Abstract
Objective
To explore and define the molecular cause(s) of a multi-generational kindred affected by Bechet’s-like mucocutaneous ulcerations and immune dysregulation.
Methods
Whole genome sequencing and confirmatory Sanger sequencing were performed. Components of the NFκB pathway were quantified by immunoblotting, and function was assessed by cytokine production (IL-6, TNF-α, IL-1β) after lipopolysaccharide (LPS) stimulation. Detailed immunophenotyping of T-cell and B-cell subsets was performed in four patients from this cohort.
Results
A novel variant in the RELA gene, p. Tyr349LeufsTer13, was identified. This variant results in premature truncation of the protein before the serine (S) 536 residue, a key phosphorylation site, resulting in enhanced degradation of the p65 protein. Immunoblotting revealed significantly decreased phosphorylated [p]p65 and pIκBα. The decrease in [p]p65 may suggest reduced heterodimer formation between p50/p65 (NFκB1/RelA). Immunophenotyping revealed decreased naïve T cells, increased memory T cells, and expanded senescent T-cell populations in one patient (P1). P1 also had substantially higher IL-6 and TNF-α levels post-stimulation compared with the other three patients.
Conclusion
Family members with this novel RELA variant have a clinical phenotype similar to other reported RELA cases with predominant chronic mucocutaneous ulceration; however, the clinical phenotype broadens to include Behçet’s syndrome and IBD. Here we describe the clinical, immunological and genetic evaluation of a large kindred to further expand identification of patients with autosomal dominant RELA deficiency, facilitating earlier diagnosis and intervention. The functional impairment of the canonical NFκB pathway suggests that this variant is causal for the clinical phenotype in these patients.
Keywords: NFκB, p65, RelA, immune dysregulation, Behçet’s syndrome, IBD
Rheumatology key messages.
Pathogenic RELA variants are associated with a varied phenotypic spectrum, with predominant mucocutaneous ulcerations.
This study expands the clinical phenotype of patients with RELA haploinsufficiency to include IBD and additional ophthalmic manifestations.
Prompt recognition of this complex immune dysregulatory phenotype may facilitate earlier diagnosis and intervention.
Introduction
The NFκB pathway is composed of a family of inducible transcription factors, which control DNA transcription, cytokine production, cell survival, and regulation of the immune response to infection. NFκB has five protein components—p65/RelA, RelB, c-Rel, p105/50 (NFκB1) and p100/p52 (NFκB2)—all of which share the Rel-homology domain (RHD) [1]. The p65/RelA protein is a key component of the canonical pathway of NFκB, and associates with p50/NFκB1for nuclear translocation and cytokine gene transcription (Fig. 1A). Signalling through the NF-κB pathway is important for maintaining self-tolerance and regulation of the immune response, besides playing a key role in control of the inflammatory response [2–7]. Defects in this pathway have been implicated in various inborn errors of immunity (IEIs). These include both immune deficiencies and autoinflammatory disorders, such as X-linked and autosomal dominant ectodermal dysplasia with immunodeficiency (IKBKG, IκBα, respectively), combined immunodeficiency secondary to pathogenic variants in IKKβ, RelB, c-Rel, NFκB1 and NFκB2, and CVID secondary to defects in NFκB1 and NFκB2 [2, 8–19].
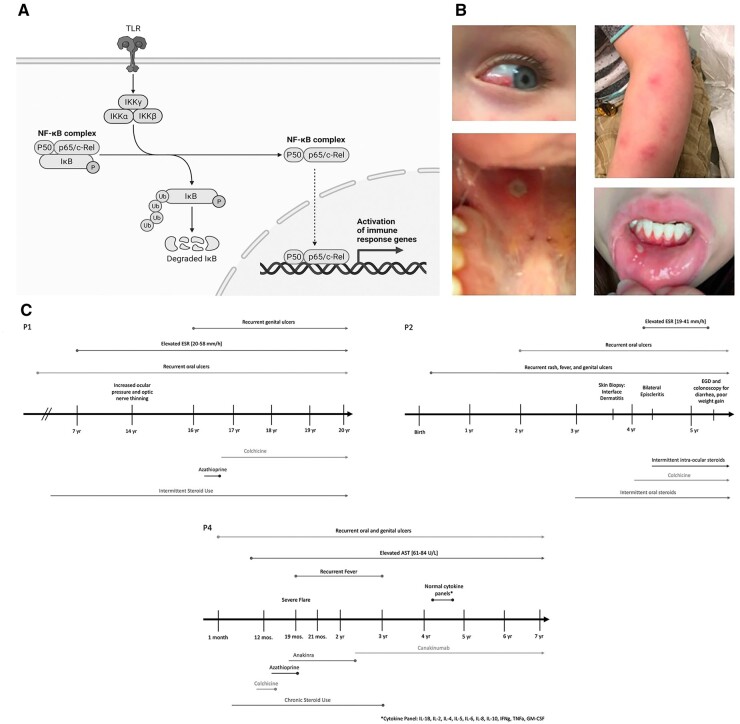
Fig. 1.
Timelines of clinical features and treatment
(A) Schematic depicting the canonical pathway of NFκB activation with p65[RelA]. Figure created with BioRender.com. (B) Photographic depiction of clinical manifestations in our cohort including mucocutaneous ulcerations in P1, along with episcleritis and rash in P2. (C) Key phenotypic and laboratory features and treatment is provided for P1, P2 and P4. Detailed clinical and laboratory history and treatment is not available for P3 as she was followed only sporadically.
Pathogenic variants in RELA have been previously reported in the last four years in at least ten patients from three different kindreds. These patients presented primarily with chronic mucocutaneous ulcerative disease. Badran et al. presented a family of four patients with a history of chronic mucocutaneous ulcerations and early onset colitis with a splice-site variant in RELA leading to RelA haploinsufficiency [20]. Adeeb et al. reported five patients within a single kindred with mucocutaneous ulcerative disease with a frameshift variant in RELA resulting in a truncated protein [21]. One of these patients had neuromyelitis optica (NMO), but did not have Behçet’s syndrome-like features [21, 22]. A single patient with a nonsense variant in RELA has been reported with ALPS (autoimmune lymphoproliferative syndrome)-like features and Evans syndrome (refractory ITP, haemolytic anaemia, neutropenia) and splenomegaly with lymphadenopathy [23, 24]. Additionally, three patients with novel RELA variants (two with the same missense variant, and one with a nonsense variant) have been reported with severe early-onset and/or familial SLE [25].
