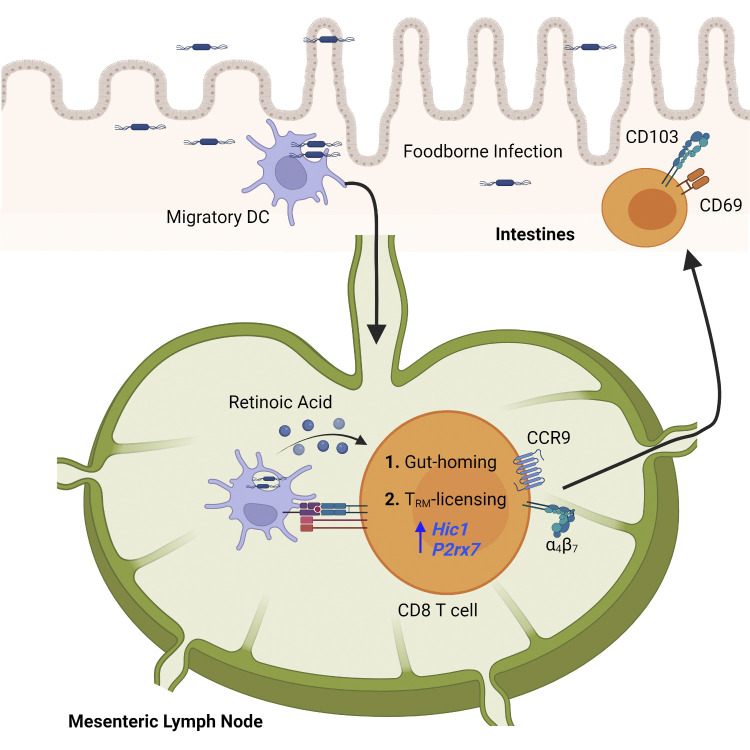
Qiu et al. demonstrate that retinoic acid signaling during T cell priming in the mesenteric lymph nodes licenses the differentiation of CD103+ CD8 tissue-resident memory T cells after entry into the small intestine, which may improve rational vaccine design.
Abstract
CD8 tissue-resident memory T (TRM) cells provide frontline protection at barrier tissues; however, mechanisms regulating TRM cell development are not completely understood. Priming dictates the migration of effector T cells to the tissue, while factors in the tissue induce in situ TRM cell differentiation. Whether priming also regulates in situ TRM cell differentiation uncoupled from migration is unclear. Here, we demonstrate that T cell priming in the mesenteric lymph nodes (MLN) regulates CD103+ TRM cell differentiation in the intestine. In contrast, T cells primed in the spleen were impaired in the ability to differentiate into CD103+ TRM cells after entry into the intestine. MLN priming initiated a CD103+ TRM cell gene signature and licensed rapid CD103+ TRM cell differentiation in response to factors in the intestine. Licensing was regulated by retinoic acid signaling and primarily driven by factors other than CCR9 expression and CCR9-mediated gut homing. Thus, the MLN is specialized to promote intestinal CD103+ CD8 TRM cell development by licensing in situ differentiation.
Graphical Abstract
Introduction
Memory CD8 T cells can be divided into three populations based on their migratory patterns: circulating central and effector memory T cells and tissue-resident memory T (TRM) cells (Gebhardt et al., 2009; Masopust et al., 2010; Mueller and Mackay, 2016; Park and Kupper, 2015; Sallusto et al., 1999; Sheridan and Lefrançois, 2011). TRM cells primarily reside in non-lymphoid tissues and are phenotypically, functionally, transcriptionally, and metabolically distinct from circulating memory T cell subsets (Kumar et al., 2017; Mackay et al., 2016; Mackay et al., 2013; Masopust et al., 2006; Pan et al., 2017; Wakim et al., 2012). CD8 TRM cells predominately express CD69, and the majority of those that reside within epithelium also express the αE integrin, CD103. CD69 promotes the residency of CD8 TRM cells in non-lymphoid tissues by inhibiting sphingosine-1-phosphate receptor-mediated egress of CD8 TRM cells from non-lymphoid tissues (Bankovich et al., 2010; Mackay et al., 2015; Shiow et al., 2006; Skon et al., 2013). CD103 binds to E-cadherin expressed in adherens junctions of epithelial cells and plays an important role in the accumulation and retention of CD8 TRM cells in barrier tissues (Casey et al., 2012; Lee et al., 2011; Mackay et al., 2013; Sheridan et al., 2014). CD8 TRM cells play a critical role in protective immunity against infections and cancer (Ganesan et al., 2017; Gebhardt et al., 2009; Jiang et al., 2012; Malik et al., 2017; Nizard et al., 2017; Sheridan et al., 2014; Shin and Iwasaki, 2012; Wu et al., 2014). They are prepositioned in the tissue to respond immediately to pathogen re-encounter and mediate protective immunity through multiple mechanisms like direct lysis of infected cells, induction of broadly active antimicrobial genes, or by activating innate immune cells and recruiting circulating memory cells through the release of cytokines IFNγ, IL-2, and TNFα (Ariotti et al., 2014; Cheuk et al., 2017; Schenkel et al., 2014; Schenkel et al., 2013). Thus, it is important to understand the mechanisms regulating the development and maintenance of CD8 TRM cells in specific tissues.
The development of CD8 TRM cells involves several steps including T cell priming in draining lymphoid organs, T cell migration to non-lymphoid tissues, and in situ TRM cell differentiation mediated by environmental signals. Priming in specific lymphoid organs instructs the migration of effector CD8 T cells to appropriate tissues, providing a critical point of regulation for the induction of proper tissue T cell responses (Park and Kupper, 2015; Sheridan and Lefrançois, 2011). Inherent factors in the tissue environment are essential for in situ CD8 TRM cell differentiation (Masopust et al., 2006). TGFβ activated by αv integrins on stromal cells in the tissue environment is crucial for inducing CD103 expression on CD8 TRM cells (Casey et al., 2012; Lee et al., 2011; Mackay et al., 2013; Mohammed et al., 2016; Qiu et al., 2021; Sheridan et al., 2014; Zhang and Bevan, 2013). Local extrinsic factors such as pathogen-induced inflammation and cognate antigen can have additional impacts on CD103+ CD8 TRM cell differentiation, although the findings are often dictated by the environment these cells reside in and the pathogen or infection route utilized (Bergsbaken and Bevan, 2015; Bergsbaken et al., 2017; Casey et al., 2012; Khan et al., 2016; Kohlmeier et al., 2007; Lee et al., 2011; Mackay et al., 2012; Muschaweckh et al., 2016; Slutter et al., 2017; Wakim et al., 2010). While the impact of local environment on CD103+ CD8 TRM cell differentiation has been extensively studied, whether CD8 T cell priming in draining lymphoid organs regulates in situ CD103+ TRM cell differentiation has not been established. The site of T cell priming is determined by the route of immunization. As the route of immunization is a critical factor in vaccine development, understanding the role of T cell priming in regulating CD103+ CD8 TRM cell differentiation will provide important insights into CD8 TRM cell biology that can be applied to improving vaccine strategies.
While priming in the mesenteric lymph nodes (MLN) has long been known to promote T cell homing to gut tissues, we report an additional distinct mechanism by which priming in the MLN promotes intestinal CD8 TRM cell formation. Priming in the MLN licensed CD8 T cells to respond to factors in the intestine to differentiate into CD103+ TRM cells. In contrast, CD8 T cells primed in the spleen were remarkably inefficient at differentiating into CD103+ TRM cells after migration into gut tissues. Moreover, priming in the MLN, rather than preconditioning during homeostasis, was the major determinant of in situ CD103+ CD8 TRM cell differentiation after migration into the intestine. We further demonstrate that MLN priming initiated CD103+ TRM cell gene signature before emigration out of the MLN, and retinoic acid (RA) signaling provided during priming licensed intestinal CD103+ TRM cell differentiation. While CCR9 contributed to CD103+ CD8 TRM cell development, the impact of RA signaling on licensing CD103+ CD8 TRM cell development appears primarily driven by factors other than CCR9 induction and gut homing. Therefore, our study establishes a distinct property of MLN priming to license CD103+ CD8 TRM cell development in the intestine.
Results
Priming in the MLN promotes CD103+ CD8 TRM precursor cell differentiation in the intestine
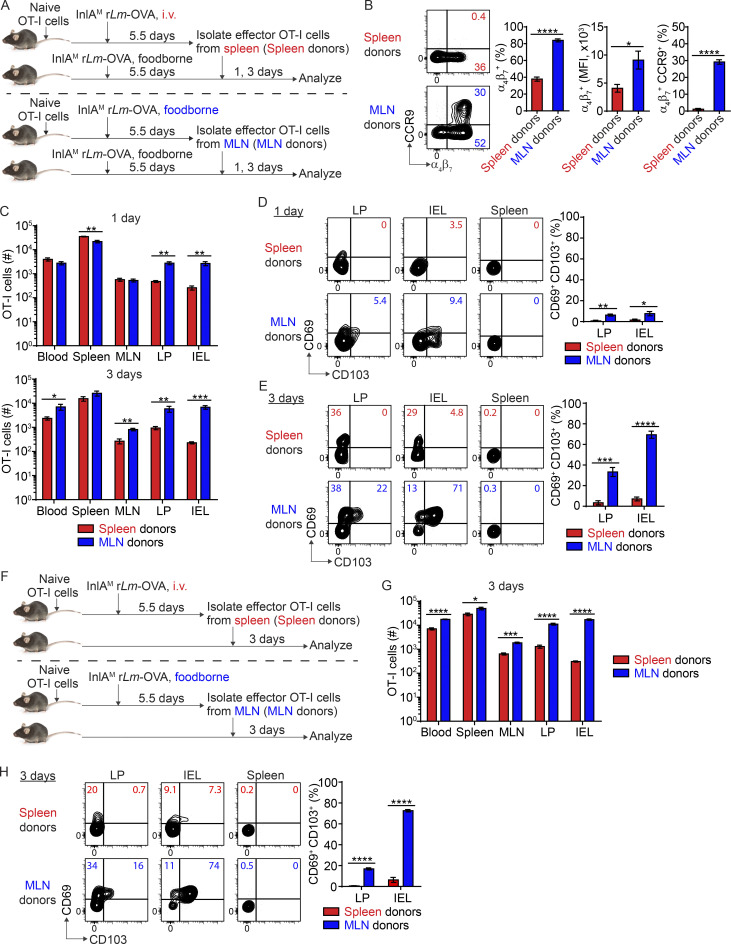
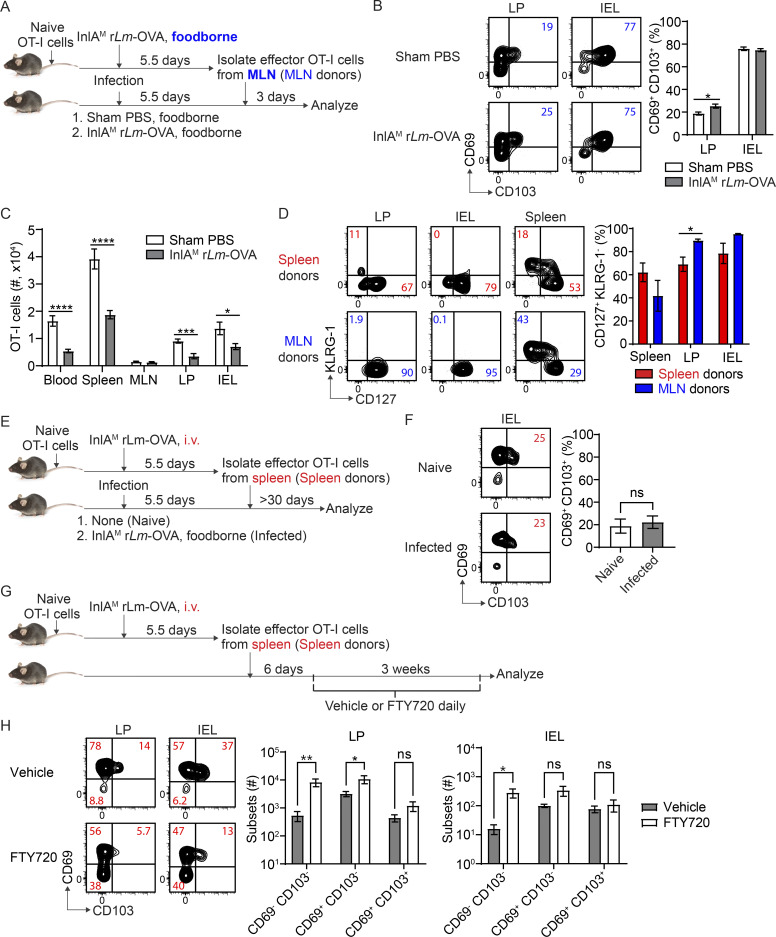
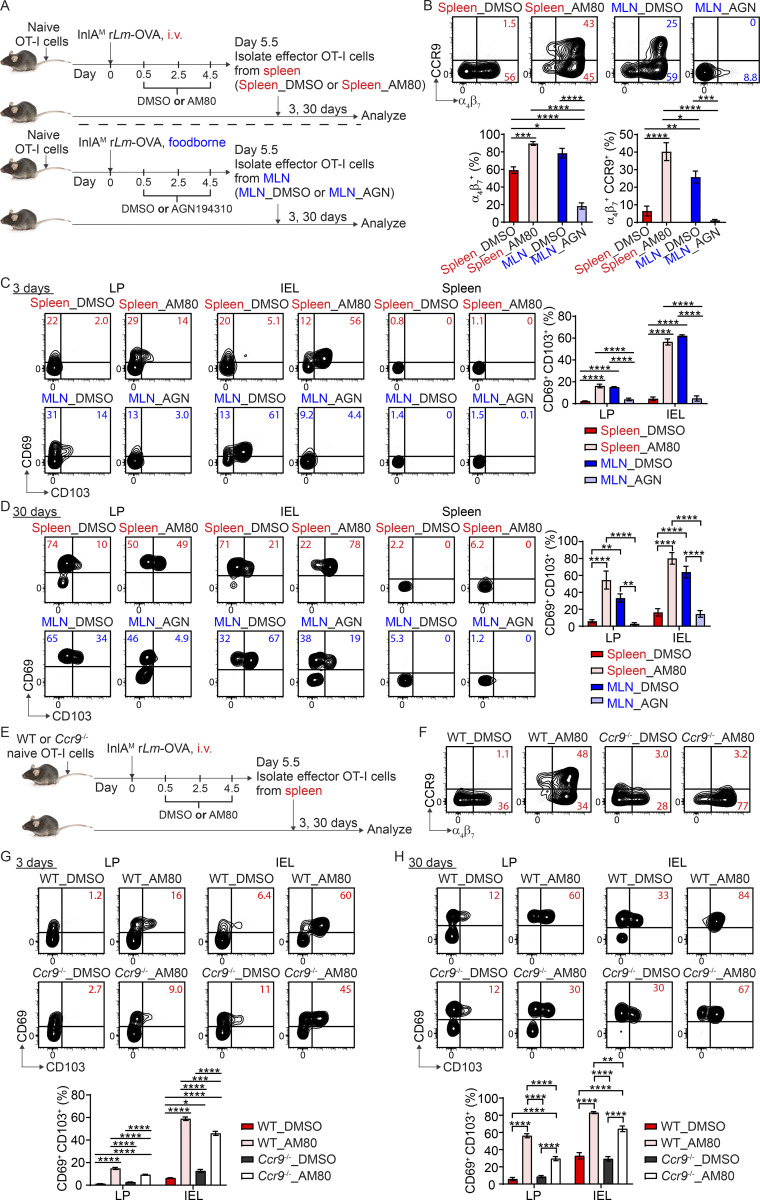
To study the impact of priming on intestinal CD103+ CD8 TRM cell differentiation, we first utilized an i.v. vs. foodborne infection model with a murinized recombinant Listeria monocytogenes strain containing a mutation in the internalin A protein (InlAM rLm) that allows efficient invasion of the murine intestinal epithelium (Wollert et al., 2007). One of the major differences between i.v. and foodborne L. monocytogenes infection is the priming site for T cells (Qiu et al., 2018). While it is well documented that the spleen is the primary priming site after i.v. L. monocytogenes infection (Khanna et al., 2007), the MLN is the draining lymph node and a major priming site after foodborne L. monocytogenes infection (Imperato et al., 2020). We employed a two-step adoptive transfer system to assess CD8 T cells primed in distinct lymphoid organs (Fig. 1 A). 1 × 104 naive OT-I cells (which express a transgenic T cell receptor that recognizes OVA257-264 presented in H2-Kb) were isolated from the spleen of OT-I Rag1−/− mice and transferred into congenic mice 1 d prior to infection. Mice were i.v. or foodborne infected with InlAM Lm expressing OVA (InlAM Lm-OVA). At 5.5 d postinfection (dpi), effector OT-I cells were isolated from the spleen of i.v.-infected mice (spleen-primed T cells) or the MLN of foodborne-infected mice (MLN-primed T cells) by FACS sorting based on congenic marker expression. 5 × 105 sorted effector OT-I cells were transferred into congenic recipient mice that were foodborne InlAM Lm-OVA infected 5.5 d previously, providing similar intestinal environments for effector cells primed at distinct lymphoid organs. 1 and 3 d after transfer, donor OT-I cells in recipient mice were analyzed. Priming in specific lymphoid organs dictates the migratory pattern of effector CD8 T cells (Park and Kupper, 2015; Sheridan and Lefrançois, 2011). In this manner, as previously reported (Johansson-Lindbom et al., 2003), priming in the MLN instructed CD8 T cells to express α4β7 and CCR9 (Fig. 1 B). CD8 T cells primed in the spleen also expressed α4β7, but to a significantly lesser extent (Fig. 1 B). Moreover, they failed to express CCR9 (Fig. 1 B). Consistent with previous studies (Masopust et al., 2010), MLN-primed T cells preferentially migrated to the lamina propria (LP) and intraepithelial lymphocytes (IEL) compartment compared with spleen-primed T cells (Fig. 1 C). These data demonstrate that priming in the MLN induced greater α4β7 and CCR9 expression on CD8 T cells and promoted their migration to the intestine.
