Abstract
Yeast vacuoles undergo fission and homotypic fusion, yielding one to three vacuoles per cell at steady state. Defects in vacuole fusion result in vacuole fragmentation. We have screened 4828 yeast strains, each with a deletion of a nonessential gene, for vacuole morphology defects. Fragmented vacuoles were found in strains deleted for genes encoding known fusion catalysts as well as 19 enzymes of lipid metabolism, 4 SNAREs, 12 GTPases and GTPase effectors, 9 additional known vacuole protein-sorting genes, 16 protein kinases, 2 phosphatases, 11 cytoskeletal proteins, and 28 genes of unknown function. Vacuole fusion and vacuole protein sorting are catalyzed by distinct, but overlapping, sets of proteins. Novel pathways of vacuole priming and docking emerged from this deletion screen. These include ergosterol biosynthesis, phosphatidylinositol (4,5)-bisphosphate turnover, and signaling from Rho GTPases to actin remodeling. These pathways are supported by the sensitivity of the late stages of vacuole fusion to inhibitors of phospholipase C, calcium channels, and actin remodeling. Using databases of yeast protein interactions, we found that many nonessential genes identified in our deletion screen interact with essential genes that are directly involved in vacuole fusion. Our screen reveals regulatory pathways of vacuole docking and provides a genomic basis for studies of this reaction.
INTRODUCTION
Membrane fusion is required for selective delivery of proteins from one organelle to another and for the maintenance of low organelle copy number. Fusion is catalyzed by a cascade of interacting proteins, including integral membrane SNAREs, chaperones such as Sec18p/NSF, Sec17p/α-SNAP and LMA1, GTPases of the Rab and Rho families, GTPase effectors, calcium channels, and calcium-responsive proteins. Certain lipids, such as phosphoinositol phosphatides, are also needed, both to recruit proteins to organelles and to generate signaling molecules. The complexity of membrane fusion has so far made it difficult to enumerate all the responsible factors and to connect them in a coherent scheme of catalysis.
Yeast vacuoles offer several advantages for studying membrane fusion (Wickner and Haas, 2000). Vacuoles are readily visualized in intact cells and undergo constant fission and fusion. Consequently, defects in fusion are readily seen as vacuole fragmentation. Large vacuoles are not required for cell growth under laboratory conditions, and thus strains with deletions of genes encoding vacuole fusion catalysts are viable. Vacuoles can be purified in large amounts and stored frozen. Vacuoles fuse during incubation in vitro, and this fusion can be assayed colorimetrically. This reaction occurs in ordered stages of priming, docking, and fusion.
Priming occurs on separate vacuoles and is needed for productive vacuole associations (Mayer et al., 1996). Priming is initiated by ATP hydrolysis by Sec18p. Priming drives the release of Sec17p, disassembly of cis-SNARE complexes (Ungermann et al., 1998), and transfer of the LMA1 cochaperone from Sec18p to the activated t-SNARE Vam3p (Xu et al., 1997, 1998). Priming requires phosphatidylinositol 4,5-bisphosphate (PI[4,5]P2; Mayer et al., 2000) and ergosterol (Kato and Wickner, 2001). Priming allows HOPS (homotypic fusion and vacuole protein sorting)/VPS class C complex, a complex of at least 6 proteins (Vps 11, 16, 18, 33, 39, and 41), to be transferred to Ypt7p (Price et al., 2000; Seals et al., 2000), catalyzing the conversion of Ypt7p to its active GTP form (Wurmser et al., 2000) and thereby initiating docking. Docking, which also requires the vacuole membrane potential ΔμH+ (Ungermann et al., 1999), Rho GTPases (Eitzen et al., 2001; Muller et al., 2001), and phosphoinositides (Mayer et al., 2000), concludes with trans-pairing of SNAREs (Ungermann et al., 1998) and calcium release from the vacuole (Peters and Mayer, 1998). Calcium-bound calmodulin binds at V0 (the integral membrane domain of the vacuolar H+-ATPase) and triggers V0-V0 association in trans (Peters et al., 2001). On protein phosphatase 1 (PP1) action (Peters et al., 1999), LMA1 is released (Xu et al., 1998) and fusion occurs. Despite this progress, few subreactions of vacuole fusion have been reconstituted with pure components, and the connections between these steps in the pathway remain obscure.
Nine of the known proteins that catalyze vacuole fusion are encoded by VAM genes. The vam mutants were identified (Wada et al., 1992) by nonselective screening for abnormal vacuole morphology. Six of the vacuole morphology (VAM) genes encode subunits of the HOPS complex (Vam1p = Vps11p, Vam9p = Vps16p, Vam8p = Vps18p, Vam5p = Vps33p, Vam6p = Vps39p, and Vam2p = Vps41p), Vam3p is the vacuolar t-SNARE, Vam7p is the vacuolar SNAP25 SNARE homologue, and Vam4p is the GTPase Ypt7p. Although each of the nine original VAM genes are allelic with a known VPS gene, the initial screen for vam mutants was not saturated. We have therefore taken a genomic approach to identify additional catalysts of vacuole fusion, exploiting a collection of 4828 yeast strains with deletions in each nonessential gene and visualizing the vacuole with the fluorescent vital dye FM4-64. The new VAM genes identified in this manner define novel pathways whose roles can be confirmed through the use of selective inhibitors of in vitro fusion of wild-type vacuoles. They reveal a striking and unexpected complexity of the priming and docking stages of homotypic vacuole fusion.
MATERIALS AND METHODS
FM4-64 and antibody to carboxypeptidase Y (CPY) were from Molecular Probes (Eugene, OR). Anti-mouse immunoglobulin G-POD was from Boehringer-Mannheim (Indianapolis, IN). Libraries of strains with deletion of each of the nonessential genes, in homozygous diploid (BY4743) and haploid (BY4739, BY4741, BY4742) backgrounds, were purchased from Research Genetics (Huntsville, AL).
Deletion Screen
Microtiter plates containing 96 yeast deletion strains were thawed, and 5–25 μl of each culture was used to inoculate 1 ml of YPD with 3 μM FM4-64 and 20 μg/ml G418. Cultures were grown for 12–36 h at 30°C with constant shaking before microscopic examination. Strains with vacuole morphology defects were streaked to single colonies and examined by at least two individuals. Microscopic examination and phenotype scoring was performed without reference to strain identity.
CPY Secretion
The CPY secretion assay was performed according to the method of Roberts et al. (1991) with minor modifications; single colonies were picked from YPD-agar plates and suspended in 200 μl of YPD, and 5 μl of each suspension was spotted onto YPD-agar plates and allowed to dry before filter overlay and incubation.
Vacuole Isolation
Vacuoles were isolated (Hass, 1995) and stored frozen (Seals et al., 2000).
Fusion Reaction
Fusion reactions (Hass, 1995) contained 3 μg each of vacuoles from BJ3505 (MATa, pep4::HIS3. prb1-Δ1.6 R, lys2–208, trp1-Δ101, ura3-52, gal2, can) and DKY6281 (MATa, leu2-3112, ura 3-52, his3-Δ200, trp1-Δ901, lys2-801, suc2-Δ9, pho8::TRP1) in reaction buffer (200 mM sorbitol, 20 mM 1,4-piperazinediethanesulfonic acid-KOH, pH 6.8, 119 mM NH4Cl, 3.9 mM MgCl2, 0.8 mM ATP [Amersham Pharmacia Biotech, Piscataway, NJ], 18.8 mM creatinine kinase [Roche Molecular Biochemicals, Summerville, NJ], 23 mM creatine phosphate [Roche Molecular Biochemicals], and 0.8 mM CoA). All reactions contained 7.8 μg of HMA, a high molecular weight fusion-enhancing fraction purified from yeast cytosol by gel filtration on Sephacryl 200HR (Amersham Pharmacia Biotech). Inhibitors (Calbiochem, San Diego, CA) were prepared as 35× concentrated stock solutions: 420 mM 2-aminoethoxydiphenyl borate (2-APB) in dimethyl sulfoxide (DMSO), 6.12 mM Ruthidium Red in PS (200 mM sorbitol, 10 mM 1,4-piperazinediethanesulfonic acid-KOH, pH 6.8), 15 mM cyclopiazonic acid in DMSO, 10.5 mM thapsigargin in DMSO, and 2 mM ET-18-OCH3 in ethanol. Antibodies to yeast vacuole proteins (α-YPT7, 33 μg/ml; α-Sec17, 150 μg/ml; α-Vam3, 75 μg/ml) were purified from the sera of rabbits immunized with the specific recombinant protein by adsorption to protein A-Sepharose. KLH-conjugated YPT7 peptide (TEAFEDDYNDAINIR) was synthesized by Biosynthesis and was added at 1 μg per reaction from a stock solution of 1 mg/ml in PS buffer.