The association of RELA haploinsufficiency and Behçet’s-like features has been previously described though there appears to be a phenotypic spectrum with an occasional patient manifesting with lymphoproliferative disease and autoimmune cytopenias. The patient cohort in this study extends the previously described clinical phenotype of mucocutaneous ulcerations and immune dysregulation.
Methods
Human subjects
The index proband (P4), his parents and two maternal aunts gave informed written consent to participate in an IRB-approved research study, IRB11-00215. The P2 proband and his parents underwent clinical exome sequencing and gave informed consent to participate in research under another IRB-approved protocol, IRB18-00662. The four patients (P1, P2, P3 and P4) also consented to an immunology study protocol, IRB#00000015, for the various functional and immunological studies described herein. All IRB protocols were approved by Nationwide Children’s Hospital and include written informed consent.
Genome sequencing (P4 proband and parents)
Genomic DNA from was extracted from peripheral blood. Genome sequencing was performed as described in Supplementary Data S1, available at Rheumatology online [26–29].
Variant analysis and interpretation
Our general approach to variant analysis and interpretation for paediatric disorders has already been described [30]. Details of variant analysis are described in Supplementary Data S1, available at Rheumatology online [31, 32].
Clinical exome sequencing (P2 and parents)
Clinical exome sequencing was performed at the Steve and Cindy Rasmussen Institute for Genomic Medicine at Nationwide Children’s Hospital, Columbus, Ohio, as previously described [33]. Details of variant analysis are described in Supplementary Data S1, available at Rheumatology online.
Sanger sequencing
Genome sequencing findings were verified by Sanger sequencing as described in the Supplementary Data S1, available at Rheumatology online.
Immunoblotting
NFκB canonical pathway analysis was performed on isolated peripheral blood mononuclear cells (PBMCs) from patients and healthy controls as described in Supplementary Data S1, available at Rheumatology online.
Immunophenotyping
Immunophenotyping of T cells and B cells was performed with multiparametric flow cytometry using antibodies (Supplementary Table S1, available at Rheumatology online) for naïve and memory T cells, activated, senescent and exhausted T cells, naïve and memory B cells, transitional B cells, and plasmablasts using fluorescently conjugated antibodies, and analysed using a CytoFLEX LX (Beckman Coulter, La Brea, CA, USA) flow cytometer. The data was further analysed with Kaluza C v.1.1.2Ⓡ (Beckman Coulter) or FCS Express v.7 (De Novo, Pasadena, CA, USA) software.
Cytokine assays
Cytokines were assessed after stimulation of peripheral blood mononuclear cells (PBMCs) from healthy controls and patients as described in Supplementary Data S1, available at Rheumatology online.
Results
Clinical cases
The four patients described below were independently seen by different providers within the Rheumatology clinic. The familial relationship of these patients was discovered subsequently after collection of genetic data and extended pedigree evaluation. Characterization of the familial variant was performed subsequent to the genetic analysis.
Patient 1 (P1) is a 20-year-old Eurasian female with recurrent oral ulcerations since infancy, followed in the Rheumatology Clinic for possible Behçet’s syndrome. At age 16 y, she developed recurrent genital ulcerations with negative testing for herpes simplex virus (HSV) and sexually transmitted diseases. She had been followed by an ophthalmologist locally for raised intra-ocular pressure and thinning of the optic nerve. Additionally, she had a history of congenital heart disease with a ventricular septal defect, and sub-valvar aortic stenosis, repaired surgically. She has received allergen immunotherapy for severe environmental allergies. Her laboratory studies have demonstrated a persistently elevated sedimentation rate (ESR) between 21–58 mm/h (reference range <20 mm/h). Clinical and laboratory screening for SLE and coeliac disease were negative. She was reluctant to try treatment with non-biologic and biologic disease modifying agents due to concern for side effects. This patient has continued to experience periodic flares of oral ulcers, which have been treated with oral corticosteroids and sucralfate (Fig. 1B). The addition of colchicine has been effective in reducing her inflammatory flares. A brief trial of azathioprine was unsuccessful and was discontinued due to reported side effects (Fig. 1C).
Patient 2 (P2) is a 5-year-old white male, who presented at age 3 y with recurrent episodes of fever, genital ulcers since infancy, and oral ulcers since age 2 y. His mother reported a personal diagnosis of Behçet’s syndrome in early childhood. P1 and P2 are related as half-siblings, sharing a maternal parent. P2 complained of leg pain with fever episodes without signs of arthritis. He developed rashes resembling erythema nodosum or panniculitis, with multiple deep, painful, erythematous papules scattered on his arms and face (Fig. 1B). Skin biopsy, performed by Dermatology, revealed interface dermatitis. At age 4 y he developed scleral inflammation, and an urgent ophthalmology evaluation provided a diagnosis of episcleritis (Fig. 1B). His laboratory studies showed a mildly elevated ESR at 19 mm/h (reference range: <13 mm/h) with disease flares. Laboratory screening studies for SLE and coeliac disease were negative. Complement components C3, C4, and quantitative immunoglobulins were normal, and HLA B51 was also negative. His disease flares improved with short courses of corticosteroids, by oral or topical route. His mother declined additional immunosuppressive therapy, such as azathioprine and TNFα-inhibitors, due to concern for side effects. At age 4 y, he developed intermittent diarrhoea with poor weight gain, raising concern for IBD. Gastroenterology evaluation, including stool calprotectin and upper and lower endoscopy, was inconclusive for IBD and intestinal Behçet’s syndrome. His disease flares have been less frequent since starting colchicine (Fig. 1C).
Patient 3 (P3) is a 32-year-old white female with history of oral and genital ulcers since age 12 y. She was initially diagnosed with Crohn’s disease given additional mucosal ulcerations seen throughout the GI tract prior to ultimately receiving a diagnosis of Behçet’s syndrome. The clinical history described below is based on patient report and clinical phenotype, as she is infrequently followed at our hospital. Approximately, from age of 15–19 y she was treated with intermittent courses of oral corticosteroids for flares of mucocutaneous ulcerations. At age 20 y, she was treated with several immunomodulatory agents, including hydroxychloroquine, colchicine and azathioprine. She was transitioned to infliximab at age 21 y due to lack of clinical improvement with prior therapies, but developed septic knee arthritis while on this drug. She was subsequently started on adalimumab, and remained on this drug for two years, with some improvement in her mucocutaneous ulcers. During this period, she required an admission to the intensive care unit for sepsis, and following discharge, voluntarily ceased all treatment. For the following six-year period, her flares were infrequent, and managed with intermittent short courses of oral corticosteroids. After developing more frequent episodes of inflammatory disease, she was started on a phosphodiesterase 4 (PDE-4) inhibitor, apremilast, which was approved for treatment of Behçet’s syndrome in 2019. She has been on this treatment for the past 2 years and appears to have excellent control of her disease.
