Abstract
Polycomb group (PcG) proteins form multimeric protein complexes which are involved in the heritable stable repression of genes. Previously, we identified two distinct human PcG protein complexes. The EED-EZH protein complex contains the EED and EZH2 PcG proteins, and the HPC-HPH PcG complex contains the HPC, HPH, BMI1, and RING1 PcG proteins. Here we show that YY1, a homolog of the Drosophila PcG protein pleiohomeotic (Pho), interacts specificially with the human PcG protein EED but not with proteins of the HPC-HPH PcG complex. Since YY1 and Pho are DNA-binding proteins, the interaction between YY1 and EED provides a direct link between the chromatin-associated EED-EZH PcG complex and the DNA of target genes. To study the functional significance of the interaction, we expressed the Xenopus homologs of EED and YY1 in Xenopus embryos. Both Xeed and XYY1 induce an ectopic neural axis but do not induce mesodermal tissues. In contrast, members of the HPC-HPH PcG complex do not induce neural tissue. The exclusive, direct neuralizing activity of both the Xeed and XYY1 proteins underlines the significance of the interaction between the two proteins. Our data also indicate a role for chromatin-associated proteins, such as PcG proteins, in Xenopus neural induction.
During embryogenesis, the fertilized egg develops into a complex organism with many differentiated cell types. Maintenance of the differentiation status of these cells requires a cellular memory system that is responsible for the stable inheritance of gene expression (10). The Polycomb group (PcG) and trithorax group (trxG) genes are part of such a memory system, and in Drosophila they have been identified as repressors (PcG) and activators (trxG), respectively (20, 33). Mutations in the PcG and trxG genes result in pleiotropic defects, of which homeotic transformations are the most apparent. The phenotypic defects in most of the PcG or trxG mutants become apparent only relatively late during Drosophila development, implying that these proteins indeed have a role in maintenance of cell differentiation. In vertebrates, similar roles for the PcG and trxG proteins in the maintenance of homeotic gene expression patterns, and consequently changes in the body plan, have been observed in PcG mutants (4, 29). However, mutations in some vertebrate PcG genes result in very early defects in development. This is exemplified by mutations in the eed (embryonic ectoderm development) gene, the vertebrate homolog of the Drosophila PcG gene extra sex combs (esc). The eed−/− mouse shows very early defects in development, resulting in gastrulation defects and lack of a node and of neural tissue (28). This indicates the involvement of PcG proteins in processes that precede maintenance of differentiation choices; it points towards involvement in embryonic inductions of tissues.
PcG proteins have been found to interact with each other to form multimeric, chromatin-associated protein complexes. Both in Drosophila and in vertebrates, various components of PcG complexes have been identified (27). Evidence has accumulated that there are at least two distinct PcG complexes. The human PcG homologs HPC2 (human Polycomb 2) (25), HPH (human Polyhomeotic) (5), BMI1 (42), and RING1 (24, 26) proteins belong to the HPC-HPH complex. The human PcG homologs EED and EZH2 (Enhancer of Zeste 2) belong to the second, EED-EZH PcG complex (30). The latter complex is associated with histone deacetylase (HDAC) activity, through a specific interaction between the EED and HDAC proteins (41).
The study of how PcG complexes regulate and interact with their target genes at the level of DNA has been hampered by the fact that most PcG proteins do not directly bind to DNA. The mouse homolog of the Drosophila PcG protein posterior sex combs, mel-18, has been shown to have DNA-binding activity (9), but not much is known about interaction partners of mel-18. Recently the Drosophila pleiohomeotic (Pho) protein has been found to share extensive homology with the vertebrate transcription factor YY1, a DNA-binding protein (1). This PcG protein could direct either the HPC-HPH or the EED-EZH PcG complex to the DNA of target genes. Here we show a specific interaction between YY1 and the EED PcG protein, providing a direct link between the DNA of target genes and the EED-EZH PcG protein complex.
To investigate the functional significance of this interaction we studied the role of the Xenopus homologs of these proteins, Xeed and XYY1, in Xenopus embryogenesis. We found that both Xeed and XYY1 directly induced neural tissue but were unable to induce mesodermal tissues. Our results indicate that the interaction between EED and YY1 is significant for early developmental decisions. They also suggest a novel role for chromatin-associated factors, such as the PcG proteins, in Xenopus neural induction.
MATERIALS AND METHODS
Two-hybrid analysis.
Two-hybrid analysis was performed as described previously (26). We cloned cDNAs encoding the EZH2, HPH1, HPC2, EED, RING1, and Bmi1 proteins in frame with the GAL4 DNA-binding domain in the pAS2 vector (Clontech). The p53 protein (pVA3) and simian virus 40 large antigen (pTD1) served as a positive interaction control. The full-length YY1 cDNA was cloned in frame with the GAL4 transactivation domain into the pGAD10 vector (Clontech). We cotransformed plasmids into the yeast Y190 strain (Clontech) and plated the transformants on medium lacking leucine, tryptophan, and histidine. After 4 days the cells were grown to an optical density at 600 nm of 1.0 to 1.2. β-Galactosidase activity was measured in permeabilized cells as described previously (5).
GST pull-down assay.
We cloned the full-length EED, EZH2, HPC2, and RING1 cDNAs into pGEX-2TK, thus creating glutathione S-transferase (GST)–EED, GST-EZH2, GST-HPC2, and GST-RING1. We immobilized these GST fusion proteins and GST protein alone on glutathione-Sepharose 4B. In vitro-translated, [35S]methionine-labeled YY1 or Pho was incubated with the GST fusion proteins in 250 μl of binding buffer (phosphate-buffered saline with 1 mM EDTA; 1 mM dithiothreitol; 2 mM phenylmethylsulfonyl fluoride [PMSF]; the protease inhibitors leupeptin, benzamidine, and aprotinin; 1% [vol/vol] Triton X-100; 0.5% NP-40; and 1 mg of bovine serum albumin per ml) for 1 h at 4°C, under constant rotation. Finally, we analyzed the washed beads on sodium dodecyl sulfate (SDS)-polyacrylamide gels, which were subjected to autoradiography.
Immunoprecipitation assay.
We prepared nuclei from human Ramos cells by 10 strokes with a glass Dounce homogenizer-pestle in a buffer containing 20 mM HEPES (pH 7.0), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, and 0.5 mM PMSF. The nuclei were pelleted by centrifugation at 1,000 × g at 4°C for 10 min. Subsequently, the nuclei were lysed in lysis buffer (250 mM NaCl, 0.1% NP-40, 50 mM HEPES [pH 7.0], 5 mM EDTA, 0.5 mM dithiothreitol, 1 mM PMSF, and the protease inhibitors leupeptin, benzamidine, and aprotinin). The lysate was sonicated two times with bursts of 15 s, and the supernatant was incubated with antibodies (indicated below) for 2 h at 4°C. Goat anti-rabbit immunoglobulin G antibody coupled to agarose beads (Sigma) was added for 30 min at 4°C. The beads were washed six times, and the immunoprecipitate (IP) was separated by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose. The blots were probed with a rabbit antibody against EED, EZH2, or HPC2 or a mouse monoclonal antibody against YY1 (SC-7872; Santa Cruz).
