Abstract
The subpellicular microtubules of the trypanosome cytoskeleton are cross-linked to each other and the plasma membrane, creating a cage-like structure. We have isolated, from Trypanosoma brucei, two related low-molecular-weight cytoskeleton-associated proteins (15- and 17-kDa), called CAP15 and CAP17, which are differentially expressed during the life cycle. Immunolabeling shows a corset-like colocalization of both CAPs and tubulin. Western blot and electron microscope analyses show CAP15 and CAP17 labeling on detergent-extracted cytoskeletons. However, the localization of both proteins is restricted to the anterior, microtubule minus, and less dynamic half of the corset. CAP15 and CAP17 share properties of microtubule-associated proteins when expressed in heterologous cells (Chinese hamster ovary and HeLa), colocalization with their microtubules, induction of microtubule bundle formation, cold resistance, and insensitivity to nocodazole. When overexpressed in T. brucei, both CAP15 and CAP17 cover the whole subpellicular corset and induce morphological disorders, cell cycle-based abnormalities, and subsequent asymmetric cytokinesis.
INTRODUCTION
The trypanosomatids are a major family of parasites responsible for important human and animal diseases (Lumsden and Evans, 1976). During its cyclical development through the mammalian host and the insect vector, the African trypanosome Trypanosoma brucei undergoes several differentiation stages defined by morphological and metabolic specificities. In all stages, the parasites share a common cell organization characterized by several organelles present as single copies within the cell (Vickerman and Preston, 1976): 1) a motile flagellum, containing a typical axoneme, that emerges from the posterior region of the cell and runs along the entire length of the cell body; 2) a typical centriolar-like organelle, the basal body, located at the base of the flagellum, which is responsible for axoneme nucleation; and 3) an unusual elongated mitochondrion whose genome is compacted in a unique structure called the kinetoplast, which is physically associated with the basal body (Robinson and Gull, 1991). Moreover, the typical cell shape of trypanosomatids is maintained by a complex subpellicular corset of cytoplasmic microtubules underlying the plasma membrane that remains intact throughout the entire cell cycle (Sherwin and Gull, 1989a).
In trypanosomes, the position, replication, and segregation of the nucleus, kinetoplast, flagellum, and basal body are highly regulated and coordinated within the cell cycle (Woodward and Gull, 1990; Kohl and Gull, 1998). Cytokinesis is initiated at the anterior end of the cell and proceeds in a linear longitudinal manner toward the posterior end. Recently, nuclear DNA synthesis inhibitors, antimicrotubule agents, and protein phosphatase inhibitors were used to investigate the regulation of the trypanosome cell cycle (Das et al., 1994; Matthews et al., 1995; Grellier et al., 1999; Ploubidou et al., 1999). It appears that cytokinesis may occur in the absence of mitosis and that basal body segregation is a critical checkpoint in cytokinesis completion.
Essential microtubule-based structures of the cytoskeleton include the flagellar axoneme, basal bodies, intranuclear mitotic spindle, and the subpellicular corset, which is a dense array consisting of up to 100 microtubules highly cross-linked by microtubule-associated proteins (MAPs). These microtubules have the same intrinsic polarity and are orientated such that the fast-growing, dynamic ends are located at the posterior end of the parasite. This posterior end is where initiation of cell elongation takes place during the growth phase of the cell cycle (Sherwin and Gull, 1989b; Robinson et al., 1995). The parasite's microtubular cytoskeleton displays unique structural and biochemical features. It is resistant to depolymerization under cold or cell fractionation conditions (Robinson et al., 1991), and its sensitivity to various microtubule-targeting drugs differs from its cytoskeleton counterpart in higher organisms (Seebeck and Gehr, 1983; MacRae and Gull, 1990). Specific studies on trypanosomal MAPs have been rare (Muller et al., 1992; Imboden et al., 1995). Nonempirical approaches have identified a few MAPs, which have subsequently been characterized and implicated in microtubule-microtubule cross-links (Balaban et al., 1989; Woods et al., 1992; Detmer et al., 1997) or microtubule-plasma membrane cross-links (Schneider et al., 1988a).
In the current study, we report the characterization of two novel, related proteins of 15- and 17-kDa isolated as stage-specific proteins and found to be associated with the subpellicullar cytoskeleton. We have investigated their respective roles and involvement in subpellicular microtubule stabilization, and subsequent cell cycle-mediated organization.
MATERIALS AND METHODS
Trypanosomes
The bloodstream forms of T. brucei (EATRO 427) were isolated from rats by ion exchange chromatography, and the procyclic forms (EATRO 1125) were cultured at 27°C in SDM-79 medium containing 10% fetal calf serum and 5 mg l−1 hemin.
Isolation of CAP15/CAP17 and Microsequencing
To enrich the 15-kDa protein (CAP15), 1010 cells (bloodstream forms) were treated with phosphate-buffered saline (PBS) (1.8 mM KH2PO4, 5 mM K2HPO4, 150 mM NaCl, pH 7.4) containing 1% (vol/vol) Triton X-100 and a cocktail of protease inhibitors (0.69 μg ml−1 pepstatin A, 0.1 μg ml−1 chymostatin, 0.43 μg ml−1 leupeptin), sonicated, and precipitated by addition of 1 volume of 10% (wt/vol) perchloric acid. After mixing for 1 h at room temperature, the cell extract was centrifuged for 30 min at 9400 × g. The supernatant was dialyzed overnight at 4°C against distilled water and lyophilized. The excess detergent was extracted by resuspending the pellets in 0.5 ml of water and 2 ml of acetone. Finally, the pellets were redissolved in 2 M guanidine-0.1% trifluoroacetic acid, loaded on a C18 reversed phase column (Vydac 218TP54; Vydac, Hesperia, CA), and the proteins separated by a linear gradient of 5% CH3CN-70% CH3CN in 0.1% trifluoracetic acid.
The 17-kDa protein (CAP17) was enriched as described (Velours et al., 1987). Procyclic cells (1010 cells) were lysed in hypotonic buffer (4.7 mM Na2HPO4, 0.3 mM KH2PO4), centrifuged, and the pellet washed twice in the same buffer, resuspended in 1 ml hypotonic buffer, and vortexed after addition of 2 ml ammonium acetate (2 M), 8 ml chloroform, and 4 ml methanol. After centrifugation for 30 min at 9400 × g, the aqueous phase was dialyzed, lyophilized, and resuspended before separation on 15% SDS-PAGE.
For microsequencing of both proteins, gel slices were obtained from Amido-Black-stained SDS-polyacrylamide gels, and peptides generated by in-gel trypsic digestion were microsequenced by J. d'Alayer (Laboratoire de Microséquençage des protéines, Institut Pasteur, Paris, France).
Cloning and Analysis of Genes Encoding CAP15/CAP17
CAP15 and CAP17 genes were identified using reverse transcription-polymerase chain reaction (PCR) approach. Single-stranded cDNA from total RNA extracted from both forms of T. brucei was synthesized using Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA). Degenerate oligonucleotides p15-2B, p17-1A, and p17-1B were used as 3′ primers and MexIII (5′-AAC GCT ATT ATT AGA ACA G-3′), corresponding to a portion of the common spliced leader (“mini-exon”) of all T. brucei mRNAs, was used as a 5′ primer. cDNA was denatured for 10 min at 95°C and the PCR amplification was carried out in the presence of 0.2 mM of each dNTP, 1 μM of both primers, and 1 unit of AmpliTaq Gold polymerase (PerkinElmer), by using the following touch down program: 30 s at 92°C, 30 s of hybridization at the following different temperatures (2 cycles at 58, 56, 54, and 52°C, followed by 25 cycles at 50°C), and 1 min at 72°C for elongation (for a total of 33 cycles) then a final 10-min extension step at 72°C was included. The p15-2B (5′-GTC ATR TCN CCC ATN ARY TC-3′), p17-1A (5′-CAT YTG YTC NGC NAR-3′), and p17-1B (5′-YTT DAT YTC NSW CAT-3′) degenerate oligonucleotides were deduced by reverse translation of peptides pep15.2 (p15-2B) and pep17.1 (p17-1A and p17-1B), as shown in Figure 1.
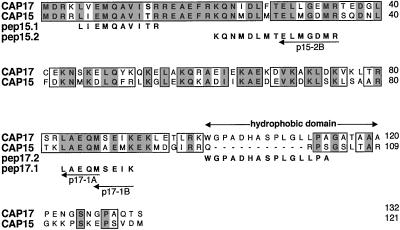
Figure 1.
