Abstract
Background
Cerebral small vessel disease (CSVD) with an insidious onset can cause overall neurological dysfunction and dementia, bringing a massive burden to society. However, the pathogenesis of CSVD is complex and reliable non-invasive biomarkers for diagnosis are still not available at present. Our study aimed to investigate abnormal exosomal miRNA patterns via microarray analysis and identify candidate biomarkers for CSVD.
Methods
We isolated exosomes from the plasma of all subjects and identified exosomes via currently universally accepted methods. The miRNAs were profiled through microarrays, and then the expression of selected differentially expressed miRNAs was validated through RT-PCR. GO and KEGG analysis predicted possible functions of differentially expressed miRNAs. Receiver operating characteristic (ROC) curve was employed to observe the diagnostic value of selective miRNAs. Finally, the relationship between the expression of miR-320e and the CSVD burden was analyzed.
Results
A total of 14 miRNAs displayed differential enrichment levels with |fold change|≥1.5 and p<0.05 through miRNA microarray analysis. The RT-PCR analysis validated that exosomal miR-320e was significantly downregulated in CSVD patients (p<0.0001). ROC curve analysis of exosomal miR-320e showed the area under the curve of 0.752. According to the multivariable analysis, miR-320e was an independent predictor of white matter hyperintensity ([aOR]= 0.452, 95% confidence interval [CI]= 0.258–0.792, p=0.006) and exhibited a negative correlation with the load of periventricular white matter hyperintensities (p=0.0021) and deep white matter hyperintensities (p=0.0018), respectively. In addition, it exhibited a negative correlation with total CSVD burden score (r=−0.276, p=0.001).
Conclusion
In our study, plasma exosomal miR-320e has a certain diagnostic value for CSVD, and a significant correlation with imaging burden of CSVD. Overall, exosomal miR-320e has the potential to be a novel biomarker for CSVD, but further research with a large sample size is necessary to assess its clinical utility.
Keywords: cerebral small vessel disease, white matter hyperintensity, diagnostic biomarker, exosomal miRNAs, miRNA microarray analysis, brain neuroimaging
Introduction
Cerebral small vessel disease (CSVD) as a whole-brain disorder involves small perforating arteries, capillaries, and small veins.1 The term describes a range of neuroimaging, pathology, and associated clinical characteristics. With the population aging, the incidence of CSVD is growing, which will aggravate the global public health burden. Currently, the diagnosis and monitoring of CSVD are mainly dependent on evaluating parenchymal lesions, which are shown on brain magnetic resonance (MR) images. It is hard to directly observe small cerebral vessels on neuroimaging, which makes the early detection and mechanism research fraught with difficulties. Circulating markers are non-invasive and easily accessible, and may be ideal tools for early detection. Studies have been conducted on circulating biomarkers of CSVD.2–4
In peripheral circulation, exosomes are vesicles derived from multivesicular vesicles wrapped by lipid bilayers with a diameter of 30–150nm.5 They can be secreted by various cells, including neurons,6 widely available in the central nervous system (CNS), and involved in many neurological functions.7 Exosomal cargos usually change under varieties of physiological and pathological conditions, reflecting the state of the originating host cell, which can redirect the functions of recipient cells after being taken up.8 Among them, microRNAs (miRNAs), as small non-coding RNAs, are vital post-transcriptional regulators of target mRNA in combination with the 3′-untranslated regions (3’UTR).9 Changes in the levels of specific exosomal miRNAs can provide insights into disease status, which plays an increasingly important diagnostic and therapeutic role in neurological disorders due to their stability and ability to traverse the blood–brain barrier, especially.10–13 Previous studies have shown that exosomal miRNAs occupy a significant position in the pathogenesis, diagnosis, and treatment of cerebral infarction.14–16 Currently, there is also a rising interest about its roles in diagnosing CSVD. Studies from Josie C. van Kralingen and Zhao W suggested exosomal miRNA-17 family members and miR-223 are associated with CSVD and CSVD-related cognitive impairment, respectively.17,18 However, there is still a limited amount of studies available, and more investigations are needed to understand the roles of exosomal miRNAs in diagnosis and treatment of CSVD.
In the present study, we are aiming at (1) describing the expression profile of circulating exosomal miRNAs in CSVD patients, (2) exploring new diagnostic markers for the diagnosis of CSVD, (3) and further analyzing their correlation with imaging markers of CSVD.
Materials and Methods
Study Participants
We recruited 150 patients with CSVD and 80 control subjects who were admitted to the Neurology Department of Affiliated Hospital of Qingdao University from July 2018 to January 2020 (230 patients in total). The controls were matched for age and gender. All enrolled individuals had subjective neurological complaints, such as dizziness, vertigo, memory loss, or suspicious symptoms of brain infarction. Among them, the patients in CSVD group were individuals with recent small subcortical infarcts (RSSI) diagnosed by brain MRI, while the control group were those without acute cerebral infarction and cerebral hemorrhage based on clinical diagnosis and imaging examination. Relevant medical histories, blood tests, and imaging data were collected for analysis. This cross-sectional study was reviewed by the Affiliated Hospital of Qingdao University ethics committee (QYFY WZLL 27395). All experiments were conducted under the Declaration of Helsinki and relevant guidelines and regulations.
Inclusion criteria for patients with CSVD: The definition of RSSI in the study is consistent with the International Standards for Reporting Vascular Changes in Neuroimaging (STRIVE recommendations).19 Exclusion criteria: (1) acute phase cerebral infarction (>20 mm in diameter); (2) >50% atherosclerotic stenosis of extracranial or intracranial arteries; (3) any possibility of cardiogenic embolism, including patients with coexisting or prior atrial fibrillation, valvular disease, acute myocardial infarction, or patent foramen ovale; (4) white matter hyperintensity (WMH) of nonvascular origin, such as WMH due to epilepsy, encephalitis, normal cranial pressure hydrocephalus, multiple sclerosis, and cerebral leukodystrophy; (5) severe liver, kidney, heart, or lung injury.
Radiological Assessment
High resolution T1-weighted imaging (T1WI), T2-weighted imaging (T2WI), T2 fluid attenuation inversion recovery imaging (T2FLAIR), diffusion-weighted imaging (DWI), and susceptibility-weighted imaging (SWI) were acquired using a 3.0T MR scanner (Discovery MR750, GE Medical Systems).
Neuroimaging assessment of CSVD was based on STRIVE recommendations. The Fazekas scale was applied to grade the load of deep white matter hyperintensities (DWMH) and periventricular white matter hyperintensities (PVWMH) (grade 0–3). The total score (0–6) was used to refer to the total white matter lesion burden, with moderate-severe cerebral white matter lesions defined by a total Fazekas score of ≥3.20,21 Based on the method of the Centre for Clinical Brain Sciences (University of Edinburgh, UK),22 we rated the total MRI burden of CSVD on an ordinal scale from 0 to 4 by counting the presence of each of the 4 MRI features. The specific assignment is a score of 1 for ≥1 lacunar infarct, 1 for ≥1 cerebral microhemorrhage, 1 for the presence of moderately to severely EPVS which are located in the basal ganglia, and 1 for either (early) confluent deep WMH (Fazekas score 2 or 3) or irregular periventricular WMH extending into the deep white matter (Fazekas score 3).
Blood Preparation and Exosomes Isolation
All subjects had fasting venous blood drawn within 24h of admission, and the blood specimens were immediately centrifuged and stratified (300 g, 4°C, 10 min). The upper layer of plasma was aspirated, diluted to 10 mL with PBS buffer, then stratified by high-speed centrifugation (2000 g, 4°C, 10 min), and the sediment at the bottom of the tube was discarded. The collected plasma was stored in a refrigerator at −80 °C pending extraction of exosomes.