Plasmid constructions.
The 1,278-bp open reading frame of Xeed was cloned into the pCS2+ vector (23). The XYY1 cDNA was cloned by PCR, using primers based on the reported sequence (21). Capped synthetic RNA was made by in vitro transcription as described previously (18). To verify that the synthesized mRNAs still retained the capacity of being translated properly, all mRNAs were translated in a rabbit reticulocyte lysate (Promega) and analyzed on an SDS–12% polyacrylamide gel.
Culture conditions and embryo manipulations.
Embryos were obtained by natural fertilization using standard procedures. Microinjection of capped synthetic RNA was performed as described before (18). Embryonic stages were determined according to the method of Nieuwkoop and Faber (15).
Histology, whole-mount immunocytochemistry, and in situ hybridization.
Embryos were fixed in Smith's fixative (2.5% acetic acid, 0.5% K2Cr2O7, 4% formaldehyde), followed by embedding in paraffin, sectioning, and hematoxylin-eosin staining. Whole-mount in situ hybridization was performed as described previously (22). As a substrate for alkaline phosphatase, BM purple AP substrate (Boehringer) was used.
RT-PCR analysis.
Capped RNA was synthesized from the plasmids as described before (18) and injected into the animal poles of two-cell embryos. Ectoderm explants were isolated from blastulae and subjected to reverse transcription (RT)-PCR at neurula stages. Primers used in the RT-PCR were as follows: EF1 alpha, 5′-CAGATTGGTGCTGGATATGC-3′ and 5′-CACTGCCTTGATGACTCCTA-3′; Muscle Actin, 5′-GCTGACAGAATGCAGAAG-3′ and 5′-TTGCTTGGAGGAGTGTGT-3′; Xnot, 5′-ATACATGGTTGGCACTGA-3′ and 5′-CTCCTACAGTTCCACATC-3′; XANF, 5′-AGCTTTCACTAGGAGCCAGA-3′ and 5′-AGGTCCAAGGCTCTATCA-3′; NCAM, 5′-GCGGGTACCTTCTAATAGTCAC-3′ and 5′-GGCTTGGCTGTGGTTCTGAAGG-3′; and NRP-1, 5′-GGGTTTCTTGGAACAAGC-3′ and 5′-ACTGTGCAGGAACACAAG-3′.
RESULTS
EED interacts with YY1 and the Drosophila PcG protein Pho.
Evidence has accumulated that PcG proteins operate as large, multimeric protein complexes (27). Previously, the existence of two distinct human PcG protein complexes, the HPC-HPH PcG complex and the EED-EZH PcG complex, was reported (5, 24, 25, 26, 27, 30). The Drosophila PcG protein Pho shares extensive homology with the vertebrate transcription factor YY1 (1). So far, it is unknown to which class of PcG proteins Pho or YY1 belongs. To screen for potential interactions between YY1 and human PcG proteins, we performed a directed two-hybrid screen, using a panel of human PcG cDNAs.
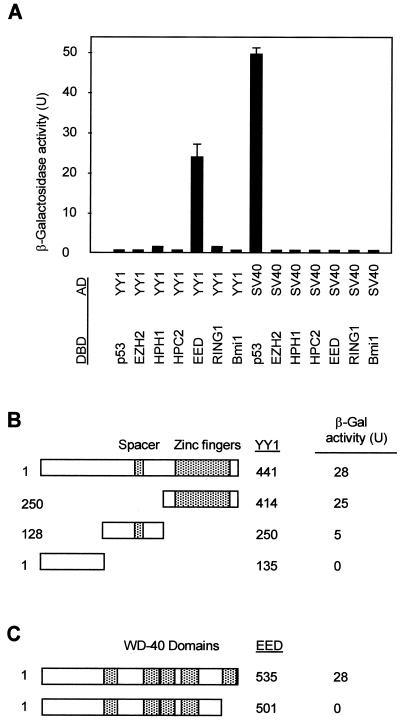
A positive interaction was found between YY1 and the EED protein (Fig. 1A), but no interaction was found between YY1 and the PcG proteins EZH2, HPH1, HPC2, RING1, and Bmi1 (Fig. 1A). This indicates that YY1 is part of the EED-EZH PcG protein complex. To define the domains that are responsible for the interaction between YY1 and EED, we subcloned different parts of YY1 and EED in frame with the GAL4 DNA-binding domain and tested whether these proteins could still interact with full-length YY1 or full-length EED. YY1 contains a C-terminal domain that contains a zinc finger binding domain that is highly conserved between YY1 and Pho (32, 38). This region is also involved in mediating repression (38). YY1 further contains a spacer region that is not conserved between YY1 and Pho (1). We found that EED interacts with the C-terminal region of YY1 (amino acids [aa] 250 to 414), hardly with aa 128 to 250, the region that overlaps the spacer region (aa 198 to 295), and not at all with the N-terminal region (aa 1 to 135) (Fig. 1B). We conclude that EED binds to the C-terminal region of YY1, which encompasses the zinc finger binding domain.
FIG. 1.
Interaction between the EED and YY1 proteins. (A) Two-hybrid analysis with YY1 and the indicated vertebrate PcG proteins showed a positive interaction between YY1 and EED. The PcG cDNAs were cloned in frame with the GAL4 DNA-binding domain (DBD), and the YY1 cDNA was cloned in frame with the GAL4 transactivation domain (AD). No interactions between the PcG proteins and simian virus 40 were observed. (B) The domain of YY1 that interacted with EED was mapped. Indicated portions of YY1 were fused to the GAL4 AD. β-Gal, β-galactosidase. (C) Deletion of the most N-terminal WD-40 domain in EED resulted in abolishment of the interaction between YY1 and EED.
EED contains five WD-40 domains, and it was previously found that all WD-40 domains of EED are necessary for the interaction between EED and EZH2 (30). A truncated EED protein that still contains the most N-terminal four WD-40 domains (aa 1 to 501) already failed to interact with YY1 (Fig. 1C). It is therefore probable that, as for the interaction between EED and EZH2, all five WD-40 domains of EED need to be present for a proper interaction between YY1 and EED.
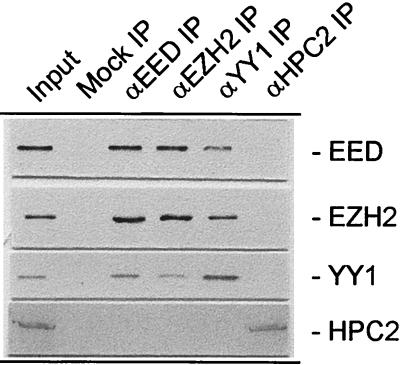
To test whether the YY1-EED interaction occurs in vivo, coimmunoprecipitation experiments were performed using antibodies against the YY1 and EED proteins. Extracts from Ramos cells in which EED and EZH2 are expressed at a high level were used (41). We detected both EED and EZH2 in the YY1 IP (Fig. 2). Conversely, YY1 was present in both the EED and EZH2 IPs (Fig. 2). Previously it was shown that HPC2 is not present in EED or EZH2 IPs (26) (Fig. 2). We have now found that HPC2 was also not present in the YY1 IP. Conversely, YY1, EED, and EZH2 were not present in the HPC2 IP (Fig. 2). Finally, no antigens were detected when the specific IP antibodies were replaced by preimmune sera (Fig. 2, mock IP), underlining the specificity of the IPs. Also, when antibodies against the human PcG proteins BMI-1 and RING1 were used instead of anti-HPC2, we did not observe YY1 in these IPs (data not shown). These data indicate that, in vivo, YY1 is associated with the EED-EZH protein complex but not with the HPC-HPH PcG complex.
