Abstract
A Schizosaccharomyces pombe spindle pole body (SPB) protein interacts in a two-hybrid system with Dlc1, which belongs to the 14-kDa Tctex-1 dynein light chain family. Green fluorescent protein-tagged Dlc1 accumulated at the SPB throughout the life cycle. During meiotic prophase, Dlc1 was present along astral microtubules and microtubule-anchoring sites on the cell cortex, reminiscent of the cytoplasmic dynein heavy chain Dhc1. In a dlc1-null mutant, Dhc1-dependent nuclear movement in meiotic prophase became irregular in its duration and direction. Dhc1 protein was displaced from the cortex anchors and the formation of microtubule bundle(s) that guide nuclear movement was impaired in the mutant. Meiotic recombination in the dlc1 mutant was reduced to levels similar to that in the dhc1 mutant. Dlc1 and Dhc1 also have roles in karyogamy and rDNA relocation during the sexual phase. Strains mutated in both the dlc1 and dhc1 loci displayed more severe defects in recombination, karyogamy, and sporulation than in either single mutant alone, suggesting that Dlc1 is involved in nuclear events that are independent of Dhc1. S. pombe contains a homolog of the 8-kDa dynein light chain, Dlc2. This class of dynein light chain, however, is not essential in either the vegetative or sexual phases.
INTRODUCTION
The fission yeast Schizosaccharomyces pombe usually propagates in a haploid state. When nutritionally starved, two cells with opposite mating types conjugate and two haploid nuclei fuse to produce a diploid nucleus in a zygote, which is immediately followed by a “zygotic” meiosis. S. pombe cells can vegetatively proliferate in a diploid state. When a diploid cell is starved, it can enter into meiosis without conjugation (azygotic meiosis). In the prophase of both types of meiosis, all telomeres cluster near the spindle pole body (SPB) to form a typical chromosomal bouquet, as occurs in many other species (Chikashige et al., 1994, 1997). Loss of a telomere-binding protein, Taz1, impairs the telomere clustering and decreases the frequency of meiotic recombination (Cooper et al., 1998; Nimmo et al., 1998). An SPB-associated protein, Kms1, is also important for the formation of the telomere cluster (Shimanuki et al., 1997; Niwa et al., 2000). kms1 mutations do not affect mitotic growth, but in the meiotic prophase the SPB tends to disintegrate into a cytoplasmic microtubule organization center (MTOC) and several dots on the nuclear membrane (Shimanuki et al., 1997; Niwa et al., 2000; Shimanuki and Niwa, unpublished data) that contain an SPB protein, Sad1 (Hagan and Yanagida, 1995). Telomeres are often colocalized with the Sad1 dots, and thus the bouquet formation is impaired in the kms1 mutant; consequently, the meiotic recombination rate is reduced (Shimanuki et al., 1997; Niwa et al., 2000). These and other studies suggest that telomere clustering is a key process for efficient chromosome pairing in fission yeast (reviewed in Hiraoka, 1998; Niwa et al., 2000).
The meiotic prophase nucleus in which telomere clustering is formed is not static. It moves along an oscillatory path with an ever-changing but generally elongated morphology (Chikashige et al., 1994). This movement, often referred to as horsetail nuclear movement, is led by the SPB and requires astral microtubules (Chikashige et al., 1994; Ding et al., 1998). Cytoplasmic dynein is involved in the nuclear movement, because the disruption of the dhc1+ gene, which encodes the dynein heavy chain, abolishes the movement (Yamamoto et al., 1999). It was recently reported that the nuclear movement is principally led by pulling microtubules that connect the SPB to microtubule-anchoring sites formed on the forward side of the cell cortex with regard to the moving SPB and that cytoplasmic dynein present at the anchoring sites is thought to generate the pulling force (Yamamoto et al., 2001). It is not known, however, how the placement of the dynein molecules is regulated at the anchoring sites.
Cytoplasmic dynein is a multisubunit complex consisting of two heavy chains that contain the motor domain and other associated proteins; that is, intermediate chains, light intermediate chains, and light chains. Various cellular processes, such as vesicular transport, spindle assembly, and nuclear migration, require cytoplasmic dynein, which probably performs its function and achieves its proper localization by selectively using the associated proteins as well as interacting factors such as the dynactin complex (Karki and Holzbaur, 1999). The light chains are represented by 8-, 14-, and 22-kDa proteins (King et al., 1996a,b); the 8- and 14-kDa light chains are also contained in axonemal dynein. The smallest subunit is highly conserved evolutionarily in several species and is required for sensory axon projection and other developmental events in Drosophila (Dick et al., 1996; Phillis et al., 1996), nuclear migration in Aspergillus (Beckwith et al., 1998), and intraflagellar retrograde transport in Chlamydomonas (Pazour et al., 1998). As for the 14-kDa light chain, the mouse gene Tctex-1, one of the candidate factors responsible for meiotic drive (Lader et al., 1989), belongs to this class of proteins (King et al., 1996b). The Drosophila homolog dtctex-1 is required for functional sperm (Caggese et al., 2001). The molecular function of the 14-kDa light chain is not understood, except that bovine Tctex-1, but not its homolog RP3, directly binds to the carboxy terminus of rhodopsin and is responsible for the cytoplasmic dynein-dependent transport of rhodopsin vesicles (Tai et al., 1999, 2001). Tctex-1 interacts with the intermediate chain (IC) as well as with other cellular factors (Mok et al., 2001). Tctex-1 and RP3 compete for binding to IC to form different types of cytoplasmic dynein complexes (Tai et al., 2001). Tctex-1 is localized to the Golgi apparatus as well as on cytoplasmic microtubules and on the nuclear membrane in fibroblasts (King et al., 1998; Tai et al., 1998). These findings suggest that the 14-kDa family of proteins has diverse functions.
Investigation of Kms1-interacting proteins by using a yeast two-hybrid system led to the identification of a protein that has homology to the 14-kDa Tctex-1 dynein light chain family proteins. The present results indicate that the Tctex-1 homolog in fission yeast is required for proper localization of Dhc1 molecules to ensure regular oscillatory nuclear movement in the meiotic prophase. We also demonstrate that it has important roles in several processes in the sexual phase, apparently independently of Dhc1.
MATERIALS AND METHODS
S. pombe Strains, Media, and Basic Methods for Genetics and Molecular Biology
The S. pombe strains used in this study are listed in Table 1. The media YE, ME, EMM-2, malt-extract broth plus galactose (MEBG), mannose synthetic medium (MSM) as well as experimental methods have been described previously (Moreno et al., 1991; Miyata et al., 1997; Okazaki et al., 2000). To observe azygotic meiosis, fresh diploid colonies were isolated for each experiment on a phloxin B-containing YE plate and were checked for efficient azygotic sporulation on an ME plate. The meiotic recombination frequency between chromosomes and a plasmid containing the ade6-L469 gene inserted into an LEU2-based vector pSP1 was measured according to Ponticelli and Smith (1989). To generate DNA fragments to be subcloned, we performed polymerase chain reaction (PCR) by using the primers listed in Table 2. The cloned PCR product sequences were confirmed by DNA sequence analysis. For site-directed mutagenesis, a QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used. Probes for filter hybridization were prepared by PCR by using a PCR digoxigenin-labeling mix (Roche Diagnostic, Mannheim, Germany). For Northern hybridization, probes prepared with primers KIP64 and KIP54, and primers PDE17 and PDE18 were used to detect dlc1+ and dhc1+ mRNAs, respectively. The hybridization signals were detected with antidigoxigenin-alkaline phosphatase conjugate (Roche Diagnostic) and CDP-Star (Tropix, Bedford, MA).
Table 1.
S. pombe strains used
| Name | Genotype |
|---|---|
| L968 | h90 wild |
| L972 | h− wild |
| SA21 | h+s wild |
| Z310-13D | h90 ura4-D18 leu1 |
| F52-3A | h90 dlc1∷ura4+ ura4 leu1 |
| Z310-10B | h90 leu1 |
| Z121-4D | h90 kms1∷ura4+ ura4 leu1 |
| F71-11D | h90 dlc1∷ura4+ kms1∷ura4+ ura4 leu1 |
| F143-6B | h90 dlc1∷ura4+ dhc1-d4∷ura4+ ura4 leu1 |
| F167-7B | h90 kms1∷ura4+ dhc1-d4∷ura4+ ura4 leu1 |
| F80-1B | h− ade8 trp1 his5 |
| F63-1C | h+s dlc1∷ura4+ ura4 leu1 |
| F84-5A | h− dlc1∷ura4+ ura4 ade8 trp1 his5 |
| F90-1D | h− dlc1∷ura4+ ura4 ura1 lys3 |
| F83-15A | h+s dlc1∷ura4+ leu1 |
| HM101 | h− ura1 lys3 |
| F78-18A | h+s fur1 ade6-M210 lys1 leu1 |
| F79-8A | h− dlc1∷ura4+ leu1 |
| F81-2D | h+ dlc1∷ura4+ fur1 ade6-M210 lys1 leu1 |
| F83-2A | h− dlc1∷ura4+ |
| Z439-2A | h+s leu1 |
| F96-2A | h+ dlc1∷ura4+ lys1 leu1 |
| F96-2C | h− dlc1∷ura4+ fur1 ade6-M210 |
| F109-2A | h+ lys1 leu1 |
| F109-2B | h− fur1 ade6-M210 |
| F95-1A | h− dlc1∷ura4+ ura1 lys3 |
| F131-1A | h+ pom1∷kanr leu1 |
| F132-1D | h+ dlc1∷ura4+ pom1∷kanr leu1 |
| F133-1A | h+ hus2-22 |
| F141-1C | h+ dlc1∷ura4+ hus2-22 |
| F154-4B | h− ade6-M26 leu1 |
| F155-1D | h+ ade6-469 leu1 |
| F156-2B | h− dlc1∷ura4+ ade6-M26 leu1 |
| F153-1A | h+ dlc1∷ura4+ ade6-469 leu1 |
| F154-7B | h90 ade6-M26 leu1 |
| F156-3A | h90 dlc1∷ura4+ ade6-M26 leu1 |
| F68-12B | h90 dhc1-d2∷ura4+ ura4 leu1 |
| F136-15B | h90 dhc1-d4∷ura4+ ura4 leu1 |
| F73-3A | h90 dlc1∷ura4+ dhc1∷GFP-LEU2 ura4 leu1 |
| MB125 | h90 dhc1∷GFP-LEU2 leu1 |
| F120-23 | h90 dlc2∷kanr leu1 |
Table 2.
