Abstract
Age-related macular degeneration (AMD) is a blinding disease characterised by dysfunction of the retinal pigmented epithelium (RPE) which culminates in disruption or loss of the neurosensory retina. Genome-wide association studies have identified >60 genetic risk factors for AMD; however, the expression profile and functional role of many of these genes remain elusive in human RPE. To facilitate functional studies of AMD-associated genes, we developed a human RPE model with integrated CRISPR interference (CRISPRi) for gene repression by generating a stable ARPE19 cell line expressing dCas9-KRAB. We performed transcriptomic analysis of the human retina to prioritise AMD-associated genes and selected TMEM97 as a candidate gene for knockdown study. Using specific sgRNAs, we showed that knockdown of TMEM97 in ARPE19 reduced reactive oxygen species (ROS) levels and exerted a protective effect against oxidative stress-induced cell death. This work provides the first functional study of TMEM97 in RPE and supports a potential role of TMEM97 in AMD pathobiology. Our study highlights the potential for using CRISPRi to study AMD genetics, and the CRISPRi RPE platform generated here provided a useful in vitro tool for functional studies of AMD-associated genes.
Keywords: age-related macular degeneration, retinal pigmented epithelium, CRISPR interference, retinal degeneration, TMEM97
1. Introduction
Age-related macular degeneration (AMD) is one of the leading causes of blindness in the western world, with a prevalence of ~8.7%, and is projected to cause vision loss in 288 million people worldwide by 2040 [1]. AMD is characterised by abnormalities of retinal pigment epithelium (RPE) at the macula region, leading to degeneration of the overlying photoreceptors and, subsequently, progressive loss of vision. Genetic and epidemiologic studies have greatly advanced our understanding of AMD pathology. Genome-wide association studies (GWAS) have successfully identified loci associated with genetic variants implicated with AMD risk and traits, with over 60 genetic variants being associated with AMD [2,3,4,5,6], including variants within the APOE, CFH, ARMS2/HTRA1 and TMEM97/VTN loci that confer risk of developing AMD. Integration with gene expression data in expression quantitative trait loci (eQTL) studies have further improved the methods to identify AMD-associated genes; however, pinpointing the exact gene variants within the loci that confer AMD risks remain challenging, which is reflective of the complexity of AMD genetics (reviewed in [7]). Understanding the function of these AMD-associated genes in the retina, as well as their expression patterns within the major retinal cells, would be important to unravel the complex genetics of AMD; however, many AMD-associated genes in the retina remain understudied. In particular, a landmark GWAS study by Frische et al. identified that the SNP rs11080055, located within the TMEM97 gene, is associated with AMD [2]. TMEM97 was recently identified as the Sigma-2 receptor [8], which plays a key role in mediating Ca2+ and K+ signalling cascades in the endoplasmic reticulum and has been implicated in cancer development and a range of neurological degenerative diseases [9]. Previous studies showed that activation of Sigma-2 receptor signalling by a number of different ligands induces apoptosis, cytotoxicity, and generation of reactive oxygen species [9,10,11]. On the other hand, Sigma-2 receptor antagonists were shown to provide a protective effect following traumatic brain injury [12]. The Sigma-2 receptor signalling pathway has emerged as an attractive therapeutic target to treat several CNS degenerative diseases, such as Alzheimer’s disease and schizophrenia [13,14]. Development of new tools to facilitate functional study of AMD associated genes in RPE, such as TMEM97, would be important to better understand the mechanisms implicated in AMD pathobiology.
The emergence of CRISPR technology provides an efficient toolset for gene editing in the retina [15]. Moreover, a recent development has highlighted the use of a nuclease-null Cas9 coupled with transcriptional repressors to modulate endogenous gene expression levels, termed CRISPR interference (CRISPRi), which offers a robust system to study gene function [16]. Notably, CRISPRi exhibits minimal off-target effects and is capable of multiplexed gene repression [17]. Several CRISPRi sgRNA libraries are also available, which allows high-throughput genome-wide screening of gene repression [18,19]. In this study, we describe the development of a CRISPRi platform in human RPE cells that would facilitate loss-of-function studies for AMD-associated genes. We report the generation of a human RPE cell line, ARPE19, that stably expresses dCas9-KRAB. This CRISPRi system utilises an inactive SpCas9 (dCas9) coupled with the transcriptional repressor domain Kruppel associated box (KRAB), which can be directed by targeting the sgRNA to induce specific gene repression [20]. We performed a transcriptomic analysis of human retina to prioritise AMD-associated genes for functional study and showed that TMEM97 was highly expressed in human RPE/choroid. Using our CRISPRi RPE system, we showed that knockdown of TMEM97 exerted a protective effect on ARPE19 against oxidative stresses. Our results support a functional role of TMEM97 in regulating oxidative stress and degeneration of RPE, which have implications in the development of new therapeutic targets to improve treatment options for AMD.