Figure 1.
Priming in the MLN promotes CD103+ CD8 TRM precursor cell differentiation in the intestine. (A) Experimental design for B–E. 1 × 104 naive OT-I cells were transferred into congenic naive first recipient mice. 1 d later, mice were i.v. or foodborne infected with InlAM Lm-OVA. At 5.5 dpi, effector OT-I cells were isolated from the spleen of i.v.-infected mice (spleen donors) or the MLN of foodborne-infected mice (MLN donors) by FACS sorting based on congenic marker expression. 5 × 105 sorted effector OT-I cells were transferred into congenic second recipient mice that were foodborne InlAM Lm-OVA infected 5.5 d previously. 1 and 3 d after transfer, donor OT-I cells in the second recipient mice were analyzed. (B) The expression of α4β7 and CCR9 by effector OT-I cells prior to adoptive transfer into the second recipient mice. (C) Numbers of donor OT-I cells in indicated tissues at 1 and 3 d after transfer. (D) The expression of CD69 and CD103 by donor OT-I cells in the LP and IEL compartment at 1 d after transfer. (E) The expression of CD69 and CD103 by donor OT-I cells in the LP and IEL compartment at 3 d after transfer. (F) Experimental design for G and H. 1 × 104 naive OT-I cells were transferred into congenic naive first recipient mice 1 d prior to i.v. or foodborne infection with InlAM Lm-OVA. At 5.5 dpi, 5 × 105 spleen donors or MLN donors were sorted and transferred into congenic naive second recipient mice. 3 d after transfer, donor OT-I cells in the second recipient mice were analyzed. (G) Numbers of donor OT-I cells in indicated tissues at 3 d after transfer. (H) The expression of CD69 and CD103 by donor OT-I cells in the LP and IEL compartment at 3 d after transfer. The data in B are pooled from five independent experiments, n = 5 experiments. For each experiment, 2–18 mice were pooled for spleen donors and 18–34 mice were pooled for MLN donors. The data in C–E and G–H are representative of three independent experiments, n = 4–5 mice/experiment. The data are expressed as mean ± SEM. Unpaired t tests were performed (B–D, G, and H). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Our previous study demonstrated that foodborne InlAM Lm infection induces rapid differentiation of CD103+ CD8 TRM precursor cells in the intestine (Sheridan et al., 2014). We next examined whether priming regulated this rapid CD103+ CD8 TRM precursor cell differentiation. 1 d after transfer, MLN-primed T cells that migrated into the intestine started to upregulate CD69 and CD103 co-expression, while spleen-primed T cells failed to coupregulate CD69 and CD103 (Fig. 1 D). Donor cells did not express CD69 or CD103 prior to transfer (Fig. S1 A), suggesting that entry into the intestines led to upregulation of CD69 and CD103 on MLN-primed but not spleen-primed CD8 T cells. 3 d after transfer, MLN-primed T cells in the LP and IEL compartment had efficiently differentiated into CD103+ TRM precursor cells (Fig. 1 E). In comparison, spleen-primed cells failed to differentiate into CD103+ TRM precursor cells in either tissue (Fig. 1 E). Additionally, MLN-primed T cells that migrated to the spleen did not differentiate into CD103+ TRM precursor cells (Fig. 1 E), suggesting that factors existing in the intestine were critical for CD103+ TRM precursor cell differentiation.
Figure S1.
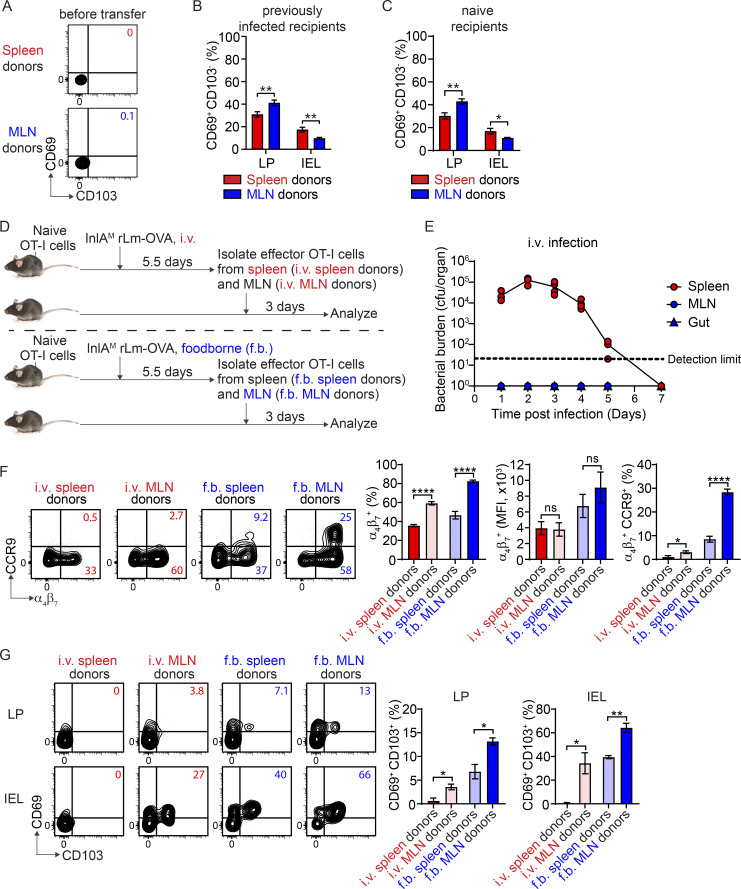
The expression of CD69 and CD103 by effector OT-I cells prior to entry into the intestine, the impact of priming on CD103− TRM precursor cell differentiation, and the impact of intrinsic environment of the MLN on CD103+ TRM precursor cell differentiation. (A) The expression of CD69 and CD103 by effector OT-I cells prior to adoptive transfer into second recipient mice. (B) The percentage of CD69+ CD103− population in indicated donor OT-I cells in the LP and IEL compartment at 3 d after transfer into second recipient mice that were foodborne InlAM Lm-OVA infected 5.5 d previously. (C) The percentage of CD69+ CD103− population in indicated donor OT-I cells in the LP and IEL compartment at 3 d after transfer into naive second recipient mice. (D) Experimental design for F and G. 1 × 104 naive OT-I cells were transferred into congenic naive first recipient mice 1 d prior to i.v. or foodborne infection with InlAM Lm-OVA. At 5.5 dpi, effector OT-I cells were sorted from the spleen and MLN of i.v. infected mice (i.v. spleen donors and i.v. MLN donors) or from the spleen and MLN of foodborne infected mice (f.b. spleen donors and f.b. MLN donors) and transferred into congenic naive second recipient mice. 3 d after transfer, donor OT-I cells in the second recipient mice were analyzed. (E) Bacterial burdens in the spleen, MLN, and small intestine (gut) at indicated days after i.v. infection with InlAM Lm-OVA. (F) The expression of α4β7 and CCR9 by i.v. spleen donors, i.v. MLN donors, f.b. spleen donors, and f.b. MLN donors prior to adoptive transfer into second recipient mice. (G) The expression of CD69 and CD103 by donor OT-I cells in the LP and IEL compartment at 3 d after transfer into second recipient mice. The data in A are representative of three independent experiments. The data in B are pooled from three independent experiments, n = 12–14 mice total. The data in C are pooled from three independent experiments, n = 12 mice total. The data in E are representative of three independent experiments, n = 4–5 mice/experiment. The data in F are pooled from five independent experiments, n = 5 experiments and for each experiment 2–18 mice were pooled for spleen donors and 18–34 mice were pooled for MLN donors. The data in G are representative of two independent experiments, n = 3 mice/experiment. The data are expressed as mean ± SEM and unpaired t tests were performed (B, C, F, and G). *, P ≤ 0.05; **, P ≤ 0.01; ****, P ≤ 0.0001.
Similar results were observed when spleen-primed and MLN-primed T cells were transferred into naive congenic recipient mice (Fig. 1, F–H), suggesting basal intestinal inherent factors alone were sufficient to drive MLN-primed but not spleen-primed T cells to rapidly differentiate into CD103+ TRM precursor cells. Thus, our data demonstrate that MLN, but not splenic priming, promotes CD8 T cell responsiveness to environmental signals present in the small intestine for CD103+ TRM precursor cell differentiation.
We next examined whether priming regulated CD103− TRM precursor cell differentiation. Regardless of whether effector T cells were transferred into previously infected recipient mice or naive mice, while MLN-primed T cells were slightly more efficient at differentiating into CD103− TRM precursor cells in the LP, they were less efficient at differentiating into CD103− TRM precursor cells in the IEL compartment (Fig. S1, B and C). Overall, the impact of priming on CD103− TRM precursor cell differentiation is less substantial and appeared compartmentalized.
To determine whether the intrinsic environment of the MLN had an impact on CD103+ TRM precursor cell differentiation, effector T cells were sorted from the spleen and MLN of i.v.-infected mice or from the spleen and MLN of foodborne-infected mice and transferred into naive congenic recipient mice for analysis 3 d later (Fig. S1 D). Since CD8 T cells are primed in the MLN but not spleen after foodborne infection (Imperato et al., 2020; Sheridan et al., 2014), effector T cells isolated from the spleen after foodborne infection are likely migrants. Additionally, L. monocytogenes did not invade the gut or MLN after i.v. infection (Fig. S1 E), suggesting that effector T cells isolated from the MLN after i.v. infection are not primed there but are migrants. Compared to foodborne MLN donor cells, foodborne spleen donor cells had reduced α4β7 and CCR9 expression (Fig. S1 F). Compared with i.v. spleen donors, i.v. MLN donors had higher α4β7 and CCR9 expression, suggesting the MLN environment enhanced gut-homing receptor expression on migrant effector T cells (Fig. S1 F). Foodborne spleen donors lost some ability to differentiate into CD103+ TRM precursor cells in the intestine compared to foodborne MLN donors (Fig. S1 G). Surprisingly, i.v. MLN donors gained the ability to differentiate into intestinal CD103+ TRM precursor cells compare to i.v. spleen donors, albeit to a lesser degree than foodborne MLN donors (Fig. S1 G). Thus, the MLN imparts an environment signal in addition to priming that promotes effector CD8 T cell differentiation into CD103+ TRM precursor cells in the intestine.
