Abstract
Dysoxylum binectariferum is an important medicinal plant distributed in the Western Ghats of India. The species has gained international importance for its anticancer component, rohitukine, a chromone alkaloid. Flavopiridol, P-276-00 and IIIM-290 are the derivatives of rohitukine in clinical trials against a wide range of cancers. Flavopiridol was recently approved as an orphan drug for chronic lymphocytic leukemia treatment. In this study, we report the isolation and characterization of rohitukine from the bark of D. binectariferum. Further, rohitukine was estimated across the Western-Ghats and the North-East regions of India. Additionally, D. binectariferum is also reported (∼45 compounds) to produce many natural derivatives of rohitukine and terpenoids, which were investigated in-silico to reveal promising CDK inhibitors. The metabolite fingerprinting of tissues of D. binectariferum was studied using HPTLC and FTIR. The distribution of major chromone alkaloid rohitukine was estimated by HPLC. Further, the pharmacological potential of D. binectariferum compounds was evaluated in-silico by discovering the potential protein targets, molecular docking, ADMET analysis and MD simulation. The isolation of rohitukine has yielded 0.6% from the bark of D. binectariferum. A higher percent of rohitukine was found in the Jog populations (0.58% & 1.28%: leaf & bark), whereas least was observed in the Phasighat population (∼0.06%: both leaf & bark). Across the geographic regions, a higher percent of rohitukine was found in the Central-southern Western Ghats, whereas lower in the northern parts of the Western Ghats and Northeast regions. The leaves produce a considerably higher percent of rohitukine and could be used as a sustainable source of rohitukine. The rohitukine analogues, along with other chromone alkaloids of D. binecatariferum were found to be more interactive with the “kinases” family of proteins, majorly “Serine/threonine-protein kinase PFTAIRE-2” (CDK15) with high confidence level (0.94–0.98). The molecular docking of these chromone alkaloids found a strong binding energy with six CDKs (−3.1 to −10.6 kcal/mol) along with a promising ADMET profile. In addition, molecular dynamic simulation found that the rohitukine complexes are virtually constant with CDK-1, 2, 9 and 15, which is substantiated with MM-PBSA free energy calculations. The chromone alkaloids, majorly rohitukine and its analogues were closely clustered with flavopiridol, P-276-00 and IIIM-290 along with other chrotacumines in the chemical phylogeny. In conclusion, D. binectariferum is a rich source of chromone alkaloids, which could lead to the discovery of more potential scaffolding for CDK inhibitors as anticancer drugs.
Keywords: Dysoxylum binectariferum, Rohitukine, HPLC, Pharmacokinetic potential, CDKs
Abbreviation
- CDK
Cyclin-dependent kinase
- CDK-15
Serine/threonine-protein kinase PFTAIRE-2
- Rh
Rohitukine
- HPTLC
High-performance thin-layer chromatography
- FTIR
Fourier-transform infrared spectroscopy
- LCMS
Liquid chromatography–mass spectrometry
- HPLC
High-performance thin-layer chromatography
- DESI-MSI
Desorption electrospray ionization -Mass Spectrometry Imaging
- NMR
Nuclear magnetic resonance
- PASS
Prediction of activity spectra for substances
- ADMET
Chemical absorption, distribution, metabolism, excretion, and toxicity
- MD simulation
Molecular dynamics simulations
- GROMACS
GROningen MAchine for Chemical Simulation
- CHARMM
Chemistry at Harvard Macromolecular Mechanics
- AMBER
Assisted Model Building with Energy Refinement
- MM-PBSA
Molecular mechanics Poisson–Boltzmann surface area
- RMSD
Root means square deviation
- Rg
Radius of gyration
- RMSF
Root means square fluctuation
- PCA
Principal component analysis
- Comp
Compound/metabolite
1. Introduction
Dysoxylum binectariferum (Roxb.) Hook. (Meliaceae) is a medicinally important species that grows on medium to large-sized trees found in tropical and subtropical climates [1]. The evergreen tree D. binectariferum is native to India, China, and other parts of Asia [2]. The pharmacologically important chemical rohitukine is a prime natural product of this species which has inspired the discovery of three anticancer clinical candidates, flavopiridol (Sanofi-Aventis), P-276-00 (Piramal Healthcare Limited, Mumbai, India), and IIIM-290 [3,4]. Rohitukine is a unique chromone alkaloid having a naringenin chromone scaffold conjugated to a ring containing one or more nitrogen atoms [5,6]. Rohitukine was first reported in Amoora rohituka [7] and later in D. binectariferum [8], D. acutangulum (Meliaceae) [9,10], Schumanniophyton magnificum and S. problematicum (Rubiaceae) [5]. Among the known species, D. binectariferum produces the highest amount of rohitukine in stem bark (3–7% by dry weight) [11]. The tree bark of D. binectariferum is the widely used source for isolation of rohitukine. However, removal of the bark threatens the species and endangers the tree's existence in the forest, which leads to extension of natural populations. This species is threatened in the Western Ghats (India), where it natively resides, due to overexploitation and habitat degradation [12]. As a result, an alternative renewable source for rohitukine isolation is desirable.
The anticancer activity of rohitukine against different cancer cell lines including leukemia, pancreatic, prostate, breast cancer and normal cell line CDKs has been evaluated [13]. Although the anticancer potential of this natural product is less explored, several synthetic and semi-synthetic analogues of this drug have been developed as a potent cyclin dependent kinase (CDKs) inhibitor [[14], [15], [16]]. Structure-activity relationship (SAR) studies of rohitukine, to enhance its anti-cancer activity, led to the development of flavopiridol [17]. Flavopiridol (alvocidib; L868275; HMR-1275; NSC 649890 of Sanofi-Aventis + NCI) is an established CDK inhibitor with broad specificity to CDK1, CDK2 and CDK4 [6,15] leading to cell cycle arrest at both G1 and G2 phases [14,18]. Flavopiridol is currently approved as an orphan drug for treatment of chronic lymphocytic leukemia. This is in Phase-II clinical trials as a monotherapy in combination with conventional chemotherapy drugs. Whereas, P-276-00 in Phase-III clinical trials for advanced refractory neoplasms and various myeloma [4,19]. The protein-ligand docking simulation is an effective computational approach to designing CDK inhibitors. Molecular docking studies showed that CDK9 is the best protein with flavopiridol interacts (−10.3 kcal/mol) efficiently. In-silico pharmacokinetic studies also predicted that hydrophobicity, oral bioavailability, and plasma protein binding were greatly enhanced in the modified flavopiridol compound [20]. There is a great possibility that molecular modifications of rohitukine and flavopiridol may lead to the discovery of more potential scaffolds for CDKs. The prediction of bioactivity of compounds is of great importance for high-throughput screening (HTS) approaches in drug discovery and chemical genomics. Many computational methods in this area focus on measuring the structural similarities and potential modification sites between chemical compounds [21].
In this present study we report rohitukine content from both leaf and bark tissues of D. binectariferum collected from the Western-Ghats and the North-East regions of India. Metabolite fingerprinting of tissues of D. binectariferum using HPTLC and FTIR. The distribution of major chromone alkaloid rohitukine was studied in various stem girth sizes and across different geographical locations. The pharmacological potential of D. binectariferum compounds were evaluated in-silico molecular docking and identified the potential protein targets. Further, the potential of drug-likeness properties of D. binectariferum compounds were analyzed by in-silico ADMET (Absorption, distribution, metabolism, excretion and toxicity) properties. Additionally, the complexes formed by the prime compound, rohitukine, were subjected to molecular dynamics (MD) simulation to analyze the stability of the docked molecules along with the MM-PBSA analysis to estimate binding free energy calculations of the rohitukine complexes.
2. Materials and methods
2.1. Plant material
The different tissues (leaves and bark) of D. binectariferum were collected from eight populations in the central Western-Ghats [Jog (14.23°N 74.81°E), Choukul (15.9°N 74.03°E), Castle rock (15.42°N 74.34°E), Kathgal (14.6°N 74.82°E), Amboli (16.96°N 73.80°E), Kargal (14.25°N 74.81°E), Thalavadi (15.69°N 74.81°E), and Bengave (14.59°N 74.59°E)] and two populations from North-East regions [Manas (26.64°N 90.91°E) and Pasighat (27.53°N 96.4°E)] of India. Additionally, fruits, seeds and seedling samples were collected and stored in a deep freezer (−80 °C) for further use. All the collected samples were dried in a hot air oven at 40–50 °C temperature for 16–20 h and powdered to 20–50 μ size. Individual extracts of samples were prepared by adding 1 g of powdered sample in 10 mL of methanol and sonicated for efficient extraction. The 0.1 g/mL extracts was used for the qualitative (HPTLC and FTIR) and quantitative (HPLC) analysis.
