Abstract
In the plant pathogen Pseudomonas syringae pv. glycinea PG4180 and other bacterial species, synthesis of the exopolysaccharide levan is catalyzed by the extracellular enzyme levansucrase. The results of Southern blotting and PCR analysis indicated the presence of three levansucrase-encoding genes in strain PG4180: lscA, lscB, and lscC. In this study, lscB and lscC were cloned from a genomic library of strain PG4180. Sequence analysis of the two lsc genes showed that they were virtually identical to each other and highly similar to the previously characterized lscA gene. lscA and lscC had a chromosomal location, whereas lscB resided on an indigenous plasmid of PG4180. Mutants with impaired expression of individual lsc genes and double mutants were generated by marker exchange mutagenesis. Determination of levansucrase activities in these mutants revealed that the lscB gene product was secreted but not that of lscA or lscC. Our results indicated that lscB and lscC but not lscA contributed to periplasmic levan synthesis of PG4180. The lscB lscC double mutant was completely defective in levan formation and could be complemented by either lscB or lscC. Our data suggested a compartment-specific localization of two lsc gene products, with LscB being the secreted, extracellular enzyme and LscC being the predominantly periplasmic levansucrase. Results of Western blot analyses indicated that lscA was not expressed and that lscA was not associated with levansucrase activities in any particular protein fraction. LscA could be detected in PG4180 only when transcribed from the vector-borne Plac promoter. PCR screening in various P. syringae strains with primers derived from the three characterized lsc genes demonstrated the presence of multiple Lsc isoenzymes in other P. syringae pathovars.
The extracellular enzyme levansucrase (Lsc) (EC 2.4.1.10) catalyzes the following three reactions: (i) synthesis of levan from sucrose by transfructosylation while releasing glucose, (ii) hydrolysis of levan to monosaccharides of fructose, and (iii) exchange of [14C]glucose in the reaction fructose-2,1-glucose plus [14C]glucose to fructose-2,1-[14C]glucose plus glucose (19). Levan is a high-molecular-weight β-(2,6)-polyfructan with extensive branching through β-(2,1) linkages. Lsc produces both linkage types.
Levansucrases have been isolated from various bacteria, such as Bacillus subtilis, Streptococcus mutans, Zymomonas mobilis, Acetobacter diazotrophicus, Erwinia amylovora, and Pseudomonas syringae (2, 9, 18, 22, 34, 41). In contrast to levansucrases from gram-positive bacteria, which differ widely in their biochemical characteristics, levansucrases from gram-negative bacteria are similar in their molecular mass and substrate-independent expression (16, 22, 23, 43).
The bacterial blight pathogen of soybean, Pseudomonas syringae pv. glycinea PG4180, causes formation of water-soaked lesions that develop into necrotic leaf spots surrounded by chlorotic halos. Like other exopolysaccharides (EPS), levan could have particular functions before or during the infection process (10, 32, 33). The polyfructan levan and alginate, an O-acetylated copolymer of β-1,4-linked d-mannuronic acid and l-glucuronic acid, are the most prominent EPS of P. syringae (12, 13, 36). Strains of P. syringae primarily produced alginate in planta and synthesized levan in culture (36). EPS are thought to enhance bacterial survival both by generating a hydrogenated matrix to minimize direct contact with plant surfaces and thereby prevent host recognition and by providing a detoxifying barrier against plant defense compounds. Additionally, EPS formation may protect bacterial cells from desiccation, concentrate minerals and nutrients, and improve attachment to surfaces during epiphytic growth.
The synthesis of EPS is a common characteristic of various plant-pathogenic bacteria, and its role in virulence has been previously reviewed in detail (1, 10, 16, 18, 32, 39). Levan may be particularly important during the early stages of infection to mask and protect the cell and support proliferation of the pathogen in host tissue (26, 33). Although levansucrases have been isolated from various bacterial species and levan formation has been used for the taxonomic classification of P. syringae (6, 42), little is known about the genetics and regulation of Lsc in P. syringae.
Previously, the biochemical characteristics of Lsc from P. syringae pv. phaseolicola were investigated (22), and two genetic loci coding for this enzyme in P. syringae pv. glycinea and P. syringae pv. phaseolicola were identified (23). In the latter study, two levansucrases from P. syringae, which differed only in their N-terminal sequences, could be expressed in Escherichia coli under control of the Plac promoter. It was shown that both enzymes were exported to the periplasm of E. coli without lethal effects but were not secreted from E. coli.
The aims of this study were the cloning and expression of two new lsc genes from P. syringae pv. glycinea PG4180 in E. coli, determination of their nucleotide sequences, analysis of the cell compartment-specific Lsc activities in PG4180 mutants with impaired expression of individual lsc genes, and analysis of their distribution in other pathovars of P. syringae.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Tables 1, 2, and 3. Pseudomonas strains were maintained on mannitol-glutamate (MG) medium (28) at 28°C. For liquid cultures, bacteria were incubated in HSC medium (37) or King's B (KB) medium (30) at 18°C by Hettwer et al. (23). E. coli DH5α was used as the cloning host (40) and was grown in Luria-Bertani (LB) broth at 37°C. Microbial growth was monitored by measuring the optical density at 600 nm (OD600). The following antibiotics (in micrograms per milliliter) were added to the media when needed: ampicillin, 50; chloramphenicol, 25; kanamycin, 25; tetracycline, 25; spectinomycin, 25; streptomycin, 25; and gentamicin, 2.
TABLE 1.
P. syringae pv. glycinea strains used in this study
| Strain | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| PG4180 | Wild type, produces levan | R. E. Mitchell (1975) |
| M1 | lscA mutant, Sm/Spr, produces levan | This study |
| M2 | lscB mutant, Gmr, produces levan | This study |
| M3 | lscA lscB mutant, Sm/Spr Gmr, produces levan | This study |
| M4 | lscC mutant, Gmr, produces levan | This study |
| M5 | lscA lscC mutant, Sm/Spr Gmr, produces levan | This study |
| M6 | lscB lscC mutant, Gmr Sm/Spr, levan deficient | This study |
Sm/Spr, streptomycin and spectinomycin resistant; Gmr, gentamicin resistant.
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant propertiesa | Source or reference |
|---|---|---|
| pBluescript II SK(+) | Ampr, cloning vector | Stratagene |
| pRK415 | Tcr, RK2-derived broad-host-range cloning vector | 29 |
| pBBR1MCS | Cmr, broad-host-range cloning vector | 31 |
| pKmobGII | Kmr, mobilizable suicide vector | 27 |
| pCAM140 | Ampr Sm/Spr, contains 2.0-kb EcoRI fragment with Sm/Spr cassette and 2.0-kb NotI fragment with promoterless uidA gene | 48 |
| pMGm | Gmr, contains 1.9-kb Gmr cassette | 35 |
| pAS-LacZ | Cmr, contains translational corS::lacZ fusion in pBBR1MCS, LacZ+ | This study |
| pHL-PhoA | Tcr, contains translational lscC::phoA fusion in pRK415, PhoA+ | This study |
| pSKL3 | Ampr, contains lscA on 3.0-kb PstI fragment (Plac>lscA), produces levan | 23 |
| pSKL3G | Ampr, contains lscA on a 3.0-kb PstI fragment (lscA>Plac), produces levan | 23 |
| pRA3.1 | Tcr, contains lscA under control of Plac on 3.0-kb PstI fragment from pSKL3 in pRK415, produces levan | This study |
| pSKL3Sm | Ampr Sm/Spr, contains lscA mutagenized by insertion of a Sm/Spr cassette in BstEII sites of pSKL3, levan deficient | 23 |
| pRA3.1-Sm | Tcr Sm/Spr, contains lscA::Sm/Spr on 5.2-kb PstI fragment from pSKL3Sm in pRK415, levan deficient | This study |
| p7C7 | Tcr, genomic library clone of PG4180 with 25- to 30-kb insert in pRK7813, contains lscB, levan deficient | This study |
| pLB7.2 | Ampr, contains 7.2-kb EcoRV fragment from p7C7 in pBluescript (lscB>Plac), levan deficient | This study |
| pLB7.2R | Ampr, contains 7.2-kb EcoRV fragment from p7C7 in pBluescript (Plac>lscB), levan deficient | This study |
| pRB7.2 | Cmr, carries lscB on 7.2-kb EcoRV fragment from pLB7.2 in pBBR1MCS, levan deficient | This study |
| pLB2.4 | Ampr, contains 2.4-kb PstI/SalI fragment from pLB7.2 derived by PCR in pBluescript (Plac>lscB), produces levan | This study |
| pLB7.2-Gm | Ampr Gmr, contains lscB mutagenized by insertion of Gmr cassette inserted in XhoI site of pLB7.2, levan deficient | This study |
| pKB7.2-Gm | Kmr Gmr, contains lscB mutagenized by insertion of Gmr cassette on 9.2-kb SalI/PstI fragment from pLB7.2-Gm in pKmobGII, levan deficient | This study |
| p5C10 | Tcr, genomic library clone of PG4180 with 25- to 30-kb insert in pRK7813, contains lscC, levan deficient | This study |
| pLC5.5 | Ampr, contains 5.5-kb PstI fragment from p5C10 in pBluescript (lscC>Plac), levan deficient | This study |
| pLC5.5-Gm | Kmr Gmr, contains lscC mutagenized by insertion of Gmr cassette in XhoI site of pLC5.5, levan deficient | This study |
| pKC5.5-Gm | Kmr Gmr, contains lscC::Gmr on 7.5-kb SalI fragment from pLC5.5-Gm in pKmobGII, levan deficient | This study |
| pLC5.5-Sm | Ampr Sm/Spr, contains lscC mutagenized by insertion of Sm/Spr cassette in XhoI site of pLC5.5, levan deficient | This study |
| pKC5.5-Sm | Kmr Sm/Spr contains lscC::Sm/Spr on 7.5-kb SalI fragment from pLC5.5-Sm in pKmobGII, levan deficient | This study |
Sm/Spr, streptomycin and spectinomycin resistant; (Plac>lscA), the promoter of the lac operon directs transcription of lscA; (lscB>Plac), the promoter of the lac operon is located downstream of lscB.
