Abstract
The spindle assembly checkpoint monitors the attachment of kinetochores to the mitotic spindle and the tension exerted on kinetochores by microtubules and delays the onset of anaphase until all the chromosomes are aligned at the metaphase plate. The target of the checkpoint control is the anaphase-promoting complex (APC)/cyclosome, a ubiquitin ligase whose activation by Cdc20 is required for separation of sister chromatids. In response to activation of the checkpoint, Mad2 binds to and inhibits Cdc20-APC. I show herein that in checkpoint-arrested cells, human Cdc20 forms two separate, inactive complexes, a lower affinity complex with Mad2 and a higher affinity complex with BubR1. Purified BubR1 binds to recombinant Cdc20 and this interaction is direct. Binding of BubR1 to Cdc20 inhibits activation of APC and this inhibition is independent of its kinase activity. Quantitative analysis indicates that BubR1 is 12-fold more potent than Mad2 as an inhibitor of Cdc20. Although at high protein concentrations BubR1 and Mad2 each is sufficient to inhibit Cdc20, BubR1 and Mad2 mutually promote each other's binding to Cdc20 and function synergistically at physiological concentrations to quantitatively inhibit Cdc20-APC. Thus, BubR1 and Mad2 act cooperatively to prevent premature separation of sister chromatids by directly inhibiting APC.
INTRODUCTION
Cell cycle progression is monitored by checkpoint mechanisms that ensure the integrity of the genome and the fidelity of chromosome segregation (Elledge, 1996). In mitosis, the spindle assembly checkpoint delays anaphase initiation until all the chromosomes are aligned at the metaphase plate, thereby ensuring the fidelity of sister chromatid separation (Amon, 1999; Fang et al., 1999). This checkpoint process monitors both the attachment of kinetochores to the mitotic spindle and the tension exerted on kinetochores by microtubules (Li and Nicklas, 1995; Rieder and Khodjakov, 1997; Shah and Cleveland, 2000; Skoufias et al., 2001). The presence of a single unattached kinetochore activates the spindle assembly checkpoint and prevents sister chromatid separation, anaphase initiation, and loss of Cdc2 kinase activity, thus providing additional time for kinetochores to be captured by the mitotic spindle and for tension to be established at kinetochores. Because unattached kinetochores are present in every prophase cell, this checkpoint also functions in every cell cycle in the normal timing of anaphase initiation (Taylor and McKeon, 1997). Genetic disruption of this checkpoint in mouse embryos causes early lethality due to mis-separation of chromosomes (Dobles et al., 2000). Thus, the spindle assembly checkpoint is an intrinsic cell cycle quality control mechanism that acts to prevent aneuploidy due to unequal chromosome separation during mitosis.
The mitotic checkpoint can be activated experimentally by microtubule-destabilizing drugs such as nocodazole. By using such drugs, genetic studies in budding yeast have identified several components of the checkpoint pathway. Mutations in any of these genes (MAD1, 2, 3; BUB1, 2, 3; and MPS1) allow for the completion of an aberrant mitosis in the presence of spindle defects and eventually lead to cell death (Hoyt et al., 1991; Li and Murray, 1991; Hardwick et al., 1996). Vertebrate homologs of the yeast checkpoint proteins have also been identified; these include Mad1, Mad2, Bub1, BubR1, and Bub3 (Chen et al., 1996; Li and Benezra, 1996; Taylor and McKeon, 1997; Chan et al., 1998, 1999; Chen et al., 1998; Fang et al., 1998b; Jin et al., 1998; Taylor et al., 1998).
The key target of the spindle assembly checkpoint is the anaphase-promoting complex (APC)/cyclosome, a ubiquitin ligase that controls sister chromatid separation and exit from mitosis (Li et al., 1997; Fang et al., 1998a,b, 1999; Hwang et al., 1998; Kallio et al., 1998; Kim et al., 1998; Wassmann and Benezra, 1998). The activity of APC is positively regulated by two regulatory proteins, Cdc20 and Cdh1/Hct1. Cdc20 and Cdh1 are activators of APC; they directly bind to APC and activate its ubiquitination activity (Schwab et al., 1997; Visintin et al., 1997; Fang et al., 1998a). At the metaphase-to-anaphase transition, Cdc20 activates APC, leading to degradation of the anaphase inhibitor securin, and separation of sister chromatids. Later, Cdh1 binds to APC, resulting in complete degradation of mitotic cyclins and exit from mitosis.
APC is also negatively regulated by two checkpoint proteins, Mad2 and BubR1. When the spindle assembly checkpoint is activated, Mad2 binds to Cdc20 and inhibits the activation of APC (Li et al., 1997; Fang et al., 1998b; Hwang et al., 1998; Kim et al., 1998). Deletion of one Mad2 allele results in premature anaphase and chromosome instability in mammalian cells, indicating an essential role of Mad2 in mitotic checkpoint control (Michel et al., 2001).
BubR1 is a protein kinase required for checkpoint control, because its inactivation by microinjection of specific antibodies abolishes the checkpoint control (Chan et al., 1998, 1999). BubR1 associates and directly phosphorylates Cdc20 (Yao et al., 2000; Skoufias et al., 2001; Tang et al., 2001). However, the functional consequence of the phosphorylation remains unknown (Wu et al., 2000). It has been shown very recently that recombinant BubR1 can directly inhibit Cdc20-APC and this inhibition is independent of its kinase activity, because a kinase-dead mutant of BubR1 is still capable of inhibiting Cdc20 (Tang et al., 2001). Furthermore, it has been reported that BubR1 forms a mitotic checkpoint complex (MCC) with Bub3, Cdc20, and Mad2 in vivo and MCC is 3000-fold more efficient than Mad2 in inhibiting Cdc20-APC (Sudakin et al., 2001).
I have independently investigated the biochemical basis of BubR1 function in mitotic checkpoint control. I found that, similar to Mad2, human BubR1 directly associates with and inhibits Cdc20, consistent with recent reports (Sudakin et al., 2001; Tang et al., 2001). Quantitative analysis indicates that BubR1 binds to Cdc20 with a higher affinity and is a more potent than Mad2 in inhibiting the activation of APC by Cdc20. Furthermore, BubR1 and Mad2 enhance each other's ability to bind to Cdc20 and they function synergistically in inhibiting APC. Thus, the two checkpoint proteins BubR1 and Mad2 act cooperatively to ensure a quantitative inhibition of APC and a complete arrest of mitotic progression.
MATERIALS AND METHODS
Expression and Purification of Recombinant Proteins
Human BubR1 and Bub3 genes were cloned by polymerase chain reaction from human fetal thymus cDNA. BubR1 and Bub3 genes were tagged with His6 by subcloning into pFastBacHTa (Invitrogen, Carlsbad, CA) and expressed in Sf9 cells. Proteins were purified by Telon beads (CLONTECH, Palo Alto, CA) and peak fractions were loaded onto a HiTrap Q column and eluted by a salt gradient. Purified recombinant proteins were then concentrated by Centriprep-10 (Amersham Biosciences, Piscataway, NJ) and dialyzed against a buffer containing 10 mM HEPES, pH 7.7, 100 mM KCl, and 1 mM dithiotreitol (DTT). Expression and purification of recombinant Mad2 and Cdc20 were described previously (Fang et al., 1998b).
Antibodies
Rabbit and mouse anti-Myc, anti-hemagglutinin (HA), and anti-cyclin B antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and used at 0.1 μg/ml for immunoprecipitation and at 1 μg/ml for Western blot analysis. Polyclonal rabbit sera were raised against the recombinant Mad1, Bub3, and BubR1 proteins. Antibodies were affinity purified with the recombinant antigens covalently coupled to Affi-Gel (Bio-Rad, Hercules, CA) or Ultralink Support (Pierce Chemical, Rockford, IL). Three separate antibodies raised against BubR1 amino acids, 1–198, 253–271, and 367–385, were used for immunoprecipitation experiments; all gave comparable results. Anti-Mad2 and anti-Cdc20 antibodies were described previously (Fang et al., 1998b).
In Vitro Binding Assays
In vitro-translated, [35S]Met-labeled, Myc-tagged, and HA-tagged proteins, 10 μl each, were incubated together for 1 h at room temperature, and then incubated with 1 μg of anti-Myc or anti-HA antibodies for 1 h. Protein A beads were added, incubated for 1 h, and washed five times with 20 volumes of XB buffer (10 mM HEPES, pH 7.7, 100 mM KCl, 0.1 mM CaCl2, and 50 mM sucrose) containing 500 mM KCl and 0.5% NP-40, and twice with XB. Immunoprecipitated proteins were then analyzed by SDS-PAGE. Alternatively, in vitro-translated proteins were incubated with 1 μg of recombinant Mad2 at room temperature for 1 h and Mad2 complexes were immunopurified by an anti-Mad2 antibody.