RESULTS
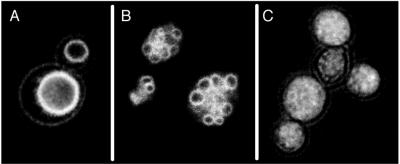
Using the fluorescent vacuolar vital stain FM4-64, we scored 4828 nonessential gene deletions in yeast for vacuole morphology. Cultures of each strain were grown at 30°C overnight in rich media, stained with FM 4-64, and screened by fluorescence light microscopy for vacuole morphology without reference to the strain identities. Wild-type yeast typically have one to three large vacuoles (Figure 1A). Abnormal vacuole morphologies were categorized as in earlier VPS screens (Rothman and Stevens, 1986; Banta et al., 1988). Class B mutants have multiple small vacuoles (Figure 1B), class C mutants have highly fragmented vacuoles (Figure 1C), class D mutants have single, grossly enlarged vacuoles, and class E mutants have an enlarged “prevacuolar” compartment surrounded by numerous membrane vesicles. Of 4828 nonessential genes, 714 deletions caused an altered vacuole morphology (see table on journal website). A subset of these were of class B and C, consistent with defects in vacuole fusion. Notably, our results match closely those previously reported for vps/vam mutants bearing distinct vacuolar fragmentation phenotypes. Of 11 previously reported vps/vam mutants (VPS5, 11, 16, 17, 18, 33, 39, 41, 43, 52, and 54) described by others as bearing either class B or C vacuoles (Bankaitis et al., 1986; Rothman et al., 1986; Weisman et al., 1990; Wada et al., 1992; Conibear and Stevens, 2000), we detected all 11 in this screen and all had either a class B or C phenotype.
Figure 1.
Vacuole morphologies. Single yeast colonies were picked from YPD-agar plates, inoculated into 1.0 ml of YPD with 3 μM FM4-64, and grown overnight at 30°C with constant shaking. Samples of 3 μl were examined by fluorescence microscopy with a rhodamine filter set on a standard microscope (Carl Zeiss, Thornwood, NY) with backlighting and photographed with T-MAX 400 Pro film (Kodak, Rochester, NY). A–C show yeast with wild-type, class B, and class C vacuoles, respectively.
Many deletion strains with a class B or C phenotype showed only a small percentage of cells with vacuole fragmentation, and some of these genes encode proteins that are unlikely to directly regulate fusion. For example, many of these genes encode nonessential transcription factors, ribosomal subunits, or nuclear pore proteins. These proteins may not be directly involved in fusion but, rather, might affect the expression of proteins that directly catalyze the fusion reaction. These considerations allowed the selection of 137 candidate VAM genes (Tables 1–9). Screening these for secretion of CPY revealed 26 new gene deletions with a moderate to strong VPS phenotype. In total, only 50 gene deletions of the 137 VAM genes, including 24 already characterized VPS genes, gave rise to a moderate or strong VPS phenotype. This suggests that there is distinction as well as overlap between pathways of traffic to the vacuole and homotypic vacuole fusion.
Table 1.
Known catalysts
| Locus | Common name(s) | Vacuolar phenotype(s) | CPY secretion | Function |
|---|---|---|---|---|
| SNAREs | ||||
| YOR106w | VAM3/PTH1 | 100%C | +++ | Syntaxin (t-SNARE) homologue |
| YGL212w | VAM7/VPS43 | 100%C | ++ | SNAP-25 homologue |
| HOPS complex | ||||
| YLR148w | VPS18/VAM8 | 100%C | +++ | HOPS subunit |
| YMR231w | VPS11/VAM1 | 100%C | +++ | HOPS subunit |
| YDL077c | VPS39/VAM6 | 100%C | +++ | HOPS subunit, Ypt7p GEF |
| YPL045w | VPS16/VAM9 | 100%C | +++ | HOPS subunit |
| YLR396c | VPS33/VAM5 | 100%C | +++ | HOPS subunit, Sec1p homologue |
| YDR080w | VPS41/VAM2 | 100%C | +++ | HOPS subunit, AP-3 coat protein |
| Vacuolar V-ATPase | ||||
| YKL080w | VMA5/VATC | 10%C 60%D | ++ | V1 sector V-ATPase subunit |
| YLR447c | VMA6 | 25%C 50%D 25%E | ++ | V0 sector V-ATPase subunit |
| YPL234c | VMA11/TFP3 | 40%C 60%D | ++ | V-ATPase proteolipid |
| YHR026w | VMA16/PPA1 | 40%C 60%D | ++ | V-ATPase proteolipid |
| YHR060w | VMA22/VPH6 | 20%C 70%D | ++ | V-ATPase assembly protein |
| YOR270c | VPH1 | 75%B 10%C | ++ | V0 sector V-ATPase subunit |
| Other known catalysts | ||||
| YML001w | YPT7/VAM4 | 100%C | ++ | Ypt/Rab GTPase |
| YNL015w | PB12 | 50%B | + | LMA1 subunit |
| YEL013w | VAC8 | 60%B 15%E | + | Docking factor required for vacuole inheritance |
Table 9.
Orphan open reading frames
| Locus | Vacuolar phenotype(s) | CPY secretion |
|---|---|---|
| YCL016c | 50%B | + |
| YDR200c | 70%B | + |
| YDR223w | 10%B 40%E | + |
| YDR433w | 80%B | + |
| YEL044w | 40%B 40%D | +++ |
| YER083c | 30%B 30%C 30%E | + |
| YGL024w | 75%B | − |
| YGL223c | 90%B | +++ |
| YJL075c | 40%B | − |
| YJL184w | 40%C 20%D | + |
| YLL002w | 60%B | + |
| YLR091w | 90%C | − |
| YLR204w | 70%D | + |
| YLR261c | 60%B 10%C | ++ |
| YLR320w | 50%B | + |
| YLR322w | 25%B 25%C | + |
| YML013c-A | 35%B 35%C | ++ |
| YMR269w | 85%B | − |
| YNL080c | 70%B | + |
| YNL281w | 70%B | + |
| YNL297c | 60%B | + |
| YOL008w | 50%B | − |
| YOL035c | 60%B | + |
| YOL050c | 70%B | + |
| YOL063c | 50%B | + |
| YOR068c | 90%B | +++ |
| YOR359w | 25%B 25%C | + |
| YPL055c | 50%B | ++ |
Three genes (YLR320w, YNL281w, and YOR068c) are adjacent to genes that are also required for normal vacuole morphology. Further analysis will be required to determine whether these deletions act by altering the expression level of an adjacent gene.
Seventeen of the genes identified in the deletion screen encode proteins that were previously known from biochemical studies to be involved in vacuole fusion (Table 1). These include the Vam3p and Vam7p SNAREs, the GTPase Ypt7p, its six HOPS effector subunits, multiple subunits of the vacuolar H+-ATPase (needed for the vacuolar membrane potential and for trans–V0-V0–pairing during fusion), the chaperone IB2, and Vac8p, a protein with armadillo repeats that is required for vacuole fusion (Veit et al., 2001; Wang et al., 2001). Many proteins that are required for vacuole fusion are also essential for cell growth and thus are not represented in the screen. These include Sec17p, Sec18p, Vti1p, Ykt6p, calmodulin, and protein phosphatase 1. Strains with a deletion of the gene for Nyv1p have normal vacuole morphology, even though this SNARE is localized to the vacuole and is found in cis- and trans-SNARE complexes (Ungermann et al., 1998). Vacuoles from nyv1Δ strains fuse poorly, and antibody to Nyv1p blocks the in vitro fusion of normal vacuoles (Nichols et al., 1997). Other SNAREs may compensate for the absence of Nyv1p function in vivo, as suggested by genetic studies of other yeast-trafficking reactions (Fischer von Mollard et al., 1997; Grote and Novick, 1999).
Membrane traffic is regulated by protein phosphorylation and dephosphorylation (Cabanoils et al., 1999; Marash and Gerst, 2001). We found that normal vacuole copy number requires 16 kinases and two protein phosphatases, in addition to two regulatory subunits of PP1 (Table 2). Several of the kinases are members of known families, such as casein kinases or mitogen-activated protein (MAP) kinase cascades, but most are categorized only as kinases by sequence homology. Other kinases and phosphatases are members of the STE pathway of mating hormone response and delivery to the vacuole. Vacuole morphology depends on two putative regulatory subunits of protein phosphatase 1 (Glc7p), an enzyme that regulates the last step of vacuole fusion (Peters et al., 1999).
Table 2.