Rapid intestinal CD103+ CD8 TRM precursor cell differentiation is independent of activation stage, intestinal homing, and KLRG-1 expression
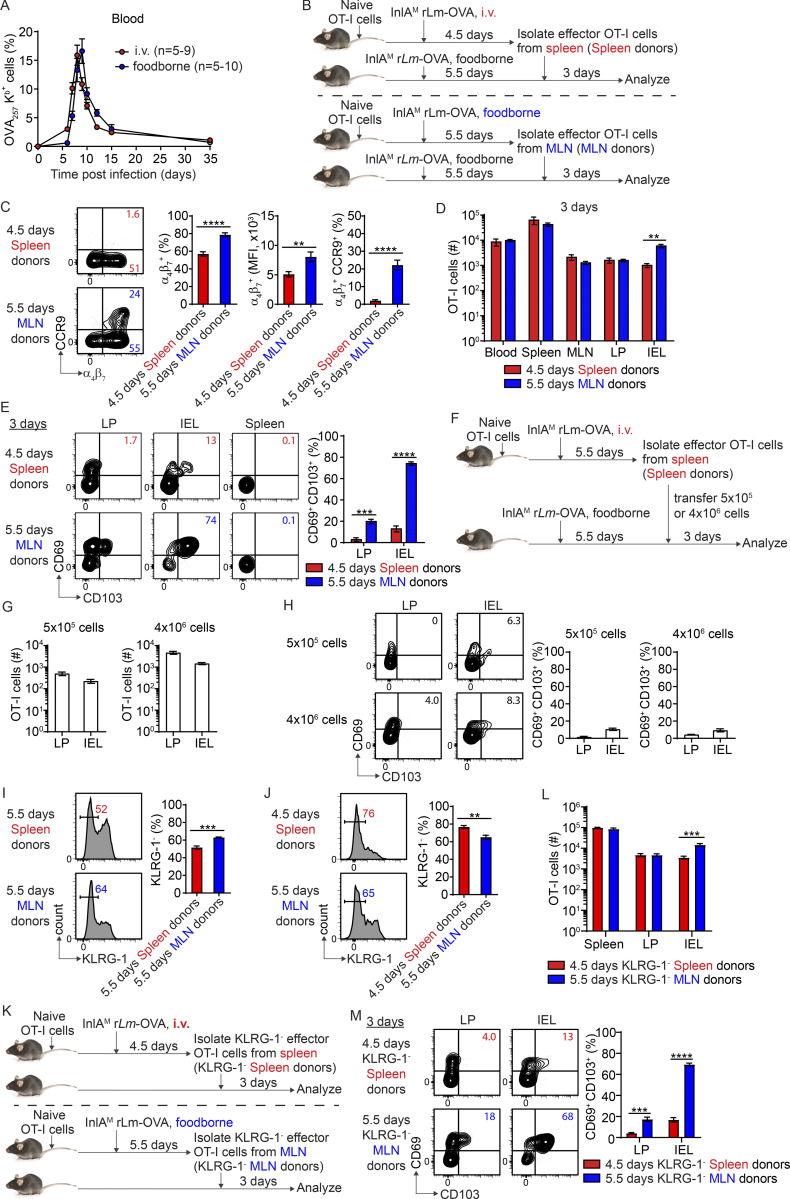
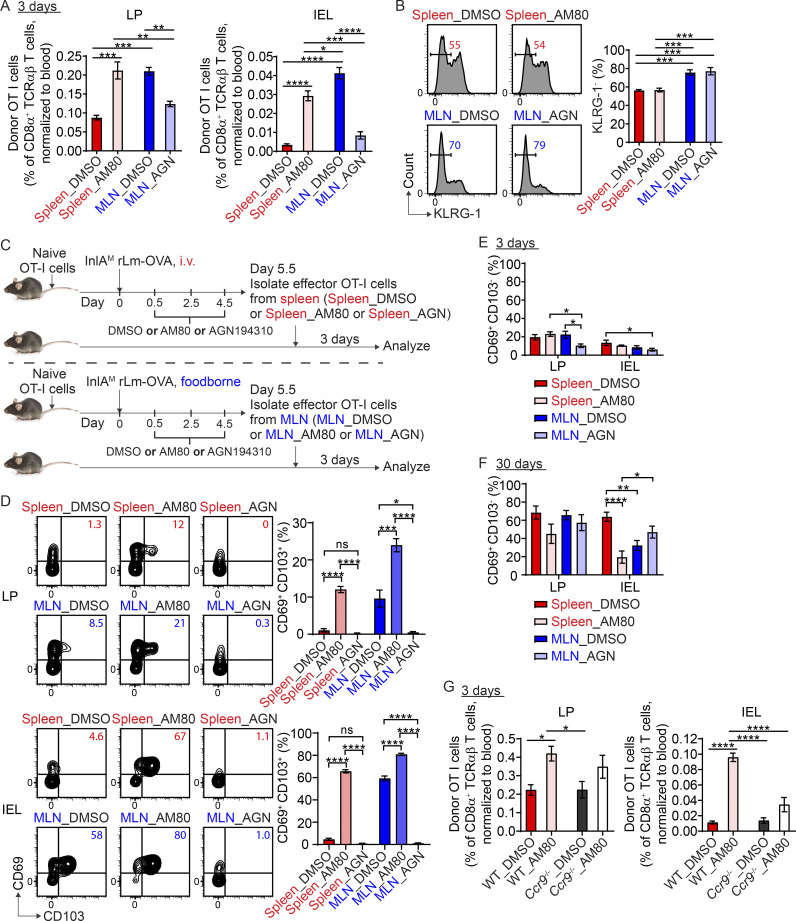
While i.v. and foodborne L. monocytogenes infection induced a similar magnitude of circulating OVA-specific CD8 T cells, the initiation and peak of the CD8 T cell response were delayed by 1 d after foodborne infection (Fig. S2 A). We sought to determine whether the difference in intestinal CD103+ TRM precursor cell differentiation between spleen-primed and MLN-primed T cells was due to a difference in activation stages at the time of transfer. To address this, effector OT-I cells were isolated from the spleen at 4.5 d after i.v. infection and from the MLN at 5.5 d after foodborne infection for adoptive transfer into mice that were foodborne-infected 5.5 d previously (Fig. S2 B), putting spleen-primed and MLN-primed T cells at a comparable activation stage based on the time to peak response. 4.5-d spleen-primed T cells did not express CCR9; however, they expressed a higher frequency of α4β7 (Fig. S2 C and Fig. 1 B) and accumulated in the LP as efficiently as 5.5-d MLN-primed T cells (Fig. S2 D), suggesting that splenic priming was as capable of inducing LP-migrant T cells as MLN priming. However, rescued LP migration did not restore the migration to the IEL compartment (Fig. S2 D), consistent with a role for CCR9 in mediating the accumulation of antigen-specific T cells in the epithelium (Johansson-Lindbom et al., 2003). Regardless, 4.5-d spleen-primed T cells failed to rapidly differentiate into CD103+ TRM precursor cells after migration into the LP or IEL compartment (Fig. S2 E), indicating that spleen-primed CD8 T cells have an inherent and limited capacity to differentiate into CD103+ TRM precursor cells in the intestinal LP that is independent of activation stage and intestinal homing ability. To confirm that the inability of spleen-primed T cells to differentiate into CD103+ TRM precursor cells was independent of their ability to accumulate in the intestine, eight times more spleen-primed T cells were transferred into recipient mice (Fig. S2 F). Although a greater number of spleen-primed T cells accumulated in the LP and IEL compartments with an increased number of transferred cells (Fig. S2 G), spleen-primed T cells remained largely unable to differentiate into CD103+ TRM precursor cells (Fig. S2 H). Thus, inability of spleen-primed T cells to differentiate into CD103+ TRM precursor cells was independent of their intestinal homing and accumulation.
Figure S2.
Rapid intestinal CD103+ CD8 TRM precursor cell differentiation is independent of activation stage, intestinal homing, and KLRG-1 expression. (A) The longitudinal OVA-specific CD8 T cell responses in the blood measured by OVA257 Kb tetramer after i.v. and foodborne infection. (B) Experimental design for C–E. 1 × 104 naive OT-I cells were transferred into congenic naive first recipient mice. 1 d later, mice were i.v. or foodborne infected with InlAM Lm-OVA. Spleen donors and MLN donors were isolated from the spleen at 4.5 d after i.v. infection and from the MLN at 5.5 d after foodborne infection, respectively. 5 × 105 sorted donor OT-I cells were transferred into congenic second recipient mice that were foodborne InlAM Lm-OVA infected 5.5 d previously. 3 d after transfer, donor OT-I cells in second recipient mice were analyzed. (C) The expression of α4β7 and CCR9 by effector OT-I cells prior to adoptive transfer into second recipient mice. (D) Numbers of donor OT-I cells in indicated tissues at 3 d after transfer. (E) The expression of CD69 and CD103 by donor OT-I cells in the LP and IEL compartment at 3 d after transfer. (F) Experimental design for G and H. 1 × 104 naive OT-I cells were transferred into congenic naive first recipient mice 1 d prior to i.v. infection with InlAM Lm-OVA. At 5.5 dpi, 5 × 105 or 4 × 106 sorted Spleen donors were transferred into congenic second recipient mice that were foodborne InlAM Lm-OVA infected 5.5 d previously. 3 d after transfer, donor OT-I cells in second recipient mice were analyzed. (G) The accumulation of donor OT-I cells in the LP and IEL compartment at 3 d after transfer. (H) The expression of CD69 and CD103 by donor OT-I cells in the LP and IEL compartment at 3 d after transfer. (I and J) The percentage of KLRG-1− population in indicated effector OT-I cells prior to adoptive transfer into second recipient mice. (K) Experimental design for L and M. 1 × 104 naive OT-I cells were transferred into congenic naive first recipient mice 1 d prior to i.v. or foodborne infection with InlAM Lm-OVA. KLRG-1− Spleen donors and MLN donors were isolated from the spleen at 4.5 d after i.v. infection and from the MLN at 5.5 d after foodborne infection respectively. 5 × 105 sorted donor OT-I cells were transferred into congenic naive second recipient mice. 3 d after transfer, donor OT-I cells in second recipient mice were analyzed. (L) Numbers of donor OT-I cells in indicated tissues at 3 d after transfer. (M) The expression of CD69 and CD103 by donor OT-I cells in the LP and IEL compartment at 3 d after transfer. The data in A are from one experiment, n = 7–10 mice per infection route and time. The data in C are pooled from three independent experiments, n = 6 mice total. The data in D and E are representative of two independent experiments, n = 4 mice/experiment. The data in G and H are from one experiment, n = 4 mice/experiment. The data in I are pooled from five independent experiments, n = 5 experiments, and for each experiment 2–18 mice were pooled for spleen donors and 18–34 mice were pooled for MLN donors. The data in J are pooled from three independent experiments, n = 6 mice total. The data in L and M are pooled from two independent experiments, n = 6 mice total. The data are expressed as mean ± SEM. Unpaired t tests were performed (C, D, E, and I–L). **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Previous studies have demonstrated that CD103+ CD8 TRM cells are derived from KLRG-1− cells (Mackay et al., 2013; Sheridan et al., 2014). It is plausible that priming in the MLN may promote KLRG-1− cells. Indeed, 5.5-d MLN-primed T cells had a slightly but significantly higher percentage of KLRG-1− cells than 5.5-d spleen-primed T cells (Fig. S2 I); however, they had a slightly but significantly lower percentage of KLRG-1− cells than 4.5-d spleen-primed T cells (Fig. S2 J). As both 4.5- and 5.5-d spleen-primed T cells failed to rapidly differentiate into CD103+ TRM precursor cells (Fig. S2 E and Fig. 1 E), the inability of spleen-primed CD8 T cells to differentiate into CD103+ TRM precursor cells did not correlate with the proportion of KLRG-1− cells among donor cells. To further confirm that the proportion of KLRG-1−cells did not impact CD103+ TRM precursor cell differentiation, KLRG-1− effector cells were isolated from the spleen at 4.5 d after i.v. infection and from the MLN at 5.5 d after foodborne infection for adoptive transfer into naive mice (Fig. S2 K). While 4.5-d KLRG-1− spleen-primed T cells accumulated in the LP as efficiently as 5.5-d KLRG-1− MLN-primed T cells (Fig. S2 L), they failed to rapidly differentiate into CD103+ TRM precursor cells after migration into the LP and IEL compartments (Fig. S2 M). These data are consistent with the notion that rapid CD103+ TRM precursor cell differentiation is independent of KLRG-1 expression.
Priming in the MLN licenses intestinal CD103+ CD8 TRM cell development
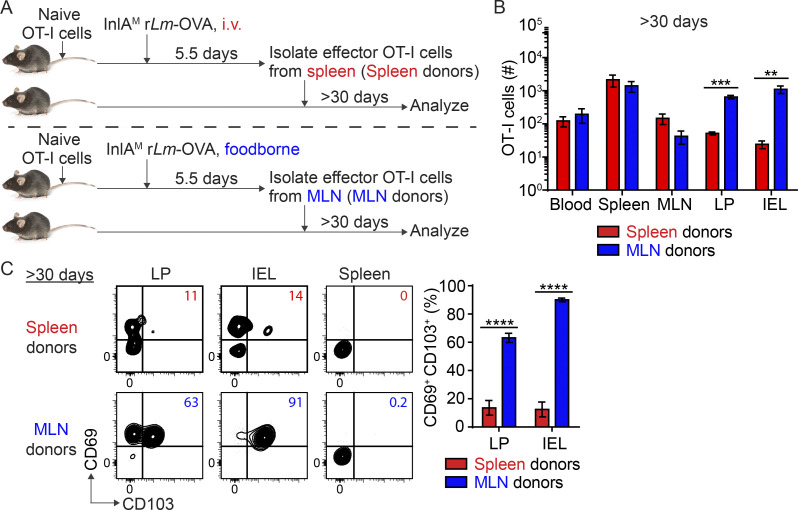
As MLN priming promoted rapid in situ CD103+ TRM precursor cell differentiation, we next evaluated its impact on CD103+ TRM cell development at memory. The basal intestinal environment was sufficient to support CD103+ TRM precursor cell differentiation (Fig. 1 H and Fig. S3, A and B), and more effector CD8 T cells could be recovered from a naive host than an infected host (Fig. S3 C), enhancing our ability to detect transferred cells at memory. As such, spleen-primed and MLN-primed CD8 T cells were adoptively transferred into naïve mice to assess their capacity to become CD103+ TRM cells (Fig. 2 A). At >30 d after transfer, spleen-primed and MLN-primed CD8 T cells were detected in all tissues examined, but significantly more MLN-primed T cells were present in the LP and IEL compartments (Fig. 2 B), suggesting MLN priming led to a significantly larger memory CD8 T cell pool in the intestine. Consistent with the enhanced CD103+ TRM precursor cell differentiation among MLN-primed cells, the majority of MLN-primed T cells in the LP and IEL compartments became CD103+ TRM cells (Fig. 2 C). In stark contrast, spleen-primed T cells remained largely incapable of differentiating into CD103+ TRM cells after 30 d (Fig. 2 C). Importantly, most spleen-primed T cells acquired a CD127+ KLRG-1− memory phenotype in the intestine at this time (Fig. S3 D), suggesting that they are capable of becoming cells with a memory phenotype but not upregulating CD103. Consistent with transfer into naive mice, spleen-primed T cells were inefficient at CD103+ TRM cell differentiation after adoptive transfer into previously infected mice (Fig. S3 F), suggesting that the local environment is insufficient to induce CD8 TRM cell differentiation in the absence of appropriate licensing. Altogether, these data demonstrated that priming in the MLN licensed CD103+ TRM cell development in the intestine.
Figure S3.
The impact of infection on intestinal CD103+ TRM cell differentiation and the residency of intestinal memory CD8 T cells derived from spleen-primed T cells. (A) Experimental design for B and C. 1 × 104 naive OT-I cells were transferred into congenic naive first recipient mice. 1 d later, mice were foodborne infected with InlAM Lm-OVA. At 5.5 dpi, effector OT-I cells were isolated from the MLN and transferred into congenic second recipient mice that were foodborne infected with InlAM Lm-OVA or treated with sham PBS 5.5 d previously. 3 d after transfer, donor OT-I cells in second recipient mice were analyzed. (B) The expression of CD69 and CD103 by donor OT-I cells in the LP and IEL compartment of indicated second recipient mice. (C) Numbers of donor OT-I cells in tissues of indicated second recipient mice. (D) The expression of CD127 and KLRG-1 by donor OT-I cells in indicated tissues at >30 d after transfer into congenic naive second recipient mice. (E) Experimental design for F. 1 × 104 naive OT-I cells were transferred into congenic naive first recipient mice 1 d prior to i.v. infection with InlAM Lm-OVA. At 5.5 dpi, effector OT-I cells were isolated from the spleen and transferred into congenic second recipient mice that were uninfected or foodborne infected with InlAM Lm-OVA 5.5 d previously. 3 d after transfer, donor OT-I cells in second recipient mice were analyzed. (F) The expression of CD69 and CD103 by donor OT-I cells in the IEL compartment of indicated second recipient mice. (G) Experimental design for H. 1 × 104 naive OT-I cells were transferred into congenic naive first recipient mice 1 d prior to i.v. infection with InlAM Lm-OVA. At 5.5 dpi, effector OT-I cells were isolated from the spleen and transferred into congenic naive second recipient mice. 6 d after transfer, second recipient mice were treated with vehicle control or FTY720 daily for 3 wk followed by analysis of donor OT-I cells. (H) The expression of CD69 and CD103 by donor OT-I cells in the LP and IEL compartment of indicated second recipient mice. The data in B and C are pooled from two independent experiments, n = 8–9 mice total. The data in D are representative of two independent experiments, n = 4 mice/experiment. The data in F are pooled from two independent experiments, n = 3–4 mice total. The data in H are pooled from two independent experiments, n = 5–6 mice total. The data are expressed as mean ± SEM. Unpaired t tests were performed (B–D, F and H). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Figure 2.