Sequence comparison of CAP15 and CAP17 (accession no. AF319943 and AF319944, respectively). Boxes define identical residues (gray) or nonconservative amino acid substitutions (white). Gaps (-) introduced to optimize the alignment demonstrate the putative hydrophobic domain present only in CAP17. The degenerate primers deduced from the microsequence data obtained for CAP15 (pep15.1 and pep15.2) and CAP17 (pep17.1 and pep17.2) are indicated by arrows.
The 402-base pair (CAP15) and 532-base pair (CAP17) PCR products were used as α-32P-labeled probes to screen a T. brucei AnTat 1 genomic DNA library constructed in the cosmid vector c2x75 as described (Bringaud et al., 1998a).
Production of Recombinant Proteins in Escherichia coli and Antibody Production
The N-terminal histidine-tagged CAP15 and CAP17 coding sequences were PCR amplified and the resulting DNA fragments were cloned into the pET3a and pET16b expression vectors (Novagen, Madison, WI), respectively. Induction of E. coli BL21 cells, transformed with pET3a-CAP15 or pET16b-CAP17 expression vectors, was performed for 2 h at 37°C with 1 mM isopropyl-β-d-thiogalactopyranoside. Cells were harvested by centrifugation, and proteins purified by nickel chelate chromatography (Novagen) according to the manufacturer's instructions. Antisera were raised in rabbits by three injections at 15-day intervals of 200 μg of each recombinant nickel-purified protein electroeluted after separation on SDS-PAGE, emulsified with complete (first injection) or incomplete Freund's adjuvant. Immunoadsorptions were performed by incubating 1:5 PBS-diluted rabbit anti-CAP15.NA or -CAP17.NA for 2 h at room temperature with recombinant CAP17 or CAP15 linked onto nickel beads, respectively. Enriched antisera for specific antibodies (anti-CAP15.A and CAP17.A) were recovered from the supernatant after centrifugation.
Western Blot
Total extracts or soluble fractions from trypanosomes or Chinese hamster ovary (CHO)-KI cells were boiled for 5 min in 2% (wt/vol) SDS. Sample preparation, migration in 15% SDS-PAGE, immunoblotting on Immobilon-P membranes (Millipore, Bedford, MA), and immunodetection by using as secondary antibody goat anti-rabbit conjugated to horseradish peroxidase (Sigma) were performed as described (Harlow and Lane, 1988). Complete or immunoadsorbed rabbit antisera were diluted 1:100 in PBS containing 0.05% (vol/vol) Tween 20 and 5% (wt/vol) nonfat milk. Blots were developed with 4-chloronaphthol or 3,3′-diaminobenzidine.
Expression in Trypanosomes, and CHO-K1 and HeLa Cells
For protein expression in CHO-KI and HeLa cells, the CAP15 coding sequence was cloned into the pTRE2 (CLONTECH, Palo Alto, CA) and pcDNA3 (Invitrogen) vectors, respectively. DNA transfections were performed using FuGENE6 reagent (Roche Molecular Biochemicals). Inducible expression of CAP15 was achieved in a CHO-K1 Tet-ON cell line according to manufacturer's specifications (CLONTECH). Transformants were selected and cloned for resistance to hygromycin B (200 μg ml−1). CAP15-expressing cells were identified by Western blot analysis after growing for 48 h with or without 2 μg ml−1 of the tetracycline analog doxycycline (Sigma). Transient expression was achieved in HeLa cells. Forty-eight hours after transfection cells were exposed for 30 min either to cold (4°C) or to 20 μM nocodazole at 37°C in culture medium.
For expression in trypanosomes, the CAP15 and CAP17 genes were cloned in the pTSA-3′proc vector (generous gift from D. Salmon and E. Pays) (Bringaud et al., 1998b). Similarly, an enhanced green fluorescent protein (EGFP)-tagged CAP15 protein (green fluorescent protein optimized for fluorescence and expression in mammalian cells) was produced for expression in parasites by using overlapping PCR. Procyclics were transformed by electroporation with BssHII-linearized recombinant vectors, as described (Bringaud et al., 1998b). Transformed cells were selected with hygromycin B and cloned by limiting dilutions in 96-well plates with conditioned medium.
Immunolocalization of CAP15 and CAP17
Cells were fixed in PBS containing 1% (vol/vol) formaldehyde for 10 min (CHO-K1 and HeLa cells grown on poly-l-lysine–coated glass slides) or 30 min (trypanosomes), and permeabilized for 10 min by adjusting the solution to 0.1% (vol/vol) Triton X-100. Glycine (0.1 M) was added for 10 min. Cells were washed once in PBS and trypanosomes were resuspended in PBS and allowed to adhere to glass slides until completely dry before incubation with antibodies. Mouse monoclonal antibodies against calf α-tubulin (Sigma) or trypanosome tubulin (TAT-1) (kind gift from K. Gull, University of Manchester, Manchester, United Kingdom) were used at 1:50 and 1:5 dilutions for CHO-K1 and HeLa cells and trypanosomes, respectively. For the observation of the mitotic spindle in immunofluorescence work, TAT1 was diluted 1:10. A mouse polyclonal immune serum specific for the flagellum of T. brucei (Giroud and Baltz, unpublished data) was diluted 1:100. Rabbit antisera raised against CAP15 and CAP17 were 1:500 (unadsorbed) and 1:100 (adsorbed) diluted, whereas secondary goat anti-mouse and goat anti-rabbit fluorescein isothiocyanate or rhodamine conjugated (Diagnostic Pasteur; Jackson Immunoresearch, West Grove, PA) were used at 1:100 dilution. All incubations were carried out for 30 min at room temperature, and all dilutions performed in PBS containing 1% (vol/vol) Triton X-100, 0.1% (wt/vol) bovine serum albumin. At the end of the immunofluorescence assay, cells were incubated for 5 min with PBS containing 1 μg ml−1 of the fluorescent DNA dye 4′,6-diamino-2-phenyl-indole (DAPI; Sigma).
For electron microscopy and immunolabeling, the cells were treated and labeled with the anti-CAP15.A as described (Sherwin and Gull, 1989b), with the following modifications. Cells were extracted for 10 min in 0.5% (vol/vol) Nonidet P-40 in PIPES buffer (100 mM PIPES, pH 6.9; 0.1 mM EGTA; 1 mM MgCl2); fixed for 10 min in 4% (wt/vol) araformaldehyde in PIPES buffer; neutralized in 0.1 M glycine for 10 min; and blocked in 1% (wt/vol) bovine serum albumin, 0.1% (vol/vol) Tween in PBS, pH 7.2, for 10 min. Electronmicroscopy grids plus the fixed cytoskeletons were floated onto affinity-purified primary antibodies (1:5 in blocking buffer) for 45 min. The cytoskeletons were washed five times (10 min each wash) in blocking buffer and incubated in secondary antibody 1:20 (10-nm gold-conjugated, anti-rabbit antibodies; British Biocell, Cardiff, United Kingdom) for 45 min. Grids were washed and fixed in 2.5% (vol/vol) glutaraldehyde for 2 min and negatively stained with 0.7% (wt/vol) aurothioglucose (Sigma).
RESULTS
Isolation of Two Low-Molecular-Weight Proteins and Cloning of Corresponding Genes
To characterize new stage-specific proteins in T. brucei, we performed fractionation assays on the procyclic and bloodstream forms. A detergent treatment (1% Triton X-100) followed by a 10% perchloric acid precipitation revealed the presence of a 15-kDa protein in the bloodstream forms, whereas a 17-kDa protein was specifically isolated from procyclic forms' extracts obtained by hypotonic lysis followed by a chloroform/methanol/amonium acetate precipitation (see MATERIALS AND METHODS). The 15- and 17-kDa protein, called CAP15 and CAP17, owing to their subcellular localization, were further purified by high-performance liquid chromatography or separated by SDS-PAGE. Genes encoding the two proteins were cloned using oligonucleotides based on their peptide sequences. Southern blot analysis of cosmids isolated from CAP15 and CAP17 screens with PCR fragments showed that CAP15 and CAP17 genes are not located on the same cosmids (our unpublished data). Comparison of the gene organization in the recombinant cosmid clones and the genomic DNA from T. brucei (AnTat 1 strain), by using CAP15- or CAP17-specific DNA probes, indicated that all the CAP15 and CAP17 genes are contained in the Cos15.1A and Cos17.5A clones, respectively (our unpublished data). Southern blot analyses indicated that the CAP17 gene is a single copy, whereas CAP15 is encoded by three identical genes tandemly arranged (our unpublished data). DNA fragments containing the CAP15 and CAP17 genes were isolated from the Cos15.1A and Cos17.5A clones, respectively, subcloned in the pUC19 vector, and sequenced.