Protein modification
| Locus | Common name(s) | Vacuolar phenotype(s) | CPY secretion | Function |
|---|---|---|---|---|
| Protein kinase related | ||||
| YAL040c | CLN3/DAF1 | 30%B 30%E | + | G1/S-cyclin, interacts with Cdc28p |
| YAR018c | KIN3/NPK1 | 15%B | + | Ser/Thr protein kinase |
| YBR059c | AKL1 | 40%B | + | Ser/Thr protein kinase |
| YBR097w | VPS15/VPT15 | 30%C 50%D | +++ | Protein kinase, interacts with Vps34p |
| YDL025c | 15%B 10%E | + | Ser/Thr protein kinase | |
| YDR507c | GIN4/ERC47 | 50%B 10%C | + | Ser/Thr protein kinase required for septin organization |
| YER123w | YCK3/CK13 | 30%B 20%C 10%D | ++ | Casein kinase I isoform |
| YFL033c | RIM15 | 30%B | + | Ser/Thr protein kinase, regulates IME2 |
| YGL215w | CLG1 | 25%B | + | Cyclin-like protein, associates with Pho85p |
| YGR188c | BUB1 | 10%B 60%D | + | Ser/Thr protein kinase, affects microtubules |
| YJL095w | BCK1 | 20%B 10%C | + | MEKK family protein kinase |
| YKL048c | ELM1/ECM41 | 80%B | + | Protein kinase-regulating pseudohyphal growth |
| YKL139w | CTK1 | 50%B 10%C | ++ | C-terminal domain kinase, α subunit |
| YLR362w | STE11 | 50%B | + | MAP kinase kinase kinase (MAPKKK) |
| YPL150w | 40%B | + | Ser/Thr protein kinase | |
| YOR061w | CKA2 | 20%B 25%C 20%D | + | Casein kinase II catalytic subunit |
| YBR028c | 15%B | + | Protein kinase with similarity to Ypk2p and Ypk1p | |
| YDR477w | SNF1/CAT1 | 50%B | + | Protein kinase, derepression of glucose-repressed genes |
| Protein phosphatase related | ||||
| YAL016w | TPD3/FUN32 | 30%B 30%C | + | PP2Ap regulatory subunit, ceramide activated |
| YDL006w | PTC1/TPD1 | 60%B | + | PP2C Ser/Thr phosphatase for MAP kinases |
| YBR050c | REG2 | 20%B 10%C | + | Possible regulatory subunit for Glc7p |
| YBL058w | SHP1 | 25%B 10%D | + | Possible regulatory subunit for Glc7p |
| Protein acetyltransferase related | ||||
| YPR131c | NAT3 | 80%B | − | Protein N-acetyltransferase |
Lipid metabolism is important for vacuole fusion (Table 3). Five steps of the ergosterol biosynthetic pathway are essential for normal vacuole copy number, although the deletion of ERG4, which encodes the final enzyme of the pathway, gave only a partial vacuole morphology defect. These observations suggest that ergosterol or its immediate precursor zymosterol is needed for vacuole fusion. Our further studies, reported elsewhere (Kato and Wickner, 2001), showed that vacuolar ergosterol is required for normal Sec18p-mediated priming and thus regulates the initial commitment to vacuole fusion.
Table 3.
Lipids
| Locus | Common name(s) | Vacuolar phenotype(s) | CPY secretion | Function |
|---|---|---|---|---|
| Phosphoinositol related | ||||
| YDR173c | IPK2/ARG82 | 50%B 10%C | ++ | Inositol polyphosphate multikinase |
| YOL065c | INP54 | 50%B | ++ | Phosphatidylinositol polyphosphate 5-phosphatase |
| YDR017c | KCS1 | 80%B | + | Inositol hexaphosphate kinase |
| YPL268w | PLC1/GSL2 | 50%B 10%C | + | Phosphoinositide-specific PLC |
| YER019w | ISC1 | 30%B 20%C | ++ | PLP C-like activity |
| YLR240w | VPS34/VPT29 | 20%C 80%D | +++ | Phosphoinositide 3-kinase, associates with Vps15p |
| YDR323c | VPS19/PEP7 | 50%C | +++ | Coordination of PI3K and Rab signaling |
| Ergosterol related | ||||
| YLR056w | ERG3/SYR1 | 60%B | + | C-5 sterol desaturase |
| YMR015c | ERG5/CYP61 | 80%B | − | Cytochrome P450 |
| YML008c | ERG6/ISE1 | 60%B | + | Zymosterol methylation |
| YNL280c | ERG24 | 30%B 20%C | − | C-14 sterol reductase |
| YER044c | ERG28 | 30%B 30%C | + | C-4 sterol demethylation |
| Sphingolipid/phospholipid related | ||||
| YLR260w | LCB5 | 40%B 40%C | − | Phosphorylation of sphingosines |
| YCR034w | FEN1/GNS1 | 75%B 10%C | − | Fatty acid elongation |
| YLR372w | SUR4/VBM1 | 100%C | − | 24–26-carbon fatty-acid conversion |
| YOR196c | LIP5 | 30%B | ++ | Lipoic acid synthetase |
| YOR221c | MCT1 | 30%B | + | Malonyl CoA:acyl carrier protein transferase |
| YKL055c | OAR1 | 30%B | + | 3-Oxoacyl-[acyl-carrier-protein] reductase |
| YBR177c | EHT1 | 20%B 10%C 10%E | + | Alcohol acyl transferase |
PI(4,5)P2 is needed for vacuole docking (Mayer et al., 2000). The phosphatidylinositol 4-kinases Stt4p and Pik1p, as well as the phosphatase SacIp, are needed for normal vacuole morphology (Audhya et al., 2000). Arf1p, which activates the PI4P 5-kinase activity of Mss4p (Donaldson and Jackson, 2000), and Glo3p, which is an Arf-GTPase–activating protein (GAP) (Dogic et al., 1999), are also required for low vacuole copy number (Table 5). Surprisingly, we found that the further metabolism of PI(4,5)P2 is required for normal vacuole morphology. Deletion of INP54, which encodes a PI(4,5)P2 5-phosphatase, causes striking vacuole fragmentation (Table 3), as do multiple deletions of the redundant PI4P phosphatases INP51, 52, and 53 (Stolz et al., 1998). The need for PI(4,5)P2 hydrolysis as well as synthesis underscores the central role of PI(4,5)P2 in docking, both as a regulatory ligand and second messenger. PI(4,5)P2 is known to regulate the mammalian Arp2/3 complex (Rohatgi et al., 2000), in accord with the observation (Table 6) that deletion of the only nonessential component of this complex, ARC18, causes vacuole fragmentation. PI(4,5)P2 can be hydrolyzed to diacylglycerol and inositol trisphosphate (IP3) by two phospholipase C (PLC) enzymes. IP3 is converted to IP(4–8) by inositol polyphosphate multikinase and inositol hexaphosphate kinase. Deletion of any of these four genes causes a strong vacuole fragmentation phenotype (Table 3).
Table 5.
G-Protein related
| Locus | Common name(s) | Vacuolar phenotype(s) | CPY secretion | Function |
|---|---|---|---|---|
| YDL192w | ARF1 | 70%B | + | GTPase involved in coat formation, activates Mss4p |
| YER122c | GLO3 | 30%B 20%C | + | Arf1p/Arf2p GAP |
| YKR001c | VPS1 | 60%B | +++ | Dynamin-like GTPase |
| YLR262c | YPT6 | 40%B 40%C | ++ | Endosomal Rab-like GTPase |
| YLR371w | ROM2 | 20%B 40%D | + | Rho1p GEF |
| YBR200w | BEM1/SRO1 | 50%B | − | Interacts with Cdc42p |
| YER155c | BEM2/SUP4 | 60%D | − | Rho1p GAP |
| YPL161c | BEM4/ROM7 | 50%B 30%E | + | Bud emergence protein, interacts with Rho GTPases |
| YOR070c | GYP1 | 50%B 25%E | + | Ypt4p/Sec4p GAP |
| YDR389w | SAC7 | 30%B | + | Rho1p GAP |
| YBR025c | 60%B 25%e | + | Uncharacterized GTPase | |
| YCR002c | CDC10 | 40%B | − | Septin, GTPase |
Table 6.
Actin/tubulin
| Locus | Common name(s) | Vacuolar phenotype(s) | CPY secretion | Function |
|---|---|---|---|---|
| Actin/polarity related | ||||
| YLR337c | VRP1/END5 | 20%B 20%C 10%D | + | Verprolin homologue, WASP-interacting protein |
| YLR370c | ARC18 | 30%B 20%D 30%E | + | ARP2/3 complex subunit |
| YDR484w | VPS52/SAC2 | 50%B 45%C | +++ | Vps52p-Vps53p-Vps54p complex subunit |
| YDR027c | VPS54/LUV1 | 45%B 50%C | +++ | Vps52p-Vps53p-Vps54p complex subunit |
| YDR129c | SAC6/ABP67 | 30%B 30%C 30%E | − | Fimbrin homologue, F-actin bundling protein |
| YLR144c | ACF2 | 30%B 30%E | − | Cortical actin assembly |
| YMR238w | DFG5 | 30%B 30%E | − | Cell polarity |
| YNL298w | CLA4/ERC10 | 45%B 30%E | + | Protein kinase required for cytokinesis |
| Microtubule related | ||||
| YCL029c | BIK1/ARM5 | 60%B 20%E | + | Microtubule-associated protein |
| YGL216c | KIP3 | 25%B 25%C | + | Kinesin-related protein |
| YOR349w | CIN1 | 25%B 25%E | + | Microtubule stability |
Proteins that mediate divalent cation transport and homeostasis also affect vacuole morphology (Table 4). Calcium is required for vacuole fusion. It is released from the vacuole lumen late in docking (Peters and Mayer, 1998), complexes with calmodulin, and regulates the formation of trans-V0:V0:Vam3p complexes (Peters et al., 2001). Additionally, three proteins related to copper transport and one sodium/proton antiporter are required for the maintenance of low vacuole copy number.