Priming in the MLN licenses intestinal CD103+ CD8 TRM cell differentiation. (A) Experimental design. 1 × 104 naive OT-I cells were transferred into congenic naive first recipient mice 1 d prior to i.v. or foodborne infection with InlAM Lm-OVA. At 5.5 dpi, 5 × 105 spleen donors or MLN donors were sorted and transferred into congenic naive second recipient mice. >30 d after transfer, donor OT-I cells in the second recipient mice were analyzed. (B) Numbers of donor OT-I cells in indicated tissues at >30 d after transfer. (C) The expression of CD69 and CD103 by donor OT-I cells in the LP and IEL compartment at >30 d after transfer. The data in B and C are representative of two independent experiments, n = 4 mice/experiment. The data are expressed as mean ± SEM. Unpaired t tests were performed (B and C). **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
While spleen-primed T cells were not efficient at differentiating into CD103+ TRM cells, they were readily detectable in the intestine at memory. We next determined whether these memory cells were resident cells or a transient migratory population. Spleen-primed T cells were adoptively transferred into naive mice (Fig. S3 G). 6 d later, after these effector cells had completely downregulated α4β7 expression, recipient mice were treated with vehicle control or FTY720 daily for 3 wk to block tissue egress of T cells (Hirai et al., 2019). The numbers of CD69− CD103− cells in the LP and IEL compartments were significantly increased in FTY720-treated mice compared to vehicle-treated mice, while the numbers of CD69+ CD103− and CD69+ CD103+ cells were largely comparable between these two groups (Fig. S3 H). These data suggest that while CD69+ populations are likely resident cells, CD69− CD103− cells represent a transient migratory population, which is consistent with previous studies (Hirai et al., 2019; Mackay et al., 2015; Takamura et al., 2016).
Priming, but not preconditioning, promotes rapid CD103+ TRM cell differentiation in the intestine
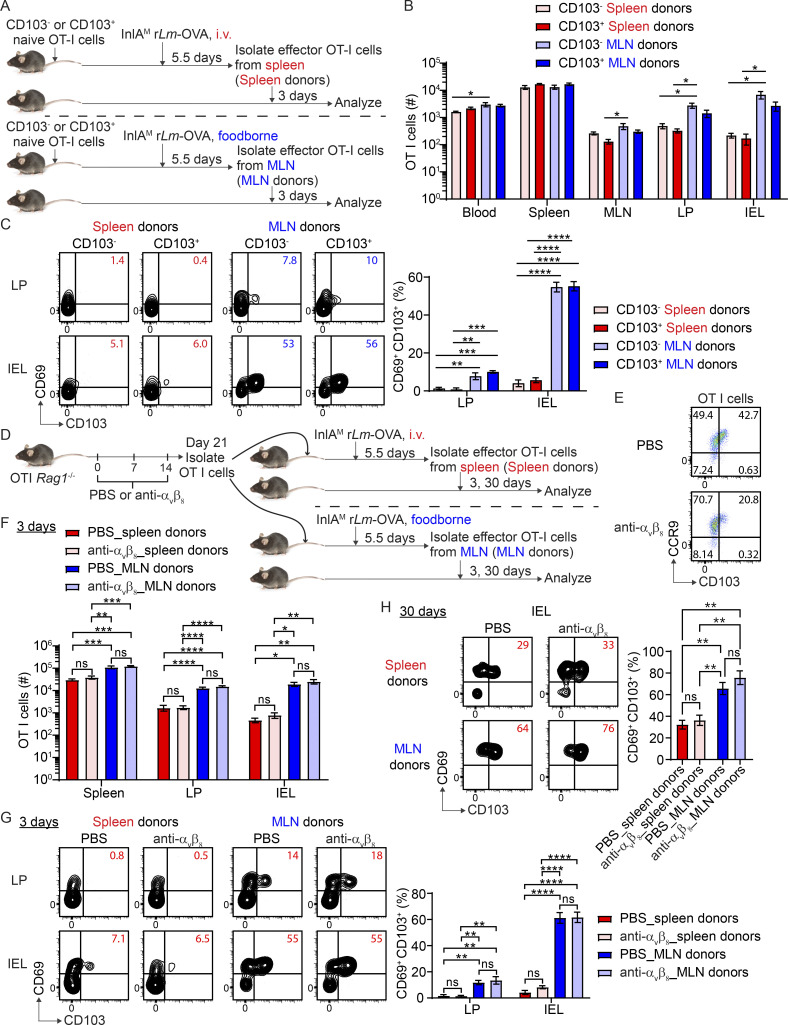
A recent study demonstrated that migratory dendritic cells (DCs) activate TGFβ through αV integrins and present it to naive CD8 T cells in lymph nodes to precondition them for CD103+ TRM cell differentiation in the skin (Mani et al., 2019). We set to determine the respective contribution of preconditioning versus priming to the differentiation of CD103+ TRM cells in the intestine. CD103 expression on naive T cells indicates TGFβ-mediated preconditioning occurred in lymph nodes (Mani et al., 2019). As such, CD103+ and CD103− naïve OT-I cells were sorted from the spleen of OT-I Rag1−/− mice and transferred into naive congenic mice (Fig. 3 A and Fig. S4 A). Recipient mice were i.v.- or foodborne-infected with InlAM Lm-OVA 1 d after transfer. At 5.5 dpi, effector OT-I cells were sorted from the spleen of i.v.-infected mice or the MLN of foodborne-infected mice and transferred into naive congenic recipient mice. Of note, CD103+ naive OT-I cells completely downregulated CD103 expression after activation (Fig. S4 B). 3 d after transfer, CD103 status of naive T cells did not significantly affect the accumulation of either spleen-primed or MLN-primed T cells in the intestine (Fig. 3 B). Interestingly, regardless of whether donor CD8 T cells were preconditioned (CD103+) or not (CD103−), as long as they were primed in the MLN, they were equally efficient in differentiating into CD103+ TRM precursor cells in the intestine (Fig. 3 C). Thus, priming, but not preconditioning, appears to be the major determinant of rapid CD103+ TRM precursor cell differentiation in the intestine. While CD103 expression depends on TGF-β signaling and is used as a surrogate marker of TGFβ-mediated preconditioning (Mani et al., 2019; Zhang and Bevan, 2012), it does not preclude that CD103− naive T cells are not preconditioned by TGF-β. Since TGF-β activation and TGFβ-mediated preconditioning in lymph nodes rely on αvβ8 expressed by migratory DCs (Mani et al., 2019; Qiu et al., 2021), anti-αvβ8 was used to block TGF-β activation and TGFβ-mediated preconditioning. Naive OT-I Rag1−/− mice were treated with PBS or anti-αvβ8 weekly for 3 wk (Dodagatta-Marri et al., 2021; Hirai et al., 2021; Fig. 3 D). Anti-αvβ8 treatment led to a 50% reduction in CD103 expression in naive T cells (Fig. 3 E). Naive OT-I cells were then isolated and transferred into congenic mice 1 d prior to infection. At 5.5 dpi, effector OT-I cells were sorted from the spleen of i.v.-infected mice or the MLN of foodborne-infected mice and transferred into naive congenic recipient mice. Anti-αvβ8 treated and untreated effector T cells accumulated similarly in the intestine of recipient mice 3 d after transfer (Fig. 3 F). More importantly, they both efficiently differentiated into intestinal CD103+ TRM precursor cells (Fig. 3 G) and CD103+ TRM cells (Fig. 3 H), if they were primed in the MLN. These results are consistent with the notion that priming, but not preconditioning, promotes CD103+ TRM cell differentiation in the intestine.
Figure 3.
Preconditioning is not a major determinant of rapid CD103+ CD8 TRM cell differentiation in the intestine. (A) Experimental design for B and C. 2 × 104 FACS sorted naive CD103+ or CD103− OT-I cells were transferred into congenic naive first recipient mice 1 d prior to i.v. or foodborne infection with InlAM Lm-OVA. At 5.5 dpi, 3 × 105 sorted spleen donors or MLN donors were transferred into congenic naive second recipient mice. 3 d after transfer, donor OT-I cells in the second recipient mice were analyzed. (B) Numbers of donor OT-I cells in indicated tissues at 3 d after transfer. (C) The expression of CD69 and CD103 by donor OT-I cells in the LP and IEL compartment at 3 d after transfer. (D) Experimental design for E–H. OT-I Rag1−/− mice were intraperitoneally injected with PBS control or 10 mg/kg anti-αvβ8 weekly for 3 wk. 1 × 104 naive OT-I cells were isolated from the spleen of PBS or anti-αvβ8 treated OT-I Rag1−/− mice and transferred into congenic naive first recipient mice 1 d prior to i.v. or foodborne infection with InlAM Lm-OVA. At 5.5 dpi, spleen donors (3 × 105 for early time point and 5 × 106 for memory time point) or MLN donors (1 × 105 for early time point and 2 × 106 for memory time point) were sorted and transferred into congenic naive second recipient mice. 3 and 30 d after transfer, donor OT-I cells in the second recipient mice were analyzed. (E) The expression of CD103 and CCR9 by naive OT-I cells after PBS and anti-αvβ8 treatment. (F) Numbers of donor OT-I cells (normalized to 5 × 105 transferred cells) in indicated tissues at 3 d after transfer. (G) The expression of CD69 and CD103 by donor OT-I cells in the LP and IEL compartment at 3 d after transfer. (H) The expression of CD69 and CD103 by donor OT-I cells in the IEL compartment 30 d after transfer. For B and C, n = 3–4 mice/experiment, and similar results were observed in two other experiments. The data in E are representative of three independent experiments. The data in F and G are pooled from two independent experiments, n = 4 mice total. The data in H are pooled from two independent experiments, n = 3–5 mice total. The data are expressed as mean ± SEM. One-way ANOVA with Tukey’s multiple comparisons test was performed (B, C, and F–H). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Figure S4.
The sorting strategy for CD103− and CD103+ naive OT-I cells, the expression of CD69 and CD103 by transferred CD103− and CD103+ naive OT-I cells after activation, and the ability to upregulate CD103 expression in response to TGF-β by Spleen and MLN donors. (A) The gating strategy for sorting CD103− and CD103+ naive OT-I cells. (B) The expression of CD69 and CD103 by spleen donors and MLN donors depicted in Fig. 3 A. (C) Experimental design for D. 1 × 104 naive OT-I cells were transferred into congenic naive recipient mice 1 d prior to i.v. or foodborne infection with InlAM Lm-OVA. 4.5 and 5.5 d spleen-primed T cells and 5.5 d MLN-primed T cells were cultured with TGF-β in vitro for 24 h followed by flow analysis of CD103 protein expression. (D) The CD103 expression by 4.5 and 5.5 d spleen-primed T cells as well as 5.5 d MLN-primed T cells. The data in A are representative of four independent experiments. The data in B are from one experiment. The data in D are representative of two independent experiments and expressed as mean ± SEM, n = 4 mice/experiment.
A CD103+ TRM cell gene signature is initiated prior to emigration from the MLN
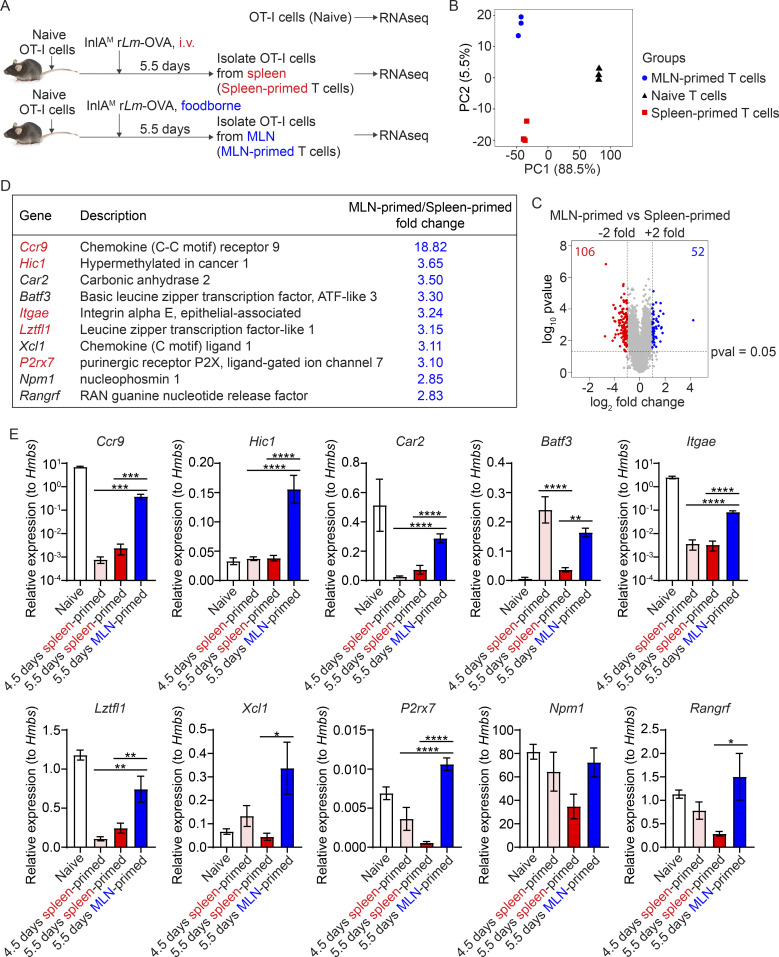
We next attempted to determine how priming regulated intestinal CD103+ TRM cell differentiation. Naive, spleen-primed, and MLN-primed OT-I cells were isolated and sequenced for transcriptional profiling (Fig. 4 A). Principle component analysis showed that the transcription profile was distinct between MLN-primed and spleen-primed T cells (Fig. 4 B). Based on the criteria for significance (P ≤ 0.05 and fold change ≥2), 158 genes were differentially expressed, of which 52 genes were upregulated and 106 genes were downregulated in MLN-primed T cells compared with spleen-primed T cells (Fig. 4 C and Table. S1). Of the top 10 most upregulated genes in the MLN-primed T cells, two were associated with the core CD103+ CD8 TRM cell signature (Itgae and Xcl1) and four were associated with the gut CD103+ CD8 TRM cell signature (Ccr9, Itgae, Xcl1, and P2rx7; Fig. 4 D; Mackay et al., 2013). Moreover, T cell-intrinsic expression of Hic 1 has been shown to promote CD103+ resident LP lymphocytes and IEL during homeostasis (Burrows et al., 2017). A recent study further identified Hic1 as a critical regulator of TRM cell differentiation in the IEL compartment (Crowl et al., 2022). Since gene expression profile changes temporally during T cell activation (Best et al., 2013), and 5.5-d spleen-primed T cells and 5.5-d MLN-primed T cells were likely at different activation stages (Fig. S2 A), we determined whether differentially expressed genes resulted from distinct priming environments or a difference in their activation stage. Quantitative PCR was performed using both 4.5- and 5.5-d spleen-primed T cells in comparison with 5.5-d MLN-primed T cells. 7 out of the 10 indicated genes (Ccr9, Hic1, Car2, Itgae, Lztfl1, Xcl1, and P2rx7) had at least twofold higher expression in 5.5-d MLN-primed T cells compared with both 4.5- and 5.5-d spleen-primed T cells (Fig. 4 E). Overall, these data suggest that MLN priming initiated a CD103+ CD8 TRM cell gene signature before effector CD8 T cells emigrated from the MLN.
Figure 4.
A CD103+ TRM cell gene signature is initiated prior to emigration from the MLN. (A) Experimental design. 1 × 104 naive OT-I cells were transferred into congenic naive recipient mice 1 d prior to i.v. or foodborne infection with InlAM Lm-OVA. At 5.5 dpi, sorted spleen donors or MLN donors were subjected to RNA extraction and RNA sequencing (RNAseq). Naive OT-I cells were used as a control. (B) Principal component analysis comparing spleen-primed T cells, MLN-primed T cells, and naive T cells. (C) Volcano plot analysis comparing spleen-primed T cells and MLN-primed T cells. (D) Top 10 most upregulated genes in MLN-primed T cells compared to spleen-primed T cells. Those in red can be induced by retinoic acid. (E) Quantitative PCR confirmation of these top 10 most upregulated genes. Relative expression to the housekeeping gene Hmbs was calculated. The data in B–D were generated using biological replicates from three independent experiments. The data in E are pooled from three independent experiments and expressed as mean ± SEM, n = 4–10 mice total. Comparisons among 4.5-d spleen-primed, 5.5-d spleen-primed, and 5.5-d MLN-primed groups were performed using one-way ANOVA with Tukey’s multiple comparisons test. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
CCR9 expression contributes to intestinal CD103+ TRM cell differentiation
Itgae, which encodes CD103, was one of the most upregulated genes in MLN-primed T cells compared with spleen-primed T cells (Fig. 4, D and E). We next examined whether this reduced Itgae gene expression in spleen-primed T cells contributed to their inability to upregulate CD103 protein expression in response to TGF-β. 4.5- and 5.5-d spleen-primed T cells and 5.5-d MLN-primed T cells were cultured with TGF-β in vitro for 24 h followed by flow analysis of CD103 protein expression (Fig. S4 C). While 5.5-d spleen-primed T cells had impaired ability to upregulate CD103 protein expression, 4.5-d spleen-primed T cells were equally efficient as 5.5 d MLN-primed T cells to upregulate CD103 protein expression in response to TGF-β in vitro (Fig. S4 D). Nevertheless, neither 4.5-d spleen-primed T cells nor 5.5-d spleen-primed T cells were able to differentiate into CD103+ TRM precursor cells in vivo (Fig. 1 E and Fig. S2 E). These data collectively suggest that reduced Itgae gene expression was not a major contributor to the inability of spleen-primed CD8 T cells to differentiate into CD103+ TRM precursor cells in the intestine.