CAP15 and CAP17 contain 121 and 132 amino acids, respectively, and are 49.2% identical and 73.5% similar, the main difference being the presence of a hydrophobic domain extending from positions 100–119 in CAP17 (Figure 1). A basic local alignment search tool search of these proteins showed no significant homology with the Swiss protein database.
Expression and Localization of CAP15 and CAP17
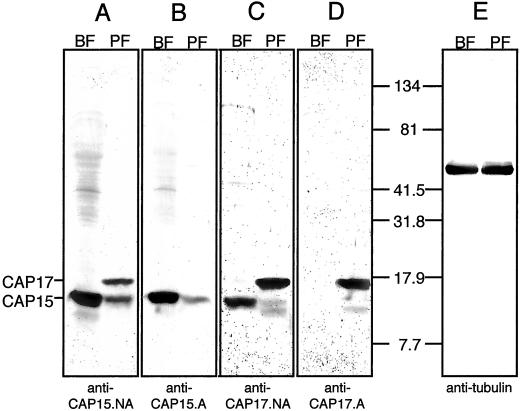
To produce antibodies against CAP15 and CAP17 (anti-CAP15.NA and anti-CAP17.NA), rabbits were injected with the full-length CAP15 or CAP17 recombinant proteins expressed in E. coli. Immunospecific sera (anti-CAP15.A and anti-CAP17.A) were obtained by immunoadsorption against the CAP17 an CAP15 recombinant proteins, respectively. Western blot analyses showed that CAP17 is present only in the procyclic forms, whereas CAP15 is expressed in both procyclic and bloodstream forms of T. brucei (Figure 2, B and D). By using a tubulin antibody as a control (Figure 2E), it appears that CAP15 is ∼10-fold more abundant in the bloodstream forms than in the procyclic forms. In addition, unadsorbed immune sera revealed that CAP17 seems to be more abundant than CAP15 in the procyclic forms (Figure 2, A and C).
Figure 2.
Western blot analysis of CAP15/CAP17 expression. Total lysates of 2 × 107 (A–D) or 2 × 106 (E) T. brucei procyclic (PF) or bloodstream (BF) forms were probed with anti-CAP15 serum (anti-CAP15.NA) (A); anti-CAP15 serum adsorbed against the recombinant CAP17 (anti-CAP15.A) (B); anti-CAP17 serum (anti-CAP17.NA) (C); and anti-CAP17 serum adsorbed against the recombinant CAP15 (anti-CAP17.A) (D). The positions of CAP15 and CAP17 are indicated on the left margin, and the molecular weight markers are indicated between D and E.
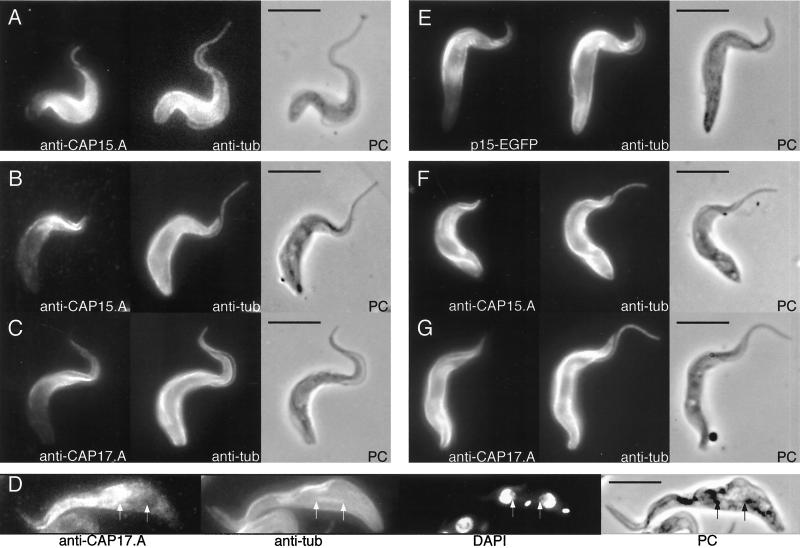
Specific immune sera were used to determine the subcellular localization of CAP15 and CAP17 by immunofluorescence. CAP15 and CAP17 show a localization comparable to anti-tubulin (TAT-1 monoclonal antibody) in the bloodstream and procyclic forms, but interestingly, it fails to label the posterior pole of the cell or the flagellum (Figure 3, A–C). The spindles of mitotic cells, which are labelled with antitubulin, are not recognized by anti-CAP17.A, indicating that CAP17 is not associated with mitotic spindles (Figure 3D). A recombinant CAP15 protein tagged with “humanized EGFP” was also expressed in procyclic cells by using the pTSA-3′proc vector. The recombinant CAP15-EGFP protein was detectable by fluorescence microscopy and shows the same localization as observed for native CAP15 and CAP17 (Figure 3E). The pTSA-3′proc vector also allowed the overexpression of both the CAP15 and CAP17 recombinant proteins in the procyclic forms of T. brucei (Figure 8A). As shown in Figure 3, F–G, the overexpressed CAP15 and CAP17 recombinant proteins colocalized with tubulin over the cell body and not the flagellum nor the mitotic spindles. These data show that both CAP15 and CAP17 colocalize with the anterior part of the subpellicular microtubules and appear to be associated with the cytoskeleton.
Figure 3.
Immunofluorescence analysis of CAP15/CAP17 expression showing labelling localized to the anterior portion of the cells. T. brucei bloodstream forms (A) or procyclic forms (B-D), and T. brucei procyclic forms overexpressing recombinant EGFP-tagged CAP15 (E), CAP15 (F), or CAP17 (G) proteins were labeled with anti-CAP15.A (A, B, and F) or anti-CAP17.A antisera (C, D, and G) and a monoclonal anti-tubulin TAT1 (A-G). The recombinant GFP-tagged CAP15 (E) was visualized by fluorescence analysis. In panel D, the cells were also stained with DAPI, and both extremities of the mitotic spindles are showed by arrows. The cell in panel D shows that, although the subpeculiar, flagellum, and spindle microtubules are labelled by anti-Cap17.NA antisera, the spindle is not labelled by anti-CAP17. Respective phase contrast (PC) images are also shown. Bar, 5 μm.
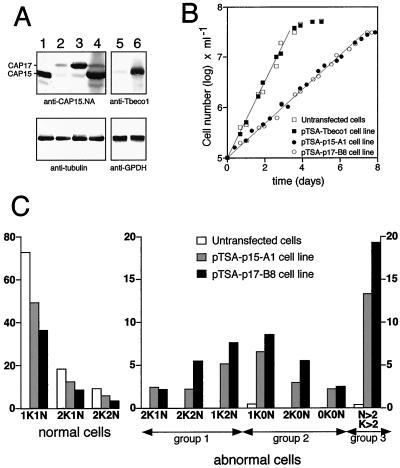
Figure 8.
CAP15 and CAP17 overexpression in procyclic cells. (A) Western blot analysis. Lysates of 107 cells (upper blot) or 2 × 106 cells (lower blot) of untransfected bloodstream (lane 1) or procyclic (lane 2) forms, and pTSA-transfected cell lines CAP17-B8 (lane 3) and CAP15-A1 (lane 4) were probed with anti-CAP15.NA serum (upper blot) or monoclonal anti-tubulin (lower blot). The position of CAP15 and CAP17 is indicated in the left margin. Lanes 5 and 6 show lysates of 107 untransfected procyclic cells and pTSA-Tbeco1 transfected cells, respectively, probed with anti-Tbeco1 (upper blot) or anti-GPDH (glycerol-3-phosphate dehydrogenase) (lower blot) immune sera. (B) Cell growth measurements. Control cells (untransfected and pTSA-Tbeco1 transfected procyclic cells) are shown with open and filled square symbols, whereas CAP15 and CAP17 over-expressing cells are indicated as open and filled circles, respectively. (C) Cell morphology analysis. Cell “type” classification was performed by immunofluorescence assays, as shown in the Figure 9, by determining both nuclear and kinetoplast cell content and by correct positioning of these two genomes. Results are indicated as percentages among the total cell population.