TABLE 3.
PCR screening for levansucrase alleles in P. syringae pathovars
| Strain | Result of PCR analyses for:
|
Reference or sourcea | |
|---|---|---|---|
| lscB | lscC | ||
| P. syringae pv. phaseolicola | |||
| NCPPB1321 | + | + | 22 |
| Psph 6/0 | − | + | B. Völksch |
| GSPB 796 | + | + | GSPB |
| P. syringae pv. glycinea | |||
| PG4180 | + | + | R. E. Mitchell |
| Psg 16/83 | + | + | 46 |
| Psg 7a/90 | + | + | 46 |
| P. syringae pv. tomato | |||
| DC3000 | + | + | D. Cuppels |
| DSM 50315 | + | + | DSM |
| GSPB 119 | + | + | GSPB |
| P. syringae pv. tabaci | |||
| GSPB 113 | + | + | GSPB |
| GSPB 117 | + | + | GSPB |
| P. syringae pv. lachrymans GSPB 77 | + | + | GSPB |
| P. syringae pv. coriandricola GSPB 1784 | + | + | GSPB |
| P. syringae pv. photiniae CFBP 11034 | + | + | CFBP |
| P. syringae pv. savastanoi | |||
| GSPB 2264 | − | + | GSPB |
| GSPB 2259 | − | + | GSPB |
| P. syringae pv. syringae | |||
| FF5 | − | + | G. W. Sundin |
| B301D | − | + | D. C. Gross |
| Pss B48 | − | + | G. W. Sundin |
| Pss 3525 | − | + | G. W. Sundin |
| P. syringae pv. myricae CFBP 11005 | − | + | CFBP |
| P. syringae pv. garcae CFBP 1634 | − | + | CFBP |
| P. syringae pv. morsprunorum | |||
| D5 | − | + | K. Naumann |
| GSPB 886 | − | + | GSPB |
| GSPB 1013 | − | + | GSPB |
| Pm 7 | − | + | A. Jones |
| P. syringae pv. persicae GSPB 1025 | − | + | GSPB |
| P. syringae pv. atropurpurea MAFF 301309 | − | + | K. Nishiyama |
| P. syringae pv. hibisci CFBP 11294 | − | − | CFBP |
| P. syringae pv. mellea CFBP 2344 | − | − | CFBP |
| P. syringae pv. striafaciens GSPB 1850 | − | − | GSPB |
| P. syringae pv. helianthi GSPB 2688 | − | − | GSPB |
| P. syringae pv. zizaniae CFBP 11040 | − | − | CFBP |
| P. syringae pv. pisi GSPB 104 | − | − | GSPB |
| P. syringae pv. apii GSPB 2153 | − | − | GSPB |
GSPB, Göttinger Sammlung phytopathogener Bakterien, Göttingen, Germany; DSM, Deutsche Sammlung für Mikroorganismen, Braunschweig, Germany; CFBP, Collection Française des Bactéries Phytopathogènes, Angers, France.
Subcellular fractionation of bacterial proteins.
Cell-free bacterial supernatants were obtained by centrifugation of cultures at 10,600 × g and subsequent filtration of the supernatants through membranes with a pore size of 0.2 μm. Supernatants were dialyzed to remove any residual glucose. Extracellular proteins were obtained from sterile bacterial supernatants by concentrating the samples 30-fold by lyophilization and then resuspended in 50 mM Tris HCl buffer. Subcellular fractionation of bacterial cells was performed by the method of Boyd et al. (5). Cells were lysed by repeated freeze-thaw cycles or by sonication. The integrity of a particular subcellular fraction was verified quantitatively by detection of two reporter enzymes, β-galactosidase (LacZ) and alkaline phosphatase (PhoA), which are representative of the cytosolic and periplasmic cell compartments, respectively. For this, two transconjugants of P. syringae pv. glycinea PG4180 that harbored plasmids pAS-LacZ and pHL-PhoA (Table 2), respectively, were used as controls. LacZ and PhoA enzyme activities were detected by the method of Rutz et al. (38). The protein concentrations in cell lysates, periplasmic fractions, and supernatants were determined by the Bradford assay (40).
Qualitative assays for levansucrase activity.
Qualitative estimation of Lsc activity in sterile bacterial supernatants and subcellular fractions was done by spotting 5 to 10 μl of samples on water-agar plates containing 5% sucrose. Enzyme activity was visualized by the formation of opalescent slime plugs after incubation at 18°C for 24 to 48 h. Zymograms with proteins from cell-free supernatants and subcellular fractions were prepared by polyacrylamide gel electrophoresis (PAGE) under nondenaturing conditions by the method of Hellio et al. (20). For this, 15-μl aliquots of native protein samples derived from concentrated supernatants or subcellular fractions of exponentially growing or stationary-phase cultures were loaded onto 10% polyacrylamide gels. Following electrophoresis, gels were incubated in sterile water containing 10% sucrose for 24 to 48 h at 18°C. Protein bands representing Lsc were detected by a whitish swelling of the gel matrix that corresponded to levan formation.
Quantitation of levansucrase activity.
Lsc activity was quantified by measuring the amount of glucose liberated during incubation with sucrose using the Gluco-quant Glucose/HK assay kit (Roche, Mannheim, Germany). A nonconcentrated 20-μl sample of glucose-free supernatant or 20 μl of a subcellular fraction was mixed with 20 μl of assay buffer (10% sucrose plus 0.09% NaCl) and 2 ml of test reagent (83 mmol of Tris per liter, 5 mmol of HEPES [pH 7.7] per liter, 4 mmol of Mg2+ per liter, 1.4 mmol of ATP per liter, 0.83 mmol of NADP per liter, 1.4 U of hexokinase per ml, and 2.5 U of glucose-6-phosphate dehydrogenase per ml). Subsequently, the reaction mixture was incubated at 25°C, and the absorbance at 365 nm (A365) was measured at 15-min intervals for 1 h. One unit of Lsc activity represents the amount of enzyme in 1 ml of bacterial culture that liberates 1 μmol of glucose per min.
Determination of the N-terminal protein sequence.
Protein extracts of bacterial supernatants were concentrated 400-fold by precipitation with 10% trichloroacetic acid and loaded onto sodium dodecyl sulfate (SDS)–10% polyacrylamide gels. Gels were stained with 0.1% Coomassie blue R250 and destained with 40% methanol and 10% acetic acid. Subsequently, gels were washed with water. Protein bands were cut out of the gel, and R. Schmid (University of Osnabrück, Osnabrück, Germany) determined the N-terminal sequence by standard procedures.
Immunodetection of levansucrase.
