Abstract
Diabetic kidney disease is the most common primary disease of end-stage kidney disease globally; however, a sensitive and accurate biomarker to predict this disease remains awaited. microRNAs are endogenous single-stranded noncoding RNAs that have intervened in different post-transcriptional regulations of various cellular biological functions. Previous literatures have reported its potential role in the pathophysiology of diabetic kidney disease, including regulation of Transforming Growth Factor-β1-mediated fibrosis, extracellular matrix and cell adhesion proteins, cellular hypertrophy, growth factor, cytokine production, and redox system activation. Urinary microRNAs have emerged as a novel, non-invasive liquid biopsy for disease diagnosis. In this review, we describe the available experimental and clinical evidence of urinary microRNA in the context of diabetic kidney disease and discuss the future application of microRNA in routine practice.
Keywords: diabetes mellitus, diabetic kidney disease, exosomes, microRNA, urinary
Diabetes mellitus (DM) is a major health threat involving 463 million persons globally [1,2]. Diabetic kidney disease (DKD) constitutes the top cause of end-stage kidney disease worldwide [3,4]. The medical burden is complicated by its strong association with cardiovascular diseases, death [5,6], as well as elevated medical financial demand [7]. Improvements in the understanding of pathophysiology and early prediction of DKD remain an unmet clinical need.
Knowledge of systems biology and advances in high-throughput sequencing technology have revolutionized the understanding of the pathophysiology of DKD and biomarker development [8]. Clinical translation of different omic biomarkers for the prediction of DKD remains limited in terms of sample size and validation cohort. An approach that precisely identifies predisposed high-risk DM patients to renal progression is urgently awaited.
Optimal biomarkers have to provide adequate sensitivity and specificity, be obtainable in a non-invasive nature and assay with friendly laboratory skills using minimal time and economical consumption. Urine represents an ideal non-invasive biomarker, mainly for genitourinary diseases, because it is easily collected in large quantities without injury to the patient. In addition to varied types of proteins, the exosomes, which can be secreted by cells of different nephron segments, may also transport protein, mRNA, and microRNA (miRNA) markers produced in conditions of kidney malfunction or structural damage [9]. From them, the urinary exosomes can provide a panoramic view of the whole urinary system.
In this narrative literature review, we examine emerging evidence from experimental and human research that suggest the potential role of urinary miRNA for clinical application in the context of DKD.
1. Introduction of microRNA
The miRNAs are important epigenetic regulators of gene expression that intervene in various cellular processes of health and disease status. Genes encoding miRNAs are situated in the noncoding region or in introns of either protein-coding genes (miR-trons) or noncoding RNA [10]. These are small, less than 70 nucleotide stem–looped structures transcribed by RNA polymerase II located in the nucleus. miRNAs rarely bind to the coding regions of mRNA or genomic DNA, including promoter regions. However, they can actively play a role as regulators of cellular crosstalk by modifying their transcriptional program [11]. Currently, there are more than 38,000 mature sequences of miRNA included in miRbase, and the entry list is still growing [12].
Most miRNAs remain stable and have a prolonged half-life, but other individual miRNAs can experience a rapid decline in certain cellular circumstances because of the existence of specific environmental stimuli or cellular factors [13]. miRNAs are ubiquitous and can be present in different compartments of the body, including blood, urine and other body fluids. To avoid the degradation of miRNA by ribonucleases, they are often packaged in the form of micro-vesicles or exosomes or carried by RNA-binding proteins [14]. The miRNAs are potentially superior biomarkers than proteins and mRNAs because of their stability in body fluids and the high reproducibility by using accurate and sensitive amplification methods. However, the isolation and quantification of miRNAs are technique and time laborious, which limits their applications in routine clinical practice.
2. miRNAs in Kidney Homeostasis
Several miRNAs are expressed primarily in the adult human kidney (such as miR-215, miR-146a and miR-886); other miRNAs (for example, miR-192, miR-194, miR-21, miR-200a, miR-204 and let-7a–g), are increased in the kidney as well as in other organs [15]. This expression is tissue-specific and can be dependent on the developmental stage. Deletion of the miR-17~92 cluster leads to defective embryogenesis, affecting progenitor cells and the development of nephrons. Mice deficient in miR-17~92 are categorized by renal hypodysplasia and cause glomerular damage and proteinuria [16].
miRNAs are involved in different renal cellular processes, glomerular haemodynamia, as well as the maintenance of fluid and electrolyte balance. Other miRNAs in the kidney can also affect renin-expressing juxtaglomerular cells, which leads to damage to these cells. Consequently, plasma renin level decreased, and it was associated with hypotension and renal fibrosis [17]. The miRNAs also can help in the osmolarity homeostasis and have effects on the process of Na+, K+ and Ca2+ regulation in the condition of hypertonic environments [18].
3. Role miRNA in DKD: Murine Experiments
Hyperglycaemia can trigger a complex interplay between metabolic and hemodynamic factors leading to the genesis of various diabetic complications, including DKD. The presence of high glucose concentration has an adverse influence on all renal cell lineages (mesangial cells, tubular cells, podocytes and endothelial cells) and can modulate the expression of miRNAs affecting cellular intercommunication and kidney tissue homeostasis [19].
Numerous studies have demonstrated the difference in the expression of circulating miRNAs throughout the progression of DKD [15]. A high glucose condition affects the expression of miR-29a and miR-29c, leading to the apoptosis of podocytes and the promotion of pro-fibrotic substances [20,21,22]. The hyperglycemia also regulates the expression of miR-25, miR-93 and miR-192, which in turn affects the redox system, vascular endothelial growth factor and tubulointerstitial fibrosis [23,24,25]. A comprehensive description of changes in the expression of 41 miRNAs in different animal studies is summarized in Table 1.
Table 1.
miRNA implicated in the diabetic animal model.