Exosome was isolated using total exosome isolation (from plasma) reagent (cat no.4484451, Thermo Fisher Scientific, Inc.). The above plasma samples were centrifuged at 2000g for 20min after being thawed at room temperature; immediately afterward, the supernatant was collected and clarified by centrifugation again at 10,000g for 20min. Add 100µl of PBS buffer solution to 200µl of clarified sample and make a mixture with 10µl of protein kinase K. Following incubation at 37°C for 10min, 60µl of exosome precipitation reagent was added to the mixture then incubated at 4 °C for 30 min. After being centrifuged at 10,000 g for 5 min at room temperature, the supernatant was discarded, and exosomes were resuspended in PBS buffer to obtain an exosome-containing solution. Store the isolated exosomes in a refrigerator at −80 °C pending RNA extraction.
Exosomes Identification
Transmission Electron Microscopy
20µl of exosome suspension was dropped onto a fixed carbon grid and left to stand for 20min at room temperature. Then excess exosome suspension was carefully blotted up with filter paper. 20µl of 2% phosphotungstic acid was dropped onto the carbon grid and left to stand for 20 s. Excess phosphotungstic acid was carefully blotted up with filter paper, and the grid was placed in a glass dish lined with filter paper. A transmission electron microscope (TEM) (JEOL, JEM 1400) was used to determine the morphology of the exosomes.
Nanoparticle Tracking Analysis
The particle concentration and size of exosome from isolated exosome suspensions was assessed using nanoparticle tracking analysis (NTA) with ZetaView PMX 110 (Particle Metrix, Germany) and corresponding ZetaView 8.04.02 software.
Western Blot Analysis
RIPA lysates containing PMSF were used to extract the total protein from the exosomes. The total protein concentration was determined using a BCA assay. The protein samples (25 µg) were added to 10% SDS-PAGE gels and then transferred to PVDF membranes by electrophoresis, which were incubated with CD9, CD63, TSG101 primary antibody (Abcam, United Kingdom) overnight. After being washed with TBST and incubated with HRP-coupled secondary antibody (Abcam, United Kingdom), the bands were finally detected by a chemiluminescence system.
MicroRNA Analysis by Expression Microarrays
To obtain a general profile of differential miRNAs from the plasma exosomes of CSVD patients, 16 CSVD patients and 16 controls were chosen randomly to create 4 CSVD pools and 4 control pools (each pool contained 4 blood samples). Differential expression of miRNAs between CSVD and control cohorts was examined by using Affymetrix GeneChip miRNA 4.0 Array. Raw data was normalized using RMA algorithm (affy package). Differential expression analysis of microarray data was performed with the limma package. The Shanghai GeneChem company (GeneChem, China) performed hybridization, scanning, and data analysis of the microarrays. Differentially expressed (DE) miRNAs were defined as expression differences meeting |fold change| ≥ 1.5 and p <0.05.
Target Prediction and Enrichment Analysis
We determined the target genes of the DE miRNAs (|fold change| ≥ 1.5 and p <0.05) using DIANA-microT-CDS (microT threshold 0.8; P value < 0.05; FDR correction; Conservative Stats) (https://diana.e-ce.uth.gr/home). DAVID (http://david.ncifcrf.gov/) an online bioinformatic tool was used for GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways enrichment analysis (P < 0.05) of these target genes.
Total RNA Extraction, Reverse Transcription, and Quantification of miRNA Expression by RT-PCR
Among the exosomal miRNAs with significant expression differences, miR-668-5p and miR-320e were chosen for the next validation based on previous literature. According to the supplier’s instructions, total RNA extraction was performed using the miRNeasy Plasm/Plasma Kit (QIAGEN). Then, the Reverse transcription of RNA was completed using the Mir-X miRNA rt-PCR SYBR kit (Takara). cDNA was then diluted to 100 μL, and the RUN6 gene was selected as the internal reference gene for quantitative RT-PCR to calculate the expression of the screened miRNAs. Finally, we calculated the relative expression of miRNAs using the 2−(∆∆Ct) method, and the data were logarithm-transformed with log10.
Statistical Analysis
We used SPSS (IBM Company, California, USA) and R software (4.0.2) for data analysis and graphing. The Shapiro–Wilk test was applied to determine the normality of the distribution of continuous variables. Mann–Whitney U-test was used if normality was not satisfied. Otherwise, the Student’s t-test was applied. Categorical data were tested by the chi-square test. The area under the curve (AUC) of ROC curve was measured to determine the diagnostic value of selected DE miRNAs. The association between miRNA level and the onset of moderate to severe WMH was assessed by binary logistic regression analysis. One-way analysis of variance (ANOVA) was used for the statistical analysis of three groups. Statistical correlation was tested by Spearman correlation. The expression of miRNAs was log-transformed before counting. Differences were considered significant at p < 0.05.
Results
The Identification of Plasma Exosomes
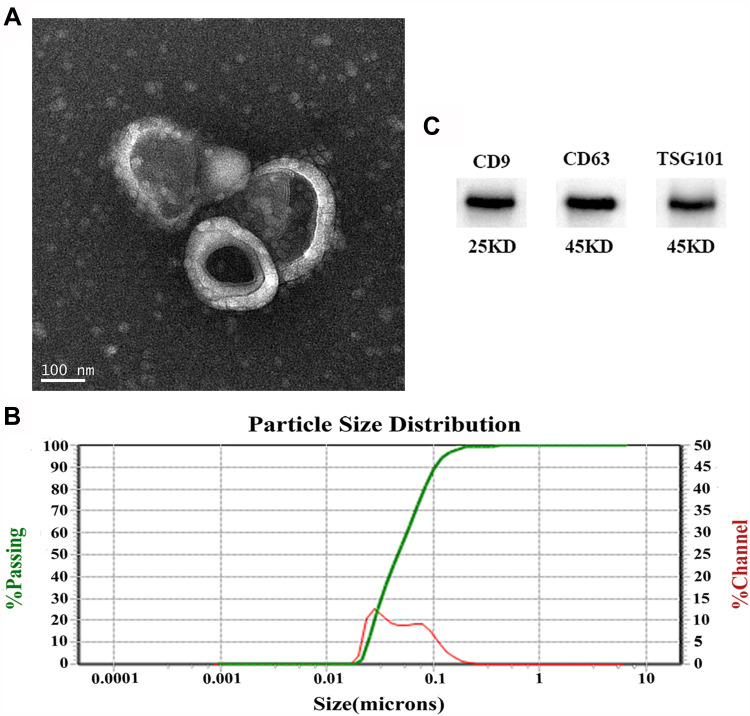
We isolated exosomes from the plasma of all subjects. The purity and integrity of the isolated exosomes were identified via TEM, NTA, and Western blotting. A typical electron microscopic image of exosomes is shown in Figure 1A, as a kind of oval or bowl-shaped microvesicles without nuclei. The exosome size and distribution were assessed using NTA (Figure 1B). Isolated exosomes ranged from 30 to 150 nm in the size distribution. Western blot analysis detected three positive protein markers associated with exosomes (CD9, CD63, and TSG101) (Figure 1C). Overall, the results of TEM, NTA, and western blotting were consistent with exosome characteristics.
Figure 1.
Characteristics of plasma-derived exosomes. (A) Representative exosome image identified by a TEM (scale bar: 100 nm). (B) Nanoparticle tracking analysis (NTA) of exosomes showed they have a diameter of 30–150 nm. (C) Western blotting identified three exosome-specific markers (CD9, CD63, and TSG101).