FIG. 2.
In vivo interaction between EED and YY1 or Pho. Extracts from human Ramos cells were immunoprecipitated using antibodies against EED, EZH2, YY1, or HPC2 or without antibody (mock IP). Western blots of the IPs were probed with antibodies against EED, EZH2, YY1, or HPC2. No antigens were detected when the specific IP antibodies were omitted from the IPs (mock IP). Input, extract from Ramos cells.
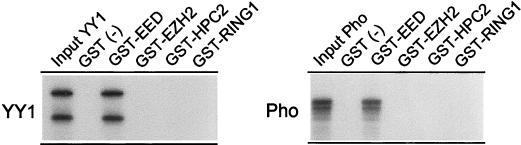
To determine whether the interaction between EED and YY1 is a direct interaction, we employed an in vitro pull-down assay. Previously described fusions of GST and EED, EZH2, and HPC2 (41) were immobilized to GST-Sepharose and incubated with [35S]methionine-labeled, in vitro-translated YY1. After extensive washing, the [35S]methionine-labeled proteins were analyzed by SDS-polyacrylamide gel electrophoresis. The YY1 protein was able to bind to immobilized GST-EED but not to GST-EZH2, GST-HPC2, GST-RING1, or immobilized GST alone (Fig. 3). We also tested whether the Drosophila PcG protein Pho interacted with GST-EED. As shown in Fig. 3, Pho also bound to immobilized GST-EED but not to GST-EZH2, GST-HPC2, GST-RING1, or immobilized GST alone. These data confirm the two-hybrid data and the coimmunoprecipitation experiments and indicate that YY1 and Pho interact directly with the EED protein but not with EZH2, HPC2, or RING1.
FIG. 3.
In vitro interaction between EED and YY1 or Pho. In vitro-translated, [35S]methionine-labeled YY1 or Pho was incubated with immobilized GST [GST (−)], GST-EED, GST-EZH2, GST-HPC2, or GST-RING1. The input was 15% of the amount that was incubated with the GST fusion proteins.
Identification and characterization of Xeed, a Xenopus homolog of the PcG protein EED.
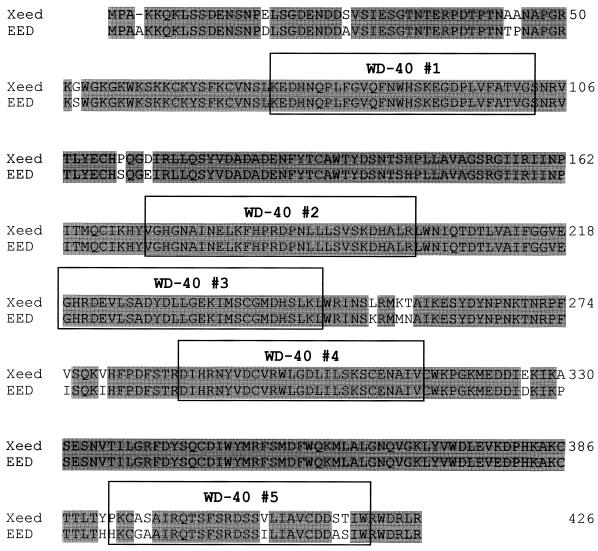
In order to test the functional significance of the EED-YY1 interaction, we wished to modify the expression levels of these proteins in Xenopus embryos. As a first step towards this goal, we needed to isolate Xenopus homologs of YY1 and EED. Since the Xenopus homolog of YY1 has been described previously (21), we concentrated our efforts on the isolation of the Xenopus homolog of EED. We screened a Xenopus oocyte cDNA library with a probe which contains the coding region of EED (30) and obtained several positive clones. The longest, a 1,324-bp cDNA clone, was further characterized (Fig. 4). Sequence analysis revealed a 1,278-bp open reading frame. A stop codon was found at the 3′ end of the clone, corresponding with a similar stop codon in the EED cDNA. Several stop codons were present approximately 15 bp upstream from the first ATG (data not shown). These data indicate that we isolated a full-length cDNA. Within the 1,278-bp coding region, the cDNA is 81% identical to EED at the nucleotide level. At the protein level the isolated cDNA is 92% identical and 96% similar to the EED protein (30) (Fig. 4). We therefore conclude that we isolated a Xenopus homolog of EED, which we call Xeed.
FIG. 4.
Predicted amino acid sequence of Xeed. The Xeed protein was compared with the human EED protein. Identical amino acids are shaded. Five WD-40 repeats are indicated with boxes.
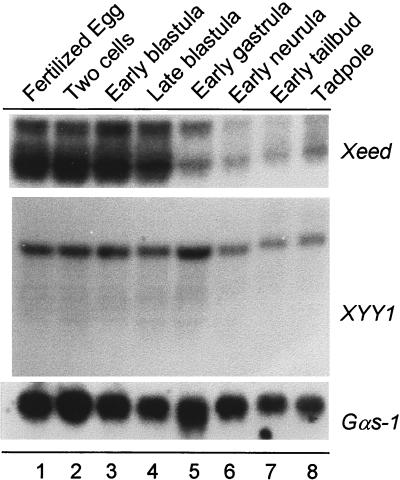
We characterized the temporal expression of the Xeed and XYY1 genes through early embryonic development of Xenopus. Total RNA from several developmental stages was analyzed by Northern blot analysis. We detected 3- and 4-kb transcripts for the Xeed gene and an approximately 5.5-kb single transcript for the XYY1 gene (Fig. 5). The highest expression level of both transcripts is found in the fertilized egg throughout blastula stages. These transcripts are of maternal origin, since transcription in the embryo is activated only after the midblastula transition. The abundance of the Xeed transcript declines during gastrula stages, and the abundance of the XYY1 transcript declines during neurula stages. However, transcription of both Xeed and XYY1 was observed throughout development, from neurula stages onward, indicating zygotic transcription. We also examined the spatial distribution of the Xeed and XYY1 transcripts in late neurula and tailbud stages by in situ hybridization. Expression was detected in tissues such as the developing neural tube (data not shown). The analysis did not reveal, however, a strongly localized expression of the transcripts in the early embryos (data not shown).
FIG. 5.
Developmental expression profiles of Xeed and XYY1. Probes for the Xeed and XYY1 genes were used for Northern analysis of total RNA (20 μg) isolated from the indicated developmental stages (15). The filter was rehybridized with a probe for Gαs-1 (19) to verify the loading and integrity of RNA in each lane.
In summary, we have isolated a highly conserved Xenopus homolog of EED, Xeed. Xeed and XYY1 have similar temporal expression profiles, and they have a strong maternal component of expression.