To analyze interactions between BubR1 and Cdc20 in a purified system, 1 μg of purified Myc-Cdc20 was incubated with 1 μg of purified BubR1 at room temperature for 1 h and the Myc–Cdc20 complex was then immunoprecipitated by 5 μg of affinity-purified anti-Myc antibody and analyzed by SDS-PAGE.
Ubiquitination Assays
To assay effects of BubR1 and Mad2 on activation of APC in reconstituted ubiquitination reactions in vitro (Fang et al., 1998a,b), 3 μl of anti-Cdc27 antibody/protein A beads was incubated with 30 μl of Xenopus interphase extracts at 4°C for 2 h. The APC beads were washed extensively and incubated, in a total volume of 9 μl, with recombinant Myc-Cdc20 (16 pmol) and various amounts of BubR1, Bub3, Mad2, and Mad2B as indicated in figure legends. APC beads were then washed twice with XB and ubiquitination assays performed in a total volume of 5 μl. The reaction mixture contains ATP or 5-adenylylimidodiphosphate (AMP-PNP), 1.25 mg/ml ubiquitin, 12.5 ng/ml labeled substrate, 200 μg/ml wheat E1, 50 μg/ml Xenopus UBCx, and 3 μl of APC beads. The substrate used was an amino-terminal fragment of Xenopus cyclin B (residues 1–102) and was labeled with 125I to a specific activity of 100 μCi/μg by using the chloramine T method. The reactions were incubated at room temperature for indicated time, quenched with SDS sample buffer, and analyzed by SDS-PAGE. Gels were scanned with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Synchronization of HeLa Cells
HeLa S3 cells (American Type Culture Collection, Manassas, VA) were grown in DMEM (Invitrogen) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and 100 μg/ml penicillin and streptomycin. For synchronization with a thymidine and nocodazole block (Fang et al., 1998b), cells were grown in the presence of 2 mM thymidine (Sigma Chemical, St. Louis, MO) for 18 h, washed with phosphate-buffered saline (PBS), and grown in fresh medium without thymidine for 4 h. Cells were then incubated with 100 ng/ml nocodazole (Sigma Chemical) for 12 h to activate the spindle assembly checkpoint. Cells were washed with PBS twice and then harvested, or transferred into fresh medium for time indicated and harvested. For double thymidine block (Fang et al., 1998a), HeLa S3 cells were grown in the presence of 2 mM thymidine for 18 h, washed with PBS, and grown in fresh medium without thymidine for 8 h. Thymidine was added again to 2 mM to block cells at G1/S. After another 18 h, cells were transferred to fresh medium and samples were harvested at time indicated. The cell cycle status of the samples was determined by fluorescence-activated cell sorting analysis of the DNA content.
Preparation and Fractionation of Cell Extracts and Immunoprecipitation
For preparation of extracts (Fang et al., 1998b), cells were lysed with seven volumes of the NP-40 lysis buffer (50 mM Tris-HCl, pH 7.7; 150 mM NaCl; 0.5% NP-40; 1 mM DTT; 10% glycerol; 0.5 μM okadaic acid; and 10 μg/ml each of leupeptin, pepstatin, and chymostatin). Lysates were then centrifuged for 30 min at 200,000 × g to make the S100 supernatants. S100 supernatant was applied to a 6-ml Resource Q column (Amersham Biosciences) equilibrated with buffer Q-A (20 mM Tris-HCl pH 7.7, 100 mM KCl, 0.1 mM CaCl2, 1 mM MgCl2, and 1 mM DTT) and eluted with a linear gradient of 0–60% buffer Q-B (Q-A + 1 M KCl) in a volume of 60 ml. Column fractions were analyzed by Western blot with anti-BubR1, Cdc20, Mad2, and APC2 antibodies. Peak fractions for BubR1 were pooled, concentrated by Centriprep-10 (Amersham Biosciences), and loaded onto a 120-ml Superose 6 column. Column fractions were then assayed with various antibodies against checkpoint proteins by Western blotting.
For immunoprecipitation, anti-BubR1, anti-Mad2, and anti-Cdc20 antibodies were covalently coupled to Affi-Prep protein A beads at a concentration of 1 mg of the antibody per milliliter of beads (Bio-Rad). The beads were washed twice with 10 volumes of 100 mM glycine pH 2.5, and neutralized with 10 mM Tris-HCl pH 7.5. Three microliters of antibody beads was incubated with 3 ml of HeLa S100 supernatants or 0.5 ml of various column fractions overnight at 4°C. Beads were washed five times with 20 volumes of XB buffer containing 500 mM KCl and 0.5% NP-40, and three times with XB alone. Proteins bound to beads were eluted with SDS sample buffer, separated by SDS-PAGE, and analyzed by Western blotting.
RESULTS
Cdc20 Forms a Stable Complex with BubR1 When Mitotic Checkpoint Is Activated
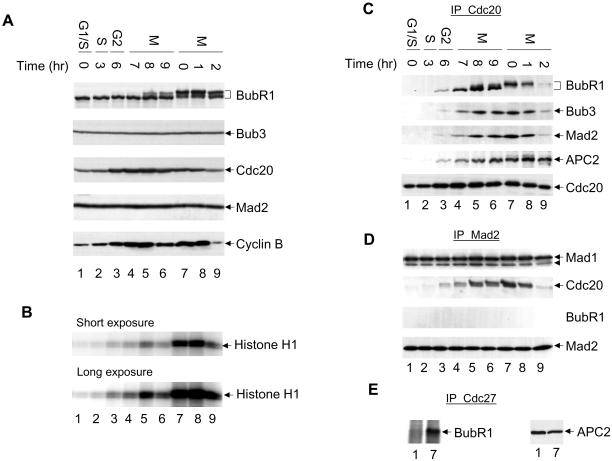
To examine the function of the BubR1 kinase in mitotic checkpoint control, I analyzed the association of BubR1 with various checkpoint proteins in the cell cycle. These experiments involve Western blot analysis as well as immunoprecipitations followed by Western blot analysis for BubR1, Bub3, Mad1, Mad2, Cdc20, and APC (Figure 1). HeLa cells were synchronized by a double thymidine block, released from their G1/S arrest, and sampled for 9 h until they began to exit from mitosis (Figure 1, A–E, lanes 1–6) (Fang et al., 1998a). In addition, HeLa cells were arrested by nocodazole treatment, which activates the mitotic checkpoint, and cell samples were collected at 0, 1, and 2 h after release from nocodazole (Figure 1, A–E, lanes 7–9) (Fang et al., 1998b). At the drug concentration used herein, nocodazole completely depolymerizes microtubules and arrests cells at prometaphase. The cell cycle stages were determined by the level of cyclin B protein, by the level of the Cdc2 kinase activity by using histone H1 as a substrate, and by fluorescence-activated cell sorting analysis (Figure 1, A and B; my unpublished data).
Figure 1.
Checkpoint complexes in the cell cycle. HeLa cells were synchronized at the G1/S boundary by a double thymidine block and cells were collected at indicated time after being released from the arrest (lanes 1–6). Alternatively, HeLa cells were synchronized at prometaphase by a thymidine-nocodazole block. Cells were collected immediately after nocodazole treatment (lane 7), or released into fresh media for 1 and 2 h (lanes 8 and 9). (A) Levels of various mitotic regulators and checkpoint proteins were determined by Western blot analyses. (B) Cdc2 kinase was immunopurified from cell lysates and assayed using histone H1 as a substrate. (C–E) Lysates of synchronized HeLa cells were immunoprecipitated with anti-Cdc20 (C), anti-Mad2 (D), or anti-Cdc27 (E) antibody, and the immunoprecipitates were analyzed by Western blotting with anti-BubR1, Bub3, Mad2, Mad1, Cdc20, and APC2 antibodies. Arrowhead in D, a partial degradation product of Mad1.
