Abstract
Here we demonstrate that multiple tetraspanin (transmembrane 4 superfamily) proteins are palmitoylated, in either the Golgi or a post-Golgi compartment. Using CD151 as a model tetraspanin, we identified and mutated intracellular N-terminal and C-terminal cysteine palmitoylation sites. Simultaneous mutations of C11, C15, C242, and C243 (each to serine) eliminated >90% of CD151 palmitoylation. Notably, palmitoylation had minimal influence on the density of tetraspanin protein complexes, did not promote tetraspanin localization into detergent-resistant microdomains, and was not required for CD151-α3β1 integrin association. However, the CD151 tetra mutant showed markedly diminished associations with other cell surface proteins, including other transmembrane 4 superfamily proteins (CD9, CD63). Thus, palmitoylation may be critical for assembly of the large network of cell surface tetraspanin-protein interactions, sometimes called the “tetraspanin web.” Also, compared with wild-type CD151, the tetra mutant was much more diffusely distributed and showed markedly diminished stability during biosynthesis. Finally, expression of the tetra-CD151 mutant profoundly altered α3 integrin-deficient kidney epithelial cells, such that they converted from a dispersed, elongated morphology to an epithelium-like cobblestone clustering. These results point to novel biochemical and biological functions for tetraspanin palmitoylation.
INTRODUCTION
There are at least 21 distinct mammalian tetraspanins, also known as transmembrane 4 superfamily proteins (Maecker et al., 1997; Todd et al., 1998; Serru et al., 2000). Various members of this widely expressed family have been implicated in infectious disease pathologies (Nakamura et al., 1995; Pileri et al., 1998), neural functions (Kopczynski et al., 1996; Banerjee et al., 1997; Dijkstra et al., 2000; Stipp and Hemler, 2000; Zemni et al., 2000), cell fusion events (sperm-egg, osteoclast, myoblast, virus; Imai and Yoshie, 1993; Tachibana and Hemler, 1999; Tanio et al., 1999; Le Naour et al., 2000; Miyado et al., 2000; Schmid et al., 2000), and signaling functions (Berditchevski and Odintsova, 1999; Odintsova et al., 2000; Hemler et al., 1996). Also, they participate in cell migration (Hemler et al., 1996; Maecker et al., 1997) and show both positive (Claas et al., 1998; Testa et al., 1999) and negative (Dong et al., 1995; Radford et al., 1995; Miyake et al., 2000) associations with tumor cell metastasis. Tetraspanin proteins appear not only on the plasma membrane but also within intracellular vesicles such as lysosomes, endosomes, and various secretory granules (Hemler et al., 1996; Maecker et al., 1997; Berditchevski and Odintsova, 1999). Tetraspanins engage in abundant lateral associations with each other and with many other types of proteins (Wright and Tomlinson, 1994; Hemler et al., 1996; Maecker et al., 1997). Within this extensive network, it has been challenging to sort out those tetraspanin interactions of primary importance. Although some tetraspanin proteins at least partially codistribute with lipid rafts (Yashiro-Ohtani et al., 2000; Claas et al., 2001), many associations with other proteins occur independently of lipid rafts. Use of detergents, such as digitonin (Serru et al., 1999) and Brij-96/97 (Imai et al., 1995; Berditchevski et al., 1996), has allowed isolation of tetraspanin complexes with a size, density, and specificity consistent with their being discrete, soluble, biochemical entities (Charrin et al., 2001; Claas et al., 2001; Stipp et al., 2001b). Compared with other tetraspanin interactions, CD151 associations with α3β1, and to a lesser extent with α6 integrins, are remarkably tight, direct, and stoichiometric (Yauch et al., 1998; Serru et al., 1999; Sterk et al., 2000; Yauch et al., 2000). Also, CD151 markedly influences both α3β1-dependent and α6β1-dependent cell morphology (Stipp and Hemler, 2000; Zhang et al., 2002).
Tetraspanins contain four transmembrane domains, short N- and C-terminal tails, a short inner loop, and small and large outer loops (Wright and Tomlinson, 1994; Maecker et al., 1997). The structure of a prototype large outer loop (from CD81) has been solved, revealing five helices, linked by intervening loops and stabilized by two disulfide bonds (Kitadokoro et al., 2001). Thus far, interactions with other proteins have been largely mapped to the large outer loop of tetraspanins (Matsumoto et al., 1993; Pileri et al., 1998; Nakamura et al., 2000). For example, tight association of α3β1 with CD151 depends of regions within the CD151 large outer loop (Yauch et al., 2000; Berditchevski et al., 2001). Tetraspanin domains responsible for weaker interactions with other proteins (i.e., those seen in Brij-96 but not in Triton X-100) have not yet been mapped.
Here we focus on tetraspanin palmitoylation. Acylation of CD9 has been reported previously (Seehafer et al., 1990). However, sites for tetraspanin acylation have not been identified, and the functional relevance has not been determined. The preferred substrate for protein S-acylation is palmitoyl CoA (Resh, 1999). For many proteins, palmitoylation contributes to membrane targeting and to localization within organized membrane microdomains (e.g., lipid rafts; Dunphy and Linder, 1998; Resh, 1999). However, for proteins containing multiple transmembrane domains, several diverse functional effects of palmitoylation are suggested, but consistent trends have not emerged (Jin et al., 1999).
We show here that multiple tetraspanin proteins are palmitoylated by a mechanism that requires a functioning Golgi apparatus. Also, we identified and mutated key palmitoylation sites in the CD151 molecule. Notably, loss of these sites had minimal effect on targeting to the cell surface plasma membrane and localization into stabilized membrane microdomains. However, loss of palmitoylation did markedly influence associations with other proteins, subcellular distribution, protein stability during biosynthesis, and cell morphology.
MATERIALS AND METHODS
Antibodies, Reagents, and Cells
Monoclonal antibodies used were anti-integrin α1, TS2/16 (Hemler et al., 1984); anti-integrin α2, A2-IIE10 (Bergelson et al., 1994); anti-integrin α3, A3-IVA5 and A3-X8 (Weitzman et al., 1993); anti-integrin α6, A6-ELE (Lee et al., 1995); anti-CD9, C9-BB (Berditchevski et al., 1996) and DU-ALL-1 (Sigma, St. Louis, MO); anti-CD63, 6H1 (Berditchevski et al., 1995); anti-CD81, M38 (Fukudome et al., 1992); anti-CD82, M104 (Fukudome et al., 1992); anti-CD151, 5C11 (Yauch et al., 1998); anti-A15/TALLA1, AZM30.4 (Azorsa et al., 1999); anti-CD71, OKT9 and HB21 (American Type Culture Collection, Manassas, VA); and anti-CD147, 8G6 (Berditchevski et al., 1997). Also utilized was a rabbit polyclonal antibody to the cytoplasmic domain of integrin α3A (Dipersio et al., 1995). The mAb to β-tubulin (Tub2.1) was from Sigma, the antibody to caveolin (C13630) was from Transduction Laboratories (Lexington, KY), and anti-fyn (FYN3) was from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal and polyclonal antibodies against green fluorescent protein (GFP) were purchased from CLONTECH (Palo Alto, CA). l-[35S]methionine/l-[35S]cysteine and [3H]palmitic acid were purchased from NEN Bioscience (Boston, MA); brefeldin A, monensin, and nocodazole were obtained from Sigma, dissolved in dimethyl sulfoxide or ethanol, and stored at −80°C at 1–2 mg/ml. Sulfo-NHS-LC-biotin and sulfo-NHS-SS-biotin biotin were obtained from Pierce (Rockford, IL). All cell lines were grown in DMEM supplemented with 10% fetal calf serum (GIBCO BRL, Rockville, MD), 10 mM HEPES, and antibiotics.
Mutagenesis and Transfection
Cysteine point mutants were generated from human CD151 cDNA by polymerase chain reaction and subcloned into EcoRI and SacII sites of the pEGFP-N1 vector (CLONTECH), yielding a C-terminal GFP fusion protein. CD151 mutants were also subcloned into Tag-3B vector via EcoRI and ApaII sites with N-terminal myc tag (Stratagene, San Diego, CA) or into pcDNA3.1 with N-terminal myc.His fusion (Invitrogen, Carlsbad, CA). For transient transfection, 293 cells were transfected using calcium phosphate. To establish stable cell lines, MCF-7, MDA-231, and NIH3T3 cells were transfected using Superfectamine (QIAGEN, Valencia, CA) and selected on 1 mg/ml G418. Much of our prior work on tetraspanins and the initial studies shown here were carried out using A431 cells. However, for technical convenience, we switched to 293 cells for transient transfections, and we utilized MCF-7 or MDA-231 cells interchangeably for stable transfections. To avoid CD151 background problems, we stably expressed human CD151 in murine NIH3T3 cells for internalization and recycling studies and in a murine α3 integrin null kidney epithelial cell line (Wang et al., 1999) for morphology studies.
Radiolabeling, Surface Labeling, Immunoprecipitation, and Immunoblotting
For 3H labeling, CD151-transfected A431, MDA-231, or 293 cells (at 80–90% confluence) were washed twice in phosphate-buffered saline (PBS), serum-starved for 3–4 h, and then pulsed for 2–3 h in medium containing 0.2–0.3 mCi/ml [3H]palmitic acid plus 5.0% dialyzed fetal bovine serum. To label surface proteins with biotin, semiconfluent cells were cooled on ice, washed three times in PBS, incubated in PBS containing sulfo-NHS-LC-biotin at 0.1 or 0.5 mg/ml for 60 min at 4°C, and then washed with cold PBS containing 300 mM glycine. To determine the half-life of CD151 proteins, semiconfluent stable MDA-231 transfectants were washed twice in PBS and starved in cysteine- and methionine-free media for 1 h. Cells were then pulsed for 1 h in methionine- and cysteine-free media containing 0.5 mCi/ml [35S]methionine/cysteine and 5.0% dialyzed fetal bovine serum. Subsequently, cells were either collected (time 0 after labeling) or chased for the indicated periods of time (see Figure 10) by replacing labeling medium with chasing medium (5.0% dialyzed fetal serum and 25× excess unlabeled l-methionine). Labeled cells were lysed in RIPA buffer (25 mM Tris, 2 mM EDTA, 150 mM NaCl, 0.1% NaN3, pH 7.2, plus 1% Triton X-100, 1% deoxycholate, and 0.1% SDS) and processed for immunoprecipitation.