Patient 4 (P4) is a 7-year-old white/Hispanic male who developed an anogenital ulcer around 1 month of age, and continued to have recurrent oral and genital non-scarring ulcers that were typically associated with fever. In addition to his mucocutaneous ulcers, he had recurrent conjunctivitis without evidence of uveitis, urticarial-like rashes, cervical lymphadenopathy and poor weight gain. He did not have a history of recurrent infections. His laboratory evaluation was notable for elevated inflammatory markers (ESR, CRP), thrombocytosis and anaemia of chronic disease. Testing was negative for HLA B51, and he had cytokine panels performed on two different occasions while on high-dose corticosteroids, which were normal. He has been treated with corticosteroids, colchicine and azathioprine with minimal response. TNF-α inhibitors were considered, but ultimately decided against given his mother’s (P3) history of failed therapy with these medications. He was started on anakinra, a recombinant IL-1 receptor antagonist, with good response, and eventually weaned off corticosteroids. He was then transitioned to canakinumab, an anti-IL-1β monoclonal antibody, which required higher than normal doses to maintain control of disease (Fig. 1C). Due to poor weight gain over the years, he had a gastrostomy tube placed to provide high caloric intake. The family history is significant for recurrent oral and vaginal ulcers in his mother (P3), as well as two maternal aunts who have Crohn’s disease of varying severity. P1 and P2 are related to P3 and P4 via maternal lineage.
Additional information for the above four patients can be found in Supplementary Table S2, available at Rheumatology online.
Genomic analysis
Genome sequencing
The index proband for this study (P4) presented to Clinical Genetics with concern for short stature and speech delay. He had a history of oral and penile ulcers and carried a clinical diagnosis of neonatal Behçet’s syndrome. His mother (P3) had a long history of Behçet’s syndrome (mouth, vaginal and GI tract sores as well as abdominal pain). Two maternal aunts had similar symptoms, though not necessarily as a formal diagnosis, and a few members of the maternal grandfather’s family have been reported to have autoimmune conditions, including SLE and Crohn’s disease (Fig. 2A). The family was referred for research testing based on the strong family history, and lack of a molecular diagnosis on targeted panel testing. The proband and his parents were enrolled for trio genome sequencing (GS). Approximately ∼240.5 gbp of raw sequencing data per individual was generated, yielding an average genome-wide read depth of 65.32×. Detailed sequencing metrics are provided in Supplementary Table S3, available at Rheumatology online.
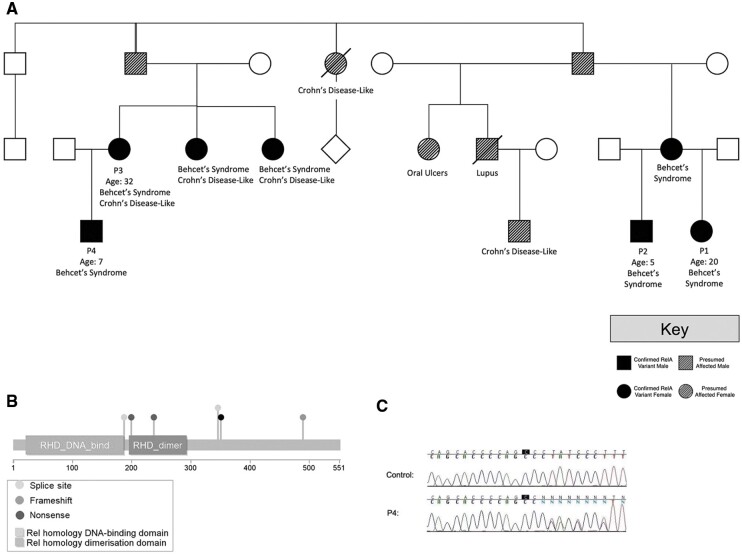
Fig. 2.
Novel RELA frameshift variant segregates with disease in a large family kindred
(A) Pedigree of the kindred with Behçet’s syndrome. Labels indicate individuals with detailed clinical phenotyping. (B) RelA protein structure with the p. Tyr349LeufsTer13 frameshift variant (black) plotted alongside other reported pathogenic variants from ClinVar and the literature. (C) Sanger sequencing chromatograms demonstrating segregation of the p. Tyr349LeufsTer13 variant in P4. See Supplementary Fig. S1 (available at Rheumatology online) for Sanger sequencing data from additional family members. Note that the presence of the (heterozygous) single-base insertion shifts the chromatogram peaks for approximately half of the pool of amplified sequences, giving rise to apparent heterozygosity at every base following the insertion (this pattern is not observed in the anonymous negative control sample).
Variant analysis and interpretation
Genome-wide copy number variant (CNV) calling with VarScan v.2.3.4 identified 15 high-confidence CNVs ranging in size from 27.5 kbp to 1.02 Mbp. Only two of these overlapped with exons of OMIM disease genes [1]: a 374.6kbp gain in VWA3B (associated with autosomal recessive spinocerebellar ataxia 22, MIM #614884) and an 861.7kbp gain of DNAI2 (associated with primary ciliary dyskinesia 9, MIM #612444). Both variants were maternally inherited, but neither condition matched the inheritance pattern or clinical features observed in this family.
For the SNV/indel analysis, multiple possible inheritance models were considered for the proband’s condition, but did not identify any compelling recessive or X-linked candidates. Analysis of maternally inherited variants under a dominant disease model uncovered 728 coding variants with MAF < 5%. Of these, 112 were rare, non-silent variants, and 20 were in genes associated with OMIM disease (Supplementary Table S2, available at Rheumatology online). Half of these were associated with autosomal recessive (AR) conditions. Of the 10 remaining candidates, only one offered a reasonable phenotypic match: a novel single-base insertion in RELA, a gene that encodes a subunit of the NFκB complex. As mentioned earlier, Badran et al. reported a splice site variant in RELA as the cause of disease in a two-generation family with autosomal dominant chronic mucocutaneous ulceration (MIM #618287) [20]. As of February 2022, the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/) contained a total of 14 RELA loss-of-function variants reported as pathogenic or likely pathogenic (Fig. 2B). One is a de novo splice site change (NM_021975.4: c.1034-2A>C) identified in a patient with mucocutaneous ulceration by Rady Children’s Hospital. Another is a de novo nonsense variant (p.Arg246*) in a patient with lymphoproliferative disease and autoimmune cytopenias [24]. The remaining three variants in ClinVar are nonsense changes identified in patients for which no phenotypic information was provided.
