Abstract
FBI-1 is a cellular POZ-domain–containing protein that binds to the HIV-1 LTR and associates with the HIV-1 transactivator protein Tat. Here we show that elevated levels of FBI-1 specifically stimulate Tat activity and that this effect is dependent on the same domain of FBI-1 that mediates Tat-FBI-1 association in vivo. FBI-1 also partially colocalizes with Tat and Tat's cellular cofactor, P-TEFb (Cdk9 and cyclin T1), at the splicing-factor–rich nuclear speckle domain. Further, a less-soluble population of FBI-1 distributes in a novel peripheral-speckle pattern of localization as well as in other nuclear regions. This distribution pattern is dependent on the FBI-1 DNA binding domain, on the presence of cellular DNA, and on active transcription. Taken together, these results suggest that FBI-1 is a cellular factor that preferentially associates with active chromatin and that can specifically stimulate Tat-activated HIV-1 transcription.
INTRODUCTION
The HIV-1 promoter can generate two types of RNA molecules (Kao et al., 1987; Ratnasabapathy et al., 1990; Sheldon et al., 1993). In the absence of the viral transactivator protein, Tat, the majority of the molecules are stable, short, nonpolyadenylated transcripts, and the minority are full-length, polyadenylated transcripts. The short transcripts correspond to RNAs with heterogeneous 3′ ends located around position +60 and contain the TAR element, the binding site for Tat. In the presence of Tat, the amount of short transcripts is decreased and the amount of full-length transcripts is dramatically increased (Tat reviewed in Karn, 1999). Tat binds to the TAR along with a cellular cofactor complex, TAK (Herrmann and Rice, 1993; Herrmann and Rice, 1995). TAK is one form of P-TEFb that contains Cdk9 and Cyclin T1 (Figure 1). P-TEFb is an elongation factor of RNA polymerase II (pol II) that phosphorylates the pol II carboxy terminal domain (CTD; reviewed in Price, 2000). CTD phosphorylation leads to the dissociation of negative elongation factors like NTEF (Price, 2000) or NELF (Yamaguchi et al., 1999) and the stimulation of pol II elongation competence. Thus, Tat-TAK associates with the TAR, which leads to the phosphorylation of the CTD, the dissociation of negative elongation factors, and the increased elongation competence of the HIV-1 transcription complexes.
Figure 1.
(A) Schematic diagram of the insert for the reporter constructs pHIV-1/TAR/Luc and pHIV-1/SLIIB/Luc containing HIV-1 sequences from −157 to +82, a region that includes 1) the HIV-1 promoter with the enhancer (ENH), the three Sp1 binding sites, and the TATA box and 2) R sequences from positions +1 to +82 with IST sites and the TAR. The SLIIB insert (B) is described in Tiley et al. (1992). (B) Schematic diagram showing the sequence of the TAR and SLIIB inserts. Tat binds to TAR along with its cellular cofactors CDK9 and Cycling T. Replacement of TAR with SLIIB allows the targeting of Rev fusion proteins such as Rev-VP16 to the R region.
In the absence of Tat, efficient synthesis of the short transcripts is dependent on a bipartite DNA element, known as the inducer of short transcripts (IST, Figure 1), located just downstream of the start site of transcription (Ratnasabapathy et al., 1990; Sheldon et al., 1993; Pendergrast et al., 1996; Pendergrast and Hernandez, 1997). IST activates transcription from the HIV-1 promoter, but the resulting RNAs are short. Thus, IST seems to stimulate the formation of transcription complexes that are not capable of efficient elongation.
A cellular factor that binds in vitro to the IST element, but not to IST mutants, has been purified and cloned (Pessler et al., 1997; Morrison et al., 1999). This factor, called FBI-1 (factor that binds to IST), is a Kruppel-type zinc finger protein that contains at its N-terminus a POZ (for poxvirus and zinc fingers) domain (Bardwell and Treisman, 1994; Albagli et al., 1995). The POZ domain is a 120-amino acid conserved motif that is often located at the N-terminus of Kruppel-type zinc finger transcription factors. FBI-1 was independently cloned as part of a screen for proteins containing POZ domains (LRF, Davies et al., 1999) and as a protein that is strongly expressed in osteoclasts (OCZF, Kukita et al., 1999). Some POZ proteins such as PLZF, BCL6, and GAF are transcription factors associated with chromatin remodeling capabilities (Dhordain et al., 1997; Hong et al., 1997; Wilkins and Lis, 1997; David et al., 1998; Huynh and Bardwell, 1998; Huynh et al., 2000). POZ proteins form homomers and heteromers via the POZ domain and often display punctate nuclear localization patterns (Dhordain et al., 1995). Indeed, FBI-1 has been shown to self-associate via both its POZ domain (Davies et al., 1999; Morrison et al., 1999) and its zinc finger (ZF) domain (Morrison et al., 1999) and to heteromultimerize with BCL-6 via POZ and ZF domains (Davies et al., 1999). In addition, overexpressed FBI-1 shows a punctate nuclear localization pattern (Davies et al., 1999), although the localization of endogenous FBI-1 has not been reported.
FBI-1 binds to IST and associates with HIV-1 Tat in coimmunoprecipitation assays, linking it to the HIV-1 promoter (Morrison et al., 1999). However FBI-1 has not been functionally linked to either short transcript production or Tat activation. Also its cellular function has not been elucidated. Here we show that elevated levels of FBI-1 significantly and specifically stimulate Tat activity and that this effect is dependent on the same domain of FBI-1 that mediates Tat-FBI-1 association. Also endogenous FBI-1 partially colocalizes with both Tat and endogenous P-TEFb (Cdk9 and Cyclin T1) within the splicing-factor–rich nuclear speckle domain, and this colocalization is dependent on the same domain necessary for Tat association and stimulation. In addition, a subpopulation of FBI-1 displays a novel peripheral-speckle pattern of localization that probably corresponds to regions of active chromatin. These results suggest a functional link between FBI-1 and Tat-activated HIV-1 transcription and that FBI-1 is preferentially associated with active chromatin in mammalian cells.
MATERIALS AND METHODS
Plasmid Constructs
All of the T7 and HA-tagged constructs expressing FBI-1 and Tat have been described previously (Morrison et al., 1999). pHIV-1/Luc was constructed by amplifying HIV-1 promoter sequences from −159 to +80 and ligating this fragment into pGL3-Basic vector (Promega, Madison, WI) cut with Asp718 and HindIII. Likewise, pHIV-1/SLIIB/Luc was constructed by amplifying HIV-1 promoter sequences from −159 to +80 from pHIV-1/R/SLIIB (Pendergrast and Hernandez, 1997) and ligating this fragment into pGL3-Basic vector cut with Asp718 and HindIII. pRL-βgl-enh+ was constructed by inserting by ligation the PCR-derived β-globin promoter sequences from H1/βgl (Ratnasabapathy et al., 1990) into the BglII-SalI–digested pRL-null vector (Promega). The resulting constructs were sequenced to ensure the fidelity of the PCR. For FBI-1-EGFP–expressing construct, EGFP coding sequences were amplified by PCR from a PBS-derived EGFP vector (a kind gift of M. Greenberg) with primers that place BglII sites at both ends of the gene and inserted by ligation into pCGN-FBI-1 digested with BglII. The resulting construct was sequenced to ensure that no point mutations were introduced during PCR and expression was checked by immunoblot (our unpublished results). FBI-1-EGFP was tested in a luciferase assay for activity and found to be able to stimulate Tat activity as well as wild-type FBI-1 (our unpublished results). pcRev, pcRev-VP16, and pcTat-Rev were kind gifts from B. Cullen.
Antibodies
The cyclin T1 and Cdk9 rabbit polyclonal antibodies used were the kind gift of D. Price (University of Iowa). They were both used at a dilution of 1:100 but similar results were seen for both at dilutions of 1:50 and 1:250 (our unpublished results). We also saw similar results with a rabbit polyclonal from Santa Cruz Biotechnology (Santa Cruz, CA) for Cdk9 (dilution 1:100) and a goat polyclonal from Santa Cruz for cyclin T1 (dilution 1:50; our unpublished results). Speckles were detected with human polyclonal ANA-S antibody (Sigma, St. Louis, MO) at a 1:100 dilution and with a mouse mAb specific for splicing factor SC35 (Sigma and also a kind gift from D. Spector), used at a 1:1000 dilution. HA-tagged proteins were detected with the mouse mAb 12CA5 (Niman et al., 1983) and T7-tagged proteins were detected with T7-Tag Antibody (Novagen, Madison, WI), both used at a dilution of 1:1000, but similar results were seen with 12CA5 as high as 1:3000 and with T7-Tag as high as 1:5000 (our unpublished results). HA-Tat was also visualized with the mAb 1975 procured from the NIH AIDS Research and Reference Reagent Program and showed aberrant nucleolar staining at multiple dilutions ranging from 1:50 to 1:1000 (our unpublished results). FBI-1 was detected with similar results using rabbit polyclonal antibodies 413 and 415, raised from two separate FBI-1 peptides (Morrison et al., 1999) at dilutions ranging from 1:100 to 1:1000 with 1:500 depicted for both. Primary antibodies were detected using the appropriate secondary antibodies conjugated to FITC or Texas Red (Jackson ImmunoChemicals, West Grove, PA; dilution 1:300).
