Abstract
Aiming to identify highly effective and selective G-quadruplex ligands as anticancer candidates, five natural compounds were investigated here, i.e., the alkaloids Canadine, D-Glaucine and Dicentrine, as well as the flavonoids Deguelin and Millettone, selected as analogs of compounds previously identified as promising G-quadruplex-targeting ligands. A preliminary screening with the G-quadruplex on the Controlled Pore Glass assay proved that, among the investigated compounds, Dicentrine is the most effective ligand of telomeric and oncogenic G-quadruplexes, also showing good G-quadruplex vs. duplex selectivity. In-depth studies in solution demonstrated the ability of Dicentrine to thermally stabilize telomeric and oncogenic G-quadruplexes without affecting the control duplex. Interestingly, it showed higher affinity for the investigated G-quadruplex structures over the control duplex (Kb~106 vs. 105 M−1), with some preference for the telomeric over the oncogenic G-quadruplex model. Molecular dynamics simulations indicated that Dicentrine preferentially binds the G-quadruplex groove or the outer G-tetrad for the telomeric and oncogenic G-quadruplexes, respectively. Finally, biological assays proved that Dicentrine is highly effective in promoting potent and selective anticancer activity by inducing cell cycle arrest through apoptosis, preferentially targeting G-quadruplex structures localized at telomeres. Taken together, these data validate Dicentrine as a putative anticancer candidate drug selectively targeting cancer-related G-quadruplex structures.
Keywords: Dicentrine, G-quadruplex, natural compounds, telomeres, oncogenes
1. Introduction
The noncanonical secondary structures of nucleic acids have attracted increasing attention as potential drug targets for their involvement in various crucial biological processes [1]. Even if DNA within cells mainly exists in the double-stranded B-form, single strands of DNA can fold into various conformations [1,2]. Among them, a growing interest is directed at G-quadruplex structures formed by single-stranded, guanine-rich sequences. G-quadruplexes consist of cyclic, planar arrays of four guanines, called G-tetrads, and sequences connecting the tracts of adjacent guanines, called loops [3]. These structures are involved in DNA damage and genome instability in addition to key cancer-related cellular pathways [3,4]. Indeed, G-quadruplexes are mainly found in telomeres and oncogene promoters, and several experiments have ascertained that their formation in DNA is modulated during the cell cycle [3,4]. Therefore, some innovative anticancer strategies rely on G-quadruplex structure stabilization induced by small molecules at telomeres and oncogene promoters in order to block telomerase activity and oncogene expression, respectively [3,5,6,7], thus developing targeted therapies potentially endowed with low-to-null side effects. To date, about 3000 different G-quadruplex ligands have been identified [8], some of which show potential anticancer activities, but only a few have progressed to clinical trials due to selectivity issues [5]. DNA in duplex form is present in chromosomes in large excess compared with G-quadruplex structures. Therefore, a stringent ability to selectively recognize G-quadruplexes discriminating duplex DNA is one of the main requirements in the design of G-quadruplex ligands as potential anticancer drugs.
In this context, we recently selected 28 natural compounds as putative G-quadruplex ligands using the docking-based virtual screening of an in-house library of ca. 1000 natural compounds and then experimentally evaluated their interaction with G-quadruplex structures [9]. Finally, 5 out of the 28 natural compounds were identified as the best G-quadruplex ligands in vitro, i.e., Bulbocapnine, Chelidonine, Ibogaine, Rotenone and Vomicine.
Aiming to discover more effective and selective G-quadruplex ligands, a focused set of structural analogs of the previously identified hit compounds was here investigated, including the natural alkaloids Canadine, D-Glaucine and Dicentrine and the flavonoids Deguelin and Millettone (Table 1). These compounds have well-recognized biological and pharmacological properties but have never been deeply investigated as putative G-quadruplex ligands or, more generally, as DNA ligands.
Here, a preliminary screening was carried out with the aid of the G-quadruplex on the Controlled Pore Glass (G4-CPG) assay [10,11] in order to assess the capacity of the examined natural compounds to interact with telomeric and oncogenic G-quadruplexes, as well as their G-quadruplex vs. duplex selectivity. The compound showing the highest affinity and selectivity for the G-quadruplexes, i.e., Dicentrine, was then studied in its interaction in solution with the DNA targets of choice using multiple biophysical techniques. Additionally, structural details of the interaction of Dicentrine with G-quadruplex structures were obtained using molecular dynamics (MD) simulations. Finally, Dicentrine was evaluated for its anticancer activity as well as its ability to target G-quadruplex structures in cells via biological assays.
Table 1.
Chemical structures and features of the natural alkaloids Canadine, D-Glaucine and Dicentrine and the flavonoids Deguelin and Millettone.
| Compound | Common Name (Library Code) | Chemical Structure | M. W. | Molecular Formula | Source | Reference |
|---|---|---|---|---|---|---|
| Alkaloids | ||||||
| 1 | Canadine (BBN142) | 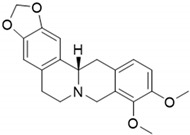 |
339.39 | C20H21NO4 | Mahonia aquifolum (Berberidaceae family) | [12] |
| 2 | D-Glaucine (BBN45) | 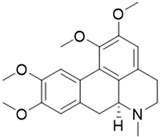 |
355.43 | C21H25NO4 | Glaucium flavum (Papaveracee family) | [13] |
| 3 | Dicentrine (BBN172) | 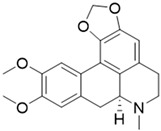 |
339.39 | C20H21NO4 | Lindera megaphylla (Lauraceae family) | [14] |
| Flavonoids | ||||||
| 4 | Deguelin (BBN238) | 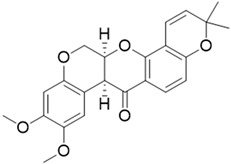 |
394.42 | C23H22O6 | Mundulea sericea(Fabaceae family) | [15] |
| 5 | Millettone (BBN271) | 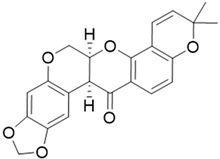 |
378.38 | C22H18O6 |
Derris trifoliata Lour (Leguminosae family) |
[16] |
2. Results and Discussion
2.1. Properties, Origin and Structural Features of the Selected Natural Compounds
The three alkaloids Canadine, D-Glaucine and Dicentrine are analogs of Bulbocapnine, an inhibitor of acetyl- and butyrylcholinesterase with dose-dependent activity [17]. Canadine is a benzylisoquinoline alkaloid that displays interesting antioxidant and antibacterial activities [18]. Correché et al. showed that Canadine has a strong antioxidant capacity against the oxidative stress induced by tert-butylhydroperoxide, a model inducer of endogenous reactive oxygen species (ROS) formation [12]. Canadine protects microsomal lipids from peroxidation, one of the reactions triggered by the formation of free radicals in cells and tissues. Furthermore, Chlebek et al. studied Canadine activity in human acetyl- and butyrylcholinesterase, involved in Alzheimer’s disease, showing an IC50 value in the low micromolar range [19].
D-Glaucine is a tetrahydroisoquinoline derivative long used as a remedy for coughs and other illnesses. D-Glaucine was identified as a relatively potent and selective inhibitor of soluble phosphodiesterase 4 (PDE4) isolated from bovine aortic muscle. PDE4 is the major isoenzyme present in human polymorphonuclear leukocytes, and its inhibition leads to increased cyclic AMP levels and the subsequent inhibition of several functional responses [20]. Moreover, D-Glaucine can inhibit the migration and invasion of human breast cancer cells [21].
Dicentrine is an aporphine-type isoquinoline alkaloid isolated from several medicinal plants, some of which are used in traditional medicine to treat cancer and other diseases [14]. Dicentrine has been shown to exert cytotoxic activity toward cancer cells by unwinding DNA and inhibiting the catalytic activity of DNA topoisomerases [22]. Konkimalla et al. have also shown that Dicentrine binds with high affinity to the erlotinib-binding site of EGFR—the epidermal growth factor receptor gene family, which evolved as important factors in cancer prognosis—by means of in silico modeling and the virtual screening approach [23]. Moreover, Dicentrine was found to stabilize a tetramolecular telomeric G-quadruplex model and to inhibit telomerase activity [24]. Furthermore, an in vitro tumor-growing assay in Severe Combined Immunodeficiency (SCID) mice showed that the intraperitoneal injection of Dicentrine at a 100 µg dose twice a week for 4 weeks significantly inhibited the tumor incidence of leukemia cell line K562 in SCID mice [14].
Deguelin and Millettone belong to the rotenoid family [16]. In particular, Deguelin is a rotenoid that shows high potential as a chemopreventive and therapeutic agent in vitro and in vivo against several types of cancer, including colon, lung and breast tumors. The anticancer effect of Deguelin is associated with cell cycle arrest and apoptosis via the downregulation of specific cell survival proteins, including Akt and mitogen-activated protein kinase. Deguelin is able to suppress HIF-1a expression and Hsp90 interaction in radioresistant lung cancer cells [15,25]. Finally, no in-depth studies on Millettone have been reported thus far.
2.2. Screening of Natural Compounds with the G4-CPG Assay
The G4-CPG assay is an affinity chromatography-based method conceived to improve the previously developed G-quadruplex on Oligo Affinity Support (G4-OAS) [11,26,27], allowing for the rapid and easy selection of G-quadruplex-specific ligands. It involves flowing solutions of putative G-quadruplex ligands through a Controlled Pore Glass (CPG) support functionalized with G-quadruplex-forming oligonucleotides. The compounds with high affinity for the G-quadruplex are retained, while those with low-to-null affinity are eluted. All the assay steps are monitored with UV measurements [10,11]. The two following cancer-related G-quadruplex-forming DNA sequences were chosen here as targets: (i) tel26, a 26-mer oligonucleotide of sequence d(TTAGGGTTAGGGTTAGGGTTAGGGTT), able to fold into a hybrid G-quadruplex, with its sequence extracted from the human 3′-telomeric overhang [28], and (ii) c-myc, a 33-mer oligonucleotide able to fold into a parallel G-quadruplex of sequence d(TGGGGAGGGTGGGGAGGGTGGGGAAGGTGGGGA), reproducing the regulatory region of the gene coding for the transcription factor C-MYC [29]. All the experiments carried out on the G-quadruplex targets were performed in parallel on a 27-mer unimolecular duplex-forming DNA sequence, hereafter named ds27, to verify if the analyzed compounds could discriminate G-quadruplex vs. duplex DNA. In particular, the duplex of choice consists of two self-complementary tracts, each containing the Dickerson sequence d(CGCGAATTCGCG), i.e., one of the best-studied models for B-DNA [30], connected by a TTT loop so as to obtain a hairpin-like structure.
The protocol used for the binding assays has been previously reported [10] and is described here in the experimental section. Briefly, the compounds were initially evaluated for their solubility at the concentration and in the washing/releasing solutions used in the G4-CPG assay, proving to be fully soluble and stable in the experimental assay conditions. For each of the five natural compounds, tests were carried out to first evaluate unspecific binding on nude CPG support and then the ability to bind G-quadruplexes- and duplex-forming DNA oligonucleotide sequences. The results of the G4-CPG assay are summarized in Table 2. No significant unspecific binding was observed for all the tested compounds, thus allowing for their further analyses of the oligonucleotide-functionalized supports. Overall, D-Glaucine, Deguelin and Millettone showed a low-to-null ability to interact with G-quadruplex- and duplex-functionalized supports. On the other hand, Canadine showed good affinity and selectivity for G-quadruplex-functionalized supports; however, it was associated with significant binding to the nude CPG. Conversely, Dicentrine showed a low affinity for the duplex-functionalized support but high affinity toward the G-quadruplex-functionalized supports, with an interesting binding preference for the hybrid tel26 G-quadruplex.
Table 2.