BubR1 is phosphorylated in mitosis, but the protein level does not seem to change in the cell cycle. Similarly, levels of Bub3, Mad1, and Mad2 do not change throughout the cell cycle (Figure 1A; my unpublished data) (Li and Benezra, 1996; Fang et al., 1998b; Chan et al., 1999; Wu et al., 2000). The level of Cdc20 protein fluctuates in the cell cycle, peaking at mitosis and dropping by about twofold as cells exit from mitosis (my unpublished data). When lysates of nocodazole-arrested cells were immunoprecipitated with an anti-Cdc20 antibody and analyzed by Western blotting with antibodies against BubR1, Bub3, Mad2, and APC2 (Figure 1C, lane 7), I found that BubR1, Bub3, Mad2, and APC all associated with Cdc20, consistent with previous reports (Fang et al., 1998b; Chan et al., 1999; Wu et al., 2000). Cdc20 associates with BubR1 and Bub3 in mitosis and dissociates as cells exit from mitosis (Figure 1C). This cell cycle profile of the association of Cdc20 with BubR1 and Bub3 is very similar to that of Cdc20 with Mad2. On the other hand, Cdc20–APC complex accumulates as cells enter mitosis (Figure 1C, lanes 3–8), but this complex lacks ubiquitin ligase activity due to inhibition by the mitotic checkpoint pathway (Fang et al., 1998b; Chan et al., 1999; Wu et al., 2000). BubR1, Bub3, and Mad2 all dissociate from Cdc20 2 h after release from nocodazole arrest. This is in sharp contrast with the association of Cdc20 and APC, which persists as cells exit from mitosis (Figure 1C, lane 9). It is during this time, 1–2 h after release from the prometaphase arrest, that APC is active and cyclin B is degraded (Figure 1A, lane 9). Quantitative analysis indicates that the level of Cdc20 only varies about twofold in the cell cycle, whereas the abundance of BubR1–Cdc20 and Mad2–Cdc20 complexes changes greater than eightfold (Figure 1; my unpublished data). Thus, formation of these checkpoint complexes is regulated. The cell cycle-dependent association of BubR1 and Bub3 with Cdc20 raises the possibility that, like Mad2, checkpoint proteins BubR1 and Bub3 may directly regulate the activity of APC in response to activation of the mitotic checkpoint.
To determine whether BubR1, Bub3, and Mad2 form a single complex with Cdc20, or several different complexes, I immunoprecipitated Mad2 with an anti-Mad2 antibody and assayed for associated proteins by Western blotting (Figure 1D). The amount of Mad2 immunoprecipitated is constant throughout the cell cycle. The checkpoint protein Mad1 binds to Mad2 and this interaction is independent of cell cycle stage. As expected, Mad2 associates with Cdc20 in mitosis. However, I failed to detect any association of Mad2 with either BubR1 or Bub3 (Figure 1D; my unpublished data). Although I cannot exclude the possibility that immunoprecipitation by the anti-Mad2 antibody dissociates a BubR1–Mad2–Cdc20 complex, this is unlikely because immunoprecipitation by three polyclonal antibodies raised against different regions of BubR1 all failed to precipitate Mad2 (my unpublished data). Thus, BubR1/Bub3 and Mad2 are likely to form two separate complexes with Cdc20.
Mad2 and Cdc20 form a ternary complex with APC and Mad2 inhibits the ubiquitin ligase activity of APC in this complex (Li et al., 1997; Fang et al., 1998b). To test whether BubR1 associates with APC, I immunopurified APC with an anti-Cdc27 antibody and found that BubR1 is associated with APC by Western blot analysis (Figure 1E, lane 7) (Chan et al., 1999). Furthermore, formation of this complex is dependent on activation of the mitotic checkpoint, because this complex is absent in G1/S cells (Figure 1E, cf. lane 1 with lane 7), suggesting that BubR1 may directly regulate the ubiquitin ligase activity of APC. Quantitative analysis indicates that, like Mad2, the majority of BubR1 forms a binary complex with Cdc20 and only a small fraction of BubR1 interacts with APC (see below; Figure 7A). Similarly, a small fraction of Bub3 also associates with APC in a checkpoint-dependent manner (my unpublished data).
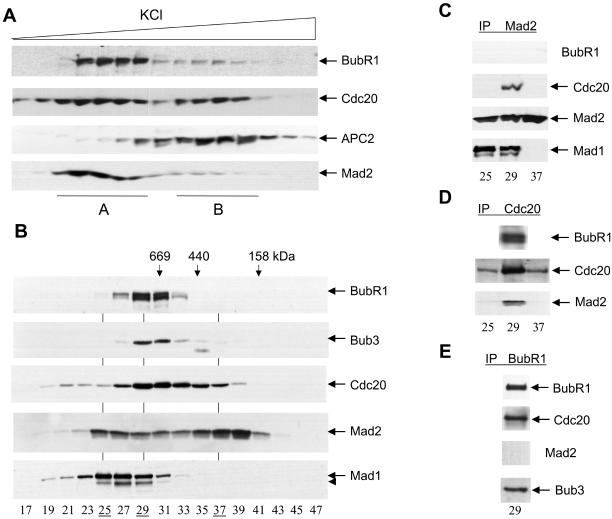
Figure 7.
Checkpoint complexes in nocodazole-arrested HeLa cells. (A) Mitotic checkpoint was activated in HeLa cells by a thymidine-nocodazole arrest. Cell lysates were fractionated through a Resource Q column and fractionation profiles of BubR1, Cdc20, APC2, and Mad2 were determined by immunoblotting. (B) Fractions from peak A were pooled, concentrated, and further fractionated by a Superose 6 column. Fractionation profiles of BubR1, Bub3, Cdc20, Mad2, and Mad1 were determined by immunoblotting. Arrowhead, a partial degradation product of Mad1. (C–E) Fractions 25, 29, and 37 from the Superose 6 column were immunoprecipitated with anti-Mad2 (C), anti-Cdc20 (D), and anti-BubR1 (E) antibodies, followed by Western blot analyses with antibodies against various checkpoint proteins.
BubR1 Directly Inhibits Cdc20
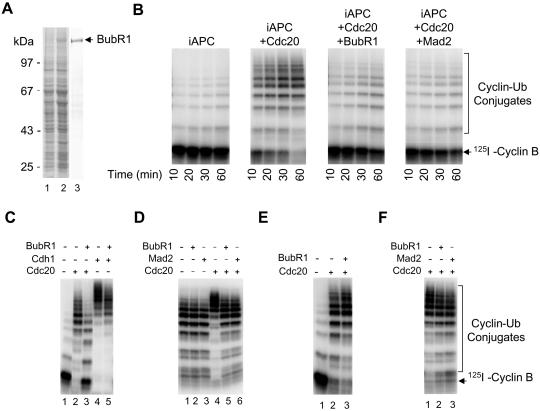
Mad2 binds to Cdc20 and inhibits its activity (Li et al., 1997; Hwang et al., 1998; Kim et al., 1998; Fang et al., 1998b). I tested whether BubR1 directly inhibits Cdc20 to prevent its activation of APC. Recombinant BubR1 was expressed in Sf9 cells and purified to homogeneity (Figure 2A). I have previously developed an efficient two-step assay to analyze the activation of APC (Fang et al., 1998a). First, interphase APC was purified by anti-Cdc27 antibody/protein A beads and then incubated with recombinant Cdc20 in the absence of ATP to allow activation to occur. Second, unbound Cdc20 was removed by several washes of APC beads and the ubiquitin ligase activity was analyzed in the presence of ATP. Interphase APC only has a basal level of activity (Figure 2B). Incubation of interphase APC with purified recombinant Cdc20 activates its ubiquitin ligase activity; radioactively labeled cyclin B was quantitatively converted to cyclin B-ubiquitin conjugates within 20 min (Figure 2B) (Fang et al., 1998a). However, incubating recombinant BubR1 with Cdc20 blocks the activation of interphase APC by Cdc20. APC incubated with BubR1 plus Cdc20 only has an activity comparable to interphase APC, consistent with a recent report (Tang et al., 2001). Similarly, when recombinant Mad2 was incubated with Cdc20, Mad2 blocks the activation of APC by Cdc20. Inhibition of the APC ligase activity by BubR1 is specific to Cdc20, because BubR1 only has a minimal effect on the activation of APC by Cdh1 (Figure 2C).
Figure 2.