Oligonucleotides used
| Name | Sequencea | Use | Sense/antisense | |
|---|---|---|---|---|
| B23 | 5′ TCCCCCGGGCCAGGATATGTTAGATAAAGCTCGA 3′ | |||
| Kms1 202 a.a. | (S) | |||
| B6 | 5′ CGCGGATCCTATCAAGAAGGCTGTACCAG 3′ | |||
| Kms1 ∼607 a.a. | (A) | |||
| pCD35 | 5′ CTTCTAGGCCTGTACGGAAGTGTTACTTCTGCTCT 3′ | |||
| dlc1+ 5′ PCR and sequence: within pCD vector | ||||
| KIP41 | 5′ CTCCGTCATATTCGGAAGCCTTCAAAACCGGC 3′ | |||
| dlc1+ 5′ PCR and sequence | (A) | |||
| KIP42 | 5′ GCAGCGTGAGCTGCGTGAACGCCTCTAGAAGG 3′ | |||
| dlc1+ 5′ PCR and sequence | (A) | |||
| KIP47 | 5′ GAAGTGGACGATTCGCTGTAAGCG 3′ | |||
| dlc1+ gene colony PCR | (S) | |||
| KIP48 | 5′ GGACGAACACGAATCGTATACACACC 3′ | |||
| dlc1+ gene colony PCR | (A) | |||
| KIP56 | 5′ CGGAATTCTAGTAGCATCTTTATGTATCGC 3′ | |||
| dlc1+ gene disruption | (S) | |||
| KIP57 | 5′ GCCCCCGGGTATTTTGATGAGTACAAGG 3′ | |||
| dlc1+ gene disruption | (A) | |||
| KIP58 | 5′ ATCCCCGGGTACCAGTTTTTGC 3′ | |||
| dlc1+ gene disruption | (S) | |||
| KIP59 | 5′ TTTCTGCTCGTTACAACGACG 3′ | |||
| dlc1+ gene disruption | (A) | |||
| ura4-4 | 5′ TGCATACATATAGCCAGTGGG 3′ | |||
| colony PCR: within ura4+ gene | (A) | |||
| ura4-3 | 5′ ATGCTCCTACAACATTACCCAC 3′ | |||
| colony PCR: within ura4+ gene | (S) | |||
| KIP64 | 5′ GGGGATCCCATATGAGCTGTCCTATAGATTCC 3′ | |||
| dlc1+ Northern probe | (S) | |||
| KIP54 | 5′ AGCGGATCCTTAAATGGAGATCCACATAATGC 3′ | |||
| dlc1+ Northern probe | (A) | |||
| KIP63 | 5′ AGGCCCCGGGCATATGGAAATGGGGTATTTTGATGAGTACAAGG 3′ | |||
| GFP-tagged Dlc1: ATG → SmaI-NdeI site | (A) | |||
| KIP64 | 5′ GGGGATCCCATATGAGCTGTCCTATAGATTCC 3′ | |||
| GFP-tagged Dlc1: ATG → SmaI-NdeI site | (S) | |||
| cGFP-5′ | 5′ GGAATTCCATATGAGTAAAGGAGAAGAACTTTTCAC 3′ | |||
| GFP-tagged Dlc1: within GFP | (S) | |||
| cGFP-3′ | 5′ ACGGCCCGGGACCTTTGTATAGTTCATCCATGC 3′ | |||
| GFP-tagged Dlc1: within GFP | (A) | |||
| PDH1 | 5′ GCTGTCCCCTTAAACCCGTTCATTGC 3′ | |||
| dhc1+ Northern probe | (S) | |||
| PDH4 | 5′ TTCCCTGAACCTGTCTTGCCTAGCAG 3′ | |||
| dhc1+ Northern probe | (A) | |||
| PDE17 | 5′ GGGATTCCATATGGCAGTCATCAAGGC 3′ | |||
| dlc2+ Northern probe and GFP-tagged Dlc2 | (S) | |||
| PDE18 | 5′ ACGCGGATCCTAACCAGACTTGAACAG 3′ | |||
| dlc2+ Northern probe and GFP-tagged Dlc2 | (A) | |||
Recognition sequences of restriction enzymes are underlined. The initiation codon or termination codons are double-underlined.
Two-Hybrid System
A commercially supplied yeast two-hybrid system (Matchmaker two-hybrid system 2; CLONTECH, Palo Alto, CA) was used according to the manufacturer's protocol. A bait plasmid containing a segment of Kms1 protein (amino acid residues 202–607) fused to the Gal4 DNA-binding domain (Gal4BD) in pAS2.1 was used to screen an S. pombe cDNA library constructed with pGAD GH (CLONTECH). Various parts of Kms1 protein as well as Dlc1 were fused to the activation domain on pACT2 by using PCR-generated fragments. In some cases, the termination codon was introduced by site-directed mutagenesis. Table 2 lists the primers used in the study.
Preparation of Anti-Dlc1 Antiserum and Western Blotting
The glutathione-S-transferase (GST)-fused Dlc1 protein was produced in Escherichia coli then purified and the GST moiety was removed. Dlc1 protein thus prepared was used to immunize rabbits. To express Dlc1 in E. coli, the open reading frame (ORF) of dlc1+ cDNA was amplified by PCR with primers KIP64 and KIP54 and inserted into the NdeI/BamHI site of pET3a (Novagen, Darmstadt, Germany). For Western blot, rabbit anti-Dlc1 antiserum and rabbit anti-Sad1 antibody (Okazaki et al., 2000) were used. Dlc1 was separated on 20% SDS-PAGE with high-molarity buffers (Okajima et al., 1993) and detected using horseradish peroxidase-conjugated donkey anti-rabbit secondary antibody (Amersham Biosciences, Piscataway, NJ) and Renaissance Western Blot Chemiluminescence Reagent Plus (PerkinElmer Life Sciences, Boston, MA). S. pombe total protein was prepared by the alkaline extraction method (Silve et al., 1991; Tange et al., 1998).
Cloning of dlc1+ and dlc2+ Genes
Using an S. pombe cosmid library (Mizukami et al., 1993), we cloned the dlc1+ gene (from cosmid c1805) and the dlc2+ gene (c926). A HincII-SpeI fragment containing the dlc1+ gene was subcloned into the ars1/LEU2-based vector pALSK+ (Tanaka et al., 2000) to generate the plasmid pSD15. A HindIII fragment containing the dlc2+ gene was also cloned in this vector. The start codon in the dlc1+ gene was determined by sequencing the PCR product amplified from a full-length cDNA library (Tanaka et al., 1999) with primers pCD35 and KIP41 or KIP42. The sequence of seven independent fragments contained cDNA with identical 5′ end, suggesting that they represented the full-length cDNA. The nucleotide sequence of the full-length dlc1+ cDNA was deposited with the accession number AF196291. We first obtained a partial cDNA sequence for Dlc2 by using degenerate oligonucleotide primers designed from the conserved amino acid sequences among the 8-kDa light chain proteins (Beckwith et al., 1998) and then cloned a full-length cDNA sequence (deposited with the accession number AF197476). There were four introns in the dlc2+ gene.
Gene Disruption
To disrupt the dlc1+ gene (Figure 1B), the upstream region from the start codon and the downstream region from the termination codon were amplified using pairs of primers, KIP56/KIP57 and KIP58/KIP59, and digested with EcoRI and SmaI and with SmaI and SpeI, respectively. These two fragments and EcoRI/SpeI-digested pBluescriptII were three-piece ligated. The resulting plasmid (pSD19) had the sequence of the SmaI site, in place of the dlc1+ coding sequence, to be inserted by the S. pombe ura4+ gene cassette. The 1.8-kb HindIII fragment of the ura4+ gene was blunt-ended and inserted into the SmaI site of pSD19 to generate the pSD26 plasmid. The BglII-SpeI fragment of pSD26 containing ura4+ was introduced into Z310-13D (h90 ura4-D18 leu1). Stable Ura+ transformants were analyzed by PCR and Southern hybridization to verify correct gene disruption. To disrupt the ORF of the dlc2+ gene with the G418-resistant marker, the PCR-based gene targeting method was used (Bähler et al., 1998).
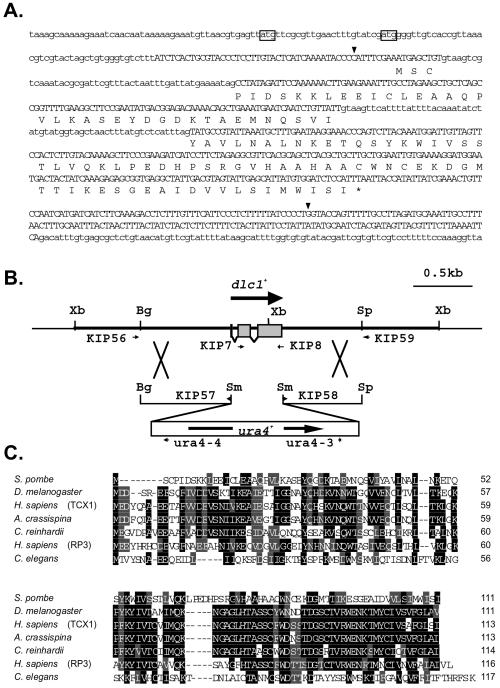
Figure 1.
dlc1+ gene and its product. (A) Nucleotides and the predicted amino acid sequences of the dlc1+ are shown. The nucleotide sequence of a full-length cDNA is shown in uppercase and the other genomic sequences, including two presumed introns, are shown in lowercase. The in-frame initiation codons upstream of the first nucleotide of the cDNA are boxed. The 5′ ends of the longest and shortest cDNAs obtained by the yeast two-hybrid screening are double underlined. The segment between the two arrowheads was replaced by the ura4+ gene cassette for gene disruption. (B) Scheme for gene disruption of the dlc1+ gene. The positions and directions of the primers are indicated by small arrows. Restriction sites shown are XbaI (Xb), BglII (Bg), SpeI (Sp), and SmaI (Sm). (C) Homology of Dlc1 with the 14-kDa dynein light chain family. Amino acid sequence of S. pombe Dlc1 is aligned with D. melanogaster Tctex1 (Y08968), Homo sapiens Tctex1 (U56255), Anthocidaris crassispina sperm flagellar outer-arm dynein LC3 (AB004251), Chlamydomonas reinhardtii inner arm dynein light chain Tctex1 (AF039437), H. sapiens rp3 (U02556), and Caenorhabditis elegans T05C12-5 ORF (Z66500). Amino acids identical among more than four proteins are shaded in black and amino acids of conservative changes are shaded in gray.
