Abstract
Spore-forming bacteria of the Bacillus genus have demonstrated potential as probiotics for human use. Bacillus clausii have been recognized as efficacious and safe agents for preventing and treating diarrhea in children and adults, with pronounced immunomodulatory properties during several in vitro and clinical studies. Herein, we characterize the novel strain of B. clausii CSI08 (Munispore®) for probiotic attributes including resistance to gastric acid and bile salts, the ability to suppress the growth of human pathogens, the capacity to assimilate wide range of carbohydrates and to produce potentially beneficial enzymes. Both spores and vegetative cells of this strain were able to adhere to a mucous-producing intestinal cell line and to attenuate the LPS- and Poly I:C-triggered pro-inflammatory cytokine gene expression in HT-29 intestinal cell line. Vegetative cells of B. clausii CSI08 were also able to elicit a robust immune response in U937-derived macrophages. Furthermore, B. clausii CSI08 demonstrated cytoprotective effects in in vitro cell culture and in vivo C. elegans models of oxidative stress. Taken together, these beneficial properties provide strong evidence for B. clausii CSI08 as a promising potential probiotic.
Keywords: Bacillus clausii, spore-based probiotics, immunomodulation, adherence, antioxidant
1. Introduction
The term probiotics acknowledged by the Agriculture Organization of the United Nations and the WHO (FAO/WHO) in 2001 and revisited later, in 2014, defines them as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” [1]. The growing body of evidence highlighting the fundamental role of microbiota in human health and disease [2] drives global interest in probiotic research, and novel approaches helping to maintain “healthier” gut microbiota are in demand. Indeed, health benefits of probiotics have been shown in multiple clinical trials and are further backed by meta-analyses and systematic reviews. Through various modalities, including interacting with the host microbiota and the host intestinal epithelium, probiotics can influence a number of physiological processes including metabolism, immunity and endocrine function amongst others [3].
Probiotic strains most cited in biomedical literature belong to the genera bifidobacteria and lactobacilli [1,4]. In this context, spore-forming bacteria of the genus Bacillus are clearly understudied [5], yet a number of bacilli strains have been shown to possess probiotic characteristics and have decades-long history of medical use [5,6]. The ability to form endospores that are resistant to heat, UV radiation, desiccation [7], can tolerate low pH and high concentrations of bile salts [8] confers Bacillus-based probiotics several unique features. Being administered in the spore state, Bacillus spp. can survive aggressive gastric conditions and reach the small intestine in large numbers where they germinate into vegetative form [9]. Spore-based probiotic formulations have a long shelf life and do not require refrigeration during storage [10,11]. Moreover, Bacillus spp. spores showed decent stability during processing and storage of functional foods, such as pastries, fruit juices and preserves, sausages, and pasta [12,13,14].
Different strains belonging to nine Bacillus species such as B. subtilis, B. coagulans, B. indicus, B. licheniformis, B. clausii have been recognized as probiotics for human use [15]. B. clausii strains are ubiquitous in nature, and have been isolated from soil [16], seawater [17], milk [18] amongst others. Importantly, isolates of B. clausii have been recovered from human feces more frequently than other Bacillus species [19].
Safety and efficacy of different probiotic strains of B. clausii have been demonstrated in a number of clinical trials conducted around the globe [20,21,22,23,24]. For example, B. clausii (O/C, N/R, SIN and T) can reduce the incidence of diarrhea, nausea, and epigastric pain associated with antibiotic treatment for H. pilory in adults [25,26]. Similarly, B clausii (A. clausii 088AE) showed their efficacy for managing diarrhea caused by broad-spectrum antibiotics in both children and adults [27]. Treatment with probiotic strains of B. clausii were also effective in reducing main clinical symptoms of acute diarrhea, including rotavirus-associated diarrhea in children [28,29,30,31]. In a recently published prospective study, B. clausii (O/C, N/R, SIN and T) was beneficial for managing symptoms of IBS in children [31]. Moreover, prophylactic administration of B. clausii to preterm infants was associated with faster achievement of full feeds [32]. In a pilot trial involving children with recurrent respiratory infections prolonged treatment with B. clausii has been proven to reduce the duration of infections [33]. B. clausii treatment has also been shown to alleviate symptoms of aphthous ulcer and oral candidiasis [34].
In vitro and preclinical studies of probiotic strains of B. clausii, as well as genome sequencing reports, revealed their key attributes, including the ability to produce antimicrobial substances and vitamins, the ability to withstand acidic pH and bile salts, and stress adaptation factors and putative adhesion proteins [35,36,37,38,39]. Interestingly, the immunomodulatory capacity of various B. clausii strains has been highlighted in a number of studies. Park et al. reported that supplementation with viable cells B. clausii KCTC 10277 BP can suppress pulmonary inflammation in ovalbumin-sensitized mice, reducing Th2 immune response and affecting hypoxia-related pathway in lung tissue [40]. B. clausii MTCC-8326 has been shown to protect murine RAW 264.7 macrophages against S. typhimurium infection supposedly by causing controlled inflammatory response and inducing several defensins and interferons [41]. Administration of B. clausii (O/C, N/R, SIN, and T) can affect expression of genes involved in inflammation, immune response, defense response etc. in duodenal mucosa of healthy individuals [42]. An in vitro study conducted on Swiss and C57 Bl/6j murine cells demonstrated the ability of the same strains to induce NOS II synthetase activity, IFN-γ production, and CD4+ T-cell proliferation [43].
MuniSpore® is a commercial preparation of the spores B. clausii CSI08. The aim of the present work was to characterize this novel strain. Firstly, we evaluated general probiotic attributes of B. clausii CSI08, such as resistance to gastrointestinal conditions, adhesion capability, enzymatic profile, and antimicrobial activity. Additionally, we examined immunomodulatory and antioxidant properties of the vegetative form of the strain in in vitro and in vivo models.
2. Materials and Methods
2.1. Bacterial Strains and Sample Preparation
Munispore®, the single-strain spore-based probiotic Bacillus clausii CSI08, was provided by Deerland Probiotics and Enzymes (US). Vegetative form of B. clausii CSI08, Escherichia coli ATCC 25922, Salmonella enteritidis ATCC 13076, Staphylococcus aureus str. RF122, Pseudomonas aeruginosa DSM 3227 were cultured routinely at 37 °C in Trypticase soy broth (TSB) or agar (Merck, Ireland), unless otherwise specified. When preparing liquid cultures, above-named bacterial strains were grown with vigorous shaking (~170 rpm). Commercial probiotic strains Lactobacillus rhamnosus GG (ATCC 53103) and Lactobacillus fermentum were grown using DeMan-Rogosa-Sharpe broth or agar (Oxoid Ltd., Basingstoke, United Kingdom) at 37 °C.
For cell culture experiments, 10 mL of overnight cultures prepared under conditions described above were transferred into 50 mL falcon tubes and centrifuged for 10 min at 4000× g. Supernatants were then removed, and bacterial pellets were washed once with 10 mL of serum-free DMEM. Washed pellets were resuspended in 10 mL of serum and additives free high glucose DMEM. To obtain cell-free supernatants bacterial preparations in DMEM were further cultivated for 18 h. After that, cultures were centrifuged for 10 min at 4000× g; pellets were discarded, and supernatants were filter-sterilized using syringe-mounted filters with 0.22 µm pore diameter. 1N NaOH was added to cell-free supernatants to bring pH to ~7.5, if necessary.
2.2. Stability of B. clausii CSI08 Spores during Pasteurization
Spore suspension was prepared by adding 50 mg of Munispore® spore powders to 100 mL of 1X PBS, pH 7.6 (Sigma-Aldrich, St. Louis, MO, USA). The suspension was mixed using a vortex for 2 min and dispensed into glass test tubes, 5 mL of suspension per each. The test tubes were treated at 45 °C, 75 °C and 90 °C in a water bath for 0.5, 1 or 3 min. Following incubation, samples were serially diluted and plated in 3M™ Petrifilm™ aerobic count plates.
2.3. Resistance of B. clausii CSI08 Spores to Simulated Gastric and Intestinal Conditions
The tolerance of B. clausii CSI08 spores to an in vitro simulated gastric and intestinal conditions were assayed following the publication by Pisano et al. [44] with some modifications. Briefly, 100 µL of spore suspensions in PBS or 100 µL of L. rhamnosus suspension in PBS were added to 900 µL of 0.3% pepsin (w/v, Sigma P6887) in NaCl pH 3 and incubated for 2 h at 37 °C in a water bath. Following incubation, spores and bacteria were pelleted by centrifuged at 3000× g for 5 min; the pellets were washed two times with 1 mL PBS and resuspended in 100 µL PBS. 900 µL of solution containing 0.1% pancreatin (w/v, Sigma P1750) and 0.3% bile salts (w/v, Sigma cat no. 48305) in PBS, pH 7.5, were added to the spores and bacteria and incubated for another 2 h at 37 °C in a water bath. After 1, 2, 3, and 4 h of incubation, samples’ aliquots were serially diluted and plated onto 3M™ Petrifilm™ aerobic count plates (B. clausii CSI08 spores) or MRS agar plates (L. rhamnosus).
2.4. Quantitative Analysis of Amino Acids and Vitamins in B. clausii CSI08 Supernatants
Overnight cultures of B. clausii CSI08 prepared in TSB were taken for analysis. Amino acids quantification was carried out using high performance liquid chromatography (HPLC) and fluorescence detection (FLD), with a precolumn derivatization step using 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate (AQC). Regarding the determination of vitamins, ultra-high performance liquid chromatography (UHPLC) coupled to a simple quadrupole mass detector (MS) was used. Full technical description of the methods can be found in Appendix A and Appendix B.
2.5. Semi-Quantitative Assays for Carbohydrate Fermentation and Hydrolytic Activities
The ability of B. clausii CSI08 to use different carbon sources was investigated with API 50CH (BioMérieux, Hampshire, UK); hydrolytic activities were determined using the API-ZYM kit system (BioMérieux, Hampshire, UK), according to the instructions provided by manufacturer.
2.6. Antimicrobial Activity of B. causii CSI08 in Liquid Media
Tests in liquid culture were performed as previously described with some modifications [45]. Briefly, 10 µL of TSB were co-inoculated with 1 × 105 CFU/mL of the test pathogen strain and 1 × 107 CFU/mL of MuniSpore®. Co-cultures were incubated at 37 °C with vigorous shaking (170 rpm) for 24 h; after that the co-cultures were serially diluted and plated. Enumeration of gram-negative bacteria was performed on MacConkey agar (Merck, Ireland). S. aureus enumeration was performed on Mannitol egg yolk polymyxin agar (MYP, Merck, Ireland). Both MacConkey and MYP agar inhibited the growth of B. clausii CSI08, enabling the detection of each pathogen.
2.7. Determination of Antioxidant Activity In Vitro
Fifty milliliters of overnight cultures of B. clausii CSI08 and L. rhamnosus prepared in TSB broth and normalized by number of bacteria (1 × 108 CFU/mL), were centrifuged at 4000× g for 15 min. Obtained pellets were washed three times with 10 mL phosphate buffered saline (PBS, Sigma-Aldrich) and resuspended in 1 mL PBS. Cell suspensions were transferred to beaded tubes (A29158, Thermofisher). Cells were lysed using BeadBug™6 homogenizer at 3500 rpm, 3 cycles 30 s each, suspensions were kept on ice between cycles for 1 min. Tubes were centrifuged at 9800× g for 15 min to remove cell debris and the supernatants were transferred to fresh microcentrifuge tubes. The level of catalase activity in cell lysates have been determined using Catalase assay kit (CAK1061, Cohesion Biosciences, UK). Total antioxidant capacity in Trolox equivalents have been measured using Total Antioxidant Capacity Assay Kit (MAK187, Sigma-Aldrich).