BubR1 directly inhibits activation of APC by Cdc20, but not by Cdh1. (A) Recombinant BubR1 was expressed in Sf9 cells (lane 2) and purified to homogeneity (lane 3). Lane 1, Sf9 cell lysates without expressing BubR1. (B) BubR1 inhibits Cdc20. Interphase APC (iAPC) was incubated with purified recombinant Cdc20, Cdc20 plus BubR1, or Cdc20 plus Mad2 and the ligase activity of APC was measured using a radioactive N-terminal fragment of cyclin B as a substrate. The recombinant Mad2 protein exists in two different forms (oligomer and monomer) and only the oligomer is an active inhibitor of APC (Fang et al., 1998b). Thus, Mad2 oligomer was used in experiments described in this article. (C) BubR1 does not inhibit Cdh1. Interphase APC (lane 1) was incubated with purified recombinant Cdc20 (lane 2), with Cdc20 plus BubR1 (lane 3), with Cdh1 (lane 4), or with Cdh1 plus BubR1 (lane 5). The ability of treated APC to ubiquitinate the radioactive N-terminal fragment of cyclin B was assayed. (D) BubR1 does not inhibit mitotic APC. Mitotic APC was incubated with a buffer (lanes 1 and 4), BubR1 (lanes 2 and 5), or with Mad2 (lanes 3 and 6) either in the presence (lanes 4–6) or absence (lanes 1–3) of Cdc20, and the ubiquitin ligase activity of APC was measured using the N-terminal fragment of cyclin B as a substrate. (E) BubR1 does not inhibit preactivated Cdc20-APC. Interphase APC beads (lane 1) were first incubated with recombinant Cdc20 (lanes 2 and 3). BubR1 was then added and incubated with the mixture of Cdc20 and APC (lane 3). The ability of treated APC to ubiquitinate the radioactive N-terminal fragment of cyclin B was assayed. (F) BubR1 and Mad2 do not directly inhibit APC. Recombinant BubR1 (lane 2) or Mad2 (lane 3) was incubated with iAPC in the presence of ATP. BubR1 and Mad2 were then removed and APC beads incubated with recombinant Cdc20. The ubiquitin ligase activity of APC was measured.
I tested whether BubR1 and Mad2 inhibit APC that had been activated by Cdc20. Active Cdc20–APC complex was immunopurified from Xenopus mitotic extracts and incubated with recombinant BubR1 and Mad2; neither BubR1 nor Mad2 inhibits the ubiquitin ligase activity of the preformed Cdc20–APC complex (Figure 2D). I have shown previously that recombinant Cdc20, when incubated with mitotic APC, superactivates its ligase activity (Fang et al., 1998a). Interestingly, both BubR1 and Mad2 prevent superactivation of mitotic APC (Figure 2D), consistent with the conclusion that both proteins inhibit unbound Cdc20, but not the preformed Cdc20–APC complex. To further confirm this conclusion, interphase APC was first activated by Cdc20, and BubR1 or Mad2 was then added and incubated with the mixture of APC and Cdc20. I found that once APC is activated, BubR1 and Mad2 have no inhibitory effect on Cdc20-APC (Figure 2E; my unpublished data). Therefore, BubR1 and Mad2 block the activation of APC by Cdc20 that has not been associated with APC, but do not inhibit the active Cdc20–APC complex. These experiments suggest that association of BubR1 and Mad2 to Cdc20 and APC may occur in a specific order and that activation of the ubiquitin ligase activity may be an irreversible process after the release of BubR1 and Mad2 from the Cdc20–APC complex at the onset of anaphase.
I next tested whether BubR1 and Mad2 have a direct inhibitory effect on APC, instead of Cdc20. Interphase APC was incubated with recombinant BubR1 or Mad2 in a kinase buffer containing ATP. The recombinant proteins were removed, and treated APC was incubated with Cdc20 and then analyzed for its ubiquitin ligase activity (Figure 2F). Prior incubation of BubR1 and Mad2 with APC only has a minimal effect on the activation by Cdc20, consistent with the conclusion that the direct target of BubR1 and Mad2 is Cdc20, not APC.
Inhibition of Cdc20 Does Not Require Its Phosphorylation
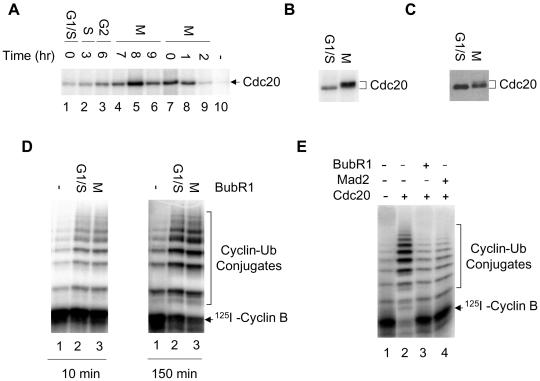
BubR1 is a kinase that phosphorylates Cdc20 (Wu et al., 2000). I purified the BubR1 immunocomplex from nocodazole-arrested HeLa cells with an anti-BubR1 antibody and found that recombinant Cdc20 is efficiently phosphorylated in the presence of radioactive γ-ATP (Figure 3A, lane 7). Thus, the BubR1 immunocomplex can phosphorylate Cdc20, although it is not clear whether BubR1 is the kinase directly responsible for phosphorylation. Furthermore, the activity of the kinase is regulated in the cell cycle; the kinase activity peaks in mitosis and drops as cells exit into G1 (Figure 3A). Interestingly, the kinase activity in normal mitotic cells is as high as, if not higher than, that in nocodazole-arrested cells (Figure 3A, cf. lane 5 with lane 8). Given that the mitotic checkpoint is only activated in a portion of cells in lane 5, I conclude that the kinase activity of the BubR1 immunocomplex is unlikely to be regulated solely by the mitotic checkpoint pathway.
Figure 3.
Phosphorylation of Cdc20 is not required for inhibition by BubR1. (A) Cell cycle-dependent phosphorylation of Cdc20 by the BubR1 immunocomplex. Synchronous HeLa cells (lanes 1–9) were prepared as described in Figure 1. BubR1 was immunopurified from cell lysates and assayed for phosphorylation of Cdc20 in the presence of radioactive γ-ATP. Lane 10, Cdc20 alone in the absence of BubR1. (B–D) Effect of phosphorylation on Cdc20 activity. BubR1 was immunopurified from double thymidine-arrested cells (G1/S, lane 2) or from thymidine-nocodazole–arrested cells (M, lane 3). BubR1 beads were incubated with recombinant Cdc20 in a kinase buffer containing radioactive γ-ATP, and phosphorylated Cdc20 was analyzed by SDS-PAGE (B). In parallel reactions, BubR1 beads were incubated with recombinant Cdc20 and nonradioactive ATP (C and D). After removal of BubR1 beads, one-tenth of Cdc20 was analyzed by Western blotting to determine its mobility (C) and the remaining Cdc20 was incubated with immunopurified iAPC. The ubiquitin ligase activity of APC was then assayed for 10 min and 150 min. Lane 1, iAPC alone. (E) Interphase APC was incubated with recombinant BubR1 (lane 3) or Mad2 (lane 4) in the presence (lanes 2–4) or absence (lane 1) of Cdc20. The ubiquitin ligase activity of APC was then analyzed in the presence of AMP-PNP, not ATP.
I analyzed whether phosphorylation of Cdc20 by the BubR1 immunocomplex affects its ability to activate APC. The active BubR1 immunocomplex was purified from a large amount of nocodazole-arrested HeLa cells (Figure 3, B–D, lane 3) and inactive complex from thymidine-arrested G1/S cells (Figure 3, lane 2). Cdc20 was incubated with the BubR1 beads in a kinase buffer containing radioactive γ-ATP and the extent of phosphorylation was analyzed by SDS-PAGE (Figure 3B). In a parallel experiment, BubR1 beads were incubated with Cdc20 plus nonradioactive ATP and the level of phosphorylation on Cdc20 analyzed by Western blotting (Figure 3C). Under the conditions used herein, recombinant Cdc20 was phosphorylated by the BubR1 immunocomplex from nocodazole-arrested cells, as indicated by a shift in its mobility. Phosphorylated Cdc20 was then incubated with inactive interphase APC and its ubiquitin ligase activity assayed. Kinetic analysis of ubiquitination reaction indicates that Cdc20 incubated with active kinase beads activates APC to the same degree as that incubated with inactive beads (Figure 3D). Thus, phosphorylation of Cdc20 by the BubR1 immunocomplex from checkpoint-arrested cells is unlikely to play a direct role in controlling Cdc20 activity. Because the BubR1 purified on antibody beads is substoichiometric to the amount of Cdc20 used, this experiment analyzes the effect of phosphorylation on Cdc20 activity, but not the effect of BubR1 binding on Cdc20 activity.
