Abstract
The aim of this work was to identify and characterize the pectolytic bacteria responsible for the emergence of bacterial soft rot on two summer cabbage hybrids (Cheers F1 and Hippo F1) grown in the Futog locality (Bačka, Vojvodina), known for the five-century-long tradition of cabbage cultivation in Serbia. Symptoms manifesting as soft lesions on outer head leaves were observed during August 2021, while the inner tissues were macerated, featuring cream to black discoloration. As the affected tissue decomposed, it exuded a specific odor. Disease incidence ranged from 15% to 25%. A total of 67 isolates producing pits on crystal violet pectate (CVP) medium were characterized for their phenotypic and genotypic features. The pathogenicity was confirmed on cabbage heads. Findings yielded by the repetitive element palindromic-polymerase chain reaction (rep-PCR) technique confirmed interspecies diversity between cabbage isolates, as well as intraspecies genetic diversity within the P. carotovorum group of isolates. Based on multilocus sequence typing (MLST) using genes dnaX, mdh, icdA, and proA, five representative isolates were identified as Pectobacterium carotovorum (Cheers F1 and Hippo F1), while two were identified as Pectobacterium versatile (Hippo F1) and Pectobacterium odoriferum (Hippo F1), respectively, indicating the presence of diverse Pectobacterium species even in combined infection in the same field. Among the obtained isolates, P. carotovorum was the most prevalent species (62.69%), while P. versatile and P. odoriferum were less represented (contributing by 19.40% and 17.91%, respectively). Multilocus sequence analysis (MLSA) performed with concatenated sequences of four housekeeping genes (proA, dnaX, icdA, and mdh) and constructed a neighbor-joining phylogenetic tree enabled insight into the phylogenetic position of the Serbian cabbage Pectobacterium isolates. Bacterium P. odoriferum was found to be the most virulent species for cabbage, followed by P. versatile, while all three species had comparable virulence with respect to potato. The results obtained in this work provide a better understanding of the spreading routes and abundance of different Pectobacterium spp. in Serbia.
Keywords: bacterial soft rot, cabbage, Pectobacterium spp., MLSA, rep-PCR, pathogenicity, virulence
1. Introduction
Cabbage (Brassica oleracea var. capitata L.) is one of the world’s most important vegetable crops, as its high yield and adaptability have facilitated worldwide distribution [1]. Owing to the abundance of health-promoting phytochemicals (glucosinolates, polyphenols, vitamins, and proteins), cabbage features in the traditional cuisine of many countries and has also been traditionally used for medicinal purposes [2]. According to the data provided by the Food and Agriculture Organization Corporate Statistical Database (FAOSTAT) for 2020, globally, about 2,414,288 ha were designated for cabbage cultivation [3].
Futog, a suburban settlement located in the province of Vojvodina (Serbia), has been known for cabbage cultivation since the 18th century [4]. However, numerous pests and pathogens currently threaten cabbage production both in Serbia and worldwide, with Pectobacterium species (fam. Enterobacteriaceae), the causative agents of bacterial soft rot disease, being particularly harmful. The symptoms of infection with these pectinolytic enterobacteria may be found in the cabbage fields during cultivation, as well as post-harvest, during transport, storage, and marketing, leading to considerable yield reduction and economic losses [5]. According to Bhat [6], bacterial soft rot causes greater total post-harvest yield loss than any other bacterial disease. Losses on cabbage caused by soft rot alone, or in combination with other storage issues, are estimated at 25–50% in a single season in the USA, with the greatest loss occurring in New York and Wisconsin [7]. However, the extent of yield loss is influenced by pathogen−host specificity, as well as numerous external factors (temperature, humidity, etc.) [8]. Although bacterial soft rot is a disease that affects only agricultural and horticultural crops, the presence of the soft rot-causing Pectobacteriaceae is not only limited to host plants but extends to the weed species growing near the infected fields, as well as water, air, and soil, thus serving as a potent source of inoculum [9]. In addition, soft rot-causing bacteria can survive in infected fleshy organs during storage, as well as in debris, on roots or other parts of host plants, and in the pupae of several insects [10]. Thus, the most likely entry routes for the pathogen involve either roots or natural openings/mechanically created wounds on the upper plant parts. Once inside the plant, the soft rot-causing bacteria begin to move and multiply in the intercellular spaces, softening the affected tissues and transforming them into a slimy mass. The slimy mass extrudes through cracks in the epidermis into the soil, or to other plants that are placed in close proximity during storage, which subsequently become infected [10]. Bacterial survival in nature is dependent on various factors. For instance, their survival in soil depends on soil pH, temperature, moisture, magnesium concentration, and calcium nutrition and mostly takes up to six months in the absence of plant debris [11]. Temperature greatly affects the survival as well as the pathogenicity of Pectobacterium species [12].
Scientific interest in Pectobacteria primarily stems from their broad host range and an emerging genetic diversity associated with dynamic evolutionary processes, mainly occurring through gene acquisition, genome rearrangement, and gene loss [13]. Achtman [14] pointed out that higher rates of divergence in Pectobacterium spp. were detected on metabolic genes compared to the ribosomal operon and posited that the observed changes may be related to the adaptation of strains to specific environmental niches. Therefore, typing and analysis of several protein-coding loci (multilocus sequence typing and analysis, MLST/MLSA) could allow the degree of phenotypic relatedness to be established more reliably than is possible by analyzing 16S rRNA or some nonprotein-coding genes [15]. In extant research, different combinations of housekeeping genes have been used for MLST and MLSA. For example, Ma et al. [16] examined seven genes (acnA, gapA, icdA, mdh, mtlD, pgi, and proA) to reconstruct the phylogeny of the tested Pectobacterium and Dickeya strains, due to (i) their proven ability to provide sufficient sequence diversity for distinguishing closely related species, (ii) their ubiquity in most enterobacteria, and (iii) the involvement of their products in diverse aspects of bacterial metabolism. Further, Moleleki et al. [17] and Pitman et al. [18] reconstructed the phylogeny of P. wasabiae strains obtained from potato grown in South Africa and New Zealand based on the gapA and mdh, and acnA and mdh genes, respectively. In their work, Waleron et al. [19] used a combination of nine housekeeping genes (gapA, gyrA, icdA, pgi, proA, recA, recN, rpoA, and rpoS) for MLSA of P. zantedeschiae strains isolated from calla lily in central Poland and Serbia. On the other hand, Marković et al. [20] used a combination of genes dnaX, proA, and mdh to identify the causal agents (P. versatile and P. carotovorum) of potato blackleg disease in Serbia. The gene dnaX was shown to reliably distinguish between Pectobacterium and Dickeya isolates in several prior studies focusing on the potato blackleg causal agents in different countries [21,22,23]. Considering the aforementioned factors (high discriminatory power and reproducibility) and previous experience gained in working with plant pathogens within the Enterobacteriaceae family, four housekeeping genes (i.e., dnaX, icdA, mdh, and proA) were utilized in the present study for typing the pectolytic isolates obtained from cabbage.
Repetitive element palindromic PCR (rep-PCR) is widely recognized as a useful technique for profiling different Pectobacterium species. In their study, Norman et al. successfully used rep-PCR with primers for BOX-, ERIC-, and REP-PCR for strain-level differentiation of P. carotovorum populations isolated from nursery retention ponds and large hypereutrophic lakes [24]. In addition to the aforementioned rep-PCR techniques (BOX-, ERIC-, and REP-PCR), for the first time, Maisuria and Nerurkar [25] also used GTG5-PCR, due to its highest discriminatory power, to differentiate P. carotovorum strains from soil and diseased fruits and vegetables. According to Zoledowska et al. [26], REP-PCR is the most useful tool for grouping P. parmentieri potato strains and is superior to BOX- and ERIC-PCR. Thus, as BOX-, ERIC-, REP-, and GTG5-PCR are among the most commonly used rep-PCR methods for DNA profiling and are proven to be sufficiently discriminative to reveal subtle differences even among strains belonging to the same species, all four methods were adopted in the present study.
According to Song et al. [27], P. carotovorum is ranked among the most common causative agents of cabbage soft rot. Nonetheless, P. aroidearum, P. brasiliense, P. odoriferum, P. polaris, and P. wasabiae species have also been reported to result in cabbage soft rot in different countries [28,29,30,31,32,33]. In Serbia, different Pectobacterium species (i.e., P. atrosepticum, P. brasiliense, P. carotovorum, P. punjabense, P. zantedeschiae, and P. versatile) have been isolated from broccoli, calla lily, carrot, celery, parsley, potato, squash, and watermelon [19,20,23,34,35,36,37,38,39].