2.8. Maintenance of Cell Lines
Human Colorectal Adenocarcinoma Cell Line HT-29 and mucous-secreting HT-29-MTX (both purchased from Sigma-Aldrich) were propagated using low glucose DMEM medium supplemented with 10% Fetal Bovine Serum, 2 mM L-glutamine, 100 U/mL penicillin, and 100 µg/mL streptomycin in a 5% CO2 atmosphere at 37 °C. All cell culture reagents were purchased from Capricorn Scientific, Ebsdorfergrund, Germany. Pro-monocytic human cell line U937 obtained from the American Type Culture Collection (ATCC, CRL-1593.2) and was routinely cultured in RPMI-1640 medium containing 10% FBS, 2 mM L-glutamine, 100 U/mL penicillin, and 100 µg/mL streptomycin under conditions indicated above. All base media and supplements were purchased from Thermofisher.
2.9. Cell Viability Assays
Cell viability assays were carried out using CyQUANT™ XTT Cell Viability kit (Invitrogen™), according to manufacturer’s instructions. Prior to experiments, HT-29 cells were seeded onto 96-well plate at a density 1 × 105 cell/well. The day after seeding, cells were washed twice with 200 µL of DPBS; 100 µL of prewashed bacteria (107 CFU/mL–108 CFU/mL) were added to the wells. Full culture medium containing 2.5% ethanol was added to the control wells (positive control). Twenty hours after exposure to bacteria HT-29 cells were washed twice with 200 µL of DPBS; 100 µL of full medium containing antibiotics were added to the wells. HT-29 cells were then stained with 0.3 mg/mL solution of XTT (sodium 3′-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis (4-methoxy6-nitro) benzene sulfonic acid hydrate) for 4 h. Absorbance was detected at 450 and 595 nm.
In a parallel experiment, HT-29 cells pretreated with B. clausii CSI08 were subjected to three rounds of washing with 200 µL of DPBS; after that HT-29 cells were exposed to H2O2 added to a final concentration of 4 mM. After 24 h of incubation staining with XTT solution was performed as described above.
2.10. Adhesion Assays
HT-29-MTX cells were seeded onto 24-well plates at a density of 5 × 105 cell/well and cultured for 21–28 days to complete maturation. Media was replaced every 2–3 days. Four hundred microliters of full media (low glucose DMEM, 10% Fetal Bovine Serum, 2 mM glutamine) without antibiotics were added to the wells allocated for bacteria; DPBS was aspirated from the wells allocated for spores after the second round of washing. One hundred microliters of pre-washed bacteria in DMEM (2.0 × 107 CFU/mL–1.2 × 108 CFU/mL) or 500 µL of spores suspensions (4.0 × 107–9.0 × 107 CFU/mL) were added to the cells, mixed by a gentle swirl, and incubated for 2.5 h at 37 °C in a 5% CO2 atmosphere. Control wells not containing mammalian cells were prepared and incubated in parallel in the same way. Upon incubation HT-29-MTX cells were washed 4 times with 0.5 mL PBS. After that, 50 µL of Trypsin/EDTA solution (CC-5012, Lonza, Switzerland) and 50 µL of PBS were added to the wells and incubated for 10 min with gentle shaking (~100 rpm) at 37 °C. Fifty microliters of Trypsin/EDTA solution were added to control wells. Consequently, 450 µL of PBS were added to the wells with bacteria or spores, contents of the wells were transferred into microcentrifuge tubes with scrapping and subjected to three rounds of vigorous shaking 30 s each. Contents of control wells were transferred into microcentrifuge tubes and subjected to one round of shaking. Serial dilutions (plus dilutions of control wells) were prepared in PBS and plated onto PetriFilm™ or MRS agar plates for quantification of B. clausii CSI08 or L. fermentum.
2.11. Anti-Inflammatory Activity Assays
HT-29 cells were seeded onto 24-well plates at a density of 5 × 105 cell/well. The day after seeding cells were washed twice with 0.5 mL DPBS; 0.4 mL of full cell culture media without antibiotics were added to the wells allotted for pretreatment with viable bacteria. Alternatively, 0.25 mL of full cell culture media containing antibiotics were added to the wells allotted for pretreatment with bacterial supernatants. One hundred microliters of pre-washed bacterial cells (108 CFU/mL) or 0.25 mL of cell free supernatants were added to the corresponding wells. After 20 h of incubation (CO2 atmosphere at 37 °C), HT-29 cells were washed twice with 0.5 mL of DPBS and 0.5 mL of full cell culture medium containing antibiotics were added to the wells. Lipopolysaccharides from E. coli O111:B4 (LPS, Sigma L4391) were added to HT-29 cells pretreated with viable bacteria and cell free supernatants to final concentration of 15 ng/mL. Poly I:C (P9582, Sigma) was added to pretreated HT-29 cells to final concentration of 10 µg/mL. Three or four hours after adding LPS or poly I:C, cell culture supernatants were removed and HT-29 cells were lysed in the wells by adding 300 µL of lysis buffer supplied with Monarch Total RNA Miniprep Kit (NEB, MA, USA).
2.12. RNA Extraction, Reverse Transcription, and Quantitative Real-Time PCR
Total RNA was extracted from cell lysates using Monarch Total RNA Miniprep Kit (NEB, MA, USA), according to manufacturer’s instructions. Then, fifty microliters of nuclease-free water were taken to elute RNA. A Qubit™ RNA broad range kit was used to quantify RNA after isolation. Next, one microgram of total RNA was taken to set up reverse transcription reactions using Luna script RT Supermix kit (NEB, MA, USA). Real-time PCR reactions were set up using Luna® Universal qPCR Master Mix (NEB, MA, USA) and appropriate primer pairs (see Table S1) at a concentration of 200 nM using 1 µL of generated cDNA per 9 µL of master mix. The reactions were performed in duplicates using the following program: initial denaturation 95 °C 5 min, denaturation 94 °C 20 s, annealing 60 °C 20 s, extension 72 °C 20 s (40 cycles). The specificity of reaction products was confirmed by melting temperature analysis (from 70 °C to 95 °C in 0.5 °C/15 s increments). Quantification of target transcripts was done using gapdh as a normalizing house-keeping gene.
2.13. Human Nulcear Factor-κB (NF-κB) p65 Transcription Factor Activity Assay
HT-29 cells were seeded onto 6-well plates at a density of 2 × 106 cell/well. The day after seeding, cells were washed twice with 2 mL of DPBS; 1.6 mL of full cell culture media without antibiotics and 0.4 mL of pre-washed as described above (see Section 2.1) B. clausii CSI08 suspension in DMEM were added to the corresponding wells. Two milliliters of media were added to the control wells. After 20 h of incubation (37 °C CO2 atmosphere), HT-29 cells were washed two times with 2 mL of DPBS and 2 mL of full media containing antibiotics were added to all the wells. LPS were added to the wells pretreated with bacteria and to the control wells to a final concentration of 15 ng/mL. HT-29 cells were collected with 1 mL of ice-cold PBS by scrapping 45 min after adding LPS. Nuclear fractions were extracted using ProteinExt® Mammalian Nuclear and Cytoplasmic Protein Extraction Kit (TransGen Biotech, Beijing, China) according to the manufacturer’s instructions with one modification. Nuclear fractions were collected with 150 µL of NPEB buffer, instead of 500 µL indicated in the supplied manual. Total protein concentration was thereafter determined using Bicinchonic Acid Protein Assay Kit (B9643, Sigma). The nuclear fractions normalized by total protein concentration (~25 µg of protein) were taken to evaluate NF-κB activity using the NF-κB activity assay kit (TFEH-p65-1, RayBiotech, GA, USA) according to the manufacturer’s instructions. Experiments were performed two times with three or four technical replicates per assay.
2.14. Macrophage Differentiation and Challenge Study
U937 cells were seeded at a concentration of 105 cells/well in 96-well plates and subjected to macrophage differentiation upon 50 ng/mL phorbol 12-myristate 13-acetate (PMA) treatment for 72–96 h. Macrophages were then stimulated with 108 CFU/mL of B. clausii CSI08 or 5 ng/mL LPS in complete RPMI-1640 medium without antibiotics for 5 h at 37 °C. Afterwards, cell supernatants were harvested and stored at −20 °C until cytokine determination. The analysis was carried out using Luminex 200™ System according to the manufacturer’s instructions with a cytokine panel from Thermofisher including IL-1β, IL-18, IL-6, TNF-α, GM-CSF, G-CSF, IL-10, IL-1RA and EGF.
2.15. C. elegans Culture Conditions and Experiments
Caenorhabditis elegans strains N2, Bristol (wild type) was obtained from the Caenorhabditis Genetics Center at the University of Minnesota and maintained at 20 °C on Nematode Growth Medium (NGM) plates with E. coli strain OP50 as normal diet for nematodes. Overnight, B. clausii CSI08 culture was grown, as described in Section 2.1, and then centrifuged for 10 min at 4000× g. Supernatants were then removed, and bacterial pellets were washed once with saline solution. C. elegans wild-type strain (N2) cultured in NGM (control fed condition), or NGM supplemented with B. clausii CSI08 at two different doses (108 and 109 cells/plate). Vitamin C (10 µg/mL) was used as positive control. Worms were incubated in these conditions and after several days were submitted to an acute oxidative stress (2 mM H2O2) according to a previously published protocol [46]. Afterwards, viability of nematodes was determined in each fed condition. Experiments were performed in duplicate. Each experiment was conducted on 5 different plates, each containing 10 worms (50 worms/assay). The antioxidant activity (worm survival) of total population was calculated. Final survival data correspond to the average of two independent assays (total population of 100 worms/condition). The effect of B. clausii CSI08 antioxidant activity was studied by comparing the survival of treated nematodes versus the control-fed nematodes.
2.16. Statistical Analysis
All data were analyzed using Prism 9 (GraphPad Software, San Diego, CA, USA). Normal distribution was determined using Shapiro-Wilk test. Samples following normal distribution were tested for significance using unpaired t test or one-way ANOVA with Tukey, Bonferroni or Dunnets post-hoc as relevant. When samples did not follow normal distribution, a Mann-Whitney U test or Kruskal-Wallis with Dunn’s post-hoc was performed.
3. Results
3.1. In Vitro Evaluation of the Probiotic Properties of B. clausii CSI08
3.1.1. Resistance to an In Vitro Simulated Gastric and Intestinal Conditions
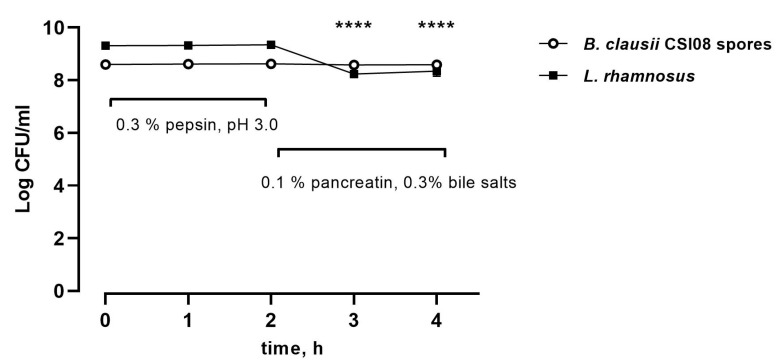
The ability of B. clausii CSI08 spores to survive during an in vitro simulated digestion process was compared to that of the commercial L. rhamnosus GG (ATCC 15103) strain. As shown in Figure 1, there was no decrease in the B. clausii CSI08 spores count after exposure to simplified gastric and small intestinal conditions. At the same time, we detected a significant drop in viable bacterial counts at the intestinal stage (0.1% pancreatin, 0.3% bile salts). These data indicate the potential ability of B. clausii CSI08 spores to efficiently survive the transit through the upper digestive tract.
Figure 1.
Survival of B. clausii CSI08 spores and L. rhamnosus ATCC 53103 at gastric and small intestinal digestion conditions simulated in vitro. The values are expressed in Log10 CFU/mL. Data represent the mean (n = 8) ± SEM of two independent experiments performed in four technical replicates. **** p < 0.0001 Survival of L. rhamnosus compared to 0 h.