Baseline Clinical Characteristics of the Enrolled population
The clinical characteristics of patients with CSVD (n=150) and controls (n=80) are exhibited in Table 1. No significant differences between CSVD patients and controls were found in age (61.77±10.11 versus 62.09±13.23), gender (71[47.3%] males versus 36[45.0%] males), race, alcohol abuse, diabetes, triglyceride, total cholesterol, high-density lipoprotein, low-density lipoprotein, fibrinogen, homocysteine, and glucose. However, there were statistically significant differences in the history of hypertension, smoking history between CSVD patients and controls. Detailed demographic data are available in Supplementary Table S1.
Table 1.
Demographic Data of CSVD Patients and Controls
| Characteristics | Control (80) | CSVD (150) | p |
|---|---|---|---|
| Age | 62.09±13.23 | 61.77±10.11 | 0.982* |
| Gender (Male/Female) | 36 (45.0%) | 71 (47.3%) | 0.735† |
| Ethnicity (Han, %) | 100% | 100% | 1† |
| Hypertension (N, %) | 46 (57.5%) | 116 (77.3%) | 0.002† |
| Diabetes mellitus (N, %) | 17 (21.2%) | 49 (32.7%) | 0.077† |
| Smoking (N, %) | 11 (13.8%) | 45 (30%) | 0.017† |
| Drinking (N, %) | 12 (15%) | 13 (9%) | 0.266† |
| TG | 1.59±0.89 | 1.50±1.02 | 0.516* |
| TC | 4.43±1.07 | 4.31±1.03 | 0.421* |
| HDL | 1.30±0.40 | 1.29±0.41 | 0.880* |
| LDL | 2.65±0.72 | 2.53±0.83 | 0.253* |
| Fibrinogen | 2.50 (2.50–2.79) | 2.73 (2.33–3.17) | 0.109‡ |
| tHcy | 11.31 (9.85–12.96) | 11.57 (10.10–14.85) | 0.217‡ |
| Glucose | 5.18 (4.63–5.76) | 5.24 (4.67–6.35) | 0.332‡ |
Notes: Significant variables (p<0.05) are bolded. *Continuous variables were compared by Student’s t-test. †Categorical data were compared by the Chi-square test. ‡Continuous variables were compared by Mann–Whitney U-test.
Abbreviations: TG, triglyceride; TC, total cholesterol; HDL, high-density lipoprotein; LDL, low-density lipoprotein; tHcy, total homocysteine; CSVD, cerebral small vessel disease.
MicroRNA Expression Profile According to the CSVD
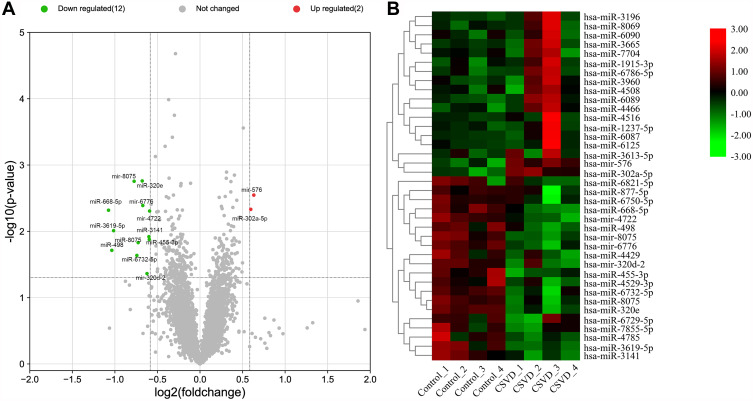
DE miRNAs were visualized by volcano plot (Figure 2A). A total of 38 miRNAs (18 upregulated and 20 downregulated) expression differences met |fold change| ≥ 1.5, of which 14 miRNAs (2 upregulated and 12 downregulated) expression differences met p < 0.05 (Table 2). The heat map exhibited hierarchical cluster expression patterns in subjects (Figure 2B).
Figure 2.
Analysis of DE miRNAs. (A) The volcano plot depicted the DE miRNAs, upregulated miRNAs and downregulated miRNAs are shown as red dots and green dots, respectively. (B) Heat maps of DE miRNAs meeting |fold change|≥1.5.
Table 2.
The Screened Differentially Expressed miRNAs in the Exosomal Samples of Patients with CSVD
| MiRNA | Fold Change | P value | Regulation |
|---|---|---|---|
| hsa-miR-320e | −1.601026847 | 0.00174 | Down |
| hsa-mir-8075 | −1.710063621 | 0.00176 | Down |
| hsa-mir-576 | 1.549797261 | 0.00284 | Up |
| hsa-mir-6776 | −1.59366401 | 0.00408 | Down |
| hsa-miR-302a-5p | 1.511890338 | 0.00466 | Up |
| hsa-miR-668-5p | −2.106565099 | 0.0048 | Down |
| hsa-mir-4722 | −1.507980215 | 0.00494 | Down |
| hsa-miR-3619-5p | −2.021634935 | 0.00976 | Down |
| hsa-miR-3141 | −1.515834765 | 0.01204 | Down |
| hsa-miR-455-3p | −1.51110876 | 0.01326 | Down |
| hsa-miR-8075 | −1.654637289 | 0.0148 | Down |
| hsa-miR-498 | −2.050939593 | 0.01934 | Down |
| hsa-miR-6732-5p | −1.672066817 | 0.0231 | Down |
| hsa-miR-320d | −1.539778146 | 0.04335 | Down |
Go and KEGG Functional Analysis of Exosomal miRNAs
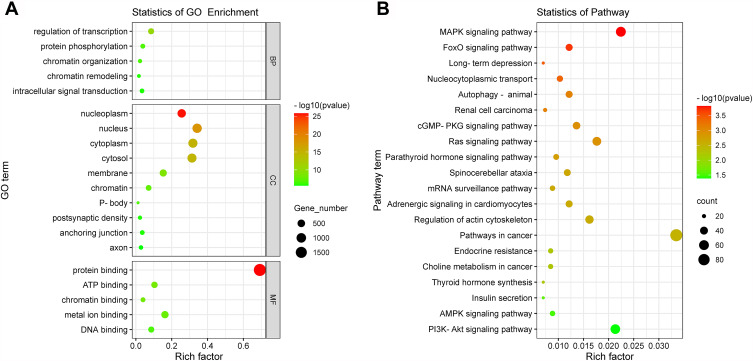
A total of 2752 DE miRNAs target genes were obtained for the subsequent functional analysis. The results are included in Supplementary Table S2. In Figure 3A, critical biological processes, molecular functions, and cellular components related to the transcription regulation and intracellular signal transduction were revealed based on GO analysis. The top 20 enriched pathways in the result of KEGG enrichment analysis are illustrated in Figure 3B, which were mainly related to the cancer, regulation of inflammatory response, metabolic, and endocrine functions. The results of enrichment analysis are shown in Supplementary Tables S3 and S4.
Figure 3.
Enrichment of target gene function of DE miRNAs. (A) Top 20 enriched GO terms including biological processes (BP), cellular components (CC), and molecular functions (MF). (B) Top 20 enriched KEGG pathways were visualized on a bubble diagram. The size of the circle shows the number of genes, and the color of the circle corresponds to the size of the P-value.