Immunolabeling
For immunolabeling, either untransfected cells or cells transfected as described (Pendergrast and Hernandez, 1997) were grown on coverslips and fixed in 2% freshly prepared formaldehyde in PBS for 10 min at room temperature (RT). Cells were permeabilized in PBS + 0.4% Triton X-100 for 10 min at RT and then washed three times for 10 min each in PBS. For the experiments depicted in Figure 4 the same results were seen when the fixing and permeabilization were performed simultaneously with 2% formaldehyde + 0.4% Triton X-100 for 10 min at RT or when 1% normal goat serum (Jackson ImmunoChemicals) was included in all of the washes (our unpublished results). To detect less-soluble protein, cells were washed once briefly with ice-cold PBS and then extracted with CSK buffer (150 mM NaCl, 300 mM sucrose, 10 mM PIPES, pH 6.8, 3 mM MgCl2, 1 mM PMSF) + 0.4% Triton X-100 for 3 min on ice and then fixed with 2% formaldehyde in PBS for 10 min at RT. After washing, the coverslips were incubated with primary antibody in PBS for 1 h at RT. Cells were then rinsed in PBS three times for 10 min at RT and then incubated with secondary antibody for 45 min at RT. Cells were then washed three times in PBS for 10 min at RT, with the final wash containing 1 μg/ml 4,6-diamidino-2-phenylindole (DAPI) where appropriate. The coverslip was then mounted onto glass sides with Vectashield (Vector Laboratories, Burlingame, CA) mounting media or with 90% glycerol in PBS with 1 mg/ml paraphenylenediamine as an antifade agent, adjusted to pH 8.0 with 0.2 M bicarbonate buffer. Cells were examined either with a Zeiss LSM 510 confocal microscope equipped with an argon-krypton laser or with a Zeiss (Thornwood, NY) Axiovert microscope equipped with epifluorescence and differential interference contrast optics. A SenSys cooled charge-coupled device camera (Photometrics, Tuscon AZ) captured images with the use of Metamorph Image software (Universal Imaging Corp., West Chester, PA). Multiple images containing several cells were collected and representative cells are shown. All images were digitally processed for presentation using Adobe PhotoShop (Adobe Systems, San Jose, CA).
Figure 4.
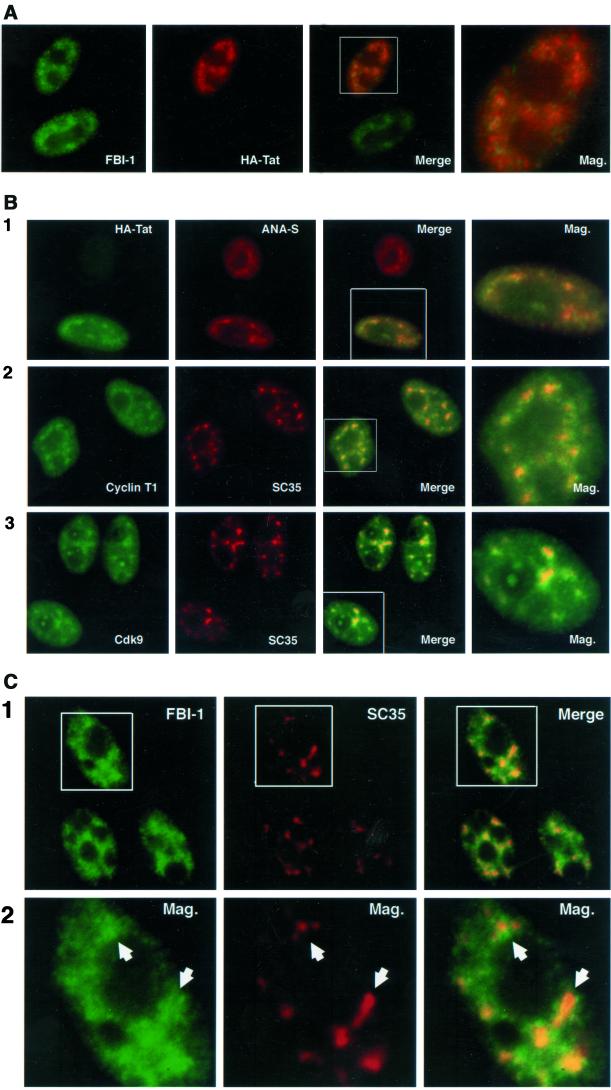
FBI-1, HIV-1 Tat, and TAK/P-TEFb localization. (A) Epifluorescent image of HeLa cells transfected with HA-Tat expressing plasmid. FBI-1 (panel 1, green) and HA-Tat (panel 2, red) colocalize (panels 3 and 4, yellow) at numerous foci within the nonnucleolar nucleoplasm. Endogenous FBI-1 was detected with a rabbit polyclonal (413) primary antibody and fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit secondary antibody (FITC-GAR). HA-Tat was detected with a mouse monoclonal (12CA5) primary antibody and a Texas Red-conjugated Goat anti-mouse secondary antibody (TR-GAM). (B) Epifluorescent image of HeLa cells transfected with 2 μg of HA-Tat expressing plasmid (panel 1) or mock transfected (2 μg of expression vector) HeLa cells (panels 2 and 3). HA-Tat (panel 1, column 1, green), endogenous cyclin T1 (panel 2, column 1, green) and Cdk9 (panel 3, column 1, green) and nuclear speckles (panels 1–3, column 2, red) colocalize (panels 1–3, column 3 and 4, yellow) at numerous irregularly shaped foci within the nonnucleolar nucleoplasm. HA-Tat was detected with 12CA5 primary antibody and an FITC-conjugated goat anti-mouse secondary (FITC-GAM). Cyclin T1 was detected with a rabbit polyclonal (anticyclin T1) antibody and FITC-GAR secondary. Cdk9 was detected with a rabbit polyclonal (anti-Cdk9) antibody and FITC-GAR secondary. Nuclear speckles were detected with a human polyclonal (ANA-S; panel 1) primary antibody or a mouse monoclonal (SC35) primary antibody and with Texas Red–conjugated goat anti-human antibody (TR-GAH) and TR-GAM, respectively. (C) Epifluorescent image of HeLa cells. Endogenous FBI-1 (green) displays a heterogeneous nuclear localization pattern characterized by numerous signal foci that surround and overlap with the nuclear speckle domain (red). Note that often there is a slight decrease in signal within a FBI-1 foci that matches the shape of the corresponding speckle (panel 2, arrowheads). FBI-1 was detected with 413 primary and FITC-GAR secondary antibodies, and nuclear speckles were detected with SC35 primary and TR-GAM secondary antibodies.
Nuclease Digestion
Cells were CSK + 0.4% Triton X-100 extracted for 3 min on ice, rinsed gently in PBS (pH 7.4), and incubated in either DNase I (100 μg/ml in PBS + 5 mM MgCl2, RNase free) + 0.5 U/μl RNasin, RNase A (100 μg/ml in PBS, DNase free), or PBS + 5 mM MgCl2 only for 30 min at 37°C. Similar results were seen for all three treatments with a 30-min RT incubation. After three washes in PBS, cells were fixed and prepared for immunolabeling as above.
In Vivo Immunoprecipitations
Three micrograms of pCGN or pCGT plasmids, either empty or expressing FBI-1 or Tat as indicated, were transfected into HeLa cells by electroporation as described (Pendergrast and Hernandez, 1997). Extract preparation, immunoprecipitations and immunoblotting were performed as described (Pendergrast and Hernandez, 1997). In all cases control immunoblots were performed to ensure equal expression and immunoprecipitation of mutant proteins. IPs and washes were performed in the presence of 75 μg/ml ethidium bromide to inhibit protein–DNA interaction.
Luciferase Assays
Two micrograms of the Firefly Luciferase reporter plasmid pHIV-1/Luc, 500 ng of the internal control reporter plasmid pRL-βgl-enh+, and either a control expression vector or a vector expressing the indicated protein were transfected into HeLa cells by electroporation as previously reported (Pendergrast and Hernandez, 1997). Similar results were also seen using 800 ng of the pRL-TK (Promega) control promoter. Twenty-four hours posttransfection the cells were harvested, and extract was prepared using Promega Passive Lysis Buffer and the Promega Dual Luciferase kit protocols. The extract was analyzed with the Bioscan Mini-Lum Luminometer (Washington, DC). Each experiment was performed three times with separate transfections, and the results shown are the average of those experiments.