CAP15 and CAP17 Are Microtubule-associated Proteins
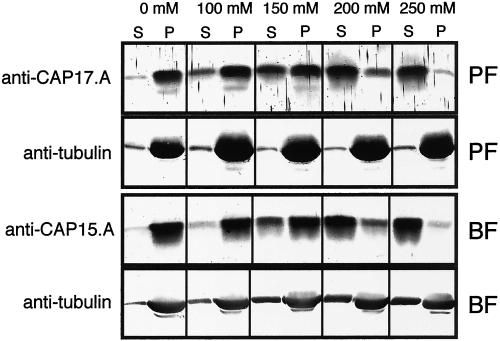
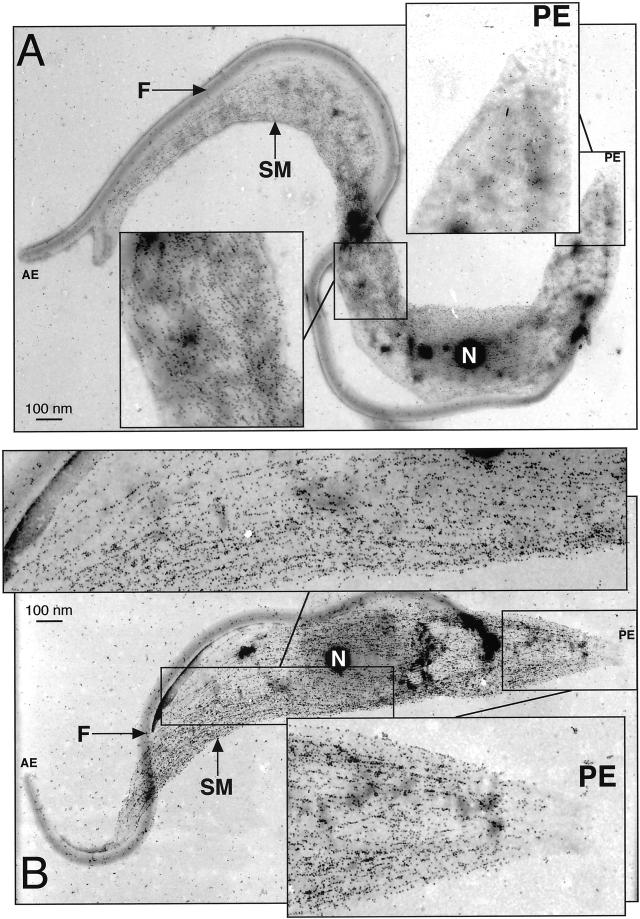
Due to its highly cross-linked nature, the trypanosome cytoskeleton can be isolated intact via a detergent extraction procedure (Robinson et al., 1991). Analysis of the detergent-soluble and -insoluble fractions shows that CAP15 and CAP17 cofractionate with tubulin in the presence of up to 100 mM NaCl (Figure 4), indicating that both proteins are associated with the cytoskeleton. The increase of the NaCl to 200 mM causes release from the cytoskeleton (Figure 4). The colocalization of tubulin and CAP15/CAP17 was confirmed by electron microscopy analysis of cytoskeletons. The anti-CAP17.A and anti-CAP15.A antibodies recognize the anterior portion of the procyclic cells as observed by immunofluorescence analysis, but not the flagellum or the cell posterior (Figure 5A). Furthermore, the same analysis conducted on CAP-overexpressing procyclic cells confirmed the presence of the recombinant CAP15 and CAP17 on the whole corset along the helical path of microtubules, which is a clear characteristic of the subpellicular microtubule corset (Figure 5B).
Figure 4.
CAP15/17 are associated with microtubules. (A) Supernatants (S) and pellets (P) from 2 × 107 T. brucei procyclic (PF) or bloodstream (BF) forms treated by detergent and various NaCl concentrations were analyzed with anti-CAP17.A or anti-CAP15.A immune sera, respectively, and monoclonal anti-tubulin. This data shows that, after 200 mM NaCl, most of the CAP proteins disassociates from the subpellicular microtubules.
Figure 5.
CAP15/17 are associated with the subpellicular corset. Electron microscope analysis of the wild-type (A) and CAP17 overexpressing (B) T. brucei procyclic forms with the anti-CAP17 immune serum. Insets represent a higher magnification (× 2.5) of the region with low (posterior end) or high levels of CAP17 in the wild-type cells, and the same regions in the CAP17 overexpressing cells, which show a high level of CAP17 labelling . The insets also show the microtubule labelling pattern of the gold particles. The gold particles clearly bind to microtubules and follow the helical path of the subpellicular microtubules along the length of the cytoskeleton. In wild-type (A) cells, the labelling is on the major portion of the cytoskeletal microtubules but show no, or extremely low, labelling at the posterior end. In CAP17, overexpressing (B) cells immunolabelling is on the whole of the subpellicular microtubules. AE, anterior end; PE, posterior end; F, flagellum; N, nucleus; SM, subpellicular microtubules.
CAP15 and CAP17 Stabilize Microtubules
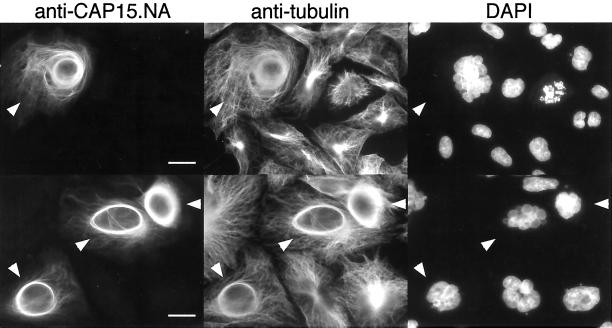
Since the recombinant CAP proteins have a tendency to agregate in solution, we investigated the role of these proteins in vivo, by expressing them in heterologous cells. CAP15 was expressed, using the pTRE2 vector, in the CHO-K1 Tet-ON cell line designed for tetracycline- or doxycycline-inducible expression. CHO-K1 Tet-ON clones expressing CAP15 were selected by Western blot analysis and approximately half of the clones tested showed a tightly controlled doxycycline-inducible expression of CAP15. A clone (C9) was used for further analyses. No signal was observed for the uninduced C9 clone analyzed by immunofluorescence with the anti-CAP15.NA antibody (our unpublished data). However, the C9 clone expressing CAP15 revealed that the protein colocalizes with the cytoskeleton network that is colabeled with an anti-tubulin antibody (Figure 6), confirming that CAP15 is a microtubule-associated protein. The level of CAP15 expression differed from cell to cell with ∼80% of the cells being devoid of CAP15, as exemplified in the Figure 6. Moreover, a perinuclear microtubule bundling was observed in about half of the CAP15 CHO-K1-expressing cells, a structure not observed in nontransfected cells (Figure 6).
Figure 6.
Immunofluorescence analysis of CHO-K1 cells expressing CAP15. CHO-K1 Tet-ON cells transfected with the pTRE2-CAP15 plasmid and induced with doxycycline were stained with anti-CAP15.NA serum, monoclonal anti-tubulin, and DAPI. This shows that CHO-K1 cells expressing CAP15 (cells labelled with white arrowheads) have bundles around the nucleus, whereas no perinuclear bundles are observed for those not expressing CAP15 (unlabelled cells). Bar, 10 μm.