Table 4.
Cations
| Locus | Common name(s) | Vacuolar phenotype(s) | CPY secretion | Function |
|---|---|---|---|---|
| Calcium related | ||||
| YAL026c | DRS2/ATC4 | 50%B | − | P-type Ca-ATPase |
| YBR131w | CCZ1 | 20%B 80%C | ++ | Required for growth in high calcium, caffeine, or zinc |
| YEL010w | SPF1/COD1 | 60%D | ++ | Putative Ca-ATPase |
| YGL167c | PMR1/DER5 | 40%B | + | P-type Ca-ATPase |
| YGR262c | – | 50%B 10%C | + | Similar to calcium/calmodulin-binding protein kinase |
| YAL058w | CNE1/FUN48 | 70%B | + | Similar to mammalian calnexin and calreticulin |
| Metal related | ||||
| YBR037c | SCO1 | 50%B | + | Putative copper transporter |
| YOL152w | FRE7 | 50%B | + | Copper regulated protein |
| YMR038c | LYS7 | 40%B | + | Copper chaperone for Sod1p |
| YDR456w | VPS44/VPL27 | 30%B 30%C 20%E | +++ | Na+/H+ antiporter |
In addition to Ypt7p (Table 1), several other GTPases and GTPase effectors were identified in our screen (Table 5). These include Arf1p and its GAP, Glo3p, which regulate PI(4,5)P2 biosynthesis (Jones et al., 2000), and exchange factors and GAPs for Rho1p, an essential GTPase that is required for normal cell polarity (Hall, 1998) and for the docking stage of vacuole fusion (Eitzen et al., 2001). Other GTPases and effectors do not have a clear link to vacuole fusion, and their effects may be direct or indirect.
Cytoskeletal proteins also govern vacuole copy number (Table 6). Intact microtubules are known to be needed for vacuole integrity in vivo (Guthrie and Wickner, 1988), although tubulin has not been shown to be needed for in vitro vacuole fusion. Actin is found on isolated vacuoles (P. Slusarewicz, A. Merz, G.E., and W.W., unpublished results) and we have recently reported that the Rho GTPases Cdc42p and Rho1p, which regulate actin polymerization, are required for vacuole docking (Eitzen et al., 2001). Cla4p, Vrp1p, which interacts directly with Bee1p, the yeast WASp, and Arc18p, a subunit of the yeast Arp2/3 complex, are part of a well-characterized regulatory cascade in yeast and mammals (Higgs and Pollard, 1999; Winter et al., 1999; Evangelista et al., 2000; Rohatgi et al., 2000). The finding that these proteins regulate vacuole copy number provides an important link in studies of the role of vacuole-bound actin.
Many VPS proteins have well-established roles in vacuole fusion (Table 1), and others with known catalytic activities are categorized accordingly. However, many VPS genes that are needed for normal vacuole morphology have no known catalytic function or have functions without obvious relationship to vacuole fusion per se (Table 7). Of the 137 genes considered in detail here, 24 were already designated VPS genes, and each of these deletion strains showed a vps phenotype of CPY secretion. Of the remaining 113 deletion strains, only 26 had a moderate or severe vps phenotype, suggesting substantial differences between homotypic vacuole fusion and heterotypic trafficking pathways to the vacuole.
Table 7.
Other VPS
| Locus | Common name(s) | Vacuolar phenotype(s) | CPY secretion | Function |
|---|---|---|---|---|
| YDR495c | VPS3/PEP6 | 30%C 60%D | +++ | Involved in vacuolar segregation |
| YOR069w | VPS5/PEP10 | 100%B | +++ | Nexin-like, involved in protein sorting |
| YAL002w | VPS8/VPT8 | 30%C 30%D 30%E | +++ | Required for Vps10p localization, Golgi retrieval |
| YOR132w | VPS17/VPT3 | 95%B | +++ | Retromer complex subunit |
| YCL008c | VPS23/STP22 | 50%D 25%B | +++ | Unknown |
| YKL041w | VPS24/VPT24 | 50%B 50%E | +++ | Protein sorting to prevacuolar compartment |
| YPL065w | VPS28/VPT28 | 60%E | +++ | May control endosomal export |
| YLR025w | VPS32/SNF7 | 30%B 30%D 30%E | +++ | Protein sorting to prevacuolar compartment |
| YJL154c | VPS35/VPT7 | 50%B | +++ | Retromer complex subunit |
Other proteins, with known or imputed roles in trafficking, are also needed for normal vacuole morphology (Table 8). Some, such as clathrin heavy and light chains or SNAREs, probably act indirectly by preventing trafficking of needed fusion catalysts to the vacuole. SNAREs may also act promiscuously in vacuole fusion, as suggested by the ability of certain SNAREs, when overexpressed, to compensate for the loss of others (Gotte and Gallwitz, 1997; Darsow et al., 1998; Tsui et al., 2001). Further studies are needed to resolve this question.
Table 8.
Other trafficking-related genes
| Locus | Common name(s) | Vacuolar phenotype(s) | CPY secretion | Function |
|---|---|---|---|---|
| YDR099w | BMH2/SCD3 | 20%B 10%C 20%E | + | 14-3-3 protein |
| YLR373c | VID22 | 30%B | + | Targeting to prevacuolar compartment |
| YKL054c | VID31 | 30%B 10%C 10%E | + | Vacuolar import and degradation |
| YGR167w | CLC1 | 50%B 40%C 10%E | ++ | Clathrin light chain, coat protein |
| YGL206c | CHC1 | 25%B 25%C | ++ | Clathrin heavy chain, coat protein |
| YHL031c | GOS1 | 70%B | + | Synaptobrevin (v-SNARE) homologue |
| YOL018c | TLG2 | 50%B | ++ | Syntaxin (t-SNARE) homologue |
| YOR036w | VPS6/PEP12 | 75%C 25%E | +++ | Syntaxin (t-SNARE) homologue |
Signaling Pathways of Vacuole Docking
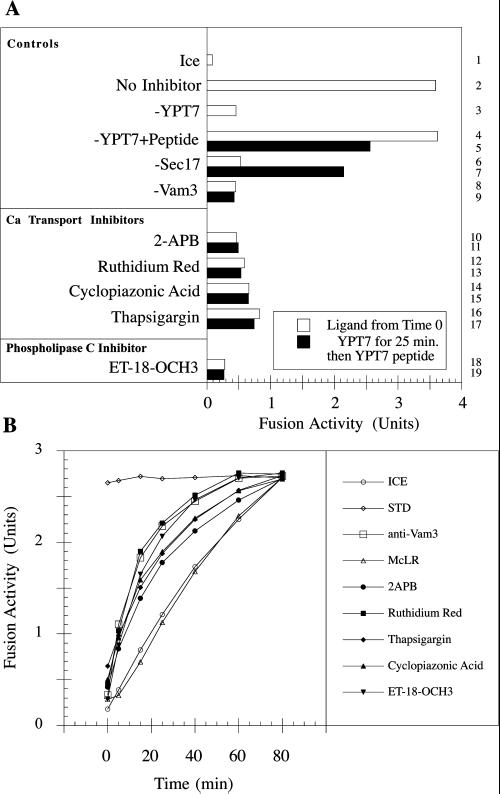
Normal vacuole morphology requires Isc1p and Plc1p, the PLC enzymes that hydrolyze PI(4,5)P2 to diacylglycerol plus IP3, and Ipk2p and Kcs1p, which further phosphorylate IP3 to IP6 and IP4-(PP)2 (Table 3; Saiardi et al., 2000). This suggests that IP3 or its derivatives may regulate later steps of vacuole docking, including the docking-dependent release of calcium from the vacuole (Belde et al., 1993). To provide an independent test of these genetic inferences, we have tested pharmacological inhibitors of PLC and of calcium channels (Figure 2). In vitro vacuole fusion is blocked by the PLC inhibitor ET-18-OCH3 (Arthur and Bittman, 1998) as well as by calcium channel inhibitors such as Ruthenium Red, cyclopiazonic acid, 2-APB, and thapsigargin (Belde et al., 1993; Takahashi et al., 1994; Thomas and Hanley, 1994; Buratti et al., 1995; Calvert and Sanders, 1995; Herrmann-Frank et al., 1996; Maruyama et al., 1997). To order their inhibitory action in the vacuole fusion pathway, we exploited the antibody to a Ypt7p peptide. This antibody blocks Ypt7p action at the start of docking but can be reversed by addition of the cognate peptide (Eitzen et al., 2001). Fusion (Figure 2A, lane 2) is blocked by anti-Ypt7p (Figure 2A, lane 3) but not by the antibody that was premixed with peptide antigen (Figure 2A, lane 4). Vacuoles that were incubated at 27°C with this antibody could be deblocked by peptide addition after 25 min (Figure 2A, lane 5) and had then acquired resistance to the antibody to Sec17p, indicative of the completion of priming (Figure 2A, lane 6, anti-Sec17p from t = 0; lane 7, anti-Ypt7p from t = 0, addition of Ypt7-peptide + anti-Sec17p at t = 25). Because Ypt7p action is essential for trans-pairing of SNAREs (Ungermann et al., 1998), the reaction did not acquire resistance to anti-Vam3p during the period of blockade by anti-Ypt7p (Figure 2A, lanes 8 and 9). Using this assay, we found that primed vacuoles remain fully sensitive to each of the PLC and calcium channel blockers (Figure 2A, lanes 10–19, filled bars), showing that these act after priming is complete. In a complementary approach, these same inhibitors were added at various times to aliquots of an ongoing fusion reaction (Figure 2B). Although each ligand inhibits completely when added from the start, the reaction acquires resistance to antibody to Vam3p as docking is completed (Ungermann et al., 1998), whereas the acquisition of resistance to microcystin LR is kinetically indistinguishable from fusion (Mayer et al., 1996). Resistance to each putative inhibitor of calcium channels or PLC is acquired only at, or shortly after, docking but well before fusion. Thus, PLC action and calcium flux occur at, or shortly after, docking.