2.2. Column chromatography isolation and characterization of rohitukine
All the samples collected were processed for the extraction of rohitukine (1) from different parts (root, stem, and leaves) of the plant following earlier reports [[22], [23], [24], [25]]. The bark samples collected were first air dried, powdered and used for pure rohitukine isolation. A known quantity of powdered sample was placed in a 30 mm × 100 mm extraction thimble (Scheider & Schuell GmbH, Dassel, Germany), and extracted with 100 mL of MeOH for 18 h in a 200 mL Soxhlet apparatus (3–5 cycles/h). The obtained extract was solvent evaporated using rotary-evaporator apparatus. The dried solid extract was then subjected to silica-column separation. The silica gel of mesh size 230–400 was used for column chromatography with an initial methanol: chloroform solvent system, then purified with a methanol: dichloromethane solvent system. The isolated compound was subjected to NMR (Bruckner AM 500 spectrometer) and IR (FTIR, Bruker, Alpha E) spectrometry for structural characterization. The molecular mass of the purified compound was confirmed by LC-MS (Shimadzu LCMS 8040). The results of IR, NMR and ESI-MS were compared to reported literature and databases (METLIN, CFM-ID) for the structural confirmation [[22], [23], [24], [25]].
2.3. HPTLC fingerprinting of different tissues
The HPTLC fingerprinting was performed on TLC silica gel plates (20 cm × 10 cm, 60 F254). The HPTLC system was equipped with Linomat 5 (CAMAG, Switzerland) automated spray-on band applicator with a 100 μL Hamilton syringe and operated using Nitrogen gas with the following settings: band length 8 mm, dosage speed 150 Nl−1, inter-band distance of 5 mm, horizontal border-distance of 15 mm, and vertical bottom-distance of 10 mm. The development of the plates was carried out in 10 min with solvent saturation of the twin-trough chamber (CAMAG, Switzerland) at ambient temperature. A solvent system consisting of a) ethyl acetate and hexane (6:3, v/v) b) methanol, chloroform and dichloromethane (4:4:1, v/v/v) was used as a mobile phase. After the mobile phase evaporation, the compounds of interest were viewed under TLC Visualizer (CAMAG, Switzerland) at two UV wavelengths (254 and 366 nm). The images were recorded and Rf values of the markers and the compounds of interest were calculated automatically by winCATS software (version 1.2.3) of CAMAG, Switzerland. All the individuals collected from various populations were subjected to HPTLC screening and additionally, the different plant parts (leaves, roots, bark, fruit pericarp, seed coat and seedling tissues) were analyzed for fingerprinting (TLC & FTIR).
2.4. Estimation of rohitukine content in different populations
The methanolic extract 0.1 g/mL was syringe filtered using 0.22 μ nylon filters, diluted to 1 mg/mL and subjected to quantitative (HPLC) analysis. The column chromatography isolated rohitukine is utilized as standard for quantification in HPLC. The RP-HPLC analysis was performed on a Shimadzu LC20AT instrument (Shimadzu, LC20AT, Japan) using the Shimadzu RP C18 column (250 mm × 4.6 mm, 0.5 m). The analyte was separated on an isocratic flow using 0.1% TFA and acetonitrile mobile phase (70:30; 1 mL/min flow rate). The column temperature was maintained at 40 °C and UV-detected at a wavelength of 254 nm with an injection volume of 5–10 μl [24,25]. Pure rohitukine isolated was used as a reference standard for HPLC analysis. The standard rohitukine was prepared with a series of con. (0.2–1.0 mg/mL) using HPLC grade MeOH and filtered using 0.45 μ syringe filters. The linear graph was obtained (y = mx) and used for quantification of rohitukine in samples. Rohitukine content was quantified for ten populations and in various tissue parts of D. binectariferum.
2.5. Computation analysis for pharmacological potential
2.5.1. Selection of ligands and protein targets
The reported 45 compounds from the species D. binecatariferum were selected as ligands [9,10,[26], [27], [28], [29]] along with preclinical drugs flavopiridol (9), IIIM-290 (10), and P-276-00 (11) as control (Fig. 1). The three-dimensional structures were retrieved from public repository PubChem (https://pubchem.ncbi.nlm.nih.gov) and some were drawn using ChemDraw Pro 12.0 software. The ligands were optimized by energy minimization using ChemDraw Pro 12.0 and were stored as.sdf format. Further, all the ligands of the SMILES were analyzed for structural similarity using ChemMine tools (http://chemmine.ucr.edu/similarity) [30]. All the ligands were subjected to “PASS targets'' (http://www.way2drug.com/passtargets/) and “SWISS targets” (http://www.swisstargetprediction.ch/) for virtual screening of possible protein targets [31,32]. The predictive accuracy was estimated as 0.94 using Leave-One-Out cross validation with the PASS targets. The score for each target is called confidence, which is a difference between probabilities for chemical compounds to interact and to not interact with the specific target. The higher confidence means the higher chance of the positive prediction being true. The output of predicted targets was obtained in distinct groups of related proteins according to ChEMBL protein classification. PASS targets of all the ligands showed the hit with a high confidence level (>0.95) majorly to “Serine/threonine-protein kinase PFTAIRE-2” (CHEMBL5856/CDK15). Further, these proteins were chosen as protein targets for docking studies.
Fig. 1.
Metabolites of D. binectariferum.
2.5.2. Validation of protein target and molecular docking
For determination of the conserved functional residues between the isoforms of CDK15, a multiple sequence alignment analysis was performed, which can be used as potential targets for the discovery of drug hits. The four isoforms of CDK15 (Q96Q40-1, −3, −4, −5 and −6) were retrieved from UniProt (https://www.uniprot.org/) in FASTA format and aligned (https://www.uniprot.org/align/) with default parameters. The default parameters: the transition matrix is Gonnet, gap opening penalty is 6 bits, gap extension is 1 bit. Clustal-Omega uses the HHaign algorithm and its default settings as the central alignment engine. The protein sequences of CDK15 were submitted to Swissmodel (https://swissmodel.expasy.org/) homology modeling software to generate 3D structures [33]. Then quality of the homology models were validated by Ramachandran plot analysis and ProQ-Protein Quality Predictor. ProQ is a neural network-based method to predict the quality of a protein model that extracts structural features such as frequency of atom-atom contacts and predicts the quality of a model, which is measured by either LGscore or MaxSub. The difference between MaxSub and LGscore is the length of target protein. The correct model should have LGscore more than 1.5 and MaxSub more than 0.1 [34].
The docking simulation was carried out on the D. binectariferum compounds with synthetically derived P276-00, flavopiridol and IIIM-290 as control against CDKs (1, 2, 4, 6, 9 and 15). The molecular docking was performed by online platform SeamDock (https://bioserv.rpbs.univ-paris-diderot.fr/services/SeamDock/) with Autodock Vina interface, Mode number (2), Energy range (5), and Exhaustiveness (8) as constant set parameters throughout the analysis. The Grid box was covered with whole protein (Blind docking) and performed docking [35,36]. The lowest energy of binding of the docked structure was considered as the best drug-receptor complex [21,37].
2.5.3. Potential drug-likeness (ADMET)
The potential of drug-likeness properties of D. binectariferum compounds were analyzed by ADMET (Absorption, distribution, metabolism, excretion and toxicity) properties by ADMETlab 2.0 in-silico tool (https://admetmesh.scbdd.com/) and SwissADME (http://www.swissadme.ch/) [38,39]. The “Absorption” property is calculated based on Caco-2 permeability, MDCK (Madin−Darby Canine Kidney cells) permeability, Pgp-inhibitor, Pgp-substrate, HIA, F-20% and F-30%. The “Distribution” property is calculated based on parameters like PPB (Plasma Protein Binding), VD (Volume Distribution), BBB (Blood-Brain Barrier) Penetration, and Fu. The “Metabolism” property is calculated based on the Cytochrome P450s enzyme system (inhibitor/substrate). The “Excretion” property is calculated based on CL (Clearance) and T½ (Half-life). The “Toxicity” property is calculated based on 11 parameters, namely, hERG Blockers, H-HT (Human Hepatotoxicity), DILI (Drug Induced Liver Injury), AMES Toxicity, Rat Oral Acute Toxicity, FDAMDD (Maximum Recommended Daily Dose), Skin Sensitization, Carcinogenicity, Eye Corrosion, Eye Irritation, Respiratory Toxicity. A visual representation of the distribution covered by different D. binectariferum metabolites and 3 preclinical synthetic derivatives (flavopiridol, IIIM-290 and P276-00) was made by employing principal component analysis (PCA) based on ADME/Tox descriptors [40].
