Abstract
Background
The interaction between the tumor-microenvironment (TME) and the cancer cells has emerged as a key player in colorectal cancer (CRC) metastasis. A small proportion of CRC cells which undergo epithelial-mesenchymal transition (EMT) facilitate the reshaping of the TME by regulating various cellular ingredients.
Methods
Immunohistochemical analysis, RNA immunoprecipitation (RIP), RNA Antisense Purification (RAP), dual luciferase assays were conducted to investigate the biological function and regulation of LINC00543 in CRC. A series in vitro and in vivo experiments were used to clarify the role of LINC00543 in CRC metastasis.
Results
Here we found that the long non-coding RNA LINC00543, was overexpressed in colorectal cancer tissues, which correlated with advanced TNM stage and poorer prognosis of CRC patients. The overexpression of LINC00543 promoted tumorigenesis and metastasis of CRC cells by enhancing EMT and remodeling the TME. Mechanistically, LINC00543 blocked the transport of pre-miR-506-3p across the nuclear-cytoplasmic transporter XPO5, thereby reducing the production of mature miR-506-3p, resulting in the increase in the expression of FOXQ1 and induction of EMT. In addition, upregulation of FOXQ1 induced the expression of CCL2 that accelerated the recruitment of macrophages and their M2 polarization.
Conclusions
Our study showed that LINC00543 enhanced EMT of CRC cells through the pre-miR-506-3p/FOXQ1 axis. This resulted in the upregulation of CCL2, leading to macrophages recruitment and M2 polarization, and ultimately stimulating the progression of CRC.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-023-04009-6.
Keywords: Epithelial-mesenchymal transition, Long non-coding RNA, Metastasis, Pre-miR-506-3p transport, Tumor microenvironment (TME)
Introduction
According to the global cancer statistics from 2020, colorectal cancer (CRC) ranked third in terms of cancer incidence and second in terms of cancer mortality [1]. Metastasis remains the major cause of death in CRC patients [2]. The tumor microenvironment (TME) is not a silent spectator, but rather an active participant that promotes tumor metastasis [3]. Tumor-associated macrophages (TAMs), one of the main cell types in the TME, are commonly activated and polarized to M1/M2 state [4]. The M2 macrophages are known to play an anti-inflammatory and cancer-promoting role during tumor progression [5, 6]. Cancer cells promote the recruitment of macrophages and their M2 polarization to build an immunosuppressive TME, which in turn facilitate cancer progression [7, 8]. Since the cross-talk between the TAMs and tumors has been largely reported, many of the recent studies have considered TAMs as promising therapeutic targets for cancer [9]. However, the underlying molecular mechanism regulating TAMs recruitment and polarization in CRC is still unclear.
Epithelial mesenchymal transition (EMT) is an important pathophysiological process in tumor progression [10] during which the cancerous cells gain the capacities of migration and invasion by losing their polarity and inter-cellular junctions [11–14]. In addition, many previous studies have uncovered that cancer cells undergoing EMT secrete various cytokines to reshape the tumor immune microenvironment [15]. In melanoma, cancer cells undergoing EMT have been reported to produce TGF-β and TSP-1 which repressed the anti-cancer immune response that accelerated cancer metastasis [16]. Hepatocellular carcinoma cells that underwent EMT secreted CCL2 to recruit TAMs that facilitated tumor metastasis [17]. Therefore, exploring how tumor cells that underwent EMT could regulate the immune microenvironment in CRC is important to identify novel therapeutic targets for treating CRC metastasis.
Long non-coding RNAs (lncRNAs) are non-protein coding RNA transcripts that play vital roles in tumor progression, and metastasis [18, 19]. With regards to CRC, lncRNAs have been recognized as important participants in the regulation of EMT and the TME. For instance, Yue, Ben et al. revealed that a positive feedback loop between the lncRNA CYTOR and the Wnt/β-Catenin axis promoted EMT and metastasis of colorectal cancer cells [20]. Wu Nan and colleagues reported that LINC00941 was directly bound to SMAD4 protein, which induced EMT and enhanced colorectal cancer metastasis [21]. Reciprocally, Liang, Zhen-Xing, and colleagues elucidated the mechanism of lncRNA RPPH1, which promoted the M2 polarization of TAMs and accelerated colorectal cancer metastasis [22]. The aberrant expression of LINC00543 was reported to be correlated with immunosuppression and poorer prognosis of CRC patients [23]. However, the exact mechanism underlying the regulation of EMT and the TME by LINC00543 in CRC is still unknown.
In this study, we found that LINC00543 was significantly elevated and markedly correlated with advanced TNM stage and poorer overall survival (OS) in colorectal cancer patients. In vitro experimental data showed that high LINC00543 expression activated EMT and boosted TAMs recruitment and M2 polarization, thereby accelerating the progression and metastasis of CRC. The above results indicated the oncogenic role of LINC00543. Further mechanistic exploration revealed that LINC00543 inhibited the transport of pre-miR-506-3p by the nuclear-cytoplasmic transporter XPO5 and downregulated the expression of miR-506-3p, thereby upregulating FOXQ1 expression and inducing EMT. In addition, upregulated FOXQ1 in tumor cells promoted the secretion of CCL2, which recruited TAMs and promoted their M2 polarization. And we verified the function of LINC00543 in regulating EMT and macrophage polarization to promote the metastasis of colorectal cancer in nude mice. The above findings revealed a novel mechanism of LINC00543 regulating the miRNA maturation process and metastasis of CRC.
Materials and methods
Patients and tissue samples
We collected primary tumor tissues and paired normal colorectal tissues (> 5 cm distance from the cancer tissue margin) from 40 CRC patients from the Zhongnan Hospital of Wuhan University (Wuhan, China). All enrolled patients were definitively diagnosed with colorectal adenocarcinoma by histopathology, and none had received neoadjuvant radiotherapy or/and chemotherapy before undergoing resection. In addition, all the enrolled patients in the group had corresponding available survival data. This study was endorsed by the Ethics Committee of Zhongnan Hospital of Wuhan University in compliance with the Declaration of Helsinki. Informed consent was obtained from all the patients before sample collection.
RNA antisense purification (RAP)
RAP analysis was performed in accordance with the manufacturer’s instructions for the RNA Antisense Purification (RAP) kit (BersinBion, Bes5103-1, China). Briefly, 4 × 107 cells per group were collected and added to the lysis solution for cell lysis. After DNA removal, the LINC00543 biotin probe, the LINC00543 segmental biotin probe, and the LINC00543 segmental antisense nucleotide fragment biotin probes were co-incubated at 37 °C for 180 min. Next, the mixture of probes and samples were added to the streptavidin magnetic beads and incubated on a vertical mixer at room temperature for about 30 min. After removing the proteins by proteinase K, the RNA was distributed for RT-PCR and agarose gel electrophoresis analysis. The sequences of the probes used for RAP are shown in the Supplementary Table S4.
RNA binding protein immunoprecipitation (RIP)
RIP analysis was performed using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, MA, USA). We first collected 2 × 107 cells that had been washed with ice-cold PBS. The configured RIP lysis solution was mixed and lysed at 0 °C for 5 min. DNA was then removed, and the nucleotide fragments of LINC00543 segments or antisense nucleotide fragments were added to hybridize at 37 °C for 3 h. The samples were then mixed with anti-XPO5 antibody and protein A/G magnetic beads and incubated on a vertical mixer at 4 °C for 12 h. The proteins were then digested after washing away the unbound material with RIP wash buffer. Finally, the RNA was extracted for subsequent experiments. The sequences of the probes used for RIP are shown in the Additional file 1: Table S4.
Flow cytometry
Macrophages and cells extracted from the tumors of nude mice were made into single-cell suspensions. The suspensions were mixed with antigen specific fluorescent antibodies and incubated on ice for 2 h (PE Mouse anti-Human CD163, APC Mouse anti-Human CD86, FITC Mouse anti-Human CD206, PerCPMouse anti-Human CD80, all from Biolegend, USA). Cells were washed twice by adding 3 ml of the flow buffer and were then resuspended in 600 μl of the flow buffer. Flow cytometry was performed using the FACSCalibur flow cytometer (BD Biosciences, USA). Flow cytometry results were analyzed using the FlowJo software (FlowJo, USA).
Statistics
The data were expressed as mean ± standard error (m ± SE) and were analyzed by two-tailed Student’s t-test. All experiments were performed in triplicate and statistical significance was defined as p < 0.05 using the GraphPad Prism software (version 6.0, GraphPad Software, USA) for Windows.