Construction of a Plasmid Expressing Green Fluorescent Protein (GFP)-tagged Dlc1
A 0.7-kb fragment in the upstream region of the start codon was amplified using primers KIP56 and KIP63, and the entire Dlc1 coding region, except for the first intron, was amplified with KIP64 and KIP59, respectively, from relevant parts of genomic DNA. These amplified fragments were digested with EcoRI and SmaI and with SmaI and SpeI, respectively, and ligated to EcoRI/SpeI digested pBluescriptII to construct plasmid pSD29. The coding sequence of GFP with improved fluorescence (Craven et al., 1998) was amplified by PCR with primers cGFP-5′ and cGFP-3′, digested with NdeI and SmaI and inserted into pSD29 digested with the same restriction enzymes. The EcoRI-SpeI fragment containing the GFP-Dlc1 fusion gene was subcloned into pALSK+ to construct plasmid pSD38.
Microscopy
Microscopic observation was performed using the Delta Vision microscope system (Applied Precision, Issaquah, WA), which allows for multicolor and three-dimensional acquisition of digitized images. For fixed cells, several Z-axis sections at 0.1-μm intervals were combined using a quick projection program. Computational removal of out-of-focus signals was not performed unless otherwise noted. For time-lapse observation of live cells, cells scraped from an agar plate were suspended in 1 μg/ml Hoechst 33342 and incubated for 5 min. Cells were collected by centrifugation, suspended in MSM medium containing 1 μg/ml Hoechst 33342, and placed onto a thin film of 2% agarose gel. The film was sandwiched with coverslips and placed on the stage. Each set of images for GFP and Hoechst 33342 were obtained with a 2-s exposure of 490 nm (neutral density filter 10%) and 0.2-s exposure of 380-nm excitations, respectively, at 25–26°C. The colors in the merged images for each wavelength in this report are all artificial.
Observation of Cells Expressing GFP-tagged Proteins
The pSD38 plasmid was introduced into S. pombe strains Z310-10B, F136-15B, Z121-4D, and F52-3A to observe GFP-Dlc1. To visualize microtubules, Z310-10B, F52-3A, and F136-15B, F143-6B were transformed with pDQ105, a multicopy plasmid that carried GFP-α-tubulin fusion gene (GFP-atb2+), which was placed under the nmt1 promoter (Ding et al., 1998) and cultivated on an EMM2 plate containing 10 μM of thiamine to allow limited expression of the fusion gene. The strain in which the dhc1+ gene was removed but expressing the GFP-tagged Dhc1 (Yamamoto et al., 1999) was crossed with appropriate strains to obtain F73-3A and MB125 (Table 1). These strains were cultivated on YE plates. Cells were induced into meiosis by placing them at high density on an ME plate for 6–12 h at 26°C.
Laser Photobleaching of GFP-labeled Microtubules
GFP-labeled microtubules were photobleached and observed on a DeltaVision microscope system in a temperature-controlled room (Haraguchi et al., 1997, 2000); experiments were performed at 26°C. For photobleaching, a nitrogen laser-pumped dye laser, Micropoint Laser System (Photonic Instrument, Arlington Heights, IL), was equipped on the DeltaVision microscope system, which was modified to remove an optical fiber illumination module, and Coumarin 440 (440-nm emission light) was used as the laser dye. The laser beam was focused ∼1 μm below the coverslip surface; the illuminated area for the photobleaching had an approximate diameter of 1 μm in the focal plane. GFP-labeled microtubules were exposed to a single pulse of the laser beam. The dynamics of the GFP-labeled microtubules was recorded on the DeltaVision microscope as described previously (Yamamoto et al., 1999, 2001). An Olympus 60×/1.4 numerical aperture PlanApo oil immersion objective lens was used for photobleaching and the following observation.
Immunofluorescence Staining and Fluorescence In Situ Hybridization
Cells were fixed with 3% formaldehyde and 0.2% glutaraldehyde. Immunostaining and in situ hybridization were performed as described previously (Okazaki et al., 2000). TAT1 mouse monoclonal anti-α-tubulin antibody and the anti-Sad1 antibody were used to stain microtubules and SPB, respectively. The secondary antibodies were Cy5-conjugated donkey anti-mouse antibody (Amersham Biosciences) and Cy3-conjugated goat anti-rabbit antibody (Jackson Immunoresearch Laboratories, West Grove, PA). DNA was stained with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI). The probes for fluorescence in situ hybridization were labeled with Cy3-conjugated or Cy5-conjugated dCTP (Amersham Biosciences). Cosmid cos212 (Funabiki et al., 1993) and plasmid YIp10.4 (Toda et al., 1984) were used for the subtelomeric repeated sequence of chromosomes I and II, and the rDNA repeat at the telomere-proximal regions of chromosome III, respectively.
RESULTS
Identification of a Protein Showing Homology with a Dynein Light Chain
We screened an S. pombe cDNA library by using the yeast two-hybrid system to identify cellular factors that interact with Kms1. Using the carboxy-terminal two-thirds of the Kms1 protein (amino acid sequence 202–607) as bait, we identified four kinds of proteins (Miki, Shimanuki and Niwa, unpublished data). The amino acid sequence of one of the proteins was similar to the 14-kDa dynein light chain family (Figure 1). It was most similar to D. melanogaster Tctex1 (25% identity and 49% similarity). Although the overall similarity to other Tctex-1 family proteins was not as high, the results obtained in the present study indicate that it is closely related to cytoplasmic dynein, hence we designated the gene dlc1+.
Characterization of dlc1+ Gene
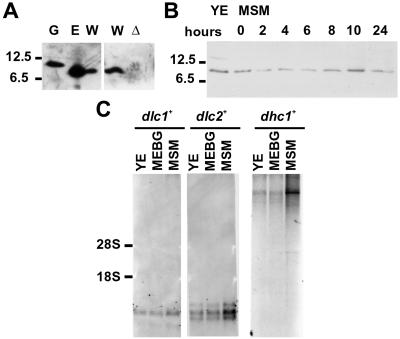
The dlc1+ gene was cloned by colony hybridization and sequenced (Figure 1A). In the genomic sequence, two additional in-frame ATG codons and therefore larger ORFs exist (Figure 1A). To determine the start codon, we sequenced full-length cDNAs obtained from another S. pombe cDNA library. None of the cDNAs contained the upstream ATG codons (Figure 1A). To verify the protein-coding sequence, the polypeptide with 111 amino acids encoded by the dlc1+ cDNAs was expressed in E. coli and compared with endogenous S. pombe Dlc1 protein in a Western blot by using anti-Dlc1 antiserum. Both proteins produced a single band that migrated to the same position (Figure 2A). The band was missing when an extract from dlc1-deleted cells (see below) was used, indicating that the antibody was specific for Dlc1 protein (Figure 2A). No extra bands were produced from extracts of cells in the sexual phase (Figure 2B). Thus, we concluded that the dlc1+ gene encodes the 111 amino acids, with a calculated molecular mass of 12.3 kDa, although it was not ruled out that a minor fraction of the gene expression might be from the larger ORFs. Note that Dlc1 protein migrated at ∼9 kDa in the SDS-polyacrylamide gel (Figure 2A). However, a bacterially produced Dlc1 with extra 16 amino acid residues (calculated molecular weight 13.6) migrated at 10.5 kDa (Figure 2A, lane G), indicating that the apparent discrepancy between the migrating position in the gel and the calculated values was probably not due to an unusual start site. The dynein heavy chain (Dhc1) is only detectable during karyogamy through early meiosis (Yamamoto et al., 1999). Consistently, the dhc1+ mRNA was massively induced during the sexual phase and the gene expression remained very low in vegetatively proliferating cells (Figure 2C). In contrast, such massive induction was not observed for the dlc1+ gene, although expression might be slightly increased during the sexual phase as shown by both Northern hybridization and Western blotting (Figure 2, B and C).
Figure 2.
Identification and expression of Dlc1. (A) Western blot analysis by using anti-Dlc1 antiserum. Purified Dlc1 protein with an additional seven amino- and nine carboxy-terminal sequences used for immunization (G), total protein of E. coli expressing the dlc1+ cDNA (E), total protein of exponentially growing wild-type fission yeast (Z310-10B) (W) or of dlc1Δ (F52-3A) cells (Δ) were separated on 20% SDS-PAGE. The positions of protein standards are shown on the left. (B) Expression level of Dlc1 protein detected by Western blot. Wild-type strains L972 and SA21 were induced to conjugate in MSM and total cell extracts from the same number of cells were prepared at the indicated times. Cells began to aggregate, conjugate, and sporulate at 6, 8, and 24 h, respectively. A control sample from exponentially growing L972 in YE was run in the left lane. (C) Northern blot hybridization with a dlc1+, dlc2+, or dhc1+ probe. Total RNA (20 μg) from wild-type homothallic strain L968 cells was loaded in each lane. RNA was extracted from cells growing exponentially in a rich medium (YE), in a poor medium (MEBG), or from cells in a conjugation medium (MSM) (38% of cells were conjugated, and among the zygotes, 30% were under the karyogamy and 70% were in the horsetail phase to meiosis I). The positions of rRNAs are shown on the left.
Disruption of dlc1+ Gene
A DNA fragment in which the whole ORF of the dlc1+ gene was replaced with a ura4+ gene cassette was used for transformation of a homothallic haploid strain (Figure 1B). Correct disruption of the gene was verified both by PCR and Southern hybridization. The dlc1-disruptant (dlc1Δ) had no discernible defect in the vegetative growth phase. The morphology and positioning of dividing nuclei were normal, based on DAPI staining. The length of the cells with the dividing nuclei was not different from that of wild-type cells. One peculiar feature we noted was that the doubling time of dlc1Δ mutants was slightly shorter than the wild-type with otherwise the same genotype, the reason for this is not known. In the sexual phase, however, the mutant had several defective phenotypes in nuclear movement, meiotic recombination, karyogamy, sporulation, and chromosome rearrangement as shown below.