Validation of Significantly Different Expression Levels of Exosomal miRNAs
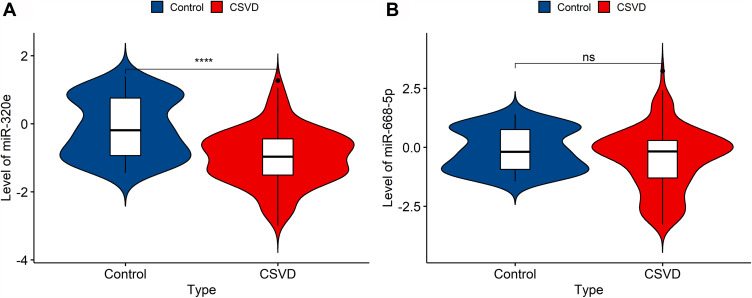
The results from miRNA microarray were validated by RT-PCR analysis. We selected miR-320e and miR-668-5p from 14 significantly different expressed miRNAs as candidate biomarkers based on microarray analysis and previous literature. The result revealed that miR-320e was significantly downregulated in plasma exosomes of CSVD patients compared to negative individuals (median, −0.968 versus −0.151; range, −2.993–1.268 versus −2.887–5.993; P<0.0001). The expression of miR-668-5p was in a downward trend but not significant (median, −0.172 versus −0.189; range, −3.282–3.237 versus −2.887–5.993; P=0.062) (Figure 4A and B).
Figure 4.
The expression of the two selective miRNAs (miR-320e, miR-668-5p) was validated by qRT-PCR. U6 snRNA was used as internal control for normalization. (A) The expression of plasma exosomal miR-320e from CSVD patients was clear downregulated (p<0.0001, Mann–Whitney test). (B) There are no significant differences in plasma exosomal miR-668-5p expression between CSVD and the control group (p=0.062, Mann–Whitney test). Violin plots showed the expression of exosomal miR-320e and miR-668-5p in patients and controls representatively. ****p < 0.0001, CSVD patients vs controls. The y-axis corresponds to miR-320e and miR-668-5p expression levels by log10 change. n = 150 in CSVD group, n = 80 in control group.
Abbreviation: ns, not statistically significant.
Diagnostic Value of Selected Exosomal miRNAs for Patient with CSVD
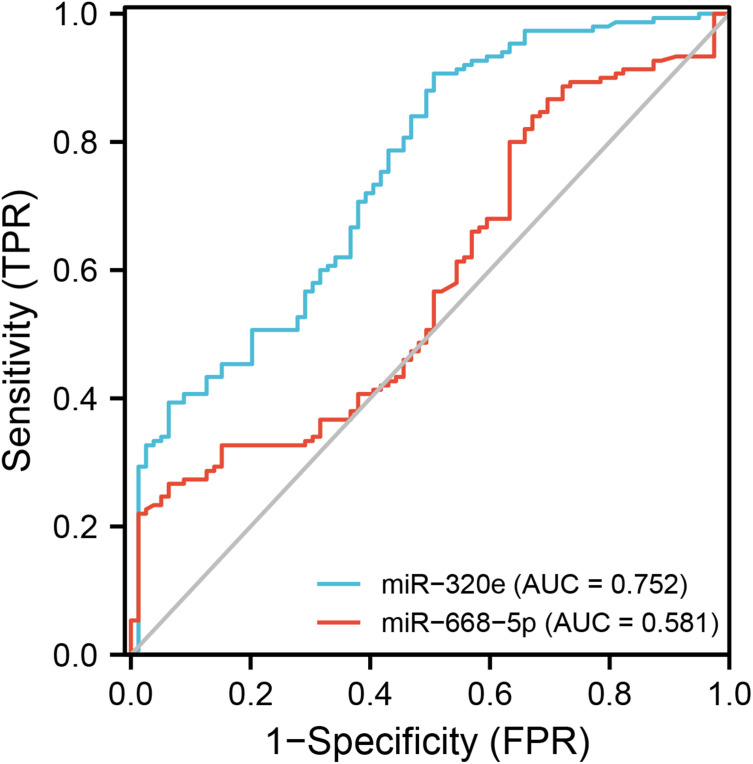
With the unmatched risk factors (hypertension and smoking history) adjusted by the binary logistic regression analysis, exosomal miR-320e was confirmed as an independent risk factor for CSVD (Table 3). We determined the sensitivity and specificity by using ROC curve analysis. Areas under the receiver-operating curve for miR-320e and miR-668-5p were 0.752 (95% CI: 0.686–0.818) and 0.581 (95% CI: 0.504–0.658), respectively (Figure 5). The best sensitivity (90.7%) and specificity (49.4%) for CSVD diagnosis when the relative expression of circulating exosomal miR-320e was taken as a cut-off value of −0.095. The result suggested a well diagnostic value of miR-320e in CSVD.
Table 3.
Exosomal miR-320e and the Risk Factors
| Items | P-value | OR | 95% CI |
|---|---|---|---|
| miR-320e | <0.001 | 0.352 | 0.239–0.519 |
| Hypertension | 0.067 | 1.871 | 0.956–3.659 |
| Smoking | 0.043 | 2.289 | 1.027–5.103 |
Notes: Exosomal miR-320e expression together with risks (Hypertension, Smoking) were analyzed in a statistical model by multivariable logistic regression.
Abbreviations: OR, odds ratio; 95% CI, 95% confidence interval.
Figure 5.
ROC curves of exosome-derived miR-320e and miR-668-5p in diagnosing CSVD.
MiR-320 and CSVD Imaging Markers
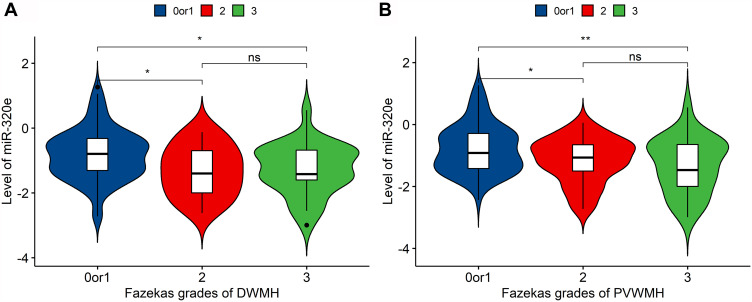
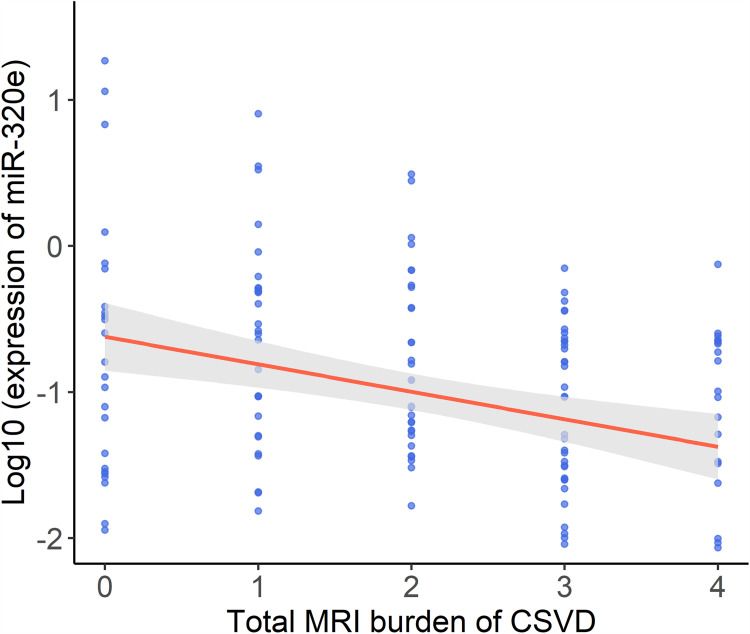
We explored the link between miR-320e and WMH severity. The patients with mild and moderate-to-severe WMH were categorized according to WMH burden as previously described. In multivariable binary logistic regression analysis, the results suggested that downregulation of miR-320e was an independent predictor of moderate-to-severe WMH (Table 4, adjusted dominance ratio [aOR]= 0.452, 95% confidence interval [CI]= 0.258–0.792, p=0.006) after adjusting for potentially confounding factors. The result in prediction of the binary logistic regression model can be seen in Supplementary Table S5. Besides, analyses showed that miR-320e was negatively correlated with Fazekas scores for DWMH (p=0.0018) and PVWMH (p=0.0021), respectively (Figure 6A and B). Furthermore, we assessed the correlation between miR-320e and the total CSVD score. The results showed a significant negative correlation between the expression of miR-320e and the total CSVD score (Figure 7, r=−0.276, p=0.001).