| Mature Sequence of miRNA | Species | Experimental Model | Expression | Number of Study |
References |
|---|---|---|---|---|---|
| hsa-let-7i-5p | Mouse | Db mice | Up | 1 | [26] |
| hsa-miR-129-1-3p | Mouse | Db mice | Down | 1 | [26] |
| hsa-miR-130a-3p | Mouse, cell | Db mice, HepG2 cells | Up | 1 | [27] |
| hsa-miR-133b | Rat | High-Fat Diet/Streptozotocin-Induced Diabetic Rat | Up | 1 | [28] |
| hsa-miR-134-5p | Mouse, cell | C57BL/KsJ db mice, Db/Db mice, podocytes | Up | 1 | [29] |
| hsa-miR-146a-5p | Mouse, cell | C57BL/6J, miR-146a−/− mice, Podocytes, HK-2 human kidney cells, Db mice, peritoneal macrophages | Up, Down | 3 | [30,31,32] |
| hsa-miR-148b-3p | Rat, cell | Proximal tubular cells | Down | 1 | [33] |
| hsa-miR-16-5p | Rat | Male Wistar rats | Up | 1 | [34] |
| hsa-miR-181a-5p | Rat, cell | HK-2 cells, Otsuka-Long-Evans-Tokushima-Fatty rats | Down | 1 | [35] |
| hsa-miR-192-5p | Mouse | Db mice, Ets-1–deficient mice | Up | 1 | [36] |
| hsa-miR-200c-3p | Mouse, cell | Db mice, M4200 cells | Down | 3 | [26,37,38] |
| hsa-miR-21-5p | Mouse, rat, cell | C57BI/6J mice, miR-21-KO mice, DBA/2J mice, Db mice, kk-ay mice, epithelial-to-mesenchymal, podocytes | Up, Down | 7 | [39,40,41,42,43,44,45] |
| hsa-miR-217-5p | Cell | Glomerular mesangial cells | Up | 1 | [46] |
| hsa-miR-218-5p | Mouse, cell | Db mice, podocytes | Up, Down | 2 | [26,47] |
| hsa-miR-23c | Rat, cell | HK-2 cells | Down | 1 | [48] |
| hsa-miR-26a-5p | Rat, cell | OLETF rats, NRK-52E cells, glomerular, mesangial cells, podocytes, proximal tubular epithelial cells | Up, Down | 2 | [49,50] |
| hsa-miR-29a-3p | Mouse, cell | Db mice, Smad3-knockout (KO) db mice, mouse embryonic fibroblasts, NRK52E cells, mesangial cells | Down | 2 | [51,52] |
| hsa-miR-29a-5p | Mouse | Male FVB mice | Down | 1 | [53] |
| hsa-miR-29c-3p | Mouse | Db mice | Down | 1 | [26] |
| hsa-miR-30a-5p | Rat | High-Fat Diet/Streptozotocin-Induced Diabetic Rat | Up | 1 | [28] |
| hsa-miR-335-5p | Mouse | Male C57BL/6J mice | Up | 1 | [54] |
| hsa-miR-33a-5p | Mouse, cell | Db mice, MMCs (CRL1927) and HEK293 cells | Down | 1 | [55] |
| hsa-miR-342-5p | Rat | High-Fat Diet/Streptozotocin-Induced Diabetic Rat | Up | 1 | [28] |
| hsa-miR-34a-5p | Mouse, cell | Db mice, mesangial cells, HK-2cells, podocytes | Down | 3 | [56,57,58] |
| hsa-miR-375-5p | Mouse | Male C57BL/6J mice | Up | 1 | [54] |
| hsa-miR-378d | Mouse | Db mice | Up | 1 | [26] |
| hsa-miR-451a | Mouse, rat | Db mice, male Wistar rats | Down | 2 | [34,59] |
| hsa-miR-7977 | Mouse | Db mice | Down | 1 | [26] |
| hsa-miR-92b-5p | Rat | Lean Zucker, obese Zucker rats | Down | 1 | [60] |
| hsa-miR-93-5p | Mouse, cell | Pod-iCreERT2 mice, podocytes | Down | 1 | [61] |
| hsa-miR-99b-5p | Mouse, rat, cell | Db mice, mesangial cells, proximal tubular epithelial cells | Down | 4 | [62,63,64,65] |
miRNAs are annotated as a mature sequence of miRNA identified from the miRbase database.
4. Urinary miRNA Research: Human Evidence
Research into miRNA has unveiled obscure puzzles of the pathophysiology of DKD. Furthermore, other investigators have interrogated the clinical utility of these miRNAs for routine practice. Emerging evidence indicates the probable roles of urinary miRNAs as predictors of DKD development or progression [66]. Urine miRNA levels provide a direct reflection of kidney tissue damage. These miRNAs originate from cells and are encapsulated into extracellular vesicles, named exosomes, and are secreted in various biological fluids, including urine. Exosomes can exert paracrine effects and serve as a mediator of intercellular communication. They are stable in biological fluid as well as in paraffin-embedded sections, rendering these exosomes suitable as “liquid biopsy” or biomarkers of specific disease conditions [67]. A number of studies have examined the relationship between urinary microRNA and blood sugar levels. They found that concentrations of various urinary extravesical miRNAs (such as miRNA-941-5p, miRNA 34c-5p and miRNA-208a-3p) were correlated with levels of glycosylated hemoglobin [68]. Table 2 lists changes in the expression of urinary miRNA associated with DKD in clinical research. We identified 141 unique urinary miRNAs associated with DKD. Peculiarly, the direction of expression of specific miRNA differs between types of DM (type 1 vs. 2) and also between distinct patient cohorts (as highlighted in bold). A possible explanation might reside in unclear mechanisms between the two types of DM. In addition, several technical concerns may affect the miRNA profiling, including methods of specimen collection (processing and storage), a fraction of extracted urine (urine, urinary extra-vesicle, extra-vesicle depleted urine fraction), analytic platform (qPCR, microarray, genome-wide profiling by small-RNA sequencing) and artifact contamination [68]. Finally, the cell source of urinary miRNA can arise from any genitourinary tract, and the function of urinary miRNA is not necessarily the same as circulating ones. Sophisticated bioinformatics analysis and network interaction maps are capable of identifying Gene Ontology processes, classification and relevant pathways of target genes in this modern era [66,69]. These approaches may help to decipher the biological functions of urinary miRNA in the pathophysiology of DKD.
Table 2.
Urinary miRNA implicated in human studies related to diabetic kidney disease.
| Mature Sequence of miRNA | Sample | DM Type | Patient Number |
Urinary Expression in DKD |
References |
|---|---|---|---|---|---|
| has-let-7a-3p | Blood/Urine | 2 | 27 | Down | [66] |
| hsa-let-7c-5p | Urine | 2 | 63 | Up | [70] |
| hsa-let-7f-1-3p | Blood/Urine | 2 | 27 | Down | [66] |
| hsa-let-7i-5p | Urine | 2 | 160 | Up | [71] |
| hsa-miR-106b-3p | Blood/Urine | 2 | 27 | Down | [66] |
| hsa-miR-10a-5p | Urine | 1 | 48 | Up vs. PMA | [68] |
| hsa-miR-10b-5p | Urine | 1 | 48 | Up vs. PMA | [68] |
| hsa-miR-1224-3p | Urine | 1 | 40 | Up | [72] |
| hsa-miR-122-5p | Urine | 1 | 48 | Up | [68] |