3.1.2. Stability of B. clausii CSI08 Spores during Pasteurization
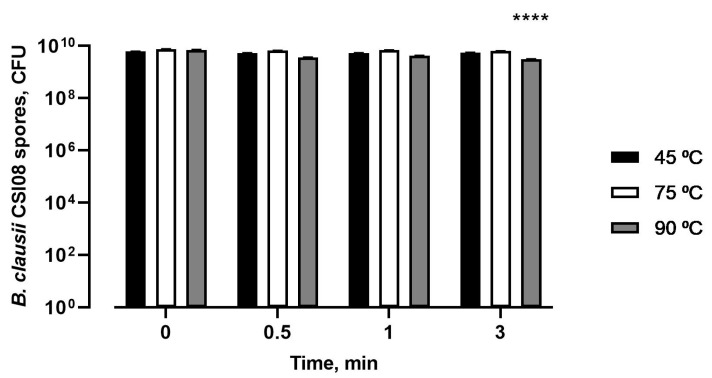
The temperature stability of B. clausii CSI08 spores was assessed at various conditions (45, 75, and 90 °C) at three different time points (0.5, 1, and 3 min) in PBS. The summarized results are shown in Figure 2. No change in viability was detected at 45 °C or 75 °C at all three time points. Yet, we recorded the reduction in spore counts after treatment at 90 °C from the 6.87 × 109 to 3.01 × 109 CFU. Nevertheless, B. clausii CSI08 spores showed good overall performance at pasteurization conditions.
Figure 2.
Thermostability of B. clausii CSI08 spores at 45, 75, and 90 °C at different time intervals. The values are expressed in CFU of spores. Data represent the mean (n = 9) ± SEM of three independent experiments performed in triplicate. Spore counts compared to 0 min. **** p < 0.0001.
3.1.3. Antimicrobial Activity
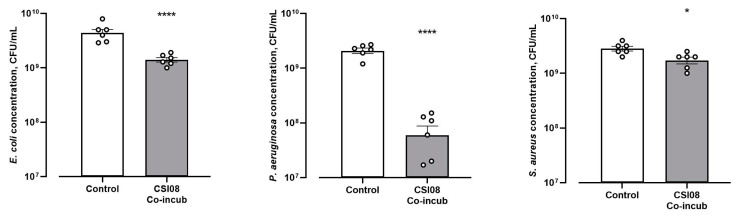
Antimicrobial activity B. clausii CSI08 in its vegetative form was evaluated for antimicrobial activity against four strains of E. coli, S. enteritidis, S. aureus, and P. aeruginosa. The results shown in Figure 3 demonstrate the reduction in bacterial counts of three tested pathogens grown in the presence of B. clausii CSI08. At the same time, under conditions indicated above B. clausii, CSI08 was unable to inhibit growth of S. enteritidis.
Figure 3.
Antimicrobial activity B. clausii CSI08 in liquid culture. Values represent average concentration ± SEM of two independent experiments with three technical replicates (n = 6). * p < 0.05, **** p < 0.0001.
3.1.4. Enzymatic Capacity of B. clausii CSI08
The carbohydrate assimilation pattern determined using the API 50 CH assay kit is shown in Table S2. B. clausii CSI08 was able to metabolize various simple carbohydrates including glycerol, L-arabinose, D-ribose, D-glucose, D-fructose, D-mannose, L-rhamnose, as well as disaccharides, including saccharose, cellobiose, trehalose, with moderate ability to metabolize polymeric amidon and glycogen.
Enzymatic profile of B. clausii CSI08 detected using the API ZYM kit is presented in Table S3. We detected activities of esterase-lipase C8 and esterase C4, hydrolyzing lipids into free glycerol and unsaturated and saturated fatty acids. Notably, B. clausii CSI08 was able to produce β-galactosidase, the enzyme that catalyzes lactose hydrolysis into glucose and galactose and, importantly, is responsible for formation of galacto-oligosaccharides (GOS) promoting the growth of Bifidobacterium and Lactobacillus species [47].
3.1.5. Amino Acids and Water-Soluble Vitamins in B. clausii CSI08 Supernatants
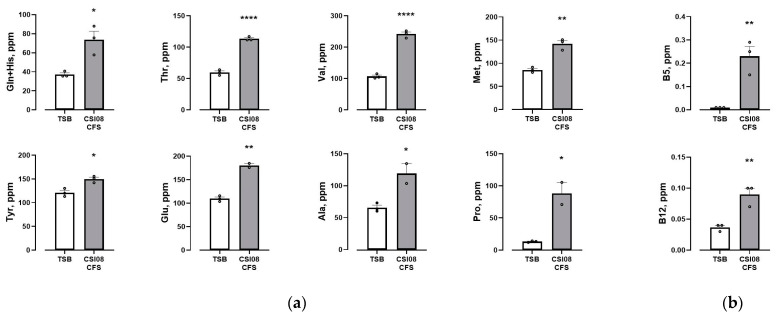
Furthermore, we performed an exploratory study to assess the ability of B. clausii CSI08 to produce amino acids and vitamins. The levels of 22 amino acids and water-soluble vitamins were determined in overnight cultures of B. clausii CSI08. The analysis showed the higher levels of glutamic acid, alanine, glutamine and histidine mixture, threonine, proline, tyrosine, valine, and methionine in bacterial supernatants compared to TSB medium (Figure 4a). These data suggest the capacity of B. clausii CSI08 to synthesize the above-mentioned amino acids during their growth in rich medium. At the same time, the elevated concentration of amino acids might be the result of a peptidase activity of the strain. We further detected high levels of pantothenic acid (B5) and cobalamin (B12) in B. clausii CSI08 supernatants (Figure 4b), which supports the assertation of the strain to produce vitamins.
Figure 4.
(a) Amino acids and (b) vitamins in B. clausii CSI08 supernatants (CFS) determined by HPLC-FLD and UHPLC-MS correspondingly. Values are means (n = 3) ± SEM. * p < 0.05, ** p < 0.01, **** p < 0.0001.
3.1.6. Assessment of Safety of B. clausii CSI08 and Adhesion to Mucous-Producing Cell Line HT-29-MTX
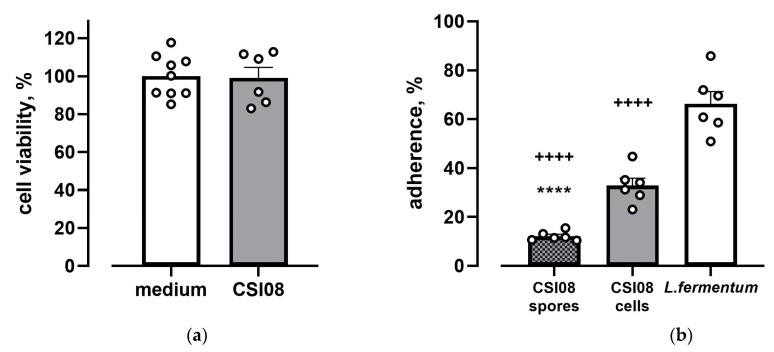
To evaluate any potential cytotoxic effect of vegetative cells B. clausii CSI08 on HT-29 intestinal cells, the XTT assay has been carried out 20 h after bacterial exposure. Approximate multiplicity of infection (MOI) in the assay constituted 1:500. As shown in Figure 5a, B. clausii CSI08 did not negatively impact the survival of intestinal epithelial cells at given MOI. The adhesion ability of B. clausii CSI08 and its spore preparations were compared to that of a commercial probiotic L. fermentum strain (Figure 5b). Both forms (cells and spores) of B. clausii were able to adhere to HT-29-MTX cells. Percentage of adhesion of the vegetative cells was approximately three times higher than that of the spores. L. fermentum was shown to adhere to the mucous-producing cells twice as efficiently as vegetative cells of B. clausii.
Figure 5.
Interaction of B. clausii CSI08 with intestinal epithelial cells. (a) B. clausii CSI08 did not reduce HT-29 viability. Data represent percentage of XTT dye conversion after 20 h of coincubation with B. clausii CSI08 compared with untreated cells (medium). Values are means ± SEM of three independent experiments. (b) Adhesion of vegetative cells and spores B. clausii CSI08 to the HT-29-MTX. Values represent percentage of adhered cells/spores after 2.5 h of incubation with HT-29-MTX followed by four rounds of washing. Values are means (n = 6) ± SEM of three independent experiments **** p < 0.0001 vs. CSI08 cells, ++++ p < 0.0001 vs. L. fermentum.
3.2. Immunomodulatory Effect of B. clausii CSI08 in Human Cell Lines
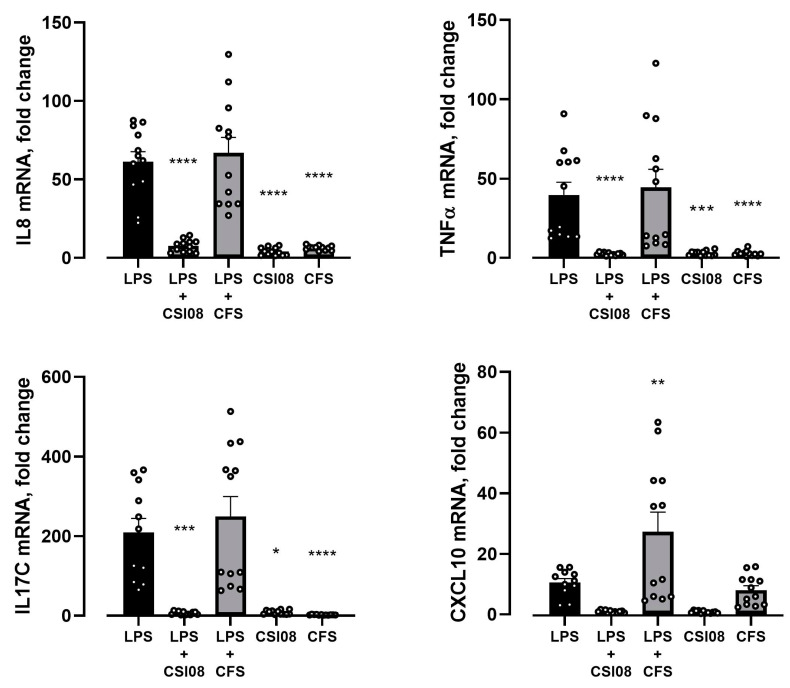
The vegetative form of B. clausii CSI08 and its cell-free supernatants were assessed for their ability to modulate pro-inflammatory response in HT-29 cell line, triggered by E. coli lipopolysaccharides (LPS) or Poly I:C, a synthetic double-stranded RNA ligand. Expression level of IL-8, TNF-α, IL-17C, CXCL10 genes have been evaluated at mRNA level using qRT-PCR. As shown in Figure 6, expression of all 4 genes was strongly induced 4 h after stimulation with LPS. Pretreatment with B. clausii CSI08, but not with its cell-free supernatants, greatly inhibited LPS-triggered pro-inflammatory response. Notably, neither the vegetative cells of B. clausii or their supernatants by themselves elicited a significant increase in IL-8, TNF-α, IL-17C, CXCL10 gene expression after co-incubation with HT-29 cells (Figure 6).
Figure 6.
B. clausii CSI08 attenuated LPS-induced pro-inflammatory response in HT-29 cell line. qPCR analysis of IL-8, TNF-α, IL-17C, and CXCL10 gene expression 4 h after exposure to 15 ng/mL LPS in HT-29 cells preincubated with B. clausii CSI08 or its cell free supernatants (CFS). * p < 0.05 vs. LPS, ** p < 0.01 vs. LPS, *** p < 0.001 vs. LPS, **** p < 0.0001 vs. LPS. The pattern of gene expression determined after co-incubation of HT-29 cells with B. clausii CSI08 and its CFS in unstimulated conditions is also shown. Results show mean ± SEM of three independent experiments.