Overexpression of Xeed, XYY1, and Pho induces an ectopic neural tube.
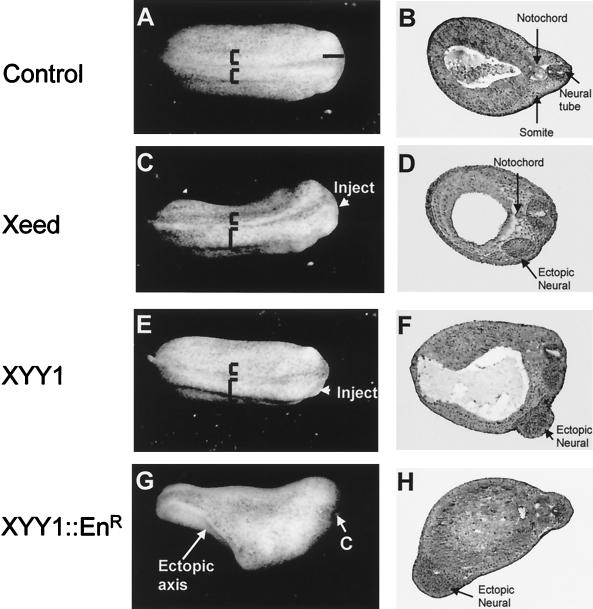
In order to test whether interference with the expression levels of Xeed and XYY1 proteins might influence early embryonic development, we injected Xeed or XYY1 mRNA into one blastomere of two-cell Xenopus embryos. Blastopore formation during gastrulation and the formation of the neural plate proceeded normally in both Xeed- and XYY1-injected embryos. However, a broadening of the neural plate at the injected side of the embryo became apparent at late neurula and tailbud stages (Fig. 6C and E). When 1 ng of either Xeed or XYY1 mRNA was injected, this phenotype was observed in approximately 30% (n = 251) of the embryos. Importantly, also when 1 ng of Pho mRNA was injected, the same phenotype was observed and with a similar frequency (data not shown). Coinjection of trace amounts of β-galactosidase mRNA showed that the broadening of the neural plate had occurred in the injected side of the embryo (data not shown). Next, we characterized the embryos by histology. This analysis revealed an ectopic neural axis at the injected side of the embryo (Fig. 6D and F). Importantly, we observed neither a notochord underlying the ectopic neural axis nor somites adjacent to the ectopic neural axis in any of the embryos we analyzed (Fig. 6D and F).
FIG. 6.
Phenotypes resulting from injection of Xeed, XYY1, and XYY1-EnR mRNA into Xenopus embryos. (A, C, E, and G) Indicated mRNAs were injected into one blastomere of two-cell-stage Xenopus embryos. The side of the embryo that was injected is indicated. The broadening of the neural plate in Xeed-injected (C) and XYY1-injected (E) embryos, compared to the uninjected embryo (A), is indicated with brackets. The ectopic axis in XYY1-EnR-injected embryos (G) is also indicated, as is the cement gland (C). (B, D, F, and H) The embryos in panels A, C, E, and G were processed for histological analysis in order to visualize the somites, notochord, and neural tissue. Shown are histological sections of uninjected (B), Xeed-injected (D), XYY1-injected (F), and XYY1-EnR-injected (H) embryos.
EED is as a transcriptional repressor (26). YY1 can also act as a transcriptional repressor, dependent on the promoter context (32). To test whether in Xenopus embryos XYY1 operates as a transcriptional repressor, we took advantage of the fact that XYY1 has a DNA-binding domain that acts independently of the domain that is involved in transcriptional regulation. We fused the DNA-binding domain of XYY1 to the Engrailed repressor (EnR) domain to create a strong transcriptional repressor (2). The resulting XYY1-EnR mRNA was injected into one blastomere of two-cell Xenopus embryos. We observed the same phenotype as with either Xeed, XYY1, or Pho, but with much higher efficiency. Injection of 50 pg of XYY1-EnR mRNA (instead of 1 ng of XYY1 mRNA) resulted in broadened neural plates in over 50% (n = 276) of the injected embryos. A significant proportion (approximately 15%) of these embryos displayed a bifurcated axis (Fig. 6G). This event was observed only rarely in Xeed- or XYY1-injected embryos. Histological analysis of these bifurcated embryos demonstrated a secondary, ectopic neural axis in the ventral regions of the XYY1-EnR-injected embryos (Fig. 6H). These secondary axes did not, however, develop into complete head structures (Fig. 6H). Importantly, as in Xeed- and XYY1-injected embryos, no notochord or somites were observed in the vicinity of the ectopic neural axis (Fig. 6H). Given the similarity of the phenotypes and the fact that the XYY1-EnR fusion protein is a strong transcriptional repressor (by virtue of the EnR domain), we conclude that the XYY1 protein also acts as a transcriptional repressor in Xenopus embryos.
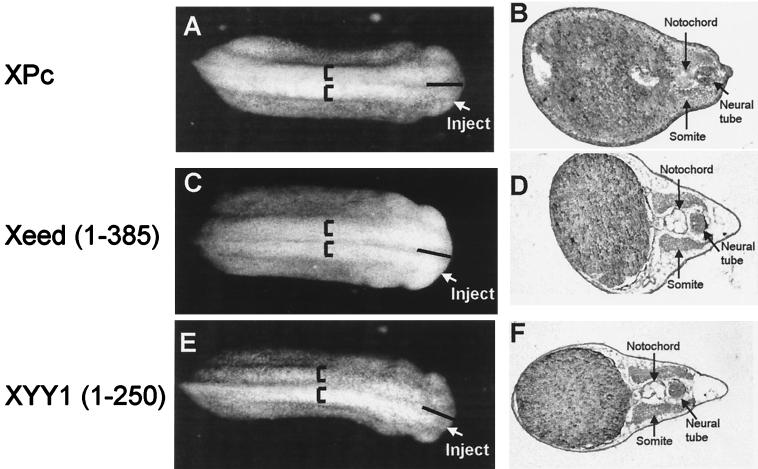
In control experiments we injected mRNAs encoding the Xenopus PcG proteins XPc (22) and Xbmi1 (22) or the RING1 protein (24) into one cell of a two-cell-stage embryo. We observed neither phenotypic effects nor changes at the histological level when XPc (Fig. 7A and B) was injected. No effects were observed with either Xbmi1 or RING1 (data not shown). Also, mutants of Xeed and XYY1 were injected. The Xeed (aa 1 to 385) mutant consisted of Xeed protein in which the most C-terminal WD-40 domain was removed. The corresponding EED mutant failed to interact with YY1 in the two-hybrid assay (Fig. 1C). The XYY1 mutant (aa 1 to 250) consisted of XYY1 protein from which the C-terminal, zinc finger binding domain was removed. The corresponding C-terminal region of YY1 was identified as mediating the binding to EED (Fig. 1B). Neither Xeed (aa 1 to 385) (Fig. 7C and D) nor XYY1 (aa 1 to 250) (Fig. 7E and F) induced phenotypic changes or changes at the histological level. Taken together, these results underline the specificity of the effects of Xeed and XYY1 on induction of neural tissue as well as underline the functional significance of the interaction between EED and YY1.