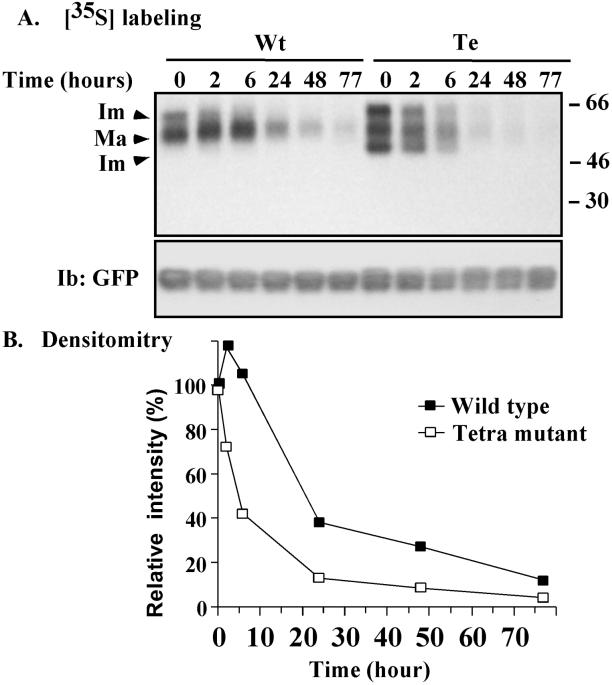
Figure 10.
Biosynthesis and degradation of CD151. (A) MDA-231 cells stably transfected with GFP-tagged CD151 were pulsed with 0.5 mCi/ml [35S]methionine for 1 h and then either lysed (time 0) or chased in medium containing 5% dialyzed serum and 25× excess cold methionine for the indicated times (2, 6, 24, 48, and 77 h) before lysis in RIPA detergent. CD151was then immunoprecipitated using anti-GFP polyclonal antibody and samples were resolved on 4–12% SDS-PAGE and transferred to a PVDF membrane. To detect [35S]methionine, the membrane was exposed to BioMax MS film using a BioMax Transcreen LE system (top). The membrane was also blotted using anti-GFP mAb to estimate CD151 loading (bottom). (B) Levels of radioactivity (for mature CD151 only) were estimated by densitometry, and the relative levels were calculated as a percentage of the labeled protein at time 0.
For immunoprecipitation, cells were lysed in RIPA or in 25 mM HEPES, 150 mM NaCl, 5 mM MgCl2, 20 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mM phenylmethylsulfonyl fluoride, 2 mM NaF, 10 mM sodium pyrophosphate, and 10 mM Na3VO4 buffer containing 1% detergent (Triton X-100, 3-[{cholamidopropyl}dimethylammonio]-1-propanesulfonic acid, Brij-99, or Brij-96). After 1 h at 4°C, insoluble material was removed by centrifugation at 16,000 × g (25 min, 4°C) and supernatants were cleared with protein A- or G-Sepharose (Roche Molecular Biochemicals, Indianapolis, IN). Specific antibodies were then incubated for 1 h at 4°C, followed by overnight incubation with protein A- or G-Sepharose (Roche Molecular Biochemicals). Immune complexes were collected by centrifugation, washed three to five times in lysis buffer, and then analyzed by SDS-PAGE (usually 10 or 12% acrylamide) under nonreducing conditions. Gels were either treated with fluorographic reagents, dried, and exposed to BioMax MR film (Kodak, Rochester, NY) for 14–21 d at −80°C or transferred onto polyvinylidene difluoride (PVDF) membrane and exposed to BioMax MS film using BioMax TRANSCREEN LE Intensifying Screen (Kodak) for 4–14 d at −80°C.
For immunoblotting, proteins resolved by SDS-PAGE were transferred to a nitrocellulose membrane and then incubated with primary antibodies and horseradish peroxidase-conjugated secondary antibody (Sigma) as described by Yauch et al. (2000) with Extravidin (Sigma) coupled to horseradish peroxidase to detect biotinylated proteins. Blots were visualized by chemiluminescence.
Sucrose Gradients, Internalization, and Recycling
Sucrose gradient analyses were carried out essentially as described by Claas et al. (2001). In brief, cell lysates were sheared through hypodermic needles (5 × 16G11/2, 10 × 26G1/2). A total of 1 ml of lysate (derived from 2 × 107 cells) was then mixed with an equal volume of 90% sucrose and overlaid with 2 ml of 35% sucrose and 1 ml of 5% sucrose (prepared in 2-[N-morpholino]ethanesulfonic acid buffer without detergent). Samples were subsequently centrifuged at 200,000 × g for 16–18 h at 4°C in an SW55 rotor (Beckman, Palo Alto, CA), and fractions of 400 μl each were collected from the top of the gradient. All lysis and gradient procedures were carried out at 4°C.
Internalization and recycling of surface CD151 proteins were assessed as described by Fabbri et al. (1999). In brief, semiconfluent NIH 3T3 cells stably transfected with myc-tagged CD151 were washed twice in PBS, cooled on ice, and labeled with 0.5 mg/ml sulfo-NHS-SS-biotin for 1 h at 4°C. Labeled cells were then washed three times in ice-cold serum-free DMEM and incubated at 37°C in serum-free DMEM for at the indicated periods of internalization time. Cells were then cooled on ice, washed, and treated with reducing solution containing 42 mM glutathione, 75 mM NaCl, 1 mM EDTA, 1% bovine serum albumin, and 75 mM NaOH. Treated cells were subsequently washed and lysed in RIPA.
For analysis of recycling, duplicate cell samples were sulfo-NHS-SS-biotin labeled and subjected to internalization for 1 h (as described above). After treatment with reducing solution, cells were further incubated at 37°C for 30 min to allow recycling to the surface. Samples were then treated with or without reducing solution before being lysed in RIPA buffer. Densitometric assessment of β-tubulin was used for normalizing the loading of samples. All densitometry was carried out using an Alpha Imager 2000 (Alpha Innotech, San Leandro, CA). Results shown in Figures 1–12 were highly reproducible in multiple experiments.
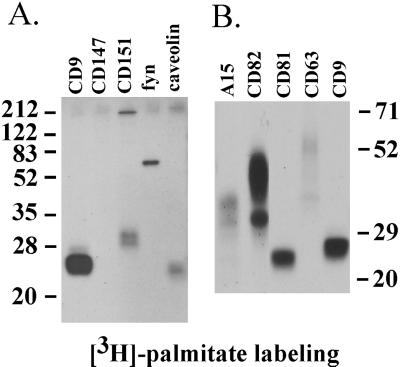
Figure 1.
Palmitoylation of tetraspanin proteins. (A) A431 cells were pulsed with [3H]palmitate and lysed in RIPA buffer, and then CD9, CD147, CD151, fyn, and caveolin were immunoprecipitated using mAbs Du-Al, 8G6, 5C11, FYN3, and C13630, respectively. (B) A15, CD82, CD81, CD63, and CD9 were immunoprecipitated using mAbs AZM30.4, M104, M38, 6H1, and Du-All-1, respectively. The A15 and CD82 samples were obtained from transiently transfected 293 cells. CD81, CD63, and CD9 samples were from A431 cells. Molecular masses are given in kilodaltons.
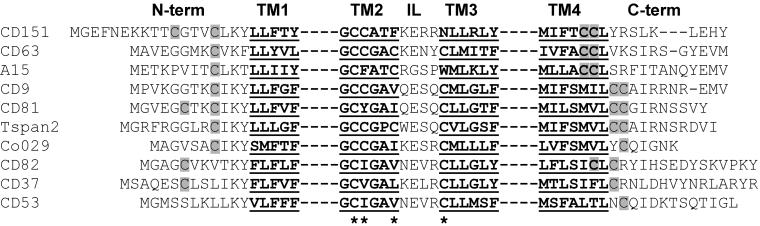
Figure 12.
Potential tetraspanin palmitoylation sites. Cysteine residues in CD151 (C11, C15, C241, C242) used as palmitoylation sites and proximal to TM1 and TM4 are compared with cysteine alignments in other tetraspanin proteins. Positions of additional membrane proximal cysteines that could be palmitoylated are marked with asterisks. Only portions of TM1, TM2, TM3, and TM4 are shown. IL, inner loop.
RESULTS
Palmitoylation of Tetraspanins and Associated Proteins
To assess palmitoylation of tetraspanin proteins, we labeled A431 cells or transiently transfected 293 cells with [3H]palmitate for 2–3 h and then prepared lysates using stringent detergent conditions (1% Triton X-100, 1.0% deoxycholate, 0.1% SDS). On immunoprecipitation, the tetraspanin proteins CD9, CD82, and CD81 showed strong 3H labeling, CD151 and A15 showed moderate labeling, and CD63 showed a lower level of labeling (Figure 1). In control experiments, known palmitoylated proteins (caveolin and fyn) were clearly labeled, whereas a cell surface protein lacking palmitoylation sites (CD147) was not labeled (Figure 1). As determined by immunoblotting and flow cytometry, all proteins analyzed are well expressed in either A431 cells or 293 cell transfectants (Yang, Claas, Kraeft, Chen, Wang, Kreidberg, and Hemler, unpublished results). High molecular weight proteins appearing in CD151 and caveolin lanes are likely because of protein multimerization.
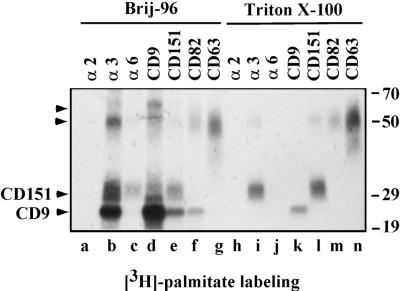
As seen in less stringent lysis conditions (1% Brij-96), palmitoylated tetraspanin proteins associated with other palmitoylated proteins. For example, 3H-labeled CD9 (Figure 2, lane d) was coprecipitated with labeled proteins resembling CD151 and CD82 (Figure 2, lanes e and f), whereas unknown labeled proteins of ∼50 and 60 kDa were also associated with CD9 (Figure 2, lane d). CD63 (Figure 2, lanes g and n) showed no obvious associations with other labeled proteins. Also, immunoprecipitation of α3 integrin yielded labeled proteins resembling CD9 and CD151, as well as unknown proteins of ∼50 and 60 kDa (Figure 2, lane b). In contrast, immunoprecipitation of α6 integrin yielded only labeled CD151 (Figure 2, lane c). In a control experiment, α2 integrin showed no association with labeled proteins (Figure 2, lanes a and h), consistent with its failure to associate with tetraspanins under these conditions (Berditchevski et al., 1996; Yauch et al., 1998). From Triton X-100 lysates, labeled CD9, CD151, CD82, and CD63 were isolated in the absence of labeled associated proteins (Figure 2, lanes k–m). The labeled protein of ∼50 kDa in the CD151 lane (Figure 2, lane l) is likely to be a CD151 dimer. Because CD151 associates tightly with α3β1 (Yauch et al., 1998), immunoprecipitation of α3 integrin yielded labeled CD151, even under Triton X-100 conditions (Figure 2, lane I).
