Abstract
The TATA sequence of the human, estrogen-responsive pS2 promoter is complexed in vivo with a rotationally and translationally positioned nucleosome (NUC T). Using a chromatin immunoprecipitation assay, we demonstrate that TATA binding protein (TBP) does not detectably interact with this genomic binding site in MCF-7 cells in the absence of transcriptional stimuli. Estrogen stimulation of these cells results in hyperacetylation of both histones H3 and H4 within the pS2 chromatin encompassing NUC T and the TATA sequence. Concurrently, TBP becomes associated with the pS2 promoter region. The relationship between histone hyperacetylation and the binding of TBP was assayed in vitro using an in vivo-assembled nucleosomal array over the pS2 promoter. With chromatin in its basal state, the binding of TBP to the pS2 TATA sequence at the edge of NUC T was severely restricted, consistent with our in vivo data. Acetylation of the core histones facilitated the binding of TBP to this nucleosomal TATA sequence. Therefore, we demonstrate that one specific, functional consequence of induced histone acetylation at a native promoter is the alleviation of nucleosome-mediated repression of the binding of TBP. Our data support a fundamental role for histone acetylation at genomic promoters in transcriptional activation by nuclear receptors and provide a general mechanism for rapid and reversible transcriptional activation from a chromatin template.
Transcription by RNA polymerase II (RNA pol II) from most eukaryotic promoters requires the evolutionarily conserved interaction of TATA binding protein (TBP) with a TATA sequence (42) in a chromatin context. Transcriptional activity, at least in yeast, correlates strongly with the degree of TBP occupancy of TATA box elements (30, 32). However, TBP can be severely inhibited from binding DNA sites located within unaltered mononucleosomes (17, 24). Positioning of a nucleosome over the TATA region appears to be a common mechanism for repressing basal transcription. In yeast, the TATA sequences of the inactive PHO5 (2), ADH2 (54), GAL80 (34, 35), and CHA1 (37) promoters are all contained within positioned nucleosomes that are disrupted during induction-dependent chromatin rearrangements. Similar mechanisms have been observed in other organisms. Both the repressed human immunodeficiency virus type 1 promoter in unstimulated human T cells (53) and the inactive beta phaseolin gene in vegetative tobacco tissues (31) contain nucleosomes that occlude their respective TATA sequences and are subsequently disrupted concomitant with transcriptional activation.
Of the multiple modifications required for conversion of chromatin from an inactive to active state, one consistent feature of active chromatin is the highly acetylated state of the core histones in the nucleosomes (13, 22, 52). Core histone acetylation influences both the interaction of specific proteins with nucleosomal DNA and the activation of gene expression (reviewed in reference 57). A broad range of transcriptional regulatory proteins possess intrinsic histone acetylase and deacetylase activity (for review, see references 49, 56, and 61). Included among the proteins that possess histone acetyltransferase (HAT) activity are several nuclear receptor coactivators that interact directly with the estrogen receptor (ER), including SRC-1/NCoA-1, ACTR/RAC3/(P/CIP), TIF2/GRIP1/NCoA-2, and CBP (also called p300), as well as the CBP-associated factor (P/CAF) (11). Of these coactivators, the HAT activity of p300 has recently been demonstrated to be critical for hormone-induced histone H4 hyperacetylation on the pS2 promoter (9). The coactivators CBP and pCAF can also acetylate nonhistone proteins, including transcription factors p53, E2F1, ELKF, GATA 1, TFIIF, and TFIIEβ and the nuclear receptor coactivator ACTR (reviewed in reference 29). While the crucial role of the acetylase activity of the nuclear receptor coactivators in hormone-induced gene regulation has been demonstrated (9), the functional consequences of acetylation of core histones remain unresolved.
Given the complexity of the structural parameters governing binding of transcription factors to nucleosomal templates, an understanding of transcriptional activation at a natural promoter requires knowledge of the native chromatin structure. To this end, we have previously mapped the chromatin structure of the human, estrogen-responsive pS2 promoter within the context of its normal genomic location in human mammary epithelial cells (48). The TATA box at −30 to −24 of the pS2 promoter is situated at the 3′ edge of a rotationally and translationally positioned nucleosome, NUC T (at nucleotides −23 to −165). NUC T remains stably positioned even upon the transcriptional induction of the gene in vivo (48). This contrasts with the previously discussed nucleosomal disruptions over other transcriptionally active TATA sequences (2, 31, 34, 35, 37, 53, 54) and suggests an alternative mechanism of alleviating nucleosomal repression of the binding of TBP. The pS2 promoter can be activated through several estrogen receptor-dependent pathways, including direct binding of steroid hormone to the receptor (6) and phosphorylation of the receptor resulting from activation of the protein kinase C (PKC) pathway (39) or the Ras mitogen-activated protein kinase cascade of the growth factor signaling pathway (28). Here we examine how induction by estradiol and the PKC pathway results in transcriptional activation of the pS2 gene on a chromatin template. First, we establish that the binding of TBP is intrinsically linked to ER-dependent transcriptional activation of the pS2 promoter. Second, we demonstrate that the core histones H3 and H4 of the nucleosomal pS2 promoter are differentially hyperacetylated in response to these transcriptional stimuli. And most significantly, we demonstrate that hyperacetylation of core histones facilitates the binding of the basal transcription factor, TBP, to the TATA sequence within the pS2 chromatin.
MATERIALS AND METHODS
Cell cultures.
CMT cells (an African green monkey kidney cell line that stably produces simian virus 40 [SV40] T antigen) and the human breast adenocarcinoma cell line MCF-7 were maintained in Dulbecco's modified Eagle's medium with 10% fetal calf serum in a 5% CO2 atmosphere at 37°C. Proliferating MCF-7 cells were grown in phenol red-free EX-CELL 320, serum-free media (JRH Biosciences) to eliminate any estrogenic activity from the media for 48 h prior to experimental treatment and chromatin immunoprecipitation (ChIP). In addition, the antiestrogen ICI 182,780 (Tocris) was added at 2 μM for this 48-h period to the unstimulated samples to fully block ER activation of the pS2 promoter. Cells were stimulated with 200 nM 17β-estradiol (Sigma) and 100 ng of tetradecanoyl phorbol acetate (TPA) (Sigma)/ml for 3 h prior to harvesting for ChIP assays.
ChIP and in vivo TBP binding assay.
Approximately 3 × 108 MCF-7 cells were used for each experimental group. ChIPs were performed essentially as previously described (5; as adapted for mammalian cell culture per instructions of Upstate Biotechnology). After the designated treatments, cells were fixed by the direct addition of formaldehyde to the culture medium to a final concentration of 1% and incubation for 10 min at 37°C. Cells were washed with ice-cold phosphate-buffered saline containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μg of aprotinin/ml, and 1 μg of pepstatin A/ml) and were lysed with 1.2 ml of sodium dodecyl sulfate (SDS) lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8.1]) containing the same protease inhibitors. This lysate was sonicated three times for 30 s using a Branson Sonifer 250 at 20% constant maximal power, resulting in an average DNA length of approximately 500 bp, as determined by electrophoresis through a 1% agarose–Tris-acetate-EDTA gel and ethidium bromide staining. After clarification, the supernatant fraction was diluted 10-fold in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.1], 167 mM NaCl) to a final volume of 12 ml. Protein levels of the 12-ml samples were quantitated with a detergent-compatible protein assay kit (Bio-Rad) per manufacturer's directions, and protein concentrations were equalized. Immunoprecipitations were performed by incubating 2 ml of sample at 4°C overnight with saturating amounts of antibodies: 10 μl of anti-acetyl-histone H4 rabbit antiserum or 10 μg of anti-acetyl-histone H3 rabbit polyclonal immunoglobulin G antibody (Upstate Biotechnology). Isolation of the immunocomplexes and further purification of the coprecipitated DNA were conducted as described previously (5). Specific sequences in the immunoprecipitates were detected by PCR, under conditions optimized for each primer set in which the yield of product was within the linear range relative to input DNA (data not shown). Identical immunoprecipitates were assayed for levels of the pS2 proximal promoter sequence, using primer set TE (5′-ATGGGCTTCATGAGCTCC and 5′-GCGACCCCGAGTCAGG), and for the control sequence, beta interferon proximal promoter sequence, using primer set F (41). Immunoprecipitations and PCRs were performed in duplicate in each experiment, and multiple independent experiments were performed to ensure reproducibility.
