Abstract
A moderately thermophilic (45 to 50°C), highly acidophilic (pH 1.5 to 2.5), chemolithotrophic Acidithiobacillus caldus strain, f, was isolated from a biooxidation process used to treat nickel ore. Trans-alternating field electrophoresis analysis of total DNA from the A. caldus cells revealed two plasmids of approximately 14 and 45 kb. The 14-kb plasmid, designated pTC-F14, was cloned and shown by replacement of the cloning vector with a kanamycin resistance gene to be capable of autonomous replication in Escherichia coli. Autonomous replication was also demonstrated in Pseudomonas putida and Agrobacterium tumefaciens LBA 4404, which suggested that pTC-F14 is a broad-host-range plasmid. Sequence analysis of the pTC-F14 replicon region revealed five open reading frames and a replicon organization like that of the broad-host-range IncQ plasmids. Three of the open reading frames encoded replication proteins which were most closely related to those of IncQ-like plasmid pTF-FC2 (amino acid sequence identities: RepA, 81%; RepB, 78%; RepC, 74%). However, the two plasmids were fully compatible and pTC-F14 represents a new IncQ-like plasmid replicon. Surprisingly, asymmetrical incompatibility was found with the less closely related IncQ plasmid R300B derivative pKE462 and the IncQ-like plasmid derivative pIE1108. Analysis of the pTC-F14 oriV region revealed five direct repeats consisting of three perfectly conserved 22-bp iterons flanked by iterons of 23 and 21 bp. Plasmid pTC-F14 had a copy number of 12 to 16 copies per chromosome in both E. coli, and A. caldus. The rep gene products of pTC-F14 and pTF-FC2 were unable to functionally complement each other's oriV regions, but replication occurred when the genes for each plasmid's own RepA, RepB, and RepC proteins were provided in trans. Two smaller open reading frames were found between the repB and repA genes of pTC-F14, which encode proteins with high amino acid sequence identity (PasA, 81%; PasB, 72%) to the plasmid addiction system of pTF-FC2. This is the second time a plasmid stability system of this type has been found on an IncQ-like plasmid.
Plasmids of Escherichia coli incompatibility group Q and related IncQ-like plasmids have a very broad host range, being capable of replication in a wide variety of gram-negative and also gram-positive bacteria (10). Although none of the IncQ and related plasmids are self-transmissible, they are efficiently mobilized by IncPα (RK2, RP4, and R68) and IncPβ (R751) plasmids (5). As a result of their host range and mobility, these plasmids are highly promiscuous. The best-studied IncQ plasmids are RSF1010 (11), R1162 (20), and R300B (2), which are identical, or nearly identical, in spite of having been isolated from different hosts (10). RSF1010 (8,486 bp) has been completely sequenced, and the replication and mobilization functions of RSF1010 and R1162 have been extensively studied (30).
The discovery of several other plasmids which have replicons with clear similarity to IncQ plasmids has been reported. These are IncQ-like plasmids pTF-FC2 (12,180 bp) (7, 8), isolated from the biomining bacterium Acidithiobacillus ferrooxidans (previously Thiobacillus ferrooxidans); pIE1107 (8,520 bp), isolated from a mixture of bacteria present in pig manure (35);and pDN1 (5,112 bp), isolated from the sheep foot rot-causing pathogen Dichelobacter nodosus (37). In addition, sequences of at least two other IncQ-related plasmids, pIE1115 and pIE1130 (both 10,687 bp), appear in sequence databases although only research on the isolation of these has been published so far (31).
IncQ replicons contain three essential replication genes (repA, repB [repB']), and repC) and an oriV region (28). The repA gene encodes a plasmid-specific helicase, repB' encodes a primase, and repC encodes a plasmid-specific DNA-binding initiation protein. The oriV region consists of 3.5 20-bp iterons and an approximately 500-bp flanking region containing G+C-rich and A+T-rich sequences and two plasmid-specific single-stranded initiation sites (ssiA and ssiB). The IncQ-related replicons have similar structures, although there are differences in some small replicon-associated proteins and the numbers and sequences of their iterons. The ability of IncQ-like plasmids to displace each other when coresident in E. coli has been tested, and it is clear that these plasmids may be divided into several distinguishable incompatibility subgroups.
Acidithiobacillus caldus (previously Thiobacillus caldus) is a sulfur-oxidizing, chemolithotrophic, obligately acidophilic (pH 1.5 to 2.5), and moderately thermophilic (45 to 50°C) bacterium (12). These bacteria, together with iron-oxidizing bacteria of the genus Leptospirillum, have been found to be the dominant organisms present in biooxidation tanks used in certain commercial processes for the recovery of gold from arsenopyrite ores (25). While investigating plasmids from strains of A. caldus isolated from biooxidation tanks, we discovered a 14-kb plasmid which was capable of replication in an E. coli polA mutant. Here we report on the characterization of this IncQ-related plasmid replicon and its biological relationship to other IncQ and IncQ-related plasmids.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strains and plasmids used in this study are shown in Table 1. E. coli cells were grown in Luria medium, and ampicillin (100 μg/ml), kanamycin (30 μg/ml), and tetracycline (20 μg/ml) were added as required. Pseudomonas putida was grown in Luria medium, and kanamycin (50 μg/ml) was added when appropriate. Agrobacterium tumefaciens LBA 4404 was grown in Luria medium which included rifampin (5 μg/ml) and, when required, kanamycin (30 μg/ml). The tetrathionate medium used to culture A. caldus strain f was made from a mineral salts solution containing the following (grams per liter): (NH4)2SO4, 3.0; KCl, 0.1; K2HPO4, 0.5; MgSO4, 0.7; H2O, 0.5; Ca(NO3)2, 0.4; H2O, 0.014; Na2SO4, 1.45. The pH was adjusted to 2.5 with H2SO4, and the mixture was autoclaved. The trace element solution used contained the following (milligrams per liter): ZnSO4, 0.7; H2O, 10.0; CuSO4, 0.5; H2O, 1.0; MnSO4, 0.4; H2O, 1.0; CoCl2, 0.6; H2O, 0.5; Cr2(SO4)3, 0.15; H2O, 0.5; Na2B4O7, 0.10; H2O, 0.5; NaMoO4, 0.2; H2O, 0.5. The mixture was autoclaved, and 1 ml was added per liter. Filter-sterilized K2S2O6 was added to a final concentration of 10 mM, and the pH was adjusted to 2.5 with H2SO4. A. caldus was grown at 37°C with constant shaking.
