Figure 1.
Overview of samples and datasets
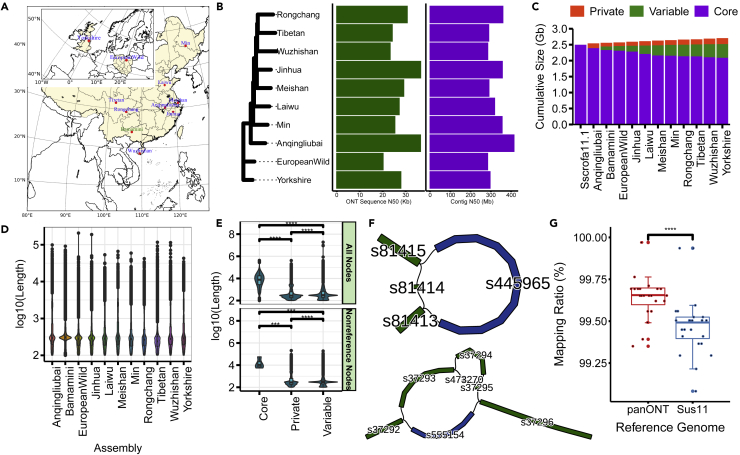
(A) Geographic distribution of the samples used in our study across Eurasia. The sequence data of Bama miniature were downloaded from public databases.
(B) Left panel: SNP-based phylogenetic tree based on Illumina sequencing of the 10 sequenced pig genomes. Middle panel: Sequence N50 of the ONT reads for each sample. Right panel: Contig N50 for each sample.
(C) The pangenome size changes with the added assembly.
(D) The length of nonreference nodes for each assembly added.
(E) The length of core, private and variable nodes for all pangenome nodes and nonreference nodes. The Wilcoxon rank-sum test was used for significance test, ∗∗∗ indicates p-value ≤0.001 and ∗∗∗∗ indicates p value ≤ 0.0001.
(F) Subgraphs of gap18 (Chr3:59,594,615-59,594,715, 100 bp) and gap4 (Chr1:214,753,360-214,753,460, 100 bp) in the reference genome Sscrofa11.1. Reference nodes are colored in green, whereas the nonreference nodes are colored in blue. Gap18 and gap4 are represented in s81414 and s37294 respectively.
(G) Improvement in mappability using the pangenome. Comparison of the mapping ratio of 24 resequencing datasets using the pangenome constructed in this study by LRS (panONT) and the pig reference genome Sscrofa11.1 (Sus11). ∗∗∗∗ indicates p value ≤ 0.0001 with two-tailed paired t test.