Measurement of the in vivo interaction of TBP with the pS2 proximal promoter was performed by modifying the ChIP protocol. Instead of anti-acetylated histone antibodies, 25 μg of anti-TBP antibodies (Santa Cruz Biotechnology) was used to immunoprecipitate cross-linked protein-DNA complexes followed by quantitative PCR analysis of the purified MCF-7 DNA fragments.
MC and core MC isolation.
CMT cells were infected with the recombinant SV40/pS2 promoter virus SVSpS2, and minichromosomes (MCs) were isolated and purified as described previously (21). Where indicated, CMT cells were treated with 400 nM trichostatin A (TSA) (Waco Pure Chemical Inc.) 3 h prior to MC isolation (21). Core MCs were isolated after incubation in 0.5 M NaCl as described previously (16), except that bovine serum albumin (BSA) (Worthington Biochemicals) was added to a concentration of 0.1 mg/ml before the final dialysis step at 4°C against 30 mM HEPES (pH 7.5), 1 mM EDTA, 0.1% Nonidet P-40 (ICN Biomedicals Inc.), 1 mM 2-mercaptoethanol, and 15% sucrose.
TBP-DNA binding reactions.
Human TBP, a glutathione transferase-tagged fusion protein (Santa Cruz Biotechnology), was added to either SVSpS2 viral DNA or to the indicated MCs. Human TFIIA, added to MCs in addition to TBP, was overexpressed in bacteria and was purified by affinity chromatography as described previously (14). The binding reactions for TBP with free DNA contained 55 mM KCl, 10 mM NaCl, 6 mM MgCl2, 0.1 mg of BSA/ml, 20 mM HEPES (pH 7.5), 20% glycerol, and 10 mM dithiothreitol. The binding reactions for TBP on MCs and core MCs contained 47 mM KCl, 10 mM NaCl, 3 mM MgCl2, 0.1 mg of BSA/ml, 20 mM HEPES (pH 7.5), 10% glycerol, 3% sucrose, and 10 mM dithiothreitol. In addition, the core MC preparation added a final concentration of 0.2 mM EDTA, 0.2 mM 2-mercaptoethanol, and 0.02% Nonidet P-40 to the reactions. All binding reactions were carried out at 30°C for 45 min in a final volume of 30 μl. Following this incubation with TBP, DNA or MCs were digested with DNase I (Worthington Biochemicals) at 0.005 or 0.05 μg/reaction mixture, respectively. The digestions were performed by adding 30 μl of DNase I in 150 mM sucrose, 80 mM KCl, 35 mM HEPES (pH 7.5), 5 mM K2HPO4, 5 mM MgCl2, 0.1 mg of BSA/ml, and 2 mM CaCl2 to the 30-μl binding reaction mixture for 2 min at 20°C. DNase I cleavage was terminated by the addition of 1 μg of EcoRI-digested genomic DNA from CMT cells and 5 μg of yeast tRNA in 60 μl of 20 mM EDTA and 20 mM Tris (pH 7.8). This mixture was placed on ice for 10 min, followed by the addition of 0.2% cetyltrimethylammonium bromide, and was incubated at 20°C for 10 min, and the DNA was precipitated by centrifugation in a microcentrifuge for 10 min at 15,000 × g. The cetyltrimethylammonium bromide was removed by resuspension of the DNA-tRNA pellet in 3 M sodium acetate, followed by ethanol precipitation. Ligation-mediated PCR (LMPCR) of 25 ng of isolated DNA was performed with primer set NTV-T, as described previously (48). The end-labeled PCR product was visualized on preflashed Reflection NEF-495 autoradiography film (Dupont) following electrophoresis through a 6% polyacrylamide–7 M urea gel (1:15, bisacrylamide:acrylamide).
Micrococcal nuclease and Western blot analysis of chromatin templates.
The nucleosomal structure of the pS2 promoter within MCs, core MCs, and genomic DNA in MCF-7 cells was monitored by micrococcal nuclease digestion and LMPCR as previously described (48). For Western blot analysis, 500 ng of MC DNA (containing approximately 500 ng of core histones) was electrophoresed through an SDS–12.5% polyacrylamide gel. Proteins were transferred onto a polyscreen polyvinylidene difluoride membrane (New England Nuclear), and the membranes were incubated with either anti-acetylated-histone H4 rabbit antiserum (1:2,000 dilution) or anti-acetylated-histone H3 rabbit polyclonal immunoglobulin G antibody (2 μg/ml) (Upstate Biotechnology) in phosphate-buffered saline containing 3% nonfat milk. Acetylated histone H4 and H3 blots were subsequently incubated with anti-rabbit horseradish peroxidase-conjugated antibody (1:3,000) (Bio-Rad) and anti-rabbit alkaline phosphatase-conjugated antibody (1:2,000), respectively. Immunoreactive species were visualized by chemiluminescence using either a Dupont NEN Renaissance kit and autoradiography or Amersham Pharmacia ECF Western blotting kit and Fluorimager (Molecular Dynamics) per manufacturers' directions. Additionally, duplicate samples not subjected to Western blotting were electrophoresed and visualized by silver staining, per directions of the manufacturer (Daiichi Co.).
RESULTS
ER activation induces hyperacetylation of histones and binding of TBP within the pS2 promoter.
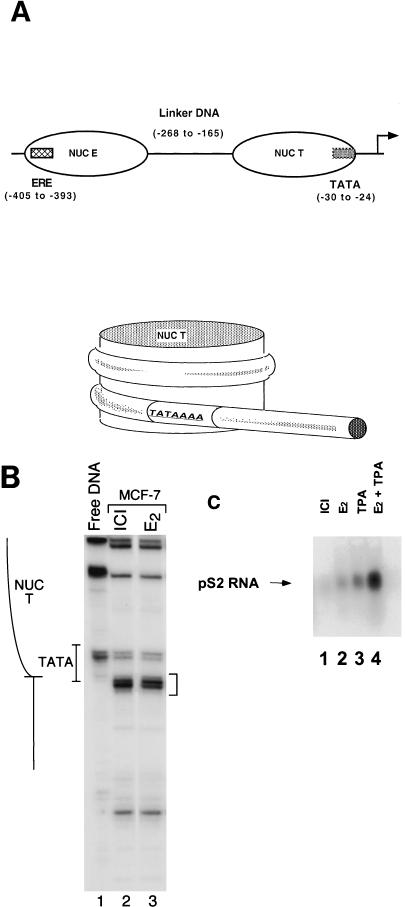
The basal transcription factor TBP can be severely inhibited from binding DNA sites located within unaltered mononucleosomes (17, 24). In the human estrogen-responsive pS2 promoter, the TATA sequence lies within a tightly positioned nucleosome (Fig. 1A) whose translational boundary is defined by nucleosome-dependent micrococcal nuclease-hypersensitive sites (48) (Fig. 1B, bracket). Strikingly, this nucleosome positioning remains unchanged upon transcriptional induction of the gene (48) (Fig. 1B, lanes 2 and 3). Given that the nucleosome-covering TATA sequence is not disrupted upon induction, we examined whether NUC T might be altered instead by modifications that would allow the binding of TBP. In particular, the biological relevance of histone acetylation of the human pS2 proximal promoter to activation with ER-dependent stimuli was investigated in the human MCF-7 cell line, a breast cancer cell line that constitutively expresses the ER. Transcriptional activation of the pS2 gene both by estradiol and by the PKC activator TPA is dependent upon the ER, and induction by these pathways is synergistic (10, 26) (Fig. 1C, lanes 2 to 4). In our experiments, levels of pS2 mRNA in the MCF-7 cells were stimulated 5-fold by estradiol, 9.5-fold by TPA, and 29-fold by a combination of the two agents. Transcriptional induction by both stimuli is inhibited by pure antiestrogens (8).
FIG. 1.
The translational positioning of the nucleosome overlapping the TATA sequence is unchanged upon induction of the pS2 promoter. (A) The upper panel is a linear representation of the pS2 proximal promoter illustrating the positions of the estrogen response element (ERE) (hatched box) and TATA sequence (grey box) relative to the positions of the two nucleosomes (NUC E and NUC T). The transcriptional start site is denoted with an arrow. The lower panel further illustrates the positioning of the TATA sequence (nucleotides −30 to −24) of the human estrogen-responsive pS2 promoter within the rotationally and translationally positioned NUC T (nucleotides −23 to −165) (48). (B) The boundary of NUC T is defined by chromatin-dependent micrococcal nuclease cleavages (bracketed on right) and is situated within the 3′ edge of the TATA sequence (48). MCF-7 cells were treated with either the antiestrogen ICI 182,780 (ICI) or 17β-estradiol (E2) before chromatin was digested in vivo by addition of micrococcal nuclease. For comparison, purified genomic DNA (Free DNA) was digested in vitro and similarly analyzed. (C) Levels of pS2 RNA were determined by Northern blot analysis following treatment of MCF-7 cells for 48 h with ICI 182,780 (ICI) or for 3 h with 17β-estradiol (E2) and/or TPA. Twenty micrograms of total isolated MCF-7 RNA per sample was electrophoresed through a 1.2% formaldehyde-agarose gel and transferred to a charged nylon membrane (NEN). The pS2 RNA was detected by hybridization with a random primed DNA probe generated from the pS2 gene sequence.