Results
LINC00543 is highly expressed in CRC tissues and is associated with poor prognosis of CRC patients
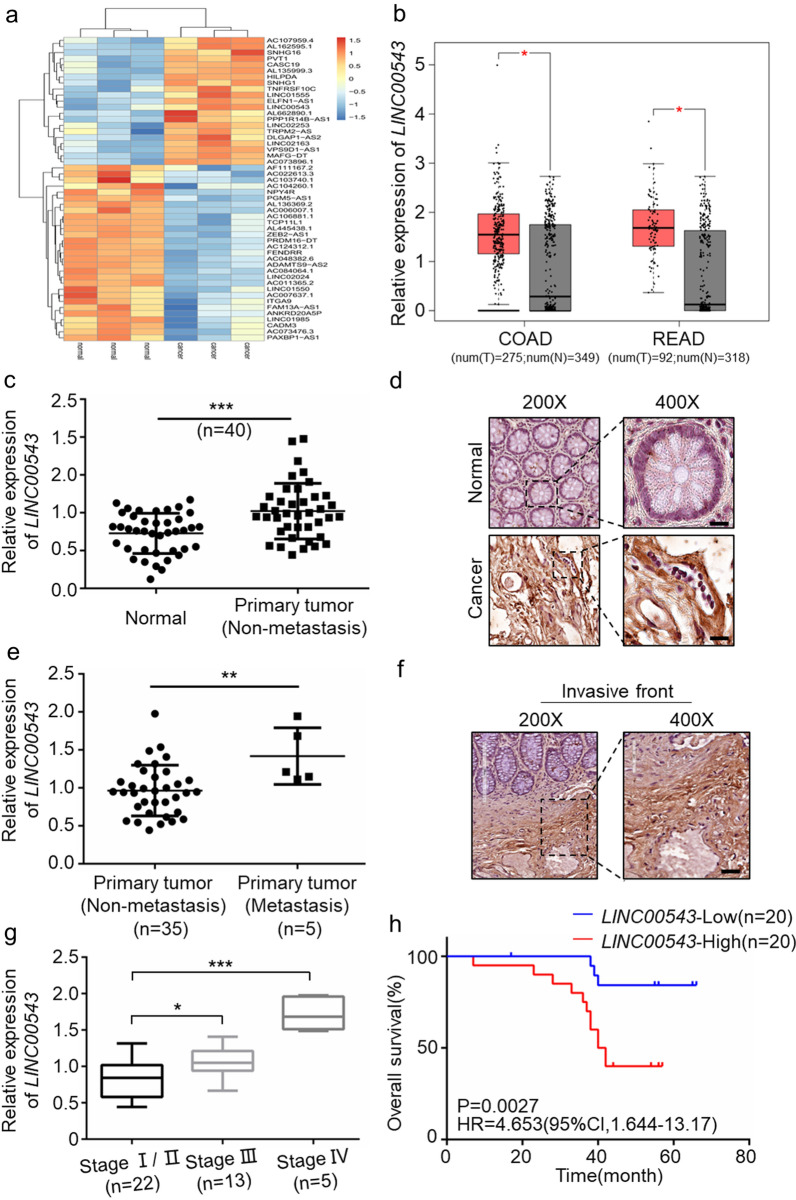
Based on the analysis of the datasets from the GEO database (GES110715) and the online analysis website GEPIA2 (http://gepia2.cancer-pku.cn/#index), we found that the expression of LINC00543 was higher in the CRC tissues as compared with the normal colorectal tissues (Fig. 1a and b). Then, we detected the expression of LINC00543 in 40 CRC tissues and their matched normal tissues, and the results were consistent with the results from the bioinformatics analysis. Besides, we found that the expression of LINC00543 was higher in the primary tumor tissues of CRC patients with distant metastasis than those without distant metastasis. Notably, the expression of LINC00543 in the invasive fronts was higher than in other regions of the primary tumor (Fig. 1c–f, Additional file 1: Fig. S1a). Moreover, we investigated the clinical data from 40 patients and found that the elevated expression of LINC00543 was correlated with advanced TNM stage and poor prognosis (Fig. 1g and h, Additional file 1: Table S1). Also, the encoding potential and location of LINC00543 were also analyzed, and the results showed that the LINC00543 gene on chromosome 13 lacked the ability to encode proteins (Additional file 1: Fig. S1b–d).
Fig. 1.
LINC00543 is an oncogene in CRC. a The expression of LINC00543 in CRC Gene Chips (GES110715). b The expression of LINC00543 through online analysis website GEPIA2. c Relative expression of LINC00543 detected by qRT-PCR in 40 CRC primary tumor tissues and their paired normal tissues normalized to 18S. d In situ hybridization (ISH) analysis of LINC00543 expression in colorectal cancer tissues and their matched normal tissues (scale bar = 100 μm). e Comparison of LINC00543 expression in samples from CRC patients with metastatic colorectal cancer and patients without metastasis. f The expression of LINC00543 in the invasive fronts (scale bar = 100 μm). g The relationship between the expression levels of LINC00543 and TNM stages in 40 CRC patients. h The database from Kaplan–Meier Plotter showed the relationship between the expression of LINC00543 and the overall survival rate. P < 0.001
The above data suggested that high LINC00543 expression was closely related to the advanced TNM stage and poor prognosis of CRC patients.
LINC00543 promotes the invasion and metastasis of CRC cells
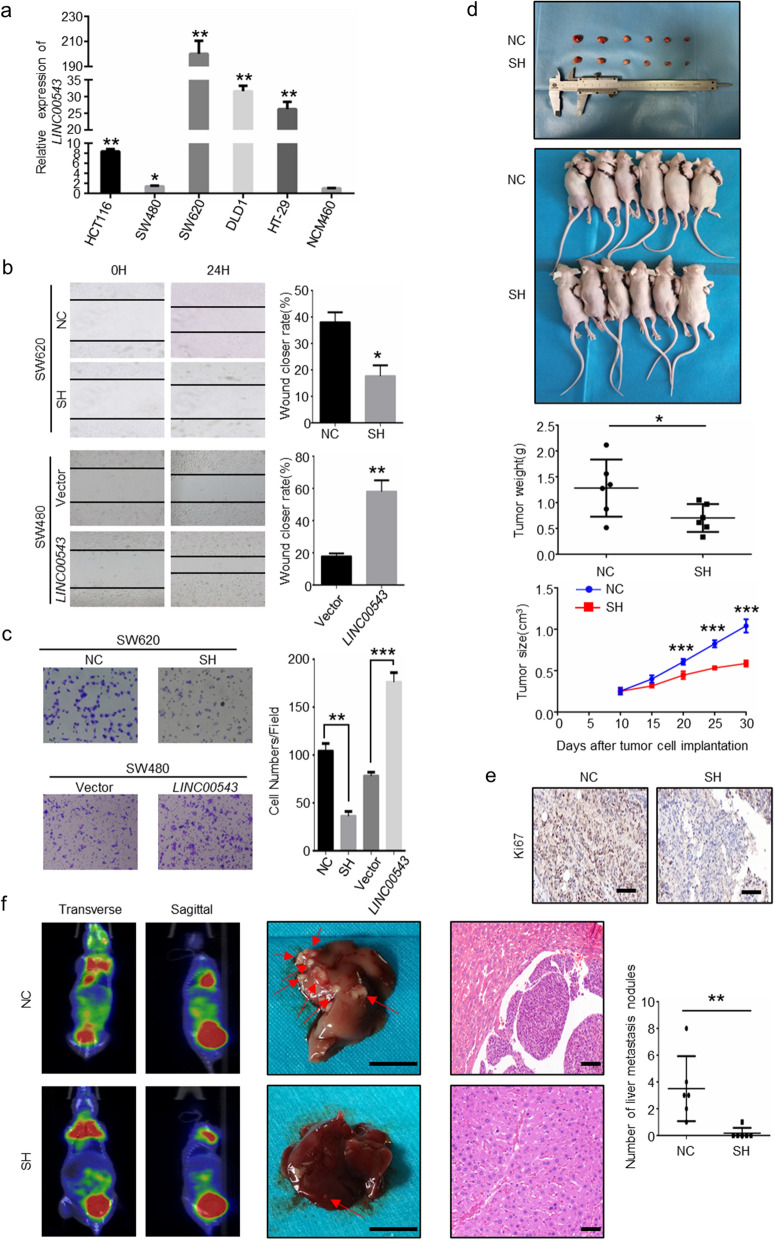
To further explore the function of LINC00543 in CRC, we evaluated the expression level of LINC00543 in a normal colorectal cell line (NCM460) and five CRC cell lines (HCT116, SW480, SW620, DLD1, HT-29). The expression of LINC00543 was significantly elevated in the CRC cell lines. Interestingly, LINC00543 expression in metastatic CRC cell line SW620, was considerably higher than that in primary CRC cell line SW480, further indicating that LINC00543 expression was associated with distant metastasis (Fig. 2a). We selected SW620 and SW480 to construct LINC00543 knockdown (SH) and overexpression (LINC00543) cell lines separately by using lentivirus and the transfection efficiency was evaluated by qRT-PCR (Additional file 1: Fig. S2a–c). The knockdown of LINC00543 suppressed the invasion and migration of CRC cells, while the overexpression of LINC00543 promoted the invasion and migration of CRC cells (Fig. 2b and c). Furthermore, the tumorigenic effect of LINC00543 was evaluated in vivo. Tumor formation and growth were inhibited upon the implantation of cell with the knockdown of LINC00543 (Fig. 2d and e, Additional file 1: Fig. S2d). Next, we evaluated the in vivo role of LINC00543 in metastasis by injecting CRC cells in the spleen to establish a liver metastatic mouse model. Compared with the NC group, mice in the SH group showed a lower rate of liver metastasis (Fig. 2f).
Fig. 2.