Ccr9 was the most upregulated gene in MLN-primed T cells compared with spleen-primed T cells (Fig. 4, D and E). A previous study demonstrated that CCR9 promotes the induction of CD103 expression on CD8 T cells following their entry into the IEL compartment (Ericsson et al., 2004). To determine the impact of CCR9 on CD103+ CD8 TRM cell differentiation in the intestine, naive CD45.1/2+ WT and CD45.2+ Ccr9−/− OT-I cells were co-transferred (1:1 ratio) into CD45.1+ recipient mice 1 d prior to foodborne or i.v. infection with InlAM rLm-OVA. At 8 dpi and memory (>30 d), WT and Ccr9−/− T cells were analyzed in recipient mice (Fig. 5 A). Consistent with previous studies that demonstrated a role of CCR9 in directing effector CD8 T cells to the LP and epithelium of the small intestine (Johansson-Lindbom et al., 2003; Stenstad et al., 2007; Wurbel et al., 2007), Ccr9−/− OT-I cells were two- and eightfold less efficient than WT OT-I cells in migrating into the LP and IEL compartment after foodborne infection, respectively (Fig. 5 B). Compared with WT OT-I cells, Ccr9−/− OT-I cells had a significantly reduced ability to differentiate into CD103+ TRM precursor cells (Fig. 5 C) and CD103+ TRM cells (Fig. 5 D) in the intestine after foodborne infection. Thus, CCR9 promoted intestinal CD103+ CD8 TRM cell differentiation. In line with the observations that spleen-primed OT-I cells did not express appreciable levels of CCR9 (Fig. 1 B and Fig. S2 C), lack of Ccr9 gene expression did not hinder competitive migration into the LP or IEL compartment or the differentiation of CD103+ CD8 TRM cells after i.v. infection (Fig. 5, B–D).
Figure 5.
CCR9 expression contributes to intestinal CD103+ TRM cell differentiation. (A) Experimental design for B–D. 1 × 104 CD45.1/2+ WT OT-I cells from OT-I Rag1−/− mice and 1 × 104 CD45.2+ Ccr9−/− OT-I cells from Ccr9−/− OT-I Rag1−/− mice were co-transferred into CD45.1+ recipient mice 1 d prior to foodborne or i.v. infection with InlAM rLm-OVA. At 8 and >30 dpi, WT and Ccr9−/− T cells were analyzed in recipient mice. (B) Numbers of WT and Ccr9−/− OT-I cells in indicated tissues at 8 dpi. (C) The expression of CD69 and CD103 by WT and Ccr9−/− OT-I cells in the LP and IEL compartment at 8 dpi. (D) The expression of CD69 and CD103 by WT and Ccr9−/− OT-I cells in the LP and IEL compartment at >30 dpi. (E) Experimental design for F–H. 1 × 104 naive OT-I cells were transferred into congenic naive first recipient mice 1 d prior to foodborne infection with InlAM Lm-OVA. At 5.5 dpi, α4β7− CCR9−, α4β7+ CCR9−, and α4β7+ CCR9+ effector OT-I cells were sorted from the MLN and transferred into congenic naive second recipient mice. 3 and 30 d after transfer, donor OT-I cells in the second recipient mice were analyzed. (F) Numbers of indicated donor OT-I cells in the LP and IEL compartment at 3 d after transfer. (G) The expression of CD69 and CD103 by indicated donor OT-I cells in the LP and IEL compartment at 3 d after transfer. (H) The expression of CD69 and CD103 by indicated donor OT-I cells in the LP and IEL compartment 30 d after transfer. (I) Experimental design for J. 1 × 104 CD45.1/2+ WT OT-I cells and 1 × 104 CD45.2+ Ccr9−/− OT-I cells were cotransferred into CD45.1+ recipient mice 1 d prior to foodborne infection with InlAM rLm-OVA. At 5.5 dpi, MLN cells were in vitro cultured in the presence or absence of TGF-β for 24 h followed by flow analysis of CD103 protein expression. (J) The expression of CD103 by WT and Ccr9−/− T cells. The data in B and C are pooled from two independent experiments, n = 6–8 mice total. The data in D are pooled from two independent experiments, n = 6–7 mice total. The data in F and G are representative of three independent experiments, n = 3 mice/experiment. The data in H are pooled from three independent experiments, n = 3 mice total. The data in J are representative of two independent experiments, n = 3 mice/experiment. Each dot in C and D depicts one individual mouse. The data in B, F, G, H, and J are expressed as mean ± SEM. One-way ANOVA with Tukey’s multiple comparisons test was performed (B, F, G, and H). Paired t tests were performed (C and D). Two-way ANOVA with Sidak’s multiple comparisons test was performed between WT and Ccr9−/− groups (J). Only comparisons between blood and other groups as well as between LP and IEL are shown (B). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
To further confirm the role of CCR9 in promoting effector CD8 T cell migration to the intestine and the subsequent CD103+ TRM cell differentiation, α4β7− CCR9−, α4β7+ CCR9−, and α4β7+ CCR9+ effector OT-I cells were sorted from the MLN after foodborne infection and transferred into naïve mice followed by analysis 3 and 30 d (memory) later (Fig. 5 E). α4β7+ CCR9− but not α4β7− CCR9− OT-I cells migrated efficiently into the LP (Fig. 5 F), substantiating a critical role for α4β7 in CD8 T cell migration to the LP (Lefrançois et al., 1999). In addition, significantly more α4β7+ CCR9+ OT-I cells accumulated in the LP and IEL compartment compared with α4β7+ CCR9− OT-I cells (Fig. 5 F), corroborating that CCR9 promoted CD8 T cell migration to the LP and especially the IEL compartment. Importantly, α4β7+ CCR9+ OT-I cells more efficiently differentiated into CD103+ TRM precursor cells at 3 d (Fig. 5 G) and CD103+ TRM cells at 30 d (Fig. 5 H) than α4β7− CCR9− and α4β7+ CCR9− OT-I cells, confirming that CCR9 contributed to the development of CD103+ TRM cells in the intestine. However, it should be noted that although not as efficient as α4β7+ CCR9+ OT-I cells, both α4β7− CCR9− and α4β7+ CCR9− OT-I cells were similarly capable of differentiating into intestinal CD103+ TRM precursor cells and CD103+ TRM cells, especially in the IEL compartment (Fig. 5, G and H), suggesting that the differentiation of CD103+ TRM cells in the intestine is regulated by both CCR9-dependent and -independent mechanisms and that α4β7 expression did not impact CD103+ TRM cell differentiation.
We next determined whether CCR9 promoted CD103+ TRM cell differentiation by directly regulating CD103 expression. Naive CD45.1/2+ WT and CD45.2+ Ccr9−/− OT-I cells were co-transferred into CD45.1+ recipient mice 1 d prior to foodborne infection with InlAM rLm-OVA. At 5.5 dpi, cells were isolated from the MLN and in vitro cultured in the presence or absence of TGF-β for 24 h followed by flow analysis of CD103 protein expression (Fig. 5 I). In response to TGF-β, Ccr9−/− OT-I cells upregulated CD103 expression as efficiently as WT OT-I cells (Fig. 5 J), suggesting that Ccr9−/− T cells were fully competent to upregulate CD103 expression in response to TGF-β in vitro. However, Ccr9−/− OT-I cells were impaired in their ability to upregulate CD103 expression and differentiate into CD103+ TRM cells in vivo (Fig. 5, C and D). Thus, these data suggest that CCR9 contributes to intestinal CD103+ TRM cell differentiation likely by localizing effector CD8 T cells to microregions in the intestine where TGF-β is abundant.
Licensing of intestinal CD103+ TRM cell differentiation occurs through RA signaling with minor contribution from CCR9
We next sought to determine the mechanism of intestinal CD103+ TRM cell licensing. At least five of the most upregulated genes in MLN-primed T cells (Ccr9, Hic 1, Itgae and Lztfl1, P2rx7) can be induced by RA (Burrows et al., 2017; Hashimoto-Hill et al., 2017; Heiss et al., 2008; Iliev et al., 2009; Iwata et al., 2004; Jiang et al., 2016; Fig. 4 D), suggesting that RA may be important for gut CD103+ TRM cell licensing. RA is metabolized from retinol (vitamin A) through a two-step oxidation process (O’Byrne and Blaner, 2013). Retinol is first oxidized to retinal by retinol dehydrogenases and retinal is subsequently oxidized to RA by retinal dehydrogenases (RALDH). MLN but not spleen DC express RALDH2 and are capable of converting retinol to RA and providing RA to instruct CD8 T cells to express α4β7 and CCR9 (Iwata et al., 2004; Svensson et al., 2008). To determine whether RA signals during T cell priming in the MLN licensed CD103+ CD8 TRM cell differentiation, mice were treated with DMSO or a pan-RA receptor (RAR) antagonist AGN194310 to block RA signaling during T cell priming in the MLN after foodborne infection (Fig. 6 A). Alternatively, mice were treated with DMSO or RARα/β agonist AM80 to stimulate RA signaling during T cell priming in the spleen after i.v. infection. Treated MLN-primed and spleen-primed T cells were sorted at 5.5 dpi and transferred into naive congenic recipient mice followed by analysis 3 and 30 d (memory) later. Consistent with the role of RA in inducing α4β7 and CCR9 expression on CD8 T cells, MLN-primed T cells treated with AGN194310 had almost completely abolished α4β7 and CCR9 expression, while spleen-primed T cells treated with AM80 had significantly increased α4β7 and CCR9 expression (Fig. 6 B). Consequently, treatment with AGN194310 led to impaired accumulation of MLN-primed T cells in the LP and IEL compartment, while treatment with AM80 resulted in enhanced accumulation of spleen-primed T cells in these compartments (Fig. S5 A). We next determined whether modulating RA signaling during T cell priming had an impact on intestinal CD103+ CD8 TRM cell differentiation. Of note, neither AGN194310 nor AM80 treatment affected KLRG-1 expression (Fig. S5 B). Strikingly, AGN194310 treatment inhibited MLN-primed T cells from differentiating into CD103+ TRM precursor cells at 3 d and CD103+ TRM cells at 30 d in the intestine (Fig. 6, C and D), suggesting RA signaling is necessary for CD103+ TRM cell licensing. Of note, AGN194310 treatment also inhibited spleen-primed T cells from differentiating into CD103+ TRM precursor cells (Fig. S5, C and D). In contrast, AM80 treatment rendered spleen-primed T cells the ability to differentiate into intestinal CD103+ TRM precursor cells at 3 d and CD103+ TRM cells at 30 d to the same level as MLN-primed T cells (Fig. 6, C and D), suggesting RA signaling is sufficient to license CD103+ TRM cell differentiation. Of note, AM80 treatment also enhanced the ability of MLN-primed T cells to rapidly differentiate into intestinal CD103+ TRM precursor cells (Fig. S5, C and D). Thus, these data provide compelling evidence that RA signals during MLN priming license intestinal CD103+ TRM cell differentiation.
Figure 6.
Licensing of rapid intestinal CD103+ TRM cell differentiation occurs through RA signaling that is primarily driven by factors other than CCR9. (A) Experimental design for B–D. 1 × 104 naive OT-I cells were transferred into congenic naive first recipient mice 1 d prior to i.v. or foodborne infection with InlAM Lm-OVA. For i.v. infection, mice were treated with DMSO control or AM80 at 0.5, 2.5, and 4.5 dpi. For foodborne infection, mice were treated with DMSO control or AGN194310 at 0.5, 2.5, and 4.5 dpi. At 5.5 dpi, effector OT-I cells were sorted from the spleen of i.v.-infected mice treated with DMSO (Spleen_DMSO) or AM80 (Spleen_AM80) or from the MLN of foodborne infected mice treated with DMSO (MLN_DMSO) or AGN194310 (MLN_AGN) and transferred into congenic naive second recipient mice. 3 and 30 d after transfer, donor OT-I cells in the second recipient mice were analyzed. (B) The expression of α4β7 and CCR9 by effector OT-I cells prior to adoptive transfer into the second recipient mice. (C) The expression of CD69 and CD103 by donor OT-I cells in the LP and IEL compartment at 3 d after transfer. (D) The expression of CD69 and CD103 by donor OT-I cells in the LP and IEL compartment 30 d after transfer. (E) Experimental design for F–H. 1 × 104 WT or Ccr9−/− OT-I cells were transferred into congenic naive first recipient mice 1 d prior to i.v. infection with InlAM Lm-OVA. Mice were treated with DMSO control or AM80 at 0.5, 2.5, and 4.5 dpi. At 5.5 dpi, WT and Ccr9−/− effector OT-I cells were sorted from the spleen of DMSO treated (WT_DMSO or Ccr9−/−_DMSO) or AM80 treated (WT_AM80 or Ccr9−/−_AM80) mice and transferred into congenic naive second recipient mice. 3 and 30 d after transfer, donor OT-I cells in the second recipient mice were analyzed. (F) The expression of α4β7 and CCR9 by effector OT-I cells prior to adoptive transfer into second recipient mice. (G) The expression of CD69 and CD103 by donor OT-I cells in the LP and IEL compartment at 3 d after transfer. (H) The expression of CD69 and CD103 by donor OT-I cells in the LP and IEL compartment 30 d after transfer. The data in B are pooled from three independent experiments, n = 5–6 mice total. The data in C are representative of two independent experiments, n = 4 mice/experiment. The data in D are pooled from four independent experiments, n = 6–8 mice total. The data in F are representative of four independent experiments. The data in G are representative of two independent experiments, n = 4 mice/experiment. The data in H are pooled from two independent experiments, n = 5 total. The data are expressed as mean ± SEM. One-way ANOVA with Tukey’s multiple comparisons test was performed (B–D, G, and H). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Figure S5.