The yeast homolog of BubR1, Mad3p, does not have a kinase domain, suggesting that the kinase activity of BubR1 may not be required for inhibition of Cdc20 (Hardwick et al., 2000). Consistent with this, the inhibitory effect of BubR1 in Figure 2 was observed when BubR1 and Cdc20 were incubated with APC in the absence of ATP and Mg2+ during the activation step. In fact, addition of EDTA to BubR1 and Cdc20 during the activation step does not affect the ability of BubR1 to inhibit Cdc20 (my unpublished data). To further confirm that the kinase activity is not involved in inhibition of Cdc20, recombinant BubR1, Mad2, and Cdc20 were incubated with immunopurified interphase APC in the absence of ATP. Recombinant proteins were then removed and the ligase activity of APC was analyzed in the presence of AMP-PNP, not ATP. AMP-PNP supports the ubiquitination reaction, not the phosphorylation reaction. The absence of ATP throughout the assay does not diminish the inhibitory effect of BubR1 or Mad2 (Figure 3E), indicating that phosphorylation of Cdc20 by BubR1 is not required for the inhibition. This is consistent with a recent report that a kinase-dead mutant of BubR1 can efficiently inhibit the activation of APC by Cdc20 (Tang et al., 2001).
BubR1 Acts Synergistically with Mad2 to Inhibit Activation of APC by Cdc20
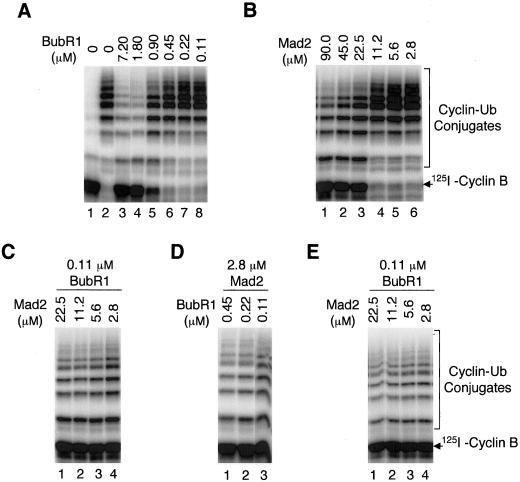
Both BubR1 and Mad2 are proteins required for mitotic checkpoint control and both can inhibit Cdc20 to prevent premature activation of APC. Why are there two inhibitors in the same pathway? Are they redundant in function? To address these questions, I examined whether these two checkpoint proteins have a synergistic effect in inhibiting activation of APC by Cdc20. First, I analyzed the dose response of BubR1 and Mad2 in inhibiting Cdc20 (Figure 4, A and B). Maximal inhibition of Cdc20 (1.7 μM) requires a stoichiometric amount of BubR1 (1.8 μM), suggesting that binding of BubR1 to Cdc20 is responsible for this inhibition (Figure 4A, lane 4). On the other hand, the amount of Mad2 (22.5 μM) required for maximal inhibition is 12-fold above the stoichiometric amount of Cdc20 (Figure 4B, lane 3). I conclude that BubR1 is 12-fold more potent than Mad2 as an inhibitor of Cdc20. Assuming that binding of Mad2 to Cdc20 is the limiting step in inhibition of APC, I estimate that the Mad2–Cdc20 complex has a dissociation constant (i.e., 50% inhibition) between 10 and 15 μM. The dissociation constant for BubR1–Cdc20 complex is at least submicromolar; the accurate value cannot be derived from the titration experiment in Figure 4A, because the concentration of Cdc20 used is above the Kd for the complex.
Figure 4.
BubR1 and Mad2 act synergistically to inhibit activation of APC by Cdc20. (A and B) Dose response of BubR1 (A) and Mad2 (B) in inhibition of Cdc20. Decreasing amounts of recombinant BubR1 and Mad2 were incubated with constant amounts of Cdc20 and iAPC. The ability of treated APC to ubiquitinate the radioactive N-terminal fragment of cyclin B was assayed. Lane 1 in A: interphase APC without Cdc20. Lane 2 in A: Cdc20-APC in the absence of BubR1. (C-E) Synergism between BubR1 and Mad2 in inhibition of Cdc20. Various amounts of recombinant BubR1 and Mad2 were incubated with constant amounts of Cdc20 and iAPC. The ability of treated APC to ubiquitinate the radioactive N-terminal fragment of cyclin B was assayed. Ubiquitination reactions were done in the presence of ATP (C and D) or in the presence of AMP-PNP (E). Data in A–D were generated in a single experiment and gels were exposed to the same extent. Therefore, APC activity in different panels is directly comparable.
Second, I examined whether BubR1 and Mad2 function synergistically in inhibiting Cdc20. At a low concentration of BubR1 (0.11 μM), BubR1 itself has no inhibitory effect on Cdc20. However, to 0.11 μM BubR1, addition of low concentrations of Mad2, ranging from 22.5 μM down to 2.8 μM, efficiently inhibits Cdc20 (Figure 4C). At 2.8 μM Mad2, a concentration below the Kd for the Mad2–Cdc20 complex, no inhibition of Cdc20 was observed if Mad2 is present by itself. In fact, efficient inhibition of Cdc20 requires an eightfold higher Mad2 concentration (Figure 4B). Similarly, efficient inhibition of Cdc20 requires a concentration 10-fold higher than 0.11 μM BubR1, if BubR1 is present alone (Figure 4A). However, combining 0.11 μM BubR1 with 2.8 μM Mad2 leads to maximal inhibition of Cdc20 (Figure 4C). Similarly, to 2.8 μM Mad2, addition of low concentrations of BubR1, ranging from 0.45 μM down to 0.11 μM, efficiently inhibits Cdc20 (Figure 4D). Thus, the BubR1 and Mad2 act synergistically in this inhibition. To determine whether the kinase activity of BubR1 is involved in its synergistic effect with Mad2, I performed my assay in the presence of AMP-PNP to prevent possible phosphorylation by BubR1, as described in Figure 3E. The same synergistic interaction between BubR1 and Mad2 was observed (Figure 4E), even when no ATP was present throughout the reaction, indicating that phosphorylation is not required for the synergy between BubR1 and Mad2.
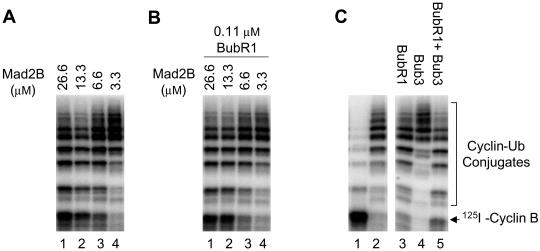
A homolog of Mad2, Mad2B, has recently been shown to be an inhibitor of APC (Chen and Fang, 2001; Pfleger et al., 2001). Similar to Mad2, Mad2B binds to activators of APC and prevents their activation of the ubiquitin ligase. However, Mad2B inhibits both Cdc20 and Cdh1 and is likely involved in transducing a cellular signal other than the mitotic checkpoint control to APC. Consistent with this, Mad2B does not interact with the checkpoint protein Mad1 (Chen and Fang, 2001). I tested whether Mad2B acts synergistically with BubR1 to inhibit Cdc20. The dose response of Mad2B in inhibition of Cdc20 is the same in the presence or absence of 0.11 μM BubR1 (Figure 5, A and B), indicating that BubR1 and Mad2B are two independent inhibitors of APC. This is consistent with the hypothesis that Mad2B does not function in the mitotic checkpoint control.
Figure 5.
Mad2B and Bub3 do not have synergistic effect with BubR1 in inhibiting Cdc20. (A) Dose response of Mad2B in inhibition of Cdc20. Decreasing amounts of recombinant Mad2B were incubated with constant amounts of Cdc20 and iAPC. The ability of treated APC to ubiquitinate the radioactive N-terminal fragment of cyclin B was assayed. (B) Various amounts of recombinant Mad2B were incubated with constant amounts of BubR1, Cdc20, and iAPC. The ability of treated APC to ubiquitinate the radioactive N-terminal fragment of cyclin B was assayed. (C) Interphase APC (lane 1) was incubated with recombinant Cdc20 (lane 2), Cdc20 plus BubR1 (lane 3), Cdc20 plus Bub3 (lane 4), or Cdc20 plus BubR1 and Bub3 (lane 5), and the ubiquitin ligase activity of treated APC was assayed.
Bub3 binds to BubR1 throughout the cell cycle (my unpublished data). In addition, Bub3 coprecipitates with Cdc20 during mitosis (Figure 1C). I examined whether Bub3 enhances the inhibitory effect of BubR1 (Figure 5C). Recombinant Bub3 alone has no effect on the activation of APC by Cdc20 (Figure 5C, lane 4). To analyze the potential synergistic inhibitory effect between BubR1 and Bub3, I selected a concentration of BubR1 at which BubR1 alone only gave a minimal inhibition of Cdc20 (Figure 5C, lane 3). Addition of an excess amount of recombinant Bub3 to BubR1 only slightly, if at all, enhances the inhibition of Cdc20 by BubR1. Titration of Bub3 protein concentration in such an experiment also failed to uncover any effect of Bub3 on inhibition of Cdc20 by BubR1 (my unpublished data). Thus, Bub3 does not act synergistically with BubR1 in inhibition of Cdc20.
