Abstract
The matrix metalloproteinase matrilysin (MMP-7) is expressed in the tumor cells of a majority of mouse intestinal and human colonic adenomas. We showed previously that matrilysin is a target gene of β-catenin–Tcf, the transcription factor complex whose activity is thought to play a crucial role in the initiation of intestinal tumorigenesis. Here we report that overexpression of a stable mutant form of β-catenin alone was not sufficient to effect expression of luciferase from a matrilysin promoter-luciferase reporter plasmid. However, cotransfection of the reporter with an expression vector encoding the PEA3 Ets transcription factor, or its close relatives ER81 and ERM, increased luciferase expression and rendered the promoter responsive to β-catenin–LEF-1 as well as to the AP-1 protein c-Jun. Other Ets proteins could not substitute for the PEA3 subfamily. Luciferase activity was induced up to 250-fold when PEA3, c-Jun, β-catenin, and LEF-1 were coexpressed. This combination of transcription factors was also sufficient to induce expression of the endogenous matrilysin gene. Furthermore, all matrilysin-expressing benign intestinal tumors of the Min mouse expressed a member of the PEA3 subfamily, as did all human colon tumor cell lines examined. These data suggest that the expression of members of the PEA3 subfamily, in conjunction with the accumulation of β-catenin in these tumors, leads to coordinate upregulation of matrilysin gene transcription, contributing to gastrointestinal tumorigenesis.
Matrilysin (MMP-7, EC 3.4.24.23), a member of the matrix metalloproteinase (MMP) family of proteins, is expressed in the malignant epithelia of the majority of human colonic adenocarcinomas (14, 41). Matrilysin transcripts also are found in the tumor epithelium of 90% of intestinal adenomas resulting from germ line-inactivating mutations in the adenomatous polyposis coli (APC) tumor suppressor gene in both humans (49) and mice (54). This pattern of expression is in contrast with the expression of most MMPs, which are confined to the surrounding stromal cells in noninvasive, benign tumors (54). The unique pattern of matrilysin expression in the neoplastic epithelia of benign polyps suggests a role in the early stages of tumor progression. Consistent with this hypothesis, in an orthotopic model of colon tumorigenesis, matrilysin expression enhances tumor formation (57) and tumor formation in the multiple intestinal neoplasia (Min) mouse is decreased by 60% when in a matrilysin-null genetic background (54).
Loss of functional APC is thought to be the most common initiating event in human colorectal cancer (27). This loss of APC activity is a result of inactivating mutations that render the APC protein incapable of targeting the proto-oncoprotein β-catenin for degradation (39). In normal epithelial cells, β-catenin is primarily localized to adherens junctions, where it interacts directly with the cell-cell adhesion molecule E-cadherin (1). However, when β-catenin is allowed to accumulate in the cytoplasm, it is efficiently transported into and retained in the nucleus (12, 22) where it acts as a transcriptional coactivator through its interaction with members of the Tcf/LEF-1 DNA binding protein family (2, 25). The transcriptional activity of the β-catenin–Tcf complex has been shown to correlate with the oncogenic potential of β-catenin protein (29). The transcription of several cognate target genes has been shown to be regulated by β-catenin–Tcf, including matrilysin (5, 9), c-myc (20), cyclin D1 (50), TCF-1 (43), and fibronectin (16).
Matrilysin is a transcriptional target of the β-catenin–Tcf complex (5, 9). In mouse and human intestinal tumors, the expression of matrilysin transcripts strongly overlaps the accumulation of β-catenin protein. Additionally, cotransfection of an expression vector encoding a stable mutant form of β-catenin with a mouse matrilysin promoter-luciferase reporter significantly upregulates luciferase expression in most colon tumor cell lines, dependent on a functional Tcf binding site in the promoter (9). Conversely, luciferase expression is reduced in these cell lines by cotransfection with an expression vector encoding the cytoplasmic domain of E-cadherin, a polypeptide that blocks association of β-catenin with Tcf factors. Taken together, these data suggest that β-catenin transactivation is necessary for matrilysin expression in intestinal tumors.
Despite the ability of β-catenin to transactivate the matrilysin promoter, other observations suggest that β-catenin accumulation is not sufficient to induce matrilysin expression. For example, rare dysplastic glandular structures of mouse intestinal tumors display high levels of nuclear β-catenin protein without concomitantly high levels of matrilysin transcripts (9). In addition, the abundance of β-catenin–Tcf in human colon tumor cell lines does not always correlate directly with the level of endogenous matrilysin gene expression (9). These findings suggest that the high levels of β-catenin protein found in gastrointestinal tumors are not sufficient to upregulate matrilysin transcription and that the activity or abundance of other transcriptional regulatory proteins common to intestinal tumors is required to effect matrilysin gene expression.
The tumor-associated expression of many MMP family members requires the activity of a variety of oncogenic transcription factors, including members of the AP-1 and Ets transcription factor families (10, 18). AP-1 and Ets binding sites are common features of the majority of MMP promoters (13). In these MMP promoters, basal promoter activity is highly dependent on an AP-1 site (5′-TGAGTCA-3′) usually located within the first 75 bp upstream of the transcription start site (3). Changes in AP-1 activity have been shown to regulate these and other promoters in response to a variety of stimuli, especially those mediated by the Ras family of small G proteins, including oncogenic activation of Ras and extracellular signaling through tyrosine kinase receptors and integrins (11, 56).
The mammalian Ets transcription factor family comprises approximately 30 individual members (53). All Ets proteins share highly related ETS DNA binding domains. The Ets family has been subdivided into subfamilies based on their sequence similarity. Subfamily members possess nearly identical ETS domains and share additional regions of sequence similarity. Ets proteins bind to recognition sites bearing a central core sequence, 5′-GGA(A/T)-3′; sequences flanking this core dictate a measure of binding specificity for individual Ets proteins (17). Ets proteins usually activate transcription, but rare members of the family repress this process. Like with members of the Jun family, the activity and expression of several Ets proteins are regulated by extracellular signals acting through the Ras pathway (53).
Ets and AP-1 factors have been shown to synergistically activate MMP transcription by interacting with their cognate binding sites in the promoters of the genes. Usually the Ets and AP-1 binding sites in these promoters are juxtaposed or in close proximity (10). In several MMP promoters, these closely spaced sites constitute a Ras- or oncogene-responsive element; mutation of either the Ets or AP-1 binding sites of such oncogene-responsive elements severely compromises the capacity of the promoter to be upregulated by oncoproteins functioning through the Ras pathway (11). Besides MMP expression, synergistic collaboration between Ets and AP-1 proteins has been shown to regulate the promoters of multiple genes associated with tumor progression (11). This apparent need for cooperation in regulating the expression of many tumor-associated genes suggests that this cooperation also is important for both Ets and AP-1 proteins to act as oncogenes.
The observation that β-catenin upregulation is insufficient under some circumstances to stimulate matrilysin gene transcription suggested that other transcription factors commonly expressed in intestinal tumor cells are involved in this process. To test this hypothesis, we used the human kidney cell line HEK293, in which β-catenin expression upregulated the activity of matrilysin Tcf site artificial promoters but not the intact human matrilysin promoter. Exploiting this characteristic, we found that expression of any of the PEA3 subfamily of Ets transcription factors rendered the matrilysin promoter responsive to β-catenin transactivation as well as to that of the AP-1 protein c-Jun. Furthermore, members of the PEA3 subfamily, particularly PEA3 and ERM, were found to be expressed frequently in mouse intestinal tumors and in every human colon tumor cell line examined. We conclude that the PEA3 subfamily acts in conjunction with β-catenin–Tcf to upregulate the transcription of the matrilysin gene during intestinal tumorigenesis.
MATERIALS AND METHODS
Cells.
HEK293 (ATCC CRL-1573), CaCo-2 (ATCC HTB-37), HCT15 (ATCC CCL-225), HCT116 (ATCC CCL-247), HT29 (ATCC HTB-38), SW480 (ATCC CCL-228), SW620 (ATCC CCL-227), and HCA7 cells (a gift of Susan Kirkland, University of London, London, United Kingdom) were maintained at 37°C in 5% CO2 in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS).
Plasmids.
Expression vectors for chicken Ets-1 (31), mouse PU.1 (28), and human TEL-1 (23) were the gifts of Scott Hiebert (Vanderbilt University, Nashville, Tenn.). Expression vectors for human ELF-1, Fli-1, GABP-α, and GABP-β (each in the pBK-CMV vector) were gifts of Barbara Graves (Huntsman Cancer Institute, Salt Lake City, Utah). Expression vectors for full-length LEF-1 (pBZ13-LEF-1) and a ΔN-LEF-1 expression vector (pFLAG hLEF-1) (58) were gifts of Elaine Fuchs (University of Chicago, Chicago, Ill.). The expression vector for Ets-2 (pSG5-Ets2) (44) was the gift of Dennis Watson (Medical University of South Carolina, Charleston). The expression vector for c-Jun (pCMX-c-Jun) (52) was the gift of Ronald Wisdom (Vanderbilt University). The pCMV-βp300-CHA expression vector was the gift of David Livingston (Dana Farber Cancer Institute, Boston, Mass.). Expression vectors for E1A and the E1A mutants 2–36E1A and E1A-928 (47) were the gift of Roland Stein (Vanderbilt University).
pCANmycPEA3 was constructed by digesting a full-length mouse PEA3 cDNA cloned into pGEM7zf (Promega) with SacI, followed by blunting with Klenow fragment and digestion with BamHI. The resulting full-length cDNA was cloned into BamHI/EcoRV-digested pCANmyc vector (Onyx Pharmaceuticals). The mouse ER81 expression vector was constructed by cloning a full-length SpeI/XhoI cDNA fragment into XbaI/XhoI-digested pCDNA3.1(−)Zeo (Invitrogen).
4×(−194Tcf)Luc and 2×(−109Tcf)Luc were constructed by synthesizing oligonucleotides with SalI-compatible overhangs on both the sense and antisense strands, annealing the oligonucleotides, and phosphorylating them with polynucleotide kinase (Promega). Phosphorylated oligonucleotides were ligated with SalI-digested TK-Luc (34). The number and orientation of inserts were determined by DNA sequencing. −194 Tcf oligonucleotides were as follows: sense, 5′-TCGACAAAAATCCTTTGAAAGACAAATACATG-3′; antisense, 5′-TCGACATGTATTTGTCTTTCAAAGGATTTTTG-3′. −109 Tcf oligonucleotides were as follows: sense, 5′-TCGACACATACTTTCAAAGTTCTGTAGACTCAG-3′; antisense, 5′-TCGACTGAGTCTACAGAACTTTGAAAGTATGTG-3′.