LINC00543 promotes the invasion and metastasis of CRC cells in vitro and in vivo. a The relative expression of LINC00543 in non-cancer cells NCM460 and CRC cells (HCT116, SW480, SW620, DLD1, HT-29). b Wound-healing assays and c transwell assays in SH cells and LINC00543 cells (magnification, × 200). d The tumor volumes in nude mice from the NC and SH groups. e The immunohistochemistry analysis of tumor from the NC and the SH groups (scale bar = 100 μm). f PET-CT images, liver anatomy (scale, 1 cm), H&E staining of the liver, and the number of liver surface metastases in spleen-injected liver metastasis models in nude mice, in the NC and SH groups (scale bar = 100 μm)
LINC00543 regulates EMT in CRC cells
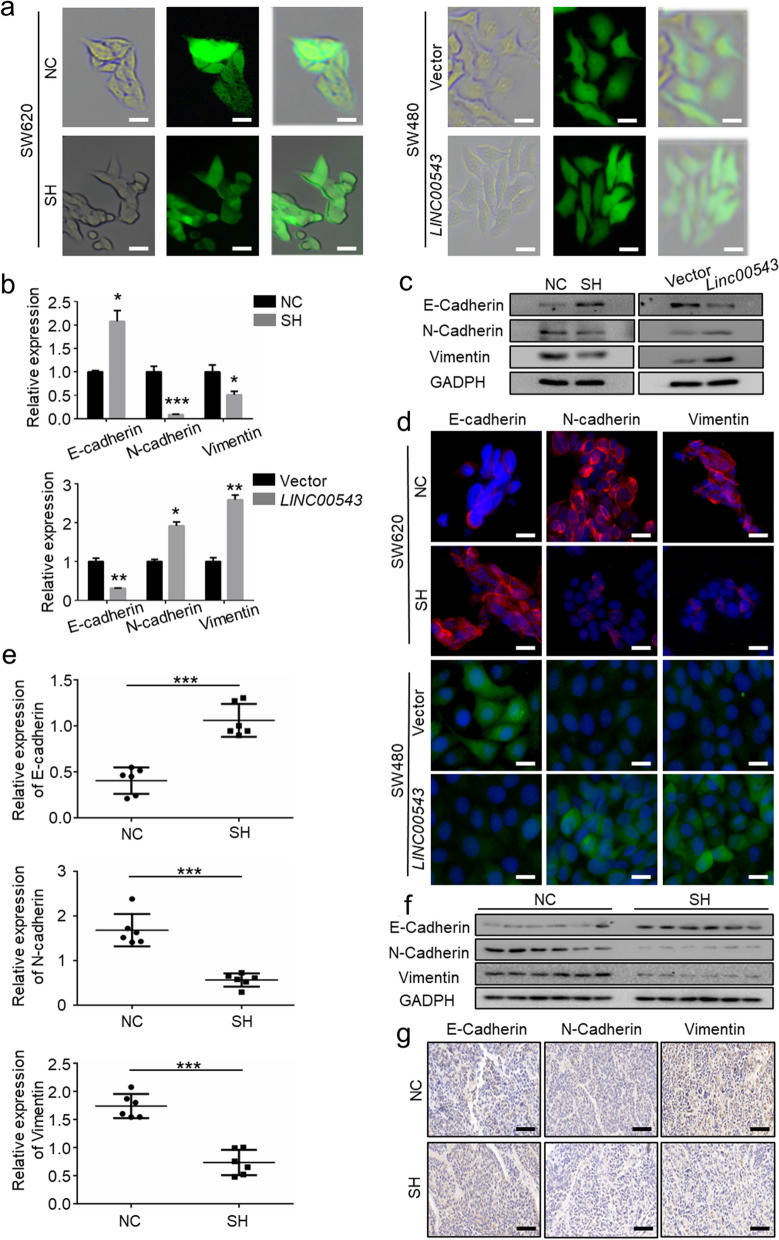
Considering that EMT is a crucial process in tumor metastasis [15], we further interrogated whether LINC00543 promoted the EMT process. We observed that in comparison with the control group, cells with the knockdown of LINC00543 lost their typical spindle-shaped morphology, while after the overexpression of the LINC00543, the cells regained their spindle shape morphology (Fig. 3a). Consistently, the expression of the epithelial cell marker E-cadherin decreased in the CRC cells overexpressing LINC00543, whereas the expression of mesenchymal cell markers N-cadherin and Vimentin were elevated. In addition, contrary results were observed in cells overexpressing the LINC00543 (Fig. 3b–d). In vivo, the knockdown of LINC00543 downregulated the expression of E-cadherin and elevated the expression of N-cadherin and Vimentin (Fig. 3e and f). These observations were consistent with the immunohistochemistry (IHC) results (Fig. 3g, Additional file 1: Fig. S3).
Fig. 3.
LINC00543 promotes EMT in CRC cells. a The morphologies of CRC cells in response to LINC00543 manipulation (scale bar = 25 μm) b qRT-PCR and c western blot analysis of E-cadherin and Vimentin in the SH and LINC00543 cells. d E-cadherin, N-cadherin, and Vimentin expression in the SH and LINC00543 cells were determined by immunofluorescence staining (scale bar = 25 μm). e and f The expression of EMT-related markers in the NC and SH group mice. g The expression of EMT-related markers in the NC and SH group mice was determined by immunohistochemistry (IHC) (scale bar = 100 μm)
LINC00543 regulates EMT by upregulating FOXQ1 in CRC cells
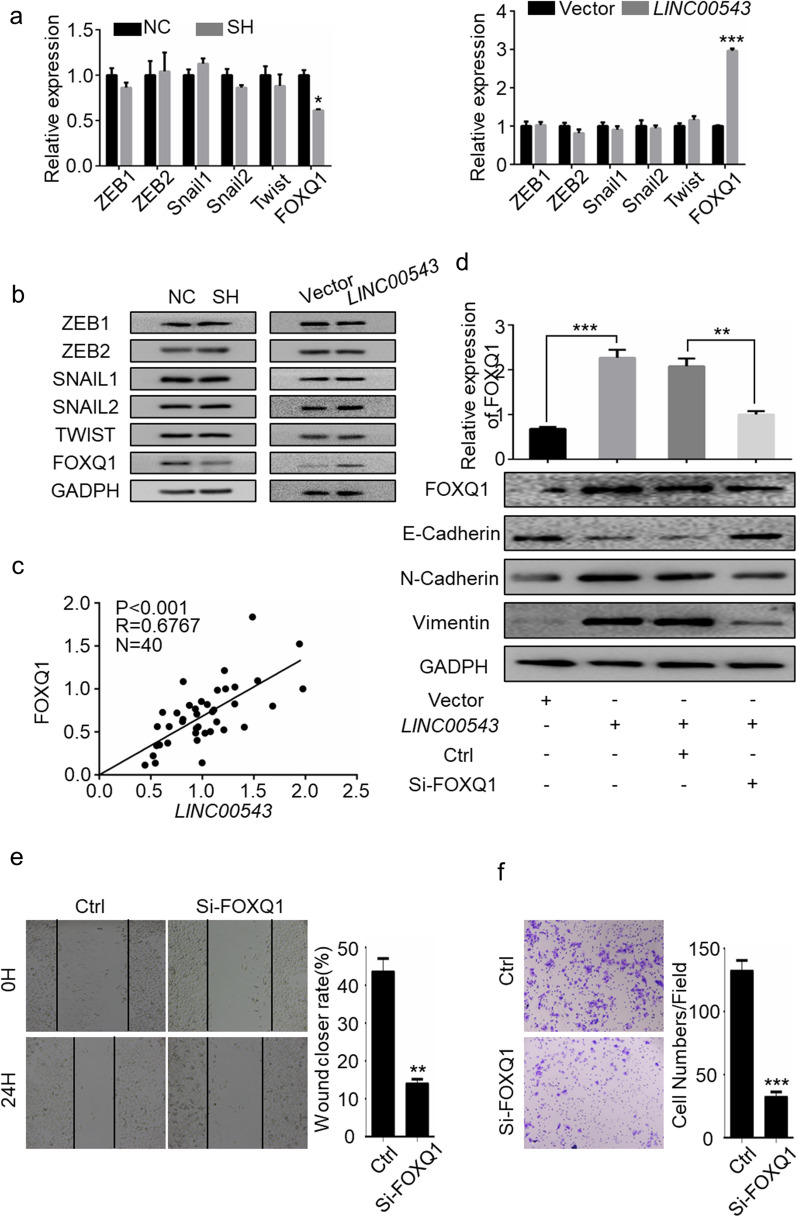
In order to further uncover the molecular mechanism of LINC00543 in EMT, we analyzed the expression of EMT-related transcription factors [24]. The results showed that the expression of FOXQ1 was significantly downregulated in cells with LINC00543 knockdown, while it was elevated in the cells overexpressing LINC00543. There were no significant changes in the expression levels of other EMT-related transcription factors (ZEB1, ZEB2, Snail1, Snail2, and Twist) (Fig. 4a and b, Additional file 1: Fig. S4). To extend the correlation of our findings, we analyzed the expression of LINC00543 and FOXQ1 in tissue specimens from 40 CRC patients and found a positive correlation between them (Fig. 4c). Moreover, the knockdown of FOXQ1 significantly attenuated the induction of EMT, invasion, and migration in CRC cells overexpressing the LINC00543 (Fig. 4d–f).
Fig. 4.