2. Results
2.1. Generation of a Stable ARPE19 Cell Line for CRISPRi
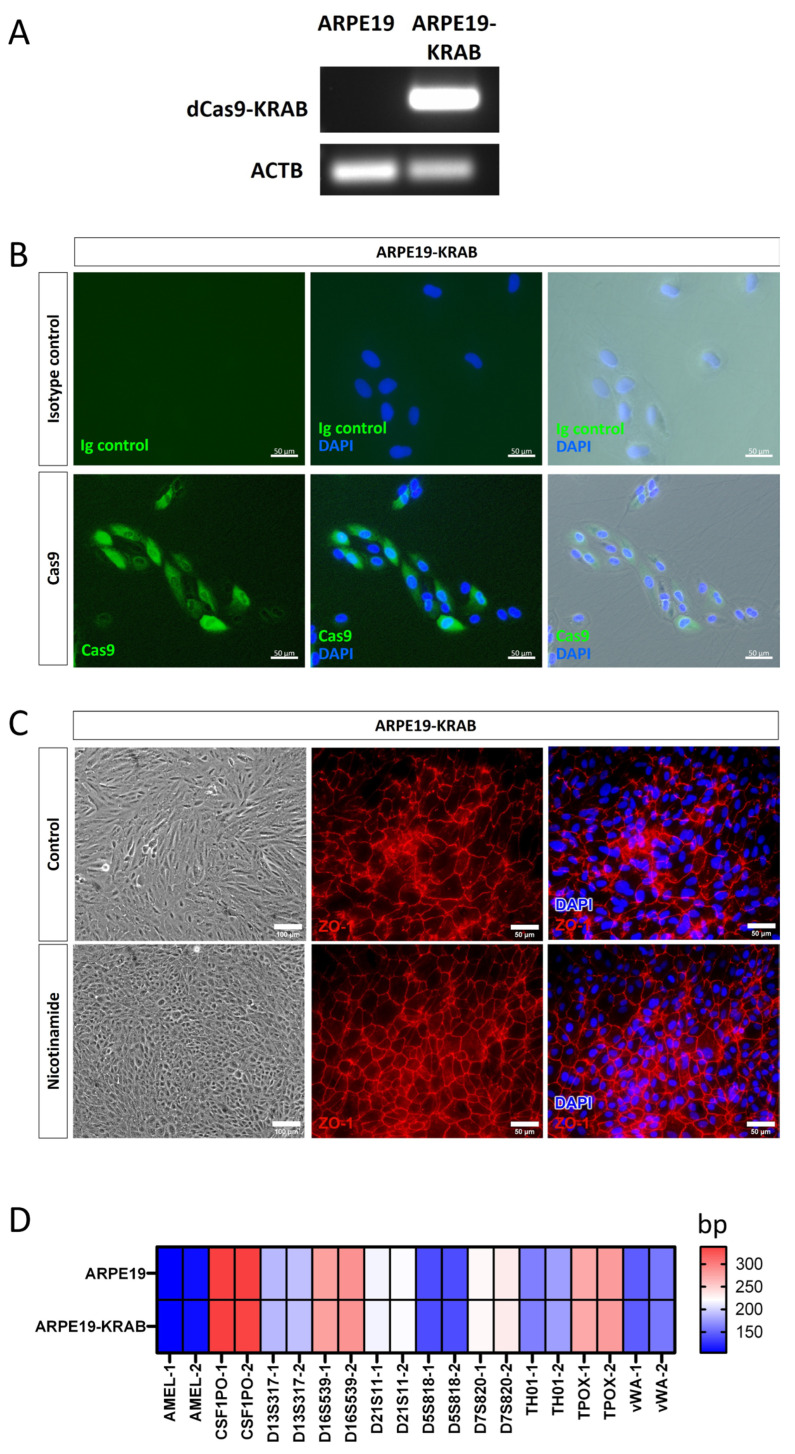
To facilitate the use of CRISPRi in retinal research, we generated a human RPE cell line that stably expresses the transcriptional repressor dCas9-KRAB under the EF1α promoter, termed ARPE19-KRAB. We performed detailed characterisation of the derived ARPE19-KRAB cells. Our results showed that ARPE19-KRAB possessed stable expression of dCas9-KRAB for at least 2 months after transduction (Figure 1A). Furthermore, immunocytochemistry analysis showed that ARPE19-KRAB express detectable levels of SpCas9 protein (Figure 1B), while no signal for SpCas9 was detected in isotype control, nor in the parental ARPE19 (Figure 1B and Figure S1). Using an established method to induce polarisation of ARPE19 with nicotinamide treatment [21], we verified that ARPE19-KRAB retained the ability to form a monolayer with organised hexagonal RPE morphology, as shown by expression of the tight junction protein ZO-1 (Figure 1C). Finally, we performed short tandem repeat (STR) analysis and confirmed that ARPE19-KRAB retained the same STR signature of the parental ARPE19 cell line (Figure 1D). Together, these results validate the quality of the derived ARPE19-KRAB as a retinal pigment epithelium platform with stable expression of a CRISPRi transcriptional repressor.
Figure 1.
Generation of a stable CRISPRi human RPE cell line expressing dCas9-KRAB. Characterization of ARPE19-KRAB cell line showing expression of (A) KRAB by RT-PCR, and (B) SpCas9 by immunocytochemistry. (C) ARPE19-KRAB was able to form polarised monolayer upon nicotinamide treatment. Left panel shows representative morphology of ARPE19-KRAB treated with control condition (10% FCS) or differentiation media with nicotinamide. Middle and right panel show immunocytochemistry of tight junction protein ZO-1. (D) Heatmap of short tandem repeat analysis of 10 polymorphic markers. Alleles 1 and 2 are designated as −1 and −2 respectively.
2.2. Gene Expression of TMEM97/VTN Locus in Human Retina
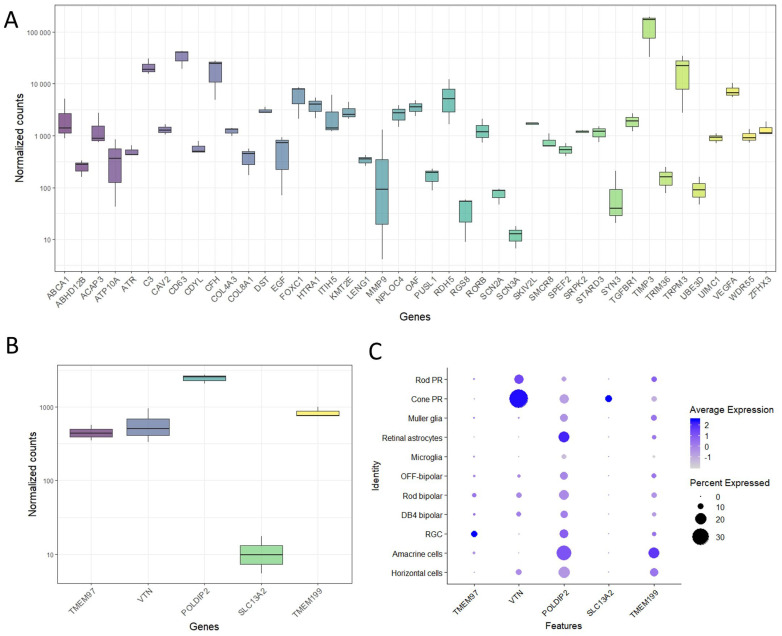
To demonstrate the value of the ARPE19-KRAB system, we set out to pick an AMD-associated gene for further loss-of-function studies using CRISPRi. We performed RNA sequencing (RNAseq) in healthy human RPE/choroid samples from 3 donors ranging from 69 to 77 years old with short retrieval time (4–6 h, Supplementary Table S1). Using a list of AMD-associated genes previously identified by a large GWAS study [2], our transcriptomic data showed that many of the AMD-associated genes are expressed in human RPE/choroid (Figure 2A).
Figure 2.
Expression of AMD-associated genes in human retina. (A) Bulk RNAseq results demonstrated expression of selected AMD-associated genes in human RPE/choroid (n = 3 donors). Expression levels of genes in the TMEM97/VTN locus in (B) human RPE/choroid as shown by bulk RNAseq, and (C) major retinal cells in human neural retina as shown by single cell-RNAseq (n = 3 donors; [22]). PR: photoreceptors; RGC; retinal ganglion cells.