BubR1 and Mad2 Mutually Enhance Their Binding to Cdc20
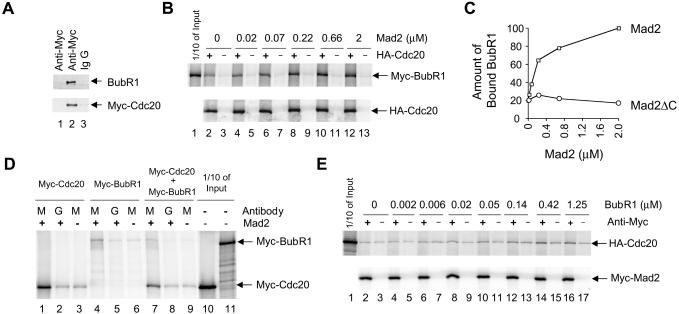
I analyzed whether BubR1 directly interacts with Cdc20 in the absence of other checkpoint proteins. Purified recombinant Myc-Cdc20 was incubated with purified recombinant BubR1 (Figure 6A). Under the protein concentration used herein, immunoprecipitation of Myc-Cdc20 by an anti-Myc antibody, but not control IgG, precipitated ∼10% of input BubR1 (Figure 6A, lanes 2 and 3; my unpublished data). This precipitation is dependent on Myc-Cdc20, because the anti-Myc antibody did not bring down BubR1 in the absence of Myc-Cdc20 (Figure 6A, lane 1). This interaction is specific because unrelated proteins, such as Mad1 or bovine serum albumin, did not coprecipitate with Myc-Cdc20 (my unpublished data). Thus, BubR1 physically interacts with Cdc20.
Figure 6.
Protein–protein interactions among BubR1, Cdc20, and Mad2. (A) Purified recombinant Cdc20 and BubR1 directly interact with each other. Recombinant Myc-Cdc20 was incubated with BubR1 and then immunoprecipitated with an anti-Myc antibody (lane 2) or with control IgG (lane 3). As a control, BubR1 was immunoprecipitated with the anti-Myc antibody in the absence of Myc-Cdc20 (lane 1). (B) Mad2 enhances interactions between BubR1 and Cdc20. Myc-BubR1 was incubated with increasing amounts of recombinant Mad2 in the presence (lanes 2, 4, 6, 8, 10, and 12) or absence (lanes 3, 5, 7, 9, 11, and 13) of labeled HA-Cdc20. Protein complexes were immunoprecipitated by an anti-HA antibody and associated Myc-BubR1 protein analyzed by SDS-PAGE. Lane 1, one-tenth of input Myc-BubR1 in the binding reaction. (C) The amount of bound Myc-BubR1 from A was quantitated, normalized with the amount of HA-Cdc20 immunoprecipitated, and plotted against Mad2 concentration (squares). The unit for the amount of bound Myc-BubR1 is arbitrary. Similarly, the interaction between Myc-BubR1 and HA-Cdc20 was analyzed in the presence of Mad2ΔC and the amount of bound BubR1 was plotted against Mad2ΔC concentration (circles). (D) Interactions among BubR1, Cdc20, and Mad2. Myc-Cdc20 (lanes 1–3), Myc-BubR1 (lanes 4–6), or Myc-Cdc20 plus Myc-BubR1 (lanes 7–9) were translated in vitro. These proteins were incubated with 4 μM recombinant Mad2 and the Mad2 complexes were immunoprecipitated by an anti-Mad2 antibody (lanes M) or by control IgG (lanes G). The amount of bound Myc-Cdc20 and Myc-BubR1 was analyzed by SDS-PAGE. Lanes 3, 6, and 9, immunoprecipitation of Myc-Cdc20 and Myc-BubR1 by the anti-Mad2 antibody in the absence of recombinant Mad2. Lanes 10 and 11, one-tenth of the input Myc-Cdc20 and Myc-BubR1 used in the binding reactions. (E) BubR1 enhances interactions between Mad2 and Cdc20. HA-Cdc20 and Myc-Mad2 were incubated with increasing amounts of recombinant BubR1. Myc-Mad2 was immunoprecipitated by an anti-Myc antibody (lanes 2, 4, 6, 8, 10, 12, 14, and 16) or by control IgG (lanes 3, 5, 7, 9, 11, 13, 15, and 17) and the amount of associated HA-Cdc20 protein was analyzed by SDS-PAGE. Lane 1, one-tenth of the input HA-Cdc20 in the binding reactions.
What is the molecular basis of the synergy between BubR1 and Mad2? Both BubR1 and Mad2 can bind to Cdc20. Thus, it is possible that binding of one protein to Cdc20 enhances the binding of the other protein and vice versa. To test this, HA-Cdc20 and Myc-BubR1 were synthesized by in vitro translation and incubated together in the presence of increasing amounts of recombinant Mad2 protein (Figure 6B). The radioactively labeled proteins synthesized in reticulocyte lysates were used due to the ease of quantitation. The amount of BubR1 and Cdc20 in the binding reaction is on the order of 10 nM (my unpublished data), and BubR1 binds specifically to Cdc20 at this concentration (Figure 6B, lanes 2 and 3). Furthermore, with increasing amounts of recombinant Mad2, increasing amounts of BubR1 were bound to Cdc20. At 2 μM Mad2, there is a fivefold increase in the amount of bound BubR1 (Figure 6, B, lanes 4–13; and C), indicating that the checkpoint protein Mad2 promotes the interaction between BubR1 and Cdc20. This enhancement requires a direct interaction between Mad2 and Cdc20, because a Mad2 mutant with the C-terminal 10 amino acids deleted, Mad2ΔC, does not interact with Cdc20 (my unpublished data) (Fang et al., 1998b) and this mutant fails to promote the interaction between Cdc20 and BubR1 (Figure 6C).
I next examined whether recombinant BubR1 promotes the interaction between Mad2 and Cdc20. Recombinant Mad2 at 4 μM interacts with in vitro-translated Myc-Cdc20, as assayed by coimmunoprecipitation (Figure 6D, lanes 1–3). However, addition of the recombinant BubR1 protein, even at a concentration of 3.3 μM, does not increase the amount of Cdc20 bound to recombinant Mad2 (my unpublished data). It is possible that the BubR1 protein may promote the binding of Mad2 to Cdc20 at lower concentrations of Mad2 and Cdc20, at which the Mad2–Cdc20 complex is unstable. Thus, I incubated together in vitro-translated Myc-Mad2 and HA-Cdc20, both on the order of 10 nM. In the absence of BubR1, the amount of HA-Cdc20 immunoprecipitated by an anti-Myc antibody is similar to that by control IgG, suggesting that HA-Cdc20 does not associate stably with Myc-Mad2 at these concentrations (Figure 6E, lanes 2 and 3). The presence of increasing amounts of recombinant BubR1 protein increases the amount of HA-Cdc20 bound to Myc-Mad2, up to fourfold in the presence of 1.25 μM BubR1 (Figure 6E, lanes 4–17), although the absolute amount of HA-Cdc20 bound to Myc-Mad2 is still relatively low. I conclude that Mad2 has a low affinity to Cdc20 and that BubR1 promotes the formation of the Mad2–Cdc20 complex under the conditions used herein.
I also examined whether BubR1 can directly interact with Mad2. Recombinant Mad2 at 4 μM was incubated with in vitro-translated Myc-BubR1. Less than 1% of input Myc-BubR1 was associated with recombinant Mad2 as assayed by coimmunoprecipitation (Figure 6D, lanes 4–6; cf. lane 4 vs. 11), consistent with the fact that Mad2 does not coimmunoprecipitate with BubR1 in checkpoint-arrested HeLa cells (Figure 1D). I next analyzed whether BubR1, Cdc20, and Mad2 form a ternary complex. Recombinant Mad2 at 4 μM was incubated with in vitro-translated Myc-BubR1 and Myc-Cdc20, both on the order of 10 nM. Again, <1% of input BubR1 was associated with recombinant Mad2 as assayed by coimmunoprecipitation (Figure 6D, lanes 7–9; cf. lane 7 vs. 11), indicating that BubR1 does not form a stable ternary complex with Cdc20 and Mad2. This is consistent with the fact that these three proteins do not form a ternary complex in vivo (Figure 1D).
Checkpoint Complexes in Nocodazole-arrested HeLa Cells
Cdc20 forms two inactive complexes in vitro, a higher affinity complex with BubR1 and a lower affinity complex with Mad2. What are the compositions of various Cdc20 complexes in checkpoint-arrested cells and what is the relative contribution of each Cdc20 complex to mitotic arrest?