| hsa-miR-126 | Urine | 2 | 92 | Up | [73] |
| hsa-miR-126-5p | Urine/blood | 1 | 147 | Down | [74] |
| hsa-miR-1275 | Urine | 2 | 6 | Up | [75] |
| hsa-miR-1307-3p | Urine | 1 | 48 | Down | [68] |
| hsa-miR-130a-3p | Urine | 1 | 48 | Up | [68] |
| hsa-miR-130a-5p | Tissue/Urine | 1 | 24 | Up | [76] |
| hsa-miR-133a-3p | Urine | 1 | 48 | Up | [68] |
| hsa-miR-133a-3p | Urine | 2 | 6 | Down | [77] |
| hsa-miR-133b | Urine | 2 | 220 | Up | [78] |
| hsa-miR-135b-5p | Urine | 2 | 160 | Up | [71] |
| hsa-miR-141-3p | Urine | 1 | 40 + 48 | Up | [68,72] |
| hsa-miR-142-3p | Urine | 1 | 48 | Up | [68] |
| hsa-miR-145-5p | Tissue/Urine | 1 | 24 | Up | [76] |
| hsa-miR-146 | Urine | 2 | 92 | Up | [73] |
| hsa-miR-148a-3p | Urine | 1 | 48 | Up | [68] |
| hsa-miR-148b-3p | Urine | 2 | 56 | Down | [79] |
| hsa-miR-150-3p | Urine | 2 | 6 | Up | [77] |
| hsa-miR-150-5p | Urine | 2 | 80 | Up | [80] |
| hsa-miR-152-3p | Urine | 1 | 48 | Up | [68] |
| hsa-miR-153-3 | Urine | 2 | 6 | Down | [77] |
| hsa-miR-155 | Urine | 2 | 92 | Up | [73] |
| hsa-miR-155-5p | Tissue/Urine | 1 | 24 | Down | [76] |
| hsa-miR-1587 | Urine | 2 | 6 | Up | [75] |
| hsa-miR-15a-5p | Urine | 2 | 80 | Down | [80] |
| hsa-miR-15b-5p | Urine | 2 | 232 + 63 + 160 | Up/Down/Down | [70,71,81] |
| hsa-miR-17-5p | Urine | 2 | 56 | Down | [79] |
| hsa-miR-17-5p | Urine | 1 | 40 | Up | [72] |
| hsa-miR-181a-5p | Urine | 1 | 48 | Up | [68] |
| hsa-miR-183-5p | Urine | 1 | 48 | Up | [68] |
| hsa-miR-188-3p | Urine | 1 | 40 | Down | [72] |
| hsa-miR-188-5p | Urine | 1 | 48 | Down | [68] |
| hsa-miR-188-5p | Urine | 2 | 6 | Up | [77] |
| hsa-miR-190a-5p | Blood/Urine | 2 | 27 | Down | [66] |
| hsa-miR-1912 | Urine | 1 | 40 | Up | [72] |
| hsa-miR-1913 | Urine | 1 | 40 | UP | [72] |
| hsa-miR-192-5p | Urine | 2 | 56 | Down | [79] |
| hsa-miR-192-5p | Urine | 1 | 48 | Up | [68] |
| hsa-miR-193b-5p | Urine | 2 | 6 | Down | [69] |
| hsa-miR-196a | Urine | 2 | 209 | UP | [82] |
| hsa-miR-197-3p | Urine | 2 | 160 | Down | [71] |
| hsa-miR-197-3p | Urine | 1 | 48 | Down | [68] |
| hsa-miR-200a-3p | Urine | 1 | 48 | Up | [68] |
| hsa-miR-200c-3p | Urine | 1 | 48 | Up | [68] |
| hsa-miR-2117 | Urine | 2 | 6 | Up | [75] |
| hsa-miR-214-3p | Urine | 1 | 40 | UP | [72] |
| hsa-miR-21-5p | Urine | 2 | 56 | Down | [79] |
| hsa-miR-21-5p | Urine/blood | 1 | 147 | Up | [74] |
| hsa-miR-216a-5p | Urine | 2 | 56 | Down | [79] |
| hsa-miR-216a-5p | Urine | 1 | 50 | Down | [83] |
| hsa-miR-217-5p | Urine | 2 | 56 | Down | [79] |
| hsa-miR-219a-3p | Urine | 2 | 6 | Up | [75] |
| hsa-miR-221-3p | Urine | 1 | 40 | Down | [72] |
| hsa-miR-222-3p | Urine | 1 | 48 + 40 | Up | [68,72] |
| hsa-miR-22-3p | Urine | 1 | 48 | Up | [68] |
| hsa-miR-23c | Urine | 2 | 6 | Up | [77] |
| hsa-miR-24-3p | Urine | 2 | 160 | Up | [71] |
| hsa-miR-27b-3p | Urine | 2 | 160 | Down | [71] |
| hsa-miR-29a-3p | Urine | 2 | 83 | NC | [84] |
| hsa-miR-29a-5p | Urine | 2 | 83 | Up | [84] |
| hsa-miR-29b-1-5p | Urine | 1 | 40 | Up | [72] |
| hsa-miR-29c-3p | Urine | 2 | 220 + 56 + 63 | Up/Up/Down | [70,78,79] |
| hsa-miR-29c-3p | Urine | 1 | 48 | Up | [68] |
| hsa-miR-29c-5p | Urine | 2 | 83 | NC | [84] |
| hsa-miR-29c-5p | Tissue/Blood/Urine | 2 | 27 + 16 | Down | [66,85] |
| hsa-miR-30a-3p | Urine | 2 | 160 | Up | [71] |
| hsa-miR-30a-5p | Urine | 1 | 48 + 27 | Up/Down | [68,86] |
| hsa-miR-30b-5p | Blood/Urine | 2 | 27 | Down | [66] |
| hsa-miR-30c-5p | Blood/Urine | 2 | 27 | Down | [66] |
| hsa-miR-30d-5p | Blood/Urine | 2 | 27 | Down | [66] |
| hsa-miR-30e-3p | Blood/Urine | 2 | 27 | Down | [66] |
| hsa-miR-3137 | Urine | 2 | 6 | Down | [69] |
| hsa-miR-31-5p | Urine | 1 | 48 | Up | [68] |
| hsa-miR-3168 | Urine | 1 | 48 | Down | [68] |
| hsa-miR-3184-3p | Urine | 1 | 48 | Up | [68] |
| hsa-miR-320b | Blood/Urine | 2 | 27 | Up | [66] |
| hsa-miR-320c | Urine | 2 | 41 | Up | [87] |
| hsa-miR-320e | Urine | 2 | 6 | Up | [77] |
| hsa-miR-323b-5p | Urine | 1 | 40 | Down | [72] |
| hsa-miR-331-3p | Blood/Urine | 2 | 27 | Down | [66] |
| hsa-miR-335-5p | Urine | 1 | 40 | Up | [72] |
| hsa-miR-339-3p | Urine | 1 | 48 | Up | [68] |
| hsa-miR-342-3p | Urine | 1 | 48 | Up | [68] |
| hsa-miR-342-5p | Urine | 2 | 220 | Up | [78] |
| hsa-miR-34a-5p | Urine | 2 | 232 | Up | [81] |
| hsa-miR-362-3p | Urine | 2 | 80 | Up | [80] |
| hsa-miR-362-5p | Urine | 1 | 48 | Down | [68] |
| hsa-miR-363-3p | Urine | 1 | 27 | Down | [86] |
| hsa-miR-3677-3p | Urine | 2 | 6 | Up | [77] |
| hsa-miR-373-5p | Urine | 1 | 40 | Down | [72] |
| hsa-miR-375-5p | Urine | 2 | 160 | Down | [71] |
| hsa-miR-377-5p | Urine | 1 | 50 | Up | [83] |
| hsa-miR-377-5p | Urine | 2 | 56 | Up | [79] |
| hsa-miR-424-3p | Urine | 1 | 48 | Down | [68] |
| hsa-miR-424-5p | Tissue/Urine | 1 | 40 + 24 + 27 | Up/Down/Down | [72,76,86] |
| hsa-miR-4270 | Urine | 2 | 6 | Up | [69] |
| hsa-miR-4286 | Urine | 1 | 48 | Down | [68] |
| hsa-miR-429 | Urine | 1 | 40 | UP | [72] |
| hsa-miR-433 | Urine | 1 | 40 | Up | [72] |
| hsa-miR-4491 | Urine | 2 | 6 | Up | [75] |
| hsa-miR-4507 | Urine | 2 | 6 | Up | [75] |
| hsa-miR-4516 | Urine | 2 | 6 | Up | [75] |
| hsa-mir-453 | Urine | 1 | 40 | Down (Up in persistent microalbuminuria) | [72] |
| hsa-miR-4534 | Urine | 2 | 6 | Up | [75] |
| hsa-miR-4687-3p | Urine | 2 | 6 | Up | [75] |
| hsa-miR-486-3p | Urine | 1 | 40 | Up | [72] |
| hsa-miR-486-5p | Urine | 1 | 27 | Up | [86] |
| hsa-miR-495-5p | Urine | 1 | 27 | Down | [86] |
| hsa-miR-498 | Urine | 2 | 6 | Up | [75] |
| hsa-miR-5007-3p | Urine | 2 | 6 | Up | [75] |
| hsa-miR-500a-5p | Urine | 2 | 160 | Down | [71] |
| hsa-miR-5088-5p | Urine | 2 | 6 | Up | [75] |
| hsa-miR-5091 | Urine | 2 | 6 | Up | [75] |
| hsa-miR-516b-5p, | Urine | 2 | 6 | Up | [75] |
| hsa-miR-520h | Urine | 1 | 40 | Down | [72] |
| hsa-miR-524-5p | Urine | 1 | 40 + 27 | Down/Down | [72,86] |
| hsa-miR-548ah-3p | Urine | 2 | 6 | Up | [77] |
| hsa-miR-548p | Urine | 2 | 6 | Up | [77] |
| hsa-miR-552-5p | Urine | 1 | 40 | Up | [72] |
| hsa-miR-589-5p | Urine | 1 | 40 | Down | [72] |
