Summary
Promoting myelination capacity of endogenous oligodendrocyte precursor cells (OPCs) is a promising therapeutic approach for CNS demyelinating disorders such as Multiple Sclerosis (MS). To aid in the discovery of myelination-promoting compounds, we generated a genome-engineered human pluripotent stem cell (hPSC) line that consists of three reporters: identification-and-purification tag, GFP, and secreted-NanoLuc, driven by the endogenous PDGFRA, PLP1, and MBP genes, respectively. Using this cell line, we established a high-throughput drug screening platform and performed a small-molecule screen, which identified at least two myelination-promoting small-molecule (Ro1138452 and SR2211) that target prostacyclin (IP) receptor and retinoic acid receptor-related orphan receptor γ (RORγ), respectively. Single-cell-transcriptomic analysis of differentiating OPCs treated with these molecules further confirmed that they promote oligodendrocyte differentiation and revealed several pathways that are potentially modulated by them. The molecules and their target pathways provide promising targets for the possible development of remyelination-based therapy for MS and other demyelinating disorders.
Subject areas: Neuroscience, Cell biology, Stem cells research
Graphical abstract
Highlights
-
•
CRISPR-engineered triple reporter hPSC line to study oligodendrocyte lineage cells
-
•
A high-throughput drug screening using human oligodendrocyte precursor cells
-
•
HTS with hOPCs identified Ro1138452 and SR2211, two myelination promoting molecules
-
•
scRNAseq on hOPCs treated with the two molecules highlights modulated pathways
Neuroscience; Cell biology; Stem cells research
Introduction
Multiple sclerosis (MS) is an immune-mediated demyelinating disease in which the immune system attacks and degrades myelin sheaths of neuronal axons in the central nervous system (CNS).1 Damage and loss of a neuron’s myelin sheath can cause apoptosis of the myelin producing oligodendrocyte cells, axonal damage and neuronal cell death, which ultimately can lead to neurological disabilities.2 All currently approved therapies for MS act by modulating the patient’s immune response. Such therapies can help to slow down the progression of the disease, but do not directly promote the remyelination that is needed to improve the long-term function and survival of damaged neurons. A number of studies have shown that following myelin damage, oligodendrocyte progenitor cells (OPCs) and neural precursor cells (NPCs) migrate toward the site of injury, where they demonstrate some ability to differentiate into mature oligodendrocytes that are capable of remyelinating the demyelinated axons.3,4 However, endogenous progenitors around a CNS lesion appear to be limited in both their mitotic competence and differentiation potential, and they progressively lose their remyelination capacity with aging.5,6 If we could employ biologically active molecules to stimulate the ability of endogenous OPCs to differentiate and produce new myelin, this could have great therapeutic value for MS and other demyelinating diseases.
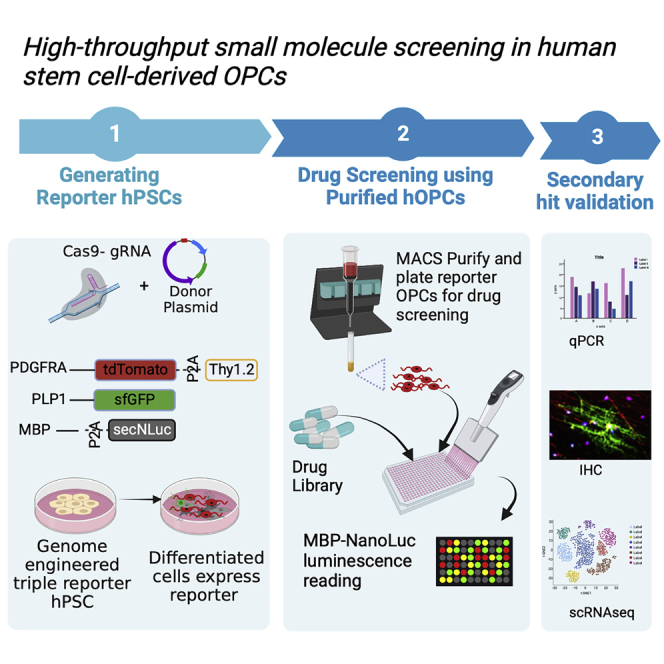
Drug screening to identify molecules that promote OL maturation and myelination has been performed using primary rodent OPCs7,8 and mouse ES-derived OPCs.9 The advent of human pluripotent stem cells (hPSC)-derived OPCs and OLs now make it possible to perform such drug screening using human cells, which presumably would be more clinically relevant to human disease. However, to our knowledge, no platforms that utilize human OPCs to screen for myelin promoting compounds have yet been reported. Use of hPSC-derived, physiologically relevant cells for drug screening to identify molecules that promote remyelination is limited by the difficulty in obtaining large quantities of pure, disease relevant cell-types such as OPCs.10 In addition, the majority of the OL maturation assays either use immunofluorescence analysis to measure the amount of MBP, or use image-based morphological analysis to detect cells that resemble oligodendrocytes.7,8,9,11,12 Although such image-based, high-content screening (HCS) assays can be powerful, morphology-based assays have limited sensitivity and they are also less scalable. Here, we used CRISPR/Cas9-based genome editing to generate a triple reporter hESC line where an identification-and-purification (IAP) tag,13 GFP, and secreted Nanoluciferase (secNluc) reporters were engineered to be driven by the endogenous PDGFRA, PLP1 and MBP genes, respectively. Once the reporter hESC is differentiated, the PDGFRA expressing OPCs can be immuno-purified using the IAP tag,13,14 and the time and efficiency of oligodendrocyte maturation can be quantified via the expression of GFP and secNluc. These qualities enabled us to use the reporter cell line to develop a highly sensitive and scalable screening platform for discovery of myelination promoting compounds that: 1) uses purified human OPCs, 2) allows time-course assays to follow real-time expression of oligodendrocyte/myelination markers, and 3) makes it possible to perform Nluc-based HTS and image-based HCS using the same cell cultures.
The hOPCs derived from this reporter system was used to screen 2500 bioactive molecules, resulting in the identification of several molecules that function at nanomolar doses to enhance maturation of oligodendrocytes from OPCs. In addition to a number of already known myelination promoting molecules, at least two compounds that have not been previously reported to improve OL differentiation and enhance myelination were identified by this screen. Image-based analysis and electrospun nanofiber-based in vitro myelination assays further validated the effect of these compounds on hOPCs-hOL maturation. In addition, we validated the efficacy of these compounds in a different hPSC-OPC reporter cell line. Furthermore, we performed scRNAseq to examine the biological processes in OPCs and OLs that are modulated by treatment with these small molecules. The identified compounds and their target pathways provide promising new potential targets for the development of remyelination-based therapies for the demyelinating diseases.
Results
Generation of a hESC triple-reporter system
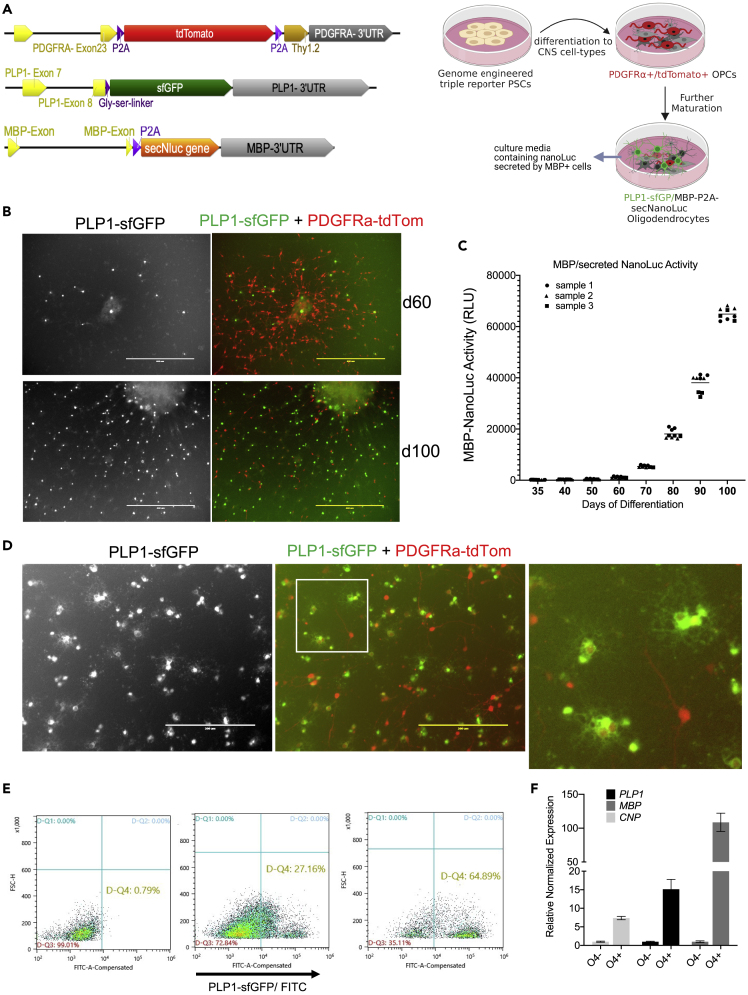
We previously engineered a hESC reporter cell line (PDTT) that allows identification and purification of hESC-derived, PDGFRA expressing oligodendrocyte lineage cells (OLLCs).13 Here, we added two more reporter sequences to the PDTT line, generating a reporter system that is suitable for high throughput screening (HTS) as well as HCS. CRISPR-Cas9-based genome editing was used to knock-in super-fold GFP (sfGFP) and secNluc reporter sequences before the stop codons of the PLP1 and MBP genes, respectively (Figure 1A). The PLP1-sfGFP construct generates a fusion reporter whereas the MBP-P2A-secNluc construct leads to two separate proteins (MBP and secNLuc) due to the presence of a self-cleaving P2A peptide.15
Figure 1.
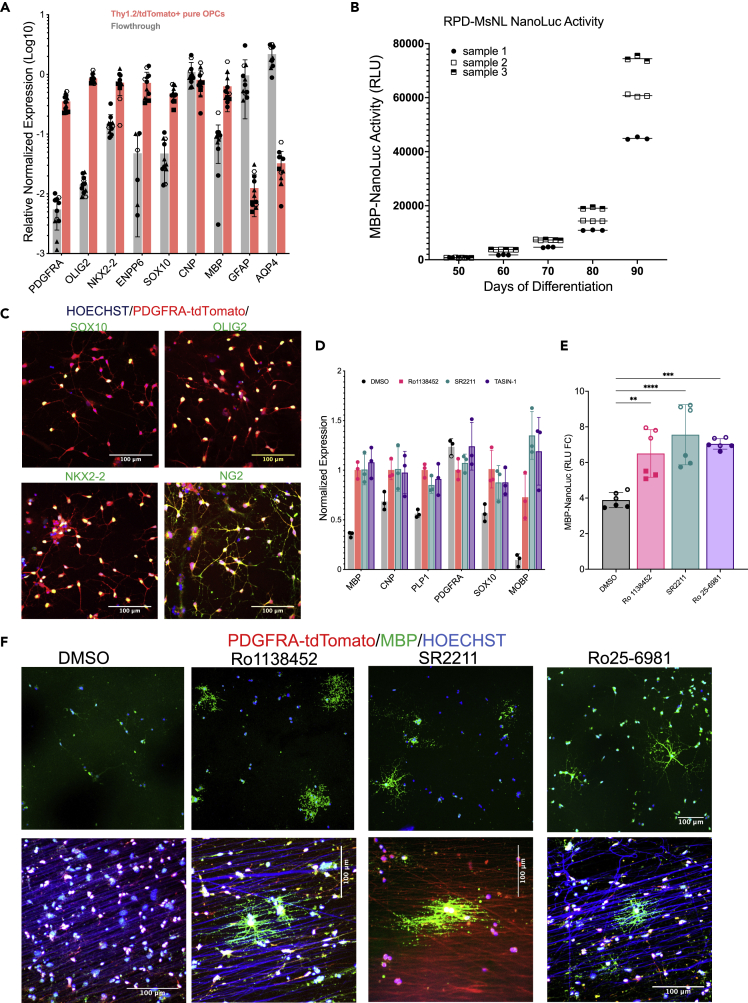
Generation and validation of an advanced hES reporter system
(A) Schematic of the endogenous PDGFRA, PLP1 and MBP locus of the Ptt-P1-MsnL reporter line after CRISPR-Cas9-based genetic knock-in of reporter sequences.
(B) Expression of the tdTomato and sfGFP fluorescent reporters in the differentiating oligodendrocyte cultures Scale bar: 400 µm
(C) secNluc activity in the cell culture medium of differentiating PtT-P1-MsNL reporter line measured with Nano-Glo assay. Culture media from different days of differentiation (xaxis) were removed for the assay. Increase in Nano-Luc activity (yaxis), which corresponds to MBP expression increases as cells mature.