Figure 2.
Tetraspanin protein complexes. A431 cells were labeled with [3H]palmitate and then lysed in the buffers containing either 1% Brij-96 (lanes a–g) or 1% Triton X-100 (lanes h–n). Equal amounts of lysate were used for each immunoprecipitation. Antibodies for individual immunoprecipitations were A2-IIE10, A3-X8, A6-ELE, Du-All-1, 5C11, M104, and 6H1 for α2, α3, α6, CD9, CD151, CD82, and CD63, respectively. Unknown proteins of ∼60 and 50 kDa (distinct from CD63) are indicated by arrowheads. At least part of the 50-kDa protein is likely to be a CD151 dimer, as confirmed by immunoblotting for CD151 (Yang, Claas, Kraeft, Chen, Wang, Kreidberg, and Hemler, unpublished results). Note: Compared with Brij-96 extraction (lane d), Triton X-100 extraction (lane k) yielded less CD9, because of destabilization of the Du-All-1 epitope. Also, more CD151 appears in the anti-α3 lane (lane b) compared with the anti-CD151 lane (lane e), because in Brij-96 lysate, the 5C11 epitope on CD151 is likely obscured by associated proteins.
Palmitoylation Is Inhibited by Brefeldin A
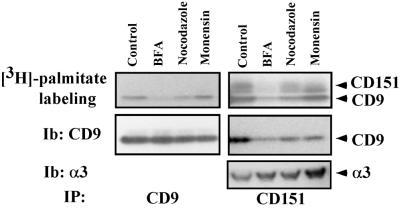
On treatment of A431 cells with the Golgi-disrupting agent brefeldin A (Dinter and Berger, 1998), palmitoylation of both CD9 and CD151 was substantially inhibited, as seen by diminished 3H labeling (Figure 3, top). In contrast, treatment with nocodazole or monensin had little effect on palmitoylation of these proteins. Levels of CD9 protein (Figure 3, bottom left) were not altered by treatment with brefeldin A, nocodazole, or monensin. Immunoprecipitation of CD151 revealed not only diminished CD151 labeling but also a decrease in associated labeled CD9 (Figure 3, top right). In addition, brefeldin A treatment caused a marked decrease in the amount of CD151-associated CD9 protein (Figure 3, middle right). In a control experiment, levels of tightly associated α3 integrin were not decreased in CD151 immunoprecipitates. Together these experiments indicate that tetraspanin palmitoylation requires trafficking through Golgi but not microtubule- or endosome-dependent trafficking. Also, the decrease in CD151-CD9 association raises the possibility that tetraspanin palmitoylation could be required for tetraspanin-tetraspanin association (see below).
Figure 3.
Palmitoylation sensitivity to brefeldin A (BFA). A431 cells were incubated with 10 μg/ml brefeldin A, 10 μg/ml nocodazole, or 20 μg/ml monensin for 1 h in DMEM in the absence of serum. Medium was then removed, and drugs were added again (same dose) in the presence of [3H]palmitate and 5% dialyzed fetal calf serum for 1 h. From Brij-96 lysates, either CD9 (left) or CD151 (right) was immunoprecipitated. Precipitated samples (IP) were analyzed for [3H]palmitate incorporation (top) or by immunoblotting (Ib) for CD9 or α3 integrin (bottom). Because CD151 tightly associates with α3 integrin, α3 was used as a marker to assess CD151 loading. SDS-PAGE was carried out on a 4–12% acrylamide gradient.
Removal of Palmitoylation by Site-directed Mutagenesis
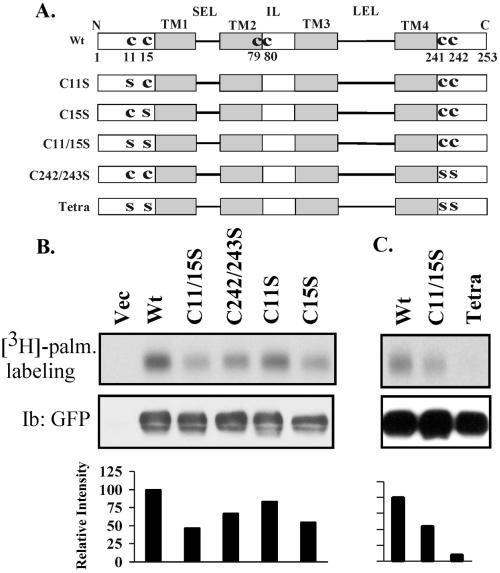
To gain more insight into the role of tetraspanin palmitoylation, we identified and mutated palmitoylation sites, using CD151 as a prototype tetraspanin protein. For transmembrane proteins, palmitoylation typically occurs on intracellular cysteine residues, proximal to transmembrane domains (Resh, 1999). Indeed, CD151 contains at least four cysteine residues (C11, C15, C241, C242) proximal to either TM1 or TM4 (Figure 4A). CD151 mutants were fused with a C-terminal GFP and transiently transfected into 293 cells. As indicated (Figure 4B), a C11/15S double mutation eliminated ∼55% of [3H]palmitate labeling, whereas C11S and C15S single mutations eliminated ∼15 and 40%, respectively. Another double mutation (C242/243S) eliminated ∼30% of the labeling. In a separate experiment, the C11/15S double mutation again eliminated nearly 50% of the labeling, whereas a C11/15/242/243S tetra mutant lost >90% of the [3H]palmitate labeling (Figure 4C). Within stable MDA-231 cell transfectants, palmitoylation was again >90% diminished in the tetra-CD151 mutant compared with wild-type CD151 (Yang, Claas, Kraeft, Chen, Wang, Kreidberg, and Hemler, unpublished results). Residual 3H labeling could be due to additional palmitoylation sites in CD151, such as C79 and C80 near the C-terminal end of TM2 (Figure 4A). However, mutations of these cysteine residues resulted in loss of CD151 expression, thus preventing evaluation of palmitoylation. In conclusion, the majority of CD151 palmitoylation is dependent on cysteine residues 11, 15, 241, and 242.
Figure 4.
CD151 palmitoylation sites. (A) Schematic diagram of candidate CD151 palmitoylation sites. Shown are the four transmembrane domains (TM1–4), short extracellular loop (SEL), inner loop (IL), large extracellular loop (LEL), and membrane-proximal cysteine residues. Various C→S (CYS→SER) mutants are also indicated. Unless otherwise indicated, all mutants contained a GFP domain fused to the carboxy terminus. Wt, wild type. (B and C) In separate experiments, 293 cells were transiently transfected with various CD151 constructs and pulsed with [3H]palmitate (palm.). After transfection (24 h), cells were lysed in RIPA buffer and immunoprecipitated using mAb 5C11. Samples were then divided equally, resolved by SDS-PAGE, and then either dried for the detection of [3H]palmitate or transferred to nitrocellulose for blotting (Ib) with GFP polyclonal antibody.
Initial Characterization of Palmitoylation-deficient CD151 Mutant
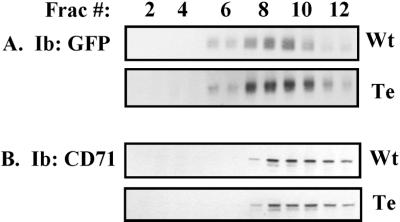
Many properties of the tetra-CD151 mutant were either unaltered or minimally altered. As described elsewhere, tetraspanin protein complexes may have a lower density than the majority of other transmembrane proteins (Claas et al., 2001; Stipp et al., 2001b). Indeed, we have utilized a sucrose density gradient (Figure 5) to confirm that wild-type CD151 has a lower density than typical transmembrane proteins, here represented by the transferrin receptor (CD71). Importantly, the density of the tetra-CD151 mutant was not markedly altered compared with wild-type CD151. Both peaks were centered in fractions 8 and 9, although the mutant did show a slightly broader peak distribution (fractions 7–11). As shown in other experiments, the tetra-CD151 mutant retained distinct epitopes recognized by mAb 5C11, 1A5, and TS151r (Yang, Claas, Kraeft, Chen, Wang, Kreidberg, and Hemler, unpublished results, and see below). In addition, the relative size of tetra-CD151-GFP was unaltered compared with wild-type CD151-GFP (Figures 4, B and C, and 6). Thus, the tetra mutant appears to remain structurally sound, despite mutation of cysteine residues 11, 15, 241 and 242.
Figure 5.
Sucrose density gradient analysis of CD151. Stably transfected MDA-231 cells containing either wild-type CD151 (Wt) or CD151-tetra mutant (Te) were lysed in 1% Brij-96. Lysates were then subjected to 5–35-45% discontinuous sucrose density gradient centrifugation and 12 fractions (Frac.) of 400 μl each were collected from the top of the gradient. (A) From each fraction, 50 μl was immunoprecipitated with anti-GFP polyclonal antibody, and samples were immunoblotted (Ib) using a monoclonal anti-GFP antibody to detect CD151-GFP. (B) From each fraction, 50 μl of lysate was resolved by SDS-PAGE, transferred to membrane, and then immunoblotted for CD71, a cell surface transmembrane protein typically found in dense fractions.