Given the expanding genetic diversity of Pectobacterium spp. and the lack of recent data on the agents causing soft rot on cabbage in Serbia, the goal of this study was to contribute to their identification and characterization based on molecular and pathogenic features.
2. Materials and Methods
2.1. Sample Collection and Pathogen Isolation
In August 2021, bacterial soft rot symptoms were noted on the summer cabbage hybrids Cheers F1 (Takii Seed, Field I (geographic coordinates 45.2507100, 19.7247660)) and Hippo F1 (Sakata Seed, Field II (geographic coordinates 45.2567200, 19.7275470)) grown in two fields located in Futog (Bačka, Vojvodina) of 0.5 ha and 1 ha size, respectively. Soft rot symptoms on outer leaves, along with deep maceration of inner head leaves accompanied by black discoloration, were found on the inspected plants (Figure 1). Due to decomposition, the affected tissue exuded a specific odor. Disease incidence in the visited fields was estimated at 15−25%.
Figure 1.
Bacterial soft rot symptoms in visited cabbage fields in Futog (Bačka, Vojvodina).
Prior to isolation, collected samples were washed under running tap water and dried on filter paper at room temperature. Isolations were performed on crystal violet pectate (CVP) media [40] from the small leaf sections (2–3 mm) that encompassed the transition zones between healthy and diseased tissue. CVP plates were incubated at 26 °C. All bacterial colonies forming characteristic cavities on the medium were selected and transferred onto nutrient agar (NA) [41] to obtain pure cultures. Isolates were long-term stored at −80 °C in lysogeny broth (LB) [42] supplemented with 30% (v/v) of sterile glycerol.
2.2. Pathogenicity on Cabbage
The pathogenicity of 67 isolates obtained in this study was evaluated using cabbage heads (unknown cultivar). Before inoculation, cabbage heads were washed under running tap water, uniformly sprayed with 70% ethanol, and dried at room temperature. Isolates used for inoculation were grown in nutrient broth (NB, HiMedia Laboratories) at 26 °C for 48 h while shaking and were adjusted to approximately 1 × 108 CFU mL−1. Inoculations were performed by puncturing holes in cabbage heads and filling them with bacterial suspensions (~200 µL). The assays were performed in two sets of three independent replicates. Inoculated cabbage heads were placed in plastic boxes which were kept under room temperature (25 ± 1 °C) and high humidity (90−100%) conditions. Sterile distilled water (SDW) was used as a negative control, while the P. carotovorum strain Pcc10, previously isolated from cabbage in Bosnia and Herzegovina [8], served as a positive control treatment.
Cabbage heads were visually observed daily in order to monitor the occurrence of soft rot symptoms and the disease progress until complete decay. Emergence of soft lesions around the holes 24 h after the inoculation of cabbage heads with the suspension of tested isolates was considered a pectolytic-positive reaction.
2.3. Genotyping Methods
2.3.1. DNA Extraction
Genomic DNA from the 67 cabbage isolates was extracted according to the hexadecyltrimethylammonium bromide (CTAB) procedure described previously by Popović et al. [43].
2.3.2. Preliminary Identification
All cabbage isolates were preliminarily identified using specific primers (F0145/E2477) designed based on the partial sequence of gene pmrA (response regulator) of P. carotovorum [44]. The sequences of the used primers are listed in Table 1. PCR amplifications were performed in a mixture (25 µL) consisting of Thermo Scientific DreamTaq PCR Master Mix (2×) (12.5 µL), nuclease-free water (Thermo ScientificTM, Waltham, MA, USA) (9.5 µL), 10 µM primers (forward/reverse) (1 µL each), and sample DNA (1 µL), according to the conditions proposed by Kettani-Halabi et al. [44]. Presence of a band in the searched position of 666 bp was checked on 1% agarose gel in relation to the positive control P. carotovorum strain Pcc10 and 200–10,000 bp SmartLadder MW-1700-10 (Eurogentec).
Table 1.
Primers used in this study.
| Primer | Sequence (5′-3′) | Reference |
|---|---|---|
| Pectobacterium-specific primers | ||
| F0145 | TACCCTGCAGATGAAATTATTGATTGTTGAAGAC | [44] |
| E2477 | TACCAAGCTTTGGTTGTTCCCCTTTGGTCA | |
| Primers for rep-PCR | ||
| ERIC1R | ATGTAAGCTCCTGGGGATTCAC | [45] |
| ERIC2 | AAGTAAGTGACTGGGGTGAGCG | |
| REP1R-I | IIIICGICGICATCIGGC | |
| REP2-I | ICGICTTATCIGGCCTAC | |
| BOXA1R | CTACGGCAAGGCGACGCTGACG | |
| GTG5 | GTGGTGGTGGTGGTG | [46] |
| Primers for MLST | ||
| dnaX-F | TATCAGGTYCTTGCCCGTAAGTGG | [22] |
| dnaX-R | TCGACATCCARCGCYTTGAGATG | |
| icdA400F | GGTGGTATCCGTTCTCTGAACG | [16] |
| icdA977R | TAGTCGCCGTTCAGGTTCATACA | |
| proAF1 | CGGYAATGCGGTGATTCTGCG | |
| proAR1 | GGGTACTGACCGCCACTTC | |
| mdh2 | GCGCGTAAGCCGGGTATGGA | [17] |
| mdh4 | CGCGGCAGCCTGGCCCATAG | |
2.3.3. Repetitive Element Palindromic PCR (Rep-PCR)
Genetic diversity among the obtained 67 cabbage isolates was evaluated using the rep-PCR fingerprinting method with two oligonucleotide primer pairs [ERIC1R/ERIC2 (ERIC-PCR) and REP1R-I/ REP2-I (REP-PCR)] and two single oligonucleotide primers [BOXA1R (BOX-PCR) and GTG5 (GTG5-PCR)] corresponding to the interspersed repetitive sequence elements. Primer sequences of the used primers are listed in Table 1. The PCR mixture (25 µL) comprised 12.5 µL of Thermo Scientific DreamTaq PCR Master Mix (2×), 9.5 µL of nuclease-free water (Thermo ScientificTM, Waltham, MA, USA), 1 µL of each (forward/reverse) primer (10 µM) and 1 µL of sample DNA. PCR amplifications for BOX-, ERIC-, and REP-PCR were performed under the conditions proposed by Louws et al. [45], while the methodology described by Versalovic et al. [46] was adopted for GTG5-PCR. After amplification, PCR products (5 µL) were mixed with DNA Gel Loading Dye (6X) (Thermo ScientificTM, Waltham, MA, USA) (2 µL) and were visualized on 2% agarose gel (FastGene®) strained with Midori Green Advance (Nippon Genetics Europe, Düren, Germany). PCR products were electrophoretically separated for 2.5 h (at 90 V and 300 mA) before being checked under a UV transilluminator. The obtained patterns were compared using PyElph 1.4 program and were subsequently used for the construction of the unweighted pair group method with arithmetic mean (UPGMA) phylogenetic tree. To easily compare the patterns obtained with each of the four used rep-PCR primers and to select all isolates that were potentially genetically different, each tree cluster, representing one DNA fingerprinting pattern, was provided with a number (DNA fingerprinting group). One isolate representing each combination of obtained DNA fingerprinting groups was randomly selected for further characterization (MLST and MLSA, and virulence assessment).
2.3.4. Multilocus Sequence Typing and Analysis (MLST/MLSA)
DNA of the seven selected representative cabbage isolates (Pc2321, Pc3821, Pc4821, Pc5421, Pv6321, Po7521, and Pc8321) was amplified with the primers produced based on the partial sequences of four housekeeping genes (dnaX (dnaX-F/dnaX-R), icdA (icdA400F/icdA977R), mdh (mdh2/mdh4), and proA (proAF1/proAR1)), encoding DNA polymerase III subunit tau, isocitrate dehydrogenase, malate dehydrogenase, and gamma-glutamyl phosphate reductase, respectively. Primer sequences of the used primers are listed in Table 1. For all reactions, PCR mixtures were created as described in the previous subsection for rep-PCR. PCR amplifications were performed under the conditions described by Sławiak et al. [22] for the gene dnaX, while those reported by Ma et al. [16] were adopted for the genes icdA and proA, and the strategy employed by Moleleki et al. [17] was utilized for the gene mdh. Presence of bands in the searched positions was checked on 1% agarose gel in relation to 200–10,000 bp SmartLadder MW-1700-10 (Eurogentec). PCR products were purified using the Qiagen QIAquick PCR Purification Kit before being sent to the Eurofins Genomics’ DNA sequencing service (Germany) for sequencing. The obtained sequences were manually checked for quality and were compared with the strains deposited in the National Center for Biotechnology Information (NCBI) database using the nucleotide BLAST (BLASTn) tool. All newly identified sequences were deposited to the NCBI database to obtain the accession numbers.