(D) A higher magnification image of the reporter expression on d100 culture shown in (B). GFP+ processes and branches resembling oligodendrocytes morphology are further magnified in the inset. Scale bar: 200 µm
(E) MACS-based enrichment of the GFP+ cells using O4 antigen microbeads. 60% GFP+ cells were achieved from d105 cells.
(F) qPCR shows enrichment of OL markers in the O4+ cells compared to the O4- cells. Data presented as mean +/- SEM
Before knocking in the P2A-secNluc reporter into the MBP locus, we wanted to confirm that the secNLuc protein product, following its P2A-mediated separation, is successfully secreted into the extracellular culture media and retains enzymatic activity. Therefore, we transfected a CMV-tdTomato-P2A-secNluc plasmid into HEK293 cells and confirmed that the culture media of the transfected cells has Nluc activity (Figures S1A–S1C).
The final reporter system that we generated (PTt-P1-MsNL) incorporates three reporters: 1) PDGFRA-P2A-tdTomato-P2A-Thy1.2 (or an IAP tag), in which, on PDGFRA expression, tdTomato as well as Thy1.2 are produced. tdTomato localizes to the cytoplasm whereas Thy1.2 localizes to the cell surface, allowing the PDGFRA expressing cells to be immunopurified via Thy1.2 antibody conjugated magnetic microbeads13; 2) PLP1-sfGFP, where expression of sfGFP is driven by PLP1; and 3) MBP-P2A-secNLuc, where NLuc protein is secreted into the cell culture media, allowing serial sampling and serial quantitative readout of MBP expression (Figures 1A and S1D–S1F).
Validation of the reporter cell line
With the PTt-P1-MsNL reporter line, similar to the PDTT cell line, PDGFRA-tdTom+ cells appear around day 45 of differentiation in our culture system.13 PLP1-GFP+ cells are visible as early as day 60. As the culture matures, less tdTomato+ cells and more GFP expressing cells are detected, and more MBP/sNLuc is also produced (Figures 1C and S2). The GFP+ cells have processes and morphology that resemble oligodendrocyte cells (Figure 1D). Moreover, the majority of GFP+ cells also express low levels of tdTomato, which indicates that these cells originated from PDGFRa expressing progenitor cells (Figures 1D and S2). We also demonstrate that an antibody against O4, an antigen specific to pre-oligodendrocyte cells, can be used to enrich PLP1-GFP+ cells from the differentiating OLLC culture (Figures 1E, 1F, and S3C). Essentially, in this advanced reporter cell-line, PDGFRA+ OLLCs can be temporally purified,13 mature OLs can be identified and enriched for using the expression of PLP1-sfGFP, and the NLuc activity, which represents MBP expression, can be quantitated over time using a small aliquot of cell culture media.
Optimization of the high throughput screening platform
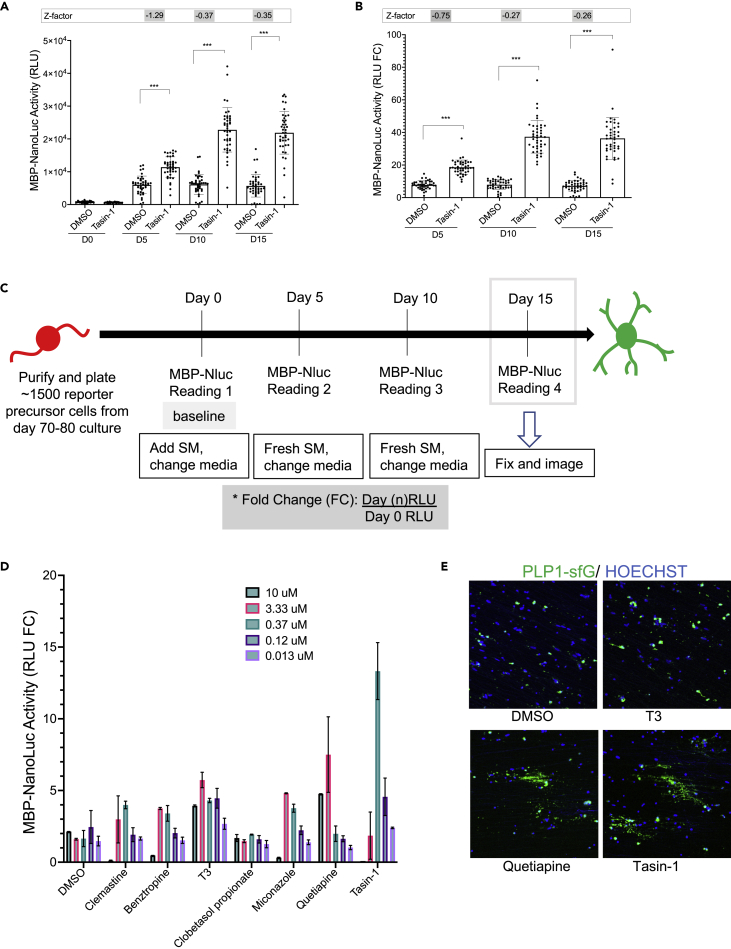
An advantage of our reporter system is that it does not require fixation or lysis for assessment, which allows us to perform time-course assays to follow real-time expression of PLP1 and MBP, oligodendrocyte/myelination marker genes. Because our main goal in generating the described reporter system was to perform HTS, we went on to optimize our assay for use in a 384-well plate format. The seeding density, NanoGlo reagent volume, culture media volumes, and the time course of the assay were optimized in preliminary studies (Figures S4A and S4B) (see the STAR Methods section). Although T3 is commonly used as positive control for OPC to OL maturation in rodent cells, we found that T3 has only a very mild effect on human OPCs (Figure 2D). Therefore, we decided to use Tasin-1 instead of T3 as a positive control and DMSO was used as negative control for optimization of our assay (Figures 2A and S4C–S4F). Tasin-1 is an inhibitor of Emopamil binding protein (EBP) and a known inducer of OL differentiation and myelination.11
Figure 2.
Drug screening assay optimization and validation
(A and B) Effect of 250 nM Tasin-1 in MBP-NLuc expression at days 5, 10, and 15 presented as (A) raw RLU value and (B) RLU fold change (FC). The Z-factor score for FC is better than the raw RLU value. Data presented as mean and compared to DMSO using one-way ANOVA (∗∗∗, P <0.0001).
(C) Timeline of the drug screening assay used for our reporter cell system.
(D) Validation of our drug screening assay using the compounds known to enhance oligodendrocyte differentiation. Data presented as mean ±SEM.
(E) purified OPCs were plated on a plate containing electrospun nanofiber in the presence of DMSO or small molecules from (D). At least two of the compounds seems to increase the myelination of the nanofiber.
To establish our screening assay (Figure S5), PDGFRA-tdTomato+ OPCs from d75-d85 differentiation cultures were purified by magnetic-activated cell sorting (MACS) with Thy1.2 microbeads and then plated into 384-well plates. We noticed a small degree of well-to-well variation in the day 0 Nluc activity (relative light unit (RLU) values), which was likely caused by small differences in the number of cells plated per well (Figure S4C). To control for this variability, instead of using absolute RLUs to compare effects of compounds on the cells, we used fold change (FC), which was calculated for each well by dividing a “time-point” RLU of a well by day 0 RLU of the same well (Figures 2A–2C). The FC value had better z’ factor values (−0.27 in FC vs −0.37 in raw RLU at d10 time-points) and better relative standard deviation (16% in FC vs 29% in raw RLU at d10 time-point) while retaining the significance of the results (Figures 2A and 2B). Therefore, RLU fold change values calculated at day 5, 10 and 15 (Figure 2C) were used as readouts for our assay.
The screening assay was validated using a number of previously reported OPC differentiation promoting compounds, and the majority of them showed a dose dependent effect on MBP-secNluc expression (Figure 2D). We have also established an in vitro myelination system where purified hOPCs are plated on electrospun nanofibers. The plated OPCs align their processed to the nanofibers as early as two days after plating, and within three weeks they mature into OLs and often myelinate the fibers as observed by PLP1-GFP+ processes (Figure S6) or by immunostaining for MBP protein.13 An increased number of PLP1-GFP+ processes aligning and potentially myelinating the nanofibers, within 10 days in culture, was observed only in the presence of a few compounds, such as Tasin-1 and Quetiapine (Figure 2E).
High throughput screen identifies compounds that increase OL maturation
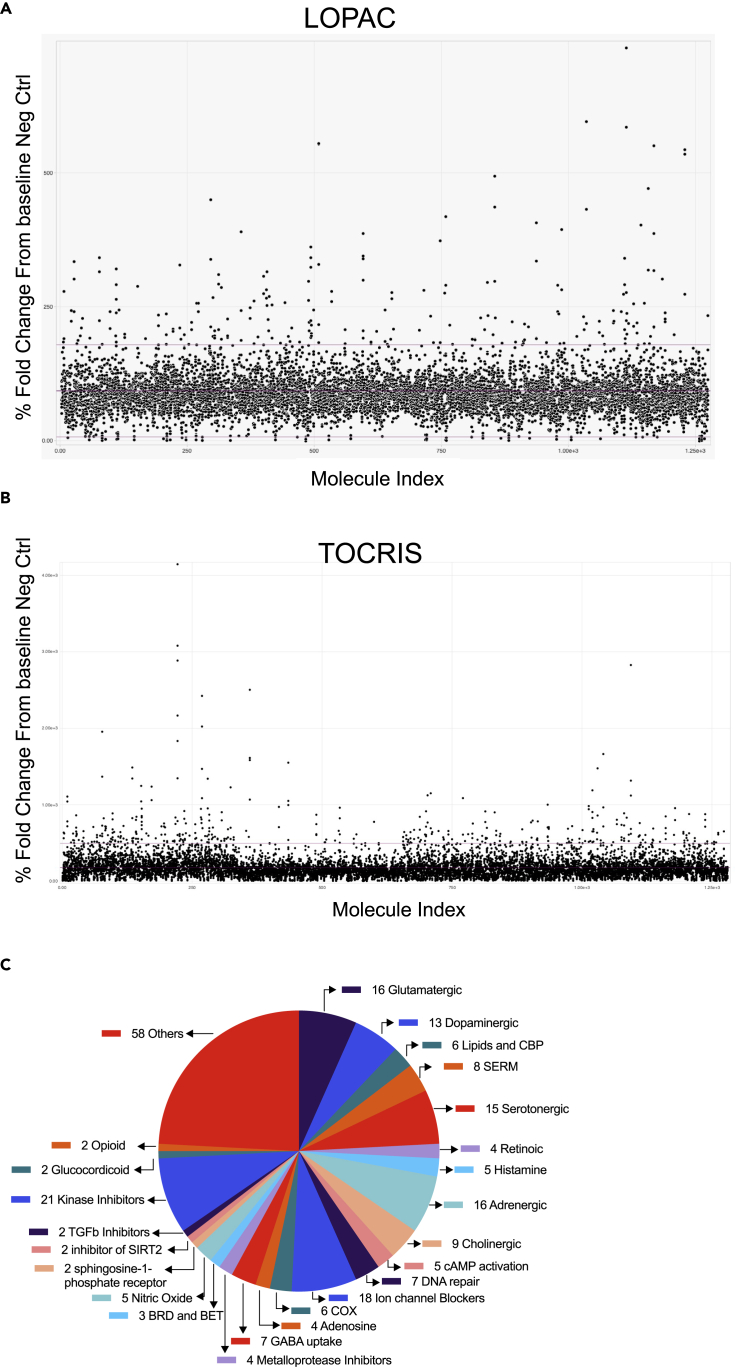
Using the above-described PT-P1-MsNL-based assay system, we screened the Library of Pharmacologically Active Compounds (LOPAC) and TOCRIS small molecule libraries (Tocriscreen Plus) (∼2500 compounds total). For an initial screen, two doses (250 nM and 1.25 μM) of the LOPAC compounds and three doses (100 nM, 500 nM, 2.5 μM) of the TOCRIS compounds were tested (Figures 3A and 3B, Table S1). A total of 240 compounds (120 from each library) that showed FC increases in secNLuc activity that were greater than two standard deviations above the DMSO vehicle control wells were retested at 7 doses (15 nM–11 μM) in duplicate, each at three time-points (5, 10 and 15 days) (Figure 3C, Table S1).
Figure 3.