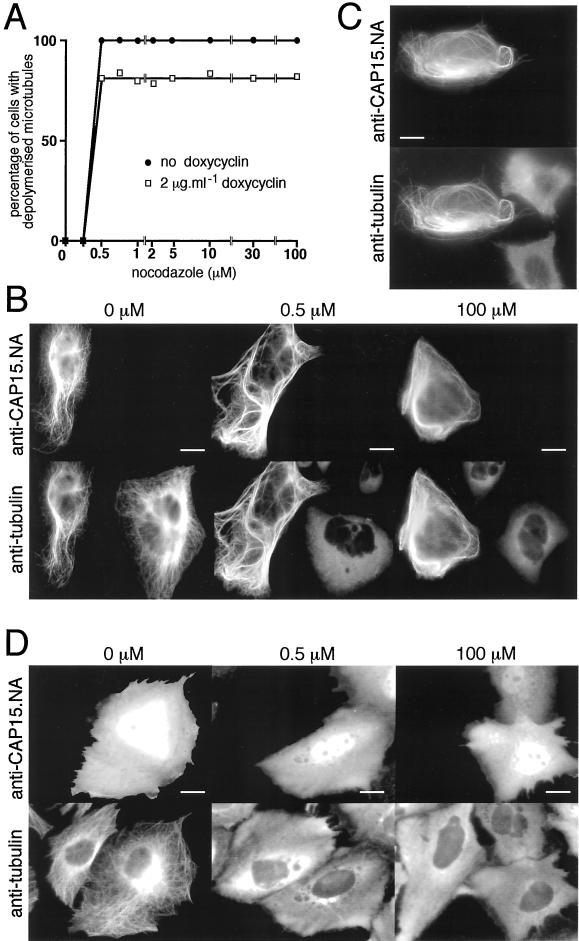
The effect of CAP15 on microtubule turnover was tested by incubating the cells with nocodazole (which inhibits microtubule formation). In untransfected cells or in the uninduced C9 clone the microtubule network was completely depolymerized after 30 min of incubation with 0.5 μM nocodazole (Figure 7A). In the presence of doxycycline (inducer of CAP15 expression) ∼20% of the C9 clone cells expressed CAP15, whereas the remaining cells did not. Under these conditions, ∼20% of the cells in the total population are insensitive to 0.5 μM nocodazole (Figure 7A). It appears that all of the nocodazole-insensitive cells express CAP15, whereas the sensitive cells do not (Figure 7B, middle), indicating that expression of CAP15 is responsible for the nocodazole insensitivity. A 200-fold increase in the nocodazole concentration (100 μM) causes only a partial effect on the CAP15-expressing cells (Figure 7B, right), because these cells still contain unaffected microtubules. The expression of CAP15 therefore appears to reduce microtubule turnover in the CHO-K1 cells, most likely by blocking depolymerization.
Figure 7.
Heterologous expression of CAP15 in CHO-K1 and HeLa cells. (A) Measurement of microtubule stability in CHO-K1 cells transfected by pTRE2-CAP15. The graph represents the percentage of cells presenting a complete depolymerization of the microtubule network as a function of nocodazole concentration. Doxycyclin-induced (▪) or uninduced (●) cells were treated for 30 min with various concentrations of nocodazole and analyzed by immunofluorescence to monitor the drug-induced microtubule depolymerization as shown in B. Approximately 20% of the doxycyclin-induced cells transfected by pTRE2-CAP15 were resistant to microtubule depolymerization after treatment with 0.5 μM nocodazole. (B) Immunofluorescence analysis of nocodazole-treated cells. Doxycyclin-induced cells were treated with 0, 0.5, or 100 μM nocodazole for 30 min and stained with anti-CAP15.NA serum and monoclonal anti-tubulin. Bar, 10 μm. (C) Cold-resistance of microtubules from HeLa cells transfected with pcDNA-CAP15. Transfected cells were treated 30 min at 4°C and stained with anti-CAP15. NA serum and monoclonal anti-tubulin. Bar = 10 μm. (D) Same experiment as in B, except that the CHO-K1 cells were transfected with the pcDNA3-CAP15nt plasmid, which encodes the CAP15 without the eight first amino acids. Bar, 10 μm. The first three panels show that the microtubule network of CHO-K1 (A and B) or HeLa (C) cells expressing CAP15 are resistant to nocodazole (A and B) and cold (C) treatments, while untransfected cells present a complete microtubule depolymerization under these conditions, indicating that CAP15 stabilizes microtubules. Panel D shows that expression in CHO-K1 cells of the truncated form of the protein does not bind to microtubules and does not protect microtubules against nocodazole treatment.
Both CAP15 and CAP17 were also transiently expressed in HeLa cells. As observed for the CAP15 CHO-K1-expressing cells (Figures 6 and 7), both CAPs colocalize with the microtubule network and protect the microtubules against nocodazole depolymerization (our unpublished data for CAP17). The microtubule network of HeLa cells is normally cold sensitive. CAP15 or CAP17 render cold-resistance to the networks of these cells (Figure 7C).
To characterize the microtubule-binding domain, we have expressed in the CHO-K1 cells a series of truncated CAP15 mutants, which start and end at amino acid positions 1–95, 57–121, 30–121, 17–121, and 8–121. All the truncated proteins were located in the cytosol and the nucleus, with no microtubule-like signal, suggesting that the entire structure is required for binding to microtubules in CHO-K1-cells. The subcellular localization of the CAP15 mutant that lacks the first seven amino acids (mutant 8–121) is presented in Figure 7D. Furthermore, the microtubule network of CHO-K1 cells expressing the 8–121 CAP15 mutant depolymerized in the presence of 0.5 μM nocodazole (Figure 7D), confirming that truncated CAP15 does not interact with the microtubules of this cell line.
Overexpression of CAP15 and CAP17 in Procyclic Forms of T. brucei
As presented above, CAP15 and CAP17 were overexpressed in procyclic forms of T. brucei (EATRO 1125 strain) by using the pTSA-3′proc vector. A Western blot analysis showed that different hygromycin-resistant clones transfected with pTSA-CAP15 or pTSA-CAP17 overexpressed similar quantities of each recombinant protein (∼10-fold higher than wild-type and similar to the level of native CAP15 in the untransfected bloodstream forms) (Figure 8A). These levels of expression affect procyclic growth. The pTSA-CAP15.A1 and pTSA-CAP17.B8 cell lines showed an increase of the cell doubling time compared with the wild-type procyclic cells (20 versus 8.5 h; Figure 8B). DAPI staining revealed that both transfected procyclic cell lines contain a large number of aberrant cells (30–50%) with abnormal nuclei and/or kinetoplast content compared with the untransfected cells (<1%) (Figure 8C). Similar results were observed for both CAP15- and CAP17-overexpressing procyclic cell lines and the data obtained with the pTSA-CAP17.B8 cell line are described herein. To determine if the phenotype observed for the CAP15 and CAP17 over-expressing procyclic cell lines is the result of CAP over-expression or a side effect of transfection or protein load, we over-expressed an unrelated T. brucei 21-kDa protein (Tbeco1) in the same cell line (EATRO 1125) using the same pTSA expression vector. This Tbeco1 over-expressing procyclic cell line is morphological identical to and behaved in terms of growth as the untransfected cells (Fig. 8B), while Tbeco1 expression was increased about 20-fold in comparison to wild type cells (Fig. 8A).
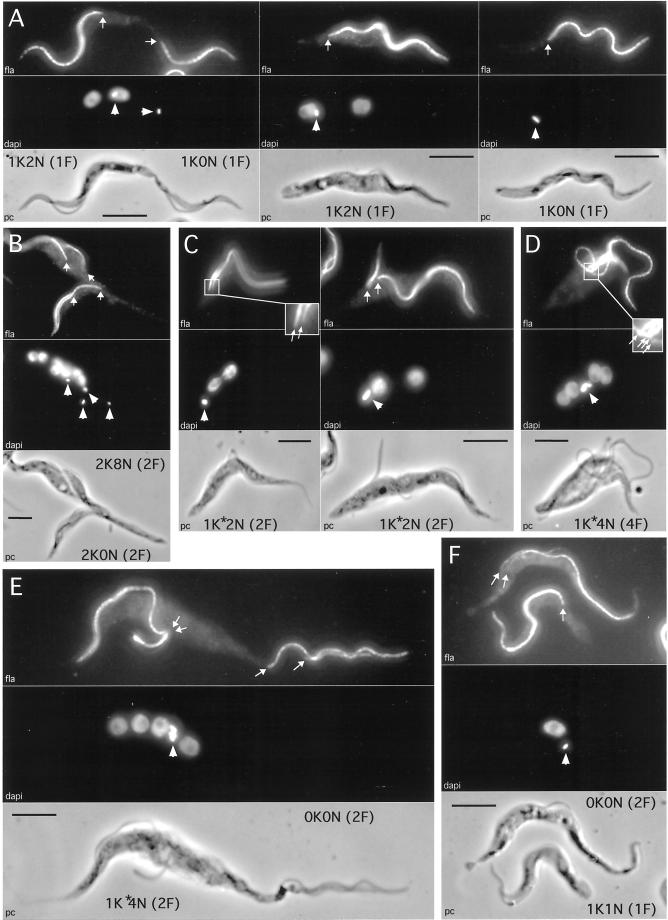
In addition to the usual 1K1N (1 kinetoplast, 1 nucleus), 2K1N, and 2K2N cells observed in wild-type populations, three groups of abnormal cell types (48.4% of the population) were detected in the pTSA-CAP17.B8 cell line (Figure 8C). The first group of cells (14.3% of the total population) had the correct number of nuclei and kinetoplasts (2K1N and 2K2N) but these organelles were mispositioned. Additionally, there were 1K2N cells (7.5%) that had an extra nucleus or alternatively had lost one kinetoplast. The second group was composed of cells without a nucleus and/or kinetoplast (15.5%). The 1K0N cells (∼8%), previously called “zoids” (Robinson et al., 1995; Ploubidou et al., 1999), were the most abundant in this group, and a significant number of cells (2.5%) had no DNA content (0K0N). The third group contained cells with more than two nuclei and/or two kinetoplasts (18.6%). Approximately one fifth of the cells composing this group contained more than 10 nuclei. All abnormal cells had at least one flagellum (as visualized using a mouse polyclonal antiserum that gives a PFR localization signal), whatever the kinetoplast or nuclei number (Figure 9).