| hsa-miR-6068 | Urine | 2 | 41 | Up | [87] |
| hsa-miR-6076 | Urine | 2 | 41 | NC | [87] |
| hsa-miR-616-5p | Urine | 1 | 27 | Up | [86] |
| hsa-miR-619-5p | Urine | 1 | 40 | Up | [72] |
| hsa-miR-628-5p | Urine | 1 | 40 | Up | [72] |
| hsa-miR-636 | Urine | 2 | 232 | Up | [81] |
| hsa-miR-638 | Urine | 1 | 40 | UP | [72] |
| hsa-miR-640 | Urine | 1 | 27 | Up | [86] |
| hsa-miR-645 | Urine | 1 | 27 | Up | [86] |
| hsa-miR-665 | Urine | 1 | 27 | Down | [86] |
| hsa-miR-6809-5p | Urine | 2 | 6 | Down | [69] |
| hsa-miR-6831-5p | Urine | 2 | 6 | Up | [69] |
| hsa-miR-760 | Urine | 2 | 6 | Up | [77] |
| hsa-miR-765 | Urine | 1 | 40 | UP | [72] |
| hsa-miR-767-3p | Urine | 1 | 27 | Down | [86] |
| hsa-miR-770-5p | Urine | 1 | 27 | Up | [86] |
| hsa-miR-7846-3p | Urine | 2 | 6 | Up | [69] |
| hsa-miR-877-3p | Urine | 2 | 80 | Up | [80] |
| hsa-miR-92a-3p | Urine | 1 | 48 + 40 | Up/Down | [68,72] |
| hsa-miR-92b-5p | Urine | 1 | 40 | UP | [72] |
| hsa-miR-93-5p | Urine | 2 | 160 | Up | [71] |
| hsa-miR-98-3p | Blood/Urine | 2 | 27 | Down | [66] |
| hsa-miR-99b-5p | Urine | 1 | 48 | Up | [68] |
| hsa-miR-99b-5p | Blood/Urine | 2 | 27 | Down | [66] |
miRNAs are annotated as a mature sequence of miRNA identified from the miRbase database. NC: not changed. miRNA in bold indicates differentially expressed direction between publications.
5. Performances of Urinary miRNAs as Disease Biomarker
A body of literature attempted to identify potential biomarkers from urine specimens in predicting the degree of kidney damage. Individual miRNAs or a cluster of miRNAs were used as possible biomarkers of DKD with satisfactory prediction performance. Li et al. found that urinary expression of let-7c-5p, miR29c-5p and miR-15b-5p could predict DKD with the area under curves (AUC) of 0.818, 0.774, and 0.818, respectively [70]. Eissa S et al. observed that the AUC of miR-15b, miR-34a, and miR-636 were 0.883, 0.917 and 0.984, respectively, for distinguishing DKD from controls. The panel composed of three miRNAs yielded an AUC of 0.912 [78,81]. The expressed levels of miR-95-3p, miR-185-5p, miR-1246, and miR-631 in urinary sediments can also yield a good accuracy (AUC, 0.863) for differentiating DKD from non-DKD or other disease conditions [88]. Collectively, all this evidence suggests the potential use of urinary miRNAs as non-invasive biomarkers for predicting DKD.
6. Conclusions
Sensitive biomarkers to guide decision-making in the management of DKD remain urgently awaited. Urinary miRNA may represent promising non-invasive, and cheap means with diagnostic or predictive implications in DKD. However, the clinical application is unsatisfactory to date because of inconsistencies between reported data. Most of the research projects reported were discovery experiments with small samples and limitation validation. In addition, the discrepancies may be in part explained by differences in procedures of urine isolation, the proportion of urine fraction (fresh urine, concentrates of extra-vesicles or extra-vesicle depleted fraction) used, heterogeneity in reporting outcome and in the degree of kidney severity (micro vs. macroalbuminuria, intermittent vs. persistent proteinuria). Furthermore, the advances in analytic methodology enabled the discovery of new miRNAs. Recently, the introduction of the sensor-based methodology using labels on magnetic beads can magnify measurement power. The incorporation of the use of spectrophotometry or electrochemistry, rather than direct visualization, can further enhance the quantification accuracy [89] with small sample sizes and limited validation. Consensus and standardization of methodologies applied to retrieve, isolate, store and measure miRNAs may enhance the reproducibility of the study results. Further validation with an extended sample size is warranted to move the field forward clinical translation of urinary miRNA in DKD.
7. Review Criteria
We conducted a narrative review of animal and human studies in published literature from 1 October 2013 until 30 July 2022. The PubMed database was used for searching the scientific literature using the following search terms: “microRNA”, “diabetic kidney disease”, “diabetic nephropathy”, and “urinary”. This narrative literature review primarily focused on original articles written in the English language and published in peer-reviewed journals. The reference lists of included articles were also hand-searched. References were managed using EndNote 20.1.
Independent researchers (CKH and YTC) managed titles and abstracts to identify potentially eligible studies for full-text review. We only included original articles and case series with over two cases in the English language for further assessment. Data extraction was performed in duplicate by two independent reviewers (CCL, KJY). When multiple articles reporting data from the same study population were identified, the most comprehensive data were used. All reported miRNAs are indexed and annotated as a mature sequence of miRNA identified from the miRbase database. Studies were required to provide quantizable data on the expression levels of miRNA, such as fold changes (down-expression, up-expression, not change or absolute value) or area under curves. We contacted the study authors regarding possible incomplete data on miRNA quantities presented in selected publications. The methodological quality of included studies was not assessed in this narrative review because large heterogenicity presented in the testing techniques (ELISA, microarray, q-PCR, sequencing, etc.) and differences in reporting outcomes.
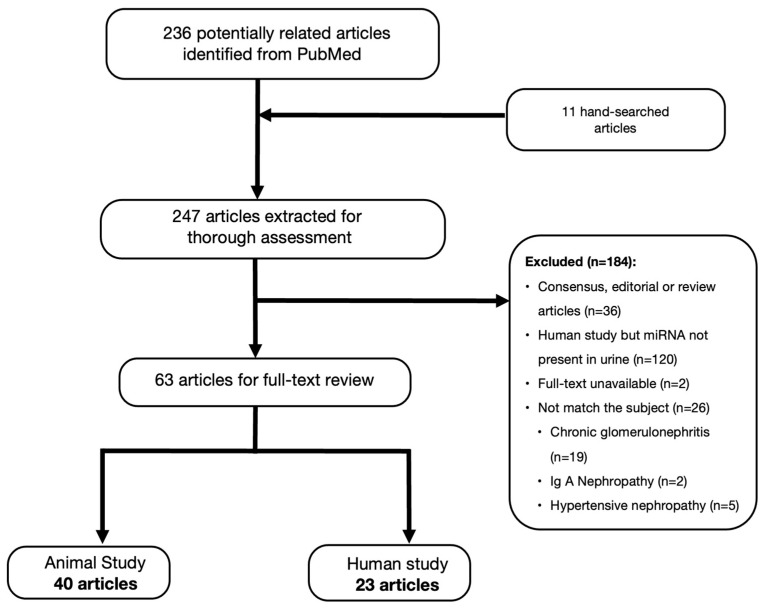
We identified 247 records from the PubMed database for the initial assessment, and only 63 articles were included for full-text review; 40 articles related to animal studies and 23 human research articles, respectively (Figure 1). This literature review included a thorough assessment of 41 miRNA from animal experiments and 141 miRNA from urine samples of DKD patients.