FOXQ1 is essential in the process of LINC00543 mediated regulation of EMT in CRC cells. a qRT-PCR and b western blot analysis of EMT-related transcription factors in the SH and LINC00543 cells. c Correlation between the expression of FOXQ1 and LINC00543 in 40 patients with colorectal cancer. d The expression of FOXQ1 and EMT markers were detected in the LINC00543 cells with Si-FOXQ1 transfected. e Wound-healing assays and f Transwell assays in LINC00543 cells transfected with Si-FOXQ1 and control; magnification, × 200
Collectively, FOXQ1 was found to be essential in mediating the effects of LINC00543 on EMT in CRC cells.
LINC00543 indirectly regulates FOXQ1 by downregulating miR-506-3p
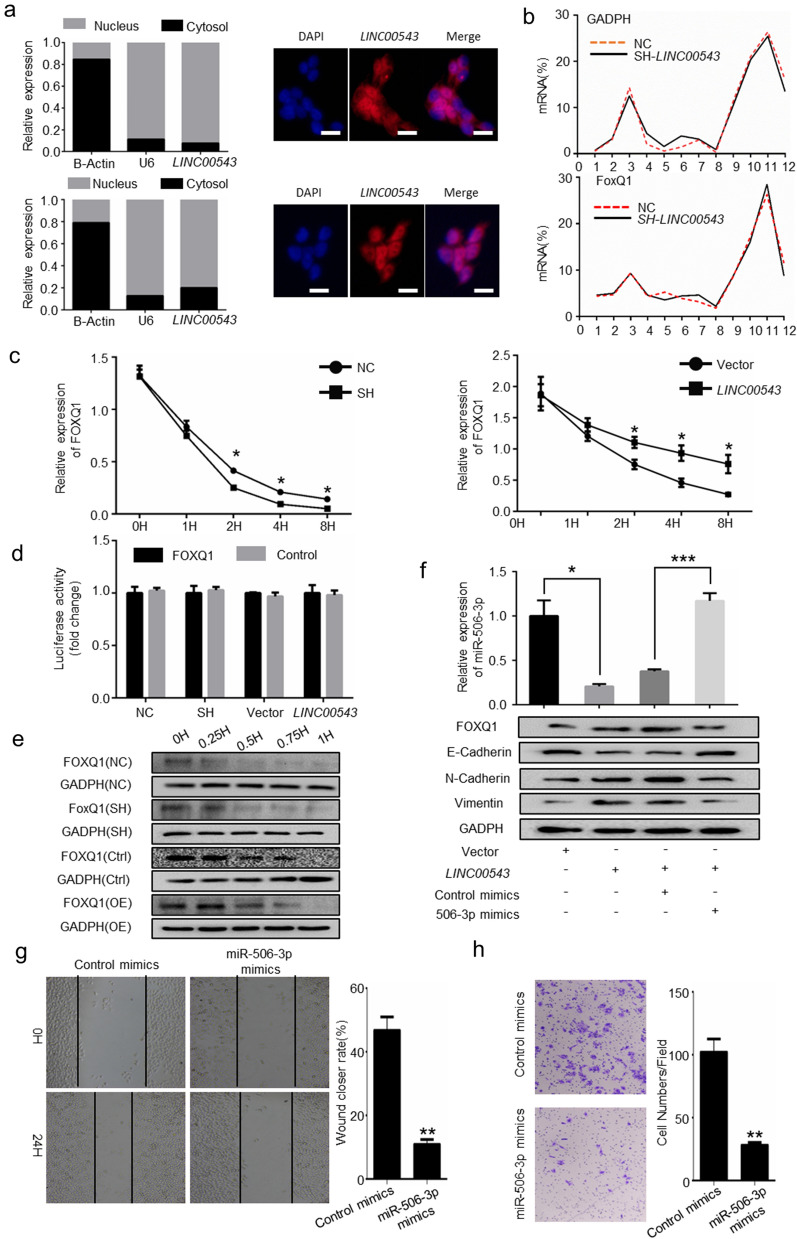
To further study the molecular mechanism of LINC00543 regulating FOXQ1, firstly, we identified the localization of LINC00543 in cells by fluorescence in situ hybridization and subcellular fraction analyses. The results showed that LINC00543 was mainly localized in the cell’s nucleus (Fig. 5a). We reasoned that LINC00543 might regulate the transcription of FOXQ1, and thus constructed a dual-luciferase reporter plasmid containing the promoter region of FOXQ1. Surprisingly, overexpression/knockdown of the LINC00543 had no effect on the transcription of FOXQ1. Then, actinomycin D assays were conducted, and the results indicated that LINC00543 suppressed the degradation of FOXQ1 mRNA. Additionally, based on the results from the polysome fractionation assay and cycloheximide-chase assay, we determined that LINC00543 regulated FOXQ1 by suppressing the degradation of FOXQ1 mRNA (Fig. 5b–e, Additional file 1: Fig. S5). Our previous studies showed that miR-506-3p regulated FOXQ1 by enhancing the degradation of FOXQ1 mRNA [25], accordingly, qRT-PCR analysis was conducted to examine the expression of miR-506-3p in CRC cells overexpressing the LINC00543. The results indicated that miR-506-3p expression was significantly downregulated in CRC cells overexpressing the LINC00543 (Fig. 5f). After transfecting miR-506-3p mimics into cells overexpressing LINC00543, we observed the positive effects of LINC00543 overexpression on EMT, invasion and migration in CRC cells were restored (Fig. 5f, g).
Fig. 5.
LINC00543/miR-506-3p/FOXQ1 regulates EMT of CRC cells. a Intracellular localization of LINC00543 was identified by nuclear/cytoplasmic fractionation experiments and FISH analysis (scale bar = 25 μm). b Polysome Fractionation analysis evaluating the effect of LINC00543 on FOXQ1 translation. c Actinomycin D analysis of the effect of LINC00543 on FOXQ1 mRNA degradation. d Dual luciferase analysis of the effect of LINC00543 on FOXQ1 DNA transcription. e Cycloheximide-chase analysis of the effect of LINC00543 on FOXQ1 protein degradation. f The expression of FOXQ1 and EMT markers were detected in LINC00543 cells upon treatment with miR-506-3p mimics. g Wound-healing assays and h transwell assays in LINC00543 cells transfected with miR-506-3p mimics and control mimics; magnification, × 200
These results showed that LINC00543 indirectly regulated FOXQ1 by downregulating miR-506-3p, thereby affecting EMT, invasion and migration of colorectal cancer cells.
LINC00543 inhibits the transportation of pre-miR-506-3p
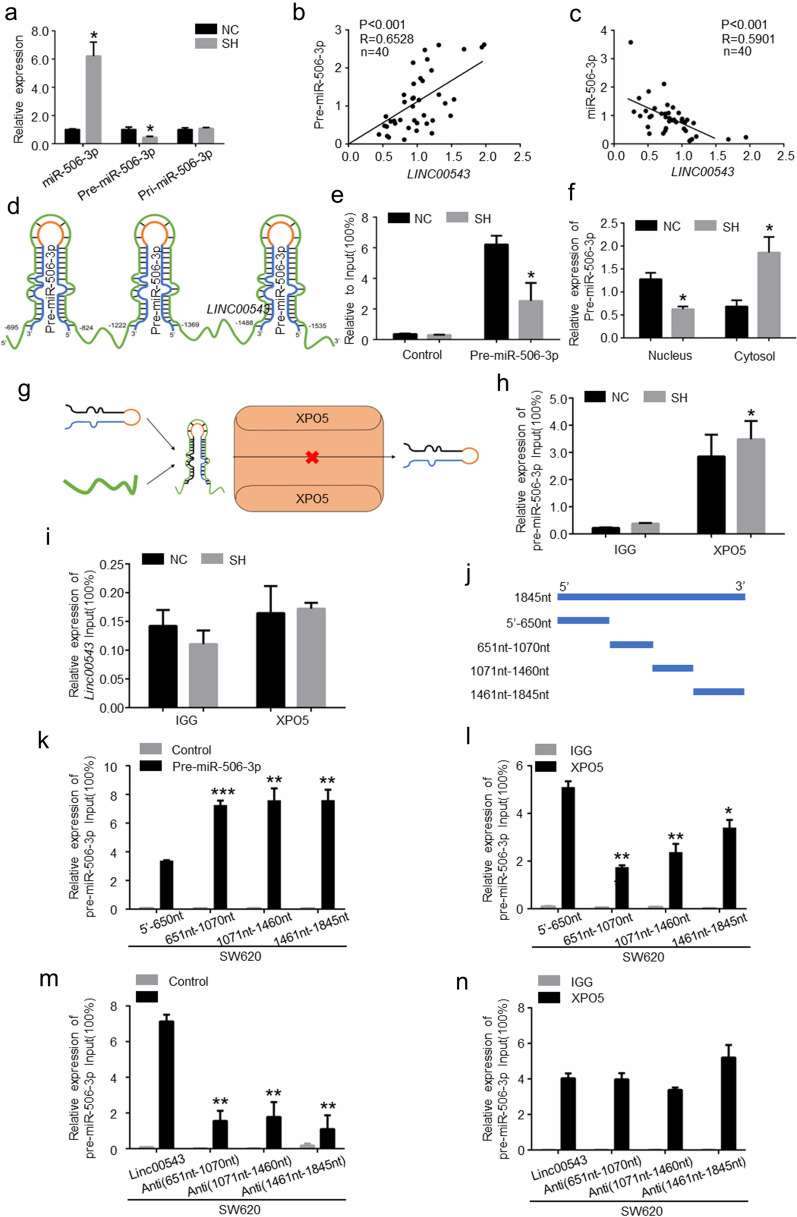
Many studies have shown that lncRNAs regulated gene expression by competing with endogenous miRNAs [26–29]. However, the online prediction website showed that there was no direct conjunction between LINC00543 and miR-506-3p (Additional file 1: Fig. S6a, b). Previous studies have reported that lncRNAs regulates the processing of pre-miRNA and pri-miRNA [30, 31]. Moreover, the expression of pre-miR-506-3p was significantly reduced in LINC00543 knockdown cells, while the opposite was applicable in LINC00543 overexpressing cells. However, the expression of pri-miR-506-3p was not significantly altered. In LINC00543 overexpressing cells, the opposite results were obtained (Fig. 6a, Additional file 1: Fig. S7a). We also confirmed that LINC00543 expression was positively correlated with pre-miR-506-3p expression and negatively correlated with miR-506-3p expression in 40 CRC patient tissues (Fig. 6b and c). We reasoned that LINC00543 blocked the processing of pre-miR-506-3p to miR-506-3p.
