Abstract
Long interspersed nuclear elements (LINEs or L1s) comprise approximately 17% of human DNA; however, only about 60 of the ∼400,000 L1s are mobile. Using a retrotransposition assay in cultured human cells, we demonstrate that L1-encoded proteins predominantly mobilize the RNA that encodes them. At much lower levels, L1-encoded proteins can act in trans to promote retrotransposition of mutant L1s and other cellular mRNAs, creating processed pseudogenes. Mutant L1 RNAs are mobilized at 0.2 to 0.9% of the retrotransposition frequency of wild-type L1s, whereas cellular RNAs are mobilized at much lower frequencies (ca. 0.01 to 0.05% of wild-type levels). Thus, we conclude that L1-encoded proteins demonstrate a profound cis preference for their encoding RNA. This mechanism could enable L1 to remain retrotransposition competent in the presence of the overwhelming number of nonfunctional L1s present in human DNA.
Retrotransposons are DNA sequences that can move (i.e., retrotranspose) to different genomic locations via an RNA intermediate. They are present in the genomes of virtually all eukaryotes and can be subdivided into two general structural classes. Long terminal repeat (LTR) retrotransposons resemble simple retroviruses but lack a functional envelope (Env) gene (2). Non-LTR retrotransposons lack LTRs and generally terminate in a polyadenylic acid [poly(A)] tail (20, 23).
L1s are the most abundant non-LTR retrotransposons in the human genome and comprise approximately 17% of nuclear DNA (42). The overwhelming majority of L1s are retrotransposition defective (RD-L1s) and cannot retrotranspose because they are 5′ truncated, internally rearranged, or mutated (23); however, an estimated 30 to 60 human L1s remain retrotransposition competent (RC-L1s) (40). RC-L1s are 6.0 kb in length and contain a 5′ untranslated region (UTR) harboring an internal promoter (43), two nonoverlapping open reading frames (open reading frame 1 [ORF1] and ORF2) (7, 41), and a 3′ UTR ending in an unorthodox poly(A) tail (20, 46). In addition, these elements are flanked by variable-length target site duplications, which are hallmarks of the retrotransposition process (20).
Non-LTR retrotransposons encode endonuclease activities, which can generate either site-specific (4, 11, 47) or relatively non-site-specific nicks in chromosomal DNA (5, 10). The liberated 3′ hydroxyl residue then acts as a primer for reverse transcription of the retrotransposon RNA by the retrotransposon-encoded reverse transcriptase (RT) by a mechanism termed target site-primed reverse transcription (TPRT) (28, 29). Thus, the processes of integration and reverse transcription are coupled for non-LTR retrotransposons.
Biochemical studies revealed that ORF1 encodes a 40-kDa RNA binding protein that colocalizes with L1 RNA in cytoplasmic ribonucleoprotein particles (RNPs) (17, 18). ORF2 encodes a multifunctional protein containing endonuclease and RT activities (10, 34) and also has a carboxyl-terminal cysteine-rich domain (C) of unknown function (9). Using an assay to monitor L1 retrotransposition in cultured human HeLa cells, we demonstrated that a wide variety of site-directed point mutations in conserved domains of the ORF1- and ORF2-encoded proteins essentially abolish L1 retrotransposition (10, 37).
L1 retrotransposition can be mutagenic and has resulted in various genetic disorders (23, 24). The characterization of mutagenic L1 insertions in humans and mice yielded the unexpected finding that each insertion is derived from a progenitor L1 containing intact ORFs (7, 19, 25, 38). Thus, despite the vast majority of RD-L1s in the genome, it appears that only RNAs derived from RC-L1s efficiently retrotranspose (i.e., the L1 proteins demonstrate an apparent cis preference) (7, 8, 37). Paradoxically, it also is proposed that the proteins encoded by RC-L1s function in trans to promote both processed pseudogene formation and the retrotransposition of certain short interspersed nuclear elements (SINEs) (1, 6, 8, 21, 23, 30, 44).
Here, we use a two-plasmid complementation assay to demonstrate that the RC-L1 proteins preferentially mobilize the transcript from which they are encoded. This cis-preference mechanism likely allows RC-L1s to persist despite the presence of overwhelming numbers of nonfunctional elements. We further show that the RC-L1 proteins can function at a low level in trans to retrotranspose both mutant L1 RNAs and cellular mRNAs, resulting in the formation of processed pseudogenes.
MATERIALS AND METHODS
The oligonucleotides used in this study were as follows: 437SNEO, 5′-CAGCCCCTGATGCTCTTCGTCC; 6664NEO, 5′CCCTTCCCGCTTCAGTGACA; 1808ASNEO, 5′-CATTGAACAAGATGGATTGCACGC; RT TESTB, 5′-CGATTTCGAACCCTGACGTC; ORF1END, 5′-TACCAGCCGCTGCAAAATCATGCC; PAI1B5′, 5′-GCCCTCACCTGCCTAGTCC; PAI1BMID, 5′-GGGAGAGAAGTTTGAAGCAC; PAI1B3′, 5′-CAGAGTGAATGTCCCCCATC; ABL5′, 5′-TTTATGGGGCAGCAGCCTGGAAAAGTACTTGGG; ABL3′, 5′-TCACTGGGTCCAGCGAGAAGGTTTTCCTTGGAGTT; IPCRPAI1B1, 5′-GATGGGGGACATTCACTCTG; IPCRPAI1B2, 5′-CTGTCACCAGCCTCCTCCG; L1PCRA, 5′-GGTTCGAAATCGATAAGCTTGG; L1IPCRB, 5′-GGACAAACCACAACTAGAATGC; JB3169, 5′-TAATACGACTCACTATAGGGGTTGACGCAAATGGGCGGTAGGCGTGTACGG; JB3165, 5′-AATTAACCCTCACTAAAGGGCAGGTTGACGCAAATGGGCGGTAGGCGTGTACGG; JB3168, 5′-TAATACGACTCACTATAGGGCAGCGGGCAGTTCGGTTTCAGGCAGGTCTTGC; and JB3167, 5′-AATAACCCTCACTAAAGGGCAGCCAGCGTCTTGTCATTGGCGAATTCGAACACGC.
Recombinant DNA plasmids.
The following recombinant plasmids contain the indicated restriction fragments of L1 DNA cloned into pCEP4 (Invitrogen) unless otherwise indicated.
pJM108/L1.3 contains a 7.2-kb NotI-BamHI fragment containing L1.3 ORF1, L1.3 ORF2, and the mneoI indicator cassette. A nonsense mutation (S119X) is present in ORF1. The mutation introduces a BclI restriction site.
pJM111/L1.3 contains a 7.2-kb NotI-BamHI fragment containing L1.3 ORF1, L1.3 ORF2, and the mneoI indicator cassette. Two missense mutations (R261A and R262A) are present in ORF1. The mutation introduces a SacII restriction site.
pJM116/L1.3 contains a 7.2-kb NotI-BamHI fragment containing L1.3 ORF1, L1.3 ORF2, and the mneoI indicator cassette. A missense mutation (H230A) is present in the endonuclease domain of ORF2. The mutation introduces an NheI restriction site.
pJM105/L1.3 contains a 7.2-kb NotI-BamHI fragment containing L1.3 ORF1, L1.3 ORF2, and the mneoI indicator cassette. A missense mutation (D702A) is present in the RT domain of ORF2. The mutation introduces a PvuII site into the plasmid.
pJM124/L1.3 contains a 7.2-kb NotI-BamHI fragment containing L1.3 ORF1, L1.3 ORF2, and the mneoI indicator cassette. The construct contains two missense mutations (R261A and R262A) in ORF1 and a missense mutation (D702A) in the RT domain of ORF2.
pJM101/L1.3 Δneo and pJM101/L1RP Δneo (and mutant derivatives) contain 6.0-kb NotI-BamHI fragments containing the complete sequence of L1.3 or L1RP, respectively. These clones lack the mneoI indicator cassette.