The 2.3-kb matrilysin promoter construct was created by cutting a 4.2-kb genomic clone of the matrilysin promoter (13) with MfeI and cloning the resultant 2.3-kb fragment into the EcoRI site of pBluescript KS. The fragment was recovered using HindIII and BamHI and cloned into the BglII and HindIII sites of pGL2Basic (Promega). The −296HMAT vector was created by digesting the 2.3-kb HMAT with KpnI/HindIII and cloning the resulting 335-bp fragment into KpnI/HindIII-digested pGL2Basic. The rat stromelysin-1 promoter construct p754TR-Luc was constructed by cloning the SmaI/BglII fragment of p754TR-CAT (13) into pGL2Basic.
GST–LEF-1 was created by PCR of the pBZ13 LEF-1 cDNA with the oligonucleotides 5′-GCCGGATCCCCAACTCTCCGGAGGA-3′ and 5′-GCGCGAATTCTCAGATGTAGGCAGCTGTCATTCTGGGA-3′ and PfuTurbo polymerase (Stratagene) and cloned into pCRScript vector (Stratagene). The LEF-1 cDNA sequence was confirmed, and cDNA was digested with BamHI and EcoRI (sites engineered into oligonucleotides) and cloned into pGEX-4T2 (Amersham Pharmacia Biotech). The glutathione S-transferase (GST)-tagged protein was purified using the manufacturer's directions, dialyzed overnight against 20 mM HEPES–20% glycerol–100 mM KCl–0.2 mM EDTA, and snap-frozen in liquid nitrogen.
Mutagenesis of the matrilysin promoter.
The matrilysin promoter was mutated by the PCR-splicing by overlap extension method (24) using −296HMAT as a template and GL1 and GL2 oligonucleotides as 5′- and 3′-end primers. Sense oligonucleotides for mutagenesis were as follows, with mutated positions underlined: for −168Ets, 5′-GTGTGCTTCTGCCAATAACGATG-3′; for −144Ets, 5′-GTAATACTTCTTCGTTTTAGTTAATG-3′; for −55Ets, 5′-CCTATTTCTACATTCGAGGC-3′; for −194Tcf, 5′-GACAGAAAAAAAAATCATTGGCGATACAAATACATTGTGTG-3′; for −109Tcf, 5′-TAACACATAATCGCCAACTTCTGTAGACTC-3′; and for mAP-1, 5′-CAAACGAGTGACCTATTTCCAC-3′. Antisense oligonucleotides were the reverse complements of the sense oligonucleotides. The double mutant Tcf site construct was created by performing PCR-splicing by overlap extension with the −194Tcf construct as a template and the −109Tcf oligonucleotides as primers. It should be noted that the inactivating mutation of Tcf sites is generally a two-nucleotide alteration (30); however, the matrilysin Tcf sites are palindromic and the 2-bp mutation does not eliminate LEF-1 binding (data not shown). The five-nucleotide alteration was made to eliminate potential Tcf interactions with the complementary strand.
Electrophoretic mobility shift assay (EMSA).
Probes were made by annealing the −194Tcf oligonucleotide (5′-GCAAAATCCTTTGAAAGACAAATCCCTCTCCTT-3′) or the −109Tcf oligonucleotide (5′-CACATACTTTCAAAGTTCTGTAGACTCCCTCTCCTT-3′) to a 10-fold excess of a primer (5′-AAGGAGAGGG-3′). Probes were labeled by primer extension with Klenow in the presence of [α-32P]dCTP for 1 h. Probes were isolated on a 5% polyacrylamide gel and eluted.
EMSA was performed by incubating 5 × 105 cpm of probe with 5 μl of purified GST–LEF-1 in EMSA buffer (20 mM HEPES, 20% glycerol, 100 mM KCl, 0.2 mM EDTA, 0.5 mM dithiothreitol) for 30 min at 37°C. The sample was then run on a 4% acrylamide–2% glycerol–0.25× Tris-borate-EDTA gel at 200 V for 3.5 h.
Transient transfections and reporter assays.
A transfection mixture was created by incubating 1 μg of firefly luciferase reporter with 10 ng of the Renilla luciferase internal control, SV40-RL (Promega), and 1 μg each of the expression vectors indicated below. The volume was brought to 200 μl using OptiMEM (Gibco-BRL), and 15 μl of Superfect (Qiagen) transfection reagent was added and mixed by pipetting. After a 15-min incubation, 1 ml of DMEM containing 10% FBS was added, the contents were mixed, and 400 μl was distributed to each of 3 wells of a 24-well plate, each well containing 1.5 × 105 HEK293 cells plated 24 h prior to transfection. Total DNA in the transfection mixture was kept constant by including the same empty vectors as those that contained the cDNAs being expressed. The transfection mixture was removed from the cells 2 to 3 h after addition and replaced with DMEM with 10% FBS.
Luciferase activity was determined using the Dual Luciferase kit (Promega) 16 to 24 h posttransfection by lysing in 50 μl of passive lysis buffer and assaying both firefly and Renilla luciferase activity in the same 30-μl aliquot of lysate. Fold induction was determined by first normalizing each firefly luciferase value to the Renilla luciferase internal control, averaging the normalized values, and dividing by the mean value of the firefly reporter cotransfected with empty vectors only. For p300 transfections, values were normalized to ratios obtained with pGL2Basic, to control for p300 effects on SV40-RL. Normalized relative light units (RLUs) were determined by normalizing each firefly luciferase value to the highest Renilla luciferase value in a given experiment by the following formula: (highest Renilla luciferase value in the experiment/Renilla luciferase value of the individual sample) × firefly luciferase value of the same individual sample. Whether using fold induction or normalized RLUs, each experiment was repeated as noted in the figure legends and the means and standard errors were calculated using Microsoft Excel.
RT-PCR.
A total of 5 × 105 HEK293 cells were plated into each well of a six-well tissue culture dish. Cells were transiently transfected as described above, except that the entire transfection mixture was added to a single well. Total RNA was isolated 24 h later using the RNeasy kit (Qiagen). Total RNA (1 μg) was reverse transcribed using 100 ng of poly(T) primer and Moloney murine leukemia virus reverse transcriptase (Gibco-BRL). PCR was performed using standard methods with 5 μl of reverse transcription (RT) mixture, Taq DNA polymerase (Promega), 1× buffer A, and 2.5 mM MgCl2. Matrilysin-specific oligonucleotides were 5′-TGGAGTGCCAGATGTTGCAG-3′ and 5′-TTTCCATATAGCTTCTGAATGCCT-3′. Data shown were obtained with 35 PCR cycles.
Immunoprecipitation.
Duplicate 35-mm plates containing 1 × 106 HEK293 cells were transfected with 1 μg of pCAN-ΔN89β-cat and 1 μg of empty pCAN-myc vector or 1 μg pCAN-ΔN89β-catenin and 1 μg of pCAN-PEA3 using Superfect reagent. After 48 h, cells were washed twice with cold 1× phosphate-buffered saline and harvested by adding 300 μl of lysis buffer (50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 5 mM MgCl2, 0.1% Nonidet P-40, 1 mM dithiothreitol, 0.1 mM sodium orthovanadate, 10 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin per ml, 10 μg of aprotinin per ml) and rocking on ice for 20 min. After lysis, duplicate samples were combined. Lysates were clarified by microcentrifugation. Clarified lysates were precleared by rocking for 4 h at 4°C after addition of 350 μg of protein A-Sepharose beads (Amersham Pharmacia Biotech) that were preswelled and stored in 100 mM NaCl–50 mM Tris-HCl (pH 7.5)–0.1% NP-40. Beads were spun out by microcentrifugation, and equal volumes of lysate were split into two fresh prechilled tubes. In one tube, 1 μg of a rabbit polyclonal anti-β-catenin antibody (C-2206; Sigma) was added with 175 μg of protein A-Sepharose. To the second tube, 175 μg of protein A-Sepharose was added as a no-primary-antibody control. Samples were rocked at 4°C for 12 h. Beads were spun out by microcentrifugation and eluates were set aside. Beads were washed once in 1 ml of lysis buffer and twice with 1 ml of wash buffer (20 mM Tris-HCl [pH 7.6], 100 mM NaCl, 1 mM EDTA, 0.1% Nonidet P-40). After the last wash, beads were spun at 12,500 rpm at 4°C and any remaining liquid was removed. Beads were then resuspended in 50 μl of 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and boiled for 10 min, as was 7 μl of the eluate from the no-antibody control to use as an input control sample. Beads were spun out and the samples were applied to an SDS–7.5% PAGE gel. The gel was transferred to NitroME nitrocellulose and blocked for 4 h in 5% milk–1× Tris-buffered saline plus Tween 20 (TBST). The blot was then incubated sequentially with 2 ng of mouse monoclonal anti-PEA3 antibody (Santa Cruz) per ml at 4°C overnight, a 1:15,000 dilution of biotinylated anti-mouse immunoglobulin G (Vector Labs) for 30 min at room temperature, and a 1:20,000 dilution of horseradish peroxidase-conjugated streptavidin (Jackson Labs) for 30 min at room temperature, each diluted in 1× TBST–5% milk; the blot was washed three times in 1× TBST between each antibody. To visualize bands, the blot was subjected to chemiluminescence using the ECL kit (Amersham Pharmacia Biotech).
In situ hybridization.
Plasmid pGEM7-MMATAH (55) was linearized with ApaI and antisense riboprobe was generated using T7 RNA polymerase (Promega) in the presence of 35S-UTP. Antisense riboprobe for mouse PEA3 was generated from pCAN-PEA3 linearized with BamHI using SP6 RNA polymerase (Promega). Antisense riboprobe for ERM was generated by linearizing a pCRII clone of nucleotides 70 to 680 with HindIII and transcribing it with T7 RNA polymerase. ER81 antisense riboprobe was generated by linearizing a pBluescript clone of a 250-bp HindIII/BamHI fragment with HindIII and transcribing it with T7 RNA polymerase.
In situ hybridization was performed on 5-μm serial sections from paraformaldehyde-fixed, paraffin-embedded Min mouse small intestinal tumors as previously described (55).