Identification of a truncating variant in RELA
The variant in this family was a novel single-base insertion in RELA predicted to cause a frameshift (NM_021975.4: c.1044dupC, p. Tyr349LeufsTer13; Fig. 2B), which reduces the amount of the protein by approximately one-third. It is absent from the gnomAD and TopMed databases, making it extremely rare. Research-based Sanger sequencing confirmed the variant was heterozygous in the proband, mother and two affected maternal aunts (Fig. 2C;Supplementary Fig. S1, available at Rheumatology online). Notably, a second male patient (proband P2) with a diagnosis of Behçet’s syndrome was seen separately in the same institution, and referred for clinical exome sequencing, which uncovered the identical frameshift variant in RELA (p.Tyr349LeufsTer13). Through pedigree comparison and sequence-based relationship inference using the KING algorithm, it was determined that the RELA-positive mothers of patients P4 and P2 were first cousins. Multiple family members in their shared lineage have clinically confirmed or family-reported features of Behçet’s syndrome (Fig. 2A).
The phenotypic similarities between our family and the one described by Badran et al. are striking. Furthermore, through international matchmaking via GeneMatcher [34], an Irish family with a similar dominant Behçet’s-like disorder caused by another truncating variant in RELA (p.His487ThrfsTer7; Fig. 2B) was identified, and later published by Adeeb et al. [21].
No rare (MAF < 0.05) coding variants were identified in NOD2 or the other 30 genes currently described to be associated with IBD/Crohn’s disease susceptibility according to OMIM (MIM #266600).
Functional analysis of the canonical NFκB pathway
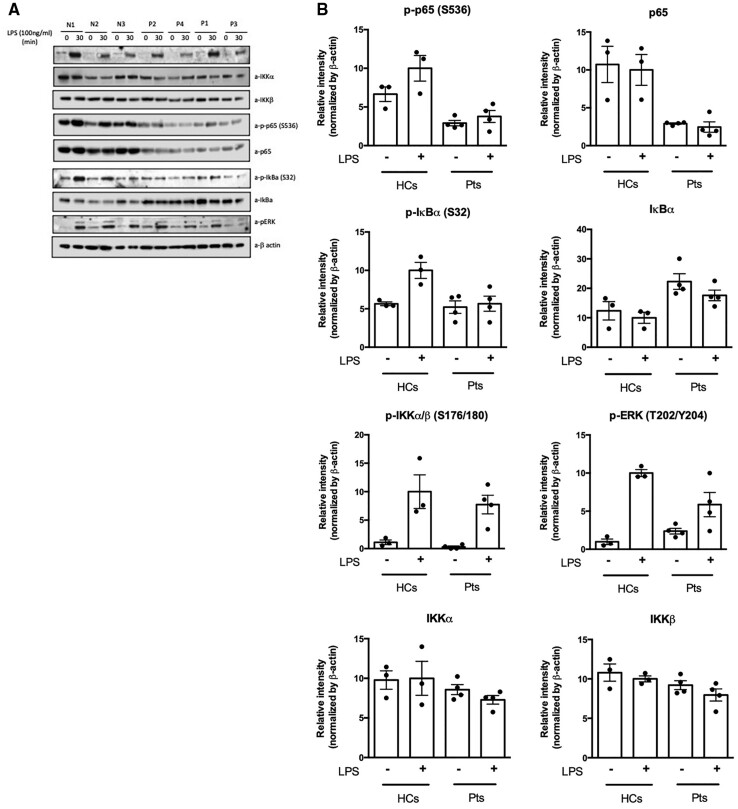
To determine whether this novel RELA variant (p.Tyr349LeufsTer13) affected downstream NFκB signalling, immunoblotting with densitometric analysis was performed before and after LPS stimulation to quantify native and phosphorylated proteins of the canonical NFκB pathway. The antibody used for assessment of native p65/RelA binds an epitope near/at Glu498 of the protein, while the patients’ variant is located upstream of this position. Therefore, the truncation of the protein due to the presence of the pathogenic variant would result in the loss of the binding epitope for the detecting antibody. This would prove that the RelA protein in the patients is truncated. After LPS stimulation, the RELA patients showed minimal phosphorylation of p65, as well as reduced levels of native p65 compared with controls, with no increment in protein expression on LPS stimulation (Fig. 3A and B). There is also substantial reduction in pERK after LPS stimulation in patients compared with controls. There does not appear to be any notable differences in protein and phosphorylation levels of IKKα and IKKβ pre- or post-LPS stimulation. The decrease in phosphorylated p65 in the patients suggests reduced heterodimer formation between p50/p65 [NFκB1/RelA], consistent with RELA haploinsufficiency.
Fig. 3.
Western Blots and densitometric quantitation of immunoblotting for native and phosphorylated proteins of the canonical NFκB pathway
Total PBMCs from healthy controls and four patients were stimulated with LPS (100 ng/mL) for 30 min. (A) The Western Blot images were acquired with a C-Digit scanner, and data were analysed using Image Studio software (Li-Cor). (B) The proteins of the NFkB canonical pathway from the Western Blot were quantified using densitometry, and the levels of proteins were normalized to β-actin. The relative phosphorylation and protein levels are shown. The values are expressed as the mean (s.d.).
Immunophenotyping analyses
Comrie et al. [24] have described immunophenotypic abnormalities in a single patient with RelA haploinsufficiency and Evans syndrome. Therefore, flow cytometric immunophenotyping of T-cell and B-cell subsets was performed in all four RELA patients (Table 1). Only one of the four RELA patients tested (P1) showed any quantitative anomalies in T-cell subsets. P1 had decreased naïve CD4+ and CD8+ CD45RA+ T cells with increased CD4+CD45RO+ memory T cells compared with the experimental healthy control, reference interval, and other RELA patients. Both P1 and P3 showed an increase in activated CD4+CD25+ T cells, with P1 also showing an expansion of CD4+CD28+CD57+ senescent T cells. B-cell phenotyping was unremarkable in all patients.
Table 1.