Proteins from crude cell extracts, concentrated supernatant extracts, or subcellular fractions were diluted, and equal amounts (2 μg/lane) were separated by SDS–10% PAGE. Electrophoresis, electroblotting on nitrocellulose membranes, and immunodetection of proteins were conducted by standard procedures (40). Polyclonal antibodies raised in rabbits against Lsc derived from P. syringae pv. phaseolicola were provided by U. Hettwer and K. Rudolph (University of Göttingen, Göttingen, Germany). The specificity of the Lsc antiserum at a dilution of 1:3,000 was evaluated with recombinant Lsc protein from E. coli and crude protein extracts of P. syringae pv. glycinea PG4180. For signal detection, secondary anti-rabbit immunoglobulin G antibodies conjugated to alkaline phosphatase (Sigma, Darmstadt, Germany) were used at a concentration of 1:7,500, and the reaction was visualized using 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium salt.
DNA procedures.
Restriction digestions, agarose gel electrophoresis, purification of DNA fragments from agarose gels, electroporation, PCR, Southern blot hybridization, and small-scale plasmid DNA preparation were performed by standard techniques (40). Subclones were generated in pBluescript II SK(+) (Stratagene, Heidelberg, Germany), pRK415 (29), or pBBR1MCS (31). Large-scale preparation of plasmid DNA from E. coli was performed by alkaline lysis and purified with Qiagen tip 100 columns (Qiagen, Hilden, Germany). Isolation of plasmid DNA from P. syringae was accomplished by the method of Kado and Liu (25). Triparental matings were done by the method of Gerhardt et al. (17). Oligonucleotide primers specific for the amplification of the genes lscA, lscB, and lscC were 5′-ATGAGTAACATCAATTAC (lscA-F), 5′-TCAGCTCAGCACCACGTTCT (lscA-R), 5′-ATGTCCACTAGCAGCTCT (lscB-F), 5′-TCAGCTTAGCGTCACGTC (lscB-R), 5′-ATGTCCACTAGCAGCTCT (lscC-F), and 5′-TCAGCTCAGTTGCACGTC (lscC-R). Oligonucleotides lscB-FC (5′-TCACTGCAGGCCCTAGCGCTGACCAAA) and lscB-RC (5′CGAGTCGACTCAGCTTAGCGTCACGTC) (PstI and SalI sites underlined) were used to amplify a 2.4-kb fragment containing lscB from plasmid pLB7.2 that was subsequently cloned into pBluescript II SK(+) to yield pLB2.4.
DNA sequencing and analysis.
Nucleotide sequencing reactions were performed by the dideoxynucleotide method (40) with the Thermo Sequenase fluorescence-labeled primer cycle sequencing kit (Amersham-Pharmacia, Freiburg, Germany). Automated DNA sequencing was accomplished with an ALF Express sequencing apparatus (Amersham-Pharmacia). Sequence data were aligned and processed with the DNASTAR version 4.1 software package (Lasergene, Madison, Wis.). DNA and protein sequence homology searches of the GenBank, EMBL, PIR, and SWISSPROT databases were performed with the University of Wisconsin Genetics Computer Group programs BLASTX, FASTEMBL, GAP, and BESTFIT. The prediction of signal peptide sequences was performed with the SignalP version 1.1 program from the Center for Biological Sequence Analysis, Department of Biotechnology, Technical University of Denmark (http: //www.cbs.dtu.dk/services/SignalP/).
Generation of lsc-deficient mutants of P. syringae pv. glycinea PG4180 by marker exchange mutagenesis.
An lscA-deficient mutant of strain PG4180 was generated by marker exchange mutagenesis as follows. The 5.0-kb PstI insert of plasmid pSKL3Sm (23), which contained lscA mutagenized by insertion of a streptomycin resistance (Smr) gene cassette that was derived from plasmid pCAM140 (48) and is expressed from its own promoter, was subcloned into the broad-host-range plasmid pRK415, resulting in pRA3.1-Sm. This plasmid was mobilized into PG4180 by triparental mating, and transconjugants were repeatedly incubated in KB medium supplemented with streptomycin. Finally, potential marker exchange mutants were isolated as Tets and Smr colonies. One of these transformants was designated M1. To obtain a PG4180 mutant deficient in lscB or lscC, a 1.9-kb gentamicin resistance (Gmr) cassette, which was derived from pMGm (35) with restriction enzyme SalI and is expressed from its own promoter, was ligated to XhoI-digested plasmids pLB7.2 and pLC5.5, respectively. The resulting 9.5- and 7.5-kb SalI inserts of plasmids pLB7.2-Gm and pLC5.5-Gm were subcloned into the mobilizable suicide vector pKmobGII (27), yielding plasmids pKB7.2-Gm and pKC5.5-Gm, respectively. Both plasmids were separately mobilized into strain PG4180 by triparental matings, thereby obtaining mutants M2 (lscB) and M4 (lscC) via homologous marker exchange mutagenesis. Double mutants M3 (lscA lscB) and M5 (lscA lscC) were obtained by separate mobilization of plasmids pKB7.2-Gm and pKC5.5-Gm into mutant M1. To generate an lscB lscC double mutant, a 2.0-kb SalI-XhoI fragment containing a streptomycin-spectinomycin resistance cassette (Smr Spr) from plasmid pCAM140 (48) was ligated to plasmid pLC5.5 that had been linearized with XhoI. The 7.5-kb SalI insert of the resulting plasmid pLC5.5-Sm was then subcloned into pKmobGII to generate plasmid pKC5.5-Sm. This plasmid was subsequently mobilized into mutant M2 by triparental mating to obtain mutant M6 (lscB lscC) via homologous recombination.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this study were deposited in GenBank and EMBL data banks under accession no. AF345638 (lscB) and AF346402 (lscC).
RESULTS
Detection of lsc genes of P. syringae pv. glycinea PG4180.
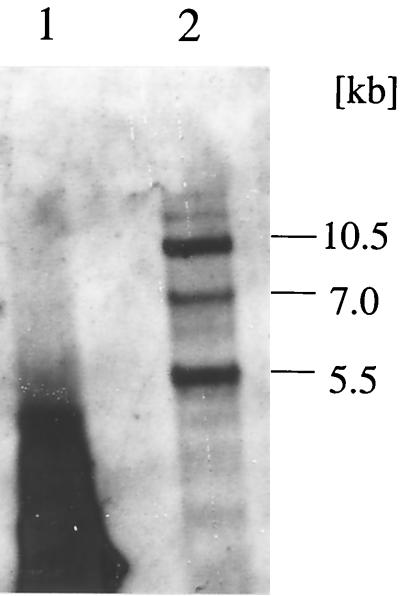
Previously, we had identified and characterized a functional lsc gene in P. syringae pv. glycinea PG4180 (23). However, after this gene had been inactivated by marker exchange mutagenesis (see below), the mutant did not exhibit a levan-deficient phenotype, suggesting the existence of at least one additional allele of this gene in strain PG4180. Southern blot hybridizations under conditions of low stringency (hybridization temperature of 55°C) with genomic DNA from PG4180 digested with the restriction enzyme SalI and a DNA probe containing the previously described lsc gene from P. syringae pv. phaseolicola NCPPB1321 (23) were performed. The probe hybridized to three fragments of 5.5, 7.0, and 10.5 kb (Fig. 1). This result and those of Southern blot analysis with other restriction enzymes indicated the presence of two additional lsc genes in PG4180. Consequently, the original lsc gene was renamed lscA, and the second and third lsc genes were designated lscB and lscC, respectively.
FIG. 1.
Detection of three lsc genes in genomic DNA from P. syringae pv. glycinea PG4180 by Southern blot hybridization. A digoxigenin-labeled PCR product of the previously characterized lsc gene from P. syringae pv. phaseolicola NCPPB1321 was used as a DNA probe under conditions of low stringency (hybridization temperature of 55°C). Lanes: 1, 1.3-kb PCR product with lsc from NCPPB1321; 2, SalI-digested genomic DNA from PG4180.
Cloning of the second and third levansucrase genes of P. syringae pv. glycinea.
According to the previously published nucleotide sequence coding for Lsc from P. syringae NCPPB1321 (23), oligonucleotide primers lscB-F and lscB-R were designed to amplify an approximately 1.3-kb PCR product from genomic DNA from strain PG4180. A genomic cosmid library of P. syringae pv. glycinea PG4180 was subsequently screened for the presence of DNA homologous to this amplified 1.3-kb DNA fragment yielding a positive signal for a cosmid that was designated p7C7. Cosmid p7C7 contained a 7.2-kb EcoRV fragment, which hybridized to the 1.3-kb PCR product from strain PG4180 used as a DNA probe in a subsequent Southern blot analysis. This fragment was subcloned into pBluescript II SK(+) in both orientations to obtain plasmids pLB7.2 and pLB7.2R. When E. coli transformants were plated on LB agar plates containing 5% sucrose, neither plasmid conferred levan formation to E. coli. A third copy of lsc was found by repeating the cosmid library screening by Southern blot hybridization with the 1.3-kb PCR product. Three individual cosmid clones contained a common 5.5-kb SalI fragment that hybridized with the DNA probe. The 5.5-kb fragment of one of those cosmids, p5C10, was subcloned into pBluescript II SK(+) to yield plasmid pLC5.5. This plasmid also did not mediate levan synthesis in the E. coli transformants.