Impaired Nuclear Movement in dlc1Δ Mutant during Meiotic Prophase
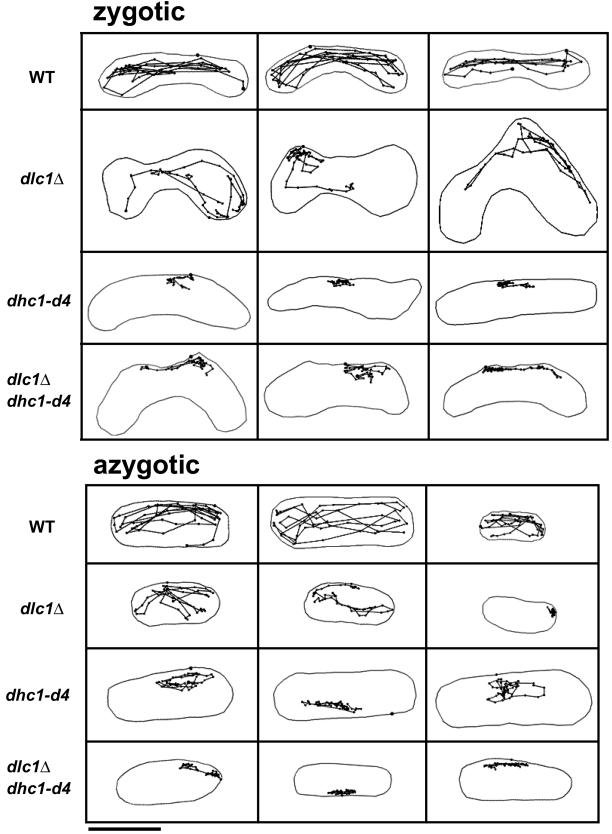
The dhc1 mutant lacks the oscillatory nuclear movement that normally occurs during prophase in wild-type meiosis (Yamamoto et al., 1999). Given the homology of Dlc1 with dynein light chain, we investigated the effect of the dlc1-null mutation on nuclear movement. Because the nuclear movement is led by the SPB that serves as the MTOC, we traced the MTOC in live cells containing GFP-labeled microtubules. The paths of the observed MTOC were projected on a plane as shown in Figure 3. Consistent with a previous report (Yamamoto et al., 1999), in both zygotic (Figure 3, top) and azygotic (Figure 3, bottom) meiosis, horsetail movement was almost absent in the dhc1-d4 mutant, although there were occasional apparently random movements.
Figure 3.
Paths of the SPB/MTOC movement during meiotic prophase. Z310-10B (WT), F52-3A (dlc1Δ), F136-15B (dhc1-d4), and F143-6B (dlc1Δ dhc1-d4) were transformed with pDQ105 and zygotic and azygotic meioses were observed in live cells. Images of GFP-α-tubulin for microtubules and of Hoechst 33342 for DNA were taken every 30 s. The position of the MTOC in each image was projected to a single plane and connected with thin lines.
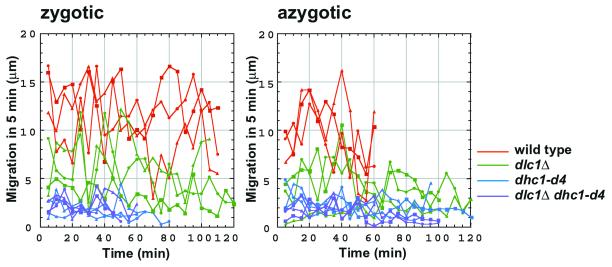
In the dlc1Δ mutant, the horsetail movement was also impaired, but apparently not as severely as in the dhc1 mutant. The profiles of the MTOC paths in dlc1 mutant cells differed from each other; nevertheless, some common features could be drawn from these profiles. First, an important feature was that when the MTOC moved, its speed often reached up to 70% of the highest speeds observed in wild type. Second, there were phases in which the MTOC slowed down or was wandering. In extreme cases, the wandering appeared to last almost throughout the prophase (Figure 3, right-most cell in azygotic meiosis). To express these features in a quantitative manner, we plotted the length of the MTOC path in 5-min intervals (Figure 4A). The average number of movements >10 μm during a 2-h observation period were 2.1 and 14.3 in the dlc1 mutant and wild type, respectively, indicating a shorter duration of the fast-moving phases in the mutant. In the dhc1 mutant, there were no movements >5 μm. Third, the regulatory mechanism for switching the direction of nuclear movement appeared to be impaired in the dlc1 mutant. The MTOC in the mutant did not follow the regular oscillatory path that occurs in wild-type cells. Another feature was that in the wandering phase, the motion of the MTOC was faster than the dhc1 mutant. Thus, abnormal behavior of the MTOC was basically similar in both zygotic and azygotic meioses.
Figure 4.
Length of the MTOC path measured at 5-min intervals from the data shown in Figure 3.
We then examined a dhc1Δ dlc1Δ double mutant to determine whether the MTOC movement in the dlc1 single mutant was dependent on dynein activity. The defect in the MTOC movement in zygotic as well as in azygotic meioses of the double mutant was indistinguishable from that in the dhc1 single mutant (Figures 3 and 4A). This result indicates the MTOC movement in the dlc1 mutant is dependent on Dhc1.
Abnormal Cytoplasmic Microtubule Arrays in Mutant
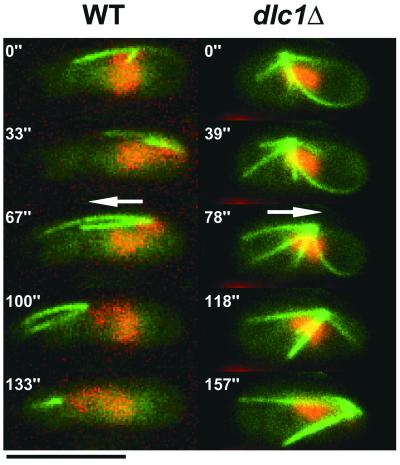
Because cytoplasmic microtubules mediate horsetail movement (Ding et al., 1998; Yamamoto et al., 2001), we observed microtubule arrays formed in the dlc1 mutant with respect to the direction of the MTOC movement, by using GFP-tagged α-tubulin. When cells with a moving SPB were arbitrarily chosen for observation, in one-third of the wild-type meioses (22/66), only microtubule(s) extending forward from the SPB were clearly visible (Figure 5), but such a conformation was never observed (0/68) in the dlc1 mutant. Instead, in 20 of 68 meioses in the mutant, prominent microtubules extending rearward with wide angles were observed but forward-extending microtubules were faint or missing (Figure 5). This type of microtubule was rarely observed (2/66) in wild type. For this observation, only one focal image was acquired at each time point (and therefore no computational image processing was applied), so that nonbundled microtubules might have been overlooked. Nevertheless, it was evident that cytoplasmic microtubule arrays formed in the dlc1 mutant were very different from those in wild type. Particularly, forward-extending microtubules, which paralleled the long axis of the cell and were apparently pulling the MTOC, were missing in the dlc1 mutant.
Figure 5.
Typical astral microtubule array formed in the prophase of a zygotic meiosis in strains with indicated genetic background. Arrows indicate the direction of SPB movement. Each image was acquired at the indicated time. Bars, 5 μm.
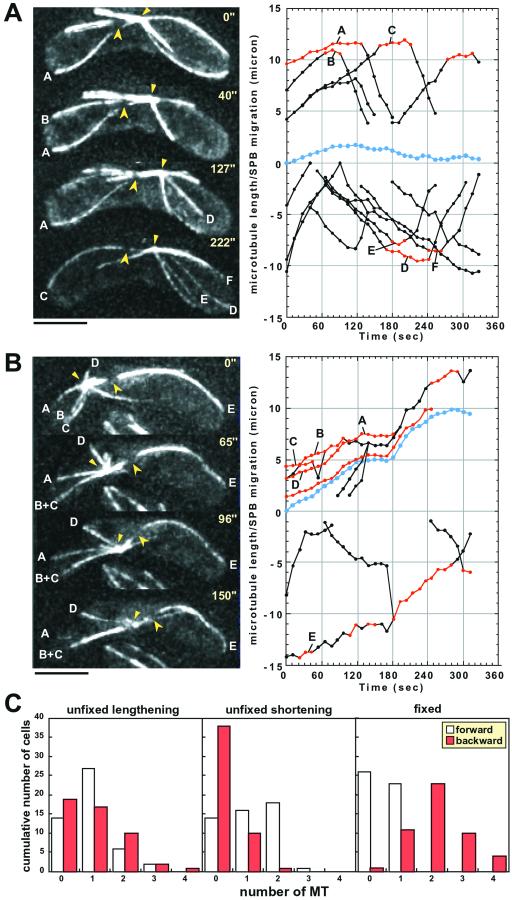
To more closely examine microtubule dynamics in the mutant, we performed a time-lapse and optical multisection observation of GFP-labeled microtubules with a minimal interval between each image acquisition. This allowed us to continuously follow the behavior of microtubule ends. Photobleaching of a microtubule segment was simultaneously performed, thus it was possible to determine at which end assembly/disassembly of microtubules took place. Previous experiments indicated that microtubule assembly and disassembly occurred only at the microtubule ends distal to the SPB both in wild type and in the dhc1 mutant (Yamamoto et al., 2001). This was also the case in the dlc1 mutant, because the distance from the bleached segment to the SPB did not change in any of the 75 cells analyzed, irrespective of whether the treated microtubules were lengthening or shortening or of the direction the SPB was moving (Figure 6). We then examined the behavior of the distal ends with regard to the direction of SPB movement. The distal ends could be fixed in position on the cell cortex or unfixed (touching or apparently not touching to the wall). Three cells with a MTOC moving at relatively high speeds (>2 μm/min) were chosen for analysis (one example is shown in Figure 6B). The number of microtubules on each side of the cell and the state of the microtubules (fixed or unfixed, lengthening or shortening) were changing with time, and the results are shown as the cumulative number of microtubules of each category obtained from a total of 49 time points (Figure 6C). As shown in Figure 6C (left), microtubules extending backward with their distal ends fixed on the cell cortex were almost always present when the MTOC was moving (Figure 6B, microtubules A–D). Such microtubules were lengthening, and therefore, appeared to be pushing the SPB. In contrast, fixed microtubules extending forward were rare. The only exceptional case in this analysis was microtubule E in Figure 6B. This microtubule was shortening more or less in conjunction with the SPB movement. It was not certain, however, whether the microtubule was engaged in pulling the SPB, judging from the orientation of the microtubule to the direction of SPB movement. For instance, in the 96-s frame in Figure 6B, the microtubule appeared to be bent due to a pushing force from the SPB. As for unfixed microtubules, they were present on both sides of the cell, although there were relatively fewer shortening microtubules on the backward side (Figure 6C, middle and right). Thus, it was likely that the nuclear movement in the dlc1 mutant was driven, at least partially, by a pushing force from the backward-extending microtubules. This appeared to also be true for slower moving MTOCs analyzed in eight cells (Figure 6A). Microtubules A and D in Figure 6A, when fixed, were lengthening in a well-coordinated manner with the SPB movement, but none were fixed and shortening. The rate of elongation of unfixed microtubules was 3.7 ± 0.9 μm/min (n = 34), which was very similar to that previously obtained for microtubules in elongation phase in wild-type cells (Yamamoto et al., 2001). On the other hand, the rate of shortening of unfixed microtubules was 6.8 ± 3.2 μm/min (n = 25). This value was also similar to that measured in wild-type cells where the SPB was not moving continuously but wandering near the ends of the cell (Yamamoto et al., 2001).