2.5.4. Molecular dynamics simulation
The evaluation of the stability of a docked protein-ligand complex was analyzed by Molecular Dynamics (MD) simulation. In the present study, the GROMACS Program package [41] and CHARMM36 force field [42] were used to simulate the molecular dynamics of the complexes in the TIP3P (Transferable Intermolecular Potential 3P) water model. The ions (Na+ or Cl−) were added to neutralize charges wherever necessary. The topology of ligand (rohitukine) was generated using the SwissParam web tool (https://www.swissparam.ch/). The systems were neutralized, and energy minimized. Then, the systems were heated from 0 K to 300 K within 100 ps in NVT (Number of particles, Volume and Temperature) ensemble with normal temperature (300 K) and another 100 ps in NPT (Number of particles, Pressure and Temperature) ensemble with normal pressure (101 kPa) [43,44]. After heating and equilibration, the docked complexes were subjected to production MD run for 10 ns after the system reached dynamic equilibrium. The simulation performed in triplicates (n = 3) and the geometric properties of the protein-ligand complexes, such as root mean square deviation (RMSD), radius of gyration (Rg), root mean square fluctuation (RMSF), and number of hydrogen bonds (H-bonds) were calculated using g_rms and g_energy programs respectively [45].
2.5.5. Binding free energies (MM-PBSA) of the complex
The free binding energies for the docked protein-ligand complexes, the rohitukine complexed with the CDKs were analyzed using the Molecular Mechanics Poisson Boltzmann Surface Area (MM-PBSA) integrated in the MMPBSA. py module of AMBER18 [46,47]. The free binding energies of the rohitukine-CDK complexes were calculated on the snapshots obtained from the 10 ns of the MD trajectories. The MM-PBSA underlines the robustness of the bonding calculation of molecular mechanical energies [47].
3. Results
3.1. HPTLC and FTIR fingerprinting of different tissues of D. binectariferum
The HPTLC fingerprint of different tissues of D. binectariferum showed clearly separated constituents without any tailing and diffuseness (Fig. 2a). The leaf and twigs observed the highest number of bands (10–15), when scanned at a wavelength of 366 nm and 5–6 bands were observed at a wavelength 254 nm. Few bands are common to all tissues (seed coat, root, fruit coat, and bark), like 3–4 major components in the range of Rf 0.16 to Rf 0.47 (Rf 0.16 = rohitukine). Whereas, some are specific to tissues, like band Rf = 0.39 was observed only in leaf and fruit coat, and Rf = 0.47 was observed in all tissues except leaf and twigs (SF 1a).
Fig. 2.
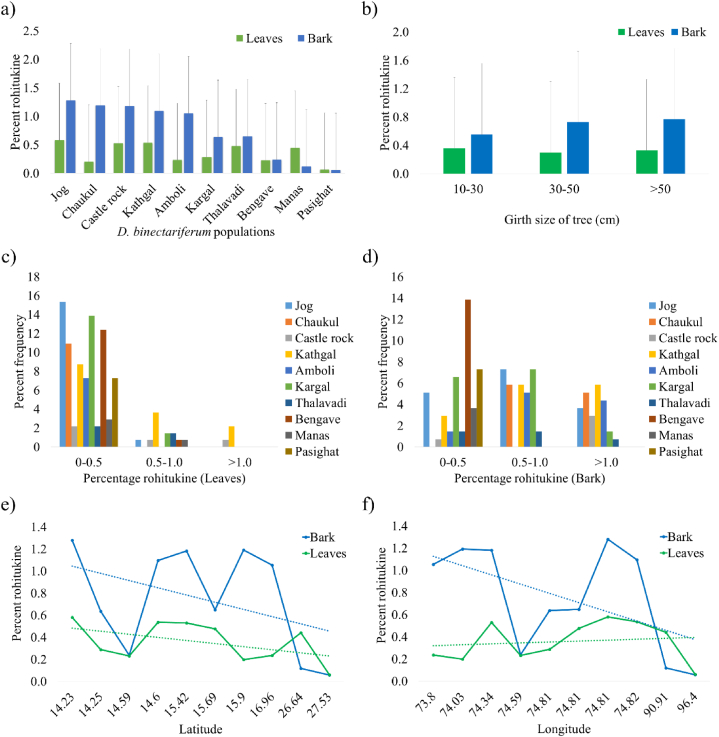
Distribution of rohitukine in leaf and bark of D. binectariferum populations. a) Average rohitukine content, b) Average rohitukine content in different girth size, c) Percent frequency of rohitukine in leaf, d) Percent frequency of rohitukine in bark, e) Rohitukine distribution across Latitude, f) Rohitukine distribution across Longitude.
Tissue specific metabolites were observed in FTIR fingerprinting (SF 1b). FTIR signals at wave number 535.1 cm−1 (C–I), 1270.3 cm−1 (C–O, aromatic ester), 1354.14 cm−1 (S O, sulfone), 1517.95 cm−1 (N–O, nitro compound) are strong and specific to bark tissue. Similarly, signals at 1032.44 cm−1 (C–N), 1383.74 cm−1 (C–H, aldehyde), 1708.89 cm−1 (C O, conjugated aldehyde) observed only in leaf and 665.98 cm−1 (C–Br, halo compound), 1047.68 cm−1 (CO–O–CO), 1567.09 cm−1 (N–H, amine) observed only in fruit coat. Additionally, common with signals at 1081.37 cm−1 (C–O, primary alcohol), 1639.56 cm−1 (C C) and many others (ST 1). FTIR fingerprinting showed twig and seed coat samples are different from the rest of other tissues. This is evident from PCA (PC1: 69.81% and PC2: 23.42%) of FTIR wave numbers of different tissues of the D. binectariferum (SF 1c & 1d). The aromatic and nitrogen containing signals were strongly observed in leaf and bark tissues (ST 1). Further, these tissues (leaf and bark) were used for qualitative and quantitative analysis of chromone alkaloids.
3.2. Isolation of rohitukine from stem bark
The Soxhlet extraction of D. binectariferum bark yielded 10% semi-solid methanolic concentrate. Further, column chromatography of methanolic concentrate (2.5 g) using silica gel obtained a pure fraction of 3–4% MeOH in ChCl3 (300 mg, yield = 0.6%) (SF 2a and 2b) (Mohana Kumara et al., 2010; Naik et al., 1988). The isolated compound assigned the structural identity and molecular formula of C16H19NO5, as deduced from mass fragmentation, 13C and 1H NMR data (SF 2c and 2d). The FTIR spectrum of isolated compound observed the strong presence of –OH (3266.59 cm−1) and –C O (1658.02 cm−1) characteristic functionalities (SF 2e) [4]. The spectrum of the isolated compound was obtained by positive ESI-MS and has characteristically yielded a major ion of m/z 306 [M+H]+. Fragmentation of the precursor ion at m/z 306.12 [M+H]+ showed a loss of 17 Da, forming the production of m/z 288 presumably due to the loss of the OH group at the piperidine ring. Further, ion at m/z 288 observed consecutive losses of 59 Da, forming the production of m/z 245. The observed characteristic ion peaks were comparable with the results of previous reports and identified as rohitukine [22,24,25] (SF 2f). The obtained rohitukine was further used for qualitative (HPTLC) and quantitative (HPLC) analysis.