Figure 9.
Immunofluorescence analysis of procyclic cells overexpressing CAP17. Cells were tested with a flagellum-specific monoclonal antibody (top picture of each panel). Positions of the proximal base of the flagella (arrows) and kinetoplasts (arrowheads) were determined by DAPI staining (middle picture of each panel). Respective phase contrast (PC) images are also shown at the bottom of each panel. The nuclear (N), kinetoplast (K), and flagellum (F) contents are indicated beside each cell. Bar, 5 μm. Panel A represents individual 1K2N (central panel) and 1K0N (right panel) cells that are the result of mispositioning of the nucleus during cell division (left panel). Panel B shows a dividing aberrant cell leading to polynucleate (8N) and anucleate (0N) cells containing two flagella and two kinetoplasts. The next two panels (C and D) illustrate dividing cells with duplicated flagellum, nucleus, and kinetoplast, which fail to segregate the kinetoplasts. The two latter panels (E and F) show cells without kinetoplast or nucleus (0K0N) during (E) and after (F) aberrant cell division.
After a few weeks of in vitro culture, the ratio of aberrant cells dropped and the doubling time reached 8.5 h, as in wild-type procyclic cells. Furthermore, immunofluorescence analysis showed that the morphologically aberrant cells expressed more CAP15/CAP17 than the “apparently morphologically normal” overexpressing cells (our unpublished data). These data suggest that the “apparently normal” cells that overexpress a low amount of recombinant protein, behave as wild-type procyclic cells, in terms of morphology and growth kinetics and subsequently enrich the population after long-term culture.
Determination of the cell cycle step(s) that is disturbed by the CAP15/CAP17 overexpression is essential to understand the role of these proteins during the trypanosome cell cycle. About half of the observed abnormal dividing cells contain the correct number of nuclei/kinetoplast but with abnormal distribution between the daughter cells (Figure 9A, left). The result of these aberrant divisions would be the formation of a cell with two nuclei (1K2N) and an anucleate cell (1K0N), belonging to the first and second group of abnormal cells, respectively (Figure 8C), as shown in Figure 9A (middle and right). The percentage of 1K2N and 1K0N cells is 7.5 versus 8.5%, respectively, suggesting that they are sibling cells. Similarly, the observation of division, which may lead to 0K0N and 2K2N daughter cells, is in agreement with the relative number of 2K2N abnormal cells and 0K0N cells (Figure 8C). In addition, 0K0N cells may also be sibling cells of other aberrant cell types, as shown in Figure 9E. In this particular case, the sibling cell contains four nuclei, indicating that abnormal polynucleated cells may go through at least one mitotic division while undergoing the processes of generating abnormal daughter cells. This latter observation is also demonstrated in Figure 9B, which shows a dividing 4K8N cell forming 2K8N and 2K0N daughter cells. Taken together, these data suggest that overexpression of CAP15/CAP17 affects cytokinesis, resulting in a misdistribution of kinetoplasts and/or nuclei in daughter cells, leading to the formation of polynucleated and anucleated cells.
Alternatively, some of the polynucleated cells may be the result of another consequence of the CAP15/CAP17 overexpression. Approximately 25% of the 1K2N cells observed, called 1K∗2N, are different from those previously shown sibling of the 1K0N cells (Figure 9A). As shown in Figure 9C, the differences are due to the position of the kinetoplast, either located between the nuclei (1K2N cells) or at the anterior end (1K∗2N cells), and the number of flagella, i.e., a single flagellum in 1K2N cells, or two in 1K∗2N cells. The 1K∗2N cells contain an apparently normal flagellum attached to the whole cell body and a free shorter one attached only in the flagellar pocket area (Figure 9C, right). Interestingly, a 1K∗2N mitotic cell showing a large duplicating nucleus and two flagella, as expected, contains a single kinetoplast instead of two kinetoplasts. This kinetoplast appears not to have fully completed DNA synthesis (Figure 9C, left). The presence of two flagella indicates that the flagellum basal body duplicated and that the cell should have segregated kinetoplasts. However, the kinetoplast did not duplicate before the cell entered mitosis, or it duplicated but failed to segregate. The observation of a single large kinetoplast in most of the 1K∗2N cells (Figure 9C, right) is in favor of the second hypothesis. The 1K∗2N cells are able to initiate a second round of kinetoplast/nucleus duplication before cytokinesis as shown by the observation of cells with four nuclei, four flagella, and a single large kinetoplast, called 1K∗4N cells (Figure 9D). The size of the kinetoplast observed in the 1K∗4N cells (Figure 9, D and E) suggests that the daughter kinetoplasts failed to segregate. As observed for the 1K∗2N cells, the 1K∗4N cells failed to initiate cytokinesis (Figure 9D) or completed the cell cycle with the formation of a dyskinetoplastic/anucleate cell (0K0N) and a 1K∗4N cell (Figure 9E). Surprisingly, two flagella remain associated with the kinetoplast mass in the 1K∗4N cell, whereas the two others moved in association with the formation of the 0K0N cell, showing that basal body/kinetoplast connections are destroyed during this type of 0K0N cell formation. All the 0K0N observed contained one or two flagella (Figure 9F).
DISCUSSION
Two related low-molecular-weight proteins, CAP15 (15- kDa) and CAP17 (17-kDa), sharing 49.2% identity, which present a microtubule-binding activity, were identified in the parasitic protozoan Trypanosoma brucei. When expressed in CHO-K1 or HeLa cells, CAP15 and/or CAP17 clearly colocalize with microtubules. The list of proteins interacting with microtubules can be subdivided into several classes. The first class comprises the so-called MAPs, also called “structural” MAPs (MAP1–4, STOP) because they bind, stabilize, destabilize, or promote the assembly of microtubules. The second class corresponds to motor proteins (kinesin and dynein), sometimes called “motor” MAPs, which generate movement along microtubules. The third and most heterogeneous class includes proteins that are not normally called MAPs but are often found associated with microtubules and may copurify with them. Some of these proteins have a role in microtubule dynamics and organization. Because acidic tubulin molecules may bind basic proteins in a nonspecific manner (Robinson et al., 1991), caution must be exercised in assigning genuine microtubule-binding activity to proteins.
CAP15 and CAP17 have the characteristics of bona fide “structural” MAPs: 1) they colocalize with microtubules in the procyclic and bloodstream forms of T. brucei; 2) CAP15 induces bundle formation in CHO-K1 cells, as observed for higher eukaryotic MAPs expressed in heterologous cells (Kosik and McConlogue, 1994; Olson et al., 1995; Yoshida et al., 1996); 3) microtubules of CAP15-expressing CHO-K1 cells and CAP15- or CAP17-expressing HeLa cells are resistant to nocodazole, indicating that microtubules are stabilized by both CAPs; 4) the cold-sensitive microtubules of the HeLa cells become cold resistant in the presence of CAP15 or CAP17, as observed for the mammalian MAPs, STOP, and F-STOP (Bosc et al., 1996; Denarier et al., 1998); and 5) no microtubule-binding activity is observed with truncated CAP15 mutants in CHO and HeLa cells.
Immunofluorescence, electron microscopy, and cell fractionation showed that CAP15 and CAP17 are associated primarily with the subpellicular corset but not the flagellum, the basal body, or the mitotic spindle. The absence of CAPs on mitotic spindle may explain why mitosis is not affected by overexpression of these proteins, whereas kinetoplast segregation and cytokinesis, which are subpellicular microtubule-mediated events (Robinson et al., 1995), are disturbed.