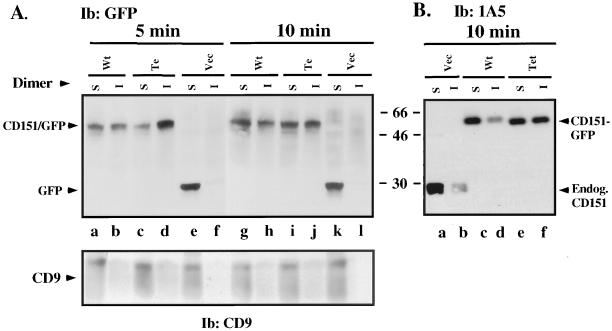
Because acylation often renders proteins more resistant to detergent extraction (Dinter and Berger, 1998; Resh, 1999), we hypothesized that wild-type CD151 might be more difficult to extract than tetra-CD151. However for wild-type and tetra CD151, initial detergent extraction experiments showed essentially identical high yields from transfected MDA-231 cells upon lysis using 1% Triton X-100, 3-[{cholamidopropyl}dimethylammonio]-1-propanesulfonic acid, or Brij-96 for at least 30 min (Yang, Claas, Kraeft, Chen, Wang, Kreidberg, and Hemler, unpublished results). When MDA-231 cells were rocked gently in 1% Brij-96 for short intervals (5–10 min), surprising differences in extractability emerged. After 5 min, nearly 50% of wild-type CD151 was extracted (Figure 6A, lanes a and b), whereas the majority of the tetra mutant remained insoluble rather than becoming more soluble (Figure 6A, lanes c and d). Differences were less obvious after 10 min, but again, wild-type CD151 showed more solubility (lane g > h) compared with the tetra mutant (lane i < j). Wild-type CD151-GFP and endogenous CD151 in MDA-231 cells were very similar in terms of showing 1% Brij-96 detergent extractability (Figure 6B, lanes a–d), indicating that the GFP tag itself is not affecting the results. More importantly, the tetra-CD151 mutant was again more resistant to 1% Brij-96 extraction for 10 min (Figure 6B, lanes e and f). For wild-type CD151 (either endogenous or GFP tagged) in 5- to 10-min extraction experiments, the mean ratio of soluble to insoluble was 1.6 ± 0.4 (N = 4). In contrast, for tetra-CD151 that ratio was 0.9 ± 0.3 (N = 3), as determined using densitometry. Another tetraspanin protein (CD9) was equally soluble in 1% Brij-96 in all cases (Figure 6A, bottom), whereas wild-type and mutant CD151 were equally insoluble in 1% Brij-99 (Yang, Claas, Kraeft, Chen, Wang, Kreidberg, and Hemler, unpublished results). The shift toward diminished detergent extractability seen for tetra-CD151 suggests that palmitoylation of this protein is not contributing to formation of stable, detergent-resistant, CD151-lipid microdomains.
Figure 6.
CD151 detergent extractability. (A) Equal numbers of semiconfluent stably transfected MDA-231 cells (mock, wt-CD151, tetra-CD151) were seeded and grown for 24 h. Cells were then washed, cooled on ice, and rocked gently in the presence of 1% Brij-96 for 5–10 min at 4°C. After removal of the soluble fraction (S), the remaining insoluble material was further extracted in RIPA for an additional 1 h at 4°C (to yield fraction I). Samples were resolved by SDS-PAGE and blotted (Ib) for CD151-GFP or GFP (using polyclonal anti-GFP) or for CD9 (using mAb C9BB). Note: The greater insolubility of CD151 compared with CD9 is possibly due to much stronger CD151 association with α3 and α6 integrins. Results shown are representative of multiple experiments. (B) MDA-231 cells were lysed in 1.0% Brij-96 for 10 min, and then soluble (S) and insoluble (I) fractions were obtained as in A. In this experiment, anti-CD151 mAb 1A5 was used to blot both endogenous CD151 and CD151-GFP proteins. Note: Expression of endogenous CD151 (lanes c–f) is greatly reduced because of the presence of exogenous CD151-GFP.
CD151 Association with Other Proteins
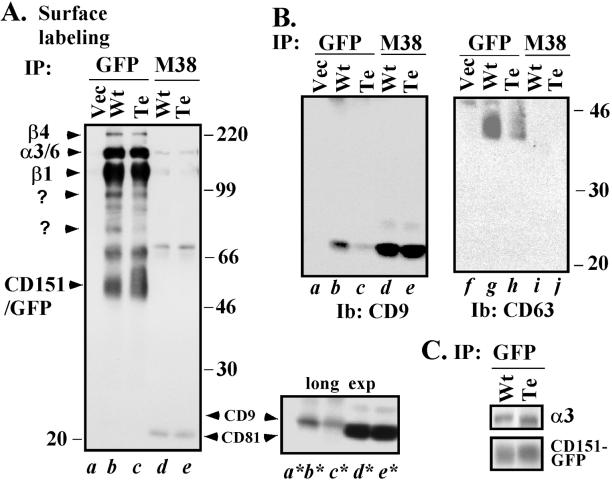
A hallmark of tetraspanins is their ability to form complexes with several other transmembrane proteins (Wright and Tomlinson, 1994; Hemler et al., 1996; Maecker et al., 1997). For example, in 1% Brij-96 lysate from MDA-431 cells, wild-type CD151 associated with cell surface biotin-labeled α3β1 and α6β4 integrins, as well as with unknown proteins of ∼95, 90, 75, and 68 kDa (Figure 7A, lane b). In contrast, tetra-CD151 showed markedly diminished association with the unknown proteins of ∼95, 90, and 75 kDa, even though tetra-CD151-GFP itself was amply present on the cell surface (Figure 7A, lane c). Extended exposure of the lower portion of Figure 7A revealed that association with surface-labeled CD9 was also markedly diminished for tetra-CD151 (compare lanes b* and c*). In a separate experiment, the tetra-CD151 mutant showed markedly diminished levels of total associated CD9 (Figure 7B, lane c) and CD63 (Figure 7B, lane h). In control experiments, patterns of proteins associated with CD81 (and levels of CD81 itself) were essentially identical whether derived from CD151 wild type or tetra transfectants (Figure 7, A, lanes d, e, d*, and e*, and B, lanes d and e).
Figure 7.
Integrity of CD151 complexes. (A) Mock or stable CD151 transfectants (MDA-231 cells) were surface labeled with biotin for 1 h at 4°C, washed, and lysed in 1.0% Brij-96 buffer. Equal amounts of lysates were subsequently immunoprecipitated (IP) using anti-GFP polyclonal antibody or anti-CD81 mAb M38. Samples were resolved by SDS-PAGE and blotted using avidin-horseradish peroxidase. The small panel to the right (bottom) represents a longer exposure (exp) of a portion of the large panel to the left. SDS-PAGE was carried out on a 5–15% acrylamide gradient. (B) Samples prepared as in A were immunoblotted for either CD63 or CD9. (C) CD151 transfectants were labeled with 0.3 mCi/ml [35S] methionine for 1 h and lysed in 1% Triton X-100, and then wt-CD151 and tetra-CD151 were immunoprecipitated (IP) using anti-GFP polyclonal antibody. After SDS-PAGE, samples were transferred to a PVDF membrane, exposed to film to detect associated α3 integrin (top), and subsequently blotted using anti-GFP mAb (bottom).
Because CD151 extracellular domains interact with α3 and α6 integrins (Yauch et al., 2000; Yang, Claas, Kraeft, Chen, Wang, Kreidberg, and Hemler, unpublished results), we predicted that loss of palmitoylation should not abolish these interactions. Indeed, the CD151-tetra mutant retained abundant association with surface-labeled α3β1 integrin in MDA-231 cell Brij-96 lysates (Figure 7A, lanes b and c). Analysis of 35S-labeled α3 coprecipitated with CD151 in RIPA detergent conditions again revealed minimal, if any, loss of association for the tetra-CD151 mutant (Figure 7C, top). Association with α6β4 was also retained, although it was ∼30% diminished, based on quantitation of β4 levels (Figure 7A, lanes b and c). We conclude that mutation of CD151 palmitoylation sites has little effect on strong CD151 associations with α3, only a marginal effect on the slightly weaker α6 integrin interaction (seen in Triton X-100), but a marked effect on associations with several other proteins.
Tetra-CD151 Mutant Shows Altered Cellular Distribution
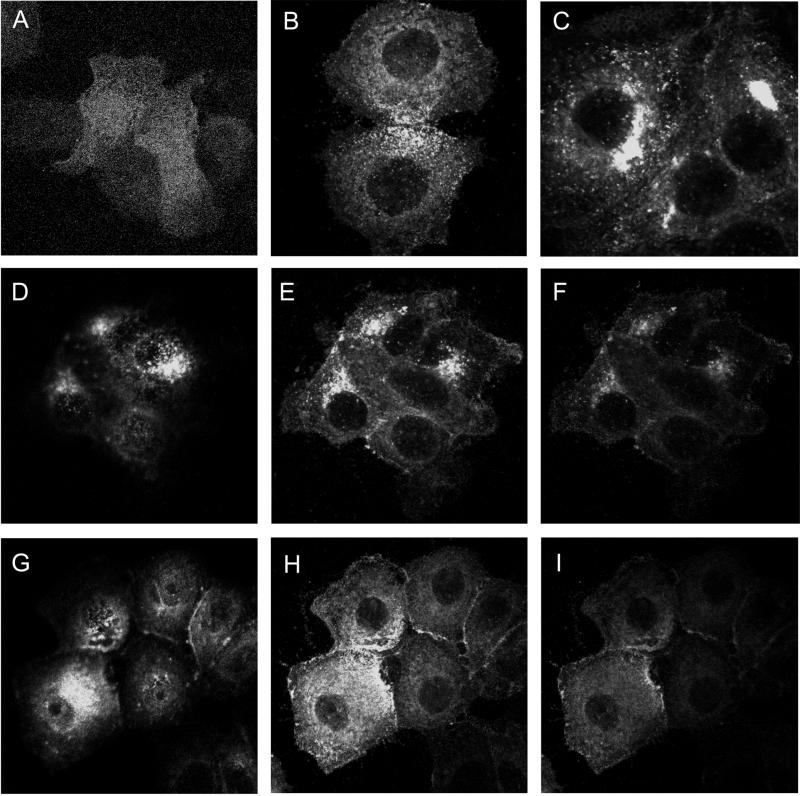
As seen by confocal microscopy, a substantial portion of wild-type CD151-GFP in MCF-7 cells appeared in perinuclear aggregates (Figure 8, D–F). In contrast, the tetra-CD151 mutant appeared to be more homogeneously dispersed throughout MCF-7 cells (Figure 8, G–I). A C15S-CD151-GFP single mutant showed a partially dispersed punctate distribution in the surface of MCF-7 cells (Figure 8B), whereas a C242/243S double mutant (Figure 8C) showed little change in CD151 distribution compared with wild type (Figure 8, D–F). In cells transfected with vector alone, GFP fluorescence was more homogenously distributed, in a pattern of small dots, typical of intracellular protein distribution. Consistent results were obtained for all focal planes examined. Similar subcellular localization differences were observed when CD151-GFPs were expressed in MDA-231 cells (Yang, Claas, Kraeft, Chen, Wang, Kreidberg, and Hemler, unpublished results). Together, these data suggest that palmitoylation of CD151, especially at the N terminus, markedly influences intracellular distribution. The presence of a GFP tag on the carboxyl termini of our CD151 proteins could itself alter CD151 distribution. However, the perinuclear staining pattern seen here for wild-type CD151-GFP strongly resembles the staining pattern seen elsewhere for endogenous CD151 in permeabilized endothelial cells (Sincock et al., 1999). Thus, CD151 distribution may not be unduly influenced by the presence of a GFP tag.
Figure 8.