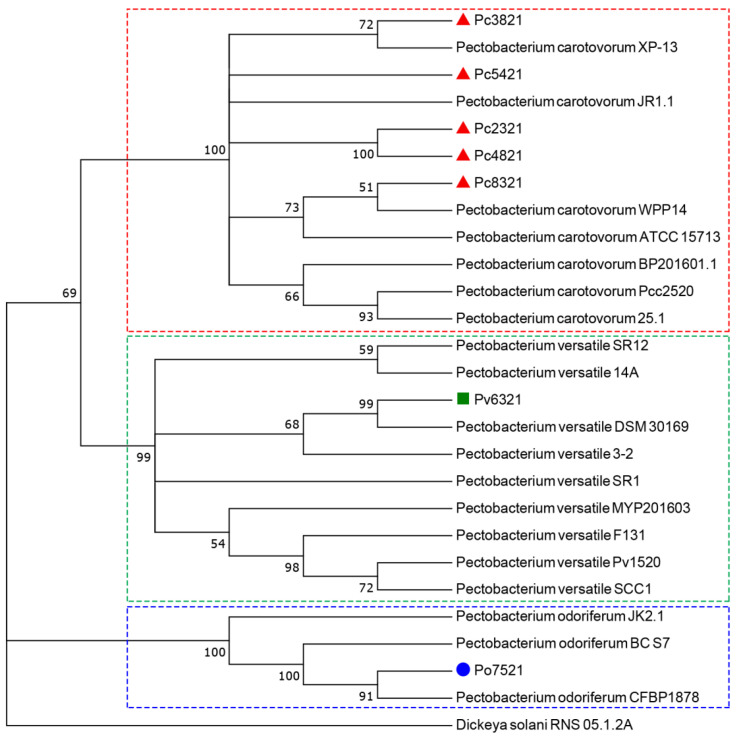
The phylogenetic position of the seven representative isolates was determined in relation to 19 strains of three Pectobacterium species, i.e., P. odoriferum (CFBP 1878, BC S7, and JK2.1), P. carotovorum (ATCC 15713, 25.1, WPP14, BP201601.1, JR1.1, XP-13, and Pcc2520), and P. versatile (14A, 3-2, SCC1, F131, DSM 30169, MYP201603, SR1, SR12, and Pv1520) isolated from different hosts (cabbage, carrot, Chinese cabbage, chicory, coleslaw, cucumber, kimchi cabbage, potato, and radish) and countries (Belarus, China, Denmark, Finland, France, Germany, Russia, Serbia, South Korea, and the USA) and retrieved from the NCBI database (Table 2). A neighbor-joining (NJ) phylogenetic tree was constructed based on the concatenated sequences (1639 nt) of genes dnaX (416 nt), icdA (479 nt), mdh (240 nt), and proA (504 nt). Before the tree construction, the sequences of all genes were aligned to the same size using the biological sequence alignment editor BioEdit 7.2 program and the ClustalW Multiple alignment tool. The tree was constructed in MEGA7 Software and was rooted with the Dickeya solani strain RNS 05.1.2A (Accession No. CP104920).
Table 2.
List of the comparative Pectobacterium spp. strains used for phylogenetic analysis with GenBank accession numbers.
| Strain a | Isolation Source | Locality | Year | GenBank Accession Numbers | |||
|---|---|---|---|---|---|---|---|
| dnaX | icdA | mdh | proA | ||||
| Pectobacterium odoriferum | |||||||
| CFBP 1878 T | Chicory | France | 1978 | MK516907 | JF926783 | JF926793 | JF926823 |
| BC S7 | Chinese cabbage | Beijing (China) | 2007 | CP009678 | CP009678 | CP009678 | CP009678 |
| JK2.1 | Kimchi cabbage | South Korea | 2016 | CP034938 | CP034938 | CP034938 | CP034938 |
| Pectobacterium carotovorum | |||||||
| ATCC 15713 T | Potato | Denmark | 1952 | MW979047 | FJ895850 | FJ895851 | FJ895853 |
| 25.1 | Cucumber | Svietlahorsk (Belarus) | 2009 | CP088019 | CP088019 | CP088019 | CP088019 |
| WPP14 | Potato | Wisconsin (USA) | 2015 | CP051652 | CP051652 | CP051652 | CP051652 |
| BP201601.1 | Potato | Boseong (South Korea) | 2016 | CP034236 | CP034236 | CP034236 | CP034236 |
| JR1.1 | Radish | South Korea | 2016 | CP034237 | CP034237 | CP034237 | CP034237 |
| XP-13 | Potato | Zhouning (China) | 2018 | CP063242 | CP063242 | CP063242 | CP063242 |
| Pcc2520 | Potato | Serbia | 2020 | MW805307 | OP751390 | OP751392 | OP751393 |
| Pectobacterium versatile | |||||||
| 14A | Potato | Minsk (Belarus) | 1978 | CP034276 | CP034276 | CP034276 | CP034276 |
| 3-2 | Potato | Minsk (Belarus) | 1979 | CP024842 | CP024842 | CP024842 | CP024842 |
| SCC1 | Potato | Finland | 1982 | CP021894 | CP021894 | CP021894 | CP021894 |
| F131 | Potato | Moscow (Russia) | 1993 | CP065030 | CP065030 | CP065030 | CP065030 |
| DSM 30169 | Cabbage | Germany | 2010 | CP065143 | CP065143 | CP065143 | CP065143 |
| MYP201603 | Potato | Miryang (South Korea) | 2013 | CP051628 | CP051628 | CP051628 | CP051628 |
| SR1 | Carrot | Iowa, Ames (USA) | 2019 | CP084656 | CP084656 | CP084656 | CP084656 |
| SR12 | Coleslaw | Iowa, Ames (USA) | 2019 | CP084654 | CP084654 | CP084654 | CP084654 |
| Pv1520 | Potato | Serbia | 2020 | MW805306 | OP751391 | MZ682621 | MZ682624 |
a Type strains are marked with the superscript T.
2.4. Virulence Assessment
The virulence potential of the seven representative cabbage isolates (Pc2321, Pc3821, Pc4821, Pc5421, Pv6321, Po7521, and Pc8321) was assessed using cabbage heads (unknown cv.) and potato tubers (cv. Arizona). Prior to inoculation, cabbage heads and potato tubers were sterilized as previously described (Section 2.2), and approximately 100 surface punctures (around 5 mm in depth) were made using a needle to allow bacteria to penetrate the tissue. Bacterial suspensions were prepared as described for the pathogenicity test (Section 2.2) and SDW was used as a negative control. Inoculations were performed by spraying cabbage heads/potato tubers as uniformly as possible, making sure to cover the entire outer surface. The experiment was conducted in three independent replicates, with three cabbage heads (nine in total per isolate) and five potato tubers (15 in total per isolate) for each replicate. Two experiments—for disease progress rating five and seven days after inoculation (d.a.i.), respectively—were performed in parallel. For both experiments, the initial weights of cabbage heads and potato tubers were recorded. To obtain statistically comparable results, sums of the initial weights for each of the tested cabbage isolates were almost equal. The inoculated cabbage heads and potato tubers were placed in plastic boxes under high humidity conditions (90−100%). The experiment was performed during summer when the room temperature was 28 ± 2 °C.
Cabbage heads and potato tubers were weighed, whereby the mean values of the initial and the final (5 and 7 d.a.i.) weights were used to calculate the disease progress curve (AUDPC) according to the following equation:
| (1) |
where yi = disease progress assessment at the ith observation, ti = time (days) at the ith observation, and n = total number of observations.
All obtained values were statistically processed using Minitab 21 Statistical Software, whereby one-way analysis of variance (one-way ANOVA) was performed, and the resulting values were compared using Tukey’s honestly significant difference (HSD) test. Values having p < 0.05 were considered statistically significant.
3. Results
3.1. Isolation, Preliminary Identification, and Pathogenicity
In this study, 67 isolates were selected from cabbage hybrids Cheers F1 (34 isolates) and Hippo F1 (33 isolates), forming characteristic cavities on CVP medium and small, whitish, irregularly shaped colonies on NA (Table 3). All isolates produced bands at 666 bp after amplification with the Pectobacterium-specific primer pair F0145/E2477 (Table 3).
Table 3.