Figure 1.
Flow chart of literature search and selection.
Acknowledgments
We thank all researchers and clinicians involved in the individual trials.
Author Contributions
Conceptualization, C.-C.C. and I.-W.W.; methodology, C.-K.H.; formal analysis, K.-J.Y.; investigation, Y.-T.C.; resources, I.-W.W.; data curation, C.-Y.C. and K.-J.Y.; writing—original draft preparation, C.-C.L.; writing—review and editing, I.-W.W.; supervision, C.-C.C. and M.-J.H.; project administration, K.-J.Y.; funding acquisition, I.-W.W. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by Chang Gung Memorial Hospital (CMRPG2C0342, CRRPG2H0124, CRRPG2H0154).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.WHO Improving Health Outcomes of People with Diabetes Mellitus: Target Setting to Reduce the Global Burden of Diabetes Mellitus by 2030. 2021. [(accessed on 30 October 2022)]. Available online: https://www.who.int/publications/m/item/improving-health-outcomes-of-people-with-diabetes-mellitus.
- 2.Federation I.D. IDF Diabetes Atlas. Ninth edition 2019. [(accessed on 13 September 2021)]. Available online: www.diabetesatlas.org.
- 3.Alicic R.Z., Cox E.J., Neumiller J.J., Tuttle K.R. Incretin drugs in diabetic kidney disease: Biological mechanisms and clinical evidence. Nat. Rev. Nephrol. 2021;17:227–244. doi: 10.1038/s41581-020-00367-2. [DOI] [PubMed] [Google Scholar]
- 4.System U.S.R.D. 2020 USRDS Annual Data Report: Epidemiology of Kidney Disease in the United States. National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases; Bethesda, MD, USA: 2020. [Google Scholar]
- 5.Webster A.C., Nagler E.V., Morton R.L., Masson P. Chronic Kidney Disease. Lancet. 2017;389:1238–1252. doi: 10.1016/S0140-6736(16)32064-5. [DOI] [PubMed] [Google Scholar]
- 6.Wen C.P., Cheng T.Y.D., Tsai M.K., Chang Y.C., Chan H.T., Tsai S.P., Chiang P.H., Hsu C.C., Sung P.K., Hsu Y.H., et al. All-cause mortality attributable to chronic kidney disease: A prospective cohort study based on 462 293 adults in Taiwan. Lancet. 2008;371:2173–2182. doi: 10.1016/S0140-6736(08)60952-6. [DOI] [PubMed] [Google Scholar]
- 7.GBD Chronic Kidney Disease Collaboration Global, regional, and national burden of chronic kidney disease, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2020;395:709–733. doi: 10.1016/S0140-6736(20)30045-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Betz B.B., Jenks S.J., Cronshaw A.D., Lamont D.J., Cairns C., Manning J.R., Goddard J., Webb D.J., Mullins J.J., Hughes J., et al. Urinary peptidomics in a rodent model of diabetic nephropathy highlights epidermal growth factor as a biomarker for renal deterioration in patients with type 2 diabetes. Kidney Int. 2016;89:1125–1135. doi: 10.1016/j.kint.2016.01.015. [DOI] [PubMed] [Google Scholar]
- 9.Alvarez M.L., Khosroheidari M., Kanchi Ravi R., Distefano J.K. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 2012;82:1024–1032. doi: 10.1038/ki.2012.256. [DOI] [PubMed] [Google Scholar]
- 10.Krutzfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 11.Trionfini P., Benigni A., Remuzzi G. microRNAs in kidney physiology and disease. Nat. Rev. Nephrol. 2015;11:23–33. doi: 10.1038/nrneph.2014.202. [DOI] [PubMed] [Google Scholar]
- 12.Kozomara A., Birgaoanu M., Griffiths-Jones S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2018;47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandrasekaran K., Karolina D.S., Sepramaniam S., Armugam A., Wintour E.M., Bertram J.F., Jeyaseelan K. Role of microRNAs in kidney homeostasis and disease. Kidney Int. 2012;81:617–627. doi: 10.1038/ki.2011.448. [DOI] [PubMed] [Google Scholar]
- 14.Sun I.O., Lerman L.O. Urinary microRNA in kidney disease: Utility and roles. Am. J. Physiol. Renal Physiol. 2019;316:F785–F793. doi: 10.1152/ajprenal.00368.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato M., Natarajan R. Diabetic nephropathy--emerging epigenetic mechanisms. Nat. Rev. Nephrol. 2014;10:517–530. doi: 10.1038/nrneph.2014.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McClelland A., Hagiwara S., Kantharidis P. Where are we in diabetic nephropathy: microRNAs and biomarkers? Curr. Opin. Nephrol. Hypertens. 2014;23:80–86. doi: 10.1097/01.mnh.0000437612.50040.ae. [DOI] [PubMed] [Google Scholar]
- 17.Carney E.F. Diabetic nephropathy: MiR-23b protects against fibrosis in diabetic nephropathy. Nat. Rev. Nephrol. 2016;12:197. doi: 10.1038/nrneph.2016.22. [DOI] [PubMed] [Google Scholar]
- 18.Trionfini P., Benigni A. microRNAs as Master Regulators of Glomerular Function in Health and Disease. J. Am. Soc. Nephrol. 2017;28:1686–1696. doi: 10.1681/ASN.2016101117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeFronzo R.A., Reeves W.B., Awad A.S. Pathophysiology of diabetic kidney disease: Impact of SGLT2 inhibitors. Nat. Rev. Nephrol. 2021;17:319–334. doi: 10.1038/s41581-021-00393-8. [DOI] [PubMed] [Google Scholar]
- 20.Rao P.V., Lu X., Standley M., Pattee P., Neelima G., Girisesh G., Dakshinamurthy K.V., Roberts C.T., Jr., Nagalla S.R. Proteomic identification of urinary biomarkers of diabetic nephropathy. Diabetes Care. 2007;30:629–637. doi: 10.2337/dc06-2056. [DOI] [PubMed] [Google Scholar]
- 21.Zhou L., Xu D.Y., Sha W.G., Shen L., Lu G.Y., Yin X., Wang M.J. High glucose induces renal tubular epithelial injury via Sirt1/NF-kappaB/microR-29/Keap1 signal pathway. J. Transl. Med. 2015;13:352. doi: 10.1186/s12967-015-0710-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang B., Komers R., Carew R., Winbanks C.E., Xu B., Herman-Edelstein M., Koh P., Thomas M., Jandeleit-Dahm K., Gregorevic P., et al. Suppression of microRNA-29 expression by TGF-beta1 promotes collagen expression and renal fibrosis. J. Am. Soc. Nephrol. 2012;23:252–265. doi: 10.1681/ASN.2011010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Putta S., Lanting L., Sun G., Lawson G., Kato M., Natarajan R. Inhibiting microRNA-192 ameliorates renal fibrosis in diabetic nephropathy. J. Am. Soc. Nephrol. 2012;23:458–469. doi: 10.1681/ASN.2011050485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu Y., Zhang Y., Wang Z., Wang L., Wei X., Zhang B., Wen Z., Fang H., Pang Q., Yi F. Regulation of NADPH oxidase activity is associated with miRNA-25-mediated NOX4 expression in experimental diabetic nephropathy. Am. J. Nephrol. 2010;32:581–589. doi: 10.1159/000322105. [DOI] [PubMed] [Google Scholar]