Summary of the binding assay data obtained for the studied natural compounds using the G4-CPG assay. The bound ligand amounts are calculated as the difference between the initially loaded amount of ligand and the unbound ligand recovered by the washing solution and are expressed as a percentage of the quantity initially loaded on each support. The errors associated with the reported percentages are within ±2%.
| Compound | Bound Ligand (%) | |||
|---|---|---|---|---|
| Nude CPG | CPG-tel26 | CPG-c-myc | CPG-ds27 | |
| Canadine | 21 | 29 | 20 | 4 |
| D-Glaucine | 0 | 2 | 1 | 0 |
| Dicentrine | 0 | 67 | 47 | 30 |
| Deguelin | 0 | 0 | 0 | 0 |
| Millettone | 8 | 8 | 8 | 8 |
In summary, within the investigated set of natural compounds, Dicentrine was the best G-quadruplex ligand in terms of target affinity and G-quadruplex vs. duplex selectivity according to the results of the G4-CPG assay, showing a two-fold higher affinity for the G-quadruplex targets than the control duplex. Therefore, it was advanced to further studies in solution to investigate its interaction with G-quadruplex structures using biophysical techniques.
2.3. Circular Dichroism Studies of Dicentrine with G-Quadruplexes and Control Duplex
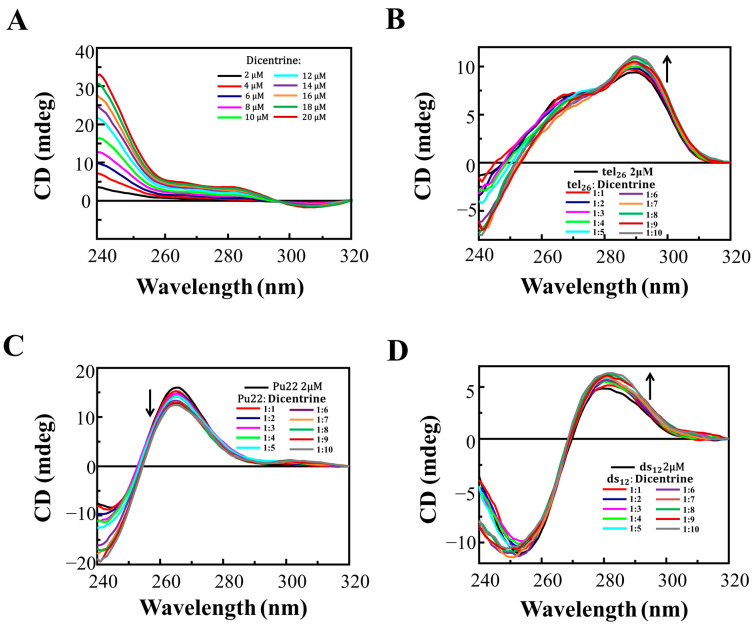
The ability of Dicentrine to interact in solution with a telomeric and an oncogenic G-quadruplex model, as well as a control duplex, was investigated with circular dichroism (CD) experiments. The human telomeric G-quadruplex tel26 [28]; the oncogenic G-quadruplex Pu22T14T23 (hereafter also named Pu22), which is a shorter variant (22-mer) of the c-myc oligonucleotide of sequence d(TGAGGGTGGGTAGGGTGGGTAA) [31,32]; and the Dickerson dodecamer d(CGCGAATTCGCG) (ds12) [33] were used as valuable G-quadruplex and duplex models for studies in solution. The G-quadruplex-forming oligonucleotides, as well as the control duplex, were prepared by overnight-annealing the proper solutions at a 2 μM DNA concentration in 5 mM KCl, 5 mM KH2PO4, 5% DMSO buffer (pH 7). According to the literature, in these conditions, (i) tel26 folds into a hybrid 2-type G-quadruplex, featured by a double hump-band with maxima centered at 265 and 290 nm [34]; (ii) Pu22 exhibits a maximum centered at 265 nm and a minimum at 245 nm, typical of a parallel G-quadruplex [32]; and (iii) ds12 shows a positive band at 280 nm, along with an intense minimum at 251 nm, characteristic of a B-DNA duplex structure [35].
These oligonucleotides were titrated with increasing amounts of Dicentrine (up to 10 molar equivalents), and the corresponding CD spectra were recorded after each addition (Figure 1). In parallel, since Dicentrine has one chiral center and shows CD signals in the same range as the investigated oligonucleotide sequences, the spectra of Dicentrine at increasing amounts were recorded in the same buffer used for the titration experiments (Figure 1A), and the contribution of Dicentrine alone was subtracted from the spectra of the titrations of oligonucleotides with this alkaloid, as reported in Figure 1B–D.
Figure 1.
(A) CD spectra of solutions of Dicentrine (from 2 to 20 µM) in 5 mM KCl, 5 mM KH2PO4, 5% DMSO buffer (pH 7) and CD spectra of 2 μM solutions of (B) tel26 G-quadruplex, (C) Pu22 G-quadruplex and (D) ds12 duplex in 5 mM KCl, 5 mM KH2PO4, 5% DMSO buffer (pH 7) in the presence of increasing amounts of Dicentrine (up to 10 equivalents). Arrows indicate the CD changes on increasing the ligand concentration.
In detail, upon titration with Dicentrine, the tel26 G-quadruplex showed an increase in the CD signal intensity of the 290 nm band (Figure 1B), whereas, for the Pu22 G-quadruplex, a decrease in the intensity of the 265 nm band was observed (Figure 1C). Finally, the titration of ds12 with Dicentrine caused a slight increase in the intensity of the 251 nm band, accompanied by a dose-dependent increase in the 280 nm band (Figure 1D).
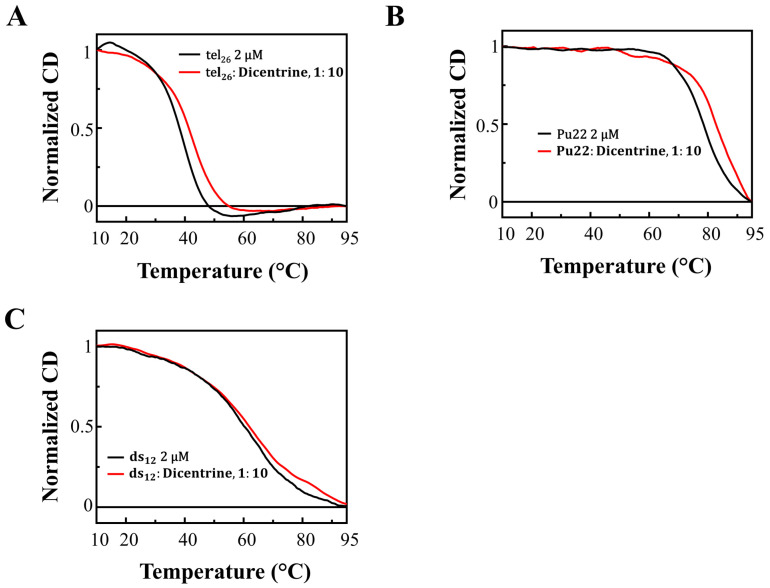
CD melting experiments were also performed on all the DNA/ligand mixtures to evaluate if the incubation with the ligand produced stabilizing effects on the G-quadruplex and duplex structures. The CD melting curves of tel26 G-quadruplex, Pu22 G-quadruplex and ds12 duplex in the absence or presence of Dicentrine (DNA/ligand 1:10 ratio) were recorded by following the changes in the wavelength of the characteristic CD intensity maximum/minimum of each DNA secondary structure (290, 265 and 251 nm for tel26, Pu22 and ds12, respectively) (Figure 2). Melting temperature (Tm) values of 40, 79 and 63 °C were found for free tel26 G-quadruplex, Pu22 G-quadruplex and ds12 duplex, respectively (Table 3). Notably, stabilizing effects on both the tel26 and Pu22 G-quadruplexes were found (ΔTm = +3 and +5 °C, respectively) (Figure 2A,B and Table 3), while no significant stabilizing effect on the ds12 duplex was observed in the presence of Dicentrine (ΔTm = +1 °C) (Figure 2C and Table 3).
Figure 2.
CD melting curves for (A) tel26 G-quadruplex, (B) Pu22 G-quadruplex and (C) ds12 duplex in the absence and presence of 10 molar equivalents of Dicentrine in 5 mM KCl, 5 mM KH2PO4, 5% DMSO buffer (pH 7), recorded at 290, 265 and 253 nm, respectively.
Table 3.
Melting temperature (Tm) values of tel26 G-quadruplex, Pu22 G-quadruplex and ds12 duplex in the absence or presence of Dicentrine (10 molar equivalents) as measured by CD melting experiments. ΔTm = Tm(DNA/ligand, 1:10)−Tm(free DNA).
| Tm ± 1 (°C) | ΔTm (°C) | |
|---|---|---|
| tel26 | 40 | - |
| tel26/Dicentrine, 1:10 | 43 | +3 |
| Pu22 | 79 | - |
| Pu22/Dicentrine, 1:10 | 84 | +5 |
| ds12 | 63 | - |
| ds12/Dicentrine, 1:10 | 64 | +1 |
Overall, the CD titration experiments proved that, upon the binding of Dicentrine, the main folds of the investigated oligonucleotides were preserved, even if slight conformational changes were observed in all cases. Moreover, CD melting experiments demonstrated that Dicentrine positively affected the stability of G-quadruplex-forming oligonucleotides, with no relevant effect on the control duplex, thus further confirming the good G-quadruplex vs. duplex selectivity of this compound, as already observed with the G4-CPG assay.
2.4. Gel Electrophoresis Studies of Dicentrine with G-Quadruplexes and Control Duplex
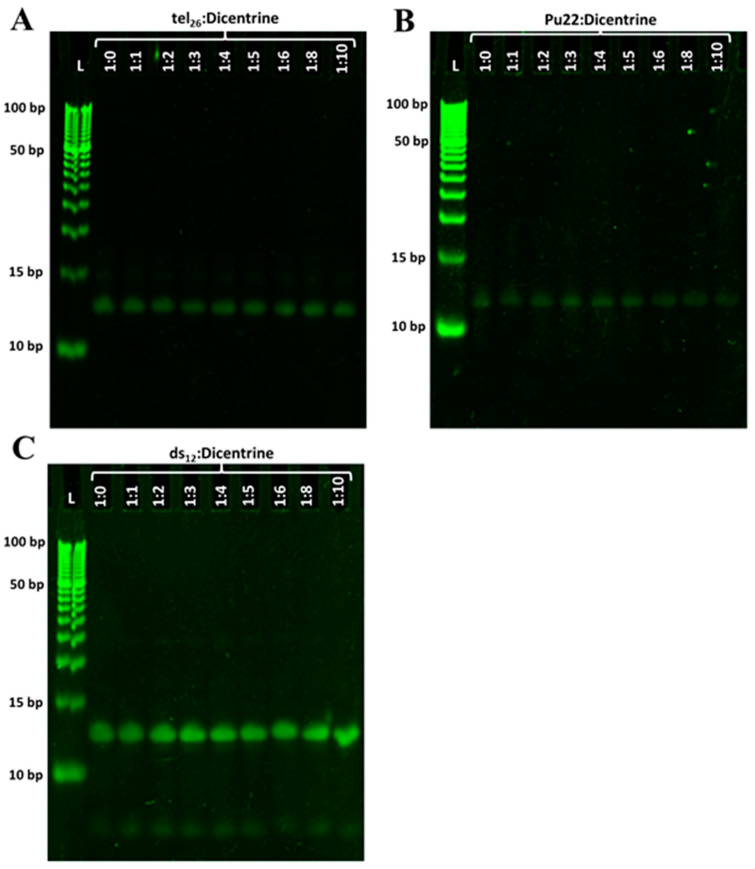
To obtain further information on the binding of Dicentrine to the G-quadruplex targets, gel electrophoresis experiments were carried out on tel26, Pu22 and ds12. Native polyacrylamide gel electrophoresis (PAGE) experiments were performed, analyzing the oligonucleotide samples at a 2 µM concentration mixed with different Dicentrine amounts (from 1:0 to 1:10 DNA/ligand ratio) in 5 mM KCl, 5 mM KH2PO4, 5% DMSO buffer (Figure 3). Furthermore, a step ladder was used as a size marker with 5 bp differences between adjacent DNA fragments.
Figure 3.
Native PAGE experiments: (A) tel26 G-quadruplex, (B) Pu22 G-quadruplex and (C) ds12 duplex samples were loaded at a 2 μM concentration in the absence and presence of different Dicentrine molar equivalents (from 1 to 10) and analyzed on 20% native PAGE. Lanes L: 5 bp DNA ladder. Gels were visualized with GelGreen™ staining.