Nucleotide sequence analysis.
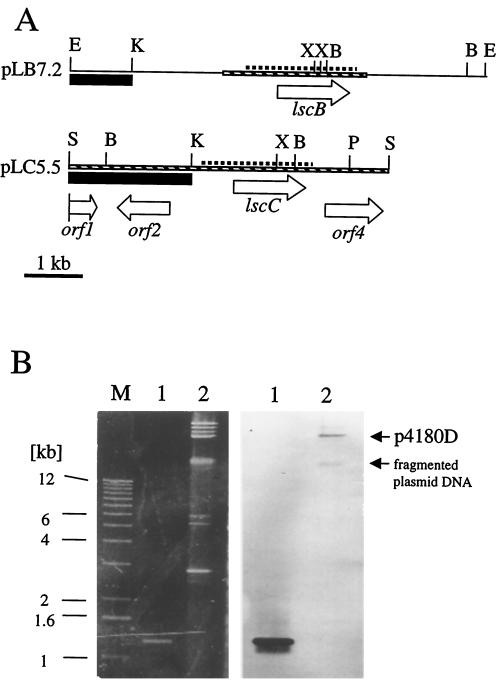
Part of the 7.2-kb insert of pLB7.2 was sequenced on the basis of four oligonucleotide primers derived from the previously described lsc gene of P. syringae pv. phaseolicola (23). The nucleotide sequence of a 1,296-bp open reading frame (ORF) designated lscB was determined and found to be 100% identical to the previously described lsc gene from P. syringae pv. phaseolicola NCPPB1321 (23). The 5,561-bp insert of plasmid pLC5.5 was completely sequenced. Three complete ORFs designated orf2, lscC, and orf4 and an N-terminally truncated ORF named orf1 (Fig. 2A) were identified following a comparison with database entries. The deduced amino acid sequences of orf2 and orf4 showed 72 and 71% similarity to an autolytic lysozyme from Xylella fastidiosa and a hypothetical protein from E. coli, respectively (accession numbers G82563 and F64902, respectively). The deduced amino acid sequence of the ORF fragment orf1 showed 89% similarity to a putative transposase from Pseudomonas sp. strain JR1 (accession number AF155505). The 1,296-bp ORF designated lscC showed 98% identity to lscB at the nucleotide sequence level. Nucleotide sequences of about 450 bp upstream of lscB and lscC were 97% identical to each other (Fig. 2A). Both lscB and lscC showed almost 99% identity at the deduced amino acid sequence level to each other. In their respective amino acid sequences, the gene products of lscB and lscC differed in only five residues distributed throughout the central and C-terminal regions (N92D, S119C, E327D, L329I, and T429Q). Both genes encode putative proteins of 47.6 kDa. Both genes also showed 86% identity to lscA at the nucleotide level and high similarity at the deduced amino acid sequence level (95%). The gene products of lscA, lscB, and lscC showed amino acid sequence similarities to various levansucrases of gram-negative and -positive bacteria comparable to those observed for lsc of P. syringae pv. phaseolicola (23).
FIG. 2.
(A) Restriction maps of the 7.2- and 5.5-kb inserts of pLB7.2 and pLC5.5, respectively. The thin hatched bars indicate the DNA for which the nucleotide sequence has been determined. White arrows symbolize identified ORFs with their designation and direction of transcription shown. Black boxes indicate DNA regions upstream of lscB and lscC used as probes for further Southern blot analyses (data not shown). Broken lines mark the highly homologous DNA regions in the inserts of both plasmids. Restriction enzyme site abbreviations: E, EcoRV; K, KpnI; X, XhoI; B, BglII; S, SalI; and P, PstI. (B) Analysis of the plasmidborne location of an lsc gene from P. syringae pv. glycinea PG4180 by Southern blot hybridization (hybridization temperature of 68°C) with lscA as a DNA probe. The positions of the approximately 60-kb plasmid p4180D and the fragmented plasmid DNA are marked by arrows. Lanes: M, molecular size markers; 1, PCR product of lscA; 2, total plasmid DNA from PG4180.
Genomic localization of lscA, lscB, and lscC.
P. syringae pv. glycinea PG4180 harbors five indigenous plasmids with molecular sizes between 45 and 100 kb which encode various virulence and fitness determinants (3, 47). To test whether any of the lsc genes might be plasmidborne, a Southern blot experiment was performed with undigested plasmid DNA from strain PG4180 and a probe containing lscA (Fig. 2B). A clear signal was detected with a band representing the approximately 60-kb plasmid p4180D (3, 46). When plasmid DNA from PG4180 was digested with the restriction enzyme SalI, a 10.5-kb fragment hybridized to the probe (data not shown). To find out which lsc gene was plasmid encoded, Southern blot analyses with DNA probes derived from DNA upstream of lscB and lscC (Fig. 2A) were conducted (data not shown). The probe from the upstream region of lscB, but not that associated with lscC, hybridized to plasmid p4180D and to the 10.5-kb SalI fragment from total plasmid DNA from PG4180. Furthermore, a 10.5-kb SalI fragment was identified in cosmid p7C7, from which lscB was subcloned (data not shown). In contrast, lscA and lscC could be located to 7.5- and 5.5-kb fragments by SalI digestion of genomic DNA (Fig. 1) and of cosmid clones from which lscA and lscC were derived (data not shown). In summary, these results suggested that lscA and lscC were located on the chromosome, whereas lscB was located on the plasmid.
Search for putative N-terminal signal peptide sequences.
The deduced amino acid sequences of all three lsc genes were analyzed for putative signal peptide sequences using the SignalP version 1.1 program. No putative signal peptidase cleavage site was found in the predicted amino acid sequence of lscA. Although a putative signal peptidase recognition site was observed in each of the deduced amino acid sequences of lscB and lscC, the SignalP program predicted them to be too close to the N terminus (eight amino acid residues downstream from the start codon) to be meaningful cleavage sites. To prove that no cleavage occurred during the translocation of Lsc through the two membranes, we determined the N-terminal amino acid sequence of a potential Lsc isoenzyme mixture from the supernatant of PG4180 cultures. Extracellular protein samples were loaded onto SDS-polyacrylamide gels, and a protein band of approximately 50 kDa was identified by zymographic detection of levan formation. The band was cut from the polyacrylamide gel, and the N terminus of the protein was sequenced. The amino acid sequence obtained was STSSSAVSQLKNSPLAGNINY, identical to the predicted N-terminal sequence derived from the nucleotide sequences of lscB and lscC. This result together with the computer prediction clearly indicated that the enzyme(s) is not proteolytically processed during translocation across the inner and outer membranes and that the transport might therefore occur via a sec-independent mechanism.
Expression and characterization of levansucrase from P. syringae in E. coli.
Originally, we planned to clone and express all three lsc genes from P. syringae PG4180 under control of the vector-based Plac promoter in E. coli to test for the functionality of each gene product. As demonstrated previously (23), lscA could be expressed under these circumstances. Because cloning of lscB in plasmid pLB7.2 or pLB7.2R resulted in undetectable Lsc expression in E. coli, we subcloned lscB as a 2.4-kb PCR product amplified from pLB7.2 into pBluescript II SK(+), yielding plasmid pLB2.4. In plasmid pLB2.4, the vector-based Plac promoter was located much closer to the translational start codon of lscB than it was in plasmid pLB7.2, and this gene was expressed (see also Fig. 5A, lane 1) leading to levan formation in E. coli (data not shown). All attempts to insert the 5.5-kb SalI fragment containing lscC into pBluescript II SK(+) in a manner that allowed Plac control of lscC failed. The lscC gene could be cloned only in the direction opposite that of the lacZ operon (Table 2), suggesting that the lack of immediate transport of its gene product to the periplasm or intrinsic characteristics of LscC might be toxic to E. coli. Lsc activities were not detected in E. coli recombinants harboring any of the three lsc genes in orientations opposite that of Plac. This indicated that none of these genes were transcribed from their native promoters in E. coli.