Fig. 6.
LINC00543 blocks the maturation of miR-506-3p. a The expression of miR-506-3p, pre-miR-506-3p, and ppi-miR-506-3p in SH cells. b The relationship between the expression of LINC00543 and Pre-miR-506-3p. c The relationship between the expression of LINC00543 and the expression of miR-506-3p. d The binding regions of LINC00543 to pre-miR-506-3p. e RAP analysis of the expression of pre-miR-506-3p in SH cells. f The expression of pre-miR-506-3p in the nucleus and the cytoplasm in SH cells. g Illustrative model showing that LINC00543 blocked the transportation of pre-miR-506-3p. h RIP analysis of the interaction between XPO5 and pre-miR-506-3p in SH cells. i RIP analysis of the interaction between XPO5 and LINC00543 in SH cells. j Schematic diagram of biotin segment probes. k RAP analysis of the interaction between four segments of LINC00543 and pre-miR-506-3p. l RIP analysis of the interaction between XPO5 and pre-miR-506-3p in cells incubated with antisense nucleotide biotin probes of the three binding regions. m and n The interaction between LINC00543, XPO5, and pre-miR-506-3p were analyzed in cells incubated with the antisense nucleotide biotin probes of the three binding regions
To further investigate the mechanism of LINC00543 acting on pre-miR-506-3p, we predicted the interaction between LINC00543 and pre-miR-506-3p through the bioinformatics program miRanda. The results showed that three regions of LINC00543 were directly bound to pre-miR-506-3p (Fig. 6d, Additional file 1: Fig. S6c). Next, we confirmed such prediction through RNA Antisense Purification (RAP) (Fig. 6e, Additional file 1: Fig. S7b). We know that lncRNAs can regulate the expression of miRNAs by blocking the transportation of pre-miRNA [32]. After the knockdown of LINC00543, the expression of pre-miR-506-3p was reduced in the nucleus but elevated in the cytoplasm (Fig. 6f), while LINC00543 overexpression increased the expression of pre-miR-506-3p in the nucleus but downregulated its expression in the cytoplasm (Additional file 1: Fig. S7c). Therefore, we speculated that the LINC00543 blocked the nuclear shuttle of Pre-miR-506–30 to the cytoplasm. Okada, Chimari et al. showed that XPO5 was a transporter of pre-miRNA from the nucleus to the cytoplasm [33] (Fig. 6g). RNA Binding Protein Immunoprecipitation showed that knocking down the LINC00543 enhanced the transport efficiency of XPO5 for Pre-miR-506-3p (Fig. 6h), and the overexpression of LINC00543 had the opposite effects (Additional file 1: Fig. S7d). However, XPO5 did not bind directly to the LINC00543 and was not regulated by it. (Fig. 6i, Additional file 1: Fig. S7e and f). The analysis of agarose electrophoresis assay showed similar results (Additional file 1: Fig. S7g). These data suggested that LINC00543 inhibited the transportation of pere-miR-506-3p by XPO5.
In order to further unveil the specific region where LINC00543 interacted with the pre-miR-506-3p, we constructed four biotin-labelled LINC00543 nucleotide fragment probes (Fig. 6j). RAP results showed that three regions (651nt-1070nt, 1071nt-1460nt, 1461nt-1845nt) of the LINC00543 directly bound to the pre-miR-506-3p, and all three regions blocked the transport of pre-miR-506-3p (Fig. 6k–n, Additional file 1: Fig. S7h-k). In conclusion, LINC00543 directly bound to pre-miR-506-3p and inhibited the transportation of pre-miR-506-3p by XPO5.
LINC00543 promotes macrophage recruitment and M2 polarization
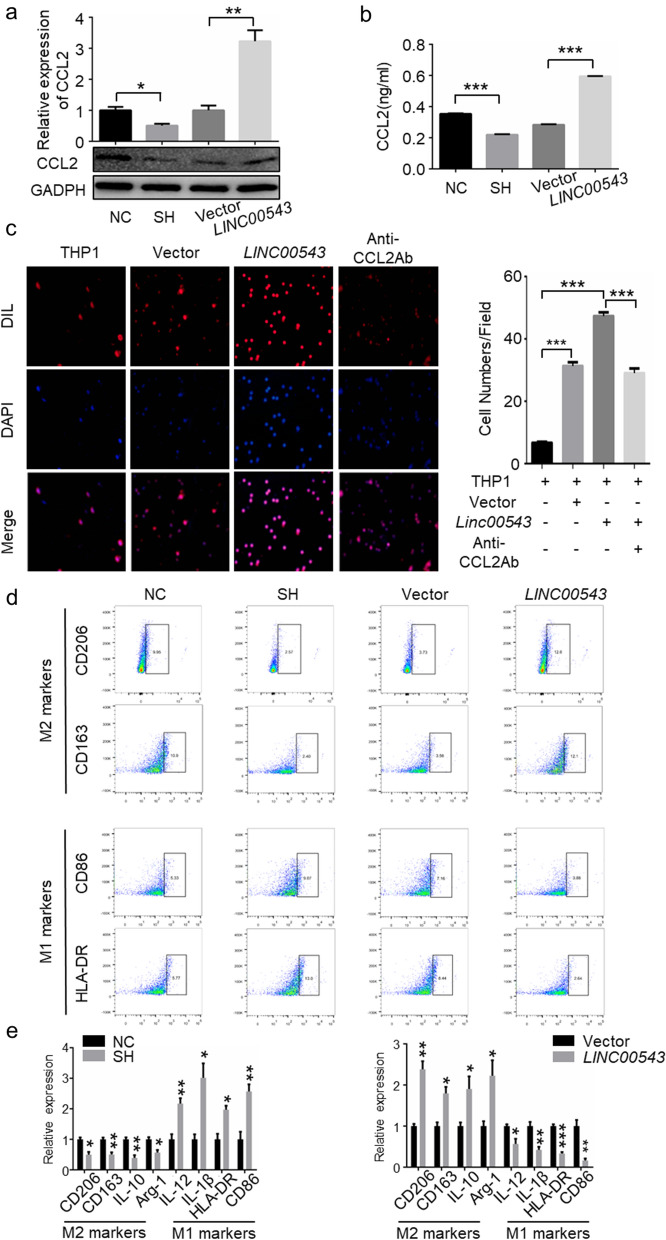
It is well known that CCL2 is a chemokine which is positively regulated by FOXQ1 to promote macrophage recruitment and M2 polarization [17, 25, 34]. We found that the knockdown of LINC00543 inhibited CCL2 expression in CRC cells and its overexpression promoted CCL2 expression (Fig. 7a). The results from ELLSA assays with the supernatant from CRC cell lines were consistent with the above findings (Fig. 7b). We co-cultured four particular cell lines (SW620-NC, SW620-SH, SW480-Vector, and SW480-LINC00543) with PMA-treated THP1 cells, respectively. It was found that the knockdown of LINC00543 inhibited the recruitment and M2 polarization of macrophages. However, the opposite results were obtained with LINC00543 overexpressing cells (Fig. 7c–e, Additional file 1: Fig. S8). Overall, LINC00543 promoted macrophage recruitment and M2 polarization by upregulating CCL2 expression.
Fig. 7.