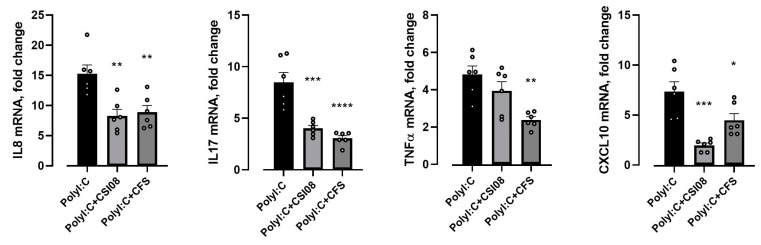
When the pro-inflammatory changes in HT-29 cell line were triggered by Poly I:C, both cells and cell-free supernatants of B. clausii CSI08 have been shown to attenuate the overexpression of IL-8, TNF-α, IL-17C, and CXCL10 genes (Figure 7).
Figure 7.
B. clausii CSI08 attenuated the Poly I:C-triggered pro-inflammatory response in HT-29 cell line. qPCR analysis of IL-8, TNF-α, IL-17C, and CXCL10 gene expression 4 h after exposure to 10 µg/mL Poly I:C in HT-29 cells preincubated with B. clausii CSI08 or its cell free supernatants (CFS). * p < 0.05, ** p < 0.01; *** p < 0.001, **** p < 0.0001. Results show mean (n = 6) ± SEM.
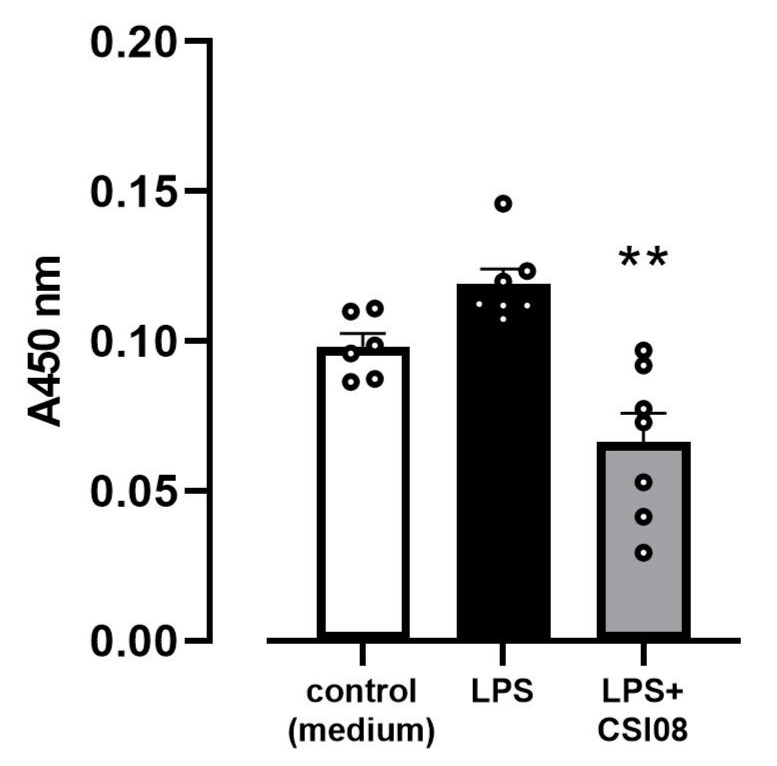
In order to confirm that inhibitory effect of B. clausii CSI08 on the expression of proinflammatory marker genes was associated with attenuation of NF-κB, we investigated the activity of the transcription factor in the nuclear fractions of HT-29 cells preincubated with viable bacteria 45 min after adding LPS. As shown in Figure 8, B. clausii CSI08 significantly counteracted the LPS-triggered activation of NF-κB.
Figure 8.
B. clausii CSI08 down-regulated the LPS-stimulated activation of NF-κB in HT-29 cell line. Adsorption values correspond to the levels of the transcription factor in the nuclear fractions of control cells, cells exposed to 15 ng/mL of LPS for 45 min, cells pretreated with B. clausii CSI08 for 20 h prior to adding LPS. Values are means ± SEM of two independent experiments. ** p < 0.01 vs. LPS.
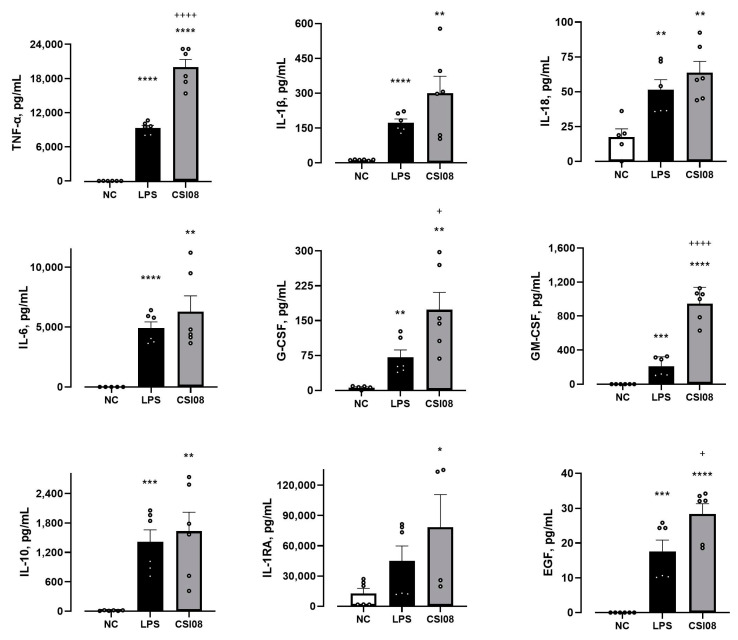
Subsequently, we investigated the effect of B. clausii CSI08 on the innate immune system, adopting the cell model of U937-derived macrophages. The latter were stimulated with 108 CFU/mL of vegetative cells B. clausii CSI08, followed by quantification of cytokines/chemokines 5 h after exposure to bacteria. The concentrations of pro-inflammatory TNF-α, IL-1β, IL-18, regulatory G-CSF, GM-CSF, IL-6, and anti-inflammatory IL-10, IL-1RA, EGF were determined. The response to E. coli lipopolysaccharides has been assessed in parallel experiments. As shown in Figure 9, B. clausii CSI08 provoked a robust cell response, resulting in secretion of high levels of all analyzed cytokines.
Figure 9.
Immunostimulatory effect of B. clausii CSI08. Cytokine levels in cell culture supernatants of U937-derived macrophages: untreated (NC) or challenged with 108 CFU/mL of vegetative cells of B. clausii CSI08 or LPS for 5 h. Values are the means ± SEM of three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001; **** p < 0.0001 LPS or CSI08 vs. NC; + p < 0.05; ++++ p < 0.0001 CSI08 vs. LPS.
3.3. Antioxidant Capacity of B. clausii CSI08 In Vitro and In Vivo
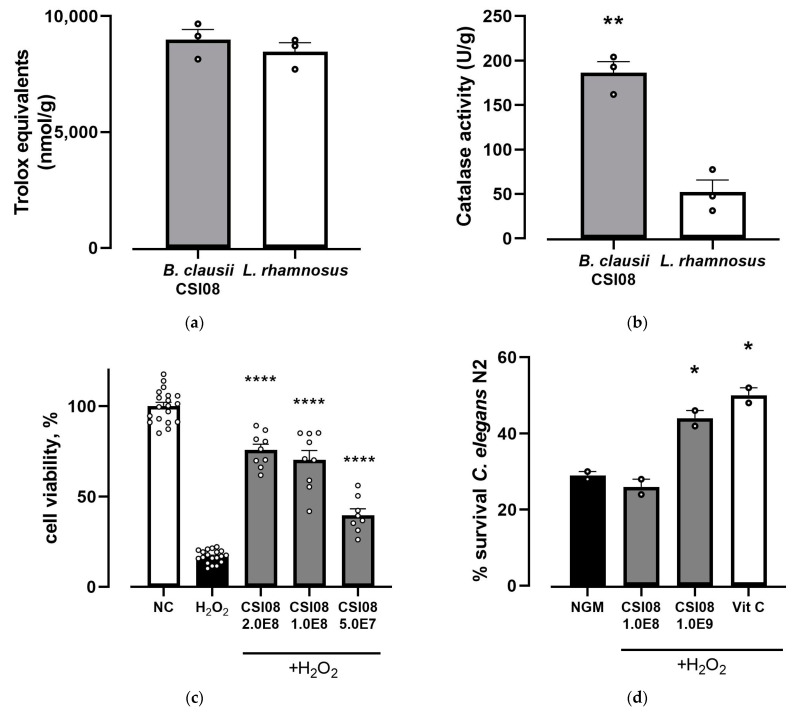
The total antioxidant capacity and the catalase activity of B. clausii CSI08 cell lysates were compared to those of L. rhamnosus GG. Strong antioxidant properties of the abovementioned strain have been acknowledged by a number of studies [48]. B. clausii CSI08 and L. rhamnosus GG exhibited a similarly high level of total antioxidant activity (Figure 10a). In contrast, the level of catalase activity was significantly higher in B. clausii CSI08 cell lysates as compared with the reference L. rhamnosus strain (Figure 10b).
Figure 10.
(a) Total antioxidant capacity of B. clausii CSI08 and L. rhamnosus GG cell lysates in Trolox equivalents (nmol/g). Presented values are means (n = 3) ± SEM. (b) B. clausii CSI08 demonstrated superior catalase activity (in U/g) as compared with L. rhamnosus GG cell lysates. Values are means (n = 3) ± SEM. ** p < 0.01. (c) Cytoprotective effect of B. clausii CSI08 on H2O2-exposed epithelium. Data represent percentage of XTT dye conversion 20 h after exposure to 4 mM H2O2 by HT-29 cells preincubated with 2.0 × 108, 1.0 × 108, 5.0 × 107 CFU of B. clausii CSI08 compared with control group (NC). The values are the means ± SEM. **** p < 0.0001 vs. H2O2. (d) Survival rate of C. elegans N2 fed with 108 and 109 CFU/mL B. clausii CSI08 followed by an acute oxidative stress caused by 2 mM H2O2. NGM—control-fed nematodes; Vitamin C as a positive control. Values presented are the average of two independent experiments (n = 100/condition). * p < 0.05.
The antioxidant potential of B. clausii CSI08 has been confirmed using the cell model of oxidative damage. The viability rate of HT-29 epithelial cells treated with hydrogen peroxide has been compared with that of the cells preincubated with B. clausii CSI08 taken at three different concentrations prior to adding H2O2. A significant reduction in viability was observed in HT-29 cells exposed to H2O2. Pretreatment with 2.0 × 108, 1.0 × 108, 5.0 × 107 CFU of B. clausii CSI08 partially restored the impaired viability of epithelial cells. Summarized results are shown in Figure 10c. To further characterize antioxidant properties of B. clausii CSI08, the in vivo C. elegans model was employed. Nematodes were fed with vegetative cells at two different concentrations and after that subjected to acute oxidative stress with hydrogen peroxide. The survival rate was compared to that of the control group nematodes (fed with nematode growth medium only). As shown in Figure 10d, B.clausii CSI08 added to the worm diet caused an increase in the survival of the C. elegans population after exposure to oxidative stress in a dose-dependent manner, being 109 cells/ml the functional dose, with a survival of 44% compared to 29% of the control fed condition.
4. Discussion
Spore-forming bacteria of the genus Bacillus represent an ample and remarkably diverse group of microorganisms. Their capacity to produce a wide variety of secondary metabolites, such as antibiotics, enzymes, vitamins, and carotenoids, created a huge potential for use in pharmaceutical, agricultural, and industrial processes [49,50,51]. Bacillus strains have been recognized as safe and efficacious probiotics, offering, in addition to classic probiotic traits, notably high stability during processing and storage [52], as well as relatively low production costs [53]. Microorganisms of B. clausii group stand out from other Bacillus species featuring a distinctive ability to modify immune response in vitro and in vivo [54]. In the present work we introduced the novel strain of B. clausii CSI08, with a focus on immunomodulatory properties of this potential probiotic.