Serbian cabbage Pectobacterium spp. isolates used in the present study, the hybrid they originate from, pectolytic activity, confirmation with Pectobacterium-specific primers, and DNA fingerprinting group affiliation.
| Isolate Code | Hybrid | Pectolytic Activity | Genus Confirmation |
DNA Fingerprinting Group | |||
|---|---|---|---|---|---|---|---|
| BOX | ERIC | GTG5 | REP | ||||
| Pc2021 | Cheers F1 | + | + | I | I | I | I |
| Pc2121 | Cheers F1 | + | + | I | I | I | I |
| Pc2221 | Cheers F1 | + | + | I | I | I | I |
| Pc2321 | Cheers F1 | + | + | I | I | I | I |
| Pc2421 | Cheers F1 | + | + | I | I | I | I |
| Pc2521 | Cheers F1 | + | + | I | I | I | I |
| Pc2621 | Cheers F1 | + | + | I | I | I | I |
| Pc2721 | Cheers F1 | + | + | I | I | I | I |
| Pc2821 | Cheers F1 | + | + | I | I | I | I |
| Pc3121 | Cheers F1 | + | + | II | II | II | II |
| Pc3221 | Cheers F1 | + | + | II | II | II | II |
| Pc3321 | Cheers F1 | + | + | II | II | II | II |
| Pc3421 | Cheers F1 | + | + | II | II | II | II |
| Pc3521 | Cheers F1 | + | + | II | II | II | II |
| Pc3621 | Cheers F1 | + | + | II | II | II | II |
| Pc3721 | Cheers F1 | + | + | II | II | II | II |
| Pc3821 | Cheers F1 | + | + | II | II | II | II |
| Pc3921 | Cheers F1 | + | + | II | II | II | II |
| Pc4221 | Cheers F1 | + | + | III | II | III | III |
| Pc4321 | Cheers F1 | + | + | III | II | III | III |
| Pc4421 | Cheers F1 | + | + | III | II | III | III |
| Pc4521 | Cheers F1 | + | + | III | II | III | III |
| Pc4621 | Cheers F1 | + | + | III | II | III | III |
| Pc4721 | Cheers F1 | + | + | III | II | III | III |
| Pc4821 | Cheers F1 | + | + | III | II | III | III |
| Pc4921 | Cheers F1 | + | + | III | II | III | III |
| Pc5021 | Cheers F1 | + | + | IV | III | IV | III |
| Pc5121 | Cheers F1 | + | + | IV | III | IV | III |
| Pc5221 | Cheers F1 | + | + | IV | III | IV | III |
| Pc5321 | Cheers F1 | + | + | IV | III | IV | III |
| Pc5421 | Cheers F1 | + | + | IV | III | IV | III |
| Pc5521 | Cheers F1 | + | + | IV | III | IV | III |
| Pc5621 | Cheers F1 | + | + | IV | III | IV | III |
| Pc5721 | Cheers F1 | + | + | IV | III | IV | III |
| Pv1021 | Hippo F1 | + | + | V | IV | V | IV |
| Pv1121 | Hippo F1 | + | + | V | IV | V | IV |
| Pv1221 | Hippo F1 | + | + | V | IV | V | IV |
| Pv1321 | Hippo F1 | + | + | V | IV | V | IV |
| Pv1421 | Hippo F1 | + | + | V | IV | V | IV |
| Pv1521 | Hippo F1 | + | + | V | IV | V | IV |
| Pv1621 | Hippo F1 | + | + | V | IV | V | IV |
| Pv6121 | Hippo F1 | + | + | V | IV | V | IV |
| Pv6221 | Hippo F1 | + | + | V | IV | V | IV |
| Pv6321 | Hippo F1 | + | + | V | IV | V | IV |
| Pv6421 | Hippo F1 | + | + | V | IV | V | IV |
| Pv6521 | Hippo F1 | + | + | V | IV | V | IV |
| Pv6621 | Hippo F1 | + | + | V | IV | V | IV |
| Po7021 | Hippo F1 | + | + | VI | V | VI | V |
| Po7121 | Hippo F1 | + | + | VI | V | VI | V |
| Po7221 | Hippo F1 | + | + | VI | V | VI | V |
| Po7321 | Hippo F1 | + | + | VI | V | VI | V |
| Po7421 | Hippo F1 | + | + | VI | V | VI | V |
| Po7521 | Hippo F1 | + | + | VI | V | VI | V |
| Po9121 | Hippo F1 | + | + | VI | V | VI | V |
| Po9221 | Hippo F1 | + | + | VI | V | VI | V |
| Po9321 | Hippo F1 | + | + | VI | V | VI | V |
| Po9421 | Hippo F1 | + | + | VI | V | VI | V |
| Po9521 | Hippo F1 | + | + | VI | V | VI | V |
| Po9621 | Hippo F1 | + | + | VI | V | VI | V |
| Pc8021 | Hippo F1 | + | + | VII | VI | VII | VI |
| Pc8121 | Hippo F1 | + | + | VII | VI | VII | VI |
| Pc8221 | Hippo F1 | + | + | VII | VI | VII | VI |
| Pc8321 | Hippo F1 | + | + | VII | VI | VII | VI |
| Pc8421 | Hippo F1 | + | + | VII | VI | VII | VI |
| Pc8521 | Hippo F1 | + | + | VII | VI | VII | VI |
| Pc8621 | Hippo F1 | + | + | VII | VI | VII | VI |
| Pc8721 | Hippo F1 | + | + | VII | VI | VII | VI |
All isolates marked in bold are used as representatives in this study.
The pathogenicity of all cabbage isolates was confirmed on cabbage heads by visually identifying irregularly shaped soft lesions (approximately 3−5 cm in diameter) around the inoculation points (holes) 1 d.a.i. The diameter of decomposing tissue enlarged daily, and the affected area spread from the outer leaves to the inner tissues, while causing tissue discoloration from cream to black. At 5 d.a.i., cabbage heads were almost completely macerated and exuded a specific odor.
3.2. Genetic Characterization
3.2.1. Rep-PCR
The UPGMA trees showing genetic diversity among the 67 tested cabbage isolates—constructed based on the obtained BOX-, ERIC-, GTG5-, and REP-PCR banding patterns—are shown in Figure S1, which also features virtual gel images of rep-PCR patterns corresponding to each group of isolates. Based on the differences found, each tree cluster was assigned a different number: BOX (I–VII), ERIC (I–VI), GTG5 (I-VII), and REP (I–VI) (Table 3). The obtained results indicate that the tested isolates are genetically diverse. BOX- and GTG5-PCR generated seven (I-VII), while ERIC- and REP-PCR generated six distinct banding patterns (I-VI). Based on the BOX- and GTG5-PCR findings, isolates were divided into the same seven groups, namely I: Pc2021–Pc2821, II: Pc3121–Pc3921, III: Pc4221–Pc4921, IV: Pc5021–Pc5721, V: Pv1021–Pv1621 and Pv6121–Pv6621, VI: Po7021–Po7521 and Po9121–Po9621, and VII: Pc8021–Pc8721 (Table 3). However, ERIC- and REP-PCR did not separate the isolates into six identical groups. ERIC-PCR implied the homogeneity of isolates Pc3121–Pc3921 and Pc4221–Pc4921 by placing them in the same tree cluster (DNA fingerprinting group II), while REP-PCR indicated homogeneity of isolates Pc4221–Pc4921 and Pc5021–Pc5721 (DNA fingerprinting group III) (Table 3). The distribution of the remaining isolates within the UPGMA groups remained the same and coincided with that obtained for BOX- and GTG5-PCR. Based on the combined results obtained with all four primers, seven isolates (Pc2321, Pc3821, Pc4821, Pc5421, Pv6321, Po7521, and Pc8321), each representing one group on the UPGMA tree (i.e., one DNA banding pattern), were randomly selected for further characterization.
3.2.2. MLST and MLSA
Based on the NCBI BLASTn analysis, five representative cabbage isolates (Pc2321, Pc3821, Pc4821, Pc5421, and Pc8321) were identified as P. carotovorum (representing the group of isolates Pc2021, Pc2121, Pc2221, Pc2321, Pc2421, Pc2521, Pc2621, Pc2721, Pc2821, Pc3121, Pc3221, Pc3321, Pc3421, Pc3521, Pc3621, Pc3721, Pc3821, Pc3921, Pc4221, Pc4321, Pc4421, Pc4521, Pc4621, Pc4721, Pc4821, Pc4921, Pc5021, Pc5121, Pc5221, Pc5321, Pc5421, Pc5521, Pc5621, Pc5721, Pc8021, Pc8121, Pc8221, Pc8321, Pc8421, Pc8521, Pc8621, and Pc8721), one representative cabbage isolate (Pv6321) was identified as P. versatile (representing the group of isolates Pv1021, Pv1121, Pv1221, Pv1321, Pv1421, Pv1521, Pv1621, Pv6121, Pv6221, Pv6321, Pv6421, Pv6521, and Pv6621), and one isolate Po7521 was identified as P. odoriferum (representing the group of isolates Po7021, Po7121, Po7221, Po7321, Po7421, Po7521, Po9121, Po9221, Po9321, Po9421, Po9521, and Po9621), with the percent identity in the 97.76–100%, 99.78–100%, and 99.57–100% range, respectively, depending on the sequenced gene (dnaX, icdA, mdh, and proA), as shown in Table 4. Accordingly, P. carotovorum was the most prevalent species (62.69%), while P. versatile and P. odoriferum were less represented (contributing by 19.40% and 17.91%, respectively). The sequences obtained for the seven representative cabbage isolates were deposited in the GenBank under the following accession numbers: OP729211-OP729217 (dnaX), OP729218-OP729224 (icdA), OP729225-OP729231 (mdh), and OP729232-OP729238 (proA).