Distribution of CD151-GFP. MCF-7 cells stably transfected with various GFP-tagged CD151 proteins were grown on glass coverslips overnight, fixed in 1.0% paraformaldehyde for 20 min at 4°C, and mounted onto glass slides using the ProLong Antifade Kit from Molecular Probes (Eugene, OR). Slides were analyzed using a confocal model LSM4 scanning microscope (Zeiss, Thornwood, NY) equipped with an external argon-krypton laser. (A) Cells expressing GFP only; (B) cells expressing C15S-CD151-GFP; (C) cells expressing C241/242S-CD151-GFP; (D–F) cells expressing wild-type CD151-GFP; (G–I) cells expressing tetra-CD151; (D and G) highest optical plane; (E and H) lower plane; (F and I) lowest plane. Expression levels for mutant and wild-type CD151 were comparable, as seen by flow cytometry (GFP fluorescence) and by immunoblotting (anti-GFP antibody).
Additional Consequences Tetra-CD151 Mutation
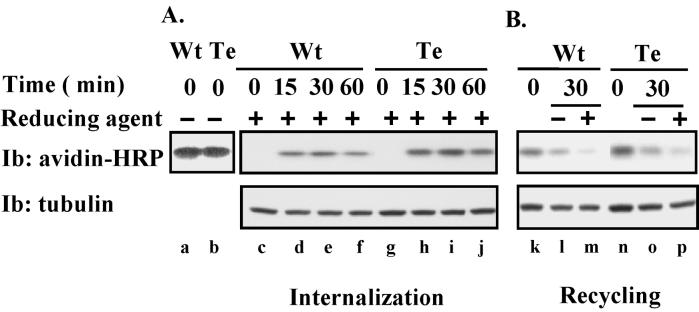
We hypothesized that altered protein complex formation (Figure 7) and altered subcellular localization (Figure 8) might be associated with changes in internalization or recycling of cell surface CD151. Hence, we labeled CD151-transfected NIH3T3 cells with sulfo-NHS-SS biotin (to create a reducible biotin-cell linkage) and determined the time course for accumulation of internalized CD151 (defined as CD151 becoming inaccessible to biotin release upon reduction). As indicated, both wild-type CD151 (Figure 9A, lanes c–f, top) and tetra-CD151 (lanes g–j) accumulated inside of cells, with peak levels observed at ∼30 min. We saw no evidence that internalization of tetra-CD151 was impaired. Similar amounts of biotin-labeled wild-type and tetra-CD151 were present at the start of the experiment (Figure 9A, lanes a and b). Immunoblots of tubulin indicate that similar amounts of lysate were sampled at each time point.
Figure 9.
Internalization and recycling of CD151. (A) NIH3T3 cells were stably transfected with Myc-tagged wild-type CD151 (Wt) or tetra-CD151 (Te). Stable transfectants were surface labeled with reducible NHS-SS-biotin at 4°C. Some labeled cells were then lysed immediately in RIPA (lanes a and b). Other cells (lanes c–j) were incubated at 37°C for various times (to allow CD151 internalization) and then treated with reducing agent to remove all biotin remaining on the cell surface. (B) NHS-SS-biotin-labeled cells were incubated at 37°C for 60 min to allow accumulation of internalized CD151 proteins and then treated with reducing agent to remove the noninternalized surface proteins. Some cells were then lysed immediately (lanes k and n). Other cells were then incubated for an additional 30 min at 37°C to allow recycling of labeled-internalized CD151 back to the cell surface. These cells were then treated either without (lanes l and o) or with (lanes m and p) reducing agent before lysis in RIPA. From all lysate samples (A and B), CD151 was immunoprecipitated (using mAb 5C11), and biotin labeling was detected by blotting with avidin-horseradish peroxidase (HRP). Each sample was also blotted (Ib) for tubulin (bottom).
Next, we considered that, if internalization was not deficient, perhaps recycling back to the cell surface might be accelerated for tetra-CD151. Hence, intracellular pools of labeled CD151 were allowed to accumulate for 60 min, and cell surface biotin was removed by treatment with reducing agent. Another 30-min incubation was then carried out, and the total labeled CD151 remaining was determined (Figure 9B, lanes l and o). In addition, levels of internal, labeled CD151 that had not recycled (not accessible to biotin removal) were also determined (Figure 9B, lanes m and p). From densitometry measurements, we estimate that wild-type CD151 underwent 78% recycling ([1 − m/l] × 100), whereas tetra-CD151 showed 69% recycling ([1 − p/o] × 100). Also, from diminution of the total biotin signal >30 min (during the recycling experiment), we estimate that wild-type CD151 was 45% degraded ([1 − l/k] × 100), whereas tetra-CD151 was 43% degraded ([1 − o/n] × 100). In conclusion, results from Figure 9 and from another independent experiments (Yang, Claas, Kraeft, Chen, Wang, Kreidberg, and Hemler, unpublished results) indicate that wild-type and tetra-CD151 on the cell surface are very similar in terms of internalization, recycling, and degradation.
Finally, we considered that differences in distribution (between tetra- and wild-type CD151) might be explained by differences in biosynthesis. To test this, stably transfected MDA-231 cells were pulse-labeled for 30 min with, and then chased with, unlabeled methionine for varying intervals. As indicated in Figure 10, A and B, the appearance of mature wild-type CD151 peaked at ∼6 h and then slowly disappeared, such that it was 50% gone in ∼22 h. In contrast, mature tetra-CD151 was already substantially diminished by 2 h, with a half-life of only ∼4–5 h. In addition, compared with wild type CD151, tetra-CD151 showed a greater accumulation of relatively transient immature forms at early time points (Figure 10A), including one form (∼47 kDa) not seen at all during wild-type CD151 biosynthesis. In a separate [35S] methionine pulse-chase experiment, wild-type-CD151-GFP and endogenous CD151 showed comparable stability in MDA-231 cells (50% gone in ∼ 24 h, Yang, Claas, Kraeft, Chen, Wang, Kreidberg, and Hemler, unpublished results). Thus, again the GFP tag itself is not markedly altering CD151 properties. In conclusion, newly synthesized, mature CD151 is less stable if it lacks palmitoylation sites.
Morphological Consequences of Altered CD151 Palmitoylation
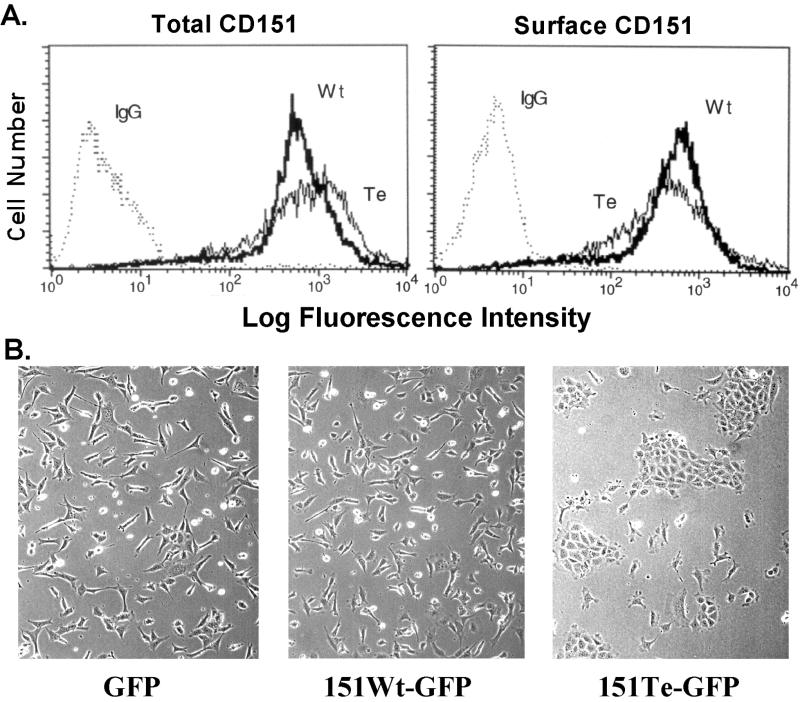
Unfortunately, high background levels of endogenous wild-type CD151 have prevented assessment of the broader biological effects of expressing palmitoylation-deficient mutant CD151 in many cell types. Because the presence of α3β1 integrin can bring more CD151 to the surface (Yauch et al., 1998), background problems are especially obvious in cells strongly positive for α3β1. To circumvent this problem, wild-type and tetra-CD151 were expressed in the B12 murine kidney epithelial cell line derived from an α3 integrin-deficient mouse (Wang et al., 1999). The tetra-CD151 and wild-type CD151 were expressed at comparable levels both in terms of total expression (Figure 11A, left) and cell surface expression (Figure 11A, right). As indicated in Figure 11B, subconfluent B12 cells expressing tetra-CD151 (right) looked very different from cells containing either GFP alone (left) or wild-type CD151-GFP (middle). In particular, the former cells displayed a characteristic epithelium-like cobblestone pattern, whereas the latter were well dispersed and more elongated. Similar results were obtained whether the B12 cells were plated on tissue culture plastic for 24 h (Figure 11B) or on laminin-5 (Yang, Claas, Kraeft, Chen, Wang, Kreidberg, and Hemler, unpublished results). However when the cells were plated for 24 h on fibronectin, an epithelial morphology was not observed and the presence of tetra-CD151 had no obvious morphological consequences (Yang, Claas, Kraeft, Chen, Wang, Kreidberg, and Hemler, unpublished results).
Figure 11.
Mutant CD151 alters cell morphology. (A) Stably transfected semiconfluent B12 kidney epithelial cells were lightly trypsinized, incubated at 37°C for 20 min, then stained on ice with either control immunoglobulin G or anti-CD151 mAb 5C11, and visualized using phycoerythrin-conjugated secondary antibody (Biosource, Camarillo, CA). Flow cytometry was carried out using a FACSCalibur (Becton-Dickinson), measuring GFP fluorescence (left) and phycoerythrin fluorescence (right), for tetra (Te) and wild-type (Wt) CD151-transfected cells. Background staining is indicated as “IgG.” Mean fluorescence intensity values for Te and Wt CD151, respectively, are 1017 and 740 (for GFP fluorescence) and 660 and 616 (for phycoerythrin fluorescence). (B) B12 cells expressing GFP alone, wild-type CD151, or tetra-CD151 were plated on tissue culture plastic for 24 h and then analyzed using a Zeiss Axiovert 135 microscope and photographed as described previously (Stipp and Hemler, 2000).