Matrilysin Western blot.
Cell lines were grown to confluence in 100-mm dishes and then incubated for 48 h in 4 ml of OptiMEM (Gibco-BRL) at 37°C and 5%CO2. Conditioned medium was then concentrated in Microcon 10 concentrators (Centricon) and quantitated using a protein assay (Bio-Rad). Twenty-five micrograms of protein from CaCo-2, HCT15, HCT116, SW480, and SW620 conditioned media and 5 μg of protein from HCA7 and HT29 conditioned media were then run on an SDS–12% PAGE gel and transferred to NitroME nitrocellulose. The blot was blocked as described above and probed with a 1:6 dilution of monoclonal rat anti-human matrilysin hybridoma supernatant (45) in 5% milk–1× TBST overnight. The blot was washed and probed as described above except that biotinylated anti-rat immunoglobulin G (Vector Labs) was used as the secondary antibody.
Northern blotting.
Probes for the PEA3 3′ untranslated region (nucleotides 2139 to 2607), ERM 3′ untranslated region (nucleotides 1404 to 1682), and the ER81 3′ untranslated region (nucleotides 1932 to 2525) were generated by PCR. Fifty nanograms of each purified probe was labeled using the random primed DNA labeling kit (Boehringer Mannheim). Total RNA was isolated using the RNeasy kit (Qiagen), and 15 μg of total RNA was run on a 1% agarose denaturing formaldehyde gel. Nucleic acids were blotted to Hybond paper (Amersham Pharmacia Biotech) by capillary transfer in 10× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The probe was hybridized to a blot in UltraHyb buffer (Amersham Pharmacia Biotech).
RESULTS
β-Catenin is insufficient to transactivate the human matrilysin promoter in HEK293 cells.
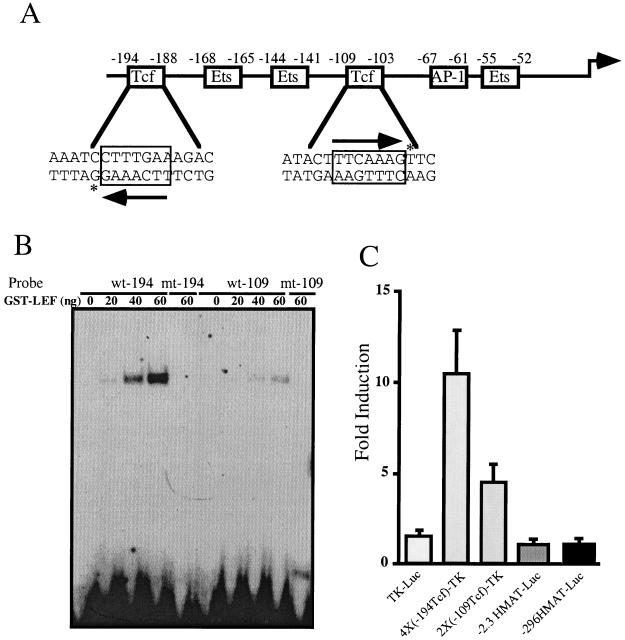
To determine whether β-catenin is sufficient to stimulate matrilysin transcription, we analyzed the responsiveness of the human promoter (13) to a stable mutant form of β-catenin (ΔN89β-cat) (38) in an immortal human embryonic kidney cell line, HEK293. The human matrilysin promoter bears two consensus Tcf binding sites (5′-[A/T] [A/T] CAAAG-3′), one in an inverted orientation between −194 and −188 (5′-CTTTGAA-3′) and another between −109 and −103 (5′-TTCAAAG-3′) (Fig. 1A). To determine whether Tcf proteins can bind to these sites, an EMSA was performed using purified GST–LEF-1 and oligonucleotides representing each site. GST–LEF-1 bound to both candidate sites but bound preferentially to the −194 site (Fig. 1B). This difference may be due to the presence of a C at position −193 compared to a T at the equivalent position in the −109 Tcf site; this nucleotide is known to affect binding of LEF-1 (33). Mutations known to diminish Tcf binding (33) were introduced into these sequences and their effect on GST–LEF-1 binding was assessed. GST–LEF-1 did not bind to either mutant site even at the highest protein concentrations tested (Fig. 1B), confirming that these mutations effectively eliminated Tcf protein binding.
FIG. 1.
The human matrilysin promoter has two functional Tcf binding sites. (A) Structure of the human matrilysin promoter. The sequence of the human matrilysin promoter has been previously reported (13) (GenBank accession no. L22525). Indicated are the sequences and positions relative to the transcriptional start site of the two consensus Tcf binding sites as well as the positions of the Ets sites and the AP-1 site. Arrows indicate the Tcf consensus sequence and orientation. Asterisks indicate the C residue that has been shown to enhance LEF-1 binding (32) in the −194 Tcf site as opposed to the T residue in the equivalent position of the −109 Tcf site. (B) EMSA of the two Tcf sites with purified GST–LEF-1. Shown are oligonucleotides representing the wild-type Tcf sites (wt-194 and wt-109) incubated with the indicated amount of GST–LEF-1. Also shown are oligonucleotides representing the mutant Tcf sites (mt-194 and mt-109) coincubated with the maximal amount of GST–LEF-1. (C) Responsiveness of the two Tcf sites to stable β-catenin. A total of 1.5 × 105 HEK293 cells were cotransfected with pCAN-ΔN89β-cat and either a thymidine kinase minimal promoter-luciferase construct (TK-luc), a promoter-reporter construct with four copies of the −194 Tcf site cloned upstream of the thymidine kinase minimal promoter [4×(−194Tcf)-TK], a promoter-reporter construct with two copies of the −109 Tcf site cloned upstream of the tk minimal promoter [2×(−109Tcf)-TK], or human matrilysin promoter reporter constructs from approximately −2300 to +35 (−2.3HMAT-Luc) or −296 to +35 (−296HMAT-Luc) cloned into pGL2Basic. Results are expressed as fold induction relative to cotransfection with an equal amount of pCANmyc empty vector. Data bars represent the means of three independent experiments, each performed in triplicate. Error bars represent standard errors.
To confirm that β-catenin was capable of activating transcription through these sites, artificial promoters were constructed that comprised multiple copies of the matrilysin Tcf sites located upstream of a minimal thymidine kinase promoter coupled to a luciferase reporter gene. These reporters were cotransfected with an expression vector encoding a stable β-catenin mutant (ΔN89β-cat) into HEK293 cells. β-Catenin stimulated expression of luciferase approximately 10-fold from a reporter bearing four copies of the −194Tcf site [4×(−194)-TK] and 4-fold from the reporter containing two copies of the −109 Tcf site [2×(−109)-TK] (Fig. 1C). Hence, under the conditions of these transfection assays, both matrilysin Tcf sites were responsive to β-catenin.
To determine whether β-catenin could stimulate reporter gene expression governed by the natural human matrilysin promoter, we used two luciferase reporters. One of these bears a human matrilysin promoter fragment from −2300 to +35 (−2.3HMAT-Luc) whereas the other comprised sequences from −296 to +35 (−296HMAT-Luc) relative to the transcription start site. Surprisingly, cotransfection of the β-catenin expression vector with either of these reporters did not stimulate luciferase expression (Fig. 1C) despite the presence of the two functional Tcf sites. Because the matrilysin promoter is activated by β-catenin in colon tumor cell lines (5, 9), we hypothesized that induction of the matrilysin promoter required other transcription factors commonly expressed in these colon carcinoma cells, but absent from HEK293 cells, to function in concert with β-catenin–Tcf.
The matrilysin promoter is selectively transactivated by PEA3 subfamily Ets transcription factors.
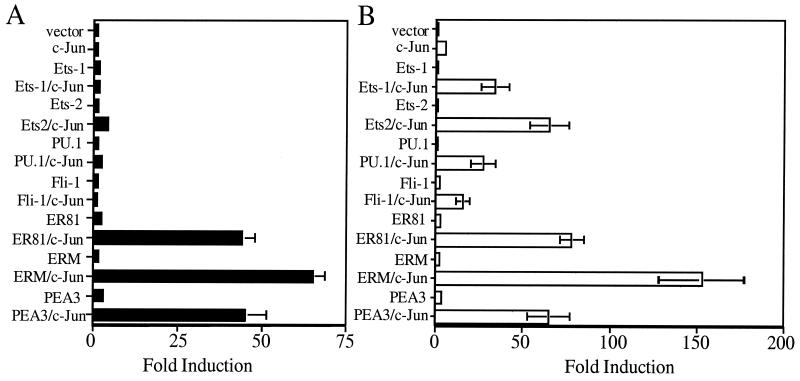
The promoters of many MMPs are responsive to AP-1 and Ets proteins (10). Indeed, these transcription factors act synergistically to activate the expression of reporter genes linked to the stromelysin-1, stromelysin-2, collagenase-1, collagenase-3, and gelatinase B promoters. Like these other MMP promoters, the matrilysin promoter has a canonical AP-1 site (5′-TGAGTCA-3′) located between −67 and −61 (3) and candidate Ets binding sites (5′-GGA[A/T]-3′) located from −55 to −52, −144 to −141, and −168 to −165 (Fig. 1A). To learn whether AP-1 can activate transcription of luciferase from the matrilysin promoter-reporter, we cotransfected the −296HMAT-Luc reporter with an expression vector encoding c-Jun, which is capable of dimerizing to constitute AP-1 activity. c-Jun did not stimulate luciferase expression from the reporter bearing the matrilysin promoter (Fig. 2A). However, c-Jun stimulated luciferase expression approximately fivefold from a reporter bearing the stromelysin-1 promoter (−754TR-1-Luc), demonstrating that c-Jun was expressed and was capable of activating transcription in these cells (Fig. 2B). The unexpected finding that the matrilysin promoter was unresponsive to c-Jun suggested that the ability of c-Jun to transactivate this promoter required additional trans-acting factors.
FIG. 2.
The matrilysin promoter is preferentially upregulated by the PEA3 subfamily of Ets transcription factors. A total of 1.5 × 105 HEK293 cells were cotransfected with either the human matrilysin promoter, −296HMAT-Luc (A), or the rat stromelysin-1 promoter, −754TR-Luc (B), and expression vectors for c-Jun and each of the Ets proteins as indicated. Data are presented as fold induction relative to cotransfection of the promoter constructs with empty expression vectors. Values were normalized to cotransfection with simian virus 40-driven Renilla luciferase and degree of induction to that of pGL2-Basic cotransfected with the same combination of expression vectors. Data bars represent the means of experiments repeated a minimum of three times, each transfection performed in triplicate. Error bars represent standard errors.