The ability to withstand low pH and the presence of high concentration of bile salts is one of the fundamental properties of probiotic strains [55]. Generally recognized as being highly resistant to extreme environmental stress, spores can struggle to maintain viability after exposure to bile salts [56]. Therefore, assessment of spores’ survival under digestion-related stress is a crucial step in a spore-based probiotic selection pipeline. B. clausii CSI08 spores demonstrated excellent resistance to the simulated gastric and small intestinal environment, indicating their potential ability to survive passage through upper gastrointestinal tract. Previous findings have demonstrated that spore formulations of the well-known B. clausii probiotic strains can tolerate pH 2 and up to 1% conjugated bile salts and, moreover, can germinate and multiplicate under mimicked human intestinal environment [39,57,58]. Similarly, we demonstrate herein that B. clausii CSI08 spores can survive simulated gastric and small intestinal conditions.
Adherence to the intestinal mucosa is another essential characteristic of potential probiotics. Stable binding to the intestinal epithelium helps bacteria and spores to avoid their quick removal by peristalsis. Even transient colonization of mucosa by probiotic strains leads to competitive exclusion of pathogens [59] and allows them to interact with the local immune system [60]. Given the critical role of mucins in adhesion [61,62], we have investigated the ability of vegetative cells and spores of B. clausii CSI08 to adhere to the mucous-producing cell line HT-29-MTX. B. clausii CSI08 in both live states showed the strong adhesive capacity, somewhat lower than that of the control L. fermentum strain. In the study published by Ahire JJ et al. [58], spores of B. clausii UBBC07 demonstrated significantly higher adhesion levels to porcine mucin compared to vegetative cells, conversely to our results. Genome sequencing analysis of the probiotic B. clausii strains [37,38] revealed the presence of genes encoding for adhesion-related proteins such as mucus-binding protein [63], enolase [64], flagellar hook-associated protein [65], and exopolysaccharide biosynthetic gene clusters [66]. Moreover, strains of B. clausii/ Enterogermina™ have been recovered from feces of healthy volunteers twelve days after single administration of probiotic preparations, which indirectly suggests the ability of B. clausii strains to temporarily colonize the intestine [67].
The enzymatic profiling conducted In the course of present study revealed β-galactosidase activity of B. clausii CSI08. Release of this enzyme, responsible for the hydrolysis of lactose, is an important feature of probiotics, since it has been involved in the relief of lactose intolerance symptoms [68]. Another significant trait of B. clausii CSI08 is the lack of detrimental β-glucuronidase, α-chymotrypsin, and β-glucosaminidase activities. Beta-glucuronidase plays a critical role in colorectal carcinogenesis, being implicated in reactivation of carcinogen metabolites [69,70], α-chymotrypsin and β-glucosaminidase, produced by gut bacteria, are among important factors in the pathogenesis of endocarditis [71].
Antimicrobial activity is another undoubtedly essential feature of potentially probiotic bacterial strains. We showed the capacity of B. clausii CSI08 to inhibit the growth of opportunistic pathogens, E. coli, S. aureus, and P. aeruginosa, during co-cultivation in liquid culture. Probiotic strains of B. clausii are known to produce substances with antimicrobial activity; some of them have been identified and characterized. Biological target of the lantibiotic clausin, first discovered in the cell-free supernatants of B. clausii O/C, are lipid intermediates essential for the biosynthesis of peptidoglycan and other bacterial cell wall polymers [43,72]. Clausin has been shown to be active against a range of Gram-positive bacteria including C. difficile [39,43]. Recently, Ahire JJ et al. demonstrated the production of class I lantibiotic clausin by B. clausii UBBC07 in the in vitro Simulator of Human Intestinal Microbial Ecosystem (SHIME) model [39]. Interestingly, production of antimicrobial substances active against Gram-negative and Gram-positive species have been observed upon whey fermentation by B. clausii [36], indicating the importance of starting substrates for synthesis of antimicrobials. Another remarkable example of indirect antimicrobial activity has been reported by Ripert G et al. in 2016 [35]. The study describes the serine protease produced during the sporulation phase by B. clausii O/C that can abolish the cytotoxic effects of toxigenic strains of C. difficile and B. cereus.
It has been shown that gut bacteria can influence the bioavailability of amino acids to the host organism [73]. Adsorption of amino acid is highly efficient in the small intestine [74], whereas in the colon amino acids are effectively metabolized by the microbiota [75]. The metabolic end products amongst others include short chain fatty acids and branched-chain fatty acids, that in turn significantly effect physiology of epithelial cells and the mucosal immune system [76]. We have demonstrated that growth of B. clausii CSI08 in rich medium was associated with an increase in concentration of eight essential amino acids. Further studies are needed to confirm the nature of the observed effect (de-novo synthesis or proteolytic activity). Furthermore, we have reported that B. clausii CSI08 can potentially synthesize pantothenic acid and cobalamin. Recent studies suggest an immunostimulatory role of pantothenic acid in the context of anticancer therapy [77]. Interestingly, vitamin B5 and its metabolite CoA, vital for metabolism of fatty acids, have been recently shown to increase mitochondrial metabolism of T cells and subsequently their anti-tumor efficacy [78]. Vitamin B12, amongst multiple biological functions, have been shown to regulate cellular immunity by increasing activity of CD8+ T lymphocytes and natural killer (NK) cells [79].
Disruption of the normal microbial community structure, environmental factors such as stress, and diet, as well as genetic factors, can trigger chronic intestinal inflammation [60]. The ability of B. clausii CSI08 to attenuate inflammatory response induced by LPS and Poly I:C, interacting with TLR4 [80] and TLR3 [81] receptor complexes correspondingly, was evaluated in the course of the present work. Pretreatment of HT-29 cells with vegetative B. clausii CSI08 attenuated the LPS-induced expression of pro-inflammatory (IL-8, TNF-α, IL-17C) genes. We also confirmed that this effect was associated with down-regulation of the canonical NF-κB transcription factor [82]. Both cells and cell-free supernatants of B. clausii CSI08 attenuated the pro-inflammatory response triggered by Poly I:C. Similarly, the protective effect of B. clausii (O/C, T, SIN and N/R) strains against Rotavirus-induced changes in Caco-2 cell line of human enterocytes have been demonstrated in the recent study published by Paparo L et al. [83]. Moreover, in a murine model of chronic colitis administration of B. clausii (O/C, T, SIN and N/R) resulted in reduction of the colonic inflammatory score [84]. Another strain of B. clausii, MTCC8326, was found to be effective in attenuating Salmonella typhimurium-associated dysbiosis and inflammation in Th2 (BALB/c)-biased mice [85].
It has been proven that certain probiotic strains can enhance innate immune response via stimulation of macrophages and dendritic cells located in the lamina propria [86,87,88]. Vegetative cells of B. clausii CSI08 were able to elicit the robust response in U937-derived macrophages, resulted in markedly increased production of pro-inflammatory (TNF-α, IL-1β, IL-18) and anti-inflammatory (IL-10, IL-1RA, EGF) cytokines. This is in agreement with Pradhan B et al. [41] who observed the induction of a pro-inflammatory response at earlier time points and an anti-inflammatory response at later time points after exposure murine RAW 264.7 macrophages to B. clausii MTCC-8326. Villéger et al. [89] have proposed the essential role of D-alanine of lipoteichoic acids for the immunomodulatory properties of probiotic strains of B. clausii. Another remarkable example of involvement of B. clausii in the immune activation, that can influence clinical outcomes in patients with pancreatic adenocarcinoma, has been described by Riquelme E et al. [90].
The antioxidant capacity of probiotics and potential health-promoting effects related to it have been widely studied in recent decades [91]. B. clausii CSI08 showed strong antioxidant activity in vitro, and, moreover, in its vegetative form exerted cytoprotective effect in cell culture and C. elegans models of oxidative stress, likely through the reduction of reactive oxygen species. Publications demonstrating the antioxidant capacity of B. clausii strains are limited and reported only in cell lines and murine studies. In a rat model of uremia, administration of B. clausii UBBC07 was associated with alleviation of oxidative response induced by acetaminophen, evidenced by increased levels of glutathione, superoxide dismutase, and catalase [92]. Similarly, in a rat trinitrobenzenesulfonic acid (TNBS)-induced colitis model treatment with B. clausii was partially protective against oxidative damage [93]. Finally, B. clausii have been shown to suppress Rotavirus-induced production of reactive oxygen species in Caco-2 cell line [83].
5. Conclusions
The study presented the novel strain of B. clausii CSI08 with prominent probiotic characteristics. Spore preparations of B. clausii CSI08/Munispore® demonstrated high level of resistance to gastric pH and high bile salts concentration. The vegetative form of the strain showed significant ability to suppress the growth of pathogenic bacteria in liquid culture, was capable of assimilating a wide range of carbohydrates and was shown to produce potentially beneficial enzymes. B. clausii CSI08 did not demonstrate cytotoxic effect towards intestinal epithelial cells and displayed moderate adhesion capacity in both live states. Additionally, we showed that spores of B. clausii CSI08 can effectively survive an in vitro simulated pasteurization process. Furthermore, B. clausii CSI08 exhibited a strong ability to attenuate LPS- and Poly I:C-triggered inflammatory response in the intestinal epithelial cell line and to enhance the innate immunity via macrophage stimulation. Moreover, this novel strain displayed excellent antioxidant characteristics when studied in vitro and in vivo. Taken together, the data herein provide strong evidence for potential therapeutic efficacy for B. clausii CSI08 as a promising probiotic strain.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11020240/s1, Table S1: Primers used in the study; Table S2: Carbohydrate assimilation profile of B. clausii CSI08 using API 50 CH system; Table S3: Enzymatic profiles of B. clausii CSI08 using the API-ZYM system.
Appendix A. HPLC-FLD Determination of Amino Acids
Materials. AccQ-Tag chemistry package for the amino acid determination was purchased from Waters Corp. (Milford, MA, USA). The 22 amino acids determined (aspartic acid, asparagine, glutamic acid, glycine, arginine, alanine, serine, glutamine, threonine, proline, cysteine, tyrosine, histidine, cystine, valine, ornithine, isoleucine, methionine, lysine, leucine, tryptophan and phenylalanine) were obtained from MilliporeSigma (Burlington, MA, USA).
Analytical procedures. Liquid chromatographic separation was performed on an Acquity Arc system (Waters, Milford, MA, USA) coupled to a fluorescence detector (FLD 2475, Waters) controlled by the Empower 3 software (v. 7.4) (Waters, Milford, MA, USA). Sample preparation includes a filtration through 0.22 µm PVDF filters followed by an aqueous dilution in the range 1/10 to 1/160, to adjust the results to the range of the standard curves. Additionally, prior to chromatographic analysis, samples were derivatized using AccQ-Tag Ultra derivatization reagent (AQC) according to the manufacturer’s instructions. Briefly, a 20 µL sample was mixed with 60 µL borate buffer (pH 8.8), then a 20 µL AccQ-tag Ultra derivatization reagent was added to each sample and shaken vigorously. Finally, they were kept at room temperature for 1 min and incubated at 55 °C for 10 min. The separation of the derivatized amino acids was carried out on an AccQ-Tag column (Nova Pak C18 4 µm; 3.9 × 150 mm, Waters, Milford, MA, USA) guarded by Nova-Pak C18 inline filter (4 µm; 3.9 × 20 mm, Waters, Milford, MA, USA). Solvent A was 10% (v/v) AccQ-Tag eluent A Concentrate in milli-Q filtered water, solvent B was HPLC-grade acetonitrile and solvent C was milli-Q filtered water. A 36 minute long gradient (0–1% 0.5 min, 1–5% 17.5 min, 5–9% 1 min, 9–17% 10.5 min, 17–60% 3.5 min and 60–0% 3 min) was established with 1 mL/min flow rate and 37 °C column temperature. The subsequent steps corresponded to the cleaning and final column equilibration stage. The injection volume was 2 µL. Fluorescence detection was carried out by λ irradiation at 250 nm excitation and 395 nm emission wavelength.
Peaks were identified by comparison to retention times of analytical standards and quantified using standard curves of the different amino acids (asparagine co-elutes with serine and glutamine with histidine). The chromatographic standards were prepared in milli-Q filtered water at a final concentration of 10 and 5 ppm, except tryptophan, which was 200 and 100 ppm. The calibration curves were prepared from the stock solutions through serial dilutions [94,95].