In particular, we focused on the AMD-associated locus TMEM97/VTN, which includes five genes: TMEM97, VTN, POLDIP2, SLC13A2 and TMEM199. Our results showed that RPE/choroid express detectable levels of TMEM97 (447 ± 35 Transcripts Per Million (TPM)), VTN (590 ± 155 TPM), POLDIP2 (2440 ± 16 TPM) and TMEM199 (863 ± 154 TPM), but not SLC13A2 (7 ± 5 TPM, Figure 2B). Next, we performed an in silico analysis using single cell RNAseq data we previously reported for adult human neural retina [22]. Our results revealed that the retinal ganglion cells possessed strong expression of TMEM97 (Figure 2C). POLDIP2 and TMEM199 were expressed ubiquitously in many retinal cell types, including photoreceptors (rods and cones), Müller glia, astrocytes, bipolar cells, retinal ganglion cells, amacrine cells, and horizontal cells, with the exception that low/no expression was detected in microglia. On the other hand, VTN and SLC13A2 were mainly expressed in the cone photoreceptors (Figure 2C). Collectively, these results show that most of the genes in TMEM97/VTN locus are expressed in both human neural retina and RPE. Given the predominant expression pattern in RPE and lack of ubiquitous expression pattern in neural retinal cells, we selected TMEM97 as a candidate gene to perform further loss-of-function study using our ARPE19-KRAB system.
2.3. Loss-of-Function Study of TMEM97 Using CRISPRi
To induce knockdown of TMEM97 expression, we designed three sgRNAs that target the proximity of the transcription start site (TSS) of TMEM97 gene (Supplementary Figure S2) using the TSS defined by Ensembl (sgRNA 1), and two sgRNAs were designed using the TSS defined by the FANTOM/CAGE promoter dataset (sgRNA 2 and 3; Supplementary Table S2 [23]). We synthesized the three sgRNAs in an expression cassette driven by a U6 promoter, as we previously described [24]. To evaluate the efficiency of the three sgRNAs to knockdown TMEM97 expression, we transiently co-transfected individual sgRNA together with a dCas9-KRAB plasmid into HEK293A cells using Lipofectamine 3000. qPCR analysis showed that sgRNA 1 and 3 did not induce knockdown of endogenous TMEM97 levels in HEK293A, while sgRNA 2 achieved the highest knockdown level and was selected for further experiments (Supplementary Figure S3).
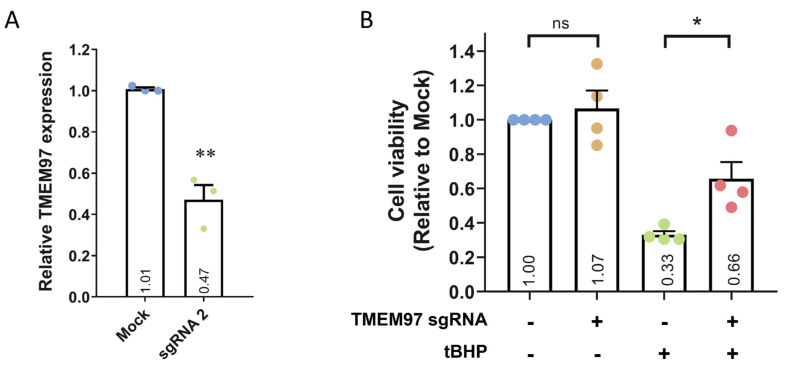
Next, we transfected sgRNA 2 into ARPE19-KRAB, which resulted in ~53% knockdown of TMEM97 after 4 days (Figure 3A and Figure S4). Moreover, transfection with a control scrambled sgRNA did not result in TMEM97 knockdown (Supplementary Figure S4). To further evaluate the specificity of this CRISPRi approach, we performed in silico analysis using Cas-OFFinder [25] to identify the off-targets of TMEM97 sgRNA 2. No off-targets were identified for sgRNA 2 for alignments with up to 2 mismatches. When we adjusted the parameters to allow for three mismatches, five potential off-targets were identified, four of which were located in deep intronic regions, while one was not close to any known genes (Supplementary Table S3). Critically, no transcription start site was located within 1 kbp upstream and 1 kbp downstream of all 5 potential off-targets, supporting the specificity of our CRISPRi approach for TMEM97 repression.
Figure 3.
CRISPRi-mediated TMEM97 knockdown and its effect on cell viability. (A) qPCR analysis of TMEM97 knockdown using CRISPRi in ARPE19-KRAB. Results are presented as the mean ± SEM of three biological repeats, each with three technical repeats. ** indicates p < 0.01. (B) Cell viability analysis of ARPE19-KRAB following TMEM97 knockdown in the presence or absence of tBHP. Results are presented as the mean ± SEM of four biological repeats, each with four to six technical repeats. ns indicates not significant; * indicates p < 0.05.
Using the ARPE19-KRAB system, we assessed if loss of TMEM97 would affect cell viability of ARPE19. Our results demonstrated that CRISPRi-mediated knockdown of TMEM97 did not affect the viability of ARPE19 cells (Figure 3B), suggesting that TMEM97 is not critical in maintaining RPE cell viability under normal conditions. Given that oxidative stress plays a key role in RPE degeneration in AMD [26], we further treated ARPE19-KRAB with tert-Butyl hydroperoxide (tBHP) to induce oxidative stress and better mimic RPE under AMD pathological conditions. Cell viability analysis showed that tBHP treatment for 24 h resulted in dramatic cell death in ARPE19-KRAB (0.33 ± 0.02-fold compared to control, Figure 3B). Interestingly, TMEM97 knockdown resulted in a significant increase in resistance to tBHP-induced cell death (0.66 ± 0.097-fold compared to control, p = 0.03).
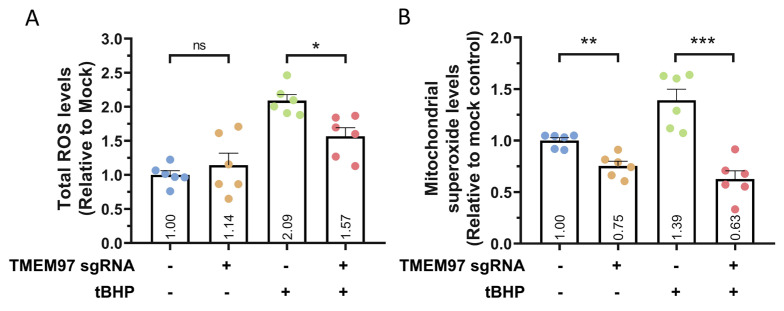
To further study the possible role of TMEM97 in modulating oxidative stress, we utilised the CellRox assay to analyse the levels of reactive oxygen species (ROS). We first induced TMEM97 knockdown in ARPE19-KRAB, followed by tBHP treatment for 24 h to induce oxidative stress. In the presence of tBHP, our results showed that TMEM97 knockdown resulted in a significant decrease in tBHP-induced total ROS levels (p = 0.02, Figure 4A). Similarly, we utilised the MitoSox assay to analyse the mitochondrial superoxide levels. Following tBHP treatment for 24 h, our results indicated that TMEM97 knockdown also reduced mitochondrial superoxide levels compared to control (p = 0.0002, Figure 4B). This provides further support for the protective effect of TMEM97 knockdown against oxidative stress-mediated cell death. In the basal condition, we also observed a 25% decrease in mitochondrial superoxide levels following TMEM97 knockdown (p = 0.001), but overall there were no statistically significant changes to the total ROS levels (p = 0.24, Figure 4). Together, these results show a novel role for TMEM97 in regulating the oxidative stress and viability of RPE and demonstrate the potential of using CRISPRi for functional studies of AMD-associated genes in RPE cells.