The effect of RA signaling modulation on effector T cell migration to the intestine, KLRG-1 expression, and TRM precursor cell differentiation. (A) The normalized percentage of indicated donor OT-I cells in the LP and IEL compartment at 3 d after transfer. (B) The percentage of KLRG-1− population in indicated effector OT-I cells prior to adoptive transfer. (C) Experimental design for D. 1 × 104 naive OT-I cells were transferred into congenic naive first recipient mice 1 d prior to i.v. or foodborne infection with InlAM Lm-OVA. Mice were treated with DMSO control, AM80, or AGN194310 at 0.5, 2.5, and 4.5 dpi. At 5.5 dpi, effector OT-I cells were sorted from the spleen of i.v.-infected or the MLN of foodborne-infected mice and transferred into congenic naive second recipient mice. 3 d after transfer, donor OT-I cells in second recipient mice were analyzed. (D) The expression of CD69 and CD103 by indicated donor OT-I cells in the LP and IEL compartment at 3 d after transfer. (E) The percentage of CD69+ CD103− population in indicated donor OT-I cells in the LP and IEL compartment at 3 d after transfer. (F) The percentage of CD69+ CD103− population in indicated donor OT-I cells in the LP and IEL compartment at 30 d after transfer. (G) The normalized percentage of indicated donor OT-I cells in the LP and IEL compartment at 3 d after transfer. The data in A are representative of two independent experiments, n = 4 mice/experiment. The data in B are pooled from three independent experiments, n = 5–6 mice total. The data in D are pooled from two independent experiments, n = 4–5 mice total. The data in E are representative of two independent experiments, n = 4 mice/experiment. The data in F are pooled from four independent experiments, n = 6–8 mice total. The data in G are representative of two independent experiments, n = 4 mice/experiment. The data are expressed as mean ± SEM. One-way ANOVA with Tukey’s multiple comparisons tests were performed (A, B, and D–G). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
We next examined whether RA signaling regulated CD103− TRM cell differentiation. While blocking RA signaling using AGN194310 significantly inhibited the differentiation of MLN-primed T cells into CD103− TRM precursor cells in the LP at 3 d (Fig. S5 E), stimulating RA signaling using AM80 significantly inhibited the differentiation of spleen-primed T cells into CD103− TRM cells in the IEL compartment at 30 d (Fig. S5 F). Overall, the impact of RA signaling on CD103− TRM cell differentiation appears less substantial and more variable.
Since RA plays a crucial role in inducing CCR9 expression on CD8 T cells, and both CCR9-dependent and -independent mechanisms contributed to CD103+ TRM cell differentiation (Fig. 5, G and H), we sought to determine the relative contribution of CCR9-dependent and -independent mechanisms to RA-licensed intestinal CD103+ TRM cell differentiation. To this end, WT and Ccr9−/− naive OT-I cells were transferred into congenic mice 1 d prior to i.v. infection (Fig. 6 E). Mice were treated with DMSO or AM80 during T cell priming. At 5.5 dpi, spleen-primed T cells were sorted and transferred into naive congenic recipient mice followed by analysis at 3 and 30 d (memory) later. As expected, while AM80 treatment drastically increased α4β7 expression in both WT and Ccr9−/− T cells, it only induced CCR9 expression in WT but not Ccr9−/− T cells (Fig. 6 F). As a result, AM80 treatment led to a significantly enhanced accumulation of WT T cells in both the LP and IEL compartment (Fig. S5 G). However, the accumulation of Ccr9−/− T cells in the LP and IEL compartment was only slightly enhanced (Fig. S5 G), consistent with the findings that CCR9 promotes CD8 T cell migration to both compartments. AM80 treatment during T cell priming rendered spleen-primed WT T cells the ability to differentiate into intestinal CD103+ TRM precursor cells at 3 d and CD103+ TRM cells at 30 d (Fig. 6, G and H). Intriguingly, AM80 treatment also enabled Ccr9−/− T cells to differentiate into intestinal CD103+ TRM precursor cells at 3 d and CD103+ TRM cells at 30 d (Fig. 6, G and H). The ability of Ccr9−/− T cells to differentiate into CD103+ TRM precursor cells and CD103+ TRM cells in the IEL compartment was at ∼75% of the level of WT T cells (Fig. 6, G and H). Thus, RA signaling licensed intestinal CD103+ TRM cell differentiation, primarily driven by factors other than CCR9 induction and CCR9-mediated gut homing.
RA signaling modulates intestinal CD103+ TRM cell formation after infection
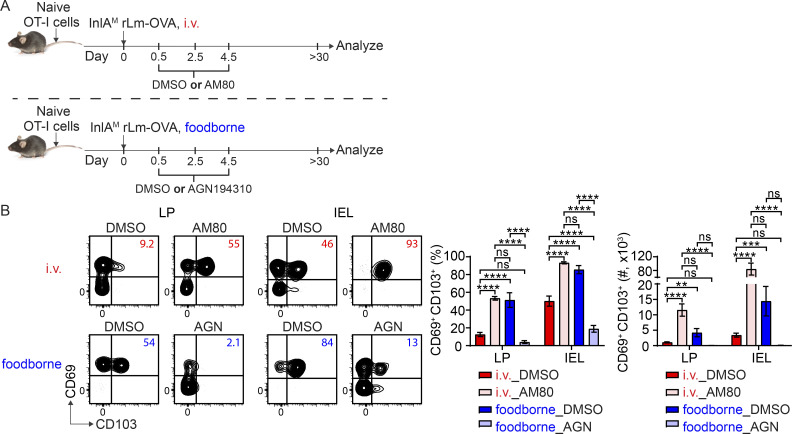
As RA signaling is sufficient and necessary for intestinal CD103+ TRM cell differentiation, we next determined whether we could modulate RA signaling to alter CD103+ TRM cell formation in the intestine without secondary transfer to determine whether this pathway has the potential for therapeutic manipulation. 1 × 104 naive OT-I cells were transferred into congenic naive recipient mice. 1 d later, mice were i.v. or foodborne infected with InlAM Lm-OVA. For i.v. infection, mice were treated with DMSO or AM80 at 0.5, 2.5, and 4.5 dpi. For foodborne infection, mice were treated with DMSO or AGN194310 at 0.5, 2.5, and 4.5 dpi. Donor OT-I cells were analyzed at >30 dpi (Fig. 7 A). Comparisons between DMSO-treated groups demonstrated that i.v. infection led to significantly less CD103+ TRM cells in the LP and IEL compartment than foodborne infection (Fig. 7 B). Importantly, stimulation of RA signaling with AM80 significantly enhanced CD103+ TRM cell formation after i.v. infection, and AGN194310 treatment that inhibits RA signaling reduced CD103+ TRM cell formation after foodborne infection (Fig. 7 B). These data indicated that CD103+ TRM cell formation could be altered by modifying RA signaling.
Figure 7.
RA signaling modulates intestinal CD103+ TRM cell formation after infection. (A) Experimental design. 1 × 104 naive OT-I cells were transferred into congenic naive recipient mice. 1 d later, mice were i.v. or foodborne infected with InlAM Lm-OVA. For i.v. infection, mice were treated with DMSO control or AM80 at 0.5, 2.5, and 4.5 dpi. For foodborne infection, mice were treated with DMSO control or AGN194310 at 0.5, 2.5, and 4.5 dpi. Donor OT-I cells were analyzed at >30 dpi. (B) The percentage and number of CD69+ CD103+ donor OT-I cells in the LP and IEL compartment at 30 dpi. The data are representative of two independent experiments, n = 6–8 mice/experiment. The data are expressed as mean ± SEM. One-way ANOVA with Tukey’s multiple comparisons test was performed. **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Discussion
Developing vaccine strategies to target induction of robust CD8 TRM cell populations is a topic of ongoing clinical interest. As such, understanding the mechanisms that regulate the development of CD8 TRM cells in their unique environments is paramount to facilitate the ability to generate and manipulate these cells for therapeutic benefits. This is especially relevant to current vaccine strategies targeting gastrointestinal infections and tumors that are primarily delivered parenterally. It is generally accepted that priming dictates the migratory pattern of effector CD8 T cells by instruction with tissue-homing receptors (Park and Kupper, 2015; Sheridan and Lefrançois, 2011). Priming in the MLN promotes the migration of effector CD8 T cells to the intestine, providing a crucial layer regulating effector CD8 T cell responses in the gut. While spleen-primed CD8 T cells are less efficient at gut homing, CD8 T cells that are primed in the spleen are still detectable in gut tissues. Here, our data established that priming in the MLN also promotes in situ CD103+ CD8 TRM cell differentiation in the intestine, providing an additional regulatory layer for the development of effector and memory CD8 T cell responses in the gut. MLN priming but not spleen priming promoted CD8 T cell differentiation into CD103+ TRM cells in the intestinal LP and epithelium. Consistent with previous studies (Masopust et al., 2006), unique cues from the intestinal environmental induced CD103+ TRM cell differentiation as MLN-primed T cells that migrated to the spleen after adoptive transfer did not become CD103+ TRM cells. Importantly, spleen-primed CD8 T cells that migrated to the intestine were inefficient at becoming CD103+ TRM cells in the intestinal environment. Thus, compelling evidence was provided to demonstrate that priming in the MLN licensed effector CD8 T cells to respond to environmental cues in the intestine to efficiently differentiate into CD103+ TRM cells. We further demonstrated that MLN priming initiated a TRM cell signature before emigration from the MLN and licensed rapid CD103+ TRM cell differentiation through RA signaling. Although CCR9 promoted CD103+ TRM cell differentiation to some extent, RA signaling primarily regulated intestinal CD103+ TRM cell differentiation through other CCR9-independent mechanisms. Therefore, our study established that RA signaling during T cell priming in the MLN promotes CD103+ TRM cell differentiation in the gut.
Our data also demonstrated that in addition to priming, the MLN could impart an environment signal to promote effector CD8 T cell differentiation into CD103+ TRM precursor cells in the intestine. However, this mechanism was not as efficient as priming in the MLN for the induction of intestinal CD103+ CD8 TRM cells. MLN stromal cells can also express high levels of RALDH and convert retinol to RA to support the induction of gut-homing T cells (Hammerschmidt et al., 2008; Molenaar et al., 2009). Therefore, it is possible that RA derived from stromal cells contributes to the intrinsic environment of the MLN that contributes to CD103+ TRM precursor cell licensing.
Previous studies demonstrated that intranasal infection of InlAM rLm is inefficient at promoting gut CD103+ TRM cell differentiation (Sheridan et al., 2014), corroborating the critical role of MLN priming in regulating intestinal CD103+ TRM cell differentiation. However, whether this is unique to the gut and gut-draining lymph nodes requires further studies. The skin environment alone appears sufficient for CD103+ TRM cell differentiation regardless of whether CD8 T cells are primed in vivo or activated in vitro (Mackay et al., 2013; Mackay et al., 2012). A recent study demonstrated that αV-expressing migratory DC activate and present TGF-β to naive CD8 T cells during homeostasis to precondition them to become skin CD103+ TRM cells upon priming (Mani et al., 2019). Spleen-primed effector CD8 T cells generated with i.v. rLm-OVA infection efficiently differentiated into CD103+ TRM cells after being pulled into the skin epidermis by topical treatment with an inflammation-inducing agent. However, our study provides strong evidence that preconditioning does not contribute to CD103+ TRM cell differentiation in the intestine. These distinctions highlight the complexity and unique biology that individual tissues contribute to TRM cell development.
TGF-β is essential for the differentiation of CD103+ CD8 TRM cells in the intestine as CD8 T cells deficient in TGF-β receptor II are not able to differentiate into intestinal CD103+ TRM cells (Casey et al., 2012; Sheridan et al., 2014; Zhang and Bevan, 2013). Our data showed that RA plays an important role in licensing effector CD8 T cell responsiveness to factors in the intestinal environment for differentiation into CD103+ TRM cells. However, how RA signaling received during MLN priming regulates CD103+ CD8 TRM cell differentiation in the intestine in response to TGF-β needs further exploration. Our data demonstrate that MLN priming provided RA signaling to license CD103+ TRM cell differentiation to a limited extent through a CCR9-dependent mechanism. In addition to directing the migration of effector CD8 T cells to the intestine, CCR9 likely further direct effector T cells that have already infiltrated the intestine to microanatomical areas where TGF-β is abundant, although the detailed mechanism requires further studies. However, a CCR9-independent mechanism appears to play the major role in licensing CD103+ TRM cell development as CCR9− MLN-primed effector CD8 T cells as well as AM80-treated spleen-primed Ccr9−/− effector CD8 T cells were capable of differentiating into CD103+ TRM cells. RA has been shown to directly induce CD103 expression on DC (Iliev et al., 2009), although this is unlikely to be the case for CD8 T cells as effector CD8 T cells that received RA signaling either through MLN-priming or RARα/β agonism did not upregulate CD103 expression after entering the spleen. RA has been shown to induce Hic1 expression on T cells, and T cell–intrinsic expression of Hic1 promotes CD103+ resident LP lymphocytes and IEL during homeostasis (Burrows et al., 2017). A recent study also showed that Hic1 is upregulated in TRM cells in the IEL compartment after infection and regulates TRM cell formation (Crowl et al., 2022). Since our data showed that Hic1 expression was upregulated in MLN-primed but not spleen-primed CD8 T cells prior to migration to the intestine, it is possible that RA signaling received during priming in the MLN promotes the differentiation of CD103+ TRM cells in the intestine through Hic1. RA is produced both in the intestinal epithelium and the MLN (Iliev et al., 2009; Iwata et al., 2004); therefore, effector CD8 T cells can receive RA signaling either during priming or after migration into the intestine to upregulate Hic1. Our data clearly showed that RA signaling received during priming is critical for intestinal CD103+ TRM cell differentiation. However, it is unclear whether RA signaling received in the intestine also contributes to TRM cell formation or homeostasis. Furthermore, AM80 treatment rendered the ability of spleen-primed CD8 T cells to differentiate into CD103+ TRM cells, indicating that spleen-primed CD8 T cells can respond to RA. However, whether their inability to differentiate into CD103+ TRM cells is due to the lack of RA signaling during priming or their inability to see RA after migrating into the intestine requires further investigation. RA has also been shown to induce P2rx7 expression (Hashimoto-Hill et al., 2017; Heiss et al., 2008), and P2rx7 promotes CD103+ CD8 TRM cell generation and maintenance by upregulating CD103 and enabling the ability of CD8 T cells to respond to TGF-β (Borges da Silva et al., 2020). Our data showed that P2rx7 was one of the top 10 most upregulated genes in MLN-primed T cells. Therefore, RA signaling received during priming in the MLN may also contribute to intestinal CD103+ TRM cell differentiation through P2rx7. A recent study showed that changes in Hic1 expression lead to changes in P2rx7 expression (Crowl et al., 2022), suggesting that RA regulation of P2rx7 expression can be both direct and indirect. In all, it is possible that RA signaling orchestrates multiple pathways collectively contributing to the differentiation of intestinal CD103+ TRM cells.
Overall, our study provides crucial insight into the mechanism regulating intestinal CD103+ CD8 TRM cell development that may shed light on rational vaccine design targeting infections and cancers of gastrointestinal tract. It emphasizes the importance of utilizing oral immunization route to target MLN priming for generating robust CD103+ CD8 TRM cell population in the intestine and provides a new regime to modulate RA signaling to tailor the development of these cells for potential therapeutic applications.
Materials and methods
Mice
C57BL/6 mice for experiments without OT-I cell transfer were purchased from The Jackson Laboratory. C57BL/6 mice expressing the congenic markers CD45.1 or CD45.2 for experiments with OT-I cell transfer were purchased from the Charles River-National Cancer Institute. OT-I Rag1−/− mice expressing CD45.1, CD45.2, or both and Ccr9−/− OT-I Rag1−/− mice expressing CD45.2 were bred and maintained at Stony Brook University under specific-pathogen-free conditions. For experiments using Ccr9−/− OT-I Rag1−/− mice, WT OT-I Rag1−/− mice were used as controls. 8–12-wk-old, age-matched female mice were used in experiments. All animal procedures were carried out in accordance with National Institutes of Health (NIH) guidelines and approved by the Stony Brook University Institutional Animal Care and Use Committee.
Bacteria
L. monocytogenes strain 10403s carrying a mutation in the internalin A protein and expressing a truncated form of OVA (InlAM rLm-OVA) have been described previously (Plumlee et al., 2013). Prior to infection, mice were deprived of food and water for 6 h. Foodborne infection was performed by providing ∼1 cm3 piece of bread inoculated with 2 × 109 cfu of InlAM rLm-OVA in PBS to individually housed mice (Chu et al., 2022). I.v. infection was performed by tail vein injection of 2 × 103 cfu of InlAM rLm-OVA in PBS.