Figure 2.
Inhibitors of docking. (A) A 44× scale (1.32 ml) fusion reaction was prepared as described in MATERIALS AND METHODS. Portions (30 μl) were dispensed into individual microfuge tubes, and each reaction was brought to a final volume of 38 μl with ligand or PS buffer as indicated. After incubation on ice for 5 min, the tubes were transferred to a 27°C water bath for 90 min and assayed for alkaline phosphatase activity. The remaining master reaction was treated with anti-YPT7 antibody for 5 min on ice and then for 15 min at 27°C. YPT7 peptide was added, and 30-μl portions were dispensed into microfuge tubes. Ligand or PS was added to a final volume of 38 μl, and the reaction was allowed to proceed for 80 min at 27°C before assay for alkaline phosphatase activity. (B) Aliquots (30 μl) from a 1.98-ml fusion reaction were transferred to ice or mixed with 1 μl of PS buffer or the indicated inhibitor at the indicated time. After a total incubation of 80 min at 27°C, all tubes were placed on ice for 5 min and then assayed for alkaline phosphatase activity.
DISCUSSION
Of the 4828 nonessential genes of yeast, ∼700 showed at least some abnormal vacuole morphology. However, most of these encode proteins of a known function or that reside in a subcellular locale, which strongly indicates that their effects are indirect. We found 137 genes that may be directly needed for normal vacuole size and copy number in growing cells and are, in this regard, VAM genes. It is clear that there is only modest overlap between the VPS and VAM genes. In one study (Banta et al., 1988), mutants in 25 of the 30 VPS genes had normal vacuole structure and thus were not VAM genes. In a later study (Raymond et al., 1992), several class A (normal vacuole appearance) vps mutants (vps13, 44, and 46) had only 21–40% secretion of CPY, whereas others (vps8, 10, 29, 30, 35, and 38) had 62–84% CPY secretion, similar to the ranges for class B or class C vps mutants. Thus, a strong vam phenotype of fragmented vacuoles is not required for a strong vps phenotype. Conversely, of the 137 VAM genes listed here, only 50 are moderate or strong VPS genes. Many gene deletions that cause striking vacuole fragmentation phenotypes do not secrete significant amounts of CPY and thus are not VPS genes. For example, none of the erg deletions have a moderate or strong vps phenotype, even though several show striking vam fragmentation phenotypes (Table 3; Kato and Wickner, 2001). Thus, there are many genes involved in regulating or catalyzing vacuole fusion that do not affect trafficking to the vacuole, and others that affect this heterotypic traffic, at vesicle budding, movement, and fusion, that do not affect homotypic vacuole fusion.
Many VAM genes, such as those needed for ergosterol biosynthesis, function at a nonvacuolar site to make a product that itself traffics to the vacuole and is more directly involved, and some of these genes will only affect fusion indirectly. Nevertheless, several criteria assure us that this set of 137 VAM genes is highly enriched in direct catalysts of the in vitro fusion reaction: 1) The screen selected each of the nonessential genes that encode proteins that are biochemically established as catalyzing vacuole fusion. 2) Functional clusters of genes were evident from the screen, for ergosterol biosynthesis (Kato and Wickner, 2001), phosphoinositide metabolism (Mayer et al., 2000), and actin regulation (Eitzen and Wickner, unpublished data), and these have been confirmed biochemically by adding specific inhibitors to the in vitro fusion reaction. 3) Genetic and proteomic relationships (Uetz et al., 2000; Ito et al., 2001; www.proteome.com) link many of these VAM genes to essential genes that directly participate in vacuole fusion. Of the 137 genes identified in our screen, 28 are in uncharacterized open reading frames and 17 encode known catalysts of vacuole fusion (Table 9). Of the remaining 92 genes, 21 can be directly tied to the reaction: two regulate protein phosphatase 1 (Table 2), six are directly involved in inositol phosphatide metabolism, five others are directly involved in ergosterol biosynthesis (Table 3), three mediate the Rho-GTPase regulation of actin (Table 6), four are effectors of Rho1p, and one activates Mss4p (Table 5).
The first novel functional gene cluster to emerge from the genetic screen was for ergosterol biosynthesis (Kato and Wickner, 2001). Striking vacuole fragmentation was seen in several ergΔ strains, and this was confirmed by direct inhibition of the in vitro fusion of wild-type vacuoles by filipin, nystatin, and amphotericin B, each a specific ligand of ergosterol. This was shown to be specific, in that addition of ergosterol or cholesterol to vacuoles overcame ergosterol deficiency or drug inhibition. Kinetic analysis showed that inhibition was seen only at the priming stage of the reaction and indeed represents a direct requirement of ergosterol for Sec18p-mediated Sec17p release (Kato and Wickner, 2001).
Roles of PI(4,5)P2
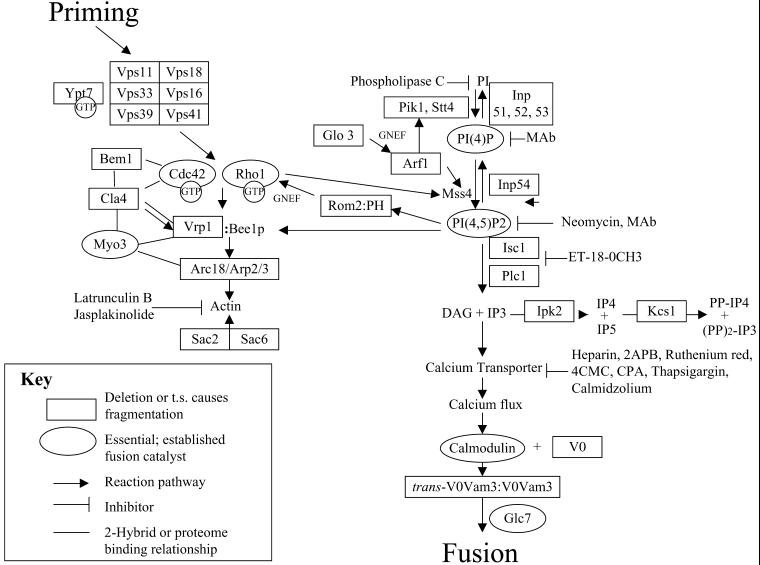
Our current VAM deletion screen has revealed that vacuole fusion is governed by highly interrelated pathways of PI(4,5)P2 synthesis, PI(4,5)P2 hydrolysis by phosphatases and phospholipases, IP3 phosphorylation, and PI(4,5)P2-dependent regulation of actin polymerization. These interrelated pathways are summarized in Figure 3. They reveal an unexpected complexity of the docking phase of vacuole fusion, and yet there is precedent for each component having a role in other fusion reactions and biochemical evidence ties these pathways together.
Figure 3.
Signaling cascades of homotypic vacuole fusion. Genes that are needed for normal vacuole morphology in vivo are in rectangles. Arrows indicate reaction pathways, in yeast and in other eukaryotes.