Appendix B. UHPLC-MS Determination of Water-Soluble Vitamins
Materials. Water-soluble vitamins (Thiamine hydrochloride, Riboflavin, Nicotinic acid, Calcium-d-pantothenate, Pyridoxine hydrochloride, Biotin, Folic acid, Cyanocobalamin and L-ascorbic acid), formic acid solution and the ammonium salt of this acid, ammonium formate, were purchased from Millipore Sigma (Burlington, MA, USA). Also, XSelect™ Premier HSST3 VanGuard™ FIT column (2.1 × 150 mm, 2.5 µm particle size) with attached guard column (2.1 × 5mm) were purchased from Waters Corp. (Milford, MA, USA). Regarding the solvents, methanol, acetone and acetonitrile, they were purchased from Scharlab (Sentmenat, Barcelona, Spain).
Analytical procedures. Liquid chromatographic separation was performed on an Acquity Arc system coupled to a single quadrupole mass detector (Acquity QDa), controlled by the Empower 3 software (v.7.4) (Waters, Milford, MA, USA). Sample preparation includes a filtration through 0.22 µm PVDF filters followed by a 1/5 aqueous dilution. Samples were analyzed as such and diluted to fit the range of the standard curves. The separation of vitamins was carried out on a XSelectTM Premier HSST3 column with a matching guard cartridge. Mobile phases A and B consisted of 0.1% formic acid 10 mM ammonium formate in milli-Q filtered water and methanol, respectively. The separation was carried out under gradient conditions (1% 6.3 min, 1–5% 0.2 min, 5–20% 4.1 min, 20–98% 4.2 min and 98% 9.2 min), at a flow rate of 0.324 mL/min. The column was kept at room temperature and the samples at 15 °C. Ten microliters were injected for analysis. Peaks were detected with a QDa mass detector (mass range 2-1250 m/z). Capillary voltage was set to 0.8 kV. The probe temperature was held at constantly 60 °C. Positive scan mode in a range of 100–700 m/z with a cone voltage of 15 V was used as a verification method. For quantification, the compounds were measured in Single Ion Recording (SIR) mode. The sampling rate was 10 points per second. The selected m/z (mass m to charge z) by vitamin was 177.0 for ascorbic acid and for group B vitamins 265.2 (B1), 124.1 (B3), 170.1 (B6), 242.2 (B5), 377.3 (B2), 245.2 (B7), 442.0 (B9) and 678.0 (B12). Standard curves of the different vitamins were used for quantification. The chromatographic standards were prepared in different solvents according to their solubility (B7 in dimethyl sulfoxide, B2 and B9 were dissolved in 0.1 M NaOH and the rest in milli-Q filtered water) at a final concentration of 75 and 50 ppm. The calibration curves were prepared from the stock solutions through serial dilutions [96].
Author Contributions
Conceptualization, E.K. and J.C.; methodology, E.K., J.C., G.G.-L., S.L., B.Á. and M.E.-L.; validation E.K., A.S., S.M., G.G.-L., S.L., B.Á. and M.E.-L.; formal analysis, E.K., A.S., S.M., G.G.-L., S.L. and M.E.-L.; investigation E.K., A.S., S.M., G.G.-L. and N.G.; resources, J.C. and K.R.; data curation, E.K., A.S., S.M. and M.E.-L.; writing—original draft preparation, E.K. and J.C.; writing—review and editing, K.R., G.G.-L., S.L. and B.Á.; visualization, E.K.; supervision, K.R., P.M. and M.T.; project administration, B.Á., P.M., J.D. and K.R.; funding acquisition, K.R. and J.D. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
Results of all analyses are included in this published article. The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study received funding from Deerland Probiotics and Enzymes/ADM. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication. All authors declare no other competing interests.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Hill C., Guarner F., Reid G., Gibson G.R., Merenstein D.J., Pot B., Morelli L., Canani R.B., Flint H.J., Salminen S., et al. Expert Consensus Document: The International Scientific Association for Probiotics and Prebiotics Consensus Statement on the Scope and Appropriate Use of the Term Probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 2.Hou K., Wu Z.-X., Chen X.-Y., Wang J.-Q., Zhang D., Xiao C., Zhu D., Koya J.B., Wei L., Li J., et al. Microbiota in Health and Diseases. Signal Transduct. Target. 2022;7:135. doi: 10.1038/s41392-022-00974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cryan J.F., O’Riordan K.J., Cowan C.S.M., Sandhu K.V., Bastiaanssen T.F.S., Boehme M., Codagnone M.G., Cussotto S., Fulling C., Golubeva A.V., et al. The Microbiota-Gut-Brain Axis. Physiol. Rev. 2019;99:1877–2013. doi: 10.1152/physrev.00018.2018. [DOI] [PubMed] [Google Scholar]
- 4.De Simone C. The Unregulated Probiotic Market. Clin. Gastroenterol. Hepatol. 2019;17:809–817. doi: 10.1016/j.cgh.2018.01.018. [DOI] [PubMed] [Google Scholar]
- 5.Elshaghabee F.M.F., Rokana N., Gulhane R.D., Sharma C., Panwar H. Bacillus As Potential Probiotics: Status, Concerns, and Future Perspectives. Front. Microbiol. 2017;8:1490. doi: 10.3389/fmicb.2017.01490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Todorov S.D., Ivanova I.V., Popov I., Weeks R., Chikindas M.L. Bacillus Spore-Forming Probiotics: Benefits with Concerns? Crit. Rev. Microbiol. 2022;48:513–530. doi: 10.1080/1040841X.2021.1983517. [DOI] [PubMed] [Google Scholar]
- 7.Nicholson W.L., Munakata N., Horneck G., Melosh H.J., Setlow P. Resistance of Bacillus Endospores to Extreme Terrestrial and Extraterrestrial Environments. Microbiol. Mol. Biol. Rev. 2000;64:548–572. doi: 10.1128/MMBR.64.3.548-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hyronimus B., Le Marrec C., Hadj Sassi A., Deschamps A. Acid and Bile Tolerance of Spore-Forming Lactic Acid Bacteria. Int. J. Food Microbiol. 2000;61:193–197. doi: 10.1016/S0168-1605(00)00366-4. [DOI] [PubMed] [Google Scholar]
- 9.Colom J., Freitas D., Simon A., Brodkorb A., Buckley M., Deaton J., Winger A.M. Presence and Germination of the Probiotic Bacillus Subtilis DE111® in the Human Small Intestinal Tract: A Randomized, Crossover, Double-Blind, and Placebo-Controlled Study. Front. Microbiol. 2021;12:2189. doi: 10.3389/fmicb.2021.715863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramlucken U., Ramchuran S.O., Moonsamy G., Jansen van Rensburg C., Thantsha M.S., Lalloo R. Production and Stability of a Multi-Strain Bacillus Based Probiotic Product for Commercial Use in Poultry. Biotechnol. Rep. 2020;29:e00575. doi: 10.1016/j.btre.2020.e00575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fajardo-Cavazos P., Nicholson W.L. Shelf Life and Simulated Gastrointestinal Tract Survival of Selected Commercial Probiotics During a Simulated Round-Trip Journey to Mars. Front. Microbiol. 2021;12:2909. doi: 10.3389/fmicb.2021.748950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majeed M., Majeed S., Nagabhushanam K., Natarajan S., Sivakumar A., Ali F. Evaluation of the Stability of Bacillus Coagulans MTCC 5856 during Processing and Storage of Functional Foods. Int. J. Food Sci. Technol. 2016;51:894–901. doi: 10.1111/ijfs.13044. [DOI] [Google Scholar]
- 13.Jafari M., Mortazavian A.M., Hosseini H., Safaei F., Mousavi Khaneghah A., Sant’Ana A.S. Probiotic Bacillus: Fate during Sausage Processing and Storage and Influence of Different Culturing Conditions on Recovery of Their Spores. Food Res. Int. 2017;95:46–51. doi: 10.1016/j.foodres.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Fares C., Menga V., Martina A., Pellegrini N., Scazzina F., Torriani S. Nutritional Profile and Cooking Quality of a New Functional Pasta Naturally Enriched in Phenolic Acids, Added with β-Glucan and Bacillus Coagulans GBI-30, 6086. J. Cereal Sci. 2015;65:260–266. doi: 10.1016/j.jcs.2015.07.017. [DOI] [Google Scholar]
- 15.Saggese A., Baccigalupi L., Ricca E. Spore Formers as Beneficial Microbes for Humans and Animals. Appl. Microbiol. 2021;1:498–509. doi: 10.3390/applmicrobiol1030032. [DOI] [Google Scholar]
- 16.Lee S.H., Park D.H. Isolation and Physiological Characterization of Bacillus clausii SKAL-16 Isolated from Wastewater. J. Microbiol. Biotechnol. 2008;18:1908–1914. doi: 10.4014/jmb.0800.175. [DOI] [PubMed] [Google Scholar]
- 17.Asha Devi N.K., Balakrishnan K., Gopal R., Padmavathy S. Bacillus clausii MB9 from the East Coast Regions of India: Isolation, Biochemical Characterization and Antimicrobial Potentials. Curr. Sci. 2008;95:627–636. [Google Scholar]
- 18.De Jonghe V., Coorevits A., De Block J., Van Coillie E., Grijspeerdt K., Herman L., De Vos P., Heyndrickx M. Toxinogenic and Spoilage Potential of Aerobic Spore-Formers Isolated from Raw Milk. Int. J. Food Microbiol. 2010;136:318–325. doi: 10.1016/j.ijfoodmicro.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 19.Hoyles L., Honda H., Logan N.A., Halket G., La Ragione R.M., McCartney A.L. Recognition of Greater Diversity of Bacillus Species and Related Bacteria in Human Faeces. Res. Microbiol. 2012;163:3–13. doi: 10.1016/j.resmic.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 20.Lahiri K., Jadhav K., Gahlowt P., Najmuddin F., Padmashree Y. Bacillus clausii As An Adjuvant Therapy In Acute Childhood Diarrhoea. IOSR J. Dent. Med. Sci. Ver. I. 2015;14:2279–2861. doi: 10.9790/0853-14517476. [DOI] [Google Scholar]
- 21.Lahiri K.R. GMA-CO Clinical Study Report: ENTER_L_01486. [(accessed on 9 January 2023)]. Sanofi-Aventis. Available online: https://www.sanofi.com/dam/jcr:04929443-77dc-4b35-bcc6-8b5859fc10d4/ENTER_L_01486_summary.pdf.
- 22.Canani R.B., Cirillo P., Terrin G., Cesarano L., Spagnuolo M.I., De Vincenzo A., Albano F., Passariello A., De Marco G., Manguso F., et al. Probiotics for Treatment of Acute Diarrhoea in Children: Randomised Clinical Trial of Five Different Preparations. Br. Med. J. 2007;335:340–342. doi: 10.1136/bmj.39272.581736.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maugo B.M. Effectiveness of Bacillus clausii in Reducing Duration of Illness in Acute Diarrhoea in Children 6–59 Months of Age Admitted with Severe Dehydration. [(accessed on 9 January 2023)]. Available online: http://erepository.uonbi.ac.ke/handle/11295/8325.