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DH5α | φ80dlacZΔM15 endA1 recA1 gyrA96 thi-1 hsdR17(rK−mK−) relA1 supE44 deoR Δ(lacZYA-argF)U169 | Promega Corp. |
| E. coli GW125a | recA polA mutant of AB1157 | 6;;1 |
| A. caldus strain f | Isolated from Billiton Process Research Bionic plant (Randburg, South Africa) | This study |
| A. tumefaciens | Nononcogenic, T-DNA deletion, vir intact | |
| LBA4404 | 13;;1 | |
| P. putida | Prototrophic | 9;;1 |
| Plasmids | ||
| pTC-F14 | Natural 14.4-kb plasmid from A. caldus strain f | This study |
| pTC-F14Cm | Cmr (Cmr gene cloned into pTC-F14) | This study |
| pIb | Apr (unique XbaI site of pTC-F14 interrupted by pBluescript) | This study |
| pK13 | Apr (unique BamHI site of pTC-F14 interrupted by pBluescript) | This study |
| pTF-FC2 | Natural 12.2-kb plasmid from A. ferrooxidans FC2 | 26;;1 |
| pKE462 | Tcr Asr, R300B replicon | 6;;1 |
| pIE1108 | Str Kmr, pIE1107 replicon with set of nonessential IncQ iterons deleted | 35;;1 |
| pDER404 | Cmr, pTF-FC2 ClaI-PstI fragment complete replicon | 26;;1 |
| pTV4164 | Apr, pTF-FC2 oriV fragment | 8;;1 |
| pTC-F101 | Kmr, pTC-F14 replicon (Fig. 1) | This study |
| pTC-F108 | Apr, pUCBM21 vector (Fig. 1) | This study |
| pTC-F109 | Apr, pGEM-T vector (Fig. 1) | This study |
| pACYC184 | Tcr Cmr, p15A replicon, cloning vector | 4;;1 |
| pBluescript (KS) | AprlacZ′, ColE1 replicon, vector | Stratagene |
| pUCBM21 | AprlacZ′, ColE1 replicon, vector | Roche Molecular Biochemicals |
| pGEM-T | Apr, T-tailed PCR product cloning vector | Promega Corp. |
| Primers | ||
| TACREPA (EcoRI) | 5′-TATTGAATTCCCCGGCAGCGCC-3′ | This study |
| TACREPAE (PstI) | 5′-TATTCTGCAGAGGGGGTGCGATAGC-3′ | This study |
| SEQORI | 5′-TATCGAGATGGCAGAGGTGCGAG-3′ | This study |
| ORIR (HindIII) | 5′-TGTCAAGCTTGGCACTCTCCTG-3′ | This study |
Ap, ampicillin resistance; Cm, chloramphenicol resistance; Km, kanamycin resistance; St, streptothricin resistance; Tc, tetracycline resistance.
DNA techniques, sequencing, and analysis.
Plasmid preparation, restriction endonuclease digestions, gel electrophoresis, ligations, and Southern blot hybridization were carried out by standard methods (29). Labeling of probes, hybridization, and detection were done by using the digoxigenin-dUTP nonradioactive DNA labeling and detection system (Roche Molecular Biochemicals). Sequencing was done by the dideoxy-chain termination method, with an ABI PRISM 377 automated DNA sequencer, and the sequence was analyzed by using a variety of software programs but mainly the personal-computer-based DNAMAN (version 4.1) package from Lynnon BioSoft. Comparison searches were performed using the gapped-BLAST program of the National Center for Biotechnology Information (1). Homology trees were constructed using the Multiple Sequence Alignment tool in DNAMAN. The PCR was carried out with the primers described in Table 1. The reaction was performed in a PCR Sprint Temperature Cycling System (Hybaid) using the Expand High Fidelity PCR System DNA polymerase (Roche Molecular Biochemicals). After an initial denaturation of 60 s at 94°C, 25 cycles of 30 s at 94°C, 30 s at 63°C (for primers TACREPA and TACREPAE) or 55°C (for primers SEQORI and ORIR), and 90 s at 72°C were performed. A final extension step of 120 s at 72°C before cooling to 4°C completed the reaction.
Harvesting of bacteria and preparation of chromosomal DNA.
Cells were recovered by centrifugation and washed three times in acidified water (pH 1.8), and extraneous sulfur species were removed by a process of low- and high-speed centrifugation. Washed cell pellets were resuspended in TE (0.01 M Tris, 0.001 M EDTA)–0.15 M NaCl (pH 7.6) buffer. Cells resuspended in TE-NaCl (pH 7.6) buffer were used for the preparation of chromosomal DNA as described by Breed et al. (3).
TAFE.