3.3. Rohitukine content in D. binectariferum populations
The 137 individuals from ten different populations of D. binectariferum leaf and bark samples were collected and quantified for rohitukine by HPLC. It was observed that, from the ten different populations, the percent rohitukine detected varied from a lowest value of 0.02% to as high as 2.37%. In the individuals analyzed from Jog population, the percent of rohitukine varied from 0.03 to 0.53%, Choukul; 0.03–2.34%, Castle rock; 0.29–1.77%, Kathgal; 0.16–2.10%, Amboli; 0.15–2.09%, Kargal; 0.02–2.37%, Thalavadi; 0.03–1.73%, Bengave; 0.04–0.62%, Manas; 0.03–0.73%, and Pasighat; 0.01–0.21% respectively (Fig. 2a). Overall, the mean percent rohitukine detected in stem bark was highest in Jog (1.28 ± 0.12%), followed by Choukul (1.19 ± 0.50%), Castle rock (1.18 ± 0.54%), Kathgal (1.10 ± 0.40%), Amboli (1.05 ± 0.53%) and others. Whereas in leaf samples, the highest percent rohitukine was observed in Jog (0.58 ± 0.18%), followed by Kathgal (0.54 ± 036%), Castle rock (0.53 ± 0.33%), Thalavadi (0.48 ± 0.10%), Manas (0.44 ± 0.17%) and others (Fig. 2a). Further, it was observed there is an increase in percent rohitukine in bark as girth size increases but not in the leaf (Fig. 2b). Across the regions of collected populations (both leaf and bark), a higher percent rohitukine was found in Central-Southern Western Ghats, whereas lower at Northern parts of the Western Ghats and Northeast regions (Fig. 2e & f).
3.4. Computational analysis of pharmacology of D. binectariferum
3.4.1. Potential target discovery of D. binectariferum metabolites
The 45 known compounds with 3 preclinical synthetic derivatives (Flavopiridol, IIIM-290 and P-276-00) were used in insilco prediction for protein targets (way2drug; “PASS targets”). All these 48 molecules are targeted to 4663 proteins (Fig. 3a). Among these, 2365 proteins belong to the “kinases” family of proteins. Out of the 45 D. binectariferum compounds, 15 compounds were majorly hit by Serine/threonine-protein kinase PFTAIRE-2 (CHEMBL5856) with a high confidence level of 0.939–0.982 and all compounds belong to chromone alkaloids (Fig. 3b). Similarly, control (9, 10 and 11) also hit with Serine/threonine-protein kinase PFTAIRE-2 with high confidence of 0.939, 0.982 and 0.978 respectively. Also, these 18 compounds hit with “Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (CHEMBL1908383)”, “Tyrosine-protein kinase Srms (CHEMBL5703)”, “Rhodopsin kinase (CHEMBL5607)”, “Serine/threonine-protein kinase TNNI3K (CHEMBL5260)”, “Mitogen-activated protein kinase kinase kinase 2 (CHEMBL5914)” and “Activin receptor type-2B (CHEMBL5466)” with confidence level >0.9 (ST 2). Further, Serine/threonine-protein kinase PFTAIRE-2 was validated and prepared for molecular docking along with other potential CDKs [1(6GU7), 2(5OO1), 4(2W96), 6(1XO2) and 9 (6GZH)].
Fig. 3.
a) Protein target prediction for D. binectariferum compounds, b) Top 5 kinases with high confidence level, c) Ramachandran plot of CDK15 homology model, and d) Quality estimate of the model.
3.4.2. Target validation
The Serine/threonine-protein kinase that acts like an antiapoptotic protein, phosphorylates BIRC5 at 'Thr-34′ and counters TRAIL/TNFSF10-induced apoptosis [48,49]. The sequence of top majorly hit protein “Serine/threonine-protein kinase PFTAIRE-2” was obtained by CHEMBL (CHEMBL5856). The sequences were subjected to BlastP against the UniPortKB/Swiss-Prot database and obtained a hit of CDK15 (Q96Q40-1) with 81.0% query coverage and 91.44% identity. The hit CDK15 (Q96Q40-1) has 5 isoforms belonging to Homo sapiens, comprising 435aa (Q96Q40-1), 400aa (Q96Q40-3), 384aa (Q96Q40-4), 429aa (Q96Q40-5), and 345aa (Q96Q40-6). The pairwise sequence alignment was obtained by collecting all isoform sequences and obtaining a similarity of 61.468% with 268 identical aa positions and 6 similar aa positions (SF 3). Ramachandran plot analysis for the all the isoforms showed that 94.14% of residues were allocated in the most favored regions in Q96Q40-1 and 93.92% in Q96Q40-3, 94.12% in Q96Q40-4, 93.90% in Q96Q40-5 and 95.15% Q96Q40-6. The Ramachandran plot statistics and quality estimation of Q96Q40-6 isoform were represented in Fig. 3c & d. The model with high Ramachandran favored score (Q96Q40-6) was further used for molecular docking studies along with other potential CDKs.
3.4.3. Molecular docking
CDK proteins are removed with water molecules and added to polar H bonds before performing molecular docking. The D. binectariferum compounds have observed a binding affinity range of −11 to −3.1 kcal/mol towards CDKs analyzed and majority compounds showing < −7.5 kcal/mol binding energy. The prime anticancer compound of D. binectariferum, rohitukine and its analogues have observed a binding affinity of −8 to −7.3 kcal/mol with all the analyzed CDKs (except Rh diacetate observed 0.8 kcal/mol with CDK15) (Fig. 4). Especially strongly with the CDK2 (−8 kcal/mol) by forming 4 hydrophobic interactions (I10(A) CB, I10(A) CD, A31(A) CB, and L131(A) CD1), 5 hydrogen bonds (I10A(O), Y15(A)OH, L83(A)O, L83(A)O, and D86(A)OD2) and 2 weak hydrogen bonds (I10(A)O, Q131(A)OE1) (Fig. 5 and ST 3). Whereas 3 preclinical synthetic derivatives (9, 10 and 11) have observed binding energy in the range of −10.5 to −7.9 kcal/mol with all the analyzed CDKs (Fig. 4c).
Fig. 4.
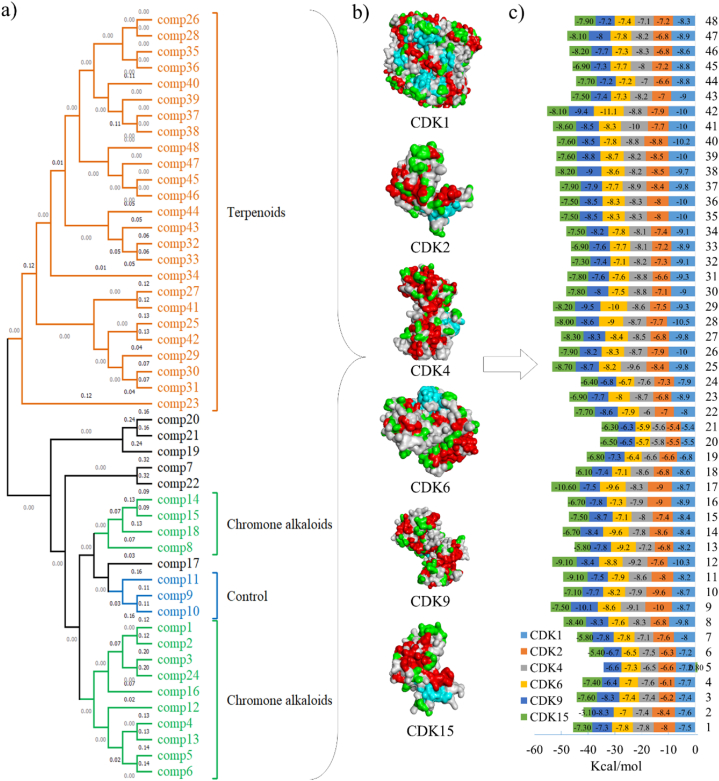
Molecular docking of D. binectariferum compounds with CDKs. a) Structural similarity of D. binectariferum compounds, b) Surface structure of CDK proteins (CDK1, 2, 4, 6, 9 and 15), c) Binding energy (Kcal/mol) of D. binectariferum compounds docked with CDKs.
Fig. 5.
2D and 3D interactions of rohitukine docked with CDKs.