RESULTS
FBI-1 Can Stimulate Tat Activity
FBI-1 binds to the HIV-1 promoter and interacts with HIV-1 Tat in coimmunoprecipitation assays (Morrison et al., 1999), suggesting that it may have some effect on Tat transactivation. To explore this possibility, we tested the effect of transient overexpression of FBI-1 on HIV-1 transcription in vivo, using a Dual Luciferase Reporter System (Promega). We cotransfected into HeLa cells: a construct containing the HIV-1 promoter upstream of a Firefly Luciferase reporter gene (Figure 1), a construct containing the β-globin promoter upstream of the Renilla Luciferase reporter gene as an internal control, and plasmids expressing HA-FBI-1 and/or HA-Tat. Twenty-four hours posttransfection we harvested the cells and assayed for Luciferase activity as a measure of transcriptional activity. Figure 2A shows the HIV-1 Firefly Luciferase activity normalized to the β-globin Renilla Luciferase activity. Cotransfection of the HA-FBI-1 expression construct lead to a strong increase in Tat activity (Figure 2A, >5-fold), whereas basal HIV-1 promoter activity was only weakly affected (2-fold). In contrast, overexpression of the transactivator Oct 1 had little effect on either basal or Tat-activated transcription. Another POZ protein, HA-BCL6, slightly repressed transcription as has been reported (Baron et al., 1997). Immunoblot analysis confirmed that Tat expression levels were unaffected by FBI-1 overexpression and that FBI-1, BCL6, and Oct 1 expression levels were comparable (our unpublished results). Similar results were obtained with HeLa cells containing an integrated HIV-1-Luciferase construct and in transient transfection experiments performed in Jurkat and 293 cells (our unpublished results). In another set of experiments, using lower amounts of plasmids expressing truncated FBI-1 mutants, we determined that both the POZ and especially the Zinc Finger (ZF) domain of FBI-1 (Figure 2B) were required for the full effect on transcription (Figure 2C). Again, immunoblot analysis confirmed that the expression plasmids for full-length FBI-1 (FL), and FBI-1 with either the POZ domain (ΔPOZ) or the ZF domain (ΔZF) deleted yielded similar amounts of protein (our unpublished results). The observation that FBI-1 more strongly activates Tat-activated transcription than basal transcription suggests that the effect is Tat specific. To determine if this is so for activated transcription, we replaced the TAR sequences in pHIV-1/Luc with a 13-base pair sequence derived from the Rev response element that constitutes an efficient RNA target for Rev binding in vivo, exactly as described by Tiley et al. (1992). The resulting construct is shown in Figure 1. We then used this construct to determine if FBI-1 could enhance the activation of Rev-VP16, a fusion protein known to activate HIV-1 transcription equipped with such a modified TAR (Tiley et al., 1992; Madore and Cullen, 1995). As expected, neither Rev nor Tat significantly stimulated transcription from this construct (Figure 2D). However expression of the Tat-Rev fusion protein significantly stimulated transcription (13-fold), and this activated transcription was strongly enhanced by coexpression of FBI-1 (>5-fold). Rev-VP16 could also activate transcription albeit at a lower level (4.8-fold) than Tat-Rev. Interestingly, FBI-1 could not significantly enhance activation by Rev-VP16. In other experiments, FBI-1 did not significantly affect activation by Rev-Rel or Rev-Sp1 (our unpublished results). These data show that increasing the amount of FBI-1 in the cell specifically enhances Tat activation of the HIV-1 promoter, suggesting that FBI-1 modulates Tat-activated HIV-1 transcription.
Figure 2.
Expressed FBI-1 FL synergizes with HIV-1 Tat at the HIV-1 LTR. (A, C, and D) HeLa cells were transiently transfected with 2 μg of the reporter plasmids HIV-1/TAR/Luc (A and C) or HIV-1/SLIIB/Luc (D), along with empty expression vector (No FBI-1) or the indicated HA-tagged expression plasmids at 1× and 3× relative amounts. The amounts of expression plasmids (100–900 ng) were calibrated to express equal amounts of proteins as judged by anti-HA immunoblot (our unpublished results). Amounts transfected were as follows: For (A) 100 ng of pcgn Tat, 200 ng (1) and 600 ng (3) pcgn FBI-1 FL, 100 ng (1) and 300 ng (3) pcgn Oct 1, 300 ng (1) and 900 ng pcgn (3) BCL6; for (C) 100 ng Tat, 200 ng (1) and 600 ng (3) pcgn FBI-1 FL and ΔZF and 150 ng (1) and 450 ng (3) pcgn ΔPOZ; for (D) 500 ng FBI-1 FL, 500 ng pcRev, 100 ng pcgn Tat, 500 ng pcTat-Rev, and 800 ng pc Rev-VP16. 500 ng of β-globin-Renilla expression vector was cotransfected as an internal control for transfection. Depicted activation levels are the results of three independent experiments, are normalized to β-globin expression, and are relative to No FBI-1, − Tat levels arbitrarily set to 1. Error bars represent SD calculated by Microsoft Excel. (B) FBI-1 and FBI-1 mutant constructs. FL FBI-1 corresponds to FBI-1 amino acids 1–584; ΔPOZ, amino acids 122–584; ZF, amino acids 377–584; ZF1, FL FBI-1 with the mutations C384A + C387A; ZF2, FL FBI-1 with the mutations C412A + C415A; ZF3, FL FBI-1 with the mutations C440A + C443A; ZF4, FL FBI-1 with the mutations C468A + C471A; ΔZF, amino acids 1–382 and 485–584.
FBI-1 Associates with Wild-type but not Activation-deficient Tat via Its ZF Domain in Vivo
In Luciferase assays, deletion of the POZ domain reduces FBI-1's stimulatory effect on Tat transactivation, and deletion of the ZF region nearly abolishes the effect. Because FBI-1 has been shown to associate in vivo with Tat, we decided to determine the region(s) of FBI-1 that are necessary for this association. To do this, we transiently expressed HA-tagged HIV-1 Tat alone or together with T7-tagged full-length or truncated derivatives of FBI-1 (Figure 2B) in HeLa cells. Protein extracts from these lysed cells were used for nondenaturing immunoprecipitations (IPs) with anti-HA antibodies, and the immunoprecipitates were checked for the presence of the T7-tagged derivatives of FBI-1 by SDS-PAGE fractionation and immunoblotting with anti-T7 antibody. As shown in Figure 3A, in the absence of coexpressed HA-Tat, anti-HA antibody immunoprecipitates did not contain T7-tagged full-length FBI-1 (FL) nor the ΔPOZ, C-terminal ZF-containing-region (ZF, Figure 2B) and ΔZF truncations, demonstrating that the anti-HA antibody does not cross-react with any of the T7 tagged FBI-1 proteins (lanes 2–5). However, when HA-Tat was coexpressed with the T7-FBI-1 derivatives, the anti-HA immunoprecipitates contained FL, ΔPOZ, and ZF FBI-1, but not ΔZF FBI-1 (lanes 6–9). An additional immunoblot confirmed that all the T7-FBI-1 derivatives were expressed at similar levels (Figure 3B). Thus, the ZF region of FBI-1 is necessary and sufficient for in vivo association with Tat.
Figure 3.
The ZF domain of FBI-1 is necessary and sufficient for Tat association in vivo. (A) Constructs expressing T7FL, T7ΔPOZ, T7ZF, or T7ΔZF and HA-Tat were expressed alone or in combination as indicated above the lanes into HeLa cells. Thirty-six hours posttransfection, extracts from transfected cells were prepared and incubated with protein A-agarose beads cross-linked to anti-HA 12CA5 monoclonal antibodies (lanes 2–9). The immunoprecipitated proteins were fractionated on a 12.5% SDS-polyacrylamide gel, transferred to nitrocellulose and blotted with anti-T7 antibody. Lane 1: one tenth the starting material for the T7FL + HA-Tat immunoprecipitation (IP; lane 6). Lanes 2–9: the IPs. The position of T7FL, T7-ΔPOZ, and T7-ZF are indicated. For lanes 6–9, an anti-HA immunoblot of a separate gel demonstrated that the HA-tagged proteins were expressed and immunoprecipitated (our unpublished results). For lanes 6–9, an anti-T7 immunoblot of a separate gel loaded with one tenth the starting material for the IP demonstrated that the mutant proteins were expressed to a similar level (B). (C) Constructs expressing T7ZF and wild-type HA-Tat, HA-18IS Tat, or HA-C30, 31A Tat were transfected alone or in combination as indicated above the lanes into HeLa cells. The experiment was performed as in (A). Lanes 1–4 show one tenth the starting materials; lanes 5–8 show the IPs.
Full-length FBI-1 does not associate efficiently with Tat mutants defective in transcriptional activation (Morrison et al., 1999). To determine if this holds true for the ZF fragment, HeLa cells were transfected with a wild-type T7-tagged ZF construct, either alone or together with wild-type or mutant HA-Tat expression constructs. The resulting lysates were then used for nondenaturing IPs with anti-HA antibody. As above, starting material and immunoprecipitated proteins were fractionated by SDS-PAGE and immunoblotted with the anti-T7 antibodies. As shown in Figure 3C, equal amounts of T7-ZF were expressed in all four transfections (lanes 1–4). A small amount of T7-ZF was detected in the anti-HA immunoprecipitate from cells transfected with T7-ZF alone, probably because of a small amount of protein sticking nonspecifically to the beads (lane 5). The coexpression of HA-Tat with T7-ZF lead to a strong increase in the amount of coimmunoprecipitated T7-ZF (lane 6). In contrast, the coexpression of T7-ZF and the transactivation-defective mutant HA-Tat derivatives HA-Tat18IS (Rice and Carlotti, 1990a, 1990b; Morrison et al., 1999) and HA-TatC30, 31A (Rice and Carlotti, 1990a; Morrison et al., 1999) did not increase immunoprecipitated ZF levels over background (lanes 7 and 8), consistent with the results previously seen with FL FBI-1 (Morrison et al., 1999). The observation that the same point mutations in Tat that prevent the interaction with FL FBI-1 also prevent interaction with FBI-1 ZF suggests that the interaction is specific and does not reflect a general stickiness of the proteins involved. In addition, because the IP's were performed in the presence of ethidium bromide, the FBI-1-Tat interaction was probably not mediated by contaminating DNA. We conclude that FBI-1 associates with Tat via its ZF domain in vivo, the very same domain required for maximal enhancement of Tat activity.