L1.3 ORF1mneoI contains a 3.8-kb NotI-BamHI fragment containing the L1.3 5′ UTR, L1.3 ORF1, and the mneoI cassette.
pPAI1amneoI contains a 2.8-kb NotI-BamHI fragment containing a 1.0-kb fragment of PAI1 cDNA and the mneoI indicator cassette. The 1.0-kb PAI1 cDNA fragment is in the antisense orientation.
pPAI1bmneoI contains a 3.8-kb NotI-BamHI fragment containing a 2.0-kb fragment of PAI1 cDNA and the mneoI indicator cassette. The increased length of the PAI1 cDNA is due to a length increase in the 3′ UTR because of the use of an alternative polyadenylation site.
pPAI1cmneoI contains a 2.8-kb NotI-BamHI fragment containing a 1.0-kb fragment of PAI1 cDNA and the mneoI indicator cassette.
pβGAL-αNLS and pβGAL-ΩNLS contain the α or Ω fragments, respectively, of the β-galactosidase gene 35 in the pRK5 mammalian expression vector. Each fragment is expressed from the cytomegalovirus (CMV) immediate-early promoter and uses the simian virus 40 (SV40) late polyadenylation signal; therefore, they are in expression contexts similar to that of the L1s used in this study.
DNA preparation and DNA sequencing.
Plasmid DNAs were purified on Qiagen Maxi or Midi prep columns according to the procedures specified by the manufacturer. DNAs for transfection experiments were checked for superhelicity by electrophoresis on 0.7% agarose-ethidium bromide gels. Only highly supercoiled preparations of DNA (>90%) were used for transfection. Genomic DNA from tissue culture cells was isolated using the Blood and Cell Midi Prep Kit (Qiagen). DNA sequencing was performed on an Applied Biosystems DNA sequencer (ABI 377) at the University of Michigan Core facilities.
Growth of HeLa cells.
HeLa cells were grown at 37°C in an atmosphere containing 7% carbon dioxide and 100% humidity in Dulbecco modified Eagle medium (DMEM)–high-glucose medium lacking pyruvate (Gibco-BRL). DMEM was supplemented with 10% fetal bovine calf serum, 0.4 mM glutamine, and 20 U of penicillin-streptomycin per ml (DMEM-complete). Cell passage and cloning of cells by limiting dilution was performed using standard techniques.
Transfection conditions.
We used a modified version of a transient-transfection protocol (45). Approximately 2 × 105 cells/ml were plated in each well of a six-well dish, and the cells were grown to about 50 to 80% confluency. The following day, duplicate dishes were cotransfected with equal amounts of a reporter plasmid (pGreen Lantern) and an L1 allele tagged with the mneoI indicator cassette. We routinely use 3 μl of Fugene-6 transfection reagent (Roche Molecular Biochemicals) and 0.5 to 1.0 μg of Qiagen prepared DNA per transfection reaction for HeLa cells plated in six-well dishes. For 175-cm2 plates, we typically plate 6 × 106 HeLa cells/dish and use 90 μl of Fugene and 30 μg of DNA per transfection reaction (45). At 72 h posttransfection, the HeLa cells in one set of tissue culture dishes were trypsinized and subjected to flow cytometry. The percentage of green fluorescent cells was used to determine the transfection efficiency of each sample (39, 45). The remaining samples were visualized to ensure that they were transfected and then were subjected to G418r selection (400 μg/ml) to score for retrotransposition. After 12 days, the media were aspirated, the cells were washed in 1× phosphate-buffered saline (PBS), and the washed cells were fixed to plates by treating with FIX solution (2% formaldehyde [of a 37% stock solution in water], 0.2% glutaraldehyde, 1× PBS) at 4°C for 30 min. The fixed cells were then stained with 0.4% Giemsa at room temperature overnight. The retrotransposition efficiency was then scored as the number of G418r foci/number of cells transfected with green fluorescent protein (GFP).
Fluorescent microscopy and fluorescence-activated cell scanning (FACS).
Fluorescence microscopy was performed using a Leica DM-IL inverted microscope with an ultra-high-pressure lamp (HBO/50W), a vertical fluorescence illuminator, and a fluorescein isothiocyanate filter set (530-nm peak excitation; Chroma). The cells were prepared for cell sorting by washing them once with 2 ml of PBS and then were removed from six-well dishes with trypsin (0.05% solution; Gibco-BRL). The suspended cells were transferred to polystyrene tubes and kept on ice until FACS analysis. Cells were analyzed with a Coulter Epics Elite tabletop analysis instrument (Beckman-Coulter) containing a blue argon laser (488 nm) and fluorescein filter sets (525-band-pass filter). Between 10,000 and 20,000 cells were analyzed per sample. Live-dead gating was performed based on the forward-scatter versus the side-scatter profile. Living cells were analyzed for fluorescence intensity, and the proportion of GFP-positive cells was determined. Mock-transfected HeLa cells were used as negative controls in these experiments. Data analysis was performed using the Coulter Elite software package.
PCR analysis.
PCR reactions were carried out in 50-μl volumes. Each reaction contained 10 U of Taq polymerase, 0.2 mM concentrations of deoxynucleoside triphosphates (dNTPs), and 200 ng of each primer in the buffer supplied by the vendor (Perkin-Elmer). In general, reactions were conducted at an annealing temperature 5°C below the Tm of the primer. One-fifth of the reaction volume was separated on 1% agarose gels containing ethidium bromide.
Inverse PCR.
The procedure described below was adapted from that of Li et al. (27). HeLa cell DNA (5 μg) derived from G418r clonal lines was digested to completion with either XbaI or SspI (New England Biolabs) in a total reaction volume of 50 μl; heating at 65°C for 30 min stopped the reactions. Restricted DNA was circularized by dilution and ligation using T4 DNA ligase (3,200 U; New England Biolabs) in a volume of 600 μl at 16°C for at least 16 h. The ligated DNA was precipitated with ethanol and dissolved in 40 μl of distilled water. Then, 2 μl of DNA was used in the primary PCR reaction in a 50-μl reaction volume containing a 20 nM concentration of each dNTP, 10 pmol of primers IPCRPAI1B1 and L1IPCRA, 1× buffer 2, and 2.5 U of enzyme mix in the Expand Long Template PCR system (Roche Molecular Biochemicals). We used a Hybaid Thermocycler programmed as follows: 95°C for 2 min, followed by 30 cycles of 94°C for 10 s, 63°C for 30 s, and 68°C for 15 min, and a final extension step at 68°C for 30 min. The amount of primary PCR was semiquantified on a 0.7% agarose-ethidium bromide gel, and 1 μl was used in a secondary PCR reaction using the same conditions, except that we used primers IPCRPAI1B2 and L1IPCRB. The secondary PCR product was separated on a 0.7% agarose-ethidium bromide gel, the product was band isolated using GeneClean (Bio 101), and the gel purified fragment was cloned into pGEM-T easy (Promega) using the manufacturer's protocols.
RNase protection analysis.