Table 4.
Univariable and Multivariable Logistic Regression Analyses Between Possible Predictors and Moderate-Severe WMH
| Items | Univariable Analysis | Multivariable Analysis | ||
|---|---|---|---|---|
| OR (95% CI) | p | aOR (95% CI) | p | |
| Age | 1.119 (1.071 to 1.170) | <0.001 | 1.093 (1.042 to 1.148) | <0.001 |
| Sex, male | 1.743 (0.911 to 3.338) | 0.094 | – | – |
| Hypertension | 1.821 (0.839 to 3.953) | 0.13 | – | – |
| Diabetes | 1.168 (0.548 to 2.335) | 0.66 | – | – |
| CHD | 4.465 (1.216 to 16.386) | 0.024 | 2.406 (0.512 to 11.314) | 0.266 |
| ALT | 0.966 (0.933 to 1.000) | 0.05 | – | – |
| AST | 1.009 (0.971 to 1.049) | 0.641 | – | – |
| TG | 0.766 (0.524 to 1.119) | 0.168 | – | – |
| TC | 0.878 (0.640 to 1.204) | 0.419 | – | – |
| HDL | 1.061 (0.487 to 2.310) | 0.882 | – | – |
| LDL | 0.885 (0.601 to 1.304) | 0.536 | – | – |
| Glucose | 0.941 (0.819 to 1.081) | 0.39 | – | – |
| BUN | 1.312 (1.035 to 1.663) | 0.025 | 1.239 (0.894 to 1.717) | 0.199 |
| Cr | 1.027 (1.003 to 1.052) | 0.029 | 0.986 (0.951 to 1.024) | 0.472 |
| Uric | 1.000 (0.996 to 1.004) | 0.884 | – | – |
| tHcy | 1.311 (1.150 to 1.495) | <0.001 | 1.308 (1.094 to 1.562) | 0.003 |
| Fibrinogen | 1.595 (0.948 to 2.685) | 0.079 | – | – |
| miR-320 | 0.414 (0.258 to 0.664) | <0.001 | 0.452 (0.258 to 0.792) | 0.006 |
Note: Significant variables (p<0.05) are bolded.
Abbreviations: CHD, coronary heart disease; AST, aspartate aminotransferase; ALT, alanine aminotransferase; TG, triglyceride; TC, total cholesterol; HDL, high-density lipoprotein; LDL, low-density lipoprotein; BUN, tlood urea nitrogen; Cr, creatinine; Uric, uric acid; tHcy, total homocysteine; OR, odds ratio; 95% CI, 95% confidence interval.
Figure 6.
Distribution of mean values of miR-320e according to severity of white matter hyperintensity. (A) miR-320e level was negatively correlated with Fazekas score of PVWMH (p=0.0021, ANOVA), n=79 when Fazekas score=0 or 1, n=46 when Fazekas score=2, n=25 when Fazekas score=3; (B) miR-320e level was negatively correlated with Fazekas score of DWMH (p=0.0018, ANOVA), n=97 when Fazekas score=0 or 1, n=24 when Fazekas score=2, n=29 when Fazekas score=3, **P < 0.01, *P < 0.05.
Abbreviations: PVWMH, periventricular white matter hyperintensity; DWMH, deep white matter hyperintensity; ns, not statistically significant.
Figure 7.
The correlation between exosomal miR-320e level and the total burden of CSVD. The scatter plot showing correlations between the two variables was calculated by Spearman correlation test (r=−0.276, p=0.001).
Discussion
Our study revealed the following findings: (1) There were 14 dysregulated plasma exosomal miRNAs (|fold change|≥1.5 and p<0.05, 2 were upregulated and 12 were downregulated) in CSVD patients compared with controls through a miRNA microarray in this study; (2) The results from miRNA array were validated by quantitative RT-PCR analysis in a large sample, and the results showed that miR-320e was significantly downregulated and have an ideal diagnostic efficacy of CSVD by plotting the ROC curve; (3) In further association analyses of image data, patients with high WMH burden tended to have a lower miR-320e expression in exosomes, and miR-320e expression was negatively correlated with total CSVD burden.
Aberrantly expressed miRNAs have been identified to be promising peripheral biomarkers for human diseases. The extraordinary value of miRNAs (especially exosomal miRNAs) in cerebrovascular disease has drawn wide attention during recent years. Exosomes are involved in the modulation of physiological and pathological processes after ischemic and contribute to brain remodeling by transferring their cargo. It has been well reported that exosomes and exosomal miRNAs, such as miR-223, miR-9, and miR124, can be used to identify acute ischemic stroke as well as predicting its outcome.16 Furthermore, the use of exosomal miRNAs derived from mesenchymal stem cells may provide a potential direction for stroke treatment. As to small vessel disease, the link of miRNAs and CSVD has also been revealed. Liao et al reported peripheral-blood miR-183 may worsen ischemia-reperfusion injury in patient with CSVD, and Kim et al found miR-378f mediate the pathological relationship between bone mineral density loss and the overall high burden of CSVD.23,24 Additionally, miR-184, let-7a, and let-7d have been reported to be beneficial in diagnosing and treating Fabry disease, a hereditary cerebral small vessel disease.25,26 Nevertheless, the study of circulating exosomal miRNAs in CSVD is still in its infancy. In the few studies published so far, exosomal miRNA-17 family and miR-223-3p were proved to be closely related to the initiation and development of CSVD.17,18 In the research, we explored exosomal miRNA expression profiles in CSVD patients by high-throughput screening. As miR-668 has been reported to participate in the homeostasis of the NO system, endothelial homeostasis and regulation of inflammatory after ischemia and tissue hypoxia, it suggests that miR-668 may be involved in cerebral small vessel function regulation.27 Besides, miR-668 may regulate the oxidative stress response and neuronal apoptosis after hypoxic injury in brain tissue by participating in endoplasmic reticulum stress and mitochondrial function regulation.28–30 In addition to extensive involvement in the inflammatory response, several animal experiments and in vitro cell experiments suggested the involvement of miR-320 in the proliferation, migration and apoptosis of mouse and human vascular smooth muscle cells, which in turn affected vascular remodeling after hypoxia.31,32 miR-320 also affects the expression of neurovascular unit homeostasis-related proteins such as AQP4 and MMP, and thus may play an important role in CSVD.33,34 Thus, we selected miR-668-5p and miR-320e for the next validation here. In the validation, the downregulated expression of the two miRNAs were revealed by RT-PCR. It should be noted that the differential expression of miR-668-5p was not significant, which may be due to insufficient sample size and single population. Therefore, a larger sample size may be required.