FBI-1 Partially Colocalizes with Tat and Tat's Cofactor, P-TEFb (Cyclin T1 and Cdk9), in the Splicing-factor–rich Nuclear Speckles
We have shown that FBI-1 associates in vivo with Tat and that elevated levels of FBI-1 stimulate Tat's activity. To further analyze the association of FBI-1 and Tat in the context of intact cell nuclei, the subcellular localization of FBI-1 and Tat were compared. HeLa cells were transfected with a construct that expresses HA-Tat, and the localization of endogenous FBI-1 and HA-Tat was examined using double-labeling immunofluorescence with anti-FBI-1 and anti-HA antibodies. The results show that the endogenous FBI-1 was localized predominantly to the nucleus with a minimal labeling in the nucleolus and the cytoplasm (Figure 4A). The nuclear distribution is heterogeneous with numerous signal foci of varying sizes and shapes in addition to more diffuse nucleoplasmic staining. This pattern was observed in the majority of cells, with two antibodies raised against different FBI-1 peptides (Morrison et al., 1999), and with different fixation methods and antibody dilutions (our unpublished results, see MATERIALS AND METHODS). HA-Tat immunolabeled with anti-HA antibody also demonstrated a speckled distribution in addition to areas of diffuse staining throughout the nucleoplasm. There was little detectable labeling in the nucleolus in the majority of cells. In addition to the anti-HA tag labeling, this pattern of Tat staining was also observed in cells expressing T7-tagged Tat (our unpublished results), suggesting that this pattern of Tat localization is not influenced by the epitope tags. A close examination of a merged image of the Tat and FBI-1 (Figure 4A, Mag.) showed that Tat and FBI-1 were concentrated in overlapping areas, predominantly at their nucleoplasmic speckles, although there was a subset of FBI-1 (green) and Tat (red) that did not overlap. Thus, endogenous FBI-1 and expressed Tat partially colocalize, consistent with a biologically relevant association.
The heterogeneous nuclear localization pattern of both proteins is reminiscent of the “nuclear speckles” domain, a subnuclear domain where many splicing factors and some transcription factors are enriched over other nuclear regions (Spector, 1993; see DISCUSSION and reviewed in Huang and Spector, 1996a). To determine whether Tat and its cellular cofactor P-TEFb (cyclin T1 and Cdk9) are part of the nuclear speckle domain, HA-Tat–transfected cells were double-labeled with anti–HA-Tat, anticyclin T1, or anti-Cdk9 and an antibody that defines nuclear speckles. To avoid secondary antibody cross-reaction, two speckle-labeling antibodies were used. The human polyclonal ANA-S (Sigma) was used in combination with the mouse monoclonal anti-HA antibody, whereas the mouse monoclonal SC35 antibody was used in combination with the rabbit polyclonal antibodies specific for cyclin T1 or Cdk9. HA-Tat predominantly colocalized with nuclear speckles as defined by the ANA-S antibody (Figure 4B, panel 1) in foci of similar shapes and sizes (panel 1, Mag.). Endogenous cyclin T1 and Cdk9 also showed predominantly nuclear localization with numerous nucleoplasmic foci (panels 2 and 3). The distribution patterns for both proteins were similar, although not identical, to the nuclear speckles distribution pattern observed when double-labeled with SC35 (panels 2 and 3, Mag.). Similar patterns for both proteins were also observed using other antibodies specific for each of these proteins (our unpublished results, see MATERIALS AND METHODS). These findings demonstrate that HA-Tat, endogenous cyclin T1, and endogenous Cdk9 are concentrated at the splicing-factor–rich nuclear speckles, in addition to being distributed at other nuclear regions.
Because Tat and FBI-1 only partially colocalized, whereas Tat largely colocalized with the splicing factor–enriched speckles, we were interested in examining the spatial relationship between FBI-1 and SC35. A double-labeling experiment with antibodies directed against endogenous FBI-1 and SC35 was performed, and the results are shown in Figure 4C. FBI-1 foci were less intense at their center than the foci of Tat, Cdk9, and cyclin T1 (compare Figure 4C, panel 1, with 4B). However, the FBI-1 signal was clearly more intense in the vicinity of the stronger SC35 foci. Interestingly, the less intensely labeled centers of FBI-1 foci often coincided with the most intensely labeled areas at the centers of the SC35 speckles (panel 2, arrowheads). Thus, contrary to Tat, cyclin T1, Cdk9, and other speckle associated proteins the FBI-1 signal was often enriched at the periphery of individual speckle foci but was only sometimes enriched at the center of the speckle.
A Less-soluble Population of FBI-1 Localizes to the Nuclear Speckle Periphery
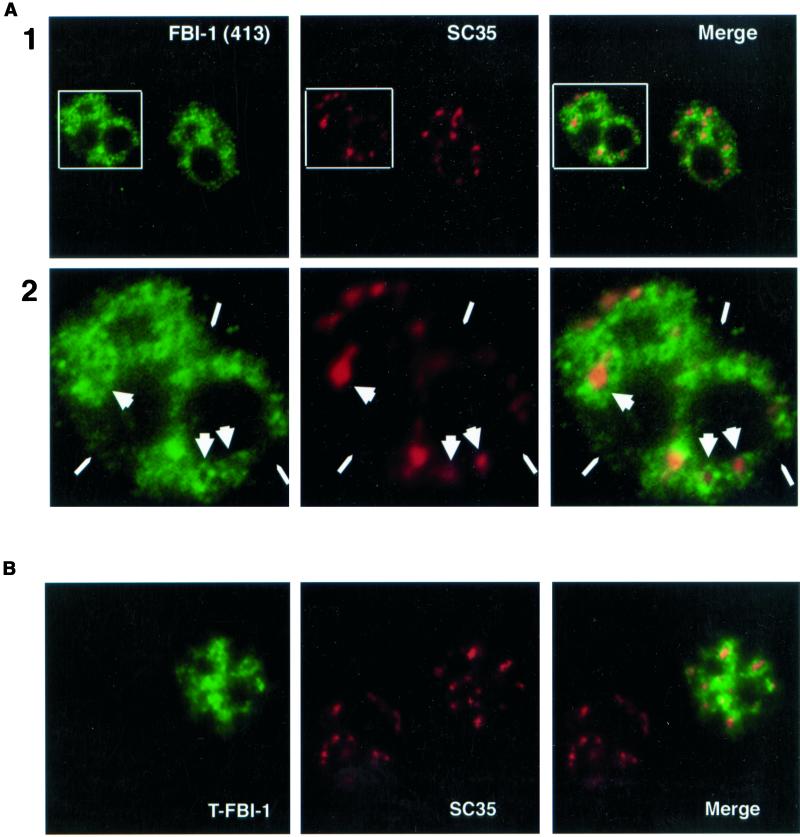
To further investigate the nature of FBI-1 distribution, we extracted HeLa cells with CSK buffer containing 0.4% Triton X-100 and 150 mM NaCl before fixation and immunostaining (see MATERIAL AND METHODS; Fu and Maniatis, 1990; Spector et al., 1991; Huang and Spector, 1996b). This gentle extraction before fixation removes the soluble fraction of nuclear components while leaving less-soluble proteins intact. The results are shown in Figure 5. As reported previously, the localization pattern obtained with the SC35 antibody was not significantly altered by the procedure, although the speckles were less extended and less connected (compare Figure 5A, column 2, with 3B, column 2). The CSK treatment more significantly altered the FBI-1 localization pattern (column 1). The diffuse nucleoplasmic signal and the labeling at the periphery of the nucleus were, for the most part, severely reduced (narrow arrowheads). However the signal in the vicinity of and at the periphery of the SC35 speckles was less affected; thus, these areas were more distinct than in nonextracted cells (compare Figure 5A, column 1, with Figure 4C, column 1). Interestingly, although extraction had little affect on the FBI-1 signal at the periphery of the speckles, the process often strongly reduced the FBI-1 signal at the center of the speckles (wide arrowheads). Thus, after extraction, FBI-1 displayed staining in the interior of the nucleus with more intense signal in the vicinity and at the periphery of SC35 speckles and less-intense signal directly overlapping the center of the speckles. This intense labeling at the periphery but not at the center of the larger SC35 speckles was similar to what was observed in untreated cells but more distinct. We call FBI-1's distribution pattern “peripheral-speckle-localization.” This pattern was different from what was observed for Tat, cyclin T1, and Cdk9, all of which showed colocalization with SC35 in either extracted cells (our unpublished results) or untreated cells (Figure 4B). To our knowledge, the peripheral-speckle-localization pattern is unique among the known speckle-associated proteins.
Figure 5.
Less-soluble FBI-1 displays peripheral-speckle localization. Epifluorescent image of prefixation CSK extracted HeLa cells. Less-soluble endogenous FBI-1 (green) shows strong signal surrounding and connecting the nuclear speckles (red) but shows a much weaker signal at the center of many speckles (large arrowheads) and from regions of the nucleus that have no speckles (narrow arrowheads). FBI-1 was detected with 413 primary antibody or with another rabbit polyclonal antibody specific for FBI-1 (415, our unpublished results) and FITC-GAR secondary antibody. Nuclear speckles were detected with SC35 primary and TR-GAM secondary antibodies. (B) Confocal image of prefixation CSK extracted Hela cells transfected with 2 μg of T7-FBI-1 (FL). T7-FBI-1 (green) signal is enhanced at the periphery of the SC35 nuclear speckles (red). T7-FBI-1 was detected with a mouse monoclonal primary antibody (T7 epitope antibody, Novagen) and FITC-GAM secondary antibody. Nuclear speckles were detected as in A.
To further analyze and resolve the Z axis relationship between the localization of FBI-1 and SC35, confocal microscopy was used to evaluate the labeling pattern of the two proteins. To achieve an observable signal for FBI-1 for the confocal studies, we transiently expressed FBI-1 tagged with the T7 epitope tag (MATERIALS AND METHODS). HeLa cells were transfected with the expression construct, and cells were extracted with CSK buffer before fixation so that the peripheral-speckle localization could be better visualized. The localization of tagged FBI-1 and SC35 were compared using immunolabeling (Figure 5B). Less soluble T7-FBI-1 was not as limited to certain regions of the nucleus as was endogenous FBI-1, probably because of overexpression; however, its signal was stronger in the area at the periphery of the speckles. Indeed, in some cases the expressed protein actually formed bright rings against the more diffuse background signal around the centers of some of the larger SC35 foci. This pattern actually makes it possible to predict, with reasonable accuracy, the position and shape of the larger and more defined speckles based on the T7-FBI-1 signal alone. A similar pattern was observed with FBI-1 tagged with EGFP (our unpublished results). Together, these results indicate that there are at least two populations of FBI-1 in the nucleus, a more soluble population that shows diffuse nucleoplasmic distribution and partial colocalization with nuclear speckles, and a less-soluble population that shows a peripheral-speckle-pattern of localization.
