Abstract
The Pseudomonas fluorescens 23F phosphonoacetate hydrolase gene (phnA) encodes a novel carbon-phosphorus bond cleavage enzyme whose expression is independent of the phosphate status of the cell. Analysis of the regions adjacent to the phosphonoacetate hydrolase structural gene (phnA) indicated the presence of five open reading frames (ORFs). These include one (phnR) whose putative product shows high levels of homology to the LysR family of positive transcriptional regulators. Its presence was shown to be necessary for induction of the hydrolase activity. 2-Phosphonopropionate was found to be an inducer (and poor substrate) for phosphonoacetate hydrolase. Unlike phosphonoacetate, which is also an inducer of phosphonoacetate hydrolase, entry of 2-phosphonopropionate into cells appeared to be dependent on the presence of a gene (phnB) that lies immediately downstream of phnA and whose putative product shows homology to the glycerol-3-phosphate transporter. RNA analysis revealed transcripts for the phnAB and phnR operons, which are transcribed divergently; the resulting mRNAs overlapped by 29 nucleotide bases at their 5′ ends. Transcripts of phnAB were detected only in cells grown in the presence of phosphonoacetate, whereas transcripts of phnR were observed in cells grown under both induced and uninduced conditions. The expression of three additional genes found in the phnA region did not appear necessary for the degradation of phosphonoacetate and 2-phosphonopropionate by either Pseudomonas putida or Escherichia coli cells.
Organophosphonates are a group of compounds of biogenic and xenobiotic origins that are characterized by possession of a direct carbon-phosphorus bond. Two different routes of C-P bond cleavage, each inducible only under conditions of phosphorus limitation, have been demonstrated when such molecules serve as the sole phosphorus source for microbial growth. “C-P lyase” is the trivial name given to an enzyme complex which catalyzes the cleavage of the C-P bond of both substituted and unsubstituted phosphonates by a mechanism which may involve redox or radical chemistry (27). By contrast, phosphonoacetaldehyde hydrolase (“phosphonatase” [EC 3.11.1.1]) is active in vitro and hydrolytically cleaves only the substituted C-P bond of phosphonoacetaldehyde to yield acetaldehyde and Pi (10). Since bacterial cleavage of the C-P bond by both C-P lyase(s) and phosphonatase is under control of the pho regulon (10) and hence occurs only under conditions of Pi limitation, organophosphonates generally fail to serve as sources of carbon for microbial growth because the excess of Pi released during the catabolism of their carbon skeletons serves to repress and/or inhibit their further mineralization. Genetic analysis of C-P bond cleavage by the C-P lyase and phosphonatase pathways has been reported (10, 18, 26), and the phosphonatase gene of Pseudomonas aeruginosa has been characterized (5).
We have recently reported the purification (15) and molecular cloning (14) of phosphonoacetate (PA) hydrolase (EC 3.11.1.2), a novel C-P cleavage enzyme from Pseudomonas fluorescens 23F which is able to mineralize PA with essentially quantitative Pi release. Expression of the enzyme is inducible in the presence of its apparently xenobiotic substrate PA and is independent of the Pi status of the cell. No mode of regulation had previously been described in which a phosphorus-containing compound is needed for induction of a gene required for its own utilization (27). To obtain a better understanding of this unique biodegradation system we now report the structural and functional analysis of those regions of the genome of P. fluorescens 23F adjacent to the phnA gene and address the function and regulation of the genes comprising the phn operon.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The PA-degrading strain P. fluorescens 23F has been described (17). Escherichia coli DH5α and Pseudomonas putida AC577 trp leu (from A. M. Chakrabarty, University of Illinois-Chicago, Chicago) were used for the analysis of cloned PA degradation genes. Preliminary experiments showed that PA and 2-phosphonopropionate could serve as only poor sources of phosphorus for P. putida AC577 without Pi release. E. coli DH5α could use PA but not 2-phosphonopropionate as a source of phosphorus, also without Pi release. Plasmid pRK2013 (7) was used as a helper plasmid. Cosmid pLAFR5 (12) was used for analysis of the expression of phn genes in E. coli and P. putida, and plasmid vectors pUC129 (12) and pUC18 were used for the analysis of the same genes in E. coli only. Promoter probe vector pKK232–8 (Pharmacia) was used for the analysis of phnA gene expression.
Media and growth conditions.
Pseudomonas and E. coli strains were propagated in a rich (2YT) medium (19) solidified when required by addition of Difco agar (1.8% [wt/vol]). For the growth and detection of recombinant plasmids, ampicillin (100 μg ml−1) or tetracycline (15 μg ml−1), IPTG (isopropyl-β-d-thiogalactopyranoside) (50 μg ml−1), and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (50 μg ml−1) were used. Mineral salts medium was free of phosphorus and contained (per liter): KCl, 0.24 g; MgSO4 · 7H2O, 0.2 g; NH4Cl, 1.0 g; CaCl2 · 2H2O, 1.0 mg; ferric ammonium citrate, 1.0 mg; HEPES buffer, 25 mM (pH 7.2); and 1 ml of trace element solution (13). Purified agar (1%) (Oxoid, Basingstoke, United Kingdom) was added when appropriate. As a source of carbon, sodium gluconate (2.0 g l−1) or sodium pyruvate (2.0 g l−1) was used. As a source of phosphorus, PA (2 mM), 2-phosphonopropionate (2PP) (2 mM), or Pi (2 mM) was used. A number of phosphonates (2 mM) were also screened for their ability to serve as sources of phosphorus. Medium for auxotrophic strains was supplemented with appropriate amino acids (50 mg l−1) and vitamins (5 mg l−1). Pseudomonas strains were routinely grown at 29°C, and E. coli was grown at 37°C.
Bacterial crosses.
Recombinant plasmids constructed using pLAFR5 were transferred into the P. putida AC577 recipient strain using a three-factor mating procedure (4) with pRK2013 as a helper plasmid.
Analysis of PA hydrolase activity.