FIG. 7.
Lack of phenotypes from injection of XPc and mutants of Xeed and XYY1. Indicated mRNAs were injected into one blastomere of two-cell-stage Xenopus embryos. The side of the embryo that was injected is indicated. Injection of neither XPc (A and B), Xeed (aa 1 to 385) (C and D), nor XYY1 (aa 1 to 250) (D and E) induced broadening of the neural plate or histologically defined neural tissue.
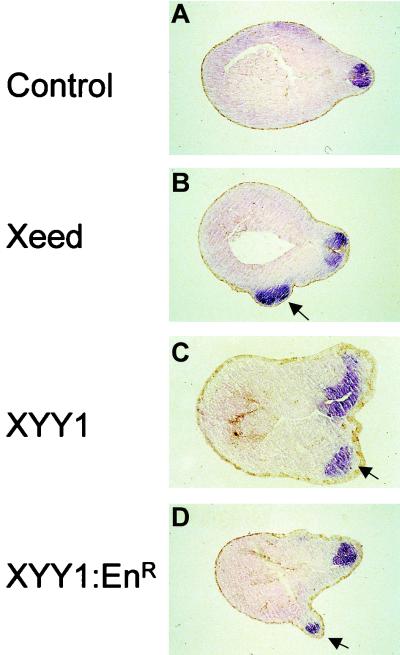
To further study the molecular characteristics of the axis that can be induced by Xeed and XYY1, we performed in situ hybridization on Xeed- and XYY1-injected embryos, using probes against neural marker genes. We chose the general neural marker NRP-1, which is expressed in the developing neural tube of stage 23 embryos (12) (Fig. 8A). We found that the ectopic tissue in Xeed- and XYY1-injected embryos was characterized by expression of NRP-1 (Fig. 8B and C). Also, in XYY1-EnR-injected embryos, NRP-1 expression in the ectopic axis was observed (Fig. 8D). These data confirm the histological analysis and demonstrate that Xeed and XYY1 induce neural tissue in the injection site. We therefore conclude that overexpression of Xeed and XYY1 induces a secondary, ectopic neural axis, but no notochord or somites.
FIG. 8.
Induction of neural tissue in Xeed- and XYY1-injected embryos. Xeed (B), XYY1 (C), and XYY1-EnR (D) mRNAs were injected into one blastomere of the two-cell-stage Xenopus embryo and cultured until stage 23. All injected embryos, as well as the uninjected control embryos (A), were subjected to whole-mount in situ hybridization using a probe that detects the neural marker NRP-1. Subsequently, the embryos were processed for histological analysis and sections were examined. The ectopic neural tissue in Xeed-, XYY1-, and XYY1-EnR-injected embryos is indicated with arrows.
Xeed, Pho, XYY1, and XYY1-EnR directly induce neural tissue in ectoderm explants.
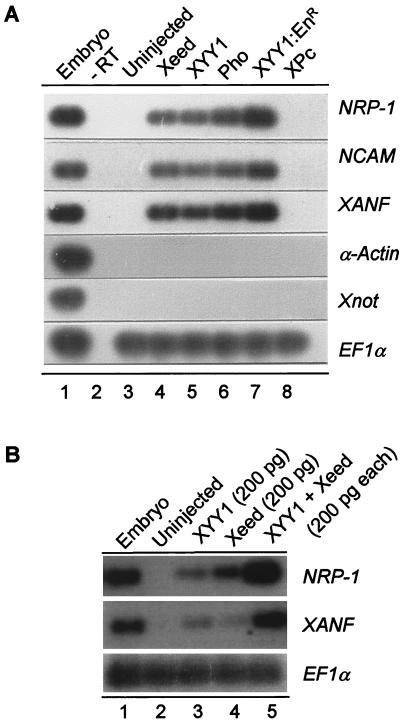
The lack of notochord or somites in the proximity of the ectopic neural axis indicated that this ectopic neural tube was the result of a direct neuralizing event. Therefore, we tested whether Xeed, Pho, XYY1, and XYY1-EnR are able to directly induce neural tissue in ectoderm tissue. We injected the respective mRNAs into the animal zone of both blastomeres of two-cell Xenopus embryos. When these embryos had reached early gastrula stage (stage 10), we dissected the ectoderm and cultured them until control embryos had reached the tailbud stage (stage 25). Total RNA was isolated, and the expression levels of the general neural markers NRP-1 (12) and NCAM (11) and the anterior neural marker XANF (45) were examined using RT-PCR. As shown in Fig. 9A, 2 ng of Xeed mRNA (lane 4), 2 ng of XYY1 mRNA (lane 5), 2 ng of Pho mRNA (lane 6), and 100 pg of XYY1-EnR mRNA (lane 7) all induced expression of the NRP-1, NCAM, and XANF neural marker genes. No expression of these neural markers was induced in control ectoderm that had been excised from early gastrula ectoderm (lane 3) and subsequently cultured to the tailbud stage. No neural markers were induced in cultured ectoderm of embryos which were injected with either 2 ng of XPc mRNA (lane 8) or 2 ng of XBmi1 mRNA (data not shown).
FIG. 9.
Direct neural induction in ectoderm by Xeed and XYY1. (A) Two nanograms of Xeed, XYY1, or Pho mRNA, 100 pg of XYY1-EnR mRNA, and 2 ng of XPc mRNA were injected into two blastomeres of two-cell Xenopus embryos and cultured to stage 10 (early gastrula). The entire ectoderm was dissected and cultured to stage 25, when RNA was isolated and analyzed by RT-PCR using primers recognizing the neural markers NRP-1, NCAM, and XANF as well as the mesodermal marker α-actin and the notochord marker Xnot. As a control, dissected stage 10 ectoderm of an uninjected embryo (lane 3) was cultured to stage 25. Uninjected, whole stage 23 embryos were analyzed as a positive control (lane 1). To verify the presence of equal amounts of RNA, EF1α was used as loading marker. −RT, no reverse transcriptase was added. (B) Two hundred picograms of Xeed (lane 3), 200 pg of XYY1 (lane 4), or 200 pg of Xeed plus 200 pg of XYY1 (lane 5) were injected into two blastomeres of two-cell Xenopus embryos and the experiment proceeded as for panel A.
We also determined whether there had been induction of mesodermal markers in the cultured ectoderm explants of the injected embryos. We monitored the expression levels of the mesodermal somite marker α-actin (13) and the notochord marker Xnot (40). No expression of these markers was found in cultured control ectoderm explants (Fig. 9A, lane 3) or in cultured explants of embryos injected with Xeed (lane 4), XYY1 (lane 5), Pho (lane 6), XYY1-EnR (lane 7), or XPc (lane 8) mRNA.
We also tested whether coinjected Xeed and XYY1 might synergize to induce neural tissue. When 200 pg of either Xeed (Fig. 9B, lane 3) or XYY1 (Fig. 9B, lane 4) mRNA was injected instead of 2 ng of mRNA (Fig. 9A), low expression levels of NRP-1 and XANF were induced. However, coinjection of 200 pg of Xeed mRNA plus 200 pg of XYY1 mRNA (Fig. 9B, lane 5) induced strong expression of NRP-1 and XANF, indicating that Xeed and XYY1 synergize to induce neural tissue.