FBI-1 Is Targeted to the Peripheral-Speckled Pattern by Its Binding to DNA
The peripheral-speckle localization of FBI-1 may be relevant to past characterization of both speckles and FBI-1, because previous studies suggest that the regions at the periphery of speckles are enriched in actively transcribing genes (see DISCUSSION). Considering the novel nature of less-soluble FBI-1's peripheral-speckle localization, we were interested in determining the mechanism by which FBI-1 is localized to these regions and the functional implications of this localization.
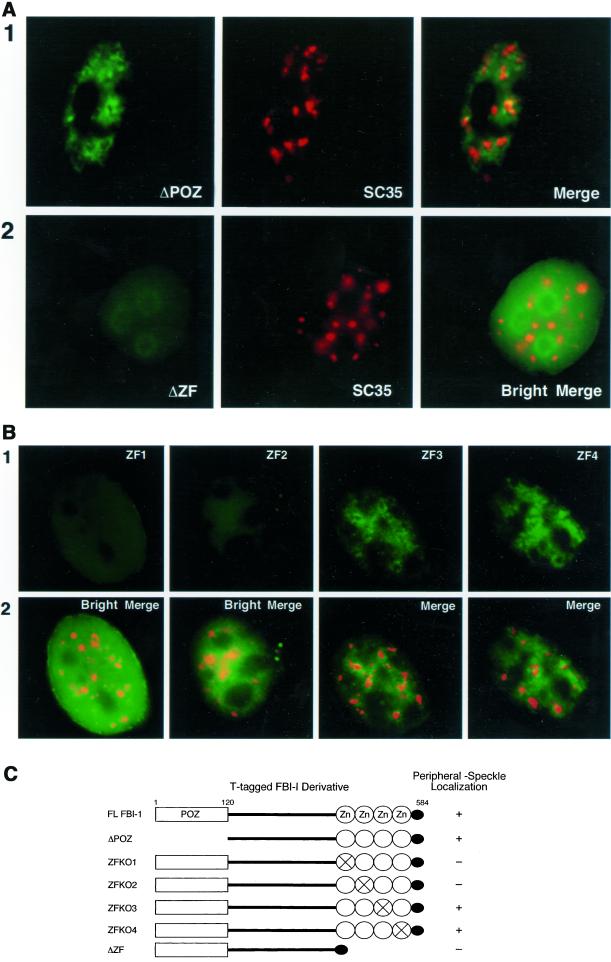
As a first step toward this goal, a series of truncation and point mutants of FBI-1 tagged with the T7 epitope were generated to determine the domain(s) of FBI-1 that are required for the peripheral-speckle localization (Figure 6C). HeLa cells were transiently transfected with mutant fusion proteins and cells were extracted with CSK buffer before fixation so that the peripheral-speckle localization pattern of FBI-1 could be more easily evaluated. Deletion of the POZ domain did not detectably altered the distribution of the mutant fusion protein compared with the wild-type fusion protein (compare Figure 6A, panel 1, with Figure 5B). However, deletion of the zinc finger (ΔZF) domain resulted in a significant alteration of the mutant fusion protein's distribution. The ZF mutant displayed a much less-intense signal after extraction (Figure 6A, panel 2, ΔZF) than the POZ deletion mutant and was more homogeneously distributed throughout the nucleoplasm with a rim around the nucleoli (panel 2, Bright Merge). To further delineate the protein domain required for the peripheral-speckle distribution, we examined the localization of mutant proteins containing point mutations in each of the four zinc fingers. Using the same mutants, we have previously shown that zinc fingers 1 and 2, but not 3 and 4, are required for FBI-1 binding to DNA (Morrison et al., 1999). As shown in Figure 6B, mutations in either zinc finger 1 or zinc finger 2 completely abolished the peripheral-speckle pattern, whereas mutations in zinc finger 3 or 4 had no effect. Thus, the same two zinc fingers that are responsible for DNA-binding are required for targeting FBI-1 to peripheral-speckle regions, suggesting that DNA binding may be responsible for the peripheral-speckle localization pattern.
Figure 6.
The DNA-binding zinc fingers 1 and 2 are required for less-soluble FBI-1's peripheral-speckle pattern of localization. Epifluorescent image of prefixation extracted HeLa cells transfected with 2 μg of the T-FBI-1 mutant-expressing plasmids ΔPOZ (A, panel 1), ΔZF (A, panel 2), ZF1 (B, column 1), ZF2 (B, column 2), ZF3 (B, column 3), and ZF4 (B, column 4). (A) Less-soluble T-FBI-1 ΔPOZ (panel 1, green) shows a peripheral-speckle pattern of localization (Merge), whereas T-FBI-1 ΔZF (panel 2, green) shows an aberrant homogeneous pattern of localization with an increased signal from the nucleoli and no increase in signal near the nuclear speckles (box 2, red and box 3). The T-tagged FBI-1 mutants were detected as T-FBI-1 in Figure 4. The nuclear speckles were detected as in Figure 5. Bright images result from 8× exposure times. (B) Less-soluble T-FBI-1 ZF3 and ZF4 (columns 3 and 4, respectively, green) also show a peripheral-speckle pattern of localization; however, T-FBI-1 ZF1 and ZF2 show aberrant homogeneous patterns of localization (columns 1 and 2, respectively, green). The T-tagged point mutants were detected as T-FBI-1 in Figure 6. Bright images result from 10× exposure times. (C) Summary of results from A and B.
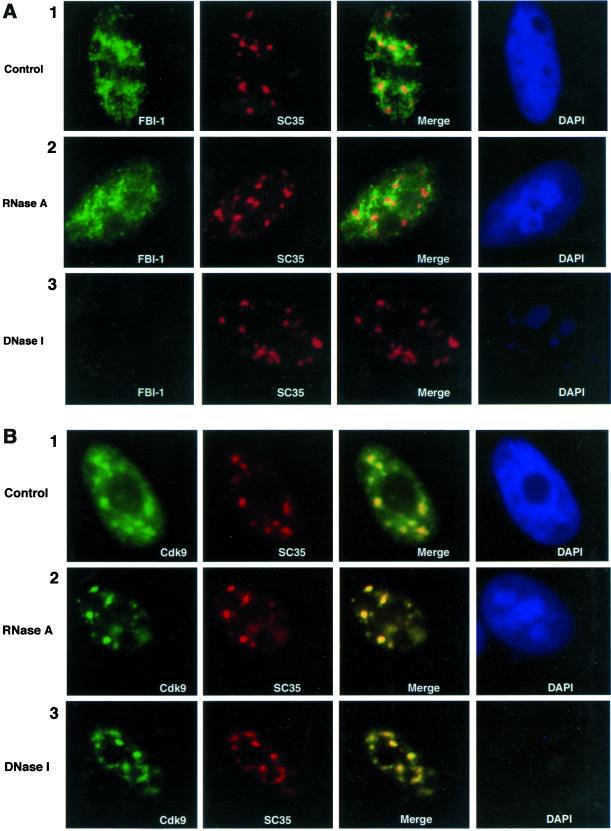
The above results, summarized in Figure 6C, suggest that FBI-1 is targeted to the peripheral-speckle pattern by binding to DNA; however, many Kruppel-type zinc finger proteins can also bind RNA and the peripheral-speckle region is enriched with RNA (Fakan and Puvion, 1980). To distinguish whether FBI-1 is localized to the peripheral-speckle regions by binding to DNA or RNA, we treated permeabilized cells with either DNase I, RNase A, or just buffer. Cells were then fixed and were double-immunolabeled with anti–FBI-1 and anti-SC35. DNase I treatment, which dramatically reduced the DNA content of the cell as seen by DAPI staining, had no effect on the SC35 nuclear speckles compared with buffer alone (Figure 7A, panels 1 and 3, column 2). RNase A treatment also had little affect on the nuclear speckles, although the treatment slightly reduced the diffuse nucleoplasmic staining of SC35 and rounded the edges of the SC35 foci (panel 2, column 2). These observations are consistent with previous studies (Spector et al., 1991). Like SC35, the peripheral-speckle pattern of endogenous FBI-1 showed little change when cells were treated with either buffer alone (panel 1) or with RNase A (panel 2). However, the nuclear labeling of FBI-1 was nearly abolished when cells were pretreated with DNase I (panel 3). These findings demonstrate that the peripheral-speckle distribution of FBI-1 is dependent on intact DNA and is independent of the presence of RNA, further supporting the hypothesis that DNA binding is responsible for its unique nuclear localization pattern.
Figure 7.
Less soluble FBI-1's peripheral-speckle pattern is dependent on cellular DNA but not on cellular RNA. Epifluorescent image of prefixation extracted HeLa cells treated for 30 min at 37°C with either RNase A (A and B, panel 2), DNase I (A and B, panel 3), or buffer (A and B, panel 1). (A) FBI-1's (green) peripheral-speckle pattern is little disturbed by treatment with buffer (panel 1) or RNase A (panel 2); however, less-soluble FBI-1 signal is drastically reduced upon DNase I treatment (panel 3, column 1), whereas the nuclear speckles (red) are undisturbed. Cdk9, and nuclear speckles were detected as in Figure 5. FBI-1 was detected as in Figure 4. (B) Less soluble Cdk9's (green) localization pattern is relatively undisturbed by all three treatments (column 1) and still colocalizes (yellow) with nuclear speckles (red). DNase I treatment results in a strong reduction of DNA content in the cell as judged by DAPI staining (A and B, blue)
As a control, we performed the same experiment concurrently on endogenous Cdk9 (Figure 7B) and cyclin T1 (our unpublished results). Both proteins showed reduced signal upon treatment with both RNase A and DNase I, but for both proteins the signal that remained colocalized extensively with SC35. Thus, of the four proteins, only the less-soluble FBI-1 was affected by DNase I treatment. Together with the mutagenesis experiments, these data strongly suggest that less-soluble FBI-1 is targeted to the peripheral-speckle pattern by binding to DNA.