The chemical matrix of D. binectariferum compounds has observed rohitukine and its analogues were closely clustered with synthetic derivatives (9, 10 and 11) in one clade along with chrotacumines (A-F) in the chemical phylogeny (Fig. 4a). The Chrotacumine E (16) and F (17) observed the lowest binding energy (−9 Kca/mol) for CDK2 and the Chrotacumine F (17) showed highest binding affinity (−10.6 kcal/mol) for CDK15. Similarly, the lactones and triterpenoids are forming another independent cluster, from which, 3-Epiocotillol (28) exhibited the strongest binding for CDK1 with the lowest binding energy (−10.5 kcal/mol), whereas eight other compounds of D. binectariferum showed ≤ −10 kcal/mol binding energy with CDK1 (Fig. 4c). Additionally, comp. (41) showed strong binding affinity (−10 kcal/mol) for CDK4, comp. (42) (−11.1 kcal/mol) for CDK6, and comp. (29) (−9.5 kcal/mol) for CDK9 (Fig. 4c). The 2D and 3D interaction of compounds with the lowest binding energy with CDKs were represented in SF 4 and ST 3.
3.4.4. Potential drug-likeness (ADMET) of D. binectariferum compounds
In the drug development pipeline, bio accessibility, gastrointestinal fate, and first-pass metabolism evaluation of phytochemicals is essential to define their pharmacokinetics. The in-silico ADMET modeling (molecular cheminformatics) properties of compounds are considered as an important condition for judging metabolic fate, potential bioactivity (drug-likeness) and toxicity. ADMET target prediction of phytochemicals could lead to identification of drug candidates. Passive diffusion is one of the prime qualities of a compound for the appropriate movement across the cell membranes. Most of the D. binectariferum compounds indicate intestinal absorption rate in the human body within the range of permeability for Caco-2 (23 compounds), MDCK (all 47), Pgp-inhibitor (20 compounds), Pgp-substrate (36 compounds), HIA (41 compounds), F 20% (39 compounds) and F 30% (16 compounds) (Fig. 6a, ST 4). Also, the distribution of drugs in plasma has a strong influence on its pharmacodynamic behavior and thus directly influences oral bioavailability. The distribution behavior of most of the D. binectariferum compounds indicate within the favorable range for PPB (21 compounds), VD (all 47 compounds), BBB Penetration (37 compounds), and Fu (22 compounds) (Fig. 6b, ST 4).
Fig. 6.
In-silico ADMET analysis a) Absorption b) Distribution c) Metabolism d) Excretion e) Toxicity f) ADMET and g) Cluster (ADMET).
The metabolism of pharmaceutical drugs is an important aspect, where its rate determines the duration and intensity of a drug's pharmacologic action. In-silico analysis of most of the D. binectariferum compounds showed the digestion in the liver. The compounds had more probability of being substrate (value > 0.5) to either of the CYPs analyzed, especially interactive with CYP2C19-sub (∼25 compounds >0.9 value) (Fig. 6c, ST 4). Whereas the CL (Clearance) and T1/2 (half-life) values of the majority of the chromone alkaloids, terpenoids are matched with the preclinical drugs, except dammarene triterpenoids were not in favorable range. The clearance and volume of distribution is an important pharmacokinetic parameter that defines the frequency of dosing of a drug. As observed, most of the compounds are indicated within the range of acceptance for CL (22 compounds) and T ½ (18 compounds) (Fig. 6d, ST 4). Also, as observed, most of the compounds are non-toxic except for “Skin Sensitization” and “Respiratory Toxicity”, where only 2 and 3 compounds did not clear the safe range respectively (Fig. 6e, ST 4). The PCA of overall “ADMET” properties comparing D. binectariferum compounds along with control (9, 10 and 11) has observed major components PC1 44.75% and PC2 of 26.16%, where the rohitukine analogues and chrotacumines are forming close cluster with the control (Fig. 6f and g). The majority of the ADMET profile of chromone alkaloids were met the criteria of drug-like and matching the control ADMET profiles (ST 4). Overall, molecules from D. binectariferum specifically, chromone alkaloids have the potential to be CDK inhibitors.
3.4.5. Molecular dynamics simulations
The binding stability of rohitukine molecules with the CDK receptors were further studied using a MD simulation in an aqueous system for a simulation time of 10 ns. For the geometric property, RMSD used to determine the average distance between the atoms of superimposed structures of a protein and a ligand over a period of time [50]. The average RMSD of the CDKs docked with rohitukine (1) seemed to remain virtually constant after 10 ns averaging (except with CDK4 and CDK6) at 0.217 ± 0.02 nm, 0.284 ± 0.06 nm, 0.940 ± 1.306 nm, 0.638 ± 1.015 nm, 0.202 ± 0.027 nm, 0.255 ± 0.034 nm for CDK1, 2, 4, 6, 9 and 15 respectively (Fig. 7a). The complex formed with CDK4 and CDK6 observed a fluctuation with larger amplitude between 4.5-9 ns and 5.5–7 ns respectively and later observed their stabilities. The radius of gyration (Rg) can be explained as the distribution of atoms of a protein around its axis (Lobanov et al., 2008). The average Rg of the CDKs docked with rohitukine (1) were 2.76 ± 0.019 nm (CDK1), 2.02 ± 0.022 nm (CDK2), 3.00 ± 0.85 nm (CDK4), 2.72 ± 0.81 nm (CDK6), 2.77 ± 0.02 nm (CDK9) and 2.07 ± 0.01 nm (CDK15) respectively (Fig. 7b).
Fig. 7.
MD simulation of rohitukine docked with CDKs a) RMSD, b) Radius of gyration (Rg), c) RMSF, and d) H-bonds interactions.
The RMSF analysis, the amino acid residues of rohitukine complexed CDK1, 2, 9 and 15 fluctuated within an average range of 0.114–0.127 Å. Whereas the amino acid residues of the rohitukine complexed CDK4 and 6 fluctuated with an average of 1.38 Å and 1.09 Å (Fig. 7c). Hydrogen bond formation with amino acid residues is the key indicator of specificity and molecular interactions between the protein and inhibitor complexes. The mean values for H-bonds formed between CDKs and rohitukine (1) were calculated for entire trajectories. The average H-bonds formed for CDK1, CDK2, CDK4, CDK6, CDK9 and CDK15 were 1.43 ± 1.21, 1.71 ± 0.57, 1.15 ± 0.66, 2.65 ± 0.77, 1.05 ± 0.72 and 0.66 ± 0.80, respectively (Fig. 7d).
3.4.6. MM-PBSA calculations
The binding free energy is a critical step of an in-silico drug designing methodology which defines the binding affinity inhibitors to the receptor. The major desirable portions of the stereoisomer binding of rohitune-CDKs complexes were the van der Waals (ΔEVDW) ranging from −20.69 (CDK4_Rh) to −38.80 kcal/mol (CDK1_Rh). Also, electrostatic energies (ΔEEL) ranging from −5.06 (CDK1_Rh) to −39.19 kcal/mol (CDK4_Rh) (ST 5). The ΔEVDW of rohitukine complexed with CDK-1, 2, 9 & 15 is comparatively lesser (−31.46 to −38.80 kcal/mol) than CDK-4 & 6 complexes (−20.67 to −27.37 kcal/mol). Similarly, the ΔEEL of rohitukine complexed with CDK-2, 4 & 15 is comparatively lesser (−21.18 to −39.19 kcal/mol) than CDK-1, 6 & 9 complexes (−5.06 to −14.50 kcal/mol) (ST 5).
4. Discussion
In the present study, the metabolic dissimilarity of different tissues of D. binectariferum was analyzed by HPTLC and FTIR fingerprinting. The nitrogen and aromatic signals were strongly observed in methanolic exacts of leaf and bark tissues (ST 1). Further, these tissues (leaf and bark) were selected for isolation of nitrogen-based compounds, chromone alkaloids. A high intensity rohitukine band was observed in methanolic exacts of bark, leaf, fruit coat and root tissues, when subjected to HPTLC fingerprinting. Further, rohitukine was isolated (0.6%) from bark of D. binectariferum and its structural characterization was done using spectral techniques [22,24,25]. The quantification of rohitukine content in ten different populations of the Western Ghats and the Northeast regions varied from 0.02% to 2.37%. Among different tissues analyzed, bark produces high rohitukine compared to leaf with the high frequency (Fig. 2c & d). Across the different geographical distribution, the Western Ghats populations produces high compared to Northeastern regions of India. In the Western Ghats, the Jog population produces high compared to other populations. There was no significant correlation between the girth of the tree and the rohitukine content. Our earlier studies reported significant amounts of rohitukine accumulation in different tissues (bark, twig, leaf and seeds) of D. binectariferum [2,23,51]. Among tissues, leaves also accumulates a considerable amount of rohitukine. Further, leaves could be used as an alternate sustainable source of rohitukine production.