Figure 6.
Microtubule dynamics in the dlc1 mutant. (A and B) Processed images at the time indicated in seconds are shown. Small arrowheads, MTOC; large arrowheads, photobleached segments. The lengths of the microtubules, marked with letters, were measured. Bars, 5 μm. In the graphs, the lengths of individual microtubules are plotted against time. Negative values indicate that microtubules are extending rightward. Segments of lines shown in red indicate that the distal tip is fixed in position on the cell cortex. Blue lines in the middle indicate the position of the MTOC, the upper side being the right side in the pictures. (C) Three zygotes with an SPB moving at a speed of >2 μm/min were chosen and the number of microtubules in each category (fixed or unfixed, lengthening or shortening) of each cell was scored at 49 time points. Cumulative number of each type of microtubule is shown in the histograms.
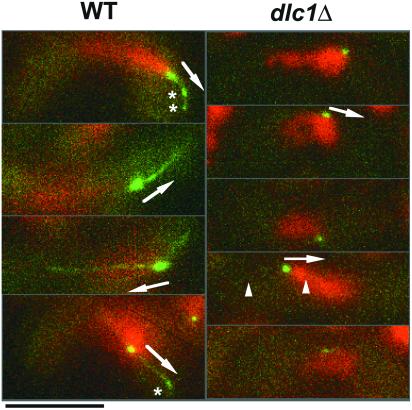
Localization of Dhc1 in dlc1 Mutant
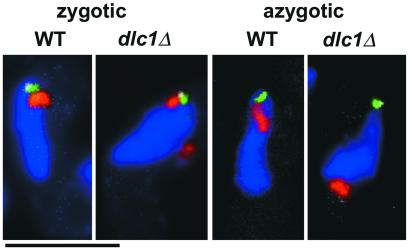
We then investigated the localization of Dhc1 in the dlc1 mutant and found that Dhc1, which was tagged with GFP (Yamamoto et al., 1999), was still present at the SPB, although the signal intensity was significantly reduced compared with wild type (Figure 7). Only occasionally was a faint fluorescent signal observed along the microtubules (Figure 7, arrowheads). Furthermore, Dhc1 signals on the cell cortex were almost invisible in the mutant. Thus, the loss of Dlc1 abolished the placement of Dhc1 to the microtubule anchors, which would be formed on the forward side of the cortex (Yamamoto et al., 1999, 2001; Figure 6, asterisks in wild type). The forward-extending microtubules in wild-type cells are linked to the cell cortex at anchoring sites and are required for normal nuclear movement (Yamamoto et al., 1999, 2001). The localization and intensity of fluorescence of GFP-Kms1 (Tange et al., 1998) were not affected by the dlc1 mutation in either vegetative or meiotic cells (our unpublished data).
Figure 7.
Reduction in localized GFP-Dhc1 in meiotic prophase nuclei in the dlc1Δ mutant. Strains MB125 (WT) and F73-3A (dlc1Δ) were observed without fixation. Representative images obtained from different cells are shown. Green, GFP-Dhc1; red, DNA. Arrows indicate the direction of nuclear movement. Asterisks indicate presumed microtubule anchors on the cell cortex; arrowheads indicate GFP signals along cytoplasmic microtubules in the dlc1 mutant. Bar, 5 μm.
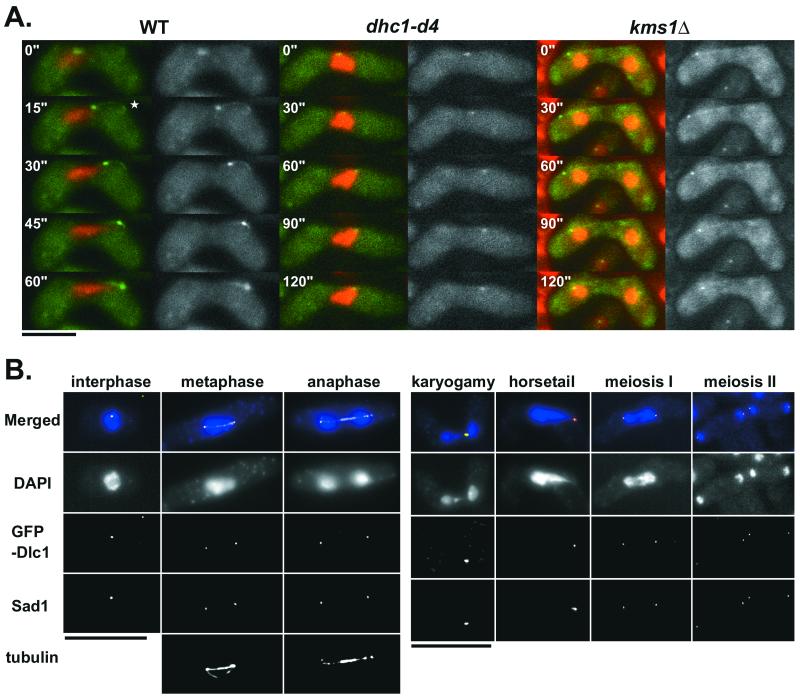
Localization of Dlc1
To analyze the localization of Dlc1, GFP was fused to the amino terminus of Dlc1. The fusion protein was expressed under the dlc1+ promoter on a multicopy plasmid. The expression level of the GFP-Dlc1 in wild-type cells was approximately twice that of the endogenous Dlc1 as estimated in a Western blot with an anti-Dlc1 antibody. This GFP-Dlc1 fusion protein was functional because the plasmid could rescue the sporulation abnormality of dlc1Δ cells (our unpublished data). In live cells expressing the fusion protein, GFP signals were most prominently observed at the leading tips of horsetail nuclei but there were also signals along cytoplasmic microtubules as well as at sites on the cell cortex (Figure 8A). The fluorescence at the leading tips overlapped with the anti-Sad1 staining (Figure 8B). Such a localization pattern was almost identical with that of GFP-Dhc1 (Yamamoto et al., 1999), except that the Dlc1 signals on the cortex were less prominent compared with Dhc1. The most remarkable difference between these proteins was that Dlc1 was present at the SPB not only during the sexual phase when Dhc1 was present (Yamamoto et al., 1999) but also during the vegetative phase as well as in a later phase of meiosis when there was little or no Dhc1 (Figure 8B). Consistently, Dlc1 in vegetative cells was localized at the SPB in the dhc1-d4 mutant at a level comparable with that in wild-type cells (our unpublished data). The vegetative localization of GFP-Dlc1 was also not affected by the kms1 mutation (our unpublished data), indicating that Dlc1 localization at the SPB during vegetative phase does not require Kms1 or Dhc1. Dlc1 was also localized at the SPB/MTOC in both dhc1 and kms1 mutants during meiotic prophase, although there might be less protein than in wild type (Figure 8A). There was very little, if any, Dlc1 on the cell cortex in the dhc1-d4 mutant. In the kms1 mutant, Dlc1 was not detected on the Sad1-containing nuclear dots that are characteristically produced in this mutant (Niwa et al., 2000; Shimanuki, unpublished data).
Figure 8.
Localization of GFP-Dlc1. (A) GFP-Dlc1 plasmid was introduced into Z310-10B (WT), F136-15B (dhc1-d4), and Z121-4D (kms1Δ), and live zygotes were observed at the indicated intervals. Chromosomes, red; GFP-Dlc1, green, also in black-and-white images on the right side of each merged image. (B) Z310-10B cells containing GFP-Dlc1 in the vegetative phase (left) and in the sexual phase (right) were fixed and stained with DAPI, anti-Sad1 antibody, and anti-tubulin TAT1 antibody simultaneously. Merged: blue (DNA), green (GFP-Dlc1), red (Sad1), and white (microtubule). Bars, 5 μm.
Reduction of Meiotic Recombination in dlc1Δ Mutant
We performed a tetrad analysis of zygotic asci to determine recombination frequencies in several chromosomal regions (Table 3). The frequencies were reduced 5.6- to 16.8-fold in the dlc1 mutant compared with those in wild-type cells. These values were comparable with those previously determined in kms1-1 and dhc1Δ mutants (Shimanuki et al., 1997; Yamamoto et al., 1999). Importantly, recombination rates between plasmids and chromosomes were only mildly affected by the dlc1 mutation as measured by the frequency of an intragenic recombination between ade6-M26 and ade6-469 (Table 4). Thus, Dlc1 appeared to have a role in the spatial arrangement of homologous chromosomes to ensure efficient meiotic recombination.
Table 3.