In our enrichment analysis of 14 DE miRNAs, KEGG pathway analysis revealed enrichment in inflammatory pathways, including some classical signaling pathway, such as MAPK-signaling pathway, AMPK-signaling pathway, PI3K-Akt signaling pathway, and autophagy pathway. Oxygen and glucose deprivation (OGD) can lead to the abnormal regulation of PI3K-AKT and MAPK signaling pathway in neurons, which mediated hypoxic-induced autophagy, and increase cell death during cerebral ischemia. PI3K/Akt and AMPK are also important inflammatory signaling proteins involved in neuroinflammation. In the brain, aberrant PI3K-AKT signaling promotes neurotoxicity and persistent inflammation by promoting pathogenic microglial responses.35 An abnormal inflammatory response after the cerebral ischemic injury is inextricably linked to structural damage and dysfunction of the neurovascular unit. Furthermore, the participation of MAPK-ERK1/2 and PI3K-AKT signaling pathways are essential to the formation and differentiation of nerve myelination.36 Also, enrichment was found in metabolism-related pathways, and it is noteworthy that bone metabolism markers (such as parathyroid hormone) were reported to be correlated with CSVD burden.23 Generally, pathway analysis suggested hypoxia and inflammation response in brain as well as metabolic disorders were significantly regulated by these DE miRNAs.
Previous studies showed that some miRNAs (including miR-320e) are expressed in human brain, making it possible to monitor CNS diseases status through quantitation of the miRNA with the help of plasma exosomes.37 In our study, consistent with microarray results, miR-320e was found to be significantly downregulated through validation, and showed an ideal diagnostic efficacy of CSVD in the ROC curve. MiR-320e is a member of the miR-320 family, which has been extensively implicated in inflammation and oxidative stress injury and identified an important role in many human diseases.38 In the past, the reduction of plasma miR-320e levels in acute stroke patients has been reported.39 Here, we first revealed the relationship of exosomal miR-320e with CSVD. miR-320 is an important inflammatory regulator in the brain. The underlying mechanisms of its effects in CSVD may be diverse. For example, miR-320 can target matrix metalloproteinase-9 (MMP-9) and reduce extracellular matrix damage, which is related to blood–brain barrier integrity in neuroinflammatory diseases.40 As miR-320 targets mitogen-activated protein kinase 1 (MAPK1), its downregulation increases the level of MAPK, which then regulates COX-2 expression by activating the ERK/NF-κB pathway and upregulates the expression of pro-inflammatory cytokines, such as IFN-γ, IL-6, and IL-17.41 There is evidence that these cytokines are associated with the progression of CSVD.42 miR-320 is also strongly correlated with metabolic disorders, affecting glucose and lipid metabolism in different cells, which may further lead to CSVD.43
WMH of vascular origin, as the most common sign of CSVD, was considered to account for 40% of all CSVD disease burden.19 It is an important manifestation of CSVD and greatly increases the risk of dementia, stroke, and disability. In our study, the further analysis revealed that miR-320e is associated with the load of WMH. The regulation of inflammatory mediated by miR-320e is speculated to contribute to the pathogenesis. Association of miR-320e with multiple sclerosis (an inflammation-associated CNS demyelination disease) suggests its possible involvement in regulating inflammatory responses in vascular white matter lesions.40 Several studies suggested that the inflammatory responses promoted by activated microglia after cerebral ischemia contributed significantly to the pathological changes of subsequent white matter lesions.44,45 TAK1 (transforming growth factor beta-activated kinase-1) as a master regulator of numerous inflammatory signaling molecules can cause proinflammatory (Microglia/macrophages) Mi/MΦ responses after cerebral ischemia and promotes ischemia-induced inflammation, brain injury and reduced white matter integrity.46 As a downstream target of miR-320e, selective inhibition of TAK1 by miR-320e may be an important way for miR-320e to regulate brain inflammation and the progress of WMH after cerebral ischemia.47 Additionally, miR-320 directly targets multiple genes involved in the pathological mechanism of WMH, including Aquaporin-4 (AQP4), E2F transcription factor 1 (E2F1), neuropilin-1 (NrP1), etc.33,48,49 Among them, AQP4 contributes to interstitial fluid clearance and lymphatic homeostasis, which is a key protein in several demyelinating diseases.50 As a cell cycle-related protein, E2F1 is important in neurogenesis and myelination, which has been shown to mediate the promoting effect of overexpressed p53 on oligodendrocyte apoptosis and white matter demyelination in a mouse model of spinal cord compression injury.51 The NrP1 protein enhances the phosphorylation of PDGFRα on (oligodendrocyte precursor cells) OPC in mouse brain microglia, which promotes PDGF-dependent OPC proliferation and subsequent myelin repair following acute demyelination.52 As a result, miR-320e may be involved in the progression of white matter lesions in CSVD by inflammatory pathway and targeting the above genes. It is noteworthy, cerebral atrophy is another an emerging feature of CSVD, and this atrophy of the gray matter is usually progressive and documented mainly in patients with acute stroke of the lacunar type.53 As a remote manifestation of CSVD, cortical atrophy can be caused by disruption of the cerebral white matter tracts due to lacunar infarcts and WMH, and is closely associated with CSVD-related cognitive impairment.1 Research into the relevance and potential mechanisms of exosomal miR-320e in cerebral atrophy would be valuable in the future.
Moreover, we found an association between miR-320e expression and the total CSVD burden score, which was suggested to better reflect the overall effect on the brain than considering 1 or 2 individual characteristics alone as imaging markers of CSVD were considered to interact with each other.22,54–56 The correlation between miR-320 and white matter lesions and total CSVD burden score may suggest its potential value in the study of depression, cognitive impairment, dementia, and gait abnormalities in patients with CSVD, as well as providing new ideas for further exploration of the quantitative detection of miRNAs to determine the severity of CSVD.
Our study has potential flaws: (i) This is a single-center retrospective observational study of a Han Chinese population, and the possibility of selection bias remains. (ii) Based on a cross-sectional study, no causal inference can be drawn. In subsequent studies, longitudinal and prospective studies are needed to elucidate the causal link between miR-320e and CSVD. (iii) It should be emphasized that the pathophysiology, prognosis and clinical features of acute small-vessel ischemic strokes are different from other types of cerebral infarcts, and lacunar infarcts are the stroke subtype with the best functional prognosis.57 Future studies including the population of other stroke subgroups are necessary to assess the specific role of exosomal miR-320e in the pathogenesis of CSVD. Future studies for larger samples and the molecular mechanisms for DE miRNAs are required for further elucidation.
Conclusion
In summary, we have expanded the pattern of dysregulated miRNAs in CSVD patients and provided clues for further study of disease mechanisms. Moreover, we identified exosomal miR-320e may serve as a valuable marker of CSVD. The association of exosomal miR-320e with the load of WMHs and overall CSVD burden provides insights into future research on CSVD, and may facilitate the diagnosis and treatment of these conditions.
Acknowledgments
We appreciate the cooperation of the patients who contributed data to the study and are thankful to all the nurses and colleagues who participated in the collection of blood samples and data. A special thank you to Dr Ma for critical review of the manuscript.
Funding Statement
This work was supported by grants from the National Natural Science Foundation of China (81971111) and (81971251).
Data Sharing Statement
The microarray data has been uploaded to the GEO database (GSE217872).
Any data generated in the analysis process can be requested from the corresponding author.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Written informed consent has been obtained from the patients to publish this paper.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by Affiliated Hospital of Qingdao University ethics committee (QYFY WZLL 27395).