The acetylation states of histones H3 and H4 within the pS2 promoter were analyzed under our experimental conditions using the ChIP assay. Sheared cellular chromatin was immunoprecipitated with antibodies raised against either a diacetylated synthetic peptide corresponding to residues 1 to 21 of histone H3 or a tetra-acetylated peptide corresponding to residues 2 to 19 of histone H4. As a control, chromatin was also mock immunoprecipitated in the absence of antibody. The DNA from these precipitates was initially analyzed by quantitative PCR utilizing primers that flank sequences containing NUC T. The background level of recovered DNA in the mock immunoprecipitates was identical, whether derived from cells treated with ICI, estradiol, or TPA (data not shown).
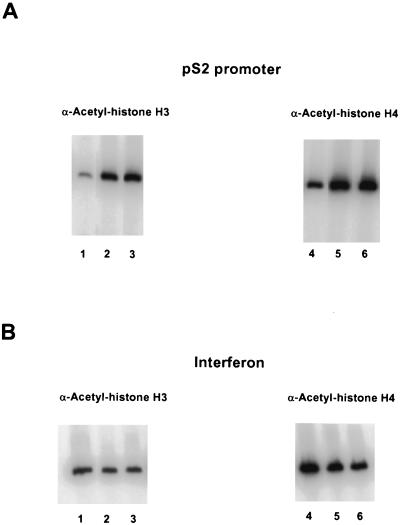
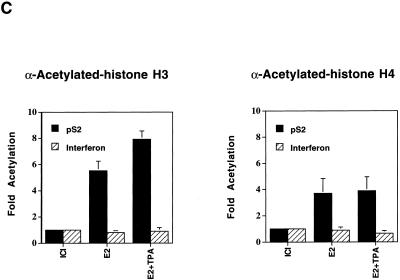
With the ChIP assays, significant changes in modification states were demonstrated upon induction of gene expression. With antibodies reactive against acetylated histone H3, we observed a significant enhancement in immunoprecipitation of the pS2 promoter region in response to treatment with estrogen, as compared to treatment with the antiestrogen ICI, and an even greater enhancement upon treatment with both estradiol and TPA (Fig. 2A and C). A somewhat distinct result was obtained with antibodies reactive against acetylated histone H4. As compared to levels of the pS2 promoter DNA associated with acetylated histone H4 in the presence of ICI, equivalent increases were observed upon treatment with estradiol either in the presence or absence of TPA (Fig. 2A and C). We note that the amount of pS2 DNA immunoprecipitated from ICI-treated samples by the H4 antibodies was much higher than the amount immunoprecipitated by the H3 antibodies. This may reflect a higher basal level of histone H4 acetylation within the pS2 proximal promoter region in its uninduced state or may reflect recognition of all acetylated forms of histone H4 by the anti-acetylated-histone H4 antibodies (33), in contrast to recognition of only highly acetylated isoforms of histone H3 by the anti-acetylated-histone H3 antibodies (4).
FIG. 2.
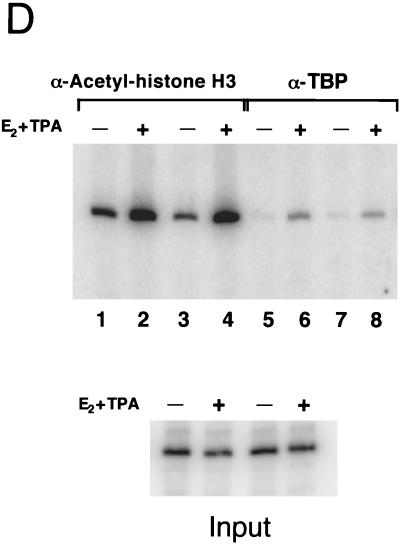
Hyperacetylation of histones H3 and H4 and binding of TBP within the pS2 proximal promoter in response to the transcriptional stimuli estradiol and TPA. (A) Chromatin fragments were immunoprecipitated with anti-acetylated-histone H3 or H4 antibody as indicated, from human mammary MCF-7 cells treated with the pure antiestrogen ICI 182,780 (lanes 1 and 4), 17β-estradiol (lanes 2 and 5), or 17β-estradiol plus TPA (lanes 3 and 6). DNA purified from these immunoprecipitations was analyzed by quantitative PCR, with the product consisting of nucleotides −440 to +18 of the pS2 promoter. (B) The same immunoprecipitates described for panel A (same lane designations) were analyzed by quantitative PCR with primer set F from the human beta interferon promoter region (−241 to −1) (41). (C) The relative levels of DNA immunoprecipitated with antibodies against acetylated histones H3 and H4 at the pS2 and beta interferon promoters were quantitated using a PhosphorImager (Molecular Dynamics). The average amounts of recovered DNA, with standard deviations, were determined from three independent sets of experiments with separate batches of cells. Levels of DNA immunoprecipitated with anti-acetylated-histone H3 antibodies from ICI-treated cells were similar (within twofold) to background levels of precipitated DNA in the absence of antibodies. Thus, the actual enhancement in association of pS2 proximal promoter DNA with highly acetylated histone H3 may be even more pronounced. (D) Genomic chromatin fragments from MCF-7 cells, treated as described for panel A with either ICI 182,780 (lanes 1, 3, 5, and 7) or 17β-estradiol plus TPA (E2 + TPA) (lanes 2, 4, 6, and 8), were immunoprecipitated either with anti-TBP antibodies (lanes 5 to 8) or with anti-acetylated-histone H3 antibodies (lanes 1 to 4). −, absence of E2 + TPA; +, presence of E2 + TPA. Duplicate experimental samples are shown to illustrate reproducibility. The relative levels of TBP binding to the proximal pS2 promoter and of immunoprecipitation with the anti-acetylated histone H3 antibody of this region were analyzed by quantitative PCR. The average increase in TBP binding with transcriptional stimuli was 4.1 ± 1.1-fold over background levels, based upon four independent experiments. Quantitative PCR analysis was performed on DNA isolated from input samples prior to immunoprecipitation to confirm equalization of initial cellular material.
In order to ensure that the enhanced histone acetylation associated with the pS2 promoter did not reflect global nucleosome modifications, we examined the acetylation state of the chromatin associated with the beta interferon proximal promoter region, encompassing its TATA sequence. Analysis of the identical chromatin immunoprecipitates in three independent experiments revealed no statistical change in the acetylation state of histones H3 or H4 associated with this control promoter upon addition of estradiol and TPA (Fig. 2B and C). This correlates with nondetectable levels of beta interferon mRNA in samples isolated from MCF-7 cells treated with ICI or estradiol and TPA, as analyzed by Northern blotting (data not shown).
In these assays, the levels of histone acetylation were determined following 3 h of exposure of the cells to the hormone. However, increases in acetylation of both histones H3 and H4 were observed as early as 15 min after treatment with estradiol (data not shown). These data are generally consistent with the recent findings of Chen and colleagues (9), although they differ in details (see Discussion).
The influence of transcriptional stimuli on the binding of the critical downstream general transcription factor, TBP, to the genomic pS2 proximal promoter was also examined using an immunoprecipitation assay. The experimental design was identical to the ChIP assay, except for the use of anti-TBP antibodies. The basal level of recovered DNA after a mock immunoprecipitation protocol was the same as the level of DNA recovered in anti-TBP immunoprecipitates from extracts of ICI-treated cells (data not shown). Thus, in the presence of the pure antiestrogen ICI 182,780, TBP did not detectably interact with this pS2 proximal promoter region, which includes the TATA sequence (Fig. 2D, lanes 5 and 7). This is a significant result, as TBP is sometimes associated with cellular promoters that remain in an inactive state (23).
Activation of ER by estradiol and TPA induced the binding of TBP to the pS2 promoter (Fig. 2D, compare lanes 5 and 7 with lanes 6 and 8). The level of binding of TBP in the induced state was fourfold higher than background levels. However, given that background levels in the absence of transcriptional stimuli were the same with or without anti-TBP antibodies, the actual extent to which transcriptional activation of the genomic pS2 promoter by ER promotes binding of TBP is undoubtedly much greater.