DISCUSSION
Initial Characterization of Tetraspanin Palmitoylation
Our results establish that all tetraspanin proteins tested (CD9, CD63, CD81, CD82, CD151, A15/Talla1) are palmitoylated. Palmitoylation of CD151 utilizes intracellular membrane proximal cysteines as determined by site-directed mutagenesis. The CD151 residues most critical for palmitoylation (C15, C241, C242) are conserved in a subset of other tetraspanin proteins (Figure 12). Two others (CD63 and A15/Talla1) have C-terminal cysteines that align precisely with C241, C242; six of nine other tetraspanins have an N-terminal cysteine aligning with C15 in CD151. Indeed, both CD63 and A15/Talla1 (which align best with CD151) are palmitoylated, although others (e.g., CD9, CD82) are also well palmitoylated. As seen previously for the palmitoylation of membrane proximal cysteines, exact positioning relative to transmembrane domains may not be that critical (Resh, 1999). Although N-terminal and C-terminal cysteines account for most of the CD151 palmitoylation, it remains possible that additional cysteines proximal to the short intracellular loop (positions marked with asterisks in Figure 12) could also contribute. Although we could not express CD151-containing C79S and C80S mutations, preliminary mutagenesis experiments revealed that cysteines proximal to transmembrane domains 2 and 3 do make a substantial contribution to CD9 palmitoylation (Yang, Claas, Kraeft, Chen, Wang, Kreidberg, and Hemler, unpublished results).
Brefeldin A inhibition results argue strongly that CD151 and CD9 must traffic through the Golgi apparatus to become palmitoylated. Indeed, brefeldin A inhibition results provide ample precedent for palmitoylation of other proteins requiring a functioning Golgi (Gonzalo and Linder, 1998; Parat and Fox, 2001). Monensin, an agent that increases the luminal pH of acidic organelles and inhibits endosomal and lysosomal trafficking (Dinter and Berger, 1998), failed to block CD151 or CD9 palmitoylation. Thus, tetraspanins are likely to acquire palmitoylation before trafficking to those organelles. Nocodazole also failed to block palmitoylation of CD9 and CD151, indicating that microtubule-dependent trafficking functions (Dinter and Berger, 1998) are not needed for tetraspanin palmitoylation.
For several reasons, [3H]palmitate labeling is likely to occur via S-acylation rather than N-acylation. First, the majority of labeling is lost upon mutation of cysteine residues. Second, palmitate is not utilized by the enzymes responsible for N-acylation (Resh, 1999). Third, whereas N-acylation occurs cotranslationally (Resh, 1999), labeling seen here is posttranslational (e.g., as evidenced by brefeldin A inhibition).
Despite the mutation of four cysteines to serine, CD151 remained structurally sound by several criteria. The CD151-tetra mutant retained at least three distinct cell surface epitopes and was unaltered with respect to size, density, internalization, or recycling. Also, the very robust association of CD151 with α3β1 integrin (Yauch et al., 1998) was retained. For that strong interaction, previously mapped to the large extracellular loop of CD151 (Yauch et al., 2000), additional contributions due to palmitoylation are apparently not necessary. Interaction of CD151 with α6β4 integrin was also largely maintained for the CD151-tetra mutant. However, with α6 integrins interacting a bit more weakly than α3β1 with CD151 (Yauch et al., 1998, 2000), a contributory role for palmitoylation begins to become evident.
Functional Consequences of CD151 Palmitoylation
Whereas CD151 interactions with α3 and α6 integrins are retained in detergents such as Triton-X100 and digitonin, interactions with other proteins are lost unless a milder detergent such as Brij-96 is utilized (Yauch et al., 1998; Serru et al., 1999). It is these weaker interactions with other tetraspanins (CD9, CD63), and with unknown proteins (95, 90, and 75 kDa), that are markedly diminished when CD151 palmitoylation sites are mutated. Thus, we suggest that tetraspanin palmitoylation may contribute to stabilization of the large assembly of tetraspanin interactions sometimes called the “tetraspanin web” (Imai and Yoshie, 1993; Rubinstein et al., 1996; Hammond et al., 1998; Hemler, 1998). Conceivably, because we are only ablating palmitoylation of CD151, but not palmitoylation of associated tetraspanins or other proteins, we could be underestimating the contribution of palmitoylation to tetraspanin complex formation. Future studies will be needed to determine whether palmitoylation of multiple components within tetraspanin complexes will have effects that are additive or possibly even synergistic. Of particular interest will be phosphatidylinositide 4-kinase, a palmitoylated enzyme (Barylko et al., 2001) that associates strongly with specific tetraspanins (Yauch and Hemler, 2000).
Mutation of cysteines to serines could have effects that extend beyond loss of palmitoylation. Thus, it is helpful to have confirmatory results from at least one experiment that does not involve mutagenesis. In this regard, treatment of A431 cells with brefeldin A caused simultaneous decreases in palmitoylation of endogenous CD151, CD9, and CD151-CD9 association. This result is consistent with the mutagenesis results and suggests that impaired protein association properties of CD151 mutants are indeed mostly due to removal of palmitoylation.
One consequence of altered CD151 connection to the tetraspan web may be an altered subcellular distribution. Localization into endosomal and lysosomal type vesicles has been previously demonstrated for CD151 and other tetraspanins (Metzelaar et al., 1991; Escola et al., 1998; Berditchevski and Odintsova, 1999; Sincock et al., 1999). Consistent with this, wild-type CD151 within intracellular vesicles in MCF-7 cells showed colocalization with rab 4, rab5, and rab 11 (endosomal markers) and CD63 (a lysosomal marker; Yang, Claas, Kraeft, Chen, Wang, Kreidberg, and Hemler, unpublished results). Our results now suggest that CD151 requires palmitoylation for endosomal/lysosomal localization. Altered tetra-CD151 localization could not be explained by altered CD151 internalization or altered recycling back to the cell surface. Instead, we suggest that loss of targeting of mutant CD151 from the Golgi to endosomal and lysosomal compartments may be due to loss of CD151 association with other proteins (such as CD63) that are required to actively assist in targeting. In addition, the absence of palmitoylation (and absence of key associated tetraspan web proteins) may make mutant CD151 more susceptible to degradation, as it begins to travel from the Golgi to other intracellular compartments; whereas it may escape degradation while trafficking to the plasma membrane. This suggestion is supported by our findings that newly synthesized, mature mutant CD151 has a markedly decreased half-life, whereas mutant CD151 on the cell surface does not show enhanced degradation. Notably, palmitoylation of lymphoma proprotein convertase (van de Loo et al., 2000) and human A1 adenosine receptor (Gao et al., 1999) may also prolong the half-life of those newly synthesized proteins.
CD151 palmitoylation deficiency also can have a dramatic influence on cell morphology. Within the murine B12 kidney epithelial cell line, mutant human CD151 caused a switch from an elongated, dispersed cell morphology toward a cobblestone, epithelial type morphology. In contrast, expression of wild-type human CD151 had little effect. We suspect that tetra-CD151 is having a dominant negative effect on endogenous murine CD151. Because no α3 is present in the B12 cells and CD151 associates well with α6β4 integrin in epithelial cells (Sterk et al., 2000), mutant CD151 may be influencing α6β4 function, possibly by altering its connection to the tetraspanin web. Our finding that tetra-CD151 altered cell morphology when cells were plated on laminin-5 (an α6β4 ligand), but not on fibronectin, supports a functional connection with α6β4. Notably, α6β4 is well established to play a key role in the transition between organized epithelium and disorganized, invasive cells (Weaver et al., 1997; Mercurio et al., 2001). Specific details regarding the mechanism of mutant CD151-induced morphological changes will be addressed in a separate manuscript.
CD151 Palmitoylation and Membrane Targeting
Palmitoylation promotes membrane targeting for many cytosolic proteins but not usually for proteins with multiple transmembrane domains (Dunphy and Linder, 1998). As we show here, palmitoylation is likewise not needed for membrane targeting of tetraspanin protein CD151. Within cellular membranes, palmitoylation of src family kinases, G proteins, and several other proteins facilitates localization into detergent insoluble, low-density complexes, sometimes called lipid rafts (Dunphy and Linder, 1998). Tetraspanins typically have a lower density than most other cellular proteins and also may associate with lipid raft-like microdomains (Yashiro-Ohtani et al., 2000; Claas et al., 2001). Thus, we hypothesized that tetraspanin palmitoylation could promote assembly of low-density lipid-containing microdomains with diminished detergent solubility. However with loss of CD151 palmitoylation, we observed minimal changes in density. Furthermore, the CD151-tetra mutant displayed detergent extractability that was diminished rather than enhanced. Thus, loss of CD151-tetra mutant associations with other proteins is clearly not due to loss of raft-like properties. Consistent with this, we observed previously that many tetraspanin (CD9 and CD81) associations with other proteins are not dependent on a raft-like microenvironment (Claas et al., 2001; Stipp et al., 2001b). Why is CD151 detergent extractability diminished with loss of palmitoylation? We speculate that, in the absence of association with other transmembrane proteins, CD151 by default may become more associated with itself, and/or more anchored to the cortical actin cytoskeleton (Berditchevski and Odintsova, 1999), and thus less extractable. In this regard, CD81 structural studies suggest that the large extracellular loop contains inherent dimerization capability (Kitadokoro et al., 2001).
Summary and Conclusions
Aside from early studies demonstrating that CD9 is palmitoylated (Seehafer et al., 1990), tetraspanin palmitoylation has not been addressed and functional consequences not established. In this first detailed study of tetraspanin palmitoylation, we have identified critical sites and begun to show that palmitoylation is indeed functionally relevant. Although not very important for conferring raft-like properties on tetraspanin complexes (e.g., lower density, detergent insolubility), palmitoylation nonetheless may have a major role in assembling the tetraspanin web. Furthermore, we provide the first evidence that tetraspanin palmitoylation may selectively influence subcellular compartmentalization, stability of newly synthesized protein, and epithelial cell morphology. Given the conserved nature of tetraspanin palmitoylation sites, many of the conclusions from this study of CD151 should be applicable to other tetraspanins.
ACKNOWLEDGMENTS
We thank Dr. Alex Kazarov (Dana-Farber Cancer Institute) for advice and assistance with cDNA constructions, and we thank Dr. Siew Heng Wong (Institute of Molecular and Cell Biology, Singapore) for advice regarding protein trafficking. This work was supported by National Institutes of Health grants CA-86712 and CA-42368 (to M.E.H.).
Abbreviations used:
- GFP
green fluorescent protein
- PBS
phosphate-buffered saline
- PVDF
polyvinylidene difluoride
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–05–0275. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–05–0275 .