- 24.De Castro J.A.A., Guno M.J.V.R., Perez M.O. Bacillus clausii as Adjunctive Treatment for Acute Community-Acquired Diarrhea among Filipino Children: A Large-Scale, Multicenter, Open-Label Study (CODDLE) Trop. Dis. Travel Med. Vaccines. 2019;5:14. doi: 10.1186/s40794-019-0089-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nista E.C., Candelli M., Cremonini F., Cazzato I.A., Zocco M.A., Franceschi F., Cammarota G., Gasbarrini G., Gasbarrini A. Bacillus clausii Therapy to Reduce Side-Effects of Anti-Helicobacter Pylori Treatment: Randomized, Double-Blind, Placebo Controlled Trial. Aliment. Pharmacol. Ther. 2004;20:1181–1188. doi: 10.1111/j.1365-2036.2004.02274.x. [DOI] [PubMed] [Google Scholar]
- 26.Plomer M., III Perez M., Greifenberg D.M. Effect of Bacillus clausii Capsules in Reducing Adverse Effects Associated with Helicobacter Pylori Eradication Therapy: A Randomized, Double-Blind, Controlled Trial. Infect. Dis. 2020;9:867–878. doi: 10.1007/s40121-020-00333-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maity C., Gupta A.K. Therapeutic Efficacy of Probiotic Alkalihalobacillus clausii 088AE in Antibiotic-Associated Diarrhea: A Randomized Controlled Trial. Heliyon. 2021;7:e07993. doi: 10.1016/j.heliyon.2021.e07993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ianiro G., Rizzatti G., Plomer M., Lopetuso L., Scaldaferri F., Franceschi F., Cammarota G., Gasbarrini A. Bacillus clausii for the Treatment of Acute Diarrhea in Children: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Nutrients. 2018;10:1074. doi: 10.3390/nu10081074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamid F., Quaium S.M.M.A., Rahman A. Comparative Study of Bacillus clausii and Multistrain Probiotics in the Management of Acute Diarrhoea in Children. Int. J. Res. Med. Sci. 2019;7:1156–1160. doi: 10.18203/2320-6012.ijrms20191317. [DOI] [Google Scholar]
- 30.Sudha M.R., Jayanthi N., Pandey D.C., Verma A.K. Bacillus clausii UBBC-07 Reduces Severity of Diarrhoea in Children under 5 Years of Age: A Double Blind Placebo Controlled Study. Benef. Microbes. 2019;10:149–154. doi: 10.3920/BM2018.0094. [DOI] [PubMed] [Google Scholar]
- 31.Acosta-Rodríguez-Bueno C.P., Abreu y Abreu A.T., Guarner F., Guno M.J.V., Pehlivanoğlu E., Perez M. Bacillus clausii for Gastrointestinal Disorders: A Narrative Literature Review. Adv. Ther. 2022;39:4854–4874. doi: 10.1007/s12325-022-02285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tewari V.V., Dubey S.K., Gupta G. Bacillus clausii for Prevention of Late-Onset Sepsis in Preterm Infants: A Randomized Controlled Trial. J. Trop. Pediatr. 2015;61:377–385. doi: 10.1093/tropej/fmv050. [DOI] [PubMed] [Google Scholar]
- 33.Marseglia G.L., Tosca M., Cirillo I., Licari A., Leone M., Marseglia A., Castellazzi A.M., Ciprandi G. Efficacy of Bacillus clausii Spores in the Prevention of Recurrent Respiratory Infections in Children: A Pilot Study. Clin. Risk. Manag. 2007;3:13–17. doi: 10.2147/tcrm.2007.3.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nirmala M., Smitha S.G., Kamath G.J. A Study to Assess The Efficacy of Local Application of Oral Probiotic in Treating Recurrent Aphthous Ulcer and Oral Candidiasis. Indian J. Otolaryngol. Head Neck Surg. 2019;71:113–117. doi: 10.1007/s12070-017-1139-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ripert G., Racedo S.M., Elie A.-M., Jacquot C., Bressollier P., Urdaci M.C. Secreted Compounds of the Probiotic Bacillus clausii Strain O/C Inhibit the Cytotoxic Effects Induced by Clostridium Difficile and Bacillus Cereus Toxins. Antimicrob. Agents Chemother. 2016;60:3445–3454. doi: 10.1128/AAC.02815-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rochín-Medina J.J., Ramírez-Medina H.K., Rangel-Peraza J.G., Pineda-Hidalgo K.V., Iribe-Arellano P. Use of Whey as a Culture Medium for Bacillus clausii for the Production of Protein Hydrolysates with Antimicrobial and Antioxidant Activity. Food Sci. Technol. Int. 2018;24:35–42. doi: 10.1177/1082013217724705. [DOI] [PubMed] [Google Scholar]
- 37.Kapse N.G., Engineer A.S., Gowdaman V., Wagh S., Dhakephalkar P.K. Genome Profiling for Health Promoting and Disease Preventing Traits Unraveled Probiotic Potential of Bacillus clausii B106. Microbiol. Biotechnol. Lett. 2018;46:334–345. doi: 10.4014/mbl.1804.04001. [DOI] [Google Scholar]
- 38.Khatri I., Sharma G., Subramanian S. Composite Genome Sequence of Bacillus clausii, a Probiotic Commercially Available as Enterogermina®, and Insights into Its Probiotic Properties. BMC Microbiol. 2019;19:307. doi: 10.1186/s12866-019-1680-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahire J.J., Kashikar M.S., Madempudi R.S. Survival and Germination of Bacillus clausii UBBC07 Spores in in Vitro Human Gastrointestinal Tract Simulation Model and Evaluation of Clausin Production. Front. Microbiol. 2020;11:1010. doi: 10.3389/fmicb.2020.01010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park H., Jung A.-Y., Chang C.-S., Kim Y.H. Bacillus clausii, a Foreshore-Derived Probiotic, Attenuates Allergic Airway Inflammation Through Downregulation of Hypoxia Signaling. J. Rhinol. 2020;27:108–116. doi: 10.18787/jr.2020.00325. [DOI] [Google Scholar]
- 41.Pradhan B., Guha D., Ray P., Das D., Aich P. Comparative Analysis of the Effects of Two Probiotic Bacterial Strains on Metabolism and Innate Immunity in the RAW 264.7 Murine Macrophage Cell Line. Probiotics Antimicrob. Prot. 2016;8:73–84. doi: 10.1007/s12602-016-9211-4. [DOI] [PubMed] [Google Scholar]
- 42.Caro S.D., Tao H., Grillo A., Franceschi F., Elia C., Zocco M.A., Gasbarrini G., Sepulveda A.R., Gasbarrini A. Bacillus clausii Effect on Gene Expression Pattern in Small Bowel Mucosa Using DNA Microarray Analysis. Eur. J. Gastroenterol. Hepatol. 2005;17:951–960. doi: 10.1097/00042737-200509000-00011. [DOI] [PubMed] [Google Scholar]
- 43.Urdaci M.C., Bressollier P., Pinchuk I. Bacillus clausii Probiotic Strains: Antimicrobial and Immunomodulatory Activities. J. Clin. Gastroenterol. 2004;38:S86–S90. doi: 10.1097/01.mcg.0000128925.06662.69. [DOI] [PubMed] [Google Scholar]
- 44.Pisano M.B., Viale S., Conti S., Fadda M.E., Deplano M., Melis M.P., Deiana M., Cosentino S. Preliminary Evaluation of Probiotic Properties of Lactobacillus Strains Isolated from Sardinian Dairy Products. Biomed. Res. Int. 2014;2014:286390. doi: 10.1155/2014/286390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marianelli C., Cifani N., Pasquali P. Evaluation of Antimicrobial Activity of Probiotic Bacteria against Salmonella Enterica Subsp. Enterica Serovar Typhimurium 1344 in a Common Medium under Different Environmental Conditions. Res. Microbiol. 2010;161:673–680. doi: 10.1016/j.resmic.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 46.Martorell P., Forment J.V., de Llanos R., Montón F., Llopis S., González N., Genovés S., Cienfuegos E., Monzó H., Ramón D. Use of Saccharomyces Cerevisiae and Caenorhabditis Elegans as Model Organisms to Study the Effect of Cocoa Polyphenols in the Resistance to Oxidative Stress. J. Agric. Food Chem. 2011;59:2077–2085. doi: 10.1021/jf104217g. [DOI] [PubMed] [Google Scholar]
- 47.Sako T., Matsumoto K., Tanaka R. Recent Progress on Research and Applications of Non-Digestible Galacto-Oligosaccharides. Int. Dairy J. 1999;9:69–80. doi: 10.1016/S0958-6946(99)00046-1. [DOI] [Google Scholar]
- 48.Seth A., Yan F., Polk D.B., Rao R.K. Probiotics Ameliorate the Hydrogen Peroxide-Induced Epithelial Barrier Disruption by a PKC- and MAP Kinase-Dependent Mechanism. Am. J. Physiol.-Gastrointest. Liver Physiol. 2008;294:G1060–G1069. doi: 10.1152/ajpgi.00202.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Danilova I., Sharipova M. The Practical Potential of Bacilli and Their Enzymes for Industrial Production. Front. Microbiol. 2020;11:1782. doi: 10.3389/fmicb.2020.01782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dame Z.T., Rahman M., Islam T. Bacilli as Sources of Agrobiotechnology: Recent Advances and Future Directions. Green Chem. Lett. Rev. 2021;14:246–271. doi: 10.1080/17518253.2021.1905080. [DOI] [Google Scholar]
- 51.Bahaddad S.A., Almalki M.H.K., Alghamdi O.A., Sohrab S.S., Yasir M., Azhar E.I., Chouayekh H. Bacillus Species as Direct-Fed Microbial Antibiotic Alternatives for Monogastric Production. Probiotics Antimicrob. Prot. 2023;15:1–16. doi: 10.1007/s12602-022-09909-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bader J., Albin A., Stahl U. Spore-Forming Bacteria and Their Utilisation as Probiotics. Benef. Microbes. 2012;3:67–75. doi: 10.3920/BM2011.0039. [DOI] [PubMed] [Google Scholar]
- 53.Abuhena M., Al-Rashid J., Azim M.F., Khan M.N.M., Kabir M.G., Barman N.C., Rasul N.M., Akter S., Huq M.A. Optimization of Industrial (3000 L) Production of Bacillus Subtilis CW-S and Its Novel Application for Minituber and Industrial-Grade Potato Cultivation. Sci. Rep. 2022;12:11153. doi: 10.1038/s41598-022-15366-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong-Chew R.M., de Castro J.-A.A., Morelli L., Perez M., Ozen M. Gut Immune Homeostasis: The Immunomodulatory Role of Bacillus clausii, from Basic to Clinical Evidence. Expert Rev. Clin. Immunol. 2022;18:717–729. doi: 10.1080/1744666X.2022.2085559. [DOI] [PubMed] [Google Scholar]
- 55.Dunne C., O’Mahony L., Murphy L., Thornton G., Morrissey D., O’Halloran S., Feeney M., Flynn S., Fitzgerald G., Daly C., et al. In Vitro Selection Criteria for Probiotic Bacteria of Human Origin: Correlation with in Vivo Findings. Am. J. Clin. Nutr. 2001;73:386s–392s. doi: 10.1093/ajcn/73.2.386s. [DOI] [PubMed] [Google Scholar]
- 56.Duc L.H., Hong H.A., Barbosa T.M., Henriques A.O., Cutting S.M. Characterization of Bacillus Probiotics Available for Human Use. Appl. Environ. Microbiol. 2004;70:2161–2171. doi: 10.1128/AEM.70.4.2161-2171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cenci G., Trotta F., Caldini G. Tolerance to Challenges Miming Gastrointestinal Transit by Spores and Vegetative Cells of Bacillus clausii. J. Appl. Microbiol. 2006;101:1208–1215. doi: 10.1111/j.1365-2672.2006.03042.x. [DOI] [PubMed] [Google Scholar]
- 58.Ahire J.J., Kashikar M.S., Madempudi R.S. Comparative Accounts of Probiotic Properties of Spore and Vegetative Cells of Bacillus clausii UBBC07 and in Silico Analysis of Probiotic Function. 3Biotech. 2021;11:116. doi: 10.1007/s13205-021-02668-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee Y.-K., Puong K.-Y., Ouwehand A.C., Salminen S. Displacement of Bacterial Pathogens from Mucus and Caco-2 Cell Surface by Lactobacilli. J. Med. Microbiol. 2003;52:925–930. doi: 10.1099/jmm.0.05009-0. [DOI] [PubMed] [Google Scholar]