Table 4.
The list of the 67 Serbian cabbage Pectobacterium spp. isolates and species affiliation, with the percent identity of the sequenced isolates based on the partial sequences of four sequenced genes (dnaX, icdA, mdh, and proA).
| Isolate Code | Hybrid | Identification According to the NCBI BLASTn (Per. Ident) | |||||
|---|---|---|---|---|---|---|---|
| Species | dnaX | icdA | mdh | proA | |||
| Pc2321 (group Pc2021–Pc2821) | Cheers F1 | P. carotovorum | 99.79% | 99.81% | 99.75% | 100% | |
| Pc3821 (group Pc3121–Pc3921) | Cheers F1 | P. carotovorum | 99.79% | 100% | 100% | 99.25% | |
| Pc4821 (group Pc4221–Pc4921) | Cheers F1 | P. carotovorum | 100% | 100% | 99.75% | 99.85% | |
| Pc5421 (group Pc5021–Pc5721) | Cheers F1 | P. carotovorum | 99.36% | 99.44% | 99.02% | 97.76% | |
| Pv6321 | (group Pv1021–Pv1621, Pv6121–Pv6621) |
Hippo F1 | P. versatile | 99.78% | 100% | 100% | 99.85% |
| Po7521 | (group Po7021–Po7521, Po9121–Po9621) | Hippo F1 | P. odoriferum | 99.79% | 100% | 100% | 99.57% |
| Pc8321 (group Pc8021–Pc8721) | Hippo F1 | P. carotovorum | 100% | 100% | 100% | 97.91% | |
The NJ phylogenetic tree generated based on the concatenated sequences of genes dnaX, icdA, mdh, and proA is presented in Figure 2. Based on these genes, the tested and comparative P. carotovorum, P. versatile, and P. odoriferum isolates/strains were separated into three clusters within the tree, each corresponding to one species. However, genetic heterogeneity (i.e., intraspecies genetic diversity) was observed within each species. Five of the seven tested cabbage P. carotovorum isolates examined in this study were separated into four groups (I: Pc2321 and Pc4821, II: Pc3821, III: Pc5421, and IV: Pc8321) within the cluster. The remaining tested P. versatile isolate Pv6321 was the most closely related to the comparative P. versatile strain DSM 30169 isolated from cabbage in Germany, while the P. odoriferum isolate Po7521 was most similar to the type P. odoriferum strain CFBP 1878 isolated from chicory in France. D. solani strain RNS 05.1.2A was placed on a monophyletic tree branch as an outgroup.
Figure 2.
The neighbor-joining phylogenetic tree constructed based on the concatenated sequences of genes dnaX, icdA, mdh, and proA for seven representative P. carotovorum
 , P. odoriferum
, P. odoriferum
 , and P. versatile
, and P. versatile
 isolates examined in this study and 19 strains of P. carotovorum, P. odoriferum, and P. versatile isolated from various hosts and countries, which were retrieved from the GenBank. The tree was rooted with the D. solani strain RNS 05.1.2A.
isolates examined in this study and 19 strains of P. carotovorum, P. odoriferum, and P. versatile isolated from various hosts and countries, which were retrieved from the GenBank. The tree was rooted with the D. solani strain RNS 05.1.2A.
3.3. Virulence Assessment
The developed disease symptoms observed seven days after the spray-inoculation of cabbage heads and potato tubers with suspensions of the tested isolates are presented in Figure 3.
Figure 3.
Examples of the bacterial soft rot symptoms on the cabbage head and potato tuber that were observed seven days after inoculation with the Serbian P. versatile strain Pv6321 and the P. carotovorum strain Pc3821, respectively.
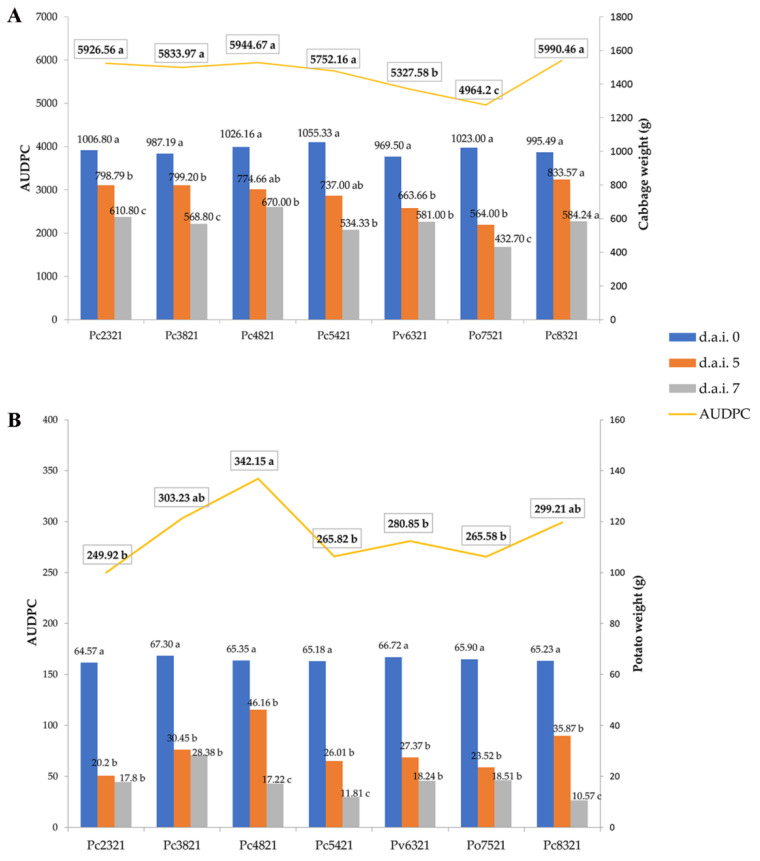
The results of the AUDPC analysis of the tested isolates on cabbage heads and potato tubers are shown in Figure 4A,B, respectively.
Figure 4.
Disease progress curves (AUDPC) showing (A) cabbage and (B) potato weight ratings (obtained 5 and 7 d.a.i.) resulting from tissue maceration due to inoculation with seven representative Pectobacterium cabbage isolates. Different letters represent statistically significant differences.
The AUDPC values pertaining to cabbage heads ranged from 4964.2 to 5990.46 for the P. odoriferum isolate Po7521 and P. carotovorum isolate Pc8321, respectively (Figure 4A). Based on the obtained AUDPC values, the P. odoriferum isolate Po7521 exhibited the highest virulence potential, followed by the P. versatile isolate Pv6321, while the P. carotovorum isolates Pc2321, Pc3821, Pc4821, Pc5421, and Pc8321 exhibited the lowest (and comparable) virulence potential. Statistically significant differences between the initial (0 d.a.i.) and the final (5 and 7 d.a.i.) weights were observed after cabbage inoculation with the P. carotovorum isolates Pc2321 and Pc3821, as well as the P. odoriferum isolate Po7521. On the other hand, the weights measured 5 and 7 d.a.i. with the P. versatile isolate Pv6321 were comparable but differed significantly from the initial weight (0 d.a.i.).
The AUDPC values pertaining to potato tubers ranged from 249.92 to 342.15 for the P. carotovorum isolates Pc2321 and Pc4821, respectively (Figure 4B). As the AUDPC values obtained for the seven representative isolates were comparable, all tested isolates appeared to be equally virulent with respect to this host. However, the weights measured 5 and 7 d.a.i. were statistically significantly lower than the initial weights when samples were inoculated with the P. carotovorum isolates Pc4821, Pc5421, and Pc8321. For the remaining two P. carotovorum isolates (Pc2321 and Pc3821), one P. versatile Pv6321, and one P. odoriferum Po7521, the weights measured 5 and 7 d.a.i. were comparable, but differed significantly from the initial values (0 d.a.i.).