A total of 106 HeLa cells were transfected with 2.5 μg of plasmid using Lipofectamine-Plus reagent as described by the manufacturer (Gibco-BRL). Approximately 52 h after transfection, transfected cells were lysed directly in 1 ml of TRIzol reagent (Gibco-BRL), and the total RNA was isolated as described by the manufacturer. The total RNA was subjected to RQ1 DNase (Promega) digestion at 37°C for 20 min. The resultant RNA was extracted with phenol-chloroform and collected by ethanol precipitation. PCR products containing T7 promoter sequences were used as template for in vitro transcription, which was carried out using T7 RNA polymerase in the presence of [α-32P]CTP using the Maxiscript in vitro transcription kit (Ambion). The primers JB3169 and JB3165 were used to generate the L1 probes, while JB3168 and JB3167 were used to generate the hyg probe (see above). JB3169 and JB3168 contain T7 promoter sequences, while JB3165 and JB3167 contain T3 promoter sequences. The RNA ladder was similarly transcribed using RNA Century Marker Plus Template Set (Ambion) as a template. Incubating the resultant samples with 2 U of DNase at 37°C for 15 min (Ambion) degraded the DNA templates. RNase protection assays were performed using the RPA III nuclease protection kit as described by the manufacturer (Ambion). Briefly, 20 μg of total RNA were hybridized to gel-purified labeled RNA probes at 42°C overnight. The hybridization products were digested using a mixture of RNase A (0.375 U) and RNase T1 (15 U) for 12 h. The remaining products were precipitated and resolved on 5% denaturing polyacrylamide gels.
RESULTS
ORF1 and ORF2 mutants are not complemented efficiently.
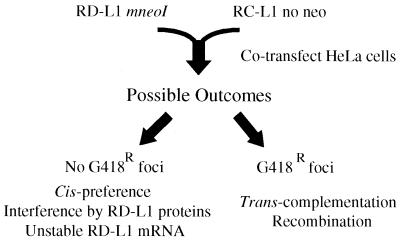
We previously demonstrated that missense mutations in conserved domains of the ORF1- and ORF2-encoded proteins greatly reduce or abolish the ability of L1 to retrotranspose (10, 37). Here, we sought to determine whether RD-L1s tagged with the mneoI indicator cassette could be complemented if they were cotransfected into HeLa cells with a RC-L1 lacking the cassette (Fig. 1). The absence of G418r foci would be consistent with a cis-preference model. However, the absence of G418r foci also could occur either if expression of the RD-L1 proteins interfered with the function of the RC-L1 proteins or if the RD-L1 RNA were unstable. By contrast, the presence of G418r foci would suggest that the RC-L1 proteins function in trans to retrotranspose RD-L1 RNAs. However, G418r foci also could arise if the RC-L1 recombined with the mneoI-tagged RD-L1 to create a recombinant L1, which could undergo subsequent retrotransposition in cis.
FIG. 1.
Rationale of the assay. Retrotransposition-defective L1s (RD-L1s) containing the mneoI indicator cassette were cotransfected into HeLa cells with retrotransposition-competent L1s (RC-L1s) lacking the cassette, and retrotransposition was determined as described in Materials and Methods. Explanations for the possible experimental outcomes are noted.
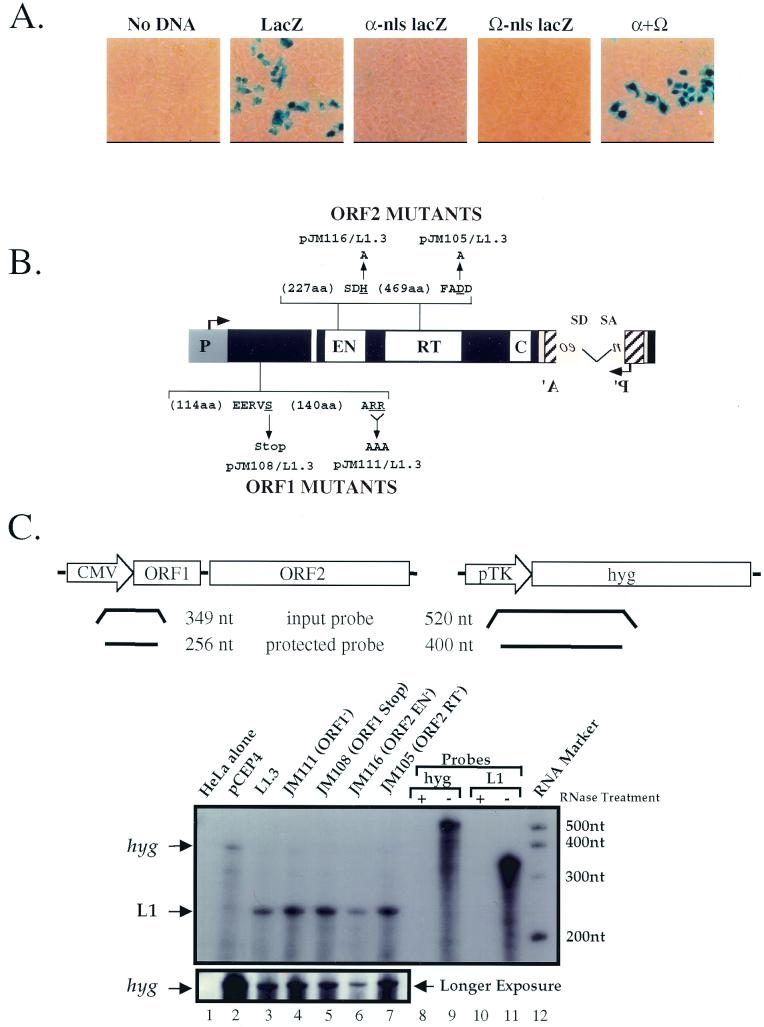
As a control for cotransfection, we used a two-plasmid system to demonstrate efficient trans complementation of β-galactosidase α and Ω fragments in HeLa cells (Fig. 2A; see Materials and Methods) (35). Thus, HeLa cells efficiently can accommodate and express proteins from two different expression vectors. Next, we conducted RT-PCR (not shown) and RNase protection assays to demonstrate that both RC-L1 and RD-L1 RNAs are expressed at similar levels (Fig. 2B and C, lanes 2 to 7). Thus, the RD-L1 mutants do not dramatically affect the stability of L1 RNA.
FIG. 2.
Controls used in this study. (A) trans complementation of β-galactosidase enzymatic activity in HeLa cells. Plasmids with the α or Ω regions of the β-galactosidase gene that contained a nuclear localization signal (nls) were transfected into HeLa cells individually or together, and β-galactosidase activity was monitored 3 days posttransfection (35). Mock transfection (no DNA) and a wild-type β-galactosidase gene (CMV β-gal; Clontech; GenBank accession no. U02451) served as negative and positive controls, respectively. (B) Mutants used in this study. Mutations in ORF1 or the endonuclease or RT domains of L1.3mneoI are indicated. The wild-type amino acids that were mutated are underlined. The arrows indicate the mutant amino acid sequence changes (e.g., ARR was changed to AAA). (C) RNA expression of representative L1 constructs. Structures of the hyg (hygromycin resistance gene) and L1 probes. Sizes of the full-length input and protected bands are indicated at the top of the figure. RNase protection assays were carried out of total RNAs prepared from HeLa cells transfected with the indicated plasmids. Probes that have undergone the RNase protection assay with (lanes 8 and 10) or without (lanes 9 and 11) the addition of RNase are shown. A longer exposure of the pCEP4-derived hyg transcripts, which serves as an internal control, is shown in the bottom panel. Consistent with earlier studies, we were unable to detect the expression of endogenous L1 transcripts in HeLa cells (43).