The FBI-1 Peripheral Speckle Pattern Is Dependent on Active Transcription
There is significant evidence that regions near speckles contain actively transcribing genes (see DISCUSSION). In combination with the results described above, this leads to the interesting hypothesis that less-soluble FBI-1 is preferentially targeted to actively transcribing DNA near to and at the periphery of nuclear speckles. To determine the relationship between FBI-1's peripheral-speckle distribution and the transcriptional activity of the cell, we examined the distribution of FBI-1 in transcriptionally inhibited cells.
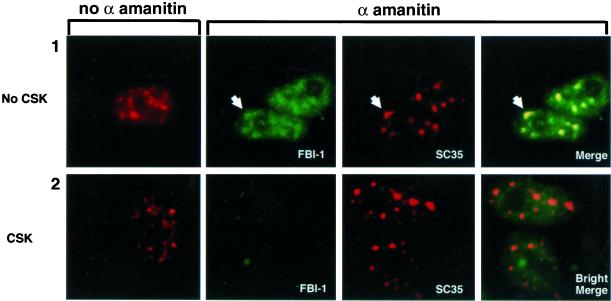
HeLa cells were treated with α-amanitin at 50 μg/ml, which strongly inhibits pol II transcription and can release factors that bind to active chromatin (Hock et al., 1998). After 5 h, we determined the localization of endogenous FBI-1 and SC35 both with and without extraction. As shown in Figure 8, α-amanitin treatment redistributed SC35 to larger and more rounded structures (compare column 1 with column 3), consistent with published studies that show that a variety of transcriptional inhibition treatments can cause such an alteration of the speckle morphology (Spector et al., 1983). Interestingly, FBI-1, which displayed partial and peripheral colocalization with SC35 speckles in the absence of α-amanitin, was also redistributed to the enlarged speckles and displayed strong colocalization with SC35 in unextracted cells (Figure 8, panel 1, for example, see large arrowhead). However, when cells were extracted almost all the FBI-1 was lost from the cells (panel 2). Moreover, the little FBI-1 that was left did not show a peripheral-speckle pattern, but rather was more uniformly distributed throughout the nucleoplasm (panel 2, Bright). Thus, the less-soluble peripheral-speckle pattern is dependent on transcription. This observation, together with the previous immunofluorescence studies suggests that less-soluble FBI-1 is targeted preferentially to actively transcribing DNA at the periphery of the speckles.
Figure 8.
Total FBI-1 is redistributed to enlarged and rounded nuclear speckles, but the less-soluble FBI-1 population and peripheral-speckle pattern is drastically reduced upon inhibition of transcription. Epifluorescent images of nonextracted (panel 1) and prefixation extracted (panel 2) HeLa cells either untreated (column 1) or treated for 5 h with 50 μg/ml of α-amanitin (columns 2–4). Total FBI-1 (panel 1, green) is redistributed into enlarged nuclear speckles (red and yellow). Note that unlike in non–α-amanitin–treated cells (Figure 4C), the FBI-1 signal foci match the shape, size, and relative intensity of the nuclear speckles even in irregularly shaped foci (arrowhead). However, less-soluble FBI-1 signal is almost completely lost (panel 2, column 2). A six times longer exposure (panel 2, column 4, Bright) shows that the little FBI-1 that is left after extraction does not show a peripheral-speckle pattern of localization. Nuclear speckles were detected as in Figure 5. FBI-1 was detected as in Figure 4.
DISCUSSION
FBI-1 and HIV-1 Tat
FBI-1 binds to the HIV-1 promoter, interacts, and partially colocalizes with the Tat-TAK/P-TEFb complex, and can increase Tat activity in a dose-dependent manner. These results suggest that FBI-1 is involved in HIV-1 transcription. But what is the role of endogenous FBI-1 in HIV-1 transcription? One possibility is that FBI-1 acts as a positive modulator of Tat-activated HIV-1 transcription by recruiting Tat/P-TEFb to the promoter via its association with Tat. In addition to the association between FBI-1 and Tat, an enhancing effect could also be due to the binding of FBI-1 to the IST element, an element that stimulates constitutive elongation-incompetent transcription in the absence of Tat and thus may maintain an open chromatin configuration. Such a promoter priming effect by a regulator has been described in the regulation of several heat shock promoters by another POZ protein, GAF (Wilkins and Lis, 1997). GAF binds to uninduced heat shock promoters and maintains an open chromatin structure, allowing the constitutive production of short transcripts and keeping the promoter primed for induction. Interestingly, a recent study shows that the Tat cofactor, P-TEFb, is recruited to activated heat shock loci and is sufficient for induction of the hsp 70 protein (Lis et al., 2000). Thus, having a POZ protein maintain a region of open/active chromatin ready for elongation induction by P-TEFb could be a general mechanism for transcriptional regulation.
Another possibility is that FBI-1 is a negative regulator of HIV-1 transcription. Although increased levels of FBI-1 enhance Tat activity in HeLa cells, we cannot exclude the possibility that the enhancement is due to the sequestering of cofactors of endogenous FBI-1 that normally repress the Tat activity. This would explain why the ΔPOZ mutant of FBI-1 still had a partial effect on Tat transcription, because deletion mutants may also act as inhibitors as long as the relevant protein binding region is intact. Consistent with this hypothesis, FBI-1 can act as a repressor when targeted to a synthetic promoter (Kukita et al., 1999). Further work will be needed to determine the mechanism of the FBI-1 effect on Tat activity.
FBI-1 Localizes to a Novel Subnuclear Domain that Includes the Nuclear Speckles
The nuclear speckles domain is defined by immunofluorescence as a subnuclear domain consisting of 10–40 “foci” of various sizes and shapes to which most splicing factors and some transcription factors localize (Spector, 1993; reviewed in Huang and Spector, 1996a; Misteli and Spector, 1998). Some of the speckles interconnect. Electron microscopic immunolabeling with anti-SC35 antibody has demonstrated that the transcription and splicing factor containing speckles observed at the light microscopic level correspond to previously characterized interchromatin granule clusters (IGC) and perichromatin fibrils (PF; Spector 1993). In vivo H3-uridine labeling and chase experiments show that perichromatin fibrils contain newly synthesized RNA, whereas the majority of interchromatin granule clusters are not labeled until several hours after chase. It therefore has been suggested that interchromatin clusters may be depots for the storage, recycling, or reassembly of transcription and splicing components, whereas the surrounding perichromatin fibrils are sites of active transcription and RNA processing (Spector, 1993; Fakan and Modak, 1973). Consistent with this hypothesis, localization studies of specific promoters have shown that many abundantly expressed genes are located near speckles and that pre-mRNA splicing factors are recruited from the speckles to sites of active transcription and splicing (Huang and Spector, 1991, 1996a; Misteli et al., 1997; Smith et al., 1999). However, in some cases, highly expressed genes may recruit large amounts of transcription and splicing factors in such a manner that the site can enlarge to resemble a speckle. The IGC has been biochemically purified and has been found to contain >70 proteins by mass spectrometry, including many splicing and some transcription factors (Mintz et al., 1999). The precise function of this nuclear domain remains to be determined.
In this study, we showed that the nuclear localization of FBI-1 overlaps with the speckled distribution of SC35, although the intensity of FBI-1 labeling tends to be stronger at the periphery and weaker at the center of the speckles. When cells were extracted with CSK before fixation, the mild extraction removed most of the FBI-1 signal that was localized at the center of individual speckles as well as most of the signal that was diffusely localized throughout the nucleoplasm. The remaining less-soluble fraction of endogenous FBI-1 displayed a novel localization pattern that often coincided with the periphery of the nuclear speckle structure. This localization pattern is representative of endogenous protein because immunolabeling using two different anti–FBI-1 antibodies or the expression of two different epitope-tagged FBI-1 proteins showed the same pattern. We call this novel pattern of localization “peripheral-speckle localization” to emphasize that FBI-1 is associated with higher affinity to the vicinity and periphery of the nuclear speckles. This association was dependent on FBI-1's DNA binding domain, the presence of DNA, and active transcription, suggesting that the localization pattern is the result of FBI-1 association with actively transcribed chromatin. These observations are consistent with FBI-1 localizing to perichromatin fibrils, which have been previously shown to surround interchromatin granule clusters, and to contain active chromatin and a large amount of newly synthesized RNA.
To our knowledge, FBI-1's peripheral-speckle pattern of localization is unique. Instructively, however, two localization patterns have been reported that share some aspects of the FBI-1 pattern. Acetylated histone H3 localizes at foci of various sizes and shapes that are excluded from the nuclear periphery, the nucleoli, and the speckles (which is to be expected as DNA is not present in the speckles; Hendzel et al., 1998). Treatment of cells with a deacetylase inhibitor, however, causes an enrichment of signal specifically around the individual speckles. Given that upon deacetylase inhibition, transcriptionally active chromatin more rapidly accumulates highly acetylated histones than the rest of the genome (Covault and Chalkley, 1980; Davie and Hendzel, 1994), the authors suggest that the regions around speckles contains active chromatin.