- 25.Long J., Wang Y., Wang W., Chang B.H., Danesh F.R. Identification of microRNA-93 as a novel regulator of vascular endothelial growth factor in hyperglycemic conditions. J. Biol. Chem. 2010;285:23457–23465. doi: 10.1074/jbc.M110.136168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du G., Xiao M., Zhang X., Wen M., Pang C., Jiang S., Sang S., Xie Y. Alpinia oxyphylla Miq. extract changes miRNA expression profiles in db-/db- mouse kidney. Biol. Res. 2017;50:9. doi: 10.1186/s40659-017-0111-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao F., Yu J., Liu B., Guo Y., Li K., Deng J., Zhang J., Wang C., Chen S., Du Y., et al. A novel function of microRNA 130a-3p in hepatic insulin sensitivity and liver steatosis. Diabetes. 2014;63:2631–2642. doi: 10.2337/db13-1689. [DOI] [PubMed] [Google Scholar]
- 28.Matboli M., Eissa S., Ibrahim D., Hegazy M.G.A., Imam S.S., Habib E.K. Caffeic Acid Attenuates Diabetic Kidney Disease via Modulation of Autophagy in a High-Fat Diet/Streptozotocin- Induced Diabetic Rat. Sci. Rep. 2017;7:2263. doi: 10.1038/s41598-017-02320-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qian X., Tan J., Liu L., Chen S., You N., Yong H., Pan M., You Q., Ding D., Lu Y. microRNA-134-5p promotes high glucose-induced podocyte apoptosis by targeting bcl-2. Am. J. Transl. Res. 2018;10:989–997. [PMC free article] [PubMed] [Google Scholar]
- 30.Bhatt K., Lanting L.L., Jia Y., Yadav S., Reddy M.A., Magilnick N., Boldin M., Natarajan R. Anti-Inflammatory Role of microRNA-146a in the Pathogenesis of Diabetic Nephropathy. J. Am. Soc. Nephrol. 2016;27:2277–2288. doi: 10.1681/ASN.2015010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee H.W., Khan S.Q., Khaliqdina S., Altintas M.M., Grahammer F., Zhao J.L., Koh K.H., Tardi N.J., Faridi M.H., Geraghty T., et al. Absence of miR-146a in Podocytes Increases Risk of Diabetic Glomerulopathy via Up-Regulation of ErbB4 and Notch-1. J. Biol. Chem. 2017;292:732–747. doi: 10.1074/jbc.M116.753822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wan R.J., Li Y.H. microRNA146a/NAPDH oxidase4 decreases reactive oxygen species generation and inflammation in a diabetic nephropathy model. Mol. Med. Rep. 2018;17:4759–4766. doi: 10.3892/mmr.2018.8407. [DOI] [PubMed] [Google Scholar]
- 33.Kuwagata S., Kume S., Chin-Kanasaki M., Araki H., Araki S., Nakazawa J., Sugaya T., Koya D., Haneda M., Maegawa H., et al. microRNA148b-3p inhibits mTORC1-dependent apoptosis in diabetes by repressing TNFR2 in proximal tubular cells. Kidney Int. 2016;90:1211–1225. doi: 10.1016/j.kint.2016.06.036. [DOI] [PubMed] [Google Scholar]
- 34.Mohan A., Singh R.S., Kumari M., Garg D., Upadhyay A., Ecelbarger C.M., Tripathy S., Tiwari S. Urinary Exosomal microRNA-451-5p Is a Potential Early Biomarker of Diabetic Nephropathy in Rats. PLoS ONE. 2016;11:e0154055. doi: 10.1371/journal.pone.0154055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu P., Guan M.P., Bi J.G., Wang D., Zheng Z.J., Xue Y.M. High glucose down-regulates microRNA-181a-5p to increase pro-fibrotic gene expression by targeting early growth response factor 1 in HK-2 cells. Cell. Signal. 2017;31:96–104. doi: 10.1016/j.cellsig.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 36.Kato M., Dang V., Wang M., Park J.T., Deshpande S., Kadam S., Mardiros A., Zhan Y., Oettgen P., Putta S., et al. TGF-beta induces acetylation of chromatin and of Ets-1 to alleviate repression of miR-192 in diabetic nephropathy. Sci. Signal. 2013;6:ra43. doi: 10.1126/scisignal.2003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dou L., Zhao T., Wang L., Huang X., Jiao J., Gao D., Zhang H., Shen T., Man Y., Wang S., et al. miR-200s contribute to interleukin-6 (IL-6)-induced insulin resistance in hepatocytes. J. Biol. Chem. 2013;288:22596–22606. doi: 10.1074/jbc.M112.423145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Z.M., Zheng H.Y., Chen L.H., Li Y.L., Wang Q., Liao C.F., Li X.W. Low expression of miR-203 promoted diabetic nephropathy via increasing TLR4. Eur. Rev. Med. Pharmacol. Sci. 2018;22:5627–5634. doi: 10.26355/eurrev_201809_15828. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z., Peng H., Chen J., Chen X., Han F., Xu X., He X., Yan N. microRNA-21 protects from mesangial cell proliferation induced by diabetic nephropathy in db/db mice. FEBS Lett. 2009;583:2009–2014. doi: 10.1016/j.febslet.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 40.Wang J., Gao Y., Ma M., Li M., Zou D., Yang J., Zhu Z., Zhao X. Effect of miR-21 on renal fibrosis by regulating MMP-9 and TIMP1 in kk-ay diabetic nephropathy mice. Cell Biochem. Biophys. 2013;67:537–546. doi: 10.1007/s12013-013-9539-2. [DOI] [PubMed] [Google Scholar]
- 41.Chen X., Zhao L., Xing Y., Lin B. Down-regulation of microRNA-21 reduces inflammation and podocyte apoptosis in diabetic nephropathy by relieving the repression of TIMP3 expression. Biomed. Pharmacother. 2018;108:7–14. doi: 10.1016/j.biopha.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 42.Zhong X., Chung A.C., Chen H.Y., Dong Y., Meng X.M., Li R., Yang W., Hou F.F., Lan H.Y. miR-21 is a key therapeutic target for renal injury in a mouse model of type 2 diabetes. Diabetologia. 2013;56:663–674. doi: 10.1007/s00125-012-2804-x. [DOI] [PubMed] [Google Scholar]
- 43.Wang J.Y., Gao Y.B., Zhang N., Zou D.W., Wang P., Zhu Z.Y., Li J.Y., Zhou S.N., Wang S.C., Wang Y.Y., et al. miR-21 overexpression enhances TGF-beta1-induced epithelial-to-mesenchymal transition by target smad7 and aggravates renal damage in diabetic nephropathy. Mol. Cell. Endocrinol. 2014;392:163–172. doi: 10.1016/j.mce.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 44.Lai J.Y., Luo J., O’Connor C., Jing X., Nair V., Ju W., Randolph A., Ben-Dov I.Z., Matar R.N., Briskin D., et al. microRNA-21 in glomerular injury. J. Am. Soc. Nephrol. 2015;26:805–816. doi: 10.1681/ASN.2013121274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J., Duan L., Tian L., Liu J., Wang S., Gao Y., Yang J. Serum miR-21 may be a Potential Diagnostic Biomarker for Diabetic Nephropathy. Exp. Clin. Endocrinol. Diabetes. 2016;124:417–423. doi: 10.1055/s-0035-1565095. [DOI] [PubMed] [Google Scholar]
- 46.Shao Y., Lv C., Wu C., Zhou Y., Wang Q. Mir-217 promotes inflammation and fibrosis in high glucose cultured rat glomerular mesangial cells via Sirt1/HIF-1alpha signaling pathway. Diabetes Metab. Res. Rev. 2016;32:534–543. doi: 10.1002/dmrr.2788. [DOI] [PubMed] [Google Scholar]
- 47.Yang H., Wang Q., Li S. microRNA-218 promotes high glucose-induced apoptosis in podocytes by targeting heme oxygenase-1. Biochem. Biophys. Res. Commun. 2016;471:582–588. doi: 10.1016/j.bbrc.2016.02.028. [DOI] [PubMed] [Google Scholar]