The capacity of Ets family transcription factors to activate expression of luciferase from the −296 HMAT reporter and, in parallel, from the −754TR-1 reporter was tested. The two reporter constructs were separately cotransfected with one of several mammalian expression vectors encoding different Ets proteins. The Ets proteins Ets-1, Ets-2, PU.1, and Fli-1 did not stimulate luciferase expression from either reporter plasmid (Fig. 2). Coexpression of these Ets proteins with c-Jun also did not stimulate expression of luciferase from the matrilysin reporter plasmid (−296HMAT) (Fig. 2A). However, each of these Ets proteins functioned synergistically with c-Jun to augment luciferase expression from the stromelysin-1 reporter (Fig. 2B). Other Ets proteins (ELF-1, GABP-α, GABP-β, and TEL-1) did not transactivate expression of luciferase from either reporter and indeed blocked the capacity of c-Jun to stimulate luciferase expression from the stromelysin-1 reporter (data not shown).
In contrast to the other Ets family members, PEA3 and its subfamily relatives ER81 and ERM modestly stimulated luciferase expression (2- to 3-fold) from the matrilysin promoter-reporter and functioned synergistically with c-Jun to transactivate this promoter (40- to 70-fold [Fig. 2A]). The PEA3 subfamily proteins also acted synergistically with c-Jun to augment luciferase expression from the stromelysin promoter (Fig. 2B). These findings starkly illustrate the functional specificity of Ets proteins for particular promoters and demonstrate that only the PEA3 subfamily members (PEA3, ER81, and ERM) can act independently and in concert with c-Jun to significantly upregulate the matrilysin promoter.
PEA3 renders the matrilysin promoter responsive to β-catenin–Tcf transactivation.
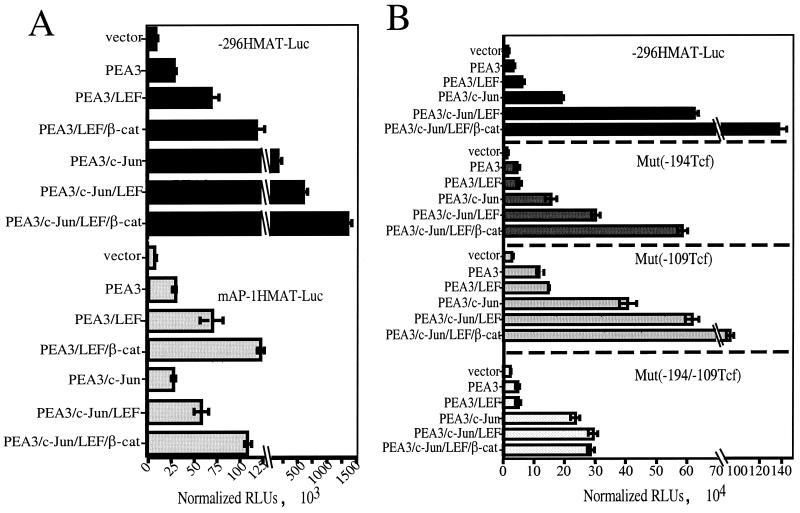
To test whether c-Jun could render the human matrilysin promoter responsive to β-catenin transactivation, we cotransfected the −296MAT-Luc reporter with expression vectors encoding c-Jun, LEF-1, or β-catenin. As anticipated from previous experiments (Fig. 1 and 2), ΔN89β-cat, LEF-1, and c-Jun individually did not significantly stimulate luciferase expression from the −296HMAT-Luc reporter (Fig. 3A). Similarly, pair-wise combinations of β-catenin, LEF-1, and c-Jun or coexpression of all three proteins did not stimulate luciferase expression from this reporter (Fig. 3A). Hence, c-Jun did not cooperate with β-catenin–LEF-1 to transactivate the matrilysin promoter.
FIG. 3.
PEA3 synergizes with both c-Jun and β-catenin–LEF-1 to upregulate matrilysin promoter activity and gene expression. (A) Synergistic activation of the human matrilysin promoter and matrilysin expression by PEA3, β-catenin (β-cat), LEF-1, and c-Jun. The −296HMAT-Luc construct was cotransfected with combinations of PEA3, c-Jun, LEF-1, and β-catenin expression vectors into 1 × 105 HEK293 cells. Data are presented as fold induction relative to cotransfection of the reporter with empty expression vectors. Raw values were normalized with the SV40-RL internal control and calculated as the degree of induction relative to that of pGL2Basic cotransfected with the same combinations of expression vectors. Data bars represent the means from 21 experiments, each performed in triplicate. Error bars represent standard errors. (Inset) HEK293 cells were transiently transfected with the expression vectors indicated, total RNA was harvested from cells 24 h later, and RT-PCR amplification was performed for matrilysin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). RT-PCR analysis is representative of three separate experiments. (B) LEF-1 and PEA3 synergy requires the β-catenin interaction domain of LEF-1. Transient transfections of HEK293 cells were performed and analyzed as described for panel A. ΔNLEF-1 is human LEF-1 with the first 36 amino acids replaced with a FLAG tag (58). Data bars represent the mean values of three experiments, each performed in triplicate. Error bars represent standard errors.
To learn whether PEA3 could render the matrilysin promoter responsive to the β-catenin–Tcf complex, we carried out similar cotransfection experiments. PEA3 modestly activated the matrilysin promoter-reporter, approximately 3-fold (Fig. 3A), and coexpression of PEA3 with either LEF-1 or β-catenin upregulated this reporter about 7-fold. Coexpression of all three proteins increased luciferase expression from the reporter 15-fold. Therefore, PEA3 cooperated with the β-catenin–LEF-1 complex to transactivate the matrilysin promoter.
In light of the capacity of PEA3 to act synergistically with either c-Jun (Fig. 2) or the β-catenin–Tcf complex, we examined the consequence of coexpressing these activators on matrilysin promoter activity. In this experiment, coexpression of c-Jun and PEA3 increased luciferase expression more than 40-fold from the −296HMAT-Luc reporter (Fig. 3A). Coexpression of PEA3 with c-Jun and either LEF-1 or β-catenin led to a dramatic increase in luciferase expression (130- or 180-fold, respectively) (Fig. 3A). Cotransfection of all four expression vectors with the −296HMAT-Luc reporter enhanced luciferase expression nearly 250-fold. These data strongly suggest that the transactivating abilities of c-Jun and the β-catenin–LEF-1 complex on the matrilysin promoter are both dependent on PEA3 activity and that these transcription factors function synergistically on this promoter.
We also tested the responsiveness of the −2.3HMAT-Luc reporter and mouse matrilysin promoter constructs to transactivation by the various proteins both individually and in combination. These reporters responded similarly to the −296HMAT-Luc reporter (data not shown). Furthermore, ER81 and ERM were fully capable of functionally substituting for PEA3 in these assays, whereas none of the other Ets proteins were capable of doing so on either the human or mouse matrilysin reporter constructs (data not shown). Taken together, these data suggest that the PEA3 subfamily Ets proteins are uniquely capable of synergizing with c-Jun and the β-catenin–LEF-1 complex to transactivate the matrilysin promoter.
The magnitude of the response of the −296HMAT reporter to transactivation by PEA3, c-Jun, and the β-catenin–LEF-1 complex prompted us to test whether this combination of transactivators could stimulate transcription of the endogenous human matrilysin gene in HEK293 cells. To this end, we transiently cotransfected HEK293 cells with the expression vectors for these transcription factors and isolated total RNA 1 day later. RT-PCR analysis of RNA from cells transfected with empty expression vectors revealed that matrilysin is not commonly expressed in HEK293 cells (Fig. 3A, inset). Coexpression of PEA3, c-Jun, and LEF-1 did not induce detectable levels of matrilysin transcript, but coexpression of β-catenin with PEA3, c-Jun, and LEF-1 did.
It was somewhat surprising to find that LEF-1 alone was capable of cooperating with PEA3 to transactivate the matrilysin promoter in HEK293 cells. LEF-1 has alternatively been described as a repressor (4, 7, 32) or transactivator that can act through both β-catenin-dependent (30) and β-catenin-independent (15) mechanisms. To test whether LEF-1 cooperation with PEA3 in HEK293 cells was dependent on its interaction with endogenous β-catenin, we used an amino-terminally truncated form of LEF-1 (ΔNLEF-1), which lacks the β-catenin interaction domain and hence functions as a dominantnegative with regard to β-catenin-dependent transactivation. Unlike LEF-1, which activated the promoter with PEA3, ΔNLEF-1 was not capable of cooperating with PEA3 to effect luciferase expression from the −296HMAT reporter (Fig. 3B). ΔNLEF-1 also compromised the ability of β-catenin to synergize with PEA3 to activate expression of this reporter. However, ΔNLEF1 did not block the capacity of PEA3 to transactivate the −296HMAT reporter, nor did it perturb the capacity of PEA3 and c-Jun to cooperate to upregulate this reporter. These findings are consistent with the contention that LEF-1 activation of the human matrilysin promoter required interaction with endogenous β-catenin and thus represented a manifestation of the cooperativity between PEA3 and β-catenin.
c-Jun and β-catenin–LEF-1 act independently to synergize with PEA3.
β-Catenin has been reported to upregulate c-Jun expression (36). Our data, in turn, show that c-Jun expression can strongly synergize with PEA3 to activate the matrilysin promoter. Together, these data suggested that β-catenin synergy with PEA3 may be indirect, resulting from an induction of endogenous c-Jun expression and subsequent synergy between c-Jun and PEA3 to transactivate the matrilysin promoter. To address this possibility, we cotransfected the HEK293 cells with the matrilysin promoter reporter containing inactivating point mutations in the AP-1 site together with expression vectors for PEA3, c-Jun, LEF-1, and β-catenin in parallel with the wild-type −296HMAT-Luc reporter (Fig. 4A).