These data confirm the histological analysis showing that Xeed, Pho, XYY1, and XYY1-EnR are able to induce ectopic neural tissue that is not accompanied by somite or notochord tissue. Importantly, this implies that the effects of these genes are due to direct neural induction within the ectoderm and are not a secondary event resulting from initial mesoderm induction.
DISCUSSION
YY1 interacts with the PcG protein EED but not with other PcG proteins.
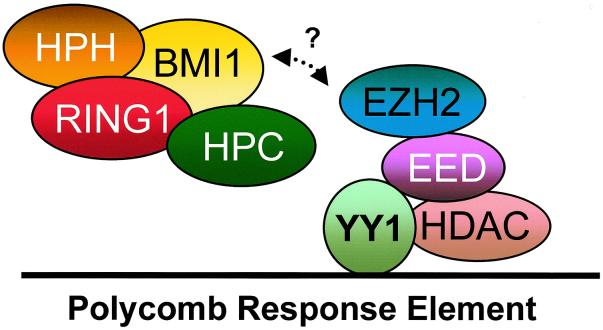
The Drosophila Pho protein (1) is, together with mel-18 (9), the only known PcG protein that displays DNA-binding properties. Since all other known PcG proteins lack this ability, this is a very important feature. It is, therefore, important to know to which PcG complex the Pho protein, or its vertebrate homolog, YY1, belongs. Potentially, YY1 could be part of either the HPC-HPH or the EED-EZH PcG protein complex. Hence, we used a panel of cDNAs cloned into two-hybrid vectors to investigate if and to which known human PcG protein YY1 binds. We show that YY1 interacts specifically with EED, the PcG protein that forms a complex with EZH2 (30) and HDAC proteins (41). We substantiated the two-hybrid interaction between EED and YY1 by performing in vivo coimmunoprecipitations and in vitro GST pull-down studies. Using both methods we demonstrated the validity of the two-hybrid interaction between EED and YY1. The identification of the interaction between EED and YY1 allows us to extend a previous model of human PcG protein complexes (27). In Fig. 10 we show our current model, in which YY1 is part of the complex that encompasses the vertebrate PcG proteins EED and EZH2. It is important to note that our present findings are consistent with the fact that HDAC activity is associated with the EED-EZH complex and not with the HPC-HPH complex (36, 41). Since it has previously been demonstrated that YY1 interacts with HDAC proteins (44), it also implies that HDAC proteins have two possibilities to interact with the EED-EZH complex: with EED (41) and with YY1 (44; also this work).
FIG. 10.
Composition of human PcG protein complexes. The HPC-HPH PcG complex contains the PcG proteins HPC2, BMI1, HPH1, HPH2, and RING1A. The EED-EZH PcG complex contains the PcG proteins EED and EZH2 and is associated with HDAC proteins. In this article we show that the homolog of the Drosophila PcG protein Pho, YY1, interacts with the EED protein and thus is either part of or associated with the EED-EZH PcG complex.
EED is a vertebrate homolog of the Drosophila PcG protein esc (6, 37). esc is distinguished from other PcG proteins in Drosophila in that it is primarily required only during embryogenesis. In a previous paper it was speculated that deacetylation of histones by HDACs and the recruitment of EED to the HDAC proteins may be among the initial repressive events during embryogenesis that eventually lead to stable and heritable PcG-mediated repression of target genes (41). Now we have found that YY1 is part of or associated with the EED-EZH PcG complex, which displays HDAC activity. Since Pho and YY1 display specific DNA-binding properties, this finding suggests a model in which YY1 directs the EED-EZH PcG complex to target genes (Fig. 10). This first step is consistent with the early developmental role of the EED-EZH complex, as has been defined genetically. It is also consistent with a role for histone deacetylation, mediated by the HDACs, which are associated with both EED and YY1, as an early event by which PcG proteins set up stable repression of target genes.
The existence of two distinct PcG protein complexes has also been observed in Drosophila. Using a two-hybrid analysis, the ESC and E(Z) proteins have been found to interact (8, 39). Furthermore, two distinct Drosophila PcG protein complexes have been characterized biochemically. One complex contains the PC, PSC, and PH proteins (31); the other contains the ESC and E(Z) proteins (14). These findings are very similar to the observations in the human system (30, 41). There are, however, significant differences between the two developmental systems. For instance, the Drosophila Pho protein lacks a domain that mediates histone deacetylation activity (1). This domain is present in the YY1 protein. It is possible that this constitutes a fundamental difference between the Drosophila and the vertebrate systems, indicating that histone deacetylation plays a less significant role in the Drosophila system. Further, Shao and coworkers did not detect the Pho protein in the PC-PH complex (31), which is in agreement with our present findings. However, the Pho protein could not be detected in the biochemically purified ESC-E(Z) complex either (14). These puzzling findings may reflect a more transient nature of interactions between Pho and other proteins, which precludes biochemical purification as part of a stable protein complex. Our observation that even an in vitro interaction between EED and the Drosophila Pho protein exists at least suggests a highly conserved nature of the interaction between EED and YY1. Also, the virtually identical phenotypes that are induced by Xeed, XYY1, and Pho in Xenopus embryos suggest that YY1 or Pho is either a stable component of or at least transiently associated with the EED-EZH PcG complex and not the HPC-HPH PcG complex.
Role of Xeed and XYY1 in the induction of neural tissue in Xenopus.
To study the functional significance of the interaction between EED and YY1, we manipulated the expression levels of the Xenopus homologs of these proteins, Xeed and XYY1. Both proteins, but no other PcG proteins, induced an ectopic neural axis in Xenopus embryos, but neither Xeed nor XYY1 was able to induce mesodermal tissue, such as muscle or notochord. Importantly, the Drosophila Pho protein induced the same phenotype. The similarity of effects underlines the significance of the EED-YY1 interaction. The fact that Pho induced the same phenotype and neural tissue in ectoderm explants also substantiates the notion that YY1 is indeed a functional homolog of the Drosophila Pho protein.
Our data point towards an early developmental role for the EED-EZH complex. Also, in homozygous eed−/− mice the earliest developmental decisions are affected, pointing towards an early role for EED in setting up vertebrate PcG-mediated repression. It may be significant that homozygous eed−/− mice lack a node and, probably as a consequence of this, also neural tissue (28). Whereas the homozygous YY1−/− mutation is embryonic lethal, in heterozygote YY1−/+ mice the formation of a proper neural tube is seriously hampered (3). Both phenotypes are complementary to the phenotypes we observed after overexpression of both Xeed and XYY1 proteins in Xenopus embryos. Although a detailed comparison between the loss-of-function data in mice and overexpression of proteins in Xenopus is not possible, the opposing effects on neural tissue are compatible with each other. The results reinforce one another and both point towards an early role of these PcG proteins in developmental decisions, such as the induction of embryonic tissues.