Adoptive transfers
1 × 104 CD45.1+ OT-I cells isolated from the spleen of OT-I Rag1−/− mice were i.v. transferred into naive CD45.2+ recipient mice 1 d prior to i.v. or foodborne infection with InlAM rLm-OVA. 5.5 d after infection, unless otherwise specified, effector OT-I cells were isolated from the spleen of i.v.-infected mice and the MLN of foodborne-infected mice based on the congenic marker CD45.1. Briefly, tissues were mashed through 70-µm cell strainers (Falcon) to obtain single-cell suspension. Cells were stained with anti-CD45.1-biotin (A20; BioLegend) followed by incubation with anti-biotin microbeads (Miltenyi Biotec) and enrichment on a MACS Separator (Miltenyi Biotec). After enrichment, cells were stained with the following fluorophore-conjugated antibodies from BioLegend or Invitrogen: anti-CD4 (GK1.5), anti-MHC II (M5/114.15.2), anti-CD8α (53–6.7), anti-CD45.1 (A20), anti-CD45.2 (104), and live/dead dye (Invitrogen) for fluorescence-activated cell sorting. Single, live, CD4− MHC II− CD8α+ CD45.2− CD45.1+ effector OT-I cells were sorted using a FACSARIA III cell sorter (BD). The purity after sorting was consistently >95%. 2-5 × 105 (for early time point) or 2-5 × 106 (for memory time point) effector OT-I cells were transferred intravenously into indicated CD45.2+ recipient mice unless otherwise indicated. Alternatively, CD45.2+ OT-I cells and CD45.1+ recipient mice were used. For WT and Ccr9−/− OT-I cotransfer experiments, 1 × 104 CD45.1/2+ WT OT-I cells from OT-I Rag1−/− mice and 1 × 104 CD45.2+ Ccr9−/− OT-I cells from Ccr9−/− OT-I Rag1−/− mice were cotransferred into CD45.1+ recipient mice.
Lymphocyte isolation
To isolate lymphocytes from the spleen and MLN, tissues were mashed through 70-µm cell strainers. Small intestine LP lymphocytes and IEL were isolated as previously described (Qiu and Sheridan, 2018; Sheridan and Lefrançois, 2012). Briefly, small intestines were cut into 1-inch long pieces after removal of Peyer’s patches and luminal content. Intestinal tissues were treated twice with 1 mM dithioerythritol (Sigma-Aldrich) solution in a shaker at 220 rpm and 37°C for 20 min. Supernatants were collected, combined, and subjected to 44%/67% Percoll (GE Healthcare) gradient for the isolation of IEL. The remaining intestine tissues were treated twice with 1.3 mM ethylenediaminetetraacetic acid (Invitrogen) solution in a shaker at 220 rpm and 37°C for 30 min to remove intestinal epithelial cells, followed by digestion with 100 U/ml of collagenase (Invitrogen) in a shaker at 300 rpm and 37°C for 45 min. Supernatants were collected after collagenase digestion and the undigested tissues were mashed through 70-µm cell strainers into the collected supernatant, which was then subjected to 44%/67% Percoll gradient for the isolation of LP lymphocytes.
Flow cytometry and antibodies
The following fluorophore-conjugated antibodies from BioLegend, Invitrogen, or BD Biosciences were used: anti-CD45 (30-F11), anti-TCRβ (H57-597), anti-CD8α (53–6.7), anti-CD11a (M17/4), anti-CD45.1 (A20), anti-CD45.2 (104), anti-CD103 (2E7), anti-CD69 (H1.2F3), anti-α4β7 (DATK32), anti-CCR9 (CW-1.2), and anti-KLRG-1 (2F1/KRLG1). Live/dead dye was purchased from Invitrogen. H-2 Kb OVA257 monomer was obtained from the NIH Tetramer Core Facility and tetramerized in-house using fluorophore-conjugated streptavidin (Invitrogen). Cells were stained with antibodies at 4°C in the dark for 20 min. If tetramer was used, staining was performed at room temperature in the dark for 1 h. After staining, cells were fixed with 2% paraformaldehyde (Electron Microscopy Sciences) for 20 min prior to acquisition on a LSRFortessa (BD) or an Aurora (Cytek). Data were analyzed with FlowJo software (Tree Star).
FTY720 treatment
FTY720 (Cayman Chemical) was dissolved in PBS with 25% DMSO. Mice were injected intraperitoneally with vehicle control or 1 mg/kg FTY720 daily for 3 wk.
Anti-αvβ8 treatment
Anti-αvβ8 (ADWA-11) was kindly provided by Dr. Dean Sheppard (University of California, San Francisco). Mice were injected intraperitoneally with PBS control or 10 mg/kg anti-αvβ8 weekly for 3 wk.
RNA sequencing and data processing
RNA was extracted using QIAGEN RNeasy Plus Micro Kit. Libraries were prepared using Illumina TruSeq Stranded mRNA library kit at the Center for Genome Innovation at UCONN Storrs, CT, USA. RNA sequencing and data analysis were performed by Macrogen. Fold changes were calculated based on log2(FPKM+1). Principle component analysis and volcano plot analysis were performed using R.
Quantitative PCR
RNA was isolated using RNeasy Plus Micro Kit (QIAGEN) and cDNA was generated using iScript Advanced cDNA Synthesis Kit (Bio-Rad). Real-time RT-PCR was performed on a Bio-Rad CFX96 using PrimePCR Assays purchased from Bio-Rad (qMmuCID0009067 for Ccr9, qMmuCED0040393 for Hic1, qMmuCID0019169 for Car2, qMmuCID0020840 for Batf3, qMmuCID0039603 for Itgae, qMmuCID0026896 for Lztfl1, qMmuCID0006409 for Xcl1, qMmuCID0040113 for P2rx7, qMmuCED0041023 for Npm1, qMmuCED0061466 for Rangrf, and qMmuCID0022816 for Hmbs).
In vitro culture
4 × 106 cells isolated from the MLN after foodborne infection or from the spleen after i.v. infection were cultured in the absence or presence of 5 ng/ml TGF-β1 (Peprotech), unless otherwise specified, in a 48-well plate with 250 µl culture medium per well for 24 h. RPMI medium 1640 (Gibco) supplemented with 10% FBS (Gibco), 1% GlutaMAX (Gibco), 10 mM Hepes (Gibco), 1 mM sodium pyruvate (Gibco), 100 units/ml penicillin (Gibco), 100 µg/ml streptomycin (Gibco), 20 µg/ml gentamicin (Gibco), and 50 µM 2-mercaptoethanal (Sigma-Aldrich) was used as culture medium.
Chemical modulation of RA signaling
Mice were injected intraperitoneally with DMSO, 5 mg/kg RARα/β agonist AM80 (Sigma-Aldrich), or 1 mg/kg pan-RAR antagonist AGN194310 every other day starting at day 0.5 until euthanization. The dosage was determined based on previous studies and our own titration experiments (Allie et al., 2013; Matsushita et al., 2011).
Bacterial burdens
Spleen and MLN were mashed through 70-µm cell strainers (Thermo Fisher Scientific) using sterile 1% saponin (Calbiochem). Gut tissues were disassociated using C Tubes and gentleMACS Dissociator (Miltenyi Biotec) in nine volumes of sterile PBS. After disassociation, 1 volume of sterile 10% saponin was added. Tissue homogenates were incubated at 4°C for 1 h before plating on BHI agar plates supplemented with 200 µg/ml streptomycin. Bacterial colonies were enumerated 24–48 h after incubation at 37°C.
Statistical analysis
All statistical analyses were performed in Prism (GraphPad Software) using two-tailed Student’s t test for comparisons of two groups and one-way ANOVA for comparisons of more than two groups. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Online supplemental material
Fig. S1 shows the expression of CD69 and CD103 by effector OT-I cells prior to entry into the intestine, the impact of priming on CD103− TRM precursor cell differentiation, and the impact of intrinsic environment of the MLN on CD103+ TRM precursor cell differentiation. Fig. S2 shows that rapid intestinal CD103+ CD8 TRM precursor cell differentiation is independent of activation stage, intestinal homing, and KLRG-1 expression. Fig. S3 demonstrates the impact of infection on intestinal CD103+ TRM cell differentiation and the residency of intestinal memory CD8 T cells derived from spleen-primed T cells. Fig. S4 demonstrates the sorting strategy for CD103− and CD103+ naive OT-I cells, the expression of CD69 and CD103 by transferred CD103− and CD103+ naive OT-I cells after activation, and the ability to upregulate CD103 expression in response to TGF-β by spleen and MLN donors. Fig. S5 shows the effect of RA signaling modulation on effector T cell migration to the intestine, KLRG-1 expression, and TRM precursor cell differentiation.
Supplementary Material
lists differentially regulated genes between spleen-primed and MLN-primed T cells.
Acknowledgments
We thank Drs. Adrianus van der Velden (Stony Brook University) and Pawan Kumar (Stony Brook University) for constructive discussion on the manuscript. We acknowledge the support of the NIH Tetramer Core Facility for providing MHC I monomers and Dr. Dean Sheppard (University of California, San Francisco, San Francisco, CA, USA) for providing the anti-αvβ8 antibody. Graphical abstract was created with Biorender.com.
This study was supported by NIH awards K12GM102778 (Z. Qiu), P01AI056172 (L. Puddington), R01AI076457 (B.S. Sheridan), R01AI172919 (B.S. Sheridan), R21AI137929 (B.S. Sheridan), and funds provided by Stony Brook University (B.S. Sheridan).
Author contributions: B.S. Sheridan and Z. Qiu directed and designed the study. Z. Qiu, C. Khairallah, T.H. Chu, J.N. Imperato, X. Lei, and G. Romanov performed the experiments. L. Puddington and A. Atakilit provided resources. Z. Qiu analyzed the data. Z. Qiu and B.S. Sheridan wrote the manuscript.
Data availability
RNA sequencing data in Fig. 4 has been deposited in Gene Expression Omnibus with the accession number GSE223983. All other data are available from the corresponding author.
References
- Allie, S.R., Zhang W., Tsai C.Y., Noelle R.J., and Usherwood E.J.. 2013. Critical role for all-trans retinoic acid for optimal effector and effector memory CD8 T cell differentiation. J. Immunol. 190:2178–2187. 10.4049/jimmunol.1201945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariotti, S., Hogenbirk M.A., Dijkgraaf F.E., Visser L.L., Hoekstra M.E., Song J.Y., Jacobs H., Haanen J.B., and Schumacher T.N.. 2014. T cell memory. Skin-resident memory CD8⁺ T cells trigger a state of tissue-wide pathogen alert. Science. 346:101–105. 10.1126/science.1254803 [DOI] [PubMed] [Google Scholar]
- Bankovich, A.J., Shiow L.R., and Cyster J.G.. 2010. CD69 suppresses sphingosine 1-phosophate receptor-1 (S1P1) function through interaction with membrane helix 4. J. Biol. Chem. 285:22328–22337. 10.1074/jbc.M110.123299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsbaken, T., and Bevan M.J.. 2015. Proinflammatory microenvironments within the intestine regulate the differentiation of tissue-resident CD8⁺ T cells responding to infection. Nat. Immunol. 16:406–414. 10.1038/ni.3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsbaken, T., Bevan M.J., and Fink P.J.. 2017. Local inflammatory cues regulate differentiation and persistence of CD8+ tissue-resident memory T cells. Cell Rep. 19:114–124. 10.1016/j.celrep.2017.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best, J.A., Blair D.A., Knell J., Yang E., Mayya V., Doedens A., Dustin M.L., Goldrath A.W., and Immunological Genome Project Consortium . 2013. Transcriptional insights into the CD8(+) T cell response to infection and memory T cell formation. Nat. Immunol. 14:404–412. 10.1038/ni.2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges da Silva, H., Peng C., Wang H., Wanhainen K.M., Ma C., Lopez S., Khoruts A., Zhang N., and Jameson S.C.. 2020. Sensing of ATP via the purinergic receptor P2RX7 promotes CD8+ trm cell generation by enhancing their sensitivity to the cytokine TGF-β. Immunity. 53:158–171.e6. 10.1016/j.immuni.2020.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows, K., Antignano F., Bramhall M., Chenery A., Scheer S., Korinek V., Underhill T.M., and Zaph C.. 2017. The transcriptional repressor HIC1 regulates intestinal immune homeostasis. Mucosal Immunol. 10:1518–1528. 10.1038/mi.2017.17 [DOI] [PubMed] [Google Scholar]
- Casey, K.A., Fraser K.A., Schenkel J.M., Moran A., Abt M.C., Beura L.K., Lucas P.J., Artis D., Wherry E.J., Hogquist K., et al. 2012. Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J. Immunol. 188:4866–4875. 10.4049/jimmunol.1200402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheuk, S., Schlums H., Gallais Sérézal I., Martini E., Chiang S.C., Marquardt N., Gibbs A., Detlofsson E., Introini A., Forkel M., et al. 2017. CD49a expression defines tissue-resident CD8+ T cells poised for cytotoxic function in human skin. Immunity. 46:287–300. 10.1016/j.immuni.2017.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, T.H., Qiu Z., and Sheridan B.S.. 2022. The use of foodborne infection to evaluate bacterial pathogenesis and host response. Methods Cell Biol. 168:299–314. 10.1016/bs.mcb.2021.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowl, J.T., Heeg M., Ferry A., Milner J.J., Omilusik K.D., Toma C., He Z., Chang J.T., and Goldrath A.W.. 2022. Tissue-resident memory CD8+ T cells possess unique transcriptional, epigenetic and functional adaptations to different tissue environments. Nat. Immunol. 23:1121–1131. 10.1038/s41590-022-01229-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodagatta-Marri, E., Ma H.Y., Liang B., Li J., Meyer D.S., Chen S.Y., Sun K.H., Ren X., Zivak B., Rosenblum M.D., et al. 2021. Integrin αvβ8 on T cells suppresses anti-tumor immunity in multiple models and is a promising target for tumor immunotherapy. Cell Rep. 36:109309. 10.1016/j.celrep.2021.109309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson, A., Svensson M., Arya A., and Agace W.W.. 2004. CCL25/CCR9 promotes the induction and function of CD103 on intestinal intraepithelial lymphocytes. Eur. J. Immunol. 34:2720–2729. 10.1002/eji.200425125 [DOI] [PubMed] [Google Scholar]
- Ganesan, A.P., Clarke J., Wood O., Garrido-Martin E.M., Chee S.J., Mellows T., Samaniego-Castruita D., Singh D., Seumois G., Alzetani A., et al. 2017. Tissue-resident memory features are linked to the magnitude of cytotoxic T cell responses in human lung cancer. Nat. Immunol. 18:940–950. 10.1038/ni.3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt, T., Wakim L.M., Eidsmo L., Reading P.C., Heath W.R., and Carbone F.R.. 2009. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat. Immunol. 10:524–530. 10.1038/ni.1718 [DOI] [PubMed] [Google Scholar]
- Hammerschmidt, S.I., Ahrendt M., Bode U., Wahl B., Kremmer E., Förster R., and Pabst O.. 2008. Stromal mesenteric lymph node cells are essential for the generation of gut-homing T cells in vivo. J. Exp. Med. 205:2483–2490. 10.1084/jem.20080039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto-Hill, S., Friesen L., Kim M., and Kim C.H.. 2017. Contraction of intestinal effector T cells by retinoic acid-induced purinergic receptor P2X7. Mucosal Immunol. 10:912–923. 10.1038/mi.2016.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss, K., Jänner N., Mähnss B., Schumacher V., Koch-Nolte F., Haag F., and Mittrücker H.W.. 2008. High sensitivity of intestinal CD8+ T cells to nucleotides indicates P2X7 as a regulator for intestinal T cell responses. J. Immunol. 181:3861–3869. 10.4049/jimmunol.181.6.3861 [DOI] [PubMed] [Google Scholar]