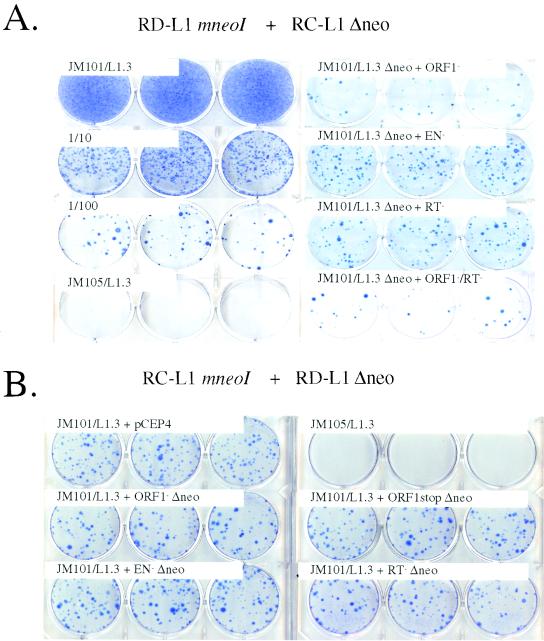
We first asked whether RD-L1s containing either a nonsense or a missense mutation in L1.3 ORF1 (JM108/L1.3 and JM111/L1.3; Fig. 2B) could be complemented if they were cotransfected into HeLa cells with equal molar amounts of an RC-L1 lacking the mneoI indicator cassette (JM101/L1.3 Δneo). As expected, the mutants could not retrotranspose by themselves (Table 1). However, upon cotransfection of the RD-L1s with JM101/L1.3 Δneo, some G418r foci were obtained (0.2 to 0.3% of the level of JM101/L1.3; Fig. 3A; Table 1), indicating that the L1.3 ORF1-encoded protein may function at a low level in trans.
TABLE 1.
Retrotransposition frequencies of the constructsa
| Group | Construct name | No. of G418r foci in HeLa cells:
|
||||
|---|---|---|---|---|---|---|
| Alone
|
Cotransfected with JM101/L1.3 Δneo
|
|||||
| n | Mean ± SEM (colonies/well) | n | Mean ± SEM (colonies/well) | % JM101/L1.3 | ||
| Wild type | JM101/L1.3 | 12 | 6,100 ± 910 | 100 | ||
| ORF1 mutants | ||||||
| Missense | JM111/L1.3 | 12 | 0 ± 0 | 21 | 16 ± 1 | 0.3 |
| Nonsense | JM108/L1.3 | 12 | 0 ± 0 | 18 | 13 ± 1 | 0.2 |
| ORF2 mutants | ||||||
| EN− | JM116/L1.3 | 12 | 2.7 ± 0.6 | 21 | 55 ± 7 | 0.9 |
| RT− | JM105/L1.3 | 12 | 0.1 ± 0.1 | 21 | 41 ± 4 | 0.7 |
| ORF1− ORF2− | JM124/L1.3 | 6 | 0 ± 0 | 6 | 12 ± 1 | 0.2 |
| ORF1 alone | ORF1mneoI | 18 | 0 ± 0 | 18 | 25 ± 4 | 0.4 |
Individual constructs tested in the retrotransposition assay are listed. The number of G418r foci obtained when the constructs were transfected into HeLa cells by themselves or cotransfected into HeLa cells with JM101/L1.3 Δneo is indicated. All of the experiments were conducted in six-well tissue culture dishes. The standard error and percentage of JM101/L1.3 retrotransposition activity are shown for each experiment. The number of independent transfections (n) is also given. EN−, endonuclease negative.
FIG. 3.
L1s retrotranspose in cis. (A) Results of the retrotransposition assay. RD-L1s containing the mneoI indicator cassette were cotransfected into 2 × 105 HeLa cells with an RC-L1 lacking the cassette (JM101/L1.3 Δneo). G418r foci were fixed and stained with Giemsa for visualization. Samples cotransfected with JM101/L1.3 Δneo and representative mutants in ORF1 (JM111/L1.3), the endonuclease or RT domains of ORF2 (JM116/L1.3 or JM105/L1.3), or a double mutant (JM124/L1.3) are shown. Cells transfected with JM101/L1.3, as well as 1/10 (2 × 104) and 1/100 (2 × 103) dilutions of transfected cells are indicated as positive controls. Cells transfected with JM105/L1.3 are shown as a negative control. (B) The coexpression of RD-L1s does not inhibit RC-L1 retrotransposition. A RC-L1 containing the mneoI indicator cassette (JM101/L1.3) was cotransfected into 2 × 104 HeLa cells with RD-L1s lacking the cassette, and retrotransposition was determined as described above. An experiment using a 1:9 (RC-L1 to RD-L1) molar ratio of transfected DNAs is shown. Cells transfected with JM101/L1.3 and an empty expression vector (CEP4) yielded G418r foci at roughly the same levels as cells that were cotransfected with JM101/L1.3 and RD-L1s lacking the indicator cassette (i.e., there was less than a 20% difference between respective samples). JM105/L1.3 was used as a negative control. Notably, RD-L1s whose transcription is driven from either the CMV promoter or the CMV promoter and L1 5′ UTR are complemented to similar extents (not shown).
Next, we repeated the experiment to determine whether RD-L1s containing point mutations in either the endonuclease (pJM116/L1.3) or the RT domain (pJM105/L1.3) of L1.3 ORF2 could be complemented in trans. Again, the mutant constructs alone could not retrotranspose efficiently (<0.04% of the level of JM101/L1.3; Table 1). However, cotransfection with JM101/L1.3 Δneo resulted in a modest increase in the number of G418r foci (ca. 0.7 to 0.9% of the level of JM101/L1.3; Fig. 3A and Table 1). Finally, we demonstrated that a construct containing missense mutations in both ORF1 and ORF2 (JM124) was complemented to the same extent as constructs containing nonsense or missense mutations in ORF1 alone (ca. 0.2% of JM101/L1.3; Fig. 3A and Table 1 [see also Table 2]).
TABLE 2.
The proteins encoded by RC-L1s can function in trans to retrotranspose cellular RNAsa
| Construct name | No. of G418r foci in HeLa cells:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Alone
|
Cotransfected with JM101/L1.3 Δneo
|
Cotransfected with JM101/L1RP Δneo
|
||||||
| n | Mean ± SEM (colonies/flask) | n | Mean ± SEM (colonies/flask) | % JM101/L1.3 | n | Mean ± SEM (colonies/flask) | % JM101/L1RP | |
| JM101/L1.3 | 2 | 53,200 ± 2,900 | 100 | |||||
| JM101/L1RP | 2 | 77,100 ± 1,200 | 100 | |||||
| JM111/L1.3 (ORF1−) | 1 | 0 ± 0 | 2 | 181 ± 25 | 0.3 | 2 | 222 ± 12 | 0.3 |
| JM105/L1.3 (RT−) | 1 | 0 ± 0 | 2 | 385 ± 7 | 0.7 | 2 | 449 ± 2 | 0.6 |
| JM124/L1.3 (ORF1−/RT−) | 3 | 0 ± 0 | 3 | 109 ± 14 | 0.2 | 3 | 147 ± 6 | 0.2 |
| ORF1mneoI | 3 | 0 ± 0 | 3 | 322 ± 30 | 0.6 | 2 | 535 ± 46 | 0.7 |
| PAI1AmneoI | 4 | 0 ± 0 | 6 | 2.3 ± 1.0 | 0.004 | 3 | 8 ± 0.9 | 0.01 |
| PAI1BmneoI | 4 | 0 ± 0 | 8 | 4.6 ± 0.4 | 0.01 | 8 | 32 ± 8 | 0.04 |
| PAI1CmneoI | 4 | 0.2 ± 0.2 | 3 | 4.0 ± 2.3 | 0.01 | 3 | 35 ± 1 | 0.05 |
Individual constructs tested in the retrotransposition assay are listed in the first column. The number of G418r foci obtained when the constructs were transfected into HeLa cells by themselves or cotransfected into HeLa cells with either JM101/L1.3 Δneo or JM101/L1RP Δneo is indicated. All of the experiments were conducted in 175-cm2 tissue culture flasks. The standard error and the percentage of wild-type L1 retrotransposition activity are shown for each experiment. The number of independent transfections (n) is indicated.