Tetrathionate-grown cells were washed twice and resuspended in SET buffer (25% sucrose, 2 mM EDTA, 50 mM Tris, pH 8) to give an optical density at 600 nm of 1. The cells were set in an equal volume of 2% LMP agarose (SeaPlaque; FMC Bioproducts) in the presence of proteinase K at 1 mg/ml. The plugs were incubated in ESP buffer (0.5 M EDTA [pH 8], 1% sodium lauryl sarcosine, proteinase K at 1 mg/ml) for 16 h at 50°C (repeated twice) to lyse the cells. Proteinase K was inactivated by incubation of the plugs in TE buffer containing 5 mM Pefabloc (Roche Molecular Biochemicals) for 16 h at 4°C. The DNA-containing plugs were washed for 30 min at 4°C in 5 volumes of distilled water, pre-equilibrated, in restriction buffer for 1 h at 4°C, and digested according to the supplier's instructions, in 3 volumes of fresh restriction buffer containing restriction enzyme. Trans-alternating field electrophoresis (TAFE) was performed with a Beckman GeneLine apparatus. DNA fragments were separated in a 1% agarose (SeaKem LE; FMC Bioproducts) gel at 150 mA and 12°C for 16 h with a pulse interval of 13 s.
Plasmid copy number determination.
Estimation of plasmid copy number was performed in a slot blot experiment by hybridizing plasmid DNA to known amounts of total DNA isolated from plasmid-containing cells and known amounts of purified plasmid DNA. By comparing the amounts of total DNA and plasmid DNA giving equivalent hybridization signals, the amount of plasmid DNA in a sample of total DNA could be estimated. As the approximate sizes of the plasmid and chromosomal DNAs are known, it was possible to calculate the number of copies of each plasmid per chromosome.
Incompatibility assay.
The ability of an incoming plasmid to displace a resident plasmid was used as the test for incompatibility. Transformation-competent, plasmid-containing E. coli DH5α host cells were transformed with a second plasmid and plated on antibiotic-containing medium which selected only for the incoming plasmid. Transformants were restreaked so as to obtain colonies derived from single cells on solid medium containing an antibiotic which again selected only for the newly acquired plasmid. Ten colonies were picked and plated onto two sets of solid medium containing a single antibiotic to separately test for the presence of the newly acquired plasmid or the plasmid which was resident at the start of the experiment. Controls to check for spontaneous loss of resident plasmids were carried out using the same procedure, except that plasmid-containing, competent E. coli cells were taken through two cycles of cell growth on solid medium without antibiotic selection before testing for retention of the resident plasmid.
Host range determination.
Electroporation of P. putida was performed with a Gene Pulser electroporation apparatus (Bio-Rad Laboratories) by using the protocol described previously (16). P. putida was grown to mid-log phase (optical density at 600 nm, 0.4) at 30°C, harvested by centrifugation, and washed once in sterile cold water. The cells were then washed twice in sterile cold 300 mM sucrose (electroporation buffer). The washed cells were resuspended in the electroporation buffer, and 100 μl of this sample was placed in a prechilled electroporation cuvette with pTC-F101 plasmid DNA. The electrical settings were as follows: voltage, 12.5 kV/cm; capacitance, 25 μF; pulse controller parallel resistance, 200 Ω. Immediately after discharge, 900 μl of Luria medium was added to the electroporated cells, which were then incubated at 30°C for 2 to 3 h prior to plating on selective medium. Triparental mating (26) was used to transform A. tumefaciens LBA 4404.
Nucleotide sequence accession number.
The nucleotide sequence determined in this study has been deposited in the GenBank database under accession number AF325537.
RESULTS
Isolation of pTC-F14 and identification of the plasmid replicon.
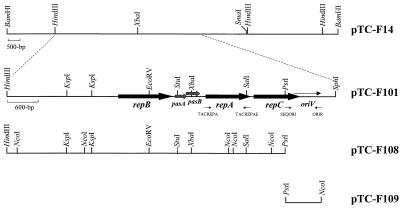
A. caldus strain f was isolated from a pilot plant used to treat a nickel concentrate situated at the Billiton Process Engineering Laboratory (Randburg, South Africa). Pulsed-field gel electrophoresis of total A. caldus DNA indicated that strain f contained at least two plasmids, one of approximately 14 kb and one of about 45 kb (data not shown). Restriction endonuclease mapping of the mixed plasmid preparation suggested that the smaller plasmid contained unique XbaI and BamHI sites. These sites were used to clone the 14-kb plasmid, called pTC-F14, into the E. coli pBluescript KS cloning vector using both the XbaI (plasmid pIb) and BamHI (plasmid pK13) sites (Fig. 1). The resulting clones were used to transform an E. coli polA mutant (GW125a), but only pK13 produced transformants. As ColE1-based cloning vectors are unable to replicate in an E. coli polA mutant strain, this result was interpreted as indicating that pTC-F14 had a replicon which was capable of independent replication in E. coli and that disruption of the XbaI site had inactivated this replicon. This interpretation was confirmed by deleting the cloning vector inserted at the pTC-F14 BamHI site and replacing it with a chloramphenicol resistance gene. The resulting construct retained the ability to replicate in both the E. coli polA mutant and polA wild-type cells. A 6.4-kb HindIII-SphI fragment spanning the XbaI site was ligated to a kanamycin resistance gene, and the resulting construct (pTC-F101) was able to replicate in E. coli, confirming that cloning at the XbaI site had disrupted the pTC-F14 replicon.
FIG. 1.
Restriction enzyme and genetic maps of the pTC-F14 replicon and subclones constructed in this study. Genes are shown as solid black or grey arrows, and the oriV region is indicated by the long, thin arrow. The positions of the primers used to amplify repA and construct pTC-F109 are shown by small arrows.
Comparative analysis of the replication region.