Disclosure
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.ter Telgte A, van Leijsen EMC, Wiegertjes K, et al. Cerebral small vessel disease: from a focal to a global perspective. Nat Rev Neurol. 2018;14:387–398. doi: 10.1038/s41582-018-0014-y [DOI] [PubMed] [Google Scholar]
- 2.Lu H, Kashani AH, Arfanakis K, et al. MarkVCID cerebral small vessel consortium: II. Neuroimaging protocols. Alzheimers Dement. 2021;17:716–725. doi: 10.1002/alz.12216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poggesi A, Pasi M, Pescini F, Pantoni L, Inzitari D. Circulating biologic markers of endothelial dysfunction in cerebral small vessel disease: a review. J Cereb Blood Flow Metab. 2016;36:72–94. doi: 10.1038/jcbfm.2015.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilcock D, Jicha G, Blacker D, et al. MarkVCID cerebral small vessel consortium: I. enrollment, clinical, fluid protocols. Alzheimers Dement. 2021;17:704–715. doi: 10.1002/alz.12215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fauré J, Lachenal G, Court M, et al. Exosomes are released by cultured cortical neurones. Mol Cell Neurosci. 2006;31:642–648. doi: 10.1016/j.mcn.2005.12.003 [DOI] [PubMed] [Google Scholar]
- 7.Shi M, Sheng L, Stewart T, Zabetian CP, Zhang J. New windows into the brain: central nervous system-derived extracellular vesicles in blood. Prog Neurobiol. 2019;175:96–106. doi: 10.1016/j.pneurobio.2019.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathieu M, Martin-Jaular L, Lavieu G, Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21:9–17. doi: 10.1038/s41556-018-0250-9 [DOI] [PubMed] [Google Scholar]
- 9.Treiber T, Treiber N, Meister G. Regulation of MicroRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. 2019;20:5–20. doi: 10.1038/s41580-018-0059-1 [DOI] [PubMed] [Google Scholar]
- 10.Chen TS, Lai RC, Lee MM, Choo ABH, Lee CN, Lim SK. Mesenchymal stem cell secretes microparticles enriched in pre-MicroRNAs. Nucleic Acids Res. 2010;38:215–224. doi: 10.1093/nar/gkp857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi H, Choi K, Kim D-H, et al. Strategies for targeted delivery of exosomes to the brain: advantages and challenges. Pharmaceutics. 2022;14:672. doi: 10.3390/pharmaceutics14030672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu W-L, Lin H-W, Lin M-R, et al. Emerging blood exosome-based biomarkers for preclinical and clinical alzheimer’s disease: a meta-analysis and systematic review. Neural Regen Res. 2022;17:2381–2390. doi: 10.4103/1673-5374.335832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang P, Rasheed M, Liang J, Wang C, Feng L, Chen Z. Emerging potential of exosomal non-coding RNA in parkinson’s disease: a review. Front Aging Neurosci. 2022;14:819836. doi: 10.3389/fnagi.2022.819836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji Q, Ji Y, Peng J, et al. Increased brain-specific MiR-9 and MiR-124 in the serum exosomes of acute ischemic stroke patients. PLoS One. 2016;11:e0163645. doi: 10.1371/journal.pone.0163645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Song Y, Huang J, et al. Increased circulating exosomal MiRNA-223 is associated with acute ischemic stroke. Front Neurol. 2017;8:57. doi: 10.3389/fneur.2017.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Y, Hu Y, Xu S, Liu F, Gao Y. Exosomal MicroRNAs as potential biomarkers and therapeutic agents for acute ischemic stroke: new expectations. Front Neurol. 2021;12:747380. doi: 10.3389/fneur.2021.747380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Kralingen JC, McFall A, Ord ENJ, et al. Altered extracellular vesicle MicroRNA expression in ischemic stroke and small vessel disease. Transl Stroke Res. 2019;10:495–508. doi: 10.1007/s12975-018-0682-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao W, Sun W, Li S, et al. Exosomal MiRNA-223-3p as potential biomarkers in patients with cerebral small vessel disease cognitive impairment. Ann Transl Med. 2021;9:1781. doi: 10.21037/atm-21-6086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wardlaw JM, Smith EE, Biessels GJ, et al. Neuroimaging standards for research into small vessel disease and its contribution to ageing and neurodegeneration. Lancet Neurol. 2013;12:822–838. doi: 10.1016/s1474-4422(13)70124-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fazekas F, Chawluk JB, Alavi A, Hurtig HI, Zimmerman RA. MR signal abnormalities at 1.5 T in Alzheimer’s dementia and normal aging. AJR Am J Roentgenol. 1987;149:351–356. doi: 10.2214/ajr.149.2.351 [DOI] [PubMed] [Google Scholar]
- 21.Wu X, Ya J, Zhou D, Ding Y, Ji X, Meng R. Pathogeneses and imaging features of cerebral white matter lesions of vascular origins. Aging Dis. 2021;12:2031–2051. doi: 10.14336/AD.2021.0414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Staals J, Makin SDJ, Doubal FN, Dennis MS, Wardlaw JM. Stroke subtype, vascular risk factors, and total MRI brain small-vessel disease burden. Neurology. 2014;83:1228–1234. doi: 10.1212/WNL.0000000000000837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J-M, Park K-Y, Kim HR, et al. Association of bone mineral density to cerebral small vessel disease burden. Neurology. 2021;96:e1290–e1300. doi: 10.1212/WNL.0000000000011526 [DOI] [PubMed] [Google Scholar]
- 24.Liao Z, Sun H, Chang Y, Chen H. The expression and clinical significance of MiRNA-183 in cerebral ischemia-reperfusion injury patients with cerebral small vessel disease. Ann Transl Med. 2020;8:1005. doi: 10.21037/atm-20-5335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maier N, Gatterer C, Haider P, et al. MiRNA let-7a and let-7d are induced by globotriaosylceramide via NF-KB activation in Fabry disease. Genes. 2021;12:1184. doi: 10.3390/genes12081184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salamon I, Biagini E, Kunderfranco P, et al. Circulating MiR-184 is a potential predictive biomarker of cardiac damage in Anderson-Fabry disease. Cell Death Dis. 2021;12:1150. doi: 10.1038/s41419-021-04438-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao Z, Gao Q, Lv X. MicroRNA-668-3p protects against oxygen-glucose deprivation in a rat H9c2 cardiomyocyte model of ischemia-reperfusion injury by targeting the Stromal Cell-Derived Factor-1 (SDF-1)/CXCR4 signaling pathway. Med Sci Monit. 2020;26:e919601. doi: 10.12659/MSM.919601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luís A, Hackl M, Jafarmadar M, et al. Circulating MiRNAs associated with ER Stress And Organ Damage In A Preclinical Model Of Trauma Hemorrhagic Shock. Front Med. 2020;7:568096. doi: 10.3389/fmed.2020.568096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei Q, Sun H, Song S, et al. MicroRNA-668 represses MTP18 to preserve mitochondrial dynamics in ischemic acute kidney injury. J Clin Invest. 2018;128:5448–5464. doi: 10.1172/JCI121859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He J, Zhang X. MiR-668 inhibitor attenuates mitochondrial membrane potential and protects against neuronal apoptosis in cerebral ischemic stroke. Folia Neuropathol. 2020;58:22–29. doi: 10.5114/fn.2020.94003 [DOI] [PubMed] [Google Scholar]