Among the subpellicular corset MAPs characterized in trypanosomes, only a few have been characterized genetically (MARP-1, MARP-2, the autoantigen 1/6, and Gb4; (Schneider et al., 1988b; Hemphill et al., 1992; Rindisbacher et al., 1993; Affolter et al., 1994; Detmer et al., 1997). All contain repetitive motifs, possibly involved in microtubule association, as shown for MARP-1 (Hemphill et al., 1992). The best-known representatives of MAPs from higher eukaryotes (MAP1, 2, and 4; tau; STOP; and F-STOP) also contain repetitive motifs (Mandelkow and Mandelkow, 1995; Denarier et al., 1998), most of them being involved in microtubule binding. Interestingly, CAP15 and CAP17 do not contain any repetitive motifs.
Balaban and Goldman (1992) previously enriched a 15-kDa microtubule-associated protein (p15) from T. brucei, which forms microtubule bundles in vitro. Herein, we show that CAP15 1) is differentially expressed in trypanosomes; 2) is located on the anterior half of the subpellicular corset; 3) stabilizes microtubules against nocodazole and cold treatments; 4) disturbs the cytoskeleton and cytokinesis when overexpressed in the procyclic forms; and 5) has a homolog (CAP17), which is also differentially expressed and presents similar characteristics. The bloodstream forms of T. brucei only express CAP15, whereas in procyclic cells, both CAPs are present. The selectable advantage of this differential expression is not clear because these proteins seem to have similar roles, i.e., they colocalize in procyclic forms and when overexpressed, both proteins induce similar abnormal phenotypes. The principal difference is the presence of a 20 amino acid, hydrophobic domain of the C-terminal region of CAP17. Although the role of this domain is unknown, we speculate that CAP17 may interact with both the subpellicular corset and the plasma membrane.
CAP15 or CAP17 overexpression in the procyclic forms leads to the production of aberrant cells similar to cells, previously called “zoids” (Robinson et al., 1995; Ploubidou et al., 1999) and polynucleated cells. The polynucleated cells contain up to several dozen nuclei and seem to be the result of multiple abnormal cell cycles. Two kinds of disorders induced by CAP overexpression were detected by a detailed analysis of the aberrant cell population. The first is the loss of mitosis/kinetoplast duplication/cytokinesis coordination, as shown by the production of “zoids” (1K0N) and their sibling 1K2N cells. The second disorder is the inhibition of kinetoplast segregation, which leads to the production of polynucleated cells containing large kinetoplast masses from which several flagella emerge. Das et al. (1994) previously showed that okadaic acid, a specific inhibitor of protein phosphatase PP1 and PP2A, yielded multinucleate trypanosomes with duplicated but unsegregated kinetoplasts. Interestingly, the 1K∗2N cells we observed and the binucleated cells generated by okadaic acid treatment, which have a single large kinetoplast, contain a second shorter flagellum, indicating that they are morphologically similar. It appears from okadaic acid treatment that loss of kinetoplast segregation blocks cytokinesis, leading to cells with two, four, and eight nuclei. The observation of a 1K∗4N containing four flagella (Figure 9D) fits with this observation, although these aberrant cells may eventually complete the cell cycle to produce an anucleate/dyskinetoplastic cell (0K0N) and a 1K∗4N cell, in this case both with two flagella, as shown in Figure 9E. Surprisingly, this process yields diskinetoplastic cells containing flagella, indicating that the physical kinetoplast/basal body connection (Robinson and Gull, 1991) can be destroyed during aberrant cell divisions (Ploubidou et al., 1999). No CAP localisation was found on the flagellum basal bodies, but since zoids and multinucleated cells are formed, we postulate that dykinetoplastic cells are an indirect consequence of perturbed cell division and not a direct association with the CAP proteins. Dyskinetoplastic trypanosomes can propagate (Riou et al., 1980), indicating that kinetoplast and probably kinetoplast/basal body connections are not essential to complete the cell cycle, but are essential for life cycle development. Clearly, anucleate cells cannot propogate.
The microtubules of the T. brucei corset are organized so that they have the same, posterior end plus, polarity (Robinson et al., 1995). This polarity has implications for the polymerization kinetics within the subpellicular array with more polymerization of individual microtubules at the posterior end of the cell. The localization of both CAPs is restricted to the anterior half of the procyclic forms, suggesting that they play a role in the stabilization of microtubules in the less polymerization-dynamic part of the corset. When overexpressed in the procyclic cells, both recombinant CAPs cover the whole subpellicular corset. This misplacing of the CAP proteins to the polymerization-dynamic region of the corset may overstabilize this microtubule array and cause cytoskeletal “chaos” during the cell cycle. Interestingly, this region contains the kinetoplast, which duplicates but fails to segregate in some of CAP-overexpressing cells. Basal body segregation has been shown to be a critical checkpoint in the cell cycle of trypanosome (Ploubidou et al., 1999). Thus, we propose that the presence of recombinant CAPs in the posterior half of the subcellular corset inhibits or delays basal body kinetoplast segregation with the resulting loss of kinetoplast duplication/cytokinesis coordination. Although our data suggests that the CAP proteins are microtubule stabilizers, we cannot rule out the model that they may also function as inactive microtubule destabilizers. In this hypothesis they may be regulated in different phases of the cell cycle or in the building of the cytoskeleton. This regulation may incur stabilizing or destabilizing effects of the CAPs. We are continuing to study CAP proteins in order to understand and address these models.
ACKNOWLEDGMENTS
We are grateful to L. Vanhamme, D. Salmon, and E. Pays for supplying the pTSA-3′proc vector; and K. Gull for supplying the TAT-1 antibody. We particularly thank D. Baltz and N. Biteau for technical help and sequencing, and M.P. Barrett and K. Gull for critical reading of the manuscript. This work was supported by the Centre National de la Recherche Scientifique, the Conseil Régional d'Aquitaine, and the GDR Centre National de la Recherche Scientifique–Parasitologie and the Ministère de l'Education Nationale de la Recherche et de la Technologie (Action Microbiologie). D.R.R. was supported by a Wellcome Trust Career Development Fellowship.
Abbreviations used:
- CAP
cytoskeletal-associated protein
- EGFP
green fluorescent protein
- MAP
microtubule-associated protein
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–06–0298. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–06–0298.