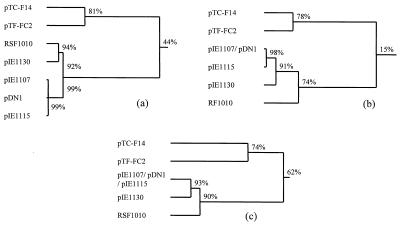
A 4-kb region incorporating the rep genes and oriV (Fig. 1) was sequenced in both directions. The G+C mole percent ratio of this region is 60%, which is typical for IncQ and IncQ-like plasmids (59 to 62%). It contains three open reading frames with high predicted amino acid sequence identity to the RepA, RepB, and RepC proteins of other IncQ-like plasmids (Fig. 2). The highest similarity was to the RepA, RepB, and RepC proteins of A. ferrooxidans plasmid pTF-FC2 (Table 2). Two small open reading frames with high amino acid sequence identity to the PasA and PasB proteins of pTF-FC2 (32) were located between the repB and repA genes (Fig. 1 and Table 2). The pas genes of pTF-FC2 have been shown to function as a plasmid addiction system which enhances plasmid stability through postsegregational killing of plasmid-free cells. The region of pTC-F14 which was required in cis for the plasmid to replicate was situated on a 716-bp fragment between the PstI and NcoI sites (Fig. 1). This oriV region contained five iterons, with the central three 22-bp iterons being perfectly conserved while flanking iterons had either a single-base-pair insertion or a single-base-pair deletion (Fig. 3). The iterons overlapped the predicted C terminus of the RepC protein. IncQ plasmids are characterized by having a G+C-rich region (24 out of 28 bp) about 25 bp downstream of the iterons (28). This is followed by an A+T-rich region (23 out of 31 bp) which is believed to be the site of the DNA melting which takes place before replication. Plasmid pTC-F14 has no equivalent G+C-rich region but does have an extended A+T-rich region (29 out of 40 bp). However, a 28-of-28-bp G+C-rich region which contains two internal 12-bp directly repeated sequences is located approximately 100 bp downstream of the extended A+T-rich region. Other features within the oriV region are two inverted repeats, one of 8 bp and one of 12 bp, with predicted stem-loop ΔG values of −0.3 and −15.2 kJ/mol, respectively. The 12-bp, but not the 8-bp, inverted repeat is therefore likely to form a stem-loop structure. Stem-loop structures have been shown to play a role in the initiation of IncQ plasmid replication (14, 15, 19, 21), but whether the 12-bp direct repeats play a role in pTC-F14 replication is unknown.
FIG. 2.
Relationships between the replicons of plasmids of the IncQ-like family based on percent amino acid sequence identities of the replication proteins. a, RepA, b, RepB; and c, RepC. Accession numbers are as follows: pTC-F14, AF325537; pTF-FC2, M35249 and M64981; pIE1107, ACZ74787; pIE1115, AJ293027; pIE1130, AJ271879; RSF1010, M28829; pDN1, ACY19120.
TABLE 2.
Replicon-associated proteins of pTC-F14 and comparison with pTF-FC2
| Protein | Translational initiation sites | pTC-F14
|
pTF-FC2
|
% Amino acid identity | ||||
|---|---|---|---|---|---|---|---|---|
| No. of amino acids | Molecular mass (Da) | pI | No. of amino acids | Molecular mass (Da) | pI | |||
| RepA | UCUGGAAAGGAGAACAGCAUG | 291 | 31,289 | 5.92 | 290 | 31,227 | 6.21 | 81.0 |
| RepB | CAGGAGAGGGCACAGGCGAUG | 352 | 40,623 | 9.73 | 352 | 40,111 | 9.77 | 78.4 |
| RepC | UACCCAGGGAGGCAAGCCAUG | 303 | 33,712 | 9.28 | 299 | 33,740 | 8.99 | 74.2 |
| PasA like | UUUGAGCAGGAGCUAAACAUG | 74 | 8,523 | 4.46 | 74 | 8,453 | 4.71 | 81.1 |
| PasB like | AGGAAGUGGAGCGCGAUCUUG | 90 | 10,483 | 10.36 | 90 | 10,307 | 10.4 | 72.2 |
Putative ribosome binding sites are in bold, and start codons are in italics.
FIG. 3.
Nucleotide sequence of the PstI-NcoI fragment (716 bp) containing the functional oriV region of pTC-F14. The amino acid sequence of the C-terminal region is shown below the nucleotide sequence. Iterons and direct repeats are shown by arrows, and inverted repeats are shown by broken arrows. The A+T-rich region is underlined, and 12-bp direct repeats are located within the G+C-rich region.
Plasmid pTC-F14 appears to have a broad host range.
As plasmid pTC-F14 was isolated from A. caldus and pTC-F14 with a chloramphenicol marker (pTC-F14Cm) was capable of replication in E. coli, it appears that, like its IncQ relatives, pTC-F14 has a broad-host-range replicon. To obtain additional evidence for the broad-host-range property of the pTC-F14 replicon, pTC-F101 (Fig. 1) was transformed into P. putida and A. tumefaciens LBA 4404 using electroporation and triparental mating, respectively. The presence of the pTC-F14 replicon was confirmed by PCR with primers specific for the repA gene of pTC-F14 (data not shown). The species identity of the transformants was confirmed by API 20E strips (bioMérieux sa, Marcy-l'Etoile, France). Furthermore, we tested for the presence of pTC-F101 in the absence of selection after 100 generations of growth in E. coli, P. putida, and A. tumefaciens. No plasmid loss was detected in E. coli DH5α or P. putida, but after 60 generations of growth in A. tumefaciens LBA 4404, the plasmid had been completely lost from the population.
Incompatibility among pTC-F14, IncQ, and IncQ-like plasmids.