C-P bond cleavage activity in cell extracts and resting cells was assayed at 30°C by measuring the amount of phosphate liberated from phosphonates supplied as substrates (17). In both cases cultures were grown in a mineral salts medium with Pi (2 mM) as phosphorus source to an optical density at 600 nm (OD600) of 1.0. Cells were then harvested, washed with NaCl (0.85%), and transferred into fresh mineral salts medium without a carbon and phosphorus source; at this point various phosphonates (2 mM) were added as potential inducers, and incubation was carried out for another 8 to 10 h. For assay of resting cells, this culture (adjusted to an OD600 of 0.5) was washed and divided into aliquots, and each potential substrate phosphonate (5 mM) was added. After 16 h of incubation at 30°C with aeration, phosphate release into culture supernatants was measured by the method of Fiske and Subbarow (8). Cell extracts were prepared as described earlier (17). As a control, cells incubated in medium lacking phosphonate supplementation were used. Lack of Pi release in phosphonate-containing sterile growth medium was confirmed. PA hydrolase activity was expressed as nanomoles of Pi liberated per minute per milligram of protein. Protein concentration was determined by the method of Bradford (2).
In all cases PA hydrolase activity was measured using cell extracts. However, to investigate the role of PhnB protein as a possible transporter, activities for both cell extracts and resting cells induced with PA were measured.
DNA techniques.
Recombinant DNA manipulations were carried out using standard protocols as described by Sambrook et al. (23). Restriction analysis of recombinant plasmids was performed using enzymes obtained from Pharmacia, according to the manufacturer's instructions. A Sephaglas BandPrep kit (Pharmacia) was used for the recovery of restriction fragments from agarose gels.
Construction of clones.
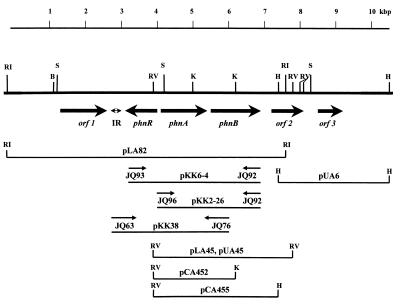
Several recombinant plasmids (pLA82, pLA45, pUA45, pCA452, and pCA455) containing the phn genes were described earlier (14) and are further characterized in this paper. Plasmids pUA6, pKK2–26, and pKK6–4 were constructed in this work. To clone the defined phn region sequences, the following oligonucleotide primers were used: JQ92, 5′-CTGAAGCTTGTGGCGCTACGATC-3′ (a reverse primer; position in the sequence 6688 to 6671 nt); JQ93, 5′-CATAAGCTTCGAACCGGTAAAACTG-3′ (2911 to 2929 nt); and JQ96, 5′-CGGAAGCTTCAAAAATGCGCAGCGC-3′ (3863 to 3881 nt). PCRs were performed on P. fluorescens 23F genomic DNA using two pairs of primers: JQ92-JQ93 and JQ92-JQ96. PfuI polymerase was used to minimize the number of possible mistakes introduced during amplification. Reactions were carried out in volumes of 50 μl with concentrations of deoxynucleoside triphosphates at 200 μM and primers at 0.15 μM each. The following temperature profile was used: denaturing at 95°C for 3 min and then 95°C for 30 s, 60°C for 30 s, and 72°C for 7 min, for 30 cycles. The HindIII sites introduced into all above listed primers (underlined) were used for the cloning of the resulting PCR fragments into pKK232–8. Primers JQ63 (5′-ACTGGCGTGCTGCTGGCA-3′ [2472 to 2489 nt]) and JQ76 (5′-CTTGTATTGCAGGGTATCAG-3′ [5404 to 5385 nt]) were used for construction of pKK38.
DNA sequencing.
Plasmids pUC129 and pUC18 containing insertions from the phnA gene region were used as templates for the Taq Dye-Deoxy Terminator Cycle sequencing kit (Applied Biosystems). DNA sequences were obtained using an automatic DNA sequencer (model 373A; Applied Biosystems). The nucleotide sequences of both strands were determined. Primer synthesis was performed by C. Stevenson (School of Biology and Biochemistry, The Queen's University of Belfast, Ireland). Initial computer analysis of the sequences was performed using the DNASIS (Hitachi) software package. The alignment of sequences was performed by using CLUSTALW software (25) with parameters set at default values.
Searches for nucleotide and amino acid sequence similarities were carried out on the National Center for Biotechnology Information web server (http://www.ncbi.nlm.nih.gov) in the nonredundant nucleotide (nr-nt) or amino acid (nr-aa) databases using the FASTA and BLAST programs (22).
RNA analysis.
P. fluorescens 23F cells were grown in mineral salts medium to an OD600 of 0.7. In the case of induction with PA, cells were harvested by centrifugation, washed, and resuspended in the same medium without a source of carbon and phosphorus. PA was added to a final concentration of 10 mM, and cells were incubated for 24 h. Cells were lysed with a solution containing 4 M guanidine thiocyanate, 0.5% (wt/vol) laurylsarcosine, and 100 mM β-mercaptoethanol. An equal volume of phenol saturated with 0.3 M sodium acetate (pH 5.5) was added, and the preparation was mixed thoroughly and heated at 80°C for 4 min. It was then cooled, and 0.1 volume of chloroform was added. Following centrifugation the aqueous phase was collected. This procedure was repeated 3 to 4 times. The RNA preparation was ethanol precipitated and dissolved in diethyl pyrocarbonate-treated water. Northern blot analysis was carried out essentially according to the method of Sambrook et al. (23). Aliquots of total RNA (20 μg) were fractionated in 0.8% or 1.2% agarose-formaldehyde gels electroblotted onto Hybond-N+ membranes (Amersham) in formaldehyde gel-running buffer for 3 h at a constant current of 150 mA using the Mini Trans-Blot cell (Bio-Rad) and were UV cross-linked to the filter. Either 5′-32P-labeled oligonucleotides or uniformly 32P-labeled single-stranded DNA fragments were used in hybridization. Primer JQ86 (5′-CGCCGTGAGTCAGGTTCAGTTCCCGGGCCG-3′) complementary to mRNA for phnR (positions 3803 through 3832 in the sequence) was 5′-32P-labeled with T4 polynucleotide kinase (Promega) by the recommendations of the suppliers. The probe used for phnA mRNA analysis was obtained as follows. First a DNA fragment (positions 3948 through 4682), corresponding to the 5′ end of the phnA gene, was amplified by PCR using primers JQ87 (5′-TTGTGTGAGAAAGATGCTCAGAATACGGTG-3′ [positions 3948 through 3977]) and JQ84 (5′-TGCCTGGCGCATGTTTGTGTTG-3′ [positions 4682 through 4661]). Residual primers, any extraneous single-stranded DNA produced by PCR, and the remaining deoxynucleoside triphosphates were removed from the PCR mixture with exonuclease I (USB) and shrimp alkaline phosphatase (USB), using the supplier's recommendations. Second, a uniformly 32P-labeled single-stranded DNA fragment was synthesized by asymmetric PCR using JQ84 as the primer and the product of the previous PCR as the template. RNA immobilized on filters was hybridized overnight with 5′-32P-labeled JQ86 or the uniformly 32P-labeled, single-stranded DNA fragment, in 6× SSC (1× SSC is 0. 15 M NaCl, 0.015 M sodium citrate) at 52°C or 68°C, respectively. Following hybridization, membranes were washed with hybridization buffer under the same conditions for 30 min and subsequently washed at room temperature in 2× SSC, 0.5× SSC, and 0.1× SSC.