Tetrad analysis of genetic linkage in dlc1Δ Spores formed in zygotic asci were analyzed for the linkage of two genetic markers indicated.
| Genetic markers | Crossa | No. of tetrads
|
Genetic distance
|
|||
|---|---|---|---|---|---|---|
| PDb | Tc | NPDd | Total | cM | ||
| ura1-lys3 | w × we | 72 | 44 | 1 | 117 | 21.4 |
| Δ × wf | 72 | 40 | 3 | 115 | 38.3 | |
| w × Δg | 72 | 28 | 0 | 100 | 14 | |
| Δ × Δh | 108 | 7 | 0 | 115 | 3 | |
| ura1-pom1 | w × wi | 94 | 27 | 2 | 123 | 15.9 |
| Δ × Δj | 117 | 2 | 0 | 123 | 2.4 | |
| ura1-hus2 | w × wk | 126 | 35 | 3 | 164 | 16.2 |
| Δ × Δl | 301 | 17 | 2 | 320 | 4.5 | |
| leu1-his5 | w × wm | 42 | 38 | 1 | 81 | 27.2 |
| Δ × wn | 57 | 47 | 6 | 110 | 37.7 | |
| w × Δo | 64 | 26 | 2 | 92 | 20.7 | |
| Δ × Δp | 114 | 4 | 0 | 118 | 1.7 | |
| trp1-ade8 | w × wm | 63 | 18 | 0 | 81 | 11.1 |
| Δ × wn | 89 | 20 | 1 | 110 | 11.8 | |
| w × Δo | 77 | 14 | 1 | 92 | 10.9 | |
| Δ × Δp | 115 | 3 | 0 | 118 | 1.3 | |
| ade6-fur1 | w × wq | 63 | 39 | 1 | 103 | 21.8 |
| w × Δr | 56 | 33 | 4 | 93 | 30.6 | |
| Δ × ws | 77 | 31 | 2 | 110 | 19.5 | |
| Δ × Δt | 95 | 9 | 0 | 104 | 4.3 | |
| Centromere linkageu | ||||||
| ade6-lys1 | w × wq | 37 | 39 | 27 | 103 | 18.9 |
| w × Δr | 33 | 31 | 24 | 88 | 17.6 | |
| Δ × ws | 39 | 31 | 37 | 107 | 14.5 | |
| Δ × Δt | 42 | 8 | 54 | 104 | 3.8 | |
w × w = wild × wild; w × Δ = wild × dlc1Δ × wild; Δ × w = dlc1Δ × wild; and Δ × Δ = dlc1Δ × dlc1Δ.
Parental ditype.
Tetratype.
Nonparental ditype.
HM101 × Z439-2A.
F95-1A × Z439-2A.
HM101 × F83-15A.
F90-1D × F83-15A.
HM101 × F131-1A.
F90-1D × F132-1D.
HM101 × F133-1A.
F90-1D × F141-1C.
F80-1B × Z439-2A.
F84-5A × Z439-2A.
F80-1B × F63-1C.
F84-5A × F63-1C.
L972 × F78-18A.
L972 × F81-2D.
F79-8A × F78-18A.
F83-2A × F81-2D.
Centromere linkage = total × 50.
Table 4.
Effect of the dlc1 mutation on allelic versus ectopic recombinationa Spores produced from zygotic meioses were examined. Three independent experiments were performed. Ade+ spores were produced from intragenic recombination between ade6-469 and ade6-M26 alleles.
| Cross | No. of Ade+ spores per 105 viable spores
|
||
|---|---|---|---|
| exp. 1 | exp. 2 | exp. 3 | |
| Allelic | |||
| Wild-typeb | 765 | 658 | 630 |
| dlc1Δc | 52 | 63 | 58 |
| No. of Ade+ spores/103 viable Leu+ spores | |||
| exp. 1 | exp. 2 | exp. 3 | |
| Ectopic | |||
| Wild-typed | 52 | 46 | 44 |
| dlc1Δe | 23 | 13 | 27 |
Both alleles were on chromosome III (allelic) or ade6-469 was on a plasmid, whereas ade6-M26 was on chromosome III (ectopic).
F154-4B × F155-1D.
F156-2B × F153-1A.
F154-7B.
F156-3A.
The effect of dhc1 mutations on recombination in zygotic meiosis was different from that in azygotic meiosis (Yamamoto et al., 1999). In wild-type cells, recombination in azygotic meiosis is generally less frequent compared with zygotic meiosis (Yamamoto et al., 1999). In dhc1Δ strains, however, it is almost reversed, because dhc1 mutations greatly interfered with recombination in zygotic meiosis, whereas the effect was mild for azygotic meiosis. We compared the two types of meiosis in the dlc1Δ mutant by using the recombination frequency between ade6 and fur1 loci with a random spore method. The dlc1 mutation had almost the same differential effect on zygotic and azygotic meioses as the dhc1 mutant (Table 5). Recombination frequency was further reduced when both of these genes were mutated (Table 5), indicating that the roles of these genes in recombination might not overlap completely. Note that the double mutant had a severe karyogamy defect, as described below, so the apparently greater reduction in recombination in zygotic meiosis might be due to the presence of spores produced from unfused nuclei.
Table 5.
Random spore analysis of genetic linkage in dlc1Δ Spores formed in zygotic and azygotic cells were analyzed for the linkage of two genetic markers.
| Genetic markers | Cross | Number of colonies
|
Genetic distance (cM)a | |||
|---|---|---|---|---|---|---|
| Ade− | Ade+ Fur− | total Fur− | Fur− | |||
| Zygotic | w × wb | 1386 | 5361 | 6747 | 20.5 | |
| dlc1Δ × dlc1Δc | 108 | 4816 | 4924 | 2.2 | ||
| dhc1-d4Δ × dhc1-d4Δd | 85 | 2679 | 2764 | 3.1 | ||
| dlc1Δ dhc1-d4Δ × dlc1Δ dhc1-d4Δe | 229 | 50842 | 5107 | 0.4 | ||
| Azygotic | w/wb | 130 | 1200 | 1330 | 9.8 | |
| dlc1Δ/dlc1Δc | 319 | 6311 | 6630 | 4.8 | ||
| dhc1-d4Δ/dhc1-d4Δd | 93 | 1054 | 1147 | 8.1 | ||
| dlc1Δ dhc1-d4Δ/dlc1Δ dhc1-d4Δe | 35 | 1435 | 1470 | 2.4 | ||
Ade+ Fur−/total Fur− × 100.
F109-2B × F109-2A.
F96-2C × F96-2A.
F170-1A × F171-1C.
F168-1A × F169-1A. Only haploid colonies were scored.
Relocation of rDNA toward SPB Was Impaired in dlc1 Mutant
We examined the effect of the dlc1Δ mutation on clustering of telomeres beneath the SPB during the meiotic prophase by fluorescence in situ hybridization by using cos212 probe for telomeres of chromosomes I and II and an rDNA probe for telomeres of chromosome III. Consistent with previous reports, in almost all wild-type nuclei, telomeres made a single cluster in both zygotic and azygotic meioses (Figure 9 and Table 6). In zygotic meiosis in the dlc1 mutant, 20% of meiotic prophase nuclei contained two separated rDNA spots, of which one located near the SPB and the other located away from it (Figure 9, left). The loss of the heavy chain results in a similar defect (Yamamoto et al., 1999). We repeated this experiment and 6 of 20 dhc1-d2 zygotes had the same abnormality in rDNA positioning. The dlc1 mutation also affected the positioning of rDNA in azygotic meiosis. In more than a quarter of the cases, a single rDNA signal was located apart from the cluster of other telomeres (Figure 9, right, and Table 6). Telomere clusters are formed through relocation of telomeres toward the SPB that occurs shortly after the induction to sexual phase, e.g., it occurs before nuclear fusion in zygotic meiosis (Chikashige et al., 1997). Thus, the defective phenotype regarding the rDNA positioning might be most plausibly explained if the relocation process is affected by the dlc1 mutation. Consistently, zygotes containing two haploid nuclei undergoing karyogamy, often contained rDNA at the tailing part of the nuclei, an abnormal configuration only rarely observed in wild-type nuclei (our unpublished data). It was not determined, however, whether the problem of telomere relocation is specific for chromosomes III.
Figure 9.
Defective rDNA relocation in the dlc1Δ mutant. Zygotic or azygotic meiotic prophase cells were stained with Cy5-rDNA (telomeres of chromosome III, red), Cy3-labeled cos212 (telomeres of chromosomes I and II, green), anti-Sad1 antibody (white), and DAPI (blue). The anti-Sad1 signal is closely associated with the cos212 signal in these figures. WT, Z310-10B; dlc1Δ, F52-3A. Bar, 10 μm.
Table 6.
Positioning of telomeres in meiotic prophase nuclei
| Type of meiosis | Telomere positioning | Wild-typea | dlc1Δa |
|---|---|---|---|
| Zygotic | All telomeres clustered | 75 | 40 |
| One of two rDNAs detachedb | 0 | 10 | |
| Azygotic | All telomeres clustered | 83 | 37 |
| rDNA at the opposite endb | 0 | 21 |
Number of cells with indicated telomere configuration. Cells with clustered cos212 probe signal (telomeres of chromosomes I and II) were scored.
See Figure 5.
Both Karyogamy and Sporulation Were Partially Defective in dlc1 Mutant
The dlc1 mutants had partially defective karyogamy and sporulation (Table 7). Because very few zygotes proceeded to meiosis I without nuclear fusion, it is likely that the higher incidence of unfused nuclei in the dlc1 mutant zygotes was indicative of a delay in the nuclear fusion process. Experiments using the mei1 mutant, which blocks entry into meiosis, supported this hypothesis (our unpublished data). The delay in the nuclear fusion process was also consistent with a defect in karyogamy in the dhc1 mutant (Yamamoto et al., 1999). We next examined a dlc1 dhc1 double mutant. In this strain both karyogamy and sporulation were impaired much more severely than in either of the single mutants (Table 7), consistent with the fact that these two genes are not involved in completely redundant functions as previously described for recombination.
Table 7.