High throughput screening for compounds that enhance oligodendrocyte differentiation
(A and B) Scatterplot of the HTS for compounds that increase activity of MBP-driven secNLuc in the culture media. Plot represents fold change of secNLuc activity versus the compound ID for (A) LOPAC and (B) TOCRIS library of small molecules.
(C) A total of 240 compounds with some effect to increase MBP-NanoLuc expression are graphed as a pie-chart according to a class they fall on. The numbers indicate the number of molecules that fall on that class.
The majority of the hit compounds identified in our screen have known targets, including dopaminergic, adrenergic, and cholinergic receptor signaling, cox, DNA repair, GABA uptake, opioid, P450/cholesterol biosynthesis, and selective estrogen receptor modulators (SERMs) (Figure 3C, Table 1) that have been identified from previous screens using rodent OPCs.16,17,18 We also identified a number of targets and molecules that have been implicated but not well studied in OL biology such as Butyryl cholinesterase19,20,21 (Tetraisopropyl pyrophosphoramide), BET bromodomain inhibitors22,23 (I-BET151 and SGC-CBP30), and an NMDA receptor blocker (Ro 25–6981 maleate).24,25,26,27 Most interestingly, we also identified two potent compounds that have not been previously reported to promote OL maturation or myelination, namely Ro1138452 and SR2211 that target the IP (prostacyclin) receptor pathway and retinoic acid receptor-related orphan receptor γ (RORγ), respectively.
Table 1.
List of top 15 hit compounds from each library
| Molecule ID | Chemical name | Action | Biological Activity |
|---|---|---|---|
| JHMI-0110651 | Oxiconazole | inhibitor | antifungal, inhibits of fungal cyctochrome P450 51 and inhibits ergosterol biosynthesis, |
| JHMI-0000208 | Fluphenazine dihydrochloride | Antagonist | D1/D2 selective dopamine receptor antagonist |
| JHMI-0002272 | Raloxifene hydrochloride | Modulator | Selective estrogen receptor modulator (SERM) |
| JHMI-0001917 | Tetraisopropyl pyrophosphoramide | Inhibitor | Selective inhibitor of butyrylcholinesterase |
| JHMI-0000185 | Clotrimazole | Inhibitor | Antifungal and Cytochrome P450 inhibitor; also, an inhibitor of the Ca2+-activated K+ channel |
| JHMI-0001683 | Ro 25–6981 hydrochloride | Antagonist | NR2B-selective NMDA antagonist |
| JHMI-0110506 | I-BET 151 dihydrochloride | Inhibitor | BET bromodomain inhibitor; also promotes differentiation of hiPSCs into megakaryocytes |
| JHMI-0000665 | CARBETAPENTANE | Ligand | Opioid and sigma1 ligand |
| JHMI-0002181 | ML-7 | Inhibitor | Selective myosin light-chain kinase (MLCK) inhibitor |
| JHMI-0002388 | (S)-(+)-Camptothecin | Inhibitor | DNA topoisomerase I inhibitor |
| JHMI-0002427 | 13-cis-retinoic acid, Isotretinoin | Regulator | Anti-inflammatory and antitumor actions mediated through RAR-beta and RAR-alpha receptors |
| JHMI-0000672 | Tamoxifen citrate | Inhibitor | An anti-estrogen, Estrogen receptor partial agonist/antagonist |
| JHMI-0001554 | Phosphoramidon disodium | Inhibitor | Inhibitor of mammalian enkephalinase and some metallo-endopeptidases; potent inhibitor of thermolysin and other bacterial metallo-endopeptidases |
| JHMI-0000623 | Tretinoin | Activator | Exerts its effects by binding to nuclear retinoic acid receptors (RARs) which directly regulate gene expression |
| JHMI-0002272 | Raloxifene hydrochloride | Modulator | Selective estrogen receptor modulator (SERM) |
| JHMI-0124771 | ZK 200775 | Antagonist | Competitive AMPA/kainate antagonist |
| JHMI-0124993 | Ro 1,138,452 hydrochloride | Antagonist | Selective prostacyclin IP receptor antagonist |
| JHMI-0001683 | Ro 25–6981 hydrochloride | Antagonist | NR2B-selective NMDA antagonist |
| JHMI-0125109 | SR 2211 | Agonist | Selective RORgamma inverse agonist |
| JHMI-0110506 | I-BET 151 dihydrochloride | Inhibitor | BET bromodomain inhibitor; also promotes differentiation of hiPSCs into megakaryocytes |
| JHMI-0125334 | SGC-CBP30 | Inhibitor | Potent CBP/p300 BRD inhibitor |
| JHMI-0125143 | ME 0328 | Inhibitor | Selective PARP-3 inhibitor |
| JHMI-0125424 | DG 172 dihydrochloride | inverse agonist | Potent PPARbeta/delta inverse agonist; promotes BMC differentiation |
| JHMI-0125008 | JHW 007 hydrochloride | Antagonist | High affinity dopamine uptake inhibitor |
| JHMI-0001736 | PNU 37883 hydrochloride | blocker | Vascular Kir6 (KATP) channel blocker |
| JHMI-0001980 | TMPH hydrochloride | Inhibitor | Neuronal nicotinic receptor antagonist |
| JHMI-0125155 | GSK 1562590 hydrochloride | Antagonist | High affinity, selective urotensin II (UT) receptor antagonist |
| JHMI-0125255 | XMD 8-87 | Inhibitor | Potent Ack1/TNK2 inhibitor |
The first 15 compounds are from the LOPAC, and the last 15 are from the TOCRIS library of molecules.
Validation of the promising hit compounds
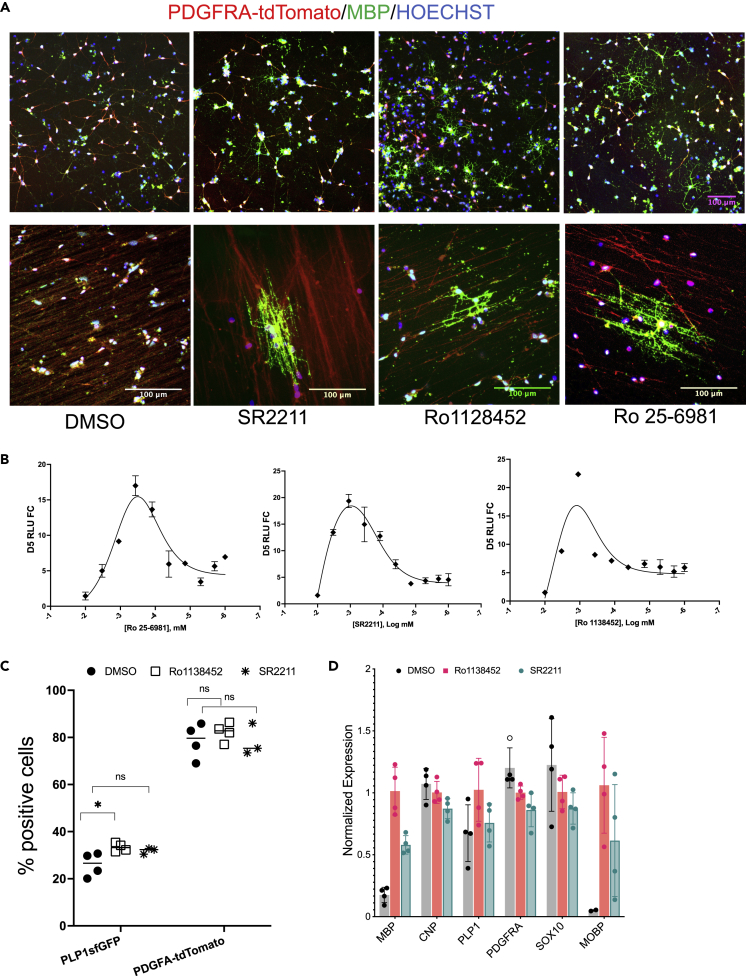
To validate the Nluc-based screening result (Figure 4B) that our potential hit compounds indeed enhance OPC maturation into OLs in the human cell system, we performed immunohistochemistry (IHC), flow and qPCR analysis of hOPCs treated with Ro1138452 and SR2211, the two most promising small molecules identified from our screen. Flow analysis showed a slight increase in percentage of PLP1-GFP positive cells (Figures 4C and S7), but no change in the PDGFRA-tdTom+ cells was detected. qPCR on OL and OPC marker genes showed increased expression of mature OL associated genes such as MBP, PLP1 and MOBP (Figure 4D). IHC using an antibody against MBP showed a higher number of MBP+ cells in the hOPCs treated with Ro1138452 and SR2211 (Figures 4A and S6C). Furthermore, in our in vitro myelination assay, hOPCs treated with these molecules for 10 days align their processes to the nanofibers and appear to myelinate them better than DMSO treated cells (Figure 4A, lower panel). Because Ro25-6981 maleate was detected as a potential hit in both LOPAC and TOCRIS library (Table 1), we examined its effect in the in vitro myelination assay. Similar to the Ro1138452 and SR2211 treatment, the Ro25-6981 treatment also appear to promote myelination of the nanofiber.
Figure 4.
Validation of the promising hit compounds
(A) Immunostaining of purified OPCs treated with different compounds for 10 days show stronger MBP staining and more MBP+ cells (top panel) and appearance of improved myelination of electrospun nanofibers ((950 nM diameter nanofibers) lower panel) compared to the DMSO treated cells. Red channel was overexposed to visualize the nanofibers in the lower panel. Immunostaining was independently repeated three times with similar results; scale bar: 100 µm.
(B) MBP-Nanoluc activity in the PTt-P1-MsNL OPCs treated with different concentration of small molecules. Yaxis is the Nanoluc reading after 5 days of drug treatment, presented as fold change to day 0. Data presented as mean ± SEM.
(C and D) Flow analysis (C) and qPCR-based quantification (D) of the OPCs treated with different small molecules show increased number of PLP1-GFP+ cells and increased PLP1 and other OL marker transcripts compared to the DMSO treated cells. In (C), comparison to DMSO is made using multiple unpaired t-test (∗, P <0.05). Data for (D) is presented as mean ±SD.
To further validate the compounds that we identified using our triple reporter system, we generated another dual reporter system (PDGFRA-P2A-tdTomato-P2A-Thy1.2 and MBP-P2A-secNluc) in an independent, male hESC line (RUES1)28 and named it RPD-MsNL (Figure 5). MACS-purified PDGFRA-tdTomato expressing OPCs generated from the RPD-MsNL hESC reporter line express other OLC markers and also produce secNluc as expected (Figures 5A, 5B, and S8). Next, we cultured the RPD-MsNL-derived OPCs in the presence of Ro1138452 and SR2211 and validated the activity of these compounds in this independent cell line via the Nluc activity, qPCR, IHC and nanofiber-based myelination assays (Figures 5C–5F).
Figure 5.
Validation of reporter OPCs generated using an independent, male hESC line (RUES1)
(A) Differentiated OPCs were MACS purified for the expression of PDGFRA-tdTomato-thy1.2, and the expression of different OPC markers between the tdTomato+ and tdTomato- (flow through) population were quantified by qPCR analysis, which shows enrichment of OPC markers in the tdTomato enriched population compared to the flow through. Three biological and three technical replicates each were used for qPCR analysis. Data are presented as mean SD. Biological and technical replicates are distinguished by filled vs clear symbols used for each data point. Source data for the qPCR are provided as a Source Data file.
(B) secNluc activity in the cell culture medium of differentiating RPD-MsNL reporter line measured with Nano-Glo assay. 20 μL of culture media from different days of differentiation (x-axis) were removed for the assay. Increase in Nano-Luc activity (y-axis), which corresponds to MBP expression increases as cells mature.
(C) Immunohistochemistry demonstrating that the MACS purified tdTomato+ cells express the OPC markers SOX10, OLIG2, NKX2.2 and NG2. Immunohistochemistry was independently repeated three times with similar results.
(D-F) qPCR (D), NanoLuc expression (E), and immunostaining (F) of the purified RPD-MsNL OPCs treated with different compounds for 10 days. (E) NanoLuc data represents two different experiments performed using different number of OPCs, represented as open (10K cells per treatment) versus filled (20k cells per treatment) symbol. (F) Stronger MBP staining, more MBP+ cells (top panel) and better myelination of the electrospun nanofibers ((950 nM diameter nanofibers) lower panel) by the compound treated OPCs compared to the DMSO treated cells is noticeable. Red channel was overexposed to visualize the nanofibers in the lower panel. Immunostaining was independently repeated three times with similar results. Data are presented as mean ± SD, scale bar: 100 µm.