The nucleosome naturally positioned over the pS2 TATA sequence severely restricts accessibility to TBP binding.
Given that core histones complexed with the genomic pS2 promoter are specifically hyperacetylated in response to transcriptional stimuli and that these same stimuli lead to increased binding of TBP, we investigated a possible functional connection at this promoter between histone acetylation and the binding of this basal transcription factor. Assembly of the preinitiation complex at most RNA pol II promoters is critically dependent on the binding of TBP with its associated factors to a TATA sequence, as RNA pol II cannot directly recognize target initiation sites (reviewed in reference 7). The binding of TBP to the TATA box within the positioned nucleosome of the human pS2 promoter was studied using a minichromosome model system. Previously, it was demonstrated that nucleotides −1100 to +10 of the pS2 promoter are sufficient to establish the positioning of NUC T, as this region of the promoter, when placed in the context of a recombinant SV40 viral genome, generates in vivo-assembled chromatin templates with structural features virtually identical to those of the genomic pS2 promoter (48). SV40 MCs are comprised of an array of the normal cellular nucleosomes, including histone H1, complexed with the viral genome. Nonhistone proteins are also bound to the MCs (for review, see references 12 and 15).
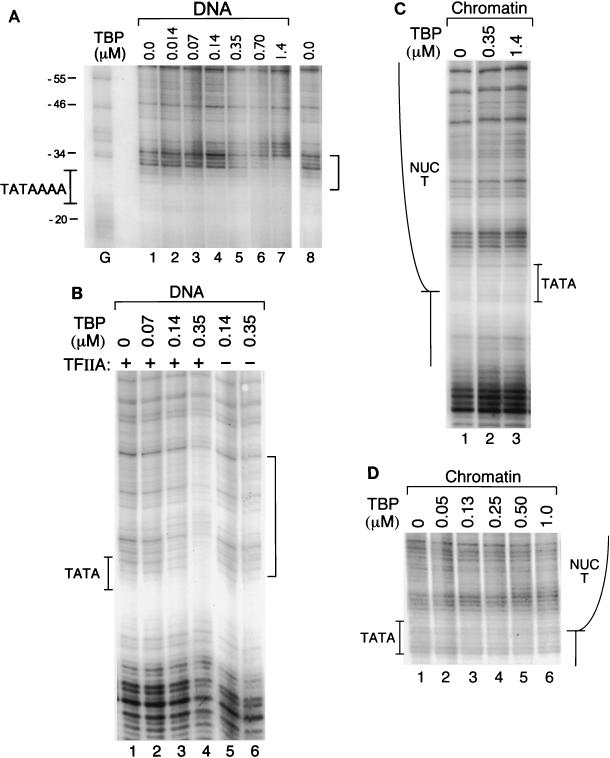
The basic characteristics of the TBP footprinting pattern and its binding avidity to the pS2 TATA sequence were initially defined on deproteinized, recombinant viral DNA (Fig. 3A). Due to the sequence specificity of DNase I, there was little cleavage of the free DNA directly within the TATA sequences (Fig. 3A, lane 1). However, a characteristic footprint was nonetheless evident upon addition of increasing amounts of TBP. The binding of TBP resulted in protection against cleavage at the 5′ edge of the TATA box from nucleotides −35 to −29 (Fig. 3A, lanes 5 to 7, bracket). DNase I footprints of TBP extending 5′ of the TATA sequence are commonly observed, such as on the adenovirus major late promoter TATA sequence, where the protected footprint extends to nucleotide −36 (62). The interaction of TBP with the TATA sequence also caused several base pairs immediately 5′ of the footprinted region (nucleotides −37 and −38) to become hypersensitive to DNase I cleavage. DNase I-hypersensitive sites have also been observed on the adenovirus major late and human heat shock protein 70 promoters, 5′ of the regions footprinted by TFIID (38). The apparent affinity of TBP for the free pS2 TATA sequence is quite weak. In these experiments, the half-maximal concentration for binding is nearly 0.6 μM TBP (0.2 μM active TBP). Thus, the apparent Kd is approximately 2 orders of magnitude lower than the reported affinity of TBP, 2.0 nM, for the adenovirus major late promoter TATA box (19, 25). This is due partly to differing sequences flanking the TATA boxes and also to binding conditions that are necessitated by the subsequent assays utilizing MCs (see below). In particular, BSA was present in our assays at concentrations that were previously shown to reduce the affinity of TBP for the adenovirus major late promoter TATA box by fivefold (25).
FIG. 3.
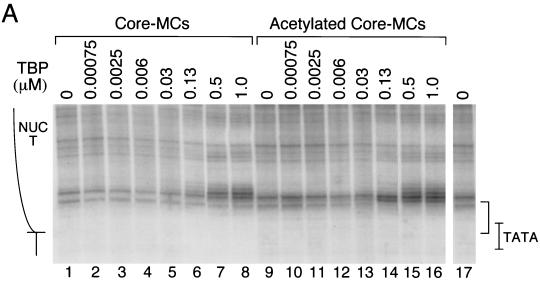
The binding of TBP to the pS2 TATA sequence is severely restricted by the in vivo chromatin structure of the promoter. (A) Purified viral DNA containing the pS2 promoter (100 ng, 1.0 nM) was incubated with increasing concentrations of human TBP, at the concentrations (in micromolars) indicated above the individual lanes. Subsequently, the reaction mixtures were digested with DNase I, and the DNA products were amplified by LMPCR (48). TBP preparations were assayed for the percentages of TBP molecules that were active for specific DNA binding as described previously (25). The preparation used for the experiments illustrated in Fig. 3D, 4A, and 5A contained essentially all active protein, while the preparation used for the experiments shown in Fig. 3A to C was approximately 30% as active. A control DNA sample was incubated in the absence of TBP (lanes 1 and 8). Dimethyl sulfate/piperidine-treated SVSpS2 viral DNA provided a G ladder (lane G); the nucleotide positions of the pS2 promoter are indicated to the left of this lane. The position of the TATA box is illustrated by the pS2 sequence TATAAAA. The binding of TBP is evidenced by a region of protection from cleavage as delineated by the bracket to the right. (B) Purified viral DNA containing the pS2 promoter was incubated with TBP at the protein concentrations indicated above the individual lanes but supplemented either with the basal transcription factor TFIIA (+) or with TFIIB (lanes 5 and 6). TFIIA and TFIIB concentrations equaled or exceeded that of TBP; TFIIA concentrations were as follows: lanes 1 and 2, 0.13 μM; lane 3, 0.25 μM; lane 4, 0.63 μM. TFIIB concentrations were as follows: lane 5, 0.14 μM; lane 6, 0.35 μM. The designations are as given for panel A. (C) MCs (230 pM) containing the pS2 promoter were isolated from CMT cells and were incubated with TBP at the concentrations noted. The position of NUC T is diagrammed on the left. The remaining designations are as noted above. (D) MCs were incubated with TBP as described for panel C, with the addition of the basal transcription factor TFIIA. TFIIA concentrations were as follows: lanes 1 to 4, 0.25 μM; lane 5, 0.50 μM; lane 6, 1.0 μM.
The general initiation factor TFIIA can enhance the binding of TBP to naked DNA when TBP binding conditions are suboptimal (25, 62), and it has therefore been included in previous studies of binding of TBP to reconstituted nucleosomal templates (17, 24). In the presence of TFIIA, the affinity of TBP for the naked pS2 TATA sequence under these binding conditions was increased but by less than 10-fold (compare Fig. 3A, lanes 5 and 6, with Fig. 3B, lane 4). On this DNA, the presence of TFIIA also enlarged the footprint significantly upstream of the TATA sequence (compare the bracketed areas in Fig. 3A and B).
When TBP alone was added in a parallel experiment to recombinant viral chromatin containing the pS2 promoter, no detectable footprint was observed even at the highest concentrations of TBP (Fig. 3C, lane 3). Thus, the binding of TBP to the pS2 TATA sequence was strongly inhibited within the context of the positioned NUC T and surrounding chromatin, despite location of the TATA sequence at the extreme edge of NUC T (Fig. 1B). Upon inclusion of TFIIA, TBP binding to the nucleosomal pS2 TATA sequence in the MC context was still only minimally detectable at the highest concentrations of TBP (Fig. 3D and 4A, lanes 1 to 6). Although the evidence for an interaction between TBP and unmodified chromatin in the presence of TFIIA was subtle, often only the appearance of DNase I hypersensitivity at nucleotides −37 and −38, this result was reproducible. TFIIA was included in all subsequent experiments.