Native PAGE experiments showed a single band for tel26 G-quadruplex in the absence of Dicentrine: the comparison with the migration of the ladder fragments confirmed a monomeric G-quadruplex fold for this oligonucleotide (Figure 3A). The addition of 1 to 10 molar equivalents of Dicentrine with respect to the DNA did not detectably affect the mobility of the oligonucleotide, suggesting that the interaction with the ligand did not alter the tel26 G-quadruplex monomeric fold (Figure 3A).
As regards the Pu22 G-quadruplex, a single band was observed in the absence of Dicentrine, and even in this case, no variation in the oligonucleotide migration was detected at the different Pu22 G-quadruplex/Dicentrine ratios explored (Figure 3B).
Finally, gel electrophoresis studies on free ds12 duplex revealed the presence of two bands: one main band with lower mobility, associated to the bimolecular duplex fold, and one less intense band with higher mobility, attributable to the single DNA strands derived from the partial unfolding of the duplex (Figure 3C). A slight increase in the single-strand band intensity was observed upon the addition of Dicentrine, suggesting a partial unwinding of the bimolecular duplex induced by the ligand, in agreement with a previous report [22].
Overall, in full agreement with the CD titration results, the PAGE experiments proved that the main conformations of the investigated DNA secondary structures were preserved upon Dicentrine binding.
2.5. Fluorescence Spectroscopy Studies of Dicentrine with G-Quadruplexes and Control Duplex
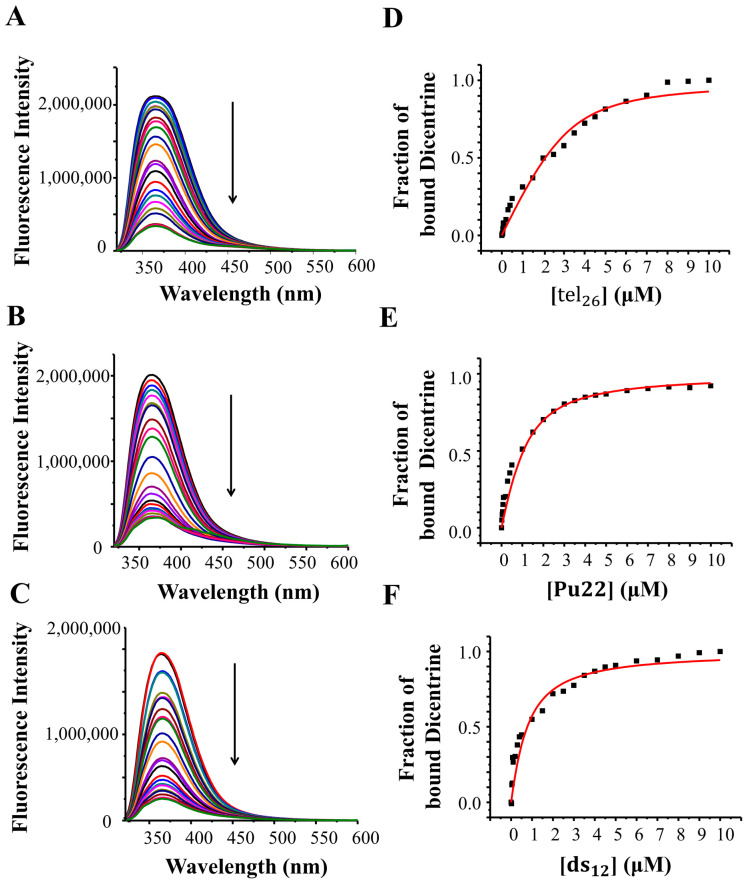
The fluorescence spectrum of a 2 μM solution of Dicentrine was recorded in 5 mM KCl, 5 mM KH2PO4, 5% DMSO buffer (pH 7). Dicentrine showed one strong emission band at 367 nm (Figure 4, black lines). Thus, the interaction of the ligand with tel26 G-quadruplex, Pu22 G-quadruplex and ds12 duplex was studied using fluorescence spectroscopy through reverse titration experiments. In detail, fluorescence titrations were carried out at a fixed concentration of the ligand (i.e., 2 μM) by adding increasing amounts of tel26, Pu22 or ds12, previously annealed in 5 mM KCl, 5 mM KH2PO4, 5% DMSO buffer (pH 7). Upon each addition, the corresponding fluorescence spectrum was recorded after stabilizing the signal. The fraction of bound ligand was calculated from the obtained fluorescence intensity values and plotted as a function of the DNA concentration. These data were then fitted with the independent and equivalent-sites model [36] in order to derive the related binding constants and stoichiometries.
Figure 4.
Left: Fluorescence emission spectra obtained by adding increasing amounts of (A) tel26 G-quadruplex, (B) Pu22 G-quadruplex and (C) ds12 duplex to 2 μM solutions of Dicentrine. Arrows indicate the fluorescence intensity changes upon increasing the DNA concentration. Right: Representative binding curves obtained by plotting the fraction of bound Dicentrine to (D) tel26 G-quadruplex, (E) Pu22 G-quadruplex and (F) ds12 duplex as a function of the DNA concentration. The black squares represent the data from a single experimental replicate; the red line represents the best fit obtained using an independent and equivalent-sites model.
Significant fluorescence quenching was observed upon the titration of Dicentrine with all the investigated oligonucleotides (Figure 4, left panels). Fitting curves related to the fluorescence data for Dicentrine with the tel26 and Pu22 G-quadruplexes and the ds12 duplex are shown in Figure 4, right panels. Notably, some preference for the hybrid tel26 G-quadruplex (Kb = (2.8 ± 1.2) × 106 M−1) compared with the parallel Pu22 G-quadruplex (Kb = (0.8 ± 0.5) × 106 M−1) was observed, in line with the affinity trend obtained with the G4-CPG assay. In addition, a good G-quadruplex vs. duplex selectivity was found, considering the significant difference of one order of magnitude in their binding constants (Kb = (3.2 ± 0.4) × 105 M−1), in full agreement with the G4-CPG assay. Binding stoichiometries of 1:1, 1:2 and 1:5 were obtained for the tel26 G-quadruplex/Dicentrine, Pu22 G-quadruplex/Dicentrine and ds12 duplex/Dicentrine systems, respectively, clearly showing that their binding to G-quadruplex structures is more specific compared with duplex DNA.
2.6. Molecular Modeling
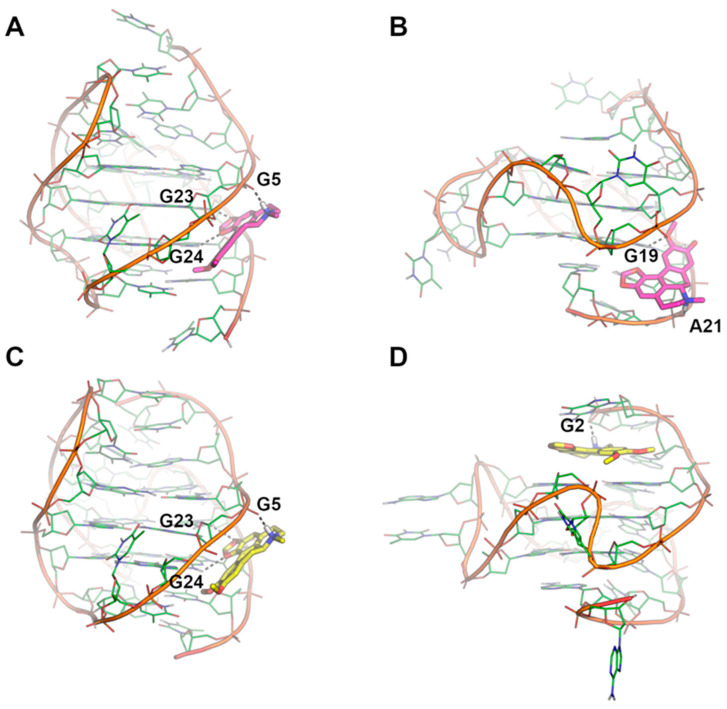
The binding mode of Dicentrine to target G-quadruplexes tel26 and Pu22 was investigated using a combination of molecular docking and molecular dynamics (MD) simulations. According to our previous study on the parent natural compounds [9], Dicentrine was docked into the groove near the 3′-end of the first NMR model of the target G-quadruplex structures in the AutoDock program [37], and the docking complexes were relaxed for 500 ns in unrestrained MD simulations carried out on explicit solvent and K+ ions (Figure 5).
Figure 5.
Binding modes of Dicentrine to tel26 and Pu22 G-quadruplexes as predicted by molecular docking and MD simulations. (A) Docking-based binding mode of Dicentrine to the first NMR model of tel26 (PDB-ID: 2JPZ) [38]. (B) Docking-based binding mode of Dicentrine to the first NMR model of Pu22 (PDB-ID: 1XAV) [39]. (C) Representative frame extracted from the MD trajectory of tel26/Dicentrine complex. (D) Representative frame extracted from the MD trajectory of Pu22/Dicentrine complex. G-quadruplexes are shown as green cartoon and lines. Dicentrine is shown using sticks and is colored magenta in the docking poses and yellow in the poses obtained by MD simulations. Polar contacts of Dicentrine are highlighted by black dashed lines; nucleotides involved in polar interactions with Dicentrine are labeled.
While molecular docking suggested that Dicentrine is potentially able to fit the groove of the target G-quadruplexes (binding energy to tel26 and Pu22 = −7.18 and −8.07 kcal/mol, respectively), differences in the binding mode to the tel26 and Pu22 G-quadruplexes were observed by MD simulations.
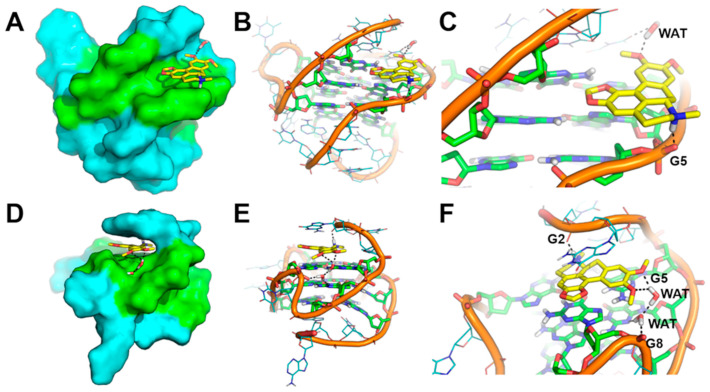
In the case of the tel26 G-quadruplex, MD simulations clearly showed that the compound binds stably and persistently to the binding site identified by docking with the groove of the G-quadruplex structure, in close proximity to G4-G6, T8 and G23-T25 (Figure 5A,C and Figure 6A–C). Within this binding site, Dicentrine interacts with the phosphate backbone of G5 from the middle G-tetrad and establishes a water-bridged H-bond with T8 from the loop (Figure 6B,C).
Figure 6.
Representative MD structures describing the interaction between Dicentrine and tel26 and Pu22 G-quadruplexes. (A) Surface representation of tel26. (B) Cartoon representation of tel26. (C) Magnification of the binding mode of Dicentrine in the groove of tel26. (D) Surface representation of Pu22. (E) Cartoon representation of Pu22. (F) Magnification of the binding mode of Dicentrine on top of the G-tetrad of Pu22. Nucleotides from G-tetrads are colored green, other nucleotides are colored cyan. Dicentrine is shown as sticks and colored yellow. Water molecules bridging the ligand and G-quadruplexes are shown as sticks. Polar contacts are highlighted by black dashed lines. Nucleotides and water molecules (WAT) involved in polar interactions with Dicentrine are labeled in panels (C,F).