PIP2 turnover, as well as synthesis, is essential for docking. The clustering of VAM genes in the pathways of PI(4,5)P2 biosynthesis and hydrolysis (Figure 3) is in accord with the finding (Mayer et al., 2000) that this lipid has a central role in docking. Each step of synthesis and degradation of PI(4,5)P2 is required for normal vacuole morphology. Thermosensitive PI 4-kinase has been shown to have fragmented vacuoles at nonpermissive temperature (Audhya et al., 2000). PI(4P) 5-kinases are activated by Arf1p (Jones et al., 2000); deletion of ARF1 or GLO3, the GAP for Arf1p, yields fragmented vacuoles (Table 5). INP54, which removes the 5-phosphate from PI(4,5)P2, is needed for normal vacuole morphology, as are the redundant phosphatases INP51, 52, or 53. These genetic findings are strengthened by the sensitivity of docking to added bacterial PLC (Mayer et al., 2000), which degrades PI. Although yeast has no obvious homologue of the mammalian IP3 receptor calcium channel proteins, the sensitivity of the reaction to inhibitors of such channels suggests a functional homologue may be responsible for docking-dependent calcium release from the vacuole (Peters and Mayer, 1998), especially because this release does not use the well-characterized vacuolar calcium transport proteins (Ungermann et al., 1999). IP3 can induce calcium release from yeast vacuoles (Belde et al., 1993).
IP3 is a biologically significant product of PI(4,5)P2 hydrolysis by PLC. Yeast have two PLC isoforms, PLC1 and ISC1, and deletion of either alters vacuole structure (Audhya et al., 2000), suggesting a role for PLC isoforms in the catalysis of vacuole fusion. PLC activity and the generation of IP3 are central for the induction of regulated calcium flux across the endoplasmic reticulum (Parker et al., 1996) and vacuolar membranes (Belde et al., 1993; Calvert et al., 1995). Because 1) calcium signaling is required for vacuole fusion, 2) the precursor of IP3, PI(4,5)P2, is required for docking (Mayer et al., 2000), and 3) deletion of either of the two PLC isoforms, PLC1 or ISC1, results in vacuole fragmentation, we used several inhibitors of PLC and of IP3-gated calcium channels to verify the participation of PLC and IP3, respectively, in vacuole fusion. Inhibitors of both PLC and of IP3 receptors inhibit vacuole fusion (Figure 2), thus supporting their involvement in the reaction. Higher-order, hyper-phosphorylated forms of inositol have been implicated in several trafficking events in the cell as well as in vacuole biogenesis. This screen showed that deletion of either of two phosphoinositol polyphosphate kinases, IPK2 and KCS1, results in fragmented vacuoles, as noted before (Saiardi et al., 2000).
Other categories of phosphatidylinositol-modifying enzymes were also revealed through this screen. These include the synaptojanin-like PI(4,5)P2 5-phosphatase-INP gene family and two high-order phosphoinositol kinases, IPK2 and KCS1. Single deletion of INP54 or double deletion of INP51 and INP52 (Soltz et al., 1998) result in fragmented vacuoles. The precise role of these phosphatases and their products are currently unknown. Other studies (Malecz et al., 2000) have suggested potential roles for inositol polyphosphate 5-phosphatases, such as those of the INP family and synaptojanin, in the regulation of the actin cytoskeleton by virtue of their association with the GTP-bound form of Rac and through genetic interactions with SAC6, a yeast fimbrin homologue essential for the assembly of normal actin structures (Adams et al., 1991). Because actin may be involved in the fusion of vacuolar membranes (Eitzen and Wickner, unpublished data), these data may represent a regulatory role for INP family-generated inositol metabolites in the fusion of vacuolar membranes.
A third functional gene cluster to emerge from this screen regulates vacuole-bound actin. Cla4p, Vrp1p, and Arc18p are elements in a well-studied cascade by which Rho GTPases regulate F-actin assembly (Higgs and Pollard, 2000; Rohatgi et al., 2000). Each of these three genes shows a two-hybrid relationship to the essential Myo3p motor protein, itself a member of this regulatory pathway (Evangelista et al., 2000; Lechler et al., 2000), and Cla4p binds to Cdc42p (Mitchell and Sprague, 2001; Mosch et al., 2001). We have recently shown that two Rho GTPases, Rho1p and Cdc42p, act after Ypt7p in the docking phase of vacuole fusion (Eitzen et al., 2001) and that actin mutations or actin-specific drugs (latrunculin B and jasplakinoloid) affect the reaction. Although the functional role of actin in vacuole docking is not known, the deletion screen provides critical insight that can be combined with biochemical approaches to establish novel pathways of the fusion reaction.
We speculate that there is an intimate relationship among actin regulation through Rho family GTPases, the need for ergosterol, phosphoinositide signaling, and calcium flux. PI(4,5)P2 (Rozelle et al., 2000) and Cdc42p directly regulate the activity of Bee1p/WASp and thus actin dynamics, whereas PLC has been shown to regulate actin polymerization and calcium release. Rho-GTPases, Arf1p, and their effectors have been shown, in conjunction with PLC, to directly modulate the activities of PI4P 5-kinases and the levels of PI(4,5)P2 in vivo (Weernink et al., 2000a,b). PI(4,5)P2 has been shown in other systems to directly activate the nucleotide exchange activities of Rho proteins (Zheng et al., 1996). Further studies have shown that PI4P 5-kinase activities, PI(4,5)P2 generation, and F-actin generation occur preferentially on membrane rafts and that these activities are sensitive to methyl-β-cyclodextrin (Rozelle et al., 2000), thus implicating sterol as a central structural catalyst for the integration of these fusion-associated events. Several important aspects of the fusion reaction can be linked to membrane microdomains known to be enriched in ergosterol in yeast. If these pathways are indeed interrelated in the context of homotypic vacuole fusion, they might signal from Ypt7p at the start of docking through activation of Rho family GTPases to the calcium flux seen at the end of docking (Figure 3).
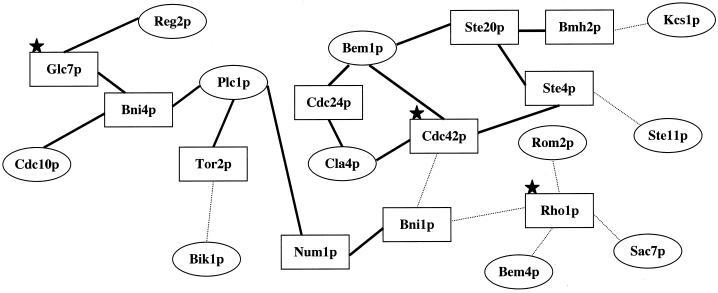
The deletion screen is limited to nonessential genes, and yet many essential genes that participate in other pathways are required for vacuole fusion. Their roles may be discovered through biochemical analysis of the in vitro fusion reaction or by their relationships to other catalysts of vacuole fusion. One example of this is shown in Figure 4. Interactions among genes may be inferred from biochemical data that is compiled at www.proteome.com (indicated by dark connector lines), typically reflecting protein copurification, directly related function, or coimmunoprecipitation from extracts (www.proteome.com), or from comprehensive two-hybrid analysis of yeast (Uetz et al., 2000; Ito et al., 2001), as indicated by thin dotted connector lines. Eleven genes identified in our deletion screen (Figure 4, ovals) have no binary connections with each other, either through two-hybrid or proteome relationships. However, these 11 genes can be connected in this manner by the addition of three essential genes, known from biochemical studies to be required for the vacuole fusion reaction (Figure 4, indicated by stars), and eight other “connector” genes (Figure 4, rectangles). Two of these eight connector genes, TOR2 and CDC24, are essential and thus were not in the deletion screen. Cdc24p is the guanine nucleotide exchange factor for Cdc42p (Zheng et al., 1994). Thus, this modest network has suggested the involvement of additional gene products. Nonessential connector genes may encode proteins that have redundant functions and thus not show a phenotype on single deletion. Gene clusters such as that shown (Figure 4) provide attractive targets for further investigation.
Figure 4.
Clustered genetic interactions. Ovals indicate genes that were detected in our screen as necessary for normal vacuole morphology. Two-hybrid and proteome relationships can connect these genes when the genes in rectangles are added. Thick lines represent experimentally determined protein-protein interactions other than two-hyrid interactions, compiled from www.proteome.com. Thin lines represent protein-protein interactions from two-hybrid analyses. Stars represent genes encoding proteins that are known from biochemical studies to be involved in the vacuole fusion reaction.
ACKNOWLEDGMENTS
The authors thank Susan Oldfield for skilled technical assistance and Drs. Andreas Mayer, Scott Emr, Tom Stevens, and Elizabeth Conibear for helpful discussions. This work was supported by a grant from the National Institute of General Medical Sciences.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc. 01–10–0512. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–10–0512.