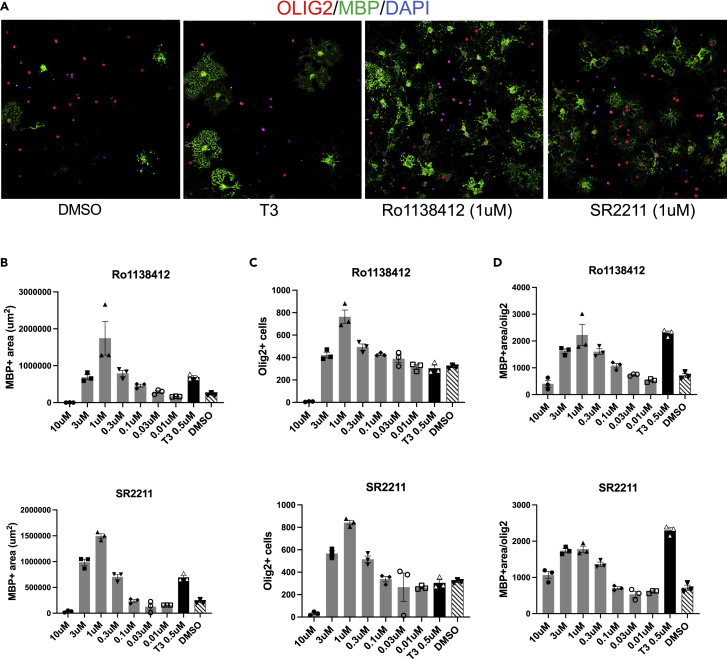
Confirmation of the hit compounds using rodent OPCs
Next, to assess if the effect of these compounds (Ro1138452 and SR2211) is specific only to human cells, we performed a validation experiment using purified primary mouse OPCs. The purified mouse OPCs treated with these molecules had significantly more MBP+ area compared to the DMSO treated culture (Figures 6A, 6B, S9A, and S9B). We also observed increased number of OLIG2+ cells in the culture treated with these molecules, which suggests that these compounds promote proliferation of the OL lineage cells in rodent OPCs (Figure 6D). No significant OPC proliferation, however, was observed in the hESC-derived OPC culture treated with these molecules. To confirm that the increased MBP+ area is not merely due to cell proliferation, we normalized MBP+ area to the number of OLIG2+ cells and show that the compounds significantly promote OPC to OL maturation in a dose dependent manner (Figures 6D, S9B, and S9C).
Figure 6.
Ro1138412 and SR2211 promote differentiation of rodent OPCs
OPCs from P7-8 mice were purified using Anti-O4 microbeads and MACS purification method, and the purified OPCs were treated with one of the following molecules: Ro1138412, SR2211, T3 as positive control or DMSO.
(A) Immunostaining for MBP protein shows significant increase in MBP positive cells and MBP+ area in the drug treated samples.
(B–D) Quantification of MBP+ area (B) OLIG2+ cell number (C), and the MBP+ area relative to OLIG2+ cells in Ro1138412 treatment (top panels) and SR2211 treatment (bottom panels) compared to T3 and DMSO treatments. Data presented as mean ± SEM.
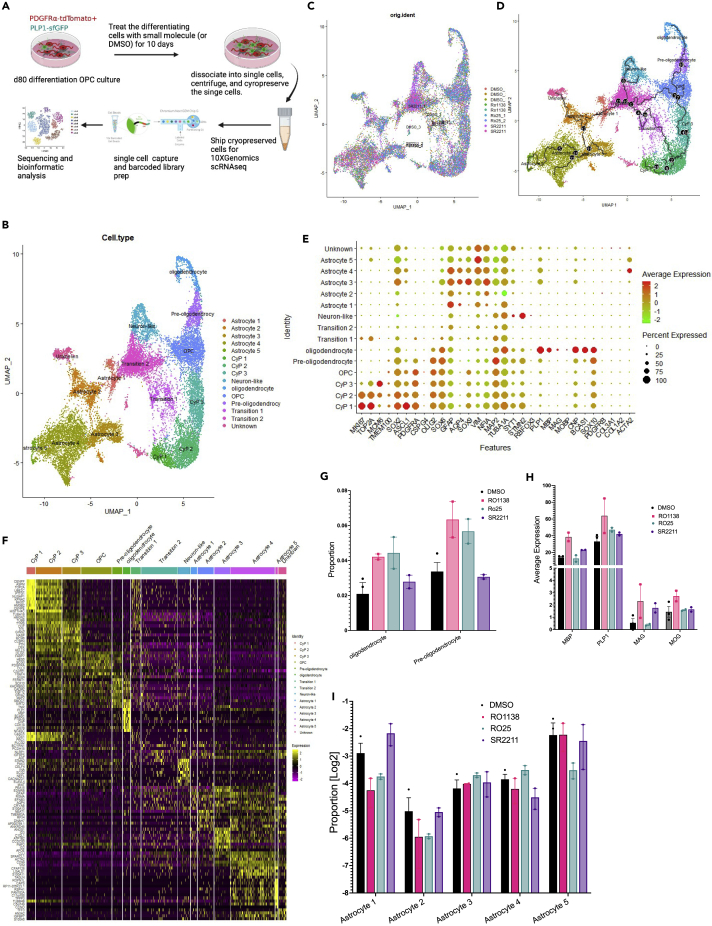
Single cell RNAseq of differentiating cultures treated with hit compounds
To better understand the transcripts and pathways modulated by the three small molecules (Ro1138452 and SR2211, and Ro25-6981 maleate), we next performed single cell transcriptomic analysis (scRNAseq) on a mixed population of differentiating OPC cultures that were treated either with DMSO or with one of the three small molecules of interest for 10 days. After removing potential doublets and cells with more than 20% mitochondrial gene content, a combined total of 18,713 cells were used for further analysis (Figures S10 and S11A, detail in STAR Methods section). Unsupervised clustering of the single cell transcripts from the combined dataset identified 15 distinct cell populations. Label prediction of the single cell clusters based on previously published datasets13,29 and expression of marker genes, predicted three cycling progenitors (CyPs), an OPC, a pre-OL, an OL, a neuron-like, a pericyte, and five astrocyte (AS) cell populations (Figures 7B and S11B, Table S2). In addition, we identified two cell populations at transitional states that did not show enrichment of any specific maker gene sets (Figures 7E and S11C). Psuedotime and trajectory analysis indicate that the cells in this transitional state are potentially transitioning from progenitor state toward either neuron-like or astrocyte cells (Figures 7D and S11D). The trajectory analysis showed a linear path from CyPs to OL differentiation; however, the astrocyte cells in our differentiation culture seem to arise either from the CyPs or, similar to our previous report,13 a sub-population of OPCs could also differentiate into astrocytes (Figure 7D).
Figure 7.
Single cell transcriptomic analysis of the differentiating PtP1MsNL OPCs treated with different small molecules
(A) schematic of the scRNAseq experiment. Day 80 differentiating OPCs were treated with small molecules for ten days before dissociation, single cell capture, and sequencing.
(B) Unsupervised clustering of the single cell transcriptional profiles of combined dataset including all the DMSO and drug treated samples. Cells are visualized with an UMAP embedding. The unbiased clustering divided the cell into 15 clusters.
(C) UMAP projection to visualize overlay of cells with different drug treatments.
(D) Trajectory analysis illustrating the maturation of mitotic progenitor cells to OL and astrocytes. Astrocytes seem to originate from early progenitors as well as OPCs.
(E) A dot plot showing average expression of a subset of known marker genes (Xaxis) across different clusters (Yaxis).
(F) heatmap of the top 10 enriched genes from each cluster. Yellow color represents increased expression.
(G and I) Proportion of cells in the oligodendrocyte and pre-oligodendrocytes clusters (G) and astrocyte clusters (I) between different drug treated samples.
(H) Average expression of oligodendrocyte markers genes in the OL cluster. Data in (G-I) are presented as mean ± SEM
We observed both the OL and Pre-OL cell proportion and average expression of OL marker genes (MBP, PLP1, MAG, MOG) in the OL cluster were slightly increased in the drug treated samples compared to the DMSO control (Figures 7G, 7H, and S10). Ro1138452 and Ro25-6981 treatment increased the OL and pre-OL cell proportion by 2-fold, but no increase was observed in the SR2211 treated samples. Ro25-6981 treatment also increased the proportion of OPC and CYP3 populations (Figure S10C), which have very high expression of OPC marker genes (Figures 7E and S11C). Average expression of the OL marker genes, particularly MBP and MAG was increased by Ro1138452 and SR2211, but not by Ro25-6981. We also observed reduction of astrocyte cell proportion in the drug treated samples. The proportion of AS1 and AS2 cell population was reduced in Ro1138452 and Ro 25–6981 treated samples, and the proportion of AS4 and AS5 was reduced in the SR2211 and Ro 25–6981 treated samples respectively (Figure 7, Table S3). These data suggest that: (1) SR2211 promotes the expression of OL marker genes but it might not significantly increase the proportion of the OL cell population, and (2) the increased proportion of OL cells in Ro1138452 and Ro25-6981 treated samples could potentially be at the expense of astrocyte lineage cells.
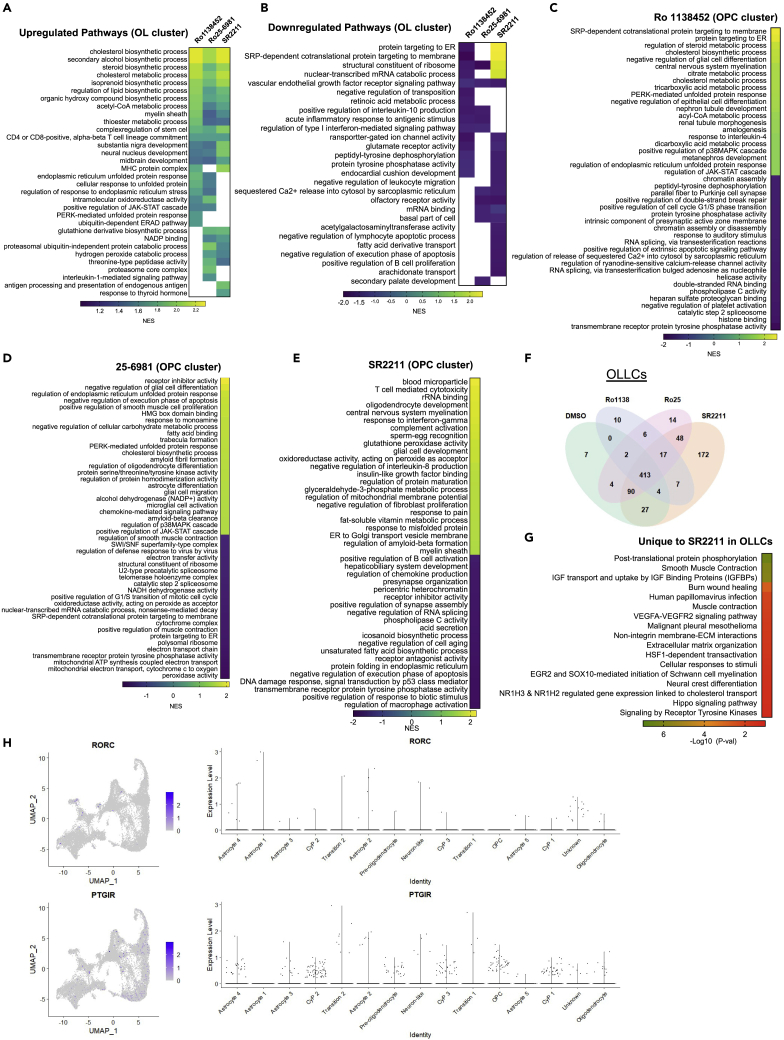
Gene set enrichment analysis reveals pathways modulated in OPCs and OLLCs by the drug treatments
We next identified the genes that are differentially expressed (DE) in the OL and OPC clusters of the drug treated samples compared to DMSO treated samples and performed gene set enrichment analysis (GSEA) on the differentially expressed genes. The GSEA revealed increased expression of genes related to sterol and cholesterol biosynthesis pathways (CBP) in the OL cluster of all the drug treated samples (Figure 8, Tables S4 and S5). CBP’s role in OL maturation myelination has been previously reported.11,30,31,32 A number of biological processes in the OL clusters that are unique to each drug treatment were also identified. Unfolded protein response (UPR) and ER stress pathways were enriched in the OL as well as OPC clusters of Ro1138452 and Ro25-6981 treated samples, but these pathways were significantly downregulated in the SR2211 treated cells (Figures 8A–8E). Processes associated with immune response such as antigen processing, regulation of Interleukin-8 and 1, complement activation, and response to interferon-gamma etc., were enriched in the SR2211 treated samples (Figure 8E). We also assessed the DE genes in OL lineage cells identified from the trajectory analysis (the Cyp > OP > pre-OL > OL trajectory) (Figure 7D) of drug treated samples when compared to the DMSO treatment. The majority of differentially expressed genes in the cells from this trajectory overlap between the samples (Figure 8F, Table S6). The highest number of unique DE genes was identified in the SR2211 treatment, and the GSEA analysis of these genes highlighted that the transport of insulin like growth factor (IGF), vascular endothelial growth factor A (VEGFA) signaling, and nuclear receptor (NR1H3 and NR1H2) mediated cholesterol transport are modulated by the SR2211 treatment (Figure 8G, Table S7).