MD simulations of Pu22/Dicentrine interactions provided very interesting results. Indeed, while docking highlighted a putative binding site within the groove of the Pu22 G-quadruplex in proximity to G19 and T20, this site proved very unstable in MD simulations and was rapidly left by Dicentrine to move into the bulk solvent. However, the remarkable propensity of Dicentrine to bind the Pu22 G-quadruplex was highlighted by the fast transfer of the compound from the solvent to the G-quadruplex, where it preferentially recognizes the G-tetrad formed by G4, G8, G13 and G17 (Figure 6D). It is worth noting that this site was not accessible in docking simulations because it is occupied by A3 in the reference NMR structure used as a rigid receptor [39]. In MD simulations, Dicentrine displaces A3 and stacks on top of the G-tetrad in a sandwich-like conformation with G2 (Figure 6D,E). Additionally, Dicentrine interacts directly with the phosphate backbone of G2 and through water-bridged connections with G5 and G8 (Figure 6F). These results corroborate the relevance of MD simulations in the investigation of the binding mode of small molecules to G-quadruplex structures.
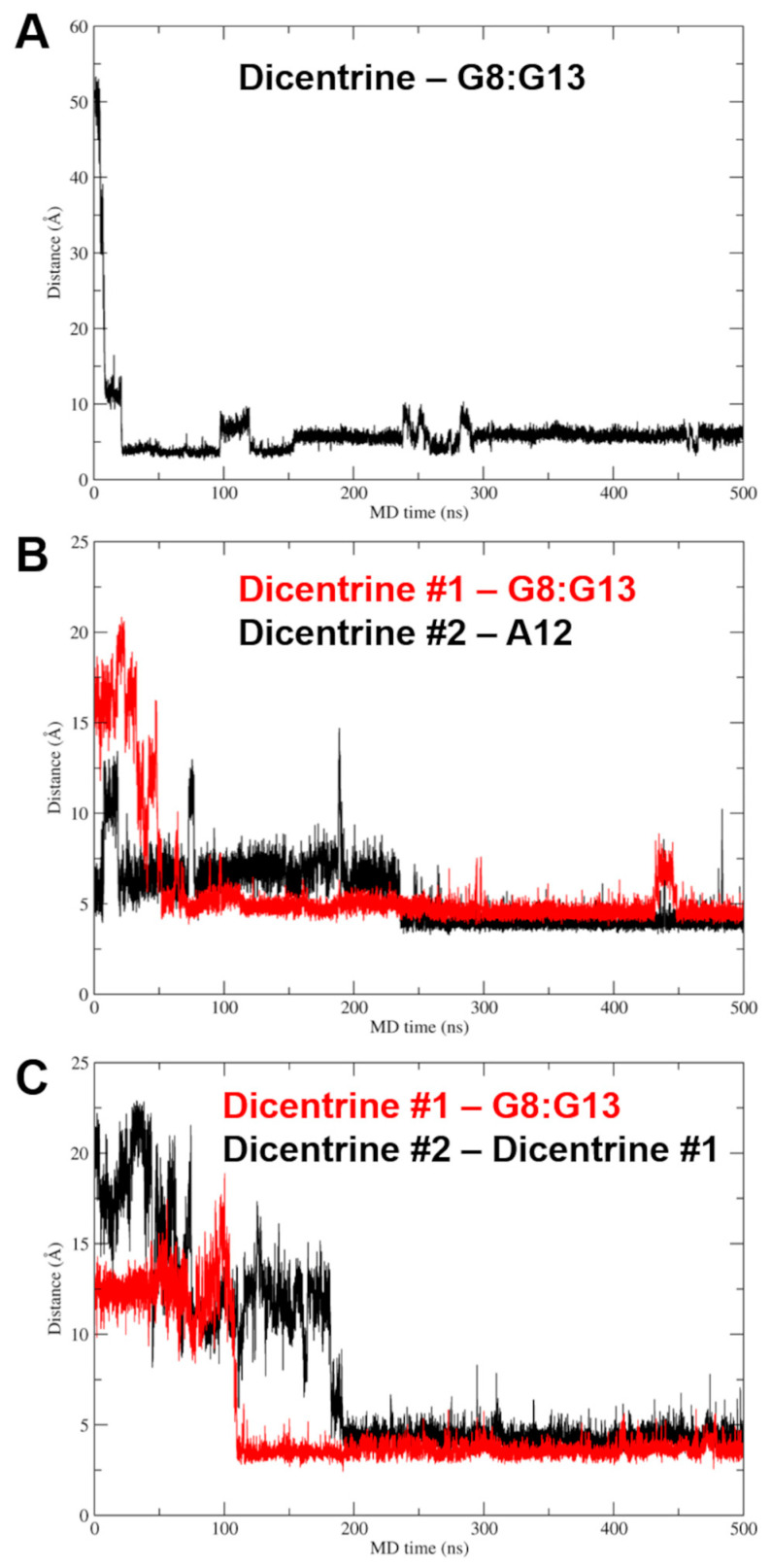
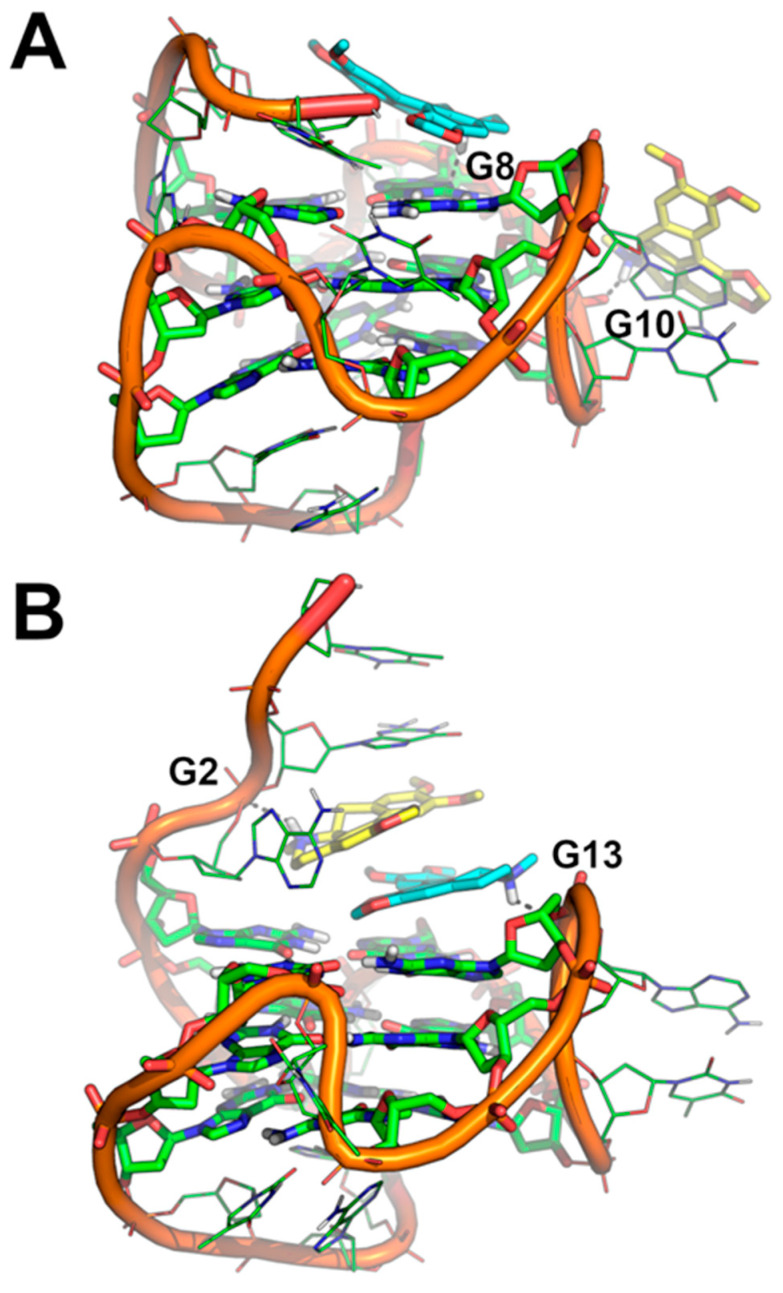
Based on the 1:2 DNA/ligand binding stoichiometry of Dicentrine to Pu22 G-quadruplex observed with fluorescence spectroscopy, intermolecular recognition was investigated using MD simulations by randomly placing two molecules of Dicentrine in the simulation box at a distance higher than 30 Å from the Pu22 G-quadruplex, as well as from each other. In agreement with the binding mode shown in Figure 6D–F, the results of these MD simulations clearly suggest that Dicentrine preferentially stacked onto the G-tetrad of the Pu22 G-quadruplex formed by G4, G8, G13 and G17, as evidenced by one of the two Dicentrine molecules in both MD replicas (Figure 7 and Figure 8). Notably, the primary binding site fully corresponds to the site observed in MD simulations carried out on the docking complex in a 1:1 stoichiometry (Figure 6D–F). On the other hand, the second Dicentrine molecule binds to different sites of the Pu22 G-quadruplex, including (i) the groove of the Pu22 G-quadruplex, including the loop T11-A12, by stacking on A12 (Figure 8A) and (ii) stacking on top of the first Dicentrine molecule on the G-tetrad at the 5′-end of the G-quadruplex in a sandwich-like conformation with G2 (Figure 8B). Interestingly, the interaction with these secondary sites seems to occur after the first Dicentrine molecule is bound, and it is apparently characterized by lower conformational stability (Figure 7B,C).
Figure 7.
Plots of the distance between the mass center of Dicentrine and target nucleotides in MD simulations. (A) Distance between Dicentrine and the G8:G13 pair in the G-tetrad in the MD trajectory of the Pu22/Dicentrine complex with 1:1 stoichiometry. (B) Distance between Dicentrine #1 and the G8:G13 pair in the G-tetrad (red) and distance between Dicentrine #2 and the A12 mass center (black) in the first MD replica on the Pu22/Dicentrine complex with 1:2 stoichiometry. (C) Distance between Dicentrine #1 and the G8:G13 pair in the G-tetrad (red) and distance between Dicentrine #2 and the Dicentrine #1 mass center (black) in the second MD replica on the Pu22/Dicentrine complex with 1:2 stoichiometry.
Figure 8.
(A,B) Binding mode of two molecules of Dicentrine to the Pu22 G-quadruplex investigated using two independent replicas of MD simulations. The representative frame extracted by the MD trajectories through cluster analysis is shown. Pu22 is represented as green cartoon and lines; residues forming the G-tetrads are shown as sticks. Dicentrine #1 and Dicentrine #2 are shown as sticks and colored cyan and yellow, respectively. Polar contacts are highlighted by black dashed lines; nucleotides involved in polar interactions with Dicentrine are labeled.
Finally, the relative binding energy of Dicentrine to the tel26 G-quadruplex and the primary and secondary sites of the Pu22 G-quadruplex identified with MD simulations was calculated using the Molecular Mechanics Generalized Born Surface Area (MM-GBSA) approach. The results clearly highlight that Dicentrine binding to the 5′-end G-tetrad of Pu22 is thermodynamically favored compared with binding to other sites, i.e., A12 or already-bound Dicentrine. The calculated delta energy of binding of Dicentrine to different systems is reported in Table 4 and corroborates the hypothesis that, considering the 1:2 DNA/ligand stoichiometry, the first molecule of Dicentrine binds with higher affinity to a primary site located on top of the G-tetrad at the 5′-end, while the second Dicentrine molecule binds to secondary sites with lower affinity and conformational stability. Notably, the delta energy of the binding of Dicentrine to the tel26 G-quadruplex is fully comparable to the one predicted for the interaction within the primary site of the Pu22 G-quadruplex, in agreement with the fluorescence data (Table 4).
Table 4.
Delta energy of the binding of Dicentrine to the Pu22 and tel26 G-quadruplexes as predicted using the MM-GBSA approach.
| Compound | Binding Energy to 5′-End G-Tetrad of Pu22 (kcal/mol ± SEM) |
Binding Energy to A12 (kcal/mol ± SEM) |
Binding Energy to Dicentrine #1 (kcal/mol ± SEM) | Binding Energy to tel26 (kcal/mol ± SEM) |
|---|---|---|---|---|
| Dicentrine #1 | −27.43 ± 4.03 (R1) a −26.85 ± 4.65 (R2) a |
- | - | −26.34 ± 3.68 |
| Dicentrine #2 | - | −16.13 ± 1.77 | −19.65 ± 2.75 | - |
a R1 = MD replica #1; R2 = MD replica #2.