Coexpression of RD-L1s does not interfere with RC-L1 retrotransposition.
Our failure to detect efficient trans complementation suggests that the RC-L1 proteins preferentially function in cis. However, it remained possible that expression of the mutant RD-L1 proteins actively interferes with the wild-type RC-L1 proteins. To exclude this possibility, we cotransfected RD-L1s lacking the mneo1 indicator gene with an RC-L1 containing the indicator gene. A drastic reduction in RC-L1 retrotransposition would be expected if dominant interference was significant.
The coexpression of various RD-L1s had little or no effect on JM101/L1.3 retrotransposition. Moreover, increasing the molar ratios of RD-L1 to RC-L1 (4:1 and 9:1, respectively) did not result in a significant reduction in the number of G418r foci (Fig. 3B). Finally, we demonstrated that the retrotransposition of an allele of JM101/L1RP tagged with an enhanced green fluorescent protein retrotransposition indicator cassette (39) was not affected by the coexpression of representative RD-L1s harboring the mneoI reporter cassette (not shown). Thus, we conclude that coexpression of mutant RD-L1 proteins does not interfere with the retrotransposition of RC-L1s.
Recombination does not account for the prevalence of G418r foci.
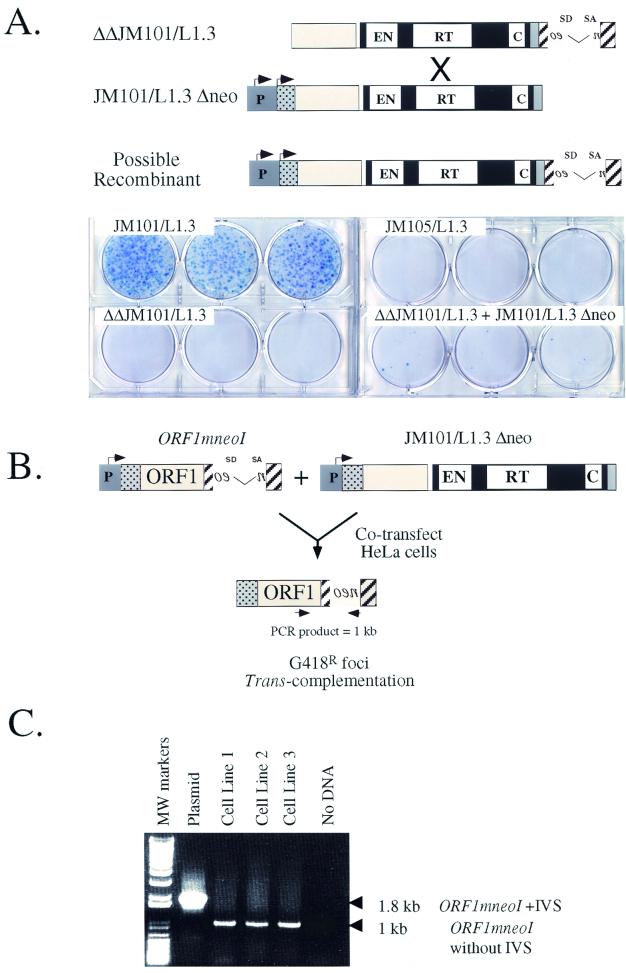
To determine whether homologous DNA recombination affected our results, a mutant allele of L1.3mneoI, which contains intact ORFs but lacks both the CMV and L1 promoters (ΔΔJM101/L1.3), was cotransfected into HeLa cells with pJM101/L1.3 Δneo. In this case, G418r foci will only result from homologous recombination between ΔΔJM101/L1.3 and pJM101/L1.3 Δneo, leading to the formation of a recombinant L1, which subsequently could undergo retrotransposition in cis (Fig. 4A).
FIG. 4.
G418r foci must arise by trans complementation. (A) The low-level rescue of RD-L1s cannot be accounted for by DNA recombination. An allele of JM101/L1.3 that lacked both the 5′ UTR and the CMV promoter (ΔΔJM101/L1.3) was transfected into HeLa cells alone or with a wild-type allele of L1.3 that lacked the mneoI indicator cassette, and retrotransposition was assayed as described in Fig. 3. The rationale for this experiment is described in the text. JM101/L1.3 and JM105/L1.3 were used as appropriate positive and negative controls. (B) Constructs used in the study. The structure of L1.3 ORF1mneoI is shown, and the rationale for the experiment is described in the text. (C) The resultant G418r foci have the predicted structure. PCR experiments using the oligonucleotides depicted in Fig. 4B (indicated by converging arrows) revealed that the retrotransposed mneoI cassette lacked the intron and was linked physically to L1.3 ORF1. The details of the experiment are provided in the text.
As expected, ΔΔJM101/L1.3 did not retrotranspose when transfected into HeLa cells alone (Fig. 4A). Moreover, cotransfection of ΔΔJM101/L1.3 with either JM101/L1.3 Δneo or point-mutated derivatives of JM101/L1.3 Δneo resulted in only rare G418r foci (<0.03% of the activity of JM101/L1.3; Fig. 4A and data not shown). Thus, homologous recombination cannot account for the G418r foci observed in Fig. 3A.
G418r foci must arise by trans complementation.
To prove that the RC-L1 proteins could function in trans, we sought to determine whether the retrotransposition of L1.3 ORF1mneoI could be stimulated by the cotransfection of JM101/L1.3 Δneo (Fig. 4B). Here, G418r foci will arise only if the JM101/L1.3 ORF2-encoded protein functions in trans to retrotranspose the L1.3 ORF1mneoI RNA. Since L1.3 ORF1mneoI completely lacks ORF2 sequences, it is difficult to envision how homology-dependent DNA recombination would recreate a recombinant L1 that could undergo subsequent retrotransposition in cis.
As expected, L1.3 ORF1mneoI was unable to retrotranspose when transfected into HeLa cells alone (Table 1). However, upon cotransfection with JM101/L1.3 Δneo, G418r foci were obtained at levels comparable to those of RD-L1s containing mutations in L1.3 ORF2 (Table 1). Next, we pooled the G418r foci obtained in these experiments and established three polyclonal cell lines. We isolated genomic DNA from each cell line and conducted PCR to determine whether the resultant retrotransposition events had the predicted structures. In each sample we detected the predicted product, indicating that ORF1 was linked physically to the retrotransposed mneoI indicator cassette (Fig. 4C). Thus, we conclude that the G418r foci obtained in these experiments arise because of trans complementation.
L1-encoded proteins can promote the retrotransposition of other cellular mRNAs.