REFERENCES
- Azorsa DO, Moog S, Cazenave JP, Lanza F. A general approach to the generation of monoclonal antibodies against members of the tetraspanin superfamily using recombinant GST fusion proteins. J Immunol Methods. 1999;229:35–48. doi: 10.1016/s0022-1759(99)00102-7. [DOI] [PubMed] [Google Scholar]
- Banerjee SA, Hadjiargyrou M, Patterson PH. An antibody to the tetraspan membrane protein CD9 promotes neurite formation in a partially α3β1 integrin-dependent manner. J Neurosci. 1997;17:2756–2765. doi: 10.1523/JNEUROSCI.17-08-02756.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barylko B, Gerber SH, Binns DD, Grichine N, Khvotchev M, Sudhof TC, Albanesi JP. A novel family of phos-phatidylinositol 4-kinases conserved from yeast to humans. J Biol Chem. 2001;276:7705–7708. doi: 10.1074/jbc.C000861200. [DOI] [PubMed] [Google Scholar]
- Berditchevski F, Bazzoni G, Hemler ME. Specific association of CD63 with the VLA-3 and VLA-6 integrins. J Biol Chem. 1995;270:17784–17790. doi: 10.1074/jbc.270.30.17784. [DOI] [PubMed] [Google Scholar]
- Berditchevski F, Chang S, Bodorova J, Hemler ME. Generation of monoclonal antibodies to integrin-associated proteins: evidence that α3β1 complexes with EMMPRIN/basigin/OX47/M6. J Biol Chem. 1997;272:29174–29180. doi: 10.1074/jbc.272.46.29174. [DOI] [PubMed] [Google Scholar]
- Berditchevski F, Gilbert E, Griffiths MR, Fitter S, Ashman L, Jenner SJ. Analysis of the CD151-alpha3beta1 integrin and CD151-tetraspanin interactions by mutagenesis. J Biol Chem. 2001;276:41165–41174. doi: 10.1074/jbc.M104041200. [DOI] [PubMed] [Google Scholar]
- Berditchevski F, Odintsova E. Characterization of integrin-tetraspanin adhesion complexes: role of tetraspanins in integrin signaling. J Cell Biol. 1999;146:477–492. doi: 10.1083/jcb.146.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berditchevski F, Zutter MM, Hemler ME. Characterization of novel complexes on the cell surface between integrins and proteins with 4 transmembranes (TM4 proteins) Mol Biol Cell. 1996;7:193–207. doi: 10.1091/mbc.7.2.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson JM, St. John NF, Kawaguchi S, Pasqualini R, Berdichevsky F, Hemler ME, Finberg RW. The I domain is essential for echovirus 1 interaction with VLA-2. Cell Adhes Commun. 1994;2:455–464. doi: 10.3109/15419069409004455. [DOI] [PubMed] [Google Scholar]
- Charrin S, Le Naour F, Oualid M, Billard M, Faure G, Hanash SM, Boucheix C, Rubinstein E. The major CD9 and CD81 molecular partner: identification and characterization of the complexes. J Biol Chem. 2001;276:14329–14337. doi: 10.1074/jbc.M011297200. [DOI] [PubMed] [Google Scholar]
- Claas C, Seiter S, Claas A, Savelyeva L, Schwab M, Zoller M. Association between rat homologue of CO-029, a metastasis-associated tetraspanin molecule and consumption coagulopathy. J Cell Biol. 1998;141:267–280. doi: 10.1083/jcb.141.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claas C, Stipp CS, Hemler ME. Evaluation of prototype TM4SF protein complexes and their relation to lipid rafts. J Biol Chem. 2001;276:7974–7984. doi: 10.1074/jbc.M008650200. [DOI] [PubMed] [Google Scholar]
- Dijkstra S, Geisert EEJ, Gispen WH, Bar PR, Joosten EA. Up-regulation of CD81 (target of the antiproliferative antibody; TAPA) by reactive microglia and astrocytes after spinal cord injury in the rat. J Comp Neurol. 2000;428:266–277. doi: 10.1002/1096-9861(20001211)428:2<266::aid-cne6>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Dinter A, Berger EG. Golgi-disturbing agents. Histochem Cell Biol. 1998;109:571–590. doi: 10.1007/s004180050256. [DOI] [PubMed] [Google Scholar]
- Dipersio CM, Shah S, Hynes RO. α3Aβ1 integrin localizes to focal contacts in response to diverse extracellular matrix proteins. J Cell Sci. 1995;108:2321–2336. doi: 10.1242/jcs.108.6.2321. [DOI] [PubMed] [Google Scholar]
- Dong J-T, Lamb PW, Rinker-Schaeffer CW, Vukanovic J, Ichikawa T, Isaacs JT, Barret JC. KAI 1, a metastasis suppressor gene for prostate cancer on human chromosome 11p11.2. Science. 1995;268:884–886. doi: 10.1126/science.7754374. [DOI] [PubMed] [Google Scholar]
- Dunphy JT, Linder ME. Signaling functions of protein palmitoylation. Biochim Biophys Acta. 1998;1436:245–261. doi: 10.1016/s0005-2760(98)00130-1. [DOI] [PubMed] [Google Scholar]
- Escola JM, Kleijmeer MJ, Stoorvogel W, Griffith JM, Yoshie O, Geuze HJ. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J Biol Chem. 1998;273:20121–20127. doi: 10.1074/jbc.273.32.20121. [DOI] [PubMed] [Google Scholar]
- Fabbri M, Fumagalli L, Bossi G, Bianchi E, Bender JR, Pardi R. A tyrosine-based sorting signal in the beta2 integrin cytoplasmic domain mediates its recycling to the plasma membrane and is required for ligand-supported migration. EMBO J. 1999;18:4915–4925. doi: 10.1093/emboj/18.18.4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukudome K, Furuse M, Imai T, Nishimura M, Takagi S, Hinuma Y, Yoshie O. Identification of membrane antigen C33 recognized by monoclonal antibodies inhibitory to human T-cell leukemia virus type 1 (HTLV-1)-induced syncytium formation: altered glycosylation of C33 antigen in HTLV-1-positive T cells. J Virol. 1992;66:1394–1401. doi: 10.1128/jvi.66.3.1394-1401.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Ni Y, Szabo G, Linden J. Palmitoylation of the recombinant human A1 adenosine receptor: enhanced proteolysis of palmitoylation-deficient mutant receptors. Biochem J. 1999;342(Pt 2):387–395. [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S, Linder ME. SNAP-25 palmitoylation and plasma membrane targeting require a functional secretory pathway. Mol Biol Cell. 1998;9:585–597. doi: 10.1091/mbc.9.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond C, Denzin LK, Pan M, Griffith JM, Geuze HJ, Cresswell P. The tetraspan protein CD82 is a resident of MHC class II compartments where it associates with HLA-DR, -DM, and -DO molecules. J Immunol. 1998;161:3282–3291. [PubMed] [Google Scholar]
- Hemler ME. Integrin-associated proteins. Curr Opin Cell Biol. 1998;10:578–585. doi: 10.1016/s0955-0674(98)80032-x. [DOI] [PubMed] [Google Scholar]
- Hemler ME, Mannion BA, Berditchevski F. Association of TM4SF proteins with integrins: relevance to cancer. Biochim Biophys Acta. 1996;1287:67–71. doi: 10.1016/0304-419x(96)00007-8. [DOI] [PubMed] [Google Scholar]
- Hemler ME, Sánchez-Madrid F, Flotte TJ, Krensky AM, Burakoff SJ, Bhan AK, Springer TA, Strominger JL. Glycoproteins of 210,000 and 130,000 m.w. on activated T cells: cell distribution and antigenic relation to components on resting cells and T cell lines. J Immunol. 1984;132:3011–3018. [PubMed] [Google Scholar]
- Imai T, Kakizaki M, Nishimura M, Yoshie O. Molecular analysis of the association of CD4 with two members of the transmembrane 4 superfamily, CD81 and CD82. J Immunol. 1995;155:1229–1239. [PubMed] [Google Scholar]
- Imai T, Yoshie O. C33 antigen and M38 antigen recognized by monoclonal antibodies inhibitory to syncytium formation by human T cell leukemia virus type 1 are both members of the transmembrane 4 superfamily and associate with each other and with CD4 and CD8 in T cells. J Immunol. 1993;151:6470–6481. [PubMed] [Google Scholar]
- Jin H, Xie Z, George SR, O'Dowd BF. Palmitoylation occurs at cysteine 347 and cysteine 351 of the dopamine D(1) receptor. Eur J Pharmacol. 1999;386:305–312. doi: 10.1016/s0014-2999(99)00727-x. [DOI] [PubMed] [Google Scholar]
- Kitadokoro K, Bordo D, Galli G, Petracca R, Falugi F, Abrignani S, Grandi G, Bolognesi M. CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs. EMBO J. 2001;20:12–18. doi: 10.1093/emboj/20.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopczynski CC, Davis GW, Goodman CS. A neural tetraspanin, encoded by late bloomer, that facilitates synapse formation. Science. 1996;271:1867–1870. doi: 10.1126/science.271.5257.1867. [DOI] [PubMed] [Google Scholar]
- Le Naour F, Rubinstein E, Jasmin C, Prenant M, Boucheix C. Severely reduced female fertility in CD9-deficient mice. Science. 2000;287:319–321. doi: 10.1126/science.287.5451.319. [DOI] [PubMed] [Google Scholar]
- Lee RT, Berditchevski F, Cheng GC, Hemler ME. Integrin-mediated collagen matrix reorganization by cultured human vascular smooth muscle cells. Circ Res. 1995;76:209–214. doi: 10.1161/01.res.76.2.209. [DOI] [PubMed] [Google Scholar]
- Maecker HT, Todd SC, Levy S. The tetraspanin superfamily: molecular facilitators. FASEB J. 1997;11:428–442. [PubMed] [Google Scholar]
- Matsumoto AK, Martin DR, Carter RH, Klickstein LB, Ahearn JM, Fearon DT. Functional dissection of the CD21/cd19/TAPA-1/Leu-13 complex of B lymphocytes. J Exp Med. 1993;178:1407–1417. doi: 10.1084/jem.178.4.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercurio AM, Rabinovitz I, Shaw LM. The alpha 6 beta 4 integrin and epithelial cell migration. Curr Opin Cell Biol. 2001;13:541–545. doi: 10.1016/s0955-0674(00)00249-0. [DOI] [PubMed] [Google Scholar]