- 48.Li X., Zeng L., Cao C., Lu C., Lian W., Han J., Zhang X., Zhang J., Tang T., Li M. Long noncoding RNA MALAT1 regulates renal tubular epithelial pyroptosis by modulated miR-23c targeting of ELAVL1 in diabetic nephropathy. Exp. Cell Res. 2017;350:327–335. doi: 10.1016/j.yexcr.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 49.Dey N., Bera A., Das F., Ghosh-Choudhury N., Kasinath B.S., Choudhury G.G. High glucose enhances microRNA-26a to activate mTORC1 for mesangial cell hypertrophy and matrix protein expression. Cell. Signal. 2015;27:1276–1285. doi: 10.1016/j.cellsig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng Z., Guan M., Jia Y., Wang D., Pang R., Lv F., Xiao Z., Wang L., Zhang H., Xue Y. The coordinated roles of miR-26a and miR-30c in regulating TGFbeta1-induced epithelial-to-mesenchymal transition in diabetic nephropathy. Sci. Rep. 2016;6:37492. doi: 10.1038/srep37492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen H.Y., Zhong X., Huang X.R., Meng X.M., You Y., Chung A.C., Lan H.Y. microRNA-29b inhibits diabetic nephropathy in db/db mice. Mol. Ther. 2014;22:842–853. doi: 10.1038/mt.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun S.F., Tang P.M.K., Feng M., Xiao J., Huang X.R., Li P., Ma R.C.W., Lan H.Y. Novel lncRNA Erbb4-IR Promotes Diabetic Kidney Injury in db/db Mice by Targeting miR-29b. Diabetes. 2018;67:731–744. doi: 10.2337/db17-0816. [DOI] [PubMed] [Google Scholar]
- 53.Lin C.L., Lee P.H., Hsu Y.C., Lei C.C., Ko J.Y., Chuang P.C., Huang Y.T., Wang S.Y., Wu S.L., Chen Y.S., et al. microRNA-29a promotion of nephrin acetylation ameliorates hyperglycemia-induced podocyte dysfunction. J. Am. Soc. Nephrol. 2014;25:1698–1709. doi: 10.1681/ASN.2013050527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Higuchi C., Nakatsuka A., Eguchi J., Teshigawara S., Kanzaki M., Katayama A., Yamaguchi S., Takahashi N., Murakami K., Ogawa D., et al. Identification of circulating miR-101, miR-375 and miR-802 as biomarkers for type 2 diabetes. Metabolism. 2015;64:489–497. doi: 10.1016/j.metabol.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 55.Tsai Y.C., Kuo P.L., Hung W.W., Wu L.Y., Wu P.H., Chang W.A., Kuo M.C., Hsu Y.L. Angpt2 Induces Mesangial Cell Apoptosis through the microRNA-33-5p-SOCS5 Loop in Diabetic Nephropathy. Mol. Ther. Nucleic Acids. 2018;13:543–555. doi: 10.1016/j.omtn.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu X.D., Zhang L.Y., Zhu T.C., Zhang R.F., Wang S.L., Bao Y. Overexpression of miR-34c inhibits high glucose-induced apoptosis in podocytes by targeting Notch signaling pathways. Int. J. Clin. Exp. Pathol. 2015;8:4525–4534. [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang L., He S., Guo S., Xie W., Xin R., Yu H., Yang F., Qiu J., Zhang D., Zhou S., et al. Down-regulation of miR-34a alleviates mesangial proliferation in vitro and glomerular hypertrophy in early diabetic nephropathy mice by targeting GAS1. J. Diabetes Complicat. 2014;28:259–264. doi: 10.1016/j.jdiacomp.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xue M., Li Y., Hu F., Jia Y.J., Zheng Z.J., Wang L., Xue Y.M. High glucose up-regulates microRNA-34a-5p to aggravate fibrosis by targeting SIRT1 in HK-2cells. Biochem. Biophys. Res. Commun. 2018;498:38–44. doi: 10.1016/j.bbrc.2017.12.048. [DOI] [PubMed] [Google Scholar]
- 59.Sun Y., Peng R., Peng H., Liu H., Wen L., Wu T., Yi H., Li A., Zhang Z. miR-451 suppresses the NF-kappaB-mediated proinflammatory molecules expression through inhibiting LMP7 in diabetic nephropathy. Mol. Cell Endocrinol. 2016;433:75–86. doi: 10.1016/j.mce.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 60.Katta A., Thulluri S., Manne N.D., Addagarla H.S., Arvapalli R., Nalabotu S.K., Gadde M., Rice K.M., Blough E.R. Overload induced heat shock proteins (HSPs), MAPK and miRNA (miR-1 and miR133a) response in insulin-resistant skeletal muscle. Cell. Physiol. Biochem. 2013;31:219–229. doi: 10.1159/000343363. [DOI] [PubMed] [Google Scholar]
- 61.Badal S.S., Wang Y., Long J., Corcoran D.L., Chang B.H., Truong L.D., Kanwar Y.S., Overbeek P.A., Danesh F.R. miR-93 regulates Msk2-mediated chromatin remodelling in diabetic nephropathy. Nat. Commun. 2016;7:12076. doi: 10.1038/ncomms12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peng R., Liu H., Peng H., Zhou J., Zha H., Chen X., Zhang L., Sun Y., Yin P., Wen L., et al. Promoter hypermethylation of let-7a-3 is relevant to its down-expression in diabetic nephropathy by targeting UHRF1. Gene. 2015;570:57–63. doi: 10.1016/j.gene.2015.05.073. [DOI] [PubMed] [Google Scholar]
- 63.Zhou J., Peng R., Li T., Luo X., Peng H., Zha H., Yin P., Wen L., Zhang Z. A potentially functional polymorphism in the regulatory region of let-7a-2 is associated with an increased risk for diabetic nephropathy. Gene. 2013;527:456–461. doi: 10.1016/j.gene.2013.06.088. [DOI] [PubMed] [Google Scholar]
- 64.Park J.T., Kato M., Lanting L., Castro N., Nam B.Y., Wang M., Kang S.W., Natarajan R. Repression of let-7 by Transforming Growth Factor-beta1-induced Lin28 up-regulates collagen expression in glomerular mesangial cells under diabetic conditions. Am. J. Physiol. Renal Physiol. 2014;307:F1390–F1403. doi: 10.1152/ajprenal.00458.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang B., Jha J.C., Hagiwara S., McClelland A.D., Jandeleit-Dahm K., Thomas M.C., Cooper M.E., Kantharidis P. Transforming growth factor-beta1-mediated renal fibrosis is dependent on the regulation of transforming growth factor receptor 1 expression by let-7b. Kidney Int. 2014;85:352–361. doi: 10.1038/ki.2013.372. [DOI] [PubMed] [Google Scholar]
- 66.Park S., Kim O.H., Lee K., Park I.B., Kim N.H., Moon S., Im J., Sharma S.P., Oh B.C., Nam S., et al. Plasma and urinary extracellular vesicle microRNAs and their related pathways in diabetic kidney disease. Genomics. 2022;114:110407. doi: 10.1016/j.ygeno.2022.110407. [DOI] [PubMed] [Google Scholar]
- 67.Sinha N., Kumar V., Puri V., Nada R., Rastogi A., Jha V., Puri S. Urinary exosomes: Potential biomarkers for diabetic nephropathy. Nephrology. 2020;25:881–887. doi: 10.1111/nep.13720. [DOI] [PubMed] [Google Scholar]