LINC00543 promotes macrophage recruitment and M2 polarization by regulating CCL2 expression. a The expression of LINC00543 in SH and LINC00543 cells. b The expression of CCL2 in the supernatant of SH and LINC00543 cells. c Transwell assays analyzing the effect of LINC00543 on CCL2 mediated macrophage recruitment; magnification, × 200. d Flow cytometry analysis of the effect of LINC00543 in macrophage polarization. e qRT-PCR analysis of M1 and M2 macrophage markers in the SH and LINC00543 cells
LINC00543 promotes tumorigenesis in vivo
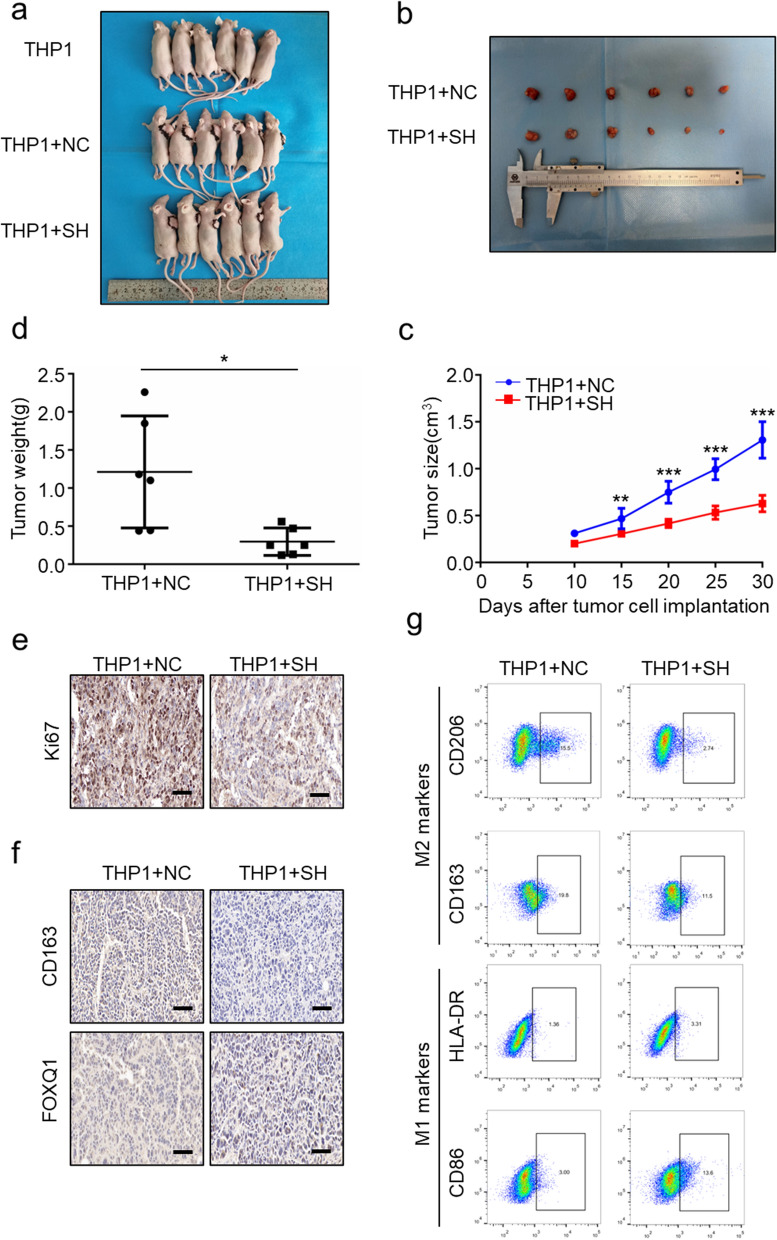
We mixed the NC group and the SH group cells with PMA-treated THP1 cells, respectively and subcutaneously injected the mixed cells into nude mice. The results showed that the knockdown of LINC00543 inhibited CRC tumorigenesis in vivo (Fig. 8a–e, Additional file 1: Fig. S9a). The IHC results indicated that the expression levels of CD163 and FOXQ1 in xenografts from the SH group mice were downregulated compared with NC group (Fig. 8f, Additional file 1: Fig. S9b). Flow cytometry analysis of the xenografts showed that the knockdown of LINC00543 suppressed the M2-like TAMs proportion (Fig. 8g). The results showed that LINC00543 regulated M2 polarization of the TAMs and promoted CRC tumorigenesis in vivo.
Fig. 8.
LINC00543 promotes the CRC tumorigenesis and the M2 polarization of macrophages in vivo. a–d The tumor volumes and weights in the THP1 group, THP1 + NC group, and THP1 + SH group. e and f The immunohistochemistry analysis results of tumors from the THP1 + N and THP1 + SH groups (scale bar = 100 μm). g The proportion of M1 and M2 macrophages in the THP1 + NC and THP1 + SH groups
Discussion
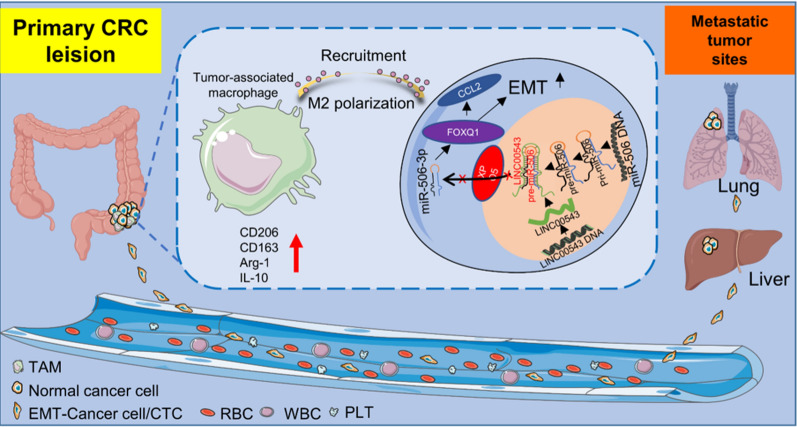
CRC is the most common gastrointestinal malignancy, and once distant metastasis occurs, the 5-year survival rate of patients decreases dramatically [35]. Therefore, it is essential to explore the metastasis associated mechanisms of CRC in greater depth and identify novel and effective targets to develop new treatment options. Increasing number of studies have shown that lncRNAs were differentially expressed in various cancers and played essential roles in cancer metastasis [36–39]. In this study, we found that LINC00543 was highly expressed in CRC tissues and was strongly associated with metastasis and poor prognosis in CRC patients. The mechanism of lncRNAs largely depends on their subcellular localization. We found LINC00543 was mainly localized in the nucleus and bound to the pre-miR-506-3p, preventing XPO5 from transporting pre-miR-506-3p from the nucleus to the cytoplasm, thereby inhibiting miR-506-3p expression and elevating FOXQ1 expression. Next, we showed that FOXQ1 promoted EMT and the secretion of CCL2 by CRC cells. Moreover, the upregulation of CCL2 secretion recruited TAMs and induced their polarization to the M2 phenotype, which ultimately promoted CRC progression (Fig. 9).
Fig. 9.
LINC00543 promotes colorectal cancer metastasis by driving epithelial-mesenchymal transition and remodeling the tumor microenvironment
EMT is a complex biological program that is extensively involved in cancer development and metastasis [10]. Many of the current studies have reported that EMT activated the stemness of gastric cancer cells and mediated chemoresistance [40, 41]. In addition, increasing number of interesting studies have reported that tumor cells undergoing EMT promoted cancer metastasis by remodeling the TME. These studies mainly revolved around the modulation of the immune cells in the TME. Among them, TAMs attracted much attention because of their indispensable role in cancer progression. The miR-195-5p/NOTCH2 axis has been reported to induce EMT in CRC cells, which promoted IL-4 secretion to enhance TAMs polarization, thereby promoting tumor progression [42]. High ZEB1 expression accelerated cervical cancer progression through CCL8-induced TAMs recruitment [43]. Here, we observed that the LINC00543/pre-miR-506-3p/FOXQ1 axis induced EMT in CRC cells which enhanced the CCL2-mediated macrophage recruitment and M2-like polarization. Our results revealed the regulation of TAMs by cancer cells that underwent EMT.
Recently, studies have shown that miRNAs influenced the EMT process mainly by regulating the expression of EMT-related transcription factors [44]. Additionally, miRNAs have been extensively reported to be regulated by lncRNAs through various mechanisms. The lncRNA MPRL is known to enhance the drug resistance of tongue squamous cell carcinoma by disrupting the processing of mature miR-483-5p [30]. The lncRNA ATB has been reported to increase the expression of ZEB1 and ZEB2 by acting as ceRNA for the miR-200 family, inducing EMT in hepatocellular carcinoma cells [45]. The lncRNA Uc.283 + A promotes cancer progression by blocking the binding of DGCR8 and pri-miR-195 [46]. In our study, we demonstrated that LINC00543 blocked the maturation of miR-506-3p by inhibiting the transport function of XPO5, which regulated FOXQ1-mediated EMT.
Our study provides solid results to support the idea that LINC00543 remodels the TME in CRC and regulates EMT in CRC by modulating the pre-miR-506-3p/FOXQ1 axis. This study extends our understanding of the molecular mechanism of colorectal cancer metastasis to help develop novel therapeutic strategies in the future.