REFERENCES
- Adams AE, Botstein D, Drubin DG. Requirement of yeast fimbrin for actin organization and morphogenesis in vivo. Nature. 1991;354:404–408. doi: 10.1038/354404a0. [DOI] [PubMed] [Google Scholar]
- Arthur G, Bittman R. The inhibition of cell signaling pathways by antitumor ether lipids. Biochim Biophys Acta. 1998;1390:85–102. doi: 10.1016/s0005-2760(97)00163-x. [DOI] [PubMed] [Google Scholar]
- Audhya A, Foti M, Emr SD. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p, and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol Biol Cell. 2000;11:2673–2689. doi: 10.1091/mbc.11.8.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis VA, Johnson LM, Emr SD. Isolation of yeast mutants defective in protein targeting to the vacuole. Proc Natl Acad Sci USA. 1986;83:9075–9079. doi: 10.1073/pnas.83.23.9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banta LM, Robinson JS, Klionsky DJ, Emr SD. Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J Cell Biol. 1988;107:1369–1383. doi: 10.1083/jcb.107.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belde PJM, Vossen JH, Borst-Pauwels GWF, Theuvenet APR. Inositol 1,4,5-trisphosphate releases Ca2+ from vacuolar membrane vesicles of Saccharomyces cerevisiae. FEBS Lett. 1993;323:113–118. doi: 10.1016/0014-5793(93)81460-h. [DOI] [PubMed] [Google Scholar]
- Buratti R, Prestipino G, Menegazzi P, Treves S, Zorzato F. Calcium dependent activation of skeletal muscle Ca2+ release channel (ryanodine receptor) by calmodulin. Biochem Biophys Res Commun. 1995;213:1082–1090. doi: 10.1006/bbrc.1995.2238. [DOI] [PubMed] [Google Scholar]
- Cabanoils J-P, Ravichandran V, Roche PA. Phosphorylation of SNAP-23 by the novel kinase SNAK regulates t-SNARE complex assembly. Mol Biol Cell. 1999;10:4033–4041. doi: 10.1091/mbc.10.12.4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert CM, Sanders D. Inositol trisphosphate-dependent and -independent Ca2+ mobilization pathways at the vacuolar membrane of Candida albicans. J Biol Chem. 1995;270:7272–7280. doi: 10.1074/jbc.270.13.7272. [DOI] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol Biol Cell. 2000;11:305–323. doi: 10.1091/mbc.11.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsow T, Burd CG, Emr SD. Acidic di-leucine motif essential for AP-3 dependent sorting and restriction of the functional specificity of the Vam3p vacuolar t-SNARE. J Cell Biol. 1998;142:913–922. doi: 10.1083/jcb.142.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogic D, de Chassey B, Pick E, Cassey D, Lefkir Y, Hemmecke S, Cosson P, Letourneur F. The ADP-ribosylation factor GTPase-activating protein Glo3p is involved in ER retrieval. Eur J Biochem. 1999;78:305–310. doi: 10.1016/s0171-9335(99)80064-8. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL. Regulators and effectors of the ARF GTPases. Curr Opin Cell Biol. 2000;12:475–482. doi: 10.1016/s0955-0674(00)00119-8. [DOI] [PubMed] [Google Scholar]
- Eitzen G, Thorngren N, Wickner W. Rho1p and Cdc42p act after Ytp7p to regulate vacuole docking. EMBO J. 2001;20:5650–5656. doi: 10.1093/emboj/20.20.5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M, Klebl BM, Tong AHY, Webb BA, Leeuw T, Leberer E, Whiteway M, Thomas DY, Boone C. A role for myosin I in actin assembly through interactions with Vrp1p, Bee1p, and the Arp2/3 complex. J Cell Biol. 2000;148:353–362. doi: 10.1083/jcb.148.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer von Mollard G, Nothwehr SF, Stevens TH. The yeast v-SNARE Vti1p mediates two vesicle transport pathways through interactions with the tSNAREs Sed5p and Pep12p. J Cell Biol. 1997;137:1511–1524. doi: 10.1083/jcb.137.7.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti M, Audhya A, Emr SD. SacI lipid phosphatase, and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol Biol Cell. 2001;12:2396–2411. doi: 10.1091/mbc.12.8.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotte M, Gallwitz D. High expression of the yeast syntaxin-related Vam3 protein suppresses the protein transport defects of a pep12 null mutant. FEBS Lett. 1997;411:48–52. doi: 10.1016/s0014-5793(97)00575-9. [DOI] [PubMed] [Google Scholar]
- Grote E, Novick PJ. Promiscuity in Rab-SNARE interactions. Mol Biol Cell. 1999;10:4149–4161. doi: 10.1091/mbc.10.12.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie B, Wickner W. Yeast vacuoles fragment when microtubules are disrupted. J Cell Biol. 1988;107:115–120. doi: 10.1083/jcb.107.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A. A quantitative assay to measure homotypic vacuole fusion in vitro. Meth Cell Sci. 1995;17:283–294. [Google Scholar]
- Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Herrmann-Frank A, Richter M, Sarkozi S, Mohr U, Lehmann-Horn F. 4-Chloro-m-cresol, a potent and specific activator of the skeletal muscle ryanodine receptor. Biochim Biophys Acta. 1996;1289:31–40. doi: 10.1016/0304-4165(95)00131-x. [DOI] [PubMed] [Google Scholar]
- Higgs HN, Pollard TD. Regulation of actin polymerization by Arp2/3 complex and WASp/Scar proteins. J Biol Chem. 1999;274:32531–32534. doi: 10.1074/jbc.274.46.32531. [DOI] [PubMed] [Google Scholar]
- Higgs HN, Pollard TD. Activation by Cdc42, and PIP(2) of Wiskott-Aldrich syndrome protein (WASp) stimulates actin nucleation by Arp 2/3 complex. J Cell Biol. 2000;18:1311–1320. doi: 10.1083/jcb.150.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DH, Morris JB, Morgan CP, Kondo H, Irvine RF, Cockcroft S. Type I phosphatidylinositol 4-phosphate 5-kinase directly interacts with ADP-ribosylation factor I and is responsible for phosphatidylinositol 4,5-bisphosphate synthesis in the Golgi compartment. J Biol Chem. 2000;275:13962–13966. doi: 10.1074/jbc.c901019199. [DOI] [PubMed] [Google Scholar]
- Kato M, Wickner W. Ergosterol is required for the Sec18/ATP-dependent priming step of homotypic vacuole fusion. EMBO J. 2001;20:4035–4040. doi: 10.1093/emboj/20.15.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler T, Shevchenko A, Li R. Direct involvement of yeast type I myosins in Cdc42-dependent actin polymerization. J Cell Biol. 2000;148:363–373. doi: 10.1083/jcb.148.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecz N, McCabe PC, Spaargaren C, Qiu R, Chuang Y, Symons M. Synaptojanin 2, a novel Rac1 effector that regulates clathrin-mediated endocytosis. Curr Biol. 2000;10:1383–1340. doi: 10.1016/s0960-9822(00)00778-8. [DOI] [PubMed] [Google Scholar]
- Marash M, Gerst JE. t-SNARE dephosphorylation promotes SNARE assembly and exocytosis in yeast. EMBO J. 2001;20:411–421. doi: 10.1093/emboj/20.3.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Kanaji T, Nakade S, Kanno T, Mikoshiba K. 2APB, 2-aminoethoxydiphenyl borate, a membrane-permeable modulator of Ins(1,4,5)P3-induced Ca2+ release. J Biochem. 1997;122:498–505. doi: 10.1093/oxfordjournals.jbchem.a021780. [DOI] [PubMed] [Google Scholar]
- Mayer A, Scheglmann D, Dove S, Glatz A, Wickner W, Haas A. Phosphatidylinositol-(4,5)-bisphosphate regulates two steps of homotypic vacuole fusion. Mol Biol Cell. 2000;11:807–817. doi: 10.1091/mbc.11.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Wickner W, Haas A. Sec18p (NSF)-driven release of Sec17p (α-SNAP) can precede docking and fusion of yeast vacuoles. Cell. 1996;85:83–94. doi: 10.1016/s0092-8674(00)81084-3. [DOI] [PubMed] [Google Scholar]
- Mitchell DA, Sprague GF., Jr The phosphotyrosyl phosphatase activator, Ncs1p (Rrd1p), functions with Cla4p to regulate the G(2)/M transition in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:488–500. doi: 10.1128/MCB.21.2.488-500.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosch HU, Kohler T, Braus GH. Different domains of the essential GTPase Cdc42p required for growth and development of Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:235–248. doi: 10.1128/MCB.21.1.235-248.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller O, Johnson DI, Mayer A. Cdc42p functions at the docking stage of yeast vacuole membrane fusion. EMBO J. 2001;20:5657–5665. doi: 10.1093/emboj/20.20.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols BJ, Ungermann C, Pelham HRB, Wickner WJ, Haas A. Homotypic vacuolar fusion mediated by t- and v- SNARES. Nature. 1997;387:199–202. doi: 10.1038/387199a0. [DOI] [PubMed] [Google Scholar]