Replicon-associated plasmid incompatibility is believed to arise from the inability of the host cell to correct fluctuations in copy number between plasmids that have elements of their replication machinery in common (23). It has previously been established that the iterons of IncQ and related plasmids are able to exert incompatibility (6, 7, 18, 24). Since the IncQ and IncQ-like plasmids share iteron sequence similarity (Fig. 4), we wished to determine which members of the IncQ plasmid family were incompatible with each other. Plasmids pIE1107 and RSF1010 were previously found to be incompatible due to the presence of nonessential iterons present on pIE1107 that are identical to IncQ iterons. When these iterons were deleted, the resulting plasmid, pIE1108, was fully compatible with RSF1010 (35). We tested the functional replicon of pTC-F14 for incompatibility with the IncQ plasmid R300B replicon (pKE462) and the replicons of IncQ-like plasmids pIE1108 and pTF-FC2 (pDER404). The replicons which, according to Rep protein sequence identity, were most closely related to each other (pIE1108 and R300B, pTC-F14 and pTF-FC2) were fully compatible in E. coli (Table 3). However, the two pairs of less well-conserved replicons were asymmetrically (unidirectionally) incompatible. When pIE1108 was the resident plasmid and either pTC-F14 or pTF-FC2 was the incoming selected plasmid, neither plasmid displaced pIE1108. When either pTC-F14 or pTF-FC2 was the resident plasmid, incoming selected pIE1108 resulted in the displacement of both pTC-F14 and pTF-FC2. A resident IncQ R300B replicon could not be displaced by incoming pTC-F14 or pTF-FC2. However, an incoming selected IncQ R300B replicon displaced resident pTC-F14 but not resident pTF-FC2. Plasmid incompatibility testing should be treated with caution when using plasmids which have a postsegregational killing system. However, in the case of both pTC-F14 and pTC-FC2, it was the plasmid which possessed the postsegregational killing system that was displaced in an E. coli host. The four plasmids could therefore be distinguished from each other based on incompatibility testing with the plasmids which were originally isolated from heterotrophic bacteria having an incompatibility advantage over those isolated from autotrophic bacteria when tested for displacement in E. coli. As regions other than the oriV region of plasmids have been isolated and shown to exert plasmid incompatibility (6, 23, 35), plasmids pTC-F108 and pTC-F109 (Fig. 1) were used to search for further incompatibility elements in the pTC-F14 replicon. Plasmid pTC-F109 containing only the pTC-F14 oriV region demonstrated incompatibility with resident pTC-F101, while pTC-F108 was compatible. Therefore, within the pTC-F14 replicon region, only oriV-associated plasmid incompatibility could be detected.
FIG. 4.
Sequences and arrangement of the iterons of the IncQ-like plasmids tested in this study. The lengths and positions of partial iterons are indicated above the arrows. Iterons of RSF1010 and pDN1 oriV and pIE1107 oriVa and oriVb are 20 bp long with 2-bp spacers (designated NN and being either AA or CC). In the case of pIE1107 oriVb, the first 2-bp spacer is missing. Iterons of pTF-FC2 and pTC-F14 are 22 bp long (except where shown otherwise), and spacers are absent.
TABLE 3.
IncQ-like plasmid displacement by other IncQ-like plasmids in E. coli
| Incoming replicon (plasmid selected) | % Resident (unselected) plasmids remaining
|
|||
|---|---|---|---|---|
| R300B (pKE462) | pIE1108 | pTF-FC2 (pDER404) | pTC-F14 (pTC-F101) | |
| R300B (pKE462) | 100 | 100 | 0 | |
| pIE1108 | 100 | 0 | 0 | |
| pTF-FC2 (pDER404) | 100 | 100 | 100 | |
| pTC-F14 (pTC101) | NEa | 100 | 100 | |
NE, plasmid not established; pTC-F14 could not be transformed into E. coli (pKE462).
Copy number of pTC-F14.
We wished to determine the copy number of pTC-F14 in both E. coli and the original A. caldus strain, f, from which it was isolated. Plasmid copy number was determined by the Southern hybridization technique using genome sizes of 4.7 Mb for E. coli (www.tigr.org) and 2.8 Mb for A. caldus. The genome size of A. caldus was estimated by digesting chromosomal DNAs from several strains with the relatively rarely cutting restriction endonucleases XbaI and DraI, followed by separation of the restriction fragments using a TAFE pulsed-field gel electrophoresis apparatus and summation of the sizes (data not shown). By using the relative sizes of the plasmid and chromosome DNAs, as well as the quantities of plasmid and total DNAs loaded onto the hybridization membrane, the copy number of pTC-F14 was calculated to be 12 to 16 copies per chromosome for both E. coli and A. caldus. This is approximately the same as the 12 to 15 copies (6) and 10 to 14 copies (32) estimated for plasmid pTF-FC2 in E. coli.
Specificity of the pTC-F14 oriV region for its own replication proteins.
Since the replication proteins of plasmids pTC-F14 and pTF-FC2 are fairly closely related (Table 2), we wished to determine whether the pTC-F14 oriV region could be complemented in trans by the pTF-FC2 plasmid replicon and vice versa. To test this, E. coli GW125a (pDER404, pTF-FC2 replicon) and E. coli GW125a (pTC-F101, pTC-F14 replicon) were transformed with pTC-F109 containing the pTC-F14 oriV region cloned into pGEM-T (Promega Corp.) vector. Since the vectors in which the oriV regions were cloned do not replicate in strain GW125a (polA) and since the oriV regions cannot replicate unless the replication proteins are provided in trans, transformants would only be obtained if replication protein complementation had occurred. Transformants were obtained for E. coli GW125a(pTC-F101) and not E. coli GW125a(pDER404). This indicated that the replication proteins of pTC-F14, but not those of pTF-FC2, were able to complement the pTC-F14 oriV region. In a reciprocal experiment, E. coli GW125a(pDER404) and E. coli GW125a(pTC-F101) were transformed with pTV4164 containing the pTF-FC2 oriV region Transformants were obtained for E. coli GW125a(pDER404) but not E. coli GW125a(pTC-F101). Therefore, each oriV region could be complemented in trans by its own replication proteins but not by those of the other plasmid. We also attempted to transform pTC-F109 and pTV4164 into E. coli GW125a(pKE462) and E. coli GW125a(pIE1108) recipients to test whether the R300B IncQ or the pIE1108 replication proteins were able to complement either the pTC-F14 or the pTF-FC2 oriV region. No transformants were obtained, which suggested that cross-complementation did not occur.