Recently, flavopiridol semisynthetic derivative of rohitukine, approved as an orphan drug for the treatment of chronic lymphocytic leukemia. The identification of alternate renewable resources for rohitukine is in high demand. Considering the importance of chromone alkaloids specifically, rohitukine, current study identified the high yielding elite lines, and these are the sustainable source for the rohitukine and its derivatives. D. binectariferum is highly threaten species in the Western Ghats regions and conserving this pharmacological relevance species is national importance to rescue the species being endangered and extinct.
Earlier studies reported the natural analogues of rohitukine such as, glycosylated, acetylated, oxidized, and methoxylated analogues by DESI MSI from different tissues of D. binectariferum [25,51]. Dysobinin (23), isolated from fruits of this tree is also reported to exhibit significant central nervous system depressant action and mild anti-inflammatory activity [26]. Dysoline (24) was isolated from the bark, chrotacumine K (4) and schumaniofioside A (22) isolated from fruits and leaves of D. binectariferum [4]. Additionally, a group of chromone alkaloids chrotacumines (A-J) were isolated from D. acutangulam, which also shares the chromone ring with the N–Me piperidine ring and ester side chains [9,10,27]. Overall, D. binectariferum is a rich source of chromone alkaloids, specifically rohitukine and its derivatives. All these alkaloids showed strong structural similarity (Fig. 4a) and share a common chromone ring with the molecular mass ranging from m/z 300–500. Further, molecular docking and ADMET studies was done for the 45 phytochemicals reported from D. binectariferum in control with anticancer rohitukine derivatives, flavopiridol, IIIM-290 and P276-00. Molecular docking is one of the in-silico assessment of interactions between proteins and drugs to understand the inhibition of targeted proteins molecules [52]. In this study, we more focused on compounds target to CDKs. CDK inhibitors are one the most important target proteins for developing anticancer therapeutics. CDKs/Cyclins are known for their vital role in regulating cell cycle, along with transcription modulation DNA damage repair, immune response, apoptosis, metabolism, and regulation of various signaling pathways through protein-kinase interactions [53]. Genetic alteration in regulatory mechanism, mutations, and overexpression of these CDKs lead to proliferation of a wide variety of human cancers [54].
The protein target prediction of 15 chromone alkaloids of D. binectariferum showed high affinity to Serine/threonine-protein kinase PFTAIRE-2 (CDK15). The CDK15 is a member of the understudied TAIRE subfamily of cyclin-dependent kinases, named after the “TAIRE” sequence motif in their cyclin binding site, and comprising of CDKs 14–18 [55]. A little is known about its function, expression, and regulation of CDK15 [56]. Additionally, these chromone alkaloids also showed highly interactive with other proteins like “Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma”, “Tyrosine-protein kinase Srms”, “Rhodopsin kinase”, “Serine/threonine-protein kinase TNNI3K”, “Mitogen-activated protein kinase kinase kinase 2” and “Activin receptor type-2B″ with confidence level >0.9 (ST 2). Most of the chromone alkaloids (except three terpenoids) exhibited high interactions with most of the CDKs (Fig. 4c). These compounds formed significant amount of hydrogen bonds within the binding site of the receptor. Few compounds (3) were exempted by Lipinski rule and many dammarene type triterpenoids (50%) by Pfizer rule/GSK rule. The highest binding energy of −11.1 kcal/mol was observed in compound 42 for CDK6, followed by Chrotacumine F (−10.6) for CDK15 and 3-Epiocotillol (−10.5) for CDK1. Additionally, the 3 preclinical synthetic derivatives (9,10, and 11) also observed significant interaction with all the analyzed CDKs and it is evident by the previous in-silico studies [20]. These rohitukine based compounds has shown earlier effective against many anticancer activities [24,57]. The IIIM-290, a rohitukine derivative showed very effective cytotoxicity against Molt-4 cells in a typical mitochondrion-mediated cell death (Mintoo et al., 2021). Also, flavopiridol shown broad specificity to CDK1, CDK2 and CDK4 [6,58].
The pharmacokinetics analysis of 45 compounds from D. binectariferum, in which chromone alkaloids showed improved ADMET profile and having similar drug-like behavior compared to control (Fig. 6f and g). Further, these chromone alkaloids can be analyzed in-vivo ADMET modeling for the development of drug. Earlier, in-vivo pharmacokinetics studies of rohitukine found be moderate binding (∼59%) to Hamster plasma, high desorption in liver with rapid oral absorption [19]. The pharmacokinetic behavior of rohitukine in hamster positively provide a basis for selection of oral anti-dyslipidemic agent and offer important lead for further structural modification or drug designing.
In molecular docking, only the ligands are flexible. However, both the protein and the ligand were found to be flexible in the MD simulation. Therefore, in the MD simulation, we were able to assess all interactions observed in molecular docking and identify potential and emerging effects on conformational changes in the ligand and enzyme binding site. Consequently, in this analysis, the lower MRSD value indicates less fluctuation during simulation, suggesting that protein stability is high [59]. As a result, the system is going to be balanced. The RMSD of rohitukine complexed with CDK-1, 2, 9 and 15 is comparatively lower (0.202–0.284 nm) than the CDK-4 and 6 complexes (0.638–0.940 nm). Similarly, the RMSF of rohitukine complexed with CDK-1, 2, 9 & 15 is comparatively lesser (0.114–0.284 Å) than CDK-4 and 6 complexes (1.098–1.383 Å). Thus, the rohitukine complexed with CDKs are more stable except CDK-4 and 6 complexes.
It is also noteworthy that MMPBSA results of rohitune-CDKs complexes substantially shown insights supporting the Bioinformatics analysis. The ΔEVDW and ΔEEL were the major contributors of the stereoisomer binding of rohitune-CDKs complexes. These interactions confirm that rohitukine is a good binding affinity towards the CDKs analyzed. The MMPBSA results of rohitune-CDKs complexes are supported by docking energies, where binding energies ranges from −7.3 to −8.0 kcal/mol (Fig. 4c). Especially, binding energies of CDK2_Rh, CDK4_Rh and CDK6_Rh are observed high (−7.8 to −8 kcal/mol) comparatively. The MMPBSA partially supports the molecular docking and MD simulation results. Further, the expedition of potential effect of rohitukine against CDKs will accelerate the pace of designing chromone based drugs against CDK proteins, which can be used to establish new therapies against different human cancer.
5. Conclusion
Plant-based drugs offered a significant contribution towards the treatment of several cancers which is second prominent cause of death globally. The flavopiridol, P276-00 and IIIM-290 obtained by synthetic derivatization of rohitukine has already succeeded as potent anticancer clinical drug candidates. Recently, flavopiridol has been approved as an orphan drug for leukemia cancer treatment. Due to increasing global demand for the CDK inhibitors more specifically, chromone alkaloids for the developing anticancer drugs. To meet the growing demand current requirement of sustainable source of chromone alkaloids, specifically rohitukine. In this study, we identified the populations/tissues produces higher quantity of rohitukine in D. binectariferum. We observed higher levels of accumulation of rohitukine in Western Ghats populations and emerge as copious for the cultivation for these resources. The demand for alternate renewable source for rohitukine might increase in near future both due to the imminent use of pre-clinical semi synthetic drugs (flavopiridol, P276-00 and IIIM-290) as anticancer and anti-HIV agent, for which leaves of D. binectariferum could be an alternate sustainable source. Apart from rohitukine, D. binectariferum produces various natural derivatives of rohitukine and potential chromone alkaloids. In the present study, integrated approach of virtual screening, molecular docking, ADMET, MD simulation studies showed chromone alkaloids as potential CDK inhibitors. To substantiate these leads of interaction of rohitukine with selected CDKs further confirmed with in vivo models. There is a great possibility that molecular modifications of chromone alkaloids may lead to the discovery of more potential scaffold for CDKs. These compounds may be further developed into promising multi-target anticancer therapeutics, that can be used for treatment of various human cancer.
Author contribution statement
Varun E: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Bhakti K, Aishwarya K, Suraj R Hosur: Performed the experiments.
Jagadish, M. R: Performed the experiments; Contributed reagents, materials, analysis tools or data.
Mohana Kumara P: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by Department of Biotechnology (DBT), Government of India (File BT/PR31331/TRM/120/227/2019).
Data availability statement
Data included in article/supp. material/referenced in article.