FIG. 4.
c-Jun and β-catenin–LEF-1 act independently to cooperate with PEA3. (A) Mutation of the AP-1 site affects only c-Jun transactivation of the matrilysin promoter. A 2-bp inactivating mutation of the AP-1 site (TGAGTCA → CGAGTGA) was introduced into the −296HMAT-Luc reporter to create mAP1HMAT-Luc. The mutant promoter was cotransfected with combinations of the PEA3, c-Jun, LEF-1, and ΔN89β-cat (β-cat) expression vectors, as indicated, in parallel with the wild-type promoter. Data are presented as RLUs normalized to cotransfected simian virus 40-driven Renilla luciferase. Data bars represent three experiments, each done in triplicate. Error bars represent standard errors. (B) Mutations of the Tcf sites affect only β-catenin–LEF-1 transactivation of the matrilysin promoter. Inactivating mutations were introduced into each single Tcf site as well as into both sites of the −296MAT-Luc reporter to create Mut(−194Tcf), Mut(−109Tcf), and Mut(−194/−109Tcf) reporters. The wild-type and mutant reporters were cotransfected in parallel with combinations of the PEA3, c-Jun, LEF-1, and ΔN89β-catenin expression vectors into HEK293 cells. Data are presented as RLUs normalized to cotransfected simian virus 40-driven Renilla luciferase. Data bars represent the means of four experiments, each performed in triplicate. Error bars represent standard errors.
Mutation of the AP-1 site had no effect on basal promoter activity compared to the control, and as in previous experiments, neither c-Jun, LEF-1, nor β-catenin alone had any effect on either reporter (data not shown). PEA3 activated both the wild-type and mutant AP-1 site promoter three- to fourfold (Fig. 4A). As expected, c-Jun coexpression with PEA3 did not activate the mutant AP-1 site reporter beyond the level observed with PEA3 alone, while c-Jun activated the wild-type promoter an additional fivefold. This is in contrast to LEF-1 and β-catenin transactivation, which was approximately equal on both wild-type and mAP-1 constructs, about two- to threefold additional activation for each factor, regardless of c-Jun expression. As would be expected if PEA3–c-Jun synergy were acting independently from PEA3–LEF-1–β-catenin synergy, the combination of c-Jun, PEA3, LEF-1, and β-catenin on the mutant AP-1 construct was approximately equal to that of PEA3, LEF-1, and β-catenin on the wild-type promoter. These results strongly argue that c-Jun upregulation by β-catenin was not involved in activating the matrilysin promoter in these transient-transfection experiments and that PEA3 and β-catenin–LEF-1 synergized directly to activate this promoter.
Because β-catenin–LEF-1 did not appear to activate the matrilysin promoter indirectly through the AP-1 site, we tested whether β-catenin–LEF-1 transactivation of the promoter was dependent upon, and limited to, the identified Tcf sites. Reporter constructs mutated at the −194 Tcf site, the −109 Tcf site, or both Tcf sites were analyzed in parallel with the wild-type reporter with respect to their responsiveness to PEA3, c-Jun, LEF-1, and β-catenin coexpression (Fig. 4B). The basal activity of each mutant reporter was higher than that of the wild-type control, particularly for the −109 Tcf site and the double Tcf site mutants; these had activities fourfold higher than that of the wild type, suggesting an inhibitory role for the resident Tcf complex, as has been observed for other promoters (9, 50).
As in previous experiments, c-Jun, LEF-1, or β-catenin expression alone had no effect on luciferase activity from these reporters (data not shown) and the reporter response to PEA3 was unaffected, being two- to fourfold in each case. Compared to PEA3 alone, LEF-1 coexpression with PEA3 activated the wild-type reporter an additional 2-fold but showed only a minor additional activation of the single Tcf site mutant reporters (<1.5-fold) and had no effect on the double Tcf site mutant reporter. c-Jun coexpression with PEA3 effectively activated both wild-type and mutant Tcf reporters 12- to 20-fold. Thus, under these conditions, mutation of the Tcf sites compromised LEF-1 transactivation without having a significant effect on c-Jun transactivation.
Similar to its effects when expressed with PEA3, LEF-1 expressed with PEA3 and c-Jun activated the wild-type reporter an additional 4-fold compared to PEA3 and c-Jun alone, while the reporters with single Tcf site mutations showed a reduced but significant response (∼2-fold) to LEF-1. The double mutant Tcf site reporter did not respond to LEF-1 under these conditions. Not surprisingly, β-catenin transactivation showed a similar dependency on the Tcf sites, activating the wild-type reporter an additional 2.5-fold when coexpressed with PEA3, c-Jun, and LEF-1 but activating the −194 Tcf site and −109 Tcf site mutants less than 2-fold and having no significant additional effect on the double Tcf site mutant reporter beyond the activation provided by the other factors. These experiments indicated that β-catenin–LEF-1 transactivation of the promoter can be partially mediated through either Tcf site. Additionally, this transactivation acted wholly through these two Tcf sites and not either through additional cryptic Tcf sites, as has been reported for the cyclin D1 promoter (50), or through upregulation of secondary trans-acting factors. Also, because β-catenin–LEF-1 activated the matrilysin reporter with an inactive AP-1 site (Fig. 4A) and c-Jun activated the matrilysin reporter with inactive Tcf sites (Fig. 4B), we conclude that c-Jun and β-catenin–LEF-1 were capable of independently cooperating with PEA3 to transactivate the matrilysin promoter.
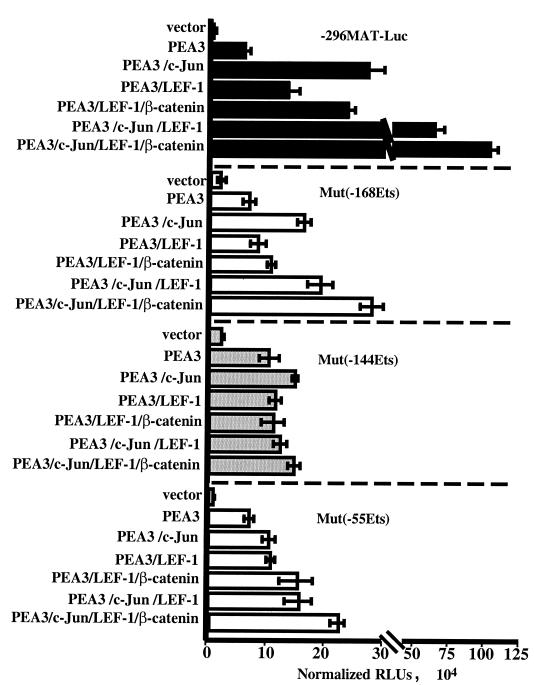
The −144 Ets site is critical for PEA3 cooperation with both c-Jun and β-catenin–LEF-1.
We have shown that the transactivating abilities of c-Jun and the β-catenin–LEF-1 complex on the matrilysin promoter were capable of functioning independently. However, both factors shared a common dependence on the activity of PEA3. To gain insight as to how PEA3 might cooperate with these transcription factors to activate the matrilysin promoter, the effects of inactivating point mutations in each putative Ets binding site (Fig. 1A) on PEA3-dependent transactivation were determined.
No Ets site mutation had a large effect on basal promoter activity, although the mutant promoters consistently tended to have a higher basal activity (Fig. 5). In these experiments, PEA3 stimulated wild-type −296HMAT-Luc 7.5-fold and each of the Ets site mutant reporters were stimulated 3- to 6-fold. Hence, no single Ets site mutation was sufficient to eliminate the PEA3 responsiveness of the matrilysin promoter.
FIG. 5.
Inactivation of the Ets sites affects PEA3 synergy with c-Jun and β-catenin–LEF-1. Inactivating point mutations of each Ets binding site were introduced into the −296MAT-Luc construct to create Mut(−168Ets), Mut(−144)Ets, and Mut(−55Ets). Wild-type and mutant reporters were cotransfected in parallel with combinations of the PEA3, c-Jun, LEF-1, and ΔN89β-catenin expression vectors into HEK293 cells. Lysates were analyzed for luciferase activity 16 to 20 h after transfection. Data are presented as RLUs normalized to cotransfected simian virus 40-driven Renilla luciferase. Data bars represent the means of three experiments, each performed in triplicate. Error bars represent standard errors.
Compared to PEA3 stimulation alone, c-Jun activated the wild-type reporter an additional 4-fold and the −168 Ets mutant an additional 2.5-fold. However, c-Jun failed to significantly coactivate either the −144 or −55 Ets mutant reporters. Thus, the Ets sites flanking the AP-1 site both appeared to be important for sensitizing the matrilysin promoter to c-Jun transactivation of the promoter.
The combination of LEF-1 and β-catenin coexpression with PEA3 activated the wild-type matrilysin reporter an additional 4.3-fold above PEA3 alone, with each protein contributing approximately 2-fold additional activation. The activation of the wild-type reporter by β-catenin–LEF-1 was also about 4-fold when coexpressed with PEA3 and c-Jun. The−168 Ets mutant reporter was not responsive to β-catenin–LEF-1 when they were coexpressed with PEA3 and showed only an additional 1.7-fold activation when coexpressed with PEA3 and c-Jun. The −144 Ets mutant reporter was completely unresponsive to β-catenin–LEF-1, regardless of c-Jun expression. The −55 Ets site mutant reporter was stimulated by β-catenin–LEF-1 more than 2-fold in both the absence and presence of c-Jun. In summary, each of the Ets sites seemed to contribute to PEA3 synergy with β-catenin–LEF-1, but the Ets sites flanked by the Tcf sites in the matrilysin promoter, −168 and −144, appeared to be especially critical for this cooperation. Interestingly, the central −144 Ets site was important for PEA3 cooperation with both β-catenin–LEF-1 and c-Jun.
Synergistic cooperation of PEA3 with β-catenin requires the activity of the transcriptional coactivator p300.
The proximity of the cooperative Ets and Tcf binding sites suggested the possibility that PEA3 and β-catenin may interact physically as well as functionally. To test this possibility, HEK293 cells were cotransfected with PEA3 and β-catenin expression vectors, and cell lysates were harvested 48 h later. β-Catenin-containing complexes were immunoprecipitated using a polyclonal anti-β-catenin antibody and, following SDS-PAGE, were immunoblotted using an anti-PEA3 monoclonal antibody. PEA3 was consistently detected in these β-catenin immunoprecipitates (Fig. 6A), suggesting that these proteins associate intracellularly. Reciprocal coimmunoprecipitations using anti-PEA3 for immunoprecipitation and anti-β-catenin antibodies for immunoblotting were difficult to interpret due to β-catenin association with both agarose and Sepharose beads, in the absence of primary antibody, under these precipitation conditions. However, the inclusion of the anti-PEA3 primary antibody consistently enriched the amount of β-catenin in the precipitated complexes three- to fivefold (data not shown).