Flow cytometric immunophenotyping of T-cell subsets in blood
| CD45RA+ | CD45RO+ | Naïve | TCM | TEM | CD28+CD57+ | CD25+ | ||
|---|---|---|---|---|---|---|---|---|
| %CD4+ T Cells | HC | 38.35 | 42 | 45 | 34 | 9 | 5.96 | 45.36 |
| P1 | 10.275 | 65.95 | 56.16 | 33.17 | 15.435 | 19.09 | 71.31 | |
| P2 | 72.8 | 15.5 | 52.8 | 29.72 | 7.4 | 4.1 | 16 | |
| P3 | 45.34 | 43.75 | 51.64 | 16.96 | 11.76 | 6.09 | 57.85 | |
| P4 | 77.82 | 14.96 | 43.25 | 23.52 | 7.12 | 7.82 | 11.38 | |
| %CD8+ T Cells | HC | 52 | 28 | 21.7 | 6 | 16 | 10.04 | 29.00 |
| P1 | 18.55 | 19.35 | 19.89 | 3.61 | 11.07 | 18.15 | 24.89 | |
| P2 | 89.2 | 1.77 | 31.81 | 1.12 | 16.83 | 3.97 | 4.75 | |
| P3 | 63.95 | 8.6 | 27.53 | 1.85 | 14.14 | 11.54 | 52.01 | |
| P4 | 92.22 | 1.98 | 31.49 | 0 | 15.27 | 11.97 | 4.87 |
P1 shows decreased naïve CD4+ and CD8+ CD45RA+ T cells and increased CD4+CD45RO+ memory T cells compared with the healthy control (HC) and other RELA patients. Both P1 and P3 showed an increase in activated CD4+CD25+ T cells, with P1 also showing an expansion of CD4+CD28+CD57+ senescent T cells. The median value (range) for each cell subset derived from 54 healthy controls is: CD4+CD45RA+=38% (14–67%), CD4+CD45RO+=42% (11–78%), CD4+CD25+=45% (20–94%), CD4+CD28+CD57+=6% (0–44%), and CD8+CD45RA+=52% (20–75%).
TCM: central memory T cells; TEM: effector memory T cells.
Cytokine assays
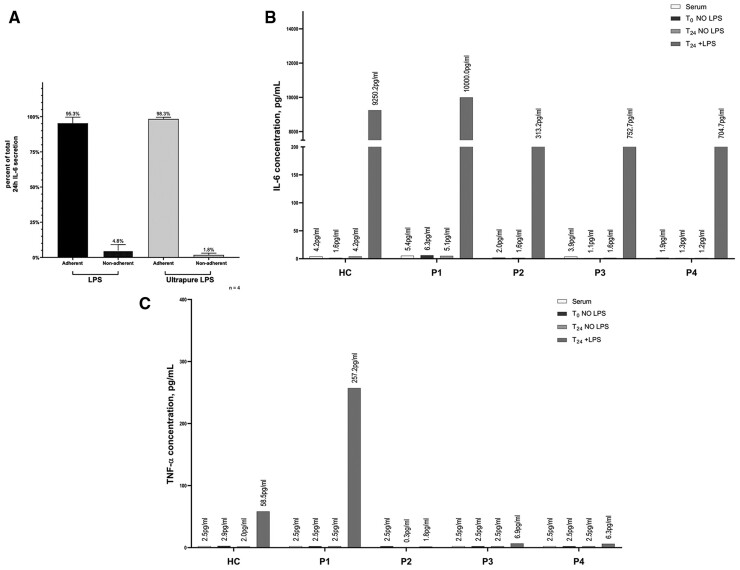
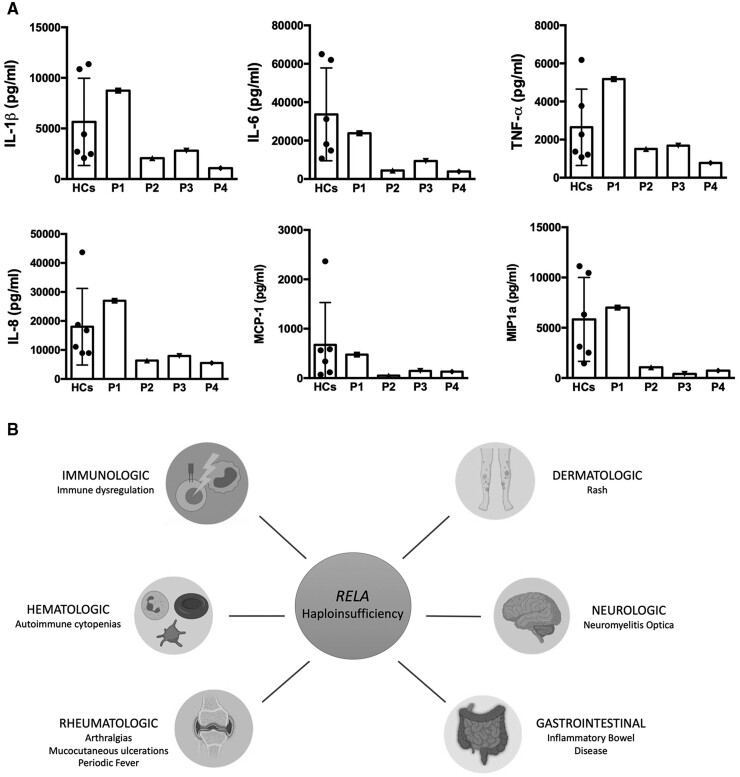
To determine whether there was dysregulation of the NFκB pathway in these patients, cytokine secretion from monocytes or PBMCs was measured before and after LPS stimulation and compared relative to healthy controls (Fig. 4). The rationale for using monocytes vs PBMCs was to ascertain the cellular contribution of the cytokines produced post-stimulation. Ultrapure and standard LPS were both tested to see if any measurable difference was noted through TLR4 signalling alone (ultrapure LPS) compared with signalling through both TLR4 and TLR2 (standard LPS) (Fig. 4A). For all tested cytokines, the majority of the secreted IL-6 was from monocytes after LPS stimulation, and not from lymphocytes (non-adherent cells), in both patients and controls. Unstimulated monocytes, as well as serum, from all four patients show negligible IL-6. However, after stimulation, P1 shows maximal IL-6 comparable to the healthy control. P2, P3 and P4 show substantially diminished IL-6 levels post-stimulation compared with P1 and the healthy experimental control (Fig. 4C). None of the patients received IL-6 or IL-6R blocking therapies. TNF-α production was highest in P1, while P2, P3 and P4 showed no TNF-α secretion post-stimulation (Fig. 4B). Individual cytokines were also assessed for each patient using total PBMCs rather than sorted monocytes (Fig. 5A), which revealed a pattern similar to what is seen in Fig. 4B and C. Three additional cytokines, IL-8, MCP-1 and MIP1α were also assessed (Fig. 5A). Cytokine and chemokine levels in P1 are within normal limits or slightly high (IL-8, MIP1α), while lower limits or reduced levels of cytokines and chemokines were detected in P2, P3 and P4.