The second protein with a localization pattern that shares a characteristic with FBI-1's peripheral-speckle pattern is the nucleosomal binding protein, HMG-17, which can alter the structure of chromatin and enhance transcription (Bustin et al., 1995). HMG-17 localizes in irregularly shaped foci that partially colocalize with active sites of transcription and with SC35 in most Hep-2 cells (Hock et al., 1998). On treatment of cells with α-amanitin, all of the HMG-17 speckles are redistributed to the enlarged SC35 speckles. Although the authors did not perform a CSK extraction in the α amanitin experiment, they did permeabilize the cells and observed a reduced signal much like the one we saw with FBI-1 in the extracted α-amanitin–treated cells. This argues that a chromatin bound protein can be released from chromatin upon inhibition of transcription by α-amanitin.
Given that the regions at the periphery of nuclear speckles contain active chromatin and that proteins can be released from chromatin upon inhibition of transcription by α-amanitin, we suggest the following model for FBI-1 distribution. FBI-1 exists in two populations. One population is soluble and resides at the center of the speckles and diffusely throughout the nucleoplasm, and the other population is less soluble and consists of FBI-1 bound to transcriptionally active, peripheral-speckle chromatin. Consistent with this notion active chromatin is relatively insoluble, possibly because of an interaction with the nuclear matrix (reviewed by Davie, 1996; Jackson, 1997). Treatment of cells with α-amanitin represses transcription, releasing the less-soluble FBI-1 from DNA that then accumulates in the enlarged speckles (Figure 8, panel 1). However, this unbound protein is now more soluble and thus is lost upon extraction (panel 2). This model predicts that FBI-1's ability to bind DNA may not be required for targeting the protein to the enlarged transcriptionally independent speckles. Indeed FBI-1 that contains a mutation that disrupts DNA binding, and peripheral speckle localization is still recruited to the enlarged speckles upon α-amanitin treatment (our unpublished results).
A question that arises from this model is what targets FBI-1 to transcriptionally active chromatin that is mostly in the peripheral-speckle regions? The areas around speckles are probably enriched in active chromatin, but there is active chromatin throughout the interior of the nucleus (Hendzel et al., 1998). Total FBI-1 is present throughout the nucleus and thus could be targeted to all active chromatin, yet most of the less-soluble, DNA-dependent, and transcriptionally dependent FBI-1 is preferentially enriched near the speckles. This may indicate that there are several stages of activated chromatin and that the peripheral-speckle chromatin represents a defined stage with some structural difference, perhaps nuclear matrix association, that makes it and the bound FBI-1 resistant to extraction.
A second question raised by this model is what is the role of FBI-1 in peripheral-speckle regions. One possibility is that FBI-1 contributes to the unique structure of this transcriptionally active chromatin region as either an activator or a repressor. Indeed, several POZ proteins have been linked to chromatin-remodeling activities (Wilkins and Lis, 1997; Dhordain et al., 1998). GAF is a transactivator protein at heat shock promoters capable of maintaining local chromatin in an open conformation, but also can bind proteins associated with deacetylases (Espinas et al., 2000). BCL6 and PLZF can bind to the Sin3 deacetylase complex and can repress transcription when targeted to heterologous promoters (Dhordain et al., 1997, 1998; Hong et al., 1997; David et al., 1998; Huynh and Bardwell, 1998; Huynh et al., 2000). Thus, FBI-1 may act as an activator or as a repressor to regulate transcriptionally active chromatin.
Tat and TAK/P-TEFb Colocalize with Nuclear Speckles
Localization of Tat tagged with two different short peptides, T7 and HA, demonstrated a heterogeneous nuclear localization pattern that predominantly colocalized with the nuclear speckle domain (Figure 4 and our unpublished results). We observed minimal nucleolar labeling. These observations of Tat distribution are in contrast to some of the published reports that Tat is predominately localized to the nucleolus by immunolabeling (Cullen et al., 1988; Siomi et al., 1990; Luznik et al., 1995). The difference may be due to the expression level of Tat in different studies.
The Tat cofactor, TAK/P-TEFb (Cyclin T1 and Cdk9), also predominantly colocalizes to the nuclear speckles (Figure 4). This finding agrees with the recent deconvolution microscopy study of TAK/P-TEFb by Herrmann and Mancini (2001). The targeting of Tat to this region may facilitate its interaction with TAK/P-TEFb and the actively transcribing HIV-1 promoter.
But why is P-TEFb found at the nuclear speckles? The phosphorylated form of pol II is not efficient at reinitiation (Gebara et al., 1997). So one possibility is that P-TEFb-pol II complexes that have finished transcribing their particular genes accumulate at nuclear speckles for reprocessing and repackaging so they can be reused at another gene. Consistent with this possibility, the phosphorylated form of RNA pol II is preferentially found at nuclear speckles, but the unphosphorylated form is not (Mortillaro et al., 1996). It will be interesting to see if other processing factors known to associate with the phosphorylated CTD such as CTD phosphatases also localize at speckles.
In summary, we demonstrated that a cellular protein, FBI-1, specifically stimulates Tat-activated HIV-1 transcription. FBI-1 associates with Tat via the zinc finger domain, and this domain is important for the stimulation of Tat. Furthermore, we showed that less-soluble FBI-1 localizes to a unique peripheral-speckled pattern in a DNA binding and transcription dependent manner, suggesting that FBI-1 preferentially associates with active chromatin.
ACKNOWLEDGMENTS
The authors thank D. Spector and members of the Spector laboratory for helpful discussions, technical assistance, and comments on the manuscript; B. Ma and Y. Sun for valuable technical assistance and R. Whitaker for assistance with cell culture; D. Price and K. Jones for antibodies and N. Zeleznik-Le, P. Dhordain, M. Greenberg, W. Herr, and R. Dalla-Favera for constructs; C. Herrmann and L. Schramm for important discussions, J. Skowronski and D. Leary for helpful comments on the manuscript, and J. Duffy, M. Ockler, and P. Renna for artwork and photography. P.S.P is supported by a Leukemia Society of America Special Fellowship Award (GN 629100). S.H. is supported by grants from National Institutes of Health National Cancer Institute grants 1 Ro1 CA 77560–01A1 and 5K01 CA74988–03to S.H.
Footnotes
DOI:10.1091/mbc.01–08–0383.
REFERENCES
- Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D. The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 1995;6:1193–1198. [PubMed] [Google Scholar]
- Bardwell VJ, Treisman R. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- Baron BW, Desai M, Baber LJ, Paras L, Zhang Q, Sadhu A, Duguay S, Nucifora G, McKeithan TW, Zeleznik-Le N. BCL6 can repress transcription from the human immunodeficiency virus type I promoter/enhancer region. Genes Chromosomes Cancer. 1997;19:14–21. [PubMed] [Google Scholar]
- Bustin M, Trieschmann L, Postnikov YV. The HMG-14/-17 chromosomal protein family: architectural elements that enhance transcription from chromatin templates. Semin Cell Biol. 1995;6:247–255. doi: 10.1006/scel.1995.0033. [DOI] [PubMed] [Google Scholar]
- Cullen BR, Hauber J, Campbell K, Sodroski JG, Haseltine WA, Rosen CA. Subcellular localization of the human immunodeficiency virus trans-acting art gene product. J Virol. 1988;62:2498–2501. doi: 10.1128/jvi.62.7.2498-2501.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covault J, Chalkley R. The identification of distinct populations of acetylated histone. J Biol Chem. 1980;255:9110–9116. [PubMed] [Google Scholar]
- David G, Alland L, Hong SH, Wong CW, DePinho RA, Dejean A. Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene. 1998;16:2549–2556. doi: 10.1038/sj.onc.1202043. [DOI] [PubMed] [Google Scholar]
- Davie JR. Histone modifications, chromatin structure, and the nuclear matrix. J Cell Biochem. 1996;62:149–157. doi: 10.1002/(sici)1097-4644(199608)62:2<149::aid-jcb2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Davie JR, Hendzel MJ. Multiple functions of dynamic histone acetylation. J Cell Biochem. 1994;55:98–105. doi: 10.1002/jcb.240550112. [DOI] [PubMed] [Google Scholar]
- Davies JM, Hawe N, Kabarowski J, Huang QH, Zhu J, Brand NJ, Leprince D, Dhordain P, Cook M, Morriss-Kay G, Zelent A. Novel BTB/POZ domain zinc-finger protein, LRF, is a potential target of the LAZ-3/BCL-6 oncogene. Oncogene. 1999;18:365–375. doi: 10.1038/sj.onc.1202332. [DOI] [PubMed] [Google Scholar]
- Dhordain P, Albagli O, Ansieau S, Koken MH, Deweindt C, Quief S, Lantoine D, Leutz A, Kerckaert JP, Leprince D. The BTB/POZ domain targets the LAZ3/BCL6 oncoprotein to nuclear dots and mediates homomerisation in vivo. Oncogene. 1995;11:2689–2697. [PubMed] [Google Scholar]