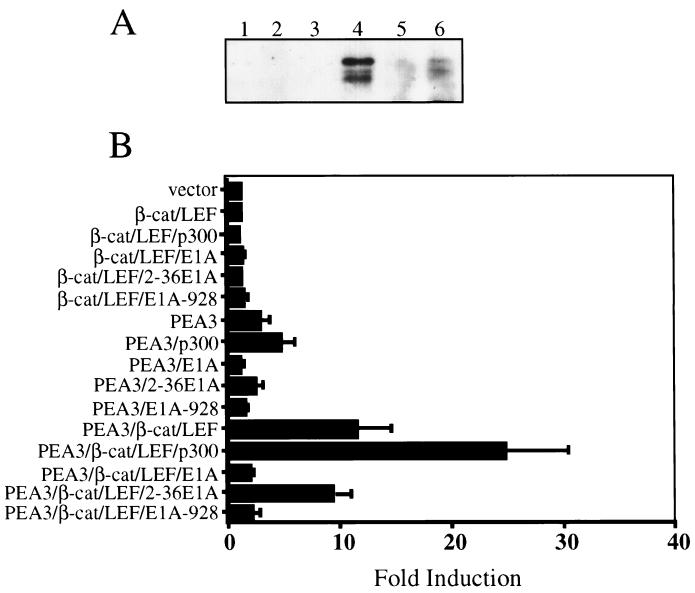
FIG. 6.
PEA3–β-catenin synergy depends upon the activity of p300. (A) PEA3 coimmunoprecipitates with β-catenin (β-cat). pCAN-ΔN89β-cat was transfected independently (lanes 1 to 3) or cotransfected with pCAN-PEA3 (lanes 4 to 6) into HEK293 cells. After 48 h, total cell lysates were immunoprecipitated with a rabbit anti-β-catenin polyclonal antibody. After being washed, the precipitated proteins were resuspended in SDS-PAGE running buffer and subjected to Western blotting with a mouse monoclonal anti-PEA3 antibody. Lanes 1 and 4, 7 μl of input lysate; lanes 2 and 5, no-antibody control; lanes 3 and 6, anti-β-catenin immunoprecipitation. (B) p300 enhances and E1A blocks PEA3 synergy with β-catenin–LEF-1. A total of 1 × 105 HEK293 cells were cotransfected with −296MAT-Luc and combinations of LEF-1, β-catenin, and PEA3 expression vectors as indicated. With each combination was included either an expression vector encoding p300, wild-type E1A (E1A), a mutant E1A with its p300 interaction domain deleted (2–36E1A), or a mutant E1A with its pRB interaction domain deleted (E1A-928). Lysates were analyzed for luciferase activity 24 h after transfection. Data are presented as fold induction relative to cotransfection of the reporter with empty vectors. Data bars represent the means of three experiments, each performed in triplicate, and represent fold induction relative to the empty-vector control. Error bars represent standard errors.
The observation that β-catenin and PEA3 protein can associate in intracellular protein complexes suggested a mechanism of transcriptional synergy wherein transcription factor complexes serve to accommodate the binding of coactivators on their target promoters (37, 40). Because many recent studies have shown that β-catenin interacts with the transcriptional coactivator p300 (21, 48), we hypothesized that PEA3–β-catenin synergy might require p300 activity. This mechanism of synergy is considered to be particularly relevant in cells where p300 is very limited, as would be the case in HEK293 cells, which express adenovirus E1A, a protein that sequesters p300 from cellular promoters. To test if PEA3 synergy with β-catenin–LEF-1 was responsive to and dependent upon p300 activity, PEA3 and β-catenin–LEF-1 were coexpressed with p300, wild-type E1A, or mutants of E1A (47). In this study, two E1A mutants were used; one cannot interact with p300 (2–36E1A), whereas the other cannot interact with the retinoblastoma gene product (E1A-928).
p300 expression was not capable of rendering the matrilysin reporter responsive to β-catenin–LEF-1 expression (Fig. 6B), but it did enhance PEA3 activation of the matrilysin promoter about twofold. In the reciprocal experiments, wild-type E1A and E1A-928 completely blocked PEA3 stimulation of the promoter, whereas the 2–36E1A mutant had a minor inhibitory effect.
p300 cooperated with PEA3–LEF-1–β-catenin activation, boosting the 11-fold activation by PEA3–LEF-1–β-catenin to almost 27-fold. As with PEA3, expression of either wild-type E1A or the E1A-928 mutant was capable of completely blocking PEA3–LEF-1–β-catenin activation of the matrilysin reporter. The 2–36E1A mutant again had a minor negative effect on the level of transactivation by PEA3–LEF-1–β-catenin.
Not surprisingly, the transactivation of the matrilysin promoter by p300 in conjunction with PEA3–β-catenin–LEF-1 required functional Ets and Tcf binding sites (data not shown), particularly the −168 and −144 Ets sites and both Tcf sites. These data support the hypothesis that synergy of PEA3 and β-catenin–LEF-1 is related to an ability to coordinately recruit p300 to the matrilysin promoter.
The PEA3 subfamily is frequently expressed in mouse intestinal tumors and human colon tumor cell lines.
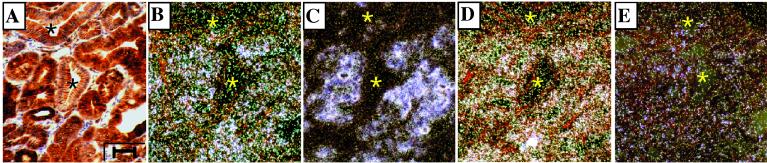
The goal of this study was to identify those trans-acting factors that act in concert with β-catenin–Tcf to activate matrilysin gene expression in vivo, particularly in intestinal tumors. If, as our data suggest, the PEA3 subfamily members are candidates to fulfill such a role, they should be commonly expressed in intestinal tumors. To determine whether this is the case, in situ hybridization was carried out on 22 small intestinal adenomas from the Min mouse. In this study, 86% (19 of 22) of the Min tumors expressed matrilysin, 77% (17 of 22) expressed PEA3, 100% (7 of 7) expressed ERM, and 86% (6 of 7) expressed ER81 (data not shown). Of the tumors examined for all members of the PEA3 subfamily, each coexpressed two or more subfamily members at elevated levels within the tumor cells (Fig. 7B, D, and E). In tumors that expressed them, PEA3 and ERM were consistently elevated in the tumor epithelium, as defined by high levels of β-catenin accumulation, compared to the nearby normal epithelium. ER81, on the other hand, was frequently found at equal levels in the tumor epithelium, normal epithelium, and, rarely, in the surrounding stroma. Therefore, while ER81 expression is frequently found in Min mouse tumors, its pattern of expression was generally distinct from that of the other two PEA3 subfamily members. Nevertheless, ER81 was selectively upregulated in the tumor epithelium in some Min tumors (3 of 7) in a manner that correlated with matrilysin expression (data not shown). Matrilysin expression was consistently found where β-catenin protein accumulation and PEA3 subfamily expression overlapped (Fig. 7).
FIG. 7.
Matrilysin expression in mouse intestinal tumors overlaps β-catenin protein accumulation and PEA3 subfamily expression. Shown are serial sections of a Min mouse tumor showing β-catenin protein immunohistochemistry (A) and in situ hybridization for PEA3 (B), matrilysin (C), ERM (D), and ER81 (E) transcripts. The asterisks mark glandular structures with junctional β-catenin localization and little to no PEA3, matrilysin, or ERM expression. The surrounding less-organized structures have accumulated β-catenin, PEA3, matrilysin, and ERM expression. ER81 expression is low and sporadic, not significantly overlapping with β-catenin, matrilysin, or the other PEA3 subfamily members. Size bar = 40 μm.
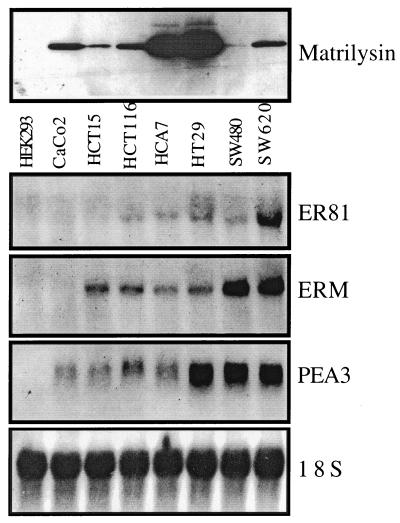
We also examined the expression of PEA3 subfamily transcripts in human colon tumor cells by Northern analysis on total RNA isolated from the CaCo-2, HCT15, HCT116, HCA7, HT29, SW480, and SW620 colon tumor lines as well as HEK293 cells (Fig. 8). Each colon tumor line has stable β-catenin (26) and expresses matrilysin protein (Fig. 8, upper panel). ER81 was found in all lines except CaCo-2 and HCT15, while ERM was found in all lines except CaCo-2. PEA3 transcripts were found in all of the colon tumor cell lines examined. Thus, matrilysin expression is common in human and mouse intestinal tumor cells that have both stable β-catenin protein and PEA3 subfamily expression.
FIG. 8.
Matrilysin and the PEA3 subfamily are expressed in every human colon tumor cell line examined. (Upper panel) Western blot of matrilysin protein secreted into the media from the HEK293 cells and the human colon tumor cell lines CaCo-2, HCT15, HCT116, HCA7, HT29, SW480, and SW620. (Lower panels) Northern analysis was performed using 15 μg of total RNA from the same cell lines and the blot was probed using 3′ untranslated region probes specific for human ER81, ERM, or PEA3. 18S rRNA is shown as a loading control.