The finding that the RC-L1 proteins could function in trans led us to ask whether other cellular mRNAs could also serve as substrates for the L1 retrotransposition machinery. Thus, we constructed a variety of plasminogen activator inhibitor 1 expression cassettes tagged with the mneoI indicator cassette [pPAI1(a-c)mneoI; see Materials and Methods]. We chose these cDNAs because they are expressed at relatively high levels in human cells (13). Indeed, the expression of each cDNA was confirmed by RT-PCR (not shown). Notably, we only used DNA sequences corresponding to the gene region of PAI1; thus, polyadenylation will occur at the SV40 pA site present in the pCEP4 expression vector (36, 37).
The resultant constructs were cotransfected into HeLa cells with either JM101/L1.3 Δneo or JM101/L1RP Δneo, a second RC-L1 that retrotransposes at a slightly higher frequency than JM101/L1.3 (25). As before, JM101/L1.3 and JM101/L1RP retrotransposed extremely efficiently, and L1.3 ORF1mneoI and RD-L1s were complemented at about 0.3 to 0.7% of the level of their respective controls (Table 2). By contrast, the PAI1(a-c)mneoI constructs were complemented at reproducibly far lower levels (ca. 0.004 to 0.05% of the respective wild-type controls; Table 2). Notably, a construct containing a von Willebrand factor expression cassette tagged with mneoI retrotransposed at a similar low frequency (not shown).
Functional domains in both the L1.3 ORF1- and ORF2-encoded proteins are required for the retrotransposition of cellular RNAs.
We determined that retrotransposition of pAI1bmneoI was dependent on both the L1.3 ORF1- and L1.3 ORF2-encoded proteins. Missense mutations in either L1.3 ORF1 or in the endonuclease or RT domains of L1.3 ORF2 were unable to stimulate the retrotransposition of pAI1bmneoI above background levels (Table 3). Moreover, cotransfection of L1.3 ORF2 alone could not stimulate pAI1bmneoI retrotransposition (Table 3). Thus, we conclude that specific functional domains in both the L1.3 ORF1- and L1.3 ORF2-encoded proteins are required for this process.
TABLE 3.
Distinct functional domains of the L1.3 ORF1- and L1.3 ORF2-encoded proteins are required to retrotranspose PAI1bmneoIa
| Construct cotransfected with PAI1bmneoI | No. of G418r foci (n = 3)
|
|
|---|---|---|
| Expt 1 | Expt 2 | |
| Alone | 2 | 0 |
| JM101/L1.3 Δneo (WT) | 17 | 16 |
| JM111/L1.3 Δneo (ORF1−) | 0 | 0 |
| JM116/L1.3 Δneo (EN−) | 0 | 1 |
| JM105/L1.3 Δneo (RT−) | 0 | 1 |
| L1.3 ORF2 Δneo | 1 | NDb |
The results from two independent experiments are shown. The constructs cotransfected with PAI1bmneoI are indicated, and the total number of G418r foci obtained in three replicate transfections is indicated. WT, wild type; EN−, endonuclease negative.
ND, not done.
The resultant integration events resemble processed pseudogenes with some unusual features.
To characterize the L1-stimulated pAI1bmneoI retrotransposition events further, we isolated genomic DNA from six clonal cell lines that were established from individual G418r foci (see Materials and Methods). Southern blot analysis demonstrated that each cell line contained a retrotransposition event, and PCR analysis indicated that the resultant retrotransposed sequences were variably 5′ truncated (data not shown [but see Fig. 5]).
FIG. 5.
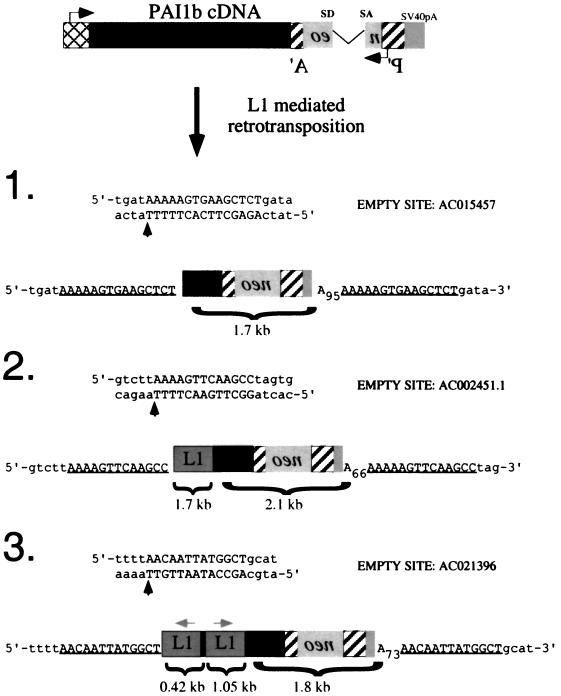
The RC-L1 proteins can generate processed pseudogenes. The structures of three pAI1bmneoI processed pseudogenes and the accession numbers of the empty sites prior to insertion of the pseudogenes are indicated. Vertical upward arrows indicate the precise insertion sites. The poly(A) tail length in each insertion is indicated in subscript; notably, polyadenylation occurred precisely at the SV40 pA site present in the CEP4 vector (36, 37). The target site duplications flanking each insertion are underlined. The black boxes represent pAI1b sequences, while the gray boxes in clones 2 and 3 represent L1 sequences that lie immediately upstream of the cDNA. The gray box between the inverted L1s indicates the pCEP4 derived plasmid sequences (see the text for additional details).
We next used inverse PCR to characterize the pAI1bmneoI retrotransposition event in clones 1 to 3 (27). Each of the resultant integration events contained the hallmarks of a retrotransposition event (Fig. 5). They were 5′ truncated, lacked the intron, ended in poly(A) tails, and were flanked by variable-length target site duplications. In each case, the cDNA integration site resembles a consensus L1 endonuclease cleavage site (5-TTTT/A) (5, 10, 21). Moreover, L1 sequences were not present at the respective “empty” target sites in HeLa DNA. Thus, we conclude that, although inefficient, the L1-encoded proteins can promote the retrotransposition of non-L1 RNAs in HeLa cells, leading to the formation of processed pseudogenes.
Notably, the structures of two of three characterized pseudogenes (clones 2 and 3; Fig. 5) were unusual and contained RC-L1 sequences immediately upstream of the 5′ truncated cDNA. In both instances, the L1-cDNA junction sequences occur in an area that lacks extensive sequence homology between the RC-L1 and pAI1b cDNAs (two or three bases, respectively; see Discussion for how these sequences may have been generated).
Pseudogene 2 contains a 1.7-kb fragment of the RC-L1 that spans bases 1723 to 3405 (7) and is in the same transcriptional orientation as the pPAI1mneoI cDNA. Pseudogene 3 contains an internally rearranged 1.5-kb fragment of the RC-L1. The 3′ fragment spans bases 4444 to 5495 (7) of the RC-L1 and is in the same transcriptional orientation as the pPAI1bmneoI cDNA. The 5′ fragment is in the opposite transcriptional orientation of the pPAI1bmneoI cDNA and contains the first 393 bp of L1, as well as 32 bp of the pCEP4 expression vector. The pCEP4 sequences are immediately upstream of the RC-L1 and begin 14 bp downstream of the CMV transcription start site. Thus, it appears that the RC-L1-derived portion of pseudogene 3 was initiated from the CMV promoter and then underwent an inversion and deletion upon its retrotransposition. Such inversion or deletion events are relatively common and may represent about 15% of all L1 retrotransposition events (14, 20).
DISCUSSION
L1s retrotranspose by cis preference.