To address these questions, I fractionated over an anion exchange column lysates of HeLa cells arrested at prometaphase by nocodazole treatment. Cdc20, BubR1, and Mad2 all bind to the anion exchange column. The endogenous Cdc20 fractionated into two peaks (Figure 7A); the first peak (A) cofractionates with the majority of BubR1 and Mad2, whereas the second peak (B) cofractionates with APC. A small amount of BubR1 and Mad2 also cofractionates with the APC peak and immunoprecipitation experiments indicated that BubR1, Mad2, and Cdc20 all associate with APC in peak B (my unpublished data). It is possible that the relatively low amount of BubR1 and Mad2 in peak B is a result of the dissociation of BubR1 and Mad2 from APC during the cell lysis and fractionation.
To determine the molecular forms of the majority of BubR1 and Mad2 proteins, I pooled fractions from peak A and further fractionated them over a gel filtration column. Both BubR1 and Bub3 elute as a sharp, symmetric peak (Figure 7B). Although Cdc20 elutes as a broad peak, the center of the peak cofractionates with the BubR1 and Bub3 proteins (fractions 29–31). In contrast, Mad2 elutes as two peaks, one centered at fractions 25–27 and the other centered at fractions 37–39. Both peaks of the Mad2 protein are offset from the peak of Cdc20, BubR1, and Bub3. In fact, the peak fractions of Cdc20, fractions 29–31, contain the least amount of Mad2 among fractions 25–39. Interestingly, the first peak of Mad2 cofractionates with the peak of the Mad1 protein, suggesting that Mad1 and Mad2 form a complex in fractions 25–27.
To analyze the molecular compositions of the Mad2 complexes, I performed immunoprecipitation experiments followed by Western blot analyses with three representative fractions from the gel filtration column (Figure 7, C–E). Fractions 25 and 37 are peak fractions for Mad2, which contain minimal amount of Cdc20 and almost no BubR1 and Bub3. Fraction 29 is the peak fraction for Cdc20, BubR1, and Bub3, but only contains a relatively low concentration of Mad2 (Figure 7B). Mad1 and Mad2 form a complex in the first, but not the second peak of Mad2; immunoprecipitation with the anti-Mad2 antibody precipitated Mad1 in fractions 25 and 29, but not in fraction 37, because there is no Mad1 in fraction 37. The Mad1–Mad2 complex does not contain Cdc20, because immunoprecipitation with two different anti-Cdc20 antibodies did not bring down Mad1, nor did immunoprecipitation with an anti-Mad1 antibody precipitate Cdc20 (my unpublished data). Mad2 in its second peak (fraction 37) does not coimmunoprecipitate with any known checkpoint protein (Figure 7C; my unpublished data).
Immunoprecipitation of Mad2 also precipitated Cdc20, but only in fraction 29, not in fractions 25 and 37 (Figure 7C). Thus, Mad2 interacts with Cdc20 in the peak fraction of Cdc20, but not in the peak fractions of Mad2. On the other hand, BubR1 does not interact with Mad2 in all three fractions tested (Figure 7C), consistent with the fact that Mad2 and BubR1 do not form a complex in crude lysates (Figure 1) (Yao et al., 2000; Skoufias et al., 2001; Tang et al., 2001).
I next analyzed the molecular forms of Cdc20 and BubR1 complexes. Cdc20 coprecipitates with both BubR1 and Mad2 in its peak fraction 29 (Figure 7D). On the other hand, immunoprecipitation with three antibodies against different regions of BubR1 coprecipitates both Bub3 and Cdc20, but not Mad2 (Figure 7E; my unpublished data). Thus, Cdc20 forms two separate complexes with BubR1 and Mad2, respectively. The BubR1–Cdc20 complex may also contain Bub3. Because the anti-BubR1 and anti-Mad2 antibodies failed to immunodeplete BubR1 and Mad2 from cell lysates and column fractions (my unpublished data), I was not able to determine the relative abundance of the Cdc20–BubR1 and Cdc20–Mad2 complexes in prometaphase cells. However, given that Mad2 in its two peaks does not coimmunoprecipitate with Cdc20 (Figure 7, C and D), I estimate that less than one-third of Mad2 is associated with Cdc20 in checkpoint-arrested HeLa cells.
DISCUSSION
I show herein that the checkpoint protein BubR1 directly binds to Cdc20 and inhibits its activation of APC, consistent with recent reports (Sudakin et al., 2001; Tang et al., 2001). Although the BubR1 immunocomplex from checkpoint-arrested cells can phosphorylate Cdc20, this inhibition is independent of the phosphorylation of Cdc20, confirming a recent observation that a kinase-dead mutant of BubR1 efficiently inhibits Cdc20 (Tang et al., 2001). Quantitative analysis indicates that BubR1 is 12-fold more potent than Mad2 as an inhibitor of Cdc20. Furthermore, BubR1 and Mad2 act synergistically in preventing premature activation of APC by Cdc20. We found that Cdc20 forms two separate inactive complexes, a higher affinity complex with BubR1 and a lower affinity complex with Mad2. Interestingly, Mad2 enhances the interaction between BubR1 and Cdc20 and this enhancement requires a direct interaction between Cdc20 and Mad2. Similarly, BubR1 also promotes the formation of the Mad2–Cdc20 complex. We conclude that the two checkpoint proteins, BubR1 and Mad2, act cooperatively to prevent premature activation of APC in response to mitotic checkpoint signals.
BubR1 Is an Inhibitor of Cdc20
As an inhibitor of Cdc20, BubR1 shares many of biochemical properties common to Mad2, such as inhibitory specificity and molecular basis of inhibition (Fang et al., 1998b). Like Mad2, BubR1-mediated inhibition is specific to Cdc20, not to Cdh1, consistent with its checkpoint function. This inhibition is mediated through a direct interaction between BubR1 and Cdc20. Human BubR1 interacts with human Cdc20 in a yeast two-hybrid assay (my unpublished data) (Wu et al., 2000) and purified recombinant BubR1 directly binds to Cdc20 in vitro. Furthermore, BubR1 is associated with Cdc20 and APC in vivo in a cell cycle-dependent manner. The cell cycle-dependent association of BubR1 with Cdc20 and APC is reminiscent of the binding of Mad2 to Cdc20 and APC (Figure 1) (Fang et al., 1998b).
Despite its similarity to Mad2, BubR1 is a novel inhibitor of Cdc20. First, unlike Mad2 (Chen et al., 1998; Jin et al., 1998), BubR1 does not interact with Mad1. It is likely that BubR1 receives and transduces a checkpoint signal different from Mad1. Second, BubR1 is a much more potent than Mad2 in inhibiting Cdc20. Dose-response analysis indicates that BubR1 can quantitatively inhibit Cdc20 at a molar concentration 12-fold lower than that required for Mad2. The BubR1–Cdc20 complex has a dissociation constant of submicromolar and the Mad2–Cdc20 complex of 10–15 μM, consistent with previously reported numbers (Tang et al., 2001).
APC Is Regulated Synergistically by Checkpoint Proteins BubR1 and Mad2
It has been shown previously that the checkpoint protein Mad2 is a stoichiometric inhibitor of APC (Fang et al., 1998b). Mad2 binds to Cdc20 and to Cdc20-APC in a checkpoint-dependent manner in vivo. In a reconstituted ubiquitination system, recombinant Mad2 directly binds to Cdc20 and inhibits the ubiquitin ligase activity of APC.
Although Mad2 is required for mitotic checkpoint control, it is not clear whether Mad2 is the only regulator of APC in the checkpoint pathway. The in vitro analysis indicates that the dissociation constant for the Mad2–Cdc20 complex is above 10 μM. On the other hand, the intracellular concentrations for Cdc20 (285 nM) and Mad2 (230 nM) in nocodazole-arrested HeLa cells are both in a submicromolar range (my unpublished data). Thus, the Mad2 protein at its physiological concentration is not sufficient to inhibit APC in response to activation of the mitotic checkpoint.
I show herein that the checkpoint protein BubR1 is another inhibitor of APC that binds to and inhibits Cdc20 (Sudakin et al., 2001; Tang et al., 2001). In checkpoint-arrested cells, the majority of BubR1 and Bub3 cofractionates and coimmunoprecipitates with Cdc20, whereas none of the two Mad2 peaks from the gel filtration chromatography cofractionates with Cdc20. In fact, less than one-third of Mad2 is associated with Cdc20 in vivo. On the other hand, the intracellular concentration of BubR1 (127 nM) in nocodazole-arrested HeLa cells is about half of that of Cdc20 and Mad2 (my unpublished data). Thus, BubR1 is likely to play an important role in control of the APC activity.