- 31.Li H, Zhao J, Liu B, et al. MicroRNA-320 targeting neuropilin 1 inhibits proliferation and migration of vascular smooth muscle cells and neointimal formation. Int J Med Sci. 2019;16:106–114. doi: 10.7150/ijms.28093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding D, Jiang H, He Y, Li X, Liu X. MiR-320-3p regulates the proliferation, migration and apoptosis of hypoxia-induced pulmonary arterial smooth muscle cells via KLF5 and HIF1α. Am J Transl Res. 2021;13:2283–2295. [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Z, Yang Z, Li X, et al. MicroRNA-320a prevent müller cells from hypoxia injury by targeting aquaporin-4. J Cell Biochem. 2020;121(12):4711–4723. doi: 10.1002/jcb.29524 [DOI] [PubMed] [Google Scholar]
- 34.Liao M, Zou S, Bao Y, et al. Matrix metalloproteinases are regulated by MicroRNA 320 in macrophages and are associated with aortic dissection. Exp Cell Res. 2018;370:98–102. doi: 10.1016/j.yexcr.2018.06.011 [DOI] [PubMed] [Google Scholar]
- 35.Chu E, Mychasiuk R, Hibbs ML, Semple BD. Dysregulated phosphoinositide 3-kinase signaling in microglia: shaping chronic neuroinflammation. J Neuroinflammation. 2021;18:276. doi: 10.1186/s12974-021-02325-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prior R, Verschoren S, Vints K, et al. HDAC3 inhibition stimulates myelination in a CMT1A mouse model. Mol Neurobiol. 2022;59:3414–3430. doi: 10.1007/s12035-022-02782-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ru X, Qu J, Li Q, et al. MiR-706 alleviates white matter injury via downregulating PKCα/MST1/NF-ΚB pathway after subarachnoid hemorrhage in mice. Exp Neurol. 2021;341:113688. doi: 10.1016/j.expneurol.2021.113688 [DOI] [PubMed] [Google Scholar]
- 38.Giorgi Silveira R, Perelló Ferrúa C, Do Amaral CC, Fernandez Garcia T, de Souza KB, Nedel F. MicroRNAs expressed in neuronal differentiation and their associated pathways: systematic review and bioinformatics analysis. Brain Res Bull. 2020;157:140–148. doi: 10.1016/j.brainresbull.2020.01.009 [DOI] [PubMed] [Google Scholar]
- 39.Wang W, Sun G, Zhang L, Shi L, Zeng Y. Circulating MicroRNAs as novel potential biomarkers for early diagnosis of acute stroke in humans. J Stroke Cerebrovasc Dis. 2014;23:2607–2613. doi: 10.1016/j.jstrokecerebrovasdis.2014.06.002 [DOI] [PubMed] [Google Scholar]
- 40.Dolati S, Aghebati-Maleki L, Ahmadi M, et al. Nanocurcumin restores aberrant MiRNA expression profile in multiple sclerosis, randomized, double-blind, placebo-controlled trial. J Cell Physiol. 2018;233:5222–5230. doi: 10.1002/jcp.26301 [DOI] [PubMed] [Google Scholar]
- 41.Cheng Z, Qiu S, Jiang L, et al. MiR-320a is downregulated in patients with myasthenia gravis and modulates inflammatory cytokines production by targeting mitogen-activated protein kinase 1. J Clin Immunol. 2013;33:567–576. doi: 10.1007/s10875-012-9834-5 [DOI] [PubMed] [Google Scholar]
- 42.Noz MP, Ter Telgte A, Wiegertjes K, et al. Trained immunity characteristics are associated with progressive cerebral small vessel disease. Stroke. 2018;49:2910–2917. doi: 10.1161/STROKEAHA.118.023192 [DOI] [PubMed] [Google Scholar]
- 43.Du H, Zhao Y, Yin Z, Wang DW, Chen C. The role of MiR-320 in glucose and lipid metabolism disorder-associated diseases. Int J Biol Sci. 2021;17:402–416. doi: 10.7150/ijbs.53419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farkas E, Donka G, de Vos RAI, Mihály A, Bari F, Luiten PGM. Experimental cerebral hypoperfusion induces white matter injury and microglial activation in the rat brain. Acta Neuropathol. 2004;108:57–64. doi: 10.1007/s00401-004-0864-9 [DOI] [PubMed] [Google Scholar]
- 45.Farkas E, Annaházi A, Institóris Á, Mihály A, Luiten PGM, Bari F. Diazoxide and dimethyl sulphoxide alleviate experimental cerebral hypoperfusion-induced white matter injury in the rat brain. Neurosci Lett. 2005;373:195–199. doi: 10.1016/j.neulet.2004.10.007 [DOI] [PubMed] [Google Scholar]
- 46.Wang R, Pu H, Ye Q, et al. Transforming growth factor beta-activated kinase 1-dependent microglial and macrophage responses aggravate long-term outcomes after ischemic stroke. Stroke. 2020;51:975–985. doi: 10.1161/STROKEAHA.119.028398 [DOI] [PubMed] [Google Scholar]
- 47.Xu C, Zhang Z, Liu N, et al. Small extracellular vesicle-mediated MiR-320e transmission promotes osteogenesis in OPLL by targeting TAK1. Nat Commun. 2022;13:2467. doi: 10.1038/s41467-022-29029-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yin M, Wang X, Yao G, et al. Transactivation of MicrornA-320 by MicroRNA-383 regulates granulosa cell functions by targeting E2F1 and SF-1 proteins. J Biol Chem. 2014;289(26):18239–18257. doi: 10.1074/jbc.M113.546044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang K, Liu Z, Tang Y, et al. LncRNA NONHSAT114552 sponges MiR-320d to promote proliferation and invasion of chordoma through upregulating NRP1. Front Pharmacol. 2021;12:773918. doi: 10.3389/fphar.2021.773918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chachaj A, Gąsiorowski K, Szuba A, Sieradzki A, Leszek J. Lymphatic system in the brain clearance mechanisms - new therapeutic perspectives for Alzheimer’s disease. Curr Neuropharmacol. 2022. doi: 10.2174/1570159X20666220411091332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma L, Yu H-J, Gan S-W, et al. P53-mediated oligodendrocyte apoptosis initiates demyelination after compressed spinal cord injury by enhancing ER-mitochondria interaction and E2F1 expression. Neurosci Lett. 2017;644:55–61. doi: 10.1016/j.neulet.2017.02.038 [DOI] [PubMed] [Google Scholar]
- 52.Sherafat A, Pfeiffer F, Reiss AM, Wood WM, Nishiyama A. Microglial neuropilin-1 promotes oligodendrocyte expansion during development and remyelination by trans-activating platelet-derived growth factor receptor. Nat Commun. 2021;12:2265. doi: 10.1038/s41467-021-22532-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grau-Olivares M, Arboix A, Junqué C, Arenaza-Urquijo EM, Rovira M, Bartrés-Faz D. Progressive gray matter atrophy in lacunar patients with vascular mild cognitive impairment. Cerebrovasc Dis. 2010;30:157–166. doi: 10.1159/000316059 [DOI] [PubMed] [Google Scholar]
- 54.Jickling GC, Chen C. Rating total cerebral small-vessel disease: does it add up? Neurology. 2014;83:1224–1225. doi: 10.1212/wnl.0000000000000843 [DOI] [PubMed] [Google Scholar]
- 55.Yilmaz P, Ikram MK, Niessen WJ, Ikram MA, Vernooij MW. Practical small vessel disease score relates to stroke, dementia, and death. Stroke. 2018;49:2857–2865. doi: 10.1161/strokeaha.118.022485 [DOI] [PubMed] [Google Scholar]
- 56.Cordonnier C, Al-Shahi Salman R, Wardlaw J. Spontaneous brain microbleeds: systematic review, subgroup analyses and standards for study design and reporting. Brain. 2007;130(8):1988–2003. doi: 10.1093/brain/awl387 [DOI] [PubMed] [Google Scholar]
- 57.Rudilosso S, Rodríguez-Vázquez A, Urra X, Arboix A. The potential impact of neuroimaging and translational research on the clinical management of lacunar stroke. IJMS. 2022;23:1497. doi: 10.3390/ijms23031497 [DOI] [PMC free article] [PubMed] [Google Scholar]