Primer extension experiments were carried out according to the method of Sambrook et al. (23). Primers JQ86 (see above) and JQ88 (5′-CAGGCGATAGGAGCGCGAGTTCACGCTGAT-3′ [positions 4090 through 4060]), which are complementary to mRNA for phnR and phnA, respectively, were employed for these purposes. 5′-32P labeling was carried out as described above. Labeled primers (500 to 1,000 × 103 cpm) were annealed with 10 to 30 μg of RNA. Hybridization was performed at 30°C in a solution containing 40 mM piperazine-N,N′-bis(2 ethanesulfonic acid) (PIPES), pH 6.4, 1 mM EDTA, 0.4 M NaCl, and 80% formamide. Primer extension was carried out with RNase H minus Moloney murine leukemia virus reverse transcriptase (Promega). The products synthesized were analyzed by electrophoresis in 6% polyacrylamide 8 M urea gels.
GC-MS determination of propionate.
Aliquots from enzyme assays containing 2PP were derivatized as the phenylphenacyl derivative of the target compound and analyzed by gas chromatography-mass spectrometry (GC-MS) with the mass spectrometer (MS) operating in the selected ion-monitoring mode. Fluoroacetate was used as an internal standard for quantitation purposes. Fluoroacetate (0.25 ml, 1 mM) was added to cell extract (1 ml), and the mixture was frozen. Following sample lyophilization, the dried powder was suspended in toluene-acetonitrile (1:1 [vol/vol], 1 ml); 1,4,7,10,13,16-hexaoxacyclooctadecane (10 mg) and 4-(bromoacetyl) biphenyl (10 mg) were added; and the mixture was heated at 75°C for 12 h. The derivatized samples were cooled and analyzed by GC-MS on a Hewlett Packard 5890 series II gas chromatograph linked to a Hewlett Packard 5971 mass selective detector controlled by a HP vectra computer using ChemStation software. The gas chromatograph was fitted with an HP Ultra 2 fused-silica wall-coated open tubular capillary column (12 m by 0.22 mm internal diameter) with 5% biphenyl–95% dimethyl polysiloxane as the bonded phase. Helium was used as the carrier gas at a flow rate of 0.8 ml min−1, and samples (1 μl) were injected at a split ratio of 80:1. After injection of samples, the oven temperature was held at 130°C for 1 min, then programmed upwards at 10°C min−1 to 300°C. The mass selective detector was operated in selected ion monitoring mode monitoring ion currents at m/z 181 and 268 atomic mass units for propionate and m/z 181 and 272 atomic mass units for fluoroacetate. To quantify propionate, the peak area for ion m/z 181 at the expected retention time of propionate was compared to a standard response curve after correcting for the response of the internal standard.
Nucleotide sequence accession number.
The nucleotide sequence reported in this work was deposited in GenBank under accession number L49465.
RESULTS
Nucleotide sequence analysis of the phnA gene region.
The construction of the P. fluorescens 23F genomic library, and cloning and sequencing of the PA-hydrolase structural gene (phnA) were reported earlier (14). Here we describe the sequencing analysis of the regions (9.5 kbp) adjacent to the phnA gene (Fig. 1 and Table 1). The nucleotide sequences of orf1, phnR, phnB, and part of orf2 were obtained using subclones produced from the pLA82 insert (Fig. 1). For sequencing of the distal part of orf2 and the whole of orf3, plasmids pUA45 and pUA6 were used (Fig. 1). The latter plasmid was produced by the cloning from the P. fluorescens 23F genomic library of the HindIII fragment overlapping with the pLA82 insert.
FIG. 1.
Physical and genetic map of the phnA gene region. Restriction sites are indicated as follows: E, EcoRI; RV, EcoRV; K, KpnI; S, SalI; H, HindIII; IR, region of imperfect inverted repeats; phnA, PA-hydrolase gene locus; phnR, putative transcriptional regulator of the PA-hydrolase gene cluster; phnB, putative transporter gene for 2PP; putative genes with unknown functions found in the region are designated as orf1, orf2, and orf3. The direction of transcription of each gene is shown by the arrows (→). The most important clones analyzed in this work are shown; the fragments containing the phnA gene were subcloned into pLAFR5 (designated as pLA plasmids), pUC129 (designated as pUA plasmids), or pUC18 (designated as pCA plasmids). Plasmid pKK2–26 was constructed by cloning a 2.8-kb PCR fragment (primers JQ96 and JQ92) into plasmid pKK232–8, and plasmid pKK6–4 was constructed by cloning a 3.78-kb PCR fragment (primers JQ93 and JQ92) into the same vector.
TABLE 1.