Declaration of interest’s statement
The authors declare no competing interests.
Acknowledgements
Authors thanks to State Forest Department, Government of Karnataka for facilitating by the kind permission for collection of samples and other field work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e13469.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Gamble J.S. Newman and Adlard; West: 1921. Flora of the Presidency of Madras. [Google Scholar]
- 2.Mahajan V., Sharma N., Kumar S., Bhardwaj V., Ali A., Khajuria R.K., Bedi Y.S., Vishwakarma R.A., Gandhi S.G. Production of rohitukine in leaves and seeds of Dysoxylum binectariferum: an alternate renewable resource. Pharm. Biol. 2015;53:446–450. doi: 10.3109/13880209.2014.923006. [DOI] [PubMed] [Google Scholar]
- 3.Houghton P.J. Chemistry and biological activity of natural and semi-synthetic chromone alkaloids. Stud. Nat. Prod. Chem. 2000;21:123–155. [Google Scholar]
- 4.Jain S.K., Meena S., Qazi A.K., Hussain A., Bhola S.K., Kshirsagar R., Pari K., Khajuria A., Hamid A., Shaanker R.U. Isolation and biological evaluation of chromone alkaloid dysoline, a new regioisomer of rohitukine from Dysoxylum binectariferum. Tetrahedron Lett. 2013;54:7140–7143. [Google Scholar]
- 5.Houghton P.J. Chromatography of the chromone and flavonoid alkaloids. J. Chromatogr. A. 2002;967:75–84. doi: 10.1016/s0021-9673(01)01555-2. [DOI] [PubMed] [Google Scholar]
- 6.Khadem S., Marles R.J. Chromone and flavonoid alkaloids: occurrence and bioactivity. Molecules. 2011;17:191–206. doi: 10.3390/molecules17010191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harmon A.D., Weiss U., V Silverton J. The structure of rohitukine, the main alkaloid of Amoora rohituka (syn. Aphanamixis polystachya)(Meliaceae) Tetrahedron Lett. 1979;20:721–724. [Google Scholar]
- 8.Naik R.G., Kattige S.L., V Bhat S., Alreja B., De Souza N.J., Rupp R.H. An antiinflammatory cum immunomodulatory piperidinylbenzopyranone from Dysoxylum binectariferum: isolation, structure and total synthesis. Tetrahedron. 1988;44:2081–2086. [Google Scholar]
- 9.Ismail I.S., Nagakura Y., Hirasawa Y., Hosoya T., Lazim M.I.M., Lajis N.H., Shiro M., Morita H., Chrotacumines A− D. Chromone alkaloids from dysoxylum acutangulum. J. Nat. Prod. 2009;72:1879–1883. doi: 10.1021/np9003849. [DOI] [PubMed] [Google Scholar]
- 10.Morita H., Nugroho A.E., Nagakura Y., Hirasawa Y., Yoshida H., Kaneda T., Shirota O., Ismail I.S., Chrotacumines G.-J. Chromone alkaloids from Dysoxylum acutangulum with osteoclast differentiation inhibitory activity. Bioorg. Med. Chem. Lett. 2014;24:2437–2439. doi: 10.1016/j.bmcl.2014.04.020. [DOI] [PubMed] [Google Scholar]
- 11.Mohana kumara P., Sreejayan N., Priti V., Ramesha B.T., Ravikanth G., Ganeshaiah K.N., Vasudeva R., Mohan J., Santhoshkumar T.R., Mishra P.D. Dysoxylum binectariferum Hook. f (Meliaceae), a rich source of rohitukine. Fitoterapia. 2010;81:145–148. doi: 10.1016/j.fitote.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 12.Sumangala R.C., Kumara P.M., Shaanker R., Vasudeva R., Ravikanth G. Development and characterization of microsatellite markers for Dysoxylum binectariferum, a medicinally important tree species in Western Ghats, India. J. Genet. 2016;93:85–88. [PubMed] [Google Scholar]
- 13.Kumar V., Guru S.K., Jain S.K., Joshi P., Gandhi S.G., Bharate S.B., Bhushan S., Bharate S.S., Vishwakarma R.A. A chromatography-free isolation of rohitukine from leaves of Dysoxylum binectariferum: evaluation for in vitro cytotoxicity, Cdk inhibition and physicochemical properties. Bioorg. Med. Chem. Lett. 2016;26:3457–3463. doi: 10.1016/j.bmcl.2016.06.046. [DOI] [PubMed] [Google Scholar]
- 14.Sedlacek H., Czech J., Naik R., Kaur G., Worland P., Losiewicz M., Parker B., Carlson B., Smith A., Senderowicz A. Flavopiridol (L86 8275; NSC 649890), a new kinase inhibitor for tumor therapy. Int. J. Oncol. 1996;9:1143–1168. doi: 10.3892/ijo.9.6.1143. [DOI] [PubMed] [Google Scholar]
- 15.K Jain S., B Bharate S., a Vishwakarma R. Cyclin-dependent kinase inhibition by flavoalkaloids. Mini Rev. Med. Chem. 2012;12:632–649. doi: 10.2174/138955712800626683. [DOI] [PubMed] [Google Scholar]
- 16.Mintoo M., Khan S., Wani A., Malik S., Bhurta D., Bharate S., Malik F., Mondhe D. A rohitukine derivative IIIM‐290 induces p53 dependent mitochondrial apoptosis in acute lymphoblastic leukemia cells. Mol. Carcinog. 2021;60:671–683. doi: 10.1002/mc.23332. [DOI] [PubMed] [Google Scholar]
- 17.Wang H.K., Flavopiridol. National Cancer Institute Curr. Opin. Investig. Drugs. 2000;2(2001):1149–1155. [PubMed] [Google Scholar]
- 18.Stadler W.M., Vogelzang N.J., Amato R., Sosman J., Taber D., Liebowitz D., Vokes E.E. Flavopiridol, a novel cyclin-dependent kinase inhibitor, in metastatic renal cancer: a University of Chicago Phase II Consortium study. J. Clin. Oncol. 2000;18:371. doi: 10.1200/JCO.2000.18.2.371. [DOI] [PubMed] [Google Scholar]
- 19.Chhonker Y.S., Chandasana H., Kumar A., Kumar D., Laxman T.S., Mishra S.K., Balaramnavar V.M., Srivastava S., Saxena A.K., Bhatta R.S. Pharmacokinetics, tissue distribution and plasma protein binding studies of rohitukine: a potent anti-hyperlipidemic agent. Drug Res. 2015;65:380–387. doi: 10.1055/s-0034-1387774. [DOI] [PubMed] [Google Scholar]
- 20.Khalipha A.B.R., Bagchi R., Hossain M.S., Mondal M., Biswas S., Ray P., Smrity S.Z., Asha U.H. 2019. A Literature Based Review on Rohitukine and Molecular Docking Studies of Flavopiridol (Rohitukine Derived) and its Derivatives against Cyclin-dependent Kinases (CDKs) for Anticancer Activity. [Google Scholar]
- 21.Nithya S., Lalasa M., Nagalakshmamma K., Archana S. Int. Conf. Comput. Bio Eng. Springer; 2019. Computational approaches in toxicity testing: an overview; pp. 255–261. [Google Scholar]
- 22.Mohana Kumara P., Zuehlke S., Priti V., Ramesha B.T., Shweta S., Ravikanth G., Vasudeva R., Santhoshkumar T.R., Spiteller M., Uma Shaanker R. Fusarium proliferatum, an endophytic fungus from Dysoxylum binectariferum Hook. f, produces rohitukine, a chromane alkaloid possessing anti-cancer activity. Antonie Leeuwenhoek. 2012;101:323–329. doi: 10.1007/s10482-011-9638-2. [DOI] [PubMed] [Google Scholar]
- 23.Mohana Kumara P., Soujanya K.N., Ravikanth G., Vasudeva R., Ganeshaiah K.N., Uma Shaanker R. Wight & Arn; Phytomedicine: 2013. Production of the Chromane Alkaloid, Rohitukine and its Attenuation in Endophytic Fungi Isolated from Dysoxylum Binectariferum Hook. F and Amoora Rohituka (Roxb) [DOI] [PubMed] [Google Scholar]
- 24.Kumara P.M., Soujanya K.N., Ravikanth G., Vasudeva R., Ganeshaiah K.N., Shaanker R.U. Rohitukine, a chromone alkaloid and a precursor of flavopiridol, is produced by endophytic fungi isolated from Dysoxylum binectariferum Hook. f and Amoora rohituka (Roxb). Wight & Arn. Phytomedicine. 2014;21:541–546. doi: 10.1016/j.phymed.2013.09.019. [DOI] [PubMed] [Google Scholar]
- 25.Mohana Kumara P., Srimany A., Arunan S., Ravikanth G., Uma Shaanker R., Pradeep T. Desorption electrospray ionization (DESI) mass spectrometric imaging of the distribution of rohitukine in the seedling of Dysoxylum binectariferum Hook. F. PLoS One. 2016;11 doi: 10.1371/journal.pone.0158099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh S., Garg H.S., Khanna N.M. Dysobinin, a new tetranortriterpene from Dysoxylum binectariferum. Phytochemistry. 1976;15:2001–2002. [Google Scholar]