- 68.Ghai V., Wu X., Bheda-Malge A., Argyropoulos C.P., Bernardo J.F., Orchard T., Galas D., Wang K. Genome-wide Profiling of Urinary Extracellular Vesicle microRNAs Associated With Diabetic Nephropathy in Type 1 Diabetes. Kidney Int. Rep. 2018;3:555–572. doi: 10.1016/j.ekir.2017.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li X., Xu R., Liu X., Xu L., Xue M., Cheng Y., Li T., Yu X., Wang Y., Li C., et al. Urinary miR-3137 and miR-4270 as potential biomarkers for diabetic kidney disease. J. Clin. Lab. Anal. 2020;34:e23549. doi: 10.1002/jcla.23549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li W., Yang S., Qiao R., Zhang J. Potential Value of Urinary Exosome-Derived let-7c-5p in the Diagnosis and Progression of Type II Diabetic Nephropathy. Clin. Lab. 2018;64:709–718. doi: 10.7754/Clin.Lab.2018.171031. [DOI] [PubMed] [Google Scholar]
- 71.Prabu P., Rome S., Sathishkumar C., Gastebois C., Meugnier E., Mohan V., Balasubramanyam M. microRNAs from urinary extracellular vesicles are non-invasive early biomarkers of diabetic nephropathy in type 2 diabetes patients with the ‘Asian Indian phenotype’. Diabetes Metab. 2019;45:276–285. doi: 10.1016/j.diabet.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 72.Argyropoulos C., Wang K., McClarty S., Huang D., Bernardo J., Ellis D., Orchard T., Galas D., Johnson J. Urinary microRNA profiling in the nephropathy of type 1 diabetes. PLoS ONE. 2013;8:e54662. doi: 10.1371/annotation/37e647d5-1781-4edf-86a8-e3b533c32ad9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.González-Palomo A.K., Pérez-Vázquez F.J., Méndez-Rodríguez K.B., Ilizaliturri-Hernández C.A., Cardona-Alvarado M.I., Flores-Nicasio M.V., Kornhauser C., Malacara J.M., Figueroa-Vega N. Profile of urinary exosomal microRNAs and their contribution to diabetic kidney disease through a predictive classification model. Nephrology. 2022;27:484–493. doi: 10.1111/nep.14039. [DOI] [PubMed] [Google Scholar]
- 74.Osipova J., Fischer D.C., Dangwal S., Volkmann I., Widera C., Schwarz K., Lorenzen J.M., Schreiver C., Jacoby U., Heimhalt M., et al. Diabetes-associated microRNAs in pediatric patients with type 1 diabetes mellitus: A cross-sectional cohort study. J. Clin. Endocrinol. Metab. 2014;99:E1661–E1665. doi: 10.1210/jc.2013-3868. [DOI] [PubMed] [Google Scholar]
- 75.Zhao Y., Shen A., Guo F., Song Y., Jing N., Ding X., Pan M., Zhang H., Wang J., Wu L., et al. Urinary Exosomal MiRNA-4534 as a Novel Diagnostic Biomarker for Diabetic Kidney Disease. Front. Endocrinol. 2020;11:590. doi: 10.3389/fendo.2020.00590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barutta F., Tricarico M., Corbelli A., Annaratone L., Pinach S., Grimaldi S., Bruno G., Cimino D., Taverna D., Deregibus M.C., et al. Urinary Exosomal microRNAs in Incipient Diabetic Nephropathy. PLoS ONE. 2013;8:e73798. doi: 10.1371/journal.pone.0073798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee W.C., Li L.C., Ng H.Y., Lin P.T., Chiou T.T., Kuo W.H., Lee C.T. Urinary Exosomal microRNA Signatures in Nephrotic, Biopsy-Proven Diabetic Nephropathy. J. Clin. Med. 2020;9:1220. doi: 10.3390/jcm9041220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Eissa S., Matboli M., Bekhet M.M. Clinical verification of a novel urinary microRNA panal: 133b, -342 and -30 as biomarkers for diabetic nephropathy identified by bioinformatics analysis. Biomed. Pharmacother. 2016;83:92–99. doi: 10.1016/j.biopha.2016.06.018. [DOI] [PubMed] [Google Scholar]
- 79.Szeto C.C., Ching-Ha K.B., Ka-Bik L., Mac-Moune L.F., Cheung-Lung C.P., Gang W., Kai-Ming C., Kam-Tao L.P. Micro-RNA expression in the urinary sediment of patients with chronic kidney diseases. Dis. Markers. 2012;33:137–144. doi: 10.1155/2012/842764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xie Y., Jia Y., Cuihua X., Hu F., Xue M., Xue Y. Urinary Exosomal microRNA Profiling in Incipient Type 2 Diabetic Kidney Disease. J. Diabetes Res. 2017;2017:6978984. doi: 10.1155/2017/6978984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Eissa S., Matboli M., Aboushahba R., Bekhet M.M., Soliman Y. Urinary exosomal microRNA panel unravels novel biomarkers for diagnosis of type 2 diabetic kidney disease. J. Diabetes Complicat. 2016;30:1585–1592. doi: 10.1016/j.jdiacomp.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 82.An Y., Zhang C., Xu F., Li W., Zeng C., Xie L., Liu Z. Increased urinary miR-196a level predicts the progression of renal injury in patients with diabetic nephropathy. Nephrol. Dial. Transpl. 2020;35:1009–1016. doi: 10.1093/ndt/gfy326. [DOI] [PubMed] [Google Scholar]
- 83.El-Samahy M.H., Adly A.A., Elhenawy Y.I., Ismail E.A., Pessar S.A., Mowafy M.E., Saad M.S., Mohammed H.H. Urinary miRNA-377 and miRNA-216a as biomarkers of nephropathy and subclinical atherosclerotic risk in pediatric patients with type 1 diabetes. J. Diabetes Complicat. 2018;32:185–192. doi: 10.1016/j.jdiacomp.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 84.Peng H., Zhong M., Zhao W., Wang C., Zhang J., Liu X., Li Y., Paudel S.D., Wang Q., Lou T. Urinary miR-29 correlates with albuminuria and carotid intima-media thickness in type 2 diabetes patients. PLoS ONE. 2013;8:e82607. doi: 10.1371/journal.pone.0082607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guo J., Li J., Zhao J., Yang S., Wang L., Cheng G., Liu D., Xiao J., Liu Z., Zhao Z. MiRNA-29c regulates the expression of inflammatory cytokines in diabetic nephropathy by targeting tristetraprolin. Sci. Rep. 2017;7:2314. doi: 10.1038/s41598-017-01027-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Argyropoulos C., Wang K., Bernardo J., Ellis D., Orchard T., Galas D., Johnson J.P. Urinary microRNA Profiling Predicts the Development of Microalbuminuria in Patients with Type 1 Diabetes. J. Clin. Med. 2015;4:1498–1517. doi: 10.3390/jcm4071498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Delic D., Eisele C., Schmid R., Baum P., Wiech F., Gerl M., Zimdahl H., Pullen S.S., Urquhart R. Urinary Exosomal miRNA Signature in Type II Diabetic Nephropathy Patients. PLoS ONE. 2016;11:e0150154. doi: 10.1371/journal.pone.0150154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Han Q., Zhang Y., Jiao T., Li Q., Ding X., Zhang D., Cai G., Zhu H. Urinary sediment microRNAs can be used as potential noninvasive biomarkers for diagnosis, reflecting the severity and prognosis of diabetic nephropathy. Nutr. Diabetes. 2021;11:24. doi: 10.1038/s41387-021-00166-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ngamdee T., Chalermwatanachai T., Siriwan C., Warachit O., Rijiravanich P., Surareungchai W. Target amplification-free detection of urinary microRNA for diabetic nephropathy diagnosis with electrocatalytic reaction. Anal. Bioanal. Chem. 2022;414:5695–5707. doi: 10.1007/s00216-022-04072-w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.