We have provided genetic evidence in support of the cis-preference model of L1 retrotransposition. Population genetic and phylogenetic analyses revealed that new L1 retrotransposition events in mice and humans likely emanated from a small number of “founder genes” (3, 12, 15, 22). Mutational and biochemical studies also have provided additional data in support of a cis preference. First, all mutagenic L1 insertions characterized in humans and mice are derived from progenitor L1s that contain intact ORFs (7, 19, 25, 37, 38). Second, cytoplasmic RNPs, which are proposed intermediates in the L1 retrotransposition pathway, are enriched for the RNAs and proteins encoded by young L1s (17, 18, 26, 31). Finally, Esnault et al. (8) recently provided experimental evidence consistent with the notion that human L1s retrotranspose by cis preference in cultured feline cells. Our finding that stable RD-L1 RNAs are complemented inefficiently provides the most compelling evidence to date that L1s predominantly retrotranspose via cis preference.
Our data are consistent with two versions of a relatively simple model for the molecular mechanism of cis preference in which L1 RNAs cotranslationally bind nascent L1 proteins. The binding of the nascent proteins to L1 RNA could be mediated by proximity. Alternatively, the RC-L1 protein(s) might only have a limited half-life in the absence of L1 RNA. In either case, these mechanisms would ensure that functional L1 RNAs were far more likely than RD-L1 RNAs or cellular mRNAs to serve as substrates for TPRT.
Processed pseudogene formation.
We demonstrated that the RC-L1 proteins also function in trans at low levels to promote the retrotransposition of other mRNAs, leading to the formation of processed pseudogenes. Interestingly, two of three characterized pAI1bmneoI processed pseudogenes were linked physically to sequences in the cotransfected RC-L1, generating chimeric L1-cDNA pseudogene structures that have not yet been found in nature. These L1-cDNA chimeras could occur if the RC-L1 and the pAI1bmneoI plasmids underwent an illegitimate (i.e., not mediated by homology) interplasmid recombination event, leading to the formation of a recombinant L1-cDNA mRNA that subsequently was retrotransposed in trans by the RC-L1 proteins. Alternatively, it is possible that L1 retrotransposition intermediates contain two RNAs and that RNA or cDNA recombination during TPRT yielded the chimeras (12, 16).
In either case, our data demonstrate that RD-L1s are trans complemented at much higher frequencies than non-L1 cDNAs. Since both the RD-L1s and cDNAs are in identical expression contexts, it is unlikely that the effect we observe is due to transcript abundance. Instead, it remains possible that the RD-L1 RNAs either colocalize with RC-L1 RNAs or that the RD-L1 RNAs contain cis-acting sequences that can recruit the RC-L1 encoded proteins (18).
It is worth comparing our results to those generated recently by Esnault et al., who demonstrated that human L1s could mediate processed pseudogene formation in a heterologous cultured feline cell system (8). We agree that the RC-L1 proteins function preferentially in cis but act at a low level in trans to promote the retrotransposition of non-L1 RNAs. Moreover, we both found that the structures of the processed pseudogenes were somewhat unusual. However, differences between our studies are noteworthy. In the experiments of Esnault et al., none of the pseudogenes that arose in the feline cells integrated at consensus L1 endonuclease cleavage sites. Indeed, two of three “pseudogenes” actually lacked poly(A) sequences. All three of our pseudogenes had all the characteristics of retrotransposition events generated via TPRT.
In addition, our data demonstrate that pseudogene formation in human cells is extremely rare (ca. 0.01 to 0.05% of the rate of L1 retrotransposition) and only is detected when our most “active” L1s are used as sources of the L1-encoded proteins. By contrast, in feline-cultured cells, the frequency of processed pseudogene formation is only 10-fold lower than the frequency of L1 retrotransposition. While it remains possible that these discrepancies reflect subtle differences that exist between our assays, it is interesting to note that both we and Dhellin et al. were unable to detect pseudogene formation when L1.2 was overexpressed in human cells (6; J. V. Moran et al., unpublished data). Thus, it appears that the feline cells are more permissive for L1-mediated processed pseudogene formation than human HeLa cells.
Finally, it is notable that the studies conducted by Esnault et al. were performed in the presence of phleomycin, a known clastogen. Thus, it remains possible that interplasmid recombination occurred more frequently in their study. Moreover, the unusual structures of the pseudogenes suggest that they may have integrated into chromosomal DNA by an L1 endonuclease-independent mechanism.
Evolutionary implications and practical considerations of the cis-preference model.
The cis-preference model would explain how a small number of autonomous L1s remain retrotransposition competent among an overwhelming abundance of nonfunctional elements. Indeed, such a mechanism would select for the retrotransposition of RC-L1 RNAs and would be consistent with the apparent patterns of concerted evolution that L1s display in different species (15, 32). It also would limit the extent to which the proteins encoded by RC-L1s could function to retrotranspose other cellular RNAs (e.g., RD-L1s and other cellular RNAs). However, it is noteworthy that particular RNAs (e.g., Alu RNAs) likely have evolved ways to usurp the cis-preference retrotransposition machinery of human RC-L1s (possible mechanisms are discussed further elsewhere [1, 23, 33]).
Finally, the finding that L1s retrotranspose by cis preference may have practical value. For example, if engineered L1s were used as transposon mutagens, there is a high likelihood that any resultant mutations would be due to the retrotransposition of the engineered L1 RNA and would not be caused by the trans mobilization of endogenous retrotransposons. Indeed, the inability of the RC-L1 proteins to efficiently mobilize cellular RNAs may prove useful when considering L1 as a potential gene delivery vehicle.
ACKNOWLEDGMENTS
We thank Anne Marie DesLauriers at the University of Michigan Flow Core for help with flow cytometry, Robert Lyons at the University of Michigan DNA Sequencing Core for help with oligonucleotide synthesis and DNA sequencing, and Ali Lotia for help with generating computer graphics. We thank David Ginsburg for providing pAI1 cDNAs. We thank Alice Telesnitsky, John Goodier, Eline Luning Prak, Tom Glaser, Dennis Hartigan-O'Connor, and current members of the Moran Lab for critical reading of the manuscript and for helpful discussions during the course of this work.
This work was supported in part by a Damon Runyon Scholar Award (J.V.M.) and National Institutes of Health grants GM60518 (J.V.M.) and CA16519 (J.D.B.).