DISCUSSION
In a previous study of β-catenin transactivation of the matrilysin promoter, it was hypothesized that nuclear β-catenin was not sufficient for matrilysin expression (9). In most colon tumor cell lines examined, the matrilysin promoter was responsive to β-catenin overexpression alone, suggesting that any other proteins necessary for matrilysin promoter activity were constitutively present in these cells. The HEK293 kidney epithelial cell line was chosen as a background distinct from colon adenocarcinoma to identify relevant transcription factors that cooperate with β-catenin to activate matrilysin gene expression. Here we have shown that the activity of the PEA3 subfamily of the Ets transcription factor family, of which PEA3 and ERM were most commonly upregulated in intestinal tumor cells, rendered the matrilysin promoter responsive to transactivation by β-catenin–Tcf as well as by the AP-1 factor c-Jun. In these cells, as well as other immortal cell lines tested, such as COS-7 and NIH 3T3 (data not shown), neither β-catenin, LEF-1, nor c-Jun had any effect on matrilysin promoter activity unless they were coexpressed with a member of the PEA3 subfamily. With PEA3 coexpression, β-catenin, LEF-1, and c-Jun synergistically transactivated the matrilysin promoter and together induced transcription from the endogenous matrilysin gene in the HEK293 cells.
Many mechanisms have been described for β-catenin–LEF-1 transactivation that lend themselves to synergy with other transcription factors. DNA bending, the original described mechanism by which LEF-1 was shown to transactivate the T-cell receptor (TCR) enhancer (15), required the binding of nearby transcription factors. The organization of the human matrilysin promoter bears a resemblance to that of the TCR enhancer in that Ets and AP-1 binding sites flank the Tcf binding sites. However, DNA bending is an unlikely mechanism for synergy on the matrilysin promoter for a number of reasons, including the following: (i) the matrilysin Tcf site sequences are not compatible with LEF-1 bending (33), (ii) LEF-1 missing its β-catenin binding region still binds and bends DNA (33) but did not synergize with PEA3 and c-Jun in our study, and (iii) mutation of the AP-1 site does not impair LEF-1 transactivation in the presence of PEA3 expression. The recent finding that β-catenin interacts with p300 (21, 48) and other reports of transcription factor synergy being mediated by stabilization of p300 on specific promoters (37, 40) led us to examine the role of p300 in PEA3–β-catenin–LEF-1 synergy. We found that p300 could indeed enhance transactivation of the matrilysin promoter and that PEA3 synergy with β-catenin–LEF-1 required endogenous p300 activity. This, combined with our observation that PEA3 protein could be coimmunoprecipitated with β-catenin, suggests that PEA3–β-catenin–LEF-1 can associate in a protein complex capable of bringing p300 to the matrilysin promoter.
Although c-Jun expression is not required for synergy between PEA3 and β-catenin–LEF-1, our studies clearly show that c-Jun is a powerful activator of the matrilysin promoter when it is coexpressed with these proteins. Although the frequency of c-Jun overexpression in intestinal tumors has been shown to be lower than that for either the PEA3 subfamily or matrilysin (35), our data likely reflect an important contribution of AP-1 complexes in general to the overall level of matrilysin expression. Indeed, in additional studies, JunB and JunD also synergized with PEA3 and β-catenin–LEF-1 to different degrees (data not shown), indicating that AP-1 activity, not just complexes containing c-Jun, can modulate the level of matrilysin transcription. On the other hand, unlike the Ets sites in the matrilysin promoter, whose inactivation has profound effects upon activation by both c-Jun and β-catenin–LEF-1, the AP-1 site affects only activation by c-Jun. Therefore, we conclude that c-Jun is not a requirement for matrilysin expression but that the AP-1 complex is an important modulator of matrilysin expression levels. Furthermore, these data draw a distinction between factors required to initiate matrilysin transcription and other factors, both positive and negative, that modulate the overall level of matrilysin production in intestinal tumor cells.
This is the first description of the expression of the PEA3 subfamily members in intestinal tumors. Their frequent expression in Min mouse tumor cells and human colon tumor cell lines suggests that the members of the PEA3 subfamily are targets of a common early alteration in a tumor-associated signaling pathway. Ets factors have been described as targets of Ras signaling (53). However, Min mouse adenomas do not have mutated Ras (46), nor do the human colon tumor cell lines HCA7 and HT29 (42). Thus, if Ras signaling is involved in PEA3 regulation in intestinal tumors, it is just as likely to be a result of extracellular signals mediated by Ras, such as epidermal growth factor (EGF) receptor signaling. EGF receptor signaling has been suggested to have relevance in human colon tumor progression (8) and has very recently been implicated in Min mouse tumor formation (51).
The matrilysin gene is not the only β-catenin–Tcf-responsive gene that has Ets binding sites in close proximity to Tcf sites. The human cyclin D1 (50), c-myc (20), and TCF-1 (43) promoters and the Xenopus fibronectin (16) and siamois (6) promoters, each of which has been shown to be regulated by Wnt or β-catenin–Tcf, all have candidate Ets binding sites within 20 bp of Tcf sites. Thus, it is possible that the close physical association of Ets and Tcf sites may have been selected for throughout evolution and is not simply a phenomenon confined to the matrilysin promoters. Indeed, while the manuscript of this article was in preparation, analysis of the Drosophila Eve enhancer revealed cooperation between Ets factors and Wnt signaling (19). We have also seen cooperation between PEA3 and β-catenin on the cyclin D1 promoter (unpublished data). Therefore, the close association of Tcf sites and Ets sites, as well as the synergistic mode of regulation, is not unique to the human and mouse matrilysin promoters and may represent a novel oncogene-responsive element common to many genes important for both development and progression of intestinal tumors.
The fact that matrilysin Tcf site artificial promoters are responsive to β-catenin overexpression, whereas the intact matrilysin promoter is not, emphasizes that not all genes with functional Tcf sites in their promoters are unconditional targets of β-catenin transactivation. Both the context of the natural promoter, as defined by other transcription factor binding sites, and the context of the cell, as defined by the expression of endogenous transcription activators and repressors, must be taken into consideration. Our results suggest that the responsiveness of the matrilysin promoter in some colon tumor cell lines is probably dependent upon both the preexisting expression of endogenous PEA3 subfamily members and the levels of endogenous β-catenin–Tcf complexes (9). Adding to the complexity of the cellular context, it has been reported that TCF-1 is both a target and an attenuator of β-catenin–Tcf transactivation (43). Hence, our work emphasizes that it is the complex interaction of the natural promoter context with the cellular context that defines what is a β-catenin target gene in any given circumstance. As a result, it is likely that there will be sets of β-catenin–Tcf target genes that will be distinct in different cell types or in the same cell type at different stages of differentiation during tissue development or tumor progression. We believe that the synergistic relationship that β-catenin exhibits with PEA3 and its relatives will be a recurring theme that will dictate the ability of β-catenin to act as a transcriptional activator of multiple genes and, by extension, its ability to act as an oncogene.
ACKNOWLEDGMENTS
We thank Jeff Fisher and Bonnie Bojovic for technical assistance. We thank Elaine Fuchs, Barbara Graves, Scott Hiebert, David Livingston, Dennis Watson, Ronald Wisdom, and Roland Stein for their plasmid gifts. DNA sequencing was performed by the Vanderbilt University sequencing facility.
This work was supported by NIH grant P30 CA68485 (Vanderbilt University sequencing facility). This work was also supported by NIH grant R01-CA 60867 (to L.M.M.), ACS Pilot Project grant IRG-58–009-41 (to H.C.C.), and by funding from the Canadian Institutes for Health Research and the Canadian Breast Cancer Research Initiative (to J.A.H.).
REFERENCES
- 1.Aberle H, Schwartz H, Kemler R. Cadherin-catenin complex: protein interactions and their implications for cadherin function. J Cell Biochem. 1996;61:514–523. doi: 10.1002/(SICI)1097-4644(19960616)61:4%3C514::AID-JCB4%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 2.Behrens J, von Kries J P, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 3.Benbow U, Brinckerhoff C E. The AP-1 site and MMP gene regulation: what is all the fuss about? Matrix Biol. 1997;15:519–526. doi: 10.1016/s0945-053x(97)90026-3. [DOI] [PubMed] [Google Scholar]
- 4.Billin A N, Thirlwell H, Ayer D E. β-Catenin–histone deacetylase interactions regulate the transition of LEF1 from a transcriptional repressor to an activator. Mol Cell Biol. 2000;20:6882–6890. doi: 10.1128/mcb.20.18.6882-6890.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brabletz T, Jung A, Dag S, Hlubek F, Kirchner T. Beta-catenin regulates the expression of the matrix metalloproteinase-7 in human colorectal cancer. Am J Pathol. 1999;155:1033–1038. doi: 10.1016/s0002-9440(10)65204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brannon M, Gomperts M, Sumoy L, Moon R T, Kimelman D. A beta-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes Dev. 1997;11:2359–2370. doi: 10.1101/gad.11.18.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavallo R A, Cox R T, Moline M M, Roose J, Polevoy G A, Clevers H, Peifer M, Bejsovec A. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- 8.Coffey R J, McCutchen C M, Graves-Deal R, Polk W H., Jr Transforming growth factors and related peptides in gastrointestinal neoplasia. J Cell Biochem Suppl. 1992;16G:111–118. doi: 10.1002/jcb.240501120. [DOI] [PubMed] [Google Scholar]
- 9.Crawford H C, Fingleton B M, Rudolph-Owen L A, Heppner Goss K J, Rubinfeld B, Polakis P, Matrisian L M. The metalloproteinase matrilysin is a target of beta-catenin transactivation in intestinal tumors. Oncogene. 1999;18:2883–2891. doi: 10.1038/sj.onc.1202627. [DOI] [PubMed] [Google Scholar]