Sequence analysis of the phnA gene region
| Gene or feature | Coding stranda | Coding region (nt) | Protein (aa/Da) | Relatedness to other proteinsb
|
||
|---|---|---|---|---|---|---|
| Protein | Identity (%) | Accession no. | ||||
| orf1 | + | 1368–2585 | 405/41681 | MhpT | 65 | P77589 |
| IR | NA | 2610–2828 | NA | NA | NA | NA |
| phnR | − | 3009–3908 | 299/32827 | GcvA | 29 | P32064 |
| AmpR | 33 | P14145 | ||||
| TrpI | 29 | P34818 | ||||
| phnA | + | 4049–5272 | 407/44237 | |||
| phnB | + | 5369–6655 | 428/47304 | GlpT | 25 | P37948 |
| orf2 | + | 7040–8173 | 377/41291 | PotF | 55 | P31133 |
| orf3 | + | 8489–9118 | 209/23775 | PmsR | 44 | P54154 |
In the nucleotide sequence, the strand coding for the PA-hydrolase gene (phnA) is designated the plus (+) strand; phnR is the minus (−) strand. NA, not applicable.
Accession numbers relate to the SWISS-PROT database; identity relates to percentage of identical amino acids in alignments.
Five open reading frames (ORFs), apart from the phnA gene, were found; they showed various degrees of similarity to existing database entries (Table 1):
(i) ORF1 is preceded by a putative ribosome-binding site (RBS) and encodes a putative protein with a molecular mass of 41,681 Da. This protein shows similarity to the hydroxyphenylpropionic acid transporter (6) and to several hypothetical metabolite transport proteins.
(ii) The ORF found immediately upstream of phnA (Fig. 1) was designated as the phnR gene. This gene is encoded by a complementary chain (the direction of transcription is opposite to that of the phnA gene) and begins 140 bp upstream of the phnA gene. This ORF is also preceded by a putative RBS and encodes a putative protein of 32,827 Da. PhnR showed significant similarity to several transcriptional regulators belonging to the LysR family. The alignment of the PhnR amino acid sequence with the amino acid sequences of three proteins belonging to the LysR family is presented in Fig. 2. Analysis of this alignment clearly demonstrates that the PhnR gene product had a high degree of similarity (identical aa residues) to other LysR proteins in its N-terminal part, the first 66 amino acid residues, and much lower similarity in the C-terminal half. This mode of similarity distribution has been shown to be typical for the LysR family of transcriptional regulators (24). Evidence for the regulation of PA-hydrolase activity by the phnR gene is presented later in this paper.
FIG. 2.
Alignment of the LysR family transcriptional regulators. PhnR, the putative transcriptional regulator encoded by the phnR gene; AmpR, the positive regulator of expression of cephalosporinase from Rhodobacter capsulatus (3); GcvA, regulatory protein for glycine cleavage enzyme system from E. coli (28); TrpI, tryptophan biosynthesis transcriptional activator from Pseudomonas syringae (1). Residues identical in all four proteins are indicated by asterisks (∗); positions where PhnR residues are identical to two proteins are indicated by dots. The amino-terminal DNA-binding domain, which was shown to be the most highly conserved in all LysR proteins (24), is underlined.
(iii) An ORF found 96 bp downstream of phnA was designated as the phnB gene. This ORF (Fig. 1) was preceded by a putative RBS. It encoded a putative protein of 428 amino acids. The amino acid sequence of this protein was shown to be similar to several transporter proteins; for example, it showed 25% of aa identity with the glycerol-3-phosphate transporter (20). The possible role of PhnB in the biodegradation of 2PP is analyzed later in this paper.
(iv) ORF2 encoded a putative protein of 377 amino acids. This protein was found to be very similar (55% aa identity) to the spermidine/putrescine-binding protein precursor from E. coli (21) and to several other similar proteins.
(v) ORF3 encoded a putative protein of 209 amino acids. Significant similarity between this protein (44% aa identity) and the peptide methionine sulfoxide reductase from Bacillus subtilis and several other sulfoxide reductases was found.
Identification of phnA and phnR transcripts and mapping of their 5′ termini.
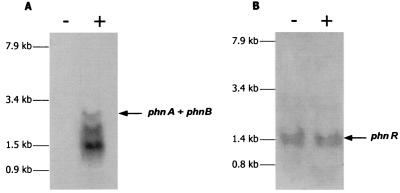
To identify the phnA and phnR transcripts, total RNA isolated from PA-induced and uninduced P. fluorescens 23F cells was fractionated by electrophoresis, transferred to nylon membranes, and hybridized with 32P-labeled probes. Transcripts for phnA were detected only in those RNA preparations obtained from induced cells (Fig. 3A). In contrast, mRNA for phnR was found in cells grown in either the presence or the absence of the inducer; it migrated as a single band, which corresponded to mRNA of ∼1.5 kb (Fig. 3B). A more complex picture was observed in the case of phnA mRNA (Fig. 3A). Three main bands were detected: an upper band of less intensity, which may correspond to the full-length phnAB transcript of ∼2.6 kb, and two bands representing transcripts of ∼2.0 and 1.4 kb.
FIG. 3.
Northern blot analysis of phnA (A) and phnR (B) mRNA. Equal amounts of total RNA (20 μg) isolated from P. fluorescens 23F cells grown in the presence (+) or in the absence (−) of PA were separated on denaturing gels, transferred onto positively charged nylon membrane, and hybridized with 32P-labeled probes specific for phnA (A) and phnR (B). (See Materials and Methods.)