FIG. 4.
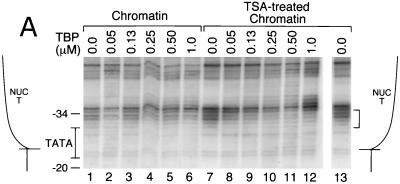
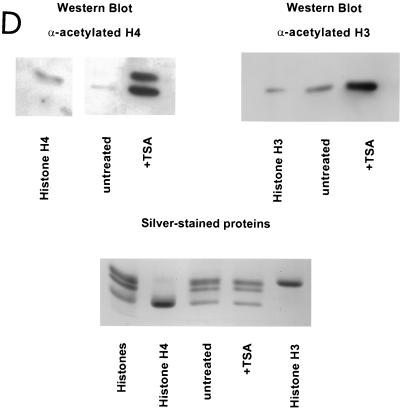
MCs isolated from TSA-treated cells support enhanced levels of TBP binding to the pS2 TATA sequences within NUC T. (A) MCs (230 pM) isolated from either untreated cells (lanes 1 to 6) or TSA-treated cells (lanes 7 to 13) were incubated with increasing concentrations of TBP for DNase I footprinting. These same MC preparations were analyzed for panel D for the acetylation state of histones H3 and H4. The concentration of TBP is indicated above each lane. TFIIA concentrations were as follows: lanes 1 to 4, 7 to 10, and 13, 0.25 μM; lanes 5 and 11, 0.50 μM; lanes 6 and 12, 1.0 μM. The position of NUC T is diagrammed on both sides of the lanes. The nucleotide positions within the pS2 promoter are numbered to the left. The region of highly specific, TBP-dependent protection from DNase I cleavage (bracket) is indicated to the right. The results shown in this figure are representative of four independent experiments. (B) The bands within the TBP-dependent footprint region (panel A, bracket) were scanned using a PhosphorImager (Molecular Dynamics). The upper graph illustrates the TBP interaction with normal chromatin (panel A, lanes 1, 3, and 5); the lower graph illustrates the TBP interaction with TSA-treated chromatin (Fig. 4A, lanes 7, 9, and 11). The positions of the nucleotides −30 to −27 (TATA) are indicated above their respective peaks. The x axis represents the relative position within the gel, while the y axis illustrates band intensity. The brackets denote the region of DNase I protection. (C) The relative DNase I accessibility of the TBP-dependent footprint region was compared for the two preparations of chromatin. The intensities of DNase I cleavage within the TBP footprint, denoted by brackets in panels A and B, were quantitated and corrected for total lane intensity by dividing this measurement by the intensity of a region within the lane unaffected by the binding of TBP (3). The accessibility without the addition of TBP was arbitrarily set at 1, with increased protection by TBP resulting in lower accessibility to DNase I. DNA sequences that became hypersensitive to DNase I were not included as part of the footprinted region. (D) Western blot analysis of the acetylation level of histones H4 and H3, from MCs isolated from either untreated or TSA-treated (+TSA) cells (preparations used for panel A). The indicated antibodies were used to probe the histone modification state. The anti-acetylated-histone H4 rabbit antiserum also interacts with acetylated histone H2B under conditions of substantial excess antibody (Upstate Biotechnology Co., personal communication). Marker lanes contained high-performance liquid chromatography-purified chicken erythrocyte histones H3 and H4. MC samples were also analyzed by staining with silver (bottom panel). The amounts of histones in the MCs from untreated versus TSA-treated cells were comparable. However, TSA treatment led to an accumulation of hyperacetylated histones H3 and H4.
Enhanced binding of TBP to the nucleosomal pS2 TATA sequence in the context of highly acetylated chromatin.
Given the extremely weak interaction of TBP with the nucleosomal pS2 TATA sequence, a role for histone acetylation on the binding of TBP to the nucleosomal pS2 promoter was tested, using highly acetylated chromatin templates. In order to isolate such chromatin, cells were infected with the recombinant SV40 virus containing the pS2 promoter and were treated subsequently with TSA. By inhibiting histone deacetylase activity, TSA causes accumulation in vivo of highly acetylated core histones (63). Western blotting analyses confirmed the higher levels of acetylation of histones H3 and H4 within MCs isolated from cells incubated with TSA (Fig. 4D).
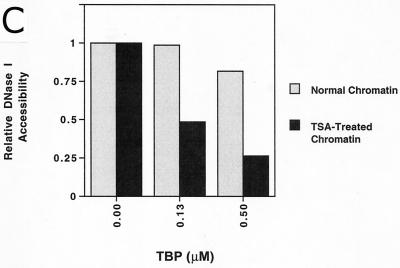
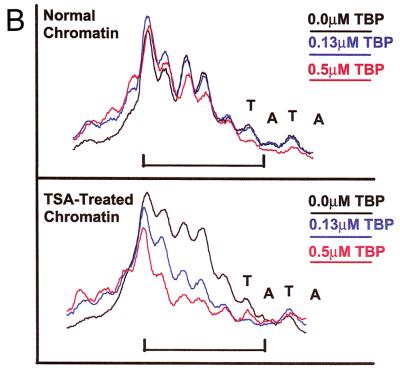
Incubation of TBP with this highly acetylated chromatin led to stable, highly specific binding at the pS2 TATA sequence (Fig. 4A, lanes 7 to 13, footprint over 5′ half of TATA box, nucleotides −30 to −34, as marked by bracket). The binding of TBP was dramatically enhanced as compared to its binding to MCs isolated from untreated cells. The differences in binding to the two chromatin preparations were quantitated both as protection from DNase I cleavage of individual bases (Fig. 4B) and as protection of the entire footprinted region relative to a region outside of the binding area (3) (Fig. 4C). At a TBP concentration (130 nM) resulting in 50% occupancy of the pS2 TATA sequence within the TSA-treated MCs (Fig. 4A, lane 9; B, blue line, TSA-treated chromatin; and C, 0.13 μM), binding to the same sequence within the unmodified MCs was undetectable (Fig. 4A, lane 3; B, blue line, normal chromatin; and C, 0.13 μM). In fact, acetylation of the MCs and the presence of TFIIA permitted levels of binding of TBP to the nucleosomal pS2 TATA sequence roughly comparable to those for binding of pure TBP to free DNA (compare Fig. 3A and 4A, noting the correction for the specific concentration of active TBP in each experiment, as indicated in the Fig. 3A legend).
Acetylation specifically of the core histones facilitates binding of TBP to NUC T.
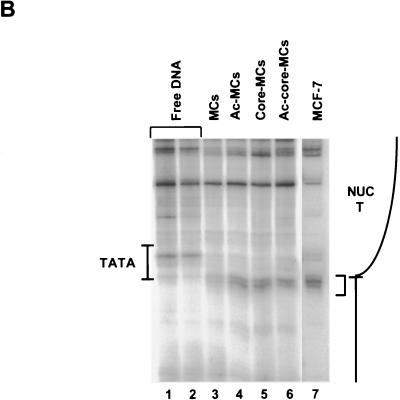
MCs assembled in vivo contain many chromatin components that may be acetylated in addition to the core histones (for example, see reference 9). In order to test definitively whether acetylation specifically of the core histones on the MCs facilitated the binding of TBP, we analyzed MCs containing only the core histones. Incubation of the native MC preparations with 0.5 M NaCl prior to purification by sucrose gradient sedimentation dissociates histone H1, along with all nonhistone proteins normally associated with native MCs, while leaving the core nucleosomes intact (16). Such core MCs were prepared from both normal and highly acetylated preparations of MCs. It was critical to verify that the in vivo translational positioning of NUC T was retained following the high-salt treatment. Therefore, both MCs and core MCs were analyzed for positioned nucleosomes by digestion with micrococcal nuclease. Nucleosome-dependent micrococcal nuclease-hypersensitive cleavages at nucleotide positions −23 and −24 are indicative of the edge of NUC T in the genomic pS2 promoter (48) (Fig. 1B and 5B, lane 7). These hypersensitive cleavages were evident in all purified MC and core MC samples of the recombinant SV40/pS2 promoter virus SVSpS2 (Fig. 5B, lanes 3 to 6) but not in the deproteinated viral DNA samples (Fig. 1B, lane 1, and 5B, lanes 1 to 2), proving that the nucleosomes remained positioned over the appropriate sequences under all conditions.
FIG. 5.