- Dhordain P, Albagli O, Lin RJ, Ansieau S, Quief S, Leutz A, Kerckaert JP, Evans RM, Leprince D. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc Natl Acad Sci USA. 1997;94:10762–10767. doi: 10.1073/pnas.94.20.10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhordain P, Lin RJ, Quief S, Lantoine D, Kerckaert JP, Evans RM, Albagli O. The LAZ3(BCL-6) oncoprotein recruits a SMRT/mSIN3A/histone deacetylase containing complex to mediate transcriptional repression. Nucleic Acids Res. 1998;26:4645–4651. doi: 10.1093/nar/26.20.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinas ML, Canudas S, Fanti L, Pimpinelli S, Casanova J, Azorin F. The GAGA factor of Drosophila interacts with SAP18, a Sin3-associated polypeptide. EMBO Rep. 2000;1:253–259. doi: 10.1093/embo-reports/kvd046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakan S, Modak SP. Localization of DNA in ultrathin tissue sections incubated with terminal deoxynucleotidyl transferase, as visualized by electronmicroscope autoradiography. Exp Cell Res. 1973;77:95–104. doi: 10.1016/0014-4827(73)90557-0. [DOI] [PubMed] [Google Scholar]
- Fakan S, Puvion E. The ultrastructural visualization of nucleolar and extranucleolar RNA synthesis and distribution. Int Rev Cytol. 1980;65:255–299. doi: 10.1016/s0074-7696(08)61962-2. [DOI] [PubMed] [Google Scholar]
- Fu XD, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Gebara MM, Sayre MH, Corden JL. Phosphorylation of the carboxy-terminal repeat domain in RNA polymerase II by cyclin-dependent kinases is sufficient to inhibit transcription. J Cell Biochem. 1997;64:390–402. [PubMed] [Google Scholar]
- Hendzel MJ, Kruhlak MJ, Bazett-Jones DP. Organization of highly acetylated chromatin around sites of heterogeneous nuclear RNA accumulation. Mol Biol Cell. 1998;9:2491–2507. doi: 10.1091/mbc.9.9.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann CH, Mancini MA. The Cdk9, and cyclin T subunits of TAK/P-TEFb localize to splicing factor-rich nuclear speckle regions. J Cell Sci. 2001;114:1491–1503. doi: 10.1242/jcs.114.8.1491. [DOI] [PubMed] [Google Scholar]
- Herrmann CH, Rice AP. Specific interaction of the human immunodeficiency virus Tat proteins with a cellular protein kinase. Virology. 1993;197:601–608. doi: 10.1006/viro.1993.1634. [DOI] [PubMed] [Google Scholar]
- Herrmann CH, Rice AP. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J Virol. 1995;69:1612–1620. doi: 10.1128/jvi.69.3.1612-1620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock R, Wilde F, Scheer U, Bustin M. Dynamic relocation of chromosomal protein HMG-17 in the nucleus is dependent on transcriptional activity. EMBO J. 1998;17:6992–7001. doi: 10.1093/emboj/17.23.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SH, David G, Wong CW, Dejean A, Privalsky ML. SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor alpha (RARalpha) and PLZF-RARalpha oncoproteins associated with acute promyelocytic leukemia. Proc Natl Acad Sci USA. 1997;94:9028–9033. doi: 10.1073/pnas.94.17.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Spector DL. Nascent pre-mRNA transcripts are associated with nuclear regions enriched in splicing factors. Genes Dev. 1991;5:22882302. doi: 10.1101/gad.5.12a.2288. [DOI] [PubMed] [Google Scholar]
- Huang S, Spector DL. Dynamic organization of pre-mRNA splicing factors. J Cell Biochem. 1996a;62:191–197. doi: 10.1002/(sici)1097-4644(199608)62:2<191::aid-jcb7>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Huang S, Spector DL. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription. J Cell Biol. 1996b;133:719–732. doi: 10.1083/jcb.133.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh KD, Bardwell VJ. The BCL-6 POZ domain and other POZ domains interact with the co- repressors N-CoR and SMRT. Oncogene. 1998;17:2473–2484. doi: 10.1038/sj.onc.1202197. [DOI] [PubMed] [Google Scholar]
- Huynh KD, Fischle W, Verdin E, Bardwell VJ. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 2000;14:1810–1823. [PMC free article] [PubMed] [Google Scholar]
- Jackson DA. Chromatin domains and nuclear compartments: establishing sites of gene expression in eukaryotic nuclei. Mol Biol Rep. 1997;24:209–220. doi: 10.1023/a:1006873614521. [DOI] [PubMed] [Google Scholar]
- Kao SY, Calman AF, Luciw PA, Peterlin BM. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- Karn J. Tackling Tat. J Mol Biol. 1999;293:235–254. doi: 10.1006/jmbi.1999.3060. [DOI] [PubMed] [Google Scholar]
- Kukita A, Kukita T, Ouchida M, Maeda H, Yatsuki H, Kohashi O. Osteoclast-derived zinc finger (OCZF) protein with POZ domain, a possible transcriptional repressor, is involved in osteoclastogenesis. Blood. 1999;94:1987–1997. [PubMed] [Google Scholar]
- Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment, and function at heat shock loci. Genes Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- Luznik L, Martone ME, Kraus G, Zhang Y, Xu Y, Ellisman MH, Wong-Staal F. Localization of human immunodeficiency virus Rev in transfected and virus-infected cells. AIDS Res Hum Retroviruses. 1995;11:795–804. doi: 10.1089/aid.1995.11.795. [DOI] [PubMed] [Google Scholar]
- Madore SJ, Cullen BR. Functional similarities between HIV-1 Tat and DNA sequence-specific transcriptional activator. Virology, 1995;206:1150–1154. doi: 10.1006/viro.1995.1041. [DOI] [PubMed] [Google Scholar]
- Mintz PJ, Patterson SD, Neuwald AF, Spahr CS, Spector DL. Purification and biochemical characterization of interchromatin granule clusters. EMBO J. 1999;18:4308–4320. doi: 10.1093/emboj/18.15.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Caceres JF, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- Misteli T, Spector DL. The cellular organization of gene expression. Curr Opin Cell Biol. 1998;10:323–331. doi: 10.1016/s0955-0674(98)80007-0. [DOI] [PubMed] [Google Scholar]
- Morrison DJ, Pendergrast PS, Stavropoulos P, Colmenares SU, Kobayashi R, Hernandez N. FBI-1, a factor that binds to the HIV-1 inducer of short transcripts (IST), is a POZ domain protein. Nucleic Acids Res. 1999;27:1251–1262. doi: 10.1093/nar/27.5.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortillaro MJ, Blencowe BJ, Wei X, Nakayasu H, Du L, Warren SL, Sharp PA, Berezney R. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc Natl Acad Sci USA. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niman HL, Houghten RA, Walker LE, Reisfeld RA, Wilson IA, Hogle JM, Lerner RA. Generation of protein-reactive antibodies by short peptides is an event of high frequency: implications for the structural basis of immune recognition. Proc Natl Acad Sci USA. 1983;80:4949–4953. doi: 10.1073/pnas.80.16.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendergrast PS, Hernandez N. RNA-targeted activators, but not DNA-targeted activators, repress the synthesis of short transcripts at the human immunodeficiency virus type 1 long terminal repeat. J Virol. 1997;71:910–917. doi: 10.1128/jvi.71.2.910-917.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendergrast PS, Morrison D, Tansey WP, Hernandez N. Mutations in the carboxy-terminal domain of TBP affect the synthesis of human immunodeficiency virus type 1 full-length and short transcripts similarly. J Virol. 1996;70:5025–5034. doi: 10.1128/jvi.70.8.5025-5034.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessler F, Pendergrast PS, Hernandez N. Purification and characterization of FBI-1, a cellular factor that binds to the human immunodeficiency virus type 1 inducer of short transcripts. Mol Cell Biol. 1997;17:3786–3798. doi: 10.1128/mcb.17.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DH. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol. 2000;20:2629–2634. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnasabapathy R, Sheldon M, Johal L, Hernandez N. The HIV-1 long terminal repeat contains an unusual element that induces the synthesis of short RNAs from various mRNA and snRNA promoters. Genes Dev. 1990;4:2061–2074. doi: 10.1101/gad.4.12a.2061. [DOI] [PubMed] [Google Scholar]
- Rice AP, Carlotti F. Mutational analysis of the conserved cysteine-rich region of the human immunodeficiency virus type 1 Tat protein. J Virol. 1990a;64:1864–1868. doi: 10.1128/jvi.64.4.1864-1868.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice AP, Carlotti F. Structural analysis of wild-type and mutant human immunodeficiency virus type 1 Tat proteins. J Virol. 1990b;64:6018–6026. doi: 10.1128/jvi.64.12.6018-6026.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon M, Ratnasabapathy R, Hernandez N. Characterization of the inducer of short transcripts, a human immunodeficiency virus type 1 transcriptional element that activates the synthesis of short RNAs. Mol Cell Biol. 1993;13:1251–1263. doi: 10.1128/mcb.13.2.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Shida H, Maki M, Hatanaka M. Effects of a highly basic region of human immunodeficiency virus Tat protein on nucleolar localization. J Virol. 1990;64:1803–1807. doi: 10.1128/jvi.64.4.1803-1807.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KP, Moen PT, Wydner KL, Coleman JR, Lawrence JB. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J Cell Biol. 1999;144:617–629. doi: 10.1083/jcb.144.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL. Nuclear organization of pre-mRNA processing. Curr Opin Cell Biol. 1993;5:442–427. doi: 10.1016/0955-0674(93)90009-f. [DOI] [PubMed] [Google Scholar]
- Spector DL, Fu XD, Maniatis T. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Schrier WH, Busch H. Immunoelectron microscopic localization of snRNPs. Biol Cell. 1983;49:1–10. doi: 10.1111/j.1768-322x.1984.tb00215.x. [DOI] [PubMed] [Google Scholar]
- Tiley LS, Modore SJ, Malim MH, Cullen BR. The BP16 transcription activation domain is functional when targeted to a promoter-proximal RNA sequence. Genes Dev. 1992;6:2077–2087. doi: 10.1101/gad.6.11.2077. [DOI] [PubMed] [Google Scholar]
- Wilkins RC, Lis JT. Dynamics of potentiation and activation: GAGA factor and its role in heat shock gene regulation. Nucleic Acids Res. 1997;25:3963–3968. doi: 10.1093/nar/25.20.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]