Figure 4.
TMEM97 knockdown resulted in reduced ROS levels. Quantification of (A) total ROS levels using CellRox assay and (B) mitochondrial superoxide levels using MitoSox assay, following TMEM97 knockdown in ARPE19-KRAB in the presence or absence of tBHP. n = 6 repeats from 2 independent experiments, error bars represented SEM; ns indicates not significant; * indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001.
3. Discussion
Functional validation of putative causal genes would foster a better understanding of AMD pathophysiology, which is important to develop better diagnosis and treatments for this disease. However, the study of AMD-associated genes in the retina using model organisms is challenging: the lack of macula in rodents represents a major limitation in modelling AMD, while primate studies are limited by the high research cost and the requirement of specialised housing facilities [27]. In this study, we present an in vitro human RPE model with CRISPRi for gene repression and demonstrate the value of our model for knockdown studies of AMD-associated genes using TMEM97 as an example. We were able to identify a sgRNA for successful knockdown using gene annotation from the FANTOM/CAGE project, which is consistent with a previous study [28]. Using this system, we were able to design a sgRNA that targets the immediate downstream region of the TSS to knockdown the expression of TMEM97. To simplify our approach, the ARPE19-KRAB platform can be combined with a sgRNA expression cassette system [24] that can be commercially synthesized, providing a rapid approach to initiate gene knockdown studies using CRISPRi. For long-term studies, our ARPE19-KRAB platform could be complemented with inducible sgRNA expression, such as doxycycline or tamoxifen-inducible systems, to precisely control the duration of gene knockdown. In addition, the ARPE19-KRAB system can also be utilised for genome-wide screening using established CRISPRi libraries [18,19], which would enable future identification of genes that contribute to AMD pathophysiology, such as dysfunctional phagocytosis or degeneration of RPE. Collectively, this study provides a valuable tool to facilitate functional studies of genetic factors that contribute to RPE dysfunction in AMD.
Using the CRISPRi system, we showed that knockdown of TMEM97 did not affect cell viability of ARPE19 under a normal physiological environment. On the other hand, in the presence of oxidative stress, TMEM97 knockdown provided protective effects against cell death in ARPE19. Accumulation of oxidative stress is known to play a key role in AMD pathogenesis and progression, which can be caused by multiple risk factors including aging, light damage, and cigarette smoking [29]. Notably, a recent transcriptome-wide association study predicted that AMD cases exhibit a higher expression level of TMEM97 compared with controls [30], suggesting that TMEM97 levels may modulate RPE degeneration caused by oxidative stresses. Consistent with this, our results provide the first evidence that TMEM97 plays an important role in regulating oxidative stress and cell survival of human RPE, supporting a potential role of TMEM97 inhibition as a therapeutic strategy for treating AMD. Future gain-of-function studies for TMEM97 would be important to further support its role in regulating oxidative stresses and cell death in RPE.
There are limitations in this study. Firstly, we used acute oxidative stress treatment to mimic the accumulated oxidative stress levels related to aging. However, other genetic variants and cellular stresses are also important in the pathogenesis of AMD, such as mitochondrial defects, lipofuscin accumulation, autophagy, and crosstalk between RPE and the immune and vascular systems [31]. Secondly, although the knockdown levels for TMEM97 achieved by our CRISPRi approach were not very high, a clear phenotype in ARPE19 could be observed following TMEM97 knockdown. Future studies to knockout TMEM97 in human RPE models, for instance using CRISPR gene editing, would support our findings and would further elucidate its role in RPE biology and AMD pathophysiology. Finally, although ARPE19 is the most commonly used RPE cell line, which retains some RPE characteristics such as the ability to phagocytose photoreceptor outer segments [32], there are limitations for its use to model native RPE. For instance, ARPE19 display reduced levels of some RPE marker genes (e.g., CRALBP and RPE65) compared to native RPE [33], changes in morphology, heterogeneity, and karyotype following prolonged culture [21,34,35]. Therefore, the CRISPRi ARPE19 model reported here may not be suitable to study AMD genes with significantly different expression levels compared to native RPE. Other in vitro RPE models have been reported, including primary RPE and RPE derived from pluripotent stem cells (reviewed in [7,36]). Future research beyond this study using in vitro RPE models, such as primary human RPE or induced pluripotent stem cell-derived RPE [7], as well as in vivo animal models for AMD [37], would be important to confirm the role of TMEM97 in regulating cell survival and oxidative stress in RPE. Further studies assessing the functional role of TMEM97 in other retinal cells, such as photoreceptors, would also be of high interest. In addition, given the potential of CRISPRi for multiplexed gene repression [38], it would provide valuable complementary information to knockdown TMEM97 together with other AMD-associated genes to determine if these genes have a synergistic contribution to retinal degeneration and the pathogenesis of AMD. In this regard, we provided a transcriptomic analysis of the human retina that can be used to prioritise AMD-associated genes, which could be further studied using our CRISPRi RPE platform.
Previous studies have demonstrated a role of the Sigma-2 receptor signalling in regulating apoptosis, cytotoxicity, and oxidative stress [9,10,11]. Our study extended these findings and showed that knockdown of TMEM97 played a protective role in RPE against oxidative stresses, providing a potential drug target to alleviate RPE degeneration in AMD. Significant progress has been made to treat neurodegenerative diseases using Sigma-2 receptor inhibitors, including clinical trials in Alzheimer’s disease [39] and schizophrenia [40]. In particular, a Sigma-2 receptor antagonist CT1812 has demonstrated a safety profile in the clinic [39] and is currently being tested as a treatment for Alzheimer’s disease in a Phase II trial (ClinicalTrials.gov: NCT03507790). Future studies that evaluate the efficacy of Sigma-2 receptor antagonists in RPE could pave the way for the development of novel treatments to improve vision in patients with AMD.
In summary, this study presents an in vitro human RPE system with integrated CRISPRi for gene repression. We show that this in vitro CRISPRi system can be used for loss-of-function studies of AMD-associated genes. Using this CRISPRi system, we reveal a novel role of TMEM97 in regulating human RPE viability against oxidative stress, providing supporting evidence for its role in AMD pathogenesis.