Supplementary Information
Additional file 1: Table S1. Relationship between LINC00543 and clinicopathological features of CRC patients(n=40). Table S2. The sequences of the primers for quantitative RT-qPCR. Table S3. The Sequences of siRNAs, Plasmid. Table S4. The Sequences of LINC00543 segmental antisense nucleotide fragment biotin probes. Fig. S1. LINC00543 is highly expressed in CRC tissues and is associated with poor prognosis of CRC patients. a. ISH analysis of the expression of LINC00543 in CRC tissues. LINC00543 is demonstrate as a long non-coding RNA without encoding proteins ability on chromosome 13 through online websites b. pubmed (https://pubmed.ncbi.nlm.nih.gov/), c. LnCAR (https://lncar.renlab.org/), and d. CPC2 (http://cpc2.gao-lab.org/). Fig. S2. LINC00543 promotes the invasion and metastasis of CRC cells. The construction of LINC00543 knockdown and overexpression stably transfected cell lines. a. The knockdown efficiencies of Si-LINC00543A, Si-LINC00543B and Si-LINC00543C in SW620 cells. b. The knockdown efficiencies of SH-LINC00543A and SH-LINC00543B in cells. c. The transfection efficiency of LINC00543 in SW480 cells. d. The Ki67 score of NC and SH cells. Fig. S3. LINC00543 regulates the EMT of CRC cells. The expression levels of E-cadherin, N-cadherin, and Vimentin in NC group and SH group nude mice. Fig. S4. LINC00543 regulates EMT by upregulating FOXQ1 in CRC cells. The expression levels of EMT-related transcription factors in SH and LINC00543 cells. Fig. S5. The Polysome analysis of SH cells. Fig. S6. LINC00543 indirectly regulates FOXQ1 by downregulating miR-506-3p. The interaction of LINC00543 with miR-506-3p and Pre-miR-506-3p were predicted through online prediction website. a. The interaction of LINC00543 with miR-506-3p was predicted through online prediction website STARBASE (https://starbase.sysu.edu.cn/). b. The interaction of LINC00543 with miR-506-3p was predicted through online prediction website LncBASE (https://diana.e-ce.uth.gr/lncbasev3/interactions). c. The LINC00543 and Pre-miR-506-3p interaction were predicted through bioinformatics tool miRanda. Fig. S7. LINC00543 inhibits the transportation of pre-miR-506-3p. a. The relative expression levels of miR-506-3p, Pre-miR-506-3p, and Pri-miR-506-3p were detected in LINC00543 cells. b. RAP were applied to analyze the interaction between LINC00543 and Pre-miR-506-3p. c. The expression of Pre-miR-506-3p in the nucleus and cytosol of LINC00543 cells. d. RIP analysis of XPO5 interaction with Pre-miR-506-3p in LINC00543 cells. e. RIP analysis of XPO5 interaction with LINC00543 in LINC00543 cells. f. Relative expression of XPO5 in SH and LINC00543 cells. g. Agarose gel electrophoresis analysis of regions of LINC00543 combination with Pre-miR-506-3p. h. RAP analysis of the interaction of four LINC00543 segments with Pre-miR-506-3p. i. RIP analysis of the interaction of Pre-miR-506-3p and LINC00543 segments in incubated of XPO5 cells. j and k. Analysis of the interactions of LINC00543, XPO5 and Pre-miR-506-3p in cells incubated with antisense nucleotide biotin probes of the 3 binding regions. Fig. S8. LINC00543 promotes macrophage recruitment and M2 polarization through regulating CCL2 expression. Transwell assays were used to verify the relationship between the expression of LINC00543 and macrophage recruitment. Fig. S9. LINC00543 promotes the tumorigenesis of CRC and the M2 polarization of macrophages in vivo. a. The expression of Ki67 in THP1+NC group and THP1+SH group. b. The expression of CD163 and foxq1 in THP1+NC group and THP1+SH group.
Acknowledgements
Not applicable
Author contributions
JZ, BX, SW participated in the design of the study; collected the clinical samples of gastric cancer; JZ, XZ, RD performed the in vitro and in vivo experiments; BZ, CF, QX provided assistance to the experiments; ZD, SH collected the clinical data; JZ, ZL, JS conducted the analysis of the data; JZ, RD, SW wrote and modified the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81572874), National Natural Science Foundation of China (82173330), Joint Fund for Translational Medicine and Interdisciplinary Research of Zhongnan Hospital of Wuhan University (ZNJC201913), the mainstay project of young and middle-aged medicine in Wuhan (WHQG202003), Technology Innovation Seed Fund (znpy2019076), Zhongnan Hospital of Wuhan University.
Availability of data and materials
All the data and material are available at the Journal of translational medicine’s website.
Declarations
Ethics approval and consent to participate
This study was approved by the Zhongnan Hospital of Wuhan University ethics committee and the patient tissue samples were conducted in accordance with the Declaration of Helsinki (2021126). All animal studies were approved by the Institutional Animal Care and Use Committee of the Zhongnan Hospital of Wuhan University.
Consent for publication
All the authors agree for the publication.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jinsen Zheng, Rongzhang Dou, and Xinyao Zhang contributed equally to this work
Contributor Information
Jinsen Zheng, Email: zjs9527@whu.edu.cn.
Rongzhang Dou, Email: dourongzhang@whu.edu.cn.
Xinyao Zhang, Email: zhangxinyao_y@163.com.
Bo Zhong, Email: zhongbo@whu.edu.cn.
Chenggang Fang, Email: fcg920767410@qq.com.
Qian Xu, Email: whuxuq@whu.edu.cn.
Ziyang Di, Email: ziyangdi@163.com.
Sihao Huang, Email: jordan_s_k@hotmail.com.
Zaihuan Lin, Email: 2014312180058@whu.edu.cn.
Jialin Song, Email: sjlsxs1995@163.com.
Shuyi Wang, Email: shuyiwang@whu.edu.cn.
Bin Xiong, Email: binxiong1961@whu.edu.cn.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Steeg PS. Targeting metastasis. Nat Rev Cancer. 2016;16(4):201–218. doi: 10.1038/nrc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmadian E, Khosroushahi AY, Eftekhari A, Farajnia S, Babaei H, Eghbal MA. Novel angiotensin receptor blocker, azilsartan induces oxidative stress and NFkB-mediated apoptosis in hepatocellular carcinoma cell line HepG2. Biomed Pharmacother Biomed Pharmacother. 2018;99:939–946. doi: 10.1016/j.biopha.2018.01.117. [DOI] [PubMed] [Google Scholar]
- 4.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11(10):889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 5.Yin Y, Liu B, Cao Y, et al. Colorectal cancer-derived small extracellular vesicles promote tumor immune evasion by upregulating PD-L1 expression in tumor-associated macrophages. Adv Sci (Weinheim, Baden-Wurttemberg, Germany) 2022;9(9):2102620. doi: 10.1002/advs.202102620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang YH, Cai K, Xu PP, et al. CREBBP/EP300 mutations promoted tumor progression in diffuse large B-cell lymphoma through altering tumor-associated macrophage polarization via FBXW7-NOTCH-CCL2/CSF1 axis. Signal Transduct Target Ther. 2021;6(1):10. doi: 10.1038/s41392-020-00437-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wen ZF, Liu H, Gao R, et al. Tumor cell-released autophagosomes (TRAPs) promote immunosuppression through induction of M2-like macrophages with increased expression of PD-L1. J Immunother Cancer. 2018;6(1):151. doi: 10.1186/s40425-018-0452-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez-Gonzalez I, Bobien A, Molnar C, et al. miR-149 suppresses breast cancer metastasis by blocking paracrine interactions with macrophages. Can Res. 2020;80(6):1330–1341. doi: 10.1158/0008-5472.CAN-19-1934. [DOI] [PubMed] [Google Scholar]
- 9.Han Y, Sun J, Yang Y, et al. TMP195 exerts antitumor effects on colorectal cancer by promoting M1 macrophages polarization. Int J Biol Sci. 2022;18(15):5653–5666. doi: 10.7150/ijbs.73264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thiery JP, Lim CT. Tumor dissemination: an EMT affair. Cancer Cell. 2013;23(3):272–273. doi: 10.1016/j.ccr.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Liu M, Zhang Y, Yang J, et al. Zinc-Dependent regulation of ZEB1 and YAP1 coactivation promotes epithelial-mesenchymal transition plasticity and metastasis in pancreatic cancer. Gastroenterol. 2021;160(5):1771–1783 e1. doi: 10.1053/j.gastro.2020.12.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li CF, Chen JY, Ho YH, et al. Snail-induced claudin-11 prompts collective migration for tumour progression. Nat Cell Biol. 2019;21(2):251–262. doi: 10.1038/s41556-018-0268-z. [DOI] [PubMed] [Google Scholar]
- 13.Cao J, Wang X, Dai T, et al. Twist promotes tumor metastasis in basal-like breast cancer by transcriptionally upregulating ROR1. Theranostics. 2018;8(10):2739–2751. doi: 10.7150/thno.21477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, Meng F, Liu G, et al. Forkhead transcription factor foxq1 promotes epithelial-mesenchymal transition and breast cancer metastasis. Can Res. 2011;71(4):1292–1301. doi: 10.1158/0008-5472.Can-10-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20(2):69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 16.Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell. 2009;15(3):195–206. doi: 10.1016/j.ccr.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 17.Xia L, Huang W, Tian D, et al. Forkhead box Q1 promotes hepatocellular carcinoma metastasis by transactivating ZEB2 and VersicanV1 expression. Hepatology (Baltimore, MD) 2014;59(3):958–973. doi: 10.1002/hep.26735. [DOI] [PubMed] [Google Scholar]
- 18.Flintoft L. Non-coding RNA: Structure and function for lncRNAs. Nat Rev Genet. 2013;14(9):598. doi: 10.1038/nrg3561. [DOI] [PubMed] [Google Scholar]