Figure 8.
Gene set enrichment analysis of the small molecule treated samples when compared to DMSO treatment
(A and B) Heatmaps of GSEA to show few highlighted pathways that are upregulated (A) or downregulated (B) in the OL cluster of different drugs treated samples.
(C–E) Up or downregulated pathways in the OPC cells treated with different small molecules. Detailed GSEA for all the clusters are listed in supplemental data X.
(F) Venn diagram showing overlap of genes enriched in the cells within OLLC trajectory (Figure 7D) of different drug treated samples.
(G) Heatmap of GSEA analysis highlighting few pathways that are enriched in the OLLCs of SR2211 treated samples. GSEA was performed with the 172 enriched genes that are unique to the SR2211 treatment.
(H) Expression of RORC and PTGIR, canonical targets of SR2211 and Ro1138452 in the single cell RNAseq dataset.
Canonical target of the SR2211 and RO1138452
The known targets of SR2211 and RO1138452 are RORγ (RORC) and prostacyclin receptor (PTGIR), respectively. Of interest, the transcriptomic analysis showed that the canonical targets of both compounds were expressed at a very low level in the differentiating cells (Figure 8H). PTGIR expression was more abundant in the OL lineage cells than the astrocytes, and higher expression was detected in the OPCs and its expression is reduced in the OL population. This suggests that the inhibition of the IP receptor pathway by Ro1138452 could promote maturation of OPCs to OL. In contrast, very few cells in the differentiating culture express RORC, which suggests that SR2211 could be promoting OL differentiation by acting through a non-canonical pathway.
Discussion
Promoting remyelination of neurons in the CNS is a promising approach for treatment of MS and other demyelinating neurodegenerations. If endogenous OPCs could be stimulated to differentiate and remyelinate affected axons, we could reduce and potentially stop disease progression, including the neurodegeneration aspects of the diseases, and might even be able to reverse already established disability in MS patients. Remyelination-based therapy could complement current immune-modulation-based treatments. Therefore, identification of lead small molecules that can promote differentiation of myelinogenic OPCs can have direct translational implications.
The systems being used currently to screen for drugs that promote oligodendrocyte maturation and myelin production are predominantly based on rodent cell model systems. For developing effective remyelination-based therapies, validating and expanding the mouse studies to a human system is essential. In addition, drug discovery performed with human cells has a better likelihood to identify leads that will translate into effective treatments. However, a verified human OPCs/OLs-based drug discovery platform has not yet been reported, probably due, in large part, owing to the difficulty and challenges in obtaining a large quantity of pure primary glial cells for such work. Because the human stem cell-derived OPCs and OL now provide a powerful and versatile source of human cell culture systems for drug discovery, we set forth to establish a drug screening platform that uses hPSC-derived OPCs and OLs.
We engineered a human embryonic stem cell-based reporter system with three reporters where two fluorescent proteins and secreted Nano luciferase (secNluc) protein are driven by OPC and OL specific promoters, and used OPCs derived from the reporter hESC to establish a drug screening platform that allows for high-throughput screening of myelination promoting drugs. With the secreted Nluc system, cells do not need to be sacrificed for analysis, allowing time-course studies and significant cost and research benefits. Using this system, we screened ∼2500 bioactive molecules in at least two doses and three time-points. 240 potential hit compounds identified from an initial screen were further tested in duplicates at 7 doses and three-time points. The extensive data on the effect of these compounds on MBP protein expression in human OLs would not have been possible without the reporter system described here. Furthermore, the reporter cell system that we have established is suitable for a larger scale, human cell-based screen of remyelination compounds.
As a validation of our screening platform, several of the compounds we identified, including muscarinic receptor antagonists (benztropine, clemastine and carbetapentane) cytochrome P450 inhibitors (clotrimazole, oxiconazole), SERMs (raloxifene, tamoxifen), dopamine receptor antagonists (fluphenazine), and ROCK inhibitors (ML9, ML7) (Table 1) were recently reported as promoting remyelination of rodent OPCs.9,11,16,33,34,35 In addition, inhibition of bromodomain containing proteins has been reported to help improve myelination in a murine system22,23 but its role in the human system has not been examined. Tetraisopropyl pyrophosphoramide, an inhibitor of butyrylcholinesterase (BChE), is another compound of interest that was identified in our screen. A role of BChE in modulating neuroinflammation, demyelination, and neuropathology in MS has been suggested21,36,37,38 and an inhibitor of BChE, rivastigmine, has been shown to suppress neuroinflammation in EAE models.37 Our study provides validation of these targets in promoting OL differentiation in human OPC and OL system.
Possibly because of the use of the highly sensitive Nluc reporter system and also because we used human OPCs for our screens, given that it is well known that human and rodent cells differ in many important ways, we were able to identify a number of targets that have not been previously reported. We show that at least two of these new compounds identified from our screen increase OL maturation in the OPCs derived from two different hESC lines. Using the in vitro myelination assay that we have established, we found that these compounds could potentially promote myelination as well.
One of the compounds that we identified, Ro1138452, is a known inhibitor of the IP receptor (encoded by PTGIR gene). PTGIR is detected in the OL lineage cells in our scRNAseq dataset. The PTGIR transcript expression was higher in the OPCs and reduced in the OL population, which suggests that the inhibition of the IP receptor pathway by Ro1138452 could potentially promote maturation of OPCs to OL. PTGIR is also reported to be expressed by PDGFRA+ OPCs and PDGFRB+ pericyte cells in the mouse CNS and also in the PDGFRA+ cells in the CNS of MS patients.39,40 Of interest, administration of Ro1138452 in the lysophosphatidylcholine (LPC) injected demyelination mouse model reduced the OPCs recruitment and impaired remyelination in the spinal cord.39 It is possible that the IP pathway is essential for OPC migration to an injury site, but once the OPCs are recruited, repression of the IP pathway promotes OL maturation and myelination. Another possibility is that the IP pathway has species-specific different functions. Conditional loss of PTGIR function in mouse OPCs versus OL, and loss of PTGIR function in hPSC-derived OPCs/OL would assist in making the species-specific distinction and would also help to understand the role of IP pathway in OPC migration, OPC to OL maturation, and myelination.
Another compound that we identified, SR2211, which has been shown to specifically inhibit RORγ,41 was a very interesting finding because of the well-known function RORγ in regulating immune homeostasis. Specifically, the RORγt isoform of RORγ is crucial for differentiation of Th17 cells and the expression of Interleukin 17 (IL-17)42,43,44 which has been implicated in the pathology of multiple sclerosis (MS).45,46 Prevention of IL-17 production both by genetic ablation RORC and by pharmacological inhibition of RORγ44 is shown to protect mice from developing experimental autoimmune encephalomyelitis (EAE), a mouse model of MS.43 However, a direct role of RORγ inhibition in promoting myelination had not been previously documented. Of note, ursolic acid (UA), which is known to promote remyelination, is also a potent RORy inhibitor, but UA is reported to act via PPARγ activation.47 Because SR2211 is known to specifically target RORγ, our data suggests that modulation of RORγ pathway could benefit inflammatory demyelination such as MS by both immune modulation and remyelination. Pro-myelinating effect of RORγ inhibition is particularly interesting because numerous reports have suggested that cholesterol biosynthesis intermediates (CBIs), especially those between lanosterol and zymosterol that promote OL maturation and myelination,11 have been reported to act as ligands to RORγ and modulate its activity.48,49,50 Therefore, it is possible that the RORγ is the not yet identified link that connects cholesterol biosynthesis to OL differentiation and myelination. On the other hand, because only a handful of cells in our differentiating OPC/OL culture express RORC, it is possible that SR2211 is working through a non-canonical pathway or by its off-target effect on RORA, PPARγ or androgen receptor.51 Alternatively, SR2211 working via a non-cell autonomous manner by targeting astrocyte cells is also possible. The ongoing loss of RORC function in human iPSC-derived OPCs/OL/astrocytes should help us better understand the role of RORγ in OL maturation and myelination, define their target pathways, and investigate their therapeutic potential.
Limitations of the study
We have established a robust human cell-based drug screening platform for the discovery of pro-myelinating compounds, performed a small-scale proof-of-principle drug screen, and identified a number of promising compounds. Current limitations of the study are that the genetic validation of the pathways targeted by these small molecules and in vivo testing of the molecules in rodent models of demyelination have not been performed. These studies would help to improve our understanding of the activity of these molecules.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-PDGFR alpha | R&D Systems | AF-307-SP |
| PE anti-human CD140a (PDGFRalpha) | BioLegend | 323505 |
| Anti-SOX10 | R&D Systems | AF2864-SP |
| Anti-NKX2.2 | DSHB | Cat# 74.5A5; RRID:AB_531794 |
| Anti-MBP | Millipore | Cat# MAB386; RRID:AB_94975 |
| Anti-OLIG2 | Millipore | Cat# AB9610, RRID:AB_570666 |
| Anti-O4 clone 81 | Millipore | MAB345 |
| Hoechst 33342 | ThermoFisher Scientific | H3570 |
| Alexa Fluor 488 goat anti-rabbit IgG | ThermoFisher Scientific | A-11034 |
| Alexa Fluor 488 goat anti-mouse IgM | ThermoFisher Scientific | A-21042 |
| Alexa Fluor 647 goat anti-rabbit IgG | ThermoFisher Scientific | A-21245 |
| Alexa Fluor 647 donkey anti-goat IgG | ThermoFisher Scientific | A-21447 |
| Bacterial and virus strains | ||
| DH10B | ThermoFisher Scientific | C404010 |
| Chemicals, peptides, and recombinant proteins | ||
| mTeSR1 | Stem Cell Technologies | 5850 |
| StemFlex | ThermoFisher Scientific | A3349401 |
| DMEM/F12 | ThermoFisher Scientific | 11320082 |
| Accutase | ThermoFisher Scientific | A11105-01 |
| Antibiotic-Antimycotic | ThermoFisher Scientific | 15240062 |
| Growth Factor Reduced BD MatrigelTM Matrix | BD Biosciences | 354230 |
| Blebbistatin | Sigma | B0560-1MG |
| Opti-MEM™ I Reduced Serum Medium | ThermoFisher Scientific | 31985088 |
| CryoStor® CS10 | Stem Cell Technologies | 7930 |
| CTLC-ABC | Immunospot | NC9711917 |
| Lipofectamine™ Stem Transfection Reagent | ThermoFisher Scientific | STEM00001 |
| Puromycin | ThermoFisher Scientific | A11138-03 |
| N2 supplement | ThermoFisher Scientific | 17502-048 |
| B27 Supplement without vitamin A | ThermoFisher Scientific | 12587-010 |
| SB431542 | Stem Cell Technologies | 72234 |
| LDN193189 | Stem Cell Technologies | 72147 |
| Smoothened agonist | Sigma | R2625 |
| All-trans retinoic acid | Millipore | 566660 |
| Recombinant human PDGF-AA | R&D Systems | 221-AA-050 |
| Recombinant human IGF-I | R&D Systems | 291-G1-200 |
| Recombinant human HGF | R&D Systems | 294-HG-025 |
| Neurotrophin 3 (NT3) | Millipore | GF031 |
| cAMP analog | Sigma | D0260 |
| 3,3,5-Triiodo-l-thyronine (T3) | Sigma | T2877 |
| l-Ascorbic acid | Sigma | A4403 |
| Insulin solution human | Sigma | 19278 |
| Biotin | Sigma | 4639 |
| Poly-L-ornithine hydrobromide | Sigma | 3655 |
| HEPES (1 M) | ThermoFisher Scientific | 15630080 |
| Laminin | MilliporeSigma | L2020-1MG |
| Critical commercial assays | ||
| MycoAlert Mycoplasma Detection kit | Lonza | LT07-218 |
| hPSC Genetic Analysis Kit | StemCell Technologies | 7550 |
| Nano-Glo® Luciferase Assay System | Promega | N1120 |
| Neural Tissue Dissociation Kit | Miltenyi | 130-092-628 |
| Deposited data | ||
| Single cell RNAseq | This study | Database: GSE223599 |
| Human single cell data | Chamling et al.,13 Jakel et al.29 | GEO Database: (GSE118257, GSE104276GSE146373), Bioproject (544731)) |
| Human reference genome NCBI build 37, GRCh37 | Genome Reference Consortium | http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/ |
| Experimental models: Cell lines | ||
| Passage 40 hESC (WA09) or H9 NIH HES # 0062 | WiCell | RRID:CVCL_9773 |
| Passage 26 hESC (RUES1) | WiCell | RRID:CVCL_B809 |
| PTt-P1-MsNL (the triple reporter) | This paper | N/A |
| Oligonucleotides | ||
| Primers for Reporter Cell line (see Table S8) | This paper | N/A |
| qPCR Primers (see Table S8) | This paper | N/A |
| Recombinant DNA | ||
| PX459.V2 | Addgene | RRID:Addgene_62988 |
| ZeroBlunt TOPO | ThermoFisher Scientific | K280020 |
| MBP-P2A-secNanoluc | This paper | N/A |
| PLP1-sfGFP | This paper | N/A |
| Software and algorithms | ||
| ImageJ | Schneider et al.52 | https://imagej.net/software/fiji/ |
| GraphPad Prism, V9 | GraphPad Software, LLC | https://www.graphpad.com |
| CFX Maestro | Biorad | 12004128 |
| Harmony software 4.9 | Perkin Elmer | HH17000010 |
| Cellomics CX7 | ThermoFisher Scientific | HCSDCX7LEDPRO |
| R package | Wu et al.53 | ClusterProfiler |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Xitiz Chamling (xchamli1@jhmi.edu).