4. Methods
4.1. Donor Retina Collection
Collection of patient samples was approved by the Human Research Ethics committee of the Royal Victorian Eye and Ear Hospital (HREC13/1151H) and carried out in accordance with the approved guidelines. Informed consent was obtained from all donors, and the experiments conformed to the principles set out in the WMA Declaration of Helsinki. Post-mortem eye globes were collected by the Lions Eye Donation Service (Royal Victorian Eye and Ear Hospital, Melbourne, Australia) for donor cornea transplantation. The remaining eye globes were used to dissect the RPE/choroid layers for this study.
4.2. Transcriptome Analysis Using RNAseq
Total RNA was extracted using the RNeasy kit and treated with DNase 1 (Qiagen, Hilden, Germany). Three donor samples of human RPE/choroid were processed for RNAseq. RNAseq was performed by the Australian Genome Research Facility. Briefly, quality of the RNA samples was checked using a bioanalyzer, and transcriptome libraries were prepared using the TruSeq Stranded mRNA kit (Illumina, San Diego, CA, USA). Subsequently, samples were processed for 100 bp single-end sequencing using an Illumina Novaseq 6000 and yielded ~38–50 million reads per sample. The human reference transcriptome GRCh38 was used as an index, and transcript level quantification was performed using Salmon v1.0 to obtain gene-level counts [41]. The tximport package was used to import and summarize the gene-level counts, using the lengthScaledTPM function [42], and was imported to DESeq2 for count normalisation and gene expression analysis [43]. Visualisation of gene expression was generated using the packages ggplot2 [44]. The transcriptome data of the human RPE/choroid in this study are available in the NCBI Gene Expression Omnibus database (GSE181550), including raw data, processed data, information of the experimental design, sequencing, and processing pipeline.
4.3. Cell Culture
HEK293A, HEK293FT, ARPE19, and ARPE19-KRAB cells were cultured in DMEM (Thermo Fisher, Waltham, MA, USA) supplemented with 10% [v/v] Fetal Bovine Serum (FBS), 1× GlutaMAX, and 0.5% Penicillin/Streptomycin (all from Thermo Fisher). All cells were grown at 37 °C and 5% CO2 and passaged using 0.25% Trypsin-EDTA (Thermo Fisher, Waltham, MA, USA).
4.4. Lentivirus Generation
7 × 106 HEK293FT cells were plated in a 10 cm dish one day prior to transfection and cultured with lentivirus packaging medium (Opti-MEM supplemented with 5% FBS and 200 µM Sodium pyruvate, all from Thermo Fisher). The lentiviruses were generated by co-transfecting the dCas9-KRAB plasmid (gifts from Kristen Brennand; #99372, Addgene, Watertown, MA, USA) with the three 3rd generation packaging vectors pMDLg/pRRE (gifts from Didier Trono, #12251, Addgene), pRSV-Rev (#12253, Addgene) pMD2.G (#12259, Addgene), using Lipofectamine 3000 (Thermo Fisher). The medium with lipofectamine 3000 was replaced with fresh media 6 h after transfection. Afterwards, the crude virus was collected at 48 and 72 h after transfection. The collected virus was filtered (0.45 µm filter, Sartorius, Göttingen, Germany) and concentrated using PEG-it overnight at 4 °C (System Biosciences, Palo Alto, CA, USA) and resuspended in cold PBS.
4.5. Generation of ARPE19-KRAB
ARPE19 cells were transduced with dCas9-KRAB lentiviruses overnight in the presence of 8 µg/mL of polybrene (Sigma-Aldrich, St. Louis, MO, USA), followed by selection with 2μg/mL puromycin (Sigma-Aldrich). We had previously performed a puromycin dosage study to determine that 2 μg/mL is the lowest puromycin concentration that can kill all ARPE19 cells. Following puromycin selection, all the transduced ARPE19 cells were further expanded to obtain the ARPE19-KRAB cell line. Polarisation induction of ARPE19-KRAB was performed as previously described [21]. On day 0, ARPE19-KRAB was cultured in MEMa media supplemented with 1% FBS, 0.5% Penicillin/Streptomycin (all from Thermo Fisher), 10 mM nicotinamide, 1% N1, 0.25 mg/mL taurine, 20 ng/mL hydrocortisone, and 0.013 ng/mL T3 (all from Sigma-Aldrich). On day 7, the sample was processed for immunocytochemistry and imaging analysis.
4.6. CRISPR Interference (CRISPRi)-Mediated Knockdown of Gene Expression
SpCas9 sgRNA expression cassettes containing a U6 promoter, sgRNA, and sgRNA scaffold were synthesized, amplified, and purified as previously described [24]. For HEK293A cells, on Day 0, 6 × 104 cells were plated down in a well on a 12-well plate. On day 1, the cells were transiently transfected with 360 ng of sgRNA expression cassette and 800 ng of dCas9-KRAB using Lipofectamine 3000. For ARPE19-KRAB cells, on Day 0, 1 × 105 cells were plated down in a well on a 12-well plate. On day 1, the cells were transiently transfected with 360 ng of sgRNA expression cassette using RNAimax (Thermo Fisher). For both HEK293A and ARPE19-KRAB cells, a mock control (no DNA transfected) was utilised as a negative control in every individual experiment. On Day 4, both HEK293A and ARPE19-KRAB samples were processed for RNA extraction and qPCR analysis to assess gene expression levels.
4.7. PCR and qPCR Analysis
Total RNA was extracted using the Illustra RNAspin kit and treated with DNase 1 (GE Healthcare, Chicago, IL, USA). cDNA synthesis was performed using a high-capacity cDNA reverse transcription kit with RNase inhibitor (Thermo Fisher). For PCR analysis, a 25 µL PCR reaction was set up using the KOD hot start DNA polymerase (Merck, Rahway, NJ, USA), following manufacturer’s instructions. The following primers were used: KRAB (FWD: CGTGAGGAGTGGAAATTGCTGG, REV: TTCCCCCTTTTCGAGCCTAAGG) and ACTB (FWD: CCCTGGCACCCAGCAC, REV: GCCGATCCACACGGAGTAC). The thermal profile used was 95 °C for 2 min; 30 cycles of 95 °C for 20 s, annealing temp for 20 s (KRAB: 65.8 °C, ACTB: 73.9 °C), 70 °C for 15 s; followed by 70 °C for 5 min. The PCR product was visualised in a 1% agarose gel. For qPCR, TaqMan gene expression assays were performed using TaqMan Fast Advanced Master Mix (Thermo Fisher) as previously described [45,46], using the following TaqMan probes for TMEM97 (Hs00299877_m1) and the housekeeping gene β-actin (Hs99999903_m1). The TaqMan assay was performed using the ABI 7500 or StepOne plus qPCR machine, following manufacturer’s instructions (Thermo Fisher). The delta delta Ct method was used to calculate and compare relative mRNA levels to control.