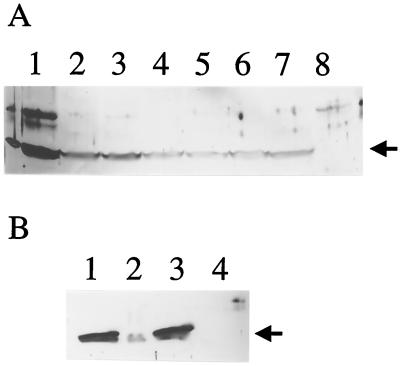
FIG. 5.
Western blot analysis of P. syringae pv. glycinea PG4180 and its lsc mutants using a polyclonal antiserum raised against Lsc from P. syringae pv. phaseolicola NCPPB1321. SDS–10% PAGE was performed with extracts or fractions. (A) Crude protein extracts. Cells were grown to an OD600 of 1.5 to 2.0 at 18°C and then subjected to total protein isolation. Lanes: 1, E. coli(pLB2.4); 2, PG4180; 3, M1 (lscA); 4, M2 (lscB); 5, M3 (lscA lscB); 6, M4 (lscC); 7, M5 (lscA lscC); and 8, M6 (lscB lscC). (B) Cell-free extracellular fractions of PG4180 and its lsc double mutants. Cells were grown to an OD600 of 1.5 to 2.0 at 18°C and then centrifuged. The supernatants were filter sterilized, concentrated 30-fold, and applied to the gel. Lanes: 1, PG4180; 2, M3 (lscA lscB); 3, M5 (lscA lscC); and 4, M6 (lscB lscC). Arrows mark the specific signals representing Lsc.
Furthermore, we analyzed the compartment-specific localization of the lscB gene product in E. coli as determined by subcellular fractionation followed by zymographic detection. As shown for lscA (23), Lsc expressed from E. coli harboring pLB2.4 was not secreted into the supernatant (data not shown). The highest level of Lsc activity was found to be present in the periplasmic fraction, thereby confirming earlier data for lscA (23). This result is in contrast to the situation in P. syringae, where Lsc is secreted to the supernatant (see below). These results suggested that the secretory pathway responsible for Lsc translocation in P. syringae is not present in E. coli.
Phenotypic analysis of lsc-deficient mutants of strain PG4180.
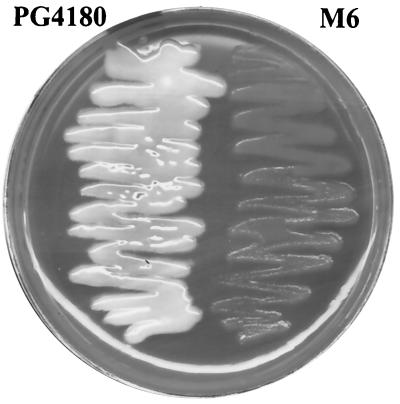
PG4180 mutants disrupted in the three lsc genes were generated. The genotypes of all mutants were verified by PCR and Southern blot analyses (data not shown), indicating that lscA::Smr, lscB::Gmr, lscC::Gmr, and lscC::Smr had replaced the native alleles in the mutants. Subsequently, all mutants were grown on MG agar plates supplemented with 5% sucrose. Mutants M1 (lscA), M2 (lscB), M3 (lscA lscB), M4 (lscC), and M5 (lscA lscC) did not exhibit a levan-deficient phenotype, suggesting that at least two lsc gene products contribute to levan formation in PG4180. When plated on MG agar containing 5% sucrose, mutant M6 (lscB lscC) did not produce levan (Fig. 3), indicating that lscB and lscC are functional, whereas lscA might not be transcribed or its gene product might exhibit an undetectable enzymatic activity.
FIG. 3.
Levan formation by P. syringae pv. glycinea PG4180 and mutant M6 (lscB lscC). Bacteria were streaked on MG agar plates containing 5% sucrose and incubated at 18°C for 7 days. PG4180, but not M6, clearly produced levan, as shown by the whitish swelling.
Compartment-specific analysis of Lsc activities in P. syringae.
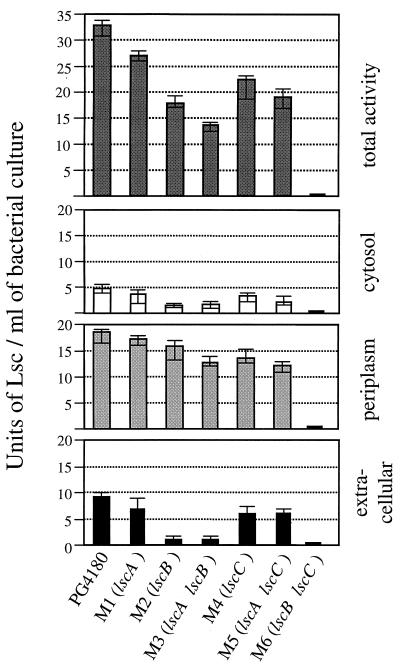
To quantitatively determine the contribution of each lsc gene product to the total Lsc activity inside and outside the cell, cultures of strain PG4180 and its lsc mutants were grown in HSC medium at 18°C until they reached an OD600 of 1.5 to 2.0. Subsequently, Lsc activities in the total cell lysate, periplasmic fraction, cytosolic fraction, and the cell-free supernatant were quantified photometrically (Fig. 4). Although visually not distinguishable from levan formation in the wild type, all mutants except M6 (lscB lscC) consistently showed a slight to moderate decrease of total Lsc activities, with values ranging from 46 to 78% of the wild-type level. No Lsc activity could be measured for the lscB lscC double mutant M6. The periplasmic portions of Lsc activities were less affected in all levan-producing mutants and represented the largest part of the total Lsc activity (Fig. 4). In contrast, cytosolic fractions contained only minor Lsc activity, suggesting that the lsc gene products were efficiently exported in PG4180. Lsc activities in the cytosolic and extracellular fractions were the lowest in the lscB mutants (M2, M3, and M6), indicating that lscB expression represents the major source of Lsc. In a comparison of supernatant samples of the three single mutants, M1 (lscA), M2 (lscB), and M4 (lscC), it appears that the lscB gene product contributes most to the extracellular Lsc activity (Fig. 4). This result was confirmed when the extracellular Lsc levels of the double mutants were compared. LscB and LscC contributed equally to the Lsc activity in the periplasm. Mutations in lscA had the least significant impact on Lsc activity regardless of the fraction studied. Additionally, we analyzed the membrane fractions of various mutants and the wild-type PG4180 for Lsc activities (data not shown). Lsc activity was negligible in those fractions, regardless of the mutant background, suggesting that Lsc is not membrane-bound.
FIG. 4.
Quantitative analysis of the compartment-specific levansucrase activities in P. syringae pv. glycinea PG4180 and its lsc mutants. Bacterial cultures were incubated in HSC medium at 18°C until they reached an OD600 of 1.5 to 2.0. Levansucrase activities (calculated per milliliter of culture) were determined photometrically in the total cell culture and in cytosolic, periplasmic, and extracellular fractions. Data represent average values from three independent experiments with three replicates each. Error bars symbolize the minimum and maximum values.
To ensure that the subcellular fractions were not significantly contaminated with proteins from other fractions, cells of strain PG4180(pAS-LacZ) and PG4180(pHL-PhoA) were incubated under the conditions described above and subjected to subcellular fractionation. Subsequently, LacZ and PhoA activities were quantified photometrically in three individual experiments, each with three replicates. The cytosolic fraction of PG4180(pAS-LacZ) showed 198 U of β-galactosidase activity, while the cognate periplasmic fraction exhibited 54 U of LacZ activity, indicating that the periplasmic fraction could have been contaminated with cytosolic LacZ by approximately 27%. When fractions of PG4180(pHL-PhoA) were analyzed for PhoA activities, 22.5 U of PhoA activity was measured in the periplasm compared to 3.3 U of PhoA activity in the cytosolic fraction, suggesting that the latter fraction was contaminated with periplasmic PhoA by 15%. Neither LacZ activity nor PhoA activity was detected in the extracellular fractions.
Immunological detection of Lsc in different cell compartments.