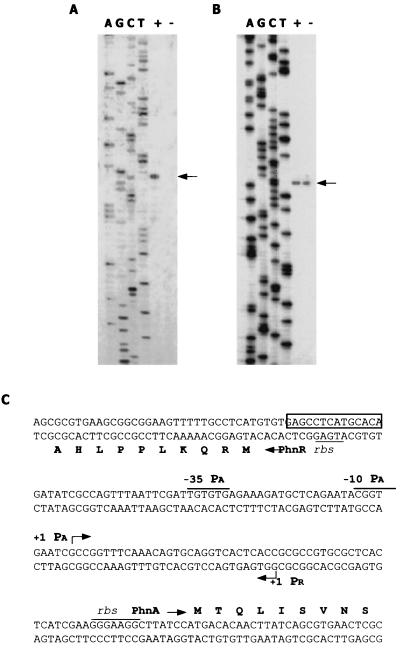
Precise mapping of the phnA and phnR 5′-transcriptional termini was carried out by primer extension analysis. In these experiments, oligonucleotides complementary to the sequence regions downstream of the 5′ ends of the phnA and phnR ORFs were used. The results of the analysis are presented in Fig. 4. In the case of the primer specific to phnA, a cDNA product was synthesized only with the RNA obtained from induced cells (Fig. 4A), whereas for phnR cDNA products were detected in both cases (Fig. 4B). These data are in perfect agreement with the results of Northern blot analysis (Fig. 3). The transcription start sites were mapped at positions 68 nt upstream of the translation start codon for the phnA ORF and 101 nt upstream of the start codon for the phnR ORF (Fig. 4C). It is also worth mentioning that a minor band was observed when mapping the phnR transcript (Fig. 4B). As transcripts usually begin with a purine (predominantly A), it is unlikely that the above-mentioned band (with an initial C) indicated a transcription start site for the phnR gene. Examination of the nucleotide sequence upstream of the transcription start site for phnA revealed −10 and −35 consensus-like hexamers typical for ς70 promoters (TACGGT and TTGTGT, respectively), followed by a putative recognition site for positive regulators belonging to the LysR family (from −70 to −58). There were no consensus-like sequences typical of ς70 or ς54 promoters in the region preceding the transcription start site for phnR.
FIG. 4.
Identification of phnA and phnR transcript initiation sites and putative regulatory sequences. Total RNA (30 μg) isolated from P. fluorescens 23F cells grown in the presence (+) or in the absence (−) of PA was annealed with 32P-labeled primers JQ88 (A), and 10 μg of the same RNA was annealed to JQ86 (B) as described in Materials and Methods. Aliquots from the primer extension reactions (lanes + and −) were separated by electrophoresis in denaturing 6% polyacrylamide gels along with the sequence ladders generated with the same oligonucleotides (lanes A, G, C, and T). The corresponding primer extension products are indicated by arrows. The nucleotide sequence of the intergenic region and the 5′ ends of phnA and phnR genes are presented (C). N-terminal amino acids translated from ORFs for both genes are shown in one-letter codes. Transcription start sites for PA and PR promoters are indicated by bent arrows. Putative −10 and −35 sequences of PA and RBS are shown. The putative recognition site for PhnR is boxed.
Substrate specificity of PA hydrolase.
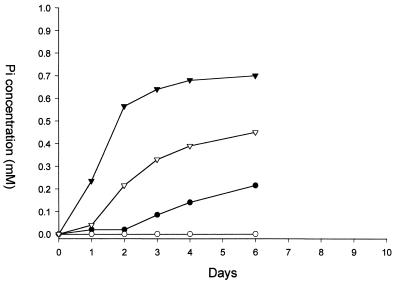
Surprisingly, only one xenobiotic (PA) was known to be degraded by the PA hydrolase encoded by the phnA gene of P. fluorescens 23F (15). To analyze the activity of this enzyme in greater detail, clones with different hybrid plasmids containing the phnA gene were examined with respect to their ability to use several structurally analogous phosphonates (2PP, 3-phosphonopropionate, 2-phosphonobutyrate, and phosphonoformate) as sources of phosphorus. It was found that P. putida AC577 containing the hybrid plasmids pLA82 and pLA45 (Fig. 1) as well as the original P. fluorescens 23F strain was able to use all tested compounds except phosphonoformate as a source of phosphorus. However, these strains released Pi only when 2PP served as a source of phosphorus (Fig. 5). This suggested that degradation of 2PP was mediated by the PhnA gene product (PA hydrolase) rather than by a classical C-P lyase since the latter enzyme is subject to repression by Pi and no net Pi release should occur as a result of its activity.
FIG. 5.
Phosphate release during growth of Pseudomonas strains on 2PP. Strains were grown in batch cultures. 2PP (2 mM) was used as a source of phosphorus and sodium gluconate (2 g l−1) as a source of carbon. Pi release was measured as described in Materials and Methods. ●, P. fluorescens 23F; ○, P. putida AC577; ▿, P. putida AC577(pLA45); ▾, P. putida AC577(pLA82).
Cleavage of the C-P bond of 2PP was detected only in cell extracts of P. putida and E. coli recombinant strains with hybrid plasmids containing the phnA gene. No activity was detected when the phnA gene was deleted. C-P cleavage activities towards 2PP were rather low, at most some 2% of the hydrolase's activity against PA (Table 2). To prove that the degradation of 2PP is determined by the enzymatic activity of the PA hydrolase encoded by the phnA gene, Pi release and accumulation of propionate were analyzed when 2PP was incubated with cell extract known to contain PA-hydrolase activity (Table 3). Although only low levels of C-P cleavage activity were found, near equimolar release of Pi and propionate from 2PP was obtained. These results confirmed that the hydrolase encoded by phnA was also active against 2PP.
TABLE 2.
Dependence of PA-hydrolase activity in P. putida AC577 and E. coli DH5α on the presence of the phnR gene
| Strain | Plasmid (substrate)a | phnR gene | PA-hydrolase activity ± SDb
|
||
|---|---|---|---|---|---|
| No inducer | PA induction | 2PP induction | |||
| P. putida | |||||
| AC577 | pLA82 (PA) | + | 0.31 ± 0.03 | 26.5 ± 1.7 | 35.25 ± 2.2 |
| AC577 | pLA82 (2PP) | + | <0.01 | 0.65 ± 0.06 | 0.35 ± 0.05 |
| AC577 | pLA45 (PA) | − | 31 ± 1.8 | 36 ± 1.4 | 39.7 ± 1.4 |
| AC577 | pLA45 (2PP) | − | 0.28 ± 0.06 | 0.34 ± 0.04 | 0.10 ± 0.01 |
| E. coli | |||||
| DH5α | pKK6-4 (PA) | + | 0.22 ± 0.02 | 2.5 ± 0.12 | 1.90 ± 0.16 |
| DH5α | pKK2-26 (PA) | − | 0.24 ± 0.03 | 0.27 ± 0.02 | 0.25 ± 0.04 |
| DH5α | PKK38 (PA) | + | 0.14 ± 0.03 | 2.1 ± 0.15 | <0.01 |
Identities of the inserts containing the phnA gene region are shown in Fig. 1.