Overall, these results provide a consistent structural explanation of the interaction of Dicentrine with the tel26 and Pu22 G-quadruplexes in solution, as observed with spectroscopic and electrophoretic techniques.
2.7. Biological Assays
Aiming to further validate Dicentrine as a putative selective antitumor candidate targeting G-quadruplex structures, in-cell assays were performed. Compared with the above experimental and in silico analyses, the use of biological models introduces an additional level of complexity, thus representing an important step in selecting candidate leads that are potentially suitable for preclinical and clinical trials.
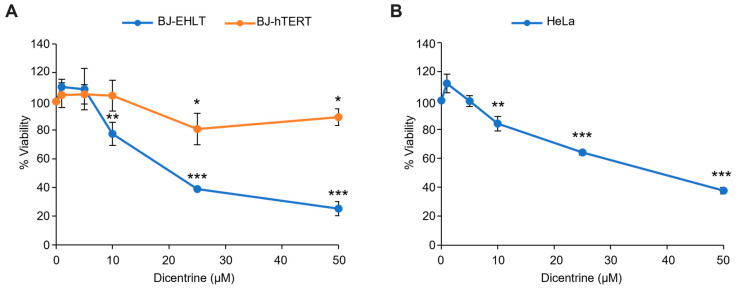
In this regard, Dicentrine was first evaluated for its properties as an antitumor agent. In detail, human-transformed BJ fibroblasts (BJ-EHLT cells) were treated with increasing doses (from 1 to 50 µM) of Dicentrine and, after 72 h of treatment, cell viability was evaluated. Interestingly, the results of the crystal violet assay (Figure 9A, blue line) evidenced the dose-dependent activity of Dicentrine, with an IC50 value of about 25 µM.
Figure 9.
Effect of Dicentrine on cell viability. (A) BJ-EHLT (blue line) and BJ-hTERT (orange line) fibroblasts or (B) HeLa cells were treated for 72 h with increasing doses of Dicentrine (1, 5, 10, 25 and 50 µM) or with DMSO (negative control). Cell viability was determined with a colorimetric crystal violet assay. Results are expressed as the percentage of viable cells over the negative control. Histograms show the mean values ± SD of three independent experiments. * p < 0.05; ** p < 0.01; *** p < 0.001.
Since anticancer drugs are much more effective if their selectivity for cancer versus normal cells is higher, viability experiments were also carried out in the normal counterpart of BJ-EHLT cells, i.e., immortalized BJ fibroblasts, BJ-hTERT. As evidenced by the growth curve, normal cells were almost insensitive to Dicentrine at all the evaluated doses, thus underlying the high selectivity of this G-quadruplex ligand against cancer cells (Figure 9A, orange line).
BJ-EHLT and BJ-hTERT, derived from the same cell line of origin (BJ fibroblasts), represent extremely valid models to test the selectivity of treatments against tumor cells. Nevertheless, the use of BJ-EHLT is frequently debated because these cells—obtained through sequential immortalization and transformation events—are considered an “artificial” tumor model. Based on these considerations, the effectiveness of Dicentrine treatment was also evaluated in HeLa, a human cervical cancer cell line. Notably, the crystal violet assay (Figure 9B), fully reproducing the results obtained in BJ-EHLT, confirmed the antitumor activity of Dicentrine (IC50 value of about 25 µM).
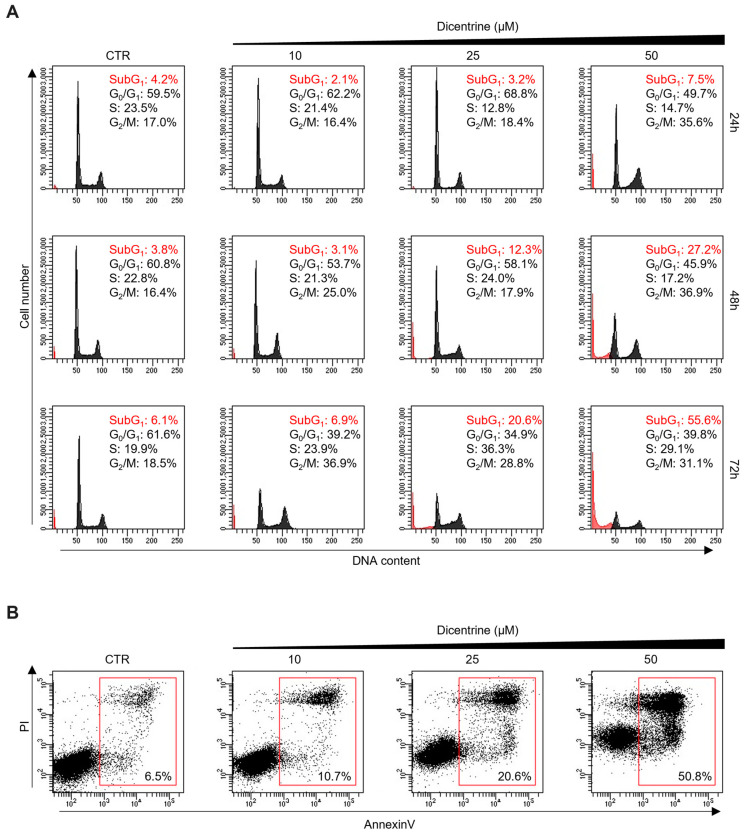
In parallel, the effect of Dicentrine on cancer cells was also evaluated using a fluorescence-associated cell sorter (FACS). Interestingly, time-course cell cycle analyses (from 24 to 72 h) showed that the treatment of HeLa cells with different doses of Dicentrine (10, 25 and 50 µM) promoted time- and dose-dependent cell cycle arrest, evidenced by the accumulation of cells in the G2/M phase (Figure 10A). In addition, FACS analyses showed the appearance of a subG1 peak, indicative of the capability of Dicentrine to induce cell apoptosis in treated cells (Figure 10A). Moreover, Annexin V analyses confirmed the capability of Dicentrine to induce apoptosis in treated HeLa cells, reaching about 20% and 50% of Annexin V-positive cells upon treatment with 25 and 50 µM of Dicentrine for 72 h, respectively (Figure 10B).
Figure 10.
Dicentrine affects the cell cycle and induces apoptosis in cancer cells. HeLa cells were treated with DMSO (negative control) or increasing doses of Dicentrine (10, 25, 50 µM) for the indicated times and then processed for flow cytometry analysis. (A) Cells were stained with propidium iodide (PI), and an analysis of the cell cycle was performed. Quantification of the percentage of cells in subG1 and G2/M phases are reported in red and in black, respectively. (B) Two-dimensional scatter plots of Annexin V analysis performed on HeLa cells untreated or treated with growing doses of Dicentrine (10, 25, 50 µM) for 72 h. Red boxes evidence the populations of Annexin V-positive cells. One representative image is shown for each experiment.
Altogether, these results highlighted the effectiveness of Dicentrine in promoting a potent and selective antitumor activity by inducing cell cycle arrest through apoptosis.
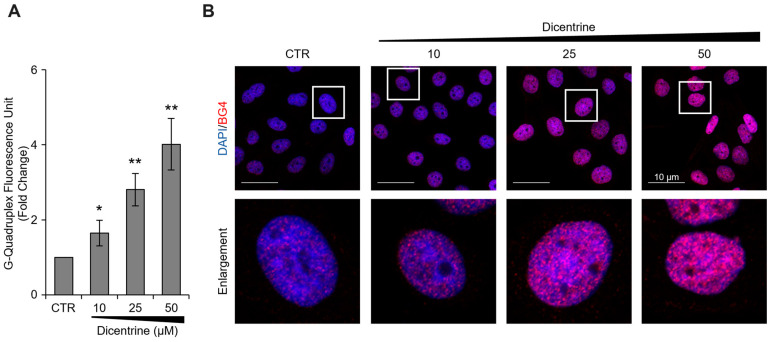
In order to investigate the mechanisms by which Dicentrine promotes apoptosis in cancer cells, additional in vitro tests were carried out. In more detail, the capability of Dicentrine to stabilize G-quadruplex structures in cells was evaluated. For these experiments, HeLa cells were treated with different doses of Dicentrine (10, 25 and 50 µM) and, after 24 h of treatment, the G-quadruplex structures—detected with a specific antibody—were evaluated using immunofluorescence (IF) microscopy. In agreement with the biophysical results, Dicentrine was found to promote, in a dose-dependent manner, the stabilization of G-quadruplex structures in tumor cells (Figure 11).
Figure 11.
Biological evaluation of the G-quadruplex-stabilizing activity of Dicentrine. Immunofluorescence analysis of G-quadruplex structures in Hela cells treated for 24 h with DMSO (negative control) or with increasing doses of Dicentrine (10, 25, 50 µM). (A) Quantitative analysis of the anti-G-quadruplex signal. For each panel, at least 25 cells/conditions were analyzed using the ImageJ software, and the results were expressed as the fold change in the fluorescence intensity over the negative control. Histograms show the mean ± SD of three independent experiments performed in triplicate. * p < 0.05; ** p < 0.01. (B) ) Upper Panels: representative images of confocal sections (63×) used for the detection of G-quadruplex structures. For each experimental condition, merged images of DAPI counterstained nuclei (blue) and G-quadruplex structures (red) are shown. Lower panels: enlargements (4×) of a single cell from each of the upper panels (white squares) are shown. Scale bars (10 µm) are reported in the figures.
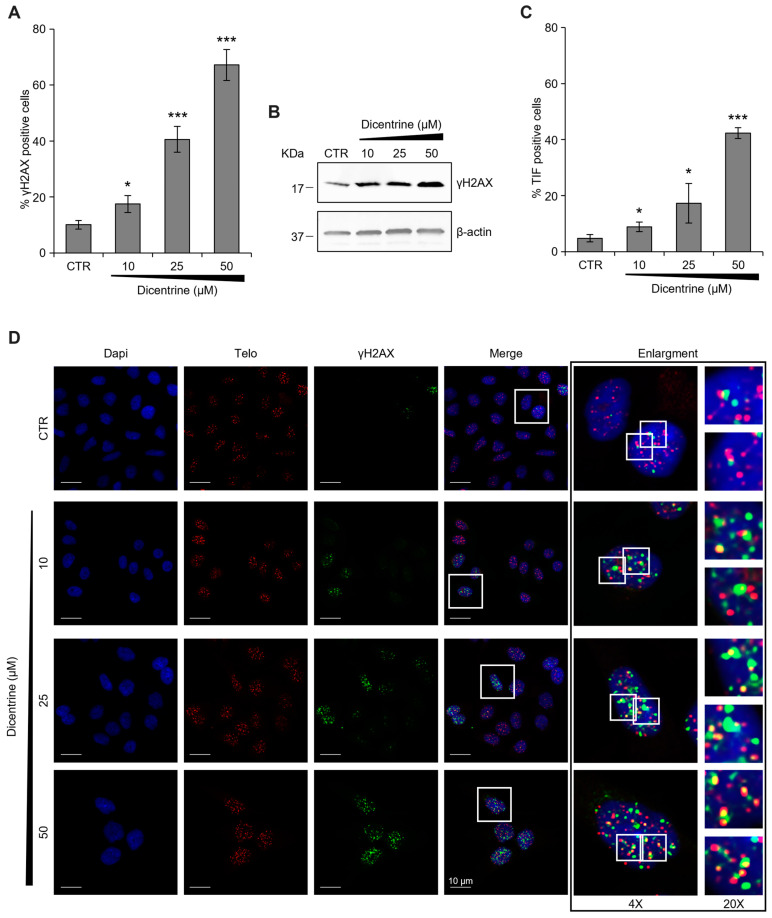
Additionally, considering that the antitumor activity of G-quadruplex ligands is mainly due to their capability to induce DNA damage, the amount of phosphorylated histone H2AX (γH2AX), a typical hallmark of DNA double-strand breaks [40], was evaluated in the cells treated with Dicentrine. Interestingly, complementary technical approaches, based on both confocal microscopy (IF) and biochemical (Western blot) analyses, evidenced a robust increase in γH2AX levels in Dicentrine-treated cells compared with their untreated counterparts (Figure 12A,B), highlighting the efficacy of the treatment in inducing DNA damage in tumor cells. For completeness, we then evaluated whether Dicentrine-induced DNA damage was localized at the telomeres. Thus, a fluorescence in situ hybridization (FISH) assay was performed, aimed at defining the number of γH2AX spots colocalizing with a fluorescent telomeric probe (Telomere Induced Foci, TIF). Interestingly, quantitative confocal analyses evidenced that Dicentrine promotes a dose-dependent increase in the percentage of TIF-positive cells (TIF ≥ 4), thus demonstrating the effective activity of this G-quadruplex ligand at the telomeres (Figure 12C,D).