To distinguish between protein secretion and enzymatic activity of Lsc, Western blot experiments were performed with total cellular protein samples and extracellular protein fractions of strain PG4180 and its lsc mutants using polyclonal antibodies raised against Lsc of P. syringae pv. phaseolicola (22). Results given in Fig. 5 demonstrate that Lsc could be detected in crude protein extracts and in the supernatant of PG4180 but not in any protein fractions of M6 (lscB lscC), suggesting that lscA is not expressed. Signals for Lsc were detected in all levan-producing mutants of PG4180 (Fig. 5A). Moreover, signals were strongly decreased or absent from the extracellular fractions of lscB mutants (M3 and M6) but were present in the lscA lscC mutant M5 (Fig. 5B), which suggests that LscB was the secreted enzyme. These results show that the compartment-specific occurrence of Lsc activities strictly correlates with the presence of the enzyme(s), allowing the possibility of a catalytic inactivation of LscC in the culture supernatant of PG4180 to be ruled out.
Complementation experiments with the levan-deficient mutant M6.
To provide evidence that the gene product of lscB is indeed secreted by strain PG4180, the 7.2-kb insert of pLB7.2 was subcloned to pBBR1MCS to yield pRB7.2. This plasmid and cosmid p5C10 carrying copies of lscB or lscC, respectively, were separately introduced to the lscB lscC mutant M6. The accumulation of Lsc in subcellular fractions of the respective transconjugants was assayed by using zymograms and Western blot analysis (data not shown). As expected, Lsc was secreted to the exterior of transconjugant M6(pRB7.2) but not to that of M6(p5C10). However, both transconjugants were visibly mucoid when streaked on MG plates containing 5% sucrose. These results supported our previous findings and suggested that even though functional lscC restored levan formation to M6, its gene product was not secreted to the supernatant but instead accumulated in the periplasm.
Analysis of the gene product of lscA.
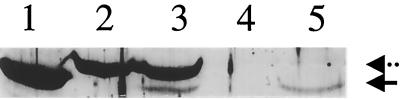
Although lscA was previously expressed in E. coli, giving rise to levan formation by E. coli (23), our current data for the levan-deficient mutant M6 (lscB lscC) suggested that lscA was not expressed in P. syringae pv. glycinea PG4180. To confirm this, the 3.1-kb PstI fragment from plasmid pSKL3 harboring a functional lscA gene was subcloned into the broad-host-range vector pRK415, yielding plasmid pRA3.1, in which lscA was transcriptionally linked to the vector-borne Plac promoter. This plasmid was then introduced into strain PG4180 and its mutant M6. Transconjugant M6(pRA3.1) exhibited a mucoid phenotype when streaked on MG plates containing 5% sucrose, indicating that the lscA gene product is functional but that its gene is not expressed from its own promoter under the in vitro conditions tested in PG4180. Subsequently, Western blot experiments were performed with PG4180, M6, and their respective transconjugants harboring plasmid pRA3.1 (Fig. 6). The immunologically detectable gene product of lscA can clearly be distinguished from those of lscB or lscC due to its smaller size. LscA could be detected when its gene was transcribed from the Plac promoter in PG4180(pRA3.1) and M6(pRA3.1) but not in PG4180 and its mutant M6 (Fig. 6), confirming the phenotypic observations.
FIG. 6.
Detection of the gene product of lscA in P. syringae by Western blot analysis following expression of lscA under control of Plac on plasmid pRA3.1. The antiserum used was raised against Lsc from P. syringae pv. phaseolicola NCPPB1321. SDS–10% PAGE was done with crude protein extracts. Cells were grown to an OD600 of 1.5 to 2.0 at 18°C and then subjected to total protein extraction. Lanes: 1, E. coli(pSKL3); 2, PG4180; 3, PG4180(pRA3.1); 4, M6; and 5, M6(pRA3.1). LscA (solid arrow) and LscB or LscC (dotted arrow) are distinguishable by their different molecular weights.
Screening for multiple levansucrase alleles in various pathovars of P. syringae
Thirty-five strains representing 21 different pathovars of P. syringae were screened for multiple lsc genes by PCR. For strains of the pathovars glycinea, tomato, phaseolicola, lachrymans, and morsprunorum, Hettwer et al. (23) had previously demonstrated the presence of lscA homologues by PCR analysis. In this study, the primer sets lscB-F–lscB-R and lscC-F–lscC-R were used to amplify 1.3-kb PCR products representing lscB and lscC, respectively (Table 3). Both signals were amplified from genomic DNA from all strains tested of the pathovars glycinea (three strains), tomato (three strains), phaseolicola (two strains), tabaci (two strains), lachrymans (one strain), coriandriocola (one strain), and photiniae (one strain) without any variability. In contrast, the PCR product of lscC but not that of lscB was amplified from strains representing pathovars atropurpurea (one strain), savastanoi (two strains), syringae (four strains), myricae (one strain), garcae (one strain), morsprunorum (four strains), and persicae (one strain), and from one strain of pathovar phaseolicola. PCR with samples of representative strains from seven pathovars (one strain for each pathovar) did not yield any detectable PCR products. These results were in part confirmed by the results of Southern blot analysis of the 1.3-kb PCR products using lscB of strain PG4180 as the DNA probe. When the respective PCR products were treated with XhoI, a restriction fragment length polymorphism was observed among samples from P. syringae pv. glycinea, phaseolicola, syringae, tomato, tabaci, lachrymans, coriandriocola, and photiniae (data not shown). These data revealed that the occurrence of multiple copies of lsc is widespread in various pathovars of P. syringae.
DISCUSSION
Herein, we provide evidence for the presence of three lsc genes in the plant pathogen P. syringae pv. glycinea PG4180 that show a high degree of similarity of their primary sequence data. While the nucleotide sequences of lscB and lscC were almost identical to each other and to lsc from P. syringae pv. phaseolicola (23), they differed from that of lscA by approximately 14%.
Screening for multiple lsc copies among P. syringae pathovars revealed that multiple occurrence of lsc genes is characteristic of pathovars other than glycinea. Our data indicated that lscB is located on the plasmid and identical to a gene coding for Lsc from P. syringae NCPPB1321 (23). This result confirmed reports on the conservation of lsc genes in plant-associated gram-negative bacteria (2, 16, 23) and implied that horizontal gene transfer might have been involved in their distribution. Subsequently, plasmid DNA from P. syringae NCPPB1321 was analyzed by Southern blot hybridization for the presence of lsc. Indeed, a signal representing a plasmid of unidentified size could be obtained, indicating that at least one lsc gene of NCPPB1321 is located on the plasmid (our unpublished observation).
So far, all levan-producing bacteria tested have contained a single gene coding for Lsc in their genomes (2, 15, 16, 21, 43–45). Redundance of lsc in the genome of P. syringae could signal its ecological importance or might hint at different functions for each of the gene products. In the fire blight pathogen, E. amylovora, levan formation was shown to significantly contribute to virulence and in planta multiplication (16). Whether levan formation plays a critical role during the infection of soybean plants by P. syringae pv. glycinea PG4180 is currently being investigated. Our preliminary results for virulence and in planta survival assays with the levan-deficient mutant M6 and the wild-type strain PG4180 suggest that M6 does not exhibit impaired pathogenicity after infiltration into soybean leaf tissue. However, when spray inoculated, M6 showed decreased survival on the leaf surface, indicating that levan formation might be required for the saprophytic fitness of P. syringae. Moreover, results of the spray inoculation experiments (and spray inoculation resembles the natural infection process much more than the infiltration technique) also suggested that there were fewer symptoms and less bacterial multiplication in the plant tissue in the levan-deficient mutant than in the wild type (data not shown). Detailed analyses of in planta levan formation and lsc expression are currently being conducted in our laboratory.
Even though all three lsc genes could be cloned in E. coli, only transcription of lscA and lscB under control of the vector-based Plac promoter gave rise to recombinant levan formation. This result is consistent with various reports showing that many P. syringae promoters are nonfunctional in members of the family Enterobacteriaceae and suggests that E. coli might lack the native regulatory system(s) for transcription of lsc genes from P. syringae. Lack of export of LscC in enterobacteria may be the primary reason for our failure to express LscC in E. coli. Intracellular accumulation of LscC might be toxic to this organism as described for sacB, the B. subtilis gene coding for Lsc (14). Furthermore, the gene products of lscA and lscB were detected in the periplasm but not in the supernatant of E. coli, suggesting a lack of the appropriate secretion machinery for Lsc in this organism.
In this report, single lscA, lscB, or lscC mutants and double mutants were generated and analyzed with respect to the contribution of each gene to the overall levan synthesis of P. syringae pv. glycinea PG4180. Interestingly, a clear levan-deficient phenotype could be observed only when lscB and lscC were simultaneously disrupted, giving rise to the assumption that the products of both genes contributed to the levan formation by PG4180. Consequently, either gene could restore levan production to mutant M6 (lscB lscC) in complementation experiments. In future experiments, whether transcription of either lscB or lscC is upregulated if the other gene is inactivated should be determined.