DISCUSSION
Plasmid pTC-F14 is the second plasmid to be isolated from acidophilic, obligately chemolithotrophic iron- or sulfur-oxidizing bacteria which has an IncQ-like replicon. All other IncQ-like plasmids have been isolated from heterotrophic bacteria found in medical and animal-associated environments. Since the ecological niche occupied by acidiphilic chemolithotrophs is very different from that occupied by mammal-associated heterotrophs, it is likely that IncQ-like replicons will be found in other ecological niches and await discovery. How many groups of IncQ replicons exist is therefore uncertain, but those so far discovered fall into two major groupings. The existence of at least two replicon groups is supported by several observations. There is a fairly deep division based on amino acid sequence comparison of the RepA, RepB, and RepC proteins (Fig. 2). In addition, one group has replicons with 20-bp iterons and 2-bp spacers while the other group has 22-bp iterons without spacers. Furthermore, plasmids pTC-F14 and pTF-FC2 also both have plasmid addition system (pas) genes between the repB and repA genes whereas none of the other IncQ and IncQ-like plasmids have similar genes within the replicon. Interestingly, pTF-FC2 is unique in having three pas genes (pasA, pasB, and pasC) whereas pTC-F14 is more typical of other plasmid addition systems in having only two. Division into two groups is even more strongly supported by the observation that RSF1010, pIE1107, pIE1115, pIE1130, and pDN1 have IncQ-like mobilization proteins while pTF-FC2 and pTC-F14 have IncP-related mobilization proteins (27; pTC-F14 partial sequence data). The relatedness between the mobilization genes of pTC-F14 and pTF-FC2 raises many questions regarding whether the mobilization system of one plasmid can transfer the other, and this will be the subject of future studies.
Based on plasmid incompatibility testing, Tietze (35) has suggested that IncQ and IncQ-like plasmids should be named differently to distinguish between the different incompatibility groups. It was proposed that the IncQ plasmids (RSF1010, R1162, and R300B) should be named IncQα plasmids and that the plasmid pIE1107 replicon (and its incompatible relatives) should be called IncQβ. Plasmid pDN1 and the recently discovered plasmid pIE1115 also belong to this group. Tietze and coworkers have recently discovered an IncQ-like plasmid, pIE1130, which belongs to a different incompatibility group which they propose to call IncQγ (31; E. Tietze and K. Smalla, Plasmid Biology 2000, p. 167). Plasmid pTC-F14 clearly belongs to an incompatibility group which is different from pTF-FC2 or any other previously reported IncQ-like plasmids. Furthermore, it is clear that there are at least two major groupings of IncQ-like plasmids, with the IncQα, IncQβ, and IncQγ plasmids belonging to one group and pTF-FC2 and pTC-F14 belonging to the other. We suggest that the former group be called IncQ-like group 1 plasmids, of which 1α, 1β, and 1γ are the currently known incompatibility subgroups. Plasmids pTF-FC2 and pTC-F14 should be called IncQ-like group 2 plasmids, and as these two plasmids are compatible, group 2 would consist of two incompatibility subgroups. We propose that the first discovered plasmid, pTF-FC2, should be designated a member of the IncQ-like 2α incompatibility subgroup, with pTC-F14 being a member of the IncQ-like 2β incompatibility subgroup.
The copy number of some iteron-containing replicons has been shown to be increased or decreased by the deletion or addition of iterons, respectively (33, 34, 36). Plasmid pTF-FC2 has three perfectly conserved 22-bp iterons, compared with the five iterons of pTC-F14, of which only the central three 22-bp iterons are perfectly conserved. We thought it possible that a reason for the increased number of iterons in pTC-F14 was to achieve a reduction in plasmid copy number with the concurrent reduction in metabolic burden to the host cell. However, the estimated copy numbers of plasmids pTC-F14 and pTF-FC2 appear to be similar and within the range of the 12 to 15 copies per chromosome reported for IncQ plasmids (10). In a study of the effects of mutations on the functionality of RSF1010 iterons, it was found that even a single-base-pair replacement in one of the iterons could result in a nonfunctional iteron-containing region and the inability of RSF1010 to replicate (22). It is therefore possible that the 23- or 21-bp iterons which flank the 22-bp iterons are nonfunctional and therefore not active in copy number control. This remains to be tested.