Figure 12.
Dicentrine induces telomere-specific DNA damage. (A) HeLa cells were treated with DMSO (negative control) or with the indicated doses of Dicentrine (10, 25, 50 µM) for 24 h. Cells were processed for telomeric FISH combined with immunofluorescence, and the amount of DNA damage was evaluated. (A) Quantitative analysis of damaged cells. Results are expressed as a percentage of γH2AX positive cells over the negative control. (B) Western blot analysis of γH2AX levels. Β-actin protein levels were used as the loading control. (C) Quantitative analysis of the percentage of TIF-positive cells. Cells with at least four γH2AX-Telo colocalizations were scored as TIF-positive. (D) Representative images of confocal sections (63×) used for the quantitative analyses reported in (A,C). DAPI counterstained nuclei (blue), telomeres (red), γH2AX (green) and merged images are shown. For each experimental condition, 4× and 20× enlargements of merged fields are reported (white boxes). Scale bars (10 µm) are shown in the figures. Histograms show the mean ± SD of three independent experiments performed in triplicate. * p < 0.05; *** p < 0.001.
Based on this robust set of collected biological data, it is possible to conclude that Dicentrine—through its capability to preferentially target G-quadruplex structures localized at the telomeres—might represent a valid lead compound in developing a new class of potent and selective anticancer drugs.
3. Materials and Methods
3.1. Chemistry
All the tested compounds are known structures belonging to the inhouse library of natural products available from the Organic Chemistry Laboratory of the Department of Chemistry and Technology of Drugs of Sapienza University of Rome, Italy. The chemical identity of noncommercial compounds was assessed by rerunning NMR experiments and proved to be in agreement with the literature data [41].
Compound 1 (Canadine, or (S)-9,10-dimethoxy-5,8,13,13a-tetrahydro-6H-[1,3]dioxolo [4,5-g]isoquinolino [3,2-a]isoquinoline) was purchased from Chemische Fabrik Karl Bucher GmbH and used without further purification.
Compound 2 (D-Glaucine, or (S)-1,2,9,10-tetramethoxy-6-methyl-5,6,6a,7-tetrahydro-4H-dibenzo[de,g]quinoline) showed NMR spectra identical to those reported in the literature [41].
Compound 3 (Dicentrine, or (S)-10,11-dimethoxy-7-methyl-6,7,7a,8-tetrahydro-5H-[1,3]dioxolo [4’,5’:4,5]benzo [1,2,3-de]benzo[g]quinoline) was purchased from Biosynth Carbosynth and used without further purification.
Compound 4 (Deguelin, or (7aS,13aS)-9,10-dimethoxy-3,3-dimethyl-13,13a-dihydro-3H-pyrano [2,3-c:6,5-f’]dichromen-7(7aH)-one) was purchased from Biosynth Carbosynth and used without further purification.
Compound 5 (Millettone, or (1S,14S)-7,7-dimethyl-2,8,18,20,24-pentaoxahexacyclo [12.11.0.03,12.04,9.015,23.017,21]pentacosa-3(12),4(9),5,10,15,17(21),22-heptaen-13-one) was purchased from BioTeck and used without further purification.
3.2. G4-CPG Assay
Stock solutions (4 mM) for each compound were prepared by dissolving a known amount of the sample in pure DMSO. A measured volume was taken from the stock solution to obtain a 60 μM compound solution in 50 mM KCl, 10% DMSO, 10% CH3CH2OH aq. solution. The detailed general procedure adopted for the assays is described as follows: weighed amounts of the nude CPG and G-quadruplex-/duplex-functionalized CPG supports (ca. 8 mg) were left in contact with 300 μL of the compound solution in a polypropylene column (4 mL volume, Alltech) equipped with a polytetrafluoroethylene frit (10 μm porosity), a stopcock and a cap [10,11]. After incubation on a vibrating shaker for 4 min, each support was washed with defined volumes of the washing solution (50 mM KCl, 10% DMSO, 10% CH3CH2OH aq. solution) or the releasing solution (2.5 M CaCl2, 15% DMSO aq. solution or pure DMSO), and all the eluted fractions were separately analyzed with UV measurements. After treatment with the releasing solution, able to induce G-quadruplexes and hairpin duplex denaturation, the supports were suspended in the washing solution and then subjected to annealing by taking them at 75 °C for 5 min and then slowly cooling them to room temperature.
The UV measurements were performed on a JASCO V-550 UV-vis spectrophotometer. A quartz cuvette with a path length of 1 cm was used. The UV quantification of the compounds was determined by measuring the absorbance relative to the λmax characteristic of each compound and referring it to the corresponding calibration curves. The errors associated with the % of bound ligands are within ±2%.
3.3. Circular Dichroism
CD spectra were recorded in a quartz cuvette with a path length of 1 cm on a Jasco J-715 spectropolarimeter equipped with a Peltier-type temperature control system (model PTC-348WI). The spectra were recorded at 20 °C in a range of 240–600 nm, with a 2 s response, a 200 nm/min scanning speed and a 2.0 nm bandwidth, and they were corrected by subtracting the background scan with buffer. All the spectra were averaged over 3 scans. The oligonucleotides d[(TTAGGG)4TT] (tel26), d(TGAGGGTGGGTAGGGTGGGTAA) (Pu22) and d(CGCGAATTCGCG) (ds12) were purchased from Biomers (www.biomers.net) as HPLC-purified compounds with a purity of > 99%. The oligonucleotides were dissolved in a 5 mM KCl, 5 mM KH2PO4, 5% DMSO buffer (pH 7) to obtain 2 μM solutions and then annealed by heating them at 95 °C for 5 min, followed by slow cooling to room temperature. CD titrations were obtained by adding increasing amounts of Dicentrine (up to 10 molar equivalents, corresponding to a 20 μM solution in ligand) to the buffer alone or to the oligonucleotide solutions. For the CD melting experiments, the ellipticity was recorded at 290 nm for tel26, 265 nm for Pu22 and 253 nm for ds12 with a temperature scan rate of 0.5 °C/min in a range of 10–95 °C.
3.4. Gel Electrophoresis
The oligonucleotide/Dicentrine mixtures (up to 1:10 ratio) and free oligonucleotide samples, together with an O’RangeRuler™ 5 bp DNA Ladder (ThermoFisher Scientific), were loaded and analyzed on native 20% polyacrylamide (19:1 acrylamide:bisacrylamide) gels. TBE 1× supplemented with 10 mM KCl was used as a running buffer. The oligonucleotide samples loaded on the gels were 2 µM in oligonucleotide concentration per strand in 5 mM KCl, 5 mM KH2PO4, 5% DMSO buffer. No migration marker was used. Samples were electrophoresed for 3 h at 100 V at room temperature. The bands were visualized using GelGreen™ staining.
3.5. Fluorescence Spectroscopy
Fluorescence spectra were recorded at 20 °C on a HORIBA JobinYvon Inc. FluoroMax®-4 spectrofluorometer equipped with an F-3004 Sample Heater/Cooler Peltier Thermocouple Drive by using a quartz cuvette with a 1 cm path length. For the fluorescence titration experiments with Dicentrine, an excitation wavelength of 307 nm was used, and the spectra were registered in a range of 320–600 nm.
Titrations were carried out at a fixed concentration (2.0 μM) of ligand. Increasing amounts of tel26, Pu22 or ds12 (up to 10 μM concentration) were added from 120 μM annealed stock solutions of each DNA sample dissolved in a 5 mM KCl, 5 mM KH2PO4, 5% DMSO buffer (pH 7). After each addition, the system was allowed to equilibrate for 10 min before recording the spectra.
The fraction of bound Dicentrine was calculated from a fluorescence intensity of 367 nm and reported in a graph as a function of the DNA concentration. The fraction of the bound ligand was determined using the equation
| α = (Y − Y0)/(Yb − Y0) |
where Y, Y0 and Yb are the values of fluorescence emission intensity at the maximum of each titrant concentration at the initial and final state of the titration. These points were fitted with an independent and equivalent-sites model [36] using the Origin 8.0 program.
The equation of the independent and equivalent-sites model is as follows:
where α is the mole fraction of the ligand in the bound form; [L]0 is the total ligand concentration; [DNA] is the added DNA concentration; n is the number of equivalent and independent sites in the DNA structure; and Kb is the binding constant.
3.6. Molecular Modeling
Molecular docking and MD simulations were carried out according to the protocol previously described [9]. Briefly, molecular docking was carried out with AutoDock4 [37] using the first NMR model of target G-quadruplexes tel26 (PDB-ID: 2JZP) [38] and c-myc (Pu22 sequence) (PDB-ID:1XAV) [39] as rigid receptors. The binding site on tel26 was centered on the groove near the 3′-end in the region surrounded by A3-G6 and G23-T26, while the binding site on Pu22 was centered on the groove near the 3′-end in the region defined by the A3-G6 and G19-A22 nucleotides. MD simulations were run with Amber18, starting from the docking complexes [42], using the General Amber Force Field for the parametrization of Dicentrine and the OL15 force field for the parametrization of the G-quadruplexes [43,44,45]. Macromolecular complexes were solvated in a rectangular box of TIP3P-type water molecules, while K+ ions were added to neutralize the total charge of the system. Production of MD trajectories was extended up to 500 ns at constant pressure and without positional restraints for every MD simulation. For MD-based intermolecular recognition studies, the initial system was built by placing the molecule(s) of Dicentrine at a distance higher than 30 Å to each other and the G-quadruplex. Two independent MD replicas of 500 ns each were run to achieve statistical significance. Analysis of MD trajectories was carried out with CPPTRAJ, while the delta energy of the binding of Dicentrine to both tel26 and Pu22 was calculated with the MM-PBSA.py software [46,47].
3.7. Biological Experiments
3.7.1. Cells and Culture Condition
Human fibroblasts (BJ) and human cervical cancer cells (HeLa) were purchased from American Type Culture Collection (ATCC, CRL-2522, CCL-2). BJ-hTERT cells were obtained by infecting primary BJ cells with a retrovirus carrying hTERT (Addgene plasmid #1773). BJ-EHLT cells were derived from the transformation of BJ fibroblasts with hTERT and SV40 early region [48]. BJ-hTERT, BJ-EHLT and HeLa were grown in Dulbecco Modified Eagle Medium (DMEM; EuroClone, Milan, Italy; ECM0728L) supplemented with 10% Fetal Bovine Serum (FBS, ThermoFisher Scientific/Gibco, Waltham, MA, USA; 10,270–106), 2 mM l-glutamine and antibiotics at 37 °C in a 5% CO2–95% air atmosphere.
3.7.2. Viability Assay (Crystal Violet)
BJ-EHLT, BJ-hTERT and HeLa cells were seeded in 24-well plates at a density of 6 × 103, 12 × 103 and 8 × 103 for each well, respectively. After 24 h, the cells were treated with different concentrations of Dicentrine (0, 1, 5, 10, 25, 50 µM) for 72 h. Upon treatment completion, the cell medium was removed, and the wells were washed twice in phosphate-buffered saline (PBS) and fixed with 4% formaldehyde for 15 min at room temperature (RT). After washing, 500 µL of crystal violet staining solution (Sigma-Aldrich, St. Louis, MO, USA) was added to each well and incubated for 30 min at RT. Finally, the plates were rinsed twice with water and air-dried, and cell pellets were dissolved in 400 µL of a 10% aqueous solution of acetic acid. In total, 200 µL of each sample was transferred to a 96-well plate, and the optical density was measured at 570 nm (OD570) with an ELISA reader (Thermo Scientific, Waltham, MA, USA). The average absorbance in each condition was used to calculate the viability expressed as a percentage of treated vs. untreated conditions.