Karyogamy and spore formation in dlc1Δ and dhc1Δ zygotes Cells were sporulated in MEB-Gal at 26°C for 24 h.
| Straina | Karyogamyb | Spore formationc | Percentage of asci containingd
|
||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | >4 spores | |||
| Zygotic | |||||||
| Wild-type | 90 | 84 | 0 | 1 | 0.5 | 99 | 0 |
| dlc1Δ | 55 | 81 | 11 | 30 | 8 | 51 | 0 |
| dhc1Δ | 44 | 71 | 9 | 20 | 10 | 58 | 3 |
| kms1Δ | nde | 69 | 22 | 33 | 11 | 33 | 1 |
| dlc1Δ dhc1Δ | 3 | 47 | 49 | 33 | 10 | 7 | 0 |
| dlc1Δ kms1Δ | nd | 38 | 76 | 20 | 3 | 1 | 0 |
| dhc1Δ kms1Δ | nd | 68 | 28 | 27 | 16 | 29 | 0.5 |
| Azygotic | |||||||
| Wild-type | 81 | 0 | 0 | 1 | 99 | 0 | |
| dlc1Δ | 43 | 7 | 15 | 6 | 72 | 0 | |
| dhc1-d4Δ | 62 | 1 | 3 | 6 | 90 | 0 | |
| dlc1Δ dhc1-d4Δ | 27 | 23 | 32 | 13 | 31 | 0 | |
Strains used were Z310-10B (wild-type), F52-3A (dlc1Δ), F68-12B (dhc1-d4), Z121-4D (kms1Δ), F71-11D (dlc1Δ kms1Δ), F143-6B (dlc1Δ dhc1-d4), and F167-7B (kms1Δ dhc1-d4).
Percentage of binucleated conjugants, but only those without a meiosis I spindle.
Percentage of sporulated zygotes in total zygotes.
More than 200 sporulated cells were counted.
Not determined.
Identification of Fission Yeast Gene Encoding a Homolog of 8-kDa Dynein Light Chain
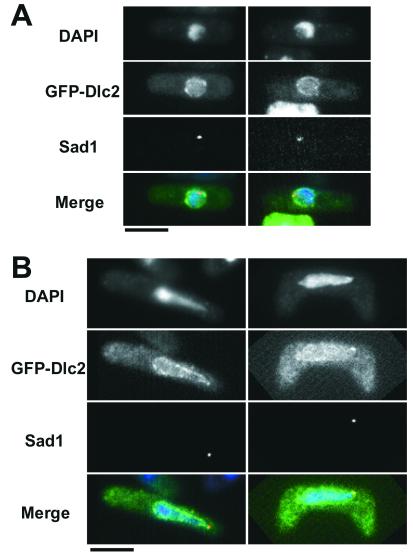
In addition to the 14-kDa light chain homolog Dlc1, fission yeast carries a gene, which we named dlc2+, that encodes a protein highly homologous to the 8-kDa dynein light chain. We cloned and sequenced the dlc2+ gene and determined the cDNA sequence as well. Dlc2 consisted of 85 amino acid residues and its predicted molecular weight was 9.8 kDa. The sequence was 78% identical with that of the 8-kDa light chain in D. melanogaster. The dlc2+ gene was transcribed during the vegetative phase and slightly induced in the sexual phase (Figure 2C). Dlc2 was tagged with GFP to examine its localization. As shown in Figure 10A, GFP-Dlc2 was enriched at the nuclear periphery. In meiotic prophase, the GFP signal was also abundant at the nuclear periphery, as well as near the SPB (Figure 10B). We then generated a dlc2-deleted allele and found that the dlc2Δ mutant did not have any notable defective phenotype in either the vegetative or the sexual phase, except that recombination frequency in zygotic meiosis was marginally reduced (for the ade6-fur1 interval, it was three-fourths of the wild-type value). Thus, the 8-kDa dynein light chain is not essential in fission yeast.
Figure 10.
Localization of GFP-Dlc2. GFP-Dlc2 plasmid was introduced into a wild-type strain (Z310-10B) and fixed for microscopic observation. Merged: blue (DNA), green (GFP-Dlc2), and red (Sad1). (A) Vegetative cells. (B) Zygotes containing a horsetail nucleus. Bar, 5 μm.
DISCUSSION
The present study identified fission yeast genes dlc1+ and dlc2+ whose products had homology to the 14- and the 8-kDa dynein light chain family proteins, respectively. We were unable to detect any defective phenotypes associated with the loss of Dlc2. We observed a subtle effect on recombination in a dlc2Δ mutant, but the exact nature of the effect remains to be determined. Thus, we conclude that despite its high sequence homology to the 8-kDa proteins, and in contrast to other organisms, Dlc2 is not essential in the normal fission yeast life cycle. The localization of Dlc2 in the meiotic prophase partly overlaps with that of Dhc1/Dlc1. Therefore, it is possible that Dlc2 in this sexual phase exists in the dynein complex, but further study is needed to elucidate the significance of the colocalization.
Dlc1 is also not essential for vegetative growth, but it is required for several sexual processes in fission yeast. First, the dlc1Δ mutation affects dynein-dependent nuclear movement in the meiotic prophase; more specifically, the direction and duration of the nuclear movement are irregular. The mutation also brings about the displacement of Dhc1 from the microtubule anchors on the cortex and the failure in the formation of “directing microtubules” that would pull the SPB (Yamamoto et al., 2001). In wild-type cells, the directing microtubules shorten from the distal ends in a coordinated manner with the SPB movement. After the SPB reaches the cell end, microtubules extend toward the opposite cell end to establish lateral binding with newly formed anchors so that the oscillatory movement will continue. Thus, the regular oscillatory nuclear movement appears to be ensured basically by the alternating assembly/disassembly of the dynein-containing anchors, although how this is achieved is not known (Yamamoto et al., 2001). Results in the present study suggest that Dlc1 is required for the positioning and/or assembly of the anchors. One possibility is that Dlc1 is a component of cytoplasmic dynein complex and mediates binding to a cargo molecule on the cell cortex directly, as in the case of rhodopsin vesicles (Tai et al., 1999), or indirectly through interacting with the IC (Mok et al., 2001). It cannot be ruled out, however, that Dlc1 has an indirect role in the localization of Dhc1; for example, Dlc1 might be required for the stability of the anchor or microtubules.
We demonstrated that the SPB movement in the dlc1 mutant was still dependent on Dhc1. Therefore, a certain form of cytoplasmic dynein that lacks Dlc1 must be participating in the nuclear movement. Because some Dhc1 was present at the SPB, we tested one possibility that the sites of assembly/disassembly were changed from the distal ends to the proximal ends of the microtubules. This possibility was not supported by the bleaching experiment. The assembly/disassembly of microtubules occurred only at the distal ends in the dlc1 mutant, similar to the dhc1 mutant and wild-type cells. The rates of elongation/shortening of unfixed microtubules were not coordinated with the SPB motion. The SPB movement in the dlc1 mutant was always accompanied by lengthening microtubules whose distal ends were fixed on the cell cortex. These facts strongly suggest that microtubules extending backward with fixed distal ends are at least partly responsible for the SPB movement in the dlc1 mutant, although it could not be ruled out that unfixed microtubules somehow contribute to generate a force to drive the SPB. Microtubules with fixed distal ends might exist in wild-type cells, but their role in nuclear movement seems to be marginal compared with the prevailing pulling microtubules. Moreover, similarly fixed microtubules are present in a dhc1 mutant and are engaged in residual nuclear movement (Yamamoto et al., 2001). This suggests that Dhc1 is not involved in the fixation of the microtubular ends. At present, it is not known how Dhc1 or dynein are involved in the nuclear movement in the dlc1 mutant. Dhc1 is noticeably present only at the SPB. It cannot be ruled out, however, that Dlc1 might be present at the fixed distal ends of microtubules on the cortex, although it must be in an amount too small to be detected. If this is the case, it is conceivable that Dhc1 enhances the elongation and/or stability of the microtubular ends and contributes to the nuclear movement in the dlc1 mutant. Other factors that could influence nuclear motion, the nucleation at the SPB and the bundling of microtubules, might also be affected by Dhc1.
Another important finding in this study is that Dlc1 is required for efficient meiotic recombination. It has been argued that the horsetail nuclear movement is required for efficient meiotic recombination because recombination is reduced in dhc1 mutants that lack nuclear movement (Yamamoto et al., 1999; Yamamoto and Hiraoka, 2001). Although Dlc1 is related to Dhc1 and its loss brings about impaired nuclear movement, it is unlikely that the reduced recombination in the dlc1 mutant is attributable solely to the problem in nuclear motion. The loss of Dlc1 function resulted in two- to threefold and 10-fold reduction in recombination frequency in azygotic and zygotic meiosis, respectively, in both wild-type and dhc1 mutant background (Table 5). Importantly, the dlc1 dhc1 double mutant completely lacked horsetail movement like in the dhc1 single mutant, indicating that Dlc1 functions for efficient recombination even in the absence of nuclear movement and its function for recombination is at least partly independent of Dhc1. It is currently unknown why the lack of Dlc1 affects recombination. As shown in the present study, its effect was mainly on homologous chromosome recombination but not on recombination between a plasmid and a chromosome; thus, it is probable that Dlc1 is required for efficient homologous chromosome pairing. Accordingly, relocation of rDNA, the marker of chromosome III telomeres, was impaired in the dlc1 mutant. It remains to be addressed, however, whether the alignment of homologous chromosomes is actually affected by the mutation and also how Dlc1 is involved in the arrangement of chromosomes during the early phase of meiosis. In addition to the role in recombination, we found that Dlc1 is involved in karyogamy and sporulation in a Dhc1-independent manner. Because the SPB appears to be the only site of Dhc1-independent localization of Dlc1, we believe that such Dhc1-independent functions of Dlc1 are executed at the SPB. This notion will be addressed experimentally in future studies.
ACKNOWLEDGMENTS
We thank Hiroyuki Tanaka and Hiroto Okayama (The University of Tokyo, Graduate School of Medicine, Tokyo, Japan) for providing the S. pombe cDNA libraries and Gerald Smith (Fred Hutchinson Cancer Research Center, Seattle, WA) for strains and plasmids. This work was supported by grants from the Kazusa DNA Research Institute Foundation and the Ministry of Education, Science and Culture of Japan.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–11–0543. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–11–0543.