Fig. 4.
Cytokine secretion after monocyte stimulation
Representative example of two healthy controls and P1 is depicted. (A) Comparison of cytokine production by adherent (monocytes) and non-adherent (lymphocytes) fractions from PBMCs after stimulation with standard LPS (stimulates through TLR2 and TLR4) and ultrapure LPS (stimulates through TLR4). (B) After stimulation, P1 shows maximal IL-6 comparable to the healthy control [HC]. (C) TNF-α production was highest in P1. The average IL-6 and TNF-α levels in eight healthy controls after 24 h stimulation with 100 ng/mL of ultrapure LPS is 9410 (4476) pg/mL and 261 (149) pg/mL, respectively.
Fig. 5.
Functional assessment of cytokine production in RelA-deficient patients and clinical overview of phenotypes
(A) Cytokine production in PBMCs. Total PBMCs from each of the four patients, and six healthy controls were stimulated for 24h with LPS, and cytokines or chemokines were assessed: IL1β, IL-6, TNFα, IL-8, MCP-1 and MIP1α. The values are expressed as the mean (s.d.) from six healthy controls. (B) Clinical spectrum of disease in patients with RELA variants. Figure created with BioRender.com.
Discussion
In this paper, we report the functional effect of a novel RELA variant, a key component of the NFκB pathway (Fig. 1A), in a large kindred with clinical phenotype of Behçet’s-like syndrome (Figs 1B, C and 2A). RELA (p65) variants along with REL (c-Rel) and NFKB1 (p50) pathogenic variants have mainly been described in the context of humoral immune defects [6]; however, this study, and others discussed herein clearly demonstrate the breadth of the clinical phenotype, including inflammatory and autoimmune manifestations.
In mouse models, RelA signalling through the NFκB pathway is critical for normal embryonic development. In RelA-deficient mice, liver degeneration and lethality secondary to increased apoptosis and necroptosis occurs around day 14–15 of embryonic development [35–37]. RelA has also been implicated in immune homeostasis, with roles in regulation of lymphocyte development and proliferation, production of specific immunoglobulin isotypes, and development and maintenance of regulatory T-cell populations [35, 37–39].
Compared with mouse studies, significant immunophenotyping abnormalities or hypogammaglobulinemia have not been reported in human cases of RelA deficiency, suggesting that, as with other inborn errors of immunity, the mouse model may not be a faithful representation of human disease. In human studies, RelA haploinsufficiency has been implicated in diminished NFκB activation in response to TNF-α stimulation. This impaired activation disrupts the balance of survival and apoptotic signals and leads to increased apoptosis of epithelial and stromal cells, resulting in a clinical phenotype of mucocutaneous ulceration as reported in several studies [20–21].
In this study, we demonstrated functional impairment of the canonical pathway of NFκB due to a novel RELA variant. The four RelA-deficient patients showed minimal phosphorylation of p65 (Fig. 3A and B), suggesting reduced heterodimer formation between p50/p65 (NFκB1/RelA). RelA and p50 heterodimers have an activating function, while p50 homodimers, which are likely to form in the presence of mutant RelA, are repressive [40]. Except for one patient (P1), quantitative skewing of the T-cell and B-cell compartments is not observed (Table 1), and most other studies reporting RELA variants have not performed extensive immunophenotyping, except for Comrie et al. [24], who report immune dysregulation with altered lymphocyte subsets, including decreased naïve T cells, increased T effector memory cells expressing CD45RA (TEMRA), and IFNγ-skewed CD4+ T cells. P1 had decreased naïve T cells, increased memory T cells, and an expanded senescent T-cell population, similar to the Comrie et al. study [24]. Interestingly, though all four patients share the same RELA variant, there are stark differences in the monocyte cytokine production with P1 demonstrating substantially higher IL-6 and TNF-α levels post-stimulation compared with the other three patients (Figs 4 and 5A). Similarly, IL-8 and MIP1α were increased in only one patient (P1) compared with the other patients (Fig. 5A). Of note, P1 was not on consistent immunosuppressive therapy at the time of the blood draws for these studies. Despite differences in immunophenotype and monocyte cytokine production, all patients share a similar clinical phenotype, which supports the pathogenic loss-of-function (LOF) of p65 (RelA), as described in other patients with different RELA variants.
The clinical presentation in this three-generation family seems to have progressively earlier-onset and increased severity in the youngest generation. This may be related to the phenomenon of genetic anticipation, which is well known in genetics, and has been described in other inborn errors of immunity, e.g. GATA2 haploinsufficiency ([41]; R.S.A., personal observation). Phenotypic overlap has been observed between RelA haploinsufficiency and A20 (TNFAIP3) haploinsufficiency [42–44], suggesting that there may be a common mechanism of TNF-α induced apoptosis in these disorders, which can also share phenotypic overlap. Given this similarity, both RELA and A20 haploinsufficiency should be considered in the differential diagnosis of patients who present with childhood-onset SLE [25, 44].
The phenotypic differences observed in this familial cohort with the same novel RELA variant may not only be related to genetic anticipation, as discussed above, but also related to the variability in nonsense-mediated decay (NMD) that has been reported with frameshift, nonsense, splicing variants or other rearrangements in several studies [45, 46].
In the case of these four patients with suspected immune dysregulation, genetic testing proved to be critical for the identification of a monogenic basis of disease. Selecting which type of genetic testing to perform depends on the clinical phenotype, family history, representation of the gene in targeted panels, gene coverage on the next-generation sequencing methodology being used, and whether there is concern for intronic pathogenic variants or pseudogenes [47]. There is no one-size-fits-all approach for every patient and every monogenic disorder of immunity.
Previously reported pathogenic variants in RELA, as well as the novel variant described here, have been associated with a varied phenotypic spectrum, with haematologic, rheumatologic, gastrointestinal, immunologic, dermatologic, ophthalmologic and neurologic manifestations (Fig. 5B). Several of these studies report patients with chronic mucocutaneous ulcerations as the predominant phenotype resulting from the RELA variants [20–21], which is consistent with the Behçet’s syndrome-like phenotype seen in the four patients described in this study. The types of RELA variants reported have included splice-site [20], frameshift [21] and nonsense [23–24].
In addition to Behçet’s syndrome-like features, the patients described here with this novel pathogenic RELA variant display a clinical phenotype that has now been expanded to include IBD and additional ophthalmic manifestations, such as episcleritis. Early identification of patients with this phenotypic spectrum is likely to facilitate the genetic and immunological analysis required for accurate diagnosis and prompt immunosuppressive intervention.