Specific activity of PA hydrolase given as Pi release ± SD in nanomoles per minute per milligram. For this analysis, cultures of P. putida and E. coli were grown and prepared as described in Materials and Methods. Cell-free extracts were then prepared, and PA-hydrolase activity was assayed. The results are expressed as an average of three experiments.
TABLE 3.
Phosphate release and accumulation of propionate in cell-free extracts of P. putida AC577(pLA82)a
| Substrate | Concn (mM) | Pi release ± SD | Propionate ± SDb |
|---|---|---|---|
| 2PP | 2 | 410 ± 12.5 | 400 ± 10.3 |
| 5 | 620 ± 21 | 560 ± 12.5 |
Pi release and propionate accumulation are given in nanomoles per milligram of protein measured in the samples after 12 h of incubation of cell-free extracts with 2PP at 30°C.
Control samples from which either 2PP or cell-free extract was omitted contained no measurable amounts of propionate after 12 h of incubation.
Regulation of PA-hydrolase activity by the phnR gene.
It has been shown above that the phnR gene encodes a protein similar to a number of positive regulators belonging to the LysR family. To further investigate a possible role for PhnR in the expression of PA hydrolase, plasmids containing either phnR and phnA or only phnA were constructed. Plasmid pLA45, in which phnR was deleted, still contained the whole of the phnA gene and 120 nt of upstream region which included a promoter sequence (Fig. 1 and 4). Plasmid pLA82 contained both phnR and phnA. Plasmids pLA82 and pLA45 (pLAFR5 vector) were transferred into P. putida AC577 using the helper plasmid pRK2013.
Phosphate release was assayed in the corresponding cell extracts using PA and 2PP as substrates after the induction of phnA with PA (2 mM) or 2PP (2 mM) or in the absence of any inducer (Table 2). These results showed that PA and 2PP were similarly effective inducers of expression of the phnA gene in clones of P. putida that contained both phnA and phnR. Uninduced activities of PA hydrolase in these clones were 30 to 100 times lower. In those clones in which the phnR gene was deleted, induction with either PA or 2PP was not required for the effective expression of the PA hydrolase (Table 2). Transcription of the phnA gene in these cases was probably achieved from the lac promoter of the pLAFR5 vector. Similar results were obtained with these plasmids in E. coli DH5α cells (data not shown). To distinguish between possible expression from the lac promoter and expression regulated by phn-regulatory elements, a promoter probe vector pKK232–8 was used for analysis of the expression of phn genes in E. coli. For the latter analysis, two clones were constructed: pKK6–4 (Fig. 1), containing the phnR, phnA, and phnB genes with a putative promoter region, and pKK2–26, which lacked most of the putative transcriptional-regulator gene sequence phnR but contained phnA and phnB genes and a putative promoter region. E. coli DH5α strains containing these plasmids were analyzed for the ability to grow in liquid medium with PA as phosphorus source. Only E. coli DH5α(pKK6–4) showed the ability to grow in this medium and release Pi. In vitro activity of the PA-hydrolase gene (phnA) was measured using cell extracts from induced and uninduced strains. The results of these experiments showed that, without induction, PA-hydrolase activity is low (0.22 to 0.25 nmol min−1 mg−1). Induction by PA led to an 11-fold increase in PA-hydrolase activity (2.5 nmol min−1 mg−1) and by 2PP to a 9-fold increase (1.9 nmol min−1 mg−1) (Table 2). Since this effect was observed only in the clone with the phnR gene, it is clear that the phnR gene is essential as a positive regulator for the PA- or 2PP-dependent induction of PA hydrolase.
Role of phnB in the utilization of 2PP.
As discussed above, the putative product of the phnB gene showed similarity to several transport proteins. To analyze the possible involvement of phnB in the transport of PA and/or 2PP into bacterial cells, the activity of PA hydrolase towards these substrates was analyzed using both cell extracts and resting cell cultures. For these experiments clones were constructed (Fig. 1) in which both the phnA and phnB genes were present (plasmid pCA455) or in which the phnB gene was deleted (plasmid pCA452). The results are summarized in Table 4. It is clear that PA-hydrolase activity against PA is present in both cell extracts and resting cell cultures induced with PA, irrespective of the presence of the phnB gene. By contrast, Pi release from 2PP was dependent on the presence of the phnB gene in the case of the resting cell cultures but not of cell extracts. These results suggest that the phnB gene product was needed for the transport of 2PP into the bacterial cell. Induction of PA-hydrolase activity by PA but not by 2PP was observed in clones with plasmid pKK38 which lacked the phnB gene (Fig. 1, Table 2). This observation supported the conclusion that the phnB gene product was involved in 2PP transport.
TABLE 4.
Phosphate release from PA and 2PP in cell-free extracts and resting cells of E. coli DH5α with different plasmids, preinduced with PA
| Plasmida | phnB gene | Phosphate release ± SD in
|
|||
|---|---|---|---|---|---|
| Cell-free extractsb (nmol min−1 mg−1) with
|
Resting cellsc with
|
||||
| PA | 2PP | PA | 2PP | ||
| PCA455 | + | 128.6 ± 1.6 | 1.03 ± 0.01 | 2.30 ± 0.03 | 0.45 ± 0.04 |
| PCA452 | − | 85.5 ± 1.2 | 0.90 ± 0.1 | 2.40 ± 0.1 | 0.05 ± 0.02 |
Identities of the inserts containing the phnA gene region are shown in Fig. 1.
Specific activity of PA hydrolase given as Pi release in nanomoles per minute per milligram. For this analysis, cultures of E. coli were grown and prepared as described in Materials and Methods. Cell-free extracts were then prepared and PA-hydrolase activity was assayed.