- Paidhungat M, Garrett S. Cdc1 and the vacuole coordinately regulate Mn2+ homeostasis in the yeast Saccharomyces cerevisiae. Genetics. 1998;148:1787–1798. doi: 10.1093/genetics/148.4.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker I, Choi J, Yao Y. Elementary events of InsP3-induced Ca2+ liberation in Xenopus oocytes: hots spots, puffs and blips. Cell Calcium. 1996;20:105–121. doi: 10.1016/s0143-4160(96)90100-1. [DOI] [PubMed] [Google Scholar]
- Peters C, Andrews PD, Stark MJR, Cesaro-Tadic S, Glatz A, Podtelejnikov A, Mann M, Mayer A. Control of the terminal step of intracellular membrane fusion by protein phosphatase 1. Science. 1999;285:1084–1087. doi: 10.1126/science.285.5430.1084. [DOI] [PubMed] [Google Scholar]
- Peters C, Bayer MJ, Buhler S, Andersen JS, Mann M, Mayer A. Trans-complex formation by proteolipid channels in the terminal phase of membrane fusion. Nature. 2001;409:581–588. doi: 10.1038/35054500. [DOI] [PubMed] [Google Scholar]
- Peters C, Mayer A. Ca2+/calmodulin signals the completion of docking and triggers a late step of vacuole fusion. Nature. 1998;396:575–580. doi: 10.1038/25133. [DOI] [PubMed] [Google Scholar]
- Price A, Seals D, Wickner W, Ungermann C. The docking stage of yeast vacuole fusion requires the transfer of proteins from a cis-SNARE complex to a Rab/Ypt protein. J Cell Biol. 2000;148:1231–1238. doi: 10.1083/jcb.148.6.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CJ, Raymond CK, Yashimiro CT, Stevens TH. Methods for studying the yeast vacuole. Methods Enzymol. 1991;194:644–661. doi: 10.1016/0076-6879(91)94047-g. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Ho HH, Kirschner MW. Mechanism of N-WASP activation by CDC42, and phosphatidylinositol 4,5-bisphosphate. J Cell Biol. 2000;150:1299–1309. doi: 10.1083/jcb.150.6.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JH, Stevens TH. Protein sorting in yeast: mutants defective in vacuole biogenesis localize vacuolar proteins into the late secretory pathway. Cell. 1986;47:1041–1051. doi: 10.1016/0092-8674(86)90819-6. [DOI] [PubMed] [Google Scholar]
- Rozelle AL, Machesky LM, Yamamoto M, Driessens MH, Insall RH, Roth MG, Luby-Phelps K, Marriott G, Hall A, Yin HL. Phosphatidylinositol 4,5-bisphosphate induces actin-based movement of raft-enriched vesicles through WASP-Arp 2/3. Curr Biol. 2000;10:311–320. doi: 10.1016/s0960-9822(00)00384-5. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Caffrey JJ, Snyder SH, Shears SB. The inositol hexakisphosphate kinase family: catalytic flexibility and function in yeast vacuole biogenesis. J Biol Chem. 2000;275:24686–24692. doi: 10.1074/jbc.M002750200. [DOI] [PubMed] [Google Scholar]
- Seals D, Eitzen G, Margolis N, Wickner W, Price A. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc Natl Acad Sci USA. 2000;97:9402–9407. doi: 10.1073/pnas.97.17.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz LE, Huynh CV, Thorner J, York JD. Identification and characterization of an essential family of inositol polyphosphate 5-phosphatases (INP51, INP52, and INP53 gene products) in the yeast Saccharomyces cerevisiae. Genetics. 1998;148:1715–1729. doi: 10.1093/genetics/148.4.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Tanzawa K, Takahashi S. Adenophostins, newly discovered metabolites of Penicillium brevicompactum, act as potent agonists of the inositol 1,4,5-trisphosphate receptor. J Biol Chem. 1994;269:369–372. [PubMed] [Google Scholar]
- Thomas D, Hanley MR. Pharmacological tools for perturbing intracellular calcium storage. Methods Enzymol. 1994;40:65–89. doi: 10.1016/s0091-679x(08)61110-3. [DOI] [PubMed] [Google Scholar]
- Tsui MMK, Tai WCS, Banfield DK. Selective formation of Sec5p-containing SNARE complexes is mediated by combinatorial binding interactions. Mol Biol Cell. 2001;12:521–538. doi: 10.1091/mbc.12.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Ungermann C, Sato K, Wickner W. Defining the functions of trans-SNARE pairs. Nature. 1998;396:543–548. doi: 10.1038/25069. [DOI] [PubMed] [Google Scholar]
- Ungermann C, Wickner W, Xu Z. Vacuole acidification is required for trans-SNARE pairing, LMA1 release, and homotypic fusion. Proc Natl Acad Sci USA. 1999;96:11194–11199. doi: 10.1073/pnas.96.20.11194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit M, Laage R, Dietrich L, Wang L, Ungermann C. Vac8p release from the SNARE complex, and its palmitoylation are coupled, and essential for vacuole fusion. EMBO J. 2001;20:3145–3155. doi: 10.1093/emboj/20.12.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y, Ohsumi Y, Anraku Y. Genes for directing vacuole morphogenesis in Saccharomyces cerevisiae. I. Isolation and characterization of two classes of vam mutants. J Biol Chem. 1992;267:18665–18670. [PubMed] [Google Scholar]
- Wang Y-X, Kauffman EJ, Duex JE, Weisman LS. Fusion of docked membranes requires the armadillo repeat protein Vac8p. J Biol Chem. 2001;276:35133–35140. doi: 10.1074/jbc.M103937200. [DOI] [PubMed] [Google Scholar]
- Weernink PA, Guo Y, Zhang C, Schmidt M, von Eichel-Streiber C, Jakobs KH. Control of cellular phosphatidylinositol 4,5-bisphosphate levels by adhesion signals, and rho GTPases in NIH 3T3 fibroblasts: involvement of both phosphatidylinositol-4-phosphate 5-kinase and phospholipase C. Eur J Biochem. 2000a;267:5237–5246. doi: 10.1046/j.1432-1327.2000.01599.x. [DOI] [PubMed] [Google Scholar]
- Weernink PA, Schulte P, Guo Y, Wetzel J, Amano M, Kaibuchi K, Haverland S, Voss M, Schmidt M, Mayr GW, Jakobs KH. Stimulation of phosphatidylinositol-4-phosphate 5-kinase by Rho-kinase. J Biol Chem. 2000b;275:10168–10174. doi: 10.1074/jbc.275.14.10168. [DOI] [PubMed] [Google Scholar]
- Weisman LS, Emr SD, Wickner WT. Mutants of Saccharomyces cerevisiae that block intervacuole vesicular traffic and vacuole division and segregation. Proc Natl Acad Sci USA. 1990;87:1076–1080. doi: 10.1073/pnas.87.3.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W, Haas A. Yeast vacuole fusion: a window on organelle trafficking mechanisms. Annu Rev Biochem. 2000;69:247–275. doi: 10.1146/annurev.biochem.69.1.247. [DOI] [PubMed] [Google Scholar]
- Winter D, Lechler T, Li R. Activation of the yeast Arp2/3 complex by Bee1p, a WASP-family protein. Curr Biol. 1999;9:501–504. doi: 10.1016/s0960-9822(99)80218-8. [DOI] [PubMed] [Google Scholar]
- Wurmser AE, Sato TK, Emr SD. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J Cell Biol. 2000;151:551–562. doi: 10.1083/jcb.151.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Mayer A, Muller E, Wickner W. A heterodimer of thioredoxin and I2B cooperates with Sec18p (NSF) to promote yeast vacuole inheritance. J Cell Biol. 1997;136:299–306. doi: 10.1083/jcb.136.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Sato K, Wickner W. LMA1 binds to vacuoles at Sec18p (NSF), transfers upon ATP hydrolysis to a t-SNARE (Vam3p) complex, and is released during fusion. Cell. 1998;93:1125–1134. doi: 10.1016/s0092-8674(00)81457-9. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Cerione R, Bender A. Control of the yeast bud-site assembly GTPase Cdc42: catalysis of guanine nucleotide exchange by Cdc24 and stimulation of GTPase activity by Bem3. J Biol Chem. 1994;269:2369–2372. [PubMed] [Google Scholar]
- Zheng Y, Glaven JA, Wu WJ, Cerione RA. Phosphatidylinositol 4,5-bisphosphate provides an alternative to guanine nucleotide exchange factors by stimulating the dissociation of GDP from Cdc42Hs. J Biol Chem. 1996;271:23815–23819. doi: 10.1074/jbc.271.39.23815. [DOI] [PubMed] [Google Scholar]