- 10.Crawford H C, Matrisian L M. Mechanisms controlling the transcription of matrix metalloproteinase genes in normal and neoplastic cells. Enzyme Protein. 1996;49:20–37. doi: 10.1159/000468614. [DOI] [PubMed] [Google Scholar]
- 11.Denhardt D T. Oncogene-initiated aberrant signaling engenders the metastatic phenotype: synergistic transcription factor interactions are targets for cancer therapy. Crit Rev Oncogen. 1996;7:261–291. doi: 10.1615/critrevoncog.v7.i3-4.70. [DOI] [PubMed] [Google Scholar]
- 12.Fagotto F, Gluck U, Gumbiner B M. Nuclear localization signal-independent and importin/karyopherin-independent nuclear import of beta-catenin. Curr Biol. 1998;8:181–190. doi: 10.1016/s0960-9822(98)70082-x. [DOI] [PubMed] [Google Scholar]
- 13.Gaire M, Magbanua Z, McDonnell S, McNeil L, Lovett D H, Matrisian L M. Structure and expression of the human gene for the matrix metalloproteinase matrilysin. J Biol Chem. 1994;269:2032–2040. [PubMed] [Google Scholar]
- 14.Giambernardi T A, Grant G M, Taylor G P, Hay R J, Maher V M, McCormick J J, Klebe R J. Overview of matrix metalloproteinase expression in cultured human cells. Matrix Biol. 1998;16:483–496. doi: 10.1016/s0945-053x(98)90019-1. [DOI] [PubMed] [Google Scholar]
- 15.Giese K, Kingsley C, Kirshner J R, Grosschedl R. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 16.Gradl D, Kuhl M, Wedlich D. The Wnt/Wg signal transducer beta-catenin controls fibronectin expression. Mol Cell Biol. 1999;19:5576–5587. doi: 10.1128/mcb.19.8.5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graves B J, Petersen J M. Specificity within the ets family of transcription factors. Adv Cancer Res. 1998;75:1–55. doi: 10.1016/s0065-230x(08)60738-1. [DOI] [PubMed] [Google Scholar]
- 18.Gutman A, Wasylyk B. Nuclear targets for transcriptional regulation by oncogenes. Trends Genet. 1991;7:49–54. doi: 10.1016/0168-9525(91)90231-E. [DOI] [PubMed] [Google Scholar]
- 19.Halfon M S, Carmena A, Gisselbrecht S, Sacherson C M, Jimenez F, Baylies M K, Michelson A M. Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell. 2000;103:63–74. doi: 10.1016/s0092-8674(00)00105-7. [DOI] [PubMed] [Google Scholar]
- 20.He T C, Sparks A B, Rago C, Hermeking H, Zawel L, Da Costa L T, Morin P J, Vogelstein B, Kinzler K W. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 21.Hecht A, Vleminckx K, Stemmler M P, Van Roy F, Kemler R. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 2000;19:1839–1850. doi: 10.1093/emboj/19.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henderson B R. Nuclear-cytoplasmic shuttling of APC regulates beta-catenin subcellular localization and turnover. Nat Cell Biol. 2000;2:653–660. doi: 10.1038/35023605. [DOI] [PubMed] [Google Scholar]
- 23.Hiebert S W, Sun W, Davis J N, Golub T, Shurtleff S, Buijs A, Downing J R, Grosveld G, Roussel M F, Gilliland D G, Lenny N, Meyers S. The t(12;21) translocation converts AML-1B from an activator to a repressor of transcription. Mol Cell Biol. 1996;16:1349–1355. doi: 10.1128/mcb.16.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 25.Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann B G, Kemler R. Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mech Dev. 1996;59:3–10. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- 26.Ilyas M, Tomlinson I P, Rowan A, Pignatelli M, Bodmer W F. Beta-catenin mutations in cell lines established from human colorectal cancers. Proc Natl Acad Sci USA. 1997;94:10330–10334. doi: 10.1073/pnas.94.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinzler K W, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 28.Klemsz J M, McKercher S R, Celada A, van Beveren C, Maki R A. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell. 1990;61:113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 29.Kolligs F T, Hu G, Dang C V, Fearon E R. Neoplastic transformation of RK3E by mutant β-catenin requires deregulation of Tcf/Lef transcription but not activation of c-myc expression. Mol Cell Biol. 1999;19:5696–5706. doi: 10.1128/mcb.19.8.5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korinek V, Barker N, Morin P J, van Wichen D, de Weger R, Kinzler K W, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 31.Leprince D, Duterque-Coquillaud M, Li R-P, Henry C, Flourens A, Debuire B, Stehelin D. Alternative splicing within the chicken c-ets-1 locus: implications for transduction within the E26 retrovirus of the c-ets proto-oncogene. J Virol. 1988;62:3233–3241. doi: 10.1128/jvi.62.9.3233-3241.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levanon D, Goldstein R E, Bernstein Y, Tang H, Goldenberg D, Stifani S, Paroush Z, Groner Y. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc Natl Acad Sci USA. 1998;95:11590–11595. doi: 10.1073/pnas.95.20.11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Love J J, Li X, Case D A, Giese K, Grosschedl R, Wright P E. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature. 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- 34.Luckow B, Schutz G. CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res. 1987;15:5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magrisso I J, Richmond R E, Carter J H, Pross C B, Gilfillen R A, Carter H W. Immunohistochemical detection of RAS, JUN, FOS, and p53 oncoprotein expression in human colorectal adenomas and carcinomas. Lab Investig. 1993;69:674–681. [PubMed] [Google Scholar]
- 36.Mann B, Gelos M, Siedow A, Hanski M L, Gratchev A, Ilyas M, Bodmer W F, Moyer M P, Riecken E O, Buhr H J, Hanski C. Target genes of beta-catenin-T cell-factor/lymphoid-enhancer-factor signaling in human colorectal carcinomas. Proc Natl Acad Sci USA. 1999;96:1603–1608. doi: 10.1073/pnas.96.4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merika M, Williams A J, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 38.Munemitsu S, Albert I, Rubinfeld B, Polakis P. Deletion of an amino-terminal sequence stabilizes β-catenin in vivo and promotes hyperphosphorylation of the adenomatous polyposis coli tumor suppressor protein. Mol Cell Biol. 1996;16:4088–4094. doi: 10.1128/mcb.16.8.4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Regulation of intracellular beta-catenin levels by the adenomatous polyposis coli (APC) tumor-suppressor protein. Proc Natl Acad Sci USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakashima K, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Kawabata M, Miyazono K, Taga T. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science. 1999;284:479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- 41.Newell K J, Witty J P, Rodgers W H, Matrisian L M. Expression and localization of matrix-degrading metalloproteinases during colorectal tumorigenesis. Mol Carcinog. 1994;10:199–206. doi: 10.1002/mc.2940100404. [DOI] [PubMed] [Google Scholar]
- 42.Rajesh D, Schell K, Verma A K. Ras mutation, irrespective of cell type and p53 status, determines a cell's destiny to undergo apoptosis by okadaic acid, an inhibitor of protein phosphatase 1 and 2A. Mol Pharmacol. 1999;56:515–525. doi: 10.1124/mol.56.3.515. [DOI] [PubMed] [Google Scholar]
- 43.Roose J, Huls G, van Beest M, Moerer P, van der Horn K, Goldschmeding R, Logtenberg T, Clevers H. Synergy between tumor suppressor APC and the beta-catenin-Tcf4 target Tcf1. Science. 1999;285:1923–1926. doi: 10.1126/science.285.5435.1923. [DOI] [PubMed] [Google Scholar]
- 44.Seth A, Robinson L, Thompson D M, Watson D K, Papas T S. Transactivation of GATA-1 promoter with ETS1, ETS2 and ERGB/Hu-FLI-1 proteins: stabilization of the ETS1 protein binding on GATA-1 promoter sequences by monoclonal antibody. Oncogene. 1993;8:1783–1790. [PubMed] [Google Scholar]
- 45.Shattuck-Brandt R L, Lamps L W, Heppner Goss K J, DuBois R N, Matrisian L M. Matrilysin and cyclooxygenase-2 are differentially expressed in intestinal and colorectal neoplasms. Mol Carcinog. 1999;24:177–187. [PubMed] [Google Scholar]
- 46.Shoemaker A R, Luongo C, Moser A R, Marton L J, Dove W F. Somatic mutational mechanisms involved in intestinal tumor formation in Min mice. Cancer Res. 1997;57:1999–2006. [PubMed] [Google Scholar]
- 47.Stein R W, Corrigan M, Yaciuk P, Whelan J, Moran E. Analysis of E1A-mediated growth regulation functions: binding of the 300-kilodalton cellular product correlates with E1A enhancer repression function and DNA synthesis-inducing activity. J Virol. 1990;64:4421–4427. doi: 10.1128/jvi.64.9.4421-4427.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takemaru K I, Moon R T. The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J Cell Biol. 2000;149:249–254. doi: 10.1083/jcb.149.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takeuchi N, Ichikawa T, Momiyama N, Hasegawa S, Nagashima Y, Miyazaki K, Koshikawa N, Mitsuhashi M, Shimada H. Matrilysin gene expression in sporadic and familial colorectal adenomas. Mol Carcinog. 1997;19:225–229. [PubMed] [Google Scholar]
- 50.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 51.Torrance C J, Jackson P E, Montgomery E, Kinzler K W, Vogelstein B, Wissner A, Nunes M, Frost P, Discafani C M. Combinatorial chemoprevention of intestinal neoplasia. Nat Med. 2000;6:1024–1028. doi: 10.1038/79534. [DOI] [PubMed] [Google Scholar]
- 52.Umesono K, Murakami K K, Thompson C C, Evans R M. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wasylyk B, Hagman J, Gutierrez-Hartmann A. Ets transcription factors: nuclear effectors of the Ras-MAP-kinase signaling pathway. Trends Biochem Sci. 1998;23:213–216. doi: 10.1016/s0968-0004(98)01211-0. [DOI] [PubMed] [Google Scholar]
- 54.Wilson C L, Heppner K J, Labosky P A, Hogan B L M, Matrisian L M. Intestinal tumorigenesis is suppressed in mice lacking the metalloproteinase matrilysin. Proc Natl Acad Sci USA. 1997;94:1402–1407. doi: 10.1073/pnas.94.4.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson C L, Heppner K J, Rudolph L A, Matrisian L M. The metalloproteinase matrilysin is preferentially expressed by epithelial cells in a tissue-restricted pattern in the mouse. Mol Biol Cell. 1995;6:851–869. doi: 10.1091/mbc.6.7.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wisdom R. AP-1: one switch for many signals. Exp Cell Res. 1999;253:180–185. doi: 10.1006/excr.1999.4685. [DOI] [PubMed] [Google Scholar]
- 57.Witty J P, McDonnell S, Newell K, Cannon P, Navre M, Tressler R, Matrisian L M. Modulation of matrilysin levels in colon carcinoma cell lines affects tumorigenicity in vivo. Cancer Res. 1994;54:4805–4812. [PubMed] [Google Scholar]
- 58.Zhou P, Byrne C, Jacobs J, Fuchs E. Lymphoid enhancer factor 1 directs hair follicle patterning and epithelial cell fate. Genes Dev. 1995;9:700–713. doi: 10.1101/gad.9.6.700. [DOI] [PubMed] [Google Scholar]