REFERENCES
- Affolter M, Hemphill A, Roditi I, Muller N, Seebeck T. The repetitive microtubule-associated proteins MARP-1 and MARP-2 of Trypanosoma brucei. J Struct Biol. 1994;112:241–251. doi: 10.1006/jsbi.1994.1024. [DOI] [PubMed] [Google Scholar]
- Balaban N, Goldman R. Isolation and characterization of a unique 15 kilodalton trypanosome subpellicular microtubule-associated protein. Cell Motil Cytoskeleton. 1992;21:138–146. doi: 10.1002/cm.970210207. [DOI] [PubMed] [Google Scholar]
- Balaban N, Waithaka HK, Njogu AR, Goldman R. Isolation of a subpellicular microtubule protein from Trypanosoma brucei that mediates cross-linking of microtubules. Cell Motil Cytoskeleton. 1989;14:393–400. doi: 10.1002/cm.970140309. [DOI] [PubMed] [Google Scholar]
- Bosc C, Cronk JD, Pirollet F, Watterson DM, Haiech J, Job D, Margolis RL. Cloning, expression, and properties of the microtubule-stabilizing protein STOP. Proc Natl Acad Sci USA. 1996;93:2125–2130. doi: 10.1073/pnas.93.5.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringaud F, Baltz D, Baltz T. Functional and molecular characterization of a glycosomal PPi-dependent enzyme in trypanosomatids: pyruvate, phosphate dikinase. Proc Natl Acad Sci USA. 1998b;95:7963–7968. doi: 10.1073/pnas.95.14.7963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringaud F, Vedrenne C, Cuvillier A, Parzy D, Baltz D, Tetaud E, Pays E, Venegas J, Merlin G, Baltz T. Conserved organization of genes in trypanosomatids. Mol Biochem Parasitol. 1998a;94:249–264. doi: 10.1016/s0166-6851(98)00080-2. [DOI] [PubMed] [Google Scholar]
- Das A, Gale M, Jr, Carter V, Parsons M. The protein phosphatase inhibitor okadaic acid induces defects in cytokinesis and organellar genome segregation in Trypanosoma brucei. J Cell Sci. 1994;107:3477–3483. doi: 10.1242/jcs.107.12.3477. [DOI] [PubMed] [Google Scholar]
- Denarier E, Fourest-Lieuvin A, Bosc C, Pirollet F, Chapel A, Margolis RL, Job D. Nonneuronal isoforms of STOP protein are responsible for microtubule cold stability in mammalian fibroblasts. Proc Natl Acad Sci USA. 1998;95:6055–6060. doi: 10.1073/pnas.95.11.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detmer E, Hemphill A, Muller N, Seebeck T. The Trypanosoma brucei autoantigen I/6 is an internally repetitive cytoskeletal protein. Eur J Cell Biol. 1997;72:378–384. [PubMed] [Google Scholar]
- Drewes G, Ebneth A, Mandelkow E-M. MAPs, MARKs and microtubule dynamics. Trends Biochem Sci. 1998;23:307–311. doi: 10.1016/s0968-0004(98)01245-6. [DOI] [PubMed] [Google Scholar]
- Grellier P, Sinou V, Garreau-de Loubresse N, Bylen E, Boulard Y, Schrevel J. Selective and reversible effects of vinca alkaloids on Trypanosoma cruzi epimastigote forms: blockage of cytokinesis without inhibition of the organelle duplication. Cell Motil Cytoskeleton. 1999;42:36–47. doi: 10.1002/(SICI)1097-0169(1999)42:1<36::AID-CM4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D, editors. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hemphill A, Affolter M, Seebeck T. A novel microtubule-binding motif identified in a high molecular weight microtubule-associated protein from Trypanosoma brucei. J Cell Biol. 1992;117:95–103. doi: 10.1083/jcb.117.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imboden M, Muller N, Hemphill A, Mattioli R, Seebeck T. Repetitive proteins from the flagellar cytoskeleton of African trypanosomes are diagnostically useful antigens. Parasitology. 1995;110:249–258. doi: 10.1017/s0031182000080835. [DOI] [PubMed] [Google Scholar]
- Kohl L, Gull K. Molecular architecture of the trypanosome cytoskeleton. Mol Biochem Parasitol. 1998;93:1–9. doi: 10.1016/s0166-6851(98)00014-0. [DOI] [PubMed] [Google Scholar]
- Kosik KS, McConlogue L. Microtubule-associated protein function: lessons from expression in Spodoptera frugiperda cells. Cell Motil Cytoskeleton. 1994;28:195–198. doi: 10.1002/cm.970280302. [DOI] [PubMed] [Google Scholar]
- Lumsden HR, Evans DA. Biology of the Kinetoplastida. New York: Academic Press; 1976. [Google Scholar]
- MacRae TH, Gull K. Purification and assembly in vitro of tubulin from Trypanosoma brucei brucei. Biochem J. 1990;265:87–93. doi: 10.1042/bj2650087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelkow E, Mandelkow E-M. Microtubules and microtubule-associated proteins. Curr Opin Cell Biol. 1995;7:72–81. doi: 10.1016/0955-0674(95)80047-6. [DOI] [PubMed] [Google Scholar]
- Matthews KR, Sherwin T, Gull K. Mitochondrial genome repositioning during the differentiation of the African trypanosome between life cycle forms is microtubule mediated. J Cell Sci. 1995;108:2231–2239. doi: 10.1242/jcs.108.6.2231. [DOI] [PubMed] [Google Scholar]
- Muller N, Hemphill A, Imboden M, Duvallet G, Dwinger RH, Seebeck T. Identification and characterization of two repetitive non-variable antigens from African trypanosomes which are recognized early during infection. Parasitology. 1992;104:111–120. doi: 10.1017/s0031182000060856. [DOI] [PubMed] [Google Scholar]
- Olson KR, McIntosh JR, Olmsted JB. Analysis of MAP 4 function in living cells using green fluorescent protein (GFP) chimeras. J Cell Biol. 1995;130:639–650. doi: 10.1083/jcb.130.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploubidou A, Robinson DR, Docherty RC, Ogbadoyi EO, Gull K. Evidence for novel cell cycle checkpoints in trypanosomes: kinetoplast segregation and cytokinesis in the absence of mitosis. J Cell Sci. 1999;112:4641–4650. doi: 10.1242/jcs.112.24.4641. [DOI] [PubMed] [Google Scholar]
- Rindisbacher L, Hemphill A, Seebeck T. A repetitive protein from Trypanosoma brucei which caps the microtubules at the posterior end of the cytoskeleton. Mol Biochem Parasitol. 1993;58:83–96. doi: 10.1016/0166-6851(93)90093-d. [DOI] [PubMed] [Google Scholar]
- Riou G, Baltz T, Gabillot M, Pautrizel R. Absence of kinetoplast DNA in a late antigenic variant of Trypanosoma equiperdum. Mol Biochem Parasitol. 1980;1:97–105. doi: 10.1016/0166-6851(80)90004-3. [DOI] [PubMed] [Google Scholar]
- Robinson DR, Beattie P, Sherwin T, Gull K. Microtubules, tubulin, and microtubule-associated proteins of trypanosomes. Methods Enzymol. 1991;196:285–299. doi: 10.1016/0076-6879(91)96027-o. [DOI] [PubMed] [Google Scholar]
- Robinson DR, Gull K. Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature. 1991;352:731–733. doi: 10.1038/352731a0. [DOI] [PubMed] [Google Scholar]
- Robinson DR, Sherwin T, Ploubidou A, Byard EH, Gull K. Microtubule polarity and dynamics in the control of organelle positioning, segregation, and cytokinesis in the trypanosome cell cycle. J Cell Biol. 1995;128:1163–1172. doi: 10.1083/jcb.128.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld J, Capdevielle J, Guillemot JC, Ferrara P. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal Biochem. 1992;203:173–179. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- Schneider A, Eichenberger W, Seebeck T. A microtubule-binding protein of Trypanosoma brucei which contains covalently bound fatty acid. J Biol Chem. 1988a;263:6472–5. [PubMed] [Google Scholar]
- Schneider A, Hemphill A, Wyler T, Seebeck T. Large microtubule-associated protein of T. brucei has tandemly repeated, near-identical sequences. Science. 1988b;241:459–462. doi: 10.1126/science.3393912. [DOI] [PubMed] [Google Scholar]
- Seebeck T, Gehr P. Trypanocidal action of neuroleptic phenothiazines in Trypanosoma brucei. Mol Biochem Parasitol. 1983;9:197–208. doi: 10.1016/0166-6851(83)90097-x. [DOI] [PubMed] [Google Scholar]
- Sherwin T, Gull K. The cell division cycle of Trypanosoma brucei brucei: timing of event markers and cytoskeletal modulations. Phil Trans R Soc Lond B Biol Sci. 1989a;323:573–588. doi: 10.1098/rstb.1989.0037. [DOI] [PubMed] [Google Scholar]
- Sherwin T, Gull K. Visualization of detyrosination along single microtubules reveals novel mechanisms of assembly during cytoskeletal duplication in trypanosomes. Cell. 1989b;57:211–21. doi: 10.1016/0092-8674(89)90959-8. [DOI] [PubMed] [Google Scholar]
- Velours J, Arselin de Chateaubodeau G, Galante M, Guerin B. Subunit 4 of ATP synthase (F0F1) from yeast mitochondria. Purification, amino-acid composition and partial N-terminal sequence. Eur J Biochem. 1987;164:579–584. doi: 10.1111/j.1432-1033.1987.tb11166.x. [DOI] [PubMed] [Google Scholar]
- Vickerman K, Preston TM. Comparative cell biology of the kinetoplastid flagellates. London: Academic Press; 1976. [Google Scholar]
- Woods A, Baines AJ, Gull K. A high molecular mass phosphoprotein defined by a novel monoclonal antibody is closely associated with the intermicrotubule cross bridges in the Trypanosoma brucei cytoskeleton. J Cell Sci. 1992;103:665–675. doi: 10.1242/jcs.103.3.665. [DOI] [PubMed] [Google Scholar]
- Woodward R, Gull K. Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei. J Cell Sci. 1990;95:49–57. doi: 10.1242/jcs.95.1.49. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Imanaka-Yoshida K, Murofushi H, Tanaka J, Ito H, Inagaki M. Microinjection of intact MAP-4 and fragments induces changes of the cytoskeleton in PtK2 cells. Cell Motil Cytoskeleton. 1996;33:252–262. doi: 10.1002/(SICI)1097-0169(1996)33:4<252::AID-CM2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]