REFERENCES
- 1.Boeke J D. LINEs and Alus—the polyA connection. Nat Genet. 1997;16:6–7. doi: 10.1038/ng0597-6. [DOI] [PubMed] [Google Scholar]
- 2.Boeke J D, Stoye J P. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 343–435. [PubMed] [Google Scholar]
- 3.Boissinot S, Chevret P, Furano A V. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol. 2000;17:915–928. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- 4.Christensen S, Pont-Kingdon G, Carroll D. Target specificity of the endonuclease from the Xenopus laevis non-lung terminal repeat retrotransposon, Tx1L. Mol Cell Biol. 2000;20:1219–1226. doi: 10.1128/mcb.20.4.1219-1226.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cost G J, Boeke J D. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry. 1998;37:18081–18093. doi: 10.1021/bi981858s. [DOI] [PubMed] [Google Scholar]
- 6.Dhellin O, Maestre J, Heidmann T. Functional differences between the human LINE retrotransposon and retroviral reverse transcriptases for in vivo mRNA reverse transcription. EMBO J. 1997;16:6590–6602. doi: 10.1093/emboj/16.21.6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dombroski B A, Mathias S L, Nanthakumar E, Scott A F, Kazazian H H., Jr Isolation of an active human transposable element. Science. 1991;254:1805–1808. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- 8.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 9.Fanning T, Singer M. The LINE-1 DNA sequences in four mammalian orders predict proteins that conserve homologies to retrovirus proteins. Nucleic Acids Res. 1987;15:2251–2260. doi: 10.1093/nar/15.5.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng Q, Moran J V, Kazazian H H, Jr, Boeke J D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 11.Feng Q, Schumann G, Boeke J D. Retrotransposon R1Bm endonuclease cleaves the target sequence. Proc Natl Acad Sci USA. 1998;95:2083–2088. doi: 10.1073/pnas.95.5.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furano A V. The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Prog Nucleic Acid Res Mol Biol. 2000;64:255–294. doi: 10.1016/s0079-6603(00)64007-2. [DOI] [PubMed] [Google Scholar]
- 13.Ginsburg D, Zeheb R, Yang A Y, Rafferty U M, Andreasen P A, Nielsen L, Dano K, Lebo R V, Gelehrter T D. cDNA cloning of human plasminogen activator-inhibitor from endothelial cells. J Clin Investig. 1986;78:1673–1680. doi: 10.1172/JCI112761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodier J L, Ostertag E M, Kazazian H H., Jr Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum Mol Genet. 2000;9:653–657. doi: 10.1093/hmg/9.4.653. [DOI] [PubMed] [Google Scholar]
- 15.Hardies S C, Martin S L, Voliva C F, Hutchison C A D, Edgell M H. An analysis of replacement and synonymous changes in the rodent L1 repeat family. Mol Biol Evol. 1986;3:109–125. doi: 10.1093/oxfordjournals.molbev.a040386. [DOI] [PubMed] [Google Scholar]
- 16.Hayward B E, Zavanelli M, Furano A V. Recombination creates novel L1 (LINE-1) elements in Rattus norvegicus. Genetics. 1997;146:641–654. doi: 10.1093/genetics/146.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hohjoh H, Singer M F. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- 18.Hohjoh H, Singer M F. Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J. 1997;16:6034–6043. doi: 10.1093/emboj/16.19.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmes S E, Dombroski B A, Krebs C M, Boehm C D, Kazazian H H., Jr A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nat Genet. 1994;7:143–148. doi: 10.1038/ng0694-143. [DOI] [PubMed] [Google Scholar]
- 20.Hutchison C A, Hardies S C, Loeb D D, Shehee W R, Edgell M H. LINES and related retroposons: long interspersed sequences in the eucaryotic genome. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: ASM Press; 1989. pp. 593–617. [Google Scholar]
- 21.Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci USA. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaplan N, Darden T, Langley C H. Evolution and extinction of transposable elements in Mendelian populations. Genetics. 1985;109:459–480. doi: 10.1093/genetics/109.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kazazian H H, Jr, Moran J V. The impact of L1 retrotransposons on the human genome. Nat Genet. 1998;19:19–24. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- 24.Kazazian H H, Jr, Wong C, Youssoufian H, Scott A F, Phillips D G, Antonarakis S E. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332:164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- 25.Kimberland M L, Divoky V, Prchal J, Schwahn U, Berger W, Kazazian H H., Jr Full-length human L1 insertions retain the capacity for high-frequency retrotransposition in cultured cells. Hum Mol Genet. 1999;8:1557–1560. doi: 10.1093/hmg/8.8.1557. [DOI] [PubMed] [Google Scholar]
- 26.Kolosha V O, Martin S L. Polymorphic sequences encoding the first open reading frame protein from LINE-1 ribonucleoprotein particles. J Biol Chem. 1995;270:2868–2873. doi: 10.1074/jbc.270.6.2868. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Shen H, Himmel K L, Dupuy A J, Largaespada D A, Nakamura T, Shaughnessy J D, Jr, Jenkins N A, Copeland N G. Leukaemia disease genes: large-scale cloning and pathway predictions. Nat Genet. 1999;23:348–353. doi: 10.1038/15531. [DOI] [PubMed] [Google Scholar]
- 28.Luan D D, Eickbush T H. RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol Cell Biol. 1995;15:3882–3891. doi: 10.1128/mcb.15.7.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luan D D, Korman M H, Jakubczak J L, Eickbush T H. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 30.Maestre J, Tchenio T, Dhellin O, Heidmann T. mRNA retroposition in human cells: processed pseudogene formation. EMBO J. 1995;14:6333–6338. doi: 10.1002/j.1460-2075.1995.tb00324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin S L. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol Cell Biol. 1991;11:4804–4807. doi: 10.1128/mcb.11.9.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin S L, Voliva C F, Hardies S C, Edgell M H, Hutchison C A D. Tempo and mode of concerted evolution in the L1 repeat family of mice. Mol Biol Evol. 1985;2:127–140. doi: 10.1093/oxfordjournals.molbev.a040340. [DOI] [PubMed] [Google Scholar]
- 33.Mathews D H, Banerjee A R, Luan D D, Eickbush T H, Turner D H. Secondary structure model of the RNA recognized by the reverse transcriptase from the R2 retrotransposable element. RNA. 1997;3:1–16. [PMC free article] [PubMed] [Google Scholar]
- 34.Mathias S L, Scott A F, Kazazian H H, Jr, Boeke J D, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 35.Mohler W A, Blau H M. Gene expression and cell fusion analyzed by lacZ complementation in mammalian cells. Proc Natl Acad Sci USA. 1996;93:12423–12427. doi: 10.1073/pnas.93.22.12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moran J V, DeBerardinis R J, Kazazian H H. Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 37.Moran J V, Holmes S E, Naas T P, DeBerardinis R J, Boeke J D, Kazazian H H., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 38.Naas T P, DeBerardinis R J, Moran J V, Ostertag E M, Kingsmore S F, Seldin M F, Hayashizaki Y, Martin S L, Kazazian H H. An actively retrotransposing, novel subfamily of mouse L1 elements. EMBO J. 1998;17:590–597. doi: 10.1093/emboj/17.2.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ostertag E M, Prak E T, DeBerardinis R J, Moran J V, Kazazian H H., Jr Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 2000;28:1418–1423. doi: 10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sassaman D M, Dombroski B A, Moran J V, Kimberland M L, Naas T P, DeBerardinis R J, Gabriel A, Swergold G D, Kazazian H H., Jr Many human L1 elements are capable of retrotransposition. Nat Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 41.Scott A F, Schmeckpeper B J, Abdelrazik M, Comey C T, O'Hara B, Rossiter J P, Cooley T, Heath P, Smith K D, Margolet L. Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics. 1987;1:113–125. doi: 10.1016/0888-7543(87)90003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smit A F. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 43.Swergold G D. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol Cell Biol. 1990;10:6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tchenio T, Segal-Bendirdjian E, Heidmann T. Generation of processed pseudogenes in murine cells. EMBO J. 1993;12:1487–1497. doi: 10.1002/j.1460-2075.1993.tb05792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei W, Morrish T A, Alisch R S, Moran J V. A transient assay reveals that cultured human cells can accommodate multiple LINE-1 retrotransposition events. Anal Biochem. 2000;284:435–438. doi: 10.1006/abio.2000.4675. [DOI] [PubMed] [Google Scholar]
- 46.Weiner A M, Deininger P L, Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- 47.Xiong Y E, Eickbush T H. Functional expression of a sequence-specific endonuclease encoded by the retrotransposon R2Bm. Cell. 1988;55:235–246. doi: 10.1016/0092-8674(88)90046-3. [DOI] [PubMed] [Google Scholar]