Activity of PA hydrolase in resting cell cultures (OD600 = 0.5) was assayed by Pi release (millimoles) after 16 h of incubation with the corresponding substrate (PA or 2PP) at 30°C. The results are expressed as an average of three experiments.
DISCUSSION
PA hydrolase has been reported to be a C-P bond cleavage enzyme of unique specificity; the purified enzyme showed no detectable activity towards 39 organophosphonate and analogous compounds tested (17, 15). The cloning of the PA-hydrolase structural gene phnA and adjacent genes has, however, allowed us to analyze expression of PA hydrolase in different genetic environments using E. coli and P. putida host strains. We have now demonstrated that the phnA gene product from P. fluorescens 23F can hydrolyze the C-P bond not only of PA but also of 2-phosphonopropionate (Table 3). Activity against 2PP was nevertheless very low, constituting no more than 2% of that on PA; the compound did not serve as sole carbon and energy source for either P. fluorescens 23F or any recombinant strain containing the cloned phnA gene. However, both PA and 2PP served as similarly effective inducers of the phnA gene in the original P. fluorescens 23F strain and in hybrid plasmids expressed in both E. coli DH5α and P. putida AC577 (Table 2). This induction was dependent on the presence in the clones of the phnR gene. phnR is located immediately upstream of the phnA gene and shows remarkable similarity to the group of transcriptional regulators belonging to the LysR family (24): (i) phnR encodes a putative protein of 299 amino acids, typical of the members of the LysR family which range from 276 to 324 residues; (ii) its direction of transcription is divergent to that of phnA, and it is located 140 bp upstream of the structural gene; (iii) since regulation of the expression of phnA is dependent on the presence of PA or 2PP, each of these may be considered as a coinducer essential for the PhnR regulator protein; (iv) the amino acid sequence of PhnR shows significant homology to other members of the LysR family; this homology is almost entirely restricted to the N-terminal part of the protein, which is characteristic of the homology distribution among LysR family regulators (Fig. 2); (v) a putative site for PhnR recognition was found in the PA promoter at position −65 (Fig. 4); this structure included dyadic symmetry and a T-N11-A motif which has been proven to be critical for binding of the NodD, NahR, and MetR transcriptional regulators (9).
Our finding was not unusual, since transcriptional regulators belonging to the LysR family are the most common group in bacteria, with more than 50 reported members that regulate a diversity of genes and complex regulons in many prokaryotic genera (24). However, the involvement of a transcriptional regulator of this type in the regulation of the expression of PA hydrolase, a unique enzyme of its class, is of particular interest; all other C-P cleavage enzymes studied have been shown to be under Pho regulon control (10).
Three ORFs, which may encode proteins involved in phosphonate utilization, were found downstream of the phnA gene. Of these, only the role of the phnB gene has been analyzed; it seems probable that its product is involved in the transportation of 2PP into the bacterial cell. The data obtained, however, do not rule out the possibility that PA can also be transported by phnB. Deletion of phnB cannot be used to prove this because E. coli DH5α can use PA but not 2PP as a source of phosphorus (A. Kulakova, data not shown). Significantly, in view of the 25% homology between phnB and the glycerol-3-phosphate transporter in B. subtilis (20) (Table 1), it is known that the antibacterial antibiotic fosfomycin (phosphonomycin; cis-1,2-epoxypropylphosphonic acid) which has close structural similarity to 2PP, is transported into bacterial cells by the glycerol-3-phosphate pathway (11). It should be noted that expression of the phnB gene has been investigated using E. coli DH5α as a host. Accordingly, extrapolation of these results to the original P. fluorescens 23F host should be made with care. However, it is significant that cloned PA hydrolase was expressed to equally high levels in P. putida and E. coli hosts (results not shown). The inducible nature of this expression (Table 2) is likely to be due to the function of the positive regulator encoded by the phnR locus.
In the course of Northern blot analysis (Fig. 3), a transcript of ∼2.6 kb was identified in RNA preparations obtained from induced cells, using a DNA probe specific to phnA. In terms of its size, this transcript could represent mRNA for phnA and phnB. Transcripts of similar lengths could be synthesized if termination of transcription occurred in the intergenic region between the phnB gene and ORF2. There were two inverted repeats in this region (positions 6742 to 6773 and 6923 to 6942). The presence of these repeats might lead to formation of stem and loop structures in RNA with free energies (ΔG25) of −8.0 kcal and −9.4 kcal, respectively. It is likely that one or both of these repeats serve as a terminator(s) of transcription.
Analysis of the expression of different clones containing the phn region (Tables 1 and 2) in P. putida and E. coli cells indicated that neither ORF2 nor ORF3 was necessary for the degradation of PA and 2PP. Furthermore the results of Northern hybridization experiments (Fig. 3) did not reveal transcripts of lengths greater than that of a dimeric transcript for phnA and phnB. Therefore, we could suppose that ORF2 and ORF3 do not belong to the phnAB operon and are transcribed independently.
It was also evident from Northern blot analysis that phnR was transcribed in the form of monocistronic mRNA. A region of tandem inverted repeats localized at the 3′ end of ORF1 (positions 2610 to 2828) might serve for termination of its transcription.
Northern blot analysis of the phnA region also revealed two transcripts of ∼2.0 and ∼1.4 kb as the major products. The former may represent the whole phnA ORF and the 5′ part of the phnB ORF, and the latter may represent phnA only. These transcripts might be produced as a result of the termination of the transcription inside the phnB gene and in the phnA and phnB intergenic region, respectively. We also can not exclude the possibility that these transcripts originated from the processing of a phnAB transcript.
In conclusion, our study has indicated a probable mechanism by which the inducible expression of PA hydrolase occurs in P. fluorescens 23F. In light of other studies in this laboratory (16) which show that 2-aminoethylphosphonic acid, phosphonomycin, and phosphonoalanine can also be degraded by environmental microorganisms in a P-insensitive manner, it is likely that the expression of a number of other C-P cleavage enzymes may prove to be similarly regulated.