How do our findings relate to other neural inducing factors that have been identified over the past few years? Most knowledge concerning pathways that mediate the earliest induction steps has been linked to secreted proteins. A common theme emerges indicating that neural induction results from antagonizing the bone morphogenetic protein signaling pathway (7, 43). Secreted proteins, such as noggin (34), are able to directly induce neural tissue in Xenopus ectoderm by antagonizing bone morphogenetic protein signaling. Further, overexpression of proteins involved in conserved signal transduction pathways, such as protein kinase C, enhances the competence of ectoderm to become induced to neural tissue, but these proteins lack the ability to directly convert ectoderm into neural tissue (16, 17, 18). In that respect, the involvement of protein kinase C in mediating neural competence resembles the role of another chromatin-associated factor. Recently it has been found that overexpression of histone H1 limits the ability of Xenopus ectoderm to become mesoderm (35). Interference with histone H1 expression does not directly induce mesoderm but changes the time window in which ectoderm can be induced to become mesoderm (35). Beside histone H1, no involvement of chromatin-associated factors in mediating embryonic induction phenomena has been described. We therefore believe that our data reveal a novel type of factors that are involved in Xenopus neural induction.
The following questions remain: which are the target genes of Xeed and XYY1, and how does the modulation of the activity of these target genes result in the induction of neural tissue? Since EED is a repressor of gene activity (30), it is likely that Xeed is also a repressor of gene activity. Furthermore, both XYY1 and XYY1-EnR directly induce neural tissue, and by virtue of the EnR domain, XYY1-EnR is a transcriptional repressor. It is, therefore, likely that the target genes of Xeed and XYY1 are repressed by these proteins and that this repression results in the induction of neural tissue. It will be of considerable interest to identify these target genes. Since the effects of Xeed and XYY1 occur early in development, these target genes may well represent a class of PcG target genes other than the known PcG target genes in Drosophila that are affected relatively late during development. Also, identification of such target genes may reveal pathways, distinct from the known ones, that are involved in mediating neural induction in Xenopus.
ACKNOWLEDGMENTS
We gratefully thank Eddy de Robertis, Peter Good, Richard Harland, Ali Hemmati-Brivanlou, David Kimelman, Paul Krieg, Doug Melton, Tim Mohun, Ralph Rupp, Colin Sharp, Jim Smith, and A. G. Zaraisky for providing us with various reagents. We further thank Thijs Hendrix for excellent frog care, Johan van der Vlag for performing the coimmunoprecipitations, Frank Raaphorst for photography, and Ralph Rupp for stimulating discussions.
This work was sponsored in part by the Human Frontier Science Program (RG0039/1999-M).
REFERENCES
- 1.Brown J L, Mucci D, Whiteley M, Dirksen M L, Kassis J A. The Drosophila Polycomb group gene pleiohomeotic encodes a sequence-specific DNA binding protein with homology to the multifunctional mammalian transcription factor YY1. Mol Cell. 1998;7:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 2.Conlon F L, Sedgwick S G, Weston K M, Smith J C. Inhibition of Xbra transcription activation causes defects in mesodermal patterning and reveals autoregulation of Xbra in dorsal mesoderm. Development. 1996;122:2427–2435. doi: 10.1242/dev.122.8.2427. [DOI] [PubMed] [Google Scholar]
- 3.Donohoe M E, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol Cell Biol. 1999;19:7237–7244. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gould A. Functions of mammalian Polycomb group and trithorax group related genes. Curr Opin Genet Dev. 1997;7:488–494. doi: 10.1016/s0959-437x(97)80075-5. [DOI] [PubMed] [Google Scholar]
- 5.Gunster J M, Satijn D P E, Hamer K M, den Blaauwen J L, de Bruijn D, Alkema M J, van Lohuizen M, van Driel R, Otte A P. Identification and characterization of interactions between the vertebrate Polycomb-group protein BMI1 and human homologs of Polyhomeotic. Mol Cell Biol. 1997;17:2326–2335. doi: 10.1128/mcb.17.4.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gutjahr T, Frei E, Spicer C, Baumgarter S, White R A H, Noll M. The Polycomb-group gene, extra sex combs, encodes a nuclear member of the WD-40 repeat family. EMBO J. 1995;14:4296–4306. doi: 10.1002/j.1460-2075.1995.tb00104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harland R M, Gerhart J C. Formation and function of Spemann's organizer. Annu Rev Cell Dev Biol. 1997;13:611–667. doi: 10.1146/annurev.cellbio.13.1.611. [DOI] [PubMed] [Google Scholar]
- 8.Jones C A, Ng J, Peterson A J, Morgan K, Simon J, Jones R S. The Drosophila esc and E(z) proteins are direct partners in Polycomb group-mediated repression. Mol Cell Biol. 1998;18:2825–2834. doi: 10.1128/mcb.18.5.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanno M, Hasegawa M, Ishida A, Isono K, Taniguchi M. mel-18, a Polycomb group-related mammalian gene, encodes a transcriptional negative regulator with tumor suppressive activity. EMBO J. 1995;14:5672–5678. doi: 10.1002/j.1460-2075.1995.tb00254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kingston R E, Bunker C A, Imbalzano A N. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 11.Kintner C R, Melton D A. Expression of Xenopus N-CAM RNA in ectoderm is an early response to neural induction. Development. 1987;99:311–325. doi: 10.1242/dev.99.3.311. [DOI] [PubMed] [Google Scholar]
- 12.Knecht A K, Good P J, Dawid I B, Harland R M. Dorsal-ventral patterning and differentiation of noggin-induced neural tissue in the absence of mesoderm. Development. 1995;121:1927–1936. doi: 10.1242/dev.121.6.1927. [DOI] [PubMed] [Google Scholar]
- 13.Mohun T J, Brennan S, Dathan N, Fairman S, Gurdon J B. Cell type-specific activation of actin genes in the early amphibian embryo. Nature. 1984;311:716–721. doi: 10.1038/311716a0. [DOI] [PubMed] [Google Scholar]
- 14.Ng J, Hart C M, Morgan K, Simon J A. A Drosophila ESC-E(Z) protein complex is distinct from other Polycomb group complexes and contains covalently modified ESC. Mol Cell Biol. 2000;20:3069–3078. doi: 10.1128/mcb.20.9.3069-3078.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nieuwkoop P D, Faber J. The normal table of Xenopus laevis (Daudin) 2nd ed. Amsterdam, The Netherlands: North Holland Publishing Co.; 1967. [Google Scholar]
- 16.Otte A P, Koster C A, Snoek G T, Durston A J. Protein kinase C mediates neural induction in Xenopus laevis. Nature. 1988;334:618–620. doi: 10.1038/334618a0. [DOI] [PubMed] [Google Scholar]