Supplementary Material
Acknowledgements
The authors acknowledge Joanna Marshall and Amber Gibbs for assistance with the cytokine assays, and the Institute of Genomic Medicine at Nationwide Children's Hospital for financial support of the genomic analyses.
Genetic and genomic analysis: D.C.K., V.J., Kr.L., T.M.M., M.M., S.E.H., S.F. and R.K.W.
NFkB functional data acquisition and analysis: H.S.K., J.R.Y., J.D., S.D.R., R.S.A.
Manuscript writing: Ke.L., D.C.K., S.A., V.S., R.S.A.
All authors read and approved the final manuscript.
Funding: This work was supported by internal department funding.
Disclosure statement: R.S.A. has been a speaker for a Beckman Coulter webinar, is Co-Chair of the ClinGen Immunology CDWG, and served on an advisory board for Enzyvant Therapeutics. The other authors have declared no conflicts of interest.
Contributor Information
Kelsey Lecerf, Division of Allergy and Immunology, Department of Pediatrics, Nationwide Children’s Hospital; Division of Allergy and Immunology, Department of Otolaryngology, The Ohio State University Wexner Medical Center.
Daniel C Koboldt, The Steve and Cindy Rasmussen Institute for Genomic Medicine, Nationwide Children’s Hospital, Columbus, OH.
Hye Sun Kuehn, Immunology Service, Department of Laboratory Medicine, NIH Clinical Center, Bethesda, MD.
Vijayakumar Jayaraman, The Steve and Cindy Rasmussen Institute for Genomic Medicine, Nationwide Children’s Hospital, Columbus, OH.
Kristy Lee, The Steve and Cindy Rasmussen Institute for Genomic Medicine, Nationwide Children’s Hospital, Columbus, OH; Department of Pathology, The Ohio State University Wexner College of Medicine, Columbus, OH.
Theresa Mihalic Mosher, The Steve and Cindy Rasmussen Institute for Genomic Medicine, Nationwide Children’s Hospital, Columbus, OH; Ambry Genetics, Aliso Viejo, CA.
Jennifer R Yonkof, Department of Pediatrics, Toledo Children’s Hospital, Toledo.
Mari Mori, Division of Genetic and Genomic Medicine.
Scott E Hickey, Division of Genetic and Genomic Medicine.
Samuel Franklin, The Steve and Cindy Rasmussen Institute for Genomic Medicine, Nationwide Children’s Hospital, Columbus, OH.
Joanne Drew, Division of Pediatric Rheumatology, Department of Pediatrics.
Shoghik Akoghlanian, Division of Pediatric Rheumatology, Department of Pediatrics.
Vidya Sivaraman, Division of Pediatric Rheumatology, Department of Pediatrics.
Sergio D Rosenzweig, Immunology Service, Department of Laboratory Medicine, NIH Clinical Center, Bethesda, MD.
Richard K Wilson, The Steve and Cindy Rasmussen Institute for Genomic Medicine, Nationwide Children’s Hospital, Columbus, OH.
Roshini S Abraham, Department of Pathology, The Ohio State University Wexner College of Medicine, Columbus, OH; Department of Pathology and Laboratory Medicine, Nationwide Children’s Hospital, Columbus, OH, USA.
Data availability statement
Data from this study can be shared on request to the corresponding author, depending on information sought, in compliance with patient protection guidelines based on institutional policy.
Supplementary data
Supplementary data are available at Rheumatology online.
References
- 1. Sen R, Baltimore D.. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell 1986;46:705–16. [DOI] [PubMed] [Google Scholar]
- 2. Scott O, Roifman CM.. NF-κB pathway and the Goldilocks principle: lessons from human disorders of immunity and inflammation. J Allergy Clin Immunol 2019;143:1688–701. [DOI] [PubMed] [Google Scholar]
- 3. Andreas N, Potthast M, Geiselhöringer AL. et al. RelB deficiency in dendritic cells protects from autoimmune inflammation due to spontaneous accumulation of tissue T regulatory cells. J Immunol 2019;203:2602–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mondor I, Schmitt-Verhulst AM, Guerder S.. RelA regulates the survival of activated effector CD8 T cells. Cell Death Differ 2005;12:1398–406. [DOI] [PubMed] [Google Scholar]
- 5. Sharfe N, Merico D, Karanxha A. et al. The effects of RelB deficiency on lymphocyte development and function. J Autoimmun 2015;65:90–100. [DOI] [PubMed] [Google Scholar]
- 6. Zhang Q, Lenardo MJ, Baltimore D.. 30 Years of NF-κB: a blossoming of relevance to human pathobiology. Cell 2017;168:37–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barnabei L, Laplantine E, Mbongo W, Rieux-Laucat F, Weil R.. NF-kB: at the borders of autoimmunity and inflammation. Front. Immunol 2021;12:716469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beaussant-Cohen S, Jaber F, Massaad MJ. et al. Combined immunodeficiency in a patient with c-Rel deficiency. J Allergy Clin Immunol 2019;144:606–8.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brue T, Quentien MH, Khetchoumian K. et al. Mutations in NFKB2 and potential genetic heterogeneity in patients with DAVID syndrome, having variable endocrine and immune deficiencies. BMC Med Genet 2014;15:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bryant VL, Tangye SG.. The expanding spectrum of NFkB1 deficiency. J Clin Immunol 2016;36:531–2. [DOI] [PubMed] [Google Scholar]
- 11. Chen K, Coonrod EM, Kumánovics A. et al. Germline mutations in NFKB2 implicate the noncanonical NF-κB pathway in the pathogenesis of common variable immunodeficiency. Am J Hum Genet 2013;93:812–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Courtois G, Smahi A, Reichenbach J. et al. A hypermorphic IκBα mutation is associated with autosomal dominant anhidrotic ectodermal dysplasia and T cell immunodeficiency. J Clin Invest 2003;112:1108–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haynes A, Kamra P, Roifman C.. Heterozygous mutations in RelB can be associated with immune dysregulation and lymphoma. LymphoSign J 2016;3:55–60. [Google Scholar]
- 14. Klemann C, Camacho-Ordonez N, Yang L. et al. Clinical and immunological phenotype of patients with primary immunodeficiency due to damaging mutations in NFKB2. Front Immunol 2019;10:297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuehn HS, Niemela JE, Sreedhara K. et al. Novel nonsense gain-of-function NFKB2 mutations associated with a combined immunodeficiency phenotype. Blood 2017;130:1553–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Merico D, Sharfe N, Hu P, Herbrick J, Roifman CM.. RelB deficiency causes combined immunodeficiency. LymphoSign J 2015;2:147–55. [Google Scholar]