Material availability
Materials are available upon request.
Experimental model and subject details
Human pluripotent stem cells (PSCs) and culture conditions
hESC WA09 and RUES1 (both WiCell), NIH-approved hESC lines (NIH approval number: NIHhESC-10-0062, and 0012), were used for this study. All hESCs related experiments were conducted by following all policies and regulations established by the Johns Hopkins University Embryonic Stem Cell Research Oversight committee. hESCs were maintained in mTesr or mTesr plus (85,850 or 100–0276, Stem Cell Technologies) on growth factor-reduced Matrigel (354,230, Corning) coated plates at 37C, 10% CO2/5% O2. hPSC colonies were passaged by dissociating with Accutase (A6964, Sigma-Aldrich). Cells were maintained in stem cell media containing 5 mM blebbistatin (B0560, Sigma-Aldrich) for the first 24 hours after passaging, to improve single cell survival. Karyotype analysis was performed using a qPCR based hPSC Genetic Analysis Kit (StemCell Technologies, #07550) and KaryoStat Karyotyping (ThermoFisher). Chromosomal Cells were routinely tested for mycoplasma contamination (MycoAlert, Lonza) and only the cells free of contamination were used for OPC differentiation.
Primary mouse OPCs
To test the effect of our two compounds on mouse OPCs, brain tissues from postnatal day 7–8 (P7-8) C57Bl/6 mice of mixed sex were collected. All animal experiments were performed at Sanofi’s animal facilities, a fully accredited Association for Assessment and Accreditation of Laboratory Animal Care facilities, by following the protocols approved by the Sanofi Institutional Animal Care and Use Committee Animals.
Method details
Cloning
A guide sequence targeting the stop codon of the MBP and PLP1 locus was designed in Deskgen.com. Guide sequence with minimal off target and very high activity score was chosen and cloned into the BbsI restriction site of the Cas9 plasmid (Cas9-P2A-Puro modified from Addgene #6298854). pSpCas9(BB)-2A-Puro (PX459) V2.0 was a gift from Feng Zhang (Addgene plasmid # 62988; http://n2t.net/addgene:62988). To clone the donor plasmid, a ∼2 kb PCR product was amplified from genomic DNA extracted from H9 ES cells and cloned into Zero Blunt TOPO cloning vector (ThermoFisher Scientific) to create an intermediate donor plasmid. Either the P2A-secNanoLuc or the sfGFP reporter DNA sequence was then introduced into the intermediate donor plasmid, precisely upstream of either the MBP or PLP1 stop codon, using Gibson assembly (New England Biolabs). NanoLuc plasmids were purchased from Promega. Plasmid maps and sequences used for cloning are included in Table S8.
Generation of reporter cell lines
Method for gene editing and report cell line generation, previously reported by our lab,13,14,52was followed. Cells were transfected using the Lipofectamine Stem (STEM00001, ThermoFisher Scientific) transfection reagent following the manufacturer’s recommended protocol. 0.35 μg Cas9 plasmid containing a gRNA sequence and 0.75 μg of donor plasmid were used for transfection. Two days after the transfection, the cells were selected with 750 ng/mL of puromycin for 24 hours, washed and cultured for 5 more days in the same dish with daily media change. After 5 days, the surviving cells were passaged at 500–1000 single cells per well of a 6 well plate for picking individual colonies and PCR-based genotyping.54 PCR was performed using the Phusion Flash mastermix (ThermoFisher Scientific) and a 2-step PCR protocol following the manufacturer’s instruction. Clones with clear homozygous knock-In were selected for the MBP-secNluc (Figures S1D andS1E), and a clone with heterozygous KI was chosen for the For PLP1-sfGFP reporter. For the PLP1-sfG KI clone, another allele of the PLP1 locus was sequenced to make sure that it had no mutation.
Oligodendrocyte differentiation protocol
Previously published and well-established oligodendrocyte differentiation protocol55 was followed with minor modifications. Briefly, hESCs were dissociated to single cells and plated on Matrigel coated plate at 100,000 cells/well of a 6-well plate and maintained in mTesr plus at 37C, 10% CO2/5% O2. For RUES1 cells 200K cells were used to start differentiation. Two days after passaging, neural differentiation and spinal cord patterning was induced through dual SMAD inhibition (SB431542, 10uM and LDN193189, 250 nM) and 100 nM all-trans RA.56 From day 8 to day 12, differentiating cells were maintained in neural induction media supplemented with RA (100 nM) and SAG (1 mM). At day 12, adherent cells were lifted and cultured in low-attachment plates to favor sphere aggregation. At day 30, spheres were plated into poly-L-ornithine/laminin-coated dishes in a media supplemented with, B27 (Thermo Fisher, 12587010), N2 supplement (Thermo Fisher, 17502048), PDGF-AA (221-AA-10, R&D systems), neurotrophin-3 (GF0308, MilliporeSigma), HGF (294-HG-025 R&D systems), Minocycline (Y0001930, Sigma) and T3 (T2877, Sigma).
Flow cytometry and MACS purification of the reporter hOPCs
For flow cytometry analysis and MACS purification, cells were dissociated into single cell suspension by incubating in accutase for ∼45 minutes. The single cell suspension was then passed through a ∼70 uM cell strainer (BD Biosciences), washed, and resuspended in Live Cell Imaging Solution (ThermoFisher Scientific) for flow analysis or MACS buffer (Miltenyi Biotec (Auburn, CA)) for MACS cell sorting. Flow analysis was performed with an SH800S Cell Sorter (Sony Biotechnology, San Jose, CA). BSC and FSC were used to select and subset live cells, and only live cells were used to quantify the number of tdTomato+ or GFP+ cells. A gate was set up using WT hES cells differentiated to day 95. MACS purification was performed by following manufacturer’s instructions with minor modifications. Cells were resuspended in MACS buffer after passing through cell strainer. A CD90.2 (THY1.2) or O4 MicroBeads were added to the single cell suspension that is resuspended in MACS buffer and incubated at room temperature for 15 minutes for cell binding. Cells were run through the LS or MS magnetic column, columns were washed 3 times with the MACS buffer, and the cells bound to the column were recovered by pushing the cells through using a syringe provided. To increase the purity of tdTomato+ population, the cells collected from the column were run through a new column without additional supplementation of MicroBeads.
Nanofiber-based myelination assay
Aligned polycaprolactone (PCL) nanofibers were electrospun on a glass coverslip according to a previously reported method.57,58 The average diameter of the fibers collected was 950 nm ± 200 nm. The nanofibers on coverslips were sterilized by soaking in 95% ethanol, and then washed three times with PBS. The nanofiber coverslips were placed on a 24 well plate and coated with PLO (50 μg/mL) for 4 hours followed by laminin (10 μg/mL) at 37°C overnight. Before cell seeding, laminin was aspirated and the coverslip was allowed to dry completely. For each well of a 24 well plate containing a nanofiber coverslip, 5000 purified OPCs were resuspended in 50 ul of PDGF media and the cells were added to the center of the coverslip. Cells were allowed to attach for 30 mins before gently adding 500 uL of the PDGF media/well to culture the cells. Media with small molecules or DMSO was replenished every 4–5 days. At the end of the experiment. media was removed, cells were fixed with 4% PFA, and used for immunofluoresce staining and imaging.
qRT-PCR, immunofluorescence staining, and microscopy
Total RNA was isolated using the RNeasy Mini Kit (QIAGEN) and reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). A 2uL PCR reaction was set up using acoustic liquid handler (ECHO 550, Labcyte) and performed with the CFX384 real-time PCR instrument (Bio-Rad). Assays included at least three technical and two biological replicates unless otherwise stated, and were run using the Ssoadvanced Universal SYBR Green Supermix (Bio-Rad).
For immunofluorescence staining, cells were fixed with 4% paraformaldehyde, and simultaneously permeabilized and blocked with 0.2% Triton X-100 + 5% BSA +5% normal goat serum (or a serum specific to the host of secondary antibody) for an hour. Cells were then incubated with appropriate dilution of primary antibodies (Anti-PDGFR alpha (1:200); PE anti-human CD140a (1:100); Anti-SOX10 (1:100); Anti-NKX2.2 (1:50); Anti-MBP (1:100); Anti-OLIG2 (1:500); Anti-O4 (1:200)) overnight followed by secondary antibodies (Alexa Fluor 488/647 (1:500)) for 2 hours. Fluorescence images were taken using either the EVOS FL Auto 2 (ThermoFisher Scientific) or Zeiss 510 confocal microscope.
High-throughput screening
MACS purified reporter OPCs from day 75–85 differentiating culture was used for screening. 1.5K cells/well in of purified OPCs were plated in PLO-laminin coated 384 well plates using the EL406 (BioTeK) peristaltic liquid dispenser. 50uL of PDGF media was used per well to plate the cells. On the day of drug treatment (on day 2 after plating the cells) 20 uL of culture media was removed from each well using an integra ViaFlow 384, and Nluc activity in the media was measured (day 0 reading).
Nluc activity was measured using NanoGlo luciferase assay reagents (Promega N1150) following manufacturer’s instruction with few modifications. 2.5 uL NanoGlo reaction mix was diluted with water (1:1 dilution) and the 5 uL diluted NanoGlo reaction mix was added per well containing 20uL of media. RLU reading was performed with a microplate reader (ClariOstar, BMG Labtech) using the preloaded settings for Nano luciferase and 0.1s exposure.
Drug dispensing was performed using ECHO 555, acoustic liquid handler. First, an intermediate plate with 10 uL of glial media in each well of a new sterile 384 well plate was prepared. Appropriate volume of small molecules was dispensed into each well using the ECHO. Since the highest dose (10 uM) was dispensed from the 10 mM stock solution, the maximum DMSO dose cells would receive was 0.01%, which is known to be well tolerated by cells in vitro. Additional 10 uL media was added to each well containing the small molecules. Using integra ViaFlow 384, 20 uL of the media containing small molecules was transferred from the intermediate plate to the plate containing cells. After adding the small molecules cells were cultured in 37 C, 5% Co2 incubator. Every 5 days, 20 uL of media was removed, Nluc activity measured and fresh media with small molecules was replenished as described above. On day 15, cells were fixed with 4% PFA, washed with PBS and either stored in 4C or immediately imaged using a High-Content Imager (Cellomics CX7).