3.7.3. Flow Cytometry
For the analysis of the cells’ progression through the cell cycle phases, HeLa cells were treated with 10, 25 and 50 µM of Dicentrine for the indicated times. Both floating and adherent cells were harvested, washed twice with PBS 1× and finally fixed in 70% CH3CH2OH at 4 °C overnight. Next, cells were washed with PBS 1×, stained with 50 mg/mL of propidium iodide (PI; Invitrogen-eBioscience) in the presence of 75 kU/mL of RNase and analyzed using FACSCelesta (BD Biosciences, San Jose, CA, USA).
The evaluation of apoptosis was carried out following the Annexin V assay versus PI staining, as previously described [49]. Briefly, both floating and adherent cells were harvested and suspended in Annexin-binding buffer (final concentration of 1 × 106 cells/mL). Thus, cells were incubated with fluorescein isothiocyanate–Annexin V (Annexin-FITC) and PI for 15 min at RT away from the light and immediately analyzed by using FACSCelesta (BD Biosciences, San Jose, CA, USA). For each condition, 20,000 events were measured, setting 488 and 525 nm as the excitation and emission wavelengths, respectively. Data obtained were analyzed with the FACS Diva software (BD Biosciences).
3.7.4. Immunofluorescence (IF) Experiments
HeLa cells were seeded on glass coverslips in a 24-well plate at a density of 2 × 104 cells/well. After 24 h of treatment with Dicentrine (0, 10, 25, 50 µM), cells were fixed in 4% formaldehyde in PBS (10 min at RT) and permeabilized with 0.25% Triton X-100 (5 min at RT). For immuno-labeling, cells were incubated with a mouse mAb that specifically recognizes DNA/RNA G-quadruplex structures (BG4 antibody; Absolute Antibody, Oxford, UK, #Ab00174-1.1). After 2 h of incubation at RT, cells were washed three times with PBS 1X and then incubated for an additional 1 h with an antibody anti-mouse IgG (H + L), F(ab’)2 Fragment (Alexa Fluor 555 Conjugate; Cell Signaling Technology, Danvers, MA, USA, #4409S). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich, D9542), and fluorescence signals were recorded by using a Zeiss Laser Scanning Microscope 510 Meta (63× magnification) (Zeiss, Jena, Germany).
IF experiments were quantified using ImageJ (version 1.53e, National Institutes of Health, NIH, Bethesda, MD, USA). At least 25 cells were screened for each condition, and the results were expressed as the fold change of the fluorescence intensity over the negative control. Histograms show the mean ± SD of three independent experiments.
3.7.5. Fluorescence In Situ Hybridization (FISH) Assays
In order to evaluate the DNA damage, immunofluorescence experiments were combined with telomeric FISH assays. After fixation and permeabilization (as reported for the IF experiments), cells were blocked with 3% BSA in PBS 1× for 1 h and incubated with the mouse mAb anti-phospho-histone H2AX (γH2AX Ser139; Millipore, 05-636) at 4 °C overnight. After two washes with 0.05% Triton X-100 in PBS 1×, the cells were incubated with the secondary antibody anti-mouse IgG (H + L), F(ab’)2 Fragment (Alexa Fluor 488 Conjugate) for 1 h at RT and then washed twice with PBS 1×. Successively, cells were fixed in 4% formaldehyde (PBS 1X) again and subjected to standard telomere DNA FISH, as previously described [50]. Nuclei were stained with DAPI. For a quantitative analysis of γH2AX positivity, at least 250 cells/condition were scored in triplicate. For Telomere-Induced Foci (TIFs) analysis, at least 25 γH2AX-positive cells on a single plane were scored. Nuclei showing at least four colocalizations of a TelC-Cy3 telomeric probe (Panagene, Daejeon, Korea) and γH2AX were considered TIF-positive. Fluorescence signals were recorded with a Zeiss Laser Scanning Microscope 510 Meta (63× magnification) (Zeiss, Jena, Germany).
3.7.6. Western Blot Analysis
Western blot analysis was performed as previously reported [51]. Briefly, HeLa cells were collected and resuspended in lysis buffer (50 mM Tris-HCl pH 7.5, 5 mM EDTA, 250 mM NaCl, 0.1% Triton) complemented with protease (Thermo Scientific, A32953) and phosphatase inhibitors (Thermo Scientific, 88667). Total proteins were fractionated using SDS-polyacrylamide gel electrophoresis and then transferred to a nitrocellulose membrane (Amersham, Arlington Heights, IL, USA). Membranes were probed with the following primary antibodies: mouse mAb anti-phospho-histone H2AX (γH2AX; Millipore, 05-636) and mouse mAb anti-β-actin (Sigma-Aldrich, A5441).
3.7.7. Statistical Analyses
Experiments were replicated three times, and the data were expressed as means ± standard deviation (SD). GraphPad Prism 6 was used for the statistical analysis, and the differences between groups were analyzed using an unpaired Student’s t-test. Differences were considered statistically significant for * p < 0.05; ** p< 0.01; *** p < 0.001.
4. Conclusions
A stringent requirement for a G-quadruplex ligand to be putatively advanced to preclinical and clinical studies is the ability to selectively recognize G-quadruplexes by discriminating duplex DNA. In this context, we recently identified five natural compounds (i.e., Bulbocapnine, Chelidonine, Ibogaine, Rotenone and Vomicine) as efficient G-quadruplexes ligands with anticancer properties through a multidisciplinary approach combining computational, biophysical and biological studies. Here, aiming to improve the effectiveness and the G-quadruplex vs. duplex DNA selectivity of the previously identified hits, a set of their structural analogs was investigated, including the natural alkaloids Canadine, D-Glaucine and Dicentrine and the flavonoids Deguelin and Millettone.
Within these natural compounds, Dicentrine emerged as the best ligand of telomeric and oncogenic G-quadruplexes in terms of affinity and G-quadruplex vs. duplex selectivity according to the results of the G4-CPG assay. Therefore, it was advanced to further studies in solution in its interaction with G-quadruplex structures to investigate its binding features in depth.
CD and PAGE experiments proved that the main folds of the investigated oligonucleotides were preserved upon the binding of Dicentrine. In parallel, CD melting experiments demonstrated that Dicentrine positively affected the stability of G-quadruplex-forming oligonucleotides, whereas no relevant effect was observed in the control duplex, further corroborating the good G-quadruplex vs. duplex selectivity of this compound, as found using the G4-CPG assay.
Significant fluorescence quenching was observed upon the titration of Dicentrine with the investigated oligonucleotides, further confirming its strong binding to telomeric and oncogenic G-quadruplexes. Moreover, some preference for the hybrid telomeric G-quadruplex (Kb = (2.8 ± 1.2) × 106 M−1) compared with the parallel oncogenic G-quadruplex (Kb = (0.8 ± 0.5) × 106 M−1) was observed, in line with the affinity trend found using the G4-CPG assay. In addition, good G-quadruplex vs. duplex selectivity was found, considering the difference of one order of magnitude in their binding constants (Kb for the model duplex = (3.2 ± 0.4) × 105 M−1)), in full agreement with the G4-CPG assay. Binding stoichiometries of 1:1, 1:2 and 1:5 were obtained for telomeric G-quadruplex/Dicentrine, oncogenic G-quadruplex/Dicentrine and control duplex/Dicentrine systems, respectively, proving that the binding to G-quadruplex structures was more specific compared with duplex DNA.
MD simulations allowed for a deeper insight into the interaction of Dicentrine with G-quadruplex structures. In the case of the telomeric G-quadruplex model, Dicentrine was proved to bind stably and persistently to the groove of the G-quadruplex structure in close proximity to G4-G6, T8 and G23-T25. On the other hand, in the case of the oncogenic G-quadruplex model, Dicentrine preferentially recognized the outer G-tetrad formed by G4, G8, G13 and G17 by displacing A3 and stacking on top of the G-tetrad in a sandwich-like conformation with G2. According to the 1:2 DNA/ligand binding stoichiometry of Dicentrine to the oncogenic G-quadruplex observed with fluorescence spectroscopy, the second Dicentrine molecule was found to bind to different sites of the G-quadruplex, including (i) the groove, including the loop T11-A12, by stacking on A12 and (ii) stacking on top of the first Dicentrine molecule on the G-tetrad at the 5′-end of the G-quadruplex in a sandwich-like conformation with G2.
Then, Dicentrine was evaluated for its anticancer activity and ability to target G-quadruplex structures in cells using biological assays. Altogether, the biological results highlighted the effectiveness of Dicentrine in promoting potent and selective antitumor activity by inducing cell cycle arrest through apoptosis. In agreement with the biophysical data, Dicentrine was found to promote the stabilization of G-quadruplex structures in cancer cells, mainly targeting and damaging telomeric DNA.
Based on all these data, Dicentrine emerged as a strong and selective ligand of G-quadruplexes, even more effective and showing higher anticancer activity than its parent compound, Bulbocapnine. Furthermore, our results allow us to hypothesize that the previously proven anticancer activity of Dicentrine in leukemia cell line K562 in SCID mice could be associated with its ability to target cancer-related G-quadruplex structures in vivo. Overall, Dicentrine can be considered a promising and selective anticancer agent whose activity against cancer cells is well correlated with its ability to selectively target genomic and mainly telomeric G-quadruplexes, discriminating duplex DNA. It thus represents a valid lead compound for the design and evaluation of suitably improved analogs to be advanced to future in vivo studies aimed at the pharmacological development of a new class of potent and selective anticancer drugs that can overcome the toxicity problems associated with current therapeutics.