- 19.Chen L, He M, Zhang M, et al. The role of non-coding RNAs in colorectal cancer, with a focus on its autophagy. Pharmacol Ther. 2021;226:107868. doi: 10.1016/j.pharmthera.2021.107868. [DOI] [PubMed] [Google Scholar]
- 20.Yue B, Liu C, Sun H, et al. A positive feed-forward loop between LncRNA-CYTOR and Wnt/β-catenin signaling promotes metastasis of colon cancer. Mol Ther J Am Soc Gene Ther. 2018;26(5):1287–1298. doi: 10.1016/j.ymthe.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu N, Jiang M, Liu H, et al. LINC00941 promotes CRC metastasis through preventing SMAD4 protein degradation and activating the TGF-β/SMAD2/3 signaling pathway. Cell Death Differ. 2021;28(1):219–232. doi: 10.1038/s41418-020-0596-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang ZX, Liu HS, Wang FW, et al. LncRNA RPPH1 promotes colorectal cancer metastasis by interacting with TUBB3 and by promoting exosomes-mediated macrophage M2 polarization. Cell Death Dis. 2019;10(11):829. doi: 10.1038/s41419-019-2077-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo JN, Xia TY, Deng SH, Xue WN, Cui BB, Liu YL. Prognostic immunity and therapeutic sensitivity analyses based on differential genomic instability-associated LncRNAs in left- and right-sided colon adenocarcinoma. Front Mol Biosci. 2021;8:668888. doi: 10.3389/fmolb.2021.668888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei C, Yang C, Wang S, et al. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol Cancer. 2019;18(1):64. doi: 10.1186/s12943-019-0976-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, Huang N, Wei S, et al. lncRNA TUG1 as a ceRNA promotes PM exposure-induced airway hyper-reactivity. J Hazard Mater. 2021;416:125878. doi: 10.1016/j.jhazmat.2021.125878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin X, Zhuang S, Chen X, et al. lncRNA ITGB8-AS1 functions as a ceRNA to promote colorectal cancer growth and migration through integrin-mediated focal adhesion signaling. Mol Ther J Am Soc Gene Ther. 2021 doi: 10.1016/j.ymthe.2021.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin D, Hu ZQ, Luo CB, et al. LINC01133 promotes hepatocellular carcinoma progression by sponging miR-199a-5p and activating annexin A2. Clin Transl Med. 2021;11(5):e409. doi: 10.1002/ctm2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu G, Ma J, Zhang J, et al. Hypoxia-induced lncHILAR promotes renal cancer metastasis via ceRNA for the miR-613/206/ 1–1-3p/Jagged-1/Notch/CXCR4 signaling pathway. Mol Ther J Am Soc Gene Ther. 2021;29(10):2979–2994. doi: 10.1016/j.ymthe.2021.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Tian T, Lv X, Pan G, et al. Long noncoding RNA MPRL promotes mitochondrial fission and cisplatin chemosensitivity via disruption of pre-miRNA processing. Clin Cancer Res. 2019;25(12):3673–3688. doi: 10.1158/1078-0432.Ccr-18-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liz J, Portela A, Soler M, et al. Regulation of pri-miRNA processing by a long noncoding RNA transcribed from an ultraconserved region. Mol Cell. 2014;55(1):138–147. doi: 10.1016/j.molcel.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 32.Yu Y, Nangia-Makker P, Farhana L, Majumdar APN. A novel mechanism of lncRNA and miRNA interaction: CCAT2 regulates miR-145 expression by suppressing its maturation process in colon cancer cells. Mol Cancer. 2017;16(1):155. doi: 10.1186/s12943-017-0725-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okada C, Yamashita E, Lee SJ, et al. A high-resolution structure of the pre-microRNA nuclear export machinery. Science (New York, NY) 2009;326(5957):1275–1279. doi: 10.1126/science.1178705. [DOI] [PubMed] [Google Scholar]
- 34.Qian BZ, Li J, Zhang H, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475(7355):222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 36.Zhang S, Zhao BS, Zhou A, et al. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31(4):591–606.e6. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z, Hou P, Fan D, et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 2017;24(1):59–71. doi: 10.1038/cdd.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang Y, Chen X, Wu Y, et al. LncRNA CASC9 promotes esophageal squamous cell carcinoma metastasis through upregulating LAMC2 expression by interacting with the CREB-binding protein. Cell Death Differ. 2018;25(11):1980–1995. doi: 10.1038/s41418-018-0084-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H, Liang L, Dong Q, et al. Long noncoding RNA miR503HG, a prognostic indicator, inhibits tumor metastasis by regulating the HNRNPA2B1/NF-κB pathway in hepatocellular carcinoma. Theranostics. 2018;8(10):2814–2829. doi: 10.7150/thno.23012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu J, Cao LL, Xu Y, et al. FOXC1 modulates stem-like cell properties and chemoresistance through hedgehog and EMT signaling in gastric adenocarcinoma. Mol Ther J Am Soc Gene Ther. 2021 doi: 10.1016/j.ymthe.2021.09.009. [DOI] [PubMed] [Google Scholar]
- 41.López-Menéndez C, Vázquez-Naharro A, Santos V, et al. E2A modulates stemness, metastasis, and therapeutic resistance of breast cancer. Can Res. 2021;81(17):4529–4544. doi: 10.1158/0008-5472.Can-20-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin X, Wang S, Sun M, et al. miR-195-5p/NOTCH2-mediated EMT modulates IL-4 secretion in colorectal cancer to affect M2-like TAM polarization. J Hematol Oncol. 2019;12(1):20. doi: 10.1186/s13045-019-0708-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Chen XJ, Deng YR, Wang ZC, et al. Hypoxia-induced ZEB1 promotes cervical cancer progression via CCL8-dependent tumour-associated macrophage recruitment. Cell Death Dis. 2019;10(7):508. doi: 10.1038/s41419-019-1748-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 45.Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 46.Lujambio A, Portela A, Liz J, et al. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene. 2010;29(48):6390–6401. doi: 10.1038/onc.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Relationship between LINC00543 and clinicopathological features of CRC patients(n=40). Table S2. The sequences of the primers for quantitative RT-qPCR. Table S3. The Sequences of siRNAs, Plasmid. Table S4. The Sequences of LINC00543 segmental antisense nucleotide fragment biotin probes. Fig. S1. LINC00543 is highly expressed in CRC tissues and is associated with poor prognosis of CRC patients. a. ISH analysis of the expression of LINC00543 in CRC tissues. LINC00543 is demonstrate as a long non-coding RNA without encoding proteins ability on chromosome 13 through online websites b. pubmed (https://pubmed.ncbi.nlm.nih.gov/), c. LnCAR (https://lncar.renlab.org/), and d. CPC2 (http://cpc2.gao-lab.org/). Fig. S2. LINC00543 promotes the invasion and metastasis of CRC cells. The construction of LINC00543 knockdown and overexpression stably transfected cell lines. a. The knockdown efficiencies of Si-LINC00543A, Si-LINC00543B and Si-LINC00543C in SW620 cells. b. The knockdown efficiencies of SH-LINC00543A and SH-LINC00543B in cells. c. The transfection efficiency of LINC00543 in SW480 cells. d. The Ki67 score of NC and SH cells. Fig. S3. LINC00543 regulates the EMT of CRC cells. The expression levels of E-cadherin, N-cadherin, and Vimentin in NC group and SH group nude mice. Fig. S4. LINC00543 regulates EMT by upregulating FOXQ1 in CRC cells. The expression levels of EMT-related transcription factors in SH and LINC00543 cells. Fig. S5. The Polysome analysis of SH cells. Fig. S6. LINC00543 indirectly regulates FOXQ1 by downregulating miR-506-3p. The interaction of LINC00543 with miR-506-3p and Pre-miR-506-3p were predicted through online prediction website. a. The interaction of LINC00543 with miR-506-3p was predicted through online prediction website STARBASE (https://starbase.sysu.edu.cn/). b. The interaction of LINC00543 with miR-506-3p was predicted through online prediction website LncBASE (https://diana.e-ce.uth.gr/lncbasev3/interactions). c. The LINC00543 and Pre-miR-506-3p interaction were predicted through bioinformatics tool miRanda. Fig. S7. LINC00543 inhibits the transportation of pre-miR-506-3p. a. The relative expression levels of miR-506-3p, Pre-miR-506-3p, and Pri-miR-506-3p were detected in LINC00543 cells. b. RAP were applied to analyze the interaction between LINC00543 and Pre-miR-506-3p. c. The expression of Pre-miR-506-3p in the nucleus and cytosol of LINC00543 cells. d. RIP analysis of XPO5 interaction with Pre-miR-506-3p in LINC00543 cells. e. RIP analysis of XPO5 interaction with LINC00543 in LINC00543 cells. f. Relative expression of XPO5 in SH and LINC00543 cells. g. Agarose gel electrophoresis analysis of regions of LINC00543 combination with Pre-miR-506-3p. h. RAP analysis of the interaction of four LINC00543 segments with Pre-miR-506-3p. i. RIP analysis of the interaction of Pre-miR-506-3p and LINC00543 segments in incubated of XPO5 cells. j and k. Analysis of the interactions of LINC00543, XPO5 and Pre-miR-506-3p in cells incubated with antisense nucleotide biotin probes of the 3 binding regions. Fig. S8. LINC00543 promotes macrophage recruitment and M2 polarization through regulating CCL2 expression. Transwell assays were used to verify the relationship between the expression of LINC00543 and macrophage recruitment. Fig. S9. LINC00543 promotes the tumorigenesis of CRC and the M2 polarization of macrophages in vivo. a. The expression of Ki67 in THP1+NC group and THP1+SH group. b. The expression of CD163 and foxq1 in THP1+NC group and THP1+SH group.
Data Availability Statement
All the data and material are available at the Journal of translational medicine’s website.