The functions of elements within the oriV regions of the IncQ plasmids have been extensively studied (see reference 28 for a review). The suggested model for initiation of replication is that the RepC proteins bind to the 20-bp iterons and this induces DNA melting and strand opening at the A+T-rich region (17). The A+T-rich region is about 60 bp from the iterons, and the two are separated by a G+C-rich region. The RepA helicase binds to the opened region and unwinds the DNA, probably mainly in the direction away from the iterons, until a pair of single-strand initiation sites (ssiA and ssiB) are reached. Each of these ssi sites has twofold symmetry, and they are situated within a region of extended twofold symmetry with a 40-bp stem and a 40-bp loop (19). The RepB primase is required to initiate replication at these ssi sites, which are arranged in such a way that replication in opposite directions takes place from each of the ssi sites by a strand displacement mechanism (14, 15, 38). Plasmid pTC-F14 clearly has the equivalent of the RepA, RepB, and RepC proteins, as well as an oriV region containing iterons, A+T-rich and G+C-rich regions, and a pair of sequences with twofold symmetry. However, several features, such as the number and sequence of the iterons, the relative positions of the A+T- and G+C-rich regions, the presence of two 12-bp direct repeats, and the lack of a region of extended twofold-symmetry, are unique to pTC-F14. Determination of whether these differences represent significant differences in the mechanism of pTC-F14 replication awaits further study. We found that the oriV regions of pTC-F14 and pTF-FC2 could be complemented only by their own replication proteins. The basis of this specificity presumably resides in the nucleotide sequences of the iterons and other features of the oriV region. For example, if each RepC protein binds specifically to its own iterons, the initiation of replication will be prevented by the lack of RepC-oriV binding. It is also possible that replicon specificity resides in the RepA or RepB protein and features of the oriV region with which they interact. We propose to investigate this by placing different combinations of rep genes expressed from non-IncQ-related replicons in trans with different features of the oriV regions to examine the basis of IncQ replication specificity.
ACKNOWLEDGMENTS
We are grateful to Erhard Tietze for providing plasmid pIE1108.
We are grateful to Billiton Process Research (Randburg, South Africa), the National Research Foundation (Pretoria, South Africa), and the University of Stellenbosch for funding support.
REFERENCES
- 1.Altshul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein data base search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barth P T, Grinter N J. Comparison of the deoxyribonucleic acid molecular weights and homologies of plasmids conferring linked resistance to streptomycin and sulfonamides. J Bacteriol. 1974;120:618–630. doi: 10.1128/jb.120.2.618-630.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breed A W, Dempers C J N, Searby G E, Gardner M N, Rawlings D E, Hansford G S. The effect of temperature on the ferrous iron oxidation kinetics of Leptospirillum ferrooxidans in continuous culture. Biotech Bioeng. 1999;65:44–53. doi: 10.1002/(sici)1097-0290(19991005)65:1<44::aid-bit6>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 4.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the p15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derbyshire K M, Willets N S. Mobilization of the non-conjugative plasmid RSF1010: a genetic analysis of its origin of transfer. Mol Gen Genet. 1987;206:154–160. doi: 10.1007/BF00326551. [DOI] [PubMed] [Google Scholar]
- 6.Dorrington R A, Rawlings D E. Identification and sequence of a broad-host-range plasmid isolated from Thiobacillus ferrooxidans. J Bacteriol. 1989;171:2735–2739. doi: 10.1128/jb.171.5.2735-2739.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorrington R A, Rawlings D E. Characterization of the minimum replicon of the broad-host-range plasmid pTF-FC2 and similarity between pTF-FC2 and the IncQ plasmids. J Bacteriol. 1990;172:5697–5705. doi: 10.1128/jb.172.10.5697-5705.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorrington R A, Bardien S, Rawlings D E. The broad-host-range plasmid pF-FC2 requires a primase-like protein for autonomous replication in Escherichia coli. Gene. 1991;108:7–14. doi: 10.1016/0378-1119(91)90481-p. [DOI] [PubMed] [Google Scholar]
- 9.Franklin F C H, Bagdasarian M, Bagdasarian M M, Timmis K N. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and the cloning of the genes for the entire regulated aromatic ring meta-cleavage pathway. Proc Natl Acad Sci USA. 1981;78:7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frey J, Bagdasarian M. The biology of IncQ plasmids. In: Thomas C M, editor. Promiscuous plasmids of Gram-negative bacteria. London, England: Academic Press; 1989. pp. 79–93. [Google Scholar]
- 11.Guerry P, van Embden J, Falkow S. Molecular nature of two nonconjugative plasmids carrying drug resistance genes. J Bacteriol. 1974;117:619–630. doi: 10.1128/jb.117.2.619-630.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hallberg K B, Lindström E B. Characterization of Thiobacillus caldus sp. nov., a moderately thermophilic acidophile. Microbiology. 1994;140:3451–3456. doi: 10.1099/13500872-140-12-3451. [DOI] [PubMed] [Google Scholar]
- 13.Hoekema A, Hirsch P R, Hooykaas P J J, Schilperoort R A. A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature. 1983;303:179–180. [Google Scholar]
- 14.Honda Y, Sakai H, Komano T, Bagdasarian M. RepB' is required in trans for the two single-strand DNA initiation signals in oriV of plasmid RSF1010. Gene. 1989;80:155–159. doi: 10.1016/0378-1119(89)90261-8. [DOI] [PubMed] [Google Scholar]