- 17. Lopez-Granados E, Keenan JE, Kinney MC. et al. A novel mutation in NFKBIA/IKBA results in a degradation-resistant N-truncated protein and is associated with ectodermal dysplasia with immunodeficiency. Hum Mutat 2008;29:861–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lorenzini T, Fliegauf M, Klammer N. et al. Characterization of the clinical and immunologic phenotype and management of 157 individuals with 56 distinct heterozygous NFKB1 mutations. J Allergy Clin Immunol 2020;146:901–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mandola AB, Sharfe N, Nagdi Z. et al. Combined immunodeficiency caused by a novel homozygous NFKB1 mutation. J Allergy Clin Immunol 2021;147:727–33.e2. [DOI] [PubMed] [Google Scholar]
- 20. Badran YR, Dedeoglu F, Leyva Castillo JM. et al. Human RELA haploinsufficiency results in autosomal-dominant chronic mucocutaneous ulceration. J Exp Med 2017;214:1937–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Adeeb F, Dorris ER, Morgan NE. et al. A novel RELA truncating mutation in familial Behçet's Disease-like mucocutaneous ulcerative condition. Arthritis Rheumatol 2021;73:490–7. [DOI] [PubMed] [Google Scholar]
- 22. Dorris E, Adeeb F, Lawless D. et al. A novel rela truncation in a 3-generation family with behcet’s disease alters the apoptotic response to inflammatory stimulants. 2019. Abstract #OP0154, Annual European Congress of Rheumatology (EULAR 2019), Madrid, Spain. [Google Scholar]
- 23. Faruqi AJ, Comrie WA, Lenardo M, Su H, Zhang Y.. RELA/p65 haploinsufficiency as a novel cause of primary immune disorder]. 2017. Abstract #59.17, American Association of Immunologists meeting, Washington DC. [Google Scholar]
- 24. Comrie WA, Faruqi AJ, Price S. et al. RELA haploinsufficiency in CD4 lymphoproliferative disease with autoimmune cytopenias. J Allergy Clin Immunol 2018;141:1507–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barnabei L, Lamrini H, Castela M. et al. Heterozygous RELA mutations cause early-onset systemic lupus erythematosus by hijacking the NF-κB pathway towards transcriptional activation of type-I Interferon genes. bioRxiv 2020;04.27.046102. doi: 10.1101/2020.04.27.046102, preprint: not peer-reviewed. [Google Scholar]
- 26. Kelly BJ, Fitch JR, Hu Y. et al. Churchill: an ultra-fast, deterministic, highly scalable and balanced parallelization strategy for the discovery of human genetic variation in clinical and population-scale genomics. Genome Biol 2015;16:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li H, Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Faust GG, Hall IM.. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics 2014;30:2503–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jun G, Wing MK, Abecasis GR, Kang HM.. An efficient and scalable analysis framework for variant extraction and refinement from population-scale DNA sequence data. Genome Res 2015;25:918–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koboldt DC, Kastury RD, Waldrop MA. et al. In-frame de novo mutation in BICD2 in two patients with muscular atrophy and arthrogryposis. Cold Spring Harb Mol Case Stud 2018;4:a003160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cingolani P, Platts A, Wang le L. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012;6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koboldt DC, Zhang Q, Larson DE. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miller CR, Lee K, Pfau RB. et al. Disease-associated mosaic variation in clinical exome sequencing: a two-year pediatric tertiary care experience. Cold Spring Harb Mol Case Stud 2020;6:a005231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sobreira N, Schiettecatte F, Valle D, Hamosh A.. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat 2015;36:928–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu C, Wu X, Zhang X. et al. Embryonic lethality and host immunity of RelA-deficient mice are mediated by both apoptosis and necroptosis. J Immunol 2018;200:271–85. [DOI] [PubMed] [Google Scholar]
- 36. Doi TS, Marino MW, Takahashi T. et al. Absence of tumor necrosis factor rescues RelA-deficient mice from embryonic lethality. Proc Natl Acad Sci USA 1999;96:2994–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Doi TS, Takahashi T, Taguchi O, Azuma T, Obata Y.. NF-kappa B RelA-deficient lymphocytes: normal development of T cells and B cells, impaired production of IgA and IgG1 and reduced proliferative responses. J Exp Med 1997;185:953–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mise-Omata S, Alles N, Fukazawa T. et al. NF-κB RELA-deficient bone marrow macrophages fail to support bone formation and to maintain the hematopoietic niche after lethal irradiation and stem cell transplantation. Int Immunol 2014;26:607–18. [DOI] [PubMed] [Google Scholar]
- 39. Ronin E, Lubrano di Ricco M, Vallion R. et al. The NF-κB RelA transcription factor is critical for regulatory T cell activation and stability . Front Immunol 2019;10:2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hoeger B, Serwas NK, Boztug K.. Human NF-κB1 haploinsufficiency and Epstein-Barr virus-induced disease-molecular mechanisms and consequences. Front Immunol 2018;8:1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Homan CC, Venugopal P, Arts P. et al. GATA2 deficiency syndrome: a decade of discovery. Hum Mutat 2021;42:1399–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou Q, Wang H, Schwartz DM. et al. Loss-of-function mutations in TNFAIP3 leading to A20 haploinsufficiency cause an early-onset autoinflammatory disease. Nat Genet 2016;48:67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maroof A, Patel DD.. TNF-α-induced protein 3 (A20): the immunological rheostat. J Allergy Clin Immunol 2018;142:401–2. [DOI] [PubMed] [Google Scholar]
- 44. Odqvist L, Jevnikar Z, Riise R. et al. Genetic variations in A20 DUB domain provide a genetic link to citrullination and neutrophil extracellular traps in systemic lupus erythematosus. Ann Rheum Dis 2019;78:1363–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nguyen LS, Wilkinson MF, Gecz J.. Nonsense-mediated mRNA decay: inter-individual variability and human disease. Neurosci Biobehav Rev 2014;46:175–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Resta N, Susca FC, Di Giacomo MC. et al. A homozygous frameshift mutation in the ESCO2 gene: evidence of intertissue and interindividual variation in Nmd efficiency. J Cell Physiol 2006;209:67–73. [DOI] [PubMed] [Google Scholar]
- 47. Lee K, Abraham RS.. Next-generation sequencing for inborn errors of immunity. Hum Immunol 2021;82:871–82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data from this study can be shared on request to the corresponding author, depending on information sought, in compliance with patient protection guidelines based on institutional policy.