4. Discussion
To the best of our knowledge, this is the first study since the pioneering work of Arsenijević and Obradović published more than 20 years ago [47], in which Erwinia carotovora subsp. carotovora was identified in the Bačka region, to provide evidence on the presence and diversity of the three bacteria (i.e., P. carotovorum, P. odoriferum, and P. versatile) causing soft rot in cabbage in Vojvodina (Serbia). The obtained results indicated presence of a combined infection in Field II, where all three identified species were confirmed on the cabbage hybrid Hippo F1. However, in Field I (hybrid Cheers F1), only P. carotovorum was detected. Analysis of the 67 cabbage isolates indicated that P. carotovorum was the most represented (62.69%), followed by P. versatile (19.40%) and P. odoriferum (17.91%). These findings are not surprising, given that P. carotovorum species is generally recognized as the main causal agent of soft rot in Brassicaceae plants [28]. This species was described on cabbage and Chinese cabbage in China, Brazil, Malaysia, Korea, and Bosnia and Herzegovina [8,48,49,50,51]. However, there is paucity of data on the presence of P. versatile and P. odoriferum on cabbage. In addition to P. carotovorum, P. versatile, and P. odoriferum, Chen et al. [31] also reported species P. aroidearum, P. brasiliense, and P. polaris on Chinese cabbage grown in different districts of Beijing (China). Presence of P. odoriferum was also reported on cabbage and Chinese cabbage in Central Poland, China, and Iran [28,48,52]. However, the lack of data on the presence of P. odoriferum and P. versatile on cabbage does not imply its absence on this host, as it is at least partly due to their recent reclassification (i.e., elevation from subspecies to the species level) within the genus Pectobacterium [53].
It is known that the genus Pectobacterium includes heterogonous strains characterized by diverse biochemical, physiological, and genetic properties even within the same species [54]. Based on the rep-PCR results, the Pectobacterium spp. isolates tested as a part of the present study were genetically heterogeneous, forming seven (BOX- and GTG5-PCR) and six (ERIC- and REP-PCR) groups on the UPGMA tree, depending on the utilized primers. In other words, the rep-PCR results indicate interspecies genetic heterogeneity, as well as intraspecies heterogeneity within P. carotovorum isolates only, which were clustered in five (BOX- and GTG5-PCR) and four (ERIC- and REP-PCR) groups. The rep-PCR (BOX-, ERIC-, and REP-PCR) analysis conducted by Alvarado et al. [49] also revealed high genetic variability among P. carotovorum strains isolated from Chinese cabbage in Pernambuco state (north-east Brazil). Based on the rep-PCR with primers for BOX-, ERIC-, and REP-PCR, Loc et al. [38] reached a similar conclusion to the one put forward in this study, positing that Pectobacterium strains of the same species tend to group closely according to their respective taxonomic designations. Considering that authors of several extant studies singled out rep-PCR as discriminative enough to reveal subtle differences between different Pectobacterium spp., as well as those among the same species, as proposed by Zoledowska et al. [26] for P. parmentieri strains from Poland, rep-PCR is a promising technique for the clarification of genetic diversity and discrimination of soft rot-causing Pectobacterium spp.
In the present study, typing of four housekeeping genes (dnaX, icdA, mdh, and proA) enabled appropriate identification of the tested cabbage isolates and a clear separation of each of the three identified species from one another. The existence of inter- and intra-species genetic heterogeneity between the three detected Pectobacterium species was again confirmed based on MLSA with concatenated sequences of the same four genes. According to Zeigler [55], among genes present in all sequenced bacterial genomes, the dnaX gene is considered one of the best candidates for assigning bacterial strains to the species level. Sławiak et al. [22] also highlighted the usefulness of gene dnaX for the identification of European potato Dickeya spp. strains. However, despite the demonstrated high resolution of the dnaX gene, the use of other protein-coding genes should not be discouraged due to the well-known claim that mutations, as the main engine of evolution, in Pectobacterium spp. mostly occur on these genes [14]. Moreover, using multiple genes allows for a greater genome coverage, which undoubtedly leads to a more reliable phylogeny reconstruction. In extant research, the remaining three genes (icdA, mdh, and proA) were successfully used for the typing of Pectobacterium spp. in different combinations with other housekeeping genes (e.g., acnA, gapA, mtlD, pgi, recA, rpoS, etc.) [16,17,18,19,20]. Important parameters when selecting these genes are (i) ubiquity in most enterobacteria, (ii) high discriminatory ability, and (iii) indispensable role in key metabolic processes [16].
In the virulence assessment assay performed as a part of this work, the lowest AUDPC values pertaining to cabbage heads (i.e., the highest virulence potential) was observed for the P. odoriferum isolate Po7521, followed by the P. versatile isolate Pv6321. On the other hand, the P. carotovorum isolates Pc2321, Pc3821, Pc4821, Pc5421, and Pc8321 were the least virulent, with a comparable virulence potential for cabbage. However, such statistical differences in aggressiveness among isolates/species were not detected on potato tubers. While these findings may be indicative of host−pathogen specificity in the case of bacteria P. odoriferum and P. versatile, more confident claims about such interactions would require more extensive studies performed on a larger number of known hosts. Bearing in mind the wide distribution, ubiquity, and polyphagous nature of P. carotovorum [56], and the resulting genetic diversity that enabled this species to survive and adapt to different ecological niches (i.e., hosts), it is likely that differences in aggressiveness will be observed between different hosts. In the study conducted by Li et al. [57], P. odoriferum strains isolated from Chinese cabbage exhibited much higher virulence potential for Chinese cabbage compared to the tested P. carotovorum and P. brasiliensis strains (measured 24 h and 30 h post-inoculation), all obtained from the same host. These authors did not observe statistically significant differences in the virulence potential between P. carotovorum and P. brasiliensis strains. However, it remains to be established whether the differences between species would be sustained for the duration of the disease progression. Moreover, based on the results reported by Waleron et al. [58], no statistically significant differences in the virulence potential on potato were observed between P. carotovorum and P. odoriferum, or between P. carotovorum and P. brasiliensis, based on the analyses performed 3 d.a.i.
This research significantly contributes to the current knowledge of the diversity of pectolytic bacteria affecting cabbage in Serbia, which has so far remained unexplored despite their great importance.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11020335/s1, Figure S1: Unweighted pair group method with arithmetic mean (UPGMA) phylogenetic trees of the 67 tested cabbage Pectobacterium spp. isolates and virtual gel images depicting rep-PCR fingerprinting patterns for each of the obtained isolate groups based on (A) BOX-PCR, (B) ERIC-PCR, (C) GTG5-PCR, and (D) REP-PCR.
Author Contributions
Conceptualization—T.P.M., A.J., and P.M. (Petar Mitrović); Methodology—T.P.M., A.J., S.M., R.I., and S.S.; Software—S.M. and R.I.; Investigation—A.J., S.M., and R.I.; Resources—T.P.M. and A.J.; Data curation—A.J., S.M., P.M. (Petar Mitrović), R.I., P.M. (Predrag Milovanović), and T.P.M.; Writing—original draft preparation—A.J. and T.P.M.; Writing—review and editing—A.J., S.M., R.I., P.M. (Predrag Milovanović), S.S., and T.P.M.; Visualization—A.J., R.I., and P.M. (Predrag Milovanović); Supervision—T.P.M. and S.S.; Funding acquisition—T.P.M., A.J., S.M., P.M. (Petar Mitrović), R.I., P.M. (Predrag Milovanović), and S.S. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Funding Statement
This research was supported by the Ministry of Education, Science and Technological Development of the Republic of Serbia, contract Nos. 451-03-68/2022-14/200053, 451-03-68/2022-14/200032, 451-03-68/2022-14/200117, 451-03-68/2022-14/200178, and 451-03-68/2022-14/200010.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Galal T.M., Khalafallah A.A., Elawa O.E., Hassan L.M. Human health risks from consuming cabbage (Brassica oleracea L. var. capitata) grown on wastewater irrigated soil. Int. J. Phytoremediation. 2018;20:1007–1016. doi: 10.1080/15226514.2018.1452186. [DOI] [PubMed] [Google Scholar]
- 2.Šamec D., Pavlović I., Salopek-Sondi B. White cabbage (Brassica oleracea var. capitata f. alba): Botanical, phytochemical and pharmacological overview. Phytochem. Rev. 2017;16:117–135. doi: 10.1007/s11101-016-9454-4. [DOI] [Google Scholar]
- 3.Food and Agriculture Organization of the United Nations FAOSTAT Statistical Database. [(accessed on 21 November 2022)]. Available online: https://www.fao.org/faostat/en/#data/QCL.