BubR1 and Mad2 mutually promote each other's binding to Cdc20 and they act synergistically in inhibition of Cdc20. This molecular synergism between BubR1 and Mad2 could result from the formation of the BubR1–Cdc20–Mad2 ternary complex. Although I did not detect such a ternary complex in immunoprecipitation experiments in cell lysates and failed to reconstitute the ternary complex in vitro, Sudakin et al. (2001) has reported in checkpoint-arrested HeLa cells a MCC complex consisting of BubR1, Bub3, Cdc20, and Mad2 in equal stoichiometry. It is possible that MCC may be a transient and unstable complex, which has escaped from detection in assays. I also note that a substoichiometric amount of BubR1 is capable of enhancing the inhibition of Cdc20 by a stoichiometric amount of Mad2, an observation that cannot be explained by the formation of a stoichiometric, stable BubR1–Cdc20–Mad2 ternary complex. I speculate that a transient association of BubR1 to Cdc20 may cause a conformation change in the Cdc20 protein, which in turn increases the affinity of Cdc20 toward Mad2.
Why does the cell need two inhibitors of APC in the same checkpoint pathway? Both BubR1 and Mad2 are localized to unattached kinetochores (Chen et al., 1996; Li and Benezra, 1996; Chan et al., 1998). Mad2 is a transient component of unattached kinetochores with a half-life of <30 s, as measured by the fluorescence recovery after photobleaching (Howell et al., 2000). It is possible that binding of Cdc20 to BubR1 recruits Cdc20 to kinetochores, where BubR1 promotes the formation of the Mad2–Cdc20 complex, which subsequently diffuses away from kinetochores and inhibits APC throughout the cell.
Alternatively, BubR1 and Mad2 may regulate the activity of APC in response to activation of different sensors in the checkpoint pathway. It has been suggested that the mitotic checkpoint pathway in vertebrates may consist of an attachment-sensitive branch and a tension-sensitive branch (Skoufias et al., 2001). Mad2 is recruited to kinetochores in response to the loss of attachment of the spindle, but not to the loss of the tension, whereas the amounts of Bub1 and BubR1 at kinetochores are increased in response to the loss of tension (Waters et al., 1998; Skoufias et al., 2001). It is possible that BubR1 and Mad2 may inhibit APC in response to different checkpoint signals.
ACKNOWLEDGMENTS
I thank James Chen for technical support during this work and am grateful to members of Fang laboratory for helpful discussion. I thank Drs. Ted Salmon and Tim Stearns for critical reading of the manuscript. G.F. is a Searle Scholar, a Kimmel Scholar in Cancer Research, and a recipient of the Beckman Young Investigator Award and a Burroughs-Wellcome Career Award in Biomedical Sciences. This work was also supported by a United States Public Health Service grant (GM62852), by a Research Scholar Grant from American Cancer Society (RSG-01-143-01) and by grants from Concern Foundation and from the Office of Technology Licensing at Stanford University.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–09–0437. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–09–0437.
Corresponding author. E-mail address: gwfang@stanford.edu.
REFERENCES
- Amon A. The spindle checkpoint. Curr Opin Genet Dev. 1999;9:69–75. doi: 10.1016/s0959-437x(99)80010-0. [DOI] [PubMed] [Google Scholar]
- Chan GK, Jablonski SA, Sudakin V, Hittle JC, Yen TJ. Human BUBR1 is a mitotic checkpoint kinase that monitors CENP-E functions at kinetochores and binds the cyclosome/APC. J Cell Biol. 1999;146:941–954. doi: 10.1083/jcb.146.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan GK, Schaar BT, Yen TJ. Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1. J Cell Biol. 1998;143:49–63. doi: 10.1083/jcb.143.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Fang G. MAD2B is an inhibitor of the anaphase-promoting complex. Genes Dev. 2001;15:1765–1770. doi: 10.1101/gad.898701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RH, Shevchenko A, Mann M, Murray AW. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J Cell Biol. 1998;143:283–295. doi: 10.1083/jcb.143.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RH, Waters JC, Salmon ED, Murray AW. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- Dobles M, Liberal V, Scott ML, Benezra R, Sorger PK. Chromosome mis-segregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 2000;101:635–645. doi: 10.1016/s0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]
- Elledge SJ. Cell cycle checkpoints-preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol Cell. 1998a;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998b;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. Mitotic proteolysis. Phil Trans R Soc Biol Sci. 1999;354:1583–1590. doi: 10.1098/rstb.1999.0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick KG, Johnston RC, Smith DL, Murray AW. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J Cell Biol. 2000;148:871–882. doi: 10.1083/jcb.148.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- Howell BJ, Hoffman DB, Fang G, Murray AW, Salmon ED. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J Cell Biol. 2000;150:1233–1250. doi: 10.1083/jcb.150.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Hwang LH, Lau LF, Smith DL, Mistrot CA, Hardwick KG, Hwang ES, Amon A, Murray AW. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- Jin DY, Spencer F, Jeang KT. Human T cell leukemia virus type 1 oncoprotein Tax targets the human mitotic checkpoint protein MAD1. Cell. 1998;93:81–91. doi: 10.1016/s0092-8674(00)81148-4. [DOI] [PubMed] [Google Scholar]
- Kallio M, Weinstein J, Daum JR, Burke DJ, Gorbsky GJ. Mammalian p55CDC mediates association of the spindle checkpoint protein Mad2 with the cyclosome/anaphase-promoting complex, and is involved in regulating anaphase onset and late mitotic events. J Cell Biol. 1998;141:1393–1406. doi: 10.1083/jcb.141.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Lin DP, Matsumoto S, Kitazono A, Matsumoto T. Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Li X, Nicklas RB. Mitotic forces control a cell-cycle checkpoint. Nature. 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- Li Y, Benezra R. Identification of a human mitotic checkpoint gene: hsMAD2. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- Li Y, Gorbea C, Mahaffey D, Rechsteiner M, Benezra R. MAD2 associates with the cyclosome/anaphase-promoting complex and inhibits its activity. Proc Natl Acad Sci USA. 1997;94:12431–12436. doi: 10.1073/pnas.94.23.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W, Dobles M, Sorger PK, Murty VVVS, Benezra R. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- Pfleger CM, Salic A, Lee E, Kirschner MW. Inhibition of Cdh1-APC by the MAD2-related protein MAD2L2: a novel mechanism for regulating Cdh1. Genes Dev. 2001;15:1759–1764. doi: 10.1101/gad.897901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Khodjakov A. Mitosis and checkpoints that control progression through mitosis in vertebrate somatic cells. Prog Cell Cycle Res. 1997;3:301–312. doi: 10.1007/978-1-4615-5371-7_24. [DOI] [PubMed] [Google Scholar]
- Schwab M, Lutum AS, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- Shah JV, Cleveland DW. Waiting for anaphase. Mad2 and the spindle assembly checkpoint. Cell. 2000;103:997–1000. doi: 10.1016/s0092-8674(00)00202-6. [DOI] [PubMed] [Google Scholar]
- Skoufias DA, Andreassen PR, Lacroix FB, Wilson L, Margolis RL. Mammalian mad2 and bub1/bubR1 recognize distinct spindle-attachment and kinetochore-tension checkpoints. Proc Natl Acad Sci USA. 2001;98:4492–4497. doi: 10.1073/pnas.081076898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Bharadwaj R, Li B, Yu H. Mad2-independent inhibition of APC-Cdc20 by the mitotic checkpoint protein BubR1. Dev Cell. 2001;1:227–237. doi: 10.1016/s1534-5807(01)00019-3. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Ha E, McKeon F. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, McKeon F. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Wassmann K, Benezra R. Mad2 transiently associates with an APC/p55Cdc complex during mitosis. Proc Natl Acad Sci USA. 1998;95:11193–11198. doi: 10.1073/pnas.95.19.11193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JC, Chen RH, Murray AW, Salmon ED. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Lan Z, Li W, Wu S, Weinstein J, Sakamoto KM, Dai W. p55CDC/hCDC20 is associated with BUBR1, and may be a downstream target of the spindle checkpoint kinase. Oncogene. 2000;19:4557–4562. doi: 10.1038/sj.onc.1203803. [DOI] [PubMed] [Google Scholar]
- Yao X, Abrieu A, Zheng Y, Sullivan KF, Cleveland DW. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat Cell Biol. 2000;2:484–491. doi: 10.1038/35019518. [DOI] [PubMed] [Google Scholar]