- 27.Izwan Mohd Lazim M., Safinar Ismail I., Shaari K., Latip J. Abd, Ali Al‐Mekhlafi N., Morita H., Chrotacumines E and F Two new chromone‐alkaloid analogs from dysoxylum acutangulum (meliaceae) leaves. Chem. Biodivers. 2013;10:1589–1596. doi: 10.1002/cbdv.201200391. [DOI] [PubMed] [Google Scholar]
- 28.Yan H.-J., Wang J.-S., Kong L.-Y. Cytotoxic steroids from the leaves of Dysoxylum binectariferum. Steroids. 2014;86:26–31. doi: 10.1016/j.steroids.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 29.Kumar V., Gupta M., Gandhi S.G., Bharate S.S., Kumar A., Vishwakarma R.A., Bharate S.B. Anti-inflammatory chromone alkaloids and glycoside from Dysoxylum binectariferum. Tetrahedron Lett. 2017;58:3974–3978. [Google Scholar]
- 30.Backman T.W.H., Cao Y., Girke T. ChemMine tools: an online service for analyzing and clustering small molecules. Nucleic Acids Res. 2011;39:W486–W491. doi: 10.1093/nar/gkr320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.V Pogodin P., Lagunin A.A., Filimonov D.A., V Poroikov V., Targets P.A.S.S. Ligand-based multi-target computational system based on a public data and naïve Bayes approach. SAR QSAR Environ. Res. 2015;26:783–793. doi: 10.1080/1062936X.2015.1078407. [DOI] [PubMed] [Google Scholar]
- 32.Daina A., Michielin O., Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47:W357–W364. doi: 10.1093/nar/gkz382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296. doi: 10.1093/nar/gky427. W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramachandran G., Ramakrishnan C., Sasisekharan V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 35.Murail S., De Vries S.J., Rey J., Moroy G., Tufféry P. SeamDock: an interactive and collaborative online docking resource to assist small compound molecular docking. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.716466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trott O., Olson A.J., Vina AutoDock. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inbathamizh L., Padmini E. Quinic acid as a potent drug candidate for prostate cancer–a comparative pharmacokinetic approach. Asian J. Pharmaceut. Clin. Res. 2013;6:106–112. [Google Scholar]
- 38.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:1–13. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiong G., Wu Z., Yi J., Fu L., Yang Z., Hsieh C., Yin M., Zeng X., Wu C., Lu A. ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021;49 doi: 10.1093/nar/gkab255. W5–W14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Durán-Iturbide N.A., Díaz-Eufracio B.I., Medina-Franco J.L. Silico ADME/Tox profiling of natural products: a focus on BIOFACQUIM. ACS Omega. 2020;5:16076–16084. doi: 10.1021/acsomega.0c01581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hess B., Kutzner C., Van Der Spoel D., Lindahl E. Gromacs 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theor. Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 42.Huang J., Rauscher S., Nawrocki G., Ran T., Feig M., de Groot B.L., Grubmüller H., MacKerell A.D. CHARMM36: an improved force field for folded and intrinsically disordered proteins. Biophys. J. 2017;112:175a–176a. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parrinello M., Rahman A. Strain fluctuations and elastic constants. J. Chem. Phys. 1982;76:2662–2666. [Google Scholar]
- 44.Berendsen H.J.C., van Postma J.P.M., Van Gunsteren W.F., DiNola A., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 45.Patel B., Patel D., Parmar K., Chauhan R., Singh D.D., Pappachan A., donovani Xprt L. Molecular characterization and evaluation of inhibitors. Biochim. Biophys. Acta, Proteins Proteomics. 2018;1866:426–441. doi: 10.1016/j.bbapap.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 46.Miller B.R., III, McGee T.D., Jr., Swails J.M., Homeyer N., Gohlke H., Roitberg A.E. MMPBSA. py: an efficient program for end-state free energy calculations. J. Chem. Theor. Comput. 2012;8:3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- 47.Case D., Ben-Shalom I.Y., Brozell S.R., Cerutti D.S., Cheatham T.E., III, Cruzeiro V.W.D., Darden T.A., Duke R.E., Ghoreishi D., Gilson M.K. AMBER 2018; university of California. San Francisco, CA, USA. 2018 http://Creativecommons.Org/Licenses/by/4.0/ © 2019 by the authors, Licens. MDPI, Basel, Switzerland. This Artic. Is an Open Access Artic. Distrib. under Terms Cond. Creat. Commons Attrib. (CC BY) Licens. 2018. [Google Scholar]
- 48.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., Yuan D., Stroe O., Wood G., Laydon A. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50:D439–D444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maiorov V.N., Crippen G.M. Significance of root-mean-square deviation in comparing three-dimensional structures of globular proteins. J. Mol. Biol. 1994;235:625–634. doi: 10.1006/jmbi.1994.1017. [DOI] [PubMed] [Google Scholar]
- 51.Kumara P.M., Srimany A., Ravikanth G., Shaanker R.U., Pradeep T. Ambient ionization mass spectrometry imaging of rohitukine, a chromone anti-cancer alkaloid, during seed development in Dysoxylum binectariferum Hook. f (Meliaceae) Phytochemistry. 2015;116:104–110. doi: 10.1016/j.phytochem.2015.02.031. [DOI] [PubMed] [Google Scholar]
- 52.De Ruyck J., Brysbaert G., Blossey R., Lensink M.F. Molecular docking as a popular tool in drug design, an in silico travel. Adv. Appl. Bioinforma. Chem. AABC. 2016;9:1. doi: 10.2147/AABC.S105289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malumbres M. Cyclin-dependent kinases. Genome Biol. 2014;15:1–10. doi: 10.1186/gb4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peyressatre M., Prével C., Pellerano M., Morris M.C. Targeting cyclin-dependent kinases in human cancers: from small molecules to peptide inhibitors. Cancers. 2015;7:179–237. doi: 10.3390/cancers7010179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferguson F.M., Doctor Z.M., Ficarro S.B., Browne C.M., Marto J.A., Johnson J.L., Yaron T.M., Cantley L.C., Kim N.D., Sim T. Discovery of covalent CDK14 inhibitors with pan-TAIRE family specificity. Cell Chem. Biol. 2019;26:804–817. doi: 10.1016/j.chembiol.2019.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mikolcevic P., Rainer J., Geley S. Orphan kinases turn eccentric: a new class of cyclin Y-activated, membrane-targeted CDKs. Cell Cycle. 2012;11:3758–3768. doi: 10.4161/cc.21592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Varshney S., Shankar K., Beg M., Balaramnavar V.M., Mishra S.K., Jagdale P., Srivastava S., Chhonker Y.S., Lakshmi V., Chaudhari B.P. Rohitukine inhibits in vitro adipogenesis arresting mitotic clonal expansion and improves dyslipidemia in vivo [S] J. Lipid Res. 2014;55:1019–1032. doi: 10.1194/jlr.M039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jain S.K., Bharate S., Vishwakarma R. Cyclin-dependent kinase inhibition by flavoalkaloids. Mini Rev. Med. Chem. 2012;12:632–649. doi: 10.2174/138955712800626683. [DOI] [PubMed] [Google Scholar]
- 59.Duran T., Minatovicz B., Bai J., Shin D., Mohammadiarani H., Chaudhuri B. Molecular dynamics simulation to uncover the mechanisms of protein instability during freezing. J. Pharmaceut. Sci. 2021;110:2457–2471. doi: 10.1016/j.xphs.2021.01.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supp. material/referenced in article.