- 4.Živanović Miljković J., Popović V., Gajić A. Land Take Processes and Challenges for Urban Agriculture: A Spatial Analysis for Novi Sad, Serbia. Land. 2022;11:769. doi: 10.3390/land11060769. [DOI] [Google Scholar]
- 5.Cui W., He P., Munir S., He P., He Y., Li X., Yang L., Wang B., Wu Y., He P. Biocontrol of soft rot of Chinese cabbage using an endophytic bacterial strain. Front. Microbiol. 2019;10:1471. doi: 10.3389/fmicb.2019.01471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhat K.A., Masood S.D., Bhat N.A., Bhat M.A., Razvi S.M., Mir M.R., Akhtar S., Wani N., Habib M. Current status of post harvest soft rot in vegetables: A review. Asian J. Plant Sci. 2010;9:200–208. doi: 10.3923/ajps.2010.200.208. [DOI] [Google Scholar]
- 7.Harter L.L., Jones L.R. Cabbage Diseases. US Government Printing Office; Washington, DC, USA: 1923. p. 20. Farmers’ Bulletin No. 1351. [Google Scholar]
- 8.Popović T., Jelušić A., Marković S., Iličić R. Characterization of Pectobacterium carotovorum subsp. carotovorum isolates from a recent outbreak on cabbage in Bosnia and Herzegovina. Pestic. Phytomedicine. 2019;34:211–222. doi: 10.2298/PIF1904211P. [DOI] [Google Scholar]
- 9.Van Gijsegem F., Toth I.K., van der Wolf J.M. Plant Diseases Caused by Dickeya and Pectobacterium Species. Springer; Cham, Switzerland: 2021. Outlook—Challenges and perspectives for management of diseases caused by Pectobacterium and Dickeya species; pp. 283–289. [DOI] [Google Scholar]
- 10.Agrios G.N. Plant Patholog. Academic Press; Cambridge, MA, USA: 2005. Plant diseases caused by prokaryotes: Bacteria and mollicutes. Bacterial Soft Rots; pp. 615–703. [DOI] [Google Scholar]
- 11.Czajkowski R., Perombelon M.C., van Veen J.A., van der Wolf J.M. Control of blackleg and tuber soft rot of potato caused by Pectobacterium and Dickeya species: A review. Plant Pathol. 2011;60:999–1013. doi: 10.1111/j.1365-3059.2011.02470.x. [DOI] [Google Scholar]
- 12.Charkowski A.O. The changing face of bacterial soft-rot diseases. Annu. Rev. Phytopathol. 2018;56:269–288. doi: 10.1146/annurev-phyto-080417-045906. [DOI] [PubMed] [Google Scholar]
- 13.Jonkheer E.M., Brankovics B., Houwers I.M., van der Wolf J.M., Bonants P.J., Vreeburg R.A., Bollema R., de Haan J.R., Berke L., Smit S., et al. The Pectobacterium pangenome, with a focus on Pectobacterium brasiliense, shows a robust core and extensive exchange of genes from a shared gene pool. BMC Genom. 2021;22:265. doi: 10.1186/s12864-021-07583-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Achtman M. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu. Rev. Microbiol. 2008;62:53–70. doi: 10.1146/annurev.micro.62.081307.162832. [DOI] [PubMed] [Google Scholar]
- 15.Nabhan S., De Boer S.H., Maiss E., Wydra K. Taxonomic relatedness between Pectobacterium carotovorum subsp. carotovorum, Pectobacterium carotovorum subsp. odoriferum and Pectobacterium carotovorum subsp. brasiliense subsp. nov. J. Appl. Microbiol. 2012;113:904–913. doi: 10.1111/j.1365-2672.2012.05383.x. [DOI] [PubMed] [Google Scholar]
- 16.Ma B., Hibbing M.E., Kim H.S., Reedy R.M., Yedidia I., Breuer J., Charkowski A.O. Host range and molecular phylogenies of the soft rot enterobacterial genera Pectobacterium and Dickeya. Phytopathology. 2007;97:1150–1163. doi: 10.1094/PHYTO-97-9-1150. [DOI] [PubMed] [Google Scholar]
- 17.Moleleki L.N., Onkendi E.M., Mongae A., Kubheka G.C. Characterisation of Pectobacterium wasabiae causing blackleg and soft rot diseases in South Africa. Eur. J. Plant Pathol. 2013;135:279–288. doi: 10.1007/s10658-012-0084-4. [DOI] [Google Scholar]
- 18.Pitman A.R., Harrow S.A., Visnovsky S.B. Genetic characterisation of Pectobacterium wasabiae causing soft rot disease of potato in New Zealand. Eur. J. Plant Pathol. 2010;126:423–435. doi: 10.1007/s10658-009-9551-y. [DOI] [Google Scholar]
- 19.Waleron M., Misztak A., Waleron M., Franczuk M., Jońca J., Wielgomas B., Mikiciński A., Popović T., Waleron K. Pectobacterium zantedeschiae sp. nov. a new species of a soft rot pathogen isolated from Calla lily (Zantedeschia spp.) Syst. Appl. Microbiol. 2019;42:275–283. doi: 10.1016/j.syapm.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 20.Marković S., Milić Komić S., Jelušić A., Iličić R., Bagi F., Stanković S., Popović T. First report of Pectobacterium versatile causing blackleg of potato in Serbia. Plant Dis. 2022;106:312. doi: 10.1094/PDIS-06-21-1128-PDN. [DOI] [Google Scholar]
- 21.Ma X., Schloop A., Swingle B., Perry K.L. Pectobacterium and Dickeya responsible for potato blackleg disease in New York State in 2016. Plant Dis. 2018;102:1834–1840. doi: 10.1094/PDIS-10-17-1595-RE. [DOI] [PubMed] [Google Scholar]
- 22.Sławiak M., van Beckhoven J.R., Speksnijder A.G., Czajkowski R., Grabe G., van der Wolf J.M. Biochemical and genetical analysis reveal a new clade of biovar 3 Dickeya spp. strains isolated from potato in Europe. Eur. J. Plant Pathol. 2009;125:245–261. doi: 10.1007/s10658-009-9479-2. [DOI] [Google Scholar]
- 23.Marković S., Stanković S., Jelušić A., Iličić R., Kosovac A., Poštić D., Popović T. Occurrence and Identification of Pectobacterium carotovorum subsp. brasiliensis and Dickeya dianthicola Causing Blackleg in some Potato Fields in Serbia. Plant Dis. 2021;105:1080–1090. doi: 10.1094/PDIS-05-20-1076-RE. [DOI] [PubMed] [Google Scholar]
- 24.Norman D.J., Yuen J.M.F., Resendiz R., Boswell J. Characterization of Erwinia populations from nursery retention ponds and lakes infecting ornamental plants in Florida. Plant Dis. 2003;87:193–196. doi: 10.1094/PDIS.2003.87.2.193. [DOI] [PubMed] [Google Scholar]
- 25.Maisuria V.B., Nerurkar A.S. Characterization and differentiation of soft rot causing Pectobacterium carotovorum of Indian origin. Eur. J. Plant Pathol. 2013;136:87–102. doi: 10.1007/s10658-012-0140-0. [DOI] [Google Scholar]
- 26.Zoledowska S., Motyka A., Zukowska D., Sledz W., Lojkowska E. Population structure and biodiversity of Pectobacterium parmentieri isolated from potato fields in temperate climate. Plant Dis. 2018;102:154–164. doi: 10.1094/PDIS-05-17-0761-RE. [DOI] [PubMed] [Google Scholar]
- 27.Song H., Lee J.Y., Lee H.W., Ha J.H. Inactivation of bacteria causing soft rot disease in fresh cut cabbage using slightly acidic electrolyzed water. Food Control. 2021;128:108217. doi: 10.1016/j.foodcont.2021.108217. [DOI] [Google Scholar]
- 28.Oskiera M., Kałużna M., Kowalska B., Smolińska U. Pectobacterium carotovorum subsp. odoriferum on cabbage and Chinese cabbage: Identification, characterization and taxonomic relatedness of bacterial soft rot causal agents. J. Plant Pathol. 2017;99:149–160. doi: 10.4454/jpp.v99i1.3831. [DOI] [Google Scholar]
- 29.Xie H., Li X.Y., Ma Y.L., Tian Y. First report of Pectobacterium aroidearum causing soft rot of Chinese cabbage in China. Plant Dis. 2018;102:674. doi: 10.1094/PDIS-07-17-1059-PDN. [DOI] [Google Scholar]
- 30.Fujimoto T., Nakayama T., Ohki T., Maoka T. First Report of Soft Rot of Cabbage Caused by Pectobacterium wasabiae in Japan. Plant Dis. 2021;105:2236. doi: 10.1094/PDIS-02-21-0238-PDN. [DOI] [Google Scholar]
- 31.Chen C., Li X., Bo Z., Du W., Fu L., Tian Y., Cui S., Shi Y., Xie H. Occurrence, Characteristics, and PCR-Based Detection of Pectobacterium polaris Causing Soft Rot of Chinese Cabbage in China. Plant Dis. 2021;105:2880–2887. doi: 10.1094/PDIS-12-20-2752-RE. [DOI] [PubMed] [Google Scholar]