Binding of TBP to NUC T is facilitated by acetylation of core histones. (A) Core MCs, containing only the core histones assembled in vivo with recombinant pS2 promoter-containing viral DNA, were prepared from MCs isolated from either untreated or TSA-treated, infected cells. The core MC preparations were incubated prior to DNase I digestion with increasing concentrations of TBP as shown. TFIIA concentrations are as follows: lanes 1 to 6, 9 to 14, and 17, 0.25 μM; lanes 7 and 15, 0.50 μM; lanes 8 and 16, 1.0 μM. Control reaction mixtures were incubated without TBP (lanes 1, 9, and 17). The designations are as given for Fig. 4A. The results shown in this figure are representative of four independent experiments. The detailed pattern of DNase I cleavages near the TATA sequence obtained here differed slightly from the pattern obtained for Fig. 3C and D and 4A. In our experience, the final step of the LMPCR procedure that generates the radiolabeled DNA products yields slight variability from experiment to experiment in the pattern of relative intensities of different bands. However, within any single experiment, consistency in the detailed DNase I cleavage patterns is observed. (B) The translational positioning of NUC T is strictly maintained on the salt-treated core MCs. MCs and core MCs, both normal and more highly acetylated (Ac) (lanes 3 to 6), were digested with micrococcal nuclease (0.05 U) prior to LMPCR amplification to establish the boundary of NUC T. In vivo micrococcal nuclease cleavage of genomic DNA within MCF-7 human breast cells, followed by LMPCR (48), demonstrates the in vivo translational positioning of NUC T (lane 7). Micrococcal nuclease cleavage (lane 1, 0.005 U; lane 2, 0.01 U) of purified recombinant viral DNA (Free DNA) is shown as controls. The enhanced cleavages present in all the chromatin preparations but not in the free DNA are delineated with a bracket. The positions of NUC T and the TATA box are indicated schematically at the sides of the lanes.
Stable binding of TBP was readily observed on the core nucleosomal templates prepared from highly acetylated MCs (Fig. 5A, bracket, lanes 9 to 17), as evidenced by complete protection from DNase I cleavage within the 5′ region of the TATA sequence. The footprint in this experiment encompassed protection of one major cleavage site near the 5′ end of the TATA sequence, as well as two minor cleavage sites in the center and on the 3′ end. In contrast, very limited protection from DNase I cleavage was observed within this region on normal core MCs when titrated with increasing amounts of TBP (Fig. 5A, lanes 1 to 8). These data indicate that acetylation of core histones is sufficient to facilitate the binding of TBP to its nucleosomal site in the pS2 promoter.
At high concentrations of TBP, enhanced sensitivity to DNase I cleavage upstream of the TATA sequence was obtained on both normal and highly acetylated core MCs (Fig. 5A, lanes 6 to 8 and 14 to 16, region above bracket). Notably, on the acetylated core MCs, significant amounts of hypersensitivity occurred at TBP concentrations higher than those sufficient to observe DNase protection. While a DNase I footprint results from continuous protection of a specific DNA sequence from nuclease throughout the entire digestion period, thereby signifying a stable interaction between a protein and DNA, hypersensitivity to cleavage by DNase I can capture a transient interaction between a protein and DNA. Thus, we infer that transient binding of TBP to the normal core MCs did occur, with the dissociation rate being too fast to permit a stable interaction at the TATA sequence.
DISCUSSION
Many reports, beginning with in vitro transcription studies of the adenovirus major late promoter, have demonstrated that when a TATA sequence is placed within a chromatin context, competition between the binding of TBP and of the core histones leads to repression of transcription by nucleosomes (36, 58, 59). Similarly, we demonstrate that on a native promoter, a highly positioned nucleosome, NUC T, represses binding of TBP to the pS2 TATA sequence in vitro within an array of positioned nucleosomes. Furthermore, we provide evidence that repression by chromatin occurs at this promoter in vivo, as the binding of TBP to the genomic pS2 TATA sequence is repressed in the absence of activated ER (Fig. 2D).
How such inhibition can be overcome is critical for understanding transcriptional activation of this type of promoter. Our data are the first to directly show that acetylation of the core histones is sufficient to permit a stable interaction between TBP and a nucleosomal TATA sequence at a native promoter. In a previous report, the presence of the amino-terminal tails of the core histones in a mononucleosome was shown to decrease the accessibility of a nucleosomal TATA box (17). These data were interpreted to suggest that histone acetylation would enhance the binding of TBP to a nucleosomal TATA sequence. However, histone acetylation and proteolytic tail removal have recently been shown to have distinct effects on nucleosomes, with histone acetylation unexpectedly increasing nucleosome stability (55). Thus, effects of acetylation must be the result of defined changes in the specific structures and conformations adopted by the histone tails rather than a general charge neutralization effect (20). In our study, the binding of TBP was not only investigated with acetylated histones but also on a naturally positioned nucleosomal array in a native promoter. This provides biochemical evidence of histone acetylation facilitating the binding of TBP in a biologically relevant context.
ER and histone acetylation.
The facilitated binding of TBP is a critical step in our proposed model of transcriptional activation of the genomic pS2 promoter by active ER. We suggest that binding of ER to the nucleosomal pS2 template recruits coactivators, which results in targeted histone acetylation of adjacent nucleosomes, including NUC T encompassing the TATA sequence. Histone acetylation of NUC T then facilitates the binding of TBP (as part of the TFIID complex) and subsequent transcription of the gene. This model proposes that the regulated accessibility of the TATA sequence is a necessary but perhaps not sufficient step for transcriptional activation. Multiple regulatory steps seem to be involved in activation of the pS2 promoter by ER. This is demonstrated by the observation that elevated levels of core histone acetylation, resulting from treatment of MCF-7 cells with the deacetylase inhibitor TSA, do not constitutively activate transcription from this promoter in vivo (43; G. F. Sewack and U. Hansen, unpublished observations). In vitro transcription studies have also demonstrated the complexity of ligand-dependent activation by ER. In a highly purified human transcription system, the TBP-associated factors (TAFIIs), the other components of TFIID, were strictly required for activation both of a synthetic DNA promoter and of a chromatin template (60). In fact, direct protein-protein interactions between ER and a variety of potential targets in the basal transcription machinery have been well documented (27, 45, 46). Therefore, transcriptional activation of the genomic pS2 promoter by ER is likely to involve multiple mechanisms, including both direct factor recruitment via protein-protein interactions and directed chromatin modification (discussed in detail below). The requirement for both functions of classical activation domains has recently been demonstrated, using synthetic yeast promoters in which the TATA sequence was assembled into a nucleosome. In this system, an artificially recruited TFIID, which essentially mimics the direct recruitment of TBP to a promoter, was insufficient to activate promoters with nucleosomal TATA elements (44).
The importance of histone acetylation in activation of transcription from estrogen-responsive promoters is supported by a variety of data. Histone acetylation is not required, however, for interaction of ER with the chromatin template, as ER binds effectively in vitro to the estrogen response element (−405 to −393) within nucleosome E of the chromatin-assembled pS2 promoter (Sewack and Hansen, unpublished). Therefore, the critical question in this regard is how binding of ER activates the promoter in its native chromatin context to facilitate binding of subsequent transcription factors. ER contains two activation domains, AF-1 and AF-2. The activity of the ligand-independent, N-terminal AF-1 is stimulated by phosphorylation of serine residues (1, 18, 28). Such phosphorylation of the AF-1 domain in ERβ by the mitogen-activated protein kinase pathway leads to the recruitment of the coactivator SRC-1 (51). Likewise, transcriptional induction by the conserved hormone-dependent AF-2 domain is dependent on the recruitment of nuclear receptor coactivators, including, as possible candidates, SRC-1, TIF2/GRIP1, ACTR/AIB1/RAC3/pCIP, CBP, and pCAF, many of which possess HAT activity (9; for review, see references 11 and 50). Formation of a complex between activated ER and coactivators containing HAT activity can be an integral component of transcriptional activation by ER from a chromatin template, as was shown directly by the inability of HAT-negative coactivators to mediate estrogen-dependent transcription (9).
We show in this study that in vivo transcriptional stimulation results in hyperacetylation of histones H3 and H4 within the region of the pS2 promoter containing NUC T. Our data are in line with a recent study demonstrating enhanced acetylation of histones H3 and H4 within the proximal promoter regions of nuclear hormone receptor-responsive genes in response to hormone treatment (9), including both estrogen-responsive genes (genes for pS2, c-myc, EB-1, and cathepsin D) and retinoic acid receptor target genes (genes for p21 and CD38). For the cathepsin D and p21 promoters, enhanced acetylation was localized, in that the acetylation states of histones H3 and H4 located 3 to 4 kb upstream of the initiation sites were unchanged by hormone treatment (9). Furthermore, hyperacetylation of histone H4 was rapid and transient at several hormone-regulated promoters, correlating with RNA pol II association on the cathepsin D promoter and expression of this gene. These data suggested a general role for core histone acetylation in the rapid transcriptional activation of nuclear receptor target genes.