The role of lscA in levan formation of P. syringae remains obscure. As shown by heterologous expression under Plac control in E. coli (23), lscA encodes a functional enzyme synthesizing the EPS levan. However, the lscB lscC mutant M6 exhibited a levan-deficient phenotype in vitro and did not show a signal for Lsc in the Western blot analysis, suggesting that the gene product of lscA is not translated and is dispensable for levan formation in P. syringae under laboratory conditions. When expressed under control of Plac, lscA restored levan production to the lscB lscC mutant M6 and LscA could be immunologically detected in M6 transconjugants. Therefore, we conclude that transcription of lscA under the tested conditions either does not occur or is too low to be detected. However, whether lscA is transcribed under natural conditions in planta is not known. Likewise, the location of lscB on an indigenous plasmid allows us to speculate that there might be multiple copies of the gene, thereby giving rise to a higher level of LscB than of LscC.
Interestingly, the enzymatic activities associated with the gene products of lscB and lscC accumulated in different compartments of strain PG4180. According to our data for the reporter enzyme controls, a significant carryover of Lsc from the cytosolic to periplasmic fraction by the applied fractionation method can be ruled out. If 27% of the cytosolic Lsc contaminated the periplasmic fraction, it would account for less than 1/18 of total periplasmic Lsc activity. Likewise, neither PhoA activity nor LacZ activity could be detected in the supernatant. In contrast, more than half of the cytosolic Lsc activity could come from the periplasmic fraction, which supports the assumption that most Lsc accumulated outside of the cytosol. LscB was found to be predominantly responsible for extracellular formation, while LscC accumulated in the periplasm. This important finding was confirmed when the lscB lscC mutant M6 was complemented in trans by lscB or lscC. Additionally, extracellular Lsc was found to possess an N-terminal sequence identical to the predicted amino acid sequences of LscB and LscC, suggesting that the export of both gene products to the periplasm and the further secretion of LscB proceed in a sec-independent manner.
The determinants of the compartment-specific accumulation of Lsc activity remain to be elucidated. LscB and LscC differ in only five amino acyl residues, and the conservative changes in amino acid residues 92, 327, 329, and 429 might not be important for the structure and physicochemical characteristics of Lsc. However, the alteration in position 119, a serine residue in LscB changed to a cysteine residue in LscC, could affect the putative number of disulfide bridges the proteins might form. LscC might contain two disulfide bridges, whereas LscB might possess only one. Such an alteration could significantly influence the overall structure of the enzyme, possibly leading to a selective transport across the outer membrane.
We determined whether lack of Lsc activity in a particular compartment was due to catalytic dysfunction or to a lack of protein transport. The results of Western blot analysis demonstrated that the presence of a given lsc gene product was strictly linked to its enzymatic activity. This result essentially ruled out the possibility that LscC might be secreted to the supernatant but might be nonfunctional in this particular environment. In future studies, experimental exchange of the particular amino acid residues in which LscB and LscC differ could reveal their potential to target their respective gene product to the extracellular space. Moreover, structural analyses of the polymeric levan products of LscB and LscC derived from their double mutants will be helpful in determining the particular role of periplasmic and extracellular levan formation for P. syringae.
Lsc activities were negligible in cytoplasmic and membrane fractions of P. syringae pv. glycinea PG4180. In contrast, high enzymatic activities were found in the periplasmic fraction and supernatant. This result suggested a putative two-step transport mechanism for Lsc via the periplasm as previously described for Lsc from A. diazotrophicus (2). However, a signal peptide sequence typical for the sec-dependent type II general secretory pathway (11) was not found in any of the predicted P. syringae lsc gene products, and the N-terminal sequence of extracellular Lsc was identical to its predicted amino acid sequence. These data indicated that the enzyme was not proteolytically processed during its translocation across the inner and outer membranes. The presence of high Lsc activities in the periplasm lowers the likelihood that Lsc translocation might occur by a classical type I or type III secretion mechanism. In these secretion pathways, proteins bypass the periplasm and are directly secreted from the cytoplasm (4, 24). Charkowski et al. (8) demonstrated that specific mutations in genes for the type III hrp secretion pathway in P. syringae caused a significant accumulation of transported proteins in the periplasm. However, this particular secretion pathway functions perfectly in PG4180 (7) and therefore cannot be the cause for this particularly interesting phenotype.
In the fire blight pathogen, E. amylovora, export of Lsc across the inner membrane was suppressed when the C terminus of this enzyme was mutated (16). The detailed mechanisms by which Lsc of P. syringae are exported from the cytoplasm and the particular signals that are necessary for the transport remain to be investigated. Molecular tools (e.g., mutants and translational fusions) to dissect the secretory pathway(s) are now available in our laboratory and are being used in ongoing studies. Ultimately, our future experiments will give new insights into the mode of Lsc export and secretion and will allow us to elucidate how the two copies of this enzyme are translocated via the bacterial membranes.
ACKNOWLEDGMENTS
We thank Frank R. Jaeckel for technical assistance in the initial work of this study and Antonio Pierik, Helge Weingart, George Sundin, and Klaus Rudolph for helpful comments.
This study was supported in part by the Deutsche Forschungsgemeinschaft (SFB 395).
REFERENCES
- 1.Alfano J R, Collmer A. Bacterial pathogens in plants: life up against the wall. Plant Cell. 1996;8:1683–1698. doi: 10.1105/tpc.8.10.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrieta J, Hernandez L, Coego A, Suarez V, Balmori E, Menendez C, Petit-Glatron M F, Chambert R, Selman-Housein G. Molecular characterization of the levansucrase gene from the endophytic sugarcane bacterium Acetobacter diazotrophicus SRT4. Microbiology. 1996;142:1077–1085. doi: 10.1099/13500872-142-5-1077. [DOI] [PubMed] [Google Scholar]
- 3.Bender C L, Young S A, Mitchell R E. Conservation of plasmid DNA sequences in coronatine-producing pathovars of Pseudomonas syringae. Appl Environ Microbiol. 1991;57:993–999. doi: 10.1128/aem.57.4.993-999.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binet R, Letoffe S, Ghigo J M, Delepelaire P, Wandersman C. Protein secretion by Gram-negative bacterial ABC exporters: a review. Gene. 1997;192:7–11. doi: 10.1016/s0378-1119(96)00829-3. [DOI] [PubMed] [Google Scholar]
- 5.Boyd D, Manoil C, Beckwith J. Determinants of membrane protein topology. Proc Natl Acad Sci USA. 1987;84:8525–8529. doi: 10.1073/pnas.84.23.8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradbury J F. Identification of cultivable bacteria from plants and plant tissue cultures by use of simple classical methods. Acta Hortic (Wageningen) 1986;225:27–37. [Google Scholar]
- 7.Budde I P, Ullrich M S. Interactions of Pseudomonas syringae pv. glycinea with host and nonhost plants in relation to temperature and phytotoxin synthesis. Mol Plant-Microbe Interact. 2000;13:951–961. doi: 10.1094/MPMI.2000.13.9.951. [DOI] [PubMed] [Google Scholar]
- 8.Charkowski A O, Huang H-C, Collmer A. Altered localization of HrpZ in Pseudomonas syringae pv. syringae hrp mutants suggests that different components of the type III secretion pathway control protein translocation across the inner and outer membranes of gram-negative bacteria. J Bacteriol. 1997;179:3866–3874. doi: 10.1128/jb.179.12.3866-3874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dedonder R. Levansucrase from Bacillus subtilis. Methods Enzymol. 1966;8:500–506. [Google Scholar]