ACKNOWLEDGMENTS
This project was supported by the Queen's University Environmental Science and Technology Research Centre (QUESTOR), the European Regional Development Fund—Technology Development Programme, and BBSRC grant 81/P11488.
We are most grateful to anonymous reviewers for their suggestions for the improvement of an earlier version of this article.
REFERENCES
- 1.Auerbach S, Gao J, Gussin G N. Nucleotide sequences of the trpI, trpB, and trpA genes of Pseudomonas syringae: positive control unique to fluorescent pseudomonads. Gene. 1993;123:25–32. doi: 10.1016/0378-1119(93)90534-a. [DOI] [PubMed] [Google Scholar]
- 2.Bradford M M. A rapid and sensitive detection method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Campbell J I, Scahill S, Gibson T, Ambler R P. The phototrophic bacterium Rhodopseudomonas capsulata sp. 108 encodes an indigenous class A beta-lactamase. Biochem J. 1989;260:803–812. doi: 10.1042/bj2600803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ditta G, Schmidhauser T, Yakobson E, Lu P, Liang X-W, Finlay D R, Guiney D, Helinski D R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985;13:149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- 5.Dumora C, Marche M, Doignon F, Aigle M, Cassaigne A, Crouzet M. First characterization of the phosphonoacetaldehyde hydrolase gene of Pseudomonas aeruginosa. Gene. 1997;197:405–412. doi: 10.1016/s0378-1119(97)00185-6. [DOI] [PubMed] [Google Scholar]
- 6.Ferrandez A, Garcia J L, Diaz E. Genetic characterization and expression in heterologous hosts of the 3–3-(hydroxyphenyl) propionate catabolic pathway of Escherichia coli K-12. J Bacteriol. 1997;170:2573–2581. doi: 10.1128/jb.179.8.2573-2581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiske C H, Subbarrow Y. The colorimetric determination of phosphorus. J Biol Chem. 1925;66:375–400. [Google Scholar]
- 9.Goethals K, VanMontagu M, Holsters M. Conserved motifs in a divergent nod box of Azorhizobium caulinodans ORS571 reveal a common structure in promoters regulated by LysR-type proteins. Proc Natl Acad Sci USA. 1992;89:1646–1650. doi: 10.1073/pnas.89.5.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang W, Metcalf W, Lee K-S, Wanner B L. Molecular cloning, mapping and regulation of Pho regulon genes for phosphonate breakdown by the phosphonatase pathway of Salmonella typhimurium LT2. J Bacteriol. 1995;177:6411–6421. doi: 10.1128/jb.177.22.6411-6421.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kahan F M, Kahan S J, Cassidy P J, Kroop H. The mechanism of action of fosfomycin. Ann N Y Acad Sci. 1974;235:364–385. doi: 10.1111/j.1749-6632.1974.tb43277.x. [DOI] [PubMed] [Google Scholar]
- 12.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 13.Krieg N R. Enrichment and isolation. In: Gerhardt P, Murray R G E, Costilow R N, Nester E W, Wood W A, Krieg N R, Phillips G B, editors. Manual of methods for general bacteriology. Washington, D.C.: American Society for Microbiology; 1981. pp. 112–142. [Google Scholar]
- 14.Kulakova A N, Kulakov L A, Quinn J P. Cloning of the phosphonoacetate hydrolase gene from Pseudomonas fluorescens 23F encoding a new type of carbon-phosphorus bond cleaving enzyme and its expression in Escherichia coli and Pseudomonas putida. Gene. 1997;195:49–53. doi: 10.1016/s0378-1119(97)00151-0. [DOI] [PubMed] [Google Scholar]
- 15.McGrath J W, Wisdom G B, McMullan G, Larkin M J, Quinn J P. The purification and properties of phosphonoacetate hydrolase, a novel carbon-phosphorus bond-cleavage enzyme from Pseudomonas fluorescens 23F. Eur J Biochem. 1995;234:225–230. doi: 10.1111/j.1432-1033.1995.225_c.x. [DOI] [PubMed] [Google Scholar]
- 16.McGrath J W, Ternan N G, Quinn J P. Utilization of organophosphonates by environmental microorganisms. Lett Appl Microbiol. 1997;24:69–73. [Google Scholar]
- 17.McMullan G, Quinn J P. In vitro characterization of a phosphate starvation-independent carbon-phosphorus bond cleavage activity in Pseudomonas fluorescens 23F. J Bacteriol. 1994;176:320–324. doi: 10.1128/jb.176.2.320-324.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metcalf W W, Wanner B L. Mutational analysis of an Escherichia coli fourteen-gene operon for phosphonate degradation using TnpoA′ elements. J Bacteriol. 1993;175:3430–3442. doi: 10.1128/jb.175.11.3430-3442.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 20.Nilsson R P, Beijer L, Rutberg B. The glpT and glpQ genes of the glycerol regulon in Bacillus subtilis. Microbiology. 1994;140:723–730. doi: 10.1099/00221287-140-4-723. [DOI] [PubMed] [Google Scholar]
- 21.Pistocchi R, Kashiwagi K, Miyamoto S, Nukui E, Sadakata Y, Kobayashi H, Igarashi K. Characteristics of the operon for a putrescine transport system that maps at 19 minutes on the Escherichia coli chromosome. J Biol Chem. 1993;268:146–152. [PubMed] [Google Scholar]
- 22.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 25.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wanner B L, Metcalf W W. Molecular studies of a 10.9-kb operon in E. coli for phosphonate uptake and biodegradation. FEMS Microbiol Lett. 1992;100:133–140. doi: 10.1111/j.1574-6968.1992.tb14031.x. [DOI] [PubMed] [Google Scholar]
- 27.Wanner B L. Molecular genetics of carbon-phosphorus bond cleavage in bacteria. Biodegradation. 1994;5:175–184. doi: 10.1007/BF00696458. [DOI] [PubMed] [Google Scholar]
- 28.Wilson R L, Stauffer G V. DNA sequence and characterization of GcvA, a LysR family regulatory protein for the Escherichia coli glycine cleavage enzyme system. J Bacteriol. 1994;176:2862–2868. doi: 10.1128/jb.176.10.2862-2868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]