4.8. Immunoblotting
Western blot analysis was performed following conventional procedures. Briefly, cell pellets were resuspended in three volumes of 1× RIPA buffer and sonicated. The lysate was mixed with 4x Laemmli Sample Buffer (Bio-Rad, Hercules, CA, USA) and 2-Mercaptoethanol (1:40, Sigma-Aldrich) and heated at 95 °C for 5 min. The lysate was loaded on the 4–15% Mini-PROTEAN TGX Precast Protein Gels (Bio-Rad) for separation and transferred to the PVDF membrane. The blots are blocked and incubated with rabbit anti-TMEM97 antibody (1:400, #OAAB22200, Aviva Systems Biology, San Diego, CA, USA) as primary antibody overnight at 4 °C. After washing, the membranes were incubated with a secondary HRP conjugated anti-rabbit IgG antibody (1:2500, GE Healthcare) and developed by chemiluminescence using ECL reagents according to the manufacturer’s instructions (Thermo Fisher).
4.9. Immunocytochemistry
Standard immunocytochemistry procedures were carried out, as we previously described [47]. Briefly, samples were fixed in 4% paraformaldehyde, followed by blocking with 10% serum (Sigma-Aldrich) and permeabilization with 0.1% Triton X-100 (Sigma-Aldrich). The samples were then immunostained with antibodies against Cas9 (#14697, Cell Signaling, Danvers, United States) or ZO-1 (#339100, Thermo Fisher), Alexa Fluor 488 secondary antibodies (Abcam, Cambridge, UK), and nuclear counterstain with DAPI (Sigma-Aldrich). Samples were imaged using a Zeiss Axio Vert.A1 fluorescent microscope. The specificity of the immunostaining was confirmed using isotype control antibodies from the appropriate species.
4.10. Cell Viability Assay
Cell viability assay was performed using a CellTiter-Glo Luminescent Cell Viability Assay (#G9241, Promega, Madison, WI, USA) following the manufacturer’s instructions. Briefly, 4 × 104 cells were seeded in a well of a 96-well plate on Day 0, followed by CRISPRi on Day 1. For some conditions, the cells were treated with 100 mM tert-Butyl Hydroperoxide (tBHP) on Day 3. On Day 4, old media was replaced with 25 μL fresh media in each well, followed by adding 25 μL CellTiter-Glo reagent before incubating at room temperature for 10 min to initiate a luminescent reaction. A volume of 40 μL of cell lysate from each well was loaded to an opaque white luminometer plate to measure luminescence using a Spark 20 M microplate reader (Tecan). The OD reading was normalised to the control condition without sgRNA and tBHP treatment, and the results are presented as fold change relative to the control condition.
4.11. Reactive Oxygen Species (ROS) Analysis
Mitochondrial superoxide and total cellular ROS levels were assessed using MitoSOX and CellROX dyes respectively (All from Thermo Fisher). Following 24 h of treatment with tBHP, cells were stained with 5 µM MitoSOX for 20 min or 5 µM CellROX for 30 min in DMEM low glucose (Thermo Fisher) at 37 °C in a humidified CO2 incubator. Cells were then washed twice with HBSS++ (Hank’s Balanced Salt Solution; Sigma-Aldrich) and imaged immediately in warm HBSS++ solution using an Olympus IX-71 microscope. Images were captured at 200 times magnification and the fluorescence intensities were analysed using ImageJ software. A total of 80 cells per group for each condition were analysed per replicate. The final fluorescence intensity was normalised to the mock control and expressed as fold change relative to mock.
4.12. Short Tandem Repeat Analysis
Genomic DNA of ARPE19 and ARPE19-KRAB were extracted using the Wizard SV Genomic DNA Purification kit (Promega), following manufacturer’s instructions. Short tandem repeat analysis was performed using the GenePrint 10 system (Promega) by the Australian Genome Research Facility.
4.13. Statistical Analysis
Statistical analysis was performed on biological repeats using paired t-test for assessment of TMEM97 knockdown levels, cell viability, total ROS levels, and mitochondrial superoxide levels (Graphpad Prism). p < 0.05 was used to establish statistical significance.
Acknowledgments
We would like to thank the Lions Eye Donation Services at Royal Victorian Eye and Ear Hospital for the collection of donor eye samples.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24043417/s1.
Author Contributions
J.-H.W., D.U.-C., S.S.C.H., A.W.H., T.L.E. and R.C.B.W. contributed to experimental design; J.-H.W., D.U.-C., J.G.L., S.M.M. and T.N. conducted the experiments; J.-H.W., D.U.-C., S.M.M., J.G.L., S.Y.L. and R.C.B.W. analysed the data; D.U.-C. and R.C.B.W. wrote the manuscript; R.C.B.W., A.W.H., S.Y.L. and T.L.E. provided the funding. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Collection of patient samples was approved by the Human Research Ethics committee of the Royal Victorian Eye and Ear Hospital (HREC13/1151H) and carried out in accordance with the approved guidelines.
Informed Consent Statement
Informed consent was obtained from all donors, and the experiments conformed to the principles set out in the WMA Declaration of Helsinki.
Data Availability Statement
The transcriptome data of the human RPE/choroid in this study are available in the NCBI Gene Expression Omnibus database (GSE181550).