In vitro mouse OPC culture, drug treatment, and imaging
Mouse OPC isolation, culture and treatment
C57Bl/6 mouse brains were collected on P7-8 and stored in Hibernate A (Thermofisher, A124750) medium until tissue digestion. The digestion was conducted using a Neural Tissue Dissociation Kit (Miltenyi, 130-092-628) and the gentalMACs Dissociator (Miltenyi) in accordance with the manufacturer’s protocol. Anti-O4 microbeads (Miltenyi, 130-094-543) and LS columns (Miltenyi, 130-042-401) were used to positively select for OPCs per the manufacturer’s protocol. The isolated cells were plated 10,000 cells per well on black 96-well plates (Perkin Elmer, 6055302), coated with 100 μg/mL Poly-D-lysine (Sigma, P6407) for at least 1 hour. Cells were cultured in DMEM/F12 (Gibco, 11330-057) with 1% N2 (Invitrogen,17502048), 100 U/ml Penicillin/100 μg/mL Streptomycin (Gibco, 15140122), 0.01% bovine serum albumin (Sigma, A9576), 5 μg/mL N-Acetyl-L-cysteine (Sigma, A8199), 10 ng/mL d-Biotin (Sigma, B4639), 2% B27 (Invitrogen, 17504044), 1 mM pyruvate (Gibco, 11360070), 5 μg/ml insulin (Sigma, I9278), 4.2 μg/mL forskolin (Sigma, F6886) and 10 ng/mL CNTF (Peprotech, 450-50). To promote proliferation, 20 ng/mL PDGF-AA (Invitrogen, PHG0035) and 1 ng/mL NT-3 (Peprotech, 450-03) were added to the medium. After two days, the proliferation medium was removed and replaced with differentiation medium (proliferation medium minus PDGF-AA and NT-3) and the Ro1138412 and SR2211 treatments were administered. Two days later the media was replenished. Cells were fixed in 4% PFA (Electron Microscopy Sciences, 15714S) three days later for a total of 5 days of treatment.
Mouse OPC immunostaining and imaging
Fixed cells were incubated with 10% normal goat serum (Vector Labs, S-1000) in PBS (Gibco, 10010023) with 0.2% Triton X-100 (Sigma, T9284; PBST) for 30 min to block nonspecific binding. The primary antibodies used were rat anti-myelin basic protein (BioRad, MCA409, 1:400 dilution) and rabbit anti-Olig2 (Millipore, AB9610 1:400). Cells were incubated with the primary antibodies in 5% normal goat serum in PBST overnight at 4C. The secondary antibodies used were AlexaFluor488 and AlexaFluor555 (1:1000, Invitrogen, A11006 and A21428), incubated for 1hr at room temperature. The cells were imaged on the Opera Phenix (Perkin Elmer). 21 images per well were taken with 4x1um thick Z Stack. Images were analyzed using Harmony software 4.9 (Perkin Elmer).
Single-cell transcriptomic analysis
Sample preparation
The reporter hESCs were differentiated to OPCs by following the protocol described above. On day 30 of differentiation, neurospheres were plated on a 6-well culture dish to facilitate OPC migration from the plated spheres. 3 wells of day 80 differentiating cultures were treated DMSO as control and two wells each were treated with different small molecules for 10 days. Media with small molecules were replenished every three days. At the end of drug treatment, cells from each well were dissociated into single cells by treating with accutase for 45 minutes (just the cells that had migrated out from the spheres were collected, the spheres remain mostly intact). Single cells were passed through 40 uM cell strainers, washed once with PBS, cryopreserved in CTLC-ABC (NC9711917, Immunospot) buffer, and stored in −80 before shipping for single cell capture.
Single cell capture
The cryopreserved cells were thawed by following the instructions provided for CTLC-ABC. Cells were washed and counted on the DeNovex cell counter. Single cell capture, barcoding, post GEM-RT cleanup, cDNA amplification, and 3′ library construction was performed using the 10X genomics’ single-cell RNA-seq technology by following their detailed instructions. Sequencing was performed on a Illumina Novaseq sequencer.
Quality control (QC) and clustering and differential gene expression
For quality control, only genes whose expression was detected in at least three cells were included in the analysis. To filter out potential doublets and poor cell samples, cells that had more than 7000 features or less than 250 features were excluded. We also excluded cells with more than 20% mitochondrial gene content. The remaining 18,714 cells that passed QC were used for all the downstream analysis. Log-normalization, principal component analysis (PCA), the Uniform Manifold Approximation and Projection (UMAP) were conducted using the Seurat package R. 2000 highly variable genes were used as input for PCA analysis. Samples from the different treatment groups were integrated using the Harmony package for R59 to remove possible batch effects. A 30-dimensional reduction by harmony with resolution 0.5 was used for the UMAP embedding and clustering. To identify differentially expressed genes from each cluster, generalized linear regression in the MAST package was performed on the genes that are expressed in at least 10% of cells in each treatment group and have a minimum of 0.25 log2 fold change. To compare gene expression in drug treated samples to that in the DMSO control samples, log fold changes and pvalue were calculated for each gene, which were further used as the inputs for GSEA analysis with the R package ClusterProfiler.53
Data integration and label transfer
To compare cells in our data with other human brain datasets, an adult human brain single-cell dataset (GSE118257) was downloaded from the GEO and only cells from healthy donors were included. Standard preprocessing was done on the reference data. The anchors between the reference data and our data were found by using the FindTransferAnchors function in Seurat60 using the default parameters. The cells in our data were classified based on the reference data using the TransferData function, and a prediction ID and a prediction score were assigned to each cell in our data. pHeatmap package in R was used to generate the prediction score heatmap.
Gene set enrichment analysis
To conduct gene set enrichment analysis, genes were ranked by the product of the sign of log2(fold change) and - log10(p-value). Ranked gene lists were used as input for gseGO function in package ClusterProfiler with minGSSize = 15, maxGSSize =500, nPerm = 10,000, and pvalue cutoff = 0.05.
Pseudotime trajectory analysis
Pseudotime trajectory analysis was performed using Monocle 3.61 Firstly, the count matrix, cell metadata, as well as dimensional reduction result stored in the Seurat object was used to create a new cds object. The trajectory graph was learned by utilizing the learn_graph function with default settings. The order_cells function was used to calculate pseudotime by setting the node in the mitotic cell cluster CyP1 as the root node of the trajectory. Differentially expressed genes along the trajectory from the CyP1 to the oligodendrocytes were identified by using a generalized linear model against the pseudotime.
Quantification and statistical analysis
All qRT-PCR data are presented as a fold change in RNA normalized to the expression of two housekeeping genes: either GAPDH and ACTB or GAPDH and CREBBP. qRT-PCR data was analyzed using CFX Maestro (Biorad) qPCR analysis software and graphed using Prism (GraphPad, V9). Statistical differences in qTR-PCR were determined by two-way ANOVA. Statistical differences in flow cytometric analysis were compared by a two-tailed t test. For high-throughput screening any compounds that showed increase in secNLuc activity that were greater than two standard deviations above the DMSO vehicle control were considered initial hit. Quantification of percent MBP+ cells in human OPCs upon treatment with the molecules was performed using imageJ, cell count.52 For the primary mouse OPCs, the cells were imaged on the Opera Phenix (Perkin Elmer), 21 images per well were analyzed and quantification performed using the built-in Harmony software 4.9 (Perkin Elmer).
Acknowledgments
Funding: Grants from the Gilbert Family Foundation (DJZ, XC), National Institutes of Health grants P30 EY001765 (DJZ, Wilmer Eye Institute), Sanofi Inc. (XC, DJZ), National Institutes of Health grants K99 EY029011 and R00EY029011 (XC), Unrestricted funds from Research to Prevent Blindness (Wilmer Eye Institute), and Generous gifts from the Guerrieri Family Foundation (DJZ).
Author contributions
W.L. assisted with experimental designs, conducted experiments, conducted bioinformatic analysis, and edited the manuscript. C.B. assisted with experimental design and high-throughput screening assay and analysis. Y.H., G.S., S.M., and J.C.D. assisted with the screening and RNA work. F.T., X.C., and Y.D. assisted in generating the stem cell reporters and differentiation of hESC-derived OPCs. W.F., C.C., and H.J., assisted with bioinformatic analysis. D.J.Z. designed the study, edited the manuscript, and provided funding support. X.C. designed the study, performed experiments, wrote the manuscript, and provided funding support.
Declaration of interests
D.J.Z. and X.C. are inventors on an intellectual property disclosure related to the technology described in the manuscript that has been filed with Johns Hopkins University, which seeks to license the technology.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: February 8, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106156.
Supplemental information
Data and code availability
-
•
Raw data for all the compounds screened in our system is uploaded as Table S1.
-
•
scRNAseq data is uploaded to NCBI gene expression repository (GSE223599).
-
•
No new codes were generated for this study.
-
•
Any additional information required to analyze the data reported in this study will be made available by the lead contact upon request.
References
- 1.Franklin R.J.M. Why does remyelination fail in multiple sclerosis? Nat. Rev. Neurosci. 2002;3:705–714. doi: 10.1038/nrn917. [DOI] [PubMed] [Google Scholar]
- 2.Pietrangelo A. Healthiline Networks, Inc; 2014. Multiple Sclerosis by the Numbers: Facts, Statisctics, and You. [Google Scholar]
- 3.Levine J.M., Reynolds R. Activation and proliferation of endogenous oligodendrocyte precursor cells during ethidium bromide-induced demyelination. Exp. Neurol. 1999;160:333–347. doi: 10.1006/exnr.1999.7224. [DOI] [PubMed] [Google Scholar]
- 4.Penderis J., Shields S.A., Franklin R.J.M. Impaired remyelination and depletion of oligodendrocyte progenitors does not occur following repeated episodes of focal demyelination in the rat central nervous system. Brain. 2003;126:1382–1391. doi: 10.1093/brain/awg126. [DOI] [PubMed] [Google Scholar]
- 5.Franklin R.J.M., Ffrench-Constant C. Remyelination in the CNS: from biology to therapy. Nat. Rev. Neurosci. 2008;9:839–855. doi: 10.1038/nrn2480. [DOI] [PubMed] [Google Scholar]
- 6.Chang A., Tourtellotte W.W., Rudick R., Trapp B.D. Premyelinating oligodendrocytes in chronic lesions of multiple sclerosis. N. Engl. J. Med. 2002;346:165–173. doi: 10.1056/NEJMoa010994. [DOI] [PubMed] [Google Scholar]
- 7.Peppard J.V., Rugg C.A., Smicker M.A., Powers E., Harnish E., Prisco J., Cirovic D., Wright P.S., August P.R., Chandross K.J. High-content phenotypic screening and triaging strategy to identify small molecules driving oligodendrocyte progenitor cell differentiation. J. Biomol. Screen. 2015;20:382–390. doi: 10.1177/1087057114559490. [DOI] [PubMed] [Google Scholar]
- 8.Mei F., Fancy S.P.J., Shen Y.A.A., Niu J., Zhao C., Presley B., Miao E., Lee S., Mayoral S.R., Redmond S.A., et al. Micropillar arrays as a high-throughput screening platform for therapeutics in multiple sclerosis. Nat. Med. 2014;20:954–960. doi: 10.1038/nm.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Najm F.J., Madhavan M., Zaremba A., Shick E., Karl R.T., Factor D.C., Miller T.E., Nevin Z.S., Kantor C., Sargent A., et al. Drug-based modulation of endogenous stem cells promotes functional remyelination in vivo. Nature. 2015;522:216–220. doi: 10.1038/nature14335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elitt M.S., Barbar L., Tesar P.J. Drug screening for human genetic diseases using iPSC models. Hum. Mol. Genet. 2018;27:R89–R98. doi: 10.1093/hmg/ddy186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hubler Z., Allimuthu D., Bederman I., Elitt M.S., Madhavan M., Allan K.C., Shick H.E., Garrison E., T Karl M., Factor D.C., et al. Accumulation of 8,9-unsaturated sterols drives oligodendrocyte formation and remyelination. Nature. 2018;560:372–376. doi: 10.1038/s41586-018-0360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mei F., Mayoral S.R., Nobuta H., Wang F., Desponts C., Lorrain D.S., Xiao L., Green A.J., Rowitch D., Whistler J., Chan J.R. Identification of the kappa-opioid receptor as a therapeutic target for oligodendrocyte remyelination. J. Neurosci. 2016;36:7925–7935. doi: 10.1523/JNEUROSCI.1493-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chamling X., Kallman A., Fang W., Berlinicke C.A., Mertz J.L., Devkota P., Pantoja I.E.M., Smith M.D., Ji Z., Chang C., et al. Single-cell transcriptomic reveals molecular diversity and developmental heterogeneity of human stem cell-derived oligodendrocyte lineage cells. Nat. Commun. 2021;12:652. doi: 10.1038/s41467-021-20892-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sluch V.M., Chamling X., Liu M.M., Berlinicke C.A., Cheng J., Mitchell K.L., Welsbie D.S., Zack D.J. Enhanced stem cell differentiation and immunopurification of genome engineered human retinal ganglion cells. Stem Cells Transl. Med. 2017;6:1972–1986. doi: 10.1002/sctm.17-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J.H., Lee S.R., Li L.H., Park H.J., Park J.H., Lee K.Y., Kim M.K., Shin B.A., Choi S.Y. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One. 2011;6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nadeem M., Sloane J.A. Targeting remyelination treatment for multiple sclerosis. World J. Neurol. 2015;5:5. doi: 10.5316/wjn.v5.i1.5. [DOI] [Google Scholar]
