchez-Herrero E, Vernos I, Marco R, Morata G. Genetic organization of the Drosophila bithorax complex. Nature. 1985;313:108–113. doi: 10.1038/313108a0. [DOI] [PubMed] [Google Scholar]
- 47.Shimell M J, Peterson A J, Burr J, Simon J A, O'Connor M B. Functional analysis of repressor binding sites in the iab-2 regulatory region of the abdominal-A homeotic gene. Dev Biol. 2000;218:38–52. doi: 10.1006/dbio.1999.9576. [DOI] [PubMed] [Google Scholar]
- 48.Shimell M J, Simon J, Bender W, O'Connor M B. Enhancer point mutation results in a homeotic transformation in Drosophila. Science. 1994;264:968–971. doi: 10.1126/science.7909957. [DOI] [PubMed] [Google Scholar]
- 49.Sigrist C J, Pirrotta V. Chromatin insulator elements block the silencing of a target gene by the Drosophila Polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics. 1997;147:209–221. doi: 10.1093/genetics/147.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simon J. Locking in stable states of gene expression: transcriptional control during Drosophila development. Curr Opin Cell Biol. 1995;7:376–385. doi: 10.1016/0955-0674(95)80093-x. [DOI] [PubMed] [Google Scholar]
- 51.Simon J, Chiang A, Bender W, Shimell M J, O'Connor M. Elements of the Drosophila bithorax complex that mediate repression by Polycomb group products. Dev Biol. 1993;158:131–144. doi: 10.1006/dbio.1993.1174. [DOI] [PubMed] [Google Scholar]
- 52.Singh H, Lebowitz J H, Baldwin A S, Jr, Sharp P A. Molecular cloning of an enhancer binding protein: isolation by screening of an expression library with a recognition site DNA. Cell. 1988;52:415–423. doi: 10.1016/s0092-8674(88)80034-5. [DOI] [PubMed] [Google Scholar]
- 53.Soeller W, Oh C E, Kornberg T B. Isolation of cDNAs encoding the Drosophila GAGA transcription factor. Mol Cell Biol. 1993;13:7961–7970. doi: 10.1128/mcb.13.12.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soeller W C, Poole S J, Kornberg T B. In vitro transcription of the Drosophila engrailed gene. Genes Dev. 1988;2:68–81. doi: 10.1101/gad.2.1.68. [DOI] [PubMed] [Google Scholar]
- 55.Strutt H, Cavalli G, Paro R. Co-localization of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. EMBO J. 1997;16:3621–3632. doi: 10.1093/emboj/16.12.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsukiyama T, Becker P B, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994;367:525–532. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- 57.White R A H, Lehmann R. A gap gene, hunchback, regulates the spatial expression of Ultrabithorax. Cell. 1986;47:311–321. doi: 10.1016/0092-8674(86)90453-8. [DOI] [PubMed] [Google Scholar]
- 58.Wu C T. Transvection, nuclear structure, and chromatin proteins. J Cell Biol. 1993;120:587–590. doi: 10.1083/jcb.120.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang C C, Bienz M. Segmental determination in Drosophila conferred by hunchback, a repressor of the homeotic gene Ultrabithorax. Proc Natl Acad Sci USA. 1992;89:7511–7515. doi: 10.1073/pnas.89.16.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]