REFERENCES
- Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Beckwith SM, Roghi CH, Liu B, Morris RN. The “8-kD” cytoplasmic dynein light chain is required for nuclear migration and for dynein heavy chain localization in Aspergillus nidulans. J Cell Biol. 1998;143:1239–1247. doi: 10.1083/jcb.143.5.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caggese C, Moschetti R, Ragone G, Barsanti P, Caizzi R. dtctex-1, the Drosophila melanogaster homolog of a putative murine t-complex distorter encoding a dynein light chain, is required for production of functional sperm. Mol Genet Genomics. 2001;265:436–444. doi: 10.1007/s004380000431. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Ding D-Q, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, Hiraoka Y. Telomere-led premeiotic chromosome movement in fission yeast. Science. 1994;264:270–273. doi: 10.1126/science.8146661. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Ding D-Q, Imai Y, Yamamoto M, Haraguchi T, Hiraoka Y. Meiotic nuclear reorganization: switching the position of centromeres and telomeres in the fission yeast Schizosaccharomyces pombe. EMBO J. 1997;16:193–202. doi: 10.1093/emboj/16.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JP, Watanabe Y, Nurse P. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature. 1998;392:828–831. doi: 10.1038/33947. [DOI] [PubMed] [Google Scholar]
- Craven RA, Griffiths DJF, Sheldrick KS, Randall RE, Hagan IM, Carr AM. Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene. 1998;221:59–68. doi: 10.1016/s0378-1119(98)00434-x. [DOI] [PubMed] [Google Scholar]
- Dick T, Ray K, Salz HK, Chia W. Cytoplasmic dynein (ddlc1) mutations cause morphogenetic defects and apoptotic cell death in Drosophila melanogaster. Mol Cell Biol. 1996;16:1966–1977. doi: 10.1128/mcb.16.5.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D-Q, Chikashige Y, Haraguchi T, Hiraoka Y. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J Cell Sci. 1998;111:701–712. doi: 10.1242/jcs.111.6.701. [DOI] [PubMed] [Google Scholar]
- Funabiki H, Hagan I, Uzawa S, Yanagida M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi T, Kaneda T, Hiraoka Y. Dynamics of chromosomes and microtubules visualized by multi-wavelength fluorescence imaging in living mammalian cells: effects of mitotic inhibitors on cell cycle progression. Genes Cells. 1997;2:369–380. doi: 10.1046/j.1365-2443.1997.1280326.x. [DOI] [PubMed] [Google Scholar]
- Haraguchi T, Koujin T, Hayakawa T, Kaneda T, Tsutsumi C, Imamoto N, Akazawa C, Sukegawa J, Yoneda Y, Hiraoka Y. Live fluorescence imaging reveals early recruitment of emerin, LBR, RanBP2, and Nup153 to reforming functional nuclear envelopes. J Cell Sci. 2000;113:779–794. doi: 10.1242/jcs.113.5.779. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y. Meiotic telomeres: a matchmaker for homologous chromosomes. Genes Cells. 1998;3:405–413. doi: 10.1046/j.1365-2443.1998.00205.x. [DOI] [PubMed] [Google Scholar]
- Karki S, Holzbaur EL. Cytoplasmic dynein and dynactin in cell division and intracellular transport. Curr Opin Cell Biol. 1999;11:45–53. doi: 10.1016/s0955-0674(99)80006-4. [DOI] [PubMed] [Google Scholar]
- King SM, Barbarese E, Dillman JF, III, Patel-King RS, Carson JH, Pfister KK. Brain cytoplasmic and flagellar outer arm dyneins share a highly conserved Mr 8,000 light chain. J Biol Chem. 1996a;271:19358–19366. doi: 10.1074/jbc.271.32.19358. [DOI] [PubMed] [Google Scholar]
- King SM, Barbarese E, Dillman JF, III, Benashski SE, Do KT, Patel-King RS, Pfister KK. Cytoplasmic dynein contains a family of differentially expressed light chains. Biochemistry. 1998;37:15033–15041. doi: 10.1021/bi9810813. [DOI] [PubMed] [Google Scholar]
- King SM, Dillman JF, III, Benashski SE, Lye RJ, Patel-King RS, Pfister KK. The mouse t-complex-encoded protein Tctex-1 is a light chain of brain cytoplasmic dynein. J Biol Chem. 1996b;271:32281–32287. doi: 10.1074/jbc.271.50.32281. [DOI] [PubMed] [Google Scholar]
- Lader E, Ha H-S, O'Neill M, Artzt K, Bennett D. tctex-1: a candidate gene family for a mouse t complex sterility locus. Cell. 1989;58:969–979. doi: 10.1016/0092-8674(89)90948-3. [DOI] [PubMed] [Google Scholar]
- Miyata M, Doi H, Miyata H, Jonson BF. Sexual co-flocculation by heterothallic cells of the fission yeast Schizosaccharomyces pombe modulated by medium constituents. Antonie Leeuwenhoek. 1997;71:207–215. doi: 10.1023/a:1000166426509. [DOI] [PubMed] [Google Scholar]
- Mizukami T, Chang WI, Garkavtsev I, Kaplan N, Lombardi D, Matsumoto T, Niwa O, Kounosu A, Yanagida M, Marr TG, Beach D. A 13 kb resolution cosmid map of the 14 Mb fission yeast genome by nonrandom sequence-tagged site mapping. Cell. 1993;73:121–132. doi: 10.1016/0092-8674(93)90165-m. [DOI] [PubMed] [Google Scholar]
- Mok YK, Lo KW, Zhang M. Structure of Tctex-1 and its interaction with cytoplasmic dynein intermediate chain. J Biol Chem. 2001;276:14067–14074. doi: 10.1074/jbc.M011358200. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of the fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Nimmo ER, Pidoux AL, Perry PE, Allshire RC. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature. 1998;392:825–828. doi: 10.1038/33941. [DOI] [PubMed] [Google Scholar]
- Niwa O, Shimanuki M, Miki F. Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. EMBO J. 2000;19:3831–3840. doi: 10.1093/emboj/19.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okajima T, Tanabe T, Yasuda T. Non-urea sodium dodecyl sulfate-polyacrylamide gel electrophoresis with high-molarity buffers for the separation of proteins and peptides. Anal Biochem. 1993;211:293–300. doi: 10.1006/abio.1993.1272. [DOI] [PubMed] [Google Scholar]
- Okazaki K, Okayama H, Niwa O. The polyubiquitin gene is essential for meiosis in fission yeast. Exp Cell Res. 2000;254:143–152. doi: 10.1006/excr.1999.4728. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Wilkerson CG, Witman GB. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT) J Cell Biol. 1998;141:979–992. doi: 10.1083/jcb.141.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillis R, Statton D, Caruccio P, Murphey RK. Mutations in the 8 kDa dynein light chain gene disrupt sensory axon projections in the Drosophila imaginal CNS. Development. 1996;122:2955–2963. doi: 10.1242/dev.122.10.2955. [DOI] [PubMed] [Google Scholar]
- Ponticelli AS, Smith GR. Meiotic recombination-deficient mutants of Schizosaccharomyces pombe. Genetics. 1989;123:45–54. doi: 10.1093/genetics/123.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimanuki M, Miki F, Ding D-Q, Chikashige Y, Hiraoka Y, Horio T, Niwa O. A novel fission yeast gene, kms1+, is required for the formation of meiotic prophase-specific nuclear architecture. Mol Gen Genet. 1997;254:238–249. doi: 10.1007/s004380050412. [DOI] [PubMed] [Google Scholar]
- Silve S, Volland C, Garnier C, Jund R, Chevallier MR, Haguenauer-Tsapis R. Membrane insertion of uracil permease, a polytopic yeast plasma membrane protein. Mol Cell Biol. 1991;11:1114–1124. doi: 10.1128/mcb.11.2.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai AW, Chuang JZ, Bode C, Wolfrum U, Sung CH. Rhodopsin's carboxy-terminal cytoplasmic tail acts as a membrane receptor for cytoplasmic dynein by binding to the dynein light chain Tctex-1. Cell. 1999;97:877–887. doi: 10.1016/s0092-8674(00)80800-4. [DOI] [PubMed] [Google Scholar]
- Tai AW, Chuang JZ, Sung CH. Localization of Tctex-1, a cytoplasmic dynein light chain, to the Golgi apparatus and evidence for dynein complex heterogeneity. J Biol Chem. 1998;273:19639–19649. doi: 10.1074/jbc.273.31.19639. [DOI] [PubMed] [Google Scholar]
- Tai AW, Chuang JZ, Sung CH. Cytoplasmic dynein regulation by subunit heterogeneity and its role in apical transport. J Cell Biol. 2001;153:1499–1509. doi: 10.1083/jcb.153.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Tanaka K, Murakami H, Okayama H. Fission yeast cdc24 is a replication factor C- and proliferating cell nuclear antigen-interacting factor essential for S-phase completion. Mol Cell Biol. 1999;19:1038–1048. doi: 10.1128/mcb.19.2.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Yonekawa T, Kawasaki Y, Kai M, Furuya K, Iwasaki M, Murakami H, Yanagida M, Okayama H. Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol Cell Biol. 2000;20:3459–3469. doi: 10.1128/mcb.20.10.3459-3469.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange Y, Horio T, Shimanuki M, Ding D-Q, Hiraoka Y, Niwa O. A novel fission yeast gene tht1+, is required for the fusion of nuclear envelopes during karyogamy. J Cell Biol. 1998;140:247–258. doi: 10.1083/jcb.140.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Nakaseko Y, Niwa O, Yanagida M. Mapping of rRNA genes by integration of hybrid plasmids in Schizosaccharomyces pombe. Curr Genet. 1984;8:93–97. doi: 10.1007/BF00420224. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Hiraoka Y. How do meiotic chromosomes meet their homologous partners?: lessons from fission yeast. BioEssays. 2001;23:526–533. doi: 10.1002/bies.1072. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Tsutsumi C, Kojima H, Oiwa K, Hiraoka Y. Dynamic behavior of microtubules during dynein-dependent nuclear migration of meiotic prophase in fission yeast. Mol Biol Cell. 2001;12:3933–3946. doi: 10.1091/mbc.12.12.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, West RR, McIntosh JR, Hiraoka Y. A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J Cell Biol. 1999;145:1233–1249. doi: 10.1083/jcb.145.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]