- 15.Honda Y, Sakai H, Hiasa H, Tanaka K, Komano T. Functional division and reconstruction of a plasmid replication origin: molecular dissection of the oriV of the broad-host-range plasmid RSF1010. Proc Natl Acad Sci USA. 1991;88:179–183. doi: 10.1073/pnas.88.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwasaki K, Uchiyama H, Yagi O, Kurabayashi T, Ishizuka K, Takamura Y. Transformation of Pseudomonas putida by electroporation. Biosci Biotech Biochem. 1994;58:851–854. doi: 10.1271/bbb.58.851. [DOI] [PubMed] [Google Scholar]
- 17.Kim Y-J, Meyer R J. An essential iteron-binding protein required for plasmid R1162 replication induces localized melting within the origin at a specific site in AT-rich DNA. J Bacteriol. 1991;173:5539–5545. doi: 10.1128/jb.173.17.5539-5545.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin L-S, Kim Y-J, Meyer R J. The 20-bp. Directly repeated DNA sequence of broad host range plasmid R1162 exerts incompatibility in vivo and inhibits R1162 DNA replication in vitro. Mol Gen Genet. 1987;208:390–397. doi: 10.1007/BF00328129. [DOI] [PubMed] [Google Scholar]
- 19.Lin L-S, Meyer R J. DNA synthesis is initiated at two positions within the origin of replication of plasmid RSF1010. Nucleic Acids Res. 1987;20:8319–8331. doi: 10.1093/nar/15.20.8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer R, Hinds M, Brasch M. Properties of R1162, a broad-host-range, high-copy-number plasmid. J Bacteriol. 1982;150:552–562. doi: 10.1128/jb.150.2.552-562.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miao D-M, Honda Y, Tanaka K, Higashi A, Nakamura T, Taguchi Y, Sakai H, Komano T, Bagdasarian M. A base-paired hairpin structure essential for the functional priming signal for DNA replication of the broad-host-range plasmid RSF1010. Nucleic Acids Res. 1993;21:4900–4903. doi: 10.1093/nar/21.21.4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miao D-M, Sakai H, Okamato S, Tanaka K, Okuda M, Honda Y, Komano T, Bagdasarian M. The interaction of RepC initiator with iterons in the replication of the broad-host-range plasmid RSF1010. Nucleic Acids Res. 1995;23:3295–3300. doi: 10.1093/nar/23.16.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novick R P. Plasmid incompatibility. Microbiol Rev. 1987;51:381–395. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Persson C, Nordstrom K. Control of replication of the broad host range plasmid RSF1010: the incompatibility determinant consists of directly repeated DNA sequences. Mol Gen Genet. 1986;203:189–192. doi: 10.1007/BF00330402. [DOI] [PubMed] [Google Scholar]
- 25.Rawlings D E, Coram N J, Gardner M N, Deane S M. Thiobacillus caldus and Leptospirillum ferrooxidans are widely distributed in continuous flow biooxidation tanks used to treat a variety of metal containing ores and concentrates. In: Amils R, Ballester A, editors. Biohydrometallurgy and the environment toward the mining of the 21st century. Part A. Amsterdam, The Netherlands: Elsevier; 1999. pp. 777–786. [Google Scholar]
- 26.Rawlings D E, Woods D R. Mobilization of Thiobacillus ferrooxidans plasmids among Escherichia coli strains. Appl Environ Microbiol. 1985;49:1323–1325. doi: 10.1128/aem.49.5.1323-1325.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rohrer J, Rawlings D E. Sequence analysis and characterization of the mobilization region of a broad host-range plasmid, pTF-FC2, isolated from Thiobacillus ferrooxidans. J Bacteriol. 1992;174:6230–6237. doi: 10.1128/jb.174.19.6230-6237.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakai H, Komano T. DNA replication of IncQ broad host-range plasmids in Gram-negative bacteria. Biosci Biotech Biochem. 1996;60:377–382. doi: 10.1271/bbb.60.377. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Scholtz P, Haring V, Wittmann-Liebold B, Ashman K, Bagdasarian M, Scherzinger E. Complete nucleotide sequence and gene organization of the broad host-range plasmid RSF1010. Gene. 1989;75:271–288. doi: 10.1016/0378-1119(89)90273-4. [DOI] [PubMed] [Google Scholar]
- 31.Smalla K, Heuer H, Götz A, Niemeyer D, Krögerrecklenfort E, Tietze E. Exogenous isolation of antibiotic resistance plasmids from piggery manure slurries reveals a high prevalence and diversity of IncQ-like plasmids. Appl Environ Microbiol. 2000;66:4854–4862. doi: 10.1128/aem.66.11.4854-4862.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith A G S, Rawlings D E. The poison-antidote system of the broad host-range Thiobacillus ferrooxidans plasmid pTF-FC2. Mol Microbiol. 1997;25:961–970. doi: 10.1046/j.1365-2958.1997.6332000.x. [DOI] [PubMed] [Google Scholar]
- 33.Thomas C M, Stalker D M, Helinski D R. Replication and incompatibility properties of segments of the origin of replication of the broad host range plasmid RK2. Mol Gen Genet. 1981;181:1–7. doi: 10.1007/BF00338996. [DOI] [PubMed] [Google Scholar]
- 34.Thomas C M, Cross M A, Hussain A A K, Smith C A. Analysis of copy number control elements in the region of the vegetative replication region of the broad host range plasmid RK2. EMBO J. 1984;3:57–63. doi: 10.1002/j.1460-2075.1984.tb01761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tietze E. Nucleotide sequence and genetic characterization of the novel IncQ-like plasmid pIE1107. Plasmid. 1998;39:165–181. doi: 10.1006/plas.1998.1343. [DOI] [PubMed] [Google Scholar]
- 36.Tsutsui H, Fujiyama A, Murotsu T, Matsubara K. Role of nine repeating sequences of the mini-F genome for expression of F-specific incompatibility phenotype and copy number control. J Bacteriol. 1983;155:337–344. doi: 10.1128/jb.155.1.337-344.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whittle G, Katz M E, Clayton E H, Cheetham B F. Identification and characterization of a native Dichelobacter nodosus plasmid, pDN1. Plasmid. 2000;43:230–234. doi: 10.1006/plas.1999.1456. [DOI] [PubMed] [Google Scholar]
- 38.Zhou H, Meyer R J. Deletion of sites for initiation of DNA synthesis in the origin of broad host-range plasmid R1162. J Mol Biol. 1990;214:685–697. doi: 10.1016/0022-2836(90)90286-u. [DOI] [PubMed] [Google Scholar]