- Metzelaar MJ, Wijngaard PLJ, Peters PJ, Sixma JJ, Nieuwenhuis HK, Clevers HC. CD63 antigen: a novel lysosomal membrane glycoprotein, cloned by a screening procedure for intracellular antigens in eukaryotic cells. J Biol Chem. 1991;266:3239–3245. [PubMed] [Google Scholar]
- Miyado K, Yamada G, Yamada S, Hasuwa H, Nakamura Y, Ryu F, Suzuki K, Kosai K, Inoue K, Ogura A, Okabe M, Mekada E. Requirement of CD9 on the egg plasma membrane for fertilization. Science. 2000;287:321–324. doi: 10.1126/science.287.5451.321. [DOI] [PubMed] [Google Scholar]
- Miyake M, Inufusa H, Adachi M, Ishida H, Hashida H, Tokuhara T, Kakehi Y. Suppression of pulmonary metastasis using adenovirally motility related protein-1 (MRP-1/CD9) gene delivery. Oncogene. 2000;19:5221–5226. doi: 10.1038/sj.onc.1203919. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Iwamoto R, Mekada E. Membrane-anchored heparin-binding EGF-like growth factor (HB-EGF) and diphtheria toxin receptor-associated protein (DRAP27)/CD9 form a complex with integrin α3β1 at cell-cell contact sites. J Cell Biol. 1995;129:1691–1705. doi: 10.1083/jcb.129.6.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Mitamura T, Takahashi T, Kobayashi T, Mekada E. Importance of the major extracellular domain of CD9 and the epidermal growth factor (E.G.F)-like domain of heparin-binding E.G.F-like growth factor for up-regulation of binding and activity. J Biol Chem. 2000;275:18284–18290. doi: 10.1074/jbc.M907971199. [DOI] [PubMed] [Google Scholar]
- Odintsova E, Sugiura T, Berditchevski F. Attenuation of EGF receptor signaling by a metastasis suppressor, the tetraspanin CD82/KAI-1. Curr Biol. 2000;10:1009–1012. doi: 10.1016/s0960-9822(00)00652-7. [DOI] [PubMed] [Google Scholar]
- Parat MO, Fox PL. Palmitoylation of caveolin-1 in endothelial cells is post-translational but irreversible. J Biol Chem. 2001;276:15776–15782. doi: 10.1074/jbc.M006722200. [DOI] [PubMed] [Google Scholar]
- Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, Abrignani S. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- Radford KJ, Mallesch J, Hersey P. Suppression of human melanoma cell growth and metastasis by the melanoma-associated antigen CD63 (ME491) Int J Cancer. 1995;62:631–635. doi: 10.1002/ijc.2910620523. [DOI] [PubMed] [Google Scholar]
- Resh MD. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim Biophys Acta. 1999;1451:1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- Rubinstein E, Le Naour F, Lagaudrière-Gesbert C, Billard M, Conjeaud H, Boucheix C. CD9, CD63, CD81, and CD82 are components of a surface tetraspan network connected to HLA-DR and VLA antigens. Eur J Immunol. 1996;26:2657–2665. doi: 10.1002/eji.1830261117. [DOI] [PubMed] [Google Scholar]
- Schmid E, Zurbriggen A, Gassen U, Rima B, ter Meulen V, Schneider-Schaulies J. Antibodies to CD9, a tetraspan transmembrane protein, inhibit canine distemper virus-induced cell-cell fusion but not virus-cell fusion. J Virol. 2000;74:7554–7561. doi: 10.1128/jvi.74.16.7554-7561.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehafer JG, Slupsky JR, Tang SC, Masellis-Smith A, Shaw AR. Myristic acid is incorporated into the two acylatable domains of the functional glycoprotein CD9 in ester, but not in amide bonds. Biochim Biophys Acta. 1990;1039:218–226. doi: 10.1016/0167-4838(90)90189-m. [DOI] [PubMed] [Google Scholar]
- Serru V, Dessen P, Boucheix C, Rubinstein E. Sequence and expression of seven new tetraspans. Biochim Biophys Acta. 2000;1478:159–163. doi: 10.1016/s0167-4838(00)00022-4. [DOI] [PubMed] [Google Scholar]
- Serru V, Naour FL, Billard M, Azorsa DO, Lanza F, Boucheix C, Rubinstein E. Selective tetraspan-integrin complexes (CD81/alpha4beta1, CD151/alpha3beta1, CD151/alpha6beta1) under conditions disrupting tetraspan interactions. Biochem J. 1999;340(Pt. 1):103–111. [PMC free article] [PubMed] [Google Scholar]
- Sincock PM, Fitter S, Parton RG, Berndt MC, Gamble JR, Ashman LK. PETA-3/CD151, a member of the transmembrane 4 superfamily, is localized to the plasma membrane and endocytic system of endothelial cells, associates with multiple integrins and modulates cell function. J Cell Sci. 1999;112:833–844. doi: 10.1242/jcs.112.6.833. [DOI] [PubMed] [Google Scholar]
- Sterk LM, Geuijen CA, Oomen LC, Calafat J, Janssen H, Sonnenberg A. The tetraspan molecule CD151, a novel constituent of hemidesmosomes, associates with the integrin alpha6beta4 and may regulate the spatial organization of hemidesmosomes. J Cell Biol. 2000;149:969–982. doi: 10.1083/jcb.149.4.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipp CS, Hemler ME. Transmembrane-4-Superfamily proteins CD151 and CD81 associate with α3β1integrin, and selectively contribute to α3β1-dependent neurite outgrowth. J Cell Sci. 2000;113:1871–1882. doi: 10.1242/jcs.113.11.1871. [DOI] [PubMed] [Google Scholar]
- Stipp CS, Kolesnikova TV, Hemler ME. EWI-2 is a major CD9 and CD81 partner, and member of a novel Ig protein subfamily. J Biol Chem. 2001a;276:40545–40554. doi: 10.1074/jbc.M107338200. [DOI] [PubMed] [Google Scholar]
- Stipp CS, Orlicky D, Hemler ME. FPRP: a major, highly stoichiometric, highly specific CD81 and CD9-associated protein. J Biol Chem. 2001b;276:4853–4862. doi: 10.1074/jbc.M009859200. [DOI] [PubMed] [Google Scholar]
- Tachibana I, Hemler ME. Role of transmembrane-4 superfamily (TM4SF) proteins CD9 and CD81 in muscle cell fusion and myotube maintenance. J Cell Biol. 1999;146:893–904. doi: 10.1083/jcb.146.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanio Y, Yamazaki H, Kunisada T, Miyake K, Hayashi SI. CD9 molecule expressed on stromal cells is involved in osteoclastogenesis. Exp Hematol. 1999;27:853–859. doi: 10.1016/s0301-472x(99)00011-9. [DOI] [PubMed] [Google Scholar]
- Testa JE, Brooks PC, Lin JM, Quigley JP. Eukaryotic expression cloning with an antimetastatic monoclonal antibody identifies a tetraspanin (PETA-3/CD151) as an effecter of human tumor cell migration and metastasis. Cancer Res. 1999;59:3812–3820. [PubMed] [Google Scholar]
- Todd SC, Doctor VS, Levy S. Sequences and expression of six new members of the tetraspanin/TM4SF family. Biochim Biophys Acta. 1998;1399:101–104. doi: 10.1016/s0167-4781(98)00087-6. [DOI] [PubMed] [Google Scholar]
- van de Loo JW, Teuchert M, Pauli I, Plets E, Van de Ven WJ, Creemers JW. Dynamic palmitoylation of lymphoma proprotein convertase prolongs its half-life, but is not essential for trans-Golgi network localization. Biochem J. 2000;352(Pt. 3):827–833. doi: 10.1042/0264-6021:3520827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Symons JM, Goldstein SL, McDonald A, Miner JH, Kreidberg JA. α3β1 integrin regulates epithelial cytoskeletal organization. J Cell Sci. 1999;112:2925–2935. doi: 10.1242/jcs.112.17.2925. [DOI] [PubMed] [Google Scholar]
- Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, Bissell MJ. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzman JB, Pasqualini R, Takada Y, Hemler ME. The function and distinctive regulation of the integrin VLA-3 in cell adhesion, spreading and homotypic cell aggregation. J Biol Chem. 1993;268:8651–8657. [PubMed] [Google Scholar]
- Wright MD, Tomlinson MG. The ins and outs of the transmembrane 4 superfamily. Immunol Today. 1994;15:588–594. doi: 10.1016/0167-5699(94)90222-4. [DOI] [PubMed] [Google Scholar]
- Yashiro-Ohtani Y, Zhou XY, Toyo-Oka K, Tai XG, Park CS, Hamaoka T, Abe R, Miyake K, Fujiwara H. Non-CD28 costimulatory molecules present in T cell rafts induce T cell costimulation by enhancing the association of TCR with rafts. J Immunol. 2000;164:1251–1259. doi: 10.4049/jimmunol.164.3.1251. [DOI] [PubMed] [Google Scholar]
- Yauch RL, Berditchevski F, Harler MB, Reichner J, Hemler ME. Highly stoichiometric, stable and specific association of integrin α3β1 with CD151 provides a major link to phosphatidylinositol 4-kinase and may regulate cell migration. Mol Biol Cell. 1998;9:2751–2765. doi: 10.1091/mbc.9.10.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch RL, Hemler ME. Specific interactions among transmembrane 4 superfamily (TM4SF) proteins and phosphatidylinositol 4-kinase. Biochem J. 2000;351:629–637. [PMC free article] [PubMed] [Google Scholar]
- Yauch RL, Kazarov AR, Desai B, Lee RT, Hemler ME. Direct extracellular contact between integrin α3β1 and TM4SF protein CD151. J Biol Chem. 2000;275:9230–9238. doi: 10.1074/jbc.275.13.9230. [DOI] [PubMed] [Google Scholar]
- Zemni R, Bienvenu T, Vinet MC, Sefiani A, Carrie A, Billuart P, McDonell N, Couvert P, Francis F, Chafey P, Fauchereau F, Friocourt G, Portes V, Cardona A, Frints S, Meindl A, Brandau O, Ronce N, Moraine C, Bokhoven H, Ropers HH, Sudbrak R, Kahn A, Fryns JP, Beldjord C, et al. A new gene involved in X-linked mental retardation identified by analysis of an X;2 balanced translocation. Nat Genet. 2000;24:167–170. doi: 10.1038/72829. [DOI] [PubMed] [Google Scholar]
- Zhang XA, Kazarov AR, Yang X, Bontrager AL, Stipp CS, Hemler ME. Function of the tetraspanin CD151–a6b1 integrin complex during cellular morphogenesis. Mol Biol Cell. 2002;13:1–11. doi: 10.1091/mbc.01-10-0481. [DOI] [PMC free article] [PubMed] [Google Scholar]