- 10.Denny T P. Involvement of bacterial polysaccharides in plant pathogenesis. Annu Rev Phytopathol. 1995;33:173–197. doi: 10.1146/annurev.py.33.090195.001133. [DOI] [PubMed] [Google Scholar]
- 11.Fekkes P, Driessen A J M. Protein targeting to the bacterial cytoplasmic membrane. Microbiol Mol Biol Rev. 1999;63:161–173. doi: 10.1128/mmbr.63.1.161-173.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fett W F, Dunn M F. Exopolysaccharides produced by phytopathogenic Pseudomonas syringae pathovars in infected leaves of susceptible hosts. Plant Physiol. 1989;89:5–9. doi: 10.1104/pp.89.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fett W F, Osman S F, Dunn M F. Characterization of exopolysaccharides produced by plant-associated fluorescent pseudomonads. Appl Environ Microbiol. 1989;55:579–583. doi: 10.1128/aem.55.3.579-583.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gay P, LeCoq D, Steinmetz M, Berkelman T, Kado C I. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol. 1985;164:918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gay P, LeCoq D, Steinmetz M, Ferrari E, Hoch J A. Cloning of the structural gene sacB, which codes for exoenzyme levansucrase of Bacillus subtilis: expression of the gene in Escherichia coli. J Bacteriol. 1983;153:1424–1431. doi: 10.1128/jb.153.3.1424-1431.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geier G, Geider K. Characterization and influence on virulence of the levansucrase gene from the fireblight pathogen Erwinia amylovora. Physiol Mol Plant Pathol. 1993;42:387–404. [Google Scholar]
- 17.Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C.: American Society for Microbiology; 1994. [Google Scholar]
- 18.Gross M, Geier G, Rudolph K, Geider K. Levan and levansucrase synthesized by the fireblight pathogen Erwinia amylovora. Physiol Mol Plant Pathol. 1992;40:371–381. [Google Scholar]
- 19.Gross M, Rudolph K. Studies on the extracellular polysaccharides (EPS) produced in vitro by Pseudomonas syringae pv. phaseolicola. II. Characterization of levan, alginate, and LPS. J Phytopathol. 1987;119:206–215. [Google Scholar]
- 20.Hellio F C, Orange N, Guespin-Michel J F. Growth temperature controls the production of a single extracellular protease by Pseudomonas fluorescens MF0, in the presence of various inducers. Res Microbiol. 1993;144:617–625. doi: 10.1016/0923-2508(93)90064-9. [DOI] [PubMed] [Google Scholar]
- 21.Hernandez L, Sotolongo M, Rosabal Y, Menendez C, Ramirez R, Caballero-Mellado J, Arrieta J. Structural levansucrase gene (lsdA) constitutes a functional locus conserved in the species Gluconacetobacter diazotrophicus. Arch Microbiol. 2000;174:120–124. doi: 10.1007/s002030000173. [DOI] [PubMed] [Google Scholar]
- 22.Hettwer U, Gross M, Rudolph K. Purification and characterization of an extracellular levansucrase from Pseudomonas syringae pv. phaseolicola. J Bacteriol. 1995;177:2834–2839. doi: 10.1128/jb.177.10.2834-2839.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hettwer U, Jaeckel F R, Boch J, Meyer M, Rudolph K, Ullrich M S. Cloning, nucleotide sequence, and expression in Escherichia coli of levansucrase genes from the plant pathogens Pseudomonas syringae pv. glycinea and P. syringae pv. phaseolicola. Appl Environ Microbiol. 1998;64:3180–3187. doi: 10.1128/aem.64.9.3180-3187.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hueck C J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasapis S, Morris E R, Gross M, Rudolph K. Solution properties of levan polysaccharide from Pseudomonas syringae pv. phaseolicola, and its possible role as a blocker of recognition during pathogenesis. Carbohydr Polym. 1994;23:55–64. [Google Scholar]
- 27.Katzen F, Becker A, Ielmini M V, Oddo C G, Ielpi L. New mobilizable vectors suitable for gene replacement in gram-negative bacteria and their use in mapping of the 3′ end of the Xanthomonas campestris pv. campestris gum operon. Appl Environ Microbiol. 1999;65:278–282. doi: 10.1128/aem.65.1.278-282.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keane P J, Kerr A, New P B. Crown gall of stone fruit. II. Identification and nomenclature of Agrobacterium isolates. Aust J Biol Sci. 1970;23:585–595. [Google Scholar]
- 29.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmid for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 30.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 31.Kovach M E, Philipps R W, Elzer P H, Roop III R M, Peterson K M. pBBR1MCS: a broad-host-range cloning vector. Biotechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- 32.Leigh J A, Coplin D L. Exopolysaccharides in plant-bacterial interactions. Annu Rev Microbiol. 1992;46:307–346. doi: 10.1146/annurev.mi.46.100192.001515. [DOI] [PubMed] [Google Scholar]
- 33.Lindow S E. Determinants of epiphytic fitness in bacteria. In: Andres J H, Hirano S S, editors. Microbial ecology of leaves. New York, N.Y: Springer-Verlag; 1991. pp. 295–314. [Google Scholar]
- 34.Lyness E W, Doelle H W. Levansucrase from Zymomonas mobilis. Biotechnol Lett. 1983;5:345–350. [Google Scholar]
- 35.Murillo J, Shen H, Gerhold D, Sharma A, Cooksey D A, Keen N T. Characterization of pPT23B, the plasmid involved in syringolide production in Pseudomonas syringae pv. tomato PT23. Plasmid. 1994;31:275–287. doi: 10.1006/plas.1994.1029. [DOI] [PubMed] [Google Scholar]
- 36.Osman S F, Fett W F, Fishman M L. Exopolysaccharides of the phytopathogen Pseudomonas syringae pv. glycinea. J Bacteriol. 1986;166:66–71. doi: 10.1128/jb.166.1.66-71.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palmer D A, Bender C L. Effects of environmental and nutritional factors on production of the polyketide phytotoxin coronatine by Pseudomonas syringae pv. glycinea. Appl Environ Microbiol. 1993;59:1619–1626. doi: 10.1128/aem.59.5.1619-1626.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rutz C, Rosenthal W, Schülein R. A single negatively charged residue affects the orientation of a membrane protein in the inner membrane of Escherichia coli only when it is located adjacent to a transmembrane domain. J Biol Chem. 1999;274:33757–33763. doi: 10.1074/jbc.274.47.33757. [DOI] [PubMed] [Google Scholar]
- 39.Saile E, McGarvey J A, Schell M A, Denny T P. Role of extracellular polysaccharides and endoglucanase in root invasion and colonization of tomato plants by Ralstonia solanacearum. Phytopathology. 1997;87:1264–1271. doi: 10.1094/PHYTO.1997.87.12.1264. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 41.Sato S, Koga T, Inoue M. Isolation and some properties of extracellular D-glucosyltransferases and D-fructosyltransferases from Streptococcus mutans serotypes c, e, and f. Carbohydr Res. 1984;134:293–304. [Google Scholar]
- 42.Schaad N W. Laboratory guide for identification of plant pathogenic bacteria. St. Paul, Minn: American Phytopathological Society; 1988. [Google Scholar]
- 43.Song K B, Joo H K, Rhee S K. Nucleotide sequence of levansucrase gene (levU) of Zymomonas mobilis ZM1 (ATCC 10988) Biochim Biophys Acta. 1993;1173:320–324. doi: 10.1016/0167-4781(93)90130-6. [DOI] [PubMed] [Google Scholar]
- 44.Song K B, Seo J W, Kim M G, Rhee S K. Levansucrase of Rahnella aquatilis ATCC33071. Gene cloning, expression, and levan formation. Ann N Y Acad Sci. 1998;864:506–511. doi: 10.1111/j.1749-6632.1998.tb10369.x. [DOI] [PubMed] [Google Scholar]
- 45.Tajima K, Tanio T, Kobayashi Y, Kohno H, Fujiwara M, Shiba T, Erata T, Munekata M, Takai M. Cloning and sequencing of the levansucrase gene from Acetobacter xylinum NCI 1005. DNA Res. 2000;7:237–242. doi: 10.1093/dnares/7.4.237. [DOI] [PubMed] [Google Scholar]
- 46.Ullrich M, Bereswill S, Völksch B, Fritsche W, Geider K. Molecular characterization of field isolates of Pseudomonas syringae pv. glycinea differing in coronatine production. J Gen Microbiol. 1993;139:1927–1937. doi: 10.1099/00221287-139-8-1927. [DOI] [PubMed] [Google Scholar]
- 47.Watanabe K, Nagahama K, Sato M. A conjugative plasmid carrying the efe gene for the ethylene-forming enzyme isolated from Pseudomonas syringae pv. glycinea. Phytopathology. 1998;88:1205–1209. doi: 10.1094/PHYTO.1998.88.11.1205. [DOI] [PubMed] [Google Scholar]
- 48.Wilson K J, Sessitsch A, Corbo J C, Giller K E, Akkermans A D L, Jefferson R A. β-Glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other gram-negative bacteria. Microbiology. 1995;141:1691–1705. doi: 10.1099/13500872-141-7-1691. [DOI] [PubMed] [Google Scholar]