- 17.Otte A P, Van Run P, Heideveld M, Van Driel R, Durston A J. Neural induction in Xenopus laevis is mediated by cross-talk between protein kinase C and cyclic AMP pathways. Cell. 1989;58:641–648. doi: 10.1016/0092-8674(89)90099-8. [DOI] [PubMed] [Google Scholar]
- 18.Otte A P, Moon R T. Protein kinase C isozymes have distinct roles in neural induction and competence in Xenopus. Cell. 1992;68:1021–1029. doi: 10.1016/0092-8674(92)90074-m. [DOI] [PubMed] [Google Scholar]
- 19.Otte A P, McGrew L L, Olate J, Nathanson N M, Moon R T. Expression and potential functions of G-protein subunits in embryos of Xenopus laevis. Development. 1992;116:141–146. doi: 10.1242/dev.116.1.141. [DOI] [PubMed] [Google Scholar]
- 20.Pirrotta V. PcG complexes and chromatin silencing. Curr Opin Genet Dev. 1997;7:249–258. doi: 10.1016/s0959-437x(97)80135-9. [DOI] [PubMed] [Google Scholar]
- 21.Pisaneschi G, Ceccotti S, Falchetti M L, Fiumicino S, Carnevali F, Beccari E. Characterization of FIII/YY1, a Xenopus laevis conserved zinc-finger protein binding to the first exon of L1 and L14 ribosomal protein genes. Biochem Biophys Res Commun. 1994;205:1236–1242. doi: 10.1006/bbrc.1994.2797. [DOI] [PubMed] [Google Scholar]
- 22.Reijnen J M, Hamer K M, den Blaauwen J L, Lambrechts C, Schoneveld I, van Driel R, Otte A P. Polycomb and bmi-1 homologs are expressed in overlapping patterns in Xenopus embryos and are able to interact with each other. Mech Dev. 1995;53:35–46. doi: 10.1016/0925-4773(95)00422-x. [DOI] [PubMed] [Google Scholar]
- 23.Rupp R A, Snider L, Weintraub H. Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev. 1994;8:1311–1323. doi: 10.1101/gad.8.11.1311. [DOI] [PubMed] [Google Scholar]
- 24.Satijn D P E, Gunster M J, van der Vlag J, Hamer K M, Schul W, Alkema M J, Saurin A J, Freemont P S, van Driel R, Otte A P. RING1 is associated with the Polycomb group protein complex and acts as a transcriptional repressor. Mol Cell Biol. 1997;17:4105–4113. doi: 10.1128/mcb.17.7.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Satijn D P E, Olson D J, Van der Vlag J, Hamer K M, Lambrechts C, Masselink H, Gunster M J, Sewalt R G A B, Van Driel R, Otte A P. Interference with the expression of a novel human Polycomb protein, HPC2, results in cellular transformation and apoptosis. Mol Cell Biol. 1997;17:6076–6086. doi: 10.1128/mcb.17.10.6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satijn D P E, Otte A P. RING1 interacts with multiple Polycomb-group proteins and displays tumorigenic activity. Mol Cell Biol. 1999;19:57–68. doi: 10.1128/mcb.19.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Satijn D P E, Otte A P. Polycomb group protein complexes, do different complexes regulate distinct target genes? Biochim Biophys Acta. 1999;1447:1–16. doi: 10.1016/s0167-4781(99)00130-x. [DOI] [PubMed] [Google Scholar]
- 28.Schumacher A, Faust C, Magnuson T. Positional cloning of a global regulator of anterior-posterior patterning in mice. Nature. 1996;383:250–253. doi: 10.1038/383250a0. [DOI] [PubMed] [Google Scholar]
- 29.Schumacher A, Magnuson T. Murine Polycomb- and trithorax-group genes regulate homeotic pathways and beyond. Trends Genet. 1997;13:167–170. [PubMed] [Google Scholar]
- 30.Sewalt R G A B, van der Vlag J, Gunster M J, Hamer K M, den Blaauwen J L, Satijn D P E, Hendrix T, van Driel R, Otte A P. Characterization of interactions between the mammalian Polycomb-group proteins Enx1/EZH2 and EED suggests the existence of different mammalian Polycomb-group protein complexes. Mol Cell Biol. 1998;18:3586–3595. doi: 10.1128/mcb.18.6.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shao Z, Raible F, Mollaaghababa R, Guyon J R, Chao-Ting W, Bender W, Kingston R E. Stabilization of chromatin structure by PCR1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 32.Shi Y, Seto E, Chang L-S, Shenk T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 33.Simon J. Locking in stable states of gene expression: transcriptional control during Drosophila development. Curr Opin Cell Biol. 1995;7:376–385. doi: 10.1016/0955-0674(95)80093-x. [DOI] [PubMed] [Google Scholar]
- 34.Smith W C, Harland R M. Expression cloning of noggin, a new dorsalizing factor localized to the Spemann organizer in Xenopus embryos. Cell. 1992;70:829–840. doi: 10.1016/0092-8674(92)90316-5. [DOI] [PubMed] [Google Scholar]
- 35.Steinbach O C, Wolffe A P, Rupp R A W. Somatic linker histones cause loss of mesodermal competence in Xenopus. Nature. 1997;389:395–399. doi: 10.1038/38755. [DOI] [PubMed] [Google Scholar]
- 36.Strouboulis J, Damjanovski S, Vermaak D, Meric F, Wolffe A P. Transcriptional repression by XPc1, a new Polycomb homolog in Xenopus laevis embryos, is independent of histone deacetylase. Mol Cell Biol. 1999;19:3958–3968. doi: 10.1128/mcb.19.6.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Struhl G, Brower D. Early role of the esc+ gene product in the determination of segments in Drosophila. Cell. 1982;31:285–292. doi: 10.1016/0092-8674(82)90428-7. [DOI] [PubMed] [Google Scholar]
- 38.Thomas M T, Seto E. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene. 1999;236:197–208. doi: 10.1016/s0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- 39.Tie F, Furuyama T, Harte P J. The Drosophila Polycomb group proteins ESC and E(Z) bind directly to each other and co-localize at multiple chromosomal sites. Development. 1998;125:3483–3496. doi: 10.1242/dev.125.17.3483. [DOI] [PubMed] [Google Scholar]
- 40.Van Dassow G, Schmidt J E, Kimelman D. Induction of the Xenopus organizer, expression and regulation of Xnot, a novel FGF and activin-regulated homeobox gene. Genes Dev. 1993;7:355–366. doi: 10.1101/gad.7.3.355. [DOI] [PubMed] [Google Scholar]
- 41.Van der Vlag J, Otte A P. Transcriptional repression mediated by the human Polycomb-group protein EED involves histone deacetylation. Nat Genet. 1999;23:474–478. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- 42.Van Lohuizen M, Verbeek S, Scheijen B, Wientjes E, van der Gulden H, Berns A. Identification of cooperating oncogenes in E(mu)-myc transgenic mice by provirus tagging. Cell. 1991;65:737–752. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- 43.Wilson P A, Hemmati-Brivanlou A. Vertebrate neural induction, inducers, inhibitors and a new synthesis. Neuron. 1997;18:699–710. doi: 10.1016/s0896-6273(00)80311-6. [DOI] [PubMed] [Google Scholar]
- 44.Yang W-M, Inouye C, Zeng Y, Bearss D, Seto E. Transcription repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zaraisky A G, Lukyanov S A, Vasiliev O L, Smirnov Y V, Belyaavsky A V, Kazanskaya O V. A novel homeobox gene expressed in the anterior neural plate of the Xenopus embryo. Dev Biol. 1992;152:373–382. doi: 10.1016/0012-1606(92)90144-6. [DOI] [PubMed] [Google Scholar]