- 32.Lee S., Vu N.-T., Oh E.-J., Rahimi-Midani A., Thi T.-N., Song Y.-R., Hwang I.-S., Choi T.-J., Oh C.-S. Biocontrol of soft rot caused by Pectobacterium odoriferum with bacteriophage phiPccP-1 in Kimchi cabbage. Microorganisms. 2021;9:779. doi: 10.3390/microorganisms9040779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smoktunowicz M., Jonca J., Stachowska A., May M., Waleron M.M., Waleron M., Waleron K. The International Trade of Ware Vegetables and Orna-Mental Plants-An Underestimated Risk of Accelerated Spreading of Phytopathogenic Bacteria in the Era of Globalisation and Ongoing Climatic Changes. Pathogens. 2022;11:728. doi: 10.3390/pathogens11070728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gavrilović V., Obradović A., Arsenijević M. Bacterial soft rot of carrot, parsley and celery. In: De Boer S.H., editor. Plant Pathogenic Bacteria. Springer; Dordrecht, The Netherlands: 2001. pp. 269–271. [Google Scholar]
- 35.Gašić K., Gavrilović V., Dolovac N., Trkulja N., Živković S., Ristić D., Obradović A. Pectobacterium carotovorum subsp. carotovorum-the causal agent of broccoli soft rot in Serbia. Pestic. Phytomedicine. 2014;29:249–255. doi: 10.2298/PIF1404249G. [DOI] [Google Scholar]
- 36.Popović T., Jelušić A., Milovanović P., Janjatović S., Budnar M., Dimkić I., Stanković S. First report of Pectobacterium atrosepticum, causing bacterial soft rot on calla lily in Serbia. Plant Dis. 2017;101:2145. doi: 10.1094/PDIS-05-17-0708-PDN. [DOI] [Google Scholar]
- 37.Zlatković N., Prokić A., Gašić K., Kuzmanović N., Ivanović M., Obradović A. First report of Pectobacterium carotovorum subsp. brasiliense causing soft rot on squash and watermelon in Serbia. Plant Dis. 2019;103:2667. doi: 10.1094/PDIS-12-18-2213-PDN. [DOI] [Google Scholar]
- 38.Loc M., Milošević D., Ivanović Ž., Ignjatov M., Budakov D., Grahovac J., Grahovac M. Genetic Diversity of Pectobacterium spp. on Potato in Serbia. Microorganisms. 2022;10:1840. doi: 10.3390/microorganisms10091840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loc M., Milošević D., Ignjatov M., Ivanović Ž., Budakov D., Grahovac J., Vlajkov V., Pajčin I., Grahovac M. First report of Pectobacterium punjabense causing potato soft rot and blackleg in Serbia. Plant Dis. 2022;106:1513. doi: 10.1094/PDIS-06-21-1199-PDN. [DOI] [PubMed] [Google Scholar]
- 40.Hélias V., Hamon P., Huchet E., Wolf J.V.D., Andrivon D. Two new effective semiselective crystal violet pectate media for isolation of Pectobacterium and Dickeya. Plant Pathol. 2012;61:339–345. doi: 10.1111/j.1365-3059.2011.02508.x. [DOI] [Google Scholar]
- 41.Fahy P.C., Persley G.J. Plant Bacterial Diseases: A diagnostic Guide. Academic Press; Sydney, Australia: 1983. 393p [Google Scholar]
- 42.Bertani G. Studies on lysogenesis I: The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Popović T., Jelušić A., Dimkić I., Stanković S., Poštić D., Aleksić G., Veljović Jovanović S. Molecular characterization of Pseudomonas syringae pv. coriandricola and biochemical changes attributable to the pathological response on its hosts carrot, parsley, and parsnip. Plant Dis. 2019;103:3072–3082. doi: 10.1094/PDIS-03-19-0674-RE. [DOI] [PubMed] [Google Scholar]
- 44.Kettani-Halabi M., Terta M., Amdan M., El Fahime E.M., Bouteau F., Ennaji M.M. An easy, simple inexpensive test for the specific detection of Pectobacterium carotovorum subsp. carotovorum based on sequence analysis of the pmrA gene. BMC Microbiol. 2013;13:176. doi: 10.1186/1471-2180-13-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Louws F.J., Fulbright D.W., Stephens C.T., De Bruijn F.J. Specific genomic fingerprints of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Appl. Environ. Microbiol. 1994;60:2286–2295. doi: 10.1128/aem.60.7.2286-2295.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Versalovic J., Schneider M., De Bruijn F.J., Lupski J.R. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 1994;5:25–40. [Google Scholar]
- 47.Arsenijević M., Obradović A. Occurrence of bacterial wilt and soft rot of seed cabbage plants (Brassica oleracea var. capitata L.) in Yugoslavia. J. Phytopathol. 1996;144:315–319. doi: 10.1111/j.1439-0434.1996.tb01535.x. [DOI] [Google Scholar]
- 48.Zhu L., Xie H., Chen S., Ma R. Rapid isolation, identification and phylogenetic analysis of Pectobacterium carotovorum ssp. J. Plant Pathol. 2010;92:479–483. [Google Scholar]
- 49.Alvarado I.C.M., Michereff S.J., Mariano R.L.R., Souza E.B., Quezado-Duval A.M., Resende L.V., Cardoso E., Mizubuti E.S.G. Characterization and variability of soft rot-causing bacteria in Chinese cabbage in North Eastern Brazil. J. Plant Pathol. 2011;93:173–181. [Google Scholar]
- 50.Nazerian E., Sijam K., Mior Ahmad Z.A., Vadamalai G. First report of cabbage soft rot caused by Pectobacterium carotovorum subsp. carotovorum in Malaysia. Plant Dis. 2011;95:491. doi: 10.1094/PDIS-09-10-0683. [DOI] [PubMed] [Google Scholar]
- 51.Park T.H., Choi B.S., Choi A.Y., Choi I.Y., Heu S., Park B.S. Genome sequence of Pectobacterium carotovorum subsp. carotovorum strain PCC21, a pathogen causing soft rot in Chinese cabbage. J. Bacteriol. 2012;194:6345–6346. doi: 10.1128/JB.01583-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rezaei R., Taghavi S.M. Phenotypic and genotypic characteristics of Iranian soft rot bacterial isolates from different hosts. Phytopathol. Mediterr. 2010;49:194–204. [Google Scholar]
- 53.Portier P., Pédron J., Taghouti G., Fischer-Le Saux M., Caullireau E., Bertrand C., Laurent A., Chawki K., Oulgazi S., Moumni M., et al. Elevation of Pectobacterium carotovorum subsp. odoriferum to species level as Pectobacterium odoriferum sp. nov., proposal of Pectobacterium brasiliense sp. nov. and Pectobacterium actinidiae sp. nov., emended description of Pectobacterium carotovorum and description of Pectobacterium versatile sp. nov., isolated from streams and symptoms on diverse plants. Int. J. Syst. Evol. Microbiol. 2019;69:3207–3216. doi: 10.1099/ijsem.0.003611. [DOI] [PubMed] [Google Scholar]
- 54.Öztürk M., Umar A.R. Occurrence, identification, and host range of Pectobacterium brasiliense causing soft rot on seed potato tubers in Turkey. J. Plant Dis. Prot. 2022:1–12. doi: 10.1007/s41348-022-00675-8. [DOI] [Google Scholar]
- 55.Zeigler D.R. Gene sequences useful for predicting relatedness of whole genomes in bacteria. Int. J. Syst. Evol. Microbiol. 2003;53:1893–1900. doi: 10.1099/ijs.0.02713-0. [DOI] [PubMed] [Google Scholar]
- 56.Cariddi C., Sanzani S.M. A severe outbreak of bacterial lettuce soft rot caused by Pectobacterium carotovorum subsp. carotovorum in Apulia (Italy) J. Plant Pathol. 2013;95:441–446. [Google Scholar]
- 57.Li X., Fu L., Chen C., Sun W., Tian Y., Xie H. Characteristics and rapid diagnosis of Pectobacterium carotovorum ssp. associated with bacterial soft rot of vegetables in China. Plant Dis. 2020;104:1158–1166. doi: 10.1094/PDIS-05-19-1033-RE. [DOI] [PubMed] [Google Scholar]
- 58.Waleron M., Waleron K., Lojkowska E. Characterization of Pectobacterium carotovorum subsp. odoriferum causing soft rot of stored vegetables. Eur. J. Plant Pathol. 2014;139:457–469. doi: 10.1007/s10658-014-0403-z. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.