Localized hyperacetylation of histones H3 and H4, centered around the transcription start site of the beta interferon promoter, also correlated with transcriptional activation of this gene (41). Given that the highly localized region containing hyperacetylated core histones also contained the TATA sequence, facilitation of binding of TBP by histone acetylation may be a common mechanism of transcriptional activation at many RNA pol II promoters.
Even though the importance of histone acetylation in transcriptional activation is becoming increasingly clear, the direct effect of specific acetylation events remains unresolved. Specialized roles for individual coactivators in the transcriptional activation process at a given promoter are suggested by the different acetylation substrate specificities of the nuclear receptor coactivators (40, 47). Our study supports regulation by multiple coactivators, in that acetylation of histone H3 and histone H4 at the pS2 promoter was differentially regulated. In particular, whereas combined treatment of cells with TPA and estradiol resulted in consistently higher levels of histone H3 acetylation than with estradiol alone, enhancement of histone H4 acetylation was unchanged by the inclusion of TPA (Fig. 1C). The time courses of acetylation of histones H3 and H4 also appeared to differ. Histone H4 was transiently acetylated on the pS2 promoter (9). The time course of histone H4 acetylation correlated with association of ACTR, p300, and CBP coactivators with the promoter but not of ER, which remained bound to the pS2 proximal promoter for at least 6 h (9). In contrast, we demonstrate that highly acetylated histone H3 remained associated with the pS2 promoter even after 3 h of estradiol stimulation (Fig. 1C). By nuclear run-on assay, the pS2 gene remains maximally active at this time, in contrast to other estrogen-stimulated promoters (6). Taken together, these results suggest a role for histone H3 acetylation in the maintenance of a transcriptionally active promoter, even though histone H4 acetylation is transient. Histone H3 acetylation could be maintained by the activity of a different coactivator. We hypothesize that either acetylation event would facilitate the binding of TBP to its nucleosomal binding site in the promoter.
In conclusion, given the lack of disruption of highly positioned nucleosomes upon induction of the pS2 promoter and the reversibility of histone acetylation in this region, we speculate that acetylation of histones within NUC T, containing the pS2 TATA sequence, provides a rapid and reversible regulatory step characteristic of nuclear receptor-controlled transcription. And finally, because the binding of TBP to a nucleosomal TATA sequence represents a general regulatory step in RNA pol II transcription (reviewed in reference 7), we propose that the acetylation of core histones to facilitate binding of TBP will provide a general mechanism of facilitating transcriptional activation at many TATA sequence-containing promoters.
ACKNOWLEDGMENTS
We thank Robert Roeder and Jeff Delong for TFIIA expression plasmids and Jeff Parvin for technical advice. We are also grateful to Tom Maniatis and Bhavin Parekh for the beta interferon PCR primers. The critical suggestions of Robert Kingston, Myles Brown, Fred Winston, Tony Imbalzano, Han-Fei Ding, Konstantin Ebralidse, and Geof Cooper are also greatly appreciated.
This work was supported by American Cancer Society grants FRA-415, BE-231, and RPG-95-005 and by NIH training grant T32-CA 09361.
REFERENCES
- 1.Ali S, Metzger D, Bornert J M, Chambon P. Modulation of transcriptional activation by ligand-dependent phosphorylation of the human oestrogen receptor A/B region. EMBO J. 1993;12:1153–1160. doi: 10.1002/j.1460-2075.1993.tb05756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almer A, Rudolph H, Hinnen A, Hörz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F, Brent R, Kingston R, Moore D, Seidman J G, Smith J, Struhl K. DNA-protein interactions. In: Janssen K, editor. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1993. p. 12.4. [Google Scholar]
- 4.Braunstein M, Sobel R E, Allis C D, Turner B M, Broach J R. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol Cell Biol. 1996;16:4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 6.Brown A M C, Jeltsch J-M, Roberts M, Chambon P. Activation of pS2 gene transcription is a primary response to estrogen in the human breast cancer cell line MCF-7. Proc Natl Acad Sci USA. 1984;81:6344–6348. doi: 10.1073/pnas.81.20.6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 8.Chalbos D, Phillips A, Galtier F, Rochefort H. Synthetic antiestrogens modulate induction of pS2 and cathepsin-D messenger ribonucleic acid by growth factors and adenosine 3′,5′-monophosphate in MCF-7 cells. Endocrinology. 1993;133:571–576. doi: 10.1210/endo.133.2.8344199. [DOI] [PubMed] [Google Scholar]
- 9.Chen H, Lin R J, Xie W, Wilpitz D, Evans R M. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 10.Cho H, Katzenellenbogen B S. Synergistic activation of estrogen receptor-mediated transcription by estradiol and protein kinase activators. Mol Endocrinol. 1993;7:441–452. doi: 10.1210/mend.7.3.7683375. [DOI] [PubMed] [Google Scholar]
- 11.Collingwood T N, Urnov F D, Wolffe A P. Nuclear receptors: coactivators, corepressors and chromatin remodeling in the control of transcription. J Mol Endocrinol. 1999;23:255–275. doi: 10.1677/jme.0.0230255. [DOI] [PubMed] [Google Scholar]
- 12.Crémisi C. Chromatin replication revealed by studies of animal cells and papovaviruses (simian virus 40 and polyoma virus) Microbiol Rev. 1979;43:297–319. doi: 10.1128/mr.43.3.297-319.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Csordas A. On the biological role of histone acetylation. Biochem J. 1990;265:23–38. doi: 10.1042/bj2650023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeJong J, Bernstein R, Roeder R G. Human general transcription factor TFIIA: characterization of a cDNA encoding the small subunit and requirement for basal and activated transcription. Proc Natl Acad Sci USA. 1995;92:3313–3317. doi: 10.1073/pnas.92.8.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DePamphilis M L, Wassarman P M. In: Organization and replication of viral DNA. Kaplan A S, editor. Boca Raton, Fla: CRC Press; 1982. pp. 37–115. [Google Scholar]
- 16.Ding H-F, Bustin M, Hansen U. Alleviation of histone H1-mediated transcriptional repression and chromatin compaction by the acidic activation region in chromosomal protein HMG-14. Mol Cell Biol. 1997;17:5843–5855. doi: 10.1128/mcb.17.10.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Godde J S, Nakatani Y, Wolffe A P. The amino-terminal tails of the core histones and the translational position of the TATA box determine TBP/TFIIA association with nucleosomal DNA. Nucleic Acids Res. 1995;23:4557–4564. doi: 10.1093/nar/23.22.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goff P L, Montano M M, Schodin D J, Katzenellenbogen B S. Phosphorylation of the human estrogen receptor. J Biol Chem. 1994;269:4458–4466. [PubMed] [Google Scholar]
- 19.Hahn S, Buratowski S, Sharp P A, Guarente L. Yeast TATA-binding protein TFIID binds to TATA elements with both consensus and nonconsensus DNA sequences. Proc Natl Acad Sci USA. 1989;86:5718–5722. doi: 10.1073/pnas.86.15.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen J C, Tse C, Wolffe A P. Structure and function of the core histone N-termini: more than meets the eye. Biochemistry. 1998;37:17637–17641. doi: 10.1021/bi982409v. [DOI] [PubMed] [Google Scholar]
- 21.Hansen U. Transcriptional and structural analyses of isolated SV40 chromatin. In: Becker P B, editor. Methods in molecular biology: chromatin protocols. Totowa, N.J: Humana Press; 1999. pp. 261–290. [DOI] [PubMed] [Google Scholar]
- 22.Hebbes T R, Thorne A W, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann A, Oelgeschläger T, Roeder R G. Considerations of transcriptional control mechanisms: do TFIID-core promoter complexes recapitulate nucleosome-like functions? Proc Natl Acad Sci USA. 1997;94:8928–8935. doi: 10.1073/pnas.94.17.8928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imbalzano A N, Kwon H, Green M R, Kingston R E. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 25.Imbalzano A N, Zaret K S, Kingston R E. Transcription factor (TF) IIB and TFIIA can independently increase the affinity of the TATA-binding protein for DNA. J Biol Chem. 1994;269:8280–8286. [PubMed] [Google Scholar]