- Hirai, T., Yang Y., Zenke Y., Li H., Chaudhri V.K., De La Cruz Diaz J.S., Zhou P.Y., Nguyen B.A., Bartholin L., Workman C.J., et al. 2021. Competition for active TGFβ cytokine allows for selective retention of antigen-specific tissue- resident memory T cells in the epidermal niche. Immunity. 54:84–98.e5. 10.1016/j.immuni.2020.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai, T., Zenke Y., Yang Y., Bartholin L., Beura L.K., Masopust D., and Kaplan D.H.. 2019. Keratinocyte-mediated activation of the cytokine TGF-β maintains skin recirculating memory CD8+ T cells. Immunity. 50:1249–1261.e5. 10.1016/j.immuni.2019.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev, I.D., Spadoni I., Mileti E., Matteoli G., Sonzogni A., Sampietro G.M., Foschi D., Caprioli F., Viale G., and Rescigno M.. 2009. Human intestinal epithelial cells promote the differentiation of tolerogenic dendritic cells. Gut. 58:1481–1489. 10.1136/gut.2008.175166 [DOI] [PubMed] [Google Scholar]
- Imperato, J.N., Xu D., Romagnoli P.A., Qiu Z., Perez P., Khairallah C., Pham Q.M., Andrusaite A., Bravo-Blas A., Milling S.W.F., et al. 2020. Mucosal CD8 T cell responses are shaped by batf3-DC after foodborne Listeria monocytogenes infection. Front. Immunol. 11:575967. 10.3389/fimmu.2020.575967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata, M., Hirakiyama A., Eshima Y., Kagechika H., Kato C., and Song S.Y.. 2004. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 21:527–538. 10.1016/j.immuni.2004.08.011 [DOI] [PubMed] [Google Scholar]
- Jiang, H., Promchan K., Lin B.R., Lockett S., Chen D., Marshall H., Badralmaa Y., and Natarajan V.. 2016. LZTFL1 upregulated by all-trans retinoic acid during CD4+ T cell activation enhances IL-5 production. J. Immunol. 196:1081–1090. 10.4049/jimmunol.1500719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, X., Clark R.A., Liu L., Wagers A.J., Fuhlbrigge R.C., and Kupper T.S.. 2012. Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin immunity. Nature. 483:227–231. 10.1038/nature10851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson-Lindbom, B., Svensson M., Wurbel M.A., Malissen B., Márquez G., and Agace W.. 2003. Selective generation of gut tropic T cells in gut-associated lymphoid tissue (GALT): Requirement for GALT dendritic cells and adjuvant. J. Exp. Med. 198:963–969. 10.1084/jem.20031244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, T.N., Mooster J.L., Kilgore A.M., Osborn J.F., and Nolz J.C.. 2016. Local antigen in nonlymphoid tissue promotes resident memory CD8+ T cell formation during viral infection. J. Exp. Med. 213:951–966. 10.1084/jem.20151855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna, K.M., McNamara J.T., and Lefrançois L.. 2007. In situ imaging of the endogenous CD8 T cell response to infection. Science. 318:116–120. 10.1126/science.1146291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmeier, J.E., Miller S.C., and Woodland D.L.. 2007. Cutting edge: Antigen is not required for the activation and maintenance of virus-specific memory CD8+ T cells in the lung airways. J. Immunol. 178:4721–4725. 10.4049/jimmunol.178.8.4721 [DOI] [PubMed] [Google Scholar]
- Kumar, B.V., Ma W., Miron M., Granot T., Guyer R.S., Carpenter D.J., Senda T., Sun X., Ho S.H., Lerner H., et al. 2017. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 20:2921–2934. 10.1016/j.celrep.2017.08.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.T., Suarez-Ramirez J.E., Wu T., Redman J.M., Bouchard K., Hadley G.A., and Cauley L.S.. 2011. Environmental and antigen receptor-derived signals support sustained surveillance of the lungs by pathogen-specific cytotoxic T lymphocytes. J. Virol. 85:4085–4094. 10.1128/JVI.02493-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrançois, L., Parker C.M., Olson S., Muller W., Wagner N., Schön M.P., and Puddington L.. 1999. The role of beta7 integrins in CD8 T cell trafficking during an antiviral immune response. J. Exp. Med. 189:1631–1638. 10.1084/jem.189.10.1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay, L.K., Braun A., Macleod B.L., Collins N., Tebartz C., Bedoui S., Carbone F.R., and Gebhardt T.. 2015. Cutting edge: CD69 interference with sphingosine-1-phosphate receptor function regulates peripheral T cell retention. J. Immunol. 194:2059–2063. 10.4049/jimmunol.1402256 [DOI] [PubMed] [Google Scholar]
- Mackay, L.K., Minnich M., Kragten N.A., Liao Y., Nota B., Seillet C., Zaid A., Man K., Preston S., Freestone D., et al. 2016. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science. 352:459–463. 10.1126/science.aad2035 [DOI] [PubMed] [Google Scholar]
- Mackay, L.K., Rahimpour A., Ma J.Z., Collins N., Stock A.T., Hafon M.L., Vega-Ramos J., Lauzurica P., Mueller S.N., Stefanovic T., et al. 2013. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat. Immunol. 14:1294–1301. 10.1038/ni.2744 [DOI] [PubMed] [Google Scholar]
- Mackay, L.K., Stock A.T., Ma J.Z., Jones C.M., Kent S.J., Mueller S.N., Heath W.R., Carbone F.R., and Gebhardt T.. 2012. Long-lived epithelial immunity by tissue-resident memory T (TRM) cells in the absence of persisting local antigen presentation. Proc. Natl. Acad. Sci. USA. 109:7037–7042. 10.1073/pnas.1202288109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, B.T., Byrne K.T., Vella J.L., Zhang P., Shabaneh T.B., Steinberg S.M., Molodtsov A.K., Bowers J.S., Angeles C.V., Paulos C.M., et al. 2017. Resident memory T cells in the skin mediate durable immunity to melanoma. Sci. Immunol. 2:eaam6346. 10.1126/sciimmunol.aam6346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani, V., Bromley S.K., Äijö T., Mora-Buch R., Carrizosa E., Warner R.D., Hamze M., Sen D.R., Chasse A.Y., Lorant A., et al. 2019. Migratory DCs activate TGF-β to precondition naïve CD8+ T cells for tissue-resident memory fate. Science. 366:366. 10.1126/science.aav5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust, D., Choo D., Vezys V., Wherry E.J., Duraiswamy J., Akondy R., Wang J., Casey K.A., Barber D.L., Kawamura K.S., et al. 2010. Dynamic T cell migration program provides resident memory within intestinal epithelium. J. Exp. Med. 207:553–564. 10.1084/jem.20090858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust, D., Vezys V., Wherry E.J., Barber D.L., and Ahmed R.. 2006. Cutting edge: Gut microenvironment promotes differentiation of a unique memory CD8 T cell population. J. Immunol. 176:2079–2083. 10.4049/jimmunol.176.4.2079 [DOI] [PubMed] [Google Scholar]
- Matsushita, H., Hijioka M., Hisatsune A., Isohama Y., Shudo K., and Katsuki H.. 2011. A retinoic acid receptor agonist Am80 rescues neurons, attenuates inflammatory reactions, and improves behavioral recovery after intracerebral hemorrhage in mice. J. Cereb. Blood Flow Metab. 31:222–234. 10.1038/jcbfm.2010.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed, J., Beura L.K., Bobr A., Astry B., Chicoine B., Kashem S.W., Welty N.E., Igyártó B.Z., Wijeyesinghe S., Thompson E.A., et al. 2016. Stromal cells control the epithelial residence of DCs and memory T cells by regulated activation of TGF-β. Nat. Immunol. 17:414–421. 10.1038/ni.3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar, R., Greuter M., van der Marel A.P., Roozendaal R., Martin S.F., Edele F., Huehn J., Förster R., O’Toole T., Jansen W., et al. 2009. Lymph node stromal cells support dendritic cell-induced gut-homing of T cells. J. Immunol. 183:6395–6402. 10.4049/jimmunol.0900311 [DOI] [PubMed] [Google Scholar]
- Mueller, S.N., and Mackay L.K.. 2016. Tissue-resident memory T cells: Local specialists in immune defence. Nat. Rev. Immunol. 16:79–89. 10.1038/nri.2015.3 [DOI] [PubMed] [Google Scholar]
- Muschaweckh, A., Buchholz V.R., Fellenzer A., Hessel C., König P.A., Tao S., Tao R., Heikenwälder M., Busch D.H., Korn T., et al. 2016. Antigen-dependent competition shapes the local repertoire of tissue-resident memory CD8+ T cells. J. Exp. Med. 213:3075–3086. 10.1084/jem.20160888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizard, M., Roussel H., Diniz M.O., Karaki S., Tran T., Voron T., Dransart E., Sandoval F., Riquet M., Rance B., et al. 2017. Induction of resident memory T cells enhances the efficacy of cancer vaccine. Nat. Commun. 8:15221. 10.1038/ncomms15221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Byrne, S.M., and Blaner W.S.. 2013. Retinol and retinyl esters: Biochemistry and physiology. J. Lipid Res. 54:1731–1743. 10.1194/jlr.R037648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Y., Tian T., Park C.O., Lofftus S.Y., Mei S., Liu X., Luo C., O’Malley J.T., Gehad A., Teague J.E., et al. 2017. Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature. 543:252–256. 10.1038/nature21379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, C.O., and Kupper T.S.. 2015. The emerging role of resident memory T cells in protective immunity and inflammatory disease. Nat. Med. 21:688–697. 10.1038/nm.3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumlee, C.R., Sheridan B.S., Cicek B.B., and Lefrançois L.. 2013. Environmental cues dictate the fate of individual CD8+ T cells responding to infection. Immunity. 39:347–356. 10.1016/j.immuni.2013.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, Z., Chu T.H., and Sheridan B.S.. 2021. TGF-β: Many paths to CD103+ CD8 T cell residency. Cells. 10:989. 10.3390/cells10050989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, Z., Khairallah C., and Sheridan B.S.. 2018. Listeria monocytogenes: A model pathogen continues to refine our knowledge of the CD8 T cell response. Pathogens. 7:55. 10.3390/pathogens7020055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, Z., and Sheridan B.S.. 2018. Isolating lymphocytes from the mouse small intestinal immune system. J. Vis. Exp. 57281. 10.3791/57281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto, F., Lenig D., Förster R., Lipp M., and Lanzavecchia A.. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 401:708–712. 10.1038/44385 [DOI] [PubMed] [Google Scholar]
- Schenkel, J.M., Fraser K.A., Beura L.K., Pauken K.E., Vezys V., and Masopust D.. 2014. T cell memory. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science. 346:98–101. 10.1126/science.1254536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel, J.M., Fraser K.A., Vezys V., and Masopust D.. 2013. Sensing and alarm function of resident memory CD8⁺ T cells. Nat. Immunol. 14:509–513. 10.1038/ni.2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan, B.S., and Lefrançois L.. 2011. Regional and mucosal memory T cells. Nat. Immunol. 12:485–491. 10.1038/ni.2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan, B.S. and Lefrançois L.. 2012. Isolation of mouse lymphocytes from small intestine tissues. Curr. Protoc. Immunol. 3:3.19.1–3.19.11. 10.1002/0471142735.im0319s99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan, B.S., Pham Q.M., Lee Y.T., Cauley L.S., Puddington L., and Lefrançois L.. 2014. Oral infection drives a distinct population of intestinal resident memory CD8(+) T cells with enhanced protective function. Immunity. 40:747–757. 10.1016/j.immuni.2014.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, H., and Iwasaki A.. 2012. A vaccine strategy that protects against genital herpes by establishing local memory T cells. Nature. 491:463–467. 10.1038/nature11522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiow, L.R., Rosen D.B., Brdicková N., Xu Y., An J., Lanier L.L., Cyster J.G., and Matloubian M.. 2006. CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature. 440:540–544. 10.1038/nature04606 [DOI] [PubMed] [Google Scholar]
- Skon, C.N., Lee J.Y., Anderson K.G., Masopust D., Hogquist K.A., and Jameson S.C.. 2013. Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nat. Immunol. 14:1285–1293. 10.1038/ni.2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slütter, B., Van Braeckel-Budimir N., Abboud G., Varga S.M., Salek-Ardakani S., and Harty J.T.. 2017. Dynamics of influenza-induced lung-resident memory T cells underlie waning heterosubtypic immunity. Sci. Immunol. 2:eaag2031. 10.1126/sciimmunol.aag2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenstad, H., Svensson M., Cucak H., Kotarsky K., and Agace W.W.. 2007. Differential homing mechanisms regulate regionalized effector CD8alphabeta+ T cell accumulation within the small intestine. Proc. Natl. Acad. Sci. USA. 104:10122–10127. 10.1073/pnas.0700269104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson, M., Johansson-Lindbom B., Zapata F., Jaensson E., Austenaa L.M., Blomhoff R., and Agace W.W.. 2008. Retinoic acid receptor signaling levels and antigen dose regulate gut homing receptor expression on CD8+ T cells. Mucosal Immunol. 1:38–48. 10.1038/mi.2007.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura, S., Yagi H., Hakata Y., Motozono C., McMaster S.R., Masumoto T., Fujisawa M., Chikaishi T., Komeda J., Itoh J., et al. 2016. Specific niches for lung-resident memory CD8+ T cells at the site of tissue regeneration enable CD69-independent maintenance. J. Exp. Med. 213:3057–3073. 10.1084/jem.20160938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakim, L.M., Woodward-Davis A., and Bevan M.J.. 2010. Memory T cells persisting within the brain after local infection show functional adaptations to their tissue of residence. Proc. Natl. Acad. Sci. USA. 107:17872–17879. 10.1073/pnas.1010201107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakim, L.M., Woodward-Davis A., Liu R., Hu Y., Villadangos J., Smyth G., and Bevan M.J.. 2012. The molecular signature of tissue resident memory CD8 T cells isolated from the brain. J. Immunol. 189:3462–3471. 10.4049/jimmunol.1201305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert, T., Pasche B., Rochon M., Deppenmeier S., van den Heuvel J., Gruber A.D., Heinz D.W., Lengeling A., and Schubert W.D.. 2007. Extending the host range of Listeria monocytogenes by rational protein design. Cell. 129:891–902. 10.1016/j.cell.2007.03.049 [DOI] [PubMed] [Google Scholar]
- Wu, T., Hu Y., Lee Y.T., Bouchard K.R., Benechet A., Khanna K., and Cauley L.S.. 2014. Lung-resident memory CD8 T cells (TRM) are indispensable for optimal cross-protection against pulmonary virus infection. J. Leukoc. Biol. 95:215–224. 10.1189/jlb.0313180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurbel, M.A., Malissen M., Guy-Grand D., Malissen B., and Campbell J.J.. 2007. Impaired accumulation of antigen-specific CD8 lymphocytes in chemokine CCL25-deficient intestinal epithelium and lamina propria. J. Immunol. 178:7598–7606. 10.4049/jimmunol.178.12.7598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, N., and Bevan M.J.. 2012. TGF-β signaling to T cells inhibits autoimmunity during lymphopenia-driven proliferation. Nat. Immunol. 13:667–673. 10.1038/ni.2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, N., and Bevan M.J.. 2013. Transforming growth factor-β signaling controls the formation and maintenance of gut-resident memory T cells by regulating migration and retention. Immunity. 39:687–696. 10.1016/j.immuni.2013.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists differentially regulated genes between spleen-primed and MLN-primed T cells.
Data Availability Statement
RNA sequencing data in Fig. 4 has been deposited in Gene Expression Omnibus with the accession number GSE223983. All other data are available from the corresponding author.