- 17.Stangel M., Kuhlmann T., Matthews P.M., Kilpatrick T.J. Achievements and obstacles of remyelinating therapies in multiple sclerosis. Nat. Rev. Neurol. 2017;13:742–754. doi: 10.1038/nrneurol.2017.139. [DOI] [PubMed] [Google Scholar]
- 18.Melchor G.S., Khan T., Reger J.F., Huang J.K. Remyelination pharmacotherapy investigations highlight diverse mechanisms underlying multiple sclerosis progression. ACS Pharmacol. Transl. Sci. 2019;2:372–386. doi: 10.1021/acsptsci.9b00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reale M., Costantini E., Di Nicola M., D'Angelo C., Franchi S., D'Aurora M., Di Bari M., Orlando V., Galizia S., Ruggieri S., et al. Butyrylcholinesterase and Acetylcholinesterase polymorphisms in Multiple Sclerosis patients: implication in peripheral inflammation. Sci. Rep. 2018;8:1319. doi: 10.1038/s41598-018-19701-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fields R.D., Dutta D.J., Belgrad J., Robnett M. Cholinergic signaling in myelination. Glia. 2017;65:687–698. doi: 10.1002/glia.23101. [DOI] [PubMed] [Google Scholar]
- 21.Darvesh S., Leblanc A.M., Macdonald I.R., Reid G.A., Bhan V., Macaulay R.J., Fisk J.D. Butyrylcholinesterase activity in multiple sclerosis neuropathology. Chem. Biol. Interact. 2010;187:425–431. doi: 10.1016/j.cbi.2010.01.037. [DOI] [PubMed] [Google Scholar]
- 22.Gacias M., Gerona-Navarro G., Plotnikov A.N., Zhang G., Zeng L., Kaur J., Moy G., Rusinova E., Rodriguez Y., Matikainen B., et al. Selective chemical modulation of gene transcription favors oligodendrocyte lineage progression. Chem. Biol. 2014;21:841–854. doi: 10.1016/j.chembiol.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiang C.M. Nonequivalent response to bromodomain-targeting BET inhibitors in oligodendrocyte cell fate decision. Chem. Biol. 2014;21:804–806. doi: 10.1016/j.chembiol.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krasnow A.M., Attwell D. NMDA receptors: power switches for oligodendrocytes. Neuron. 2016;91:3–5. doi: 10.1016/j.neuron.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 25.Káradóttir R., Cavelier P., Bergersen L.H., Attwell D. NMDA receptors are expressed in oligodendrocytes and activated in ischaemia. Nature. 2005;438:1162–1166. doi: 10.1038/nature04302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Biase L.M., Kang S.H., Baxi E.G., Fukaya M., Pucak M.L., Mishina M., Calabresi P.A., Bergles D.E. NMDA receptor signaling in oligodendrocyte progenitors is not required for oligodendrogenesis and myelination. J. Neurosci. 2011;31:12650–12662. doi: 10.1523/JNEUROSCI.2455-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elitt M.S., Shick H.E., Madhavan M., Allan K.C., Clayton B.L.L., Weng C., Miller T.E., Factor D.C., Barbar L., Nawash B.S., et al. Chemical screening identifies enhancers of mutant oligodendrocyte survival and unmasks a distinct pathological phase in pelizaeus-merzbacher disease. Stem Cell Rep. 2018;11:711–726. doi: 10.1016/j.stemcr.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Douvaras P., Wang J., Zimmer M., Hanchuk S., O'Bara M.A., Sadiq S., Sim F.J., Goldman J., Fossati V. Efficient generation of myelinating oligodendrocytes from primary progressive multiple sclerosis patients by induced pluripotent stem cells. Stem Cell Rep. 2014;3:250–259. doi: 10.1016/j.stemcr.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jäkel S., Agirre E., Mendanha Falcão A., van Bruggen D., Lee K.W., Knuesel I., Malhotra D., Ffrench-Constant C., Williams A., Castelo-Branco G. Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature. 2019;566:543–547. doi: 10.1038/s41586-019-0903-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ačimovič J., Goyal S., Košir R., Goličnik M., Perše M., Belič A., Urlep Ž., Guengerich F.P., Rozman D. Cytochrome P450 metabolism of the post-lanosterol intermediates explains enigmas of cholesterol synthesis. Sci. Rep. 2016;6:28462. doi: 10.1038/srep28462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hubler Z., Friedrich R.M., Sax J.L., Allimuthu D., Gao F., Rivera-León A.M., Pleshinger M.J., Bederman I., Adams D.J. Modulation of lanosterol synthase drives 24,25-epoxysterol synthesis and oligodendrocyte formation. Cell Chem. Biol. 2021;28:866–875.e5. doi: 10.1016/j.chembiol.2021.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khandker L., Jeffries M.A., Chang Y.J., Mather M.L., Evangelou A.V., Bourne J.N., Tafreshi A.K., Ornelas I.M., Bozdagi-Gunal O., Macklin W.B., Wood T.L. Cholesterol biosynthesis defines oligodendrocyte precursor heterogeneity between brain and spinal cord. Cell Rep. 2022;38:110423. doi: 10.1016/j.celrep.2022.110423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du C., Duan Y., Wei W., Cai Y., Chai H., Lv J., Du X., Zhu J., Xie X. Kappa opioid receptor activation alleviates experimental autoimmune encephalomyelitis and promotes oligodendrocyte-mediated remyelination. Nat. Commun. 2016;7:11120. doi: 10.1038/ncomms11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dietz K.C., Polanco J.J., Pol S.U., Sim F.J. Targeting human oligodendrocyte progenitors for myelin repair. Exp. Neurol. 2016;283:489–500. doi: 10.1016/j.expneurol.2016.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lariosa-Willingham K.D., Rosler E.S., Tung J.S., Dugas J.C., Collins T.L., Leonoudakis D. Development of a high throughput drug screening assay to identify compounds that protect oligodendrocyte viability and differentiation under inflammatory conditions. BMC Res. Notes. 2016;9:444. doi: 10.1186/s13104-016-2219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lumsden C.E. Fundamental problems in the pathology of multiple sclerosis and allied demyelinating diseases. Br. Med. J. 1951;1:1035–1043. doi: 10.1136/bmj.1.4714.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lumsden C.E. Quantitative studies on lipolytic enzyme activity in degenerating and regenerating nerve. Q. J. Exp. Physiol. Cogn. Med. Sci. 1952;37:45–57. doi: 10.1113/expphysiol.1952.sp000979. [DOI] [PubMed] [Google Scholar]
- 38.Kim S.U., Oh T., Johnson D.D. Developmental changes of acetylcholinesterase and pseudocholinesterase in organotypic cultures of spinal cord. Exp. Neurol. 1972;35:274–281. doi: 10.1016/0014-4886(72)90153-7. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi C., Muramatsu R., Fujimura H., Mochizuki H., Yamashita T. Prostacyclin promotes oligodendrocyte precursor recruitment and remyelination after spinal cord demyelination. Cell Death Dis. 2013;4:e795. doi: 10.1038/cddis.2013.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muramatsu R., Kuroda M., Matoba K., Lin H., Takahashi C., Koyama Y., Yamashita T. Prostacyclin prevents pericyte loss and demyelination induced by lysophosphatidylcholine in the central nervous system. J. Biol. Chem. 2015;290:11515–11525. doi: 10.1074/jbc.M114.587253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar N., Lyda B., Chang M.R., Lauer J.L., Solt L.A., Burris T.P., Kamenecka T.M., Griffin P.R. Identification of SR2211: a potent synthetic RORgamma-selective modulator. ACS Chem. Biol. 2012;7:672–677. doi: 10.1021/cb200496y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruan Q., Kameswaran V., Zhang Y., Zheng S., Sun J., Wang J., DeVirgiliis J., Liou H.C., Beg A.A., Chen Y.H. The Th17 immune response is controlled by the Rel-RORgamma-RORgamma T transcriptional axis. J. Exp. Med. 2011;208:2321–2333. doi: 10.1084/jem.20110462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang X.O., Pappu B.P., Nurieva R., Akimzhanov A., Kang H.S., Chung Y., Ma L., Shah B., Panopoulos A.D., Schluns K.S., et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Solt L.A., Kumar N., Nuhant P., Wang Y., Lauer J.L., Liu J., Istrate M.A., Kamenecka T.M., Roush W.R., Vidović D., et al. Suppression of TH17 differentiation and autoimmunity by a synthetic ROR ligand. Nature. 2011;472:491–494. doi: 10.1038/nature10075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang Z., Wang C., Zepp J., Wu L., Sun K., Zhao J., Chandrasekharan U., DiCorleto P.E., Trapp B.D., Ransohoff R.M., Li X. Act1 mediates IL-17-induced EAE pathogenesis selectively in NG2+ glial cells. Nat. Neurosci. 2013;16:1401–1408. doi: 10.1038/nn.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang C., Zhang C.J., Martin B.N., Bulek K., Kang Z., Zhao J., Bian G., Carman J.A., Gao J., Dongre A., et al. IL-17 induced NOTCH1 activation in oligodendrocyte progenitor cells enhances proliferation and inflammatory gene expression. Nat. Commun. 2017;8:15508. doi: 10.1038/ncomms15508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y., Li X., Ciric B., Curtis M.T., Chen W.J., Rostami A., Zhang G.X. A dual effect of ursolic acid to the treatment of multiple sclerosis through both immunomodulation and direct remyelination. Proc. Natl. Acad. Sci. USA. 2020;117:9082–9093. doi: 10.1073/pnas.2000208117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santori F.R., Huang P., van de Pavert S.A., Douglass E.F., Jr., Leaver D.J., Haubrich B.A., Keber R., Lorbek G., Konijn T., Rosales B.N., et al. Identification of natural RORgamma ligands that regulate the development of lymphoid cells. Cell Metab. 2015;21:286–298. doi: 10.1016/j.cmet.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jetten A.M., Takeda Y., Slominski A., Kang H.S. Retinoic acid-related Orphan Receptor gamma (RORgamma): connecting sterol metabolism to regulation of the immune system and autoimmune disease. Curr. Opin. Toxicol. 2018;8:66–80. doi: 10.1016/j.cotox.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jetten A.M., Cook D.N. Inverse) agonists of retinoic acid-related orphan receptor gamma: regulation of immune responses, inflammation, and autoimmune disease. Annu. Rev. Pharmacol. Toxicol. 2020;60:371–390. doi: 10.1146/annurev-pharmtox-010919-023711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J., Zou J.X., Xue X., Cai D., Zhang Y., Duan Z., Xiang Q., Yang J.C., Louie M.C., Borowsky A.D., et al. ROR-gamma drives androgen receptor expression and represents a therapeutic target in castration-resistant prostate cancer. Nat. Med. 2016;22:488–496. doi: 10.1038/nm.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sluch V.M., Chamling X., Wenger C., Duan Y., Rice D.S., Zack D.J. Highly efficient scarless knock-in of reporter genes into human and mouse pluripotent stem cells via transient antibiotic selection. PLoS One. 2018;13:e0201683. doi: 10.1371/journal.pone.0201683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Douvaras P., Fossati V. Generation and isolation of oligodendrocyte progenitor cells from human pluripotent stem cells. Nat. Protoc. 2015;10:1143–1154. doi: 10.1038/nprot.2015.075. [DOI] [PubMed] [Google Scholar]
- 56.Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ren Y.J., Zhang S., Mi R., Liu Q., Zeng X., Rao M., Hoke A., Mao H.Q. Enhanced differentiation of human neural crest stem cells towards the Schwann cell lineage by aligned electrospun fiber matrix. Acta Biomater. 2013;9:7727–7736. doi: 10.1016/j.actbio.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 58.Sluch V.M., Davis C.-h.O., Ranganathan V., Kerr J.M., Krick K., Martin R., Berlinicke C.A., Marsh-Armstrong N., Diamond J.S., Mao H.-Q., Zack D.J. Differentiation of human ESCs to retinal ganglion cells using a CRISPR engineered reporter cell line. Sci. Rep. 2015;5:16595. doi: 10.1038/srep16595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P.R., Raychaudhuri S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trapnell C. 2016. Monocole: differential expression and time-series analysis for single-cell RNA-Se.http://cole-trapnell-lab.github.io/monocle-release/articles/v2.2.0 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Raw data for all the compounds screened in our system is uploaded as Table S1.
-
•
scRNAseq data is uploaded to NCBI gene expression repository (GSE223599).
-
•
No new codes were generated for this study.
-
•
Any additional information required to analyze the data reported in this study will be made available by the lead contact upon request.