- 26.Ingar-Trowbridge D M, Pimentel M, Parker M G, McLachlan J A, Korach K S. Peptide growth factor cross-talk with the estrogen receptor requires the A/B domain and occurs independently of protein kinase C or estradiol. Endocrinology. 1996;137:1735–1744. doi: 10.1210/endo.137.5.8612509. [DOI] [PubMed] [Google Scholar]
- 27.Jacq X, Brou C, Lutz Y, Davidson I, Chambon P, Tora L. Human TAFII30 is present in a distinct TFIID complex and is required for transcriptional activation by the estrogen receptor. Cell. 1994;79:107–117. doi: 10.1016/0092-8674(94)90404-9. [DOI] [PubMed] [Google Scholar]
- 28.Kato S, Endoh H, Masuhiro Y, Kitamoto T, Uchiyama S, Sasaki H, Masushige S, Gotoh Y, Nishida E, Kashime H, Metzger D, Chambon P. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270:1491–1494. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- 29.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 31.Li G, Chandler S P, Wolffe A P, Hall T C. Architectural specificity in chromatin structure at the TATA box in vivo: nucleosome displacement upon B-phaseolin gene activation. Proc Natl Acad Sci USA. 1998;95:4772–4777. doi: 10.1073/pnas.95.8.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X-Y, Virasius A, Zhu X, Green M. Enhancement of TBP binding by activators and general transcription factors. Nature. 1999;399:605–609. doi: 10.1038/21232. [DOI] [PubMed] [Google Scholar]
- 33.Lin R, Leone J W, Cook R G, Allis C D. Antibodies specific to acetylated histones document the existence of deposition- and transcription-related histone acetylation in Tetrahymena. J Cell Biol. 1989;108:1577–1588. doi: 10.1083/jcb.108.5.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lohr D. Chromatin structure and regulation of the eukaryotic regulatory gene GAL80. Proc Natl Acad Sci USA. 1993;90:10628–10632. doi: 10.1073/pnas.90.22.10628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lohr D, Lopez J. GAL4/GAL80-dependent nucleosome disruption/disposition on the upstream regions of the yeast GAL1–10 and GAL80 genes. J Biol Chem. 1995;270:27671–27678. doi: 10.1074/jbc.270.46.27671. [DOI] [PubMed] [Google Scholar]
- 36.Meisterernst M, Horikoshi M, Roeder R G. Recombinant yeast TFIID, a general transcription factor, mediates activation by the gene-specific factor USF in a chromatin assembly assay. Proc Natl Acad Sci USA. 1990;87:9153–9157. doi: 10.1073/pnas.87.23.9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moreira J M, Holmberg S. Nucleosome structure of the yeast CHA1 promoter: analysis of activation-dependent chromatin remodeling of an RNA-polymerase-II-transcribed gene in TBP and RNA pol II mutants defective in vivo in response to acidic activators. EMBO J. 1998;17:6028–6038. doi: 10.1093/emboj/17.20.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakajima N, Horikoshi M, Roeder R G. Factors involved in specific transcription by mammalian RNA polymerase II: purification, genetic specificity, and TATA box-promoter interactions of TFIID. Mol Cell Biol. 1988;8:4028–4040. doi: 10.1128/mcb.8.10.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nunez A-M, Berry M, Imler J-L, Chambon P. The 5′ flanking region of the pS2 gene contains a complex enhancer region responsive to oestrogens, epidermal growth factor, a tumour promoter (TPA), the c-Ha-ras oncoprotein and the c-jun protein. EMBO J. 1989;8:823–829. doi: 10.1002/j.1460-2075.1989.tb03443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 41.Parekh B S, Maniatis T. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the INF-B promoter. Mol Cell. 1999;3:125–129. doi: 10.1016/s1097-2765(00)80181-1. [DOI] [PubMed] [Google Scholar]
- 42.Patikoglou G A, Kim J L, Sun L, Yang S-H, Kodadek T, Burley S K. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 1999;13:3217–3230. doi: 10.1101/gad.13.24.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruh M F, Tian S, Cox L K, Ruh T S. The effects of histone acetylation on estrogen responsiveness in MCF-7 cells. Endocrine. 1999;11:157–164. doi: 10.1385/ENDO:11:2:157. [DOI] [PubMed] [Google Scholar]
- 44.Ryan M P, Stafford G A, Yu L, Morse R H. Artificially recruited TATA-binding protein fails to remodel chromatin and does not activate three promoters that require chromatin remodeling. Mol Cell Biol. 2000;20:5847–5857. doi: 10.1128/mcb.20.16.5847-5857.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sabbah M, Kang K, Tora L, Redeuilh G. Oestrogen receptor facilitates the formation of preinitiation complex assembly: involvement of the general transcription factor TFIIB. Biochem J. 1998;336:639–646. doi: 10.1042/bj3360639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sadovsky Y, Webb P, Lopez G, Baxter J D, Fitzpatrick P M, Gizang-Ginsberg E, Cavailles V, Parker M G, Kushner P J. Transcriptional activators differ in their responses to overexpression of TATA-box-binding protein. Mol Cell Biol. 1995;15:1554–1563. doi: 10.1128/mcb.15.3.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schiltz R L, Mizzen C A, Vassilev A, Cook R G, Allis C D, Nakatani Y. Overlapping but distinct patterns of histone acetylation by the human co-activators p300 and PCAF within nucleosomal substrates. J Biol Chem. 1999;274:1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- 48.Sewack G F, Hansen U. Nucleosome positioning and transcription-associated chromatin alterations on the human estrogen-responsive pS2 promoter. J Biol Chem. 1997;272:31118–31129. doi: 10.1074/jbc.272.49.31118. [DOI] [PubMed] [Google Scholar]
- 49.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 50.Torchia J, Glass C, Rosenfeld M G. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 51.Tremblay A, Tremblay G B, Labrie F, Giguere V. Ligand-independent recruitment of SRC-1 to estrogen receptor β through phosphorylation of activator function AF-1. Mol Cell. 1999;3:513–519. doi: 10.1016/s1097-2765(00)80479-7. [DOI] [PubMed] [Google Scholar]
- 52.Turner B M. Decoding the nucleosome. Cell. 1993;75:5–8. [PubMed] [Google Scholar]
- 53.Verdin E, Paras P, Jr, Lint C V. Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcription activation. EMBO J. 1993;12:3249–3259. doi: 10.1002/j.1460-2075.1993.tb05994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verdone L, Camilloni G, Mauro E D, Caserta M. Chromatin remodeling during Saccharomyces cerevisiae ADH2 gene activation. Mol Cell Biol. 1996;16:1978–1988. doi: 10.1128/mcb.16.5.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Widlund H R, Vitolo J M, Thiriet C, Hayes J J. DNA sequence-dependent contributions of core histone tails to nucleosome stability: differential effects of acetylation and proteolytic tail removal. Biochemistry. 2000;39:3835–3841. doi: 10.1021/bi991957l. [DOI] [PubMed] [Google Scholar]
- 56.Wolffe A P, Pruss D. Targeting chromatin disruption: transcription regulators that acetylate histones. Cell. 1996;84:817–819. doi: 10.1016/s0092-8674(00)81059-4. [DOI] [PubMed] [Google Scholar]
- 57.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 58.Workman J L, Roeder R G. Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell. 1987;51:613–622. doi: 10.1016/0092-8674(87)90130-9. [DOI] [PubMed] [Google Scholar]
- 59.Workman J L, Roeder R G, Kingston R E. An upstream transcription factor, USF (MLTF), facilitates the formation of preinitiation complexes during in vitro chromatin assembly. EMBO J. 1990;9:1299–1308. doi: 10.1002/j.1460-2075.1990.tb08239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu S-Y, Thomas M C, Hou S Y, Likhite V, Chiang C-M. Isolation of mouse TFIID and functional characterization of TBP and TFIID in mediating estrogen receptor and chromatin transcription. J Biol Chem. 1999;274:23480–23490. doi: 10.1074/jbc.274.33.23480. [DOI] [PubMed] [Google Scholar]
- 61.Wu C. Chromatin remodeling and the control of gene expression. J Biol Chem. 1997;272:28171–28174. doi: 10.1074/jbc.272.45.28171. [DOI] [PubMed] [Google Scholar]
- 62.Yokomori K, Zeidler M P, Chen J-L, Verrijzer C P, Mlodzik M. Drosophila TFIIA directs cooperative DNA binding with TBP and mediates transcriptional activation. Genes Dev. 1994;8:2313–2323. doi: 10.1101/gad.8.19.2313. [DOI] [PubMed] [Google Scholar]
- 63.Yoshida M, Kijima M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]