Author Contributions
Conceptualization, C.P., F.G., M.M. and D.M. (Daniela Montesarchio); methodology, C.P., P.Z. and M.M.; formal analysis, C.P., P.Z. and M.M.; investigation, C.P., F.G., D.M. (Domenica Musumeci), D.Q., P.Z., S.I., C.D. and M.M.; original draft preparation, C.P., F.G., P.Z. and M.M.; review and editing, all authors; visualization, C.P. and M.M.; supervision, F.G., A.B., M.M. and D.M. (Daniela Montesarchio); funding acquisition: A.B., B.B., M.M. and D.M. (Daniela Montesarchio). All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
C.P. was supported by an AIRC fellowship for Italy. The research leading to these results received funding from AIRC under grant IG 2020—ID. 25046—P.I. Montesarchio Daniela. A.B. thanks AIRC for the financial support (grant IG2018 n. 21579 to A.B.). M.M. wishes to acknowledge the financial support by Piano di Sostegno alla Ricerca USiena 2021 F-CUR (Natiplex project).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Nakanishi C., Seimiya H. G-quadruplex in cancer biology and drug discovery. Biochem. Biophys. Res. Commun. 2020;531:45–50. doi: 10.1016/j.bbrc.2020.03.178. [DOI] [PubMed] [Google Scholar]
- 2.Lerner L.K., Sale J.E. Replication of G-quadruplex DNA. Genes. 2019;10:95. doi: 10.3390/genes10020095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Varshney D., Spiegel J., Zyner K., Tannahill D., Balasubramanian S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020;21:459–474. doi: 10.1038/s41580-020-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biffi G., Tannahill D., McCafferty J., Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kosiol N., Juranek S., Brossart P., Heine A., Paeschke K. G-quadruplexes: A promising target for cancer therapy. Mol. Cancer. 2021;20:40. doi: 10.1186/s12943-021-01328-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Platella C., Capasso D., Riccardi C., Musumeci D., Dellagreca M., Montesarchio D. Natural compounds from: Juncus plants interacting with telomeric and oncogene G-quadruplex structures as potential anticancer agents. Org. Biomol. Chem. 2021;19:9953–9965. doi: 10.1039/D1OB01995C. [DOI] [PubMed] [Google Scholar]
- 7.Platella C., Mazzini S., Napolitano E., Mattio L.M., Beretta G.L., Zaffaroni N., Pinto A., Montesarchio D., Dallavalle S. Plant-derived stilbenoids as DNA-binding agents: From monomers to dimers. Chem. Eur. J. 2021;27:8832–8845. doi: 10.1002/chem.202101229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Criscuolo A., Napolitano E., Riccardi C., Musumeci D., Platella C., Montesarchio D. Insights into the small molecule targeting of biologically relevant G-quadruplexes: An overview of NMR and crystal structures. Pharmaceutics. 2022;14:2361. doi: 10.3390/pharmaceutics14112361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Platella C., Ghirga F., Zizza P., Pompili L., Marzano S., Pagano B., Quaglio D., Vergine V., Cammarone S., Botta B., et al. Identification of effective anticancer G-quadruplex-targeting chemotypes through the exploration of a high diversity library of natural compounds. Pharmaceutics. 2021;13:1611. doi: 10.3390/pharmaceutics13101611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Platella C., Musumeci D., Arciello A., Doria F., Freccero M., Randazzo A., Amato J., Pagano B., Montesarchio D. Controlled Pore Glass-based oligonucleotide affinity support: Towards High Throughput Screening methods for the identification of conformation-selective G-quadruplex ligands. Anal. Chim. Acta. 2018;1030:133–141. doi: 10.1016/j.aca.2018.04.071. [DOI] [PubMed] [Google Scholar]
- 11.Platella C., Napolitano E., Riccardi C., Musumeci D., Montesarchio D. Affinity chromatography-based assays for the screening of potential ligands selective for G-quadruplex structures. ChemistryOpen. 2022;11:e202200090. doi: 10.1002/open.202200090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Correché E.R., Andujar S.A., Kurdelas R.R., Lechón M.J.G., Freile M.L., Enriz R.D. Antioxidant and cytotoxic activities of canadine: Biological effects and structural aspects. Bioorg. Med. Chem. 2008;16:3641–3651. doi: 10.1016/j.bmc.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 13.Iwahori A., Hirota Y., Sampe R. NII-Electronic Library Service. Chem. Pharm. Bull. 1970;43:2091. [Google Scholar]
- 14.Huang R.L., Chen C.C., Huang Y.L., Ou J.C., Hu C.P., Chen C.F., Chang C. Anti-tumor effects of d-Dicentrine from the root of Lindera megaphylla. Planta Med. 1998;64:212–215. doi: 10.1055/s-2006-957411. [DOI] [PubMed] [Google Scholar]
- 15.Thamilselvan V., Menon M., Thamilselvan S. Anticancer efficacy of deguelin in human prostate cancer cells targeting glycogen synthase kinase-3 β/β-catenin pathway. Int. J. Cancer. 2011;129:2916–2927. doi: 10.1002/ijc.25949. [DOI] [PubMed] [Google Scholar]
- 16.Ito C., Itoigawa M., Kojima N., Tan H.T.W., Takayasu J., Tokuda H., Nishino H., Furukawa H. Cancer chemopreventive activity of rotenoids from Derris trifoliata. Planta Med. 2004;70:585–588. doi: 10.1055/s-2004-815447. [DOI] [PubMed] [Google Scholar]
- 17.Adsersen A., Kjølbye A., Dall O., Jäger A.K. Acetylcholinesterase and butyrylcholinesterase inhibitory compounds from Corydalis cava Schweigg. & Kort. J. Ethnopharmacol. 2007;113:179–182. doi: 10.1016/j.jep.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 18.Scazzocchio F., Cometa M.F., Tomassini L., Palmery M. Antibacterial activity of Hydrastis canadensis extract and its major isolated alkaloids. Planta Med. 2001;67:561–564. doi: 10.1055/s-2001-16493. [DOI] [PubMed] [Google Scholar]
- 19.Chlebek J., Korábečný J., Doležal R., Štěpánková Š., Pérez D., Hošťálková A., Opletal L., Cahlíková L., Macáková K., Kučera T., et al. In vitro and in silico acetylcholinesterase inhibitory activity of thalictricavine and canadine and their predicted penetration across the Blood-Brain Barrier. Molecules. 2019;24:1340. doi: 10.3390/molecules24071340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cortijo J., Villagrasa V., Pons R., Berto L., Martí-Cabrera M., Martinez-Losa M., Domenech T., Beleta J., Morcillo E.J. Bronchodilator and anti-inflammatory activities of glaucine: In vitro studies in human airway smooth muscle and polymorphonuclear leukocytes. Br. J. Pharmacol. 1999;127:1641–1651. doi: 10.1038/sj.bjp.0702702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang H., Jang S.W., Pak J.H., Shim S. Glaucine inhibits breast cancer cell migration and invasion by inhibiting MMP-9 gene expression through the suppression of NF-κB activation. Mol. Cell. Biochem. 2015;403:85–94. doi: 10.1007/s11010-015-2339-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woo S.H., Sun N.J., Cassady J.M., Snapka R.M. Topoisomerase II inhibition by aporphine alkaloids. Biochem. Pharmacol. 1999;57:1141–1145. doi: 10.1016/S0006-2952(99)00018-0. [DOI] [PubMed] [Google Scholar]
- 23.Konkimalla V.B., Efferth T. Inhibition of epidermal growth factor receptor over-expressing cancer cells by the aphorphine-type isoquinoline alkaloid, dicentrine. Biochem. Pharmacol. 2010;79:1092–1099. doi: 10.1016/j.bcp.2009.11.025. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y., Zhang H., Jin X., Xiang J.F., Tang Y.L. Inhibitory effect of dicentrine on human lung cancer cell line A549 proliferation and on telomerase activity. Chin. J. Pharmacol. Toxicol. 2011;25:543–546. doi: 10.3867/j.issn.1000-3002.2011.06.004. [DOI] [Google Scholar]
- 25.Oh S.H., Woo J.K., Jin Q., Kang H.J., Jeong J.W., Kim K.W., Waun K.H., Lee H.Y. Identification of novel antiangiogenic anticancer activities of deguelin targeting hypoxia-inducible factor-1 alpha. Int. J. Cancer. 2008;122:5–14. doi: 10.1002/ijc.23075. [DOI] [PubMed] [Google Scholar]
- 26.Musumeci D., Amato J., Zizza P., Platella C., Cosconati S., Cingolani C., Biroccio A., Novellino E., Randazzo A., Giancola C., et al. Tandem application of ligand-based virtual screening and G4-OAS assay to identify novel G-quadruplex-targeting chemotypes. Biochim. Biophys. Acta—Gen. Subj. 2017;1861:1341–1352. doi: 10.1016/j.bbagen.2017.01.024. [DOI] [PubMed] [Google Scholar]
- 27.Musumeci D., Amato J., Randazzo A., Novellino E., Giancola C., Montesarchio D., Pagano B. G-quadruplex on Oligo Affinity Support (G4-OAS): An easy affinity chromatography-based assay for the screening of G-quadruplex ligands. Anal. Chem. 2014;86:4126–4130. doi: 10.1021/ac500444m. [DOI] [PubMed] [Google Scholar]
- 28.Ambrus A., Chen D., Dai J., Bialis T., Jones R.A., Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun D., Hurley L.H. The importance of negative superhelicit in inducing the formation of G-quadruplex. J. Med. Chem. 2009;52:2863–2874. doi: 10.1021/jm900055s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drew H.R., Wing R.M., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R.E. Structure of a B-DNA dodecamer: Conformation and dynamics. Proc. Natl. Acad. Sci. USA. 1981;78:2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dai J., Carver M., Hurley L.H., Yang D. Solution structure of a 2:1 quindoline-c-MYC G-quadruplex: Insights into G-quadruplex-interactive small molecule drug design. J. Am. Chem. Soc. 2011;133:17673–17680. doi: 10.1021/ja205646q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazzini S., Gargallo R., Musso L., De Santis F., Aviñó A., Scaglioni L., Eritja R., Di Nicola M., Zunino F., Amatulli A., et al. Stabilization of c-KIT G-quadruplex DNA structures by the RNA polymerase I inhibitors BMH-21 and BA-41. Int. J. Mol. Sci. 2019;20:4927. doi: 10.3390/ijms20194927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaroszewski J.W., Clausen V., Cohen J.S., Dahl O. NMR investigations of duplex stability of phosphorothioate and phosphorodithioate DNA analogues modified in both strands. Nucleic Acids Res. 1996;24:829–834. doi: 10.1093/nar/24.5.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amato J., Platella C., Iachettini S., Zizza P., Musumeci D., Cosconati S., Pagano A., Novellino E., Biroccio A., Randazzo A., et al. Tailoring a lead-like compound targeting multiple G-quadruplex structures. Eur. J. Med. Chem. 2019;163:295–306. doi: 10.1016/j.ejmech.2018.11.058. [DOI] [PubMed] [Google Scholar]
- 35.Petraccone L. Higher-Order Quadruplex Structures. Top Curr. Chem. 2013;330:23–46. doi: 10.1007/128_2012_350. [DOI] [PubMed] [Google Scholar]
- 36.Giancola C., Pagano B. Energetics of ligand binding to G-quadruplexes. Top Curr. Chem. 2013;330:211–242. doi: 10.1007/128_2012_347. [DOI] [PubMed] [Google Scholar]
- 37.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dai J., Carver M., Punchihewa C., Jones R.A., Yang D. Structure of the hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: Insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007;35:4927–4940. doi: 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ambrus A., Chen D., Dai J., Jones R.A., Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 40.Francisco S., Francisco S. γ-H2AX—A novel biomarker for DNA double-strand breaks. In Vivo. 2008;22:305–309. [PubMed] [Google Scholar]
- 41.García Díaz J., Tuenter E., Escalona Arranz J.C., Llauradó Maury G., Cos P., Pieters L. Antimicrobial activity of leaf extracts and isolated constituents of Croton linearis. J. Ethnopharmacol. 2019;236:250–257. doi: 10.1016/j.jep.2019.01.049. [DOI] [PubMed] [Google Scholar]
- 42.Salomon-Ferrer R., Case D.A., Walker R.C. An overview of the Amber biomolecular simulation package. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2013;3:198–210. doi: 10.1002/wcms.1121. [DOI] [Google Scholar]
- 43.Galindo-Murillo R., Robertson J.C., Zgarbová M., Šponer J., Otyepka M., Jurečka P., Cheatham T.E. Assessing the current state of Amber force field modifications for DNA. J. Chem. Theory Comput. 2016;12:4114–4127. doi: 10.1021/acs.jctc.6b00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zgarbová M., Šponer J., Otyepka M., Cheatham T.E., Galindo-Murillo R., Jurečka P. Refinement of the sugar-phosphate backbone torsion beta for AMBER force fields improves the description of Z- and B-DNA. J. Chem. Theory Comput. 2015;11:5723–5736. doi: 10.1021/acs.jctc.5b00716. [DOI] [PubMed] [Google Scholar]
- 45.Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general Amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 46.Roe D.R., Cheatham T.E. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9:3084–3097. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 47.Miller B.R., McGee T.D., Swails J.M., Homeyer N., Gohlke H., Roitberg A.E. MMPBSA.py: An efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012;8:3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- 48.Chen W., Hahn W.C. SV40 early region oncoproteins and human cell transformation. Histol. Histopathol. 2003;18:541–550. doi: 10.14670/HH-18.541. [DOI] [PubMed] [Google Scholar]
- 49.Biroccio A., Benassi B., Filomeni G., Amodei S., Marchini S., Chiorino G., Rotilio G., Zupi G., Ciriolo M.R. Glutathione influences c-Myc-induced apoptosis in M14 human melanoma Cells. J. Biol. Chem. 2002;277:43763–43770. doi: 10.1074/jbc.M207684200. [DOI] [PubMed] [Google Scholar]
- 50.Petti E., Buemi V., Zappone A., Schillaci O., Veneziano Broccia P., Dinami R., Matteoni S., Benetti R., Schoeftner S. SFPQ and NONO suppress RNA:DNA-hybrid-related telomere instability. Nat. Commun. 2019;10:1001. doi: 10.1038/s41467-019-08863-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iachettini S., Trisciuoglio D., Rotili D., Lucidi A., Salvati E., Zizza P., Di Leo L., Del Bufalo D., Ciriolo M.R., Leonetti C., et al. Pharmacological activation of SIRT6 triggers lethal autophagy in human cancer cells. Cell Death Dis. 2018;9:996. doi: 10.1038/s41419-018-1065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.