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the University of Melbourne (R.C.B.W), the Centre for Eye Research Australia (R.C.B.W), and the Stafford Fox Medical Research Foundation (S.Y.L). D.U.-C and T.N. are supported by the Melbourne Research Scholarship. The Centre for Eye Research Australia and St Vincent Institute of Medical Research receive operational infrastructure support from the Victorian Government.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Wong W.L., Su X., Li X., Cheung C.M.G., Klein R., Cheng C.-Y., Wong T.Y. Global Prevalence of Age-Related Macular Degeneration and Disease Burden Projection for 2020 and 2040: A Systematic Review and Meta-Analysis. Lancet Glob. Health. 2014;2:e106–e116. doi: 10.1016/S2214-109X(13)70145-1. [DOI] [PubMed] [Google Scholar]
- 2.Fritsche L.G., Igl W., Bailey J.N.C., Grassmann F., Sengupta S., Bragg-Gresham J.L., Burdon K.P., Hebbring S.J., Wen C., Gorski M., et al. A Large Genome-Wide Association Study of Age-Related Macular Degeneration Highlights Contributions of Rare and Common Variants. Nat. Genet. 2016;48:134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neale B.M., Fagerness J., Reynolds R., Sobrin L., Parker M., Raychaudhuri S., Tan P.L., Oh E.C., Merriam J.E., Souied E., et al. Genome-Wide Association Study of Advanced Age-Related Macular Degeneration Identifies a Role of the Hepatic Lipase Gene (LIPC) Proc. Natl. Acad. Sci. USA. 2010;107:7395–7400. doi: 10.1073/pnas.0912019107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Logue M.W., Schu M., Vardarajan B.N., Farrell J., Lunetta K.L., Jun G., Baldwin C.T., Deangelis M.M., Farrer L.A. Search for Age-Related Macular Degeneration Risk Variants in Alzheimer Disease Genes and Pathways. Neurobiol. Aging. 2014;35:1510.e7–1510.e18. doi: 10.1016/j.neurobiolaging.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu Y., Bhangale T.R., Fagerness J., Ripke S., Thorleifsson G., Tan P.L., Souied E.H., Richardson A.J., Merriam J.E., Buitendijk G.H.S., et al. Common Variants near FRK/COL10A1 and VEGFA Are Associated with Advanced Age-Related Macular Degeneration. Hum. Mol. Genet. 2011;20:3699–3709. doi: 10.1093/hmg/ddr270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng C.-Y., Yamashiro K., Chen L.J., Ahn J., Huang L., Huang L., Cheung C.M.G., Miyake M., Cackett P.D., Yeo I.Y., et al. New Loci and Coding Variants Confer Risk for Age-Related Macular Degeneration in East Asians. Nat. Commun. 2015;6:6063. doi: 10.1038/ncomms7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen T., Urrutia-Cabrera D., Liou R.H.-C., Luu C.D., Guymer R., Wong R.C.-B. New Technologies to Study Functional Genomics of Age-Related Macular Degeneration. Front. Cell Dev. Biol. 2020;8:604220. doi: 10.3389/fcell.2020.604220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alon A., Schmidt H.R., Wood M.D., Sahn J.J., Martin S.F., Kruse A.C. Identification of the Gene That Codes for the σ2 Receptor. Proc. Natl. Acad. Sci. USA. 2017;114:7160–7165. doi: 10.1073/pnas.1705154114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oyer H.M., Sanders C.M., Kim F.J. Small-Molecule Modulators of Sigma1 and Sigma2/TMEM97 in the Context of Cancer: Foundational Concepts and Emerging Themes. Front. Pharmacol. 2019;10:1141. doi: 10.3389/fphar.2019.01141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu C.-C., Yu C.-F., Wang S.-C., Li H.-Y., Lin C.-M., Wang H.-H., Abate C., Chiang C.-S. Sigma-2 receptor/TMEM97 Agonist PB221 as an Alternative Drug for Brain Tumor. BMC Cancer. 2019;19:473. doi: 10.1186/s12885-019-5700-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicholson H.E., Alsharif W.F., Comeau A.B., Mesangeau C., Intagliata S., Mottinelli M., McCurdy C.R., Bowen W.D. Divergent Cytotoxic and Metabolically Stimulative Functions of Sigma-2 Receptors: Structure-Activity Relationships of 6-Acetyl-3-(4-(4-(4-Fluorophenyl)piperazin-1-Yl)butyl)benzod]oxazol-2(3H)-One (SN79) Derivatives. J. Pharmacol. Exp. Ther. 2019;368:272–281. doi: 10.1124/jpet.118.253484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vázquez-Rosa E., Watson M.R., Sahn J.J., Hodges T.R., Schroeder R.E., Cintrón-Pérez C.J., Shin M.-K., Yin T.C., Emery J.L., Martin S.F., et al. Neuroprotective Efficacy of a Sigma 2 Receptor/TMEM97 Modulator (DKR-1677) after Traumatic Brain Injury. ACS Chem. Neurosci. 2019;10:1595–1602. doi: 10.1021/acschemneuro.8b00543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Izzo N.J., Colom-Cadena M., Riad A.A., Xu J., Singh M., Abate C., Cahill M.A., Spires-Jones T.L., Bowen W.D., Mach R.H., et al. Proceedings from the Fourth International Symposium on σ-2 Receptors: Role in Health and Disease. eNeuro. 2020;7:ENEURO.0317-20.2020. doi: 10.1523/ENEURO.0317-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terada K., Migita K., Matsushima Y., Kamei C. Sigma-2 Receptor as a Potential Therapeutic Target for Treating Central Nervous System Disorders. Neural Regen. Res. 2019;14:1893–1894. doi: 10.4103/1673-5374.259609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hung S.S.C., McCaughey T., Swann O., Pébay A., Hewitt A.W. Genome Engineering in Ophthalmology: Application of CRISPR/Cas to the Treatment of Eye Disease. Prog. Retin. Eye Res. 2016;53:1–20. doi: 10.1016/j.preteyeres.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 16.Dominguez A.A., Lim W.A., Qi L.S. Beyond Editing: Repurposing CRISPR-Cas9 for Precision Genome Regulation and Interrogation. Nat. Rev. Mol. Cell Biol. 2016;17:5–15. doi: 10.1038/nrm.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Housden B.E., Perrimon N. Comparing CRISPR and RNAi-Based Screening Technologies. Nat. Biotechnol. 2016;34:621–623. doi: 10.1038/nbt.3599. [DOI] [PubMed] [Google Scholar]
- 18.Gilbert L.A., Horlbeck M.A., Adamson B., Villalta J.E., Chen Y., Whitehead E.H., Guimaraes C., Panning B., Ploegh H.L., Bassik M.C., et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanson K.R., Hanna R.E., Hegde M., Donovan K.F., Strand C., Sullender M.E., Vaimberg E.W., Goodale A., Root D.E., Piccioni F., et al. Optimized Libraries for CRISPR-Cas9 Genetic Screens with Multiple Modalities. Nat. Commun. 2018;9:5416. doi: 10.1038/s41467-018-07901-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E., Stern-Ginossar N., Brandman O., Whitehead E.H., Doudna J.A., et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hazim R.A., Volland S., Yen A., Burgess B.L., Williams D.S. Rapid Differentiation of the Human RPE Cell Line, ARPE-19, Induced by Nicotinamide. Exp. Eye Res. 2019;179:18–24. doi: 10.1016/j.exer.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lukowski S.W., Lo C.Y., Sharov A.A., Nguyen Q., Fang L., Hung S.S., Zhu L., Zhang T., Grünert U., Nguyen T., et al. A Single-Cell Transcriptome Atlas of the Adult Human Retina. EMBO J. 2019;38:e100811. doi: 10.15252/embj.2018100811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R., et al. Optimized sgRNA Design to Maximize Activity and Minimize off-Target Effects of CRISPR-Cas9. Nat. Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang L., Hung S.S.C., Yek J., El Wazan L., Nguyen T., Khan S., Lim S.Y., Hewitt A.W., Wong R.C.B. A Simple Cloning-Free Method to Efficiently Induce Gene Expression Using CRISPR/Cas9. Mol. Ther. Nucleic Acids. 2019;14:184–191.24. doi: 10.1016/j.omtn.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bae S., Park J., Kim J.-S. Cas-OFFinder: A Fast and Versatile Algorithm That Searches for Potential off-Target Sites of Cas9 RNA-Guided Endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Datta S., Cano M., Ebrahimi K., Wang L., Handa J.T. The Impact of Oxidative Stress and Inflammation on RPE Degeneration in Non-Neovascular AMD. Prog. Retin. Eye Res. 2017;60:201–218. doi: 10.1016/j.preteyeres.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bringmann A., Syrbe S., Görner K., Kacza J., Francke M., Wiedemann P., Reichenbach A. The Primate Fovea: Structure, Function and Development. Prog. Retin. Eye Res. 2018;66:49–84. doi: 10.1016/j.preteyeres.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 28.Radzisheuskaya A., Shlyueva D., Müller I., Helin K. Optimizing sgRNA Position Markedly Improves the Efficiency of CRISPR/dCas9-Mediated Transcriptional Repression. Nucleic Acids Res. 2016;44:e141. doi: 10.1093/nar/gkw583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellezza I. Oxidative Stress in Age-Related Macular Degeneration: Nrf2 as Therapeutic Target. Front. Pharmacol. 2018;9:1280. doi: 10.3389/fphar.2018.01280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strunz T., Lauwen S., Kiel C., International AMD Genomics Consortium (IAMDGC) den Hollander A., Weber B.H.F. A Transcriptome-Wide Association Study Based on 27 Tissues Identifies 106 Genes Potentially Relevant for Disease Pathology in Age-Related Macular Degeneration. Sci. Rep. 2020;10:1584. doi: 10.1038/s41598-020-58510-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ambati J., Fowler B.J. Mechanisms of Age-Related Macular Degeneration. Neuron. 2012;75:26–39. doi: 10.1016/j.neuron.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finnemann S.C., Bonilha V.L., Marmorstein A.D., Rodriguez-Boulan E. Phagocytosis of Rod Outer Segments by Retinal Pigment Epithelial Cells Requires alpha(v)beta5 Integrin for Binding but Not for Internalization. Proc. Natl. Acad. Sci. USA. 1997;94:12932–12937. doi: 10.1073/pnas.94.24.12932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ablonczy Z., Dahrouj M., Tang P.H., Liu Y., Sambamurti K., Marmorstein A.D., Crosson C.E. Human Retinal Pigment Epithelium Cells as Functional Models for the RPE in Vivo. Investig. Ophthalmol. Vis. Sci. 2011;52:8614–8620. doi: 10.1167/iovs.11-8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo Y., Zhuo Y., Fukuhara M., Rizzolo L.J. Effects of Culture Conditions on Heterogeneity and the Apical Junctional Complex of the ARPE-19 Cell Line. Investig. Ophthalmol. Vis. Sci. 2006;47:3644–3655. doi: 10.1167/iovs.06-0166. [DOI] [PubMed] [Google Scholar]
- 35.Schnichels S., Paquet-Durand F., Löscher M., Tsai T., Hurst J., Joachim S.C., Klettner A. Retina in a Dish: Cell Cultures, Retinal Explants and Animal Models for Common Diseases of the Retina. Prog. Retin. Eye Res. 2021;81:100880. doi: 10.1016/j.preteyeres.2020.100880. [DOI] [PubMed] [Google Scholar]
- 36.Alfonsetti M., Castelli V., d’Angelo M., Benedetti E., Allegretti M., Barboni B., Cimini A. Looking for In Vitro Models for Retinal Diseases. Int. J. Mol. Sci. 2021;22:10334. doi: 10.3390/ijms221910334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rastoin O., Pagès G., Dufies M. Experimental Models in Neovascular Age Related Macular Degeneration. Int. J. Mol. Sci. 2020;21:4627. doi: 10.3390/ijms21134627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCarty N.S., Graham A.E., Studená L., Ledesma-Amaro R. Multiplexed CRISPR Technologies for Gene Editing and Transcriptional Regulation. Nat. Commun. 2020;11:1281. doi: 10.1038/s41467-020-15053-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grundman M., Morgan R., Lickliter J.D., Schneider L.S., DeKosky S., Izzo N.J., Guttendorf R., Higgin M., Pribyl J., Mozzoni K., et al. A Phase 1 Clinical Trial of the Sigma-2 Receptor Complex Allosteric Antagonist CT1812, a Novel Therapeutic Candidate for Alzheimer’s Disease. Alzheimers. Dement. 2019;5:20–26. doi: 10.1016/j.trci.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davidson M., Saoud J., Staner C., Noel N., Luthringer E., Werner S., Reilly J., Schaffhauser J.-Y., Rabinowitz J., Weiser M., et al. Efficacy and Safety of MIN-101: A 12-Week Randomized, Double-Blind, Placebo-Controlled Trial of a New Drug in Development for the Treatment of Negative Symptoms in Schizophrenia. Am. J. Psychiatry. 2017;174:1195–1202. doi: 10.1176/appi.ajp.2017.17010122. [DOI] [PubMed] [Google Scholar]
- 41.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soneson C., Love M.I., Robinson M.D. Differential Analyses for RNA-Seq: Transcript-Level Estimates Improve Gene-Level Inferences. F1000Research. 2015;4:1521. doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love M.I., Huber W., Anders S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Villanueva R.A.M., Chen Z.J. ggplot2: Elegant Graphics for Data Analysis (2nd Ed.) Meas.: Interdiscip. Res. Perspect. 2019;17:160–167. doi: 10.1080/15366367.2019.1565254. [DOI] [Google Scholar]
- 45.Hung S.S.C., Van Bergen N.J., Jackson S., Liang H., Mackey D.A., Hernández D., Lim S.Y., Hewitt A.W., Trounce I., Pébay A., et al. Study of Mitochondrial Respiratory Defects on Reprogramming to Human Induced Pluripotent Stem Cells. Aging. 2016;8:945–957. doi: 10.18632/aging.100950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crombie D.E., Curl C.L., Raaijmakers A.J., Sivakumaran P., Kulkarni T., Wong R.C., Minami I., Evans-Galea M.V., Lim S.Y., Delbridge L., et al. Friedreich’s Ataxia Induced Pluripotent Stem Cell-Derived Cardiomyocytes Display Electrophysiological Abnormalities and Calcium Handling Deficiency. Aging. 2017;9:1440–1452. doi: 10.18632/aging.101247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hung S.S.C., Wong R.C.B., Sharov A.A., Nakatake Y., Yu H., Ko M.S.H. Repression of Global Protein Synthesis by Eif1a-like Genes That Are Expressed Specifically in the Two-Cell Embryos and the Transient Zscan4-Positive State of Embryonic Stem Cells. DNA Res. 2013;20:391–402. doi: 10.1093/dnares/dst018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptome data of the human RPE/choroid in this study are available in the NCBI Gene Expression Omnibus database (GSE181550).