- 60.Kamada N., Chen G.Y., Inohara N., Núñez G. Control of Pathogens and Pathobionts by the Gut Microbiota. Nat. Immunol. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Laparra J.M., Sanz Y. Comparison of in Vitro Models to Study Bacterial Adhesion to the Intestinal Epithelium. Lett. Appl. Microbiol. 2009;49:695–701. doi: 10.1111/j.1472-765X.2009.02729.x. [DOI] [PubMed] [Google Scholar]
- 62.Gagnon M., Zihler Berner A., Chervet N., Chassard C., Lacroix C. Comparison of the Caco-2, HT-29 and the Mucus-Secreting HT29-MTX Intestinal Cell Models to Investigate Salmonella Adhesion and Invasion. J. Microbiol. Methods. 2013;94:274–279. doi: 10.1016/j.mimet.2013.06.027. [DOI] [PubMed] [Google Scholar]
- 63.Navarre W.W., Schneewind O. Surface Proteins of Gram-Positive Bacteria and Mechanisms of Their Targeting to the Cell Wall Envelope. Microbiol. Mol. Biol. Rev. 1999;63:174–229. doi: 10.1128/MMBR.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marcos C.M., de Fátima da Silva J., de Oliveira H.C., Moraes da Silva R.A., Mendes-Giannini M.J.S., Fusco-Almeida A.M. Surface-Expressed Enolase Contributes to the Adhesion of Paracoccidioides Brasiliensis to Host Cells. FEMS Yeast Res. 2012;12:557–570. doi: 10.1111/j.1567-1364.2012.00806.x. [DOI] [PubMed] [Google Scholar]
- 65.Haiko J., Westerlund-Wikström B. The Role of the Bacterial Flagellum in Adhesion and Virulence. Biology. 2013;2:1242–1267. doi: 10.3390/biology2041242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.RUAS-MADIEDO P., GUEIMONDE M., MARGOLLES A., de los REYES-GAVILÁN C.G., SALMINEN S. Exopolysaccharides Produced by Probiotic Strains Modify the Adhesion of Probiotics and Enteropathogens to Human Intestinal Mucus. J. Food Prot. 2006;69:2011–2015. doi: 10.4315/0362-028X-69.8.2011. [DOI] [PubMed] [Google Scholar]
- 67.Ghelardi E., Celandroni F., Salvetti S., Gueye S.A., Lupetti A., Senesi S. Survival and Persistence of Bacillus clausii in the Human Gastrointestinal Tract Following Oral Administration as Spore-Based Probiotic Formulation. J. Appl. Microbiol. 2015;119:552–559. doi: 10.1111/jam.12848. [DOI] [PubMed] [Google Scholar]
- 68.Jiang T., Mustapha A., Savaiano D.A. Improvement of Lactose Digestion in Humans by Ingestion of Unfermented Milk Containing Bifidobacterium Longum. J. Dairy Sci. 1996;79:750–757. doi: 10.3168/jds.S0022-0302(96)76422-6. [DOI] [PubMed] [Google Scholar]
- 69.Parodi P.W. The Role of Intestinal Bacteria in the Causation and Prevention of Cancer: Modulation by Diet and Probiotics. Aust. J. Dairy Technol. 1999;54:103–121. [Google Scholar]
- 70.Cheng K.-W., Tseng C.-H., Chen I.-J., Huang B.-C., Liu H.-J., Ho K.-W., Lin W.-W., Chuang C.-H., Huang M.-Y., Leu Y.-L., et al. Inhibition of Gut Microbial β-Glucuronidase Effectively Prevents Carcinogen-Induced Microbial Dysbiosis and Intestinal Tumorigenesis. Pharmacol. Res. 2022;177:106115. doi: 10.1016/j.phrs.2022.106115. [DOI] [PubMed] [Google Scholar]
- 71.Oakey H.J., Harty D.W.S., Knox K.W. Enzyme Production by Lactobacilli and the Potential Link with Infective Endocarditis. J. Appl. Bacteriol. 1995;78:142–148. doi: 10.1111/j.1365-2672.1995.tb02834.x. [DOI] [PubMed] [Google Scholar]
- 72.Bouhss A., Al-Dabbagh B., Vincent M., Odaert B., Aumont-Nicaise M., Bressolier P., Desmadril M., Mengin-Lecreulx D., Urdaci M.C., Gallay J. Specific Interactions of Clausin, a New Lantibiotic, with Lipid Precursors of the Bacterial Cell Wall. Biophys. J. 2009;97:1390–1397. doi: 10.1016/j.bpj.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Macfarlane G.T., Allison C., Gibson S.A., Cummings J.H. Contribution of the Microflora to Proteolysis in the Human Large Intestine. J. Appl. Bacteriol. 1988;64:37–46. doi: 10.1111/j.1365-2672.1988.tb02427.x. [DOI] [PubMed] [Google Scholar]
- 74.Evenepoel P., Claus D., Geypens B., Hiele M., Geboes K., Rutgeerts P., Ghoos Y. Amount and Fate of Egg Protein Escaping Assimilation in the Small Intestine of Humans. Am. J. Physiol.-Gastrointest. Liver Physiol. 1999;277:G935–G943. doi: 10.1152/ajpgi.1999.277.5.G935. [DOI] [PubMed] [Google Scholar]
- 75.Davila A.-M., Blachier F., Gotteland M., Andriamihaja M., Benetti P.-H., Sanz Y., Tomé D. Intestinal Luminal Nitrogen Metabolism: Role of the Gut Microbiota and Consequences for the Host. Pharmacol. Res. 2013;68:95–107. doi: 10.1016/j.phrs.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 76.Blachier F., Mariotti F., Huneau J.F., Tomé D. Effects of Amino Acid-Derived Luminal Metabolites on the Colonic Epithelium and Physiopathological Consequences. Amino Acids. 2007;33:547–562. doi: 10.1007/s00726-006-0477-9. [DOI] [PubMed] [Google Scholar]
- 77.Bourgin M., Kepp O., Kroemer G. Immunostimulatory Effects of Vitamin B5 Improve Anticancer Immunotherapy. OncoImmunology. 2022;11:2031500. doi: 10.1080/2162402X.2022.2031500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Paul M.S., Saibil S.D., Han S., Israni-Winger K., Lien S.C., Laister R.C., Sayad A., Penny S., Amaria R.N., Haydu L.E., et al. Coenzyme A Fuels T Cell Anti-Tumor Immunity. Cell Metab. 2021;33:2415–2427.e6. doi: 10.1016/j.cmet.2021.11.010. [DOI] [PubMed] [Google Scholar]
- 79.Tamura J., Kubota K., Murakami H., Sawamura M., Matsushima T., Tamura T., Saitoh T., Kurabayshi H., Naruse T. Immunomodulation by Vitamin B12: Augmentation of CD8+ T Lymphocytes and Natural Killer (NK) Cell Activity in Vitamin B12-Deficient Patients by Methyl-B12 Treatment. Clin. Exp. Immunol. 1999;116:28–32. doi: 10.1046/j.1365-2249.1999.00870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Poltorak A., He X., Smirnova I., Liu M.-Y., Huffel C.V., Du X., Birdwell D., Alejos E., Silva M., Galanos C., et al. Defective LPS Signaling in C3H/HeJ and C57BL/10ScCr Mice: Mutations in Tlr4 Gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 81.Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A. Recognition of Double-Stranded RNA and Activation of NF-ΚB by Toll-like Receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 82.Gilmore T.D. Introduction to NF-ΚB: Players, Pathways, Perspectives. Oncogene. 2006;25:6680–6684. doi: 10.1038/sj.onc.1209954. [DOI] [PubMed] [Google Scholar]
- 83.Paparo L., Tripodi L., Bruno C., Pisapia L., Damiano C., Pastore L., Berni Canani R. Protective Action of Bacillus clausii Probiotic Strains in an in Vitro Model of Rotavirus Infection. Sci. Rep. 2020;10:12636. doi: 10.1038/s41598-020-69533-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Scaldaferri F., Graziani C., Mora V., Petito V., Lopetuso L.R., Puca P., Ianiro G., Napolitano D., Quaranta G., Masucci L., et al. Bacillus clausii (O/C, SIN, N/R, T) Improves Acute Mild Colitis in Mice While In-Vivo Modulating Gut Microbiota. Ann. Gastroenterol. Dig. Syst. 2021;4:1035. [Google Scholar]
- 85.Pradhan B., Guha D., Naik A.K., Banerjee A., Tambat S., Chawla S., Senapati S., Aich P. Probiotics L. acidophilus and B. clausii Modulate Gut Microbiota in Th1- and Th2-Biased Mice to Ameliorate Salmonella Typhimurium-Induced Diarrhea. Probiotics Antimicrob. Prot. 2019;11:887–904. doi: 10.1007/s12602-018-9436-5. [DOI] [PubMed] [Google Scholar]
- 86.Shida K., Nanno M., Nagata S. Flexible Cytokine Production by Macrophages and T Cells in Response to Probiotic Bacteria: A Possible Mechanism by Which Probiotics Exert Multifunctional Immune Regulatory Activities. Gut Microbes. 2011;2:109–114. doi: 10.4161/gmic.2.2.15661. [DOI] [PubMed] [Google Scholar]
- 87.Rocha-Ramírez L.M., Pérez-Solano R.A., Castañón-Alonso S.L., Moreno Guerrero S.S., Ramírez Pacheco A., García Garibay M., Eslava C. Probiotic Lactobacillus Strains Stimulate the Inflammatory Response and Activate Human Macrophages. J. Immunol. Res. 2017;2017:4607491. doi: 10.1155/2017/4607491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maldonado Galdeano C., Cazorla S.I., Lemme Dumit J.M., Vélez E., Perdigón G. Beneficial Effects of Probiotic Consumption on the Immune System. Ann. Nutr. Metab. 2019;74:115–124. doi: 10.1159/000496426. [DOI] [PubMed] [Google Scholar]
- 89.Villéger R., Saad N., Grenier K., Falourd X., Foucat L., Urdaci M.C., Bressollier P., Ouk T.-S. Characterization of Lipoteichoic Acid Structures from Three Probiotic Bacillus Strains: Involvement of d-Alanine in Their Biological Activity. Antonie Leeuwenhoek. 2014;106:693–706. doi: 10.1007/s10482-014-0239-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Riquelme E., Zhang Y., Zhang L., Montiel M., Zoltan M., Dong W., Quesada P., Sahin I., Chandra V., Lucas A.S., et al. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell. 2019;178:795–806.e12. doi: 10.1016/j.cell.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang Y., Wu Y., Wang Y., Xu H., Mei X., Yu D., Wang Y., Li W. Antioxidant Properties of Probiotic Bacteria. Nutrients. 2017;9:521. doi: 10.3390/nu9050521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Patel C., Patel P., Acharya S. Therapeutic Prospective of a Spore-Forming Probiotic—Bacillus clausii UBBC07 Against Acetaminophen-Induced Uremia in Rats. Probiotics Antimicrob. Prot. 2020;12:253–258. doi: 10.1007/s12602-019-09540-x. [DOI] [PubMed] [Google Scholar]
- 93.Özkoç M., Can B., Şentürk H., Burukoğlu Dönmez D., Kanbak G. Possible Curative Effects of Boric Acid and Bacillus clausii Treatments on TNBS-Induced Ulcerative Colitis in Rats. Biol. Trace Elem. Res. 2022:1–15. doi: 10.1007/s12011-022-03215-5. [DOI] [PubMed] [Google Scholar]
- 94.Guba A., Bába O., Tőzsér J., Csősz É., Kalló G. Fast and Sensitive Quantification of AccQ-Tag Derivatized Amino Acids and Biogenic Amines by UHPLC-UV Analysis from Complex Biological Samples. Metabolites. 2022;12:272. doi: 10.3390/metabo12030272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jaworska M., Stańczyk M., Wilk M., Kłaczkow G., Anuszewska E., Barzał J., Rzepecki P. New Approach for Amino Acid Profiling in Human Plasma by Selective Fluorescence Derivatization. Amino Acids. 2012;43:1653–1661. doi: 10.1007/s00726-012-1243-9. [DOI] [PubMed] [Google Scholar]
- 96.Benvenuti M.E. Selective Quantitative Determination of Water Soluble Vitamins in Various Food Matrices Using the ACQUITY UPLC H-Class System and ACQUITY QDa Mass Detector. Waters Corporation; Milford, MA, USA: 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Results of all analyses are included in this published article. The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.